Background: BRCA1 and PALB2 interact with each other to promote homologous recombination and DNA double strand break repair.
Results: Mice with abrogated PALB2-BRCA1 interaction show male fertility defect.
Conclusion: PALB2 and BRCA1 function together to ensure normal male meiosis.
Significance: This work demonstrates the importance of the PALB2-BRCA1 interaction in vivo and reveals a novel role of PALB2 in sex chromosome synapsis.
Keywords: BRCA1, DNA Damage Response, Homologous Recombination, Meiosis, Spermatogenesis, BRCA2, FANCN, Fanconi Anemia, PALB2, Male Infertility
Abstract
PALB2 links BRCA1 and BRCA2 in homologous recombinational repair of DNA double strand breaks (DSBs). Mono-allelic mutations in PALB2 increase the risk of breast, pancreatic, and other cancers, and biallelic mutations cause Fanconi anemia (FA). Like Brca1 and Brca2, systemic knock-out of Palb2 in mice results in embryonic lethality. In this study, we generated a hypomorphic Palb2 allele expressing a mutant PALB2 protein unable to bind BRCA1. Consistent with an FA-like phenotype, cells from the mutant mice showed hypersensitivity and chromosomal breakage when treated with mitomycin C, a DNA interstrand crosslinker. Moreover, mutant males showed reduced fertility due to impaired meiosis and increased apoptosis in germ cells. Interestingly, mutant meiocytes showed a significant defect in sex chromosome synapsis, which likely contributed to the germ cell loss and fertility defect. Our results underscore the in vivo importance of the PALB2-BRCA1 complex formation in DSB repair and male meiosis.
Introduction
PALB2, also known as FANCN, controls the function of BRCA2 (FANCD1) and links BRCA1 and BRCA2 in homologous recombination (HR)3-based repair of DNA double strand breaks (DSBs) (1, 2). Like BRCA1 and BRCA2, PALB2 itself is a tumor suppressor gene mutated in breast, ovarian, pancreatic, and other cancers (2–4). Like BRCA2, mono-allelic mutations in PALB2 increase the risk of adult cancers, whereas bi-allelic germline mutations cause Fanconi anemia (FA) (5, 6). FA is a rare, recessive syndrome characterized by congenital and developmental abnormalities, progressive bone marrow failure, and increased tumor incidence (7, 8). In addition, infertility is also a common feature among male FA patients (8). At the cellular level, the hallmark of FA is chromosomal breakage and hypersensitivity when treated with DNA interstrand crosslinkers (ICLs), which generate lesions that require HR for repair.
BRCA1, BRCA2, and PALB2 play a critical role in HR by promoting the recruitment of RAD51, the recombination enzyme, to DNA damage sites (5, 9–11). In particular, BRCA2, which can simultaneously bind up to 6–7 molecules of RAD51 (12), is directly responsible for much of RAD51 localization to damage sites, and its formation of the nucleoprotein filament essential for HR initiation (12–16). BRCA2, in turn, is stabilized in the nucleus and recruited to DNA damage sites by PALB2, the partner and localizer of BRCA2 (17). Interestingly, PALB2 also directly interacts with BRCA1, thereby connecting BRCA1 and BRCA2 in the HR pathway (18–20). The recruitment of BRCA1 to DSB sites is independent of PALB2 and BRCA2, while loss of BRCA1 greatly diminishes the recruitment of PALB2 and, in turn, that of BRCA2 and RAD51 (19, 20). Thus, there exists a BRCA1-PALB2-BRCA2-RAD51 axis for HR initiation, although each of the links may be subject to additional regulation.
Similar to Brca1 and Brca2, systemic knock-out of Palb2 leads to early embryonic lethality (21–23), and conditional ablation in the mammary gland results in breast cancer development (23, 24). To circumvent the embryonic lethality of complete Palb2 knock-out and further study PALB2 function in vivo, we generated a knockin mutation in the N terminus of mouse PALB2 that abrogates its interaction with BRCA1. Cells derived from the mutant animals showed hypersensitivity and greatly elevated chromosomal instability when treated with an ICL. Although the mutant mice were viable and developed normally, males homozygous for the mutation displayed reduced fertility due to germ cell loss in the testes.
EXPERIMENTAL PROCEDURES
Generation of the Palb2CC6 Knockin Strain
The targeting construct was based on an existing conditional knock-out construct recently described (23). The construct contains loxP sites flanking exons 2 and 3 of the mouse Palb2 gene. The CC6 mutation was introduced into exon 2 by site-directed mutagenesis. The resulting plasmid was used for gene targeting, which was performed by the Transgenic/Knock-out Mouse Shared Resource of the Rutgers Cancer Institute of New Jersey, using V6.5 ES cells (F1 hybrid of C57BL/6 and 129/Sv). Genomic DNA was extracted from neomycin-resistant ES cell clones, digested with BamH1 or BsaB1 and subjected to Southern blotting analysis using 5′ and 3′ probes. Verified ES cells were injected into blastocysts of C57BL/6 mice to generate chimeras, which were then crossed with FLP deleter mice (129S4/SvJaeSor-Gt(ROSA)26Sortm1(FLP1)Dym/J, stock number 003946, The Jackson Laboratory) to achieve germline transmission and remove the drug (neomycin) selection cassette. The final strain contains the knockin mutation along with two loxP sites and one Frt site on a mixed 129/B6 background. Genotyping was conducted as described (23). All mouse work was approved by the Institutional Animal Care and Use Committee (IACUC) of the Rutgers Robert Wood Johnson Medical School (New Brunswick, NJ) under the protocol numbers I08-073-9 and I11-029-5.
Cell Culture
293T and DR-U2OS cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1× penicillin-streptomycin (Pen-Strep). B cells were isolated from spleens of 8-week-old males using anti-CD43 Microbeads (Miltenyi Biotech) and stimulated with 25 μg/ml LPS (Sigma) and 50 units/ml IL-4 (Invitrogen) as described (25). The cells were cultured in RPMI 1640 supplemented with 10% heat-inactivated FBS, 1× Pen-Strep, 1% glutamine, 1X non-essential amino acids, 10 mm HEPES, 1% sodium pyruvate, and 53 μm β-mercaptoethanol. Primary mouse embryonic fibroblasts (MEFs) were generated from E13.5 embryos according to standard protocol and cultured in DMEM with 15% FBS and 1× Pen-Strep. All cells were cultured at 37 °C in a humidified incubator with 5% CO2.
Plasmid Constructs
The CC mutant series of human PALB2 expression constructs were based on pOZ-FH-C-PALB2Δ4, which expresses an internal deletion mutant of PALB2 lacking the segment encoded by exon 4 (aa 71–561) but still able to bind BRCA1 and BRCA2 (5). The CC1–11 mutations were generated by site-directed mutagenesis following the QuikChange protocol (Agilent Technologies). The pOZ vectors express various versions of PALB2 proteins double-tagged with FLAG and HA epitopes at the C terminus.
cDNA Transfection, Immunoprecipitation (IP), and Western Blotting
cDNA constructs were transfected into 293T cells in 6-well plates using FuGENE® 6 HD (Roche). Cells were plated at a density of 5 × 105 cells per well the day before transfection, and 2 μg of plasmid and 4 μl of transfection reagent were used for each well. Cells were collected 30 h after transfection and lysed with NETNG-300 (300 mm NaCl, 1 mm EDTA, 20 mm Tris-HCl, 0.5% Non-Idet P-40, and 10% glycerol) containing Complete® protease inhibitor mixture (Roche). The double-tagged proteins were IPed overnight with anti-FLAG M2-agarose beads (Sigma). For Western blotting analyses, proteins were resolved on 4–12% Tris-glycine gels, transferred onto nitrocellulose membranes and probed with relevant antibodies. Immobilon Western Chemiluminescent HRP Substrate (Millipore) was used to develop the blots. The PALB2 antibodies M11 and NB3 used were raised in rabbits against aa 601–880 of human PALB2 and aa 1–200 of mouse PALB2, respectively, and affinity purified. Other antibodies used were HA (HA7, Sigma), BRCA1 (Ab-4, EMD Biosciences), BRCA2 (Ab-1, EMD Biosciences), RAD51 (H-92, Santa Cruz Biotechnology), α-tubulin (T9026, Sigma), and β-Actin (AC-15, Santa Cruz Biotechnology).
