Abstract
Clostridium jeddahense strain JCDT (= CSUR P693 = DSM 27834) is the type strain of C. jeddahense sp. nov. This strain, whose genome is described here, was isolated from the fecal flora of an obese 24 year-old Saudian male (BMI=52 kg/m2). Clostridium jeddahense strain JCDT is an obligate Gram-positive bacillus. Here we describe the features of this organism, together with the complete genome sequence and annotation. The 3,613,503 bp long genome (1 chromosome, no plasmid) exhibits a G+C content of 51.95% and contains 3,462 protein-coding and 53 RNA genes, including 4 rRNA genes.
Keywords: Clostridium jeddahense, genome, culturomics, taxonogenomics
Introduction
Clostridium jeddahense strain JCDT (=CSUR P693 = DSM 27834), is the type strain of Clostridium jeddahense sp. nov. This bacterium is a Gram-positive, anaerobic, spore-forming indole, positive bacillus that was isolated from the stool of an obese 24 year-old Saudian individual, as a part of a culturomics study as previously reported.
The usual parameters used to delineate a bacterial species include 16S rDNA sequence identity and phylogeny [1,2], genomic G + C content diversity, and DNA–DNA hybridization (DDH) [3,4]. Nevertheless, some limitations appeared notably because the cutoff values vary dramatically between species and genera [5]. The introduction of high-throughput sequencing techniques made genomic data for many bacterial species available [6]. We recently proposed a new method (taxono-genomics), which includes genomic data in a polyphasic approach to describe new bacterial species [6]. This strategy combines phenotypic characteristics, including MALDI-TOF MS spectrum, and genomic analysis [7-37].
Here, we present a summary classification and a set of features for C. jeddahense sp. nov. strain JCDT (=CSUR P693 = DSM 27834), together with the description of the complete genome sequencing and annotation. These characteristics support the circumscription of the species C. jeddahense.
The genus Clostridium was created in 1880 [38] and consists of obligate anaerobic rod-shaped bacilli able to produce endospores [38]. More than 200 species have been described to date (http://www.bacterio.cict.fr/c/clostridium.html). Members of the genus Clostridium are mostly environmental bacteria or associated with the commensal digestive flora of mammals. However, several are major human pathogens, including C. botulinum, C. difficile and C. tetani [38].
Classification and features
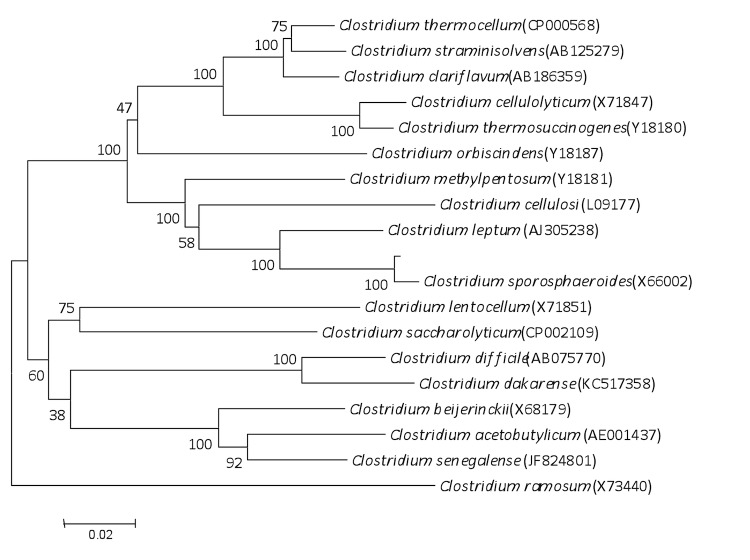
A stool sample was collected from an obese 24-year-old male Saudian volunteer patient living in Jeddah. The patient gave an informed and signed consent, and the agreement of the Ethical Committee of the King Abdulaziz University, King Fahd medical Research Centre, Saudi Arabia, and the local ethics committee of the IFR48 (Marseille, France) were obtained under agreement number 014-CEGMR-2-ETH-P and 09-022 respectively. The fecal specimen was preserved at -80°C after collection and sent to Marseille. Strain JCDT (Table 1) was isolated in July 2013 by anaerobic cultivation on 5% sheep blood-enriched Columbia agar (BioMerieux, Marcy l’Etoile, France) after a 5-day preincubation on blood culture bottle with rumen fluid. This strain exhibited a 97.3% nucleotide sequence similarity with Clostridium sporosphaeroides strain DSM 1294 (Figure 1). This value was lower than the 98.7% 16S rRNA gene sequence similarity threshold recommended by Stackebrandt and Ebers to delineate a new species without carrying out DNA-DNA hybridization [2] and was in the 78. 4 to 98.9% range of 16S rRNA identity values observed among 41 Clostridium species with validly published names [52].
Table 1. Classification and general features of Clostridium jeddahense strain JCDT according to the MIGS recommendations [39].
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [40] | |
| Phylum Firmicutes | TAS [41-43] | ||
| Class Clostridia | TAS [44,45] | ||
| Order Clostridiales | TAS [46,47] | ||
| Family Clostridiaceae | TAS [46,48] | ||
| Genus Clostridium | IDA [46,49,50] | ||
| Species Clostridium jeddahense | IDA | ||
| Type strain JCDT | IDA | ||
| Gram stain | Positive | IDA | |
| Cell shape | Rod | IDA | |
| Motility | Motile | IDA | |
| Sporulation | Sporulating | IDA | |
| Temperature range | Mesophile | IDA | |
| Optimum temperature | 37°C | IDA | |
| MIGS-6.3 | Salinity | Unknown | IDA |
| MIGS-22 | Oxygen requirement | Anaerobic | IDA |
| Carbon source | Unknown | IDA | |
| Energy source | Unknown | IDA | |
| MIGS-6 | Habitat | Human gut | IDA |
| MIGS-15 | Biotic relationship | Free living | IDA |
| MIGS-14 | Pathogenicity Biosafety level Isolation |
Unknown 2 Human feces |
|
| MIGS-4 | Geographic location | Jeddah, Saudi Arabia | IDA |
| MIGS-5 | Sample collection time | July 2013 | IDA |
| MIGS-4.1 | Latitude | 21.422487 | IDA |
| MIGS-4.1 | Longitude | 39.856184 | IDA |
| MIGS-4.3 | Depth | Surface | IDA |
| MIGS-4.4 | Altitude | 0 m above sea level | IDA |
Evidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [51]. If the evidence is IDA, then the property was directly observed for a live isolate by one of the authors or an expert mentioned in the acknowledgements.
