Abstract
Rubrobacter radiotolerans strain RSPS-4 is a slightly thermophilic member of the phylum “Actinobacteria” isolated from a hot spring in São Pedro do Sul, Portugal. This aerobic and halotolerant bacterium is also extremely resistant to gamma and UV radiation, which are the main reasons for the interest in sequencing its genome. Here, we present the complete genome sequence of strain RSPS-4 as well as its assembly and annotation. We also compare the gene sequence of this organism with that of the type strain of the species R. radiotolerans isolated from a hot spring in Japan. The genome of strain RSPS-4 comprises one circular chromosome of 2,875,491 bp with a G+C content of 66.91%, and 3 circular plasmids of 190,889 bp, 149,806 bp and 51,047 bp, harboring 3,214 predicted protein coding genes, 46 tRNA genes and a single rRNA operon.
Keywords: Rubrobacter radiotolerans, radiation-resistance, gram positive, genome sequence, 454 sequencing
Introduction
Rubrobacter radiotolerans strain RSPS-4 is a slightly thermophilic actinobaterium isolated from a hot spring in central Portugal [1]. Species of the genus Rubrobacter are extremely resistant to ionizing radiation [1-4]. The type strain of Rubrobacter radiotolerans P-1T (DSM 5868T, JCM 2153T) from Japan and strain RSPS-4 are two of the most resistant organisms to ionizing radiation, more so than the archetypal Deinococcus rediodurans strain R1 (DSM 20539T, JCM 16871T) [5]. Both R. radiotolerans strains have a sigmoid shaped survival curve on a dose-response irradiation curve up to 25 kGy and a shoulder dose of 5.7 kGy, with a 37% survival of 7.6 and 9.0 for the type strain of R. radiotolerans and strain RSPS-4 [1].
Microbial resistance to ionizing radiation is rather intriguing since gamma radiation is restricted to locations contaminated with nuclear waste, while natural environments with high doses of gamma radiation are not known in the biosphere. One hypothesis is that radiation resistance is related to desiccation resistance in some organisms that do not produce spores [6]. While the species of Deinococcus are the preeminent extremely radiation-resistant organisms [7], other radiation-resistant microbes are isolated from very diverse environments and are classified in taxa that belong to different phyla and domains, suggesting that this characteristic evolved independently and in response to other environmental challenges, possibly desiccation or reactive oxidative stress (ROS). Radiation-resistant organisms have been described from archaea such as Thermococcus gammatolerans [8] to bacteria, such as Deinococcus [9] and Truepera [10], both of which belong to the phylum Deinococcus-Thermus, Actinobacteria of the genera Rubrobacter [1] and Kineococcus [11], Proteobacteria of the genera Methylobacterium [12] and Acinetobacter, Sphingobacteria of the genus Hymenobacter [9] and cyanobacteria of the genus Chroococcidiopsis [13].
Deinococcus radiodurans has been the most studied bacterium for radiation resistance mechanisms [14-20], however, no single key factor has been identified to explain its resistance and it is now hypothesized that the ability to recover from high doses of irradiation results from a combination of several mechanisms. DNA repair systems were the first studied resistance mechanisms, but D. radiodurans contains a set of enzymatic repair systems similar to radiation-sensitive bacteria such as Shewanella oneidensis and Pseudomonas putida [15,21] and such systems are thus insufficient per se to justify the almost error-free reassembly of the irradiated D. radiodurans genome [22]. Recent findings shifted the focus from DNA repair to antioxidant protein protection as a main resistance mechanism [15,23]. Several studies showed that irradiated cells maintained enzymes protected from oxidative stress and thus available for an efficient repair of DNA lesions [24-27].
Here, we describe the complete sequencing and annotation of the genome of R. radiotolerans RSPS-4, identify genes involved in the main repair pathways for DNA, the synthesis of compatible solutes that could be involved in the protection of enzymes and in the response to oxidative stress, and analyze the differences between strain RSPS-4 and the type strain P-1T (DSM 5868T, JCM 2153T). This genome will help understand the genetic basis for radiation resistance mechanisms and provide data for broader comparative studies with other radiation resistance bacteria.
Classification and features
Rubrobacter belongs to the phylum “Actinobacteria” formerly known as the high G+C Gram-positive bacteria. The genus comprises four species, three of which, were isolated from thermal environments, namely Rubrobacter xylanophilus recovered from the thermally polluted runoff from a carpet factory in the United Kingdom [2]; Rubrobacter radiotolerans [3] isolated from a hot spring in Japan after gamma-irradiation of the water sample [28] and Rubrobacter taiwanensis isolated from non-irradiated samples from Lu-Shan hot spring in the central region of Taiwan [4]. The mesophilic species Rubrobacter bracarensis, was isolated from a green biofilm covering the biodeteriorated interior walls of a Church at Vilar de Frades, in Portugal [29], but is not known to be gamma-, UV- or desiccation-resistant.
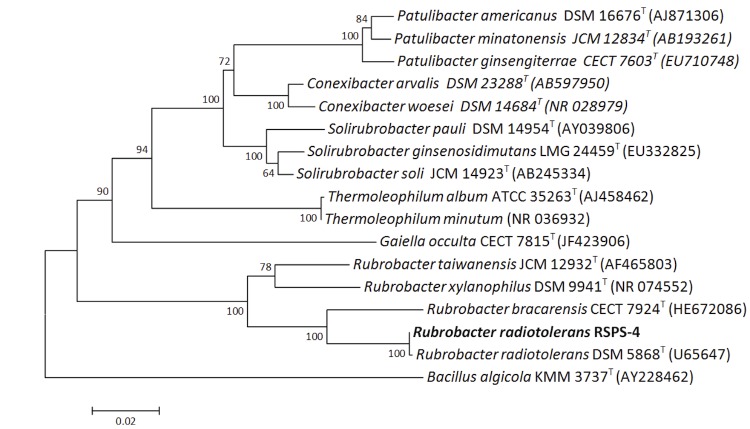
Phylogenetic analysis of 16S rRNA gene sequences indicates that the species of the genus Rubrobacter belong to the monogeneric family Rubrobacteraceae (Figure 1), which along with the genera of the families Solirubrobacteriaceae, Thermoleophilaceae, Conexibacteraceae, Patulibacteraceae and Gaiellaceae form deep-branching lineages of the subclass Rubrobacteridae of the phylum “Actinobacteria” [34]. The species R. radiotolerans is most closely related to R. bracarensis [29].
Figure 1.
Phylogenetic tree showing the position of Rubrobacter radiotolerans strain RSPS-4 with other organisms within the subclass Rubrobacteridae. The tree was inferred from 1,301 aligned characters of the 16S rRNA sequences of different species using the Neighbor-Joining method [30]; bootstrap values are based on 1,000 replicates [31]. The evolutionary distances were calculated using the Jukes-Cantor method [32]. Analysis was carried out with MEGA6 [33]. Bacillus algicola KMM 3737T (AY228462) was used as an outgroup.
