Abstract
Anoxybacillus flavithermus subsp. yunnanensis is the only strictly thermophilic bacterium that is able to tolerate a broad range of toxic solvents at its optimal temperature of 55-60°C. The type strain E13T was isolated from water-sediment slurries collected from a hot spring. This study presents the draft genome sequence of A. flavithermus subsp. yunnanensis E13T and its annotation. The 2,838,393bp long genome (67 contigs) contains 3,035 protein-coding genes and 85 RNA genes, including 10 rRNA genes, and no plasmids. The genome information has been used to compare with the genomes from A. flavithermus subsp. flavithermus strains.
Keywords: Anoxybacillus flavithermus subsp. yunnanensis, genome, solvent tolerance, thermophile
Introduction
Solvent-tolerant bacteria are a relatively new group of extremophilic microorganisms. They are able to overcome the toxic and destructive effects of organic solvents due to their unique adaptive mechanisms. Most of the reported solvent-tolerant bacteria are mesophilic bacteria that have an optimal temperature of between 25-37°C [1]. So far, Anoxybacillus flavithermus subsp. yunnanensis is the only strictly thermophilic bacterial species known to tolerate a broad range of solvents at its optimal temperature of 55-60°C [2,3]. The strains show unusual physiological features in the presence of solvents, such as a higher cell yield [2], an observable incrassation of electron-transparent intracellular material and a distorted cytoplasm [3]. However, mechanisms of solvent tolerance in thermophilic species have not been proposed.
The type strain E13T (=CCTCC AB2010187T =KCTC 13759T) and the additional strain PGDY12 were isolated from water-sediment slurries collected from a hot spring in Yunnan Province of China in our lab, and are most closely related to A. flavithermus subsp. flavithermus, first discovered in a hot spring in New Zealand [4]. At present, a total of 19 species and two subspecies of Anoxybacillus with validly published names have been reported [5]. None of these Anoxybacillus strains is reported to tolerate solvents except A. flavithermus subsp. yunnanensis. To understand the molecular basis of the ability to tolerate solvents under high temperature conditions, we sequenced and annotated a draft genome of the type strain E13T of A. flavithermus subsp. yunnanensis.
Classification and features
A. flavithermus subsp. yunnanensis E13T (Table 1) was isolated in 2008 by static cultivation in rich Luria-Bertani (LB) medium supplemented with 10% ethanol [2]. This strain is a facultatively aerobic, Gram-positive, motile, spore-forming rod that is capable of utilizing a wide range of carbon sources, such as arabinose, cellobiose, galactose, maltose, trehalose and xylose. The strain E13T not only exhibited a remarkable ability to grow in ethanol concentrations reaching 13% at 55°C, but can also tolerate highly toxic solvents including toluene, benzene, xylene, chloroform and cyclohexane. Because A. flavithermus subsp. yunnanensis is the only strictly thermophilic bacterium that is able to tolerate toxic solvents, the effect of temperature on solvent tolerance has not yet been studied. The reports of the effect of temperature on ethanol (a much less toxic solvent) tolerance indicated that ethanol tolerance decreased with increasing temperature [20,21]. The comparison of the growth of strain E13T at different temperatures showed that a temperature increase of 20°C, from 45 to 65°C, resulted in a decrease of the critical inhibitory toluene concentration from 0.56 to 0.31%. A similar sharp decrease occurred in the cases of benzene, xylene, chloroform and cyclohexane. The results suggested that temperature plays a vitally important role in determining solvent tolerance in bacteria, which may explain why such thermophilic bacteria are rare in nature.
Table 1. Classification and general features of A. flavithermus subsp. yunnanensis E13T according to the MIGS recommendations [6].
