Background: Lipase maturation factor 1 (Lmf1) plays an important role in plasma lipid metabolism, but its regulation remains uncharacterized.
Results: Endoplasmic reticulum (ER) stress induces Lmf1 expression in cell lines and mouse liver. Atf6α deficiency abolishes, whereas active Atf6α stimulates this response.
Conclusion: Lmf1 is an unfolded protein response (UPR) target through Atf6α signaling.
Significance: Lmf1 regulation by the UPR suggests a possible role in ER homeostasis.
Keywords: Chaperone, Endoplasmic Reticulum (ER), Endoplasmic Reticulum Stress (ER Stress), Lipase, Unfolded Protein Response (UPR), Activating Transcription Factor 6, Lipase Maturation Factor 1
Abstract
Lipase maturation factor 1 (Lmf1) is a critical determinant of plasma lipid metabolism, as demonstrated by severe hypertriglyceridemia associated with its mutations in mice and human subjects. Lmf1 is a chaperone localized to the endoplasmic reticulum (ER) and required for the post-translational maturation and activation of several vascular lipases. Despite its importance in plasma lipid homeostasis, the regulation of Lmf1 remains unexplored. We report here that Lmf1 expression is induced by ER stress in various cell lines and in tunicamycin (TM)-injected mice. Using genetic deficiencies in mouse embryonic fibroblasts and mouse liver, we identified the Atf6α arm of the unfolded protein response as being responsible for the up-regulation of Lmf1 in ER stress. Experiments with luciferase reporter constructs indicated that ER stress activates the Lmf1 promoter through a GC-rich DNA sequence 264 bp upstream of the transcriptional start site. We demonstrated that Atf6α is sufficient to induce the Lmf1 promoter in the absence of ER stress, and this effect is mediated by the TM-responsive cis-regulatory element. Conversely, Atf6α deficiency induced by genetic ablation or a dominant-negative form of Atf6α abolished TM stimulation of the Lmf1 promoter. In conclusion, our results indicate that Lmf1 is an unfolded protein response target gene, and Atf6α signaling is sufficient and necessary for activation of the Lmf1 promoter. Importantly, the induction of Lmf1 by ER stress appears to be a general phenomenon not restricted to lipase-expressing cells, which suggests a lipase-independent cellular role for this protein in ER homeostasis.
Introduction
Lipoprotein lipase, hepatic lipase, and endothelial lipase are members of the vascular lipase protein family and are involved in plasma lipid metabolism (1). Through their lipolytic activities against triglycerides and phospholipids associated with lipoprotein particles in the circulation, these enzymes play critical roles in the regulation of plasma lipid levels, tissue lipid utilization, and cardiovascular disease risk.
The biosynthesis of vascular lipases takes place in the endoplasmic reticulum (ER)2 of parenchymal cells within lipase-expressing tissues such as adipose muscle, heart (2–4). The conversion of nascent lipase polypeptide chain into catalytically active enzyme requires post-translational maturation that involves glycosylation, glycan processing, and protein folding (5). The maturation of lipases is facilitated by both major chaperone systems operating within the ER, the calnexin/calreticulin and 78-kDa glucose-regulated protein (Grp78)/Grp94 systems (6, 7). In addition to general chaperones, lipase maturation critically depends on a client-specific chaperone residing in the ER membrane, lipase maturation factor 1 (Lmf1) (8). Lmf1 was first identified as the protein affected by a naturally occurring mutation in the mouse, combined lipase deficiency (cld) (9). Homozygous cld mutant mice suffer from massive hypertriglyceridemia and neonatal lethality owing to greatly reduced plasma lipoprotein lipase and hepatic lipase activities (10). Subsequent studies demonstrated that endothelial lipase activity is also diminished in the absence of Lmf1 (11). Although lipase proteins are expressed at normal levels in cld cells, they remain inactive, form high-molecular weight aggregates within the ER, and are degraded (12). Similar to cld mice, LMF1 deficiency causes abnormalities in lipid metabolism in humans as highlighted by the identification of loss-of-function mutations in patients with combined lipase deficiency and hypertriglyceridemia (8, 13).
Prompted by the dramatic hyperlipidemia phenotype in cld mutant mice, previous studies characterized Lmf1 exclusively in the context of lipase maturation. Nonetheless, Lmf1 is ubiquitously expressed in cells and tissues independent of the presence of lipases (8). Moreover, several naturally occurring splice variants of Lmf1 lack the domain that is critical for lipase maturation (14). Based on these observations, it has been hypothesized that in addition to its established function in the post-translational maturation of lipases, Lmf1 may play a wider role in ER homeostasis (15).
Homeostasis in the ER is maintained through the unfolded protein response (UPR), a network of signaling pathways coordinating the cellular response to perturbations in protein biosynthesis within the organelle (16). In response to ER stress, the UPR activates processes that reduce protein load within the ER through inhibition of protein translation and transcriptional activation of genes involved in protein folding and ER-associated protein degradation. When chronic or excessive ER stress cannot be resolved by these adaptive mechanisms, the UPR triggers apoptotic signaling and cell death (17).
UPR signaling is initiated by three signal transducers anchored in the ER membrane, protein kinase R-like ER kinase (Perk), inositol-requiring transmembrane kinase and endoribonuclease 1α (Ire1α), and activating transcription factor 6α (Atf6α) (16). Perk signaling leads to phosphorylation and inhibition of eukaryotic initiation factor 2α (eIF2α), resulting in transient translational attenuation of most mRNAs. In addition, the Perk pathway also coordinates transcriptional activation of genes related to protein biosynthesis and redox regulation through the up-regulation of the Atf4 and C/EBP homologous protein (Chop) transcription factors (18, 19). Activation of Ire1α promotes splicing of Xbp1 mRNA to produce a spliced Xbp1 transcript (sXbp1), which allows expression of a transcription factor involved in the up-regulation of ER-associated protein degradation genes (20). Upon ER stress, Atf6α is transported to the Golgi complex, where site-specific proteolytic cleavage generates a transcription factor competent for nuclear translocation (nAtf6α) (21). Within the nucleus, nAtf6α activates transcription through binding to ER stress response elements within promoters of chaperones and ER-associated protein degradation components (22, 23). Impaired protein folding, secretion, and degradation in Atf6α−/− cells and tissues demonstrate the importance of Atf6α signaling during ER stress (22).
The transcriptional regulation of Lmf1 expression has remained unexplored so far. In the present study, we address this issue and provide evidence that Lmf1 is a UPR target gene. We demonstrate that ER stress activation of Lmf1 requires Atf6α signaling in vitro and in vivo and identify a cis-acting element that mediates this effect within the Lmf1 promoter. Our results point to a conserved pathway linking ER stress to Lmf1 expression in diverse cell types, thus raising the possibility that Lmf1 has a more general function in ER homeostasis beyond its established role in the maturation of lipases.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Viability Assays
Immortalized Ire1α−/−, Perk−/− mouse embryonic fibroblasts (MEFs) with or without lentiviral reconstitution of Ire1α and Perk expression (24) were obtained from Dr. Fumihiko Urano (Washington University School of Medicine). Primary Atf6α−/− MEFs were generated as described (22, 24) and maintained in DMEM supplemented with 10% fetal bovine serum. To restore expression of Atf6α in Atf6α−/− primary MEF, cells were transiently transfected with a constitutively active nuclear form of Atf6α (nAtf6α) containing the N-terminal 373 amino acids of the protein (25) using the Amaxa Nucleofector protocol for MEF (Lonza) according to the manufacturer's instructions. 3T3-L1 fibroblasts were obtained from ATCC and maintained in DMEM supplemented with 10% calf serum and transfected with polyethylenimine. For a single well of a 48-well plate, 0.5 μg of plasmid DNA and 1 μl of 1 mg/ml polyethylenimine was preincubated for 20 min and added to cells in the absence of antibiotics. Six hours later, cells were washed with PBS, and complete medium was added. INS1–832/13 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum as described (26). Cell viability was assessed with the alamarBlue Reagent (Invitrogen) according to the manufacturer's instructions.
