Background: The potential role of Dim1 in RANKL-mediated induction of osteoclast differentiation is unknown.
Results: Dim1 directly interacts with c-Fos to diminish the expression of NFATc1 target genes that are essential for RANKL-induced osteoclatogenesis.
Conclusion: Dim1 functions as a negative regulator of RANKL-induced osteoclast differentiation.
Significance: Understanding of the inhibitory role of Dim1 in RANKL-induced osteoclastogenesis might provide therapeutic benefits for treating various types of bone diseases.
Keywords: Chromatin, FBJ Murine Osteosarcoma Viral Oncogene Homolog (FOS), Gene Expression, NFAT Transcription Factor, Osteoclast, Dim1, NFATc1, Osteoclastogenesis, RANKL, c-Fos
Abstract
Bone remodeling is a continuous process of osteoblastic bone formation and osteoclastic bone resorption to maintain normal bone mass. NFATc1 is the master regulator of osteoclastogensis and transcriptionally activated by c-Fos and NF-κB in response to receptor activator of NF-κB ligand (RANKL) treatment. Defective entry into mitosis 1 (Dim1) is a nuclear protein that is implicated in pre-mRNA splicing and cell cycle progression, but the possible role of Dim1 in regulating other cellular processes remains unknown. Here, we demonstrate that Dim1 attenuates RANKL-induced osteoclastogenesis by targeting NFATc1 signaling pathway. Expression levels of Dim1 and NFATc1 are significantly increased during the formation of multinucleated osteoclasts. RNAi-mediated knockdown of Dim1 markedly enhances the expression of NFATc1 and its target genes, leading to the increase of RANKL-induced osteoclastogenesis in bone marrow-derived macrophages. Conversely, ectopic expression of Dim1 decreases RANKL-induced osteoclast differentiation by silencing NFATc1 and its target genes, further linking Dim1 to the dynamic regulation of osteoclastogenesis. Consistent with this notion, ChIP and interaction studies show that Dim1 directly associates with c-Fos and prevents c-Fos from binding to the NFATc1 promoter, resulting in targeted inactivation of the NFATc1 gene. Therefore, our studies reveal an unrecognized role for Dim1 as a master modulator of osteoclast differentiation, as well as the molecular mechanism underlying its repressive action toward osteoclastogensis.
Introduction
New bone formation and maintenance in vertebrate animals are carried out by the coordinated action of osteoblasts and osteoclasts. Osteoblasts are mesenchymal stem cells that form bone matrix, whereas osteoclasts are large multinucleated cells whose function is to resorb bone matrix (1–3). The excess formation or activity of osteoclasts in humans leads to many pathological bone diseases, including osteoporosis, rheumatoid arthritis, Paget's disease, and tumor bone metastases (4–6). Osteoclast differentiation is triggered by the stimulation of receptor activator of NF-κB ligand (RANKL4; also called TRANCE, TNSF11, OPGL, and ODF), which is expressed as a membrane-bound protein in osteoblasts and provides osteoclast-specific differentiation signals (7–10). RANKL binds to its cognate receptor RANK on pre-osteoclast cell membrane and initiates multiple signal transduction pathways to turn on the expression of several transcription factors such as NF-κB, c-Fos, and NFATc1 in precursor cells (11). Activation of RANKL-RANK signaling leads to the recruitment of the adaptor molecule TRAF6, followed by the activation and nuclear translocation of NF-κB (12, 13). When NF-κB is activated, it triggers c-Fos signals, which then induce the expression of NFATc1 (14). After the initial induction, NFATc1 autoamplifies its expression by binding to its own promoter, induces the expression of a number of genes, and activates differentiation of osteoclast, leading to bone resorption and remodeling (15–19).
Dim1 is a small protein with the molecular mass of 15 kDa and was initially identified in fission yeast as an essential protein for cell cycle regulation as well as for chromosome segregation during mitosis (20). Dim1 is extraordinarily well conserved throughout the eukaryotic kingdom and is present in numerous species including mammals, plants, and yeasts, with ∼80% sequence identity throughout the entire length of 142 amino acids (21, 22). Dim1 has been reported to interact with components of pre-mRNA splicing machinery such hnRNP-F, hnRNP-H, and the RNA-binding protein Npw38/PQBP-1 (23). Under some circumstances, Dim1 has effects on cell cycle progression via its role in the control of pre-mRNA splicing of proteins responsible for accurate cell cycle progression. The three-dimensional structure of Dim1p has been determined by both nuclear magnetic resonance (21) and x-ray crystallography (24). The results indicate that Dim1 contains a common thioredoxin-like fold characterized by a four-stranded β-sheet comprising pairs of parallel and antiparallel strands flanked by three α-helices with a C-terminal extension. In addition to its function as a cell cycle regulator, Dim1 seems to participate in regulating other cellular reactions, such as host-virus interaction and viral replication, although the details remain largely unknown at present (25).
Here, we demonstrate that Dim1 functions as a negative regulator of RANKL-induced osteoclast differentiation. Specifically, Dim1 directly interacts with c-Fos and suppresses c-Fos transcriptional activity toward the NFATc1 gene, thereby diminishing the expression of NFATc1 target genes and inhibiting RANKL-induced osteoclatogenesis. Consistent with these results, Dim1-deficient osteoclast precursors exhibit increased expression of NFATc1 target genes and show up-regulation of RANKL-mediated induction of osteoclast differentiation. Our studies uncover a novel role for Dim1 in regulating RANKL-induced osteoclastogenesis and may provide the basis for novel therapeutic approaches to various types of bone diseases.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies
For mammalian expression of Dim1, FLAG-Dim1 cDNA was amplified by PCR and inserted into of retroviral expression vector pMX. To generate GST-Dim1, Dim1 cDNA was ligated into pGEX4T-1 vector. Antibodies for c-Fos, Dim1, HA, NFATc1, and NF-κB p65 were from Santa Cruz Biotechnology; antibodies for actin and FLAG were from Sigma; antibody for Mitf was from Active Motif.
