Abstract
We report the genome sequence of a healthcare-associated MRSA type ST239 clone isolated from a patient with septicemia in Malaysia. This clone typifies the characteristics of ST239 lineage, including resistance to multiple antibiotics and antiseptics.
Keywords: Staphylococcus aureus, MRSA, Malaysia, Genomics
Introduction
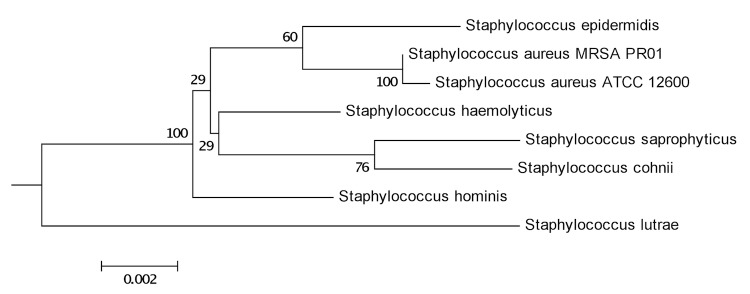
Antibiotic resistance in S. aureus is a major concern, as an increasing number of infections are caused by methicillin-resistant S. aureus (MRSA). Figure 1 shows the phylogenetic position of S. aureus in relation to other staphylococci. In Malaysia, the incidence of MRSA-related infections is a cause of concern in hospitals country-wide. Health-associated MRSA (HA-MRSA) has been dominated by a few lineages in Southeast Asia, particularly ST239. Sequence type 239 is an international healthcare-associated (HA) MRSA lineage prevalent in Asia, South America and Eastern Europe, which includes EMRSA-1, -4, -7, and -11 and the Brazilian, Portuguese, Hungarian, and Viennese clones. Strains of type ST239 are typically resistant to multiple classes of antibiotics and antiseptics such as β-lactam antibiotics.
Figure 1.
Phylogenetic tree highlighting the position of Staphylococcus aureus strain PR01 relative to other type strains within the Staphylococcaceae. The strains and their corresponding GenBank accession numbers for 16S rRNA genes are: S. aureus strain ATCC 12600, L36472; S. saprophyticus strain ATCC 15305, AP008934; S. epidermidis strain ATCC 14990, D83363; S. hominis strain DSM 20328, X66101; S. haemolyticus strain CCM2737, X66100; and S. cohnii strain ATCC 49330, AB009936. The tree uses sequences aligned by the RDP aligner, and uses the Jukes-Cantor corrected distance model to construct a distance matrix based on alignment model positions without the use of alignment inserts, and uses a minimum comparable position of 200. The tree is built with RDP Tree Builder, which uses Weighbor [1] with an alphabet size of 4 and length size of 1000. The building of the tree also involves a bootstrapping process repeated 100 times to generate a majority consensus tree [2]. Staphylococcus lutrae (X84731) was used as an outgroup.
Classification and features
We have chosen a representative of an MRSA strain, termed MRSA PR01 isolated from a patient with septicemia, isolated from a hospital in Kuala Lumpur. Table 1 indicates general information gathered on MRSA PR01. The MRSA PR01 strain has been identified as sequence type 239 (ST239) by multilocus sequence typing (MLST). Initial disc susceptibility tests showed that the strain is resistant to β-lactam antibiotics oxacillin, ampicillin, cefuroxime, ceftriaxone, gentamicin, erythromycin, ciprofloxacin and co-trimoxazole.
Table 1. Classification and general features of Staphylococcus aureus MRSA PR01.