Gel Filtration
Mouse PALB2 proteins tagged with FLAG-HA epitopes at the C terminus were expressed in 293T cells by transient transfection as described above. 30 h after transfection, cells were collected and lysed in NETNG250 (250 mm NaCl, 1 mm EDTA, 20 mm Tris-HCl, 0.5% Non-Idet P-40, 10% glycerol) with 5 mm NaF. Insoluble material was removed by high speed centrifugation (16,000 rpm for 30 min) at 4 °C. 2 mg of each extract was analyzed on an FPLC AKTA Purifier (GE Healthcare) with a Superpose 6, 10/300 GL Tricorn column pre-equilibrated with NETNG250 buffer containing 5 mm NaF. 0.5 ml fractions were collected, and the proteins were precipitated with acetone and analyzed by Western blotting.
Homologous Recombination Assay
The DR-U2OS cell line stably integrated with a single copy of the DR-GFP repair substrate DNA has been described elsewhere (17, 26). To measure homologous recombination (HR) activity of various PALB2 proteins, DR-U2OS cells were first transfected with a PALB2 siRNA to deplete endogenous protein. 30 h after transfection, cells were split into 6-well plates at 180,000 cells per well and allowed to adapt overnight. Cells were then co-transfected with 1.5 μg of I-SceI expression vector (pCBASce) and 0.5 μg of pOZC vector or various PALB2Δ4 constructs. GFP-positive cells resulting from HR-mediated repair of the I-SceI-induced DSBs were scored by flow cytometry 48 h after the second transfection, using a Beckman Coulter Cytomics FC 500 flow cytometer. The sequences of the control and PALB2 siRNAs are 5′-UUCGAACGUGUCACGUCAAdTdT and 5′-UCAUUUGGAUGUCAAGAAAdTdT, respectively.
B Cell Chromosome Spreads
For analysis of metaphase chromosomes, mitomycin C (MMC, Sigma Cat. M4287) was added to activated B cells at a final concentration of 250 nm for 16 h, after which cells were arrested with 100 ng/ml colcemid (Sigma) for 1 h. This was followed by treatment with hypotonic solution (0.075 m KCl) and fixation with 3:1 methanol/acetic acid. Telomere-FISH analysis was performed with Cy3-labeled telomere peptide nucleic acid probe (Panagene). 50–55 images were analyzed per sample.
Immunofluorescence
To detect RAD51 foci in activated B cells before and after MMC treatment, concentrated cell suspensions were seeded onto poly-l-lysine-coated coverslips, fixed and stained with anti-RAD51 (H-92, Santa Cruz Biotechnology) as described (27).
Cell Viability Assay
Activated B cells were seeded into 96-well plates at a density of 4,000 cells per well. MMC was added, and cells were incubated with the drug for 48 h. Cell viability was assessed with CellTiterGlo (Promega) according to the manufacturer's instructions. Luminescence signal was read on the GloMax®-96 Microplate luminometer (Promega).
β-Galactosidase Staining
Staining for acidic β-galactosidase was carried out using the Senescence β-Galactosidase Staining Kit (Cell Signaling Tech.) according to manufacturer's instructions. MEFs were analyzed in passage 3. The assay was allowed to proceed for 72 h before positive cells were scored.
Meiotic Spreads
Prophase I spermatocyte surface spreads were prepared from testes of 8-week-old males (28, 29). Decapsulated testes were processed in testis cell isolation medium (104 mm NaCl, 45 mm KCl, 1.2 mm MgSO4, 0.6 mm KH2PO4, 0.1% (w/v) glucose, 6 mm sodium lactate, 1 mm sodium pyruvate, pH adjusted to 7.3 with 1N NaOH) to isolate the seminiferous tubules. Single cells in 0.1 m sucrose solution at various stages of prophase I were spread onto slides coated with 1% paraformaldehyde/0.1% Triton X-100 in a moist chamber, then dried for 3.5 h. The slides were subsequently rinsed with Photo-Flo 200 solution (1:250, Kodak 146 4510) and air-dried. Antibodies used were SYCP3 (Abcam, 15093; Santa Cruz Biotechnology, sc-74569), RAD51 (EMD Biosciences, PC130), DMC1 (Santa Cruz, sc-22768), γ-H2AX (Millipore, 05636), and MLH1 (BD Biosciences, 51-1327GR). Nuclei were sub-staged (30) based on SYCP3 staining as leptotene (short stretches of axis with no synapsis), early zygotene (longer stretches of axis and some synapsis), late zygotene (the majority of axes are synapsed) and pachytene (completely synapsed axes with intensely staining sex body chromatin).
Tissue Collection and Analyses
Testes were collected from 8-week-old male mice immediately after euthanization by CO2 asphyxiation. Testes were fixed overnight either in Bouin's or phosphate-buffered formalin, transferred to 70% ethanol, embedded in paraffin, and cross-sectioned at 3-μm thickness. Sections were stained with hematoxylin and eosin (H&E) and examined by light microscopy.
Immunohistochemistry
Tissue sections were de-waxed in xylene and hydrated in a graded series of ethanol. For staining with γ-H2AX, after a brief wash with PBS, antigen retrieval was carried out with 10 mm sodium citrate buffer, pH 6.0 at 95 °C for 30 min after which the slides were cooled down to room temperature. Sections were incubated in 3% H2O2 for 10 min, blocked with 10% goat serum in PBS for 1 h at room temperature. Incubation with γ-H2AX antibody (Millipore, 05-636) was carried out overnight at 4 °C in a humidified chamber at a dilution of 1:200 in 5% goat serum. Secondary antibody incubations were carried out using Dako LSAB2 System-HRP (Dako, K0675) following the manufacturer's protocol. Bound antibodies were visualized with 3,3′-diaminobenzidine followed by washes with distilled water. Sections were counter-stained with hematoxylin and dehydrated in a graded series of ethanol, followed by xylene before mounting with CytoSeal-60 (Thermo Scientific).
Terminal Deoxynucleotidyltransferase dUTP Nick-end Labeling (TUNEL) Assay
Staining was performed using the DeadEndTM Fluorometric TUNEL System (Promega) according to the manufacturer's instructions on 3-μm sections.
Statistical Analyses
Statistical analyses were performed using ANOVA or two-tailed Student's t test with GraphPad Prism 6. Significance is denoted as the following: ns, p > 0.05; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001 and ****, p ≤ 0.0001.
RESULTS
Mutational Analysis of the PALB2 Coiled-coil Motif
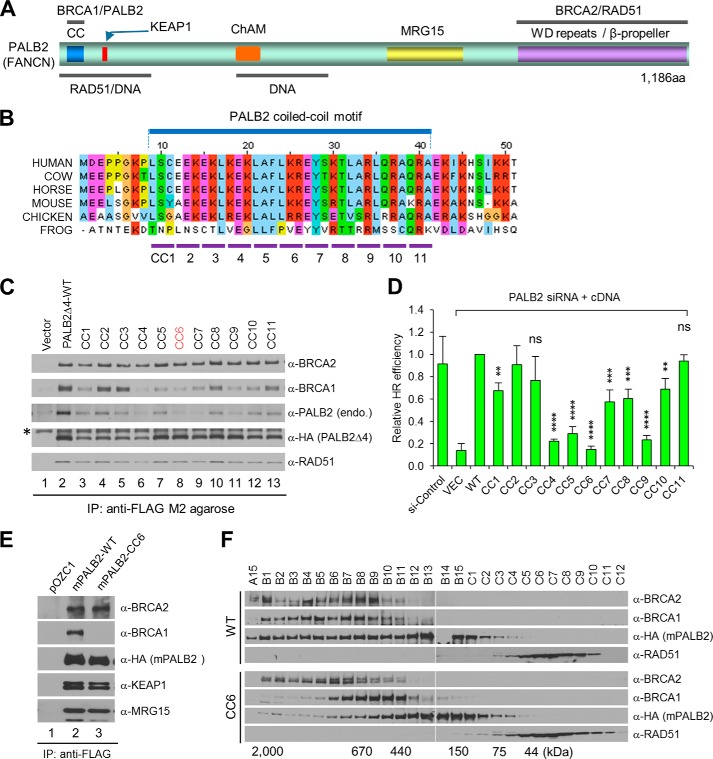
PALB2 contains a highly conserved coiled-coil (CC) motif at the N terminus and an equally conserved WD repeat domain at the C terminus (Fig. 1A). The CC motif directly interacts with BRCA1 (18–20), and the WD domain forms a seven-bladed β–propeller structure that tightly binds BRCA2 (31). PALB2 also directly interacts with KEAP1, an oxidative/electrophilic stress sensor, via a conserved “ETGE” motif at the N terminus (27). Additionally, PALB2 has been reported to bind RAD51, MRG15, and DNA, although the exact binding sites have not been well characterized (32–35). Interestingly, the CC motif can also mediate the formation of a PALB2 dimer and/or oligomer (36, 37). A PALB2 protein deleted of the motif is monomeric in vitro and shows higher DNA binding affinity and increased activity in promoting RAD51-mediated strand invasion, suggesting that PALB2 self-association may regulate its activity (37). However, cell- or animal-based data supporting this notion are lacking, and it is currently unknown whether PALB2 binding to BRCA1 is separable from its self-association.
