Abstract
Burkholderia phymatum is a soil bacterium able to develop a nitrogen-fixing symbiosis with species of the legume genus Mimosa, and is frequently found associated specifically with Mimosa pudica. The type strain of the species, STM 815T, was isolated from a root nodule in French Guiana in 2000. The strain is an aerobic, motile, non-spore forming, Gram-negative rod, and is a highly competitive strain for nodulation compared to other Mimosa symbionts, as it also nodulates a broad range of other legume genera and species. The 8,676,562 bp genome is composed of two chromosomes (3,479,187 and 2,697,374 bp), a megaplasmid (1,904,893 bp) and a plasmid hosting the symbiotic functions (595,108 bp).
Keywords: Burkholderia, symbiosis, Mimosa, rhizobia, nitrogen fixation
Introduction
Rhizobia are a functional class of bacteria able to enter into nitrogen-fixing symbioses with legumes. The bacterial symbiont induces the formation of nodules on the roots of the plant where they differentiate into nitrogen-fixing bacteroids. Bacteria then allocate combined nitrogen to the plant, which in return provides the bacteria with energy derived from photosynthesis. This symbiosis confers agricultural advantages to the legumes by reducing the need for fertilization and allows them to be pioneer plants on degraded or contaminated soils.
Rhizobia are polyphyletic and are placed within two classes of Proteobacteria, the Alphaproteobacteria and the Betaproteobacteria. They are closely related to non-symbiotic species, including important human, animal or plant pathogens or saprophytes. Most research has focused on the α-rhizobia, since the β-rhizobia were only recently discovered [1,2]. The α-rhizobia include 10 genera (Sinorhizobium, Mesorhizobium, Rhizobium, Methylobacterium, Devosia, Azorhizobium, Bradyrhizobium, Ochrobactrum, Bosea and Phyllobacterium) and have a worldwide distribution associated with a diversity of legume species (from herbs to trees). To date, the β-rhizobia include only two genera, Burkholderia and Cupriavidus (ex Ralstonia), and a dozen species (for review [3], updated in [4]). They are found preferentially associated with Mimosa species (at least 68 nodulated species, and especially M. pudica, M. pigra, and M. bimucronata) in Asia, Australia, and Central and South America [5,6]. Based on a comparison of house-keeping and nodulation gene phylogenies, Burkholderia species have been postulated to be ancestral symbionts of South American Mimosa and Piptadenia species [4,5]. Here we describe the genome sequence of one of the first described β-rhizobia, the type strain of Burkholderia phymatum, STM815T.