The Rubrobacter radiotolerans strain RSPS-4 was isolated from a hot spring runoff at São Pedro do Sul, in central Portugal, after irradiation of the water sample with a cobalt-60 source at a dose of 8.06 kGy and a rate of 1.5kGyh-1 [1]. The hot spring runoff had a temperature of 50ºC and a pH of 8.9. The strain grew optimally at 45ºC in medium containing no added NaCl but was able to grow in medium containing 6% NaCl (w/v), degraded hide powder azure, gelatine, hippurate, arbutin and esculin, was cytochrome oxidase, catalase and β-galactosidase positive and produced nitrite from nitrate. The fatty acid composition of RSPS-4 was dominated by the unique 12-methyl-16:0 (70% of the total) with lower amounts of 4-methyl-18:0 (15.7%-17.8% of the total fatty acids). The 16S rRNA gene sequence of strain RSPS-4 is similar to the sequence of the type strain of R. radiotolerans DSM 5868T [1], only differing in 2 positions. Classification and general features of Rubrobacter radiotolerans RSPS-4 are shown in Table 1. The strain was deposited at the Spanish Type Culture Collection (CETC) with the code CECT 8386.
Table 1. Classification and general features of Rubrobacter radiotolerans RSPS-4 according to the MIGS recommendations [35].
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Domain Bacteria | TAS [36] | ||
| Phylum Actinobacteria | TAS [37] | ||
| Class Actinobacteria | TAS [38] | ||
| Subclass Rubrobacteridae | TAS [36-40] | ||
| Current classification | Order Rubrobacterales | TAS [38,40,41] | |
| Family Rubrobacteraceae | TAS [38-40] | ||
| Genus Rubrobacter | TAS [42,43] | ||
| Species Rubrobacter radiotolerans | TAS [42,43] | ||
| Strain RSPS-4 | |||
| Gram stain | Positive | TAS [1,3] | |
| Cell shape and pigmentation | Pleomorphic rod shaped; red pigmented, Pink colonies | TAS [1,3] | |
| Motility | Non-motile | ||
| Sporulation | Does not produce spores | TAS [3] | |
| Temperature range | 30-55ºC | TAS [1] | |
| Optimum temperature | 45ºC | TAS [1] | |
| Carbon source | Organic carbon compounds | ||
| Energy source | Organic carbon compounds | ||
| Terminal electron receptor | O2, nitrate | ||
| MIGS-6 | Habitat | Hot springs | TAS [1,3] |
| MIGS-6.3 | Salinity | <Than 0.2% NaCl | TAS [1] |
| MIGS-22 | Oxygen | Aerobic | TAS [1,3] |
| MIGS-15 | Biotic relationship | Free living | TAS [1,3] |
| MIGS-14 | Pathogenicity | None | |
| MIGS-4 | Geographic location | São Pedro do Sul | TAS [1] |
| MIGS-5 | Sample collection time | 1999 | TAS [1] |
| MIGS-4.1 | Latitude | 40° 44' 22.09'' N | |
| MIGS-4.2 | Longitude | 8° 5' 32.47'' W | TAS [1] |
| MIGS-4.3 | Depth | Surface hot spring |
Evidence codes - TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from of the Gene Ontology project [44].
Genome sequencing information
Genome Project History
R. radiotolerans strain RSPS-4 was selected for sequencing based on its extremely high resistance to ionizing radiation. The complete genome sequence is available from GenBank. Sequencing was performed at Roche Diagnostics GmbH, Penzberg, finishing and annotation were performed by Biocant. A summary of the project is shown in Table 2.
Table 2. Genome sequence project information.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing Quality | Finished |
| MIGS-28 | Libraries Used | 1 GS DNA Standard Library for 454 Pyrosequencing |
| MIGS-29 | Sequencing Platforms | 454 GS20 and 3500/3500XL Genetic Analyzer |
| MIGS-31.2 | Fold Coverage | 23× Pyrosequence and Sanger |
| MIGS-30 | Assemblers | 454 Roche Newbler Assembler, Phrap and Consed. |
| MIGS-32 | Gene Calling Method | Prodigal |
Growth conditions and DNA isolation
Rubrobacter radiotolerans strain RSPS-4 was grown on Thermus medium at 45ºC as described elsewhere [1]. DNA isolation was performed from a 1g pellet of a 24h, exponential growth phase culture using the Wizard Genomic DNA Purification Kit (Promega, Madison, USA) following the standard protocol, without modifications, for genomic DNA isolation of Gram positive bacteria, as recommended by the manufacturer.
Genome sequencing and assembly
The genome of strain RSPS-4 was sequenced in the GS20 sequencing platform (Roche -454 Life Sciences) at Roche Diagnostics GmbH, Penzberg, Germany. A total of 3 PicoTiterPlates generated 889,098 pyrosequencing reads with an average length of 106 bp. These were assembled into 70 contigs, with an N50 of 120,613, using the Newbler assembler (Roche). The Phred/Phrap/Consed software package [45,46] was used for sequence assembly and quality assessment in the subsequent finishing procedures. The gaps were closed by an Optimized Multiplex PCR approach [47], where 156 Sanger sequences were produced and added to the 454 reads to produce a hybrid assembly with Phrap. Together, 454 and Sanger sequences provided a 23× coverage of the genome.
Genome annotation
Structural and functional annotation has been performed using PGP (Prokaryotic Genome Prediction) an in-house developed pipeline. PGP used tRNAscan-SE [48], RNAMMer [49] and PILERCR [50] to predict non-coding genes and miscellaneous features. These features were then masked and the CDS predicted with Prodigal [51]. Furthermore, PGP automatically corrected the start position of each CDS based on the Geneprimp [52] algorithm. Functional annotation was carried out under PGP in InterProScan [53] against PFAM [54], TIGRFAM [55], Hamap [56], PIRSF [57], PRINTS [58], SMART [59], SUPERFAMILY [60], ProSite [61] databases and RPS-BLAST against COG database [62] The product name of each CDS was assigned using TIGRFAM, COG and PFAM annotation [63]. Those CDS that were not annotated with these databases were assigned as hypothetical proteins. The automatic annotation was followed by a round of manual curation to eliminate obvious overlaps by visualization of the obtained functional annotation with Artemis [64]. Possible mis-assemblies and mis-annotations were corrected by comparison to the genome of Rubrobacter radiotolerans DSM 5868T, assembly of 2011, produced by the US Department of Energy Joint Genome Institute. The genome sequence was completed in 2013 and presented for public access in 2014.
