Background: The regulation of ACOT12 by ADP and ATP is thought to occur through oligomerization.
Results: The structures of apo-ACOT12 and ADP-bound ACOT12 reveal new insights into regulation.
Conclusion: ACOT12 is a homotrimer and neither ADP nor ATP alter the oligomeric state of the protein.
Significance: These findings provide the first structural insights in the regulation of this enzyme family.
Keywords: Acetyl Coenzyme A (Acetyl-CoA), ADP, Allosteric Regulation, ATP, Metabolism, ACOT12, START Domain, Thioesterase
Abstract
Acetyl-CoA plays a fundamental role in cell signaling and metabolic pathways, with its cellular levels tightly controlled through reciprocal regulation of enzymes that mediate its synthesis and catabolism. ACOT12, the primary acetyl-CoA thioesterase in the liver of human, mouse, and rat, is responsible for cleavage of the thioester bond within acetyl-CoA, producing acetate and coenzyme A for a range of cellular processes. The enzyme is regulated by ADP and ATP, which is believed to be mediated through the ligand-induced oligomerization of the thioesterase domains, whereby ATP induces active dimers and tetramers, whereas apo- and ADP-bound ACOT12 are monomeric and inactive. Here, using a range of structural and biophysical techniques, it is demonstrated that ACOT12 is a trimer rather than a tetramer and that neither ADP nor ATP exert their regulatory effects by altering the oligomeric status of the enzyme. Rather, the binding site and mechanism of ADP regulation have been determined to occur through two novel regulatory regions, one involving a large loop that links the thioesterase domains (Phe154-Thr178), defined here as RegLoop1, and a second region involving the C terminus of thioesterase domain 2 (Gln304-Gly326), designated RegLoop2. Mutagenesis confirmed that Arg312 and Arg313 are crucial for this mode of regulation, and novel interactions with the START domain are presented together with insights into domain swapping within eukaryotic thioesterases for substrate recognition. In summary, these experiments provide the first structural insights into the regulation of this enzyme family, revealing an alternate hypothesis likely to be conserved throughout evolution.
Introduction
Acetyl-CoA is one of the most important cellular metabolites, playing a central role in both the metabolism of carbohydrates, lipids, and proteins, as well as a range of signaling pathways. Its levels in the mitochondria are allosterically regulated through control of the pyruvate dehydrogenase complex and enzymes involved in β-oxidation that mediate its production and reciprocally regulated by enzymes of the citric acid cycle and ketogenic pathways that are responsible for its oxidative degradation to produce ATP or ketone bodies (1–4). In the cytoplasm, acetyl-CoA plays important roles in fatty acid synthesis and in the production of key signaling molecules, including acetyl-choline and the HMG-CoA/mevalonic acid pathway that lead to synthesis of isoprenoids and cholesterol (5). Here, acetyl-CoA levels are regulated by key enzymes, including ATP-citrate lyase (6), cytosolic acetyl-CoA synthetase (7), and acetyl-CoA thioesterase 12 (ACOT12)3 (8, 9).
ACOT12, known also as StarD15 or cytoplasmic acetyl-CoA hydrolase, is the major cytoplasmic acetyl-CoA thioesterase expressed in the liver of mouse (10), rat (11), and human (12). The enzyme is highly conserved, with >82% sequence identity within these species, and all mammalian homologues are comprised of two N-terminal, tandem hotdog domains, and a C-terminal steroidogenic acute regulatory protein-related lipid transfer (START) domain. Thioesterase activity resides in the double hotdog thioesterase domains which typically self-associate into dimers, trimers, tetramers, or hexamers depending on the thioesterase family (13–16). Significantly, oligomerization of ACOT12 has been suggested to play a key role in its regulation, where the apo- and ADP-bound forms are believed to exist as inactive monomers, while ATP is thought to induce the enzyme into an active dimers and tetramers (14, 17, 18). The C-terminal START domain in ACOT12 remains to be fully characterized, but related domains have been shown to act in lipid binding, transferring and sensing domains for phospholipids, sterols, and sphingolipids (19, 20). In humans, there are 15 proteins containing START domains, termed StarD1-StarD15, 8 of which are part of multidomain proteins (20), however interactions between START and interdomain partners are yet to be elucidated. ACOT12 is the acyl-CoA thioesterase responsible for hydrolysis of acetyl-CoA in the liver of mouse, rat, and human (10–12). Acetyl-CoA is an important component in the biogenic synthesis of signaling molecules, and together with the role of the liver in managing and supplying acetyl-CoA and its metabolic derivatives to other organs, highlights the importance of ACOT12 and need for a detailed understanding of its regulation. ACOT12 has shown to be reciprocally regulated by ADP and ATP, with the current dogma for the mechanism of regulation, believed to occur through induced oligomerization of the thioesterase domains, whereby ATP promote these domains into active dimer and tetramer forms, whereas apo- and ADP-bound ACOT12 are inactive monomers (10–12, 17).
Here, we describe the crystal structures of the thioesterase domains of ACOT12 both in the apo- and ADP-bound forms to provide the first structural basis for thioesterase regulation. Using a range of complementary biophysical techniques, we demonstrate that neither ADP nor ATP regulate the oligomerization of the enzyme but instead influence the mobility of two flexible regions critical for thioesterase activity (21). Rather than forming a tetramer, we show, using crystallography, small angle x-ray scattering (SAXS), and size exclusion chromatography (SEC), that the enzyme forms a trimer of double hotdog protomers, revealing a domain-swapping event between eukaryote thioesterases for substrate recognition. Finally, we present the first view of the interactions between the START and juxtapositional thioesterase domains to provide a detailed account of thioesterase regulation, which is likely to be conserved throughout evolution.
MATERIALS AND METHODS
Expression and Purification
The thioesterase domains of human ACOT12 (residues 7–336; accession no. NC_000005.9) were cloned into the pNIC-Bsa4 expression vector at the Structural Genomics Consortium (Stockholm, Sweden). The protein was recombinantly expressed as a His-tagged fusion protein in Escherichia coli BL21(DE3) pLysS or BL21(DE3) gold pRARE2 cells by the addition of 1 mm isopropyl β-d-1-thiogalactopyranoside at an OD of 0.6, and growth continued for 21 h at 291–297 K. Cells were harvested by centrifugation and resuspended in His buffer A containing 50 mm phosphate buffer, pH 8.0, 300 mm NaCl, and 20 mm imidazole. Purification was carried out in very similar fashion in both laboratories and here we describe the one leading to structures 4MOC and 4MOB. Affinity chromatography (HisTrap HP, GE Healthcare) was performed using His buffer A to load and remove unbound proteins, and a gradient elution up to 500 mm imidazole over 10 column volumes used to elute the enzyme. Fractions containing recombinant ACOT12 were pooled and treated with TEV protease (15 μg/ml) for 12 h at 276 K to remove the His-affinity tag (leaving 2 vector encoded amino acids, Ser and Met). The protein was further purified by SEC (S200, 26/60 column, GE Healthcare) in buffer containing 50 mm Tris, pH 7.4, 125 mm NaCl, and then concentrated to 21 mg/ml using an Amicon centrifugal filter. The protein for the apo-ACOT12 structure (PDB code 3B7K) was concentrated to 16.3 mg/ml in a final buffer containing 20 mm HEPES, 300 mm NaCl, 10% glycerol, 0.5 mm TCEP, pH 7.5. The purity was assessed to be >95% by SDS-PAGE and stored as 50-μl aliquots at 253 K.