Figure 1.
A consensus phylogenetic tree highlighting the position of Clostridium jeddahense strain JCDT relative to other type strains within the Clostridum genus. GenBank accession numbers are indicated in parentheses. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using the maximum-likelihood method in the MEGA software package. Numbers at the nodes are the percentages of bootstrap values from 500 replicates that support the node. Clostridium ramosum was used as outgroup. The scale bar represents a 2% nucleotide sequence divergence.
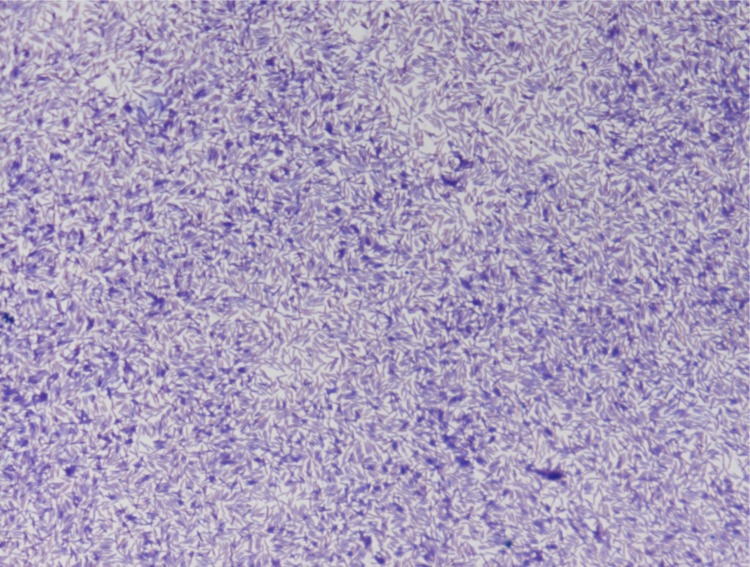
Four growth temperatures (25, 30, 37, 45°C) were tested; growth occurred between 25 and 37°C, but optimal growth was observed at 37°C, 24 hours after inoculation. No growth occurred at 45°C. Colonies were translucent and approximately 0.2 to 0.3 mm in diameter on 5% sheep blood-enriched Columbia agar (BioMerieux). Growth of the strain was tested on the same agar under anaerobic and microaerophilic conditions using GENbag anaer and GENbag microaer systems, respectively (BioMerieux), and in aerobic conditions, with or without 5% CO2. Growth was observed only anaerobically. No growth occurred in aerobic or microaerophilic conditions. Gram staining showed Gram-positive rods able to form spores (Figure 2). A motility test was positive. Cells grown on agar exhibit a mean diameter of 1 µm and a mean length of 1.22 µm in electron microscopy (Figure 3).
Figure 2.
Gram stain of Clostridium jeddahense strain JCDT
Figure 3.
Transmission electron micrograph of C. jeddahense strain JCDT, taken using a Morgani 268D (Philips) at an operating voltage of 60kV.The scale bar represents 500 nm.
Strain JCDT exhibited neither catalase nor oxidase activity (Table 2). Using an API Rapid ID 32A strip (BioMerieux), positive reactions were obtained for indole production, alkaline phosphatase, arginine arylamidase, proline arylamidase, alanine arylamidase, glycine arylamidase, histidine arylamidase, glutamyl glutamic acid arylamidase and serine arylamidase. Negative reactions were obtained for arginine dihydrolase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, α-arabinosidase, N-acetyl-β-glucosaminidase, glutamic acid decarboxylase, α-fucosidase, nitrate reduction, leucyl glycine arylamidase, fermentation of mannose and raffinose, urease, β-galactosidase-6-phosphatase, β-glucuronidase, phenylalanine arylamidase, leucine arylamidase, pyroglutamic acid arylamidase and tyrosine arylamidase. Using an API 50CH strip (Biomerieux), strain JCDT was asaccharolytic.
Table 2. Differential characteristics of Clostridium jeddahense JCDT, C. senegalense JC122 [11], C. dakarense FF1 [34], C. beijerinckii NCIMB 8052, C. difficile B1, C. cellulolyticum H10, C. leptum DSM 753 and C. sporosphaeroides DSM 1294 [53]†.
| Properties | C. jeddahense |
C. sporosphaeroides |
C. cellulolyticum |
C. leptum |
C. senegalense |
C. dakarense |
C. beijerinckii |
C. difficile |
|---|---|---|---|---|---|---|---|---|
| Cell diameter (µm) | 1.0 | 0.5-0.6 | 1.5 | 0.6-0.8 | 1.1 | 1.2 | 1.7 | 3.0 |
| Oxygen requirement | Strictly anaerobic |
Strictly anaerobic |
Strictly anaerobic |
Strictly anaerobic | Strictly anaerobic |
Strictly anaerobic |
Strictly anaerobic |
Strictly anaerobic |
| Gram stain | Positive | Positive | Positive | Positive | Positive | Positive | Variable | Variable |
| Motility | Motile | Non Motile | Motile | Non Motile | Motile | Motile | Motile | Motile |
| Endospore formation | + | + | + | + | + | + | + | + |
| Indole | + | - | - | Na | - | + | Na | Na |
| Production of | ||||||||
| Alkaline phosphatase | + | Na | Na | Na | - | + | Na | Na |
| Catalase | - | Na | - | Na | - | - | - | Na |
| Oxidase | - | Na | Na | Na | - | - | Na | Na |
| Nitrate reductase | - | - | - | Na | - | - | - | - |
| Urease | - | - | - | Na | - | - | - | Na |
| β-galactosidase | - | Na | Na | Na | - | - | Na | Na |
| N-acetyl-glucosamine | - | Na | Na | Na | + | Na | Na | |
| Acid from | ||||||||
| L-Arabinose | - | - | Na | Na | Na | - | + | - |
| Ribose | - | - | Na | W | Na | - | - | - |
| Mannose | - | - | Na | - | Na | - | + | + |
| Mannitol | - | - | Na | - | Na | - | + | + |
| Sucrose | - | -Na | Na | W | Na | - | + | + |
| D-glucose | - | Na | Na | Na | Na | + | + | Na |
| D-fructose | - | Na | Na | Na | Na | - | + | + |
| D-maltose | - | Na | Na | Na | Na | + | + | - |
| D-lactose | - | Na | Na | Na | Na | - | + | - |
| G+C content (%) | 52 | 41 | 51 | 26.8 | 27.98 | 28 | 28 | |
| Habitat | Human gut | Environment | Compost | Human gut | Human gut | Human gut | Human gut | Human gut |
†(Na = data not available; w = weak, v = variable reaction)
C. jeddahense is susceptible to amoxicillin, amoxicillin-clavulanate, imipenem, metronidazole, doxycycline, rifampicin, vancomycin but resistant to ceftriaxone, ciprofloxacin and trimethoprim-sulfamethoxazole. The comparisons with other Clostridium species are summarized in Table 2.