Osteoclast Formation
Bone marrow (BM) cells were collected by flushing femurs and tibias from 6- to 8-week-old C57BL/6 mice. Cells were cultured in α-minimum essential medium (α-MEM) supplemented with 10% FBS and M-CSF (5 ng/ml) for 16 h. Non-adherent cells were harvested and cultured with M-CSF (30 ng/ml) for 3 days. Floating cells were removed and adherent cells were used as bone marrow-derived macrophage (BMM) cells. To generate osteoclasts, BMMs were cultured in the presence of M-CSF (30 ng/ml) and RANKL (50 ng/ml). On day 3, the cells were fixed and stained for tartrate-resistant acid phosphatase (TRAP) using the acid phosphatase, leukocyte kit (Sigma). TRAP-positive multinucleated cells containing three or more nuclei are counted as osteoclasts under a light microscope.
Protein-Protein Interactions
For in vitro pulldown assays, whole cell lysates from 293T cells expressing Mitf, c-Fos, and NF-κB p65 were incubated with GST-Dim1 (2 μg) immobilized on glutathione-Sepharose beads in 750 μl of binding buffer (20 mm HEPES-KOH, pH 7.9, 0.5 mm EDTA, 200 mm NaCl, 1 mm dithiothreitol, 10% glycerol, and 0.1% Nonidet P-40) for 16 h at 4 °C. After washing beads three times with washing buffer (20 mm HEPES-KOH, pH 7.9, 0.5 mm EDTA, 250 mm NaCl, 1 mm dithiothreitol, 10% glycerol, and 0.1% Nonidet P-40), bound proteins were detected by immunoblotting. For in vivo interaction studies, RAW 264.7 cells were stably infected with retroviral vectors encoding FLAG-Dim1. Cell lysates were subjected to anti-FLAG immunoprecipitation, and the bound proteins were analyzed by immunoblotting.
Lentiviral-mediated RNA Interference
For shRNA-based knockdown, DNA oligonucleotides encoding shRNA specific for Dim1 mRNA (5′- CAAGCAAGAAATGGTTGACAT-3′) were annealed and ligated into the lentiviral expression vector pLKO.1 (Addgene). Lentivirus particles were generated in 293T cells by co-transfecting plasmids encoding VSV-G, NL-BH, and the shRNA. For Dim1 knockdown, BMM cells were infected with these viruses and selected with puromycin (2 μg/ml) for 3 days. After selection, BMM cells were cultured for additional 3 days in the presence of M-CSF (30 ng/ml) and RANKL (50 ng/ml).
Retroviral-mediated Gene Transfer
To generate retroviral particles, pMX-FLAG-Dim1 was transfected into the packaging cell line Plat-E. Viral soup was collected from cultured media 2 days after transfection. BMM cells were infected with viral soup and selected with puromycin (2 μg/ml) for 3 days. After selection, cells were cultured with M-CSF (30 ng/ml) and RANKL (50 ng/ml) for 3 days.
Cell Proliferation Assays
Cell proliferation was assessed by the 3–4,5-(dimethyl-thyazol-2-yl)-2,5-diphenyltetrazolium (MTT) assay. In brief, BMM cells were seeded in 24-well tissue culture plates at a density of 2 × 104 and treated with the MTT labeling reagent (0.5 mg/ml) at 37 °C for 1 h. The blue MTT formazan precipitate was dissolved with the MTT solvent (0.2 ml) and measured at a wavelength of 570 nm using a microplate reader (Bio-Rad).
Reporter Gene Assays
RAW 264.7 cells were plated in 12-well plates at 50% confluence and transfected with reporter plasmids and expression vectors for c-Fos, NF-κB p65, and/or Dim1 in the presence or absence of RANKL (30 ng/ml) for 24 h. Cells were lysed in Reporter Lysis buffer (Promega) and assayed for luciferase activity using Plate Chameleon (Hidex).
Microarray and qRT-PCR
BMM cells were treated with M-CSF and RANKL for 0 and 3 days. Total RNA was isolated and analyzed by gene expression microarray using the MouseRef-8 Expression BeadChip (version 2.0). Differential gene expression analysis was carried out using the ArrayPipe software. Genes that are up-regulated with RANKL treatment in BMM cells were functionally analyzed in the context of gene ontology and molecular networks by using Ingenuity Pathway Analysis (IPA) software. For quantitative reverse transcription (qRT)-PCR analysis, total RNA was isolated as for microarray and subjected to RT reactions (26). Assays were normalized to β-actin mRNA levels. The following primers were used for qRT-PCR to quantify target gene expression: β-actin (5′-GCAAGTGCTTCTAGGCGGAC-3′ and 5′-AAGAAAGGGTGTAAAACGCAGC-3′), c-fos (5′-CCAGTCAAGAGCATCAGCAA-3′ and 5′-AAGTAGTGCAGCCCGGAGTA-3′), Ctsk (5′-ACGGAGGCATTGACTCTGAAGATG-3′ and 5′-GGAAGCACCAACGAGAGGAGAAAT-3′), Dim1 (5′-CATCGCCGAAAAGGTTAAAA-3′ and 5′-GGCCCAGTTGATCTTGTTGT-3′), integrin-β3 (5′-GAATGAATGCGCAGCACAGAGC-3′ and 5′-ACAGAGACTGGACCGAAACCAC-3′), Nfatc1 (5′-CTCGAAAGACAGCACTGGAGCAT-3′ and 5′-CGGCTGCCTTCCGTCTCATAG-3′), OSCAR (5′-CTGCTGGTAACGGATCAGCTCCCCAGA-3′ and 5′-CCAAGGAGCCAGAACCTTCGAAACT-3′), NF-κB p65 (5′-GGAGTTCCAGTACTTGCC-3′ and 5′-GTCCTTTTGCGCTTCTCT-3′), and GAPDH (5′-GGTCCTCAGTGTAGCCCAAG-3′ and 5′-AATGTGTCCGTCGTGGATCT-3′).
