Background: Increased transcription by mammalian amino acid response (AAR) has been linked primarily to the GCN2/eIF2/ATF4 pathway.
Results: Some cells contain a GCN2- and ATF4-independent AAR pathway that is MEK-dependent.
Conclusion: A novel MEK pathway activates a specific subset of AA-responsive genes.
Significance: The mammalian AAR network is more complex than previously thought, extending beyond the GCN2/eIF2/ATF4 pathway.
Keywords: Amino Acid, Gene Transcription, Hepatocellular Carcinoma, Mitogen-activated Protein Kinase (MAPK), Transcription, ATF4, HepG2 Cells, Amino Acid Response, Asparagine Synthetase
Abstract
Amino acid (AA) limitation in mammalian cells triggers a collection of signaling cascades jointly referred to as the AA response (AAR). In human HepG2 hepatocellular carcinoma, the early growth response 1 (EGR1) gene was induced by either AA deprivation or endoplasmic reticulum stress. AAR-dependent EGR1 activation was discovered to be independent of the well characterized GCN2-ATF4 pathway and instead dependent on MEK-ERK signaling, one of the MAPK pathways. ChIP showed that constitutively bound ELK1 at the EGR1 proximal promoter region was phosphorylated after AAR activation. Increased p-ELK1 binding was associated with increased de novo recruitment of RNA polymerase II to the EGR1 promoter. EGR1 transcription was not induced in HEK293T cells lacking endogenous MEK activity, but overexpression of exogenous constitutively active MEK in HEK293T cells resulted in increased basal and AAR-induced EGR1 expression. ChIP analysis of the human vascular endothelial growth factor A (VEGF-A) gene, a known EGR1-responsive gene, revealed moderate increases in AAR-induced EGR1 binding within the proximal promoter and highly inducible binding to a site within the first intron. Collectively, these data document a novel AA-activated MEK-ERK-ELK1 signaling mechanism.
Introduction
Mammals detect and respond to inadequacies in protein or amino acid (AA)2 content via a collection of signaling cascades jointly referred to as the AA response (AAR) (reviewed in Refs. 1 and 2). The best characterized among these networks involves activation of the general control nonderepressible 2 (GCN2) kinase, which senses increased intracellular uncharged tRNA levels. Once activated, GCN2 phosphorylates the eukaryotic translation initiation factor (eIF2α), triggering a global suppression of protein translation initiation, but an increase in the translation of selected mRNAs including that for activating transcription factor 4 (ATF4) (3). Endoplasmic reticulum (ER) stress also triggers ATF4 synthesis through activation of a separate eIF2 kinase called PKR-like endoplasmic reticulum kinase (PERK) (4, 5). ATF4 mediates an increase in transcription from a number of genes that contain an enhancer sequence composed of a half-site for C/EBP and a half-site for the ATF family of transcription factors (6, 7); consequently, these sequences are referred to as a C/EBP-ATF response element (CARE). Most, but not all (8), functional CARE sites respond to ATF4, regardless of which eIF2 kinase was activated, so CARE sites can exhibit ER stress element activity or AAR element activity (reviewed in Ref. 1). The products of these CARE-containing genes control a wide range of physiological processes and microarray analyses of human HepG2-C3A (9) or HepG2 hepatocellular carcinoma cells (10), as well as mouse embryonic fibroblasts (MEF) (11), has revealed that AA limitation activates hundreds of genes, a majority of which have functions other than within protein or AA homeostasis. Interestingly, the list of genes induced during the AAR in HepG2 cells included several immediate-early response genes such as selected members of the FOS-JUN and the early growth response (EGR) transcription factor families (10). Follow-up studies established that cJUN is a novel AAR-inducible gene, for which the induction is ATF4-independent, and instead involves autoactivation in response to signaling from the JNK arm of MAPK signaling (12). The elevated cJUN expression impacts the regulation of other AAR targets, including ATF3 (13).
EGR1, which encodes a zinc finger transcription factor, is an important immediate-early response gene activated by a broad range of extracellular stimuli. EGR1 impacts cell growth, proliferation, differentiation, and apoptosis (reviewed in Ref. 14). The signaling pathways that control EGR1 transcription vary depending on the initial stimulus and target tissue, but many studies have established that phosphorylation of constitutively bound E twenty-six-like factor (ELK1) in response to MEK-ERK signaling is an important mechanism (14, 15). ELK1 belongs to the ternary complex factor subfamily of the ETS (E twenty-six) superfamily of transcription factors (16, 17). Once increased in its expression, EGR1 regulates the transcription of target genes by binding to GC-rich sequences (14, 18). Egr1 knock-out mice, though viable, exhibit impaired liver regeneration following partial hepatectomy, and Egr1 has been proposed as a central regulator of cell cycle progression during hepatocellular regeneration following injury (19). Thus, control of hepatic EGR1 expression by AA limitation or ER stress may be a critical factor in liver physiology.
The present study documents that the AAR-initiated induction of EGR1 transcription is not mediated by the well documented GCN2-eIF2-ATF4 signaling pathway, but instead by AA-responsive MEK-ERK signaling. ERK-dependent phosphorylation of ELK1, constitutively bound to the EGR1 gene is associated with increased transcription and a marked elevation of EGR1 expression. Therefore, these results provide evidence for the existence of an AA-controlled MEK signaling pathway that terminates with phosphorylation of ELK1. The AA-dependent transcription via p-ELK1 reveals a new family of transcription factors, the ETS family, within the AAR. Correspondingly, transcription is induced through ETS genomic enhancer sequences previously unknown to have AAR element activity. Furthermore, the induction of immediate-early response genes in AA-deprived tumor cells provides a possible link between protein/AA nutrition and cell growth in the transformed state.
EXPERIMENTAL PROCEDURES
Cell Culture
All of the cell lines used in these studies were cultured in DMEM (pH 7.4; Mediatech, Herndon, VA), supplemented with 1× nonessential AA, 2 mm glutamine, 100 μg/ml streptomycin sulfate, 100 units/ml penicillin G, 0.25 μg/ml amphotericin B, and 10% (v/v) fetal bovine serum. The HEK293T-ATF4 cell line was created by Ord et al. (20) after virally transforming HEK293T cells with a tetracycline (Tet)-inducible construct that contains the ATF4 coding region. The HEK293T-ATF4 DMEM was the same as above but was also supplemented with 10% (v/v) tetracycline-free fetal bovine serum, 25 μg/ml Zeocin, and 2.5 μg/ml blasticidin. All cells were maintained at 37 °C in an atmosphere of 5% CO2 and 95% air and maintained in growth phase at 60–70% confluence. Approximately 12 h prior to treatments, cells were replenished with fresh DMEM to ensure more complete nutrition when experiments were initiated. For the HEK293T-ATF4 cells, overexpression of ATF4 in the absence of other possible AAR signals was induced by adding tetracycline at the concentrations and times indicated. For activation of the AAR, cells were incubated in either DMEM lacking histidine (catalog number D9801-02; United States Biological, Swampscott, MA) or complete DMEM containing 2 mm histidinol (HisOH), an amino alcohol that triggers the AAR. HisOH competitively inhibits histidinyl tRNA synthetase, causing an increase in uncharged tRNAHis and thereby inducing the AAR (21). Replicating experiments with either DMEM-histidine or DMEM + HisOH yielded no qualitative differences.
Inhibitor Assays
The MEK inhibitor PD98059 (Sigma-Aldrich) was diluted in DMSO. The initial concentrations tested were chosen based on previous studies (22) and then optimized as described in the text. All cell lines were pretreated with an equal volume of DMSO (control) or PD98059 for 1 h prior to activation of the AAR for the indicated times in the continued presence of inhibitor.
Transient Transfection
HEK293T cells (0.5 × 106 cells/60-mm dish) were plated in DMEM 24 h before transfection to achieve 30–40% confluence. The cells were transiently transfected with plasmids expressing full-length ATF4 cDNA, a constitutively active MEK1 (MEKCA, kindly provided by Dr. Xingming Deng), or as a control, green fluorescent protein (GFP-pcDNA3.1) at 5 μg/60-mm dish, using a calcium phosphate protocol (23). The constitutively active MEKCA was created by the mutations S218E and S222D, two phospho-serine residues in the activation loop of MEK1 (24). The cells to be transfected were incubated with the plasmids overnight, washed twice with PBS, replenished with complete DMEM, and incubated for another 36 h prior to activation of the AAR. HC-04 human hepatocytes (MRA-156, MR4; ATCC, Manassas, VA) were transiently transfected using calcium phosphate.
Knockdown of Selected Proteins
The GIPZ-shRNA plasmid constructs against GCN2 (catalog number RHS4430-101133792) or a nonsilencing Control (catalog number RHS4346) were purchased from Open Biosystems (Huntsville, AL). The shRNA constructs were packaged in HEK293T cells with the Trans-Lentiviral shRNA packaging kit (catalog number TLP5912) following the manufacturer's protocol. HepG2 cells were incubated at 37 °C for 6 h with lentiviral particles containing the shRNA construct. The infected cells were cultured with fresh culture medium for 48 h before puromycin selection (2.5 μg/ml) for at least 14 days. After the initial puromycin selection, individual clones were isolated by serial dilution and screened for the reduction in ATF4 induction following activation of the AAR.
The siRNA si-ERK1 (L-003592-00-0005), siERK2 (L-003555-00-0005), siJNK1 (L-03514-00-0005), and siJNK2 (L-003505-00-0005) were purchased from Dharmacon/Thermoscientific and are unmodified siGENOME SMARTpool constructs. Transient siRNA transfections with 50 nm for each member to yield a total of 100 nm for siERK1+siERK2 or siJNK1+siJNK2 were performed in 12-well plates according to the manufacturer's protocol using DharmaFECT4 transfection reagent (T-2004-02) 72 h prior to activating the AAR. The same amount of siRNA with a scrambled sequence (D-001810-02-05) was used as the siControl.
RNA Isolation and Real Time Quantitative PCR (qPCR)
Total RNA was isolated with the TRIzol reagent (Invitrogen) following the manufacturer's directions. Transcription activity (hnRNA) and steady state mRNA levels were assayed by qPCR, as described previously (25). A 1-μg aliquot of total RNA was used to synthesize first strand cDNA with the qScript cDNA synthesis kit (Quanta Biosciences, Gaithersburg, MD). For qPCR, each cDNA sample was diluted 10× with TE buffer, 2 μl of this diluted solution was mixed with 10 μl of SYBR Green master mixture, and 5 pmol of forward and reverse primers were added in a total volume of 20 μl. The mixture was subjected to 40 cycles at 95 °C for 15 s and 60 °C for 60 s. The primers used are listed in Table 1. After qPCR, melting curves were acquired by stepwise increase from 55 to 95 °C to ensure that only a single product was amplified in the reaction. GAPDH was used as an internal control, and all calculations were based on the difference of threshold cycle number of the analyzed gene relative to the GAPDH mRNA content in the same sample.
TABLE 1.