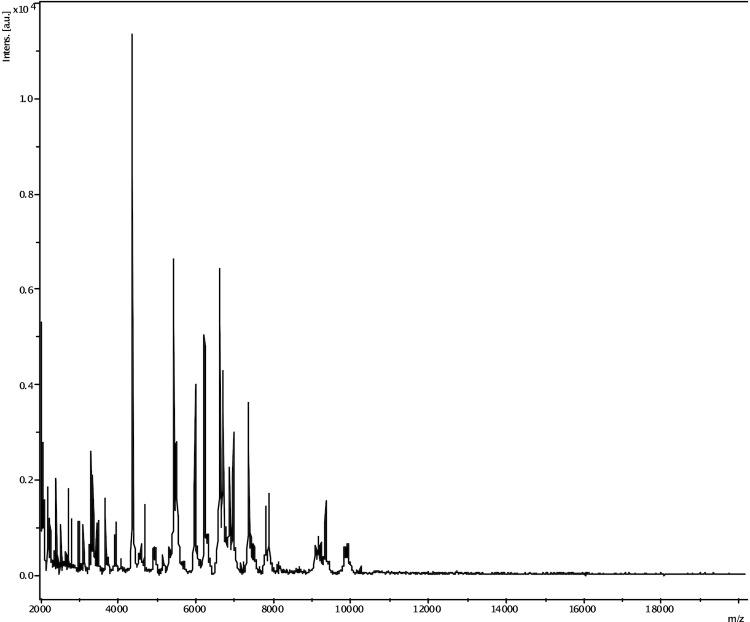
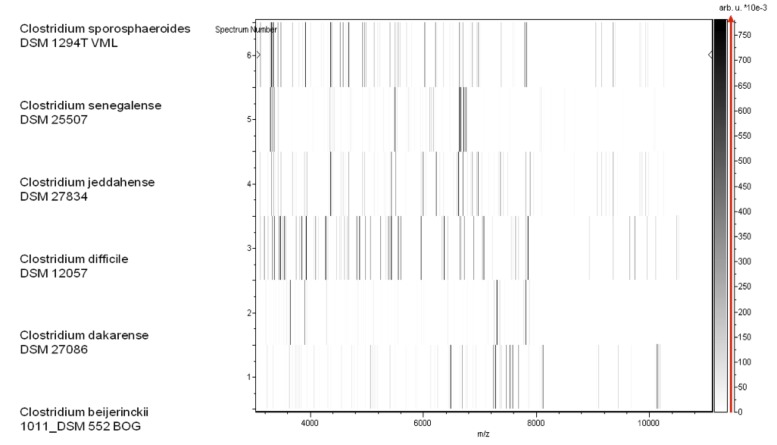
Matrix-assisted laser-desorption/ionization time-of-flight (MALDI-TOF) MS protein analysis was carried out as previously described [54]. Briefly, a pipette tip was used to pick one isolated bacterial colony from a culture agar plate and spread it as a thin film on a MTP 384 MALDI-TOF target plate (Bruker Daltonics, Leipzig, Germany). Twelve distinct deposits from twelve isolated colonies were performed for strain JCDT. Each smear was overlaid with 2 µL of matrix solution (saturated solution of alpha-cyano-4-hydroxycinnamic acid) in 50% acetonitrile, 2.5% tri-fluoracetic acid, and allowed to dry for 5 minutes. Measurements were performed with a Microflex spectrometer (Bruker). Spectra were recorded in the positive linear mode for the mass range of 2,000 to 20,000 Da (parameter settings: ion source 1 (ISI), 20kV; IS2, 18.5 kV; lens, 7 kV). A spectrum was obtained after 675 shots with variable laser power. The time of acquisition was between 30 seconds and 1 minute per spot. The twelve JCDT spectra were imported into the MALDI BioTyper software (version 2.0, Bruker) and analyzed by standard pattern matching (with default parameter settings) against the main spectra of 3,769 bacteria, including 228 spectra from 96 Clostridium species. The method of identification included the m/z from 3,000 to 15,000 Da. For every spectrum, a maximum of 100 peaks were compared with spectra in database. The resulting score enabled the identification of tested species, or not: a score ≥ 2 with a validly published species enabled identification at the species level, a score ≥ 1.7 but < 2 enabled identification at the genus level, and a score < 1.7 did not enable any identification. No significant MALDI-TOF score was obtained for strain JCDT against the Bruker database, suggesting that our isolate was not a member of a known species. We added the spectrum from strain JCDT to our database (Figure 4). Finally, the gel view showed the spectral differences with other members of the genus Clostridium (Figure 5).
Figure 4.
Reference mass spectrum from C. jeddahense strain JCDT. Spectra from 12 individual colonies were compared and a reference spectrum was generated.
Figure 5.
Gel view comparing C. jeddahense strain JCDT to other Clostridium species. The gel view displays the raw spectra of loaded spectrum files arranged as a pseudo-electrophoretic gel. The x-axis records the m/z value. The left y-axis displays the running spectrum number originating from subsequent spectra loading. The peak intensity is expressed by a grey scale scheme code. The grey scale bar on the right y-axis indicates the relation between the shade of grey a peak is displayed with and the peak intensity in arbitrary units. Species names are shown on the left.