ChIP
Mock-depleted or Dim1-depleted BMM cells, either treated or not treated with RANKL, were cross-linked with 1% formaldehyde for 10 min and processed for ChIP. All samples were run in triplicate, and results were averaged. Sequences of the primers used for quantitative real time PCR are as follows: AP-1 binding site (5′-CCGGGACGCCCATGCAATCTGTTAGTAATT-3′ and 5′-GCGGGTGCCCTGAGAAAGCTACTCTCCCTT-3′) and distal region (5′-TCTGAGAGGGAGTGGCTGAT-3′ and 5′-CCTGGCTGGTTTGAGTTGAT-3′).
Accession Number
The NCBI GEO accession number for microarray data reported in this paper is GSE57468.
RESULTS
RANKL-induced Osteoclastogenesis Coincides with High Expression of Dim1 and NFATc1
Osteoclastogenesis is a complex process that reflects numerous changes in gene expression and cellular pathways. We used BMM cells to study the global transcription network during RANKL-induced osteoclastogenesis. Our initial TRAP staining confirmed that a significant number of multinucleated osteoclast-like cells were formed from BMM cells when cultured in the presence of soluble RANKL (Fig. 1A). To screen for altered gene expression during osteoclastogenesis, RANKL-treated BMM cells were subjected to gene expression profiling using the MouseRef-8 Expression microarrays (version 2.0). With a fold-change cutoff of >2.0, the expression profiling showed that 1073 genes were activated and 1070 genes were repressed (Fig. 1B and supplemental Table S1) in RANKL-treated cells compared with the untreated cells. Many of the genes up-regulated upon RANKL treatment are known to be involved in osteoclastogenic signaling events and osteoclast function (supplemental Table S2). Furthermore, our data showed that a group of genes including NFATc1 and its target genes that are essential for bone remodeling were differentially expressed in the RANKL-treated cells (supplemental Table S1).
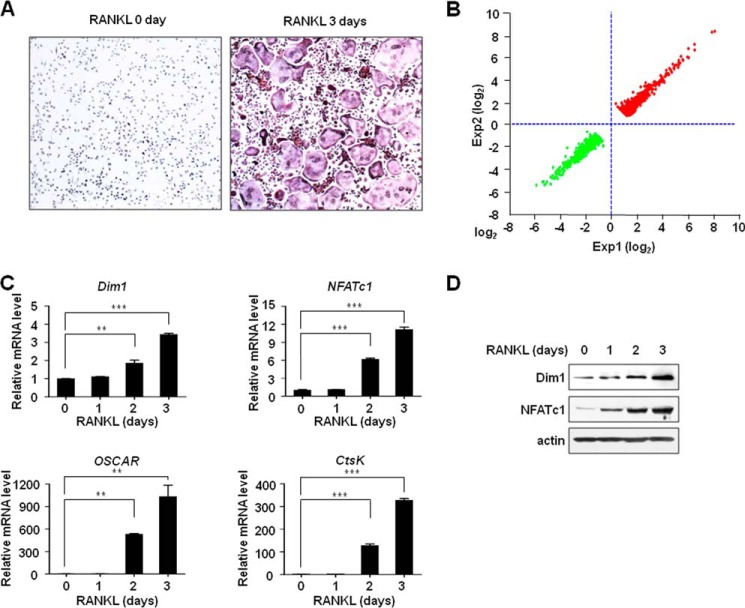
FIGURE 1.
Dim1 and NFATc1 activation in response to RANKL-induced osteoclastogenesis. A, osteoclastogenesis was induced by treating BMM cells with M-CSF (30 ng/ml) plus RANKL (50 ng/ml) for 3 days. Cells were fixed, stained for TRAP, and photographed under a light microscope. B, BMM cells were treated with RANKL for 0 and 3 days, and total RNA was extracted and analyzed by cDNA microarray. Genes that were up-regulated (1073 genes) or down-regulated (1070 genes) in RANLK-treated cells versus the day 0 control were selected (fold change > 2.0) and were sorted by fold changes. C, BMM cells were cultured for 0, 1, 2, and 3 days in the presence of M-CSF (30 ng/ml) and RANKL (50 ng/ml). Total RNA was isolated from cell lysates and analyzed by qRT-PCR. mRNA levels were normalized against an internal β-actin control for each time point, and values at the zero time point are set to 1. The results shown are mean values from three independent experiments for each time point. **, p < 0.01; ***, p < 0.001. D, whole cell lysates were prepared from M-CSF/RANKL-treated BMM cells as described in C and analyzed by immunoblot with Dim1 and NFATc1 antibodies. β-Actin was probed as a loading control.
Unexpectedly, our profiling data also indicated that Dim1 expression was up-regulated in RANKL-treated cells relative to its untreated controls (Table S1). Although Dim1 has been mainly characterized as an essential factor in the control of pre-RNA splicing in human cells, these results suggest that Dim1 may play a role in the regulation of osteoclastogenesis. To explore this possibility and validate the microarray data, we analyzed the expression of Dim1, NFATc1, and NFATc1 target genes, which showed the consistent response to RANKL treatment in the gene expression data, by qRT-PCR (Fig. 1C). The results correlated well with those obtained from the gene expression microarrays and demonstrated that RANKL-induced osteoclastogenesis is linked to transcriptional activation of these genes over a period of 3 days. In determining the protein levels of Dim1 and NFATc1 by immunoblot, we again found that RANKL-induced osteoclastogenesis is associated with the up-regulation of Dim1 and NFATc1 expression (Fig. 1D).
