Background: The stable presentation of HLA is necessary for effective T-cell activity in infectious diseases.
Results: Conformational fluctuation analysis revealed a minor state of HLA.
Conclusion: The minor state avoids the disintegration of HLA by tightly packing toward the peptide. The minor population correlates with the elongated presentation of HLA on the cell surface.
Significance: We revealed the stabilization mechanism for HLA.
Keywords: Major Histocompatibility Complex (MHC), Nuclear Magnetic Resonance (NMR), Protein Dynamic, Protein Stability, Thermodynamics
Abstract
In immune-mediated control of pathogens, human leukocyte antigen (HLA) class I presents various antigenic peptides to CD8+ T-cells. Long-lived peptide presentation is important for efficient antigen-specific T-cell activation. Presentation time depends on the peptide sequence and the stability of the peptide-HLA complex (pHLA). However, the determinant of peptide-dependent pHLA stability remains elusive. Here, to reveal the pHLA stabilization mechanism, we examined the crystal structures of an HLA class I allomorph in complex with HIV-derived peptides and evaluated site-specific conformational fluctuations using NMR. Although the crystal structures of various pHLAs were almost identical independent of the peptides, fluctuation analyses identified a peptide-dependent minor state that would be more tightly packed toward the peptide. The minor population correlated well with the thermostability and cell surface presentation of pHLA, indicating that this newly identified minor state is important for stabilizing the pHLA and facilitating T-cell recognition.
Introduction
Human leukocyte antigen (HLA)4 class I presents a wide variety of self and non-self peptides to T-cell receptors (TCRs), resulting in activation of antigen-specific CD8+ T lymphocytes. HLA-restricted T-cells are essential components of effective host immune responses to infection with many viruses, including HIV-1 (1, 2), and they also play important roles in autoimmune diseases and cancer immune surveillance (3, 4). The potency of T-cell antiviral activity is not equal at the single-cell level (5) and depends on the cell antigen specificity, which highlights the potential importance of antigen peptides bound to HLAs for generating effective antiviral T-cells (6). Variations in peptide presentation cause drastic differences in the progression of a number of diseases, such as AIDS and ankylosing spondylitis (7, 8).
HLA class I forms a ternary complex composed of a highly polymorphic heavy chain, a monomorphic β2-microglobulin, and an antigen peptide typically composed of 8–11 amino acids. A number of crystal structures of peptide-HLA (pHLA) complexes have been reported, especially for HLA-A*02, HLA-B*27, and HLA-B*35 in complex with various self and non-self peptides (9–11). The heavy chain contains the peptide binding domain, with six pockets (A-F pockets) that vary in structure among different HLA class I allomorphs. For most HLA allomorphs, two of the pockets play a dominant role in determining peptide specificity (12, 13). The antigenic surface formed by the bound peptide and the peptide binding domain of the heavy chain is unique to each pHLA structure. Such structural differences play a major role in recruiting antigen-specific TCRs and resulting T-cell activation. On the other hand, intriguingly, the overall structures of the heavy chains are virtually identical irrespective of the bound peptides (11). At most, all-atom root-mean-square deviations (r.m.s.d.) between the heavy chains in each HLA class I allomorph are 1.1 Å.
In stark contrast, the thermostability profiles of pHLAs differ drastically depending on the bound peptides. The melting temperatures of several HLA allomorphs in complex with different peptides range from 42.7 to 71.8 °C (14–16). Similarly, the antigenic properties of pHLAs also depend on the bound peptides. We previously reported that HLA-B*35:01-restricted cytotoxic T lymphocytes specific for two HIV-1 Nef peptides, VPLRPMTY (VY8) and RPQVPLRPMTY (RY11), showed substantially different antiviral activities despite their sequence similarity and the comparable binding affinity between each pHLA and cognate TCR (15). HLA-B*35:01 exhibited a higher melting temperature, longer cell surface presentation, and more potent susceptibility to killing by cytotoxic T lymphocytes when in complex with VY8 (HLA/VY8) than when in complex with RY11 (HLA/RY11). This relationship was further demonstrated by HLA-B*35:01 in complex with various variant peptides of VY8 and RY11, where the surface expression levels of pHLAs were controlled to be constant (15). Although the interaction mode between the pHLA and TCR is a key factor in determining the potency of T-cell responses (17, 18), these results suggest that pHLA stability itself has a significant impact on the mounting of efficient T-cell responses. However, the determinant of the peptide-dependent stabilization mechanism remains elusive. Observed differences in thermostability and antigenicity are difficult to explain by static structural differences between the peptide binding domain and the bound peptide itself. Rather, it is thought that the conformational dynamics of the pHLA affect its thermostability and antigenic properties.
In the present study we determined the crystal structure of HLA-B*35:01 in complex with RY11 and also examined conformational fluctuations of three HLA-B*35:01 complexes exhibiting different thermostability and cytotoxic T lymphocyte activity profiles using relaxation dispersion NMR spectroscopy (19, 20). Although the crystal structure was nearly identical to previously determined structures of HLA-B*35:01 in complex with different bound peptides, the NMR analyses identified peptide-dependent fluctuations and a transiently formed minor state at the interface of the peptide binding domain. Our data revealed that the minor state is responsible for peptide-dependent stabilization of HLA-B*35:01.
EXPERIMENTAL PROCEDURES
Sample Preparation
For crystallographic studies, residues 1–274 of the HLA-B*35:01 heavy chain were cloned into the plasmid pET21a (+) (Invitrogen) and expressed in BL21 (DE3) cells. The heavy chain was refolded with the HIV-1 RY11 peptide and β2-microglobulin and purified as described elsewhere (21). The collected protein fractions were concentrated to 10 mg/ml.
For NMR measurements, 2H and 15N or 2H, 5N, and 13C uniformly labeled heavy chain and 2H-labeled β2-microglobulin were expressed as inclusion bodies in BL21 (DE3) cells grown in M9 minimal medium. The protein-containing cells were lysed, and the pellets were separately dissolved in 6 m guanidine-HCl buffer. The HLA-binding peptides VY8, VY8(P5A), VY8(L3A), and RY11(P8A) were purchased from GenScript. The HLA complex was refolded in the presence of β2-microglobulin and one of the peptides and then purified as described previously (15).
Crystallography Methods
HLA/RY11 was crystallized via the hanging-drop vapor diffusion method at 18 °C. The crystals grew in a reservoir solution of 0.1 m MES monohydrate (pH 6.5) and 12% (w/v) polyethylene glycol 20,000. All crystals were flash-cooled in liquid nitrogen after a brief soaking in reservoir solution with the addition of 17% (v/v) glycerol. The x-ray diffraction data were collected at Shanghai Synchrotron Radiation Facility using beamline 17U. All data were processed with HKL2000 software (22).
The HLA/RY11 structure was solved by molecular replacement using Phaser (23) from the CCP4 program suite (24) with the structure of HLA/VY8 (PDB code 1A1N) serving as a search model. The initial model was refined using REFMAC5 (25), and extensive model building was performed using COOT (26). Further rounds of refinement were carried out using the phenix.refine program implemented in the PHENIX package (27), with energy minimization, group and individual B-factor refinement, bulk solvent modeling, and TLS refinement. The stereochemical quality of the final model was assessed with the PROCHECK program (28). The formula for R-factors is as stated in Brünger et al. (29). The structure was deposited in the PDB under accession number 4LNR. The mutated structures HLA/VY8(P5A), HLA/VY8(L3A), and HLA/RY11(P8A) were modeled from the structure of HLA/VY8 or HLA/RY11 using the PyMOL Molecular Graphics System, Version 1.6.0.0 (Schrödinger, LLC.) and YASARA force field (30).
