Background: Phytochromes are dimeric bili-proteins central to photoperception by plants and microorganisms.
Results: An informative perspective on photoconversion from the dark-adapted to the photoactivated state was provided by crystallographic and electron microscopic analyses of a bacterial version.
Conclusion: Light-induced conformational changes in the bilin induce a large scale reorientation of the sister output modules.
Significance: The structures offer a model for signal transmission by phytochromes.
Keywords: Electron Microscopy (EM), Photomorphogenesis, Photoreceptor, Phototransduction, Plant Biochemistry, Protein Structure, X-ray Crystallography, Bilin, Photoconversion, Phytochrome
Abstract
Phytochromes are multidomain photoswitches that drive light perception in plants and microorganisms by coupling photoreversible isomerization of their bilin chromophore to various signaling cascades. How changes in bilin conformation affect output by these photoreceptors remains poorly resolved and might include several species-specific routes. Here, we present detailed three-dimensional models of the photosensing module and a picture of an entire dimeric photoreceptor through structural analysis of the Deinococcus radiodurans phytochrome BphP assembled with biliverdin (BV). A 1.16-Å resolution crystal structure of the bilin-binding pocket in the dark-adapted red light-absorbing state illuminated the intricate network of bilin/protein/water interactions and confirmed the protonation and ZZZssa conformation of BV. Structural and spectroscopic comparisons with the photochemically compromised D207A mutant revealed that substitutions of Asp-207 allow inclusion of cyclic porphyrins in addition to BV. A crystal structure of the entire photosensing module showed a head-to-head, twisted dimeric arrangement with bowed helical spines and a hairpin protrusion connecting the cGMP phosphodiesterase/adenylyl cyclase/FhlA (GAF) and phytochrome-specific (PHY) domains. A key conserved hairpin feature is its anti-parallel, two β-strand stem, which we show by mutagenesis to be critical for BphP photochemistry. Comparisons of single particle electron microscopic images of the full-length BphP dimer in the red light-absorbing state and the photoactivated far-red light-absorbing state revealed a large scale reorientation of the PHY domain relative to the GAF domain, which alters the position of the downstream histidine kinase output module. Together, our data support a toggle model whereby bilin photoisomerization alters GAF/PHY domain interactions through conformational modification of the hairpin, which regulates signaling by impacting the relationship between sister output modules.
Introduction
The phytochrome (Phy)3 superfamily encompasses a structurally diverse collection of photochromic photoreceptors in plants and microorganisms that use a bilin (or open-chain tetrapyrrole) chromophore for light detection (1–3). They typically exist in two forms, a dark-adapted red light-absorbing state (Pr) that is biologically inactive and a metastable far-red light-absorbing state (Pfr) that is biologically active. The ability to reversibly photointerconvert between Pr and Pfr with red and far-red light, respectively, and revert thermally from Pfr to Pr allows Phys to act as photoswitches in various signaling cascades capable of measuring light quantity, duration, and spectral quality. In plants, Phys are the major regulators of plant morphogenesis, including a number of agronomically important processes such as seed germination, chloroplast development, flowering time, and shade avoidance (4). In microorganisms, Phys have been connected to pigmentation, phototaxis, sporulation, and the chromatic acclimation of the photosynthetic machinery to the available light spectrum (1, 5, 6).
Phys are homodimeric with each polypeptide containing an N-terminal photosensing module (PSM) that houses the bilin followed by a C-terminal output module (OPM) that transmits a chemical or conformational signal upon light activation of the photoreceptor (2, 3). In canonical Phys, the PSM sequentially contains a Period/Arnt/Single-minded (PAS) domain of unknown function, a signature cGMP phosphodiesterase/adenylyl cyclase/FhlA (GAF) domain that cradles the bilin through numerous electrostatic and hydrophobic interactions, and a Phy-specific (PHY) domain that stabilizes the photoactivated Pfr state and presumably relays the light signal to the downstream OPM.
Using a lyase activity intrinsic to the GAF domain, the bilin is bound covalently via a thioether linkage between the C3 side chain of the A pyrrole ring and a conserved cysteine present either upstream of the PAS domain (proteobacteria and fungi (7, 8)) or within the GAF domain (cyanobacteria and plants (9–12)). Whereas proteobacterial and fungal Phys use the immediate breakdown product of heme, biliverdin IXα (BV), as their chromophore, cyanobacterial and plant Phys use the more reduced bilins, phycocyanobilin and phytochromobilin, respectively. Photoconversion of Pr to Pfr appears to involve a 15Z to 15E isomerization of the C15=C16 double bond connecting the C and D pyrrole rings that flips the D ring (13, 14), but structural and photochemical studies indicate that other routes might exist for some Phy subtypes (15–17). The OPM arrangement most commonly employed includes a histidine kinase (HK) or related domain, implying that these Phys work in two-component phosphotransfer cascades, but domains with predicted adenylyl cyclase, diguanylate cyclase, guanylate phosphatase, methyl-chemotaxis accepting, and methyl binding activities have also been found, demonstrating that the Phy system has been co-opted to control a variety of signaling outcomes (1, 18, 19).
In addition to canonical Phys, numerous proteobacteria and cyanobacteria express variants with novel architectures and photochemical properties. These include Phys that use Pfr as the dark-adapted state (bathy-Phys), thus requiring far-red light for photoactivation, Phys lacking the PAS domain (PAS-less), and Phys missing the PAS and PHY domains with the GAF domain sometimes arranged in repeated units, each with its own photochromic bilin (1, 3, 20, 21). This diversity is particularly evident in cyanobacteria, where a collection of Phy variants called cyanobacteriochromes are expressed with remarkably diverse end states, including violet/blue, violet/orange, blue/green, green/red, and red/green light-absorbing pairs (6, 22).
How photoisomerization of the bilin impinges on the Phy protein to ultimately impact signaling is poorly understood. To address this question, a number of crystallographic and solution NMR three-dimensional structures have been generated for the GAF domain from cyanobacteriochromes and PAS-less Phys (15, 23–26), and PSM fragments from canonical and bathy-Phys (10, 11, 19, 21, 27–30). Structures of the PSM in the dark-adapted state revealed a multidomain arrangement with the sister GAF domains most often providing contacts for a head-to-head dimer architecture. Notably, the PAS and GAF domains are tightly tethered by a novel figure-of-eight knot created by a loop extending from the GAF domain that lassos the N-terminal sequence upstream from the PAS domain (27). The GAF and PHY domains are in turn linked by a long helical spine (29, 31) and further connected noncovalently by an unusual hairpin (or tongue) feature that projects from the PHY domain to make specific electrostatic and hydrophobic contacts with the GAF domain surface near the chromophore (10, 11, 19, 21, 29). The bilin as Pr assumes a 5Zsyn, 10Zsyn, 15Zanti (ZZZssa) configuration within the GAF domain crevice. It is tightly fixed by an intricate network of conserved contacts, including electrostatic interactions between the B and C ring side-chain propionates and a collection of arginines, serines, and/or lysines, and a hydrogen bond lattice involving the A-C pyrrole ring nitrogens, a central pyrrole water, and neighboring His and Asp residues, the latter of which sits within a centrally positioned, invariant Asp-Ile-Pro (DIP) motif (32). Mutagenic studies have revealed the importance of many of these interactions to bilin incorporation and the photochemical properties of Phys (11, 15, 21, 25, 29, 33).
By comparing the structures of canonical Phys with representative bathy-Phys and cyanobacteriochromes (11, 19, 21), the development of paired solution NMR structures of GAF domains in their dark-adapted and photoactivated states (15, 23) and temperature-scanning crystallography (14), a “toggle” model for the initial photoconversion events has emerged (11, 21, 31). Following the light-induced flip of the D pyrrole ring, the bilin slides within the GAF domain crevice to induce a cascade of bilin/protein and protein/protein contact alterations that ultimately melts the connection between the PHY-domain hairpin and the GAF-domain surface proximal to the chromophore, including one involving the DIP aspartate. The hairpin β-strand stem swivels and assumes a helical conformation, which then reassociates with the GAF domain by a different complement of contacts to presumably alter the distance between and/or orientation of the PHY domain relative to the GAF domain. Ultimately, these changes impact the relative arrangement of the paired OPM effector domains in the Phy dimer.
Whereas current three-dimensional models and mutagenic studies support the conformational changes at the GAF/PHY domain interface (11, 19, 21, 30, 34), structural information is currently lacking to support light-induced rearrangements of the sister OPMs. To provide such support, we present here a collection of structural images for the bacterial Phy BphP from Deinococcus radiodurans with a canonical PAS-GAF-PHY domain PSM followed by a HK domain OPM (35). Included are high resolution views of the bilin and its protein/water contacts for wild-type DrBphP and its photochemically compromised D207A variant (33), and a well resolved view of the entire PSM in the Pr state, all generated by x-ray crystallography. Using these structures for orientation, we also developed single particle EM (SPEM) images of the full-length DrBphP dimer in both the Pr and Pfr states. Comparison of these images together with a recent low resolution crystal structure of the DrBphP PSM as a Pr/Pfr mixture (30) support a model whereby changes in hairpin conformation reorients the sister OPMs by large scale repositioning of the connecting PHY domains. Given the conservation of the participating features among Phys, it is likely that this toggle mechanism is relevant to others within the superfamily, especially those with canonical PAS-GAF-PHY-OPM modular arrangements.
EXPERIMENTAL PROCEDURES
DrBphP Protein Expression and Purification
D. radiodurans BphP apoproteins bearing an N-terminal T7 tag (MASMTGGQQMGRGS) and a C-terminal hexahistidine tag were expressed in Escherichia coli BL21(DE3) cells using the pET21b plasmid (Novagen, Madison, WI) as a full-length form (775 amino acids), encompassing just the PSM (residues 1–501) or encompassing just the PAS-GAF domain region (residues 1–321 and including the Y307S mutation (32)) as described (27, 33). Site-directed mutations were introduced using the QuikChange method (Stratagene, La Jolla, CA) and appropriate primers (33). Cultures were grown at 25 °C in terrific broth and harvested by centrifugation 4 h after inducing apoprotein expression with isopropyl β-d-1-thiogalactopyranoside. The cell pellets were frozen in liquid nitrogen and stored at −80 °C.
Frozen pellets were resuspended in lysis buffer (10% glycerol, 20 mm HEPES-NaOH (pH 7.8), 500 mm NaCl, 0.05% Tween 20, 30 mm imidazole, 1 mm phenylmethanesulfonyl fluoride, and 1 mm 2-mercaptoethanol), sonicated, and clarified by centrifugation. The apoproteins were enriched from the clarified lysates by nickel-nitrilotriacetic acid (Qiagen, Valencia, CA) chromatography, using the lysis buffer for washing and lysis buffer supplemented with 300 mm imidazole for elution. The DrBphP-containing fractions were incubated overnight with a molar excess of BV (Frontiers Scientific, Logan, UT), and the free BV was then removed by filtration through a 50-ml G-25 Sephadex column (GE Healthcare) equilibrated in the ion-exchange buffer (10% glycerol, 20 mm HEPES-NaOH (pH 7.8), 20 mm NaCl, and 10 mm 2-mercaptoethanol). The chromoproteins were further purified by ion-exchange chromatography using a Q-Sepharose HP column (GE Healthcare) equilibrated in ion-exchange buffer and employing a 20–1000 mm linear NaCl gradient for elution. DrBphP-containing fractions were exchanged into 50 mm HEPES-KOH (pH 7.8) and 150 mm KCl at 25 °C using a 50-ml G-25 Sephadex column (GE Healthcare), frozen as 30-μl droplets in liquid nitrogen, and stored at −80 °C. Covalent binding of BV to the DrBphP apoproteins was monitored by zinc-induced fluorescence of the chromoproteins following SDS-PAGE (33).