Dim1 Exerts Repressive Effects on NFATc1 and Its Target Genes during RANKL-induced Osteoclastogenesis
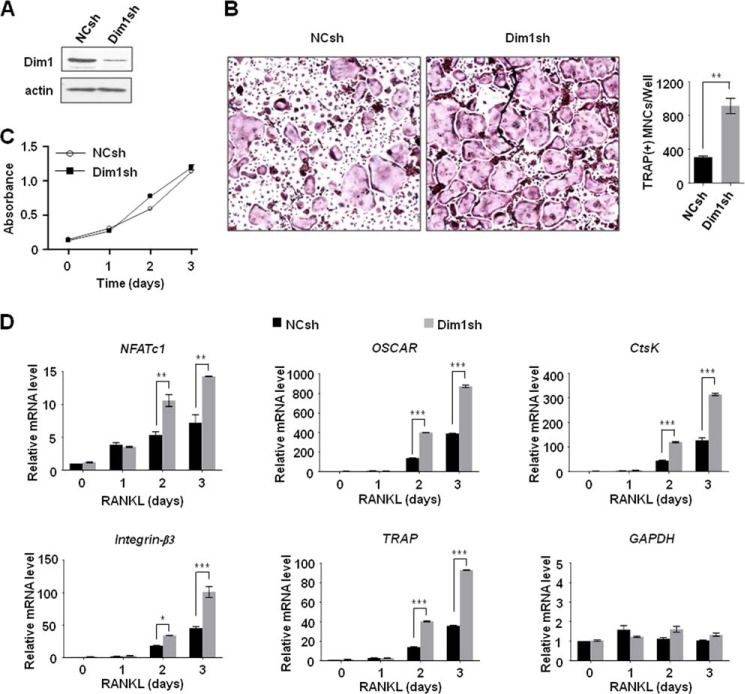
One of the key observations made in our gene expression profiling is that high-level expression of Dim1 coincides with transcriptional activation of NFATc1 and its target genes, which are critical for RANKL-induced osteoclastogenesis. To assess whether Dim1 directly influences osteoclastogenesis, we first depleted Dim1 in BMM cells and checked its effects on osteoclastic cell growth and the NFATc1 transcription pathway. Immunoblotting confirmed that infection of BMM cells with lentivirus expressing Dim1 shRNA efficiently silenced Dim1 expression (Fig. 2A). Because osteoclastogenesis enhances the expression of Dim1, we expected that Dim1 knockdown would negatively affect NFATc1 transactivation and thus RANKL-induced osteoclastogenesis. Contrary to these expectations, we found that osteoclast development was significantly accelerated in Dim1-depleted BMM cells in the presence of RANKL (Fig. 2B). Moreover, the finding that the proliferation of osteoclast precursors was not affected by Dim1 knockdown suggests that Dim1 inhibits the differentiation, not the proliferation, of pre-osteoclasts (Fig. 2C).
FIGURE 2.
Stimulatory effects of Dim1 knockdown on osteoclastogenesis and NFATc1 expression. A, BMM cells were infected with lentiviruses expressing either control shRNA (NC) or Dim1 shRNA for 3 days. The efficiency of depletion was determined by immunoblot with Dim1 antibody. β-Actin was used as a loading control. B, mock- or Dim1-depleted BMM cells were treated with M-CSF (30 ng/ml) and RANKL (50 ng/ml) for 3 days. Cells were fixed and stained for TRAP (left panel), and the number of TRAP-positive multinuclear cells were counted (right panel). **, p < 0.01 (Student's t test). C, mock- or Dim1-depleted BMM cells were treated with M-CSF for 3 days, and cell proliferation was measured by MTT assay. Results represent the mean ± S.D. of three experiments performed in triplicate over a 3-day period. D, mock- or Dim1-depleted BMM cells were treated with M-CSF and RANKL for 0, 1, 2, and 3 days. Total RNA was prepared from cells, and qRT-PCR was performed using primers specific for Oscar, Ctsk, integrin-β3, TRAP, and GAPDH genes. The mRNA levels from each reaction were normalized to β-actin control. The results shown are mean values from three independent experiments, and values derived from β-actin at the 0 time point in mock-transfected cells are arbitrarily set to 1. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
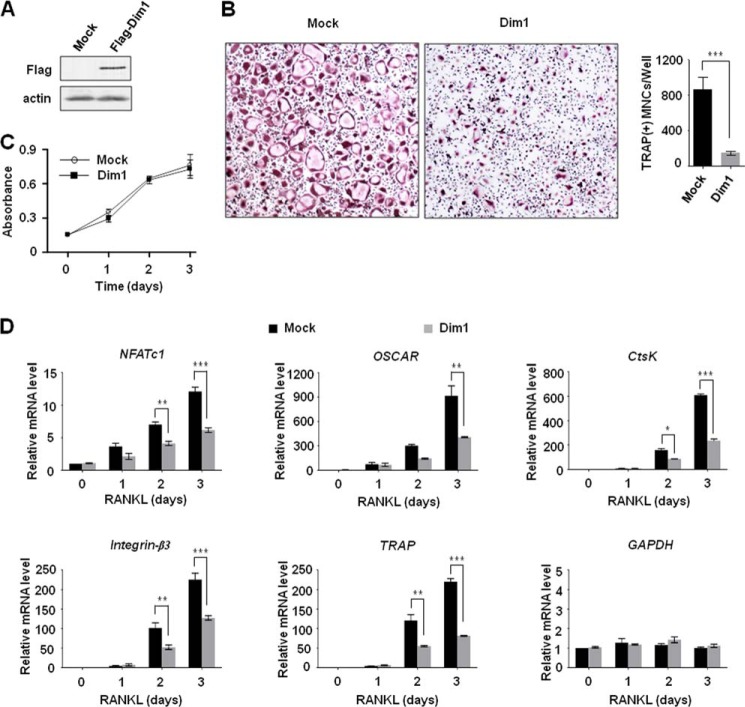
We next determined whether Dim1 would participate in regulating the expression of NFATc1 and its target genes in the context of RANKL-induced osteoclast differentiation by qRT-PCR. Consistent with the results of osteoclastogenesis assays in Fig. 2B, knockdown of Dim1 resulted in higher expression of NFATc1 over 3 days of RANKL treatment (Fig. 2D, NFATc1). Further analyses revealed a similar increase in the expression of the four NFATc1 target genes, which were identified from our gene expression profiling, in Dim1-depleted BMM cells during osteoclast differentiation (Fig. 2D). In consonance with our observations from knockdown experiments, ectopic expression of Dim1 suppressed RANKL-induced osteoclast formation in BMM cells by ∼20%, but did not affect the proliferation of BMM cells (Fig. 3, A–C). Expression of NFATc1 was also repressed by Dim1 expression, and the observed repression was paralleled by inactivation of the four NFATc1 target genes (Fig. 3D). These observations are consistent with the hypothesis that Dim1-mediated suppression of RANKL-induced osteoclastogenesis involves transcriptional alterations of NFATc1 and its target genes.