NMR Methods
Three-dimensional transverse relaxation-optimized spectroscopy (TROSY) spectra of HNCO, HN(CA)CO, HNCA, HN(CO)CA, HNCACB, HN(CO)CACB, CC(CO)NH, and 15N-NOESY were measured on an AVANCE DMX750 spectrometer (Bruker BioSpin) for sequential assignments of the backbone 1H, 13C, and 15N chemical shifts (31) of the HLA complexes at 0.8–0.9 mm in NMR buffer (95% H2O, 5% D2O, 20 mm PBS (pH 7.40, 143 mm NaCl, 2 mm NaN3). NMR data were processed using the program NMRPipe (32), and signals were assigned using the programs KUJIRA (33) and NMRView (34). 15N R2 relaxation rates were measured on AVANCE DRX600 and AVANCE DMX750 spectrometers (Bruker BioSpin) using the TROSY-type constant-time Carr-Purcell-Meiboom-Gill pulse sequence (19, 20) at 10, 15, 20, and 25 °C for HLA/VY8(P5A) and at 20 °C for HLA/RY11(P8A) and HLA/VY8(L3A). The spectra were processed, and effective R2 rates were calculated as described previously (35). Relaxation dispersion curves were fitted to a two-state exchange model (major ↔ minor) using the program GLOVE (36). The relaxation dispersion curves were initially fitted for each individual residue with fitting parameters of the population-average intrinsic relaxation rate (R20), the exchange rate constant (kex = kmajor → minor + kminor → major), the chemical shift difference between states (Δω), and the product of the populations of the major and minor states (pmajorpminor). The population of the minor state was calculated according to the formula pminor = (1 − (1 − 4pmajorpminor)1/2)/2. The kinetic rates kmajor → minor and kminor → major were calculated as kmajor → minor = pminorkex and kminor → major = pmajorkex, respectively. Data for which R2 changed by <1 s−1 over the entire range of τcp or for which an F test showed less than a 99% confidence limit were excluded from further analysis. Because the fluctuating residues were concentrated in the peptide binding domain, all relaxation dispersions, except those for a few linker residues, were fitted globally with global parameters of kex and pmajorpminor.
van't Hoff Fitting of Fluctuation Parameters
The equilibrium constant of the fluctuation between the major and minor states was calculated as Keq = kminor → major/kmajor → minor, and lnKeq was plotted as a function of the inverse of the absolute temperature, 1/T × 103. The data were fitted to the nonlinear van't Hoff equation (26), RTlnKeq = ΔH − TΔS + ΔCp(T − T0) − TΔCpln(T/T0), where T0 = 293.15, and R is the gas constant. The terms ΔCp, ΔH, and ΔS represent the changes in heat capacity, enthalpy, and entropy, respectively.
Calculation of ΔCp from the Buried Surface Area (BSA) Value
The BSA of the peptide and heavy chain interface was calculated using the Protein Interfaces, Surfaces, and Assemblies (PISA) service of the European Bioinformatics Institute (37).
Circular Dichroism Analysis
For circular dichroism measurements, the change in ellipticity (θ) with heating from 4 to 90 °C was monitored at 222 nm using a J-820 spectropolarimeter (JASCO), with a sample-cell volume of ∼0.4 ml in a quartz cell with an optical path length of 2 mm at the rate of 1 °C·min−1. The concentration of the sample was set between 0.1 and 0.2 mg/ml. The experiment was conducted single time for each sample. The Tm value in the circular dichroism analysis was calculated using the software provided by the manufacturer (JASCO).
RESULTS
Comparison of the Crystal Structures of Peptide-HLA-B*35:01 Complexes Exhibiting Different Thermostabilities
To address the correlation between the structure and thermostability of various peptide-HLA-B*35:01 complexes, we determined the crystal structure of HLA/RY11 at a resolution of 2.0 Å (Fig. 1a; Table 1). The structure was compared with previously determined crystal structures of HLA-B*35:01 (Table 2), especially HLA/VY8, as HLA/VY8 is more stable than HLA/RY11 despite the similar sequences of the bound peptides. The overall structure of HLA/RY11 was nearly identical to all of the 21 crystal structures of HLA-B*35:01, including HLA/VY8 (11), with all-atom r.m.s.d. of 0.22–1.09 Å for the heavy chain. However, the structures of the bound VY8 and RY11 peptides differed, with the middle of RY11 being bulged, as is the case with long HLA-binding peptides (Fig. 1b) (38, 39). Nevertheless, both peptides were bound to HLA-B*35:01 in the typical manner in which the B and F pockets prefer peptides with a proline residue at position 2 and either a tyrosine, phenylalanine, methionine, or leucine residue at the carboxyl terminus (40, 41). The remaining pockets (A, C, D, and E) exhibit little preference with respect to the amino acid sequence of the bound peptide (12, 13). The second proline and the carboxyl-terminal tyrosine in both VY8 and RY11 form hydrogen bonds with Tyr-99 in the B pocket and Ser-116 in the F pocket, respectively (Fig. 1b). Curiously, HLA/RY11 is less thermostable, and the RY11 peptide is easy to dissociate from HLA (15), but the BSA of the HLA-peptide contact surface of HLA/RY11 was similar to that of HLA/VY8 (1780 Å2 versus 1710 Å2). Although RY11 has a larger B-factor than the other part of HLA/RY11, no significant difference in B-factor was found in other parts of HLA/RY11 and HLA/VY8. It is known that pHLA with bulged peptides tends to have low thermostability. However, it is unclear from the crystal structures alone how the large thermostability difference could arise by the difference in the peptide.
FIGURE 1.
Similarity of the crystal structures of HLA/VY8 and HLA/RY11. a, the crystal structures of HLA/VY8 (red; PDB code 1A1N) and HLA/RY11 (blue; PDB code 4LNR) superposed using the HLA heavy chain. The structure of β2-microglobulin is shown in gray. b, closed view around the peptide-binding domain in the structures of HLA/VY8 (red) and HLA/RY11 (blue). The regions around the B and F pockets are circled, and hydrogen bonds important for peptide recognition involving Tyr-99 and Ser-116 are represented as dashed lines.
TABLE 1.
Data collection and refinement statistics (molecular replacement)
| HLA/RY11 | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 50.80, 81.42, 108.97 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| Resolution (Å) | 50.00-2.00 (2.00-2.07)a |
| Rsym or Rmerge | 0.062 (0.331) |
| I/σI | 36.3 (8.1) |
| Completeness (%) | 97.3 (96.2) |
| Redundancy | 10.2 (9.8) |
| Refinement | |
| Resolution (Å) | 24.00-2.00 |
| No. reflections | 29915 |
| Rwork/Rfree | 0.1887/0.2084 |
| No. atoms | |
| Protein | 3178 |
| Ligand/ion | 0 |
| Water | 443 |
| B-factors | |
| Protein | 29.1 |
| Ligand/ion | 0 |
| Water | 37.9 |
| r.m.s.d. | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.734 |
a Values in parentheses are for the highest resolution shell.