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Domain Bacteria | TAS [7] | ||
| Phylum Firmicutes | TAS [8-10] | ||
| Class Bacilli | TAS [11,12] | ||
| Current classification | Order Bacillales | TAS [13,14] | |
| Family Bacillaceae | TAS [13,15] | ||
| Genus Anoxybacillus | TAS [16,17] | ||
| Species Anoxybacillus flavithermus | TAS [16] | ||
| Subspecies Anoxybacillus flavithermus subsp. yunnanensis | TAS [2,18] | ||
| Type strain E13T | TAS [2] | ||
| Gram stain | positive | TAS [2] | |
| Cell shape | rod | TAS [2] | |
| Motility | motile | TAS [2] | |
| Sporulation | sporulating | TAS [2] | |
| Temperature range | 30-66°C | TAS [2] | |
| Optimum temperature | 60°C | TAS [2] | |
| Carbon source | carbohydrates | TAS [2] | |
| Energy source | heterotrophic | TAS [2] | |
| MIGS-6 | Habitat | hot spring | TAS [2] |
| MIGS-6.3 | Salinity | optimum 0.3% (w/v) NaCl | TAS [2] |
| MIGS-22 | Oxygen requirement | facultative anaerobe | TAS [2] |
| MIGS-15 | Biotic relationship | free-living | TAS [2] |
| MIGS-14 | Pathogenicity | non-pathogenic | NAS |
| MIGS-4 | Geographic location | Yunnan, China | TAS [2] |
| MIGS-5 | Sample collection time | 2008 | IDA |
| MIGS-4.1 | Latitude | N 4°56.5951’ | IDA |
| MIGS-4.2 | Longitude | W 98°26.2032’ | IDA |
| MIGS-4.3 | Depth | water-sediment slurry (shallow) | IDA |
| MIGS-4.4 | Altitude | 1,457 m above sea level | NAS |
aEvidence codes - IDA: Inferred from Direct Assay (first time in publication); TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [19]. If the evidence code is IDA, then the property was directly observed by one of the authors or an expert mentioned in the acknowledgements.
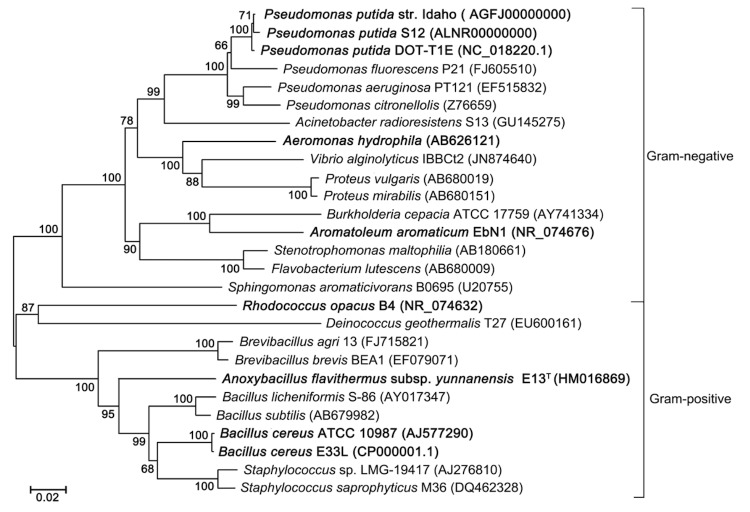
Currently, more than 30 solvent-tolerant mesophilic bacteria have been reported, and 8 genomes are available in GenBank. The phylogenetic position of A. flavithermus subsp. yunnanensis E13T among these typical solvent-tolerant bacteria is shown in Figure 1. This strain is most closely related to Bacillus species. The genomes of B. cereus strain E33L and strain ATCC 10987 might provide valuable guidance in a genetic analysis of the solvent tolerance of A. flavithermus subsp. yunnanensis E13T.
Figure 1.
Phylogenetic tree highlighting the position of A. flavithermus subsp. yunnanensis E13T relative to other typical solvent-tolerant bacteria. The 16S rRNA sequences were aligned using ClustalX2, and phylogenetic inferences obtained using the neighbor-joining method with the MEGA program. Species and GenBank accession numbers are indicated. Bootstrap values based on 1,000 replicates show the robustness of the branching. Scale bar represents 0.02 substitutions per nucleotide position. Strains with genome sequencing projects registered in GenBank are shown in bold.
Genome sequencing information
Genome project history
The organism was selected based on its unique characteristics as a solvent-tolerant thermophile and in order to investigate new mechanisms of solvent tolerance. The genome was sequenced at BGI-Shenzhen (Shenzhen, China) and deposited in Genbank under the accession number AVGH00000000. The version described in this paper is version AVGH01000000. To our knowledge, it was the first genome of A. flavithermus subsp. yunnanensis, the 8th genome of an Anoxybacillus species and the 9th genome of solvent-tolerant bacteria to be sequenced. A summary of the project information associated with MIGS version 2.0 compliance [6] is shown in Table 2.