FIGURE 3.
Repressive effects of Dim1 expression on osteoclastogenesis and NFATc1 transactivation. A, BMM cells were infected with retroviruses expressing FLAG-Dim1 or control empty viruses for 3 days. Whole cell lysates from infected cells were analyzed by immunoblot for the expression of FLAG-Dim1. β-Actin was used as a loading control. B, Dim1 expression virus- or control virus-infected BMM cells were treated with M-CSF and RANKL for 3 days, and osteoclastogenesis was examined by TRAP staining (left) and counting the number of TRAP-positive multinuclear osteoclasts (right). **, p < 0.01 (Student's t test). C, BMM cells were infected with Dim1 or control viruses and treated with M-CSF, and the proliferative potential of the cells was measured by MTT assays at the indicated times. Data shown are the averages and S.D. of three independent experiments. D, Dim1 virus- or control virus-infected BMM cells were treated with M-CSF and RANKL for 0, 1, 2, and 3 days, and the expression levels of the four representative NFATc1 target genes (Ctsk, integrin-β3, Oscar, and Trap) were assessed by qRT-PCR as in Fig. 2D. Results represent the means ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Dim1 Down-regulates NFATc1 Expression by Antagonizing the Recruitment and Function of c-Fos
RANKL-induced transcriptional activation of the Nfatc1 gene during osteoclastogenesis is dependent on both c-Fos and NF-κB pathways (26–30). Previous studies identified several factors negatively regulating RANKL-induced osteoclastogenesis by interfering with the DNA-binding activities of c-Fos and NF-κB, thereby reducing the expression of NFATc1 (31, 32). Considering the transrepression activities of Dim1 toward NFATc1 expression, we were interested in investigating whether the observed function of Dim1 in attenuating osteoclastogenesis is the consequence of inhibiting the expression of these upstream signaling components. Therefore, we examined the relative levels of c-Fos and NF-κB p65 subunit in control and Dim1-depleted osteoclastic cells by qRT-PCR. Knocking down Dim1 led to a distinct increase in NFATc1 transcription during osteoclast formation (Fig. 4A, NFATc1), but transcription of c-Fos and NF-κB p65 was only weakly affected by Dim1 knockdown (c-fos and p65). Similarly, we did not see any obvious antagonistic regulation of c-fos and NF-κB p65 genes by Dim1 expression during RANKL-induced osteoclastogenesis (Fig. 4B). Consistent with these results, reporter gene assays showed that Dim1 selectively suppresses NFATc1 transcription without affecting the transcription of c-Fos and NF-κB p65 (Fig. 4C). This observation strongly suggests that it is the function of c-Fos/NF-κB, not their expression, which is disrupted by Dim1 for NFATc1 suppression. To check this possibility, we first sought to determine whether the impact of Dim1 on NFATc1 expression is due to its direct interaction with c-Fos and/or NF-κB. GST-fused Dim1 was immobilized to glutathione-Sepharose beads, and incubated with cell lysates of 293T cells expressing Mitf, c-Fos, or NF-κB p65. The same binding assays were performed with GST control. As illustrated in Fig. 5A, c-Fos efficiently interacted with Dim1, but Mitf and NF-κB p65 showed no detectable binding to Dim1. The observed interaction between Dim1 and c-Fos appears to be specific as GST alone showed no interaction with c-Fos. To further validate these in vitro binding results, extracts from RAW 264.7 cells infected with virus expressing FLAG-Dim1 were immunoprecipitated using anti-FLAG antibody. Consistent with the in vitro data, ectopic Dim1 was coimmunoprecipitated with endogenous c-Fos, but failed to show any coprecipitation of Mitf and NF-κB p65 (Fig. 5B).
FIGURE 4.
Minimal effects of Dim1 on the expression of c-Fos and NF-κB p65. A, total RNA was prepared from mock- or Dim1-depleted BMMs after treating with M-CSF and RANKL for 0, 1, 2, and 3 days. The mRNA levels of c-fos, NF-κB p65, and Nfatc1 genes were quantified by qRT-PCR. B, qRT-PCR was performed as described in A, but using total RNA isolated from Dim1 expression virus- or control virus-infected BMM cells. C, RAW 264.7 macrophage cells were transiently transfected with c-Fos, NF-κB p65, or NFATc1 reporter together with Dim1 expression vector. 24 h post-transfection, cells were treated with RANKL (50 ng/ml) for 16 h, and luciferase activity was measured using Dual-Luciferase assay kit (Promega). Each bar represents the means ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
FIGURE 5.
