Abstract
Background
Brown planthopper (BPH, Nilaparvata lugens Stål), is the most destructive phloem-feeding insect pest of rice (Oryza sativa). The BPH-resistance gene BPH15 has been proved to be effective in controlling the pest and widely applied in rice breeding programs. Nevertheless, molecular mechanism of the resistance remain unclear. In this study, we narrowed down the position of BPH15 on chromosome 4 and investigated the transcriptome of BPH15 rice after BPH attacked.
Results
We analyzed 13,000 BC2F2 plants of cross between susceptible rice TN1 and the recombinant inbred line RI93 that carrying the BPH15 gene from original resistant donor B5. BPH15 was mapped to a 0.0269 cM region on chromosome 4, which is 210-kb in the reference genome of Nipponbare. Sequencing bacterial artificial chromosome (BAC) clones that span the BPH15 region revealed that the physical size of BPH15 region in resistant rice B5 is 580-kb, much bigger than the corresponding region in the reference genome of Nipponbare. There were 87 predicted genes in the BPH15 region in resistant rice. The expression profiles of predicted genes were analyzed. Four jacalin-related lectin proteins genes and one LRR protein gene were found constitutively expressed in resistant parent and considered the candidate genes of BPH15. The transcriptomes of resistant BPH15 introgression line and the susceptible recipient line were analyzed using high-throughput RNA sequencing. In total, 2,914 differentially expressed genes (DEGs) were identified. BPH-responsive transcript profiles were distinct between resistant and susceptible plants and between the early stage (6 h after infestation, HAI) and late stage (48 HAI). The key defense mechanism was related to jasmonate signaling, ethylene signaling, receptor kinase, MAPK cascades, Ca2+ signaling, PR genes, transcription factors, and protein posttranslational modifications.
Conclusions
Our work combined BAC and RNA sequencing to identify candidate genes of BPH15 and revealed the resistance mechanism that it mediated. These results increase our understanding of plant–insect interactions and can be used to protect against this destructive agricultural pest.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2164-15-674) contains supplementary material, which is available to authorized users.
Keywords: Brown planthopper, BPH15, Recombination coldspot, Candidate gene, RNA sequencing, Transcriptome, Defense mechanism
Background
The brown planthopper (BPH; Nilaparvata lugens Stål) is a typical phloem-feeding insect and a major pest of rice (Oryza sativa). At this time, 24 BPH-resistance genes have been identified in rice, 20 of which are located on chromosomes [1]. Resistance genes BPH14 and BPH15 were introgressed from wild rice Oryza officinalis [2]. These two genes showed significant resistance to BPH and have been broadly employed in rice breeding programs [3, 4]. Recently, BPH14 was isolated using a map-based cloning strategy, and it was found to encode a coiled-coil, nucleotide-binding, and leucine-rich repeat (CC-NB-LRR) protein that activates the SA signaling pathway [5]. In rice breeding, BPH15 shows a greater resistance effect than BPH14 and BPH18 when introgressed into the elite indica rice 9311 and hybrid rice [6]. BPH15 is located on the short arm of chromosome 4, where 4 BPH-resistance genes are clustered [7, 8].
Plant responses to insect attack are correlated to the mode of feeding [9, 10]. Gene expression profiles suggested that defense mechanisms against BPH differ from those against chewing insects. Genes involved in macromolecule degradation and plant defenses were found to be upregulated, whereas those involved in photosynthesis and cell growth were downregulated after BPH infestation [11]. Quantitative proteomics results revealed that proteins involved in JA synthesis, oxidative stress response, β-glucanases, and kinases showed significant changes in expression in response to BPH feeding [12]. Nitric oxide was used by plants as a signaling molecule and plays a role in the rice tolerance response to BPH feeding [13]. Callose deposition on the sieve plates occluded the sieve tubes and inhibited continuous feeding by BPH in resistant lines; thus, the death of BPH on resistant lines was the result of starvation and not poisoning [14]. Nevertheless, complete transcriptional analysis of the BPH response genes remains unavailable, and more comprehensive differential expression profiles are required to better understand the molecular mechanism of BPH resistance in rice.
To clone BPH15 and increase our understanding of the molecular mechanism of resistance, we backcrossed the resistant plant carrying the BPH15 locus to susceptible rice and developed the backcrossed populations. BPH15 was located in a recombination cold spot of 580-kb. High-throughput RNA sequencing represents the latest and most suitable tool for characterizing the transcriptome [15]. We then applied deep RNA sequencing to investigate the transcriptomes of BPH15 introgression line and susceptible recipient line. In the sequenced BPH15 region, four jacalin-related lectin (JRL) domain proteins and a LRR family protein were considered candidate BPH15 genes. The molecular mechanism of resistance to BPH was also explored by comparing differentially expressed genes (DEGs) between resistant and susceptible rice.
Results
BPH resistance gene BPH15is located in a recombination cold-spot region
We previously mapped the major BPH-resistance gene on the short arm of chromosome 4 between molecular markers RG1 and RG2 using the F2 population of cross between RI93, a recombinant inbred line that carrying the BPH15 gene from original resistant donor B5, and susceptible rice TN1 (Figure 1A) [7]. To fine-map the gene, the resistant rice line YHY15 carrying the BPH15 locus was selected from the F2 population and backcrossed to TN1 to develop mapping populations (Additional file 1). After genotyping BC1F2 and phenotyping the BC1F3 populations, BPH15 was mapped between markers RM261 and S16 (Figure 1A). We further screened 13,000 BC2F2 plants for recombination between RM261 and S16, and 54 recombination events were identified. The average genome-wide recombination rate (the ratio of total genetic map length in centimorgans divided by the genome size in base pairs) is about 0.004 cM/kb in rice [16]. Chromosome intervals between RM261 and S16 showed a much lower recombination rate (0.0005 cM/kb). After genotyping the recombinant plants using newly developed markers (Additional file 2) and phenotyping the fixed recombinant plants, BPH15 was mapped to a 0.0269 cM interval defined by g12140-2 and T6 (7 recombinants in 13,000 plants) (Figure 1B; Additional file 3-1). A high recombination rate was observed just outside of the BPH15 region. Specifically, the 0.7-kb fragment from marker T6 to 20M14 is a hot spot with a genetic recombination rate of 0.049 cM/kb, much higher than the whole genome level (Figure 1C). Afterward, we identified 61 recombinants from 10,000 BC4F2 plants, similar to BC2F2 plants, and no crossover was found in the BPH15 region (Additional file 3-2). Based on these results, BPH15 is located in a recombination cold-spot region, and identifying candidate BPH15 genes using conventional analysis of recombination is difficult.
Figure 1.
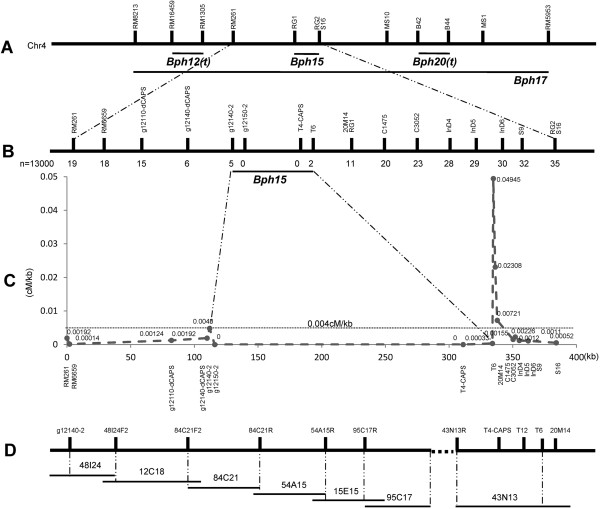
Fine mapping of the BPH15 locus. A, The marker positions of four previously mapped BPH resistant genes. B, Screened recombinant information in the BC2F2 family. Numbers under the linkage map indicate the number of recombinants detected between the marker and BPH15. BPH15 was mapped to the region between markers g12140-2 and T6. C, Recombination frequencies between each adjacent DNA marker. The dashed line represents genome average 0.004 cM/kb value. D, Physical map assembled by PCR-screened BAC clones. The dashed line represents the gap where no BAC clones overlapped.
High level of sequence diversity in the BPH15region
The physical distance between the markers g12140-2 and T6 is approximately 210-kb in the Nipponbare genome. To detect the actual length and identify the genes in the region spanning the BPH15 locus, we constructed a genomic BAC library of long insertion fragments for the resistant rice B5, the original donor of BPH15. The markers g12140-2 and T6 were used to screen BAC clones by amplification of the BAC DNA pools for an initial chromosome walking. Using five steps of walking, the physical map of the BPH15 region was assembled, and the shortest path consisted of seven BAC clones (Figure 1D). A gap existed between the BAC 95C17 and BAC 43N13. The seven BAC clones were sequenced and included a 700-kb region. Finally, we mapped the BPH15 locus between g12140-2 and newly developed marker T12 (Additional file 3-1; Figure 1D), and the physical distance was at least 580-kb according to sequenced BAC clones. Eighty-seven genes within this 580-kb region were annotated using online FGENESH software (from F8 to F94) and 70 genes were TE-related genes (Additional file 4-1). There were 31 annotated genes in the 210-kb Nipponbare sequence according to MSU 7.0 and 21 genes were TE-related genes (Additional file 4-2). Excluding several functional genes (Figure 2), the sequences in this region of the two rice genotypes are highly diverse. Based on these results, the sequence of this region in the resistant rice genome differed significantly from the corresponding region in the reference genome of Nipponbare; i.e., the 210-kb fragment in Nipponbare was replaced by a much larger fragment containing several repeat sequences in resistant rice. Thus, developing codominant molecular markers for this region was difficult. The high level of sequence diversity in the BPH15 region explained the heavy suppression of recombination in this region.