FIGURE 1.
Mutational analysis of the PALB2 coiled-coil motif. A, schematic of PALB2 domain structures and binding sites for its interacting partners and DNA. ChAM stands for chromatin-association motif. B, sequence alignment of the N-terminal residues in PALB2 from various species. Shown below are the residues from the coiled-coil motif (aa 9–41) that are mutated in each of the CC mutants. C, effects of CC mutations on PALB2 binding to BRCA1 and its self-association. The WT and CC mutant PALB2Δ4 proteins were transiently expressed in 293T cells, and their expression levels and binding partners were subjected to immunoprecipitation (IP)-Western analysis using indicated antibodies. The asterisk indicates a nonspecific band reacting to the HA antibody. Note: the endogenous PALB2 protein is sufficiently larger than the PALB2Δ4 protein so that it can be separated adequately from the latter. D, effect of CC mutations on the HR activity of PALB2. DR-U2OS cells were first depleted of endogenous PALB2 and then co-transfected with an I-SceI endonuclease expression vector and each of the mutant constructs. GFP-positive cells were quantitated by flow cytometry. Error bars represent standard deviations (SDs) from three independent experiments. ns, p > 0.05; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001 and ****, p ≤ 0.0001. E, effect of CC6 mutation on the binding of mouse PALB2 to BRCA1 and other interacting partners. The same procedure was performed as in C. F, gel filtration elution profiles of the WT and CC6 mutant mouse PALB2 proteins. See text for details.
To determine the residues in the PALB2 CC motif required for BRCA1binding and self-association, we conducted alanine-scanning mutagenesis in which every three consecutive amino acids of the 33-residue motif were mutated into alanines, creating 11 mutants (CC1 to CC11; Fig. 1B). To facilitate the analysis of PALB2 self-association, the mutations were generated in an abbreviated version of PALB2 wherein the region encoded by exon 4 (aa71–561) was deleted. This shortened PALB2 protein was previously identified as a natural revertant in an FA-N cell line and shows full HR activity (5). The ability of the mutant proteins to bind endogenous BRCA1 and PALB2 was assessed by transient expression followed by immunoprecipitation (IP)-Western blotting analysis. Mutants CC1-CC4 and, to lesser extents, CC7, CC10 and CC11, showed lower expression levels. The remaining mutant proteins were expressed at levels close to that of the wild-type protein (Fig. 1C). Mutant proteins CC2 and CC3 appeared to largely retain BRCA1 binding capacity, whereas all the other mutants displayed reduced binding affinity, albeit to varying degrees. As to dimer/oligomer formation, all mutants showed reduced ability to associate with the endogenous PALB2 protein. Notably, the degrees to which the different mutations affected PALB2 self-binding and BRCA1 binding did not completely correlate, suggesting that the binding modes of PALB2-BRCA1 and PALB2-PALB2 association are similar but not identical. As expected, all mutant proteins were able to bind BRCA2 and RAD51.
To test the functionality of the mutants in HR, we performed a “protein replacement” assay, wherein endogenous PALB2 is first depleted by an siRNA and then PALB2 mutants are expressed from siRNA-resistant cDNAs, using a DR-U2OS cell line harboring a single copy of a GFP-based HR reporter (26). Mutants CC4, CC5, CC6, and CC9 resulted in a three- to 5-fold reduction of HR activity, whereas other mutants retained over 50% of the function (Fig. 1D). Since monomeric PALB2 has been shown to possess higher activity in DNA binding and RAD51-mediated strand invasion in vitro (37) and a monomeric PALB2 recruited to DNA sites by a fused BRCT domain is functional in HR in the cell (38), it appears that loss of BRCA1 binding is likely the cause of the reduced HR activities of the mutant proteins.
Generation of a Palb2CC6 Knock-in Mouse Strain
To study the functional relevance of the PALB2-BRCA1 interaction in vivo, we decided to knock in the CC6 mutation in the mouse Palb2 locus. This mutation was chosen because it resulted in a protein that was relatively stable yet showed severely diminished BRCA1 binding and the lowest HR activity among the mutations described above. To ensure that the mutation would produce expected functional deficiencies in the mouse, we introduced it into the mouse Palb2 cDNA, overexpressed the protein and tested its abilities to bind BRCA1 and to dimerize or oligomerize by IP-Western and gel filtration, respectively. Indeed, the mutation selectively abrogated the binding of mouse PALB2 to BRCA1, without affecting its interaction with BRCA2, KEAP1, and MRG15 (Fig. 1E). Gel filtration analysis showed that wild-type PALB2 eluted in fractions corresponding to molecular weights from 150 kDa to ∼2 MDa (Fig. 1F), indicating the formation of dimers and oligomers of different sizes. In contrast, the mutant protein eluted in what appeared to be a single but broad peak with a molecular mass range of 150 to 440 kDa, indicating that it is still able to form a dimer and perhaps also a trimer. Besides, in cells overexpressing the mutant PALB2, the elution profiles of BRCA1 and BRCA2 also appeared to be altered.
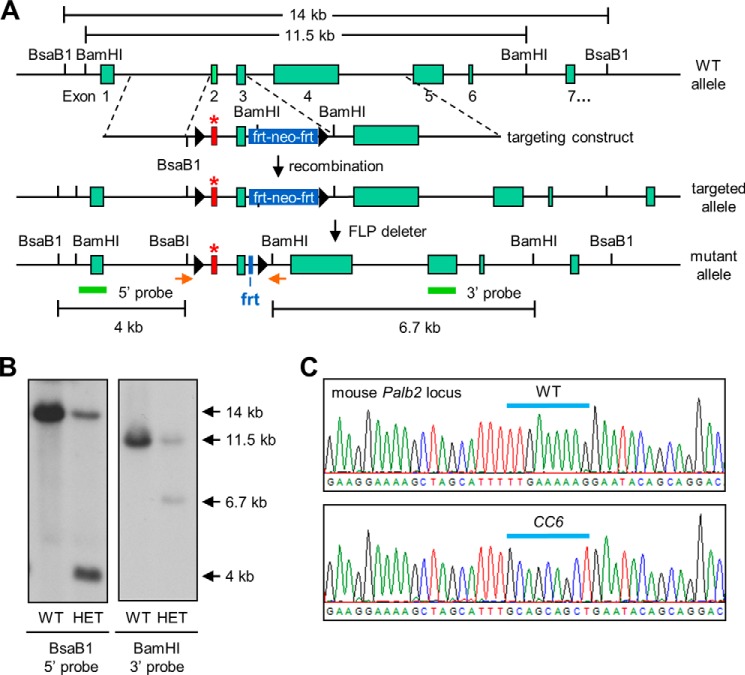
The CC6 mutation was knocked into the mouse Palb2 locus as illustrated in Fig. 2A. Southern blotting confirmed the proper integration of the targeting cassette into the Palb2 locus (Fig. 2B). Following the removal of the neo cassette by crossing the chimera with a FLP-deleter mouse, the final targeted allele is otherwise intact except for intronic loxP and frt sites. These extraneous sequences do not have any effects in the context of the wild-type Palb2 (23, 24). Palb2CC6/CC6 homozygous mutant mice were obtained at normal Mendelian frequency and showed normal development. The homozygous mutation was confirmed by sequencing genomic DNA from the mice (Fig. 2C). Since the ES cells (V6.5) used were derived from 129Sv/C57BL/6 F1 hybrid mice and the FLP-deleter had a 129S4/SvJaeSor background, the mutant mice are on a mixed genetic background.