DrBphP Crystallography
Crystallization trials employed the sitting drop vapor diffusion method. Crystallization environments for the PAS-GAF fragment alone or in combination with the D207A mutation were identified by optimizing conditions employed previously for the PAS-GAF fragment (32). The final crystallization condition for the PAS-GAF chromoprotein was 10% poly(ethylene)glycol (PEG) 3350, 19% isopropyl alcohol, 5% glycerol, and 100 mm citric acid/NaOH (pH 5.6), and for the PAS-GAF(D207A) chromoprotein was 19% PEG 3350, 19% isopropyl alcohol, 5% glycerol, and 100 mm citric acid/NaOH (pH 5.6). Crystals for both PAS-GAF chromoproteins were flash-cooled in liquid nitrogen in the latter buffer. Crystallization conditions for the DrBphP PSM construction were identified using the Hampton index screen (Hampton Research, Aliso Viejo, CA) in combination with various additives (e.g. ethylene glycol, glycerol, and isopropyl alcohol). The optimized condition was 18% PEG 3350, 5% tacsimate at pH 6.5 (Hampton Research), 2% ethylene glycol, and 100 mm PIPES-NaOH (pH 6.5). Before flash-cooling in liquid nitrogen, crystals were soaked in a buffer with the same components, except that the tacsimate and ethylene glycol concentrations were increased to 6 and 23%, respectively.
X-ray Crystallography and Structure Refinement
X-ray diffraction datasets for the DrBphP PSM and the PAS-GAF chromoproteins were collected at the Advanced Photon Source GM/CA Collaborative Access Team 23-ID-D and the LS Collaborative Access Team 21-ID-F beamlines at Argonne National Laboratory, whereas the diffraction datasets for the D207A mutant were collected at the Protein Crystallography Research Resource at the National Synchrotron Light Source X29 beamline at Brookhaven National Laboratory. All datasets were indexed, integrated, and scaled using HKL2000 (36). Initial phases were calculated by PHASER (37) using the 1.45-Å DrBphP PAS-GAF structure (PDB ID code 2O9C) generated by Wagner et al. (32) as the search model. Manual model building was conducted with COOT (38), and structures were refined with PHENIX (37). Model validation was conducted with MOLPROBITY (39), and superpositions were calculated with LSQKAB (40). Structural figures were prepared with the PyMOL Molecular Graphics System.
SPEM of DrBphP
Negative-staining EM visualization employed full-length DrBphP fixed onto carbon films. The films were prepared by evaporating a thin layer of carbon onto a piece of freshly cleaved mica using an Edwards (Sanborn, NY) vacuum evaporator (< 10−5 torr). The carbon was peeled off by floating the mica on the surface of deionized water, and the resulting film was then deposited onto 300 mesh copper grids. The carbon-coated grids were glow discharged in a 100-millitorr argon atmosphere for 1 min. We prepared EM grids for Pr (4–5 μl samples) under dim green safelight. Excess solution was removed by blotting the edge of the grids with filter paper, and the grids were washed with water followed by staining twice with 4 μl of 2% (w/v) phosphotungstic acid (pH 7.4) for 30 s, and then air-dried. To prepare EM grids for the Pfr state, protein samples were illuminated with red light to photoequilibrium and then stained while continuing the red light irradiation at low fluence until the grids were completely dry. Placement of the DrBphP samples onto the grids and subsequent fixation took ∼3 min for each sample.
The negatively stained EM grids were imaged in a JEOL JEM-2010F transmission electron microscope (JEOL Ltd., Tokyo, Japan) operating at 200 kV. Micrographs were recorded in a low dose mode (15 e−/Å2) at ×50,000 microscope magnification in a Gatan (Pleasanton, CA) UltraScan 4000 CCD camera (4096 × 4096 pixels), which corresponded to 2.12-Å/pixel sampling at the specimen level. Particle selection and image processing were conducted using the EMAN (41) and EMAN2 software packages (42). Raw particle images were selected in a semi-automatic manner with e2boxer.py in EMAN2. The selected particles were manually inspected to remove “bad” particles that were partially disassembled, having low contrast, and/or contacting other particles.
For three-dimensional reconstructions of DrBphP in the Pr state, the contrast transfer function was first determined with raw images and corrected by flipping the phases in EMAN2. Reference-free two-dimensional classifications of the phase-flipped data sets were developed by e2refine2d.py. The resulting averaged images were then used to generate a starting model by e2initialmodel.py. The model was low-pass filtered and used for further three-dimensional refinement against phase-flipped particle images (∼18,500) by e2refine.py. Three-dimensional reconstruction of DrBphP in the photoactivated state used a dataset of ∼12,000 particles. Because two distinct types of averaged particles were observed, one with all expected domains present and the other apparently missing the HK domains, we used e2refinemulti.py to refine two models. One model (Pfr) included 7200 particles, and the other model (Pfr′) included 4700 particles. Finally, the two models were refined independently against their corresponding data sets by e2refine_easy.py. All image processing and three-dimensional reconstructions were done on an 8-CPU Dell Linux workstation. The resolutions of the final three-dimensional maps were estimated by the gold-standard Fourier shell correlation method at a threshold of 0.143. Surface rendering of the three-dimensional density maps and atomic model docking were performed in Chimera (43).
Spectroscopic and Thermal Reversion Measurements
Samples utilized for the acquisition of Pr and Pfr absorption spectra, and Pfr → Pr thermal reversion were dissolved in 150 mm KCl and 50 mm HEPES-KOH (pH 7.8) that was titrated to pH at the temperature of interest. Spectra were recorded using a Cary60 spectrophotometer (Agilent Technologies, Santa Clara, CA). Red light was provided by a 660-nm peak output LED (Digi-Key, Thief River Falls, MN) combined with a 660-nm interference filter (10-nm half-bandwidth). Absorption spectra of DrBphP polypeptides assembled in vitro with BV or protoporphyrin IXα (PPIX) (Sigma) followed by denaturation were recorded after dissolution in 8 m urea (pH 2.0). Fluorescence excitation and emission spectra were recorded using a QuantaMaster Model C-60/2000 spectrofluorimeter (Photon Technologies International, Edison, NJ).
Protein Sequence Alignments
Amino acid sequence alignments were generated by Clustal Omega under the default settings and displayed with BioEdit.
RESULTS
DrBphP Constructions
Based on the utility of DrBphP for the structural analysis of Phys and for defining their photoconversion mechanism through an array of biophysical approaches (27, 31–33, 44, 45), we further exploited this Phy to better understand how conformational changes in the chromophore are transduced from the PSM to the downstream OPM. Our ultimate goal was to generate well resolved three-dimensional images of the Pr and Pfr states to confirm and extend the toggle model for photoconversion (11, 21, 31). These studies involved full-length dimeric DrBphP and its PSM (residues 1–501) and PAS-GAF (residues 1–321) fragments expressed recombinantly with N-terminal T7 and C-terminal hexahistidine tags, purified, and assembled with BV in vitro. The PAS-GAF fragment also included the Y307A mutation that was shown previously to aid crystallization by eliminating a surface conflict at the dimerization interface (32).
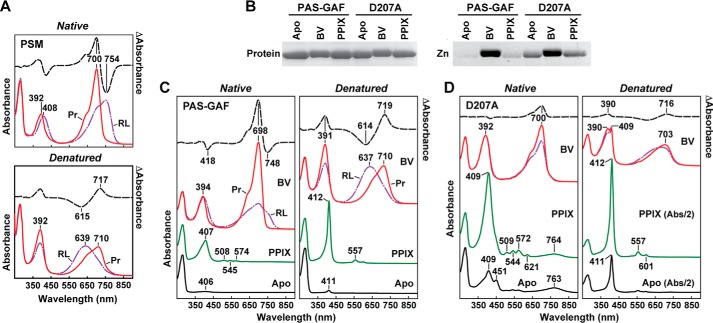
All proteins were highly stable in solution, efficiently bound BV covalently after purification, and displayed near normal absorption for the dark-adapted Pr state (Fig. 1, A and B). The PSM photoconverted normally to Pfr in red light with its Pr and Pfr absorption following denaturation in acidic urea consistent with the 15Z and 15E isomers of BV, respectively (see Fig. 4A). In contrast, the PAS-GAF truncation was substantially impaired photochemically and generated a mixed population containing Pfr and a bleached species even under saturating red light (Fig. 1B), consistent with the importance of the PHY domain to full photochromicity (18). The mixed species generated from the PAS-GAF fragment after red light was also substantially less stable than the Pfr state of full-length DrBphP and its PSM as judged by its substantially faster rate of thermal reversion back to Pr (t/2 = 6 min at 45 °C for PAS-GAF versus 652/607 min for the PSM/full-length chromoproteins (Fig. 1C)). Whereas reversion kinetics for PSM fragments with a functional PHY domain were best fit to a two-exponential function, those for the PAS-GAF fragment required only one exponential.
FIGURE 1.
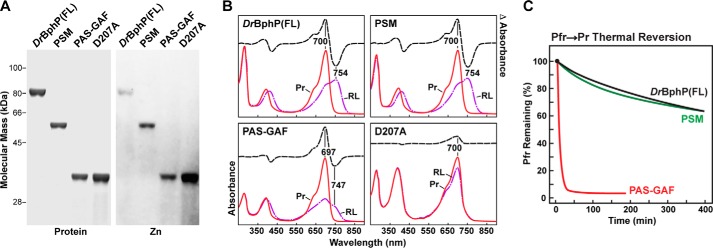
BV binding and photochemical properties of full-length (FL) DrBphP and its PSM and PAS-GAF fragments. A, SDS-PAGE of recombinant full-length, PSM, PAS-GAF, and D207A chromoproteins either stained for protein with Coomassie Blue (Protein) or assayed for the bound bilin by zinc-induced fluorescence (Zn). B, UV-visible absorption spectra as Pr or following saturating red-light irradiation (RL, mostly Pfr) and red light-minus-darkness difference spectra (upper dashed lines). The scales for the difference spectra were reduced by 0.7 relative to the absorption spectra. Difference absorption maxima and minima are indicated. C, Pfr → Pr thermal reversion kinetics at 45 °C monitored at 750 nm following excitation with saturating red light.
FIGURE 4.
UV-visible absorption and fluorescent properties of the D207A mutation assembled with BV and PPIX. A, absorption spectra before and after acidic denaturation of the PSM of DrBphP assembled with BV. Absorption and difference spectra were recorded for the Pr state or after saturating red-light (RL) irradiation (mostly Pfr) in nondenaturing buffer or after dissolution in 8 m urea (pH 2.0). Absorbance maxima are indicated. Difference spectra (dashed lines) were scaled to 70% of absorption spectra. B, covalent binding of BV and PPIX to the wild-type PAS-GAF fragment and the D207A mutant. Following in vitro assembly, the samples were subjected to SDS-PAGE and either stained for protein with Coomassie Blue (Protein) or assayed for the bound BV/PPIX by zinc-induced fluorescence (Zn). C and D, absorption spectra before and after acidic denaturation of the PAS-GAF fragment from wild-type DrBphP (C) and the D207A mutant (D) assembled with BV or PPIX. Absorption and difference spectra were recorded as in A. Apo, apoprotein before bilin assembly. The scales of the absorption spectra for the denatured PPIX and Apo samples from the D207A mutant were reduced 2-fold for clarity.
High Resolution Model of the PAS-GAF Region from DrBphP
To better define the bilin/protein contacts and the positions of ordered waters within the GAF domain, we generated a 1.16-Å resolution crystal structure of the PAS-GAF fragment as Pr by optimizing previous crystallization conditions (32). Here, radiation damage to the crystals was minimized using a 5-μm x-ray beam and continuous vector translation across the length of the crystal during data collection.4 The resulting crystal structure (PDB code 4Q0H; Table 1) superposed well with the prior 1.45-Å resolution PAS-GAF structure by Wagner et al. (32), having a root mean-squared deviation (r.m.s.d.) of 0.078 Å over 264 Cα atoms, and it likewise was arranged as a head-to-head dimer. The dimer interface was provided by the GAF domain helical bundle created by the α1, α2, and α5 helices, along with a contribution from strand β3 of the PAS domain. BV was clearly arranged in a ZZZssa configuration with the D pyrrole ring tilted by 51° as compared with the relatively planar A-C rings (Fig. 2A). However, diffuse electron density for BV was evident at the A-ring C3 side chain near the thioether linkage, which generated negative Fo − Fc difference map features even at ∼5 σ. These observations implied high mobility of this region, an incomplete or transient thioether linkage, or extreme sensitivity of this bond to ionizing radiation (27, 28).