Real-time PCR
RNA and cDNA was prepared as described previously (39). Quantitative real-time PCR was performed using the QuantiTect SYBR Green PCR kit (Qiagen) on a 7500 real-time PCR System (Applied Biosystems). Relative mRNA values were calculated using the standard curve method. Expression of several housekeeping genes, including GAPDH, 36B4, actin, and Tbp, was typically assessed in experiments and the gene(s) showing the least variation among samples was used as an internal control for normalization. Primer sequences are available upon request.
Western Blotting
Cell lysates were prepared in homogenization buffer (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 1% Nonidet P-40), and particulate matter was removed by centrifugation at 20,000 × g for 10 min at 4 °C. Protein content was determined using the bicinchoninic assay (Pierce), and equal total protein amounts were separated on 7% Tris acetate SDS-polyacrylamide gels (Invitrogen). After electrotransfer, PVDF membranes were blocked with 5% BSA for 1 h, followed by overnight incubations with primary antibodies at 4 °C and secondary antibodies for 1 h. Lmf1 was detected with a polyclonal rabbit antibody raised against a C-terminal peptide and HRP-conjugated anti-rabbit IgG (Pierce). For normalization, hsp90 was detected with anti-hsp90 antibody (sc-7947, Santa Cruz Biotechnology) and HRP-conjugated anti-rabbit IgG. Immunoreactive bands were visualized using the West Femto chemiluminescent substrate (Pierce).
Mouse Experiments
Wild-type C57BL/6J and Atf6α−/− mice backcrossed (>10 generations) to C57BL/6J were maintained on standard laboratory chow in the University of Iowa specific pathogen-free facility on a 12:12 h light cycle. Four-month-old mice of both sexes were intraperitoneally injected with 1 mg/kg body weight of tunicamycin (TM), and livers were harvested for RNA isolation 48 h after injection. All animal experiments were approved by the Institutional Animal Care and Use Committees at Cedars-Sinai Medical Center and the University of Iowa.
Luciferase Reporter Assays
A bacterial artificial chromosome clone (RP24-180G21) carrying the mouse Lmf1 gene was used to PCR-amplify various fragments of the Lmf1 promoter with primers flanked by KpnI and XhoI restriction sites. Primer sequences are available upon request. Lmf1 promoter fragments were cloned between KpnI and XhoI sites of the pGL3-Basic promoterless firefly (FF) luciferase reporter plasmid (Promega). Point-mutations in promoter fragments were generated with the QuikChange site-directed mutagenesis kit (Stratagene). For luciferase assays, cells were plated in 48-well plates and transfected with a mixture of pGL3-promoter-FF and phRL(SV) control plasmid (Promega) expressing Renilla luciferase in a 100:1 ratio. Six hours after transfection, cells were treated with TM or thapsigargin for 16 h, and normalized FF luciferase activities were determined in cell lysates using the Dual-Luciferase reporter assay system and a GloMax-96 microplate luminometer (Promega). In some experiments, the transfection mixture also contained Atf6 expression vectors, or empty vector, at a ratio of pGL3-promoter-FF/phRL(SV)/Atf6 = 4:1.
Statistical Analysis
Results are shown as means ± S.D. or S.E. as indicated in figure legends. Statistical analyses were performed with SigmaPlot 11 software. Two-tailed unpaired Student's t test was used to compare two groups of data and one-way analysis of variance (ANOVA) followed by Tukey's test was applied for the analysis of multiple group comparisons. Differences were considered statistically significant at p values <0.05.
RESULTS
Lmf1 Is Induced by ER Stress
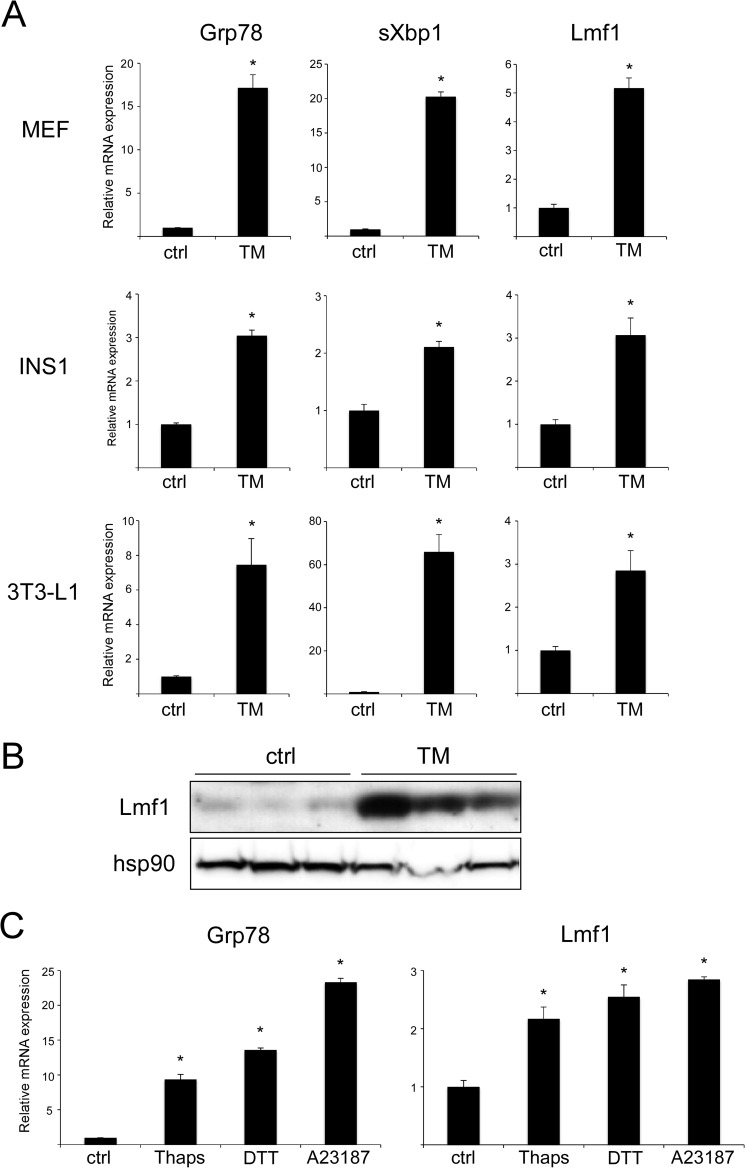
As Lmf1 functions as a chaperone within the ER, we investigated whether its expression is induced under conditions of ER stress. We triggered ER stress in MEFs with TM, a drug that inhibits N-glycosylation within the ER and induces the UPR. As expected, expression of the ER stress markers Grp78 and sXbp1 was robustly elevated after 16 h of treatment (Fig. 1A). Furthermore, TM treatment significantly increased (∼5-fold) Lmf1 transcript and protein levels (Fig. 1, A and B). The TM effect on Lmf1 expression was not limited to MEFs, as similar results were obtained in INS1 insulinoma cells and 3T3-L1 fibroblasts (Fig. 1A). To assess whether Lmf1 expression is affected by ER stress induced by other mechanisms, 3T3-L1 cells were treated with thapsigargin and A23187, which perturb ER Ca2+ homeostasis, and DTT, which alters ER redox state. All three treatments induced Grp78 as well as Lmf1 expression (Fig. 1C), indicating that Lmf1 is a target of the ER stress response program.
FIGURE 1.