Nucleotide sequence accession numbers
The genome sequence for Rubrobacter radiotolerans strain RSPS-4 was deposited at GenBank under the accession number XXXXXXX (Chromosome), XXXXXXX (Plasmid 1), XXXXXX (Plasmid 2) and XXXXXX (Plasmid 3).
Genome properties
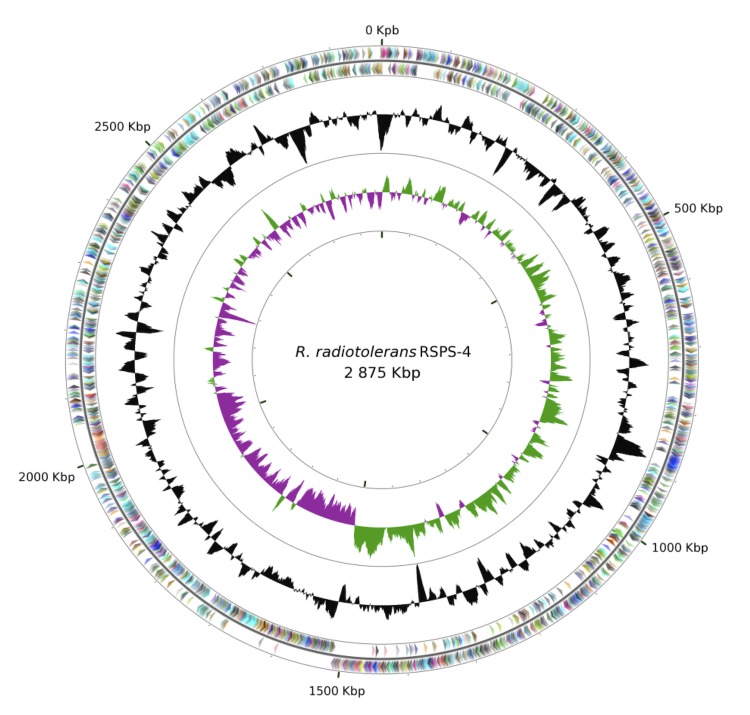
The Rubrobacter radiotolerans strain RSPS-4 genome consists of one circular chromosome of 2,875,491 bp (G+C content 66.9%) (Figure 2) and three plasmids, with 190,889 bp (G+C content 65.66%), 149,806 bp (G+C content 66.48%) and 51,047 bp (G+C content 63.17%) (Figure 3). Of the 3,214 protein-coding genes predicted, 2,772 are located in the chromosome, 191 in plasmid 1, 137 in plasmid 2, and 59 in plasmid 3. The majority of the protein-coding genes, (2,646, 82%) were assigned a putative function while the remainder were annotated as hypothetical proteins. 46 tRNAs encoding all the 20 amino acids and 1 rRNA operon were annotated. The replication origin was set to the first nucleotide of the dnaA gene. Genome properties are summarized in Table 3, Table S1 and Figure 2 and the distribution of genes into COGs functional categories is presented in Table 4.
Figure 2.
Circular representation of the chromosome of R. radiotolerans RSPS-4. From outside in, the outer two circles show genes on forward strand and reverse strand, colored by COG categories, the third circle shows the G+C% content plot (colored in black), and the inner circle the GC skew (green and purple). Graphics were created on the CGViewer Server [65].
Figure 3.
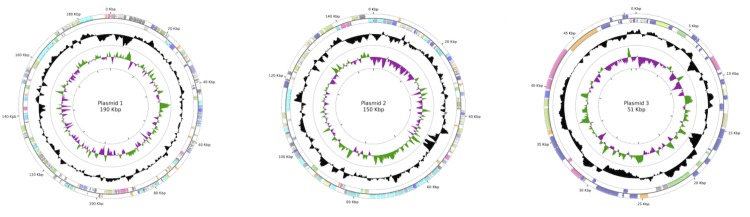
Circular representation of the plasmids of R. radiotolerans RSPS-4. From outside in, the outer two circles show genes on forward strand and reverse strand, colored by COG categories, the third circle shows the G+C% content plot (colored in black), and the inner circle the GC skew (green and purple). Graphics were created on the CGViewer Server [65].
Table 3. Genome Statistics.
| Attribute | Value | % of total |
|---|---|---|
| Genome Size (bp) | 3,267,233 | 100 |
| DNA Coding region (bp) | 2,993,158 | 91.6 |
| Chromosomal DNA G+C content (bp) | 2,181,219 | 66.7 |
| Extrachromossomal elements | 3 | |
| Total genes | 3260 | 100 |
| RNA genes | 46 | 1.42 |
| rRNA operons | 1 | |
| Protein-coding genes | 3214 | 98.6 |
| Genes with function prediction | 2646 | 81.7 |
| Genes assigned to COGs | 2565 | 79.1 |
| CRISPR repeats | 2 |
Table 4. Number of genes associated with the general COG functional categories.
| Code | Value | %age | Description |
|---|---|---|---|
| J | 139 | 4.46 | Translation, ribosomal structure and biogenesis |
| A | - | - | RNA processing and modification |
| K | 143 | 4.59 | Transcription |
| L | 118 | 3.79 | Replication, recombination and repair |
| B | 3 | 0.10 | Chromatin structure and dynamics |
| D | 30 | 0.96 | Cell cycle control, mitosis and meiosis |
| Y | - | - | Nuclear structure |
| V | 46 | 1.48 | Defense mechanisms |
| T | 121 | 3.89 | Signal transduction mechanisms |
| M | 143 | 4.59 | Cell wall/membrane biogenesis |
| N | 19 | 0.61 | Cell motility |
| Z | - | - | Cytoskeleton |
| W | - | - | Extracellular structures |
| U | 26 | 0.83 | Intracellular trafficking and secretion |
| O | 88 | 2.83 | Posttranslational modification, protein turnover, chaperones |
| C | 225 | 7.23 | Energy production and conversion |
| G | 162 | 5.20 | Carbohydrate transport and metabolism |
| E | 224 | 7.19 | Amino acid transport and metabolism |
| F | 75 | 2.41 | Nucleotide transport and metabolism |
| H | 131 | 4.21 | Coenzyme transport and metabolism |
| I | 128 | 4.11 | Lipid transport and metabolism |
| P | 136 | 4.37 | Inorganic ion transport and metabolism |
| Q | 37 | 1.19 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 326 | 10.47 | General function prediction only |
| S | 245 | 7.87 | Function unknown |
| - | 649 | 20.84 | Not in COGs |
Insights from the genome sequence
The genome sequence of strain RSPS-4 was analyzed for genes involved in recovery from ionizing radiation: DNA repair, oxidative stress response and compatible solute production. The key enzymes for the main DNA repair mechanisms were present in the genome, except for the non-homologous end joining, suggesting a shared set of DNA repair enzymes with other radiation-resistant bacteria. The pathways for oxidative stress response were also investigated but not all key enzymes were identified, suggesting that further studies are required in this bacterium to fully understand oxidative stress response and its role in radiation resistance. RSPS-4 possessed the four described pathways for trehalose production. Finally, the RSPS-4 genome sequence was compared to that of R. radiotolerans DSM 5868T type strain. The two strains practically shared the same nucleotide sequence, the same gene order, orientation and synteny, however strain DSM 5868T had extra segments, probably corresponding to putative prophages.