Figure 2.
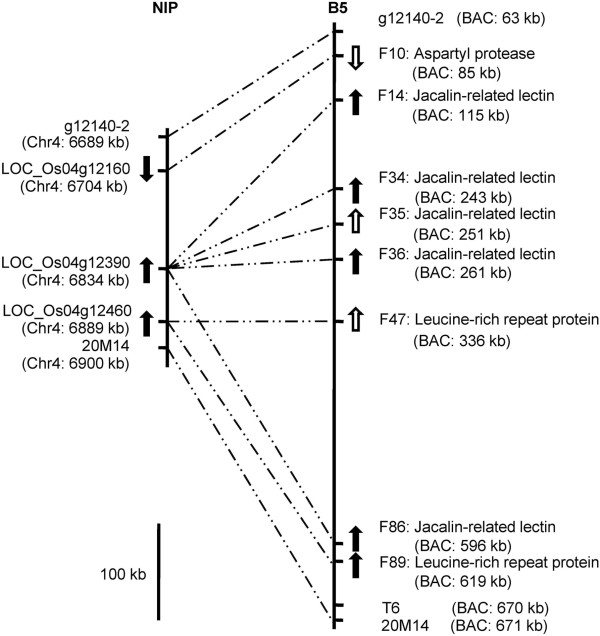
The BPH15 region relationship between Nipponbare and BPH-resistant rice B5. The solid and hollow arrows represent expressed and unexpressed genes and their direction, respectively. Markers g12140-2, T6 and 20M14 relate to the region in Figure 1D. The notes in brackets represent gene location on the chromosome or assembled 700-kb BAC sequences. LOC_Os04g12160: aspartic proteinase nepenthesin-2 precursor; LOC_Os04g12390: transposon protein, containing a jacalin lectin domain; LOC_Os04g12460: leucine rich repeat family protein. The genes location are listed in Additional file 4.
BPH-responsive transcript profiles are distinct in resistant BPH15introgression line and susceptible recipient line
To identify the BPH15 candidate genes and understand the molecular mechanism of resistance, the expression profiles of BPH15 introgression line (R) and susceptible recipient line (S) were determined using deep RNA sequencing. RNAs extracted from rice samples at the early stage (6 h after infestation, HAI), late stage (48 HAI) and control (0 HAI) of two rice lines were sequenced. As a result, 198 million paired-end sequence reads of 100 bp in length were generated in six samples. After removing low-quality reads, a total of 150 million high-quality clean reads remained, of which 90.48–92.15% were aligned to the reference genome using TopHat (Table 1).
Table 1.
Statistics of sequencing reads and alignment to the reference genome
| Samples | Raw data reads | Raw data base (bp) | High-quality reads | High-quality base (bp) | Percentage of alignment (%) |
|---|---|---|---|---|---|
| S0 | 37,061,382 | 3,706,138,200 | 28,008,690 | 2,357,352,788 | 91.84 |
| S6 | 24,715,554 | 2,471,555,400 | 18,742,423 | 1,581,929,269 | 91.38 |
| S48 | 32,339,240 | 3,233,924,000 | 24,661,359 | 2,081,307,813 | 90.48 |
| R0 | 39,251,668 | 3,925,166,800 | 29,661,746 | 2,489,101,005 | 92.15 |
| R6 | 35,214,634 | 3,521,463,400 | 26,576,329 | 2,234,567,794 | 91.39 |
| R48 | 29,490,134 | 2,949,013,400 | 22,533,122 | 1,903,312,211 | 91.84 |
| all | 198,072,612 | 19,807,261,200 | 150,183,669 | 12,647,570,880 | - |
Percentage of alignment = high quality reads aligned to genome/high-quality reads.
One fundamental use of transcriptome sequencing is analysis of differentially expressed genes (DEGs) between samples [15]. In our study, we defined DEGs as the transcripts showing at least a 1.5-fold change of the FPKM (fragments per kilobase of exon per million fragments mapped) (log2FC ≥ 0.585 or log2FC ≤ –0.585) and P-value < 0.05. In total, 2,914 DEGs were detected among seven comparisons: S0_S6, S0_S48, R0_R6, R0_R48, S0_R0, S6_R6, and S48_R48 (Additional file 5). In susceptible rice, 615 and 1966 DEGs were found in S0_S6 and S0_S48 comparisons, respectively. In contrast, in resistant rice, the DEGs numbers were 451 and 651 in R0_R6 and R0_R48, respectively (Figure 3).
Figure 3.
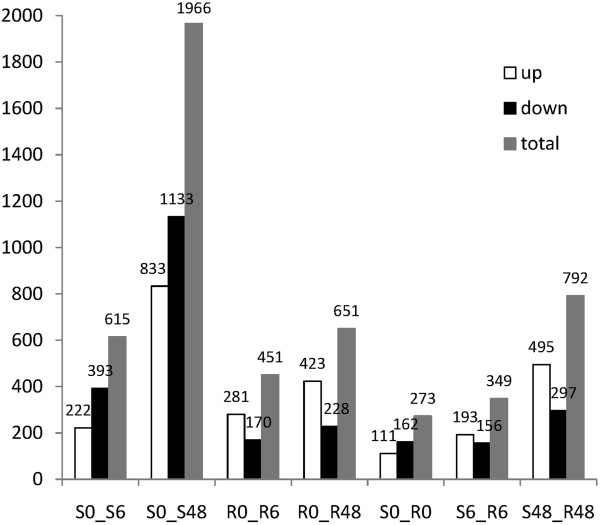
Contrast between upregulated and downregulated DEGs in all comparisons.
DEGs in susceptible and resistant rice at 6 HAI and 48 HAI were hierarchically clustered, and the heat map is shown in Figure 4A. The majority of DEGs had similar expression patterns among four comparisons, showing consistent upregulation or downregulation, although not all P-values for the four comparisons were below 0.05. To compare the two rice genotypes, DEGs exclusively at 48 HAI were selected and hierarchically clustered. Most of these DEGs had lower amplitude of variation in the resistant genotype compared to the susceptible genotype (Figure 4B). The k-means clustering analysis results also supported the conclusion (Additional file 6).
Figure 4.
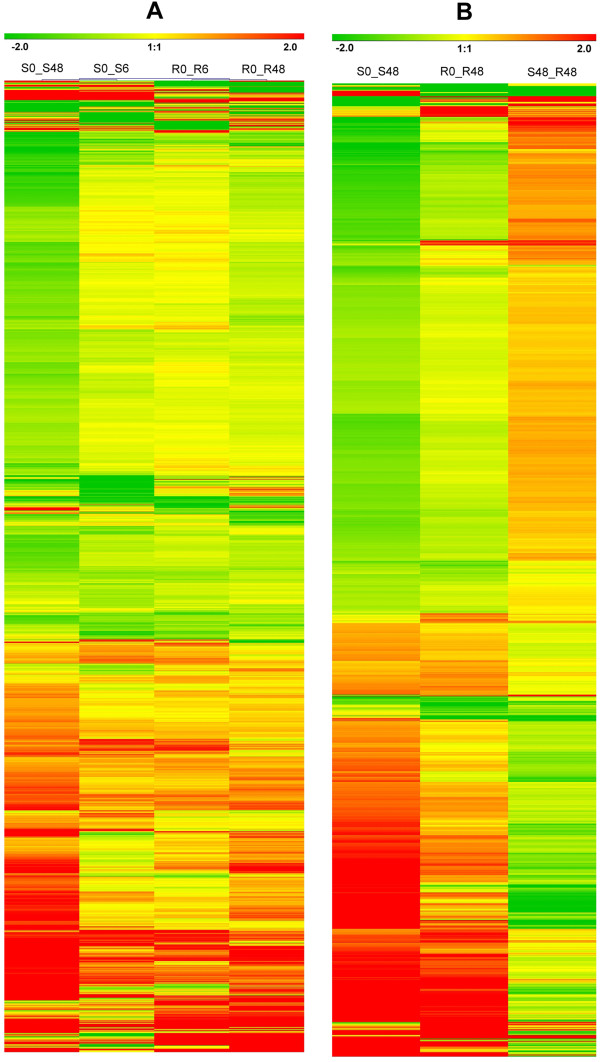
Hierarchical clustering analysis of DEGs based on the log ratio of FPKM data. The color key represents FPKM normalized log2 transformed counts. Red indicates upregulated DEGs and green denotes downregulated DEGs. Each column shows a comparison and each row represents a gene. A, DEGs of S and R. B, DEGs at 48 HAI.
High-throughput technologies such as microarray and sequencing methods generate enormous amounts of data, but individual functional annotation of all DEGs remains challenging. Pathway-based analysis to characterize the interaction between genes increases our understanding of the biological function of DEGs [17]. DEGs assigned to MapMan pathways and important classifications are listed in Additional file 7 and provided in Table 2. To verify the RNA-Seq results, the expression of 23 DEGs was analyzed by quantitative polymerase chain reaction (qPCR) with gene-specific primers (Additional file 2). Additional file 8 provided detailed RNA-seq fold-change values for every DEG and their qPCR results of three biological replicates. The qPCR results were consistent with RNA-seq data, since the genes displayed similar fold-changes with a correlation ratio of R2 = 0.971 (Additional file 9).
Table 2.