TABLE 1.
Dr-BphP X-ray crystallographic data collection and refinement statistics
| Crystal | PAS-GAF | PAS-GAF(D207A) | PSM |
|---|---|---|---|
| Data collectiona | |||
| Space group | C2 | C2 | P6422 |
| Cell dimensions (Å) | 89.5, 51.6, 80.6 | 87.5, 51.3, 80.4 | 149.0, 149.0, 148.9 |
| 90° × 116.3° × 90° | 90°, 115.8°, 90° | 90°, 90°, 120° | |
| Resolution (Å) | 50–1.16 (1.18–1.16) | 36.5–1.75 (1.78–1.75) | 50–2.75 (2.80–2.75) |
| Rsymb | 0.061 (0.631) | 0.057 (0.535) | 0.101 (0.978) |
| I/σ(I) | 22.8 (2.02) | 22.5 (2.68) | 21.9 (2.48) |
| Completeness (%) | 98.3 (95.7) | 99.3 (99.7) | 99.8 (100) |
| Redundancy | 4.2 (3.6) | 5.9 (5.5) | 11.6 (11.8) |
| Refinement | |||
| Resolution (Å) | 36.1–1.16 | 36.2–1.74 | 46.3–2.75 |
| No. of reflections | 111,918 | 32,501 | 25,901 |
| Rwork/Rfreec | 0.131/0.157 | 0.161/0.195 | 0.202/0.224 |
| Atoms | 3873 | 2945 | 3881 |
| Protein | 3395 | 2620 | 3786 |
| Ligandd | 57 | 51 | 43 |
| Water | 388 | 270 | 52 |
| Average B-factors | 25.2 | 33.4 | 92.6 |
| Protein | 23.8 | 32.5 | 93.2 |
| Ligand | 21.1 | 28.8 | 67.1 |
| Water | 37.5 | 43.0 | 69.5 |
| r.m.s.d. | |||
| Bond lengths (Å) | 0.011 | 0.017 | 0.008 |
| Bond angles (°) | 1.4 | 1.7 | 0.91 |
| Steric clashes | |||
| Clash score | 2.8 | 2.6 | 3.0 |
| Ramachandran (%) | |||
| Favored | 99.8 | 99.4 | 98.2 |
| Outliers | 0 | 0 | 0 |
| PDB codes | 4Q0H | 4Q0I | 4Q0J |
a Outer shell values are in parentheses.
b Rsym = ΣhΣI|Ii(h) − 〈I(h)〉|/ΣhΣiIi(h), where Ii(h) is the intensity of an individual measurement of the reflection and 〈I(h)〉 is the mean intensity of the reflection.
c Test set sizes for Rfree calculations were 1.8, 5.07, and 5.0% for PAS-GAF, D207A, and PSM datasets, respectively, and were selected randomly. Each Rfactor = Σh‖Fobs| − |Fcalc‖/Σh|Fobs|, where Fobs and Fcalc are the observed and calculated structure-factor amplitudes, respectively.
d For PAS-GAF, these included atoms from 1 BV, 3 isopropyl alcohol, and 1 chloride molecule; for the D207A mutant, 1 BV molecule, and 2 isopropyl alcohol molecules; and for the PSM, one BV molecule.
FIGURE 2.
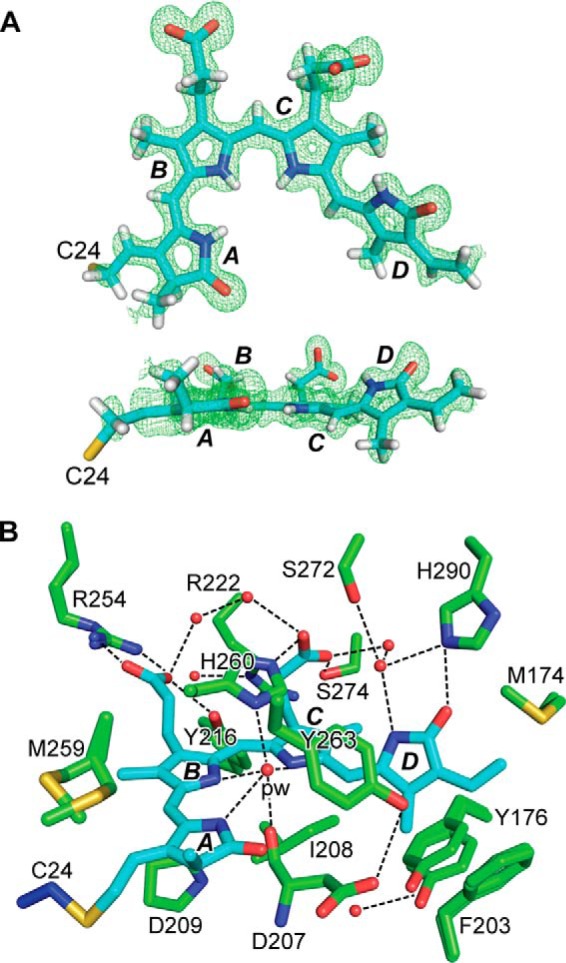
Crystal structure of the PAS-GAF fragment from D. radiodurans BphP as Pr at 1.16 Å resolution. A, ZZZssa conformation of BV in the PAS-GAF structure (PDB code 4Q0H) displayed in front and side orientations superimposed on the Fo − Fc electron density map generated by omitting BV and contouring to 3 σ. The sulfur atom in Cys-24 (yellow) that participates in the thioether linkage to BV is included, and pyrrole rings A–D are labeled. Carbons, nitrogens, oxygens, and hydrogens are colored cyan, blue, red, and white, respectively. B, architecture of the bilin-binding pocket illustrating relevant hydrophobic and electrostatic interactions that fix BV in the Pr state. Alternative conformations of Tyr-176, Phe-203, Arg-254, and Met-259, and the positions of several well ordered waters (red spheres) are included. The pyrrole water (pw) is labeled. The coloring scheme is as in A, except that the GAF domain and Cys-24 carbons are colored in green and blue, respectively. All atoms for Asp-207 are shown to illustrate its interaction with the pyrrole water. For all other residues, only the side-chain atoms are displayed. Dashed lines locate hydrogen bond contacts.
The improved resolution revealed a number of fine features in the resulting electron density map of the PAS-GAF region as Pr, including alternate conformations of several residues adjacent to BV (e.g. Tyr-176, Phe-203, Met-259, and Arg-254), the exact positioning of several waters, well fixed B and C ring propionate side chains, and structural support for the protonation of all four pyrrole rings of BV (Fig. 2, A and B). From this structure, characterization of the comprehensive hydrogen-bonding network between BV and the GAF domain crevice was possible, with the pyrrole water and Asp-207 as its focal points (Fig. 2B). In general, the amino acid side chains associated with BV were well ordered (Fig. 2B). However, Tyr-176 exhibited flared electron density that could be well described by simulating a slight rocking motion at the main chain, and the Tyr-263 atomic positions produced a negative Fo − Fc map defect similar in magnitude to that found at the thioether linkage. The difference density for Tyr-263 might reflect an alternative conformation of low occupancy that could have been induced by the pH at crystallization or by removing the PHY domain that normally abuts this residue (see below). These data also permitted refinement of anisotropic atomic displacement parameters to facilitate visualization of atomic movements. Notably, displacements of the BV carbons/nitrogens had the largest magnitudes roughly perpendicular to the plane of the A-C pyrrole rings, which appeared to correlate with displacements of protein and solvent atoms in the binding pocket.
Structure of the DrBphP PAS-GAF(D207A) Mutant
Not surprisingly, substitutions of Asp-207 have severe consequences on DrBphP photoconversion given its central position at the bilin/GAF domain interface and its likely role in the deprotonation/protonation cycle of the bilin during photoconversion (33, 46), with even subtle substitutions generating photochemically impaired and often highly fluorescent variants (33, 47, 48). To better understand the molecular consequences of these substitutions, we generated a crystal structure of the PAS-GAF fragment harboring the D207A substitution to 1.75 Å resolution (PDB code 4Q0I; Table 1). As shown in Fig. 1, the D207A polypeptide efficiently bound BV to generate a Pr-like state with a substantially increased Soret absorption peak at 390 nm relative to the main peak at 700 nm and a diminished absorption band at 700 nm in comparison with the wild-type PAS-GAF fragment. It also failed to accumulate a far-red light-absorbing Pfr-type species in red light but instead showed slight bleaching during prolonged irradiations.
The overall structure of the D207A mutant was highly congruent with the wild-type PAS-GAF structure described above with an r.m.s.d. of 0.20 Å for the core 250 Cα atoms (minus flexible regions) and an r.m.s.d. of 0.16 Å for the 142 Cα atoms of the GAF domains. Whereas the main chain position of residue 207 remained consistent whether it was occupied by alanine or aspartate, the position of Tyr-263 was significantly impacted (Fig. 3D), suggesting that its placement is governed in part by interactions with residue 207. This conclusion is supported by the work of Mailliet et al. (49) with Synechocystis Cph1, which showed a similar disposition of residue 263 in the Y263F mutant that precludes hydrogen bonding with the aspartate. Difference electron density maps around the chromophore gave strong indications of alternate conformations/species for BV and Tyr-263, and possibly a non-bilin adduct (Fig. 3A). In particular, much of the signal from the hydrophobic side of the D pyrrole ring (i.e. C17, C18, and the C18 vinyl side chain) was absent, and a defect in the difference map appeared between the D ring and the Tyr-263 side chain. Insertion of the ZZZssa configuration of BV at full occupancy largely accounted for the difference map defects at the B and C ring positions but led to negative difference map peaks adjacent to the hydrophobic half of the D ring and failed to account for the density between the D ring and Tyr263.4
FIGURE 3.
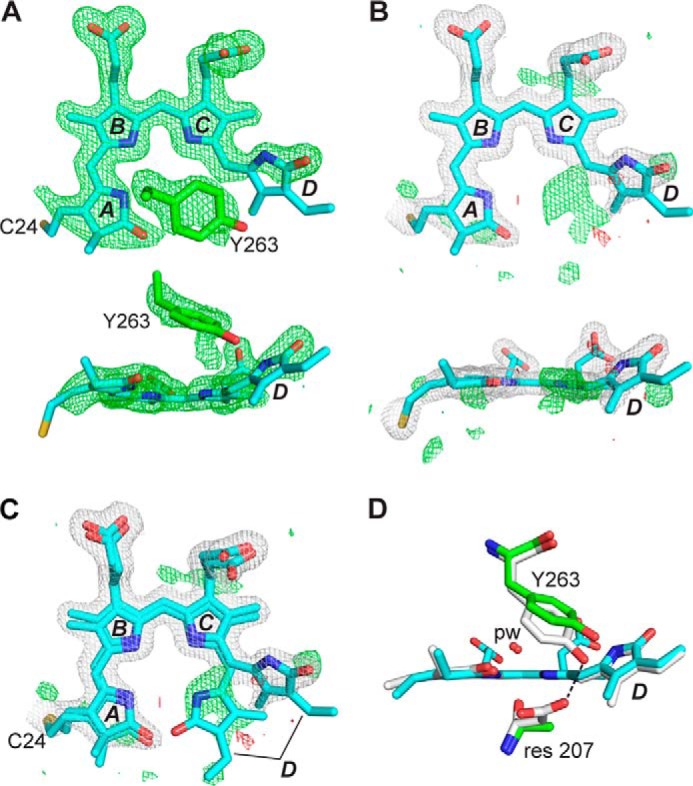
Crystal structure of the DrBphP PAS-GAF fragment as Pr and bearing the D207A mutation. A, structure of BV at 1.75 Å resolution (PDB code 4Q0I) displayed in front and side orientations superimposed on the Fo − Fc electron density map generated by omitting BV and Tyr-263 and contouring to 3 σ. The final refined position of Tyr-263 is also shown. The sulfur atom in Cys-24 (yellow) that participates in the thioether linkage to BV is included, and pyrrole rings A–D are labeled. Carbons, nitrogens, and oxygens are colored cyan, blue, and red, respectively. B, BV is shown as in A, superposed with the final 2Fo − Fc (white) and Fo − Fc (positive, green; negative, red) electron density maps that were contoured at 1 σ and 3 σ, respectively. Atoms of the A and D pyrroles were refined at 70% occupancy, and atoms of the B and C pyrroles were refined at 100% occupancy. C, refined positions of alternate conformations of BV having the ZZZssa and ZZZsss configurations were superposed on the electron density maps from B. D, positions of Tyr-263 for wild-type PAS-GAF fragment and the D207A mutant were shown after superposition of the GAF domains. ZZZssa BV configuration, pyrrole water (pw), and residue (res) 207 positions are shown. Coloring scheme is the same as in A–C, except that wild-type carbons are white. Dashed line locates a hydrogen bond contact.