FIGURE 2.
Generation of Palb2CC6/CC6 knockin mice. A, schematic of the mouse Palb2 locus and the procedure used to generate the knockin strain. The structure of the gene targeting construct used is shown below the WT Palb2 locus. The mutated exon 2 is shown in red, the frt-neomycin-frt drug selection cassette in blue, and the sequences used as 5′ and 3′ probes as green bars. LoxP sites are shown as black arrows. B, Southern blot results of representative ES recombinant clones. Genomic DNAs were digested with BsaB1 or BamH1 and then subjected to Southern blotting analysis, using 5′ and 3′ probes, respectively. Compared with the wild-type (WT) clone, the correct (heterozygous) recombinant clone (HET) showed an additional 4 kb band upon BsaB1 digestion and 6.7 kb band upon BamH1 cleavage. C, sequencing results of PCR-amplified mouse genomic DNA from WT (top) and MUT (bottom) mice, confirming the presence of the CC6 knockin mutation and absence of the WT allele in MUT mice. The positions of the PCR primers are shown as orange arrows in A.
ICL Repair Defects and Premature Senescence of Palb2CC6/CC6 Mutant Cells
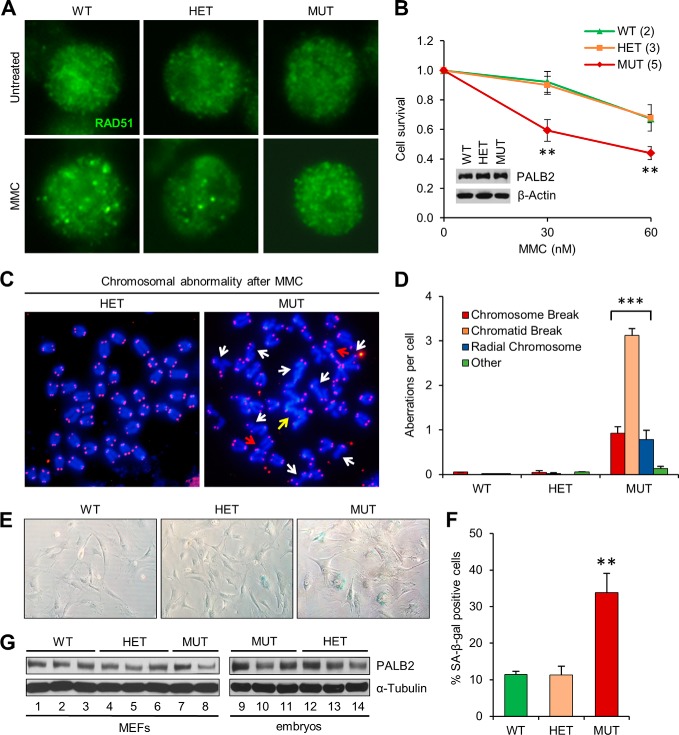
To test the DNA repair activity of the endogenous mutant PALB2 protein, we generated primary B lymphocytes from wild-type (WT), heterozygous (HET), and homozygous mutant (MUT) mice. Activated B cells were challenged with MMC, and RAD51 foci formation was examined as an indicator of repair activity. Bright RAD51 foci formed in the WT cells after MMC treatment, whereas the foci were smaller and less distinct in MUT cells, and. HET cells showed an intermediate phenotype (Fig. 3A). Moreover, MUT cells were more sensitive to MMC when compared with WT or HET cells (Fig. 3B). PALB2 protein abundance was similar in cells of the different genotypes (Fig. 3B), indicating that the mutation does not affect the intrinsic stability of the protein. The smaller RAD51 foci in the mutant cells and their MMC sensitivity suggest that the mutant protein is unable to promote optimal assembly of RAD51 into recombination-competent structures.
FIGURE 3.
ICL repair defect and premature senescence of Palb2CC6/CC6 mutant cells. A, RAD51 foci formation in activated B cells before and after mitomycin C (MMC) treatment. Cells derived from spleens of males of the indicated genotypes were treated with 150 nm MMC for 6 h and then fixed and analyzed by immunofluorescence (IF). B, MMC sensitivity of the mutant B cells. Activated B cells were treated with indicated concentrations of MMC for 48 h, and cell viability was determined with CellTiterGlo. Data presented are the average of three independent experiments. Immunoblot (inset) shows similar PALB2 protein levels among cells with the different genotypes. C and D, MMC-induced chromosome aberrations in mutant B cells. Cells were treated with 250 nm MMC for 16 h before harvest. Panel C shows representative metaphase spreads of cells following MMC treatment. White arrows denote chromatid breakage, red arrows denote chromosome breaks, and the yellow arrow indicates a radial chromosome. Panel D shows the quantification of different types of aberrations in cells of each genotype. At least 50 metaphase spreads were analyzed per genotype. E and F, increased cellular senescence of mutant MEFs at passage 3. Panel E shows representative images of β-gal stained cells, and F shows quantification of senescent cells in the MEFs. n ≥ 5 embryos per genotype were analyzed. G, Western blots showing PALB2 levels in MEFs (left) and E13.5 whole embryos (right). Error bars represent errors of the mean (S.E.). **, p ≤ 0.01; ***, p ≤ 0.001.
Bi-allelic germline mutations in PALB2 result in FA, the standard diagnosis of which is increased chromosomal breakage in response to ICL-inducing agents such as diepoxybutane (DEB) and MMC (8). To test if this hallmark feature of FA was recapitulated in our model, we prepared metaphase spreads from activated B cells before and after MMC treatment. In the absence of exogenous DNA damage, both the MUT and HET cells, like the WT cells, showed virtually no chromosomal abnormalities (data not shown). However, after MMC treatment, the MUT cells displayed greatly increased structural aberrations, including chromosome and chromatid breaks and radial chromosomes (Fig. 3, C and D). In contrast, chromosomes in WT and HET cells remained intact at the doses used.
We also attempted to test the above parameters using mouse embryonic fibroblasts (MEFs). However, MEFs from MUT embryos grew slower and showed premature senescence when compared with cells derived from either WT or HET embryos (Fig. 3E). Overall, there was an ∼3-fold increase in the percentage of cells expressing senescence-associated β-galactosidase (SA-β-gal) in the MUT MEFs by passage 3 (Fig. 3F). The protein did not appear to be grossly unstable since early-passage MEFs and whole embryos expressed similar amounts of protein across the three genotypes (Fig. 3G). Therefore, the premature senescence of the MUT cells was not due to an insufficient amount of the mutant protein.