Pathway classification by MapMan
| Pathways | S-all | R-all | S_R-all | |||
|---|---|---|---|---|---|---|
| U | D | U | D | U | D | |
| Ethylene | 21 | 7 | 14 | 3 | 4 | 8 |
| Jasmonate | 4 | 2 | 2 | 0 | 0 | 0 |
| Salicylic acid | 4 | 0 | 2 | 0 | 0 | 0 |
| Receptor kinase | 20 | 5 | 9 | 3 | 5 | 4 |
| Ca2+ signaling | 14 | 8 | 6 | 3 | 1 | 3 |
| Biotic stress | 35 | 8 | 13 | 2 | 4 | 20 |
| Wounding | 4 | 1 | 4 | 0 | 1 | 1 |
| AP2/EREBP TF | 15 | 3 | 9 | 0 | 1 | 8 |
| bHLH TF | 6 | 1 | 7 | 0 | 0 | 1 |
| Zinc finger family TF | 12 | 1 | 5 | 1 | 4 | 4 |
| WRKY domain TF | 10 | 1 | 7 | 0 | 2 | 1 |
| Protein synthesis, targeting and folding | 6 | 148 | 3 | 30 | 71 | 9 |
| Protein degradation-ubiquitin | 26 | 4 | 17 | 1 | 5 | 9 |
| Protein posttranslational modification | 25 | 12 | 12 | 0 | 3 | 8 |
| Bowman–Birk-type bran trypsin inhibitor | 7 | 1 | 6 | 1 | 0 | 5 |
| Glucan endo-1,3-β-glucosidase | 11 | 2 | 6 | 0 | 1 | 9 |
| Photosynthesis | 8 | 97 | 4 | 27 | 38 | 5 |
| Tetrapyrrole synthesis | 0 | 23 | 0 | 10 | 14 | 1 |
| Major CHO synthesis | 2 | 6 | 2 | 2 | 2 | 0 |
| Major CHO degradation | 6 | 3 | 3 | 1 | 3 | 2 |
| Lipid metabolism | 5 | 51 | 6 | 5 | 26 | 5 |
| TCA, mitochondrial electron transport | 2 | 22 | 3 | 1 | 1 | 3 |
S-all: total DEG number in comparisons of S0_S6 and S0_S48; R-all: DEG number in comparisons of R0_R6 and R0_R48; S_R-all: DEG number in comparisons of S0_R0, S6_R6 and S48_R48.U: upregulated; D: downregulated.
Hormone signaling pathways play pivotal roles in plant defense [18, 19]. In this study, the majority of ethylene synthesis genes and ethylene signal transduction genes, such as ACO (LOC_Os02g53180) and ERF (LOC_Os02g43790), were upregulated in two rice genotypes, but the number of DEGs in resistant rice was less than that in susceptible rice (Additional file 7; Table 3). This result suggested that BPH feeding activated the ET signaling pathway in susceptible rice. Jasmonate synthesis genes, such as lipoxygenase LOX (LOC_Os08g39850), allene oxidase synthase AOS2 (LOC_Os03g12500), and 12-oxophytodienoate reductase OPR1 (LOC_Os06g11210) were upregulated in susceptible rice, which suggests that BPH attack induces the JA signaling pathway (Additional file 7; Table 3). Four SA carboxyl methyltransferase (SAMT) genes were upregulated in susceptible rice (Additional file 7), which can reduce SA content by forming MeSA from SA in the plants [20]. Other SA synthesis and signaling genes were not identified in DEGs. We measured the SA content in leaf sheath surrounding the stem from plants exposed to BPH for 0, 3, 6, 24, and 48 h using gas chromatography–mass spectrometry (GC-MS; Additional file 10). No significant difference in SA levels was observed between the BPH-infested and control plants or between resistant and susceptible plants, except that SA levels in 48 HAI susceptible rice were significantly lower than in resistant rice. The lower SA content may be caused by upregulated SAMT genes expression. These results indicated that the SA signaling pathway may not be activated in BPH15-mediated resistance or during the basal defense of susceptible rice.
Table 3.
Representative pathway genes
| Pathways | Representative genes | Transcript | S0_S6 | S0_S48 | R0_R6 | R0_R48 |
|---|---|---|---|---|---|---|
| Ethylene | ACO | LOC_Os02g53180.1 | - | U | - | - |
| ERF | LOC_Os02g43790.1 | - | U | - | - | |
| Jasmonate | LOX | LOC_Os08g39850.1 | - | U | - | - |
| AOS2 | LOC_Os03g12500.1 | - | U | - | - | |
| OPR1 | LOC_Os06g11210.1 | - | U | - | - | |
| Salicylic acid | SAMT | LOC_Os11g15040.1 | U | U | - | U |
| Receptor kinase | Receptor-like protein kinase 5 | LOC_Os02g13510.1 | - | U | - | U |
| BAK1 | LOC_Os03g32580.1 | - | U | - | - | |
| XA21 | LOC_Os11g36180.1 | - | - | - | U | |
| MAP kinase | MAPK | LOC_Os03g17700.1 | - | U | - | - |
| Ca2+ signaling | OsCML15 | LOC_Os05g31620.1 | - | U | - | - |
| Calmodulin-binding protein | LOC_Os12g36910.1 | - | U | - | U | |
| Biotic stress | PR1a | LOC_Os07g03710.1 | - | U | - | - |
| PR1b | LOC_Os01g28450.1 | - | U | - | U | |
| PR3 | LOC_Os03g30470.1 | - | U | - | - | |
| PR4b | LOC_Os11g37960.1 | - | U | - | - | |
| PR9 | LOC_Os07g48020.1 | - | U | - | U | |
| PR10 | LOC_Os12g36880.1 | - | U | U | U | |
| Wounding | WI12 | LOC_Os03g18770.1 | U | - | - | - |
| TF | EREBP | LOC_Os03g08500.1 | - | U | - | - |
| Basic helix–loop–helix protein | LOC_Os09g31300.1 | - | U | - | U | |
| C2H2 zinc finger protein | LOC_Os05g37190.1 | - | U | - | U | |
| C3H zinc finger protein | LOC_Os07g38090.1 | - | - | - | - | |
| WRKY domain TF | LOC_Os01g14440.1 | - | U | - | - | |
| Pr. synthesis | tRNA synthetase | LOC_Os01g54020.2 | - | D | - | - |
| 40S ribosomal protein | LOC_Os03g18570.1 | - | D | - | - | |
| 60S ribosomal protein | LOC_Os05g06310.1 | - | D | - | - | |
| Translation initiation factor | LOC_Os05g49970.2 | - | D | - | - | |
| Pr. targeting | Mitochondrial import translocase | LOC_Os02g48610.2 | - | D | - | - |
| Pr. folding | Chaperonin | LOC_Os06g09679.1 | - | D | - | D |
| Pr. degradation | Ubiquitin family protein | LOC_Os02g06640.1 | - | U | - | - |
| OsFBL7 | LOC_Os02g10700.1 | U | U | - | - | |
| Pr. modification | Protein phosphatase | LOC_Os02g13100.1 | - | U | - | - |
| Calcium-dependent protein kinases | LOC_Os07g05620.2 | - | U | - | U | |
| CRK5 | LOC_Os04g56430.1 | - | U | - | - | |
| CRK6 | LOC_Os03g16960.1 | - | U | - | - | |
| Serine/threonine protein kinase | LOC_Os02g01730.1 | - | U | - | - | |
| Trypsin inhibitor | BBTI5 | LOC_Os01g03360.1 | U | U | U | - |
| Glucosidase | GNS1 | LOC_Os05g31140.1 | - | U | - | - |
| GNS4 | LOC_Os01g71670.1 | - | U | - | U | |
| GNS5 | LOC_Os01g71340.1 | - | U | - | U | |
| PS light reaction | PS I reaction center subunit | LOC_Os03g56670.1 | - | D | - | - |
| PS II reaction center protein | LOC_Os01g71190.1 | D | D | - | D | |
| ATP synthase | LOC_Os07g32880.1 | - | D | - | - | |
| PS Calvin cycle | Ribulose bisphosphate carboxylase | LOC_Os12g17600.1 | - | D | - | - |
| Tetrapyrrole | Aminolevulinic acid dehydratase | LOC_Os06g49110.1 | - | D | - | - |
| CHO synthesis | Starch synthase | LOC_Os04g53310.1 | D | - | D | - |
| CHO degradation | β-amylase | LOC_Os03g22790.1 | U | U | U | - |
| Lipid synthesis | Acetyl-CoA carboxylase | LOC_Os05g22940.1 | - | D | - | - |
| Glycerol-3-phosphate acyltransferase | LOC_Os01g63580.1 | - | D | - | - | |
| TCA | Citrate synthase | LOC_Os01g19450.1 | - | D | - | - |
In total, 35 and 13 proteins responding to biotic stress were upregulated in susceptible and resistant rice, respectively, such as SCP-like extracellular protein PR1a (LOC_Os07g03710), PR1b (LOC_Os01g28450), chitinase family protein PR3 (LOC_Os03g30470), wound-induced protein PR4b (LOC_Os11g37960), PR9 (LOC_Os07g48020), and pathogenesis-related Betv1 family protein PR10a (LOC_Os12g36880) (Tables 2 and 3). However, the number of DEGs in resistant rice was lower than in susceptible rice (Additional file 7). Herbivore-induced callose deposition on the sieve plates of rice is an important mechanism for host resistance. β-1,3 glucan hydrolase genes are activated and cause unplugging of the sieve tube occlusions in susceptible plants [14]. The majority of this gene family were upregulated in susceptible and resistant rice, such as GNS1 (LOC_Os05g31140), GNS4 (LOC_Os01g71670), and GNS5 (LOC_Os01g71340), but the number of DEGs was lower in resistant rice (Additional file 7; Tables 2 and 3). In total, 148 of 154 protein synthesis, protein targeting, and protein folding related genes were downregulated in susceptible rice, but only 30 were downregulated in resistant rice (Tables 2 and 3). Thus, the reprogramming of protein synthesis and secretion machinery was significantly affected in the susceptible lines but not in the resistant lines, which was suggestive of significant damage in susceptible rice.