TABLE 2.
Structural comparison of HLA-B*35:01
| PDB ID | r.m.s.d.a | Peptide sequence | Comment | References |
|---|---|---|---|---|
| 4LNR | 0 | RPQVPLRPMTY | HLA/RY11 | |
| 0.597 | RPQVPLRAMTY | HLA/RY11 (P8A)-modeled structure | ||
| 1A1N | 0.358 | VPLRPMTY | HLA/VY8 | 11 |
| 0.566 | VPARPMTY | HLA/VY8 (L3A)-modeled structure | ||
| 0.524 | VPLRAMTY | HLA/VY8 (P5A)-modeled structure | ||
| 2CIK | 0.36 | KPIVVLHGY | 52 | |
| 1ZHK | 0.365 | LPEPLPQGQLTAY | 60 | |
| 2FYY | 0.208 | HPVGEADYFEY | 55 | |
| 2H6P | 0.384 | KPIVVLHGY | 61 | |
| 3LKP | 0.299 | LPFDKSTIM | 53 | |
| 3LKN | 0.37 | LPFERATIM | 53 | |
| 3LKS | 0.367 | LPFEKSTVM | 53 | |
| 3LKQ | 0.347 | LPFDKTTIM | 53 | |
| 1ZSD | 0.167 | EPLPQGQLTAY | 59 | |
| 1XH3 | 0.153 | LPAVVGLSPGEQEY | 38 | |
| 3LKO | 0.243 | LPFDRTTIM | 53 | |
| 2AXG | 0.186 | APQPAPENAY | 57 | |
| 3LKR | 0.252 | LPFERATVM | 53 | |
| 3MV8 | 0.552 | HPVGEADYFEY | Complex with TCR | 54 |
| 3MV9 | 0.856 | HPVGEADYFEY | Complex with TCR | 54 |
| 3MV7 | 0.82 | HPVGEADYFEY | Complex with TCR | 54 |
| 2NX5 | 0.946 | EPLPQGQLTAY | Complex with TCR | 58 |
| 1A9B | 1.063 | LPPLDITPY | 56 | |
| 1A9E | 1.029 | LPPLDITPY | 56 | |
| 1CG9 | 1.064 | LPPLDITPY |
a The r.m.s.d. of the all atom of heavy chain was calculated using PyMOL Molecular Graphics System, Version 1.6.0.0. (Schrödinger, LLC.) against the structure of HLA/RY11.
Analysis of the Conformational Dynamics of HLA-B*35:01 in Solution
We attempted to analyze the conformational dynamics of HLA/VY8 and HLA/RY11 in solution by relaxation dispersion NMR spectroscopy. However, disintegration and aggregate formation hampered the relaxation dispersion measurements. The aggregation was most likely due to insufficient stability of the pHLAs under the sample condition and NMR measurements (∼1 week) employed. Therefore, we used a VY8 peptide variant containing an Ala substituted for Pro-5 (VY8(P5A)), as this variant is known to improve thermostability while preserving TCR specificity (15). There was no significant interaction observed between Pro5 and the heavy chain, whereas the change in BSA resulting from the mutation was very small (ΔBSA = 1% of the total interaction surface). The HSQC spectrum of HLA/VY8(P5A) was virtually identical to that of HLA/VY8 (Fig. 2). The secondary structure of HLA/VY8(P5A) estimated from the 13Cα, 13Cβ, 13CO, and 15N chemical shifts closely matched that of the crystal structure of HLA/VY8.
FIGURE 2.
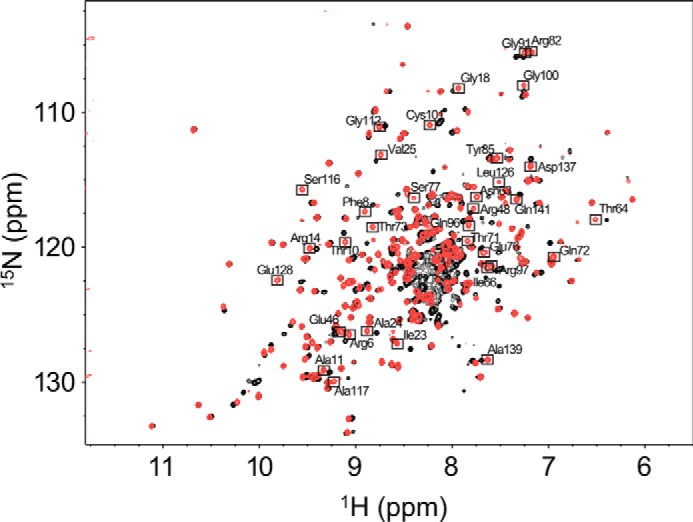
HSQC spectra of HLA/VY8 and HLA/VY8(P5A). The 1H,15N TROSY HSQC spectra of HLA/VY8 and HLA/VY8(P5A) are shown in black and red, respectively. The resonances of HLA/VY8(P5A) that do not overlap with those of HLA/VY8 are boxed with the assignments.
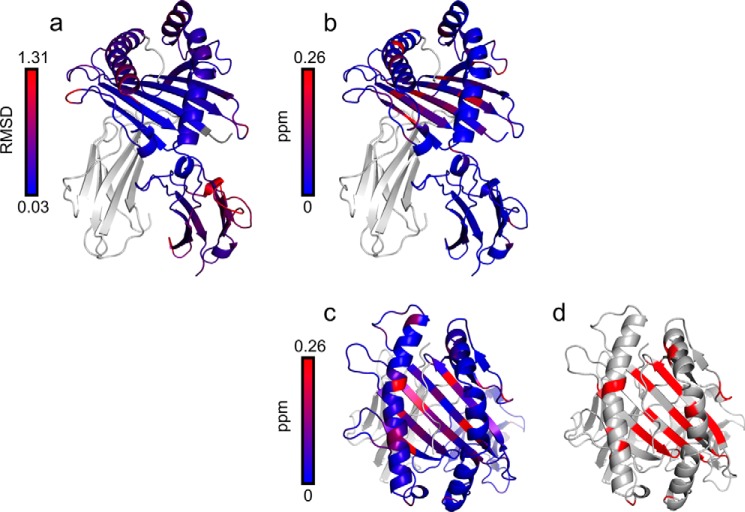
The relaxation dispersion experiment identified fluctuating residues in the peptide binding domain of HLA/VY8(P5A) at 20 °C (Fig. 3a). No fluctuation was observed in the Ig-like C1-type domain. We fitted all the relaxation dispersions globally to a two-state exchange model (major ↔ minor), yielding an exchange rate (kex = kmajor → minor + kminor → major) of 766 ± 41 s−1 and a minor population (pminor) of 4.37 ± 0.09%. Analysis of the chemical shift differences (Δω, representing the amplitude of the conformational change between the major and minor states) showed large-amplitude conformational changes in the B and F pockets compared with the other fluctuating regions (Fig. 3a). It is of note that the fluctuating residues included Tyr-99 in the B pocket and Ser-116 in the F pocket, both of which are important for peptide recognition by HLA-B*35:01.