Genome sequencing information
Genome project history
The organism was selected for sequencing on the basis of its phylogenetic position and 16S rDNA similarity to members of the genus Clostridium, and is part of a study of the human digestive flora aiming at isolating all bacterial species in human feces [55]. It was the 101st genome of a Clostridium species and the first genome of C. jeddahense sp. nov. The GenBank accession number is CBYL00000000. The assembly consists of 104 contigs. Table 3 shows the project information and its association with MIGS version 2.0 compliance [39].
Table 3. Project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | High-quality draft |
| MIGS-28 | Libraries used | Paired end and Mate pair |
| MIGS-29 | Sequencing platform | MySeq Illumina |
| MIGS-31.2 | Fold coverage | 94.91× |
| MIGS-30 | Assemblers | Newbler |
| MIGS-32 | Gene calling method | PRODIGAL |
| Genbank Date of Release | February 12, 2014 | |
| Genbank project ID | CBYL00000000 | |
| MIGS-13 | Project relevance | Study of the human gut microbiome |
Growth conditions and DNA isolation
C. jeddahense sp. nov., strain JCDT (= CSUR P693 = DSM 27834) was grown on 5% sheep blood-enriched Columbia agar (BioMerieux) at 37°C in anaerobic atmosphere. Bacteria grown on three Petri dishes were harvested and resuspended in 4x100µL of TE buffer. Then, 200 µL of this suspension was diluted in 1ml TE buffer for lysis treatment that included a 30- minute incubation with 2.5 µg/µL lysozyme at 37°C, followed by an overnight incubation with 20 µg/µL proteinase K at 37°C. Extracted DNA was then purified using 3 successive phenol-chloroform extractions and ethanol precipitation at -20°C overnight. After centrifugation, the DNA was resuspended in 160 µL TE buffer.
Genome sequencing and assembly
Genomic DNA of Clostridium jeddahense was sequenced on a MiSeq sequencer (Illumina, Inc, San Diego CA 92121, USA) with 2 applications: paired end and mate pair. The paired end and the mate pair strategies were barcoded in order to be mixed respectively with 14 other genomic projects constructed according the Nextera XT library kit (Illumina) and 11 others projects with the nextera Mate pair kit (Illumina).
The gDNA was quantified by a Qubit assay with the high sensitivity kit (Life technologies, Carlsbad, CA, USA) to 11.1 ng/µL and dilution was performed such that 1ng of each strain’s gDNA was used to construct the paired end library. The “tagmentation” step fragmented and tagged the DNA .Then limited cycle PCR amplification completed the tag adapters and introduced dual-index barcodes. After purification on Ampure beads (Life Technolgies, Carlsbad, CA, USA), the libraries were normalized on specific beads according to the Nextera XT protocol (Illumina). Normalized libraries are pooled into a single library for sequencing on the MiSeq. The pooled single strand library was loaded onto the reagent cartridge and then onto the instrument along with the flow cell. Automated cluster generation and paired-end sequencing with dual index reads was performed in a single 39-hour run at a 2x250 bp read length. Within this pooled run, the index representation was determined to be 7.3%. Total information of 5.3 Gbases was obtained from a 574 K/mm2 density with 95.4% (11,188,000 clusters) of the clusters passing quality control (QC) filters. From the genome sequencing process, the 753,292 produced Illumina reads for Clostridium jeddahense were filtered according to the read qualities.
The mate pair library was constructed from 1 µg of genomic DNA using the Nextera Mate Pair Illumina guide. The genomic DNA sample is simultaneously fragmented and tagged with a mate pair junction adapter. The profile of the fragmentation was validated on an Agilent 2100 BioAnalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA) with a DNA7500 labchip. The DNA fragments range in size from 1 kb up to 11 kb with a mean size of 7kb. No size selection was performed and 600 ng tagmented fragments were circularized. The larger circularized DNA molecules were physically sheared to smaller sized fragments with a mean size of 625 bp on the Covaris device S2 in microtubes (Woburn, MA, USA) .The library’s profile and the quantitation were visualized on a High Sensitivity Bioanalyzer LabChip. The libraries were normalized to 2 nM and pooled. After a denaturation step and dilution at 10 pM the pool of libraries was loaded onto the reagent cartridge and then onto the instrument along with the flow cell. Automated cluster generation and sequencing run was performed in a single 39-hour run at a 2x250 bp read length.
Total information of 3.9 Gb was obtained from a 399 K/mm2 density with 97.9% (7,840,000 clusters) of the clusters passing quality control (QC) filters. Within this pooled run, the index representation for Clostridium jeddahense was determined to be 6.54%.
From this genome sequencing process, the 501,426 produced Illumina reads for Clostridium jeddahense were filtered according to the read qualities.
Genome annotation
Open Reading Frames (ORFs) were predicted using Prodigal [56] with default parameters. However, the predicted ORFs were excluded if they spanned a sequencing gap region. The predicted bacterial protein sequences were searched against the GenBank [57] and Clusters of Orthologous Groups (COG) databases using BLASTP. The tRNAs and rRNAs were predicted using the tRNAScan-SE [58] and RNAmmer [59] tools, respectively. Signal peptides and numbers of transmembrane helices were predicted using SignalP [60] and TMHMM [61], respectively. Mobile genetic elements were predicted using PHAST [62] and RAST [63]. ORFans were identified if their BLASTP E-value was lower than 1e-03 for alignment length greater than 80 amino acids. If alignment lengths were smaller than 80 amino acids, we used an E-value of 1e-05. Such parameter thresholds have already been used in previous work to define ORFans. Artemis [64] and DNA Plotter [65] were used for data management and visualization of genomic features, respectively. The Mauve alignment tool (version 2.3.1) was used for multiple genomic sequence alignment [66].
To estimate the mean level of nucleotide sequence similarity at the genome level between C. jeddahense and 7 other members of the genus Clostridium, we used the Average Genomic Identity Of gene Sequences (AGIOS) home-made software [6]. Briefly, this software combines the Proteinortho software [67] for detecting orthologous proteins between pairs of genomes, then retrieves the corresponding genes and determines the mean percentage of nucleotide sequence identity among orthologous ORFs using the Needleman-Wunsch global alignment algorithm. C. jeddahense strain JCDT was compared to C. senegalense strain JC122, C. dakarense strain FF1, Clostridium beijerinckii strain NCIMB 8052, C. difficile strain B1, Clostridium cellulolyticum strain H10, Clostridium leptum strain DSM 753, and Clostridium sporosphaeroides strain DSM 1294 (see Table 6B).