PCR primers
| Primer specificity | Primer sequences (human)a |
|---|---|
| ASNS, mRNA | FP 5′-GCAGCTGAAAGAAGCCCAAGT-3′ |
| RP 5′-TGTCTTCCATGCCAATTGCA-3′ | |
| GCN2, mRNA | FP 5′-GAAATGGTAAACATCGGGCAAACTC-3′ |
| RP 5′-TTCACAAGAGCCAGGAGAATCTTCAC-3′ | |
| GAPDH, mRNA | FP 5′-TTGGTATCGTGGAAGGACTC-3′ |
| RP 5′-ACAGTCTTCTGGGTGGCAGT-3′ | |
| VEGF-A, mRNA | FP 5′-ACTGAGGAGTCCAACATCAC-3′ |
| RP 5′-CTTGTCTTGCTCTATCTTTC-3′ | |
| EGR1, mRNA | FP 5′-AGAAGGACAAGAAAGCAGACAAAAGTGT-3′ |
| RP 5′-GGGGACGGGTAGGAAGAGAG-3′ | |
| EGR1, transcription activity | FP 5′-CTACGAGCACCTGACCGCAGG-3′ |
| RP 5′-ACAGGACGCCAGGATGGTGG-3′ | |
| EGR1 P1, ChIP assay | FP 5′-CCCCGTCTCAGAAAGAATAAAAACATTA-3′ |
| RP 5′-CCTTGTGTCTGAATGTCCATTTTGC-3′ | |
| EGR1 P2, ChIP assay | FP 5′-CCTCTTTCGGATTCCCGCAG-3′ |
| RP 5′-GGTCCTTGTGGTGAGGGGTCA-3′ | |
| EGR1 P3, ChIP assay | FP 5′-GAGGGAGCGAGGGAGCAACC-3′ |
| RP 5′-CTCCAAATAAGGTGCTGCCCAAA-3′ | |
| EGR1 P4, ChIP assay | FP 5′-CATATTAGGGCTTCCTGCTTCCCATA-3′ |
| RP 5′-CCGCCTCTATTTGAAGGGTCTGG-3′ | |
| EGR1 P5, ChIP assay | FP 5′-GTCACGACGGAGGCGGACC-3′ |
| RP 5′-CGGCGGCTCCCCAAGTTC-3′ | |
| EGR1 P6, ChIP assay | FP 5′-CGCAGAGGACCGAGCTTTTGT-3′ |
| RP 5′-GCAGCCCCGCTCATCAAAA-3′ | |
| EGR1 P7, ChIP assay | FP 5′-GGGGATTCTCCGTATTTGCGTC-3′ |
| RP 5′-GGCTACCATTGACTCCCGAGGT-3′ | |
| EGR1 P8, ChIP assay | FP 5′-GTCCCAGCTCATCAAACCCAGC-3′ |
| RP 5′-AGAAGCGGCGATCACAGGACTC-3′ | |
| VEGF-A upstream, ChIP assay | FP 5′-ACTTTCCTGCTCCCTCCTCGC-3′ |
| RP 5′-CCACCAAGGTTCACAGCCTG-3′ | |
| VEGF-A promoter, ChIP assay | FP 5′-CGCTCGGTGCTGGAATTTGATA-3′ |
| RP 5′-TGGGGAATGGCAAGCAAAAA-3′ | |
| VEGF-A intron 1, ChIP assay | FP 5′-GCTGTCACTGCCACTCGGTCTC-3′ |
| RP 5′-GCAGCAATCCACCCCAAAAC-3′ | |
| VEGF-A intron 4, ChIP assay | FP 5′-GTGAGGATGTAGTCACGGATTC-3′ |
| RP 5′-CCAAAGGTCACATAGCGGGA-3′ | |
| JNK1, mRNA | FP 5′-CCATTTCAGAATCAGACTCATGCCA-3′ |
| RP 5′-TGTGGTGTGAAAACATTCAAAAGGC-3′ | |
| JNK2, mRNA | FP 5′-GGGATTGTTGTGCTGCATTTGATAC-3′ |
| RP 5′-TGGTTCTGAAAAGGACGGCTTAGTTT-3′ | |
| ERK1, mRNA | FP 5′-CGCTTCCGCCATGAGAATGTC-3′ |
| RP 5′-CAGGTCAGTCTCCATCAGGTCCTG-3′ | |
| ERK2, mRNA | FP 5′-CGTGTTGCAGATCCAGACCATGAT-3′ |
| RP 5′-TGGACTTGGTGTAGCCCTTGGAA-3′ | |
| ATF3, mRNA | FP 5′-GAGCGGAGCCTGGAGCAAAA-3′ |
| RP 5′-GGGGACGATGGCAGAAGCACT-3′ |
a FP, forward primer; RP, reverse primer.
Microarray Analysis
Total RNA was isolated using the Qiagen RNeasy kit (Qiagen), including DNase I treatment before the final elution to eliminate any DNA contamination. The integrity of total RNA was monitored with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). A 100-ng aliquot of total RNA from four independent incubations for each of four conditions (DMEM versus DMEM lacking histidine, each with or without 2.5 μm PD98059) was amplified using the GeneChip® WT PLUS reagent kit (Affymetrix, Santa Clara, CA) following the manufacturer's instructions, and then 5.5 μg of cDNA was fragmented and terminally labeled. Labeled targets were hybridized to Affymetrix GeneChip® Human Transcriptome Array 2.0 for 16 h at 45 °C and washed according to Affymetrix standard protocols. For gene expression analysis, arrays were normalized using RMA as implemented in Partek Genomics Suite 6.6 (Partek Incorporated, St. Louis, MO). Analysis of variance was used to detect differentially expressed genes. The microarray data have been deposited in the NCBI Gene Expression Omnibus (accession number GSE58869).
Protein Isolation and Immunoblotting
Whole cell protein was extracted with a RIPA buffer (50 mm Tris-HCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 150 mm NaCl, 2 mm EDTA, containing Pierce protease and phosphatase inhibitor mini-tablets (Thermo Scientific, Waltham, MA). Immunoblotting was performed as described previously (26). The membrane was then incubated with one of the following antibodies: rabbit anti-ATF4 polyclonal antibody (27), mouse anti-phospho ERK mouse monoclonal antibody (sc-7383), rabbit anti-total ERK polyclonal antibody (sc-94), rabbit anti-EGR1 polyclonal antibody (sc-189), and mouse anti-GAPDH monoclonal antibody (sc-32233) from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). The rabbit anti-β-actin polyclonal antibody (A2066) was from Sigma-Aldrich. The rabbit anti-JNK polyclonal antibody (9252s) was from Cell Signaling Technology (Danvers, MA). The bound secondary antibody was detected using an enhanced chemiluminescence kit (32106; Thermo Scientific) and then exposing the blot to Classic Blue Autoradiography Film BX (MIDSCI, St. Louis, MO).
Chromatin Immunoprecipitation
ChIP analysis was performed according to a previously published protocol (26). HepG2 cells were seeded at a density of 1.5 × 107/150-mm dish with DMEM and cultured for ∼36 h, which includes a transfer to fresh DMEM during the final 12 h prior to AAR induction. Immunoprecipitation was performed with one of the following antibodies: rabbit anti-ATF4 polyclonal antibody described previously (27), rabbit anti-RNA polymerase II polyclonal antibody (sc-899), rabbit anti-EGR1 polyclonal antibody (sc-189), rabbit anti-serum response factor (SRF, sc-335), and, as a nonspecific negative control, a normal rabbit IgG (sc-2027) purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The total ELK1 (9182) and S103 phospho-SRF (4261) antibodies were purchased from Cell Signaling, whereas the S383 phospho-ELK1 antibody was obtained from Abcam (32799; Boston, MA). Immunoprecipitated DNA was analyzed with qPCR as described above, using primers listed in Table 1. The ChIP results are presented as the ratio to input DNA.
Statistical Analysis
Each experiment contained three or more individual samples to detect experimental variation, and each experiment was repeated one or more times with separate batches of cells to ensure reproducibility between experiments. The data are expressed as the averages ± standard deviations within an individual experiment containing three or four replicates, and the results, analyzed using Student's t test, with p ≤ 0.05 were considered statistically significant.
RESULTS
AAR-Induced EGR1 Expression
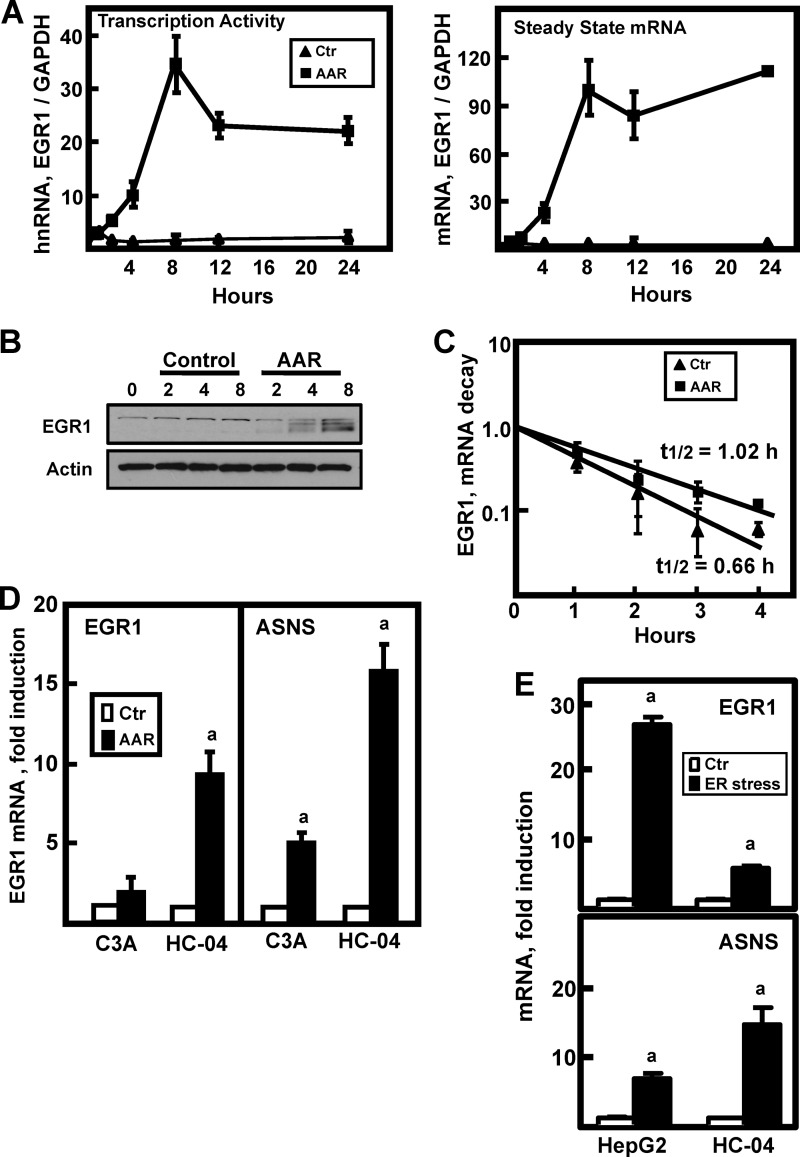
A recent microarray expression analysis performed by our laboratory revealed the EGR1 gene to be the most highly induced gene following activation of the AAR in HepG2 hepatoma cells (10). This observation is consistent with an earlier demonstration of EGR1 induction in MEF (11). To investigate the mechanism of the EGR1 induction in HepG2 cells, both transcription activity and steady state mRNA content was assayed from 1 to 24 h (Fig. 1A). The results showed that transcription from the EGR1 gene was increased within 2 h of AAR activation and was enhanced by more than 30-fold at 8 h. Steady state mRNA content paralleled the transcription activity (Fig. 1A), and cellular EGR1 protein content was also increased (Fig. 1B). The half-life of the EGR1 mRNA was modestly stabilized by AAR activation (Fig. 1C), but transcription appeared to account for most of the increased mRNA. Although the EGR1 induction was also evident in nontransformed, immortalized HC-04 human hepatocytes (Fig. 1D), compared with the HepG2 cells, it was considerably smaller in magnitude (compare values in Fig. 1A “steady state mRNA” to Fig. 1D). To determine whether this difference was linked to cellular transformation, additional human hepatoma cell lines were examined. The induction of EGR1 mRNA in Hep3B cells was less than 2-fold, and in LH86 cells it was ∼3.5-fold (data not shown). Interestingly, among the human hepatoma cells that did not exhibit significant EGR1 induction was the HepG2-derived subclone, C3A cells. The C3A subclone was selected from a parental HepG2 culture based on more “hepatocyte-like” properties (American Type Cell Culture). Although striking, this contrast between HepG2 and C3A cells is consistent with the expression array data of Lee et al. (9), who did not detect significant induction of EGR1 in the C3A line in response to the AA limitation.