Reduced Fertility and Testicular Abnormalities of Palb2CC6/CC6 Males
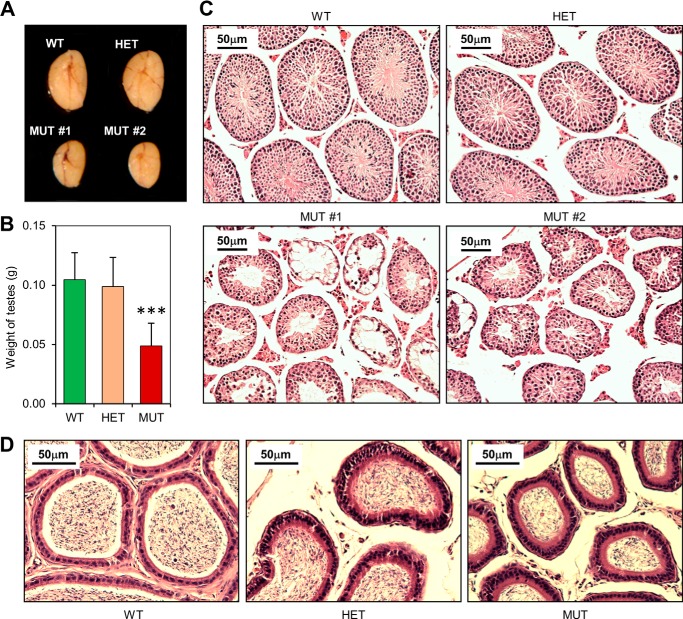
While female MUT mice were fertile, most MUT males were not. When MUT males did produce litters, only 1 or 2 pups per litter were observed (compared with an average litter size of 8 for WT and HET males). Consistent with the fertility defect, testes of MUT males were substantially smaller (Fig. 4A). The average weight of MUT testes was ∼50% of those of WT and HET males (Fig. 4B). Significant variability was observed, as some MUT testes were <30% the weight of WT testes and one of the ∼50 MUT testes harvested during the course of the study had a nearly normal size.
FIGURE 4.
Testicular abnormalities of the Palb2CC6/CC6mutant mice. A, representative images of testes from WT, HET, and MUT males at 8 weeks of age. Testes from two different MUT males are shown. B, quantification of the weights of the testes from n≥14 males for each genotype. C, H&E-stained cross-sections of the testes in A. Note the smaller sizes of the seminiferous tubules in the mutant testes. Error bars represent S.E.. ***, p ≤ 0.001. D, representative H&E-stained cross sections of epididymides of 8-week-old males.
Histological analyses were carried out to characterize the nature of the testicular defect in MUT males, using two MUT testes that represent the more typical size (Fig. 4A). The cross-section of a mouse testis reveals a number of seminiferous tubules, each with several germ cell types undergoing continuous cycles of proliferation and differentiation (39, 40). Spermatogonial stem cells at the periphery of each tubule undergo a series of mitotic divisions and eventually differentiate into spermatocytes, which then enter meiosis. The two divisions of meiosis finally yield haploid, round spermatids that mature into elongated spermatids, which further mature into spermatozoa before being released into the central lumen. Since the cells at the tubule periphery are at an earlier stage of development than the ones occupying the central portion and all the tubules in adult testes are not synchronized, a cross-section of a sexually mature normal testis will generally contain seminiferous tubules containing germ cells at all stages of spermatogenic development.
Hematoxylin and eosin (H&E)-stained cross-sections of WT or HET testes revealed well-developed seminiferous tubules, each displaying a clear progression in germ cell maturation, and almost all tubules contained mature spermatozoa (Fig. 4C). Consistent with the reduced testis size, tubules in the MUT testes were distinctly smaller (Fig. 4C). In one MUT testis (#1), large numbers of tubules showed a highly disorganized internal structure with apparent germ cell loss and no elongated spermatids, whereas in the other MUT testis (#2), the majority of tubules showed seemingly normal cellularity and contained spermatozoa. Still, even when spermatozoa were seen in the mutant tubules, they were generally fewer in number. As expected, MUT epididymis contained much reduced number of sperms (Fig. 4D). Thus, the reduced fertility of the MUT males was due to a defect in spermatogenesis.
DNA Repair Defect and Increased Apoptosis in the Mutant Testes
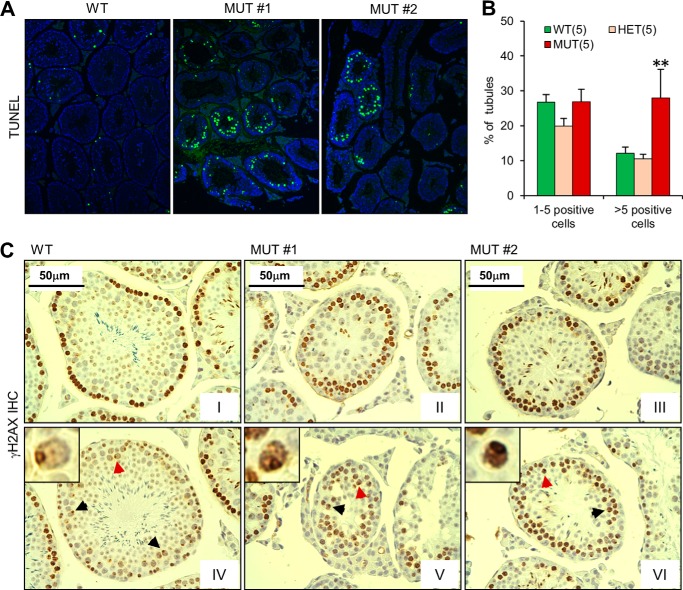
To test whether the lacunae formation and reduced sperm generation in the mutant seminiferous tubules were a result of apoptosis of germ cells, we performed the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay (Fig. 5A). The proportion of seminiferous tubules containing 1–5 TUNEL-positive germ cells was similar across the three genotypes. However, the proportion of seminiferous tubules containing >5 TUNEL-positive germ cells was about 2.5-fold higher in the MUT mutant testes than in either WT or HET testes (Fig. 5B). Considering that a significant number of tubules in the MUT testes were nearly devoid of germ cells, or apparently “post-apoptotic,” and therefore did not possibly reveal TUNEL-positive cells, the actual difference in apoptotic cell number could be larger.
FIGURE 5.
Defective DNA repair and increased apoptosis in the Palb2CC6/CC6mutant mice. A and B, apoptosis of germ cells in the testes. Panel A shows representative TUNEL staining patterns of the WT and MUT testes shown in Fig. 4. Panel B shows the quantification of seminiferous tubules with 1–5 or more than 5 apoptotic cells from 5 mice for each genotype. Error bars represent S.E.. **, p ≤ 0.01. C, IHC analysis of γH2AX in the WT and MUT testes in A. Two different images from each testis are shown. Arrows indicate representative pachytene cells. Insets show high-power views of the nuclei indicated by the red arrows.
During meiosis, the meiosis-specific transesterase SPO11 generates numerous DSBs along the chromosomes, which are subsequently repaired by recombination between homologous chromosomes (41, 42). To test whether the CC6 mutation in PALB2 affects the repair of SPO11-induced DSBs, we analyzed γH2AX levels in the testes by immunohistochemistry (IHC) (Fig. 5C). The first meiotic division has a lengthy prophase that is sub-divided into leptonema, zygonema, pachynema, diplonema, and diakinesis. The DSBs are induced in leptonema and persist into zygonema, and are accompanied by high levels of γH2AX formation. By pachynema, the DSBs are mostly repaired, and γH2AX only decorates the “sex body” consisting of the X and Y chromosomes (43). In WT seminiferous tubules, strong γH2AX staining was observed in cells located at the most outer layer, which corresponds to spermatogonia and leptotene (and perhaps some zygotene) spermatocytes (Fig. 5, C–I); strong staining of inner layer meiocytes was seen in only a small number of tubules, and pachytene cells showed weak staining except on the XY body (Fig. 5C-IV, inset). A similar observation was made in HET testes (data not shown). In the MUT testes, early meiocytes with strong pan-nuclear γH2AX staining were observed, as were pachytene cells with sex body staining, indicating meiotic progression. However, cells with strong γH2AX staining were often found at the second or even third layer in the tubules (Fig. 5C-II-III), suggesting that some meiotic cells had a defect in repair of SPO11-induced DSBs. Based on their location, morphology and staining pattern, most of these cells appear to be in the zygotene stage. Moreover, pachytene cells, i.e. those with darkly stained sex body, showing strong pan-nuclear staining were also occasionally seen (Fig. 5C, arrows and insets in panels V and VI).