Table 2. Project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | High-quality Draft |
| MIGS-28 | Libraries used | One 454 shotgun library and two paired-end Illumina Hiseq 2000 libraries |
| MIGS-29 | Sequencing platforms | 454 GS FLX Titanium, Illumina HiSeq 2000 sequencing platform |
| MIGS-31.2 | Fold coverage | 52.5×454 Titanium, 368.5×Illumina |
| MIGS-30 | Assemblers | Newbler version 2.6 |
| MIGS-32 | Gene calling method | Glimmer 3.02 |
| Genbank ID | AVGH00000000 | |
| Genbank Date of Release | August 01, 2014 | |
| GOLD ID | Gi0037576 | |
| MIGS-13 | Project relevance | Strictly thermophilic and organic solvent-tolerant strain |
Growth conditions and DNA isolation
A. flavithermus subsp. yunnanensis strain E13T was grown in LB medium at 60°C for 8 h. The cells were harvested by centrifugation at 12,000 g, and washed twice with distilled water. Genomic DNA from the strain E13T was extracted with a Genomic DNA Mini Preparation Kit (Beyotime, Shanghai, China) according to the method for extracting genomic DNA from Gram-positive bacteria. The quality and concentration of the genomic DNA were measured by spectrophotometric analysis using a biophotometer (Eppendorf BioPhotometer Plus, Eppendorf, Germany).
Genome sequencing and assembly
The genome of A. flavithermus subsp. yunnanensis was sequenced using a combination of 454 GS FLX Titanium (Roche) with a shotgun library (1.8-kb insert size), and Illumina Hiseq2000 sequencing platform with two paired-end libraries (0.5 and 6-kb insert size). The 454 shotgun library was constructed with 500 ng of DNA as described by the manufacturer with the GS Rapid library Prep kit (Roche), and the details of Illumina paired-end library construction and sequencing can be found at the Illumina web site. For the genome, we constructed and sequenced a 454 shotgun library which generated 352,901 reads totaling 148.6 Mb, and 2 Illumina paired-end libraries which generated 1,182 Mb raw data. The final assembly was based on 148.6 Mb of 454 draft data, which provides an average 52.5× coverage of the genome and 1,043 Mb of Illumina draft data, which provides an average 368.5× coverage of the genome. These sequences were assembled using Newbler software with 90% identity and 40 bp as overlap. The resulting 67 contigs were scaffolded via read-pairing relationships with SSPACE [22] using all available libraries of high quality reads. The final assembly identified 67 contigs arranged in 24 scaffolds and generated a genome size of 2,838,393 bp.
Genome annotation
Genes were predicted by merging the results obtained from the RAST (Rapid Annotation using Subsystem Technology) server [23] and the Glimmer modeling software package [24]. The predicted coding sequences (CDSs) were translated and used to search the National Center for Biotechnology Information (NCBI) nonredundant database, KEGG, Clusters of Orthologous Groups (COG), Swiss-Prot and TrEMBL databases. The tool RNAmmer [25] was used to find rRNA genes, whereas tRNA genes were found by using the tool tRNAscanSE [26]. Other non-coding RNAs were identified by searching the genome for Rfam profiles using INFERNAL (v0.81) [27]. Signal peptides and numbers of transmembrane helices were predicted using SignalP [28] and TMHMM [29], respectively.
Genome properties
The genome is 2,838,393 bp long (1 chromosome, no plasmids) with a 41.4% G+C content (Figure 2 and Table 3). Of the 3,120 predicted genes, 3,035 were protein-coding genes, and 85 were RNAs. In addition, ten rRNA genes (two 16S rRNA, one 23S rRNA and seven 5S rRNA) and 75 predicted tRNA genes were identified in the genome. A total of 2,267 genes (72.66%) were assigned a putative function. The remaining genes were annotated as hypothetical proteins. The properties and the statistics of the genome are summarized in Table 3. The distribution of genes into COGs and KEGG functional categories is presented in Table 4.