FIGURE 1.
Activation of the EGR1 gene by the AAR. A, HepG2 cells were incubated in DMEM (Ctr) or DMEM lacking histidine (AAR) to activate the AAR. RNA was isolated at the times indicated, and EGR1 transcription activity (by assaying hnRNA) or steady state mRNA levels were measured using qPCR as described under “Experimental Procedures.” The data were plotted as the ratio of the respective EGR1 RNA to the GAPDH control, and normalized relative to the DMEM t = 0 value. The results are presented as the averages ± standard deviations of three or more samples and are representative of multiple independent experiments. B, after incubating HepG2 cells in DMEM (Control) or DMEM lacking histidine (AAR) for the times indicated, whole cell extracts were subjected to immunoblot analysis for EGR1 or actin. A representative blot of multiple experiments is shown. C, to test for a possible effect of the AAR on EGR1 mRNA stabilization, HepG2 cells were incubated in DMEM + 2 mm HisOH (AAR) for 4 h to activate the AAR, and then the cells were transferred to fresh DMEM (Ctr) or DMEM + 2 mm HisOH (AAR) with both media containing 5 μm actinomycin D. After the transfer, RNA was isolated at the times indicated, and then EGR1 and GAPDH mRNA were measured by qPCR. The data are presented as a semi-log plot and the half-life calculated using the equation t½ = −0.693/k. D, a subclone of HepG2 cells, C3A cells, and nontransformed, immortalized HC-04 human hepatocytes were incubated for 6 h in DMEM (Ctr) or DMEM + 2 mm HisOH (AAR) to activate the AAR. RNA was isolated, and the steady state ASNS and EGR1 mRNA levels were analyzed. The data are normalized relative to the DMEM values to illustrate the relative fold induction. E, HepG2 hepatoma cells or HC-04 hepatocytes were incubated in DMEM (Ctr) or DMEM containing 100 nm thapsigargin (ER stress) for 6 h to trigger ER stress and activate the unfolded protein response pathways. RNA was isolated, and EGR1, ASNS, and GAPDH mRNA levels were measured. The data were plotted as the ratio of EGR1 or ASNS mRNA to the GAPDH control and normalized relative to the DMEM value. For all of the RNA data, the results are shown as the averages ± standard deviations of three or four assays per experiment and are representative of multiple independent experiments. An a indicates that the AAR value is different from the DMEM control at p ≤ 0.05.
Given the common link of eIF2 phosphorylation and ATF4 production by the GCN2 (AAR) and PKR-like endoplasmic reticulum kinase (unfolded protein response, UPR) kinases, there is considerable overlap between the subset of genes induced by the AAR and by ER stress (28). The effect of ER stress on EGR1 expression in HepG2 and HC-04 cells was analyzed after treatment with thapsigargin for 6 h to perturb ER calcium levels. Notably, ER stress induced EGR1 expression in both cell types, but the relative relationship between them was similar to that observed for the AAR; the increase in HepG2 cells was much greater than that in HC-04 cells (Fig. 1E). The relative induction of the ATF4-dependent ASNS gene was just the opposite; the thapsigargin-dependent increase was ∼3-fold greater in the HC-04 cells.
EGR1 Induction by the AAR Is Independent of GCN2-ATF4
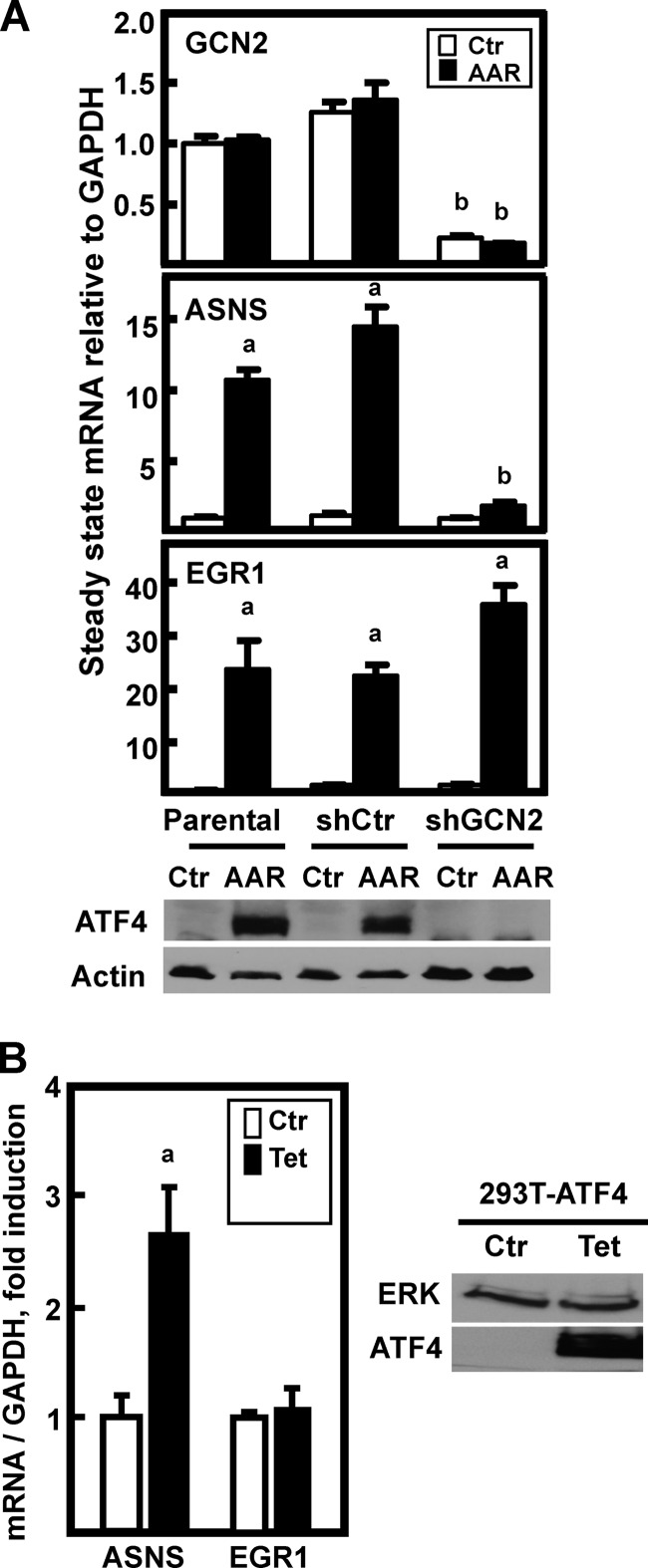
It is becoming clear that the AAR is a collection of signal transduction pathways (reviewed in Refs. 1, 2, and 29). The best studied of these is the GCN2-ATF4 arm of the AAR, but several AAR-induced genes have been shown to be GCN2-independent (11) and/or ATF4-independent (12, 30). To test whether or not EGR1 induction was dependent on GCN2-ATF4 signaling, GCN2 expression was suppressed by isolating clonal cell lines of HepG2 that stably express an shRNA against GCN2 or an unrelated shControl sequence. Averaging the data from five shControl and seven shGCN2 clonal cell lines, the shControl caused little or no difference in GCN2, ASNS, or EGR1 mRNA expression, whereas the shGCN2 clones averaged ∼80% knockdown of GCN2 mRNA (not shown). A single representative clone for each was chosen to illustrate as examples (Fig. 2A). GCN2 mRNA content itself was independent of AAR activity. ATF4 immunoblotting revealed strong suppression of ATF4 protein induction within the shGCN2 clonal cell line. The AAR induction of ASNS expression, known to be tightly linked to the GCN2-dependent increase in ATF4 protein (31), was largely blocked by a reduction in GCN2, whereas the induction of EGR1 mRNA was slightly, but reproducibly, enhanced (Fig. 2A). These observations in HepG2 cells extend the observation of Deval et al. (11), who showed AAR induction of Egr1 in Gcn2-deficient MEF cells.
FIGURE 2.
Induction of EGR1 mRNA expression in HepG2 cells is not dependent on GCN2-ATF4. A, HepG2 cells stably expressing an shRNA control sequence (shCtr) or against GCN2 (shGCN2) were prepared as described under “Experimental Procedures.” These cells, along with parental HepG2 cells, were incubated for 8 h in DMEM (Ctr) or DMEM lacking histidine (AAR) to activate the AAR prior to isolation of RNA. The mRNA levels of GCN2, ASNS, EGR1, and GAPDH were measured by qPCR, and the data are plotted as the ratio of the indicated mRNA to the GAPDH control relative to the DMEM value for the parental HepG2 cells. The results shown are the averages ± standard deviations of three or four assays within a representative experiment. An a indicates that the AAR value is different from the DMEM control at p ≤ 0.05, and a b indicates that the shGCN2 value is statistically different from the corresponding shControl value at p ≤ 0.05. For the blots shown in the bottom panel of A, corresponding whole cell extracts were probed for ATF4 protein and actin as a loading control. B, HEK293T-ATF4 cells were incubated in DMEM (Ctr) or DMEM + 0.01 μg/ml Tet for 8 h. RNA and a whole cell protein extract were isolated. ASNS, EGR1, and GAPDH mRNA levels were measured by qPCR, and the results are shown as the averages ± standard deviations of three or four assays and are representative of multiple independent experiments. An a indicates that the AAR value is different from the DMEM value at p ≤ 0.05. The whole cell extracts were subjected to immunoblot analysis for ATF4 and total ERK as a loading control.
To further test the relationship between ATF4 and EGR1 induction, we used human embryonic kidney HEK293T cells that had been selected by Ord et al. (20) for stable expression of a Tet-inducible ATF4 cDNA. These “HEK293T-ATF4” cells were monitored for ASNS and EGR1 mRNA after treatment with Tet to induce ATF4 alone in the absence of other possible AAR signaling events (Fig. 2B). As we have reported previously (32), Tet-induced ATF4 caused a significant increase in ASNS expression. However, despite immunoblot evidence for a large increase in ATF4 protein in response to the Tet, no effect of ATF4 overexpression alone was observed on the EGR1 mRNA level. Collectively, these data indicate that the AAR-dependent induction of EGR1 is independent of the GCN2-ATF4 pathway.