Table 6B. Genomic comparison of C. jeddahense with 7 other Clostridium species†.
| C. jeddahense |
C. sporosphaeroides |
C. cellulolyticum |
C. dakarense |
C. difficile |
C. leptum |
C. senegalense |
C. beijerincki |
|
|---|---|---|---|---|---|---|---|---|
| C. jeddahense | 3,462 | 1,573 | 876 | 816 | 847 | 1,030 | 770 | 1,044 |
| C. sporosphaeroides | 91.97 | 2,951 | 854 | 776 | 819 | 1,016 | 745 | 1,015 |
| C. cellulolyticum | 61.60 | 60.62 | 3,923 | 806 | 851 | 754 | 814 | 946 |
| C. dakarense | 57.30 | 56.34 | 65.70 | 5,020 | 1,271 | 665 | 1,110 | 1,142 |
| C. difficile | 57.56 | 56.80 | 65.65 | 77.74 | 3,390 | 714 | 1,098 | 1,171 |
| C. leptum | 67.98 | 68.07 | 61.94 | 58.55 | 58.84 | 3,591 | 651 | 780 |
| C. senegalense | 57.52 | 56.73 | 65.71 | 70.18 | 69.41 | 58.72 | 3,704 | 1,125 |
| C. beijerincki | 58.63 | 57.95 | 65.93 | 68.98 | 68.48 | 59.58 | 71.37 | 3,818 |
†Numbers of orthologous proteins shared between genomes (above diagonal), AGIOS values (below diagonal) and numbers of proteins per genome (bold numbers)
Genome properties
The genome is 3,613,503 bp long (1 chromosome, but no plasmid) with a 51.95% G+C content (Figure 6 and Table 4). Of the 3,515 predicted genes, 3,462 were protein-coding genes and 53 were RNAs, including 4 rRNAs. A total of 2,193 genes (62.38%) were assigned a putative function and 81 genes were identified as ORFans (2.3%). The properties and statistics of the genome are summarized in Tables 4 and 5. The distribution of genes into COG functional categories is presented in Table 5.
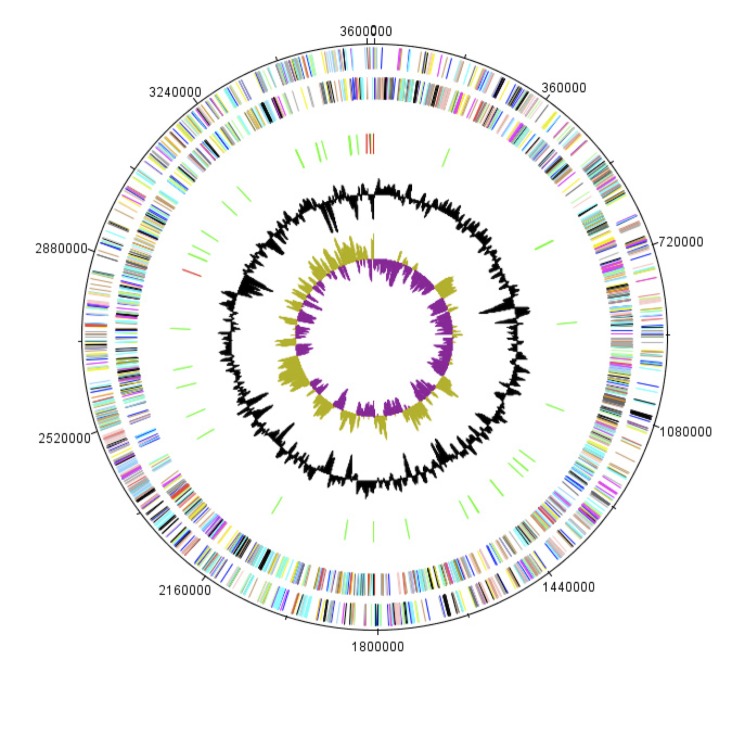
Figure 6.
Graphical circular map of the chromosome. From the outside in: open reading frames oriented in the forward (colored by COG categories) direction, open reading frames oriented in the reverse (colored by COG categories) direction, RNA operon (red), and tRNAs (green), GC content plot, and GC skew (purple: negative values, olive: positive values).
Table 4. Nucleotide content and gene count levels of the genome.
| Attribute | Value | % of totala |
|---|---|---|
| Genome size (bp) | 3,613,503 | |
| DNA G+C content (bp) | 1,877,214 | 51.95 |
| DNA Coding region (bp) | 3,152,277 | 87.23 |
| Number of replicons | 1 | |
| Extra chromosomal element | 0 | |
| Total genes | 3,515 | 100 |
| RNA genes | 53 | 1.51 |
| Protein-coding genes | 3,462 | 98.49 |
| Genes with function prediction | 2,193 | 87.19 |
| Genes assigned to COGs | 2,515 | 71.55 |
| Genes with peptide signals | 135 | 3.84 |
| Genes with transmembrane helices | 887 | 25.23 |
a The total is based on either the size of the genome in base pairs or the total number of protein-coding genes in the annotated genome
Table 5. Number of genes associated with the 25 general COG functional categories.