DEGs assigned to photosynthesis (PS), tricarboxylic acid cycle (TCA), mitochondrial electron transport/ATP synthesis, major carbohydrate (CHO) metabolism and lipid metabolism were highly downregulated in susceptible rice. However, these genes had low amplitude of variation in resistant rice (Additional file 7; Table 2). So, these genes were upregulated in resistant rice as compared between with those in suseptible rice (Figure 5). The PS comprised genes coding for proteins involved in the photosynthetic electron transport chain, photorespiration, Calvin cycle, and Photosystem I and II complexes. We observed a general downregulation of genes associated with photosynthetic processes at 6 HAI and 48 HAI in both susceptible and resistant rice, but with a higher number of DEGs in susceptible genotype at 48 HAI. BPH response included genes involved in the primary metabolism, mainly related to carbohydrate and lipid metabolism. These genes were predominantly downregulated in susceptible rice after BPH infestation, but the carbohydrate degradation genes were upregulated (Table 3). Our data indicated that BPH attack generally represses photosynthesis-related genes in susceptible rice leaves, as well as those involved in primary metabolism.
Figure 5.
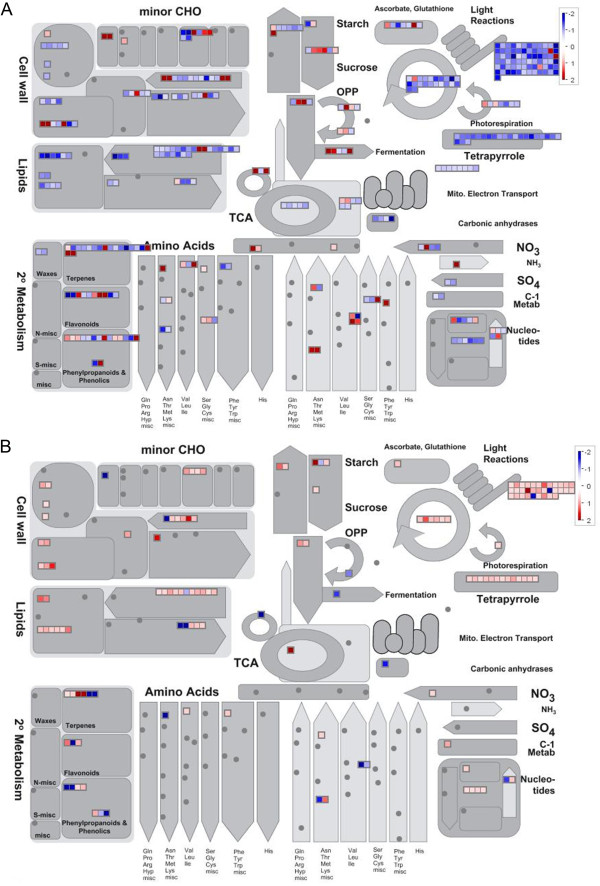
MapMan overview of metabolism. Individual genes are represented by small squares. The color key represents FPKM normalized log2 transformed counts. Red indicates upregulation and dark blue denotes downregulation. A: S0_S48 DEG; B: S48_R48 DEG.
BPH-responsive transcript profiles are distinct in early and late feeding stages of two rice genotypes
As shown in Figure 3, the number of DEGs at 6 HAI was less than 48 HAI in both the resistant and susceptible rice. This demonstrated that both rice genotypes experienced weak damage in the early stages of BPH feeding, and resistant rice showed a relatively normal physiological status compared to susceptible rice. In addition, the amplitude of variation at 6 HAI was less than that at 48 HAI in both rice genotypes (Figure 4A; Additional file 6).
To understand key biological processes involved in the rice response to BPH, we used singular enrichment analysis of agriGO to identify enriched GO terms. All compared DEGs showed significant GO terms, excluding S0_R0 and S6_R6 (Additional file 11). As a result, 40 significant GO terms were found in total DEGs in susceptible line (union of S0_S6 and S0_S48), 25 in resistant line (union of R0_R6 and R0_R48), and 35 in comparison of the two lines (union of S0_R0, S6_R6, and S48_R48) (Additional files 12, 13 and 14). Investigation of the rice transcriptome following BPH feeding revealed the activation of a wide and complex response, and the transcriptional reconfiguration involved a broad range of biological processes. The 40 GO terms were mainly distributed in several categories, including photosynthesis, primary metabolism, secondary metabolism, response to stimulus, cell wall, and ribosome. GO terms of important biological functions were compared for significance in early and late feeding stages of two rice genotypes (Table 4). Common GO terms significant in all comparisons and unions were response to stimulus, response to abiotic stimulus, response to stress, cell wall, plastid, and thylakoid. The 48 HAI-specific GO terms were response to biotic stimulus, photosynthesis, generation of precursor metabolites and energy, ribosome, and intracellular organelle. Several GO terms, including macromolecule biosynthetic process, translation, gene expression, secondary metabolic process, and biosynthetic process, were significant only in 48 HAI-susceptible rice and the corresponding S48_R48. All GO terms were significant in 48 HAI-susceptible rice, and showed smaller P-values. Overall, during BPH infestation, the most rapid gene expression adjustments were observed in the categories of response to stimulus and cell wall; the DEGs involved in photosynthesis, response to biotic stimulus, and ribosome gradually increased and became significant at late feeding stage.
Table 4.
Significant GO terms
| GO accession | GO terms | R0_R6 | S0_S6 | R0_R48 | R-all | S0_S48 | S-all | S48_R48 | S_R-all |
|---|---|---|---|---|---|---|---|---|---|
| GO:0050896 | Response to stimulus | 7.90E-07 | 3.50E-08 | 1.00E-08 | 1.80E-11 | 2.10E-14 | 5.50E-17 | 2.10E-06 | 2.40E-09 |
| GO:0009628 | Response to abiotic stimulus | 1.10E-06 | 3.10E-08 | 7.90E-08 | 4.10E-10 | 1.10E-20 | 6.00E-24 | 1.10E-12 | 3.40E-16 |
| GO:0006950 | Response to stress | 7.90E-06 | 1.10E-07 | 7.10E-08 | 1.60E-10 | 1.40E-12 | 5.80E-16 | 6.30E-07 | 7.70E-10 |
| GO:0005618 | Cell wall | 2.60E-06 | 1.10E-06 | 1.50E-03 | 8.60E-08 | 2.90E-12 | 3.70E-15 | 3.10E-08 | 6.60E-09 |
| GO:0009536 | Plastid | 3.80E-06 | 1.60E-07 | 3.40E-10 | 8.40E-12 | 4.20E-83 | 3.90E-79 | 1.80E-31 | 1.20E-28 |
| GO:0009579 | Thylakoid | 1.90E-05 | 2.50E-12 | 4.30E-20 | 1.70E-18 | 1.00E-102 | 1.60E-97 | 8.70E-52 | 1.10E-47 |
| GO:0015979 | Photosynthesis | - | - | 8.90E-09 | 2.20E-07 | 4.90E-42 | 8.90E-41 | 5.00E-21 | 1.30E-17 |
| GO:0009607 | Response to biotic stimulus | - | - | 4.70E-07 | 5.20E-08 | 2.40E-13 | 4.80E-13 | 5.70E-08 | 4.70E-08 |
| GO:0006091 | Generation of precursor metabolites and energy | - | - | 4.80E-04 | 8.40E-04 | 4.50E-21 | 2.30E-20 | 4.00E-11 | 3.30E-11 |
| GO:0005840 | Ribosome | - | - | 2.00E-06 | 2.70E-04 | 2.60E-24 | 9.50E-23 | 4.70E-16 | 4.20E-16 |
| GO:0043229 | Intracellular organelle | - | - | 6.10E-04 | 6.30E-04 | 3.90E-23 | 4.20E-21 | 2.30E-09 | 6.10E-09 |
| GO:0009059 | Macromolecule biosynthetic process | - | - | - | - | 3.50E-15 | 9.40E-13 | 6.00E-13 | 1.40E-12 |
| GO:0006412 | Translation | - | - | - | - | 3.50E-15 | 9.40E-13 | 6.00E-13 | 1.40E-12 |
| GO:0010467 | Gene expression | - | - | - | - | 3.00E-10 | 2.10E-08 | 6.00E-10 | 1.10E-09 |
| GO:0019748 | Secondary metabolic process | - | - | - | - | 2.30E-04 | 1.40E-05 | 4.60E-04 | 1.50E-06 |
| GO:0009058 | Biosynthetic process | - | - | - | - | 2.30E-04 | 4.20E-04 | 1.20E-03 | 3.30E-04 |
R-all: union of R0_R6 and R0_R48; S-all: union of S0_S6 and S0_S48; S_R-all: union of S0_R0, S6_R6, and S48_R48.
Common defense-related genes in two rice genotypes
Receptor kinase, kinase cascades, and Ca2+ signaling-related genes are important components of signal transduction and play roles in transmitting resistance signals to downstream response genes [21]. Twenty and nine receptor kinases in susceptible and resistant rice were upregulated, respectively (Table 2). This included LRR family protein receptor-like protein kinase 5 (LOC_Os02g13510), brassinosteroid insensitive 1-associated receptor kinase 1 precursor BAK1 (LOC_Os03g32580), and receptor kinase XA21 (LOC_Os11g36180) (Table 3). These upregulated receptor kinase genes suggested that signal perception was activated after BPH feeding. The intracellular concentration of Ca2+, an important second messenger, typically increases in response to biotic or abiotic stress. In our studies, the majority of Ca2+ signaling-related genes were upregulated, such as calmodulin-related calcium sensor protein OsCML15 (LOC_Os05g31620) and calmodulin-binding protein (LOC_Os12g36910) (Table 3), which suggests that the rice defense response to BPH involved Ca2+ influx. Two MAP kinase genes LOC_Os03g17700 and LOC_Os06g48590 were upregulated (Additional file 7). Protein posttranslational modification genes were also upregulated, such as cysteine-rich receptor-like protein kinase CRK5 (LOC_Os04g56430), CRK6 (LOC_Os03g16960), calcium/calmodulin dependent protein kinases (LOC_Os07g05620), serine/threonine protein kinase (LOC_Os02g01730), and protein phosphatase (LOC_Os02g13100) (Table 3). CRK5 and CRK6 were detected based on a proteomic approach to evaluate BPH15 [12]. These upregulated genes suggest that a resistance signal is transmitted to downstream regulatory networks after rice perceives BPH.