DNA repair and associated systems
Gamma radiation induces the production of highly reactive oxygen radicals (ROS) in cells by ionizing water from several macromolecules. In DNA, ROS induce base modifications, DNA single-strand and double-strand breaks [66]. The latter are considered the most dangerous for survival and most difficult to repair. The R. radiotolerans RSPS-4 genome was analyzed for DNA repair pathways by searching genes involved in homologous recombination, single strand annealing (SSA), extended synthesis-dependent strand annealing or non-homologous end joining [67,68].
RSPS-4 strain encodes a set of essential genes for homologous recombination. All genes for the RecFOR pathway were detected, recF (RradSPS_0004), recR (RradSPS_0466), recO (RradSPS_1511), and recJ (RradSPS_0780). RecA, the protein responsible for strand invasion and exchange, was encoded as a single copy (RradSPS_1428). Genes encoding the branch migration and resolution of Holliday junction proteins RuvA (RradSPS_1317), RuvB (RradSPS_1318), RuvC (RradSPS_1316), and RecG (RradSPS_1377) were detected, as were the homologs of the genes encoding the SbcD (mre11) (RradSPS_2355), and SbcC (Rad50) (RradSPS_2356) proteins. The gene coding for the RecX (RradSPS_1429) protein, which acts as a negative regulator of RecA, was also present.
The genes encoding proteins involved in extended synthesis dependent strand annealing were also present in the strain RSPS-4 genome: PolA (RradSPS_0202), which participates in the initial DNA synthesis-step, RecA, that ensures the maturation of the linear intermediates into full-size circular chromosomes and RadA (RradSPS_1965), a protein involved in the stabilization or processing of branched DNA molecules [69] and in the priming step in DNA strand elongation [66].
Homologs of the Ku-like complex proteins and DNA ligase IV complex, the most relevant proteins involved in non-homologous end-joining pathway were not found in RSPS-4 strain, suggesting that this repair pathway may not be functional.
Mismatch repair (MMR) is probably functional in RSPS-4 with gene copies of mutL (RradSPS_0036 and _0159), and mutS (RradSPS_0158). The absence of the gene encoding the MutH endonuclease, which participates in the recognition of GATC methylated sequences discriminating the DNA strand to be repaired, should not compromise the efficiency of this pathway, as this protein is also absent from several bacterial genomes [70]. No dcm or dam site specific methylase genes were detected, suggesting that strain RSPS-4 uses different proteins for strand recognition and incision to complete MMR. Homologs of xseA and xseB, encoding the subunits of a MMR exonuclease, were not detected.
A particular feature of the RSPS-4 DNA repair pathways was the absence of the LexA autoprotease, the repressor that controls the expression of the SOS regulon in E. coli. The gene could not be identified in the RSPS-4 genome, based on a nucleotide sequence search using the two lexA genes described for the type strain DSM 5868T. Furthermore, the absence of this gene was validated by PCR amplification, using the genes of R. radiotolerans strain DSM 5868T as template to design specific lexA primers. The absence of this gene has been observed in bacteria from many genera, such as Mycoplasma, Chlamydia, Borrelia, Helicobacter, Coxiella, and the Cyanobacteria [71-73], and suggests alternative mechanisms to repair DNA damage in bacteria could be employed. In Lactococcus lactis, a LexA–independent SOS response is led by the HdiR protein [74]. Moreover, other mechanisms may act alone or synergistically with the SOS response in DNA damage repair. In the radio-resistant D. radiodurans, the two LexA homologs undergo RecA-dependent cleavage after DNA damage, however, no regulon under the control of LexA1 or LexA2 proteins has been identified to date [66]. In this organism, recA induction following gamma irradiation is not controlled by LexA1 or LexA2 but depends on a Deinococcus specific regulatory protein IrrE, also designated PprI, a positive effector that enhances the expression of some DNA repair genes following exposure to radiation [66].
In deinococci, additional genes have been identified for DNA repair of irradiation damage, however these were absent from strain RSPS-4. No homologs of irrE or pprA were found [75], suggesting that Rubrobacter uses other pathways for the same mechanism. The ddrA, ddrB, ddrC and ddrD genes [76] are also absent. Their absence from the Kineococcus radiotolerans genome [77] suggests their specificity to the deinococci.
Reactive oxygen species detoxification
Reactive oxygen species (ROS) including hydrogen peroxide, superoxide and hydroxyl radicals are toxic to cells due to their ability to damage DNA and specially proteins containing iron-sulfur clusters or sculpture atoms. In order to prevent the damage induced by ROS, cells have several mechanisms of response to oxidative stress. Strain RSPS-4 has a single gene for a manganese containing catalase (RradSPS_2184), whilst katA, katE and katG were not detected [15,78]. Superoxide dismutase was also encoded as a single gene identified as sodA (RradSPS_0327), and was also dependent on manganese [79]. Peroxiredoxins, which reduce H2O2 to water, were encoded by six copies of alkyl hydroperoxide reductase subunit C/ Thiol specific antioxidant (AhpC/TSA) (RradSPS_0148, _0515,_ 988, _1124, _2530, _2650) while peroxidases such as BsaA were not detected. A single gene for thioredoxin reductase trxR (RradSPS_0074) [78] was detected in the genome, as well as a gene for the trxA (RradSPS_0519) and two for grxA redoxins (RradSPS_1230 and _3087), involved in redox balance [80]. A single gene for thiosulfate transferase (RradSPS_0885) [78] was also found in the genome. Although regulatory genes of the oxyR family were not detected, lysR (RradSPS_1021, _1060, _1715, _1856 and _2024), which activates the transcription of genes involved in peroxide metabolism and protection in D. radiodurans (katG, ahpC, ahpF, and dps) were detected [80].
Manganese has been proposed to be an important mechanism for oxidative stress response. This ion is suggested to protect cytosolic proteins from ROS by replacing Fe2+ and other divalent cations such as Mg2+ or Cu2+ as cofactors, and by forming ROS-scavenging complexes with various metabolites that preserve enzyme quaternary structures [23]. In strain RSPS-4, the catalase and the superoxide dismutase genes are predicted to encode manganese-containing enzymes and two ABC-type Mn2+/Zn2+ transport systems were present in the genome sequence (RradSPS_1136, _1137 and _1138; RradSPS_2222, _2223 and _2224) [80].