Crystallization
Sparse matrix, hanging drop vapor diffusion was used to screen crystallization conditions over a number of commercially available screens, in both the presence and absence of ADP (PEG/Ion, PEG/Ion 2, Crystal Screen, and Crystal Screen 2 from Hampton Research, JCSG+, and PACT from Molecular Dimensions). Briefly, 1.5 μl of the protein sample was combined with an equal volume of reservoir solution and suspended above the requisite reservoir solution. Apo-ACOT12 structure was crystallized with 5 mm acetyl-CoA with 23% PEG 3350, 350 mm sodium thiocyanate. For improved apo-ACOT12 structure, small hexagonal crystals were obtained from slightly modified condition without acetyl-CoA (22% PEG 3350, 400 mm sodium thiocyanate). For ACOT12-ADP, small cuboidal crystals were obtained from PEG/Ion 42 condition (20% PEG 3350, 200 mm potassium phosphate dibasic). These initial crystallization conditions were optimized by screening around different PEG and salt concentrations until large, single, diffraction quality crystals were achieved. The final crystallization mixture contained PEG 3350, 150 mm potassium phosphate dibasic, and 1 mm ADP.
X-ray Data Collection and Structure Determination
Cryocrystallography was undertaken during x-ray diffraction data collection to minimize radiation damage. Briefly, crystals were soaked in reservoir solution containing 15% glycerol or 5% butanediol (PDB code 3B7K) and flash-cooled in liquid nitrogen at 100 K. Diffraction data were collected at the Australian Synchrotron on the MX1 and MX2 crystallography beamlines and at European Synchrotron Radiation Facility on ID14-1. Diffraction images were indexed, merged, and scaled using iMOSFLM (22) and AIMLESS (23) or XDS (24). Phases were determined by molecular replacement with Molrep (25) using a dimer generated from Bacillus halodurans ACOT structure (PDB code 1VPM) or chain A from ACOT7 (PDB code 2Q2B). Molecular replacement was followed by iterative rounds of model building with Coot (26) and refinement using PHENIX Refine (27) or REFMAC5 (28). Structures have been deposited to the PDB and were assigned codes 3B7K (apo-ACOT12; 2.7 Å), 4MOC (apo-ACOT12; 2.5 Å), and 4MOB (ACOT12-ADP).
Mutagenesis
Mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene). Each reaction mixture consisted of 39.3 μl of double distilled H2O, 5 μl of 10× reaction buffer, 1 μl of dNTP mix, 1.25 μl each of forward and reverse primers (100 μm), 1 μl of PfuUltra HF polymerase and 1.2 μl of ACOT12 plasmid. Mixtures were heated to 95 °C for 30 s followed by 16 cycles of 95 °C for 30 s, 55 °C for 1 min, and 68 °C for 7 min. The amplified products were then treated with the restriction enzyme DpnI (1 μl of 10 units/μl) for 1 h at 37 °C to digest the remaining nonmutated parental DNA, and then 1 μl of treated DNA was transformed into XL1-Blue supercompetent cells. The fidelity of the clones was confirmed by DNA sequencing.
Activity Assays
Enzyme activity of ACOT12 was measured spectrophotometrically using a coupled color change reaction with 5,5-dithiobis-(2-nitrobenzoate) as described previously (29–32). Each 100-μl assay volume contained 10 mm 5,5-dithiobis-(2-nitrobenzoate) in 100 mm sodium phosphate buffer, pH 8.0, 6 μg of protein, acetyl-CoA at various concentrations as specified, and regulatory molecules (ADP, ATP, GDP, GTP, citric acid, and NADH) at a final concentration of 10 mm. Release of CoA at 412 nm was measured over 20 min at 21 °C in a 96-well plate, and activity was calculated using extinction coefficient Σ412 = 13,600 m−1 cm−1 (29). Graphpad Prism (version 6) was used to plot the data and calculate enzyme kinetics.
Small Angle X-ray Scattering
SAXS measurements were performed on ACOT12 in the absence and presence of ADP and ATP at the SAXS beamline of the Australian Synchrotron. Each SAXS measurement represents the average of ten 1-s exposures and were performed on a dilution series of protein from 5 to 0.1 mg/ml. To minimize radiation damage, samples flowed through a 1.5-mm quartz capillary at a rate of 4 μl/s. A Pilatus 1 m detector (Dektris) with a 1.6-m camera length was used with an x-ray energy of 12 KeV, giving a Q range from 0.01 to 0.5 Å−1. Averaging of images, subtraction of blanks, and radial integration was performed using the beamline control software ScatterBrain (Australian Synchrotron), and all measurements were made at 25 °C. Scattering intensities I(q) for sample and buffer were recorded as a function of scattering vector q. The scattering profiles were calculated after subtracting the scattering contributions of buffer. Theoretical scattering curves of crystallographic atomic models were computed using CRYSOL (33).