Classification and features
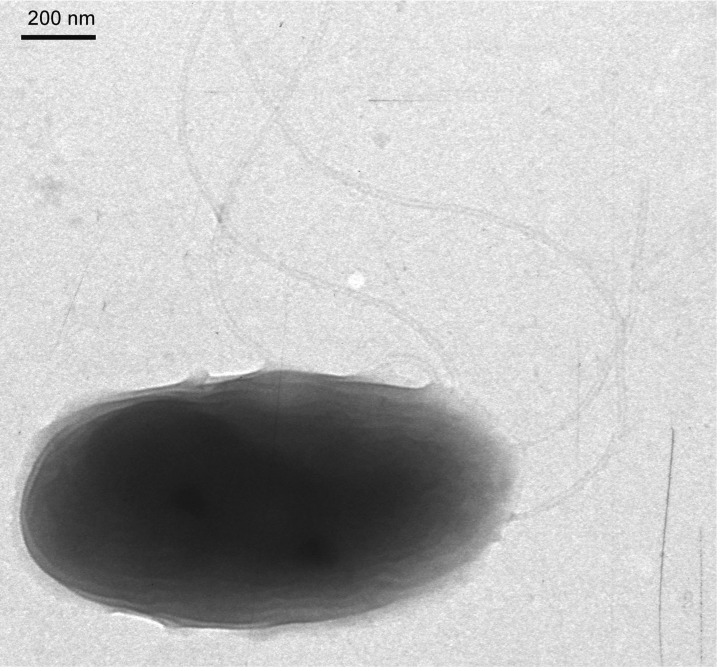
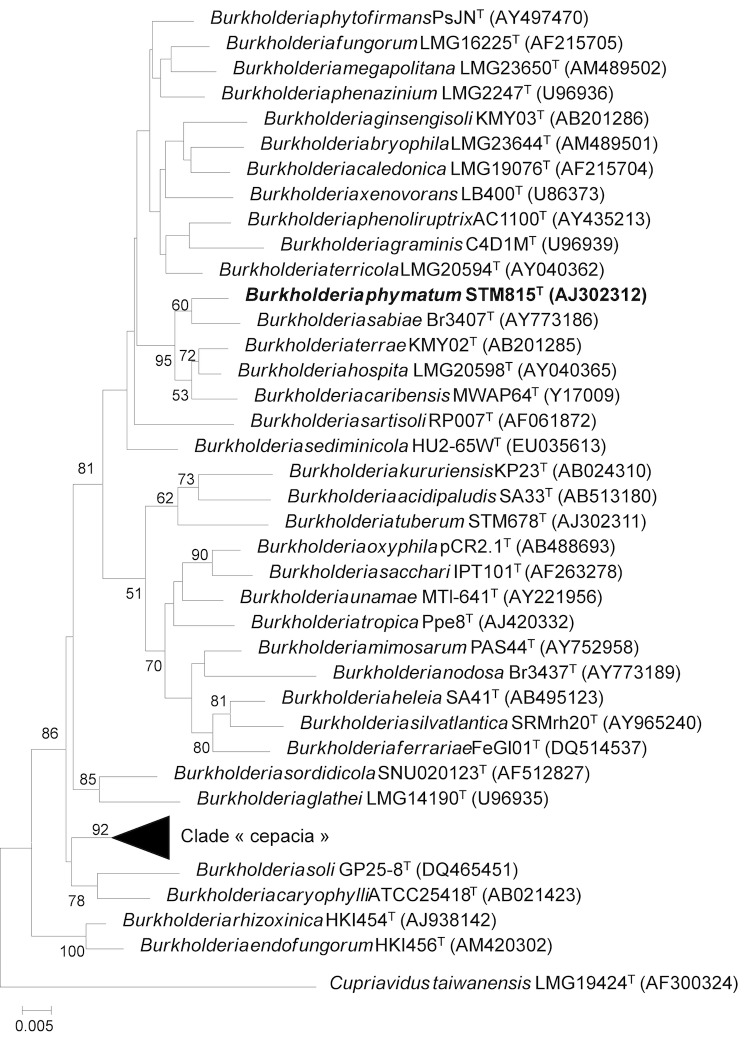
Burkholderia phymatum STM815T is a motile, Gram-negative rod (Figure 1) in the order Burkholderiales of the class Betaproteobacteria. It is fast growing, forming colonies within 3-4 days when grown on yeast-mannitol agar (YMA [7],) at 28°C. It is one of the first described members of the β-rhizobia. The strain STM815T, which is the type strain of the species, was isolated from nodules of Machaerium lunatum in French Guiana in 2000 [1], and the species, B. phymatum, was described based on this single isolate [8]. However, the species has subsequently been shown not to nodulate Machaerium species [9], but it can nodulate species in the large genus Mimosa [9,10]. Indeed, the symbiotic abilities of STM815T have been demonstrated on numerous Mimosa species, and this strain is now considered to be an efficient symbiont of a broad range of legumes, particularly in Mimosa and related genera in the sub-family Mimosoideae [9]. Strain STM815T is also able to fix nitrogen in free-living conditions [9]. Many isolates of B. phymatum have been sampled from Mimosa pudica in French Guiana [10], Papua New Guinea [9], China [11] and India [12]. Phylogenetic analyses of core and symbiotic genes have illustrated the ancestral status of Burkholderia species in symbioses with Mimosa [4,5]. Burkholderia phymatum STM815T is now considered to be a model system for studying the adaptive processes of Burkholderia in symbioses with legumes, in comparison with α-rhizobia. The B. phymatum species is phylogenetically related to symbiotic and non-pathogenic species, and is distant from the “cepacia” clade of Burkholderia (which contains many pathogenic species) (Figure 2, Table 1).
Figure 1.
Transmission electron microscopy of B. phymatum STM815 (credit: Geoffrey Elliott).
Figure 2.
Phylogenetic tree highlighting the position of Burkholderia phymatum strain STM815T relative to other type strains within the genus Burkholderia. The 16S rDNA sequences from type strains were obtained from the ribosomal database project [13], aligned with muscle 3.6, and a neighbor-joining tree was built from a Kimura-2P corrected distance matrix using BioNJ on the www.phylogeny.fr server [14]. Numbers at nodes are % bootstraps from 1000 replicates (shown only if >50%). Accession numbers of 16S rDNA are indicated between parentheses for each strain. C. taiwanensis LMG19424T was used as outgroup.
Table 1. Classification and general features of Burkholderia phymatum STM815 according to MIGS recommendations [15].