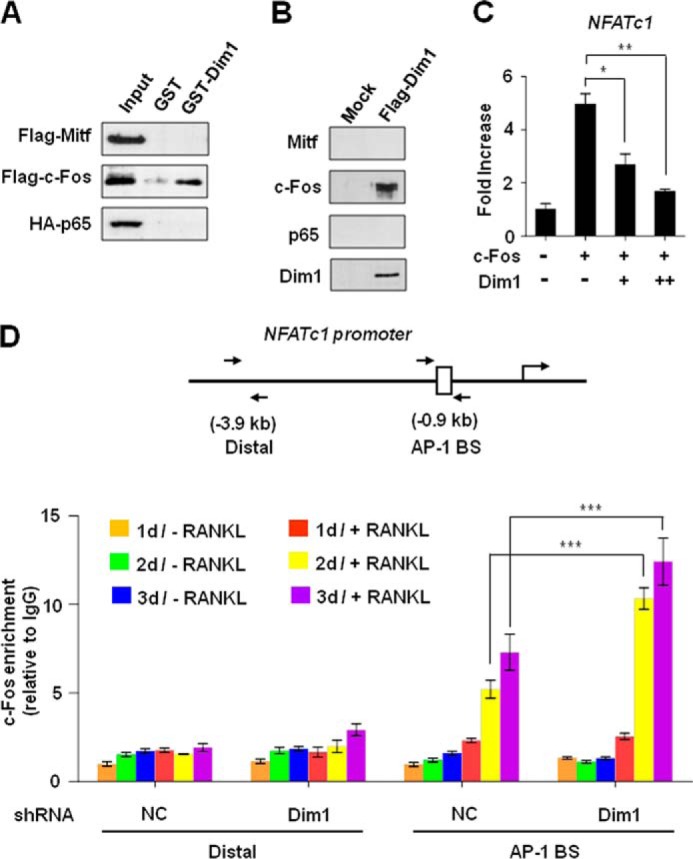
Blocking c-Fos recruitment to NFATc1 promoter by Dim1. A, FLAG-Mitf, FLAG-c-Fos, or HA-NF-κB p65 expression vectors were transfected into 293T cells. 2 days after transfection, cell lysates were prepared and incubated with GST or GST-Dim1 immobilized on glutathione beads. Bound proteins were resolved on 10% SDS-PAGE and analyzed by immunoblot with anti-FLAG or anti-HA antibody. Results shown are representative of at least three independent experiments. B, RAW 264.7 cells were infected with control or Dim1 expression viruses, and whole cell lysates were prepared and immunoprecipitated with anti-FLAG antibody. The precipitates were analyzed by immunoblot with the indicated antibodies. C, 293T cells were transiently co-transfected with the reporter plasmid NFATc1-Luc along with c-Fos and Dim1 expression vectors. Cells were assayed for luciferase activity 24 h after transfection. D, mock-depleted or Dim1-depleted BMMs were treated with or without RANKL for 1, 2, and 3 days, and ChIP assays of Nfatc1 gene were performed using c-Fos antibody. Precipitation efficiencies relative to non-enriched input samples were determined by quantitative real time PCR with primers specific for the AP-1 binding site and distal region. Each bar represents the means ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
In an effort to directly assess c-Fos-targeted repressive action of Dim1, we next tested whether Dim1 can inhibit transcriptional activation of transiently transfected reporter plasmids by c-Fos. The expression of the reporter gene bearing c-Fos binding sites was highly activated by transient expression of c-Fos. Because co-expression of Dim1 generated a substantial inactivation of c-Fos-mediated reporter gene transcription (Fig. 5C), our findings suggest that the repressive function of Dim1 is highly dependent on c-Fos. To gain further insight into the mechanism by which Dim1 modulates the expression of Nfatc1 gene, ChIP assays were performed in control and Dim1-depleted cells using c-Fos antibody. In control cells, a distinct localization of c-Fos at the proximal promoter of Nfatc1 gene was detected in response to RANKL stimulation (Fig. 5D). However, a clear increase in c-Fos occupancy was observed at the promoter region upon knocking down Dim1 by shRNA. Taken together, these findings support the view that Dim1 suppresses Nfatc1 gene transcription by affecting the recruitment of c-Fos to NFATc1 promoter, rather than affecting the expression of c-Fos, during osteoclastogenesis.
DISCUSSION
In the present study, we focused on potential roles of Dim1 in regulating the initial stage of the differentiation process in which quiescent osteoclast precursors are induced to become osteoclasts by RANKL stimulation. Our study indicates that Dim1 plays a critical role in controlling the expression of NFATc1, a pivotal component for osteoclast development, in osteoclast precursors by interfering with the transactivation function of c-Fos, which is an essential transcription factor for NFATc1 activation (26). Whereas Dim1 has no apparent effect on c-Fos expression, it seems to antagonize Nfatc1 gene transactivation through negatively regulating c-Fos localization at NFATc1 locus. This idea is supported by the fact that Dim1 knockdown caused an increase in c-Fos binding to the NFATc1 promoter. The strong correlation between the cellular levels of Dim1 and the efficiency of Nfatc1 gene silencing we have observed in Figs. 2 and 3 also fits well with the repression of NFATc1 target genes. This striking finding suggests that direct disruption of transcription integrity via the interaction between Dim1 and c-Fos is specific and significant in the regulation of bone remodeling. This is the first study indicating that Dim1 is a key molecule in determining the differentiation rate of osteoclasts as an upstream effector of RANKL signaling. Our study failed to show the binding of Dim1 to p65 subunit of NF-κB, suggesting that Dim1 is mainly targeting c-Fos to affect osteoclast differentiation. Our results, however, do not rule out the possibility that other signaling transduction pathways may also offer a molecular basis of Dim1-triggered repression of osteoclast differentiation. Thus, to better understand how much the Dim1-mediated regulatory mechanism contributes to osteoclast formation, it will be important to identify other osteoclast-specific genes whose transcription is particularly sensitive to Dim1 expression in future investigations.