RESULTS
Structure Determination of ACOT12
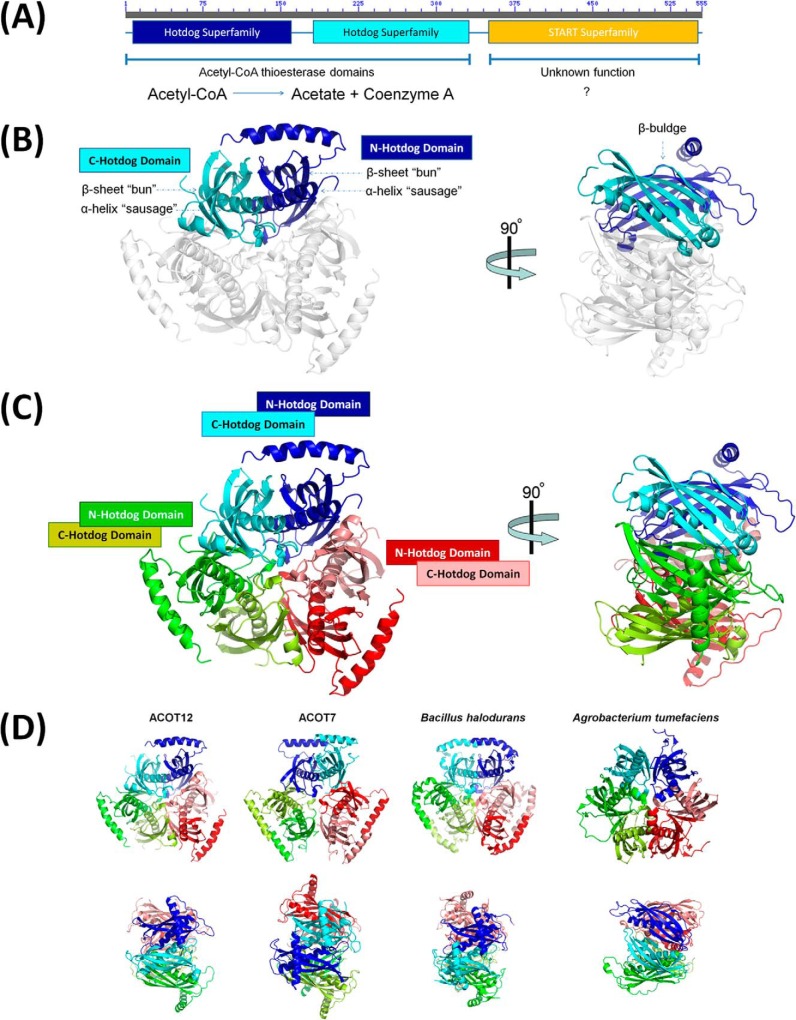
As mentioned previously, ACOT12 is of major importance, and there is a particular need for a detailed understanding of its regulation. To address this, because the structural basis for induced oligomerization and regulation of thioesterases is yet to be elucidated, we set out to determine the high resolution structures of the ACOT12 thioesterase domains in the absence and presence of ADP to better understand this regulatory mechanism. The crystals of the apo-enzyme were initially solved at 2.7 Å resolution. The asymmetric unit contained a trimer, which did not fit the oligomerization described in the literature. A large loop on the outer face of monomer (residues 153–179) did not have electron density, and density was also missing for the C-terminal part of the construct. We therefore decided to try to improve the crystal quality and try to stabilize these regions in the protein. A new crystal form of the apo-enzyme was discovered where one monomer was present in the asymmetric unit and the crystals diffracting to slightly higher resolution (2.5 Å; outer shell mean(I)/sd(I), 2.3; Rpim, 0.33; Table 1). This crystal structure provided clear contiguous electron density enabling 267 residues to be modeled (R/Rfree, 0.20/0.25 and good stereochemistry; Table 1), with the exception of two regions corresponding to residues 154–178 and 304–336 similar to the original structure. Notably, both of these regions are visible in the ADP-bound ACOT12 described below. The apo structures are very similar (average monomer r.m.s.d. of 0.4 Å for Cα atoms) with the major differences at the conformations of the surface exposed residues and in the 10 amino acids preceding the disordered C-terminal part (modeled only to one chain in the trimeric structure). In the following we will discuss the higher resolution structure unless otherwise stated. The ACOT12 monomer contained two hotdog domains, each comprised of a 5 stranded β-sheet enclosing a central α-helix, dimerized with the two central α-helices face-to-face, and β-sheets associating through β2 surrounding the central α-helices (Fig. 1, A and B).
TABLE 1.
Data collection and refinement statistics for ACOT12 and ACOT12:ADP
Values in brackets describe the highest resolution shell.
| ACOT12 (PDB code 3B7K) | ACOT12 (PDB code 4MOC) | ACOT12:ADP (PDB code 4MOB) | |
|---|---|---|---|
| Wavelength (Å) | 0.93400 | 0.9537 | 0.9537 |
| Resolution range (Å) | 2.70–20 (2.70–2.80) | 2.50–31.38 (2.50–2.60) | 2.40–34.71 (2.40–2.51) |
| Space group | C2221 | P321 | I23 |
| Unit cell | 82.70, 126.05, and 185.34 Å; 90, 90, and 90° | 123.55, 123.55, and 49.75 Å, 90, 90, and 120° | 138.85 Å; 90° |
| Total reflections | 191,938 (20,062) | 116,336 (11,446) | 409,008 (46,051) |
| Unique reflections | 26,807 (2737) | 14,338 (1505) | 17,555 (2114) |
| Multiplicity | 7.2 (7.3) | 8.1 (7.6) | 23.3 (21.8) |
| Completeness (%) | 99.3 (100) | 93.5 (87.6) | 100 (100) |
| Mean I/σ(I) | 12.5 (3.4) | 14.9 (2.3) | 18.9 (2.6) |
| Wilson B-factor (Å2) | 69.80 | 42.25 | 43.61 |
| Rpim | 0.116 (0.598)a | 0.037 (0.329) | 0.030 (0.300) |
| Rwork | 0.23 | 0.20 | 0.16 |
| Rfree | 0.28 | 0.25 | 0.21 |
| No. of atoms | 2240 | 2671 | |
| Macromolecules | 5917 | 2169 | 2576 |
| Ligands | 144 | 48 | 72 |
| Water | 0 | 71 | 95 |
| Protein residues | 769 | 273 | 320 |
| r.m.s.d. | |||
| Bonds (Å) | 0.008 | 0.004 | 0.010 |
| Angles | 1.11° | 0.86° | 1.23° |
| Ramachandran favored (%) | 94.81 | 95.56 | 96.86 |
| Ramachandran allowed (%) | 4.92 | 4.44 | 3.14 |
| Ramachandran outliers (%) | 0.27 | 0.00 | 0.00 |
a Rmerge value.
FIGURE 1.
Structure of ACOT12. A, domain architecture of ACOT12 comprised of two hotdog domains and C-terminal START domain. B, structure of the ACOT12 double hotdog domain protomer shown in schematic representation, with each hotdog domain in dark and light shades of the same color for each protomer. A 90° rotation highlights the presence of a β-bulge induced at each dimer interface. C, structure of the ACOT12 quaternary arrangement comprised of a trimer of hotdog dimers. D, structures of thioesterases containing the same hexameric arrangement built from either three double hotdog dimers (eukaryotes) or six single hotdog monomers (prokaryotes). Shown are structures of the thioesterase domains from B. halodurans (PDB code 1VPM (21)) a single hotdog fold thioesterase that forms a hexamer; the full length structure of ACOT7 (PDB codes 2Q2B and 2V1O (34)) and the structure of the double hotdog thioesterase from A. tumefaciens (PDB code 2GVH), which forms a back-to-back protomer.
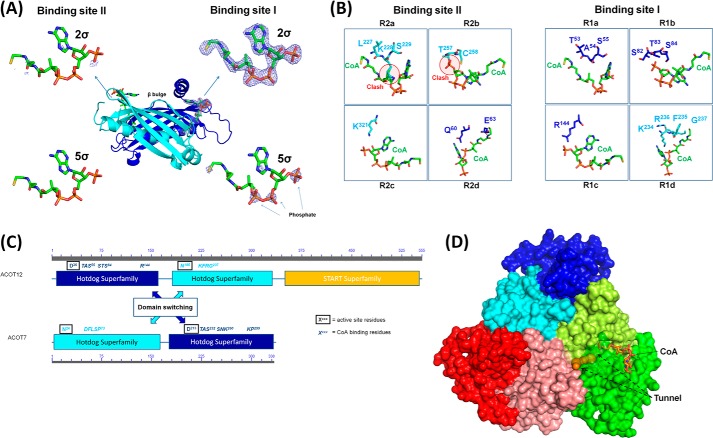
Coenzyme A (CoA), not added to the crystallization conditions, was identified at one of two symmetry related active sites, occupying the interface of the hotdog dimers and inducing a β-bulge (Fig. 2, A and B). That only half of the potential CoA binding sites contained ligand is similar to that of mouse ACOT7 (34), and emerging to be a distinguishing feature between the double hotdog thioesterases found in eukaryotes, and the single thioesterase domains of prokaryotic thioesterases, since the former contain two non-identical tandem hotdog domains creating symmetry related, but chemically distinct binding sites, while the latter contain only a single thioesterase domain and identical CoA binding sites (see (34) and discussion for full description). Since the structural basis for the preference of CoA-binding in eukaryotic thioesterases has yet to be determined, we superimposed CoA into the non-binding site, and compared these two pockets. Binding site one (containing CoA in our structure), bound through H-bonding and salt bridge interactions involving residues clustered into four regions (R): R1a (TAS55), R1b (STS84), R1c (R144); and R1d (KFRG237), while superimposition of the CoA molecule onto binding site two, revealed major clashes with K228 and T257, corresponding to residues from the analogous regions R2a and R2b (Fig. 2) respectively.