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Domain Bacteria | TAS [16] | ||
| Phylum Proteobacteria | TAS [17] | ||
| Class Betaproteobacteria | TAS [18,19] | ||
| Current classification | Order Burkholderiales | TAS [18,20] | |
| Family Burkholderiaceae | TAS [18,21] | ||
| Genus Burkholderia | TAS [22-24] | ||
| Species Burkholderia phymatum | TAS [8,25] | ||
| Type strain STM815 | |||
| Gram stain | negative | TAS [8] | |
| Cell shape | straight rods | TAS [8] | |
| Motility | motile | ||
| Sporulation | non-sporulating | TAS [8] | |
| Temperature range | mesophile, no growth at 42°C | TAS [8] | |
| Optimum temperature | 28°C | TAS [8] | |
| Carbon source | D-glucose, L-arabinose, D-mannose, D-mannitol, N-acteyl-D-glucosamine, D-gluconate, caprate, D-galactose, citric acid, D-galacturonate acid, methyl-pyruvate, L-aspartic acid, L-glutamic acid, L-asparagine, D,L-lactic acid |
TAS [8] TAS [8] IDA IDA IDA IDA |
|
| Energy source | chemoorganotroph | TAS [8] | |
| MIGS-6 | Habitat | Soil, nodule, host | TAS [1] |
| MIGS-6.3 | Salinity | Not reported | |
| MIGS-22 | Oxygen | Aerobic | TAS [8] |
| MIGS-15 | Biotic relationship | Free living, Symbiotic | TAS [1,9] |
| MIGS-14 | Pathogenicity | None | |
| MIGS-4 | Geographic location | Root nodule of Machaerium lunatum in French Guiana (Paracou) | TAS [1] |
| MIGS-5 | Sample collection time | 2000 | TAS [1] |
| MIGS-4.1 | Latitude | 5°15’N | TAS [1] |
| MIGS-4.2 | Longitude | 52°55’W | TAS [1] |
| MIGS-4.3 | Depth | Not reported | |
| MIGS-4.4 | Altitude | 32 m | TAS [1] |
a) Evidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project [26].
Symbiotaxonomy
Burkholderia phymatum STM815T forms nodules (Nod+) and fixes N2 (Fix+) with a broad range of Mimosa species [6,9] as well as with other genera in the tribe Mimoseae in the Mimosoideae legumes sub-family [9]. Nodulation data were compiled in Table 2.
Table 2. Mimosoid legumes tested for nodulation with Burkholderia phymatum STM815.
| Tribe / Genus | Species | Nodulation by STM815 |
|---|---|---|
| Tribe Mimoseae* | ||
| Acacia | farnesiana, karroo, nilotica var. kraussiana, nilotica var. leiocarpa, pennatula, schaffneri, seyal, tortilis | F |
| Anadenanthera | pavonina, colubrina | F |
| Desmanthus | bicornutus, fruticosus, virgatus | O |
| Dichrostachys | cinerea, microcephala | O |
| Leucaena | collinsii, cuspidata, pulverulenta, trichodes | N |
| confertiflora, esculenta, greggii, retusa, salvadorensis | O | |
| leucocephala, multicapitula | F | |
| Microlobius | foetidus | O |
| Mimosa | aculeaticarpa1, luisana1, setosissima4 | O |
| acutistipula1, albida1, albolanata4, artemisiana1, bimucronata1, caesalpiniifolia1, camporum1, cordistipula4, debilis4, diplotricha1, foliolosa4, flocculosa1, hexandra1, himalayana1, invisa1, latispinosa1, ophtalmocentra1, pigra1, polydactyla1, pudica1, somnians1, tenuiflora, setosa4, ursina4, velloziana4, xanthocentra4 | F | |
| adenocarpa1, affinis1, bahamensis1, blanchetii1, borealis1, callithrix4, claussenii4, decorticans4, delicatula1, densa4, dysocarpa1, melanocarpa4, menabeensis1, polyantha1, scabrella1, uruguensis1 | I | |
| Neptunia | dimorphantha, gracilis, majore, monosperma, plena | O |
| oleracea | N | |
| Parapiptadenia | rigida | N |
| Piptadenia | gonoacantha, stipulacea, viridiflora2 | F |
| Pityrocarpa3 | moniliformis, obliqua | F |
| Prosopis | africana, farcta, glandulosa, velutina | O |
| chilensis, pubescens | N | |
| juliflora | F | |
| Schleinitzia | novo-guineensis | O |
| Stryphnodendron | coriaceum, guianensis, pulcherrimum | O |
| Tribe Ingeae† | ||
| Acacia (Ac) | senegal | N |
| Acacia (P) | dealbata | O |
| mangium | N | |
| Albizia | adenocephala, kalkora, niopoides | O |
| julibrissin | N | |
| Calliandra | houstiana var. acapulcens, houstiana var. anomala, houstiana var. calothyrsus, juzepczukii, trinervia | F |
| physocalyx, rubescens | N | |
| Chloroleucon | tortum | O |
| Enterolobium | cyclocarpum | O |
| Faidherbia | albida | N |
| Pithecellobium | dulce | F |
| Samanea | saman | O |
| Zapoteca | tetragona | O |
Legend: O = no nodules formed; N = outgrowths on roots, superficially similar to nodules but ineffective; I = nodules formed are inefficient; F = nitrogen fixing nodules formed (these may not all be fully effective, but plants gave acetylene reduction values at least twice that of non-nodulated control plants).