Compatible solutes and stress protection
Two compatible solutes, namely trehalose and mannosylglycerate, accumulate to high levels in R. xylanophilus and R. radiotolerans RSPS-4 [81,82], but the accumulation of these osmolytes is not dependent on the salt concentration of the growth medium as it is in so many halotorerant and halophilic organisms. Instead, the accumulation of these compatible solutes is constitutive indicating that the accumulation of these compatible solutes is most likely in response to any of several stress conditions that may affect the survival of the cells [81]. The genome sequence of R. radiotolerans RSPS-4 and R. radiotolerans DSM 5868T possess many of the key genes for the synthesis of trehalose and mannosylglycerate. Strain RSPS-4 and DSM 5868T possess identical mannosylphospglycerate synthases that are homologs of R. xylanophilus (mpgS, EU847586.1) (RradSPS_0500; Rrad_0501). The synthesis of trehalose could proceed via four pathways namely through the TpS/TpP (RradSPS_0264 and _0265; Rrad_0265 and _0266), the TreS (RradSPS_0192; Rrad_0194), the TreT (RradSPS_0753; Rrad_0753) or the TreY/TreZ pathways (RradSPS_0196 and _0195; Rrad_0198 and _0197), homologs of which are found in both R. radiotolerans strains. The TpS/TpP and the TreT pathways were examined in R. xylanophilus and both were involved in the synthesis of trehalose [82]. Trehalose is considered to be a solute involved in the protection of many biological structures under different stress conditions and is considered to be especially important under extreme desiccation [83].
Comparisons with other fully sequenced genomes
The genome sequence of R. radiotolerans RSPS-4 was compared to the genome of Rubrobacter radiotolerans DSM 5868T. The genome structure was similar in the two strains, a major chromosome and 3 plasmids. However, the genome of strain DSM 5868T, with 3,398,074 bp and 3266 genes, is larger by 130,841 bp than the genome of the RSPS-4 genome. Alignment of the two genome nucleotide sequences with the tool MISHIMA [84] revealed the two genome sequences practically shared the same nucleotide sequence, the same order, orientation and synteny. The main differences resided in chromosome 1 and plasmid 1. In chromosome 1, strain DSM 5868T had an extra 87,086 bp segment of 70 genes (Table S2). This segment encodes a putative prophage, flanked by an integrase in the first position (Rrad_2453), and transposases at the other end (Rrad_2527 and _2528). Plasmid 1 of DSM 5868T contained a segment of 55 genes that was only observed in this strain (Table S3). This 56,691 bp segment was flanked at one end by a resolvase (Rrad_2844) but no enzyme related to mobile elements was observed at the other end. Strain RSPS-4 also had a genome segment that was absent from the DSM 5868T strain. This was a segment of 11 genes, corresponding to 11,639 bp, in plasmid 1 (Table S4). This segment harbors two transposases (RradSPS_2871 and _2872) and a recombinase (RradSPS_2873), although they are positioned in the middle of the segment and may be the remnant of the insertion of mobile elements.
Apart from the proteins related to mobile elements, the two additional segments present in the type strain have several transcriptional regulators (Table S2 and S3). LexA was present in two copies, one in the chromosomal segment (Rrad_2461) and the other in plasmid 1 (Rrad_3065). The location of this gene within a mobile region may explain its absence from the RSPS-4 strain. Other transcriptional regulators were identified in these genome segments; the transcriptional regulator of the XRE family (Rrad_2455 and _3061), which acts as a repressor–like protein of several phages [85], ArsR (Rrad_2469), which participates in the stress response to heavy metals [86] and LuxR (Rrad_2505 and _2856) and TetR (Rrad_2866 and _2871), which are involved in diverse pathways such as those encoding virulence factors and antibiotics biosynthesis [87], the control of multidrug efflux pumps, and the response to osmotic stress or toxic chemicals [88]. The extra segments present in DSM 5868T genome were populated by hypothetical or conserved hypothetical genes, 32 in the chromosome 1, and 29 in plasmid 1 (Table S2 and S3). The segments additionally encoded transporters involved in heavy metal homeostasis (such as heavy metal translocating P-type ATPase, (Rrad_2468)), proteins involved in lipid biosynthesis (such as 4'-phosphopantetheinyl transferase), proteins involved in the type II/III secretion system (Rrad_2515, _2516 and _2853) and several enzymes involved in the respiratory chain (Rrad_2470-2477) (Table S2). Most genes in the extra segments of type strain were actually also encoded in other regions of the genome, suggesting that the duplication of certain genes may provide the DSM 5868T strain with increased ability to respond to stress conditions.
Conclusion
The complete genome sequence and annotation of Rubrobacter radiotolerans strain RSPS-4 was hereby presented. The genome comprises 1 chromosome and 3 circular plasmids which together represent an organism of approximately 3.2 Mb. The genome sequence encodes for several key genes involved in the mechanisms of DNA repair, in response to oxidative stress and in the production of compatible solutes. However, some of the described pathways are not complete and await further studies to fully understand the mechanisms behind the RSPS-4 extreme resistance to radiation.