Induction of EGR1 Is a MEK-dependent Mechanism
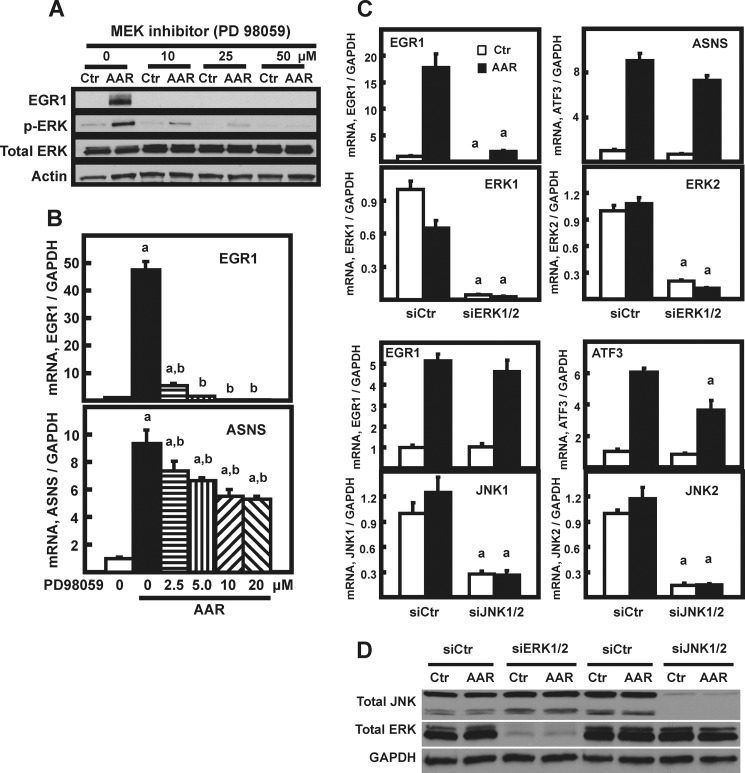
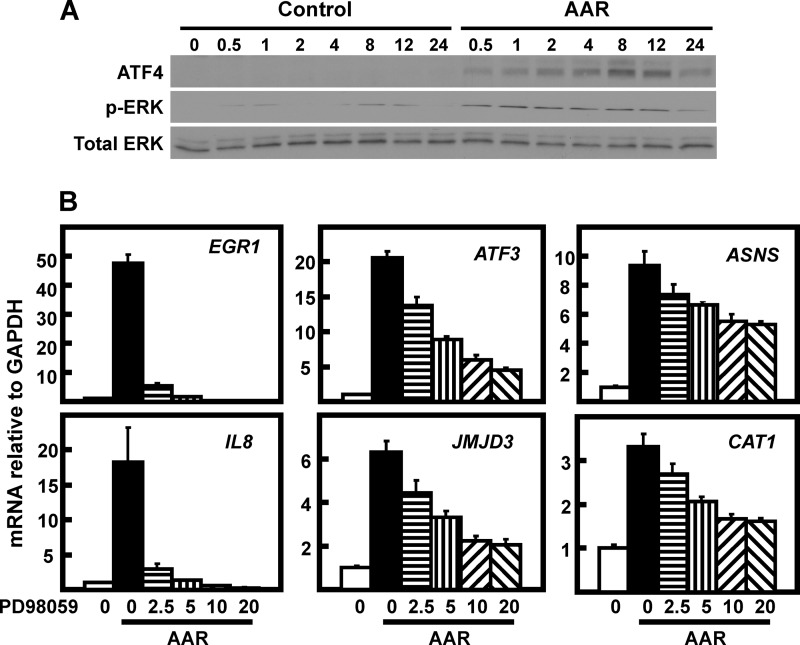
Transcriptional induction of the EGR1 gene by stimuli other than the AAR is known to be downstream of the three principle MAPK pathways, with the specific MAPK arm determined by the cell type or the activating stress (reviewed in Ref. 14). To determine whether one or more of the MAPK pathways was involved in AAR-induced EGR1 expression, they were initially screened by small molecule inhibition. Both basal and AAR-induced expression of EGR1 was completely blocked by inhibition of MEK with PD98059, whereas JNK inhibition caused a partial suppression and inhibition of p38 signaling actually enhanced EGR1 expression (data not shown). Although the initial screening experiments used 10–50 μm PD98059 (Fig. 3A), it was determined subsequently that a PD98059 concentration as low as 2.5 μm was sufficient to block ∼90% of the EGR1 mRNA induction but caused only a 20% decrease for ASNS (Fig. 3B). The sensitivity of the EGR1 induction to chemical inhibition of MEK activity was confirmed by testing U0126 (data not shown).
FIGURE 3.
AAR induction of EGR1 in HepG2 cells is linked to MEK signaling. A, HepG2 were pretreated for 1 h with the indicated concentration of MEK inhibitor (PD98059). The control cells were incubated in an equal volume of DMSO. The cells were then maintained in the presence of the inhibitor but transferred to either DMEM (Ctr) or DMEM + HisOH (AAR) for an additional 8 h to activate the AAR. Then whole cell protein extracts were collected and analyzed by immunoblotting for changes in EGR1, p-ERK, total ERK, and actin protein content. B, HepG2 cells were incubated in DMEM (open bar) or DMEM lacking histidine (AAR) and with the indicated concentration of PD98059 MEK inhibitor, as described for A. RNA was analyzed by qPCR for EGR1, ASNS, and GAPDH mRNA content. The data are plotted as the averages ± standard deviations for triplicate samples and are representative of multiple independent experiments. An a indicates that the AAR value is different from the DMEM control at p ≤ 0.05, and a b indicates that the value for inhibitor-treated cells is statistically different from the histidine-deprived cells in the absence of inhibitor (AAR 0 μm value) at p ≤ 0.05. C, HepG2 cells were transiently transfected with siRNA oligonucleotides against JNK1/2 or ERK1/2 to test the effect on AAR-induced EGR1 expression. To knock down both ERK1 and ERK2 simultaneously (likewise for JNK1 and JNK2), 50 nm of each siRNA was mixed to give a final concentration of 100 nm. At 72 h after transfection, the cells were incubated for 8 h in DMEM (Ctr) or DMEM lacking histidine (AAR) to induce the AAR. The mRNA for ASNS, ATF3, ERK1, ERK2, JNK1, JNK2, EGR1, and GAPDH were measured by qPCR. The data are presented as the averages ± standard deviations for at least three samples. An “a” indicates that the siERK1/2 or siJNK1/2 value is different from the corresponding siControl value at p ≤ 0.05. D, the knockdown of ERK1/2 and JNK1/2 were confirmed by immunoblotting samples from cells treated as described for C.
To assess their relative contribution, ERK1/2 and JNK1/2 were subjected to knockdown by treatment of HepG2 cells with siRNA oligonucleotides (Fig. 3, C and D). The results show that knockdown of ERK1/2 caused a significant reduction in the basal (DMEM) and AAR-induced level of EGR1 expression. As a negative control, there was little or no decrease in the induction of the largely ATF4-dependent ASNS gene. Knockdown of JNK1/2 did not significantly affect EGR1 mRNA content. As a positive control for siJNK1/2 action, ATF3 was partially suppressed in its induction (Fig. 3C), consistent with published results (13, 30). These data provide additional documentation for the specificity of AA signaling to the EGR1 gene through the MEK-ERK pathway.
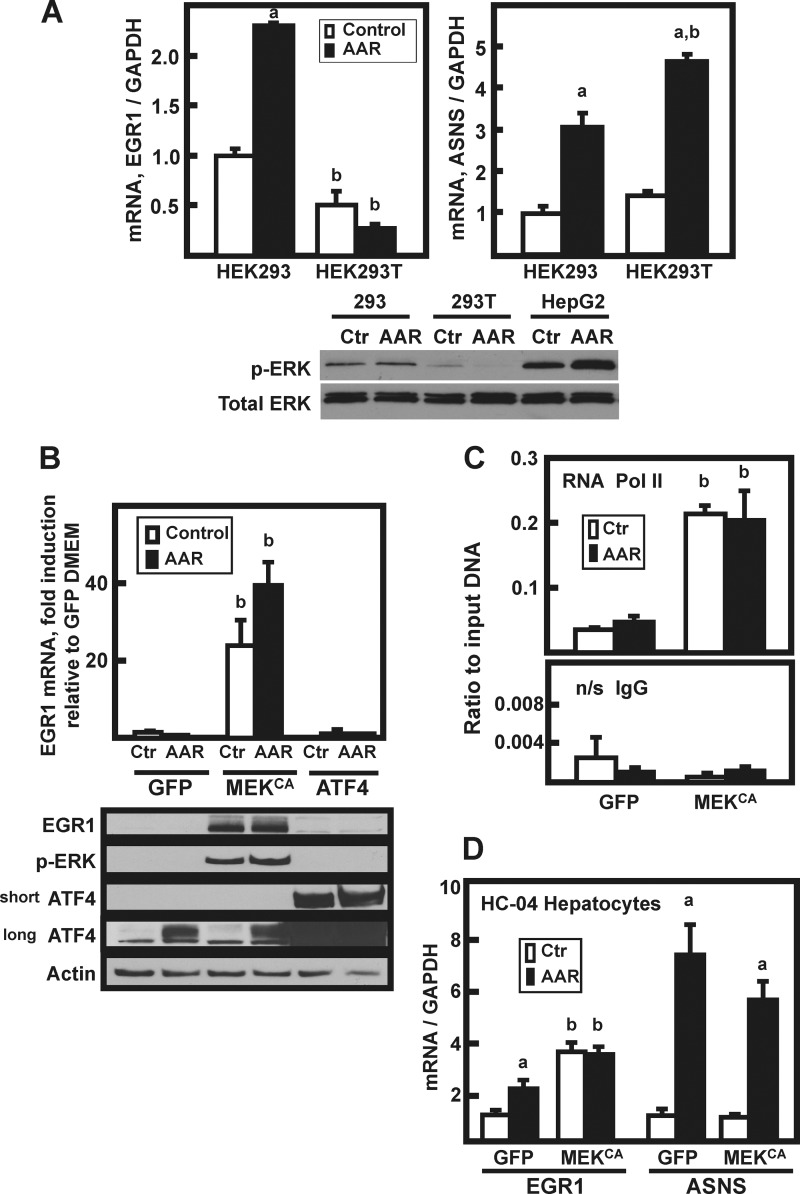
A comparison of p-ERK production in HEK293 cells that do not express T antigen with the T antigen-expressing HEK293T cells revealed that non-T-containing HEK293 cells exhibit a low level of MEK activity, whereas little or no detectable MEK activity occurs in the HEK293T cell line (Fig. 4A, immunoblot panel). The AAR associated p-ERK increase in HepG2 hepatoma cells is shown as a reference. Whereas ASNS mRNA induction was similar in both HEK293 and HEK293T cells (3–5-fold), EGR1 induction occurred only in the MEK-containing HEK293 cells (Fig. 4A), and even that was modest in magnitude compared with HepG2 cells (Fig. 1A). By making the mutations S218E and S222D at two phosphorylated serine residues in the activation loop of MEK1, a constitutively active MEK1 construct (MEKCA) was created by Mansour et al. (24). We have shown previously that exogenously expressed MEKCA strongly drives ERK phosphorylation in HEK293T cells (12, 22). When HEK293T cells were transiently transfected with plasmids encoding ATF4 or as a negative control green fluorescent protein, there was little or no detectable increase in p-ERK protein or EGR1, with or without AAR activation (Fig. 4B). However, exogenous expression of MEKCA caused an increase in p-ERK level and a 25-fold EGR1 mRNA induction in cells that were incubated in DMEM control medium. Activation of the AAR caused a further enhancement of the MEKCA action. Consistent with these results, a strong increase in EGR1 protein levels was also observed in the HEK293T cells expressing the MEKCA (Fig. 4B). Consistent with transcriptional activation, in the MEKCA expressing cells ChIP analysis for Pol II revealed increased recruitment to the EGR1 promoter (Fig. 4C). Neither AAR-induced endogenous ATF4 nor highly overexpressed exogenous ATF4 resulted in a significant increase in EGR1 mRNA or protein expression (Fig. 4B). These transient overexpression data are consistent with the results of Fig. 2B showing that Tet-inducible ATF4 does not increase EGR1 mRNA content.
FIGURE 4.