Transcription factors (TFs) play an important role in the defense response [22]. We detected approximately 200 TF DEGs in two rice genotypes, and more were found in susceptible rice than resistant rice (Tables 2 and 3; Additional file 7). EREBP binds to the GCC box, a conserved ethylene responsive promoter element found in many defense-related genes [23]. A systematic expression analysis of rice WRKY revealed a large number of WRKY DNA-binding proteins involved in the transcriptional activation of defense-related genes in response to rice pathogens [24]. Overexpression of the OsWRKY89 gene enhanced resistance to the rice blast fungus and white-backed planthopper [25].
Protein degradation-related genes were upregulated in both rice genotypes (Table 2). The protein degradation system is thought to be responsible for selective degradation of proteins folded incorrectly as a result of stress [26]. In response to abiotic stress, wound-responsive proteins WI12 (LOC_Os03g18770) were upregulated. Wound-response pathways were also detected in BPH-feeding rice [27]. Eight Bowman–Birk-type bran trypsin inhibitor precursor genes play a role in resistance to BPH in BPH14-containing rice [5], and we also found that they were upregulated in this research (Additional file 7; Table 3).
Candidate genes of BPH15
We searched for expressed genes within the 87 genes predicted in the 580-kb BPH15 region using RNA sequencing data of resistant rice. We identified five jacalin-related lectins (F14, 34, 35, 36, and 86) and two LRR family proteins (F47 and 89) (Additional file 4-1; Figure 2), most of which showed higher FPKM values than other predicted genes. These genes had similar FPKM values and their expression levels were not significantly different at three time points (Additional file 4-1). These genes were examined using reverse transcription-PCR (RT-PCR), and we found that four jacalin-related lectins (F14, 34, 36, and 86) and a LRR family protein (F89) were constitutively expressed in resistant rice before and after BPH feeding (Figure 2). Other functional genes (one aspartyl protease family protein and six receptor-like protein kinases), which were only detected in a few raw fragments (Additional file 4-1), were not detected in RT-PCR. The majority of transposon protein and retrotransposon protein predicted genes were also detected in a few raw fragments. We then detected all potentially expressed genes (FGENESH and RiceGAAS predicted genes) and found that no other genes were expressed. Note that the four jacalin-related lectin genes showed similar BLASTp results in the Nipponbare genome, and the corresponding gene LOC_Os04g12390 also contained a jacalin domain. However, the gene was annotated as an En/Spm subclass transposon protein in the MSU Rice Genome Annotation Project Release 7.0 (Additional file 4-1). We inferred that these orthologous genes originated from the same ancestral gene (Figure 2). The LRR protein F89 expressed in resistant rice corresponded to LOC_Os04g12460, which also predicted as a LRR gene. Based on these results, the candidate gene BPH15 represents one or a few of these expressed genes, which is being verified based on complementation tests.
Discussion
The wild relatives of crops contain numerous genes of economic importance that are critical for genetic improvement of crops and understanding the mechanism of traits under control by these genes [28]. The use of genes from wild relatives to improve crop performance is well established [29]. Genetic improvement of crop plants by developing introgressed lines originating in wild relatives usually results in a reduction in recombination rates within introgressed segments [30]. By using molecular markers, a lot of introgression of chromosome segments from O. officinalis to O. sativa had been observed [31–33], though the mechanism of introgression is poorly understood. Important traits such as insect and disease resistance have been transferred into cultivated rice [32, 34]. BPH resistance gene BPH15 was introgressed from wild rice O. officinalis [35]. In this study, BPH15 was located in a recombination cold spot, which is similar to the powdery mildew resistance gene Mla in barley [36].
The C genome of O. officinalis is estimated to be 651 Mb, which is larger than the 430 Mb of the A genome of cultivated rice O. sativa. This 580-kb region of BPH15, which contains numerous repeat sequences, in the resistant rice genome corresponds to the 210-kb fragment in the Nipponbare genome. Excluding several functional genes (Figure 2), the sequences in this region of the two genotypes are highly diverse. More TE-related genes appear in the BPH15 region of resistant rice than in the corresponding region of Nipponbare (Additional file 4-1). The important roles of retrotransposons to modify genome size, remodel genome structure, and displace gene functions in the plant genome have been observed in several studies, which indicates that retrotransposons are an important driving force in genome evolution [37]. Previous studies demonstrated that retroelement insertions contributed to the C genome expansion of O. officinalis [38] [Oryza Map Alignment Project (OMAP), http://www.omap.org], which might cause divergence in the BPH15 region between O. officinalis and O. sativa. The diversity in the intergenic, repetitive DNA regions should be responsible for the low chromosome pairing and recombination between O. sativa and O. officinalis [33]. In this experiment, introgression of the highly diverse 580-kb segment is the primary cause of the low recombinant rate, making it difficult to isolate the BPH15 gene using positional cloning. These difficulties were also encountered in other processes of gene cloning. The recombination repression of nematode resistance gene Mi was also thought to be a consequence of the alien origin of the DNA segment [39]. Another reason for suppressed recombination in the BPH15 region is that the locus is located near the centromere. A study found that chromosomal recombination at the centromere core and surrounding regions on six chromosomes was completely suppressed [16], and a substantial reduction in recombination was observed in the regions of the short arm and the pericentromeric region of chromosome 4 [40].
We also observed a recombination hot spot located just outside the BPH15 segment, where the local recombination rate is much higher than the whole genome average value in the BC2F2 family. Recombination hot spots in many species show significant relationships with gene density, GC content, and specific gene functional categories [41]. The 0.7-kb recombination hot-spot between markers T6 and 20M14, located just on the right side of the 580-kb replacement fragment, shows average base composition of the overall chromosome (data not shown). Based on these observations, the recombination hot-spot region showed no correlation with its base properties but position (close to the large replacement fragment). This is important for future studies on the position relationship between recombination hot spots and cold spots to analyze the mechanism of activation and inactivation of recombination.
Identifying candidate genes in an uncharacterized genomic region with no recombination is difficult. In our study, BAC clone sequencing of the BPH15 region and deep RNA sequencing of resistant rice were combined to analyze candidate resistance genes. Eighty-seven genes annotated in this 580-kb region exist between marker g12140-2 and T12, and most of them are TE-related genes. Moreover, only four jacalin-related lectins and a LRR domain protein were expressed in the resistant rice. Many plant lectins have anti-insect potential, some have strong insecticidal properties. Transgenic rice plants expressing lectins showed resistance to BPH and other insects [42]. Hessian fly-responsive gene 1 (Hfr-1) is a novel jacalin-like lectin gene from wheat (Triticum aestivum) plants that responds to infestation by Hessian fly (Mayetiola destructor) larvae [43]. Lectins, are also known to play important roles in defense responses against pathogens. A jacalin-related lectin-like gene (TaJRLL1) in wheat is a component of the plant defense system [44]. The mannose-binding lectin gene CaMBL1 from pepper plays a key role in the regulation of plant cell death and defense responses through the induction of downstream defense-related genes and SA accumulation after the recognition of microbial pathogens [45]. Some lectin receptor-like kinases have been implicated in rice resistance to pathogens and herbivores [46, 47]. As the four jacalin-related lectin genes in the BPH15 region expressed in the resistant rice, we speculated that one or multiple of these lectin genes are BPH15 candidates and function in resistance to BPH. The jacalin-related lectin genes clustered in BPH15 region might evolve from ancient duplications driven by TE elements, as what has been shown in other resistance gene evolution in plant [48]. Another candidate gene is the LRR domain protein in the BPH15 region. The majority of resistance (R) genes that have been cloned belong to the NB-LRR family and the first cloned BPH-resistance gene BPH14 is a NB-LRR member [5, 49, 50]. Therefore, the five candidate genes are currently being verified using complementation tests.