References
- 1.Ferreira AC, Nobre MF, Moore E, Rainey FA, Battista JR, da Costa MS. Characterization and radiation resistance of new isolates of Rubrobacter radiotolerans and Rubrobacter xylanophilus. Extremophiles 1999; 3:235-238 10.1007/s007920050121 [DOI] [PubMed] [Google Scholar]
- 2.Carreto L, Moore E, Nobre MF, Wait RRPW, Sharp RJ, Da Costa MS. Rubrobacter xylanophilus sp. nov., a New Thermophilic Species Isolated from a Thermally Polluted Effluent. Int J Syst Bacteriol 1996; 46:460-465 10.1099/00207713-46-2-460 [DOI] [Google Scholar]
- 3.Suzuki K, Collins MD, Iijima E, Komagata K. Chemotaxonomic characterization of a radiotolerant bacterium, Arthrobacter radiotolerans: Description of Rubrobacter radiotolerans gen. nov., comb. nov. FEMS Microbiol Lett 1988; 52:33-39 10.1111/j.1574-6968.1988.tb02568.x [DOI] [Google Scholar]
- 4.Chen MY, Wu SH, Lin GH, Lu CP, Lin YT, Chang WC, Tsay SS. Rubrobacter taiwanensis sp. nov., a novel thermophilic, radiation-resistant species isolated from hot springs. Int J Syst Evol Microbiol 2004; 54:1849-1855 10.1099/ijs.0.63109-0 [DOI] [PubMed] [Google Scholar]
- 5.Brooks BW, Murray RGE. Nomenclature for “Micrococcus radiodurans” and Other Radiation-Resistant Cocci: Deinococcaceae fam. nov. and Deinococcus gen. nov., Including Five Species. Int J Syst Bacteriol 1981; 31:353-360 10.1099/00207713-31-3-353 [DOI] [Google Scholar]
- 6.Mattimore V, Battista JR. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J Bacteriol 1996; 178:633-637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Battista JR, Rainey FA. The genus Deinococcus In: Bergey’s Manual of Systematic Bacteriology (Boone, D.R. and Castenholz, R.W, Eds).Vol 1. 2nd ed. New york: Springer; 2001:369–403. [Google Scholar]
- 8.Jolivet E, L’Haridon S, Corre E, Forterre P, Prieur D. Thermococcus gammatolerans sp. nov., a hyperthermophilic archaeon from a deep-sea hydrothermal vent that resists ionizing radiation. Int J Syst Evol Microbiol 2003; 53:847-851 10.1099/ijs.0.02503-0 [DOI] [PubMed] [Google Scholar]
- 9.Anderson AW, Nordon HC, Cain RF, Parrish G, Duggan G. Studies on a radio-resistant micrococcus. I. Isolation, morphology, cultural characteristics, and resistance to gamma radiation. Food Technol 1956; •••:575-578 [Google Scholar]
- 10.Albuquerque L, Simões C, Nobre MF, Pino NM, Battista JR, Silva MT, Rainey FA, da Costa MS. Truepera radiovictrix gen. nov., sp. nov., a new radiation resistant species and the proposal of Trueperaceae fam. nov. FEMS Microbiol Lett 2005; 247:161-169 10.1016/j.femsle.2005.05.002 [DOI] [PubMed] [Google Scholar]
- 11.Phillips RW, Wiegel J, Berry CJ, Fliermans C, Peacock AD, White DC, Shimkets LJ. Kineococcus radiotolerans sp. nov., a radiation-resistant, gram-positive bacterium. Int J Syst Evol Microbiol 2002; 52:933-938 10.1099/ijs.0.02029-0 [DOI] [PubMed] [Google Scholar]
- 12.Green PN, Bousfield IJ. Emendation of Methylobacterium Patt, Cole, and Hanson 1976; Methylobacterium rhodinum (Heumann 1962) comb. nov. corrig.; Methylobacterium radiotolerans (Ito and Iizuka 1971) comb. nov. corrig.; and Methylobacterium mesophilicum (Austin and Goodfellow 1979) comb. nov. Int J Syst Bacteriol 1983; 33:875-877 10.1099/00207713-33-4-875 [DOI] [Google Scholar]
- 13.Billi D, Friedmann EI, Hofer KG, Caiola MG, Ocampo-Friedmann R. Ionizing-Radiation Resistance in the Desiccation-Tolerant Cyanobacterium Chroococcidiopsis. Appl Environ Microbiol 2000; 66:1489-1492 10.1128/AEM.66.4.1489-1492.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White O, Eisen JA, Heidelberg JF, Hickey EK, Peterson JD, Dodson RJ, Haft DH, Gwinn ML, Nelson WC, Richardson DL, et al. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 1999; 286:1571-1577 10.1126/science.286.5444.1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makarova KS, Aravind L, Wolf YI, Tatusov RL, Minton KW, Koonin EV, Daly MJ. Genome of the Extremely Radiation-Resistant Bacterium Deinococcus radiodurans Viewed from the Perspective of Comparative Genomics. Microbiol Mol Biol Rev 2001; 65:44-79 10.1128/MMBR.65.1.44-79.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipton MS, Pasa-Tolic' L, Anderson GA, Anderson DJ, Auberry DL, Battista JR, Daly MJ, Fredrickson J, Hixson KK, Kostandarithes H, et al. Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc Natl Acad Sci USA 2002; 99:11049-11054 10.1073/pnas.172170199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox MM, Battista JR. Deinococcus radiodurans — the consummate survivor. Nat Rev Microbiol 2005; 3:882-892 10.1038/nrmicro1264 [DOI] [PubMed] [Google Scholar]
- 18.Misra HS, Khairnar NP, Kota S, Shrivastava S, Joshi VP, Apte SK. An exonuclease I-sensitive DNA repair pathway in Deinococcus radiodurans: a major determinant of radiation resistance. Mol Microbiol 2006; 59:1308-1316 10.1111/j.1365-2958.2005.05005.x [DOI] [PubMed] [Google Scholar]
- 19.Zahradka K, Slade D, Bailone A, Sommer S, Averbeck D, Petranovic M, Lindner AB, Radman M. Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 2006; 443:569-573 [DOI] [PubMed] [Google Scholar]
- 20.Slade D, Lindner AB, Paul G, Radman M. Recombination and Replication in DNA Repair of Heavily Irradiated Deinococcus radiodurans. Cell 2009; 136:1044-1055 10.1016/j.cell.2009.01.018 [DOI] [PubMed] [Google Scholar]
- 21.Daly MJ, Gaidamakova EK, Matrosova VY, Vasilenko A, Zhai M, Leapman RD, Lai B, Ravel B, Li SM, Kemner KM, Fredrickson JK. Protein Oxidation Implicated as the Primary Determinant of Bacterial Radioresistance. PLoS Biol 2007; 5:e92 10.1371/journal.pbio.0050092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Battista JR, Earl AM, Park MJ. Why is Deinococcus radiodurans so resistant to ionizing radiation? Trends Microbiol 1999; 7:362-365 10.1016/S0966-842X(99)01566-8 [DOI] [PubMed] [Google Scholar]
- 23.Daly MJ. Death by protein damage in irradiated cells. DNA Repair (Amst) 2012; 11:12-21 10.1016/j.dnarep.2011.10.024 [DOI] [PubMed] [Google Scholar]
- 24.Daly MJ, Gaidamakova EK, Matrosova VY, Kiang JG, Fukumoto R, Lee DY, Wehr NB, Viteri GA, Berlett BS, Levine RL. Small-molecule antioxidant proteome-shields in Deinococcus radiodurans. PLoS ONE 2010; 5:e12570 10.1371/journal.pone.0012570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fredrickson JK, Li SM, Gaidamakova EK, Matrosova VY, Zhai M, Sulloway HM, Scholten JC, Brown MG, Balkwill DL, Daly MJ. Protein oxidation: key to bacterial desiccation resistance? ISME J 2008; 2:393-403 10.1038/ismej.2007.116 [DOI] [PubMed] [Google Scholar]