FIGURE 3.
Conformational fluctuations of HLA/VY8(P5A). a, chemical shift differences (Δω) between the major and minor states plotted on the crystal structure of HLA/VY8 as a continuous color scheme from gray to red. The positions around Tyr-99 and Ser-116, which are important for peptide recognition, are circled. b, relaxation dispersion profiles for Ser-116 and Tyr-99 recorded at 14.1 tesla (black) and 17.6 tesla (red). c, the van't Hoff plot for the fluctuations of pHLA between the major and minor states. The best-fit curve is shown as a solid line. The determinant factor (R2) is 0.99.
To further characterize the minor state, we also measured relaxation dispersions of HLA/VY8(P5A) at 10, 15, and 25 °C. The exchange rates obtained at these temperatures were converted to the equilibrium constants (Keq = kminor → major/kmajor → minor) and fitted to the van't Hoff equation (Fig. 3b). This analysis provided a change in the heat capacity (ΔCp) of −0.88 ± 0.46 kcal·mol−1·K−1, a change in enthalpy (ΔH) of −261 ± 138 kcal·mol−1, and a change in entropy (ΔS) of −6.16 ± 3.22 cal·mol−1·K−1. It is generally appreciated that a negative ΔCp value for a protein complex is strong evidence that dehydration has occurred due to interaction and/or induced fit (42, 43). In the case of HLA/VY8(P5A), only −0.25 kcal·mol−1·K−1 of the total ΔCp value was calculated as being due to interaction with the peptide. Therefore, the remainder of the ΔCp value (−0.63 kcal·mol−1·K−1) can be ascribed to an induced-fit process. Because the secondary structure predicted by the chemical shift matched that of the crystal structure well, the fluctuating regions would already have formed a well folded conformation in the major state. Therefore, the minor state most likely forms a more packed dehydrated conformation. Note that the peptide binding domain is composed of many fluctuating hydrophobic residues (Fig. 4), and dehydration in a hydrophobic area generally contributes to the formation of a strong interaction (43). Furthermore, the negative ΔH and ΔS values are both consistent with a more packed dehydrated conformation because they can be interpreted as indicating the formation of new electrostatic- or hydrogen-bonding interactions and an increasingly ordered state, respectively (44). Therefore, it is likely that the interaction between the peptide binding domain and the peptide becomes stronger in the dehydrated minor state.
FIGURE 4.
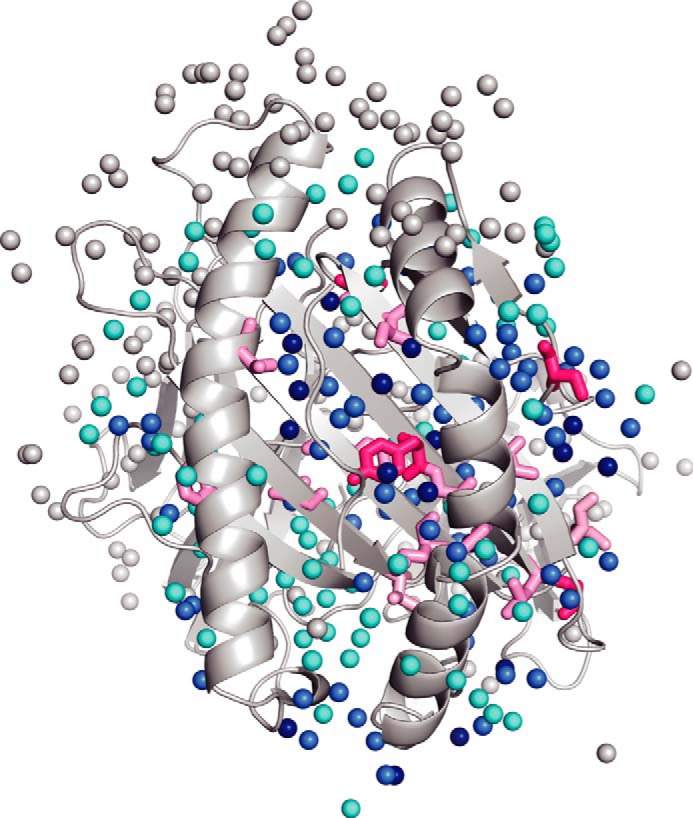
The water molecules around the fluctuating residues. Water molecules are shown as spheres in the HLA/VY8 structure. The fluctuating hydrophobic residues are shown as sticks in the same color scheme as in Fig. 2. Dark blue, blue, and cyan waters are located within 3.2, 6.4, and 9.6 Å from the fluctuating residues, respectively.
Together with the nearly identical crystal structures of the pHLAs, these results led us to hypothesize that the minor state is important for the stabilization of the pHLA to prevent disintegration of the complex. We also hypothesized that the loosely packed major conformation enables binding of a variety of peptides without inducing substantial structural changes in HLA.
Comparison of the Minor State among Different HLA-B*35:01 Complexes
We postulated that pHLAs of differing thermostability have minor states that differ in abundance and/or nature. To test this hypothesis, we analyzed the conformational dynamics of HLA-B*35:01 in complex with other peptide variants: Ala at p8 of RY11 and Ala at p3 of VY8, designated as RY11(P8A) and VY8(L3A), respectively. The peptide variants differentially increased the stability of the pHLAs in the previous study while preserving the specificity of the cognate TCR (15). We avoided introducing mutations into the fluctuating peptide binding domain of the HLA heavy chain because such mutations may cause unwanted effects, such as changing the peptide sequence preference.
The HSQC spectra of both HLA/RY11(P8A) and HLA/VY8(L3A) were similar to that of HLA/VY8(P5A) (Fig. 5). The resonances of the Ig-like C1-type domain of HLA/RY11(P8A) and HLA/VY8(L3A) superimposed well on those of HLA/VY8(P5A), whereas many residues in the peptide binding domains of HLA/RY11(P8A) and HLA/VY8(L3A) showed slightly different chemical shifts compared with those of HLA/VY8(P5A) (Figs. 5 and 6). Almost all the fluctuating residues in HLA/VY8(P5A) have different chemical shifts of HLA/RY11(P8A) and HLA/VY8(L3A) (Fig. 7). This suggests that the NMR signals were observed at the population-averaged chemical shifts of the major and minor state. Interestingly, most of the shifted resonances of HLA/VY8(L3A) appeared in between those of HLA/VY8(P5A) and HLA/RY11(P8A), suggesting that the minor populations are different among the pHLAs. The residues showing chemical shift differences between the pHLAs were spread over a large area of the peptide binding domain and included residues that have no direct contacts with the peptide.
FIGURE 5.

Similarity in the solution structures of HLA/VY8(P5A), HLA/VY8(L3A), and HLA/RY11(P8A). 1H,15N TROSY HSQC spectra of HLA/VY8(P5A), HLA/VY8(L3A), and HLA/RY11(P8A) are shown in red, purple, and blue, respectively. Residues showing relatively large chemical shift differences in the peptide binding groove are shown in the inset.
FIGURE 6.