In the WT and HET testes, the few apoptotic cells were predominantly at the basal layer of the seminiferous tubule, where spermatogonia and pre-leptotene/leptotene spermatocytes reside. In the MUT testes, however, apoptotic cells were mostly found in the inner layers corresponding to zygotene or later-stage spermatocytes (Fig. 5A). Taken together, our data suggest that the mutant PALB2 protein is unable to sustain adequate DSB repair during meiosis prophase I, resulting in spermatocytes loss via apoptosis during zygonema and later stages.
DMC1/RAD51 and MLH1 Foci Formation in the Mutant Testes
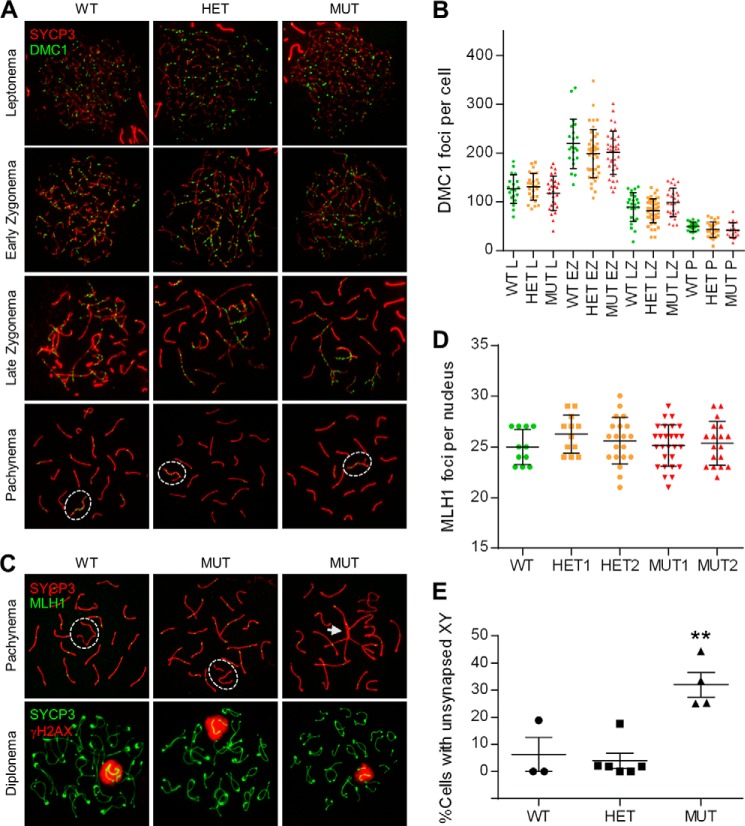
In mammals, defects in recombination and chromosome synapsis are associated with infertility that often manifest more severely in males than in females (44). RAD51 and its meiotic counterpart, DMC1, are components of recombination foci along leptotene and zygotene chromosomal axes. DMC1/RAD51 foci number reaches a peak in zygonema and then decreases as DSB repair progresses (30). By the end of pachynema, very few foci adorn the compacted chromosomes. The late-synapsing X and Y chromosomes, on the contrary, display relatively more DMC1/RAD51 foci at pachynema (45). To assess the ability of the mutant PALB2 to support meiotic recombination, we performed immunolabeling for DMC1 and RAD51 on meiotic spreads along with SYCP3, a structural component of the axis of the synaptonemal complex (SC). DMC1 foci appeared to form and disappear normally in all sub-stages of prophase I (Fig. 6, A and B), and a similar observation was made for RAD51 (data not shown).
FIGURE 6.
DMC1/RAD51 foci formation, synaptonemal complex (SC) assembly and chromosome synapsis in spermatocytes. A, SC assembly and DMC1 focus formation in normal and mutant spermatocytes. SC assembly was followed through the various stages of prophase I by IF staining for SYCP3 (red), and DMC1 focus formation (green) was analyzed to gauge recombination activity. B, quantification of DMC1 foci at various stages of prophase I. The 5 males analyzed were littermates (note that MUT1 and MUT2 are different from MUT #1 and MUT #2 in Figs. 4 and 5). Each dot represents the total foci count from a single nucleus. Horizontal lines denote the means and vertical lines the standard errors of the mean (SEs). C, double immunolabeling of SYCP3/MLH1 (top panels) and SYCP3/γH2AX (bottom panels) at pachynema and diplonema, respectively. Dashed white ovals delineate the sex chromosomes, which are synapsed at the pseudo-autosomal region (PAR) in the WT but asynapsed in the mutant cells. The white arrow marks a “tangle” involving the sex chromosomes and several autosomes. D, quantification of MLH1 foci at pachynema. E, quantification of XY asynapsis in the pachytene spermatocytes. At least 3 adult males per genotype were analyzed with ≥50 nuclei scored per animal. Error bars represent S.E.; **, p ≤ 0.001.
To further understand the meiotic defects in the MUT testes, we examined chromosome synapsis in the spermatocytes. Synapsis occurs in zygonema. By pachynema, every autosome is juxtaposed alongside its homologue to form 19 fully-assembled autosomal SCs; the X and Y sex chromosomes, which are sequestered in the sex body and mostly non-homologous, synapse only at the homologous pseudo-autosomal region (46). By examining SYCP3 through the sub-phases in prophase I, it was apparent that SC formation and autosomal synapses can occur normally in mutant meiocytes (Fig. 6A). MLH1 foci, which mark the sites of crossing-over between homologous chromosomes during mid-late pachytene stage, also appeared to form normally in the mutant spermatocytes (Fig. 6, C and D). However, in some mutant nuclei, there were “tangles” involving multiple chromosomes (Fig. 6C, white arrow). Importantly as well, a significant fraction (∼32%) of spermatocytes in the MUT testes displayed asynapsis of sex chromosomes in pachynema, which persisted into diplonema (Fig. 6, C and E). Formation of the sex body, as revealed by γH2AX staining, however, was not affected in the MUT spermatocytes (Fig. 6C, lower panel), suggesting that sex chromosome inactivation was normal.
DISCUSSION
In this study, we engineered a knockin mutation in mouse Palb2 that diminishes the PALB2-BRCA1 interaction. In contrast to the embryonic lethality phenotype caused by Palb2 knock-out, mice homozygous for the knockin mutation were viable and did not show obvious congenital or developmental defects. Importantly, the mice displayed ICL-induced chromosomal abnormalities and male fertility defects, which are also the hallmark characteristics of most FA gene knock-out animals (47, 48). These FA-like phenotypes underscore the functional importance of PALB2-BRCA1 communication in vivo.
PALB2 directly interacts with both BRCA1 and BRCA2. While the binding mode between PALB2 and BRCA2 has been revealed by crystal structure (31) and the control of BRCA2 by PALB2 is relatively well defined, how BRCA1 binds and regulates PALB2 is less clear. The matter is further complicated by the fact that the PALB2 CC motif responsible for BRCA1 binding also mediates its own homo-dimerization and oligomerization (36, 37). Deletion or mutations of this motif have been shown to severely affect HR activity (18–20). Based on a hypothesized model of a two-helix bundle formation between the corresponding CC motifs of PALB2 and BRCA1, Sy et al. mutated hydrophobic residues predicted to be responsible for the complex formation (18). The L21A, Y28A, and L35A mutants indeed showed greatly reduced BRCA1 binding capacity and strong HR defects. Consistent with the above finding, mutations CC5, CC7, and CC9 in the present study, each encompassing one of the above residues, significantly affected HR activity, although the effect of the CC7 mutation containing Y28A was moderate (Fig. 1D). Interestingly, our mutation CC6 (24LKK26→AAA) caused an even stronger reduction of HR activity, suggesting an involvement of additional residues and/or alternative binding modes between PALB2 and BRCA1.