MEK signaling is both necessary and sufficient for induction of EGR1. A, HEK293 or HEK293T cells were incubated in DMEM (Control or Ctr) or DMEM lacking histidine (AAR) for 8 h, and then EGR1, ASNS, and GAPDH mRNA content was analyzed by qPCR. The data are plotted as the averages ± standard deviations of triplicate samples and are representative of multiple independent experiments. An a indicates that the AAR value is different from the DMEM control at p ≤ 0.05, and a b indicates that the HEK293T value is statistically different from the corresponding value obtained with HEK293 cells p ≤ 0.05. Whole cell extracts were also analyzed by immunoblotting for total ERK and p-ERK protein content. B, HepG2 cells were transiently transfected with plasmids expressing GFP, constitutively active MEK (MEKCA), or ATF4, and 36 h later the cells were incubated for 8 h in DMEM (Ctr) or DMEM + HisOH (AAR). RNA and protein extracts were prepared, and EGR1 mRNA content was analyzed by qPCR. The data are plotted as the averages ± standard deviations of triplicate samples and are representative of multiple independent experiments. A b indicates that the value is different from the corresponding GFP-transfected control (GFP) value at p ≤ 0.05. The whole cell extracts were analyzed by immunoblotting for changes in EGR1, p-ERK, ATF4, and actin protein content. Two different exposures (“short” or “long”) of the ATF4 blot are shown to illustrate the relative abundance of endogenous versus overexpressed protein. C, HepG2 cells were transiently transfected with (GFP) or constitutively active MEK (MEKCA), and the AAR was induced as described for B. Following activation of the AAR, the cells were subjected to ChIP analysis for RNA Pol II binding to the EGR1 promoter. The primer sequences used are listed in Table 1. A nonspecific IgG antibody was used as the negative control. The results are given as the averages ± standard deviations for at least three samples, and a b denotes a significant difference of p ≤ 0.05 relative to the corresponding GFP-transfected control cell value. D, nontransformed HC-04 human hepatocytes were transiently transfected with GFP or constitutively active MEK (MEKCA) and treated with HisOH, as described for HepG2 cells in B. The mRNA levels of EGR1, ASNS, and GAPDH were measured by qPCR, and the data were normalized relative to the GFP DMEM value for either EGR1 or ASNS. The results are given as the averages ± standard deviations for at least three samples. An a indicates that the AAR value is different from the DMEM control at p ≤ 0.05, and a b indicates a significant difference of p ≤ 0.05 relative to the corresponding GFP-transfected control cell value.
As shown above, the magnitude of EGR1 induction in nontransformed HC-04 human hepatocyte was considerably less than in HepG2 cells (Fig. 1D), and immunoblotting of HC-04 cell extracts showed low levels of p-ERK (data not shown). To test the effect of ectopically increasing MEK activity in a second cell type, HC-04 cells were transfected with MEKCA, and then ASNS and EGR1 mRNA expression was monitored (Fig. 4D). As observed for HEK293T cells, artificially increasing the HC-04 MEK activity resulted in an induction of basal EGR1 expression with little or no effect on ASNS. A minor difference between the HC-04 and HEK293T cells was that when MEKCA was expressed in the HC-04 cells, activation of the AAR in the MEK-expressing cell produced no further EGR1 induction. Collectively, these results indicate that increased MEK activity alone is sufficient for the EGR1 mRNA and protein induction during the AAR, and conversely, ATF4 is neither sufficient nor necessary.
Relative Contribution of MEK-independent and -dependent Pathways
To compare the kinetics of induction and duration of the AA responsive MEK and GCN2 pathways, the abundance of their respective products, p-ERK and ATF4, was analyzed from 0 to 24 h after incubation of HepG2 cells in DMEM ± histidine (Fig. 5A). The data show that the initiation of the response is quite similar with the effectors of both pathways increased in abundance within 30 min. However, the peak and signal maintenance was slightly different, with p-ERK remaining at similar level from 0.5 to 12 h, whereas ATF4 exhibited a sharper peak at 8–12 h. Both pathways declined by 24 h. These results indicate that the rapid induction of the GCN2 and the MEK pathways is similar, but the period of greatest activation differs to some degree.
FIGURE 5.
Relative contribution of the GCN2-ATF4 and MEK pathways to AA signaling. A, after incubating HepG2 cells in DMEM (Control) or DMEM lacking histidine (AAR) for the times indicated, whole cell extracts were subjected to immunoblot analysis for ATF4, p-ERK, or total ERK. The total ERK also serves as the loading control. A representative blot is shown. B, HepG2 were pretreated for 1 h with the indicated concentration (0–20 μm) of MEK inhibitor (PD98059). The control cells were incubated in an equal volume of DMSO. The cells were then maintained in the presence of the inhibitor but transferred to either DMEM (0) or DMEM lacking histidine (AAR) for an additional 8 h to activate the AAR. RNA was analyzed by qPCR for the indicated mRNA content. The data shown for ASNS are the same as those shown in Fig. 3C. The data are plotted as the averages ± standard deviations for triplicate samples and are representative of multiple independent experiments.
To determine whether the MEK-dependent pathway complements the ATF4 pathway for specific genes, the PD98059 sensitivity of several AA-responsive genes was tested (Fig. 5B). The genes tested could be grouped into three general categories. Induction of EGR1 and IL-8 showed the greatest MEK dependence, and each was inhibited by more than 90% at 20 μm PD98059. Expression of ATF3 and JMJD3 was somewhat less MEK-driven, but approximately two-thirds of their AAR induction was dependent on MEK. This result for ATF3 is compatible with the data of Deval et al. (11) showing that AAR induction of Atf3 is only reduced by 50% in Gcn2-deficient fibroblasts. Consistent with their known dependence on ATF4 (26, 33), ASNS and CAT1 were the least affected by MEK inhibition, with less than a 50% decline. The plateau of inhibition after 10 μm PD98059 for the four partially sensitive genes demonstrates that a portion of their induction is truly independent of MEK. Based on their known dependence on ATF4 from previously published studies, this residual induction is assumed to be the result of GCN2-ATF4 signaling. These data illustrate that the MEK and ATF4 pathways complement one another and can coincidently contribute to the AA control of a specific gene.
To extend the results shown in Fig. 5B across the genome, an expression microarray was used to determine the sensitivity of AAR-induced genes to MEK inhibition. HepG2 cells were incubated for 8 h in DMEM with or without histidine and in the presence or absence of 2.5 μm PD98059 (n = 4 for each condition). RNA was collected and analyzed by microarray using the Affymetrix Human Transcriptome 2.0 Array chip.
The data were analyzed using bioinformatic approaches that we have used previously (10). Using the criteria of a change greater than 2-fold and a p ≤ 0.001, 502 activated and 465 repressed genes were identified. Of the 502 activated genes, 305 and 126 exhibited a MEK dependence of >25% and >50%, respectively, and of the 465 repressed genes, 323 and 78 showed >25% and >50% MEK dependence, respectively. Table 2 lists the top 15 AAR-induced genes regardless of mechanism, whereas Table 3 lists the top 15 AAR-induced genes (of 2-fold or more) that exhibited the greatest degree of MEK dependence. Within the two groups of genes, only EGR1, cFOS, and EREG are in common, indicating that MEK-ERK signaling represents a unique and novel AAR pathway.
TABLE 2.
Genes most highly induced by the AAR regardless of mechanism
The 15 genes most highly induced by the AAR regardless of mechanism are shown. The expression microarray data are from HepG2 cells incubated for 8 h in DMEM ± histidine with or without 2.5 μm PD98059.
| Probe set ID | Gene symbol | p value (−His/DMEM) | Fold change (−His/DMEM) |
|---|---|---|---|
| PSR05009822.hg.1 | EGR1 | 2.50E-13 | 23.8496 |
| JUC14003021.hg.1 | FOS | 2.37E-07 | 11.7154 |
| PSR01016127.hg.1 | SLC22A15 | 4.94E-08 | 10.7615 |
| PSR05001385.hg.1 | SUB1 | 1.92E-10 | 9.99431 |
| JUC06005364.hg.1 | NCOA7 | 6.34E-08 | 9.89601 |
| JUC04002867.hg.1 | EREG | 2.39E-08 | 9.82107 |
| PSR01027796.hg.1 | ATF3 | 1.13E-10 | 9.71156 |
| JUC09000186.hg.1 | VLDLR | 5.11E-08 | 9.60593 |
| JUC19005801.hg.1 | PPP1R15A | 1.28E-12 | 9.24786 |
| PSR12006935.hg.1 | SHMT2 | 1.69E-16 | 8.12226 |
| PSR15003833.hg.1 | TMOD2 | 2.01E-09 | 7.07384 |
| PSR12006973.hg.1 | INHBE | 5.90E-12 | 7.0701 |
| JUC07012293.hg.1 | ASNS | 1.32E-07 | 6.78668 |
| JUC01023344.hg.1 | ZZZ3 | 1.46E-06 | 6.78157 |
| JUC01003020.hg.1 | SESN2 | 1.33E-10 | 6.72953 |
TABLE 3.
AAR-induced Genes with the Greatest Degree of MEK Dependence
The 15 AAR-induced genes with the greatest degree of MEK dependence are shown. The expression microarray data are from HepG2 cells incubated for 8 h in DMEM ± histidine with or without 2.5 μm PD98059.
| Probe set ID | Gene symbol | Fold change (−His/DMEM) | p value (−His/DMEM) | Percentage of MEK dependence |
|---|---|---|---|---|
| JUC14003021.hg.1 | FOS | 11.7154 | 2.37E-07 | 88.6184 |
| PSR05009822.hg.1 | EGR1 | 23.8496 | 2.50E-13 | 88.3077 |
| PSR04005637.hg.1 | IL8 | 5.50397 | 9.82E-08 | 86.2509 |
| PSR10009587.hg.1 | DUSP5 | 2.44081 | 1.12E-09 | 82.6263 |
| PSR07004534.hg.1 | IGFBP1 | 2.11808 | 7.68E-05 | 80.4794 |
| JUC02011397.hg.1 | CCL20 | 2.73831 | 6.44E-10 | 80.0432 |
| JUC18005299.hg.1 | SERPINB8 | 2.29491 | 0.00177338 | 79.9067 |
| PSR09013309.hg.1 | AQP3 | 3.58114 | 3.36E-08 | 78.3926 |
| PSR22011280.hg.1 | LIF | 3.97938 | 2.96E-11 | 77.2063 |
| JUC01008446.hg.1 | MAB21L3 | 3.2181 | 1.47E-09 | 76.3792 |
| JUC03014269.hg.1 | FILIP1L | 2.22555 | 0.00718454 | 75.6935 |
| PSR05025990.hg.1 | SPRY4 | 3.24396 | 2.34E-09 | 75.0695 |
| JUC08008916.hg.1 | HEY1 | 3.2598 | 1.08E-08 | 75.0352 |
| JUC04002867.hg.1 | EREG | 9.82107 | 2.39E-08 | 74.9477 |
| JUC06000413.hg.1 | EDN1 | 2.84431 | 4.59E-06 | 74.6727 |
Assuming that the GCN2-ATF4 pathway mediates the majority of the MEK-independent regulation, these results illustrate that GCN2 signaling is the primary mechanism for AA control in HepG2 cells. However, it is just as clear that MEK-dependent signaling represents a substantial contribution to the global AAR and that many genes respond to both pathways.