The chemical shift differences of HLA/VY8(L3A) and HLA/RY11(P8A) from HLA/VY8(P5A). The chemical shift differences ((ΔδH)2+(ΔδN/5)2)1/2 between HLA/VY8(P5A) and HLA/RY11(P8A) and between HLA/VY8(P5A) and HLA/VY8(P3A) are shown as blue bars in the upper and lower graphs, respectively. The gray background shows the unassigned residues in HLA/VY8(L3A) and HLA/RY11(P8A). The yellow and red boxes in the middle indicate the secondary structural components observed in the crystal structure of HLA/VY8.
FIGURE 7.
Comparison of the r.m.s.d. differences and chemical shift differences. a, the r.m.s.d. for each residue in the crystal structures of HLA/VY8 and HLA/RY11 are shown. b and c, the chemical shift differences between HLA/VY8(P5A) and HLA/VY8(L3A) are shown from different angles. d, the residues showing relaxation dispersions in HLA/VY8(P5A) and HLA/VY8(P3A) are shown in red.
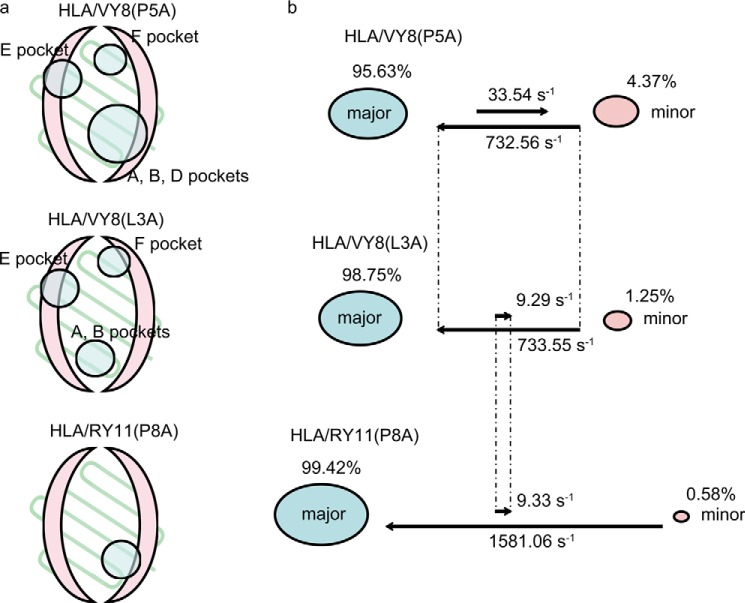
The relaxation dispersion experiments with HLA/RY11(P8A) and HLA/VY8(L3A) showed that both pHLAs fluctuate in the peptide binding domain, as was the case for HLA/VY8(P5A) (Fig. 8). However, fewer residues fluctuated in HLA/RY11(P8A) and HLA/VY8(L3A) than in HLA/VY8(P5A). For HLA/VY8(L3A), the fluctuating residues were located more distal from the center of the peptide binding domain than were those in HLA/VY8(P5A). For HLA/RY11(P8A), only a small number of residues near the A and B pockets fluctuated. It is notable that Tyr-99 and Ser-116, which are responsible for peptide binding, fluctuated only in HLA/VY8(P5A) (Fig. 5), suggesting that fluctuations at these residues play an important role in the peptide-dependent minor state. Fitting of relaxation dispersion curves yielded pminor of 1.25 ± 0.05% and kex of 742 ± 48 s−1 for HLA/VY8(L3A) and pminor of 0.58 ± 0.02% and kex of 1590 ± 96 s−1 for HLA/RY11(P8A). The minor populations of both HLA/RY11(P8A) and HLA/VY8(L3A) were smaller than that of HLA/VY8(P5A) (4.37%). The kinetic rate constants kmajor → minor and kminor → major had an interesting correlation among the pHLAs (Fig. 9). HLA/RY11(P8A) and HLA/VY8(L3A) had similar kmajor → minor rates (9.29 ± 0.07 and 9.33 ± 0.07 s−1, respectively) which were slower than that of HLA/VY8(P5A) (33.54 ± 0.06 s−1). In contrast, HLA/VY8(L3A) and HLA/VY8(P5A) had similar kminor → major rates (733.55 ± 0.06 and 732.56 ± 0.07 s−1, respectively), which were slower than that of HLA/RY11(P8A) (1581.06 ± 0.07 s−1). Thus, HLA/VY8(L3A) exhibits the characteristics of HLA/VY8(P5A) regarding the minor-to-major transition and the characteristics of HLA/RY11(P8A) regarding the major-to-minor transition. A comparison of the fluctuating regions among the three pHLAs suggested that the kminor → major rate is determined by the residues in the E and F pockets fluctuating in HLA/VY8(L3A) and HLA/VY8(P5A) and that the kmajor → minor rate is determined by the residues in the D pocket fluctuating only in HLA/VY8(P5A) (Fig. 9). Analysis of the relaxation dispersions also revealed chemical shift differences (Δω) between the major and minor states of HLA/RY11(P8A) and HLA/VY8(L3A). Almost all of the Δω values of HLA/RY11(P8A) and HLA/VY8(L3A) were larger than those of HLA/VY8(P5A) (Figs. 3a and 8), suggesting that the conformational changes from the major to the minor states are larger for HLA/RY11(P8A) and HLA/VY8(L3A) than for HLA/VY8(P5A).
FIGURE 8.
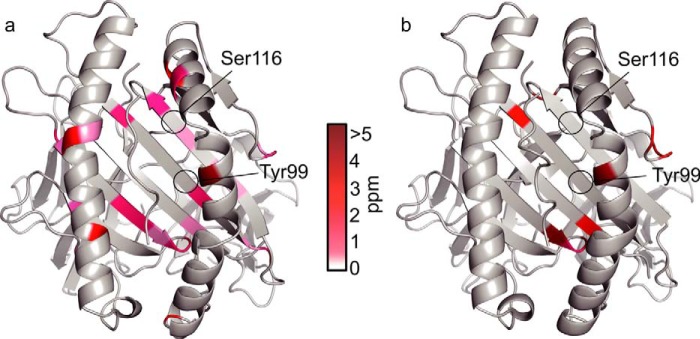
Conformational fluctuations of HLA/VY8(L3A) and HLA/RY11(P8A). Chemical shift differences (Δω) are plotted on the crystal structures of HLA/VY8 (a) and HLA/RY11 (b) in the same manner as in Fig. 3a.
FIGURE 9.
Fluctuating regions in three pHLA complexes. a, fluctuating regions in HLA/VY8(P5A), HLA/VY8(L3A), and HLA/RY11(P8A) are shown as schematic drawings. The α-helices and β-strands in the peptide binding domain are shown in pink and light green, respectively. The fluctuating regions are circled with the name of the pockets. b, the kinetic rates and the populations of the major and minor states are visualized for the three pHLA complexes.
Analysis of the Correlation between Conformational Dynamics, Thermostability, and pHLA Decay on the Cell Surface
We conducted thermostability analyses of HLA/VY8(L3A), HLA/VY8(P5A), and HLA/RY11(P8A) using circular dichroism. The melting temperatures were 50.1 °C for HLA/RY11(P8A), 57.7 °C for HLA/VY8(L3A), and 61.4 °C for HLA/VY8(P5A). Theses values are different from our previous determined values (15). The differences were caused most likely because we used a different instrument and newly prepared samples. Although the values were different, we observed the Tm tendency (HLA/VY8(P5A) > HLA/RY11(P8A)) being preserved. Interestingly, as the melting temperatures of the three pHLAs increased, so did the sizes of the minor populations determined by the relaxation dispersion experiments (Fig. 10). In other words, the pHLAs with larger major populations tended to disintegrate more rapidly. These results strongly support our hypothesis that the minor state is responsible for pHLA thermostability and that the peptides are loosely bound in the major state.