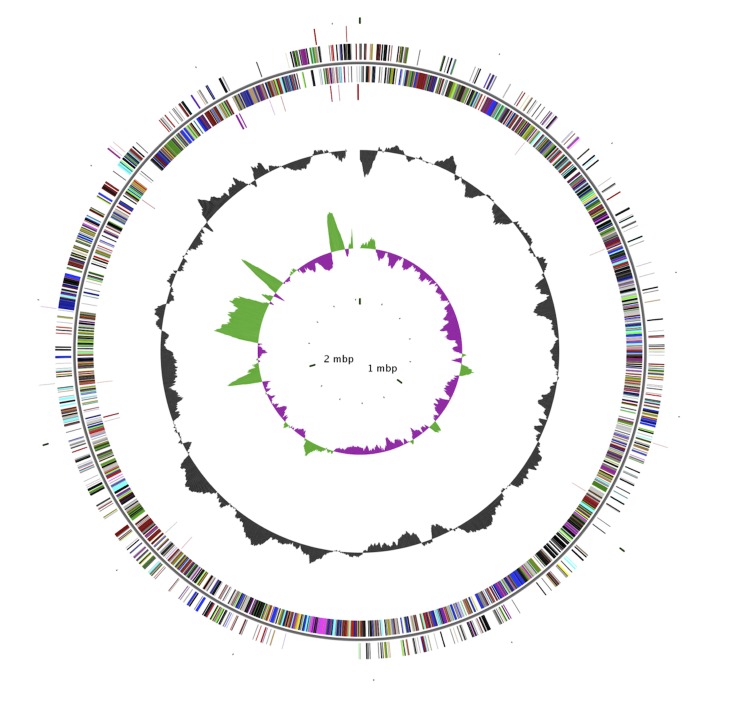
Figure 2.
Graphical circular map of the chromosome. From the outside to the center: RNA genes (tRNA red, rRNAs purple and sRNA black) on the forward strand, genes on the forward strand (colored by COG categories), genes on the reverse strand, RNA genes on the reverse strand, G+C content, and GC skew (purple negative values, olive positive values).
Table 3. Nucleotide content and gene count levels of the genome.
| Attribute | Value | % of totala |
|---|---|---|
| Genome size (bp) | 2,838,393 | 100 |
| DNA G+C content (bp) | 1,176,230 | 41.44 |
| DNA Coding region (bp) | 2,555,544 | 90.03 |
| Total genes | 3,120 | 100 |
| RNA genes | 85 | 2.72 |
| Protein-coding genes | 3,035 | 97.28 |
| Genes with protein function prediction | 2,267 | 72.66 |
| Genes assigns to KEGG pathways | 1,936 | 62.05 |
| Genes assigned to KEGG Orthology | 1,012 | 32.43 |
| Genes assigned to COGs | 1,886 | 60.44 |
| Genes with signal peptides | 99 | 3.17 |
| Genes with transmembrane helices | 716 | 22.94 |
aThe total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome
Table 4. Number of genes associated with the 25 general COG functional categories.
| Code | Value | %agea | Description |
|---|---|---|---|
| J | 148 | 4.88 | Translation |
| A | 0 | 0.00 | RNA processing and modification |
| K | 131 | 4.32 | Transcription |
| L | 153 | 5.04 | Replication, recombination and repair |
| B | 1 | 0.03 | Chromatin structure and dynamics |
| D | 27 | 0.89 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0.00 | Nuclear structure |
| V | 17 | 0.56 | Defense mechanisms |
| T | 93 | 3.06 | Signal transduction mechanisms |
| M | 85 | 2.80 | Cell wall/membrane biogenesis |
| N | 52 | 1.71 | Cell motility |
| Z | 0 | 0.00 | Cytoskeleton |
| W | 0 | 0.00 | Extracellular structures |
| U | 34 | 1.12 | Intracellular trafficking and secretion |
| O | 91 | 2.99 | Posttranslational modification, protein turnover, chaperones |
| C | 137 | 4.51 | Energy production and conversion |
| G | 144 | 4.74 | Carbohydrate transport and metabolism |
| E | 200 | 6.59 | Amino acid transport and metabolism |
| F | 63 | 2.08 | Nucleotide transport and metabolism |
| H | 103 | 3.39 | Coenzyme transport and metabolism |
| I | 79 | 2.60 | Lipid transport and metabolism |
| P | 122 | 4.02 | Inorganic ion transport and metabolism |
| Q | 32 | 1.05 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 231 | 7.61 | General function prediction only |
| S | 169 | 5.57 | Function unknown |
| - | 1,234 | 40.66 | Not in COGs |
a) The total is based on the total number of protein coding genes in the annotated genome.