Effect of Serum
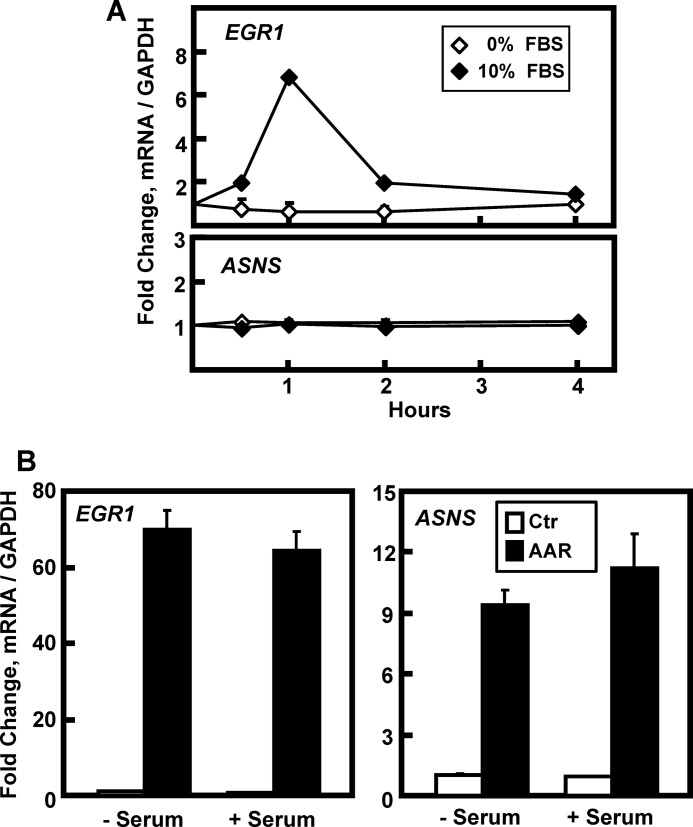
The fact that EGR1 is a known immediate-early response gene and the AA-responsive induction of ELK1 phosphorylation raised the possibility that the culture medium serum might contribute to this regulatory mechanism. To confirm the well recognized “serum response” in HepG2 cells, they were incubated for 16 h in DMEM lacking FBS and then transferred to fresh DMEM ± 10% FBS for 0–4 h (Fig. 6A). The serum-dependent increase in EGR1 mRNA expression was observed at 1 h and returned to the basal state by 4 h. ASNS was used as a negative control to show that the serum effect was gene-specific. As shown in Fig. 1A, the induction of EGR1 by the AAR occurs primarily after 4 h. However, to test whether or not the presence of serum may have an effect on the AA response beyond the initial rapid serum response, HepG2 cells were incubated for 16 h in the absence of FBS and then transferred to fresh DMEM ± 2 mm HisOH with or without 10% FBS for 6 h (Fig. 6B). As the data illustrate, despite the absence of serum for a total of 22 h, there was little or no effect on the EGR1 mRNA induction.
FIGURE 6.
Serum is not required for AAR induction of EGR1. HepG2 cells were incubated in DMEM lacking serum for 16 h and then transferred to DMEM with or without 10% FBS for 0–4 h (A) or to DMEM (Ctr) or DMEM + HisOH (AAR) in the presence or absence of 10% FBS for 6 h (B). The mRNA levels of EGR1, ASNS, and GAPDH were measured by qPCR, and the data were normalized relative to DMEM value for either EGR1 or ASNS. The results are given as the averages ± standard deviations for at least three samples.
Transcription Factors That Signal AA Limitation to the EGR1 Gene
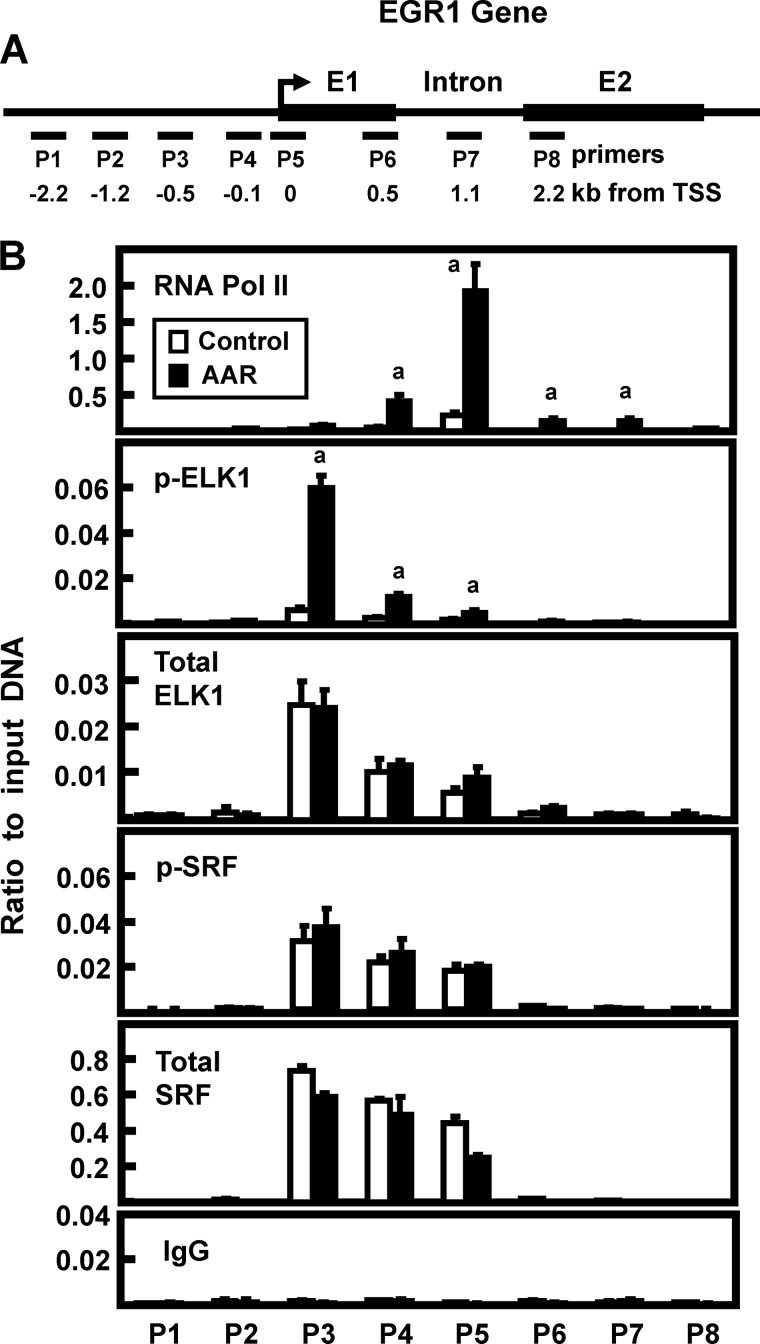
SRF binding to genomic serum response elements (SRE) and ETS-like factor 1 (ELK1) binding to ETS sites often exhibit cooperative binding (reviewed in Refs. 16 and 17). It has been documented that a series of SRE-ETS sites upstream of the EGR1 transcription start site (TSS) mediate increased transcription in a stimuli- and cell-specific manner (14, 34). Although the total amount of SRF and ELK1 binding does not typically change with gene activation, increased phosphorylation of ETS-bound ELK1 and/or phosphorylation of SRE-bound SRF results in enhanced transcriptional activity (14, 34–36). To determine whether ELK1 or SRF is involved in AA signaling in HepG2 cells, ChIP of the EGR1 gene locus was analyzed using primer sets that spanned ∼2.2 kb on either side of the TSS (Fig. 7A). Pol II showed basal association at the promoter region, and additional recruitment occurred in response to the AAR (Fig. 7B). Consistent with the proposed model for SRE-ETS function, the total abundance of both SRF and ELK1 binding was unaffected by the AAR. In contrast, the amount of p-ELK1 association with the ETS-containing promoter region was significantly increased in response to AAR activation (Fig. 7B). However, the amount of p-SRF was unchanged, consistent with the lack of a serum effect on the AAR induction of EGR1. For ChIP negative controls, binding of total SRF, total ELK1, p-SRF, and p-ELK1 at other regions of the gene were negligible, and the association of nonspecific IgG was low across the entire region tested. ChIP analysis was performed using antibodies specific for ERK or p-ERK, and no binding above the level of the IgG background was detected (data not shown).
FIGURE 7.
Transcription factor binding to the EGR1 gene in response to the AAR. A, the locations of primers (labeled P1–P8) used to analyze the human EGR1 gene are illustrated relative to the transcription start site (arrow) and the coding region of the gene. The primer sequences are listed in Table 1. B, HepG2 cells were incubated in DMEM (Control) or DMEM lacking histidine (AAR) for 8 h, and then the cells were subjected to ChIP analysis with antibodies specific for RNA Pol II, total SRF, p-SRF, total ELK1, p-ELK1, and a nonspecific IgG as a negative control. The data are plotted as the ratio to the input DNA and are the averages ± standard deviations for at least three samples. The data shown are representative of multiple independent experiments. An a denotes a significant difference of p ≤ 0.05 relative to the corresponding DMEM control value for that particular primer set.
Elevated EGR1 Contributes to AAR-enhanced Gene Expression in HepG2 Cells
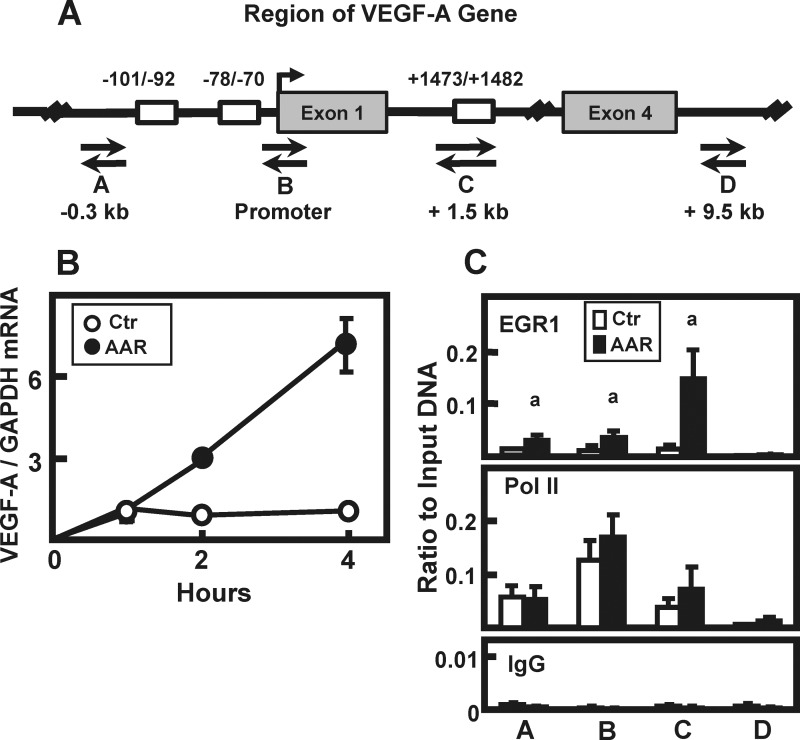
The vascular endothelial growth factor A (VEGF-A) gene is activated by the AAR (37, 38), and it is known that transcription from the VEGF-A gene can be induced in HepG2 cells through EGR1 binding to proximal promoter sites (39). Scanning the VEGF-A gene revealed the presence of at least two potential EGR1 binding sites in the promoter region (nucleotides −101/−92 and nucleotides −78/−70) and a third site (nucleotides +1473/+1482) in the first intron (Fig. 8A). Induction of VEGF-A was confirmed in HepG2 cells incubated in medium containing 2 mm HisOH for 0–4 h (Fig. 8B). To determine whether this AAR-dependent induction was associated with EGR1, ChIP analysis was performed to monitor EGR1 and Pol II binding to the VEGF-A gene. As shown in Fig. 8A, primers were generated to analyze four distinct genomic regions: (a) 0.3 kb upstream of the VEGF-A TSS, (b) at the TSS, (c) 1.5 kb downstream of the TSS in intron 1, and (d) as a negative control, 9.5 kb downstream in intron 4 (see Table 1 for primer sequences). Nonspecific IgG binding, as a negative control, was negligible at all regions tested (Fig. 8C). Pol II association was the greatest at the TSS, and although there was a reproducible slight trend of increased Pol II levels at the TSS and +1.5 kb sites following AAR activation, the change was not statistically significant, suggesting that minimal additional Pol II recruitment may be necessary for increased transcription (Fig. 8C). Activation of the AAR enhanced EGR1 recruitment to all three potential EGR1 binding regions within the VEGF-A gene, with the highest increase occurring at the intronic site. The lack of EGR1 signal within intron 4 served as a “background” control for the EGR1 antibody. Therefore, consistent with the AAR-dependent induction of VEGF-A expression, the ChIP results show that EGR1 is recruited to the VEGF-A gene in an AA-dependent manner.
FIGURE 8.