Keeping the balance between bone formation and resorption is tightly regulated by various hormones and cytokines in local microenvironments. Aberrant regulation of osteoclast formation and function has been implicated in bone loss with concomitant suppression of bone growth and repair, which can weaken the skeleton and increase the risk of fracture. The controlled manipulation of osteoclastogenesis can be used to arrest such conditions, protect against accelerated bone loss, and preserve bone mass. The association of Dim1 with bone disorders has not been reported so far. Therefore, our present study raises the possibility that Dim1 could be a novel therapeutic tool for osteoclastogenic disorders because of its preferential expression on osteoclast cells as well as its significant inhibitory role in differentiation into mature osteoclasts. In light of this view, Dim1-mediated inactivation of NFATc1 expression described in this study will be very useful for the purpose of gaining further insight on the process to osteoclast formation and treating patients suffering from bone loss or skeletal complications of disease. In addition, osteoclast pathway is known to be regulated by a combination of gene specific and general transcription factors, which collectively maintain normal osteoclastic resorption within an acceptable level. In this respect, investigating how these multiple osteoclastic transcription factors functionally interact with Dim1 will increase our understanding of anti-osteoclastogenic function of Dim1.
In conclusion, we investigated the possible effects of Dim1 on osteoclastogenesis using defined experimental systems. Unexpectedly, our initial characterization of Dim1 demonstrated that Dim1 plays a critical role in controlling RANKL-induced osteoclast differentiation. We extended these findings by showing that c-Fos is the target for the observed function of Dim1 as an inhibitor of RANKL-induced osteoclastogenesis. Thus, our findings argue that the distinct interaction of Dim1 with c-Fos has a critical function in controlling RANKL-induced osteoclast differentiation. It remains a challenge to understand how Dim1 cooperates with other signaling factors that have been reported to be involved in osteoclast differentiation. Identification of additional factors as well as targets for Dim1 will definitely be required for deeper understanding of the molecular basis of disease associated with increased bone resorption.
Supplementary Material

This article contains supplemental Tables S1 and S2.
The NCBI GEO accession number for microarray data reported in this paper is GSE57468.
- RANKL
- receptor activator of NF-κB ligand
- Dim1
- defective entry into mitosis 1
- BM
- bone marrow
- BMM
- bone marrow-derived macrophage
- TRAP
- tartrate-resistant acid phosphatase
- MTT
- 3–4,5-(dimethyl-thyazol-2-yl)-2,5-diphenyltetrazolium
- qRT
- quantitative reverse transcription.
REFERENCES
- 1. Karsenty G., Wagner E. F. (2002) Reaching a genetic and molecular understanding of skeletal development. Dev. Cell 2, 389–406 [DOI] [PubMed] [Google Scholar]
- 2. Zaidi M. (2007) Skeletal remodeling in health and disease. Nat. Med. 13, 791–801 [DOI] [PubMed] [Google Scholar]
- 3. Zelzer E., Olsen B. R. (2003) The genetic basis for skeletal diseases. Nature 423, 343–348 [DOI] [PubMed] [Google Scholar]
- 4. Novack D. V., Teitelbaum S. L. (2008) The osteoclast: friend or foe? Annu. Rev. Pathol. 3, 457–484 [DOI] [PubMed] [Google Scholar]
- 5. Rodan G. A., Martin T. J. (2000) Therapeutic approaches to bone diseases. Science 289, 1508–1514 [DOI] [PubMed] [Google Scholar]
- 6. Tanaka S., Nakamura K., Takahasi N., Suda T. (2005) Role of RANKL in physiological and pathological bone resorption and therapeutics targeting the RANKL-RANK signaling system. Immunol. Rev. 208, 30–49 [DOI] [PubMed] [Google Scholar]
- 7. Baron R. (2004) Arming the osteoclast. Nat. Med. 10, 458–460 [DOI] [PubMed] [Google Scholar]
- 8. Nakashima T., Hayashi M., Fukunaga T., Kurata K., Oh-Hora M., Feng J. Q., Bonewald L. F., Kodama T., Wutz A., Wagner E. F., Penninger J. M., Takayanagi H. (2011) Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nat. Med. 17, 1231–1234 [DOI] [PubMed] [Google Scholar]
- 9. Lacey D. L., Timms E., Tan H. L., Kelley M. J., Dunstan C. R., Burgess T., Elliott R., Colombero A., Elliott G., Scully S., Hsu H., Sullivan J., Hawkins N., Davy E., Capparelli C., Eli A., Qian Y. X., Kaufman S., Sarosi I., Shalhoub V., Senaldi G., Guo J., Delaney J., Boyle W. J. (1998) Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93, 165–176 [DOI] [PubMed] [Google Scholar]
- 10. Yasuda H., Shima N., Nakagawa N., Yamaguchi K., Kinosaki M., Mochizuki S., Tomoyasu A., Yano K., Goto M., Murakami A., Tsuda E., Morinaga T., Higashio K., Udagawa N., Takahashi N., Suda T. (1998) Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc. Natl. Acad. Sci. U.S.A. 95, 3597–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Teitelbaum S. L. (2000) Bone resorption by osteoclasts. Science 289, 1504–1508 [DOI] [PubMed] [Google Scholar]
- 12. Asagiri M., Takayanagi H. (2007) The molecular understanding of osteoclast differentiation. Bone 40, 251–264 [DOI] [PubMed] [Google Scholar]