Lmf1 expression is induced by ER stress. A, MEFs, INS1 cells and 3T3-L1 fibroblasts were exposed, respectively, to 0.5, 2.5, and 5 μg/ml TM for 16 h followed by real-time PCR analysis of Grp78, sXbp1, and Lmf1 gene expression. B, Western blot analysis of Lmf1 and hsp90 in 3T3-L1 fibroblasts exposed to 5 μg/ml TM for 24 h. C, real-time PCR analysis of Grp78 and Lmf1 expression in 3T3-L1 fibroblasts treated with 10 nm thapsigargin (Thaps), 5 mm DTT or 50 μm A23187 for 16 h. Results are expressed as mean ± S.D., n = 3 per group. *, p < 0.05 versus control (ctrl).
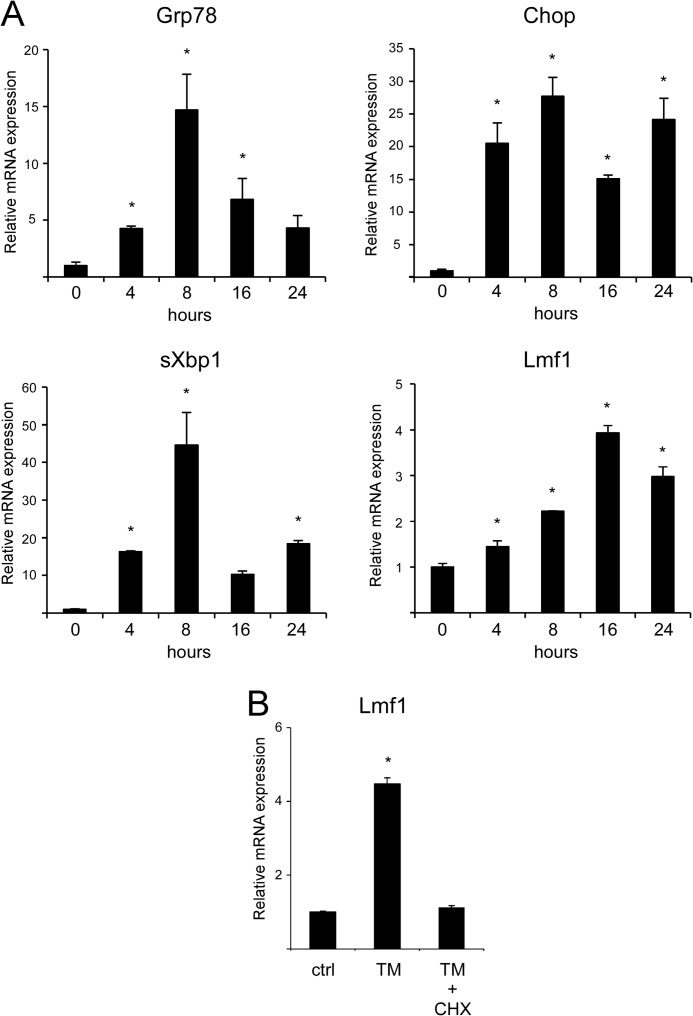
Next, we investigated the time course of Lmf1 induction by ER stress. The ER stress markers Grp78, Chop, and sXbp1 reached maximal expression within 8 h of TM exposure, after which their transcript levels decreased, consistent with previous reports (Fig. 2A) (27, 28). In contrast, the induction of Lmf1 occurred later with highest expression observed 16 h after TM treatment (Fig. 2A). Qualitatively similar results were obtained in INS1 cells, where Grp78, Chop, and sXbp1 were maximally induced after 4–8 h of TM treatment, whereas Lmf1 peaked at 16 h (data not shown). Delayed induction of Lmf1 relative to direct targets of UPR signaling suggested that the transcriptional response of Lmf1 may require new protein synthesis. Consistent with this idea, the translation inhibitor cycloheximide completely abolished TM-induced Lmf1 expression (Fig. 2B).
FIGURE 2.
Lmf1 induction occurs late during ER stress and depends on new protein synthesis. A, time course of ER stress-induced gene expression was analyzed in TM-treated (5 μg/ml) 3T3-L1 fibroblasts by real-time PCR. B, 3T3-L1 cells were treated with TM (5 μg/ml) in the presence and absence of 10 μg/ml cycloheximide (CHX) for 16 h followed by real-time PCR analysis of Lmf1 expression. Results are expressed as mean ± S.D., n = 3 per group. *, p < 0.05 versus control (ctrl).
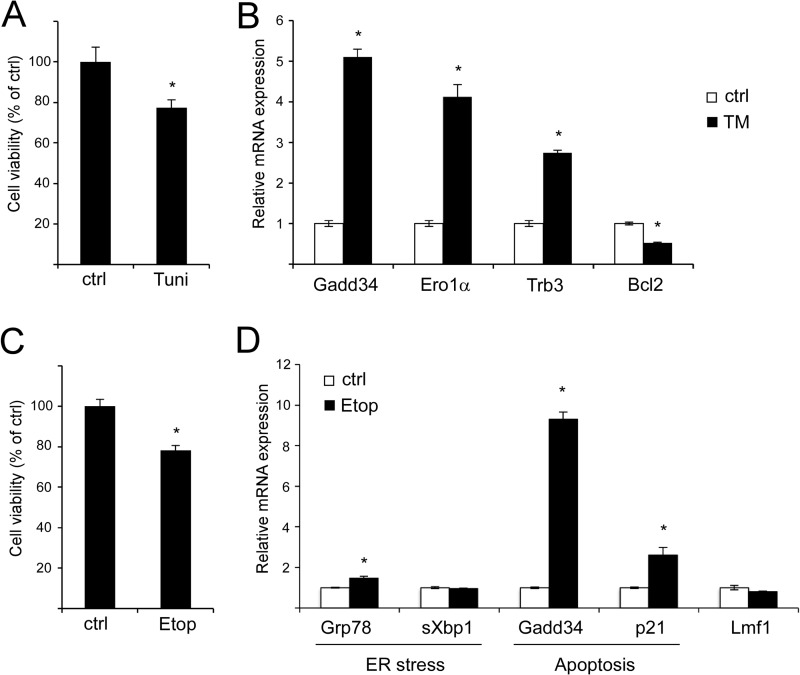
Prolonged and unmitigated ER stress leads to the activation of apoptotic signaling cascades and promotes apoptosis (29). Thus, the relatively high dose of TM (5 μg/ml) and long exposure (16 h) required for maximal Lmf1 induction raised the possibility that the apoptotic program may have been initiated at the time point of maximal Lmf1 expression. Indeed, after 16 h of TM treatment cell viability was reduced (Fig. 3A), and the expression of proapoptotic (Gadd34, Ero1α, Trb3) and anti-apoptotic (Bcl2) genes was elevated and diminished, respectively (Fig. 3B). To discriminate between ER stress versus apoptotic signaling as the underlying mechanism of Lmf1 induction, cells were treated with etoposide, a topoisomerase inhibitor known to trigger apoptosis through genotoxic stress. Etoposide treatment for 48 h resulted in reduced cell viability (Fig. 3C) and elevated expression of the proapoptotic markers the Gadd34 and p21 (Fig. 3D). As expected, etoposide failed to induce ER stress as indicated by only mildly elevated and unaffected expression of Grp78 and sXbp1, respectively (Fig. 3D). Importantly, Lmf1 expression remained unchanged under these conditions. Taken together, our results suggest that Lmf1 expression is induced by ER stress signaling through a pathway dependent on new protein synthesis.
FIGURE 3.
Prolonged TM treatment of 3T3-L1 cells induces cell death and apoptotic gene expression, but Lmf1 expression is unaffected by etoposide-induced apoptosis. A, confluent cell layers were exposed to 5 μg/ml TM or vehicle (control; ctrl), and cell viability was assessed by the analysis of total cellular protein 16 h later. B, pro- (Gadd34, Ero1α, Trb3) and anti-apoptotic (Bcl2) gene expression was determined after 16 h of TM treatment. C, confluent cell layers were exposed to 50 mm etoposide (Etop) or vehicle (control; ctrl), and cell viability was assessed by the analysis of total cellular protein 48 h later. D, expression of ER stress markers, proapoptotic genes, and Lmf1 was determined by real-time PCR analysis. Relative gene expression is normalized to vehicle-treated (control) samples, and results are expressed as mean ± S.D., n = 3 per group. *, p < 0.05 versus control (ctrl). Tuni, tunicamycin.