AAR-induced EGR1 binds to the VEGF-A gene. A, a region of the human VEGF-A gene is depicted (not drawn to scale) to show the location of possible EGR1 binding sequences (open boxes) with the nucleotide numbers representing the locations of these sites relative to the transcription start site (arrow). Four sets of primers (labeled A–D) were designed to scan specific regions of the gene by ChIP analysis. B, HepG2 cells were incubated in DMEM (Ctr) or DMEM + HisOH (AAR) to activate the AAR. RNA was isolated at the times indicated, and steady state VEGF-A and GAPDH mRNA levels were measured using qPCR. The VEGF-A/GAPDH ratio was calculated, and the data were normalized relative to the DMEM t = 0 value. The results are represented as the averages ± standard deviations of assays in triplicate and are representative of multiple independent experiments. C, HepG2 cells were cultured in DMEM (Ctr) or DMEM + HisOH (AAR) for 4 h, and then the binding of EGR1 or RNA Pol II at the VEGF-A gene was analyzed by ChIP (see A for primer locations and Table 1 for primer sequences). Within each ChIP experiment, a nonspecific IgG was used as negative control. The data, plotted as the ratio to input DNA, are represented as the averages ± standard deviations for at least three samples. An a indicates a significant difference of p ≤ 0.05 relative to the DMEM control value for that specific primer set.
DISCUSSION
The results in this study provide several novel observations on AA-responsiveness in HepG2 hepatoma cells: (a) the data extend published array studies indicating that EGR1 expression is highly induced in HepG2 human hepatoma cells; this increase is largely the result of enhanced transcription; (b) the EGR1 induction is independent of the well described GCN2-ATF4 pathway for AA signaling but instead requires MEK-ERK signaling; (c) MEK-ERK activation was both necessary and sufficient for the AAR induction of EGR1 mRNA content; (d) the AAR-dependent induction of EGR1 transcription was associated with increased recruitment of Pol II and increased phosphorylation of ELK1, which is constitutively bound at the proximal promoter; (e) the results indicate that the current list of AA-responsive transcription factors and their associated genomic sequences that exhibit AAR element activity must be expanded to include the factor ELK1 and its consensus ETS core binding sequence; (f) EGR1 is required for full activation of other AAR responsive genes; for example, AAR-inducible EGR1 binding was observed at the proximal promoter and at a region located within intron 1 of the VEGF-A gene.
The GCN2 eIF2 kinase is activated by uncharged tRNA and thus serves as an AA-responsive sensor of protein/AA limitation (40–42). Although the GCN2-ATF4 pathway appears to be ubiquitous in mammalian cells, there is evidence for GCN2- and ATF4-independent AA signaling mechanisms as well (11, 12). Knockdown of GCN2, and consequently ATF4, caused a strong suppression of AAR-dependent ASNS induction, consistent with a tight linkage to ATF4-driven transcription. In contrast, EGR1 mRNA content was slightly enhanced by GCN2-ATF4 knockdown. Furthermore, ectopic overexpression of ATF4 in MEK-deficient HEK293T cells, by either transient or Tet-inducible stable methods, produced significant activation of ASNS but no increase in EGR1 expression. The AAR induction of EGR1 was extremely sensitive to MEK inhibition as illustrated by the fact that 2.5 μm PD98059 blocked ∼90% of the increase, whereas that same concentration only suppressed ASNS mRNA content by ∼20%. This small reduction in ASNS may be related to the known, but not fully characterized cross-talk between the GCN2-ATF4 and MEK pathways (22, 43). Previous results from our laboratory using cultured Gcn2-deficient MEF cells (22) and the in vivo data of Bunpo et al. (43) investigating liver tissue in Gcn2 knock-out mice showed that the induction of p-ERK in response to AA limitation was decreased in the absence of Gcn2. However, beyond the species and cell type differences, interpretation of the mouse data are complicated by the fact that in both the MEF cells and the liver tissue the basal level of p-ERK, that is, in the absence of AA limitation, was elevated in the Gcn2-deficient cells/tissue. Conversely, we observed an increase in the induction of MAPK-dependent cJUN induction after ATF4 knockdown (12), and the present results revealed the same trend for EGR1 induction following knockdown of GCN2. Interestingly, Koumenis and co-workers (44) showed that the GCN2-ATF4 pathway is necessary for optimal growth of some tumors, and yet, we have not observed a decline in HepG2 proliferation after knockdown of either GCN2 or ATF4. It is possible that even a modest enhancement of the immediate-early response genes, such as cFOS, cJUN, and EGR1, permits HepG2 cells to continue proliferating during the AAR. Additional experimentation is required to better understand the interrelationships between the GCN2-ATF4 pathway and the MAPK pathways.
The ETS proteins make up a large family of transcription factors that recognize the core sequence of GGA(A/T) embedded within a larger sequence of ∼10 bp (16, 17). ELK1 is a member of the ternary complex factor subfamily of the ETS superfamily. Often ELK1 forms cooperative protein complexes with SRF bound to SRE sites, and thus, co-localization of ETS and SRE enhancer sites is a common but not obligatory motif. Indeed, whether or not SRF is present in conjunction with ELK1 influences the functional consequences of ELK1 binding (17). The present results document that hypophosphorylated SRF and ELK1 are bound to the EGR1 promoter region prior to AA limitation. Although the total amount of SRF and ELK1 binding does not change in response to AA limitation, the level of p-ELK1 is increased. In contrast, the association of p-SRF was not increased by the AAR, distinguishing this regulatory mechanism from the serum response and suggesting that the functional complex induced by the AAR and assembled at the EGR1 promoter does not include a p-SRF/p-ELK1 heterodimer. The phosphorylation of ELK1 is associated with increased recruitment of RNA Pol II and increased transcription activity from the EGR1 gene. Prior to the present research, analysis of many AA-responsive genes revealed that members of the ATF, FOS/JUN, and C/EBP subfamilies of the bZIP transcription factor superfamily were responsible for regulated AAR transcription (reviewed in Refs. 2 and 29). However, the current results demonstrate that the paradigm must be expanded to include ELK1 and perhaps other ETS members, for those cell types in which the MEK-ERK pathway is highly activated in response to AA limitation.
Following AAR induction of EGR1, the factor is recruited to the VEGF-A promoter, as well as to a previously unknown EGR1-binding site within intron 1. This observation is consistent with reports that EGR1 promotes the in vivo motility and invasion of hepatocellular carcinoma cells (45). Furthermore, one mechanism by which EGR1 may aid hepatocellular carcinoma proliferation in the face of nutrient limitation may be via induction of VEGF-A. For example, Lee and Kim (39) documented EGR1 binding to the VEGF-A proximal promoter during hepatocyte growth factor-mediated proliferation of HepG2 and Hep3B hepatoma cells. Likewise, Liu et al. (46) reported that hepatocellular carcinoma progression is critically dependent on VEGF-A-driven endothelial cell proliferation in the tumor-associated vasculature. In fact, therapies targeting VEGF-A receptor signaling are being employed for the management of aggressive and advanced stage hepatocellular carcinoma (47). Relative to normal mouse liver tissue, diethylnitrosoamine-induced mouse liver tumors have elevated expression of many MEK- and ATF4-dependent AAR target genes, including EGR1 (11.6-fold), ASNS (6.3-fold), cFOS (4.0-fold), and ATF3 (6.6-fold) (48), consistent with the hypothesis that tumors are often nutrient-deprived.
Asparagine starvation, via delivery of bacterial asparaginase, has been a critical component of multidrug therapy for childhood acute lymphoblastic leukemia for several decades (49). More recently, additional examples of successful therapeutic suppression of pathways that promote tumor cell survival during nutrient-restrictive conditions are beginning to be exploited clinically (50, 51). As highlighted by our microarray expression analysis, the AAR in HepG2 cells induces several key activities, including several FOS-JUN members, EGR1, and VEGF-A, that may enable tumor cells to remain poised for growth under conditions restrictive for normal cells (10). The GCN2-ATF4 pathway activation by dietary AA restriction provides resistance to the inflammatory stress and ischemia associated with surgery (52) and also confers pro-survival and proliferative capabilities to tumor cells undergoing nutrient limitation (44). Fu et al. (12) showed that AAR-induced ERK and JNK activity in transformed cells contributed to the autoactivation of the cJUN gene, coincident with a 2-fold greater maintenance of proliferation for HepG2 and Huh7 hepatocellular carcinoma cells compared with growth of nontransformed HC-04 human hepatocytes under the same AAR conditions. The present results suggest that the MEK-ERK-ELK1 pathway leading to activation of EGR1 may be another critical component required for the relative robustness of many tumor cells under AA-deprived conditions. Future research on the impact of nutrient stress, and AA limitation in particular, on tumor proliferation will be an exciting avenue for providing a better understanding of the relationship between nutrition and cancer.
Acknowledgments
We thank Fan Zhang for help preparing immunoblots. We also thank Dr. Tonis Örd (Estonian Biocenter, Tartu, Estonia) for supplying the HEK293T-ATF4 cells. We thank the Malaria Research and Reference Reagent Resource Center (MR4) for the HC-04 human hepatocyte cell line that was originally submitted by Jetsumon Sattabongkot Prachumsri (Walter Reed Army Institute for Research).
This work was supported, in whole or in part, by National Institutes of Health Grant DK92062 and DK94729 (to M. S. K.).
- AA
- amino acid
- AAR
- AA response
- ASNS
- asparagine synthetase
- ATF
- activating transcription factor
- CARE
- C/EBP-ATF response elements
- EGR1
- early growth response 1
- ELK1
- E-twenty-six (ETS)-like factor 1
- GCN2
- general control nonderepressible 2
- HisOH
- histidinol
- MEKCA
- constitutively active MEK
- qPCR
- real time quantitative PCR
- SRE
- serum response element
- SRF
- serum response factor
- TSS
- transcription start site
- MEF
- mouse embryonic fibroblast
- ER
- endoplasmic reticulum
- Tet
- tetracycline
- Pol
- polymerase.