We have previously shown that acute depletion of BRCA1 resulted in loss of (endogenous) PALB2 focus formation in human cells (19). However, the above noted L21A, Y28A, and L35A mutants of PALB2 that are defective in BRCA1 binding were found to form ionizing radiation-induced nuclear foci to varying degrees (18), suggesting that PALB2 can form damage-induced foci without binding to BRCA1. The discrepancy between the results may be explained by a model wherein BRCA1 facilitates PALB2 recruitment to DNA damage sites by both direct and indirect mechanisms. First, BRCA1 may directly recruit PALB2 via their protein-protein interaction. Alternatively, BRCA1 may facilitate PALB2 recruitment by promoting end resection, effectively creating a landing pad for PALB2 to dock via its inherent DNA binding and RPA-displacing activity (34, 35). A further possibility is that BRCA1 prevents 53BP1 accumulation at DSBs (25), thereby eliminating a potential inhibitory effect of 53BP1 on PALB2's access to DNA damage sites. Depletion of BRCA1 would interfere with all of these mechanisms, leading to severe loss of PALB2 foci formation, whereas the PALB2 mutants may still form foci, since BRCA1 is present and functional upstream. Importantly, the PALB2-L21A and Y28A mutants showing nearly normal levels of focus formation had much reduced HR activity (18), suggesting that the foci may be different in nature and nonfunctional or less functional without being first guided by BRCA1. Alternatively, PALB2 may need to interact with BRCA1 after its recruitment to be productive, as proposed by Sy et al. (18).
Mutation or depletion of PALB2 leads to a practically complete loss of RAD51 focus formation, indicating that PALB2 is critical for RAD51 recruitment to DNA damage sites (5, 18). Under normal conditions, the requirement of PALB2 for RAD51 recruitment is likely to be due to its role in localizing BRCA2 to break sites. Yet, given strong in vitro evidence that PALB2 can directly bind RAD51 and promote its strand-invading activity (34, 35), a direct, BRCA2-independent control of RAD51 by PALB2 may also contribute to RAD51 foci assembly. In this study, the observed RAD51 focus formation in the mutant B cells was likely due to a reduced but still significant recruitment of the CC6 mutant protein. However, the foci were not only smaller but also less functional, as the mutant cells were shown to be more sensitive to MMC (Fig. 3, A and B), again suggesting that BRCA1 may impart certain instructions to PALB2 (and thus to BRCA2 and RAD51) to promote optimal assembly of RAD51/DMC1 recombination structures for HR to proceed normally.
In the mutant spermatocytes, surprisingly, the formation and clearance of DMC1/RAD51 foci were normal at all stages of meiotic prophase I (Fig. 6, A and B and data not shown). Nonetheless, additional analysis revealed what appeared to be a prolonged zygotene stage in the mutant meiocytes with the cells staining strongly for γH2AX (Fig. 5C). Furthermore, some mutant cells showed strong pan-nuclear γH2AX staining in the pachytene stage. These findings imply that recombination did not proceed normally and that the chromosomes had accumulated some form(s) of unrepaired DNA damage, echoing the above notion that a direct communication between PALB2 and BRCA1 is important for the assembly of productive recombination structures. Our findings are reminiscent of earlier reports that spermatocytes in Brca1−/− (Trp53−/−) testes failed to progress (through the zygotene stage) into the pachytene stage and that Brca1Δ11/Δ11 spermatocytes showed abnormal, pan-nuclear γH2AX staining in late zygotene and pachytene stages (49, 50), further suggesting that the meiotic function of BRCA1 may be, at least in part, mediated by PALB2.
Interestingly, ∼30% of mutant spermatocytes were found to contain unsynapsed sex chromosomes during prophase I (Fig. 6, C and E). Sex chromosome asynapsis can lead to an arrest of meiotic progression and eventually apoptosis during the mid-pachytene stage (51). Indeed, substantially increased apoptosis were observed in the mutant testes (Fig. 5, B and C). Thus, it appears that the reduced fertility of the mutant males is caused by germ cell attrition resulting from a combination of unrepaired DNA breaks and XY asynapsis. Taken together, our results reveal new insights into the in vivo role of the PALB2-BRCA1 complex formation in DNA repair and male meiosis.
Acknowledgments
We thank Florent le Masson for valuable technical assistance, Scott Keeney for critical comments on the manuscript, and Reena Shakya for helpful discussions. We thank the Transgenic and Knockout Mouse Shared Resource of the Rutgers Cancer Institute of New Jersey, which is supported by an NCI CCSG Grant (P30CA072720) and by a Robert Wood Johnson Foundation grant to the Child Health Institute of New Jersey (Grant 67038).
This work was supported, in whole or in part, by the National Institutes of Health (R01CA138804, to B. X., R01GM105421, to M. J. and Scott Keeney, and R00CA160574, to S. F. B.), the American Cancer Society (RSG #TBG-119822, to B. X.).
- HR
- homologous recombination
- PALB2
- partner and localizer of BRCA2
- DSB
- double strand break
- CC
- coiled-coil
- FA
- Fanconi anemia
- MMC
- mitomycin C
- FISH
- fluorescent in situ hybridization
- TUNEL
- terminal deoxynucleotidyltransferase dUTP nick end labeling.
REFERENCES
- 1. Moynahan M. E., Jasin M. (2010) Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nature Reviews 11, 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tischkowitz M., Xia B. (2010) PALB2/FANCN: recombining cancer and Fanconi anemia. Cancer Res. 70, 7353–7359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Casadei S., Norquist B. M., Walsh T., Stray S., Mandell J. B., Lee M. K., Stamatoyannopoulos J. A., King M. C. (2011) Contribution of inherited mutations in the BRCA2-interacting protein PALB2 to familial breast cancer. Cancer Res. 71, 2222–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walsh T., Casadei S., Lee M. K., Pennil C. C., Nord A. S., Thornton A. M., Roeb W., Agnew K. J., Stray S. M., Wickramanayake A., Norquist B., Pennington K. P., Garcia R. L., King M. C., Swisher E. M. (2011) Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 108, 18032–18037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xia B., Dorsman J. C., Ameziane N., de Vries Y., Rooimans M. A., Sheng Q., Pals G., Errami A., Gluckman E., Llera J., Wang W., Livingston D. M., Joenje H., de Winter J. P. (2007) Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat. Genet. 39, 159–161 [DOI] [PubMed] [Google Scholar]
- 6. Reid S., Schindler D., Hanenberg H., Barker K., Hanks S., Kalb R., Neveling K., Kelly P., Seal S., Freund M., Wurm M., Batish S. D., Lach F. P., Yetgin S., Neitzel H., Ariffin H., Tischkowitz M., Mathew C. G., Auerbach A. D., Rahman N. (2007) Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat. Genet. 39, 162–164 [DOI] [PubMed] [Google Scholar]
- 7. Kee Y., D'Andrea A. D. (2012) Molecular pathogenesis and clinical management of Fanconi anemia. J. Clin. Investig. 122, 3799–3806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Auerbach A. D. (2009) Fanconi anemia and its diagnosis. Mutation Res. 668, 4–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bhattacharyya A., Ear U. S., Koller B. H., Weichselbaum R. R., Bishop D. K. (2000) The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J. Biol. Chem. 275, 23899–23903 [DOI] [PubMed] [Google Scholar]
- 10. Yuan S. S., Lee S. Y., Chen G., Song M., Tomlinson G. E., Lee E. Y. (1999) BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 59, 3547–3551 [PubMed] [Google Scholar]
- 11. Farmer H., McCabe N., Lord C. J., Tutt A. N., Johnson D. A., Richardson T. B., Santarosa M., Dillon K. J., Hickson I., Knights C., Martin N. M., Jackson S. P., Smith G. C., Ashworth A. (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 [DOI] [PubMed] [Google Scholar]
- 12. Liu J., Doty T., Gibson B., Heyer W. D. (2010) Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1260–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jensen R. B., Carreira A., Kowalczykowski S. C. (2010) Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467, 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang H., Li Q., Fan J., Holloman W. K., Pavletich N. P. (2005) The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature 433, 653–657 [DOI] [PubMed] [Google Scholar]