Atf6α Is Required for ER Stress-induced Lmf1 Expression
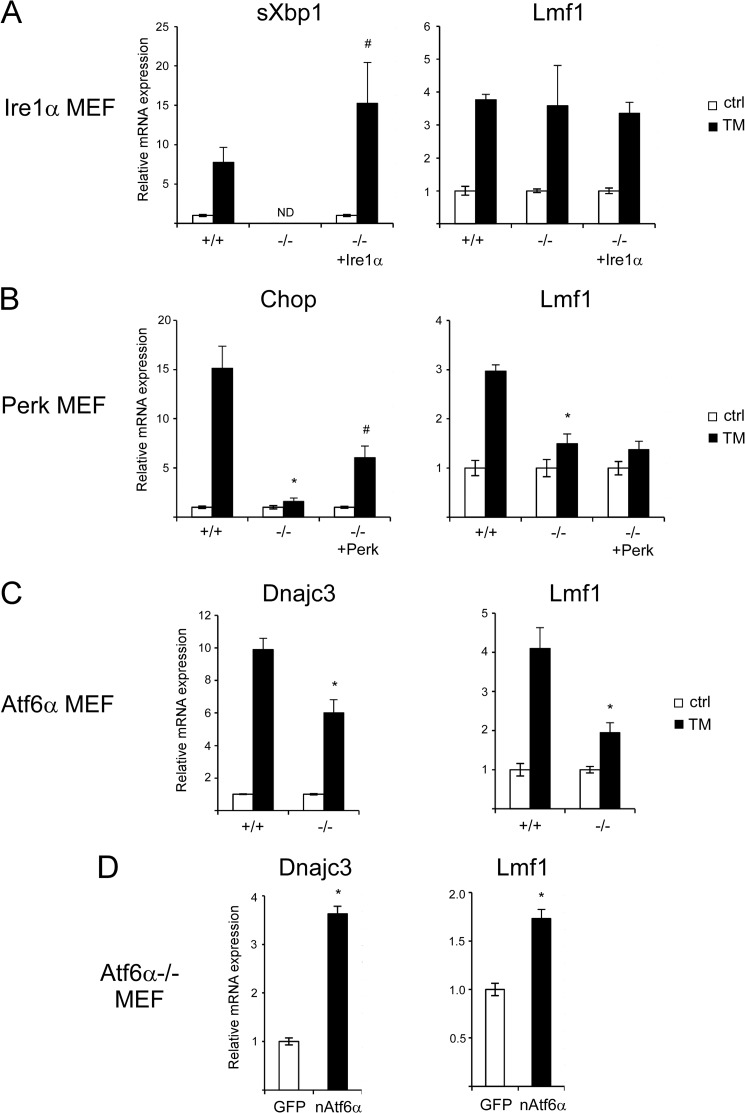
To identify the UPR signaling branch(es) mediating the effect of TM on Lmf1 expression, we characterized transcriptional responses in MEFs deficient in each of the three principal signal transducers. Consistent with the critical role of Ire1α in Xbp1 mRNA splicing, basal and TM-induced expression of sXbp1 was completely abolished in Ire1α−/− MEFs and restored after reconstitution of these cells with Ire1α (Fig. 4A, left panel). In contrast, Ire1α deficiency had no effect on Lmf1, indicating that the Ire1α signaling pathway is not involved in TM induction of Lmf1 (Fig. 4A, right panel).
FIGURE 4.
ER stress-induced Lmf1 expression is dependent on Atf6α. A, wild-type (+/+), Ire1α-deficient (−/−), and Ire1α-deficient MEFs transduced with lentivirus (LV) expressing Ire1α (−/− + Ire1α) were exposed to vehicle (control; ctrl) or 0.5 μg/ml TM, and gene expression was analyzed by real-time PCR 16 h later. B, wild-type (+/+), Perk-deficient (−/−), and Perk-deficient MEFs transduced with LV-Perk (−/− + Perk) were analyzed as described above. C, wild-type (+/+) and Atf6α-deficient (−/−) MEFs were analyzed as described in A. D, Atf6α-deficient MEFs were transfected with GFP or nuclear Atf6α (nAtf6α), and gene expression was analyzed 24 h later. Results are normalized to vehicle treatment (control; ctrl) of the same genotype (A–C) or GFP transfection (D) and expressed as mean ± S.D., n = 3 per group. *, p < 0.05 versus +/+ cells treated with TM (A–C) or GFP transfection (D); #, p < 0.05 versus −/− cells treated with TM. ND, not detectable.
As expected, TM-induced expression of Chop, a transcriptional target of the Perk signaling pathway (30), was greatly reduced in Perk−/− MEFs compared with wild-type cells and partially restored when Perk deficiency was rescued (Fig. 4B, left panel). Similarly, the induction of Lmf1 was diminished in Perk−/− MEFs, but re-expression of Perk had no effect (Fig. 4B, right panel). We interpret the reduced Lmf1 response observed in the Perk−/− cell line as an epiphenomenon unrelated to Perk deficiency and suggest that Perk signaling is unlikely to be involved in TM-induced regulation of Lmf1. Nonetheless, we cannot exclude the possibility that the Perk pathway may also contribute to Lmf1 expression.
To assess the role of Atf6α signaling on Lmf1 expression, Atf6α−/− and Atf6α+/+ primary MEFs were compared. Consistent with previous studies (22), TM-stimulated up-regulation of Dnajc3 was diminished in Atf6α−/− MEFs (Fig. 4C, left panel). The expression of Lmf1 was similarly affected with ∼50% suppression of TM-induction in Atf6α-deficient cells (Fig. 4C, right panel). Importantly, rescue with nuclear Atf6α (nAtf6α) increased both Dnajc3 and Lmf1 expression in Atf6α−/− cells (Fig. 4D). Based on these results, we conclude that TM-induction of Lmf1 is mediated, at least in part, by the Atf6α signaling pathway. Although Atf6α is activated through a post-translational mechanism (21), Lmf1 induction requires new protein synthesis (Fig. 2B), suggesting that the effect of Atf6α on Lmf1 expression is likely to be indirect.