FIGURE 2.
The CoA binding motif of ACOT12. A, the two potential binding sites for CoA on ACOT12, with binding site I occupied by CoA as demonstrated by density at both 2 and 5σ of Fo − Fc annealed omit maps and, in binding site II, a clear lack of density. B, the binding sites for CoA involving four separate binding regions; R1a (TAS55), R1b (STS84), R1c (Arg144), and R1d (KRFG237) for binding site I, whereas the binding site II contains steric clashes inhibiting CoA from occupying this site. C, location of CoA binding and active site residues on ACOT12 and ACOT7, demonstrating domain swapping between these two thioesterases for these critical functional regions. D, surface representation of ACOT12 with CoA shown as sticks and the substrate-binding tunnel is shown as spheres determined using Caver (38).
Interestingly, despite the low sequence identity between ACOT7 and ACOT12 of ∼30%, the CoA binding pockets are well conserved, but located on opposite domains, indicating that a domain switch for substrate binding has occurred (Fig. 2C). In both cases, the CoA binding site is distinguished by a TAS binding motif in R1a, while the non-CoA binding site contains a less favorable binding pocket and steric clashes. This will prove to be a useful feature in the prediction of CoA binding sites in eukaryotic thioesterases. In similar fashion to the CoA binding site, the active site residues for ACOT12 also reside on the opposite domains to those for ACOT7, and comprised of D36 and N195 in ACOT12, equivalent to the ACOT7 active site (N24, D213) (Fig. 2C) (34). A tunnel protruding from the active site to the surface of the protein can also be clearly discerned (Fig. 2D).
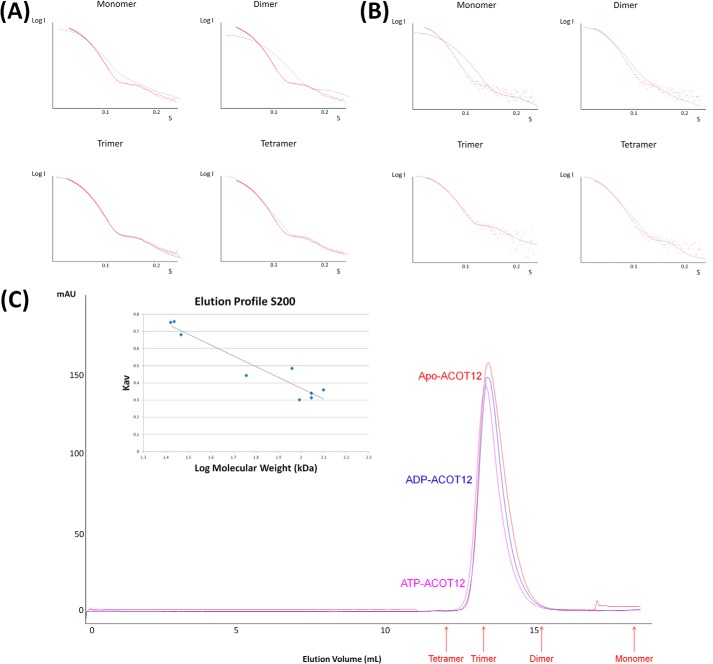
While only one ACOT12 molecule was present in the asymmetric unit of the crystal, the unit cell was comprised of six ACOT12 protomers arranged as two trimers. Consistent with this observation, the biological unit assigned by the Proteins Interfaces Structures and Assemblies (PISA) server, was a trimer of the monomeric double hotdog protomer (Fig. 1C). This trimer is similar to the one found in the asymmetric unit of the other crystal form and it is also consistent with the quaternary structure of other related thioesterases, including mouse ACOT7 (34) and the bacterial thioesterase from Bacillus halodurans (21) (Fig. 1D). To confirm the biological assembly observed in our crystal structure, both SAXS and SEC were performed. SAXS data confirmed that ACOT12 exists as a trimer in solution (Fig. 3A; χ of 62, 25, 1.3, and 14.4 for monomer, dimer, trimer and tetramer respectively), and was consistent with SEC data (Fig. 3C), confirming that ACOT12, in the absence of ADP, exists as a trimer, and thus, structurally similar to ACOT7 and bacterial thioesterases.
FIGURE 3.
SAXS and SEC data confirm the quaternary structure observed from crystallographic data. A, ACOT12 scattering data (red dots) superimposed on the theoretical scattering curves (blue line) for an ACOT12 monomer, dimer, trimer, and tetramer using CRYSOL with χ values of 62, 25, 1.3, and 14.4, respectively. B, ACOT12-ADP scattering data with the theoretical scattering curves for ACOT12-ADP monomer, dimer, trimer, and tetramer superimposed with χ values of 25, 7, 1.8, and 5.5. C, SEC elution profiles for apo-ACOT12, ACOT12-ADP, and ACOT12-ATP plotted against known molecular weight standards to assess the native molecular weight demonstrate that neither ADP nor ATP disrupt oligomerization, consistent with the SAXS and crystallographic data.