- 26.Krisko A, Radman M. Protein damage and death by radiation in Escherichia coli and Deinococcus radiodurans. Proc Natl Acad Sci USA 2010; 107:14373-14377 10.1073/pnas.1009312107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slade D, Radman M. Oxidative stress resistance in Deinococcus radiodurans. Microbiol. Mol. Biol. Rev. MMBR 2011; 75:133-191 10.1128/MMBR.00015-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshinaka T, Yano K, Yamaguchi H. Isolation of highly radioresistant bacterium Arthrobacter radiotolerans nov. sp. Agric Biol Chem 1973; 37:2269-2275 10.1271/bbb1961.37.2269 [DOI] [Google Scholar]
- 29.Jurado V, Miller AZ, Alias-Villegas C, Laiz L, Saiz-Jimenez C. Rubrobacter bracarensis sp. nov., a novel member of the genus Rubrobacter isolated from a biodeteriorated monument. Syst Appl Microbiol 2012; 35:306-309 10.1016/j.syapm.2012.04.007 [DOI] [PubMed] [Google Scholar]
- 30.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4:406-425 [DOI] [PubMed] [Google Scholar]
- 31.Felsenstein J. Confidence limits on Phylogenies:an approach using bootstrap. Evolution 1985; 39:783-791 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- 32.Jukes T, Cantor CR. Evolution of protein molecules. Mamm. Protein Metab. 1969; 3:21-132 10.1016/B978-1-4832-3211-9.50009-7 [DOI] [Google Scholar]
- 33.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 2011; 28:2731-2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albuquerque L, França L, Rainey FA, Schumann P, Nobre MF, da Costa MS. Gaiella occulta gen. nov., sp. nov., a novel representative of a deep branching phylogenetic lineage within the class Actinobacteria and proposal of Gaiellaceae fam. nov. and Gaiellales ord. nov. Syst Appl Microbiol 2011; 34:595-599 10.1016/j.syapm.2011.07.001 [DOI] [PubMed] [Google Scholar]
- 35.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garrity GM, Holt JG. The Road Map to the Manual. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 119-169. [Google Scholar]
- 38.Stackebrandt E, Rainey FA, Ward-Rainey NL. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol 1997; 47:479-491 10.1099/00207713-47-2-479 [DOI] [Google Scholar]
- 39.Stackebrandt E. Will we ever understand? The undescribable diversity of the prokaryotes. Acta Microbiol Immunol Hung 2004; 51:449-462 10.1556/AMicr.51.2004.4.5 [DOI] [PubMed] [Google Scholar]
- 40.Zhi XY, Li WJ, Stackebrandt E. An update of the structure and 16S rRNA gene sequence-based definition of higher ranks of the class Actinobacteria, with the proposal of two new suborders and four new families and emended descriptions of the existing higher taxa. Int J Syst Evol Microbiol 2009; 59:589-608 10.1099/ijs.0.65780-0 [DOI] [PubMed] [Google Scholar]
- 41.Reddy GSN, Garcia-Pichel F. Description of Patulibacter americanus sp. nov., isolated from biological soil crusts, emended description of the genus Patulibacter Takahashi et al. 2006 and proposal of Solirubrobacterales ord. nov. and Thermoleophilales ord. nov. Int J Syst Evol Microbiol 2009; 59:87-94 10.1099/ijs.0.64185-0 [DOI] [PubMed] [Google Scholar]
- 42.Validation of the publication of new names and new combinations previously effectively published outside the IJSB. List No. 28. Int J Syst Bacteriol 1989; 39:93-94 10.1099/00207713-39-1-93 [DOI] [Google Scholar]
- 43.Suzuki K, Collins MD, Iijima E, Komagata K. Chemotaxonomic characterization of a radiotolerant bacterium Arthrobacter radiotolerans: describtion of Rubrobacter radiotolerans gen nov., comb. nov. FEMS Microbiol Lett 1988; 52:33-39 10.1111/j.1574-6968.1988.tb02568.x [DOI] [Google Scholar]
- 44.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gordon D, Abajian C, Green P. Consed: a graphical tool for sequence finishing. Genome Res 1998; 8:195-202 10.1101/gr.8.3.195 [DOI] [PubMed] [Google Scholar]
- 46.Gordon D, Desmarais C, Green P. Automated Finishing with Autofinish. Genome Res 2001; 11:614-625 10.1101/gr.171401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tettelin H, Radune D, Kasif S, Khouri H, Salzberg SL. Optimized Multiplex PCR: Efficiently Closing a Whole-Genome Shotgun Sequencing Project. Genomics 1999; 62:500-507 10.1006/geno.1999.6048 [DOI] [PubMed] [Google Scholar]
- 48.Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 2005; 33:W686-W689 10.1093/nar/gki366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lagesen K, Hallin P, Rødland EA, Stærfeldt HH, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 2007; 35:3100-3108 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Edgar RC. Search and Clustering Orders of Magnitude Faster Than BLAST. Bioinformatics 2010; 26:2460-2461 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 51.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010; 11:119 10.1186/1471-2105-11-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, Kyrpides NC. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods 2010; 7:455-457 10.1038/nmeth.1457 [DOI] [PubMed] [Google Scholar]
- 53.Zdobnov EM, Apweiler R. InterProScan--an integration platform for the signature-recognition methods in InterPro. Bioinformatics 2001; 17:847-848 10.1093/bioinformatics/17.9.847 [DOI] [PubMed] [Google Scholar]
- 54.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res 2008; 36:D281-D288 10.1093/nar/gkm960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haft DH, Selengut JD, White O. The TIGRFAMs database of protein families. Nucleic Acids Res 2003; 31:371-373 10.1093/nar/gkg128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pedruzzi I, Rivoire C, Auchincloss AH, Coudert E, Keller G, de Castro E, Baratin D, Cuche BA, Bougueleret L, Poux S. HAMAP in 2013, new developments in the protein family classification and annotation system. Nucleic Acids Res 2013; 41:D584-D589 10.1093/nar/gks1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu CH, Nikolskaya A, Huang H, Yeh LS, Natale DA, Vinayaka CR, Hu ZZ, Mazumder R, Kumar S, Kourtesis P. PIRSF: family classification system at the Protein Information Resource. Nucleic Acids Res 2004; 32:D112-D114 10.1093/nar/gkh097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Attwood TK, Coletta A, Muirhead G, et al. The PRINTS database: a fine-grained protein sequence annotation and analysis resource--its status in 2012. Database J. Biol. Databases Curation 2012;2012:bas019. [DOI] [PMC free article] [PubMed]
- 59.Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res 2012; 40:D302-D305 10.1093/nar/gkr931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson D, Pethica R, Zhou Y, Talbot C, Vogel C, Madera M, Chothia C, Gough J. SUPERFAMILY--sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res 2009; 37:D380-D386 10.1093/nar/gkn762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res 2010; 38:D161-D166 10.1093/nar/gkp885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science 1997; 278:631-637 10.1126/science.278.5338.631 [DOI] [PubMed] [Google Scholar]