However, if the B and C pyrrole rings were modeled at full occupancy and the A and D ring occupancy was reduced to 70%, most negative difference map features were reduced to noise levels (Fig. 3B). The difference density peaks that remained suggest two possible origins. The D207A mutant harbored steady state levels of both the ZZZssa and ZZZsss configurations of BV, with the anti to syn rotation of C14 swiveling the D ring into the unaccounted for density, or a cyclized bilin such as PPIX entered the binding pocket as seen previously by Wagner et al. (33) for a D207H variant. Both these scenarios are tenable and are perhaps facilitated by the absence of the more bulky Asp residue at position 207 and the increased distance between residues Ala-207 and Tyr-263 in the mutant that might open the binding crevice sufficiently to accept cyclic porphyrins. Modeling PPIX into the pocket instead of BV eliminated some of the negative density defects at the D ring but created new defects especially at the pyrrole water.4
In support for acceptance of a PPIX-type bilin by D207A, we compared the absorption spectra of the wild type PAS-GAF and D207A apoproteins when assembled with PPIX or BV in both native and acidic denaturing conditions (8 m urea, pH 2.0). As shown in Fig. 4B, the wild-type PAS-GAF fragment effectively bound BV but marginally bound PPIX covalently, as judged by zinc-induced fluorescence of the polypeptides following SDS-PAGE, whereas the D207A mutant could better accept PPIX. This improved PPIX binding could then be visualized by absorption spectra of the D207A mutant following denaturation. Here, the strong Soret peak at 412 nm and additional maxima at 509, 544, 572, and 621 nm, which are characteristic of PPIX, became obvious for D207A (Fig. 4D). Surprisingly, this Soret peak for PPIX was also apparent in the spectra of denatured D207A samples assembled with BV in vitro and in D207A apoprotein samples prior to assembly. Together, these observations imply that the D207A apoprotein also incorporated PPIX from a pool available during its recombinant expression in E. coli. This occupancy was then reflected in the crystal structures by added electron density for a cyclic porphyrin seen for the D207A chromoprotein assembled with BV (Fig. 3C). Consistent with this scenario, fluorescence spectroscopy of the D207A apoprotein assembled with BV also detected two fluorescent species,5 one matching the absorption and emission spectra of incorporated PPIX and a second matching those for BV (47, 48).
X-ray Crystal Structure of the PSM from DrBphP
To better understand the modular arrangement of DrBphP and to help interpret subsequent SPEM images, we generated a crystal structure of the dimeric PSM (Fig. 5). Comprehensive trials identified several conditions for crystal growth, which were optimized to enable collection of an x-ray diffraction dataset to 2.75 Å resolution (PDB code 4Q0J; Table 1). The PAS-GAF-PHY arrangement of the individual polypeptides in the resulting structure (Fig. 5A) matched reasonably well with those previously described for Synechocystis Cph1 (10), Pseudomonas aeruginosa BphP (29), and Rhodopseudomonas plaustris BphP1 (19). Its extracted PAS and GAF domains also superposed well with the 1.16-Å resolution model of the PAS-GAF fragment determined alone as evidenced by an r.m.s.d. of 0.606 Å for 284 Cα atoms (Fig. 5C).
FIGURE 5.
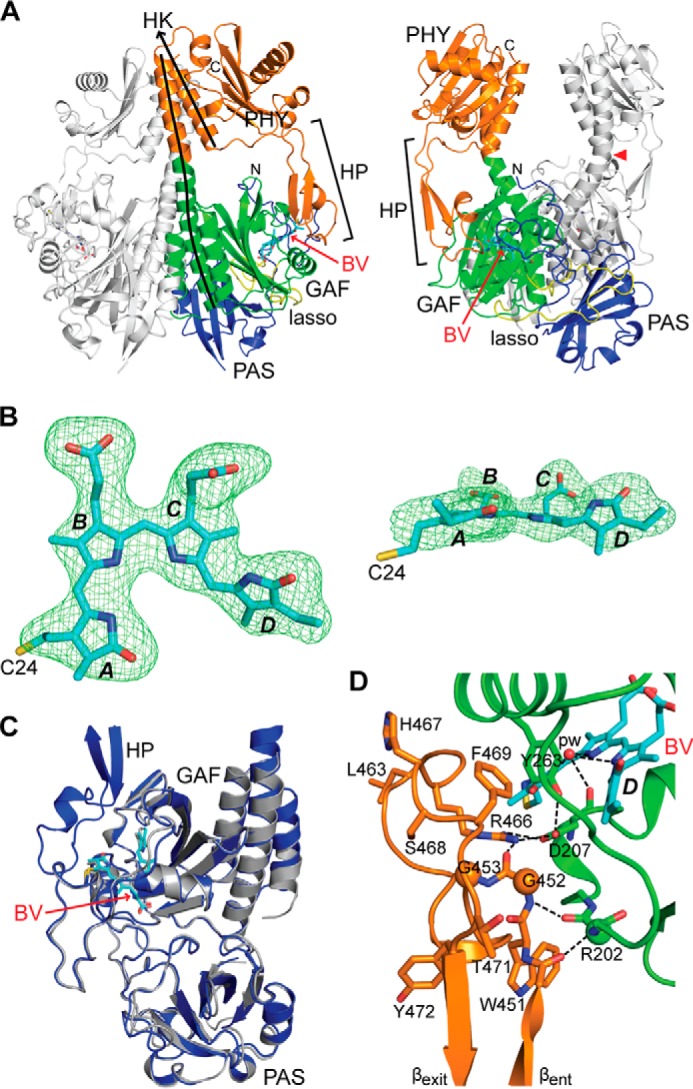
Crystal structure of the PSM dimer from DrBphP as Pr at 2.75 Å resolution. A, ribbon diagram of the PSM dimer (PDB code 4Q0J) in front and side views. For one subunit, the PAS, GAF, and PHY domains and the knot lasso are colored in blue, green, orange, and yellow, respectively. BV is located by the arrow and colored in cyan with the nitrogens and oxygens colored in blue and red, respectively. The hairpin (HP) extending from the PHY domain is located by the bracket. The lines track the helical spine of one monomer. Ala-326 at the apogee of the bowed helical spine is marked by a red arrowhead. HK, histidine kinase domain; N, N terminus; C, C terminus. B, ZZZssa conformation of BV displayed in front and side orientations superposed on the Fo − Fc electron density map generated by omitting BV and contouring to 3 σ. The sulfur atom in Cys-24 (yellow) that participates in the thioether linkage to BV is included, and pyrrole rings A–D are labeled. Carbons, nitrogens, and oxygens are colored cyan, blue, and red, respectively. C, superposition of the high resolution PAS-GAF structure (PDB code 4Q0H, gray) onto the PSM structure (blue). D, close-up view of the hairpin region (orange) extending toward and contacting the GAF domain close to the chromophore. The side chains of key amino acids are included, some of which are analyzed in Figs. 9 and 10. Dashed lines locate hydrogen bond contacts. The PSM structure is colored as in A. βent and βexit label the entrance (N-terminal) and exit (C-terminal) β-strands in the hairpin.
As with the PAS-GAF structures (this work and Refs. 27, 32), the DrBphP PSM crystallized as a head-to-head dimer with 2-fold symmetry corresponding to the crystallographic axis and dimerization contacts contributed by the GAF and PAS domains (Fig. 5A). However, the angle between sister subunits differed substantially between the PSM and PAS-GAF structures, suggesting that the dimer interface serves as a flexible joint that can accommodate substantial motion. In this case, the angle between helices α5 of the GAF domains at the pivot point near residue 304 was 21° greater for the PSM construction than for the wild-type PAS-GAF construction without the Y307S substitution (27). Modeling also revealed that the paired PAS domains would eventually provide a barrier to this rotation, suggesting that one function of this feature is to limit angular movement at the dimerization interface, and thus facilitate directional propagation of conformational signals to the OPM.
The sister subunits in the PSM twist about the 2-fold rotational axis in a left-handed fashion partially due to a remarkable bow in the helical spines (Fig. 5A). An outward splaying of each helical spine began at the exiting helix α5 of the GAF domain with the return generated by a prominent kink involving Ala-326 at the helical junction between the GAF and PHY domains. Despite this bend, no contact was evident between sister PHY domains; in fact, the closest approach was at the α6 helix that transitions from the PHY domain to the OPM. Here, atoms of the Asp-496 side chains of sister subunits came within ∼8 Å of each other (Fig. 5A). Prior SPEM maps of the full-length DrBphP dimer detected a central hole separating the expected positions of the paired PHY domains (31). When our bowed PSM structure was docked with this EM image as a rigid body, this cavity was largely explained by the curved helical spines (see below).
The BV-binding pockets of the PAS-GAF and PSM constructs were remarkably similar, having an r.m.s.d. of 0.26 Å for 127 Cα atoms, and likewise the PSM cradled BV in the ZZZssa configuration (Fig. 5B). This robust congruence of GAF domain folds strongly implies that the PHY domain and its hairpin have little influence on the overall structure of the PAS-GAF region, at least for the Pr state. Moreover, rotamers and side chain positions of residues adjacent to the BV were highly consistent between the constructions, suggesting that structural analyses of the PAS-GAF fragment should provide pertinent insights into bilin transitions between the Pr and Pfr states and how the bilin would respond to site-directed mutations or chemical modification.
Conformation of the DrBphP Hairpin in the Pr State
An intriguing feature in available PSM structures is the hairpin (or tongue) that extends from the PHY domain to contact the GAF domain near the bilin-binding crevice. Electron density for the DrBphP PSM was continuous throughout the hairpin sequence, thus permitting modeling of the entire feature. Like hairpins from other Phys that employ Pr as the dark-adapted state (Fig. 6) (10, 11, 21), the DrBphP hairpin as Pr includes an anti-parallel two β-strand stem that binds to the GAF domain through contacts mediated by conserved residues at the end of the stem (Fig. 5D). Prior analyses (11, 21) identified the Trp-Gly-Gly (WGG, residues 451–453 in DrBphP), Pro-Arg-Xaa-Ser-Phe (PRXSF, residues 465–469), and bulky hydrophobic Xaa-Glu (HbXE, residues 472–474) motifs as influential hairpin/GAF domain contacts (see Fig. 9A). These motifs are present in DrBphP and appear to generate the same associations, including Arg-466 that forms a salt bridge with Asp-207, and hydrophobic interactions involving Trp-451, Gly-452, Pro-456, Pro-465, and Phe-469 (Fig. 5D). In addition, we detected hydrogen bonds between the main-chain carbonyl and amide of Ala-450 and Gly-452, respectively, and Arg-202 in strand β3 of the GAF domain, which further cement the hairpin/GAF association while extending the GAF domain β-sheet (Fig. 5D).
FIGURE 6.