| Code | Value | % agea | Description |
|---|---|---|---|
| J | 154 | 4.45 | Translation |
| A | 0 | 0 | RNA processing and modification |
| K | 296 | 8.55 | Transcription |
| L | 138 | 3.98 | Replication, recombination and repair |
| B | 1 | 0.03 | Chromatin structure and dynamics |
| D | 24 | 0.69 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0 | Nuclear structure |
| V | 73 | 2.11 | Defense mechanisms |
| T | 156 | 4.5 | Signal transduction mechanisms |
| M | 116 | 3.35 | Cell wall/membrane biogenesis |
| N | 62 | 1.79 | Cell motility |
| Z | 0 | 0 | Cytoskeleton |
| W | 0 | 0 | Extracellular structures |
| U | 48 | 1.38 | Intracellular trafficking and secretion |
| O | 66 | 1.9 | Posttranslational modification, protein turnover, chaperones |
| C | 154 | 4.45 | Energy production and conversion |
| G | 237 | 6.84 | Carbohydrate transport and metabolism |
| E | 328 | 9.47 | Amino acid transport and metabolism |
| F | 56 | 1.61 | Nucleotide transport and metabolism |
| H | 92 | 2.66 | Coenzyme transport and metabolism |
| I | 85 | 2.45 | Lipid transport and metabolism |
| P | 164 | 4.74 | Inorganic ion transport and metabolism |
| Q | 53 | 1.53 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 346 | 10 | General function prediction only |
| S | 195 | 5.63 | Function unknown |
| - | 947 | 27.35 | Not in COGs |
a The total is based on the total number of protein-coding genes in the annotated genome
Genome comparison with other Clostridium genomes
We compared the genomes of C. jeddahense JCDT, C. sporosphaeroides DSM 1294, C. leptum DSM 753, C. beijerincki NCIMB 8052, C. cellulolyticum H10, C. difficile B1, C. senegalense DSM 25507, C. dakarense DSM 27086 (Table 6A).
Table 6A. Genomic comparison of C. jeddahense with 7 other Clostridium species†.
| Species | Strain | Genome accession number |
Genome size (Mb) | G+C content |
|---|---|---|---|---|
| C. jeddahense | JCDT | CBYL00000000 | 3.61 | 51.95 |
| C. sporosphaeroides | DSM 1294 | ARTA01000000 | 3.17 | 53.5 |
| C. cellulolyticum | H10 | NC_011898 | 4.07 | 37.4 |
| C. dakarense | DSM 27086 | CBTZ010000000 | 3.73 | 27.98 |
| C. difficile | B1 | NC_017179 | 4.46 | 28.4 |
| C. leptum | DSM 753 | ABCB02000000 | 3.27 | 50.2 |
| C. senegalense | DSM 25507 | CAEV01000001 | 3.89 | 26.8 |
| C. beijerincki | NCIMB 8052 | NC_009617 | 6.0 | 29.0 |
† A: Species, Strain, GenBank accession number, genome size and G+C content of all compared genomes
The draft genome of C. jeddahense (3.61 Mb) is larger than C. sporosphaeroides and C. leptum (3.17 and 3.27 Mb respectively) but smaller than C. beijerincki, C. cellulolyticum, C. difficile, C. senegalense and C. dakarense (6.0, 4.07, 4.46, 3.89, 3.73 Mb respectively). It exhibits a higher G+C content than all other compared genome except C. sporosphaeroides (53.5%). C. jeddahense has a higher gene content (3,462) than C. sporosphaeroides, C. difficile (2,951 and 3,390 respectively) but smaller than C. leptum, C. beijerincki, C. cellulolyticum, C. senegalense and C. dakarense (3,591, 3,818, 3,923, 3,704, 5,020 respectively). C. jeddahense shared 1,573, 876, 816, 847, 1,030, 770 and 1,044 orthologous genes with C. sporosphaeroides, C. cellulolyticum, C. dakarense, C. difficile, C. leptum, C. senegalense and C. beijerincki respectively.
When we compared C. jeddahense with other species, AGIOS values ranged from 57.52 with C. senegalense to 91.97% with C. sporosphaeroides. Although the AGIOS value was elevated between C. jeddahense and C. sporosphaeroides, we believe that the remarkable phenotypic differences, including motility, indole production (Table 2), and protein profile (Figure 7), enable the classification of C. jeddahense as a new species.
Figure 7.
Distribution of predicted genes of C. jeddahense and 7 other Clostridium species into COG categories. C.jdm= C. jeddahense, C.spo= C. sporosphaeroides, C. lep= C. leptum, C.bej = C. beijerinckii, C. cel = C. cellulolyticum, C. diff = C. difficile, C. sen = C. senegalense, C. dak = C. dakarense.
Conclusion
On the basis of phenotypic, phylogenetic and genomic analyses (taxono-genomics), we formally propose the creation of Clostridium jeddahense sp. nov. that contains strain JCDT. This strain was isolated from the fecal flora of an obese 24 year-old Saudian individual living in Jeddah.
Description of C. jeddahense sp. nov.
Clostridium jeddahense (jed.dah..en’.se L.gen. neutr. n. combination of Jeddah, the city in Saudi Arabia where the specimen was obtained from an obese Saudian patient sample.) Transparent colonies were 0.2 to 0.3 mm in diameter on blood-enriched agar. C. jeddahense is a Gram-positive, obligate anaerobic, endospore-forming bacterium with a mean diameter of 1 µm. Optimal growth on axenic medium was observed at 37°C.
C. jeddahense is catalase negative and oxidase negative. Alkaline phosphatase, arginine arylamidase, proline arylamidase, alanine arylamidase, glycine arylamidase, histidine arylamidase, glutamyl glutamic acid arylamidase and serine arylamidase activities were positive. Arginine dihydrolase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, α-arabinosidase, N-acetyl-β-glucosaminidase, glutamic acid decarboxylase, α-fucosidase, reduction of nitrate, leucyl glycine arylamidase, fermentation of mannose and raffinose, urease, β-galactosidase-6-phosphatase, β-glucuronidase, phenylalanine arylamidase, leucine arylamidase, pyroglutamic acid arylamidase and tyrosine arylamidase activities were negative. Asaccharolytic. Positive for indole. Cells are susceptible to amoxicillin, amoxicillin-clavulanate, imipenem, metronidazole, doxycycline, rifampicin, vancomycin but resistant to ceftriaxone, ciprofloxacin and trimethoprim-sulfamethoxazole.
The G+C content of the genome is 51.95%. The 16S rDNA and genome sequences are deposited in GenBank under accession numbers HG726040 and CBYL00000000, respectively. The type strain is JCDT (= CSUR P693 = DSM 27834).
Acknowledgements
This work was funded by the Deanship of Scientific Research (DSR), King Abdulaziz University, under grant No. (1-141/1433 HiCi). The authors, therefore, acknowledge technical and financial support of KAU. The authors thank the Xegen Company (www.xegen.fr) for automating the genomic annotation process.