FIGURE 10.

Correlation between minor populations, stability, and T-cell activity decay rate. The left, middle, and right graphs show the minor populations, melting temperatures, and T-cell activity decay rates of the three pHLAs. The formula y = a × exp(−b × t) was used to calculate the T-cell activity decay rate with previously reported data (15), where t represents the time for activity measurement, and log b was determined as the decay rate.
We then calculated the pHLA decay rates on the antigen-presenting cell surface measured by the cognate T-cell responses determined from previously published data (15). Interestingly, the rates of pHLA decay on the antigen-presenting cell surface also correlated well with the minor populations. That is, the larger the minor populations, the longer the pHLAs were presented on the cell surface (Fig. 10).
DISCUSSION
In this study we report the conformational fluctuations that are responsible for the stabilization of peptide-HLA-B*35:01 complexes. In solution, pHLAs predominantly form a bound peptide-independent conformation that would be similar to the crystal structure except that they form a peptide-dependent dehydrated minor conformation at the peptide binding domain. The minor populations of the pHLAs correlated well with the length of time antigenic peptides are presented on the cell surface, which is important for T-cell recognition. It is, therefore, likely that the conformational fluctuations that we observed are part of the molecular basis of pHLA antigenicity.
A number of crystal structures of HLA class I allomorphs in complex with various peptides have been reported, but the structures of each HLA heavy chain are virtually identical (Table 2). In contrast, thermodynamic analyses using differential scanning calorimetry, circular dichroism (14, 15), infrared spectrometry (45), and hydrogen/deuterium exchange mass spectroscopy (46) have indicated that pHLAs are flexible in solution. These studies indicated that the overall flexibility of pHLAs differs depending on the bound peptides, suggesting the importance of the fluctuation of HLA on its function (47). However, they did not show how pHLAs are stabilized by the bound peptides.
Here, we characterized for the first time peptide-dependent site-specific pHLA fluctuations. Our results enabled us to explain observed peptide-dependent differences in the stability of various pHLAs. For example, Tyr-99 and Ser-116, which are important for peptide recognition by HLA-B*35:01, fluctuated only in the most stable pHLA analyzed in this study (HLA/VY8(P5A)). This result suggests that peptide recognition by a pHLA and further stabilization of the pHLA are dynamically regulated by conformational fluctuations.
With respect to the process of pHLA disintegration, previous molecular dynamic simulation studies indicated that in the absence of a bound peptide, HLA-B*27, becomes partially unfolded on a nanosecond time scale (48). Winkler et al. (49) showed that the disintegration of HLA-B*27 is initiated by dissociation of the peptide or β2-microglobulin, with disintegration rates of 10−4-10−7 s−1, depending on the peptide. Consistent with these data, we previously reported that peptide-HLA-B*35:01 complexes, including HLA/VY8(L3A), HLA/VY8(P5A), and HLA/RY11(P8A), disintegrate with rate constants of 1 × 10−6-4 × 10−6 s−1 (15). In contrast, the rate constants of the major-to-minor transition observed in this study (9–34 s−1) were much faster than the disintegration rate constants. Taken together, these data suggest that a pHLA rapidly forms the more packed minor conformation (constituting only up to 5% of the population) before it enters the disintegration pathway.
How does the mutation incorporated into the position where the side chain of the residue points out of the binding pocket affect the fluctuation of the residues even at the bottom of the peptide-binding groove? As the mutation changed the chemical shifts of the wide area of the heavy chain including the regions distal to the peptide (Fig. 2), the fluctuation change in the peptide by the mutation may be transmitted to the heavy chain through the intermolecular interactions between the peptide and the heavy chain and through the intramolecular interactions in the heavy chain.
What is the physiological role of the pHLA major state? Antigenic peptides are selected and loaded onto HLA class I proteins at the endoplasmic reticulum (ER) by various ER-resident chaperones in the so-called “peptide-loading complex” (50). As a component of this large complex, HLA would need to assume a similar conformation to accommodate a peptide. Our data suggest that the major state of HLA seems to accommodate an antigenic peptide more loosely compared with the well packed minor conformation, as if it allows the loading of various structurally different peptides in a similar structure. We propose such a two-step “transient induced-fit model” for the loading of a peptide to HLA. In the first step, the major state loads a peptide loosely, without the need for conformational adjustment, and in the second step, conformational tuning to the well packed minor state enables the pHLA to avoid rapid disintegration (Fig. 11). It would be intriguing to examine whether such dynamic conformational transitions of pHLA play a role in the endoplasmic reticulum peptide-loading complex as well as in TCR binding on the cell surface.
FIGURE 11.

The pHLA transient induced-fit model. A schematic illustration of pHLA fluctuation is shown. The HLA heavy chain is shown in pink, and the peptide and β2-microglobulin are represented as a trapezoid and a circle, respectively. Water molecules are shown as light blue circles.
The limited use of an HLA class I allomorph in this study may be of concern. It is not sure if a similar correlation between the minor population and the thermostability will generally be observed in other HLAs. The analysis of various HLA class I allomorphs in complex with different peptides of immunological interest using the same NMR approach is, therefore, warranted. Elucidating the stabilization mechanisms of various pHLAs will help facilitate the rational design of more stable therapeutically useful pHLAs (51).
Acknowledgments
We thank Dr. Jose M. M. Caaveiro for critically reading the manuscript and Hao Cheng for protein preparation.
This work was supported by the Japanese Society for the Promotion of Science Research Grant 11J06193.
The atomic coordinates and structure factors (code 4LNR) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The accession number for the chemical shift data (19825) has been deposited in the Biological Magnetic Resonance Data Bank.
- HLA
- human leukocyte antigen
- TCR
- T-cell receptor
- pHLA
- peptide-HLA
- r.m.s.d.
- root mean square deviation
- TROSY
- transverse relaxation-optimized spectroscopy
- BSA
- buried surface area
- HSQC
- heteronuclear single quantum correlation(s).