RNA sequencing of the BPH15 introgression line and the susceptible recipient line provided transcriptome data on the mechanism of resistance conferred by BPH15. In our study, several rice genes have been associated with the BPH response for the first time, which provides information on signal transduction pathways and defense responses elicited by the BPH in rice. We analyzed plants at 6 HAI and 48 HAI, before the development of visible symptoms, and compared the expression profiles between early and late stages of infestation. The BPH extracts large volumes of phloem sap to attain adequate sugar, which should influence expression of genes involved in carbon assimilation and mobilization. The transcriptional downregulation of photosynthetic and primary metabolism related genes appears to be a universal adaptive response of plants to phloem-feeding insects and represents a shift in resource allocation from growth to basal defense [19]. In susceptible rice, BPH can significantly reduce photosynthetic rates in host plants, but resistant plants show few symptoms of damage and grow normally after 2 days of feeding [11]. In this experiment, photosynthesis, TCA, CHO metabolism, lipid metabolism, and protein synthesis related genes were downregulated in susceptible rice. However, resistant rice containing BPH15 shows lower expression changes, suggesting that the resistant rice has a stronger tolerance than susceptible rice. The early stage and late stage showed significantly different expression profiles. The majority of DEGs show a less significant change and fewer DEGs are observed at 6 HAI, which may be due to minor damage during the short BPH feeding time. Significant GO terms of photosynthesis, generation of precursor metabolites and energy, ribosome, and intracellular organelle only appeared during the late stages. Previous comparative analyses of expression profiles of proteins in BPH15 rice leaf sheaths in response to infestation by the BPH found that in response to stress caused by pest invasion, plants develop a basal defense, which appeared stronger in the susceptible lines compared with resistant lines [12]. In this report, the amplitude of variation in resistance rice was lower than that in susceptible rice (Figure 4), in which the upregulated genes in susceptible rice may function as a basal defense. The majority of upregulated ethylene signals, receptor kinases, biotic response PR genes, and transcription factor genes in susceptible rice showed larger fold-changes than in resistant rice (Additional file 7; Table 2). BPH14 and Mi-1 both activate an SA-dependent resistance pathway after BPH and nematode feeding [5, 51]. However, the SA pathway may not be present in the BPH15 resistance mechanism according to our sequencing data and SA content measured using GC-MS. The key defense mechanism found in our study was related to jasmonate signaling, ethylene signaling, receptor kinase, MAPK cascades, Ca2+ signaling, PR genes, TFs, and protein posttranslational modifications. Receptor kinase XA21 (LOC_Os11g36180) was upregulated only in resistant rice (Additional file 7). These exclusively upregulated DEGs in resistant rice increase our understanding of the molecular mechanism of resistance in BPH15. In addition, the candidate genes (JRL and LRR genes) may participate in the unique defense mechanism of BPH15. These lectin proteins may have insecticidal properties or deterrent activity to the BPH. The JRL and LRR genes may also perceive BPH feeding as a receptor to transduce defense signals to downstream genes. However, this hypothesis requires further confirmation and candidate gene complementation tests.
Conclusions
The BPH-resistance gene BPH15 was mapped to a recombination cold-spot region. The high level of sequence diversity in the BPH15 region explained the heavy suppression of recombination in this region. We found that BPH-responsive transcript profiles were distinct between resistant and susceptible plants and between the early stage and late stage. Susceptible rice showed more DEGs and larger amplitude of variation than resistant rice after BPH feeding. More genes were regulated at the late stage than those at the early stage. The key defense mechanism in resistant BPH15 and susceptible recipient rice was related to jasmonate signaling, ethylene signaling, receptor kinase, MAPK cascades, Ca2+ signaling, PR genes, transcription factors, and protein posttranslational modifications. Four jacalin-related lectin proteins genes and one LRR protein gene predicted in the BPH15 region were expressed constitutively and considered candidate BPH15 genes. These results increase our understanding of plant–insect interactions and can be used to protect against this destructive agricultural pest.
Methods
Fine mapping of BPH15
YHY15, a resistant rice line containing the BPH15 locus from RI93/TN1 F2 population [7], was backcrossed to the susceptible rice TN1 to develop populations to fine-map the gene (Additional file 1). The BC1F2 plants were used for genotype analysis and BC1F3 lines harvested from each of the BC1F2 plants were assayed for BPH resistance. To fine-map BPH15, 13,000 BC2F2 plants were screened to obtain recombinants between PCR markers RM261 and S16. The selected recombinant plants were self-pollinated and used to select fixed recombinants (BC2F3), which showed homozygous resistant and susceptible genotypes on both sides of the crossover. The seeds of fixed recombinants (BC2F4) were used to phenotype using the seedling bulk test method. Twenty seeds of each line were sown in a 20-cm-long row with 2.5 cm between rows in a plastic box. YHY15 and TN1 were randomly sown among the BC2F4 plants as controls. At the third-leaf stage, the seedlings were infested with BPH nymphs at a level of eight insects per seedling. When all of the TN1 seedlings had died (scored as 9), each seedling was given a score of 0, 1, 3, 5, 7, or 9 according to Huang et al. [35]. The resistance score of each BC2F3 plant was then inferred from the scores of the seedlings in the corresponding BC2F4 plants. At the same time, additional molecular markers (CAPS, dCAPS, InDel, and SSR) were developed in the region to genotype the recombinants plants. Briefly, primer pairs were designed for single-copy regions in Nippobare and used to amplify YHY15 and TN1. Products were cloned into the T-vector for sequencing, and the polymorphisms between YHY15 and TN1 were used to develop molecular markers. BPH15 was located in a smaller region according to the genotype and phenotype of recombinants plants. To identify more recombinant lines, BC4F2 was also screened in the region.
BAC sequencing of the BPH15region
A genomic BAC library for resistant rice B5, the original donor of BPH15, was constructed. The library contained 36,864 clones with an average insert size of 130-kb. The coverage of the library was about 10 genome equivalents, which increases the probability of isolating unique rice genes or sequences in the library. BAC DNA pools were prepared and candidate clones were screened using PCR-based analysis [52]. Initial screening was performed using primers for the distal markers g12140-2 and T6. BAC ends were used to develop single-copy markers, rescreen the library, and extend the contig by chromosome walking. The low-copy sequences were efficiently identified using low-pass DNA sequencing of the BAC when the BAC end sequences were repetitive and could not be used for the walking step. Finally, overlapping BAC clones were assembled to the contig and sequenced by conventional BAC subclone library shotgun sequencing, assembling, and gap-filling. New markers were developed from BAC sequences to genotype the recombinant lines. The assembled sequence was annotated using the FGENESH (http://linux1.softberry.com/berry.phtml) and RiceGAAS (Rice Genome Automated Annotation System, http://ricegaas.dna.affrc.go.jp/) annotation systems.
Preparation of RNA-Seq libraries
The BPH-resistant rice 9311(15), introgression lines containing BPH15 [6] and susceptible recurrent parent 9311 were grown in pots (20 cm in diameter and 20 cm in height) with 30 plants per pot in a greenhouse, which was controlled to have 30 ± 2°C/14 h light (07:00–21:00) and 28 ± 2°C/10 h dark (21:00–07:00) cycles. At the three-leaf stage of rice (12 days after sowing), third-instar nymphs of BPH insects (biotype 1) were introduced to the rice plants at a density of eight insects per seedling. The stems were harvested after BPH infestation for 0 (uninfested control), 6, and 48 h. The six samples were named R0, R6, and R48 for the resistant genotype, and S0, S6, and S48 for the susceptible genotype in which the number represents the HAI. All time points begun at different times and stopped at the same time. The stems (5 cm in length) of 30 rice plants of each treatment were collected as a combined sample, quickly frozen in liquid nitrogen, and kept at –80°C until further analysis.
Total RNAs were prepared using RNAiso Plus according to the manufacturer’s protocol (TaKaRa Code: D9108A). All subsequent procedures, including mRNA purification, cDNA preparation, end repair of cDNA, adaptor ligation, and cDNA amplification were performed according to the manufacturer’s protocols accompanying the mRNA-Seq Sample Preparation Kit (Illumina). Each library had an insert size of 150 bp, and paired end sequences of 100 bp on each end (2*100 bp) were generated via Illumina HiSeq2000.
Analysis of differentially expressed genes
The most recent rice genome and gene information (MSU Release 7.0) were downloaded from the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu). The raw reads were cleaned by removing adaptor sequences, empty reads, low-quality sequences, and short reads. The remaining high-quality reads were aligned to the Oryza sativa genome by TopHat [53] (version: 2.0.6 http://tophat.cbcb.umd.edu). Cufflinks [54] (version: 2.0.2 http://cufflinks.cbcb.umd.edu) was used to calculate the FPKM value of every transcript. The P-value of different expression was calculated using Fisher’s exact test. We used P < 0.05 and the absolute value of log2FC ≥ 0.585 as the threshold to judge the significance of each gene expression difference. Cluster analysis and heat maps were performed with the Genesis software based on the hierarchical and k-means clustering method [55] (version: 1.7.6 http://genome.tugraz.at). GO analysis was performed using a Web-based tool agriGO [56] (http://bioinfo.cau.edu.cn/agriGO). For pathway analysis, we mapped all DEGs using the MapMan package [57] with the Osa_MSU_v7 mapping file and latest pathways downloaded from the official Web site (http://mapman.gabipd.org/web/guest/mapman).
Real-time qPCR
Twenty-three genes of MapMan pathway classifications were selected for validation using real-time qPCR. Primer sets were designed with the Primer Premier 5 software. The qPCR was performed with the Sso Advanced SYBR Green Supermix and CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad) following the manufacturer’s instructions. The results were analyzed using CFX Manager Software 2.1. EF-Tu (LOC_Os03g08020.1) was used as an internal control to standardize the results according to sequencing data. All results had three biological replicates and three technical replicates.
Reverse transcription-PCR
The RNAs of BPH15 introgression line and recipient line (0 and 48 HAI) were reverse transcribed to first strand cDNA. The expression of predicted genes in BPH15 region was confirmed using RT-PCR. All primers were designed according to specific fragment of the genes. The PCR products of RT–PCR were verified by sequencing.
Measurement of SA content using GC-MS
Four-week-old resistant and susceptible rice were infested with BPH nymphs for 0, 3, 6, 24, and 48 h. The leaf sheaths surrounding the stem were separated and frozen immediately in liquid nitrogen. The SA content was determined using a modified vapor-phase extraction method [58, 59]. Briefly, 100 mg of leaf sheath was ground to a fine powder in liquid nitrogen. After the addition of internal standards (2H6-SA, 300 ng), samples were extracted using a mixture of acetone and 50 mM citric acid (v/v = 7/3), and ethyl acetate. The supernatant was then dried using N2 and subsequently methylated with trimethylsilyldiazomethane. After stopping the methylation reaction with acetic acid in hexane, the samples were subjected to a vapor-phase extraction procedure using a volatile collector trap packed with Tenax absorbent and eluted with n-hexane. The eluted samples were then analyzed using GC-MS equipped with an AS3000 auto sampler (Trace GC Ultra/ISQ, Thermo Fisher Scientific). Compounds were separated on an Rtx-5MS (30 m × 0.25 mm × 0.25 μm) column held at 50°C for 1 min after injection, after which the temperature was increased by 10°C min–1 to 180°C (4 min) and by 15°C min–1 to 280°C (5 min), with helium as the carrier gas (constant flow rate 1 mL min–1). Quantification of SA was completed by correlating the peak area (extracted ion) of the compound with that of the corresponding internal standard. Three independent biological replicates were sampled and all samples were measured three times with similar results.