*This is taken to include Acacia subgenus Acacia, now thought to be closely related to tribe Mimoseae and given the generic name Vachellia by some.
†This is taken to include Acacia, subgenera Aculeiferum (Ac) and Phyllodineae (P). The species listed below are now also included in genera Senegalia and Acacia respectively. Species from other genera in former Acacia have not been studied here.
1 Nodulation data from [9]; 2 This species has been transferred to an as yet unnamed genus by [27]; 3 This genus was formerly in Piptadenia [27]; 4 Nodulation data from [6]. Nodulation data for other legumes are from unpublished data from E.K. James and L. Moulin.
Genome sequencing information
Genome project history
The genome was selected by a consortium of researchers led by M. Riley, to be sequenced by the DOE Joint Genome Institute as part of the “Recommendations for Sequencing Targets in Support of the Science Missions of the Office of Biological and Environmental Research”. Initially, the strain was chosen to enrich genome data in the Burkholderia genus for comparative genomics. The genome was selected for genome determination because strain STM815T is a legume symbiont, as compared to the large number of genome sequences available for opportunistic and human-pathogens. The genome sequence was completed in 2007 and presented for public access on April 2008. Automatic annotation was performed using the JGI-Oak Ridge National Laboratory annotation pipeline [28]. Additional automatic and manual sequence annotation, as well as comparative genome analysis, were performed using the MicroScope platform at Genoscope [29]. Table 3 presents the project information and its association with MIGS version 2.0 compliance [30].
Table 3. Project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Complete |
| MIGS-28 | Libraries used | 3 kb, 8 kb and 40 kb (fosmid) |
| MIGS-29 | Sequencing platforms | Sanger |
| MIGS-31.2 | Fold coverage | 11.2 |
| MIGS-30 | Assemblers | Phred/Phrap/Consed |
| MIGS-32 | Gene calling method | DOE-JGI tools |
| Genome Database release | December 12, 2008 | |
| Genbank ID | CP001043 - CP001046 | |
| Genbank Date of Release | April 22, 2008 | |
| NCBI BioProject ID | PRJNA17409 | |
| GOLD ID | Gc00775 | |
| Project relevance | biotechnological |
Growth conditions and DNA isolation
The strain was grown in 50 ml of broth Yeast-mannitol medium (YM [7],) and DNA isolation was performed using a CTAB (Cetyl trimethyl ammonium bromide) bacterial genomic DNA isolation method [31].
Genome sequencing and assembly
The genome of Burkholderia phymatum STM815T was sequenced by Sanger technology at the Joint Genome Institute (JGI) using a combination of 3 kb, 8 kb and 40 kb (fosmid) DNA libraries. All general aspects of library construction and sequencing performed at the JGI can be found at the DOE JGI website [32].
Draft assemblies were based on 115,329 total reads and resulted in approximately 11.2× coverage of the genome. The Phred/Phrap/Consed software package was used for sequence assembly and quality assessment [33-35]. Gaps between contigs were closed by custom primer walks on gap spanning clones or PCR products. A total of 1,282 additional reactions were necessary to close gaps and to raise the quality of the finished sequence. The completed genome sequences of B. phymatum STM815T contain 115,487 reads, achieving an average of 11.2-fold sequence coverage per base with an error rate less than 1 in 100,000.
Genome annotation
Automatic annotation was performed using the Integrated Microbial Genomes (IMG) platform [36] developed by the Joint Genome Institute, Walnut Creek, CA, USA [28]. Additional automatic and manual sequence annotation, as well as comparative genome analysis, were performed using the MicroScope platform at Genoscope [29]. Gene calling in Microscope resulted in the prediction of 940 additional protein coding sequences compared to the 7,496 detected at IMG. These additional genes were mostly short coding sequences considered as gene remnants or fragmented CDS, so that genome statistics presented here are from the IMG platform.