- 63.Mavromatis K, Ivanova NN, Chen IMA, Szeto E, Markowitz VM, Kyrpides NC. The DOE-JGI Standard Operating Procedure for the Annotations of Microbial Genomes. Stand Genomic Sci 2009; 1:63-67 10.4056/sigs.632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics 2000; 16:944-945 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- 65.Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res 2008; 36:W181-W184 10.1093/nar/gkn179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Confalonieri F, Sommer S. Bacterial and archaeal resistance to ionizing radiation. J Phys Conf Ser 2011; 261:012005 10.1088/1742-6596/261/1/012005 [DOI] [Google Scholar]
- 67.Hefferin ML, Tomkinson AE. Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair (Amst) 2005; 4:639-648 10.1016/j.dnarep.2004.12.005 [DOI] [PubMed] [Google Scholar]
- 68.Blasius M, Hübscher U, Sommer S. Deinococcus radiodurans: What Belongs to the Survival Kit? Crit Rev Biochem Mol Biol 2008; 43:221-238 10.1080/10409230802122274 [DOI] [PubMed] [Google Scholar]
- 69.Beam CE, Saveson CJ, Lovett ST. Role for radA/sms in Recombination Intermediate Processing in Escherichia coli. J Bacteriol 2002; 184:6836-6844 10.1128/JB.184.24.6836-6844.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nowosielska A, Marinus MG. DNA mismatch repair-induced double-strand breaks. DNA Repair (Amst) 2008; 7:48-56 10.1016/j.dnarep.2007.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li S, Xu M, Su Z. Computational analysis of LexA regulons in Cyanobacteria. BMC Genomics 2010; 11:527 10.1186/1471-2164-11-527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Campoy S, Fontes M, Padmanabhan S, Cortés P, Llagostera M, Barbé J. LexA-independent DNA damage-mediated induction of gene expression in Myxococcus xanthus. Mol Microbiol 2003; 49:769-781 10.1046/j.1365-2958.2003.03592.x [DOI] [PubMed] [Google Scholar]
- 73.Mertens K, Lantsheer L, Ennis DG, Samuel JE. Constitutive SOS expression and damage-inducible AddAB-mediated recombinational repair systems for Coxiella burnetii as potential adaptations for survival within macrophages. Mol Microbiol 2008; 69:1411-1426 10.1111/j.1365-2958.2008.06373.x [DOI] [PubMed] [Google Scholar]
- 74.Kelley WL. Lex marks the spot: the virulent side of SOS and a closer look at the LexA regulon. Mol Microbiol 2006; 62:1228-1238 10.1111/j.1365-2958.2006.05444.x [DOI] [PubMed] [Google Scholar]
- 75.Narumi I, Satoh K, Cui S, Funayama T, Kitayama S, Watanabe H. PprA: a novel protein from Deinococcus radiodurans that stimulates DNA ligation. Mol Microbiol 2004; 54:278-285 10.1111/j.1365-2958.2004.04272.x [DOI] [PubMed] [Google Scholar]
- 76.Tanaka M, Earl AM, Howell HA, Park MJ, Eisen JA, Peterson SN, Battista JR. Analysis of Deinococcus radiodurans’s Transcriptional Response to Ionizing Radiation and Desiccation Reveals Novel Proteins That Contribute to Extreme Radioresistance. Genetics 2004; 168:21-33 10.1534/genetics.104.029249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bagwell CE, Bhat S, Hawkins GM, Smith BW, Biswas T, Hoover TR, Saunders E, Han CS, Tsodikov OV, Shimkets LJ. Survival in Nuclear Waste, Extreme Resistance, and Potential Applications Gleaned from the Genome Sequence of Kineococcus radiotolerans SRS30216. PLoS ONE 2008; 3:e3878 10.1371/journal.pone.0003878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Basu B, Apte SK. Gamma Radiation-induced Proteome of Deinococcus radiodurans Primarily Targets DNA Repair and Oxidative Stress Alleviation. Mol Cell Proteomics 2012; 11:M111.011734. [DOI] [PMC free article] [PubMed]
- 79.Terato H, Suzuki K, Nishioka N, Okamoto A, Shimazaki-Tokuyama Y, Inoue Y, Saito T. Characterization and Radio-resistant Function of Manganese Superoxide Dismutase of Rubrobacter radiotolerans. J Radiat Res (Tokyo) 2011; 52:735-742 10.1269/jrr.11105 [DOI] [PubMed] [Google Scholar]
- 80.Yuan M, Chen M, Zhang W, Lu W, Wang J, Yang M, Zhao P, Tang R, Li X, Hao Y, et al. Genome Sequence and Transcriptome Analysis of the Radioresistant Bacterium Deinococcus gobiensis: Insights into the Extreme Environmental Adaptations. PLoS ONE 2012; 7:e34458 10.1371/journal.pone.0034458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Empadinhas N, Mendes V, Simões C, Santos MS, Mingote A, Lamosa P, Santos H, Costa MS. Organic solutes in Rubrobacter xylanophilus: the first example of di-myo-inositol-phosphate in a thermophile. Extrem. Extremophiles 2007; 11:667-673 10.1007/s00792-007-0084-z [DOI] [PubMed] [Google Scholar]
- 82.Nobre A, Alarico S, Fernandes C, Empadinhas N, da Costa MS. A Unique Combination of Genetic Systems for the Synthesis of Trehalose in Rubrobacter xylanophilus: Properties of a Rare Actinobacterial TreT. J Bacteriol 2008; 190:7939-7946 10.1128/JB.01055-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Santos H, da Costa MS. Compatible solutes of organisms that live in hot saline environments. Environ Microbiol 2002; 4:501-509 10.1046/j.1462-2920.2002.00335.x [DOI] [PubMed] [Google Scholar]
- 84.Kryukov K, Saitou N. MISHIMA - a new method for high speed multiple alignment of nucleotide sequences of bacterial genome scale data. BMC Bioinformatics 2010; 11:142 10.1186/1471-2105-11-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maddocks SE, Oyston PCF. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 2008; 154:3609-3623 10.1099/mic.0.2008/022772-0 [DOI] [PubMed] [Google Scholar]
- 86.Murphy JN, Saltikov CW. The ArsR repressor mediates arsenite-dependent regulation of arsenate respiration and detoxification operons of Shewanella sp. strain ANA-3. J Bacteriol 2009; 191:6722-6731 10.1128/JB.00801-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen J, Xie J. Role and regulation of bacterial LuxR-like regulators. J Cell Biochem 2011; 112:2694-2702 10.1002/jcb.23219 [DOI] [PubMed] [Google Scholar]
- 88.Ramos JL, Martínez-Bueno M, Molina-Henares AJ, Terán W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. MMBR 2005; 69:326-356 10.1128/MMBR.69.2.326-356.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Collins MD, Hutson RA, Grant IR, Patterson MF. Phylogenetic characterization of a novel radiation-resistant bacterium from irradiated pork: description of Hymenobacter actinosclerus sp. nov. Int J Syst Evol Microbiol 2000; 50:731-734 10.1099/00207713-50-2-731 [DOI] [PubMed] [Google Scholar]