Availability of supporting data
The data sets supporting the results of this article are included within the article and its additional files. The RNA-Seq raw data were submitted to Short Read Archive at NCBI with accession number SRP039374.
Electronic supplementary material
Additional file 1: Pedigree flow chart. (PDF 172 KB)
Additional file 2: Primers used in this study. (XLS 35 KB)
Additional file 3: Recombinants genotype and phenotype. (XLS 56 KB)
Additional file 4: Cufflink analysis of sequenced BAC clones and annotation MSU release 7.0 of Nipponbare between markers g12140-2 and 20M14. (XLS 66 KB)
Additional file 5: Detailed information for all 2,914 DEGs. (XLS 1 MB)
Additional file 6: K-means clustering analysis of DEGs based on log ratio of FPKM data. (PDF 401 KB)
Additional file 7: The DEG assign to MapMan major pathways. (XLS 289 KB)
Additional file 8: Details on the qPCR data. (XLS 40 KB)
Additional file 9: RNA-seq concordance with real-time qPCR results. (PDF 99 KB)
Additional file 10: Measurement of salicylic acid content using gas chromatography–mass spectrometry. (PDF 164 KB)
Additional file 11: DEG enrichment of GO terms. (XLS 46 KB)
Additional file 12: Graphical result of significant GO terms of S-all. (PDF 2 MB)
Additional file 13: Graphical result of significant GO terms of R-all. (PDF 472 KB)
Additional file 14: Graphical result of significant GO terms of S_R-all. (PDF 774 KB)
Acknowledgements
This work was supported by National Natural Science Foundation of China (Grant No. 31230060), the National 863 Project of China (Grant No. 2012AA10A303) and the National 973 Project of China (Grant No. 2013CBA01400).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
WL, GH designed the research, and wrote the paper. WL analyzed the experiment results, prepared figures and tables. WL and BD mapped the gene. WL, XS, YZ and YP collected samples, measured SA, and conducted qPCR analysis. LZ prepared experiment regent. YH developed the BPH15 introgression line. All authors read and approved the final manuscript.
Contributor Information
Wentang Lv, Email: lvwentang@gmail.com.
Ba Du, Email: Duba20050906@163.com.
Xinxin Shangguan, Email: Shangguanxin0409@163.com.
Yan Zhao, Email: yanz@whu.edu.cn.
Yufang Pan, Email: panyufanghao@126.com.
Lili Zhu, Email: zhulili58@sina.com.
Yuqing He, Email: yqhe@mail.hzau.edu.cn.
Guangcun He, Email: gche@whu.edu.cn.
References
- 1.Cheng X, Zhu L, He G. Towards understanding of molecular interactions between rice and the brown planthopper. Mol Plant. 2013;6(3):621–634. doi: 10.1093/mp/sst030. [DOI] [PubMed] [Google Scholar]
- 2.Wang B, Huang Z, Shu L, Ren X, Li X, He G. Mapping of two new brown planthopper resistance genes from wild rice. Chin Sci Bull. 2001;46(13):1092–1095. doi: 10.1007/BF02900685. [DOI] [Google Scholar]
- 3.Hu J, Li X, Wu C, Yang C, Hua H, Gao G, Xiao J, He Y. Pyramiding and evaluation of the brown planthopper resistance genes Bph14 and Bph15 in hybrid rice. Mol Breed. 2012;29(1):61–69. doi: 10.1007/s11032-010-9526-x. [DOI] [Google Scholar]
- 4.Li J, Xia M, Qi H, He G, Wan B, Zha Z. Marker-assisted selection for brown planthopper (Nilaparvata lugens Stål) resistance genes Bph14 and Bph15 in rice. Sci Agric Sin. 2006;10:028. [Google Scholar]
- 5.Du B, Zhang W, Liu B, Hu J, Wei Z, Shi Z, He R, Zhu L, Chen R, Han B, He G. Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc Natl Acad Sci U S A. 2009;106(52):22163–22168. doi: 10.1073/pnas.0912139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu J, Cheng M, Gao G, Zhang Q, Xiao J, He Y. Pyramiding and evaluation of three dominant brown planthopper resistance genes in the elite indica rice 9311 and its hybrids. Pest Manag Sci. 2013;69(7):802–808. doi: 10.1002/ps.3437. [DOI] [PubMed] [Google Scholar]
- 7.Yang H, You A, Yang Z, Zhang F, He R, Zhu L, He G. High-resolution genetic mapping at the Bph15 locus for brown planthopper resistance in rice (Oryza sativa L.) Theor Appl Genet. 2004;110(1):182–191. doi: 10.1007/s00122-004-1844-0. [DOI] [PubMed] [Google Scholar]
- 8.Fujita D, Kohli A, Horgan FG. Rice resistance to planthoppers and leafhoppers. Crit Rev Plant Sci. 2013;32(3):162–191. doi: 10.1080/07352689.2012.735986. [DOI] [Google Scholar]
- 9.Walling LL. The myriad plant responses to herbivores. J Plant Growth Regul. 2000;19(2):195–216. doi: 10.1007/s003440000026. [DOI] [PubMed] [Google Scholar]
- 10.Walling LL. Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol. 2008;146(3):859–866. doi: 10.1104/pp.107.113142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Wang X, Yuan H, Chen R, Zhu L, He R, He G. Responses of two contrasting genotypes of rice to brown planthopper. Mol Plant Microbe Interact. 2007;21(1):122–132. doi: 10.1094/MPMI-21-1-0122. [DOI] [PubMed] [Google Scholar]
- 12.Wei Z, Hu W, Lin Q, Cheng X, Tong M, Zhu L, Chen R, He G. Understanding rice plant resistance to the brown planthopper (Nilaparvata lugens): a proteomic approach. Proteomics. 2009;9(10):2798–2808. doi: 10.1002/pmic.200800840. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, He J, Jiang L, Wu H, Xiao Y, Liu Y, Li G, Du Y, Liu C, Wan J. Nitric oxide production is associated with response to brown planthopper infestation in rice. J Plant Physiol. 2011;168(8):739–745. doi: 10.1016/j.jplph.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 14.Hao P, Liu C, Wang Y, Chen R, Tang M, Du B, Zhu L, He G. Herbivore-induced callose deposition on the sieve plates of rice: an important mechanism for host resistance. Plant Physiol. 2008;146(4):1810–1820. doi: 10.1104/pp.107.111484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J, Mizuno H, Hayashi-Tsugane M, Ito Y, Chiden Y, Fujisawa M, Katagiri S, Saji S, Yoshiki S, Karasawa W, Yoshihara R, Hayashi A, Kobayashi H, Ito K, Hamada M, Okamoto M, Ikeno M, Ichikawa Y, Katayose Y, Yano M, Matsumoto T, Sasaki T. Physical maps and recombination frequency of six rice chromosomes. Plant J. 2003;36(5):720–730. doi: 10.1046/j.1365-313X.2003.01903.x. [DOI] [PubMed] [Google Scholar]
- 17.Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30(1):42–46. doi: 10.1093/nar/30.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zarate SI, Kempema LA, Walling LL. Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol. 2007;143(2):866–875. doi: 10.1104/pp.106.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson GA, Goggin FL. Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J Exp Bot. 2006;57(4):755–766. doi: 10.1093/jxb/erj135. [DOI] [PubMed] [Google Scholar]
- 20.Koo Y, Kim M, Kim E, Song J, Jung C, Moon J-K, Kim J-H, Seo H, Song S, Kim J-K, Lee J, Cheong J-J, Choi Y. Overexpression of salicylic acid carboxyl methyltransferase reduces salicylic acid-mediated pathogen resistance in Arabidopsis thaliana. Plant Mol Biol. 2007;64(1–2):1–15. doi: 10.1007/s11103-006-9123-x. [DOI] [PubMed] [Google Scholar]
- 21.Wu J, Baldwin IT. New insights into plant responses to the attack from insect herbivores. Annu Rev Genet. 2010;44(1):1–24. doi: 10.1146/annurev-genet-102209-163500. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Guo H, Li H, Zhang H, Miao X. Identification of transcription factors potential related to brown planthopper resistance in rice via microarray expression profiling. BMC Genomics. 2012;13(1):687. doi: 10.1186/1471-2164-13-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 2000;12(3):393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryu H-S, Han M, Lee S-K, Cho J-I, Ryoo N, Heu S, Lee Y-H, Bhoo S, Wang G-L, Hahn T-R, Jeon J-S. A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep. 2006;25(8):836–847. doi: 10.1007/s00299-006-0138-1. [DOI] [PubMed] [Google Scholar]
- 25.Wang H, Hao J, Chen X, Hao Z, Wang X, Lou Y, Peng Y, Guo Z. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol. 2007;65(6):799–815. doi: 10.1007/s11103-007-9244-x. [DOI] [PubMed] [Google Scholar]
- 26.Garbarino JE, Oosumi T, Belknap WR. Isolation of a polyubiquitin promoter and its expression in transgenic potato plants. Plant Physiol. 1995;109(4):1371–1378. doi: 10.1104/pp.109.4.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang F, Zhu L, He G. Differential gene expression in response to brown planthopper feeding in rice. J Plant Physiol. 2004;161(1):53–62. doi: 10.1078/0176-1617-01179. [DOI] [PubMed] [Google Scholar]