References
- 1.Tindall BJ, Rosselló-Móra R, Busse HJ, Ludwig W, Kämpfer P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int J Syst Evol Microbiol 2010; 60:249-266 10.1099/ijs.0.016949-0 [DOI] [PubMed] [Google Scholar]
- 2.Stackebrandt E, Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today 2006; 33:152-155 [Google Scholar]
- 3.Wayne LG, Brenner DJ, Colwell PR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematic. Int J Syst Bacteriol 1987; 37:463-464 10.1099/00207713-37-4-463 [DOI] [Google Scholar]
- 4.Rossello-Mora R. DNA-DNA Reassociation Methods Applied to Microbial Taxonomy and Their Critical Evaluation. In: Stackebrandt E (ed), Molecular Identification, Systematics, and population Structure of Prokaryotes. Springer, Berlin, 2006; p. 23-50. [Google Scholar]
- 5.Welker M, Moore ER. Applications of whole-cell matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry in systematic microbiology. Syst Appl Microbiol 2011; 34:2-11 10.1016/j.syapm.2010.11.013 [DOI] [PubMed] [Google Scholar]
- 6.Ramasamy D, Mishra AK, Lagier JC, Padhmanabhan R, Rossi-Tamisier M, Sentausa E, Raoult D, Fournier PE. A polyphasic strategy incorporating genomic data for the taxonomic description of new bacterial species. Int J Syst Evol Microbiol 2014; (In press). 10.1099/ijs.0.057091-0 [DOI] [PubMed] [Google Scholar]
- 7.Kokcha S, Mishra AK, Lagier JC, Million M, Leroy Q, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Bacillus timonensis sp. nov. Stand Genomic Sci 2012; 6:346-355 10.4056/sigs.2776064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lagier JC, El Karkouri K, Nguyen TT, Armougom F, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Anaerococcus senegalensis sp. nov. Stand Genomic Sci 2012; 6:116-125 10.4056/sigs.2415480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mishra AK, Gimenez G, Lagier JC, Robert C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Alistipes senegalensis sp. nov. Stand Genomic Sci 2012; 6:304-314 10.4056/sigs.2625821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagier JC, Armougom F, Mishra AK, Ngyuen TT, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Alistipes timonensis sp. nov. Stand Genomic Sci 2012; 6:315-324 10.4056/sigs.2685971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mishra AK, Lagier JC, Robert C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Clostridium senegalense sp. nov. Stand Genomic Sci 2012; 6:386-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mishra AK, Lagier JC, Robert C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Peptoniphilus timonensis sp. nov. Stand Genomic Sci 2012; 7:1-11 10.4056/sigs.2956294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mishra AK, Lagier JC, Rivet R, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Paenibacillus senegalensis sp. nov. Stand Genomic Sci 2012; 7:70-81 10.4056/sigs.3056450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagier JC, Gimenez G, Robert C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Herbaspirillum massiliense sp. nov. Stand Genomic Sci 2012; 7:200-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kokcha S, Ramasamy D, Lagier JC, Robert C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Brevibacterium senegalense sp. nov. Stand Genomic Sci 2012; 7:233-245 10.4056/sigs.3256677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramasamy D, Kokcha S, Lagier JC, N’Guyen TT, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Aeromicrobium massilense sp. nov. Stand Genomic Sci 2012; 7:246-257 10.4056/sigs.3306717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagier JC, Ramasamy D, Rivet R, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Cellulomonas massiliensis sp. nov. Stand Genomic Sci 2012; 7:258-270 10.4056/sigs.3316719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagier JC, Karkouri K, Rivet R, Couderc C, Raoult D, Fournier PE. Non contiguous-finished genome sequence and description of Senegalemassilia anaerobia gen. nov., sp. nov. Stand Genomic Sci 2013; 7:343-356 10.4056/sigs.3246665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra AK, Hugon P, Nguyen TT, Robert C, Couderc C, Raoult D, Fournier PE. Non contiguous-finished genome sequence and description of Peptoniphilus obesi sp. nov. Stand Genomic Sci 2013; 7:357-369 10.4056/sigs.32766871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishra AK, Lagier JC, Nguyen TT, Raoult D, Fournier PE. Non contiguous-finished genome sequence and description of Peptoniphilus senegalensis sp. nov. Stand Genomic Sci 2013; 7:357-369 10.4056/sigs.32766871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lagier JC, Karkouri K, Mishra AK, Robert C, Raoult D, Fournier PE. Non contiguous-finished genome sequence and description of Enterobacter massiliensis sp. nov. Stand Genomic Sci 2013; 7:399-412 10.4056/sigs.3396830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hugon P, Ramasamy D, Rivet R, Raoult D, Fournier PE. Non contiguous-finished genome sequence and description of Alistipes obesi sp. nov. Stand Genomic Sci 2013; 7:427-439 10.4056/sigs.3336746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hugon P, Mishra AK, Nguyen TT, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Brevibacillus massiliensis sp. nov. Stand Genomic Sci 2013; 8:1-14 10.4056/sigs.3466975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishra AK, Hugon P, Nguyen TT, Raoult D, Fournier PE. Non contiguous-finished genome sequence and description of Enorma massiliensis gen. nov., sp. nov., a new member of the Family Coriobacteriaceae. Stand Genomic Sci 2013; 8:290-305 10.4056/sigs.3426906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramasamy D, Lagier JC, Gorlas A, Raoult D, Fournier PE. Non contiguous-finished genome sequence and description of Bacillus massiliosenegalensis sp. Nov. Stand Genomic Sci 2013; 8:264-278 10.4056/sigs.3496989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramasamy D, Lagier JC, Nguyen TT, Raoult D, Fournier PE. Non contiguous-finished genome sequence and description of Dielma fastidiosa gen. nov., sp. nov., a new member of the Family Erysipelotrichaceae. Stand Genomic Sci 2013; 8:336-351 10.4056/sigs.3567059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mishra AK, Pfleiderer A, Lagier JC, Robert C, Raoult D, Fournier PE. Non contiguous-finished genome sequence and description of Bacillus massilioanorexius sp. nov. Stand Genomic Sci 2013; 8:465-479 10.4056/sigs.4087826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hugon P, Ramasamy D, Robert C, Couderc C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Kallipyga massiliensis gen. nov., sp. nov., a