REFERENCES
- 1. Goulder P. J., Walker B. D. (2012) HIV and HLA class I: an evolving relationship. Immunity 37, 426–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Streeck H., Nixon D. F. (2010) T cell immunity in acute HIV-1 infection. J. Infect. Dis. 202, S302–S308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Garcia-Lora A., Algarra I., Garrido F. (2003) MHC class I antigens, immune surveillance, and tumor immune escape. J. Cell. Physiol. 195, 346–355 [DOI] [PubMed] [Google Scholar]
- 4. Singer D. S., Mozes E., Kirshner S., Kohn L. D. (1997) Role of MHC class I molecules in autoimmune disease. Crit. Rev. Immunol. 17, 463–468 [PubMed] [Google Scholar]
- 5. Goulder P. J., Altfeld M. A., Rosenberg E. S., Nguyen T., Tang Y., Eldridge R. L., Addo M. M., He S., Mukherjee J. S., Phillips M. N., Bunce M., Kalams S. A., Sekaly R. P., Walker B. D., Brander C. (2001) Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J. Exp. Med. 193, 181–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ueno T., Idegami Y., Motozono C., Oka S., Takiguchi M. (2007) Altering effects of antigenic variations in HIV-1 on antiviral effectiveness of HIV-specific CTLs. J. Immunol. 178, 5513–5523 [DOI] [PubMed] [Google Scholar]
- 7. Pereyra F., et al. (2010) The major genetic determinants of HIV control affect HLA class I peptide presentation. Science 330, 1551–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tam L. S., Gu J., Yu D. (2010) Pathogenesis of ankylosing spondylitis. Nat. Rev. Rheumatol. 6, 399–405 [DOI] [PubMed] [Google Scholar]
- 9. Madden D. R., Gorga J. C., Strominger J. L., Wiley D. C. (1992) The three-dimensional structure of HLA-B27 at 2.1Å resolution suggests a general mechanism for tight peptide binding to MHC. Cell 70, 1035–1048 [DOI] [PubMed] [Google Scholar]
- 10. Silver M. L., Parker K. C., Wiley D. C. (1991) Reconstitution by MHC-restricted peptides of HLA-A2 heavy chain with β2-microglobulin in vitro. Nature 350, 619–622 [DOI] [PubMed] [Google Scholar]
- 11. Smith K. J., Reid S. W., Stuart D. I., McMichael A. J., Jones E. Y., Bell J. I. (1996) An altered position of the α2 helix of MHC class I is revealed by the crystal structure of HLA-B*3501. Immunity 4, 203–213 [DOI] [PubMed] [Google Scholar]
- 12. Escobar H., Crockett D. K., Reyes-Vargas E., Baena A., Rockwood A. L., Jensen P. E., Delgado J. C. (2008) Large scale mass spectrometric profiling of peptides eluted from HLA molecules reveals N-terminal-extended peptide motifs. J. Immunol. 181, 4874–4882 [DOI] [PubMed] [Google Scholar]
- 13. Schönbach C., Ibe M., Shiga H., Takamiya Y., Miwa K., Nokihara K., Takiguchi M. (1995) Fine tuning of peptide binding to HLA-B*3501 molecules by nonanchor residues. J. Immunol. 154, 5951–5958 [PubMed] [Google Scholar]
- 14. Hillig R. C., Hülsmeyer M., Saenger W., Welfle K., Misselwitz R., Welfle H., Kozerski C., Volz A., Uchanska-Ziegler B., Ziegler A. (2004) Thermodynamic and structural analysis of peptide- and allele-dependent properties of two HLA-B27 subtypes exhibiting differential disease association. J. Biol. Chem. 279, 652–663 [DOI] [PubMed] [Google Scholar]
- 15. Motozono C., Yanaka S., Tsumoto K., Takiguchi M., Ueno T. (2009) Impact of intrinsic cooperative thermodynamics of peptide-MHC complexes on antiviral activity of HIV-specific CTL. J. Immunol. 182, 5528–5536 [DOI] [PubMed] [Google Scholar]
- 16. Walpole N. G., Kjer-Nielsen L., Kostenko L., McCluskey J., Brooks A. G., Rossjohn J., Clements C. S. (2010) The structure and stability of the monomorphic HLA-G are influenced by the nature of the bound peptide. J. Mol. Biol. 397, 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bowerman N. A., Crofts T. S., Chlewicki L., Do P., Baker B. M., Christopher Garcia K., Kranz D. M. (2009) Engineering the binding properties of the T cell receptor:peptide:MHC ternary complex that governs T cell activity. Mol. Immunol. 46, 3000–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Riquelme E., Carreño L. J., González P. A., Kalergis A. M. (2009) The duration of TCR/MHC interactions regulates CTL effector function and tumor-killing capacity. Eur. J. immunol. 39, 2259–2269 [DOI] [PubMed] [Google Scholar]
- 19. Loria J. P., Rance M., Palmer A. G., 3rd (1999) A relaxation-compensated Carr-Purcell-Meiboom-Gill sequence for characterizing chemical exchange by NMR spectroscopy. J. Am. Chem. Soc. 121, 2331–2332 [Google Scholar]
- 20. Tollinger M., Skrynnikov N. R., Mulder F. A., Forman-Kay J. D., Kay L. E. (2001) Slow dynamics in folded and unfolded states of an SH3 domain. J. Am. Chem. Soc. 123, 11341–11352 [DOI] [PubMed] [Google Scholar]
- 21. Shi Y., Qi J., Iwamoto A., Gao G. F. (2011) Plasticity of human CD8αα binding to peptide-HLA-A*2402. Mol. Immunol. 48, 2198–2202 [DOI] [PubMed] [Google Scholar]
- 22. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 23. Read R. J. (2001) Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 57, 1373–1382 [DOI] [PubMed] [Google Scholar]
- 24. Collaborative Computational Project (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 25. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 26. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 27. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Laskowski R. A., Macarthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 29. Brünger A. T. (1997) Free R value: Cross-validation in crystallography. Methods Enzymol. 277, 366–396 [DOI] [PubMed] [Google Scholar]
- 30. Krieger E., Joo K., Lee J., Lee J., Raman S., Thompson J., Tyka M., Baker D., Karplus K. (2009) Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins 77, 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ikura M., Kay L. E., Bax A. (1990) A novel approach for sequential assignment of 1H, 13C, and 15N spectra of proteins: heteronuclear triple-resonance three-dimensional NMR spectroscopy. Application to calmodulin. Biochemistry 29, 4659–4667 [DOI] [PubMed] [Google Scholar]
- 32. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 33. Kobayashi N., Iwahara J., Koshiba S., Tomizawa T., Tochio N., Güntert P., Kigawa T., Yokoyama S. (2007) KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J. Biomol. NMR 39, 31–52 [DOI] [PubMed] [Google Scholar]
- 34. Johnson B. A., Blevins R. A. (1994) NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 35. Sugase K., Dyson H. J., Wright P. E. (2007) Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 447, 1021–1025 [DOI] [PubMed] [Google Scholar]
- 36. Sugase K., Konuma T., Lansing J. C., Wright P. E. (2013) Fast and accurate fitting of relaxation dispersion data using the flexible software package GLOVE. J. Biomol. NMR 56, 275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 38. Probst-Kepper M., Hecht H. J., Herrmann H., Janke V., Ocklenburg F., Klempnauer J., van den Eynde B. J., Weiss S. (2004) Conformational restraints and flexibility of 14-meric peptides in complex with HLA-B*3501. J. Immunol. 173, 5610–5616 [DOI] [PubMed] [Google Scholar]