Genome properties
The genome includes two chromosomes and two plasmids, for a total size of 8,676,562 bp (62.3% GC content). Chromosome 1 is 3.48 Mb in size (63.0% GC), chromosome 2 is 2.69 Mb (62.3% GC), plasmid 1 is 1.90 Mb (62.0% GC) and plasmid 2 0.59 Mb (59.2% GC). For chromosomes 1 and 2, 3,140 and 2,358 genes were predicted, respectively. For plasmid 1 and 2, 1,627 and 449 genes were predicted, respectively. A total 7,496 of protein coding genes were predicted, of which 5,601 were assigned to a putative function with the remaining annotated as hypothetical proteins. 5,630 protein coding genes belong to COG families in this genome. The properties and the statistics of the genome are summarized in Tables 4-6, and circular maps of each replicon are shown in Figure 3 (chromosomes) and Figure 4 (plasmids). Plasmid 2 was identified as the symbiotic plasmid of STM815, as it carried nod, nif and fix genes directly involved in symbiosis as well as several other genes coding for proteins indirectly linked to symbiotic interactions with plants. Among them were found genes coding for the biosynthesis of phytohormones such as indol acetic acid (iaaHM), ACC deaminase (acdS), and genes involved in the biosynthesis of rhizobitoxine (rtxAC-like). A Type 4 secretion system was also identified on this plasmid, while no type 3 system could be detected in the whole genome.
Table 4. Summary of genome: two chromosomes and two plasmids.
| Label | Size (Mb) | Topology | INSDC identifier | Refseq identifier |
|---|---|---|---|---|
| Chromosome 1 | 3.479189 | Circular | NC_010622.1 | CP001043.1 |
| Chromosome 2 | 2.697376 | Circular | NC_010627.1 | CP001044.1 |
| Plasmid 1 | 1.904895 | Circular | NC_010623.1 | CP001045.1 |
| Plasmid 2 | 0.595110 | Circular | NC_010625.1 | CP001046.1 |
Table 6. Number of genes associated with the 25 general COG functional categories.
| Code | Value | %agea | Description |
|---|---|---|---|
| J | 195 | 3.02 | Translation |
| A | 1 | 0.02 | RNA processing and modification |
| K | 643 | 10.00 | Transcription |
| L | 235 | 3.65 | Replication, recombination and repair |
| B | 2 | 0.03 | Chromatin structure and dynamics |
| D | 37 | 0.58 | Cell cycle control, mitosis and meiosis |
| V | 68 | 1.06 | Defense mechanisms |
| T | 397 | 6.17 | Signal transduction mechanisms |
| M | 396 | 6.16 | Cell wall/membrane biogenesis |
| N | 113 | 1.76 | Cell motility |
| W | 1 | 0.02 | Extracellular structures |
| U | 139 | 2.16 | Intracellular trafficking and secretion |
| O | 213 | 3.31 | Posttranslational modification, protein turnover, chaperones |
| C | 503 | 7.82 | Energy production and conversion |
| G | 486 | 7.55 | Carbohydrate transport and metabolism |
| E | 663 | 10.31 | Amino acid transport and metabolism |
| F | 101 | 1.57 | Nucleotide transport and metabolism |
| H | 223 | 3.47 | Coenzyme transport and metabolism |
| I | 290 | 4.51 | Lipid transport and metabolism |
| P | 287 | 4.46 | Inorganic ion transport and metabolism |
| Q | 200 | 3.11 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 719 | 11.18 | General function prediction only |
| S | 522 | 8.11 | Function unknown |
| - | 1944 | 25.67 | Not in COGs |
a) The total is based on the total number of protein coding genes in the annotated genome.
Figure 3.
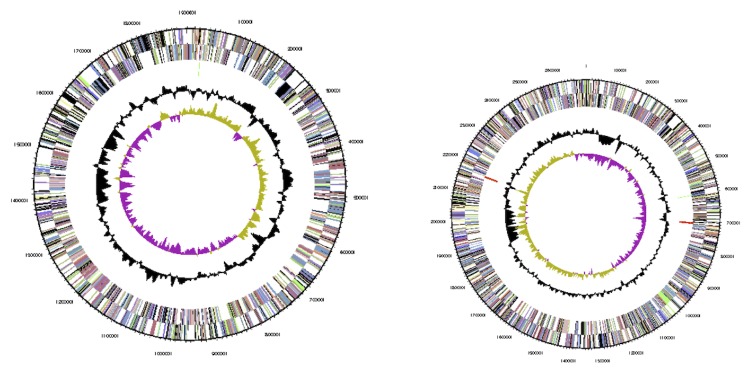
Circular maps of Chromosome 1 (left) and Chromosome 2 (right) of B. phymatum STM815T. From outside to center: Genes on forward strand (color by COG categories as denoted by the IMG platform), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, sRNAs red, other RNAs black), GC content, GC skew. Replicons are not drawn to scale.
Figure 4.