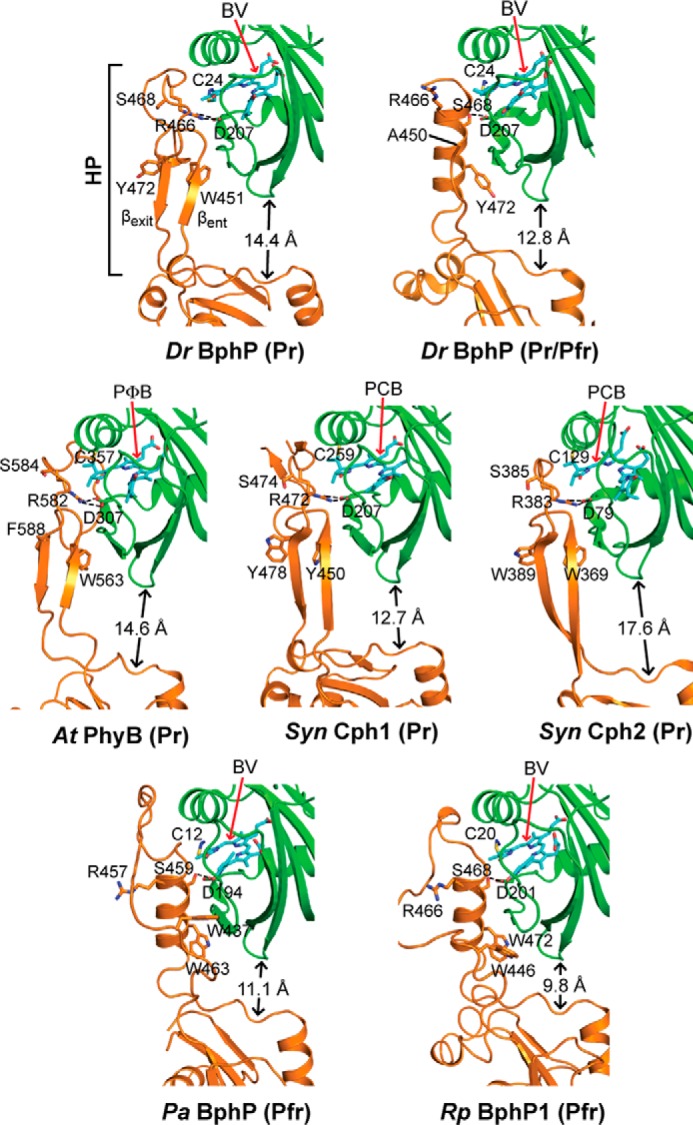
Conformation of the PHY domain hairpin from representative Phys and its interaction with the GAF domain. PSM structures were extracted from D. radiodurans BphP as Pr (PDB code 4Q0J; this report) and a mixed Pr/Pfr state (4O01 (30)), Synechocystis Cph1 as Pr (2VEA (10)), and Cph2 as Pr (4BWI (21)), A. thaliana PhyB as Pr (4OUR (11)), P. aeruginosa BphP as Pfr (3C2W (29)), and R. palustris BphP1 (4GW9 (19)) as Pfr. The GAF and PHY domains are colored in green and orange, respectively, and the bilin is colored in cyan (arrow), the type of which is indicated. PCB, phycocyanobilin. PΦB, phytochromobilin. Side chains are shown for relevant amino acids. Dashed lines indicate hydrogen bond contacts between the DIP (Asp-Ile-Pro) motif aspartate in the GAF domain and either the conserved arginine or serine in the hairpin stem. The distance separating the GAF and PHY domain globular regions is indicated. The distances were measured from the loop separating the β1 and β2 strands of the GAF domain and the α-carbon of a conserved tryptophan (Trp-483 in DrBphP) just proximal to the exiting α-helix of the PHY domain. HP, hairpin. βent and βexit label the entrance (N-terminal) and exit (C-terminal) β-strands in the hairpin.
FIGURE 9.
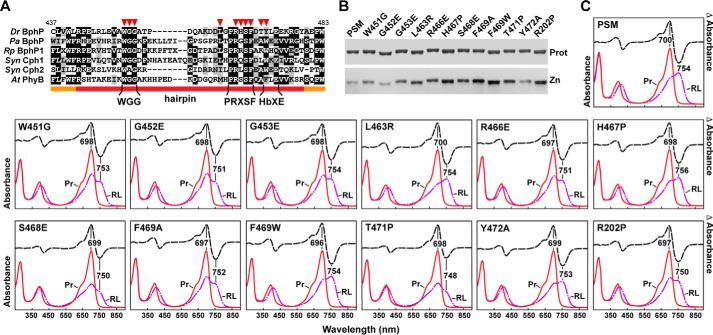
Assembly and photochemical properties of amino acid substitutions affecting the hairpin/GAF domain contact. A, amino acid sequence alignment of hairpin region from the representative Phys illustrated in Fig. 6. The hairpin and bracketing PHY domain sequences are located by the orange and red lines, respectively. The WGG, PRXSF, and HbXE motifs in the hairpin β-strand stem are highlighted. Specific amino acids analyzed by site-directed mutagenesis are indicated by the red arrowheads. Identical and similar amino acids are shown by the black and gray boxes, respectively. The numbering is for D. radiodurans BphP. B, SDS-PAGE of the recombinant chromoproteins either stained for protein with Coomassie Blue (Prot) or assayed for the bound bilin by zinc-induced fluorescence (Zn). C, UV-visible absorption spectra as Pr or following saturating red-light irradiation (RL, mostly Pfr), and red light-minus-darkness difference spectra (upper lines). The scales for the difference spectra were reduced by 0.7 relative to the absorption spectra. Difference absorption maxima and minima are indicated.
SPEM Images of the Pr and Pfr Dimers
To investigate how photoactivation reconfigures the DrBphP structure, particularly in the OPM region, we performed negative-staining EM and single particle reconstructions on full-length wild-type DrBphP in both the dark-adapted Pr and photoactivated Pfr states (Fig. 7). To ensure that we avoided Pfr contamination in the Pr samples and retained much of the Pfr state in the photoactivated samples, the preparations were rapidly fixed in darkness or soon after the saturating red-light irradiation, respectively. We found that both states assembled as dimers (Fig. 7, A and B), in agreement with prior size determinations by size exclusion chromatography (32). The SPEM map of DrBphP in the Pr state was similar to the previously reported negatively stained and cryo-EM density maps of the same state, including retention of the central hole and a more diffuse C-terminal bundle reflecting the paired HK OPMs (31). Accuracy of the Pr map was provided by docking the crystal structure of the DrBphP PSM dimer solved in this study as a single rigid body. As shown in Fig. 7C, this PSM model fit well into the SPEM map, with the superposition further demonstrating that the bowed dimer interface in the PSM crystal is physiologically relevant and not artifactually induced by crystal packing.
FIGURE 7.
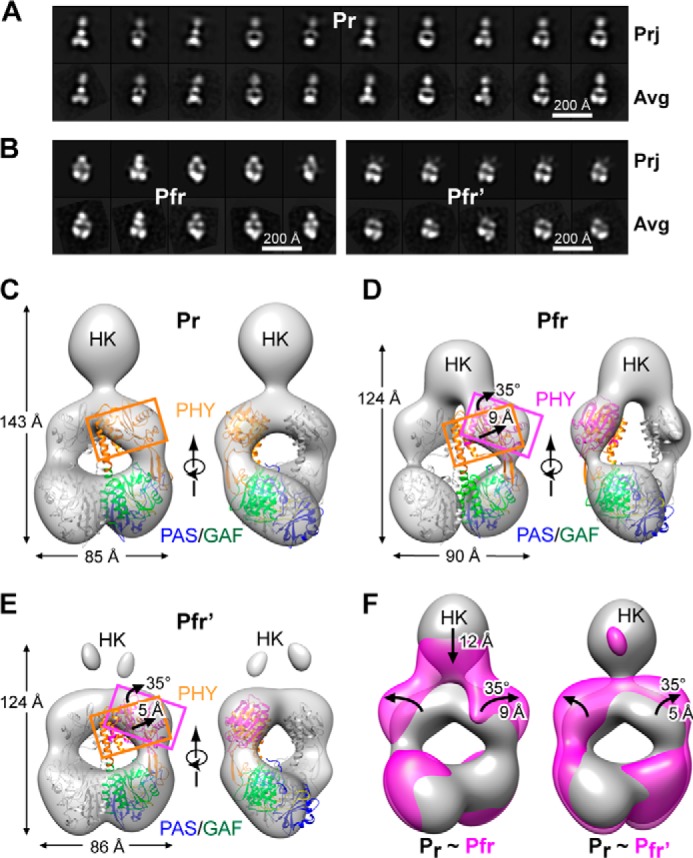
SPEM images of the DrBphP dimer in the Pr and Pfr states. A and B, selected reference-free two-dimensional class averages (bottom row, Avg)) generated after negative staining of Pr (A) and of samples subjected to saturating red-light irradiation (mostly Pfr) (B) in comparisons with selected two-dimensional projections (top row, Prj) of the respective three-dimensional EM maps. Left panel B shows Pfr particles with visible HK domains, and the right panel shows particles with HK domains largely invisible. C, SPEM map of DrBphP in the Pr state shown in the surface-rendered front and side views. D and E, SPEM maps of DrBphP in the Pfr (D) and Pfr′ conformers (E). The crystal structure of the PSM dimer as Pr (PDB code 4Q0J, this report) docked as a rigid body was included in C–E. One monomer is in gray, and the other monomer is shown in blue, green, and orange for the PAS, GAF, and PHY domains, respectively. PHY domain fits the density well when it is isolated from the crystal structure and allowed to rotate by 35° clockwise and move outward by ∼9 Å for Pfr (D) or by 5 Å for Pfr′ (E). F, superposition of the SPEM maps of Pr (gray) with Pfr (magenta, left) or Pfr′ (magenta, right). The downward shift of the sister HK domains, and the rotation and outward movement of the PHY domains in the Pfr or Pfr′ models are indicated by the black arrows.
Interestingly, we observed two distinct populations of particles in the EM image collections from the photoactivated samples as follows: one with all expected domains visible and the other largely missing density for the HK OPM, which we have called Pfr and Pfr′, respectively (Fig. 7B, left and right panels). From analysis of 12,200 particle images, ∼59% (7200) were classified as Pfr and 39% (4700) classified as Pfr′. We subsequently obtained SPEM reconstructions for the Pr, Pfr, and Pfr′ states at 24, 26, and 27 Å resolution, respectively (Fig. 7, C–E). Close comparisons of the Pfr/Pfr′ models showed a clear difference from the Pr model, which was detected as an increased diameter of the central hole and a broadening across the paired PHY domains along with a shortening from the PAS to HK domains. The sum was a markedly altered aspect ratio, which changed from l/w = 1.68 for Pr to l/w = 1.38 for Pfr (Fig. 7, A and B).
A clear discrepancy in the EM densities surrounding the PHY regions as Pfr/Pfr′ was also evident when the PSM dimeric crystal structure in the Pr state was docked as a single rigid body. This mismatch could be corrected by rotating the PHY domain by ∼35° and displacing it outward by 9 and 5 Å for the Pfr and Pfr′ maps, respectively, as compared with Pr (Fig. 7, D and E). In contrast, no differences were detected at the PAS-GAF regions between Pr and the Pfr/Pfr′ states. Collectively, the superposed SPEM models revealed a large scale conformational change during photoactivation, which involves separation of the sister PHY domains by rotation and outward movement relative to the PAS-GAF region.
In addition to density differences at the PSM regions, even larger differences between Pr and Pfr/Pfr′ were evident for the density corresponding to the HK OPMs. In the Pfr SPEM model, the associated HK domains were evident but appeared to be displaced downward toward the PSM by ∼12 Å (Fig. 7, D and F). In contrast, the Pfr′ model revealed little density for the HK domains, suggesting that the DrBphP OPM pair in this population had largely separated upon photoconversion and became more mobile (Fig. 7, E and F). It was possible that the Pfr′ species represents DrBphP polypeptides actually missing HK by proteolytic cleavage during photoconversion and sample preparation. However, SDS-PAGE of the red light-irradiated samples detected only a single molecular species at the expected size of the full-length DrBphP polypeptide.6
Following completion of our work, Takala et al. (30) reported low resolution but informative crystallographic structures of the DrBphP PSM dimer before and after photoactivation with the photoactivated structure appearing to represent a mixed population of Pr/Pfr species based on an absorption spectrum of the crystal. Importantly, the sister PHY domains of the Pr/Pfr model were splayed out as expected from our SPEM models of Pfr, and the hairpin stem region best fit a helical configuration as predicted from the ensemble of crystal structures for Phys in the dark-adapted and photoactivated states (see Fig. 6). When we docked this photoactivated structure onto the SPEM models as a rigid body, a strong fit was observed once we rotated the PHY domain counterclockwise by 25° and 34° for the Pfr and Pfr′ models, respectively (Fig. 8). This discordance likely reflected the absence of the OPM in the photoactivated PSM structure that would presumably constrain rearrangement of the sister PHY domains via additional dimerization contacts.