REFERENCES
- 1. Kilberg M. S., Shan J., Su N. (2009) ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol. Metab. 20, 436–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chaveroux C., Lambert-Langlais S., Cherasse Y., Averous J., Parry L., Carraro V., Jousse C., Maurin A. C., Bruhat A., Fafournoux P. (2010) Molecular mechanisms involved in the adaptation to amino acid limitation in mammals. Biochimie 92, 736–745 [DOI] [PubMed] [Google Scholar]
- 3. Vattem K. M., Wek R. C. (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harding H. P., Zhang Y., Ron D. (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271–274 [DOI] [PubMed] [Google Scholar]
- 5. Harding H. P., Zhang Y., Bertolotti A., Zeng H., Ron D. (2000) Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol. Cell 5, 897–904 [DOI] [PubMed] [Google Scholar]
- 6. Wolfgang C. D., Chen B. P., Martindale J. L., Holbrook N. J., Hai T. (1997) gadd153/Chop10, a potential target gene of the transcriptional repressor ATF3. Mol. Cell. Biol. 17, 6700–6707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fawcett T. W., Martindale J. L., Guyton K. Z., Hai T., Holbrook N. J. (1999) Complexes containing activating transcription factor (ATF)/cAMP-responsive-element-binding protein (CREB) interact with the CCAAT enhancer-binding protein (C/EBP)-ATF composite site to regulate Gadd153 expression during the stress response. Biochem. J. 339, 135–141 [PMC free article] [PubMed] [Google Scholar]
- 8. Gjymishka A., Palii S. S., Shan J., Kilberg M. S. (2008) Despite increased ATF4 binding at the C/EBP-ATF composite site following activation of the unfolded protein response, system A transporter 2 (SNAT2) transcription activity is repressed in HepG2 cells. J. Biol. Chem. 283, 27736–27747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee J. I., Dominy J. E., Jr., Sikalidis A. K., Hirschberger L. L., Wang W., Stipanuk M. H. (2008) HepG2/C3A cells respond to cysteine-deprivation by induction of the amino acid deprivation/integrated stress response pathway. Physiol. Genomics 33, 218–229 [DOI] [PubMed] [Google Scholar]
- 10. Shan J., Lopez M. C., Baker H. V., Kilberg M. S. (2010) Expression profiling after activation of the amino acid deprivation response in HepG2 human hepatoma cells. Physiol. Genomics 41, 315–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deval C., Chaveroux C., Maurin A. C., Cherasse Y., Parry L., Carraro V., Milenkovic D., Ferrara M., Bruhat A., Jousse C., Fafournoux P. (2009) Amino acid limitation regulates the expression of genes involved in several specific biological processes through GCN2-dependent and GCN2-independent pathways. FEBS J. 276, 707–718 [DOI] [PubMed] [Google Scholar]
- 12. Fu L., Balasubramanian M., Shan J., Dudenhausen E. E., Kilberg M. S. (2011) Auto-activation of c-JUN gene by amino acid deprivation of hepatocellular carcinoma cells reveals a novel c-JUN-mediated signaling pathway. J. Biol. Chem. 286, 36724–36738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fu L., Kilberg M. S. (2013) Elevated cJUN expression and an ATF/CRE site within the ATF3 promoter contribute to activation of ATF3 transcription by the amino acid response. Physiol. Genomics 45, 127–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pagel J. I., Deindl E. (2011) Early growth response 1: a transcription factor in the crossfire of signal transduction cascades. Indian J. Biochem. Biophys. 48, 226–235 [PubMed] [Google Scholar]
- 15. Hodge C., Liao J., Stofega M., Guan K., Carter-Su C., Schwartz J. (1998) Growth hormone stimulates phosphorylation and activation of elk-1 and expression of c-fos, egr-1, and junB through activation of extracellular signal-regulated kinases 1 and 2. J. Biol. Chem. 273, 31327–31336 [DOI] [PubMed] [Google Scholar]
- 16. Hollenhorst P. C., McIntosh L. P., Graves B. J. (2011) Genomic and biochemical insights into the specificity of ETS transcription factors. Annu. Rev. Biochem. 80, 437–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Odrowaz Z., Sharrocks A. D. (2012) ELK1 uses different DNA binding modes to regulate functionally distinct classes of target genes. PLoS Genet. 8, e1002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cao X. M., Koski R. A., Gashler A., McKiernan M., Morris C. F., Gaffney R., Hay R. V., Sukhatme V. P. (1990) Identification and characterization of the Egr-1 gene product, a DNA-binding zinc finger protein induced by differentiation and growth signals. Mol. Cell. Biol. 10, 1931–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liao Y., Shikapwashya O. N., Shteyer E., Dieckgraefe B. K., Hruz P. W., Rudnick D. A. (2004) Delayed hepatocellular mitotic progression and impaired liver regeneration in early growth response-1-deficient mice. J. Biol. Chem. 279, 43107–43116 [DOI] [PubMed] [Google Scholar]
- 20. Ord D., Meerits K., Ord T. (2007) TRB3 protects cells against the growth inhibitory and cytotoxic effect of ATF4. Exp. Cell Res. 313, 3556–3567 [DOI] [PubMed] [Google Scholar]
- 21. Thiaville M. M., Dudenhausen E. E., Zhong C., Pan Y. X., Kilberg M. S. (2008) Deprivation of protein or amino acid induces C/EBPβ synthesis and binding to amino acid response elements, but its action is not an absolute requirement for enhanced transcription. Biochem. J. 410, 473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thiaville M. M., Pan Y. X., Gjymishka A., Zhong C., Kaufman R. J., Kilberg M. S. (2008) MEK signaling is required for phosphorylation of eIF2α following amino acid limitation of HepG2 human hepatoma cells. J. Biol. Chem. 283, 10848–10857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pear W. S., Nolan G. P., Scott M. L., Baltimore D. (1993) Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. U.S.A. 90, 8392–8396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mansour S. J., Matten W. T., Hermann A. S., Candia J. M., Rong S., Fukasawa K., Vande Woude G. F., Ahn N. G. (1994) Transformation of mammalian cells by constitutively active MAP kinase kinase. Science 265, 966–970 [DOI] [PubMed] [Google Scholar]
- 25. Shan J., Fu L., Balasubramanian M. N., Anthony T., Kilberg M. S. (2012) ATF4-dependent regulation of the JMJD3 gene during amino acid deprivation can be rescued in Atf4-deficient cells by inhibition of deacetylation. J. Biol. Chem. 287, 36393–36403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen H., Pan Y. X., Dudenhausen E. E., Kilberg M. S. (2004) Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive bZIP transcription factors as well as localized histone acetylation. J. Biol. Chem. 279, 50829–50839 [DOI] [PubMed] [Google Scholar]
- 27. Su N., Kilberg M. S. (2008) C/EBP homology protein (CHOP) interacts with activating transcription factor 4 (ATF4) and negatively regulates the stress-dependent induction of the asparagine synthetase gene. J. Biol. Chem. 283, 35106–35117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 29. Kilberg M. S., Balasubramanian M., Fu L., Shan J. (2012) The transcription factor network associated with the amino acid response in mammalian cells. Adv. Nutr. 3, 295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chaveroux C., Jousse C., Cherasse Y., Maurin A. C., Parry L., Carraro V., Derijard B., Bruhat A., Fafournoux P. (2009) Identification of a novel amino acid response pathway triggering ATF2 phosphorylation in mammals. Mol. Cell. Biol. 29, 6515–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Siu F., Bain P. J., LeBlanc-Chaffin R., Chen H., Kilberg M. S. (2002) ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 277, 24120–24127 [DOI] [PubMed] [Google Scholar]
- 32. Shan J., Ord D., Ord T., Kilberg M. S. (2009) Elevated ATF4 expression, in the absence of other signals, is sufficient for transcriptonal induction via CCAAT enhancer-binding protein-activating transcription factor response elements. J. Biol. Chem. 284, 21241–21248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lopez A. B., Wang C., Huang C. C., Yaman I., Li Y., Chakravarty K., Johnson P. F., Chiang C. M., Snider M. D., Wek R. C., Hatzoglou M. (2007) A feedback transcriptional mechanism controls the level of the arginine/lysine transporter cat-1 during amino acid starvation. Biochem. J. 402, 163–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen C. C., Lee W. R., Safe S. (2004) Egr-1 is activated by 17β-estradiol in MCF-7 cells by mitogen-activated protein kinase-dependent phosphorylation of ELK-1. J. Cell. Biochem. 93, 1063–1074 [DOI] [PubMed] [Google Scholar]
- 35. O'Donnell A., Odrowaz Z., Sharrocks A. D. (2012) Immediate-early gene activation by the MAPK pathways: what do and don't we know? Biochem. Soc. Trans. 40, 58–66 [DOI] [PubMed] [Google Scholar]
- 36. Yang S. H., Sharrocks A. D., Whitmarsh A. J. (2013) MAP kinase signalling cascades and transcriptional regulation. Gene 513, 1–13 [DOI] [PubMed] [Google Scholar]
- 37. Abcouwer S. F., Marjon P. L., Loper R. K., Vander Jagt D. L. (2002) Response of VEGF expression to amino acid deprivation and inducers of endoplasmic reticulum stress. Invest. Ophthalmol. Vis. Sci. 43, 2791–2798 [PubMed] [Google Scholar]
- 38. Pan Y. X., Chen H., Thiaville M. M., Kilberg M. S. (2007) Activation of the ATF3 gene through a co-ordinated amino acid-sensing response programme that controls transcriptional regulation of responsive genes following amino acid limitation. Biochem. J. 401, 299–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee K. H., Kim J. R. (2009) Hepatocyte growth factor induced up-regulations of VEGF through Egr-1 in hepatocellular carcinoma cells. Clin. Exp. Metastasis 26, 685–692 [DOI] [PubMed] [Google Scholar]
- 40. Zhang P., McGrath B. C., Reinert J., Olsen D. S., Lei L., Gill S., Wek S. A., Vattem K. M., Wek R. C., Kimball S. R., Jefferson L. S., Cavener D. R. (2002) The GCN2 eIF2α kinase is required for adaptation to amino acid deprivation in mice. Mol. Cell. Biol. 22, 6681–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kimball S. R., Anthony T. G., Cavener D. R., Jefferson L. S. (eds) (2005) Nutrient Signaling through Mammalian GCN2, Springer-Verlag, New York [Google Scholar]
- 42. Bunpo P., Cundiff J. K., Reinert R. B., Wek R. C., Aldrich C. J., Anthony T. G. (2010) The eIF2 kinase GCN2 is essential for the murine immune system to adapt to amino acid deprivation by asparaginase. J. Nutr. 140, 2020–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bunpo P., Dudley A., Cundiff J. K., Cavener D. R., Wek R. C., Anthony T. G. (2009) The GCN2 protein kinase is required to activate amino acid deprivation responses in mice treated with the anti-cancer agent, l-asparaginase. J. Biol. Chem. 284, 32742–32749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ye J., Kumanova M., Hart L. S., Sloane K., Zhang H., De Panis D. N., Bobrovnikova-Marjon E., Diehl J. A., Ron D., Koumenis C. (2010) The GCN2-ATF4 pathway is critical for tumor cell survival and proliferation in response to nutrient deprivation. EMBO J. 29, 2082–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ozen E., Gozukizil A., Erdal E., Uren A., Bottaro D. P., Atabey N. (2012) Heparin inhibits Hepatocyte Growth Factor induced motility and invasion of hepatocellular carcinoma cells through early growth response protein 1. PLoS One 7, e42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu L. P., Liang H. F., Chen X. P., Zhang W. G., Yang S. L., Xu T., Ren L. (2010) The role of NF-κB in Hepatitis b virus X protein-mediated upregulation of VEGF and MMPs. Cancer Invest. 28, 443–451 [DOI] [PubMed] [Google Scholar]
- 47. Cheng A. L., Guan Z., Chen Z., Tsao C. J., Qin S., Kim J. S., Yang T. S., Tak W. Y., Pan H., Yu S., Xu J., Fang F., Zou J., Lentini G., Voliotis D., Kang Y. K. (2012) Efficacy and safety of sorafenib in patients with advanced hepatocellular carcinoma according to baseline status: subset analyses of the phase III Sorafenib Asia-Pacific trial. Eur. J. Cancer 48, 1452–1465 [DOI] [PubMed] [Google Scholar]
- 48. DeZwaan-McCabe D., Riordan J. D., Arensdorf A. M., Icardi M. S., Dupuy A. J., Rutkowski D. T. (2013) The stress-regulated transcription factor CHOP promotes hepatic inflammatory gene expression, fibrosis, and oncogenesis. PLoS Genet. 9, e1003937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pui C. H., Relling M. V., Downing J. R. (2004) Acute lymphoblastic leukemia. N. Engl. J. Med. 350, 1535–1548 [DOI] [PubMed] [Google Scholar]
- 50. Lee C., Raffaghello L., Brandhorst S., Safdie F. M., Bianchi G., Martin-Montalvo A., Pistoia V., Wei M., Hwang S., Merlino A., Emionite L., de Cabo R., Longo V. D. (2012) Fasting cycles retard growth of tumors and sensitize a range of cancer cell types to chemotherapy. Sci. Transl. Med. 4, 124ra127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Scrable H. (2012) Impersonalized medicine. Sci. Transl. Med. 4, 124ps126. [DOI] [PubMed] [Google Scholar]
- 52. Peng W., Robertson L., Gallinetti J., Mejia P., Vose S., Charlip A., Chu T., Mitchell J. R. (2012) Surgical stress resistance induced by single amino acid deprivation requires Gcn2 in mice. Sci. Transl. Med. 4, 118ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]