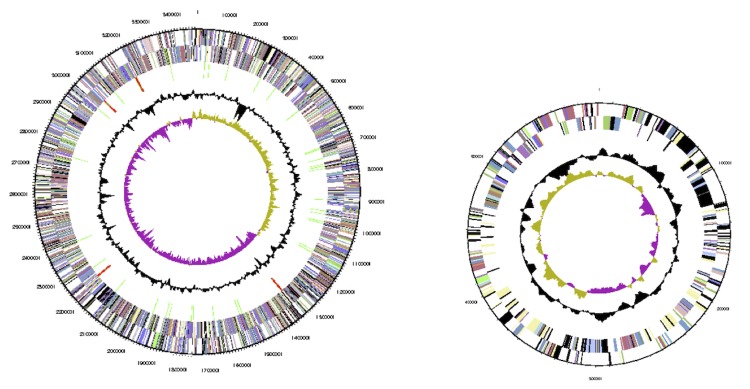
Circular maps of Plasmid 1 (left) and Plasmid 2 (right) of B. phymatum STM815T. From outside to center: Genes on forward strand (color by COG categories as denoted by the IMG platform), Genes on reverse strand (color by COG categories), RNA genes (tRNAs green, sRNAs red, other RNAs black), GC content, GC skew. Replicons are not drawn to scale.
Table 5. Nucleotide content and gene count levels of the genome.
| Attribute | Value | % of totala |
|---|---|---|
| Genome size (bp) | 8676562 | 100.00% |
| DNA coding region (bp) | 7328930 | 84.47% |
| DNA G+C content (bp) | 5404839 | 62.29% |
| Total genesb | 7574 | 100.00% |
| RNA genes | 78 | 1.03% |
| Protein-coding genes | 7496 | 98.93% |
| Genes assigned to COGs | 5630 | 74.33% |
| Genes with signal peptides | 701 | 9.26% |
| Genes with transmembrane helices | 1709 | 22.56% |
a) The total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome.
b) Also includes 39 pseudogenes.
Comparison of Burkholderia phymatum STM815T with other fully sequenced genomes of Burkholderia
Venn diagram (family number)
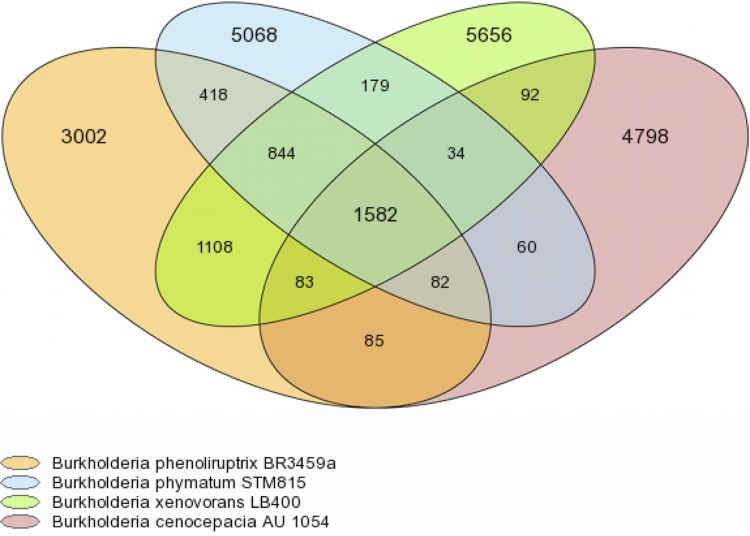
Gene families specific to, or shared by, Burkholderia phymatum STM815T and 3 other Burkholderia species, were determined using MICFAM [Figure 5]. This tool is based on MicroScope gene families [39] which are computed using an algorithm implemented in the SiLiX software [40]: a single linkage clustering algorithm of homologous genes sharing an amino-acid alignment coverage and identity above a defined threshold. This algorithm operates on the “The friends of my friends are my friends” principle of gene comparison. If two genes are homologous, they are clustered. Moreover, if one of the genes is already clustered with another one, the three genes are clustered into the same MICFAM.
Figure 5.
B. phymatum STM815T was compared to 3 others Burkholderia strains from similar and different ecological niches: a legume symbiont (B. phenoliruptrix BR3459a, a Mimosa flocculosa nodule symbiont from Brazil [37,38]; a soil bacterium (B. xenovorans LB400) and a human opportunistic pathogen (B. cenocepacia AU1054). The core genomes of all four bacteria yielded 1,582 gene families. Each bacterium had more gene families specific to its species, (from 3,002 to 5,656 depending on strain) than shared ones (1,582 core gene families). There were 418 gene families specific to the two Mimosa symbionts (STM815 and BR3459a), including symbiosis-related genes (nod genes) and nitrogen fixation genes (nif, fix), glutamine transporters, biosynthesis genes of the phytohormone indol acetic acid (IAA), and hydrogenase genes (hup, hyp).
Conclusion
Burkholderia phymatum STM815T possesses a large genome composed of two chromosomes and two plasmids, one of which encodes the symbiotic functions. Further studies on the genome of this bacterium will help elucidate the high nodulation competitiveness [41], broad host range and symbiotic efficiency of this strain.