new member of the family Clostridiales Incertae Sedis XI. Stand Genomic Sci 2013; 8:500-515 10.4056/sigs.4047997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Padhmanabhan R, Lagier JC, Dangui NPM, Michelle C, Couderc C, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Megasphaera massiliensis. Stand Genomic Sci 2013; 8:525-538 10.4056/sigs.4077819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mishra AK, Edouard S, Dangui NPM, Lagier JC, Caputo A, Blanc-Tailleur C, Ravaux I, Raoult D, Fournier PE. Non-contiguous finished genome sequence and description of Nosocomiicoccus massiliensis sp. nov. Stand Genomic Sci 2013; 9:205-219 10.4056/sigs.4378121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra AK, Lagier JC, Robert C, Raoult D, Fournier PE. Genome sequence and description of Timonella senegalensis gen. nov., sp. nov., a new member of the suborder Micrococcineae. Stand Genomic Sci 2013; 8:318-335 10.4056/sigs.3476977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keita MB, Diene SM, Robert C, Raoult D, Fournier PE. Non contiguous-finished genome sequence and description of Bacillus massiliogorillae sp. nov. Stand Genomic Sci 2013; 9:93-105 10.4056/sigs.4388124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mediannikov O, El Karkouri K, Robert C, Fournier PE, Raoult D. Non contiguous-finished genome sequence and description of Bartonella florenciae sp. nov. Stand Genomic Sci 2013; 9:185-196 10.4056/sigs.4358060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo CI, Mishra AK, Padhmanabhan R, Samb Ba B, Gassama Sow A, Robert C, Couderc C, Faye N, Raoult D, Fournier PE, Fenollar F. Non contiguous-finished genome sequence and description of Clostridium dakarense sp. nov. Stand Genomic Sci 2013; 9:14-27 10.4056/sigs.4097825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishra AK, Hugon P, Robert C, Raoult D, Fournier PE. Non contiguous-finished genome sequence and description of Peptoniphilus grossensis sp. nov. Stand Genomic Sci 2012; 7:320-330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mediannikov O, El Karkouri K, Diatta G, Robert C, Fournier PE, Raoult D. Non contiguous-finished genome sequence and description of Bartonella senegalensis sp. nov. Stand Genomic Sci 2013; 8:279-289 10.4056/sigs.3807472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roux V, Million M, Robert C, Magne A, Raoult D. Non-contiguous finished genome sequence and description of Oceanobacillus massiliensis sp. nov. Stand Genomic Sci 2013; •••:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wells CL, Wilkins TD. (1996). Clostridia: Spore forming Anaerobic Bacilli In: Baron's Medical Microbiology (Baron S et al., eds.) (4th ed.). University of Texas Medical Branch. [PubMed] [Google Scholar]
- 39.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1. Springer, New York 2001; 119-169. [Google Scholar]
- 42.Gibbons NE, Murray RGE. Proposals Concerning the Higher Taxa of Bacteria. Int J Syst Bacteriol 1978; 28:1-6 10.1099/00207713-28-1-1 [DOI] [Google Scholar]
- 43.Murray RGE. The Higher Taxa, or, a Place for Everything...? In: Holt JG (ed), Bergey's Manual of Systematic Bacteriology, First Edition, Volume 1, The Williams and Wilkins Co., Baltimore, 1984, p. 31-34. [Google Scholar]
- 44.List of new names and new combinations previously effectively, but not validly, published. List no. 132. Int J Syst Evol Microbiol 2010; 60:469-472 10.1099/ijs.0.022855-0 [DOI] [PubMed] [Google Scholar]
- 45.Rainey FA. Class II. Clostridia class nov. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 3, Springer-Verlag, New York, 2009, p. 736. [Google Scholar]
- 46.Skerman VBD, Sneath PHA. Approved list of bacterial names. Int J Syst Bact 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [Google Scholar]
- 47.Prevot AR. Dictionnaire des bactéries pathogens. In: Hauduroy P, Ehringer G, Guillot G, Magrou J, Prevot AR, Rosset, Urbain A (eds). Paris, Masson, 1953, p.1-692. [Google Scholar]
- 48.Pribram E. Klassification der Schizomyceten. Klassifikation der Schizomyceten (Bakterien), Franz Deuticke, Leipzig, 1933, p. 1-143. [Google Scholar]
- 49.Prazmowski A. "Untersuchung über die Entwickelungsgeschichte und Fermentwirking einiger Bakterien-Arten." Ph.D. Dissertation, University of Leipzig, Germany, 1880, p. 366-371. [Google Scholar]
- 50.Smith LDS, Hobbs G. Genus III. Clostridium Prazmowski 1880, 23. In: Buchanan RE, Gibbons NE (eds), Bergey's Manual of Determinative Bacteriology, Eighth Edition, The Williams and Wilkins Co., Baltimore, 1974, p. 551-572. [Google Scholar]
- 51.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.16S Yourself database. http://www.mediterranee-infection.com/article.php?larub=152&titre=16s-yourself
- 53.Logan NA, De Vos P. Genus I. Clostiridum Prazmowski 1880. In: (Eds?) De Vos P, Garrity D, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB. Bergey’a manual of Systematic Bacteriology. Volume 3: The Firmicutes, Springer, 738-828. [Google Scholar]
- 54.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 2009; 49:543-551 10.1086/600885 [DOI] [PubMed] [Google Scholar]
- 55.Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect 2012; 18:1185-1193 [DOI] [PubMed] [Google Scholar]
- 56.Prodigal. http://prodigal.ornl.gov/
- 57.Benson DA, Karsch-Mizrachi I, Clark K, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res 2012; 40:D48-D53 10.1093/nar/gkr1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955-964 10.1093/nar/25.5.0955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lagesen K, Hallin P, Rodland EA, Staerfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100-3108 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 2004; 340:783-795 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]
- 61.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001; 305:567-580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 62.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic Acids Res 2011; 39:W347-W352 10.1093/nar/gkr485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008; 9:75 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics 2000; 16:944-945 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- 65.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 2009; 25:119-120 10.1093/bioinformatics/btn578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 2004; 14:1394-1403 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lechner M, Findeib S, Steiner L, Marz M, Stadler PF, Prohaska SJ. Proteinortho: Detection of (Co-)orthologs in large-scale analysis. BMC Bioinformatics 2011; 12:124 10.1186/1471-2105-12-124 [DOI] [PMC free article] [PubMed] [Google Scholar]