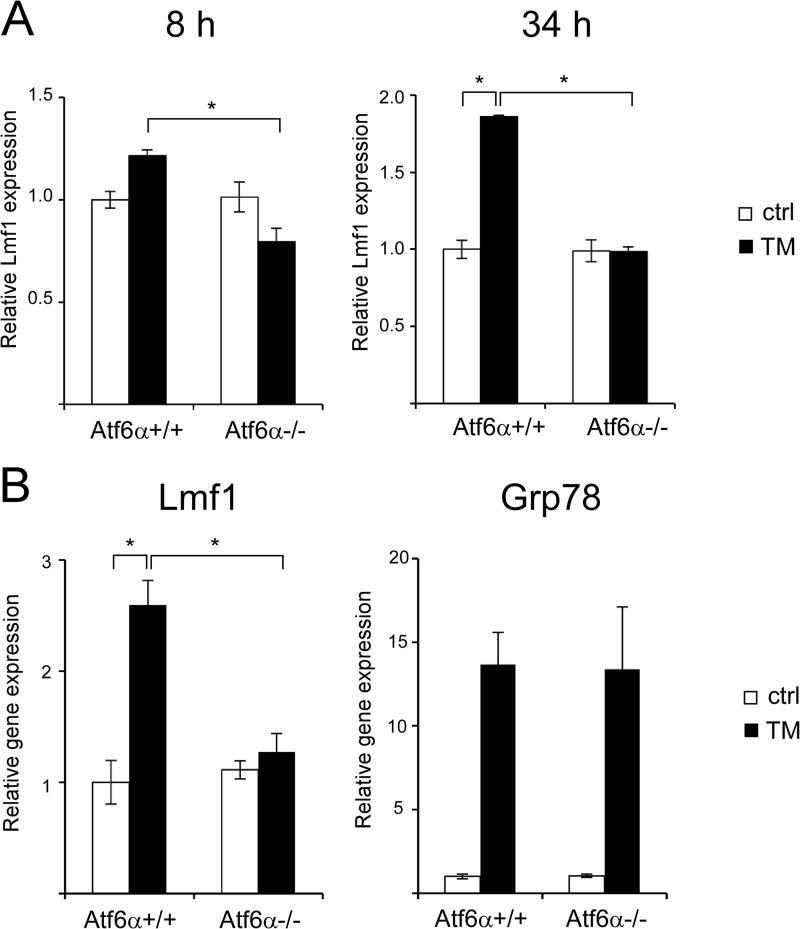
Lmf1 Is Induced in Vivo by ER Stress in an Atf6α-dependent Manner
To investigate the in vivo relevance of the above results, we analyzed mice injected with a sublethal dose (1 mg/kg body weight) of TM, an established model of hepatic ER stress (31–33). Using this model, we previously applied microarray analysis to document temporal changes in global gene expression patterns associated with ER stress in the liver of wild-type and Atf6α−/− mice (28, 34). Analysis of these data sets revealed that TM treatment led to modest but detectable induction of Lmf1 8 h after drug injection and produced a more robust increase a day later (Fig. 5A). Importantly, Atf6α deficiency abolished TM-induced Lmf1 expression at both time points. To confirm these results, we injected TM in an independent cohort of Atf6α+/+ and Atf6α−/− mice and analyzed hepatic Lmf1 expression by quantitative real-time PCR 48 h later. Consistent with earlier observations, Lmf1 expression was significantly elevated by TM treatment and this induction was completely abrogated in Atf6α−/− mice (Fig. 5B). In conclusion, these results demonstrate that Lmf1 is induced by ER stress in vivo and signaling through the Atf6α pathway is required for this effect.
FIGURE 5.
ER stress induces Lmf1 expression in vivo in an Atf6α-dependent manner. A, Atf6α+/+ and Atf6α−/− mice were intraperitoneally (i.p.) injected with 2 or 1 mg/kg TM or vehicle (control; ctrl), and total RNA was isolated from liver 8 or 34 h later, respectively. Lmf1 expression, based on microarray data from Refs. 28 and 34, is shown. B, Atf6α+/+ and Atf6α−/− mice were i.p.-injected with 1 mg/kg TM or vehicle (control; ctrl), and hepatic Lmf1 and Grp78 expression was analyzed 48 h later by real-time PCR. Relative gene expression is normalized to vehicle-treated wild-type samples, and results are expressed as mean ± S.E., n = 3 per group. *, p < 0.05 between indicated groups (ANOVA).
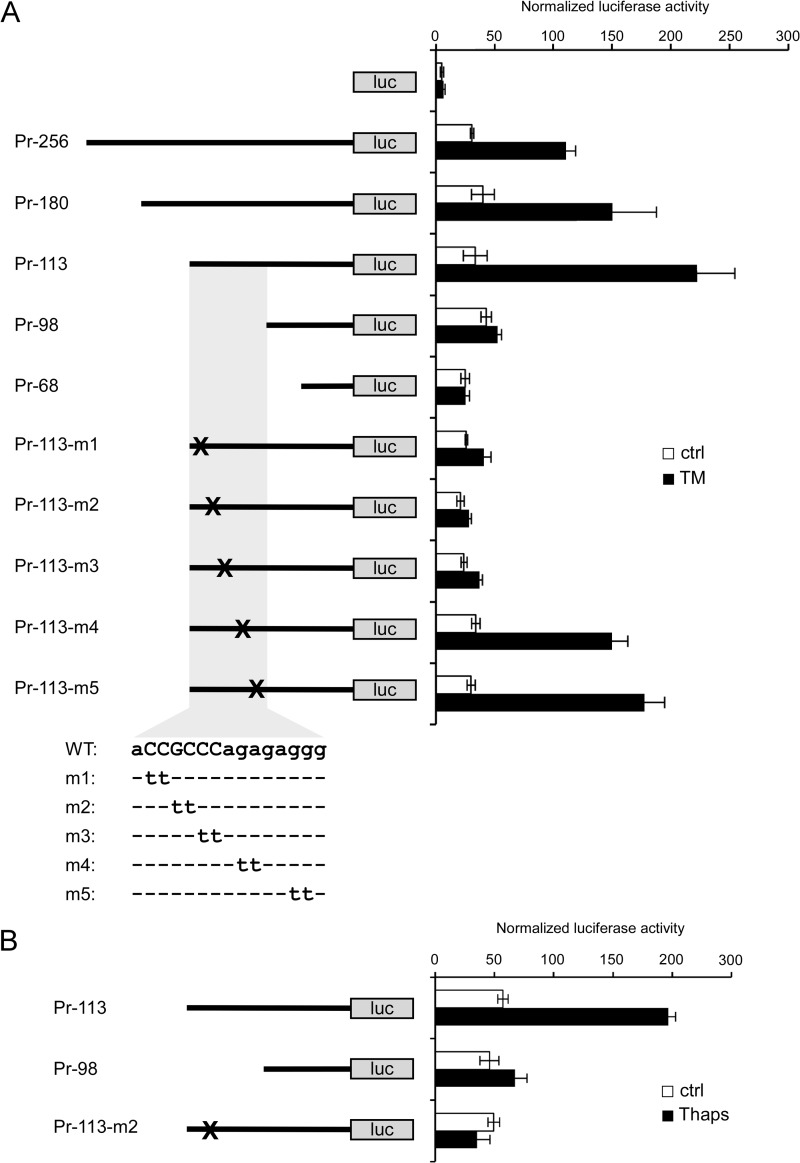
ER Stress Response Is Mediated by GC-rich Sequence in the Lmf1 Promoter
The broad effects of ER stress on the transcriptome are mediated through both transcriptional and post-transcriptional mechanisms (35). To investigate whether ER stress-induced Lmf1 expression is due to transcriptional changes mediated by the Lmf1 promoter, luciferase reporter constructs were generated and analyzed in 3T3-L1 fibroblasts. In untreated cells, a construct containing −267 to +39 of the promoter region (Pr-256) increased luciferase activity 6-fold relative to a promoterless plasmid indicating the presence of the Lmf1 core promoter in this region (Fig. 6A). Importantly, Pr-256 responded to TM treatment and exhibited ∼3-fold increased luciferase activity relative to untreated cells, an effect similar in magnitude to that observed on endogenous Lmf1 mRNA expression (Fig. 1A). This suggests that a transcriptional mechanism is largely responsible for ER stress-induced Lmf1 expression. To localize the cis-acting DNA element mediating the effect of TM on the promoter, we generated a series of 5′-terminal deletions from Pr-256. Analysis of these constructs localized the response element to the −113 to −96 region (Fig. 6A). Subsequent site-directed mutagenesis experiments revealed that mutations of the CCGCCC sequence in this region abolish TM induction of the promoter (Fig. 6A). To extend these results to ER stress triggered by perturbations in Ca2+ homeostasis, we performed similar experiments with thapsigargin. Consistent with the results of TM treatment, deletion or mutation of the GC box also abolished thapsigargin-responsiveness of the promoter (Fig. 6B). We conclude that ER stress-induced Lmf1 expression is mediated by a transcriptional mechanism through a GC box within the proximal promoter.
FIGURE 6.