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Current classification | Domain Bacteria Phylum Firmicutes Class Bacilli Order Bacillales Family Staphylococcaceae Genus Staphylococcus Species Staphylococcus aureus Type strain MRSA PR01 |
[3] [4-7] [8,9] [6,10] [9,11] [6,12] [6,12] TAS |
|
| Gram stain | Positive | TAS | |
| Cell shape | Coccus | TAS | |
| Motility | Non-motile | TAS | |
| Sporulation | Non-sporulating | TAS | |
| Temperature range | Mesophile | TAS | |
| Optimum temperature | 30-37°C | TAS | |
| Carbon source | Glucose | TAS | |
| Energy source | Chemoorganotrophic | ||
| Terminal electron receptor | |||
| MIGS-6 | Habitat | Human respiratory tract, skin | TAS |
| MIGS-6.3 | Salinity | ||
| MIGS-22 | Oxygen | Facultative anaerobe | TAS |
| MIGS-15 | Biotic relationship | ||
| MIGS-14 | Pathogenicity | Opportunistic pathogen | TAS |
| MIGS-4 | Geographic location | Malaysia | IDA |
| MIGS-5 | Sample collection time | May 2009 | IDA |
| MIGS-4.1 | Latitude | 4.1936°N | IDA |
| MIGS-4.2 | Longitude | 103.7249°E | IDA |
| MIGS-4.3 | Depth | Not reported | IDA |
| MIGS-4.4 | Altitude | Not reported | IDA |
a) Evidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [13].
Genome sequencing information
Genome project history
This organism was selected for sequencing as a representative of MRSA infection in a local Malaysian hospital. The genome sequences of this organism were deposited in GenBank (WGS database). Sequencing, finishing and annotation were performed at the Integrative Pharmacogenomics Centre (PROMISE), UiTM. Table 2 presents the project information and its association with MIGS version 2.0 compliance [14].
Table 2. Project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Non-contiguous Finished |
| MIGS-28 | Libraries used | One 350bp Illumina GAIIx genomic library |
| MIGS-29 | Sequencing platforms | Illumina GAIIx, Sanger |
| MIGS-31.2 | Fold coverage | >200× |
| MIGS-30 | Assemblers | CLCBio Genomics Workbench |
| MIGS-32 | Gene calling method | Glimmer and GeneMark |
| Genome Database release | DDBJ/EMBL/Genbank/ | |
| Genbank ID | ANPO01000000 | |
| Genbank Date of Release | January 11, 2014 | |
| GOLD ID | ||
| Project relevance | Medical, Tree of life |
Growth conditions and DNA isolation
MRSA PR01 was grown overnight under aerobic conditions in Tryptic Soy Broth at 37°C. DNA extraction was performed using MasterPure™ Gram Positive DNA Purification Kit (Epicentre, Madison, USA) as per manufacturer's instructions. The concentration and purity of resultant DNA was assessed by UV spectrophotometry (Nanodrop, Thermo Scientific). 5 µg of genomic DNA (A260/280 = 1.88) was used for library preparation.
Genome sequencing and assembly
The genome sequence was obtained using 104 Mb of paired-end (300 bp spacing) data from the Illumina GAIIx platform (Illumina, San Diego, CA) with 36-bp reads. Sequence data were assembled using CLCBio Genomics Workbench (CLC bio, Aarhus, Denmark). One hundred and ninety-five contigs (N50: 13,272 bp) were generated, and were overlaid with the reference sequence Mu50 using OSLay. Fourteen supercontigs were generated as a result. Gaps were closed using Sanger sequencing.