FIGURE 8.
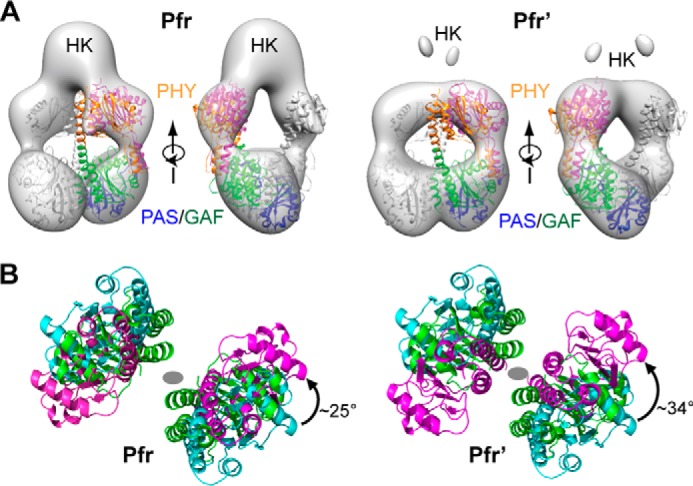
Docking of the crystal structure of the PSM as Pfr with the EM models of the BphP dimer as Pfr. A, alignment of the crystal structure of the PSM from a Pr/Pfr mixture with the SPEM models of the Pfr and Pfr′ states shown in Fig. 7. The low resolution Pr/Pfr crystal structure (PDB code 4O01 (30)) was placed into the EM density by first fitting the PAS-GAF regions alone and then docking as a rigid body the entire PSM via its alignment with the fitted PAS-GAF region. Best fit was achieved when the PHY domain crystal structure was allowed to rotate independently of the PAS-GAF domains. One monomer is in gray, and the other monomer is shown in blue, green, and orange for the PAS, GAF, and PHY domains, respectively. B, rotation of the PHY domain relative to the PAS-GAF region for improved fit of the PSM crystal structure into the EM models of the full-length DrBphP dimer. PHY domains colored in green, cyan, and magenta were isolated from the docked PSM dimer as Pr/Pfr (PDB code 4O01). The viewing direction is from the OPM toward the PSM along the 2-fold axis of the DrBphP dimer. The gray oval marks the 2-fold axis position in each panel. The PHY domain (magenta) was rotated counter-clockwise by 25 and 34° to better fit the crystal structure into the Pfr and Pfr′ EM maps, respectively.
Role of the Hairpin on DrBphP Photochemistry
The most parsimonious explanation for the differences between the Pr and Pfr/Pfr′ SPEM structures is that the paired PHY domains flared outward during photoconversion relative to the static PAS-GAF regions, which then loosen the contacts between the sister OPMs (Fig. 7F). From structural comparisons of several canonical Phy and bathy-Phy PSM structures (Fig. 6) combined with mutagenic studies, an underpinning “tryptophan switch” mechanism has been proposed to drive this motion (21), whereby the β-strand stem of the hairpin releases from the GAF domain and melts to allow swiveling of the polypeptide chain positions and assumption of an α-helical conformation (11, 21, 30). The distance between the GAF and the PHY domains decreases as a result of this helical contraction (see Figs. 6 and 7), which could pull the sister PHY domains away from each other by lessening the prominent helical spine kink in the DrBphP PSM structure (Fig. 5A) and/or by changing the relative angle of sister subunits at the dimer interface.
To test this toggle model, we examined hairpin function in detail by site-directed mutagenesis of the PSM. A special focus was on characterizing the photochemical effects of modifications to residues within the WGG, PRXSF, and HbXE motifs that show strong conservation within Phys having PAS-GAF-PHY architectures and were considered important for the tryptophan switch (Fig. 9A), and Arg-202 that participates in a heretofore unknown hairpin stem/GAF domain hydrogen bond interaction (Fig. 5D). The impacts of other GAF domain residues at the hairpin interface on DrBphP photochemistry (e.g. Asp-207 and Tyr-263) were described previously (33). In two positions (His-467 and Thr-471) we substituted prolines that, based on Ramachandran angles, could disrupt formation of the Pfr helix without compromising the native Pr β-strand structure.
To aid in interpretation, the PAS-GAF fragment of DrBphP was used as the expected baseline for PSM mutants with completely compromised hairpin/GAF interactions. As described above, the most apparent consequences of removing the PHY domain were a partially impaired photoconversion of Pr to Pfr and a substantially enhanced thermal reversion rate (100-fold faster (Fig. 1C)), implying that the hairpin mainly helps lock DrBphP in the photoactivated state. Like the PAS-GAF fragment, all the mutants tested here were soluble, covalently bound BV efficiently, and generated a relatively normal Pr absorption spectrum at 25 °C (Fig. 9, B and C). Thermal reversion was monitored at both at 25 and 45 °C, with the higher temperature needed to increase this relaxation to a practical rate for those mutants with highly stable Pfr states (Fig. 10). Interestingly, no mutants significantly impaired the absorption or reversion properties of the PSM beyond those seen for the PAS-GAF fragment, implying that none worked dominantly.
FIGURE 10.
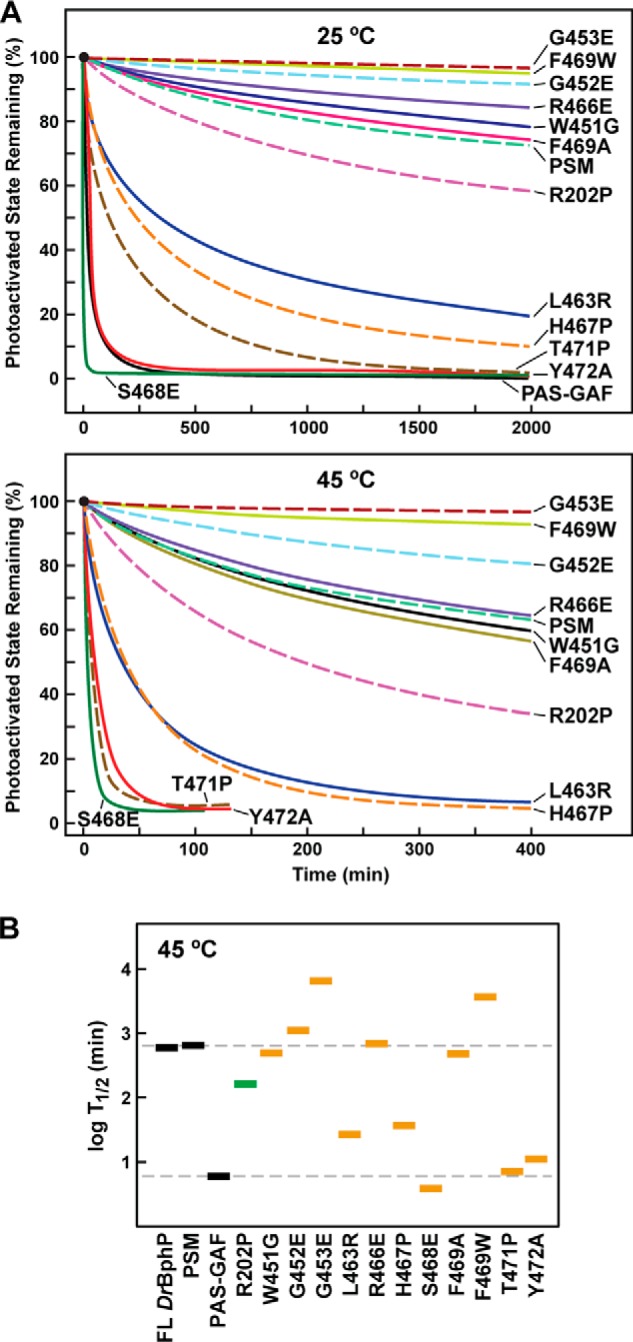
Thermal reversion rates for DrBphP PSM mutants affecting the hairpin/GAF domain contact. A, kinetics of Pfr → Pr thermal reversion for the mutant chromoproteins at 25 and 45 °C. B, half-life of Pfr for each mutant at 45 °C. Gray boxes represent the full-length (FL) chromoprotein and the PSM and PAS-GAF fragments. Green box represents the R202P mutant. Orange boxes represent various PHY-domain hairpin mutations. The dashed lines indicate the reversion rates for the PSM and PAS-GAF truncations. See Fig. 9 for the BV assembly and absorption spectra of each mutant.
Our working predictions were that mutants that destabilize/stabilize the hairpin/GAF contact as Pr would suppress/accelerate thermal reversion of Pfr back to Pr, and mutants that destabilize/stabilize the reconfigured contact as Pfr would accelerate/suppress reversion, respectively. Most substitutions met these expectations. For example, Ser-468 within the PRXSF motif is proposed to form a key hydrogen bond with Asp-207 upon swiveling the hairpin to its helical Pfr configuration. Its replacement with glutamate to subsequently repel Asp-207 compromised photoconversion to baseline levels (i.e. similar to the PAS-GAF fragment) and generated one of the fastest thermal reversion rates detected (t/2 of 4 min at 45 °C, versus 652 min for the wild-type PSM or a 160-fold increased rate (Figs. 9C and 10)). Substitution of the adjacent His-467 to a proline was designed to disrupt the Pfr helix and thus potentially destabilize Pfr. In agreement, the H467P chromoprotein had relatively normal Pfr absorption but a substantially faster Pfr → Pr thermal reversion rate (19-fold faster). Similarly, Thr-471 and Tyr-472, which are proposed to rotate during hairpin swivel with the tyrosine participating in the tryptophan switch (21), also had strongly accelerated thermal reversion rates (93- and 60-fold faster). Although the Y472A substitution, which removes the hydrophobic residue in the HbXE motif (Fig. 9A), had a relatively normal Pfr absorption spectrum after saturating red light irradiation, the adjacent T471P substitution, which was designed to disrupt the Pfr hairpin helix, was partially compromised (Figs. 9C and 10).
In contrast, a glutamate substitution of Gly-452 or Gly-453 in the WGG motif and the tryptophan substitution of Phe-469 in the PRXSF motif substantially suppressed thermal reversion with little impact on the absorption spectrum of Pfr, suggesting that these mutants help lock Pfr in place or inhibit its return to Pr. Notably, the counterpart F469A substitution had reduced Pfr accumulation, indicating that a bulky hydrophobic residue is important in this position. Finally, the Leu-463 to arginine substitution was designed to stabilize the Pr state by potentially adding a hydrogen bond link between it and a pair of glutamate residues in the nearby PAS domain (Glu-18 and Glu-22). This mutant appeared to increase the amount of Pfr at photoequilibrium in red light as judged by the reduced shoulder at 700 nm in the “Pfr” absorption spectrum that represents residual Pr (Fig. 9C). As predicted, the Pfr state of the L463R mutant also was substantially more unstable as judged by a 24-fold faster rate of thermal reversion (Fig. 10).
However, considering hairpin mutants that were predicted to enhance Pfr stability, to our surprise none met the level of Pfr stabilization predicted from comparison with our recent studies on Arabidopsis PhyB (11, 50). Most of these DrBphP hairpin mutants could be categorized as residues that do not participate directly in Pfr formation, as judged by Pfr state crystal structures, but they were expected to slow thermal reversion by compromising one of the many contacts in the hairpin/GAF interface of the Pr state (e.g. R466E, W451G, G452E, G453E, and R202P) (Figs. 5D and 10). The R202P substitution actually increased the thermal reversion rate, which is consistent with the apparent role of its guandinium group in Pfr stabilization (30). Arg-466 in the PRXSF motif forms a hydrogen bond with Asp-207 and is thus well positioned to respond to the flip of the D pyrrole ring impinging on Asp-207. Remarkably, the drastic glutamate substitution of this arginine (R466E) had a relatively normal Pfr absorption and showed only a mild increase in Pfr stability, suggesting that this contact does not have a major role in stabilizing either Pr or Pfr (Figs. 9C and 10). Similarly, substitution of Trp-451 in the WGG motif had little impact on the Pr → Pfr photoconversion and stability of the Pfr state. Thus, although the tryptophan switch may occur during photoconversion, regaining contact between Trp-451 and the GAF domain surface is not essential for recovery to the Pr state.