Structural elucidation of ACOT12 bound to ADP
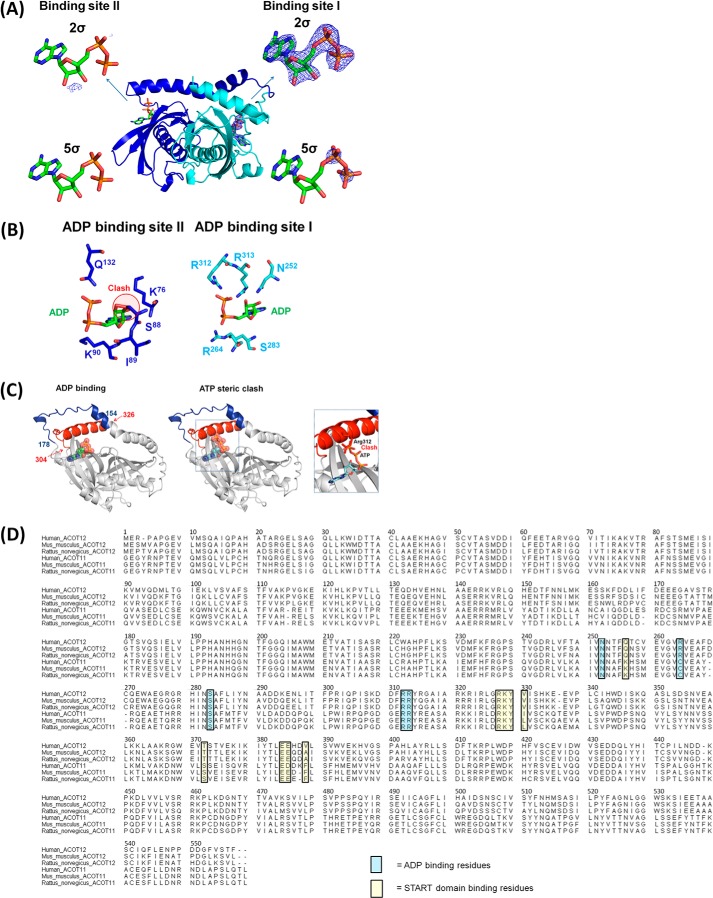
The lack of structural information pertaining to thioesterase regulation by alloesteric inhibitors represents a major gap in our understanding of this enzyme family, and since the structural elucidation of ACOT12 in the non-ADP form appeared to exist as a trimer of the double hotdog protomers, we set out to determine how allosteric regulation may perturb oligomerization. Attempts to crystallize ACOT12 bound to ADP in the conditions identified for apo-ACOT12 were unsuccessful, suggesting that the enzyme was adopting a significantly different conformation. Crystallization conditions producing crystals with one monomer in the asymmetric unit, diffracting to 2.4 Å resolution, enabled elucidation of a structure similar to the non-ADP form, with the two hotdog domains dimerized in the same face-to-face configuration (Fig. 4), and CoA located in the same position. One, well defined ADP molecule was identified (Fig. 5), located at the base of the β-sheet, and bound at a pocket involving residues Asn252, Arg264, Ser283, Arg312, and Arg313 (Fig. 5, A and B). These residues are well conserved (Fig. 5D), and similarly to ACOT12 bound to CoA, a symmetry related, but non-identical ADP binding site did not contain ADP (Fig. 5A), due to the presence of steric clashes (Fig. 5B).
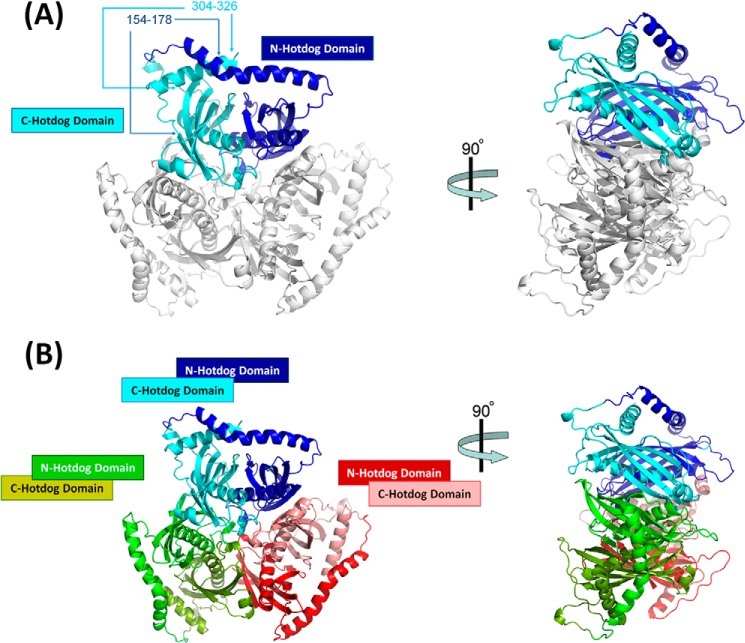
FIGURE 4.
Structure of ACOT12-ADP. A, the flexible regions 154–178 and 304–326 are visible in the ACOT12:ADP structure, revealing a 50° kink in the helix joining the two hotdog domains (loop I) at His150, causing residues 151–179 to link over the C-terminal α-helix. B, the hexamer arrangement of the ACOT12-ADP demonstrates its similarity to apo-ACOT12.
FIGURE 5.
ACOT12 binding site of ADP. A, two potential binding sites, with binding site I having clear density at both 2 and 5σ in simulated annealed omit maps and binding site II exhibiting negligible density corresponding to ADP. B, residues involved in binding ADP at the two potential binding sites, with binding site I utilizing Arg312 and Arg313 to interact with the phosphate of ADP, whereas at binding site II, ADP is inhibited from binding due to the presence of Ser88 (equivalent to Gly in binding site I). C, superposition of ATP on the ADP site reveals the γ-phosphate of ATP clashes with Arg312 preventing the C-terminal helix from being locked into place. D, sequence alignment of human, mouse, and rat ACOT12 and ACOT11. The ADP and START domain binding residues are highlighted in blue and yellow, respectively.
Unexpectedly, the quaternary structure was almost identical to the CoA bound-ACOT12 structure, exhibiting the same trimer of double hotdog protomer domains (Fig. 4). It is unlikely that the quaternary structure can be due to crystal artifacts due to its similarity to the apo-ACOT12 that was crystallized in two different space groups (r.m.s.d. of trimer of 0.423 Å2), and its similarity with other thioesterases. Nevertheless, we set out to also confirm the ACOT12-ADP quaternary structure in solution, and found that it was not altered by the presence or absence of ADP (χ of 25, 7, 1.8 and 5.5 for monomer, dimer, trimer and tetramer respectively) in both SAXS and SEC experiments (Fig 3BC).
That the ADP binding site is located on the surface of the protein and distinct from all inter-domain interfaces, and, that these interfaces were not altered in either structure, suggests that it is unlikely for ADP to be exerting its regulatory action by influencing the oligomeric state of the enzyme (confirmed in our structural and solution analysis above). Thus, to reveal the structural basis of ACOT12 regulation, a full comparative analysis of both the apo- and ADP-bound ACOT12 structures was undertaken. The most striking difference was that the previously flexible, unresolved regions in the CoA-bound-ACOT12 structure (residues 154–178, and 304–326), were now well ordered and clearly visible in the ADP-bound structure. These large regions were ordered due to interactions between the ADP phosphate moieties with Arg312 and Arg313, located within the second of these flexible loops, which in turn, were bound through a large array of interactions with the first flexible loop, specifically involving Glu160 and Glu173 bound to Arg322 and Arg302 respectively; Leu167, Phe169 and Val176 interacting with Asn304; and Ser161 bonding with Arg315. Secondly, ADP induces a 50° kink in the helix joining the two hotdog domains at His150, causing residues 151–179 to link over the C-terminal α-helix (residues 308 to 326) locking it in place (Fig. 4A). Repulsion between positively charged Arg264 and Arg312+Arg313 is removed when negative phosphate binds forming a sandwich stabilizing the whole loop system to this conformation. These charges are not present in binding site II (Fig. 5B). Since flexibility of the C-terminal α-helix is critical for activity (21), ADP binding in this pocket through Arg312 and Arg313, and the subsequent locking by loop 1 represents an effective mechanism through which ADP inhibits enzyme activity, that is distinct from the current ligand induced oligomerization model.