Acknowledgements
This work was performed under the auspices of the US Department of Energy's Office of Science, Biological and Environmental Research Program and the University of California, Lawrence Livermore National Laboratory under Contract No. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under Contract No. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract No. DE-AC02-06NA25396, and French National Agency of Research (ANR) (Project “BETASYM” ANR-09-JCJC-0046).
Abbreviations:
- EMBL- European Molecular Biology Laboratory
NCBI- National Center for Biotechnology Information (Bethesda, MD, USA), RDP- Ribosomal Database Project (East Lansing, MI, USA), COG- Cluster of Orthologous Genes
References
- 1.Moulin L. Munive a, Dreyfus B, Boivin-Masson C. Nodulation of legumes by members of the beta-subclass of Proteobacteria. Nature 2001; 411:948-950 10.1038/35082070 [DOI] [PubMed] [Google Scholar]
- 2.Chen WM, Laevens S, Lee TM, Coenye T, De Vos P, Mergeay M, Vandamme P. Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int J Syst Evol Microbiol 2001; 51:1729-1735 10.1099/00207713-51-5-1729 [DOI] [PubMed] [Google Scholar]
- 3.Gyaneshwar P, Hirsch AM, Moulin L, Chen WM, Elliott GN, Bontemps C, Estrada-de Los Santos P, Gross E, Dos Reis FB, Sprent JI. Legume-nodulating betaproteobacteria: diversity, host range, and future prospects. Molecular plant-microbe interactions. Mol Plant Microbe Interact 2011; 24:1276-1288 10.1094/MPMI-06-11-0172 [DOI] [PubMed] [Google Scholar]
- 4.Bournaud C, de Faria SM, dos Santos JM, Tisseyre P, Silva M, Chaintreuil C, Gross E, James EK, Prin Y, Moulin L. Burkholderia species are the most common and preferred nodulating symbionts of the piptadenia group (tribe mimoseae). PLoS ONE 2013; 8:e63478 10.1371/journal.pone.0063478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bontemps C, Elliott GN, Simon MF, Dos Reis Júnior FB, Gross E, Lawton RC, Neto NE, de Fátima Loureiro M, De Faria SM, Sprent JI, et al. Burkholderia species are ancient symbionts of legumes. Mol Ecol 2010; 19:44-52 10.1111/j.1365-294X.2009.04458.x [DOI] [PubMed] [Google Scholar]
- 6.dos Reis FB, Jr, Simon MF, Gross E, Boddey RM, Elliott GN, Neto NE, Loureiro Mde F, de Queiroz LP, Scotti MR, Chen WM, et al. Nodulation and nitrogen fixation by Mimosa spp. in the Cerrado and Caatinga biomes of Brazil. New Phytol 2010; 186:934-946 10.1111/j.1469-8137.2010.03267.x [DOI] [PubMed] [Google Scholar]
- 7.Vincent J. A manual for the pratical study of root-nodule bacteria. I.B.P. Han. Ltd, Oxford: Blackwell Scientific Publications; 1970:vol 15. [Google Scholar]
- 8.Vandamme P, Goris J, Chen WM, De Vos P, Willems A. Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst Appl Microbiol 2002; 25:507-512 10.1078/07232020260517634 [DOI] [PubMed] [Google Scholar]
- 9.Elliott GN, Chen WM, Chou JH, Wang HC, Sheu SY, Perin L, Reis VM, Moulin L, Simon MF, Bontemps C, et al. Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol 2007; 173:168-180 10.1111/j.1469-8137.2006.01894.x [DOI] [PubMed] [Google Scholar]
- 10.Mishra RP, Tisseyre P, Melkonian R, Chaintreuil C, Miché L, Klonowska A, Gonzalez S, Bena G, Laguerre G, Moulin L, et al. Genetic diversity of Mimosa pudica rhizobial symbionts in soils of French Guiana: Investigating the origin and diversity of Burkholderia phymatum and other beta-rhizobia. FEMS Microbiol Ecol 2012; 79:487-503 10.1111/j.1574-6941.2011.01235.x [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Wei S, Wang F, James EK, Guo X, Zagar C, Xia LG, Dong X, Wang YP. Burkholderia and Cupriavidus spp. are the preferred symbionts of Mimosa spp. in southern China. FEMS Microbiol Ecol 2012; 80:417-426 10.1111/j.1574-6941.2012.01310.x [DOI] [PubMed] [Google Scholar]
- 12.Gehlot HS, Tak N, Kaushik M, Mitra S, Chen WM, Poweleit N, Panwar D, Poonar N, Parihar R, Tak A, et al. An invasive Mimosa in India does not adopt the symbionts of its native relatives. Ann Bot (Lond) 2013; 112:179-196 10.1093/aob/mct112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 2009; 37:D141-D145 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 2008; 36:W465-W469 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1. [Google Scholar]