Collectively, the mutant analyses support the conformational changes in the PHY domain hairpin and the key role(s) of most prominent residues as proposed in the toggle model. However, its action might be the collective sum of multiple hairpin/GAF domain interactions and not solely dependent on the tryptophan switch nor light-induced rupture of the Arg-466/Asp-207 salt bridge.
DISCUSSION
Attempts to understand how Phys work mechanistically have focused in the past few years on developing an atomic level appreciation of these bili-proteins and on defining the conformational changes induced by photoconversion from Pr to Pfr. Here, we present one of the most in-depth views currently available from structural analyses of D. radiodurans BphP using both x-ray crystallography and SPEM. The highest resolution crystal structure of the PAS-GAF region to date (1.16 Å) provides a detailed view into the bilin-binding pocket as Pr. This structure revealed individual atomic motions within the GAF domain that were highly correlated between the chromophore and surrounding residues and provided structural evidence for full protonation of the four pyrrole nitrogens (33, 46).
When the PAS-GAF structure was compared with the photochemically compromised D207A variant, it appears that the high fluorescence of this mutant is derived from structural variability in several surrounding residues, including Tyr-263, in addition to replacement of the aspartate. These changes might compromise light-driven photoisomerization of the bilin as well as open up the GAF domain cavity for other tetrapyrroles. In particular, modeling suggests that the bilin-binding pocket is tolerant to more cyclic bilins by either allowing a PPIX-type species to occupy the binding crevice or permitting rotation of BV to assume a ZZZsss configuration. This propensity likely underpins the different fluorescence excitation and emission spectra for the various Asp-207 mutant fluorophores reported previously (33, 47, 48). In our situation, the inclusion of a PPIX-type adduct likely occurred during recombinant synthesis. Of potential use as a novel fluorophore, we note that the PAS-GAF D207A mutant of DrBphP assembled with PPIX might be an order of magnitude more fluorescent than when assembled with BV.5
Solution of the PSM structure by x-ray crystallography permitted modeling of the full hairpin motif and its parent PHY domain. It revealed a greatly bowed helical spine and verified the anti-parallel two β-strand hairpin stem thought to be a signature of canonical Phys that use Pr as the dark-adapted state (11, 21). Contacts between the hairpin and the GAF domain include residues within the stem as well as the intervening loop and involve many of the same conserved hairpin residues that participate in the Synechocystis Cph1 and Cph2 and Arabidopsis PhyB structures (10, 11, 21). As first described for Arabidopsis PhyB (11), the hairpin of DrBphP also extends the GAF domain β-sheet from strand β3 through a tenuous parallel β-strand connection. This interface is stabilized through multiple contacts, some of which involve residues in the conserved PRXSF and WGG motifs and Arg-202 in the GAF domain.
Superposition of the PAS-GAF region from the PSM structure with that determined for the PAS-GAF fragment alone revealed remarkable congruence, indicating that the PHY domain has little impact on the PAS-GAF conformation. However, the PHY domain clearly influences photochemistry of the PSM as evidenced here by the partially compromised absorption spectrum of the PAS-GAF fragment after saturating red light and its substantially more rapid Pfr → Pr thermal reversion (Fig. 1, B and C). Thus, it is possible that the PAS-GAF region does not engage the PHY domain during the initial steps in photoconversion (e.g. photoisomerization) but needs it during the later stages of the photocycle that eventually generates and maintains Pfr.
To date, the molecular functions of the PAS domain remain obscure. For the PSM of DrBphP, it participates modestly in dimer formation while possibly limiting the range of motion available at the PAS-GAF dimer interface. More globally, it is conceivable that the PAS domain through this restriction helps focus the conformational changes underpinning signal transmission from the bilin toward the PHY domain and the OPM. The figure-of-eight knot might aid in this focus by reinforcing the connection between the PAS and GAF domains.
From numerous studies, a toggle model has emerged to explain how light absorption by the bilin is mechanically transmitted to the output module, thereby controlling signaling by these photoreceptors (for various aspects of the model, see Refs. 11, 14, 21, 23, 30, 31, 34). The initial step in canonical Phys is thought to be a red light-induced Z to E isomerization of the C15=C16 bond, which flips the D pyrrole ring (13, 14). As the D pyrrole ring is amphipathic and is surrounded by a complementary “amphipathic” environment in the Pr state, its flip promotes sliding of the entire bilin to a new energetically favorable position within the pocket. At this Pfr position, the bilin is partially anchored through an interaction between the D pyrrole ring nitrogen and the carboxylate group of Asp-207 in DrBphP and new contacts between the GAF domain and the B and C ring propionates. The new bilin position induces altered conformations for the aromatic side chains within the pocket (Tyr-176 and Phy-203 in DrBphP) and crowding of Tyr-263. In a currently obscure manner, these changes (and likely others) serve to melt the β-strand interaction between the hairpin stem and the GAF domain, swivel the hairpin, and reform a new set of interactions between the GAF domain and a now helical hairpin stem (30). Presumably, the end result is to reorient the PHY domain relative to the GAF domain, which then reverberates downstream to the OPM. For DrBphP, the net effect is to presumably impact phosphotransfer between the sister HK domains or from the HK domains to the corresponding response regulator BphR.
Although previous studies support the initial molecular events in the toggle model, how conformational changes within the PSM impact the OPM have not been documented. Here, using SPEM of the Pr and Pfr states of full-length DrBphP, we have observed these changes as seen by large scale repositioning of the sister HK domains relative to the PSM. Docking the PSM crystal structure resolved here into the SPEM maps detected a shortening and broadening of the Pfr dimer caused by separation of the sister PHY domains at their shoulders, leading to a downward displacement of the HK domains by as much as 12 Å (Fig. 7F). Based on the bowed helical spine for the Pr state, this separation could be induced by straightening the bow upon photoconversion at the Ala-326 kink separating the GAF and PHY helices (see Fig. 5A). Importantly, because the HK domain immediately follows the PHY in the primary sequence, the outward movement of the PHY domain during photoconversion to Pfr should reposition the HK domains downward and distort or possibly melt the dimer interface near the HK domain catalytic sites (Fig. 11). The resulting weakened HK-HK interaction might underline our detection of two Pfr species as follows: the Pfr conformer in which the HK domains remain in contact and the Pfr′ conformer in which the HK domains have become more dynamic (Fig. 11). Given that full photoconversion of Pr to Pfr is not possible due to spectra overlap, an intriguing alternative scenario is that the Pfr and Pfr′ conformers reflect photoactivated DrBphP dimers in which the Pfr conformer represents the Pr/Pfr heterodimer and the Pfr′ conformer represents the Pfr/Pfr homodimer.
FIGURE 11.
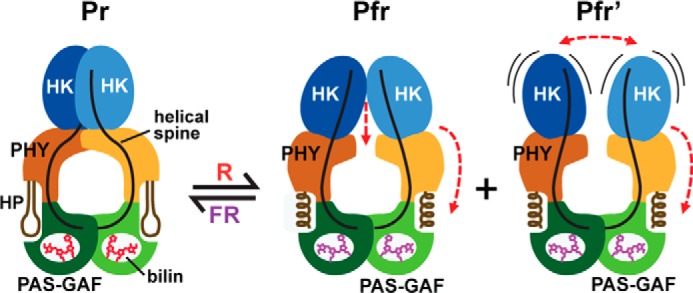
Model for DrBphP photoconversion. Model involves the reorientation of the sister PHY domains relative to the GAF domains through light-induced conformational changes in the PHY domain hairpin from β-strand to helical and altered contacts with the GAF domain. These modifications reorient and/or increase the mobility of the sister HK domains through alterations of the bowed helical spines to ultimately impact phosphotransferase activity of the dimer. HP, hairpin. Pfr and Pfr′ represent the two photoactivated states observed by SPEM.
It is also important to note that our SPEM modeling of the PAS-GAF region was nearly indistinguishable for the Pr and Pfr/Pfr′ states, strongly suggesting that light-induced conformational changes in this region are subtle and probably limited to the chemical level. Consequently, the PHY domain might act as an amplifier, which converts small input changes in the PAS-GAF region into a large physical separation that forces the downstream HK domains to either alter their relative orientations but remain in contact or possibly separate.
We presume that the conformational changes seen in the SPEM models are driven at least in part by structural rearrangements in the hairpin. Its participation was originally based on comparisons between hairpin structures of canonical Phys in the dark-adapted Pr state with those for two bathy-Phys as Pfr followed by mutational analyses (11, 21, 34), and more recently it was reinforced by the low resolution paired Pr and irradiated structures of the DrBphP PSM (30). After melting of the hairpin-GAF domain connection, the hairpin refolds from its β strand-dominated structure in Pr to a helical Pfr-type form. This change is coincident with positional swapping of the N- and C-terminal portions of the hairpin with respect to the GAF domain through a switch in the electrostatic contact between Asp-207 of the DIP motif and Arg-466 to the adjacent Ser-468 in the hairpin PRXSF motif (see Fig. 9A).
Given the observations of this light-induced hairpin swivel and interconversion between β-strand and helical stems (30), we provide functional support by analysis of a suite of site-directed mutants affecting this region, especially within the WGG, PRXSF, and HbXE motifs (see Fig. 9A and Ref. 21). As examples, a number of hairpin mutants predicted to strengthen the hairpin stem/GAF domain contacts as Pr or weaken the contact as Pfr had pronounced effects on the absorption spectrum of the photoactivated samples and dampened the stability of Pfr as seen by markedly faster rates of Pfr → Pr thermal reversion (L463R, H467P, S468E, T471P, and Y472A). Conversely, mutants (G453E and F469W) that might weaken the contact between the Pr hairpin and the GAF domain substantially slowed Pfr → Pr thermal reversion.
The main surprise from the mutant analyses of DrBphP was the relatively small stabilizing effect on the Pfr state for the G452E substitution, which shows strong consequences in Arabidopsis PhyB (11). Also notable was the negligible effect for the R466E substitution, which we expected to have strong effects given its electrostatic contact with Asp-207. Alanine replacement at a comparable position to Arg-466 in Synechocystis Cph1 (R472A) also had little effect on its photochemistry (10), whereas a similar alanine replacement in Arabidopsis PhyB (R582A) strongly compromised thermal reversion, but the resulting mutant photoreceptor was still competent in signaling in vivo (11, 50). Other mutations that seemingly would add stability to the Pfr state by virtue of their ability to hamper reattachment of the Pr-type hairpin (e.g. R303P, R466E, and W452G) also elicited only small effects. In any event, it is clear that full appreciation of the hairpin on signal transmission will likely require assays that measure the influence of individual amino acids on signal transmission by the downstream OPM (e.g. HK phosphotransfer).
Remarkably, we could still influence the Pfr → Pr thermal reversion rate of the DrBphP PSM by ∼1000-fold by single amino acid substitutions in the hairpin (e.g. G453E versus S468E (Fig. 10)), thus illustrating the strong impact this feature has on Pfr relaxation even without making direct contact with the bilin. The predominant energy barrier to thermal reversion is likely set by the probability of π-bond rupture at the C15=C16 bond that would permit rotation of the D pyrrole ring back to the 15Zs configuration. As this rotation likely coincides with back-sliding of the bilin within the GAF domain crevice to its Pr position, the hairpin might impact this movement by affecting the nearby amino acids (Asp-207 and Tyr-263) and the freedom of Tyr-176 and Phe-203 that are predicted to move in concert with the D ring during its isomerization-induced backflip (11). Clearly, higher resolution structures of the PSM in its pure Pfr state and the full-length structures of the Pr and Pfr dimers are now needed to fully appreciate how the hairpin strongly impacts the Pfr stability of Phys and how Phys reversibly transform the OPM between its dark-adapted and photoactivated states.