ER stress induces Lmf1 promoter activity through GC-rich sequence. 3T3-L1 fibroblasts were transfected with Lmf1 promoter-luciferase constructs and treated with 5 μg/ml TM (A) or 10 nm thapsigargin (Thaps; B) for 16 h. Sequence of relevant promoter region (gray shading) is shown, and critical nucleotides are capitalized. Firefly luciferase (luc) activities were normalized by Renilla luciferase transfection control and are expressed as mean ± S.D.; n = 4 per group.
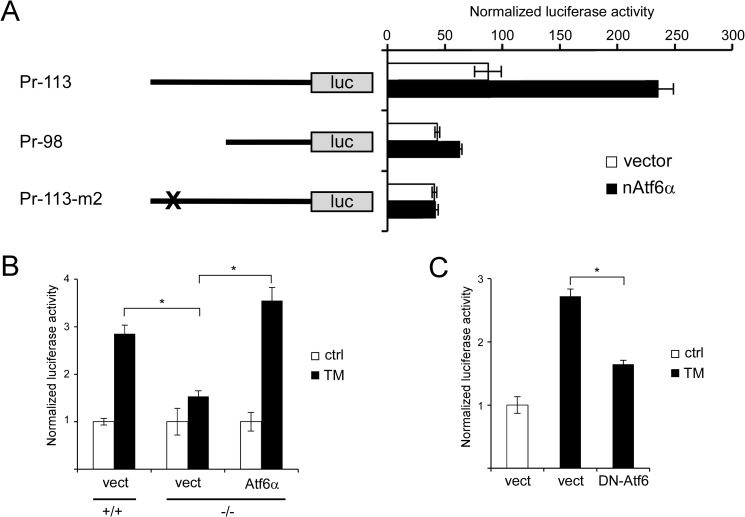
Atf6α Is Sufficient and Necessary for Lmf1 Promoter Activation
Our results in Atf6α-deficient MEFs and mice suggested that Atf6α was required for TM-induced expression of endogenous Lmf1 (Figs. 3 and 4). To investigate whether Atf6α signaling is involved in the activation of the Lmf1 promoter, we co-transfected nAtf6α with the Pr-113 luciferase construct in 3T3-L1 cells. Consistent with earlier results, nAtf6α up-regulated the Lmf1 proximal promoter (Fig. 7A). Moreover, this effect was abolished by deletion (Pr-98) or mutation (Pr-113-m2) of the GC-box involved in the TM-response (Fig. 7A). These results indicate that Atf6α signaling is sufficient for the transcriptional activation of Lmf1 and the effects of TM and Atf6α are mediated by the same cis-acting sequence within the Lmf1 promoter.
FIGURE 7.
Atf6α is sufficient and necessary for Lmf1 promoter activation. A, 3T3-L1 fibroblasts were co-transfected with Lmf1 promoter-luciferase constructs and vector or nAtf6α, and luciferase activity was measured 24 h later. B, wild-type (+/+) and Atf6α-deficient (−/−) MEFs were co-transfected with Pr-113 reporter construct and Atf6α or vector (vect) followed by treatment with 0.5 μg/ml TM. Luciferase activity was measured after 16 h exposure to TM. C, 3T3-L1 fibroblasts co-transfected with Pr-113 and dominant-negative Atf6 (DN-Atf6) or vector (vect) were treated with 5 μg/ml TM and assayed for luciferase (luc) activity 16 h later. Renilla luciferase-normalized firefly luciferase activities are shown. Results are expressed as mean ± S.D., n = 4 per group. *, p < 0.05 between indicated groups (ANOVA). ctrl, control.
To investigate whether Atf6α is required for TM-induction of the Lmf1 promoter, we performed loss-of-function experiments. Genetic deficiency of Atf6α in MEFs significantly reduced the effect of TM treatment on the Pr-113 reporter construct, and the TM response could be restored by reconstitution of Atf6α−/− cells with Atf6α (Fig. 7B). Furthermore, a dominant negative form of Atf6α (DN-Atf6) lacking the transactivation domain (25) diminished Lmf1 promoter activation by TM in 3T3-L1 cells (Fig. 7C). In conclusion, these results demonstrate that TM-induced activation of the Lmf1 promoter is mediated by Atf6α.
DISCUSSION
Although the role of Lmf1 in the posttranslational maturation of lipases is well established, the regulation of Lmf1 expression remains poorly characterized. We initially hypothesized that, as a posttranslational effector of lipase activities, Lmf1 may be regulated by metabolic cues. However, we and others (36) found that Lmf1 expression was unaffected by feeding status.3 The rationale for the present study was the hypothesis that Lmf1 regulation may be related to its function as an ER chaperone. Indeed, we demonstrate here that Lmf1 expression is induced by the UPR triggered by diverse mechanisms, including perturbed protein glycosylation, redox state, and Ca2+ signaling. Importantly, ER stress-induced Lmf1 expression was observed in diverse cell lines as well as in the liver of TM-injected mice. Thus, our results indicate that induction of Lmf1 gene expression is a general feature of the ER stress response in vitro and in vivo.
Induction of the UPR results in widespread transcriptional changes affecting hundreds of genes related to ER homeostasis, cell survival, and other physiological processes (22, 28, 32). In general terms, acute ER stress triggers early transcriptional changes that mitigate stress and promote survival, whereas prolonged and excessive stress leads to the activation of proapoptotic pathways in later stages of the UPR program. In this context, maximal induction of Lmf1 is a late event occurring 8–12 h after that of Grp78 and sXbp1 and requires relatively high concentrations of TM. Furthermore, peak Lmf1 expression coincides with high expression of Chop, a key transcriptional regulator of ER stress-induced apoptosis (17), and the induction of its proapoptotic (Gadd34, Trb3, Ero1α) and suppression of prosurvival (Bcl2) targets. Thus, Lmf1 is induced under proapoptotic conditions in TM-treated 3T3-L1 cells, which raises the question of whether Lmf1 is a bona fide target of the UPR or apoptotic signaling. To discriminate between these possibilities, we used etoposide-induced genotoxic stress to trigger apoptosis independent of ER stress. Under these conditions, Lmf1 expression remained unaffected, indicating that the intrinsic (i.e. mitochondrial) apoptosis pathway does not induce Lmf1. Collectively, our results suggest that UPR signaling is necessary for ER stress-induced regulation of Lmf1.
Having established the role of UPR in Lmf1 regulation, we demonstrated that the Atf6α signaling branch plays a major role in TM induction of Lmf1 both in vitro and in vivo. We localized the cis-acting element mediating the effect of ER stress on the Lmf1 promoter and demonstrated that Atf6α is necessary and sufficient for the induction of Lmf1 by TM through this DNA sequence. Although Atf6α is a transcription factor, multiple lines of evidence suggest that the involvement of Atf6α in the regulation of Lmf1 is indirect. First, the kinetics of TM-induced Lmf1 expression is significantly delayed compared with direct targets of Atf6α such as Dnajc3, Erdj3, and Erp72 (34). Moreover, whereas stress-induced activation of Atf6α is a posttranslational process, TM-induced Lmf1 expression depends on new protein synthesis, as demonstrated by the inhibitory effect of cycloheximide. Finally, the GC-rich sequence mediating the effect of TM within the Lmf1 promoter does not resemble the canonical Atf6α binding sites ERSE and ERSE-II (37). Indeed, interactions between Atf6 and the GC box could not be detected in electrophoretic mobility shift assays.3 Thus, the induction of Lmf1 expression during ER stress is likely to be mediated by a putative GC box-binding transcription factor whose activity is regulated by Atf6α. Using overexpression and knockdown approaches, we functionally evaluated several candidate GC motif-binding transcription factors previously implicated in UPR signaling, including Sp1, E2F1, and YY1 (38–40). However, we were unable to demonstrate modulation of the Lmf1 promoter through the TM-responsive GC box by any of the transcription factors tested in initial experiments (data not shown). Thus, further studies will be required to identify the molecular mechanisms involved in UPR-induced regulation of Lmf1.