Genome annotation
Genome properties
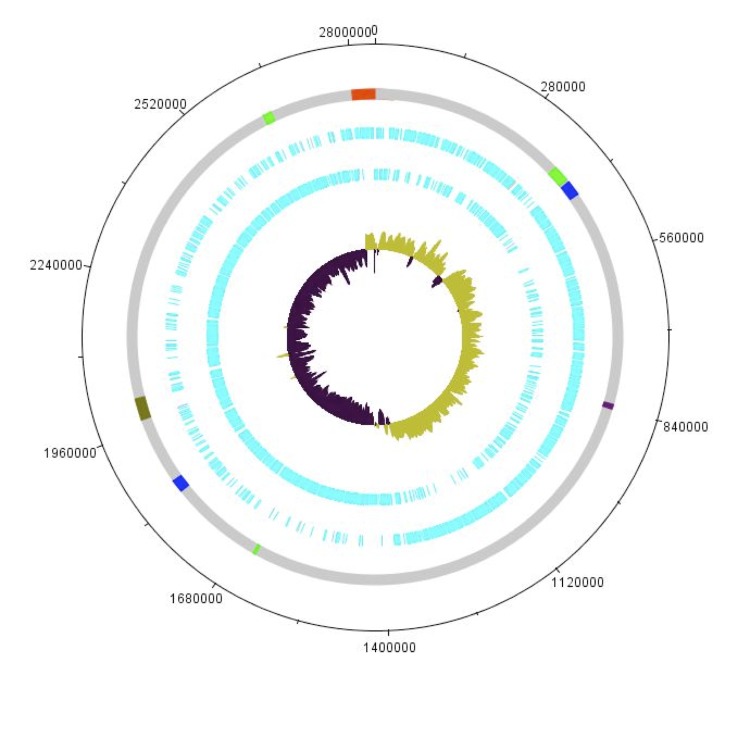
The MRSA PR01 genome consists of a 2,725,110-bp circular chromosome with a GC content of 32.6% (Table 3). The MRSA PR01 genome contains 2668 CDs with 19 rRNA features (). A total of 1722 (64.5%) of protein coding genes were assigned to COGs, and a breakdown of the functional assignment of COG-assigned genes is shown in Table 4. Plasmid sequences were only partially sequenced. Figure 2 depicts genomic regions of interest found in the preliminary analysis of the MRSA PR01 genome.
Table 3. Nucleotide content and gene count levels of the MRSA PR01 genome.
| Attribute | Value | % of totala |
|---|---|---|
| Genome size (bp) | 2,725,110 | |
| DNA G+C content (bp) | 888,386 | 32.6 |
| Total genes | 2687 | |
| RNA genes | 19 | 0.7 |
| Protein-coding genes | 2668 | 99.3 |
| Genes assigned to COGs | 1722 | 64.5 |
a) The total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome.
Table 4. Number of genes associated with the 25 general COG functional categories.
| Code | Value | %agea | Description |
|---|---|---|---|
| J | 140 | 5.247 | Translation |
| A | - | - | RNA processing and modification |
| K | 127 | 4.760 | Transcription |
| L | 126 | 4.723 | Replication, recombination and repair |
| B | - | - | Chromatin structure and dynamics |
| D | 23 | 0.862 | Cell cycle control, mitosis and meiosis |
| Y | - | - | Nuclear structure |
| V | - | - | Defense mechanisms |
| T | 47 | 1.762 | Signal transduction mechanisms |
| M | 91 | 3.411 | Cell wall/membrane biogenesis |
| N | 4 | 0.150 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 0 | 0 | Extracellular structures |
| U | 0 | 0 | Intracellular trafficking and secretion |
| O | 72 | 2.699 | Posttranslational modification, protein turnover, chaperones |
| C | 106 | 3.973 | Energy production and conversion |
| G | 129 | 4.835 | Carbohydrate transport and metabolism |
| E | 186 | 6.972 | Amino acid transport and metabolism |
| F | 68 | 2.549 | Nucleotide transport and metabolism |
| H | 83 | 3.111 | Coenzyme transport and metabolism |
| I | 62 | 2.324 | Lipid transport and metabolism |
| P | 123 | 4.610 | Inorganic ion transport and metabolism |
| Q | 23 | 0.862 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 193 | 7.234 | General function prediction only |
| S | 119 | 4.460 | Function unknown |
| - | 946 | 35.457 | Not in COGs |
a) The total is based on the total number of protein coding genes in the annotated genome.
Figure 2.
Visual representation of the MRSA PR01 genome. From outer to inner tracks: Scale (in bases); annotated CDSs colored according to predicted function (refer to legend); forward strand CDS; reverse strand CDS; GC skew.
Initial analysis of the genome revealed several key features. This genome has a typical SCCmec type III cassette, containing cadmium resistance genes. SCCmec type III is a composite element that is comprised of SCCmec and SCCmercury. In the MRSA PR01 genome, like others, this region harbors ccrC, pI258 and Tn554 as well as the genes involved in cadmium resistance. The MRSA PR01 genome contains two pathogenicity islands, and several resistance features were identified such as the qacA gene, which confers resistance to antiseptics such as cationic biocides, quaternary ammonium salts, and diamidines via an export-mediated mechanism, and the norA gene which confers resistance to hydrophilic quinolones such as norfloxacin and ciprofloxacin. There were 9 regions defined as prophage regions by PHAST [17] with one complete prophage region.