Comparison with other Anoxybacillus flavithermus genomes
As of this moment, six genome sequences from Anoxybacillus species are available in GenBank database, including four A. flavithermus subsp. flavithermus strains, one A. kamchatkensis strain and one Anoxybacillus sp. strain. Only A. flavithermus subsp. flavithermus strain WK1 and strain TNO-09.006 have complete genome sequences [30,31]. Here we compare the genome sequence of A. flavithermus subsp. yunnanensis E13T with those of the four A. flavithermus subsp. flavithermus strains. The draft genome of A. flavithermus subsp. yunnanensis E13T is similar in size to that of A. flavithermus subsp. flavithermus strain WK1 (2.83 vs 2.84 Mb, respectively), but larger than that of strain TNO-09.006, strain AK1 and strain NBRC 109594 (2.65, 2.63 and 2.77 Mb, respectively). The G+C content of A. flavithermus subsp. yunnanensis E13T is similar to those of A. flavithermus subsp. flavithermus strain WK1, strain TNO-09.006 and strain NBRC 109594 (41.4, 41.7, 41.8 and 41.7%, respectively), but slightly less than that of strain AK1 (42.7%). The gene content of A. flavithermus subsp. yunnanensis E13T is greater than those of A. flavithermus subsp. flavithermus strain WK1, strain TNO-09.006, strain AK1 and strain NBRC 109594 (3,120, 2,954, 2,819, 2,799 and 2,963 genes, respectively). In addition, A. flavithermus subsp. yunnanensis E13T shared a mean genome sequence similarity of 90% (range 80-99%), 90% (79-100%), 86% (73-99%) and 91% (71-100%) with A. flavithermus subsp. flavithermus strain WK1, strain TNO-09.006, strain AK1 and strain NBRC 109594, respectively.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31270081, 31140061, 30800010).
References
- 1.Kongpol A, Kato J, Vangnai AS. Isolation and characterization of Deinococcus geothermalis T27, a slightly thermophilic and organic solvent-tolerant bacterium able to survive in the presence of high concentrations of ethyl acetate. FEMS Microbiol Lett 2008; 286:227-235 10.1111/j.1574-6968.2008.01273.x [DOI] [PubMed] [Google Scholar]
- 2.Dai J, Liu Y, Lei Y, Gao Y, Han F, Xiao Y, Peng H. A new subspecies of Anoxybacillus flavithermus ssp. yunnanensis ssp. nov. with very high ethanol tolerance. FEMS Microbiol Lett 2011; 320:72-78 10.1111/j.1574-6968.2011.02294.x [DOI] [PubMed] [Google Scholar]
- 3.Gao Y, Dai J, Peng H, Liu Y, Xu T. Isolation and characterization of a novel organic solvent-tolerant Anoxybacillus sp. PGDY12, a thermophilic Gram-positive bacterium. J Appl Microbiol 2011; 110:472-478 10.1111/j.1365-2672.2010.04903.x [DOI] [PubMed] [Google Scholar]
- 4.Heinen W, Lauwers AM, Mulders JWM. Bacillus flavothermus, a newly isolated facultative thermophile. Antonie van Leeuwenhoek 1982; 48:265-272 10.1007/BF00400386 [DOI] [PubMed] [Google Scholar]
- 5.Goh KM, Kahar UM, Chai YY, Chong CS, Chai KP, Ranjani V, Md R, Chan KG. Recent discoveries and applications of Anoxybacillus. Appl Microbiol Biotechnol 2013; 97:1475-1488 10.1007/s00253-012-4663-2 [DOI] [PubMed] [Google Scholar]
- 6.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archae, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 119–169. [Google Scholar]
- 9.Murray RGE. The Higher Taxa, or, a Place for Everything...? In: Holt JG (ed), Bergey's Manual of Systematic Bacteriology, First Edition, Volume 1, The Williams and Wilkins Co., Baltimore, 1984, p. 31–34. [Google Scholar]
- 10.Gibbons NE, Murray RGE. Proposals Concerning the Higher Taxa of Bacteria. Int J Syst Bacteriol 1978; 28:1-6 10.1099/00207713-28-1-1 [DOI] [Google Scholar]
- 11.List of new names and new combinations previously effectively, but not validly, published. List no. 132. Int J Syst Evol Microbiol 2010; 60:469-472 10.1099/ijs.0.022855-0 [DOI] [PubMed] [Google Scholar]