Model for enhanced activity in the presence of ATP
Since ATP is known to exert the opposite regulatory effect of ADP on ACOT12, a common phenomenon observed in many metabolic enzymes since ADP and ATP are opposing signals for the energy status of the cell, we assessed whether our model could account for reciprocal ATP activation of ACOT12. Superposition of ATP onto ADP reveals that the γ-phosphate on ATP clashes with the C-terminal helix (Fig. 5C). This would prevent it from being tethered and locked in place by loop one, thus, promoting the mobility of these domains (rather than immobilizing them), and providing a structural basis for the switch between inhibition and activation by ADP and ATP respectively. Indeed, these structural predictions are supported by our activity and mutagenesis analysis below, while furthermore, the alternative induced oligomerization mechanism cannot support the differences between ADP and ATP.
Enzyme activity and regulation by ADP/ATP confirmed through mutagenesis
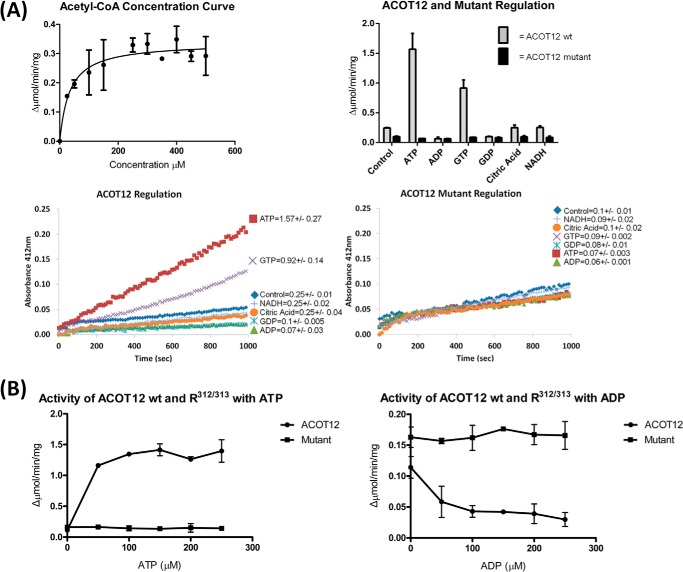
To probe the activity and regulation of ACOT12 based on our structural observations, we first assessed the activity of the ACOT12 and responsiveness to the ADP and ATP previously reported in the literature (14, 17, 18). We found the wtACOT12 cleaved acetyl-CoA (100 μm) at a rate of 0.25 μmol/min/mg, comparable to that described previously (11), and was sensitive to ADP (0.07 μmol/min/mg) and ATP (1.6 μmol/min/mg) (Fig. 6). Since flexibility of the C-terminal α-helix has previously been shown to be essential for thioesterase activity (21), and that Arg312 Arg313 was key to this interaction, we designed a Glu312 Glu313 ACOT12 double mutant to test whether nucleotide regulation could be abolished. As predicted by our structural model, the mutant was no longer susceptible to nucleotide regulation (Fig. 6) due to the repulsive charges introduced by these Glu residues (Fig. 6).
FIGURE 6.
ACOT12 activity and regulation activity. A, the ACOT12 thioesterase activity against acetyl-CoA concentration was plotted in GraphPad Prism using Michaelis-Menten kinetics (left panel). The activity of ACOT12 at 100 μm acetyl-CoA is enhanced by the presence of ATP and decreased by ADP, but this regulatory action is abolished in the Arg312/313 mutant, for which ATP and ADP (or GTP or GDP) show no significant differences (bottom panels, and summarized in right panel). The results shown in the bottom panel are from a single typical experiment repeated two or more times, with pooled data shown in the right panel representing the mean of activity (±) S.D. B, activity curves investigating the activity of ACOT12 and the Arg312/313 mutant over a range of ATP and ADP concentrations. Using the log concentrations of regulator the IC50 = 0.07290 mm for ADP and EC50 = 0.00297 mm for ATP were determined using Graphpad Prism.
Structural insights into thioesterase and START domain interactions
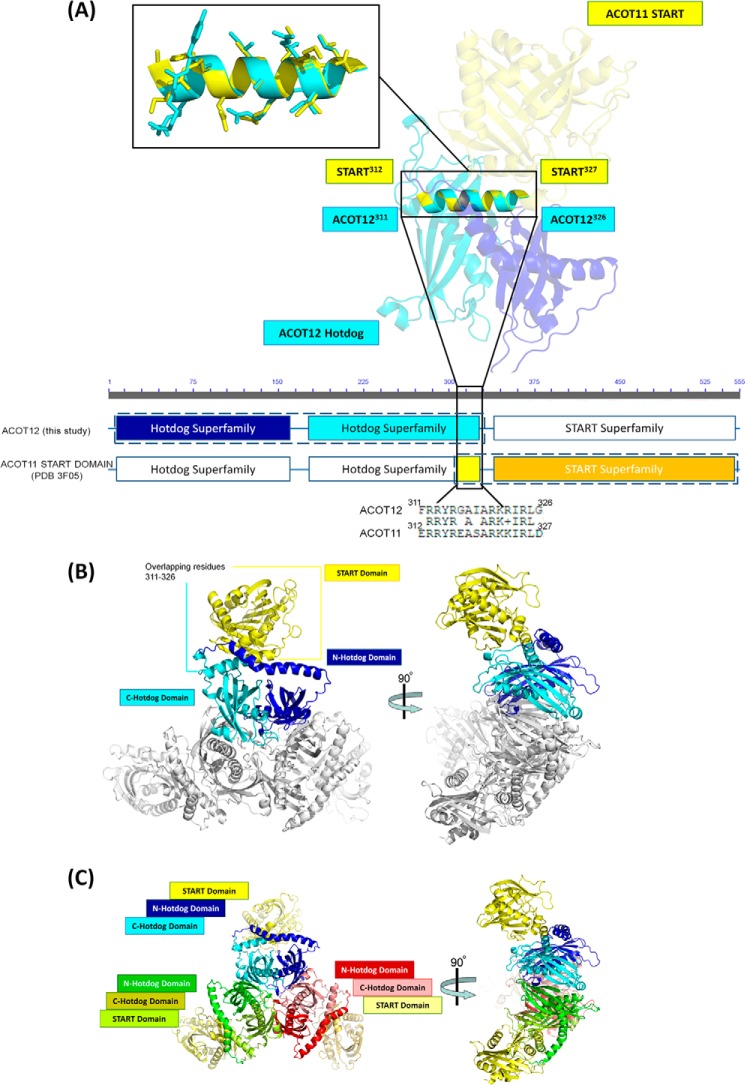
The human genome encodes 15 START-domain containing proteins, many of which are fused with other multi-domain proteins including thioesterases, the metastatic lymph node 64 protein (MLN64), and the Rho-GTPase-activating-protein domain, however association between START and multi-domain proteins has yet to be structurally resolved (20). Interestingly, the structure of our ADP-bound ACOT12 revealed a 15 residue overlap with the previously published structure of the START domain from the related homologue, ACOT11 (35). Superposition of these overlapping domains is presented in Fig. 7A. Residues 311–326 of our ACOT12-ADP structure overlapped with residues 312–327 of the START domain from ACOT11 (containing 55% sequence identity), and were superimposed with an r.m.s.d. of 0.308 Å2 for Cα (Fig. 7), providing a model with no steric clashes and a binding interface of approximately ∼1000 Å2 between the C-terminal thioesterase domain and the START. Interactions at the interface, many of which were conserved among human, mouse, and rat ACOT12 and ACOT11 proteins (Fig. 5D), include Gln256 of the C-terminal thioesterase domain (Ctt) bound to Ser372 of the START domain; Lys328 Ctt with Glu384 START; Tyr329 Ctt with Asp385 START, Tyr329 Ctt with Phe388 START, Arg327 Ctt with Ile330 START, and Tyr329 Ctt with Ile330 START. Significantly, the interaction between the thioesterase and START domains is distinct from the ADP-binding pocket, and therefore, it is unlikely that the START domain plays a role in regulation of the ACOT12 activity. Interestingly, START domain regulation has been reported by previous researchers for both ACOT12 (14) and ACOT11 (36), the mechanism of which is still unknown and will be the subject of future research.