- 15. Thorslund T., McIlwraith M. J., Compton S. A., Lekomtsev S., Petronczki M., Griffith J. D., West S. C. (2010) The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1263–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Holloman W. K. (2011) Unraveling the mechanism of BRCA2 in homologous recombination. Nat. Struct. Mol. Biol. 18, 748–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xia B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F. J., Livingston D. M. (2006) Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell 22, 719–729 [DOI] [PubMed] [Google Scholar]
- 18. Sy S. M., Huen M. S., Chen J. (2009) PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc. Natl. Acad. Sci. U.S.A. 106, 7155–7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang F., Ma J., Wu J., Ye L., Cai H., Xia B., Yu X. (2009) PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr. Biol. 19, 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang F., Fan Q., Ren K., Andreassen P. R. (2009) PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol. Cancer Res. 7, 1110–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rantakari P., Nikkilä J., Jokela H., Ola R., Pylkäs K., Lagerbohm H., Sainio K., Poutanen M., Winqvist R. (2010) Inactivation of Palb2 gene leads to mesoderm differentiation defect and early embryonic lethality in mice. Hum. Mol. Genet. 19, 3021–3029 [DOI] [PubMed] [Google Scholar]
- 22. Bouwman P., Drost R., Klijn C., Pieterse M., van der Gulden H., Song J. Y., Szuhai K., Jonkers J. (2011) Loss of p53 partially rescues embryonic development of Palb2 knockout mice but does not foster haploinsufficiency of Palb2 in tumour suppression. J. Pathol. 224, 10–21 [DOI] [PubMed] [Google Scholar]
- 23. Bowman-Colin C., Xia B., Bunting S., Klijn C., Drost R., Bouwman P., Fineman L., Chen X., Culhane A. C., Cai H., Rodig S. J., Bronson R. T., Jonkers J., Nussenzweig A., Kanellopoulou C., Livingston D. M. (2013) Palb2 synergizes with Trp53 to suppress mammary tumor formation in a model of inherited breast cancer. Proc. Natl. Acad. Sci. U.S.A. 110, 8632–8637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huo Y., Cai H., Teplova I., Bowman-Colin C., Chen G., Price S., Barnard N., Ganesan S., Karantza V., White E., Xia B. (2013) Autophagy opposes p53-mediated tumor barrier to facilitate tumorigenesis in a model of PALB2-associated hereditary breast cancer. Cancer Disc. 3, 894–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bunting S. F., Callén E., Wong N., Chen H. T., Polato F., Gunn A., Bothmer A., Feldhahn N., Fernandez-Capetillo O., Cao L., Xu X., Deng C. X., Finkel T., Nussenzweig M., Stark J. M., Nussenzweig A. (2010) 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nakanishi K., Yang Y. G., Pierce A. J., Taniguchi T., Digweed M., D'Andrea A. D., Wang Z. Q., Jasin M. (2005) Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc. Natl. Acad. Sci. U.S.A. 102, 1110–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma J., Cai H., Wu T., Sobhian B., Huo Y., Alcivar A., Mehta M., Cheung K. L., Ganesan S., Kong A. N., Zhang D. D., Xia B. (2012) PALB2 interacts with KEAP1 to promote NRF2 nuclear accumulation and function. Mol. Cell. Biol. 32, 1506–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barchi M., Roig I., Di Giacomo M., de Rooij D. G., Keeney S., Jasin M. (2008) ATM promotes the obligate XY crossover and both crossover control and chromosome axis integrity on autosomes. PLoS Genet. 4, e1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Heyting C., Dietrich A. J. (1991) Meiotic chromosome preparation and protein labeling. Methods Cell Biol. 35, 177–202 [DOI] [PubMed] [Google Scholar]
- 30. Cole F., Kauppi L., Lange J., Roig I., Wang R., Keeney S., Jasin M. (2012) Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat. Cell Biol. 14, 424–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oliver A. W., Swift S., Lord C. J., Ashworth A., Pearl L. H. (2009) Structural basis for recruitment of BRCA2 by PALB2. EMBO Reports 10, 990–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sy S. M., Huen M. S., Chen J. (2009) MRG15 is a novel PALB2-interacting factor involved in homologous recombination. J. Biol. Chem. 284, 21127–21131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hayakawa T., Zhang F., Hayakawa N., Ohtani Y., Shinmyozu K., Nakayama J., Andreassen P. R. (2010) MRG15 binds directly to PALB2 and stimulates homology-directed repair of chromosomal breaks. J. Cell Sci. 123, 1124–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Buisson R., Dion-Côté A. M., Coulombe Y., Launay H., Cai H., Stasiak A. Z., Stasiak A., Xia B., Masson J. Y. (2010) Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 17, 1247–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dray E., Etchin J., Wiese C., Saro D., Williams G. J., Hammel M., Yu X., Galkin V. E., Liu D., Tsai M. S., Sy S. M., Schild D., Egelman E., Chen J., Sung P. (2010) Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Struct. Mol. Biol. 17, 1255–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sy S. M., Huen M. S., Zhu Y., Chen J. (2009) PALB2 regulates recombinational repair through chromatin association and oligomerization. J. Biol. Chem. 284, 18302–18310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Buisson R., Masson J. Y. (2012) PALB2 self-interaction controls homologous recombination. Nucleic Acids Res. 40, 10312–10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang F., Bick G., Park J. Y., Andreassen P. R. (2012) MDC1 and RNF8 function in a pathway that directs BRCA1-dependent localization of PALB2 required for homologous recombination. J. Cell Sci. 125, 6049–6057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ahmed E. A., de Rooij D. G. (2009) Staging of mouse seminiferous tubule cross-sections. Methods Mol. Biol. 558, 263–277 [DOI] [PubMed] [Google Scholar]
- 40. Oakberg E. F. (1956) Duration of spermatogenesis in the mouse and timing of stages of the cycle of the seminiferous epithelium. Am. J. Anat. 99, 507–516 [DOI] [PubMed] [Google Scholar]
- 41. Cole F., Keeney S., Jasin M. (2010) Evolutionary conservation of meiotic DSB proteins: more than just Spo11. Genes Dev. 24, 1201–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hunter N. (2007) in Topics in Current Genetics, Molecular Genetics of Recombination, Meiotic Recombination. Springer-Verlag, Heidelberg [Google Scholar]
- 43. Mahadevaiah S. K., Turner J. M., Baudat F., Rogakou E. P., de Boer P., Blanco-Rodríguez J., Jasin M., Keeney S., Bonner W. M., Burgoyne P. S. (2001) Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genet. 27, 271–276 [DOI] [PubMed] [Google Scholar]
- 44. Kolas N. K., Marcon E., Crackower M. A., Höög C., Penninger J. M., Spyropoulos B., Moens P. B. (2005) Mutant meiotic chromosome core components in mice can cause apparent sexual dimorphic endpoints at prophase or X-Y defective male-specific sterility. Chromosoma 114, 92–102 [DOI] [PubMed] [Google Scholar]
- 45. Kauppi L., Barchi M., Lange J., Baudat F., Jasin M., Keeney S. (2013) Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev. 27, 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kauppi L., Barchi M., Baudat F., Romanienko P. J., Keeney S., Jasin M. (2011) Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 331, 916–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bakker S. T., de Winter J. P., te Riele H. (2013) Learning from a paradox: recent insights into Fanconi anaemia through studying mouse models. Disease Models Mechanisms 6, 40–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tischkowitz M., Winqvist R. (2011) Using mouse models to investigate the biological and physiological consequences of defects in the Fanconi anaemia/breast cancer DNA repair signalling pathway. J. Pathol. 224, 301–305 [DOI] [PubMed] [Google Scholar]
- 49. Xu X., Aprelikova O., Moens P., Deng C. X., Furth P. A. (2003) Impaired meiotic DNA-damage repair and lack of crossing-over during spermatogenesis in BRCA1 full-length isoform deficient mice. Development 130, 2001–2012 [DOI] [PubMed] [Google Scholar]
- 50. Cressman V. L., Backlund D. C., Avrutskaya A. V., Leadon S. A., Godfrey V., Koller B. H. (1999) Growth retardation, DNA repair defects, and lack of spermatogenesis in BRCA1-deficient mice. Mol. Cell. Biol. 19, 7061–7075; Partial Retraction (2006) Mol. Cell. Biol. 26, 9571 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 51. Burgoyne P. S., Mahadevaiah S. K., Turner J. M. (2009) The consequences of asynapsis for mammalian meiosis. Nature Reviews. Genetics 10, 207–216 [DOI] [PubMed] [Google Scholar]