- 18.Validation List No 107. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 2006; 56:1-6 10.1099/ijs.0.64188-0 [DOI] [PubMed] [Google Scholar]
- 19.Garrity GM, Bell JA, Lilburn T. Class II. Betaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 575. [Google Scholar]
- 20.Garrity GM, Bell JA, Lilburn T. Order I. Burkholderiales ord. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 575. [Google Scholar]
- 21.Garrity GM, Bell JA, Lilburn T. Family I. Burkholderiaceae fam. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 575. [Google Scholar]
- 22.Validation of the publication of new names and new combinations previously effectively published outside the IJSB. List No. 45. Int J Syst Bacteriol 1993; 43:398-399 10.1099/00207713-43-2-398 [DOI] [PubMed] [Google Scholar]
- 23.Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol Immunol 1992; 36:1251-1275 10.1111/j.1348-0421.1992.tb02129.x [DOI] [PubMed] [Google Scholar]
- 24.Gillis M, Van TV, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez MP. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol 1995; 45:274-289 10.1099/00207713-45-2-274 [DOI] [Google Scholar]
- 25.Validation of publication of new names and new combinations previously effectively published outside the IJSEM. Validation List No. 91. Int J Syst Evol Microbiol 2003; 53:627-628 10.1099/ijs.0.02771-0 [DOI] [PubMed] [Google Scholar]
- 26.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jobson RW, Luckow M. Phylogenetic Study of the Genus Piptadenia (Mimosoideae: Leguminosae) using Plastid trnL-F and trnK / matK Sequence Data. Syst Bot 2007; 32:569-575 10.1600/036364407782250544 [DOI] [Google Scholar]
- 28.Mavromatis K, Chu K, Ivanova N, Hooper SD, Markowitz VM, Kyrpides NC. Gene context analysis in the Integrated Microbial Genomes (IMG) data management system. PLoS ONE 2009; 4:e7979 10.1371/journal.pone.0007979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vallenet D, Engelen S, Mornico D, et al. MicroScope: a platform for microbial genome annotation and comparative genomics. Database : the journal of biological databases and curation 2009;2009:bap021. [DOI] [PMC free article] [PubMed]
- 30.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DOE Joint Genome Institute user home. http://my.jgi.doe.gov/general/index.html
- 32.DOE Joint Genome Institute http://www.jgi.doe.gov
- 33.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 1998; 8:186-194 10.1101/gr.8.3.175 [DOI] [PubMed] [Google Scholar]
- 34.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 1998; 8:175-185 10.1101/gr.8.3.175 [DOI] [PubMed] [Google Scholar]
- 35.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res 1998; 8:195-202 10.1101/gr.8.3.195 [DOI] [PubMed] [Google Scholar]
- 36.Integrated Microbial Genomes (IMG) platform. http://img.jgi.doe.gov
- 37.de Oliveira Cunha C, Goda Zuleta LF, Paula de Almeida LG, Prioli Ciapina L, Lustrino Borges W, Pitard RM, Baldani JI, Straliotto R, de Faria SM, Hungria M, et al. Complete Genome Sequence of Burkholderia phenoliruptrix BR3459a (CLA1), a Heat-Tolerant, Nitrogen-Fixing Symbiont of Mimosa flocculosa. J Bacteriol 2012; 194:6675-6676 10.1128/JB.01821-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen WM, de Faria SM, Straliotto R, Pitard RM, Simões-Araùjo JL, Chou JH, Chou YJ, Barrios E, Prescott AR, Elliott GN, et al. Proof that Burkholderia Strains Form Effective Symbioses with Legumes: a Study of Novel Mimosa-Nodulating Strains from South America. Appl Environ Microbiol 2005; 71:7461-7471 10.1128/AEM.71.11.7461-7471.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miele V, Penel S, Duret L. Ultra-fast sequence clustering from similarity networks with SiLiX. BMC Bioinformatics 2011; 12:116 10.1186/1471-2105-12-116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.SiLiX http://lbbe.univ-lyon1.fr/-SiLiX-.html
- 41.Melkonian R, Moulin L, Béna G, Tisseyre P, Chaintreuil C, Heulin K, Rezkallah N, Klonowska A, Gonzalez S, Simon M, et al. The geographical patterns of symbiont diversity in the invasive legume Mimosa pudica can be explained by the competitiveness of its symbionts and by the host genotype. Environ Microbiol. 2013 doi: 10.1111/1462-2920.12286. [DOI] [PubMed] [Google Scholar]