FIGURE 7.
The ACOT12 full-length model. A, structural alignment and superposition of ACOT12 with ACOT11 through overlapping residues 311–326. B, the ACOT11 START domain, shown in yellow, superimposed onto the ACOT12-ADP structure using the overlapping residues 311–326. C, the hexameric model of full-length ACOT12 containing all three START domains. Superimpositions were performed using Coot (26).
DISCUSSION
Here, we describe the first ADP-bound structure within this enzyme family to reveal a novel regulatory mechanism and the molecular basis for thioesterase regulation. In both ADP-bound and unbound structures, the enzyme is a trimer of double hotdog protomers, confirmed through crystallographic, SAXS, and SEC analysis. Previous to this report, the regulation of ACOT12 was believed to be regulated by ADP and ATP through differential oligomerization (11, 14, 17). These observations were performed using size exclusion chromatography and have, to date, not been confirmed with any other biophysical methods. Our structural and biophysical data do not support such a mechanism because the nucleotide-binding site does not appear to disrupt (or promote) the binding of the quaternary interfaces or influence the START domain interactions. Rather, ADP tethers two novel regulatory regions, 154–178 and 304–326, designated here as RegLoop1 and RegLoop2, through specific interactions involving Arg312 and Arg313. RegLoop1 further immobilizes RegLoop2 through an extensive array of interactions, preventing flexibility of the C-terminal α-helix, which has shown to be essential for enzyme activity (21). ATP appears to reciprocally regulate the enzyme by preventing immobilization of RegLoop2 due to a steric clash with the γ-phosphate.
The presence of CoA in both of our structures infers that the ligand remained bound to the protein during the purification process. That only half of the potential CoA-binding sites contained CoA was strikingly similar to ACOT7; however, that the two enzymes contained the same active site configuration but on opposite domains was quite unexpected. This has implications for the regulatory mechanism described here; because CoA is binding on the opposite domains, the locking mechanism would no longer be possible in ACOT7, and indeed, this has been confirmed in our activity assays, where ADP, ATP, GDP, and GTP do not alter ACOT7 activity. Thus, this mechanism of regulation is likely to only be applicable in thioesterase family members where the CoA binds in the same pocket as that described here for ACOT12. Interestingly, domain swapping is likely to emerge as a common feature in this enzyme family and extend beyond the human homologues described here. For example, most bacteria contain only a single hotdog domain in this family of enzymes yet maintain the same overall quaternary structure (Fig. 1D). However, an exception to this rule, the thioesterase of Argobacterium tumefaciens (Fig. 1D) contains a double hotdog protomer, where the hotdog domains bind back to back rather than face to face, yet remarkably still achieve the same quaternary structure (Fig. 1D). The structural basis for determining how (and why) these thioesterases can achieve the same overall quaternary structure through different protomer arrangements will only be fully elucidated with additional structural characterization within this enzyme family.
At the CoA site, a β-bulge was also observed at the β-sheet interface, becoming a signature feature of CoA-bound thioesterases (21, 37). This was recently documented in the analysis of CoA bound and unbound forms of acyl-CoA thioesterase from B. halodurans, where the CoA was shown to induce this β-bulge and interaction with the C-terminal α-helices of the hotdog domains. This is now confirmed in human ACOT12 and other higher order thioesterases (PDB codes 1YLI and 2V1O), suggesting that this is likely to be a highly conserved structural regulatory element within this thioesterase family.
Overall, our structural insights into the regulation of ACOT12 demonstrate a novel mechanism of regulation, distinct from that which has been proposed previously. Due to the highly conserved nature of the thioesterase fold, it is likely that other thioesterases (with the same CoA domain binding configuration) can be regulated by ADP/ATP via a similar mechanism to that described here.
Acknowledgments
We thank the Australian Synchrotron for valuable assistance during data collection. Crystallographic experiments were also performed on the ID14-1 beamline at the European Synchrotron Radiation Facility (Grenoble, France). We are grateful to Local Contact at European Synchrotron Radiation Facility for providing assistance in using beamline ID14-1.
The atomic coordinates and structure factors (codes 4MOC and 4MOB) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- ACOT12
- acetyl-CoA thioesterase 12
- START
- steroidogenic acute regulatory protein-related lipid transfer
- SAXS
- small angle x-ray scattering
- SEC
- size exclusion chromatography
- r.m.s.d.
- root mean square deviation
- Ctt
- C-terminal thioesterase domain.