In addition to its established role in proteostasis, UPR signaling has also been implicated in the regulation of lipid metabolism (41). Canonical signaling through Ire1α affects lipogenesis and very low-density lipoprotein (VLDL) secretion (42–44), whereas non-canonical UPR signaling involving the cAMP response element-binding protein, hepatocyte-specific regulates lipolysis through the transcriptional regulation of apolipoproteins that activate or inhibit lipoprotein lipase activity (45). As Lmf1 overexpression results in elevated tissue lipoprotein lipase activity (46), UPR-induced Lmf1 expression may represent a novel mechanism through which ER stress modulates lipolysis and tissue lipid uptake. However, our study demonstrates that ER stress-induced Lmf1 regulation also occurs in a lipase-independent cellular context. Indeed, fibroblasts used in our experiments are not known to express Lmf1-dependent lipases (47, 48). Thus, regulation of Lmf1 by the UPR appears to be a general cellular mechanism, which suggests a broader role for Lmf1 in ER homeostasis. For example, the chaperone function of Lmf1 may not be restricted to lipases and may affect a wider range of secretory proteins. In addition, Lmf1 may be involved in other ER-associated functions such as protein trafficking, Ca2+ homeostasis, or protein degradation. Consistent with these possibilities, Lmf1 exhibits a ubiquitous tissue expression pattern that is independent of lipases (8). Moreover, naturally occurring Lmf1 isoforms that lack the domain involved in lipase maturation have been identified (49). Thus, our results warrant future studies to explore the lipase-independent cellular functions of Lmf1.
In conclusion, we identified Lmf1 as a transcriptional target of the UPR and demonstrated a critical role for Atf6α signaling in ER stress-induced Lmf1 regulation. The present work represents the first study of Lmf1 in a lipase-independent context and raises the possibility that, in addition to its established function in lipase maturation, Lmf1 may have a novel role in ER homeostasis.
Acknowledgments
We thank C. Newgard for INS1-832/13 cells, F. Urano for Ire1α−/− and Perk−/− MEFs, and C. Glembotski for the DN-Atf6 construct.
This work was supported in part by National Institutes of Health Grants HL094709 (to I. N. M.), DK084058 (to D. T. R.), DK088227, DK042394, HL052173 (to R. J. K.), and HL28481 (to M. P.) and a University of Iowa Cardiovascular Center Institutional Training Grant (to D. D.-M.)
H. Z. Mao, N. Ehrhardt, C. Bedoya, J. A. Gomez, D. DeZwann-McCabe, I. N. Mungrue, R. J. Kaufman, D. T. Rutkowski, and M. Péterfy, unpublished observations.
- ER
- endoplasmic reticulum
- Atf6α
- activating transcription factor 6α
- nAtf6α
- constitutively active nuclear Atf6α
- Chop
- C/EBP homologous protein
- cld
- combined lipase deficiency mutation
- Grp78
- 78-kDa glucose-regulated protein
- Ire1α
- endoribonuclease 1α
- Lmf1
- lipase maturation factor 1
- MEF
- mouse embryonic fibroblast
- Perk
- protein kinase R-like ER kinase
- TM
- tunicamycin
- UPR
- unfolded protein response
- ANOVA
- one-way analysis of variance.
REFERENCES
- 1. Young S. G., Zechner R. (2013) Biochemistry and pathophysiology of intravascular and intracellular lipolysis. Genes Dev. 27, 459–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ben-Zeev O., Doolittle M. H. (2004) Maturation of hepatic lipase. Formation of functional enzyme in the endoplasmic reticulum is the rate-limiting step in its secretion. J. Biol. Chem. 279, 6171–6181 [DOI] [PubMed] [Google Scholar]
- 3. Ben-Zeev O., Doolittle M. H., Davis R. C., Elovson J., Schotz M. C. (1992) Maturation of lipoprotein lipase. Expression of full catalytic activity requires glucose trimming but not translocation to the cis-Golgi compartment. J. Biol. Chem. 267, 6219–6227 [PubMed] [Google Scholar]
- 4. Ben-Zeev O., Mao H. Z., Doolittle M. H. (2002) Maturation of lipoprotein lipase in the endoplasmic reticulum. Concurrent formation of functional dimers and inactive aggregates. J. Biol. Chem. 277, 10727–10738 [DOI] [PubMed] [Google Scholar]
- 5. Doolittle M. H., Péterfy M. (2010) Mechanisms of lipase maturation. Clin. Lipidol. 5, 71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meunier L., Usherwood Y. K., Chung K. T., Hendershot L. M. (2002) A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol. Biol. Cell 13, 4456–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Doolittle M. H., Ben-Zeev O., Bassilian S., Whitelegge J. P., Péterfy M., Wong H. (2009) Hepatic lipase maturation: a partial proteome of interacting factors. J. Lipid Res. 50, 1173–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Péterfy M., Ben-Zeev O., Mao H. Z., Weissglas-Volkov D., Aouizerat B. E., Pullinger C. R., Frost P. H., Kane J. P., Malloy M. J., Reue K., Pajukanta P., Doolittle M. H. (2007) Mutations in LMF1 cause combined lipase deficiency and severe hypertriglyceridemia. Nat. Genet. 39, 1483–1487 [DOI] [PubMed] [Google Scholar]
- 9. Paterniti J. R., Jr., Brown W. V., Ginsberg H. N., Artzt K. (1983) Combined lipase deficiency (cld): a lethal mutation on chromosome 17 of the mouse. Science 221, 167–169 [DOI] [PubMed] [Google Scholar]
- 10. Davis R. C., Ben-Zeev O., Martin D., Doolittle M. H. (1990) Combined lipase deficiency in the mouse. Evidence of impaired lipase processing and secretion. J. Biol. Chem. 265, 17960–17966 [PubMed] [Google Scholar]
- 11. Ben-Zeev O., Hosseini M., Lai C. M., Ehrhardt N., Wong H., Cefalù A. B., Noto D., Averna M. R., Doolittle M. H., Péterfy M. (2011) Lipase maturation factor 1 is required for endothelial lipase activity. J. Lipid Res. 52, 1162–1169 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12. Briquet-Laugier V., Ben-Zeev O., White A., Doolittle M. H. (1999) cld and lec23 are disparate mutations that affect maturation of lipoprotein lipase in the endoplasmic reticulum. J. Lipid Res. 40, 2044–2058 [PubMed] [Google Scholar]
- 13. Cefalù A. B., Noto D., Arpi M. L., Yin F., Spina R., Hilden H., Barbagallo C. M., Carroccio A., Tarugi P., Squatrito S., Vigneri R., Taskinen M. R., Péterfy M., Averna M. R. (2009) Novel LMF1 nonsense mutation in a patient with severe hypertriglyceridemia. J. Clin. Endocrinol. Metab. 94, 4584–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Doolittle M. H., Neher S. B., Ben-Zeev O., Ling-Liao J., Gallagher C. M., Hosseini M., Yin F., Wong H., Walter P., Péterfy M. (2009) Lipase maturation factor LMF1, membrane topology and interaction with lipase proteins in the endoplasmic reticulum. J. Biol. Chem. 284, 33623–33633 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15. Péterfy M. (2012) Lipase maturation factor 1: A lipase chaperone involved in lipid metabolism. Biochim. Biophys. Acta 1821, 790–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ron D., Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 17. Tabas I., Ron D. (2011) Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 13, 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Harding H. P., Zhang Y., Zeng H., Novoa I., Lu P. D., Calfon M., Sadri N., Yun C., Popko B., Paules R., Stojdl D. F., Bell J. C., Hettmann T., Leiden J. M., Ron D. (2003) An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 11, 619–633 [DOI] [PubMed] [Google Scholar]