- 12.Ludwig W, Schleifer KH, Whitman WB. Class I. Bacilli class nov. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 3, Springer-Verlag, New York, 2009, p. 19–20. [Google Scholar]
- 13.Skerman VBD, Sneath PHA. Approved list of bacterial names. Int J Syst Bact 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [Google Scholar]
- 14.Prévot AR. Dictionnaire des bactéries pathogens. In: Hauduroy P, Ehringer G, Guillot G, Magrou J, Prevot AR, Rosset, Urbain A (eds). Paris, Masson, 1953, p.1–692. [Google Scholar]
- 15.Fischer A. Untersuchungen über bakterien. Jahrbücher für Wissenschaftliche Botanik 1895; 27:1-163 [Google Scholar]
- 16.Pikuta E, Lysenko A, Chuvilskaya N, Mendrock U, Hippe H, Suzina N, Nikitin D, Osipov G, Laurinavichius K. Anoxybacillus pushchinensis gen. nov., sp. nov., a novel anaerobic, alkaliphilic, moderately thermophilic bacterium from manure, and description of Anoxybacillus flavithermus comb. nov. Int J Syst Evol Microbiol 2000; 50:2109-2117 10.1099/00207713-50-6-2109 [DOI] [PubMed] [Google Scholar]
- 17.Pikuta E, Cleland D, Tang J. Aerobic growth of Anoxybacillus pushchinoensis K1: emended descriptions of A. pushchinoensis and the genus Anoxybacillus. Int J Syst Evol Microbiol 2003; 53:1561-1562 10.1099/ijs.0.02643-0 [DOI] [PubMed] [Google Scholar]
- 18.List of new names and new combinations previously effectively, but not validly, published – Validation List No. 141. Int J Syst Evol Microbiol 2011; 61:2025-2026 10.1099/ijs.0.036640-0 [DOI] [PubMed] [Google Scholar]
- 19.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baskaran S, Ahn HJ, Lynd LR. Investigation of ethanol tolerance of Clostridium thermosaccharolyticum in continuous culture. Biotechnol Prog 1995; 11:276-281 10.1021/bp00033a006 [DOI] [Google Scholar]
- 21.Georgieva TI, Skiadas IV, Ahring BK. Effect of temperature on ethanol tolerance of a thermophilic anaerobic ethanol producer Thermoanaerobacter A10: modeling and simulation. Biotechnol Bioeng 2007; 98:1161-1170 10.1002/bit.21536 [DOI] [PubMed] [Google Scholar]
- 22.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011; 27:578-579 10.1093/bioinformatics/btq683 [DOI] [PubMed] [Google Scholar]
- 23.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, et al. The RAST server: Rapid Annotations using Subsystems Technology. BMC Genomics 2008; 9:75 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007; 23:673-679 10.1093/bioinformatics/btm009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagesen K, Hallin P, Rødland EA, Stærfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100-3108 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955-964 10.1093/nar/25.5.0955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res 2005; 33:D121-D124 10.1093/nar/gki081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bendtsen JD, Nielsen H, Von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 2004; 340:783-795 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]
- 29.Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001; 305:567-580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 30.Saw JH, Mountain BW, Feng L, Omelchenko MV, Hou S, Saito JA, Stott MB, Li D, Zhao G, Wu J, et al. Encapsulated in silica: genome, proteome and physiology of the thermophilic bacterium Anoxybacillus flavithermus WK1. Genome Biol 2008; 9:R161 10.1186/gb-2008-9-11-r161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caspers MPM, Boekhorst J, Abee T, Siezen RJ, Kort R. Complete genome sequence of Anoxybacillus flavithermus TNO-09.006, a thermophilic sporeformer associated with a dairy-processing environment. Genome Announc 2013; 1(1). [DOI] [PMC free article] [PubMed]