Acknowledgments
We thank Drs. Jeremiah Wagner, Junrui Zhang, and Andrew Ulijasz for the various DrBphP plasmids and advice on protein purification. We also appreciate the help of Drs. George Phillips, Jr., and Craig A. Bingman for technical advice and use of their crystallization facilities, and the Biophysics Instrumentation Facility at the University of Wisconsin at Madison for access to fluorimetry instrumentation.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI070285 (to H. L.). This work was also supported by National Science Foundation Grant MCB-1329956 (to R. D. V.) and Brookhaven National Laboratory Laboratory-directed Research Project 10-16.
The atomic coordinates and structure factors (codes 4Q0H, 4Q0I, and 4Q0J) have been deposited in the Protein Data Bank (http://wwpdb.org/).
E. S. Burgie and R. D. Vierstra, unpublished data.
J. M Walker, E. S. Burgie, and R. D. Vierstra, unpublished data.
T. Wang and H. Li, unpublished data.
- Phy
- phytochrome
- PSM
- photosensory module
- OPM
- output module
- GAF
- cGMP phosphodiesterase/adenylyl cyclase/FhlA
- PAS
- Period/Arnt/Single-minded
- PHY
- Phy-specific
- Pr
- red light-absorbing state
- Pfr
- far-red light-absorbing state
- BV
- biliverdin
- HK
- histidine kinase
- SPEM
- single particle electron microscopy
- r.m.s.d.
- root mean squared deviation
- PPIX
- protoporphyrin IXα
- PDB
- Protein Data Bank
- DrBphP
- D. radiodurans bacteriophytochrome.
REFERENCES
- 1. Auldridge M. E., Forest K. T. (2011) Bacterial phytochromes: more than meets the light. Crit. Rev. Biochem. Mol. Biol. 46, 67–88 [DOI] [PubMed] [Google Scholar]
- 2. Rockwell N. C., Su Y. S., Lagarias J. C. (2006) Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vierstra R. D., Zhang J. (2011) Phytochrome signaling: solving the Gordian knot with microbial relatives. Trends Plant Sci. 16, 417–426 [DOI] [PubMed] [Google Scholar]
- 4. Franklin K. A., Quail P. H. (2010) Phytochrome functions in Arabidopsis development. J. Exp. Bot. 61, 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gutu A., Kehoe D. M. (2012) Emerging perspectives on the mechanisms, regulation, and distribution of light color acclimation in cyanobacteria. Mol. Plant 5, 1–13 [DOI] [PubMed] [Google Scholar]
- 6. Bussell A. N., Kehoe D. M. (2014) in The Cell Biology of Cyanobacteria (Flores E., Herrero A., eds) pp. 149–170, Caister Academic Press, Poole, UK [Google Scholar]
- 7. Lamparter T., Carrascal M., Michael N., Martinez E., Rottwinkel G., Abian J. (2004) The biliverdin chromophore binds covalently to a conserved cysteine residue in the N terminus of Agrobacterium phytochrome Agp1. Biochemistry 43, 3659–3669 [DOI] [PubMed] [Google Scholar]
- 8. Froehlich A. C., Noh B., Vierstra R. D., Loros J., Dunlap J. C. (2005) Genetic and molecular analysis of phytochromes from the filamentous fungus Neurospora crassa. Eukaryot. Cell 4, 2140–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yeh K. C., Wu S. H., Murphy J. T., Lagarias J. C. (1997) A cyanobacterial phytochrome two-component light sensory system. Science 277, 1505–1508 [DOI] [PubMed] [Google Scholar]
- 10. Essen L. O., Mailliet J., Hughes J. (2008) The structure of a complete phytochrome sensory module in the Pr ground state. Proc. Natl. Acad. Sci. U.S.A. 105, 14709–14714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Burgie E. S., Bussell A. N., Walker J. M., Dubiel K., Vierstra R. D. (2014) Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc. Natl. Acad. Sci. U.S.A. 111, 10179–10184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lagarias J. C., Rapoport H. (1980) Chromopeptides from phytochrome. The structure and linkage of the Pr form of the phytochrome chromophore. J. Am. Chem. Soc. 102, 4821–4828 [Google Scholar]
- 13. Song C., Psakis G., Lang C., Mailliet J., Gärtner W., Hughes J., Matysik J. (2011) Two ground state isoforms and a chromophore D-ring photoflip triggering extensive intramolecular changes in a canonical phytochrome. Proc. Natl. Acad. Sci. U.S.A. 108, 3842–3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang X., Ren Z., Kuk J., Moffat K. (2011) Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome. Nature 479, 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ulijasz A. T., Cornilescu G., Cornilescu C. C., Zhang J., Rivera M., Markley J. L., Vierstra R. D. (2010) Structural basis for the photoconversion of a phytochrome to the activated Pfr form. Nature 463, 250–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rockwell N. C., Duanmu D., Martin S. S., Bachy C., Price D. C., Bhattacharya D., Worden A. Z., Lagarias J. C. (2014) Eukaryotic algal phytochromes span the visible spectrum. Proc. Natl. Acad. Sci. U.S.A. 111, 3871–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hirose Y., Rockwell N. C., Nishiyama K., Narikawa R., Ukaji Y., Inomata K., Lagarias J. C., Ikeuchi M. (2013) Green/red cyanobacteriochromes regulate complementary chromatic acclimation via a protochromic photocycle. Proc. Natl. Acad. Sci. U.S.A. 110, 4974–4979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Karniol B., Wagner J. R., Walker J. M., Vierstra R. D. (2005) Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors. Biochem. J. 392, 103–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bellini D., Papiz M. Z. (2012) Structure of a bacteriophytochrome and light-stimulated protomer swapping with a gene repressor. Structure 20, 1436–1446 [DOI] [PubMed] [Google Scholar]
- 20. Bussell A. N., Kehoe D. M. (2013) Control of a four-color sensing photoreceptor by a two-color sensing photoreceptor reveals complex light regulation in cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 110, 12834–12839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Anders K., Daminelli-Widany G., Mroginski M. A., von Stetten D., Essen L. O. (2013) Structure of the cyanobacterial phytochrome 2 photosensor implies a tryptophan switch for phytochrome signaling. J. Biol. Chem. 288, 35714–35725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rockwell N. C., Ohlendorf R., Möglich A. (2013) Cyanobacteriochromes in full color and three dimensions. Proc. Natl. Acad. Sci. U.S.A. 110, 806–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cornilescu C. C., Cornilescu G., Burgie E. S., Markley J. L., Ulijasz A. T., Vierstra R. D. (2014) Dynamic structural changes underpin photoconversion of a blue/green cyanobacteriochrome between its dark and photoactivated states. J. Biol. Chem. 289, 3055–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Narikawa R., Ishizuka T., Muraki N., Shiba T., Kurisu G., Ikeuchi M. (2013) Structures of cyanobacteriochromes from phototaxis regulators AnPixJ and TePixJ reveal general and specific photoconversion mechanism. Proc. Natl. Acad. Sci. U.S.A. 110, 918–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Burgie E. S., Walker J. M., Phillips G. N., Jr., Vierstra R. D. (2013) A photo-labile thioether linkage to phycoviolobilin provides the foundation for the blue/green photocycles in DXCF-cyanobacteriochromes. Structure 21, 88–97 [DOI] [PubMed] [Google Scholar]
- 26. Cornilescu G., Ulijasz A. T., Cornilescu C. C., Markley J. L., Vierstra R. D. (2008) Solution structure of a cyanobacterial phytochrome GAF domain in the red-light-absorbing ground state. J. Mol. Biol. 383, 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wagner J. R., Brunzelle J. S., Forest K. T., Vierstra R. D. (2005) A light-sensing knot revealed by the structure of the chromophore-binding domain of phytochrome. Nature 438, 325–331 [DOI] [PubMed] [Google Scholar]
- 28. Yang X., Stojkovic E. A., Kuk J., Moffat K. (2007) Crystal structure of the chromophore binding domain of an unusual bacteriophytochrome, RpBphP3, reveals residues that modulate photoconversion. Proc. Natl. Acad. Sci. U.S.A. 104, 12571–12576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang X., Kuk J., Moffat K. (2008) Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proc. Natl. Acad. Sci. U.S.A. 105, 14715–14720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takala H., Björling A., Berntsson O., Lehtivuori H., Niebling S., Hoernke M., Kosheleva I., Henning R., Menzel A., Ihalainen J. A., Westenhoff S. (2014) Signal amplification and transduction in phytochrome photosensors. Nature 509, 245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li H., Zhang J., Vierstra R. D. (2010) Quaternary organization of a phytochrome dimer as revealed by cryoelectron microscopy. Proc. Natl. Acad. Sci. U.S.A. 107, 10872–10977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wagner J. R., Zhang J., Brunzelle J. S., Vierstra R. D., Forest K. T. (2007) High resolution structure of Deinococcus bacteriophytochrome yields new insights into phytochrome architecture and evolution. J. Biol. Chem. 282, 12298–12309 [DOI] [PubMed] [Google Scholar]
- 33. Wagner J. R., Zhang J., von Stetten D., Günther M., Murgida D. H., Mroginski M. A., Walker J. M., Forest K. T., Hildebrandt P., Vierstra R. D. (2008) Mutational analysis of Deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes. J. Biol. Chem. 283, 12212–12226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yang X., Kuk J., Moffat K. (2009) Conformational differences between the Pfr and Pr states of Pseudomonas aeruginosa bacteriophytochrome. Proc. Natl. Acad. Sci. U.S.A. 106, 15639–15644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Davis S. J., Vener A. V., Vierstra R. D. (1999) Bacteriophytochromes: phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science 286, 2517–2520 [DOI] [PubMed] [Google Scholar]
- 36. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. A 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 37. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Emsley P., Cowtan K. (2004) COOT: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 39. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kabsch W. (1976) A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A 32, 922–923 [Google Scholar]
- 41. Ludtke S. J., Baldwin P. R., Chiu W. (1999) EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 42. Tang G., Peng L., Baldwin P. R., Mann D. S., Jiang W., Rees I., Ludtke S. J. (2007) EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 [DOI] [PubMed] [Google Scholar]
- 43. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 44. Bhoo S. H., Davis S. J., Walker J., Karniol B., Vierstra R. D. (2001) Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature 414, 776–779 [DOI] [PubMed] [Google Scholar]
- 45. von Stetten D., Günther M., Scheerer P., Murgida D. H., Mroginski M. A., Krauss N., Lamparter T., Zhang J., Anstrom D. M., Vierstra R. D., Forest K. T., Hildebrandt P. (2008) Chromophore heterogeneity and photoconversion in phytochrome crystals and solution studied by resonance Raman spectroscopy. Angew. Chem. Int. Ed. Engl. 47, 4753–4755 [DOI] [PubMed] [Google Scholar]
- 46. von Stetten D., Seibeck S., Michael N., Scheerer P., Mroginski M. A., Murgida D. H., Krauss N., Heyn M. P., Hildebrandt P., Borucki B., Lamparter T. (2007) Highly conserved residues Asp-197 and His-250 in Agp1 phytochrome control the proton affinity of the chromophore and Pfr formation. J. Biol. Chem. 282, 2116–2123 [DOI] [PubMed] [Google Scholar]
- 47. Auldridge M. E., Satyshur K. A., Anstrom D. M., Forest K. T. (2012) Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein. J. Biol. Chem. 287, 7000–7009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shu X., Royant A., Lin M. Z., Aguilera T. A., Lev-Ram V., Steinbach P. A., Tsien R. Y. (2009) Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science 324, 804–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mailliet J., Psakis G., Feilke K., Sineshchekov V., Essen L. O., Hughes J. (2011) Spectroscopy and a high-resolution crystal structure of Tyr263 mutants of cyanobacterial phytochrome Cph1. J. Mol. Biol. 413, 115–127 [DOI] [PubMed] [Google Scholar]
- 50. Zhang J., Stankey R. J., Vierstra R. D. (2013) Structure-guided engineering of plant phytochrome B with altered photochemistry and light signaling. Plant Physiol. 161, 1445–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]