- 13. Gohda J., Akiyama T., Koga T., Takayanagi H., Tanaka S., Inoue J. (2005) RANK-mediated amplification of TRAF6 signaling leads to NFATc1 induction during osteoclastogenesis. EMBO J. 24, 790–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamashita T., Yao Z., Li F., Zhang Q., Badell I. R., Schwarz E. M., Takeshita S., Wagner E. F., Noda M., Matsuo K., Xing L., Boyce B. F. (2007) NF-κB p50 and p52 regulate receptor activator of NF-κB ligand (RANKL) and tumor necrosis factor-induced osteoclast precursor differentiation by activating c-Fos and NFATc1. J. Biol. Chem. 282, 18245–18253 [DOI] [PubMed] [Google Scholar]
- 15. Ishida N., Hayashi K., Hoshijima M., Ogawa T., Koga S., Miyatake Y., Kumegawa M., Kimura T., Takeya T. (2002) Large scale gene expression analysis of osteoclastogenesis in vitro and elucidation of NFAT2 as a key regulator. J. Biol. Chem. 277, 41147–41156 [DOI] [PubMed] [Google Scholar]
- 16. Yagi M., Miyamoto T., Sawatani Y., Iwamoto K., Hosogane N., Fujita N., Morita K., Ninomiya K., Suzuki T., Miyamoto K., Oike Y., Takeya M., Toyama Y., Suda T. (2005) DC-STAMP is essential for cell-cell fusion in osteoclasts and foreign body giant cells. J. Exp. Med. 202, 345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim Y., Sato K., Asagiri M., Morita I., Soma K., Takayanagi H. (2005) Contribution of nuclear factor of activated T cells c1 to the transcriptional control of immunoreceptor osteoclast-associated receptor but not triggering receptor expressed by myeloid cells-2 during osteoclastogenesis. J. Biol. Chem. 280, 32905–32913 [DOI] [PubMed] [Google Scholar]
- 18. Asagiri M., Sato K., Usami T., Ochi S., Nishina H., Yoshida H., Morita I., Wagner E. F., Mak T. W., Serfling E., Takayanagi H. (2005) Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J. Exp. Med. 202, 1261–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen S., Pan M. (2013) NFAT signaling and bone homeostasis. Hematol. Thromb. Dis. 10.4172/2329-8790.1000102 [DOI] [Google Scholar]
- 20. Berry L. D., Feoktistova A., Wright M. D., Gould K. L. (1999) The Schizosaccharomyces pombe dim1(+) gene interacts with the anaphase-promoting complex or cyclosome (APC/C) component lid1(+) and is required for APC/C function. Mol. Cell. Biol. 19, 2535–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reuter K., Nottrott S., Fabrizio P., Lührmann R., Ficner R. (1999) Identification, characterization and crystal structure analysis of the human spliceosomal U5 snRNP-specific 15 kD protein. J. Mol. Biol. 294, 515–525 [DOI] [PubMed] [Google Scholar]
- 22. Zhang Y. Z., Gould K. L., Dunbrack R. L., Cheng H., Roder H., Golemis E. A. (1999) The evolutionarily conserved Dim1 protein defines a novel branch of the thioredoxin fold superfamily. Physiol. Genomics 1, 109–118 [DOI] [PubMed] [Google Scholar]
- 23. Zhang Y., Lindblom T., Chang A., Sudol M., Sluder A. E., Golemis E. A. (2000) Evidence that dim1 associates with proteins involved in pre-mRNA splicing, and delineation of residues essential for dim1 interactions with hnRNP F and Npw38/PQBP-1. Gene. 257, 33–43 [DOI] [PubMed] [Google Scholar]
- 24. Simeoni F., Arvai A., Bello P., Gondeau C., Hopfner K. P., Neyroz P., Heitz F., Tainer J., Divita G. (2005) Biochemical characterization and crystal structure of a Dim1 family-associated protein: Dim2. Biochemistry 44, 11997–12008 [DOI] [PubMed] [Google Scholar]
- 25. Mehrabadi M., Hussain M., Asgari S. (2013) Cloning and characterization of a Dim1-like mitosis gene of Spodoptera frugiperda cells (Sf9) induced by Autographa californica multiple nucleopolyhedrovirus. J. Invertebr. Pathol. 113, 152–159 [DOI] [PubMed] [Google Scholar]
- 26. Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J., Wagner E. F., Mak T. W., Kodama T., Taniguchi T. (2002) Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell 3, 889–901 [DOI] [PubMed] [Google Scholar]
- 27. Nishikawa K., Nakashima T., Hayashi M., Fukunaga T., Kato S., Kodama T., Takahashi S., Calame K., Takayanagi H. (2010) Blimp1-mediated repression of negative regulators is required for osteoclast differentiation. Proc. Natl. Acad. Sci. U.S.A. 107, 3117–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao B., Takami M., Yamada A., Wang X., Koga T., Hu X., Tamura T., Ozato K., Choi Y., Ivashkiv L. B., Takayanagi H., Kamijo R. (2009) Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat. Med. 15, 1066–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matsuo K., Galson D. L., Zhao C., Peng L., Laplace C., Wang K. Z., Bachler M. A., Amano H., Aburatani H., Ishikawa H., Wagner E. F. (2004) Nuclear factor of activated T-cells (NFAT) rescues osteoclastogenesis in precursors lacking c-Fos. J. Biol. Chem. 279, 26475–26480 [DOI] [PubMed] [Google Scholar]
- 30. Vaira S., Alhawagri M., Anwisye I., Kitaura H., Faccio R., Novack D. V. (2008) RelA/p65 promotes osteoclast differentiation by blocking a RANKL-induced apoptotic JNK pathway in mice. J. Clin. Invest. 118, 2088–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim K., Kim J. H., Lee J., Jin H. M., Kook H., Kim K. K., Lee S. Y., Kim N. (2007) MafB negatively regulates RANKL-mediated osteoclast differentiation. Blood 109, 3253–3259 [DOI] [PubMed] [Google Scholar]
- 32. Zhao B., Ivashkiv L. B. (2011) Negative regulation of osteoclastogenesis and bone resorption by cytokines and transcriptional repressors. Arthritis Res. Ther. 13, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.