REFERENCES
- 1. Bartlett K., Eaton S. (2004) Mitochondrial β-oxidation. Eur. J. Biochem. 271, 462–469 [DOI] [PubMed] [Google Scholar]
- 2. Wieland O., Weiss L., Eger-Neufeldt I. (1964) Enzymatic regulation of liver acetyl-CoA metabolism in relation to ketogenesis. Adv. Enzyme Regul. 2, 85–99 [DOI] [PubMed] [Google Scholar]
- 3. Williamson D. H., Bates M. W., Krebs H. A. (1968) Activity and intracellular distribution of enzymes of ketone-body metabolism in rat liver. Biochem. J. 108, 353–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wakil S. J., Abu-Elheiga L. A. (2009) Fatty acid metabolism: target for metabolic syndrome. J. Lipid Res. 50, S138–S143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown M. S., Goldstein J. L. (1980) Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J. Lipid Res. 21, 505–517 [PubMed] [Google Scholar]
- 6. Bauer D. E., Hatzivassiliou G., Zhao F., Andreadis C., Thompson C. B. (2005) ATP citrate lyase is an important component of cell growth and transformation. Oncogene 24, 6314–6322 [DOI] [PubMed] [Google Scholar]
- 7. Fujino T., Kondo J., Ishikawa M., Morikawa K., Yamamoto T. T. (2001) Acetyl-CoA synthetase 2, a mitochondrial matrix enzyme involved in the oxidation of acetate. J. Biol. Chem. 276, 11420–11426 [DOI] [PubMed] [Google Scholar]
- 8. Westin M. A., Hunt M. C., Alexson S. E. (2008) Short-and medium-chain carnitine acyltransferases and acyl-CoA thioesterases in mouse provide complementary systems for transport of β-oxidation products out of peroxisomes. Cell. Mol. Life Sci. 65, 982–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Matsunaga T., Isohashi F., Nakanishi Y., Sakamoto Y. (1985) Physiological changes in the activities of extramitochondrial acetyl-CoA hydrolase in the liver of rats under various metabolic conditions. Eur. J. Biochem. 152, 331–336 [DOI] [PubMed] [Google Scholar]
- 10. Suematsu N., Okamoto K., Isohashi F. (2002) Mouse cytosolic acetyl-CoA hydrolase, a novel candidate for a key enzyme involved in fat metabolism: cDNA cloning, sequencing and functional expression. Acta Biochim. Pol. 49, 937–945 [PubMed] [Google Scholar]
- 11. Suematsu N., Okamoto K., Shibata K., Nakanishi Y., Isohashi F. (2001) Molecular cloning and functional expression of rat liver cytosolic acetyl-CoA hydrolase. Eur. J. Biochem. 268, 2700–2709 [DOI] [PubMed] [Google Scholar]
- 12. Suematsu N., Isohashi F. (2006) Molecular cloning and functional expression of human cytosolic acetyl-CoA hydrolase. Acta Biochim. Pol. 53, 553–561 [PubMed] [Google Scholar]
- 13. Pidugu L. S., Maity K., Ramaswamy K., Surolia N., Suguna K. (2009) Analysis of proteins with the 'hot dog' fold: Prediction of function and identification of catalytic residues of hypothetical proteins. BMC Struct. Biol. 9, 37–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Horibata Y., Ando H., Itoh M., Sugimoto H. (2013) Enzymatic and transcriptional regulation of the cytoplasmic acetyl-CoA hydrolase ACOT12. J. Lipid Res. 54, 2049–2059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cantu D. C., Chen Y., Lemons M. L., Reilly P. J. (2011) ThYme: a database for thioester-active enzymes. Nucleic Acids Res. 39, D342–D346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cantu D. C., Chen Y., Reilly P. J. (2010) Thioesterases: a new perspective based on their primary and tertiary structures. Protein Sci. 19, 1281–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Isohashi F., Nakanishi Y., Sakamoto Y. (1983) Effects of nucleotides on a cold labile acetyl-coenzyme A hydrolase from the supernatant fraction of rat liver. Biochemistry 22, 584–590 [DOI] [PubMed] [Google Scholar]
- 18. Söling H. D., Rescher C. (1985) On the regulation of cold-labile cytosolic and of mitochondrial acetyl-CoA hydrolase in rat liver. Eur. J. Biochem. 147, 111–117 [DOI] [PubMed] [Google Scholar]
- 19. Soccio R. E., Breslow J. L. (2003) StAR-related lipid transfer (START) proteins: mediators of intracellular lipid metabolism. J. Biol. Chem. 278, 22183–22186 [DOI] [PubMed] [Google Scholar]
- 20. Alpy F., Tomasetto C. (2005) Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J. Cell Sci. 118, 2791–2801 [DOI] [PubMed] [Google Scholar]
- 21. Marfori M., Kobe B., Forwood J. K. (2011) Ligand-induced conformational changes within a hexameric Acyl-CoA thioesterase. J. Biol. Chem. 286, 35643–35649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rosano C., Zuccotti S., Stefani M., Bucciantini M., Ramponi G., Bolognesi M. (2002) Crystallization and preliminary X-ray characterization of the acylphosphatase-like domain from the Escherichia coli hydrogenase maturation factor HypF. Acta Crystallogr. D Biol. Crystallogr. 58, 524–525 [DOI] [PubMed] [Google Scholar]
- 23. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 24. Kabsch W. (2010) Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vagin A. A., Isupov M. N. (2001) Spherically averaged phased translation function and its application to the search for molecules and fragments in electron-density maps. Acta Crystallogr. D Biol. Crystallogr. 57, 1451–1456 [DOI] [PubMed] [Google Scholar]
- 26. Emsley P., Lohkamp B., Scott W., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Afonine P. V., Grosse-Kunstleve R. W., Echols N., Headd J. J., Moriarty N. W., Mustyakimov M., Terwilliger T. C., Urzhumtsev A., Zwart P. H., Adams P. D. (2012) Towards automated crystallographic structure refinement with phenix. refine. Acta Crystallogr. D Biol. Crystallogr. 68, 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vagin A. A., Steiner R. A., Lebedev A. A., Potterton L., McNicholas S., Long F., Murshudov G. N. (2004) REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 29. Yamada J., Furihata T., Tamura H., Watanabe T., Suga T. (1996) Long-chain acyl-CoA hydrolase from rat brain cytosol: purification, characterization, and immunohistochemical localization. Arch. Biochem. Biophys. 326, 106–114 [DOI] [PubMed] [Google Scholar]
- 30. Tilton G. B., Shockey J. M. (2004) Biochemical and molecular characterization of ACH2, an acyl-CoA thioesterase from Arabidopsis thaliana. J. Biol. Chem. 279, 7487–7494 [DOI] [PubMed] [Google Scholar]
- 31. Wei J., Kang H. W., Cohen D. E. (2009) Thioesterase superfamily member 2 (Them2)/acyl-CoA thioesterase 13 (Acot13): a homotetrameric hotdog fold thioesterase with selectivity for long-chain fatty acyl-CoAs. Biochem. J. 421, 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hunt M. C., Solaas K., Kase B. F., Alexson S. E. (2002) Characterization of an acyl-CoA thioesterase that functions as a major regulator of peroxisomal lipid metabolism. J. Biol. Chem. 277, 1128–1138 [DOI] [PubMed] [Google Scholar]
- 33. Svergun D., Barberato C., Koch M. (1995) CRYSOL-a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 [Google Scholar]
- 34. Forwood J. K., Thakur A. S., Guncar G., Marfori M., Mouradov D., Meng W., Robinson J., Huber T., Kellie S., Martin J. L. (2007) Structural basis for recruitment of tandem hotdog domains in acyl-CoA thioesterase 7 and its role in inflammation. Proc. Natl. Acad. Sci. U.S.A. 104, 10382–10387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thorsell A. G., Lee W. H., Persson C., Siponen M. I., Nilsson M., Busam R. D., Kotenyova T., Schüler H., Lehtiö L. (2011) Comparative structural analysis of lipid binding START domains. PLoS One 6, e19521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Han S., Cohen D. E. (2012) Functional characterization of thioesterase superfamily member 1/Acyl-CoA thioesterase 11: implications for metabolic regulation. J. Lipid Res. 53, 2620–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Willis M. A., Zhuang Z., Song F., Howard A., Dunaway-Mariano D., Herzberg O. (2008) Structure of YciA from Haemophilus influenzae (HI0827), a Hexameric Broad Specificity Acyl-Coenzyme A Thioesterase. Biochemistry 47, 2797–2805 [DOI] [PubMed] [Google Scholar]
- 38. Chovancova E., Pavelka A., Benes P., Strnad O., Brezovsky J., Kozlikova B., Gora A., Sustr V., Klvana M., Medek P. (2012) CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput. Biol. 8, e1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]