- 19. Han J., Back S. H., Hur J., Lin Y. H., Gildersleeve R., Shan J., Yuan C. L., Krokowski D., Wang S., Hatzoglou M., Kilberg M. S., Sartor M. A., Kaufman R. J. (2013) ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee A. H., Iwakoshi N. N., Glimcher L. H. (2003) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell Biol. 23, 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haze K., Yoshida H., Yanagi H., Yura T., Mori K. (1999) Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10, 3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu J., Rutkowski D. T., Dubois M., Swathirajan J., Saunders T., Wang J., Song B., Yau G. D., Kaufman R. J. (2007) ATF6α optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev. Cell 13, 351–364 [DOI] [PubMed] [Google Scholar]
- 23. Adachi Y., Yamamoto K., Okada T., Yoshida H., Harada A., Mori K. (2008) ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct. Funct. 33, 75–89 [DOI] [PubMed] [Google Scholar]
- 24. Ghosh R., Lipson K. L., Sargent K. E., Mercurio A. M., Hunt J. S., Ron D., Urano F. (2010) Transcriptional regulation of VEGF-A by the unfolded protein response pathway. PloS One 5, e9575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thuerauf D. J., Morrison L., Glembotski C. C. (2004) Opposing roles for ATF6α and ATF6β in endoplasmic reticulum stress response gene induction. J. Biol. Chem. 279, 21078–21084 [DOI] [PubMed] [Google Scholar]
- 26. Hohmeier H. E., Mulder H., Chen G., Henkel-Rieger R., Prentki M., Newgard C. B. (2000) Isolation of INS-1-derived cell lines with robust ATP-sensitive K+ channel-dependent and -independent glucose-stimulated insulin secretion. Diabetes 49, 424–430 [DOI] [PubMed] [Google Scholar]
- 27. Lin J. H., Li H., Yasumura D., Cohen H. R., Zhang C., Panning B., Shokat K. M., Lavail M. M., Walter P. (2007) IRE1 signaling affects cell fate during the unfolded protein response. Science 318, 944–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rutkowski D. T., Wu J., Back S. H., Callaghan M. U., Ferris S. P., Iqbal J., Clark R., Miao H., Hassler J. R., Fornek J., Katze M. G., Hussain M. M., Song B., Swathirajan J., Wang J., Yau G. D., Kaufman R. J. (2008) UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev. Cell 15, 829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu C., Bailly-Maitre B., Reed J. C. (2005) Endoplasmic reticulum stress: cell life and death decisions. J. Clin. Invest. 115, 2656–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 31. Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R. T., Remotti H., Stevens J. L., Ron D. (1998) CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 12, 982–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marciniak S. J., Yun C. Y., Oyadomari S., Novoa I., Zhang Y., Jungreis R., Nagata K., Harding H. P., Ron D. (2004) CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 18, 3066–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hetz C., Bernasconi P., Fisher J., Lee A. H., Bassik M. C., Antonsson B., Brandt G. S., Iwakoshi N. N., Schinzel A., Glimcher L. H., Korsmeyer S. J. (2006) Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1α. Science 312, 572–576 [DOI] [PubMed] [Google Scholar]
- 34. Arensdorf A. M., Dezwaan McCabe D., Kaufman R. J., Rutkowski D. T. (2013) Temporal clustering of gene expression links the metabolic transcription factor HNF4α to the ER stress-dependent gene regulatory network. Front. Genet. 4, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Arensdorf A. M., Diedrichs D., Rutkowski D. T. (2013) Regulation of the transcriptome by ER stress: non-canonical mechanisms and physiological consequences. Front. Genet. 4, 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kroupa O., Vorrsjö E., Stienstra R., Mattijssen F., Nilsson S. K., Sukonina V., Kersten S., Olivecrona G., Olivecrona T. (2012) Linking nutritional regulation of Angptl4, Gpihbp1, and Lmf1 to lipoprotein lipase activity in rodent adipose tissue. BMC Physiol. 12, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yamamoto K., Yoshida H., Kokame K., Kaufman R. J., Mori K. (2004) Differential contributions of ATF6 and XBP1 to the activation of endoplasmic reticulum stress-responsive cis-acting elements ERSE, UPRE and ERSE-II. J. Biochem. 136, 343–350 [DOI] [PubMed] [Google Scholar]
- 38. Abdelrahim M., Liu S., Safe S. (2005) Induction of endoplasmic reticulum-induced stress genes in Panc-1 pancreatic cancer cells is dependent on Sp proteins. J. Biol. Chem. 280, 16508–16513 [DOI] [PubMed] [Google Scholar]
- 39. Racek T., Buhlmann S., Rüst F., Knoll S., Alla V., Pützer B. M. (2008) Transcriptional repression of the prosurvival endoplasmic reticulum chaperone GRP78/BIP by E2F1. J. Biol. Chem. 283, 34305–34314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baumeister P., Luo S., Skarnes W. C., Sui G., Seto E., Shi Y., Lee A. S. (2005) Endoplasmic reticulum stress induction of the Grp78/BiP promoter: activating mechanisms mediated by YY1 and its interactive chromatin modifiers. Mol. Cell Biol. 25, 4529–4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang S., Kaufman R. J. (2014) How does protein misfolding in the endoplasmic reticulum affect lipid metabolism in the liver? Curr. Opin. Lipidol. 25, 125–132 [DOI] [PubMed] [Google Scholar]
- 42. Lee A. H., Scapa E. F., Cohen D. E., Glimcher L. H. (2008) Regulation of hepatic lipogenesis by the transcription factor XBP1. Science 320, 1492–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. So J. S., Hur K. Y., Tarrio M., Ruda V., Frank-Kamenetsky M., Fitzgerald K., Koteliansky V., Lichtman A. H., Iwawaki T., Glimcher L. H., Lee A. H. (2012) Silencing of lipid metabolism genes through IRE1α-mediated mRNA decay lowers plasma lipids in mice. Cell Metab. 16, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang S., Chen Z., Lam V., Han J., Hassler J., Finck B. N., Davidson N. O., Kaufman R. J. (2012) IRE1α-XBP1s induces PDI expression to increase MTP activity for hepatic VLDL assembly and lipid homeostasis. Cell Metab. 16, 473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang C., Wang G., Zheng Z., Maddipati K. R., Zhang X., Dyson G., Williams P., Duncan S. A., Kaufman R. J., Zhang K. (2012) Endoplasmic reticulum-tethered transcription factor cAMP responsive element-binding protein, hepatocyte specific, regulates hepatic lipogenesis, fatty acid oxidation, and lipolysis upon metabolic stress in mice. Hepatology 55, 1070–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hosseini M., Ehrhardt N., Weissglas-Volkov D., Lai C. M., Mao H. Z., Liao J. L., Nikkola E., Bensadoun A., Taskinen M. R., Doolittle M. H., Pajukanta P., Péterfy M. (2012) Transgenic expression and genetic variation of Lmf1 affect LPL activity in mice and humans. Arterioscler. Thromb. Vasc. Biol. 32, 1204–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cornelius P., MacDougald O. A., Lane M. D. (1994) Regulation of adipocyte development. Annu. Rev. Nutr. 14, 99–129 [DOI] [PubMed] [Google Scholar]
- 48. Hirata K., Dichek H. L., Cioffi J. A., Choi S. Y., Leeper N. J., Quintana L., Kronmal G. S., Cooper A. D., Quertermous T. (1999) Cloning of a unique lipase from endothelial cells extends the lipase gene family. J. Biol. Chem. 274, 14170–14175 [DOI] [PubMed] [Google Scholar]
- 49. Doolittle M. H., Ehrhardt N., Péterfy M. (2010) Lipase maturation factor 1: structure and role in lipase folding and assembly. Curr. Opin. Lipidol. 21, 198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]