- 39. Tynan F. E., Burrows S. R., Buckle A. M., Clements C. S., Borg N. A., Miles J. J., Beddoe T., Whisstock J. C., Wilce M. C., Silins S. L., Burrows J. M., Kjer-Nielsen L., Kostenko L., Purcell A. W., McCluskey J., Rossjohn J. (2005) T cell receptor recognition of a “super-bulged” major histocompatibility complex class I-bound peptide. Nat. Immunol. 6, 1114–1122 [DOI] [PubMed] [Google Scholar]
- 40. Rammensee H. G., Friede T., Stevanoviíc S. (1995) MHC ligands and peptide motifs: first listing. Immunogenetics 41, 178–228 [DOI] [PubMed] [Google Scholar]
- 41. Rammensee H., Bachmann J., Emmerich N. P., Bachor O. A., Stevanović S. (1999) SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50, 213–219 [DOI] [PubMed] [Google Scholar]
- 42. Gómez J., Hilser V. J., Xie D., Freire E. (1995) The heat capacity of proteins. Proteins 22, 404–412 [DOI] [PubMed] [Google Scholar]
- 43. Prabhu N. V., Sharp K. A. (2005) Heat capacity in proteins. Annu. Rev. Phys. Chem. 56, 521–548 [DOI] [PubMed] [Google Scholar]
- 44. Todd M. J., Luque I., Velázquez-Campoy A., Freire E. (2000) Thermodynamic basis of resistance to HIV-1 protease inhibition: calorimetric analysis of the V82F/I84V active site resistant mutant. Biochemistry 39, 11876–11883 [DOI] [PubMed] [Google Scholar]
- 45. Hülsmeyer M., Hillig R. C., Volz A., Rühl M., Schröder W., Saenger W., Ziegler A., Uchanska-Ziegler B. (2002) HLA-B27 subtypes differentially associated with disease exhibit subtle structural alterations. J. Biol. Chem. 277, 47844–47853 [DOI] [PubMed] [Google Scholar]
- 46. Hawse W. F., Gloor B. E., Ayres C. M., Kho K., Nuter E., Baker B. M. (2013) Peptide modulation of class histocompatibility complex protein molecular flexibility and the implications for immune recognition. J. Biol. Chem. 288, 24372–24381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Baker B. M., Scott D. R., Blevins S. J., Hawse W. F. (2012) Structural and dynamic control of T-cell receptor specificity, cross-reactivity, and binding mechanism. Immunol. Rev. 250, 10–31 [DOI] [PubMed] [Google Scholar]
- 48. Narzi D., Becker C. M., Fiorillo M. T., Uchanska-Ziegler B., Ziegler A., Böckmann R. A. (2012) Dynamical characterization of two differentially disease associated MHC class I proteins in complex with viral and self-peptides. J. Mol. Biol. 415, 429–442 [DOI] [PubMed] [Google Scholar]
- 49. Winkler K., Winter A., Rueckert C., Uchanska-Ziegler B., Alexiev U. (2007) Natural MHC class I polymorphism controls the pathway of peptide dissociation from HLA-B27 complexes. Biophys. J. 93, 2743–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Garbi N., Tanaka S., van den Broek M., Momburg F., Hämmerling G. J. (2005) Accessory molecules in the assembly of major histocompatibility complex class I/peptide complexes: how essential are they for CD8(+) T-cell immune responses? Immunol. Rev. 207, 77–88 [DOI] [PubMed] [Google Scholar]
- 51. Oelke M., Maus M. V., Didiano D., June C. H., Mackensen A., Schneck J. P. (2003) Ex vivo induction and expansion of antigen-specific cytotoxic T cells by HLA-Ig-coated artificial antigen-presenting cells. Nat. Med. 9, 619–624 [DOI] [PubMed] [Google Scholar]
- 52. Hourigan C. S., Harkiolaki M., Peterson N. A., Bell J. I., Jones E. Y., O'Callaghan C. A. (2006) The structure of the human allo-ligand HLA-B*3501 in complex with a cytochrome p450 peptide: steric hindrance influences TCR allo-recognition. Eur. J. Immunol. 36, 3288–3293 [DOI] [PubMed] [Google Scholar]
- 53. Gras S., Kedzierski L., Valkenburg S. A., Laurie K., Liu Y. C., Denholm J. T., Richards M. J., Rimmelzwaan G. F., Kelso A., Doherty P. C., Turner S. J., Rossjohn J., Kedzierska K. (2010) Cross-reactive CD8+ T-cell immunity between the pandemic H1N1–2009 and H1N1–1918 influenza A viruses. Proc. Natl. Acad. Sci. U.S.A. 107, 12599–12604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gras S., Chen Z., Miles J.J., Liu Y.C., Bell M.J., Sullivan L.C., Kjer-Nielsen L., Brennan R.M., Burrows J.M., Neller M. A., Khanna R., Purcell A. W., Brooks A. G., McCluskey J., Rossjohn J., Burrows S. R. (2010) Allelic polymorphism in the T cell receptor and its impact on immune responses. J. Exp. Med. 207, 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Miles J. J., Borg N. A., Brennan R. M., Tynan F. E., Kjer-Nielsen L., Silins S. L., Bell M. J., Burrows J. M., McCluskey J., Rossjohn J., Burrows S. R. (2006) TCRα genes direct MHC restriction in the potent human T cell response to a class I-bound viral epitope. J. Immunol. 177, 6804–6814 [DOI] [PubMed] [Google Scholar]
- 56. Menssen R., Orth P., Ziegler A., Saenger W. (1999) Decamer-like conformation of a nona-peptide bound to HLA-B*3501 due to non-standard positioning of the C terminus. J. Mol. Biol. 285, 645–653 [DOI] [PubMed] [Google Scholar]
- 57. Tynan F. E., Elhassen D., Purcell A. W., Burrows J. M., Borg N. A., Miles J. J., Williamson N. A., Green K. J., Tellam J., Kjer-Nielsen L., McCluskey J., Rossjohn J., Burrows S. R. (2005) The immunogenicity of a viral cytotoxic T cell epitope is controlled by its MHC-bound conformation. J. Exp. Med. 202, 1249–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tynan F. E., Reid H. H., Kjer-Nielsen L., Miles J. J., Wilce M. C., Kostenko L., Borg N. A., Williamson N. A., Beddoe T., Purcell A. W., Burrows S.R., McCluskey J., Rossjohn J. (2007) T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat. Immunol. 8, 268–276 [DOI] [PubMed] [Google Scholar]
- 59. Miles J. J., Elhassen D., Borg N. A., Silins S. L., Tynan F. E., Burrows J. M., Purcell A. W., Kjer-Nielsen L., Rossjohn J., Burrows S. R., McCluskey J. (2005) CTL recognition of a bulged viral peptide involves biased TCR selection. J. Immunol. 175, 3826–3834 [DOI] [PubMed] [Google Scholar]
- 60. Tynan F. E., Borg N. A., Miles J. J., Beddoe T., El-Hassen D., Silins S. L., van Zuylen W. J., Purcell A. W., Kjer-Nielsen L., McCluskey J., Burrows S.R., Rossjohn J. (2005) High resolution structures of highly bulged viral epitopes bound to major histocompatibility complex class I. Implications for T-cell receptor engagement and T-cell immunodominance. J. Biol. Chem. 280, 23900–23909 [DOI] [PubMed] [Google Scholar]
- 61. Archbold J. K., Macdonald W. A., Miles J. J., Brennan R. M., Kjer-Nielsen L., McCluskey J., Burrows S. R., Rossjohn J. (2006) Alloreactivity between disparate cognate and allogeneic pMHC-I complexes is the result of highly focused, peptide-dependent structural mimicry. J. Biol. Chem. 281, 34324–34332 [DOI] [PubMed] [Google Scholar]