Conclusion
This study is the first to report on the whole genome sequence of a Malaysian MRSA isolate. Preliminary analysis of the genome has highlighted the genetic determinants that are responsible for the organism to adapt easily to selective pressures. Further research is being conducted to provide insight on the adaptive power of this healthcare-associated strain to attain high resistance to antibiotics.
Nucleotide sequence accession numbers. This Whole Genome Shotgun project has been deposited at DDBJ/EMBL/GenBank under the accession ANPO00000000. The version described in this paper is the first version, ANPO01000000.
Acknowledgements
We would like to thank BioEasy Sdn. Bhd. and Illumina for providing technical advice. This project was supported by a grant from the Ministry of Higher Education Malaysia (Grant no. 600-RMI/ST/FRGS 5/3/Fst (58/2010))
Abbreviations
- MRSA
Methicillin-resistant Staphylococcus aureus
References
- 1.Bruno WJ, Socci ND, Halpern AL. Weighted neighbor joining: a likelihood-based approach to distance-based phylogeny reconstruction. Mol Biol Evol 2000; 17:189-197 10.1093/oxfordjournals.molbev.a026231 [DOI] [PubMed] [Google Scholar]
- 2.Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM. The ribosomal database project (RDP-II): introducing my RDP space and quality controlled public data. Nucleic Acids Res 2007; 35:D169-D172 10.1093/nar/gkl889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 119-169. [Google Scholar]
- 5.Murray RGE. The Higher Taxa, or, a Place for Everything...? In: Holt JG (ed), Bergey's Manual of Systematic Bacteriology, First Edition, Volume 1, The Williams and Wilkins Co., Baltimore, 1984, p. 31-34. [Google Scholar]
- 6.Skerman VBD, McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [PubMed] [Google Scholar]
- 7.Gibbons NE, Murray RGE. Proposals Concerning the Higher Taxa of Bacteria. Int J Syst Bacteriol 1978; 28:1-6 10.1099/00207713-28-1-1 [DOI] [Google Scholar]
- 8.Ludwig W, Schleifer KH, Whitman WB. Class I. Bacilli class nov. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 3, Springer-Verlag, New York, 2009, p. 19-20. [Google Scholar]
- 9.List Editor List of new names and new combinations previously effectively, but not validly, published. List no. 132. Int J Syst Evol Microbiol 2010; 60:469-472 10.1099/ijs.0.022855-0 [DOI] [PubMed] [Google Scholar]
- 10.Prévot AR. In: Hauderoy P, Ehringer G, Guillot G, Magrou. J., Prévot AR, Rosset D, Urbain A (eds), Dictionnaire des Bactéries Pathogènes, Second Edition, Masson et Cie, Paris, 1953, p. 1-692. [Google Scholar]
- 11.Schleifer KH, Bell JA. Family VIII. Staphylococcaceae fam. nov. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 3, Springer-Verlag, New York, 2009, p. 392. [Google Scholar]
- 12.Judicial Commission Opinion 17. Conservation of the Generic name Staphylococcus Rosenbach, Designation of Staphylococcus aureus Rosenbach as the Nomenclatural Type of the Genus Staphylococcus aureus Rosenbach, and Designation of the Neotype culture of Staphylococcus aureus Rosenbach. Int Bull Bacteriol Nomencl Taxon 1958; 8:153-154 [Google Scholar]
- 13.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Domselaar GH, Stothard P, Shrivastava S, Cruz JA, Guo A, Dong X, Lu P, Szafron D, Greiner R, Wishart DS. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res 2005;33(Web Server issue):W455-9. [DOI] [PMC free article] [PubMed]
- 16.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics 2008; 9:75 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: A Fast Phage Search Tool. Nucleic Acids Res 2011; 39(suppl 2):W347-52 10.1093/nar/gkr485 [DOI] [PMC free article] [PubMed] [Google Scholar]