- 28.Tanksley SD, McCouch SR. Seed banks and molecular maps: unlocking genetic potential from the wild. Science. 1997;277(5329):1063–1066. doi: 10.1126/science.277.5329.1063. [DOI] [PubMed] [Google Scholar]
- 29.Hajjar R, Hodgkin T. The use of wild relatives in crop improvement: a survey of developments over the last 20 years. Euphytica. 2007;156(1–2):1–13. doi: 10.1007/s10681-007-9363-0. [DOI] [Google Scholar]
- 30.Canady MA, Ji Y, Chetelat RT. Homeologous recombination in Solanum lycopersicoides introgression lines of cultivated tomato. Genetics. 2006;174(4):1775–1788. doi: 10.1534/genetics.106.065144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jena KK, Kochert G, Khush GS. RFLP analysis of rice (Oryza sativa L.) introgression lines. Theor Appl Genet. 1992;84(5–6):608–616. doi: 10.1007/BF00224159. [DOI] [PubMed] [Google Scholar]
- 32.Brar DS, Khush GS. Alien introgression in rice. Plant Mol Biol. 1997;35(1–2):35–47. doi: 10.1023/A:1005825519998. [DOI] [PubMed] [Google Scholar]
- 33.Jin H, Tan G, Brar DS, Tang M, Li G, Zhu L, He G. Molecular and cytogenetic characterization of an Oryza officinalis–O. sativa chromosome 4 addition line and its progenies. Plant Mol Biol. 2006;62(4–5):769–777. doi: 10.1007/s11103-006-9056-4. [DOI] [PubMed] [Google Scholar]
- 34.Jena KK. The species of the genus Oryza and transfer of useful genes from wild species into cultivated rice, O. sativa. Breed Sci. 2010;60(5):518–523. doi: 10.1270/jsbbs.60.518. [DOI] [Google Scholar]
- 35.Huang Z, He G, Shu L, Li X, Zhang Q. Identification and mapping of two brown planthopper resistance genes in rice. Theor Appl Genet. 2001;102(6–7):929–934. doi: 10.1007/s001220000455. [DOI] [Google Scholar]
- 36.Wei F, Gobelman-Werner K, Morroll SM, Kurth J, Mao L, Wing R, Leister D, Schulze-Lefert P, Wise RP. The Mla (Powdery Mildew) resistance cluster is associated with three NBS-LRR gene families and suppressed recombination within a 240-kb DNA interval on chromosome 5S (1HS) of Barley. Genetics. 1999;153(4):1929–1948. doi: 10.1093/genetics/153.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dooner HK, He L. Maize genome structure variation: interplay between retrotransposon polymorphisms and genic recombination. Plant Cell. 2008;20(2):249–258. doi: 10.1105/tpc.107.057596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng Q, Huang T, Zhao Q, Zhu J, Lin Z, Han B. Analysis of collinear regions of Oryza AA and CC genomes. J Genet Genomics. 2009;36(11):667–677. doi: 10.1016/S1673-8527(08)60159-9. [DOI] [PubMed] [Google Scholar]
- 39.Kaloshian I, Yaghoobi J, Liharska T, Hontelez J, Hanson D, Hogan P, Jesse T, Wijbrandi J, Simons G, Vos P, Zabel P, Williamson VM. Genetic and physical localization of the root-knot nematode resistance locus Mi in tomato. Mol Gen Genet. 1998;257(3):376–385. doi: 10.1007/s004380050660. [DOI] [PubMed] [Google Scholar]
- 40.Zhao Q, Zhang Y, Cheng Z, Chen M, Wang S, Feng Q, Huang Y, Li Y, Tang Y, Zhou B, Chen Z, Yu S, Zhu J, Hu X, Mu J, Ying K, Hao P, Zhang L, Lu Y, Zhang LS, Liu Y, Yu Z, Fan D, Weng Q, Chen L, Lu T, Liu X, Jia P, Sun T, Wu Y, et al. A fine physical map of the rice chromosome 4. Genome Res. 2002;12(5):817–823. doi: 10.1101/gr.48902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paape T, Zhou P, Branca A, Briskine R, Young N, Tiffin P. Fine-scale population recombination rates, hotspots, and correlates of recombination in the Medicago truncatula genome. Genome Biol Evol. 2012;4(5):726–737. doi: 10.1093/gbe/evs046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michiels K, Van Damme EJM, Smagghe G. Plant-insect interactions: what can we learn from plant lectins? Arch Insect Biochem Physiol. 2010;73(4):193–212. doi: 10.1002/arch.20351. [DOI] [PubMed] [Google Scholar]
- 43.Subramanyam S, Smith DF, Clemens JC, Webb MA, Sardesai N, Williams CE. Functional characterization of HFR1, a high-mannose n-glycan-specific wheat lectin induced by hessian fly larvae. Plant Physiol. 2008;147(3):1412–1426. doi: 10.1104/pp.108.116145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang Y, Song M, Wei Z, Tong J, Zhang L, Xiao L, Ma Z, Wang Y. A jacalin-related lectin-like gene in wheat is a component of the plant defence system. J Exp Bot. 2011;62(15):5471–5483. doi: 10.1093/jxb/err226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hwang IS, Hwang BK. The pepper mannose-binding lectin gene CaMBL1 is required to regulate cell death and defense responses to microbial pathogens. Plant Physiol. 2011;155(1):447–463. doi: 10.1104/pp.110.164848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X, Shang J, Chen D, Lei C, Zou Y, Zhai W, Liu G, Xu J, Ling Z, Cao G, Ma B, Wang Y, Zhao X, Li S, Zhu L. A β-lectin receptor kinase gene conferring rice blast resistance. Plant J. 2006;46(5):794–804. doi: 10.1111/j.1365-313X.2006.02739.x. [DOI] [PubMed] [Google Scholar]
- 47.Cheng X, Wu Y, Guo J, Du B, Chen R, Zhu L, He G. A rice lectin receptor-like kinase that is involved in innate immune responses also contributes to seed germination. Plant J. 2013;76(4):687–698. doi: 10.1111/tpj.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michelmore RW, Meyers BC. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 1998;8(11):1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- 49.Rossi M, Goggin FL, Milligan SB, Kaloshian I, Ullman DE, Williamson VM. The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc Natl Acad Sci U S A. 1998;95(17):9750–9754. doi: 10.1073/pnas.95.17.9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pauquet J, Burget E, Hagen L, Chovelon V, Menn A, Valot N, Desloire S, Caboche M, Rousselle P, Pitrat M. Progress in Cucurbit Genetics and Breeding Research. Proceedings of Cucurbitaceae 2004, the 8th EUCARPIA Meeting on Cucurbit Genetics and Breeding, Olomouc, Czech Republic, 12-17 July, 2004: Palacký University in Olomouc. 2004. Map-based cloning of the Vat gene from melon conferring resistance to both aphid colonization and aphid transmission of several viruses; pp. 325–329. [Google Scholar]
- 51.Branch C, Hwang C-F, Navarre DA, Williamson VM. Salicylic acid is part of the Mi-1-mediated defense response to root-knot nematode in tomato. Mol Plant Microbe Interact. 2004;17(4):351–356. doi: 10.1094/MPMI.2004.17.4.351. [DOI] [PubMed] [Google Scholar]
- 52.Xu J, Yang D, Domingo J, Ni J, Huang N. Screening for overlapping bacterial artificial chromosome clones by PCR analysis with an arbitrary primer. Proc Natl Acad Sci U S A. 1998;95(10):5661–5666. doi: 10.1073/pnas.95.10.5661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18(1):207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 56.Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38(suppl 2):W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. Mapman: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37(6):914–939. doi: 10.1111/j.1365-313X.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- 58.Schmelz EA, Engelberth J, Tumlinson JH, Block A, Alborn HT. The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J. 2004;39(5):790–808. doi: 10.1111/j.1365-313X.2004.02168.x. [DOI] [PubMed] [Google Scholar]
- 59.Song Y, Xu J, Liang X, Su Y, Xie L, Zeng R. Simultaneous quantification of jasmonic and salicylic acids in tomato plants by gas chromatography. Acta Agri Univ Jiangxi. 2010;32:1056–1060. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Pedigree flow chart. (PDF 172 KB)
Additional file 2: Primers used in this study. (XLS 35 KB)
Additional file 3: Recombinants genotype and phenotype. (XLS 56 KB)
Additional file 4: Cufflink analysis of sequenced BAC clones and annotation MSU release 7.0 of Nipponbare between markers g12140-2 and 20M14. (XLS 66 KB)
Additional file 5: Detailed information for all 2,914 DEGs. (XLS 1 MB)
Additional file 6: K-means clustering analysis of DEGs based on log ratio of FPKM data. (PDF 401 KB)
Additional file 7: The DEG assign to MapMan major pathways. (XLS 289 KB)
Additional file 8: Details on the qPCR data. (XLS 40 KB)
Additional file 9: RNA-seq concordance with real-time qPCR results. (PDF 99 KB)
Additional file 10: Measurement of salicylic acid content using gas chromatography–mass spectrometry. (PDF 164 KB)
Additional file 11: DEG enrichment of GO terms. (XLS 46 KB)
Additional file 12: Graphical result of significant GO terms of S-all. (PDF 2 MB)
Additional file 13: Graphical result of significant GO terms of R-all. (PDF 472 KB)
Additional file 14: Graphical result of significant GO terms of S_R-all. (PDF 774 KB)


