Abstract
Members of genus Skermanella were described as Gram-negative, motile, aerobic, rod-shaped, obligate-heterotrophic bacteria and unable to fix nitrogen. In this study, the genome sequence of Skermanella stibiiresistens SB22T is reported. Phylogenetic analysis using core proteins confirmed the phylogenetic assignment based on 16S rRNA gene sequences. Strain SB22T has all the proteins for complete glycolysis, tricarboxylic acid cycle and pentose phosphate pathway. The RuBisCO encoding genes cbbL1S1 and nitrogenase delta subunit gene anfG are absent, consistent with its inability to fix carbon and nitrogen, respectively. In addition, the genome possesses a series of flagellar assembly and chemotaxis genes to ensure its motility.
Keywords: Skermanella stibiiresistens, genome sequence, metabolism, flagella, chemotaxis
Introduction
The type species for the genus Skermanella is Skermanella parooens ACM 2042T, which was originally proposed as Conglomeromonas largomobilis subsp. parooensis by Skerman et al. in 1983 [1]. Later, it was transferred to the genus Skermanella (family Rhodospirillaceae) on the basis of phylogenetic evidence and phenotypic characteristics, especially the inability to fix nitrogen [2,3]. At present, this genus comprises four validly published species, Skermanella parooensis [3], Skermanella aerolata [4], Skermanella xinjiangensis [5] and Skermanella stibiiresistens [6], which were isolated from fresh water, air, sandy soil and a coal mine, respectively.
Skermanella was characterized as a Gram-negative, non-spore-forming bacterium with unicellular and multicellular phases of growth, an obligate chemo-organotroph and facultative anaerobe, unable to fix nitrogen, and with a high DNA G+C content. To the best of our knowledge, genome information for Skermanella members is still not available. In this study, we present the draft genome sequence of a Skermanella type strain S. stibiiresistens SB22T and compare it with the members of the related genera Azospirillum and Rhodospirillum.
Classification and features
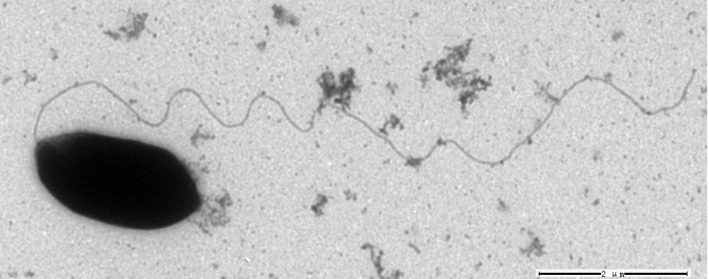
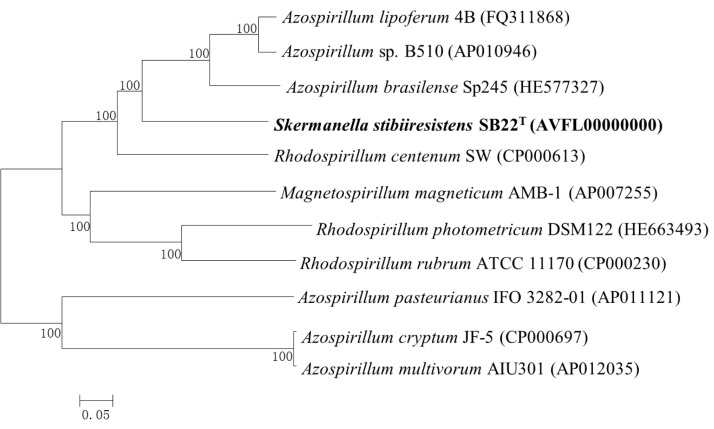
S. stibiiresistens SB22T is Gram-negative, motile, rod-shaped with single polar flagella and non-sporulating (Figure 1). Ten related strains with complete genome sequences belong to Azospirillum (3 strains), Rhodospirillum (3 strains), Acidiphilium (2 strains), Magnetospirillum (1 strain) and Acetobacter (1 strain). A total of 515 conserved proteins were identified among them using cluster algorithm tool MCL (http://micans.org/mcl/) with default values and a neighbor joining (NJ) tree was built based on this set. The phylogenetic tree showed that strain SB22T was closely related to the genera Azospirillum and Rhodospirillum, which was consistent with the taxonomy previously determined by 16S rRNA gene sequence analysis [6]. Classification and general features of S. stibiiresistens SB22T are shown in Table 1. Figure 2 shows the phylogenetic neighborhood of S. stibiiresistens SB22T in a core-protein based tree.
Figure 1.
A transmission micrograph of S. stibiiresistens SB22T, made using a Hitachi H-7000FA transmission electron microscope operating at 100 kV. The scale bar represents 2 μm.
Table 1. Classification and general features of S. stibiiresistens SB22T according to the MIGS recommendations [7].
| MIGS ID | Property | Term | Evidence code | |
|---|---|---|---|---|
| Current classification | Domain Bacteria | TAS [8] | ||
| Phylum Proteobacteria | TAS [9] | |||
| Class Alphaproteobacteria | TAS [10,11] | |||
| Order Rhodospirillales | TAS [12,13] | |||
| Family Rhodospirillaceae | TAS [12,13] | |||
| Genus Skermanella | TAS [3,4,6] | |||
| Species Skermanella stibiiresistens | TAS [6] | |||
| Type strain SB22T | TAS [6] | |||
| Gram stain | Negative | TAS [6] | ||
| Cell shape | Rod-shaped | TAS [6] | ||
| Motility | Motile | TAS [6] | ||
| Sporulation | Non-sporulating | TAS [6] | ||
| Optimum temperature | 4-37ºC | TAS [6] | ||
| Carbon source | D-glucose, D-ribose, rhamnose, L-proline, salicin, inositol, DL-lactate, L-alanine, malic acid, potassium 2-ketogluconate and 3-hydroxybutyric acid | TAS [6] | ||
| Energy source | Chemoorganotroph | TAS [6] | ||
| Terminal electron receptor | Molecular oxygen | TAS [6] | ||
| MIGS-6.2 | pH | 5-9 | TAS [6] | |
| MIGS-22 | Oxygen | Aerobic | TAS [6] | |
| MIGS-15 | Biotic relationship | Free-living | NAS | |
| MIGS-14 | Pathogenicity | Non-pathogenic | NAS | |
| MIGS-4 | Geographic location | Jixi coal mine of Jixi City, Heilongjiang Province, northeast China | TAS [6] | |
| MIGS-5 | Sample collection time | 2011 | TAS [6] | |
| MIGS-4.1 MIGS-4.2 |
Latitude Longitude |
N45°18' E130°57' |
TAS [6] TAS [6] |
|
| MIGS-4.3 | Depth | Surface sandy soil | TAS [6] | |
| MIGS-4.4 | Altitude | Not reported | ||
Evidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement; NAS: Non-traceable Author Statement. These evidence codes are from the Gene Ontology project [14]. If the evidence is IDA, then the property was directly observed for a live isolate by one of the authors or an expert mentioned in the acknowledgements.
Figure 2.
A NJ phylogenetic tree highlighting the position of S. stibiiresistens SB22T with other completely sequenced strains that belong to the same family (Rhodospirillaceae). All protein FASTA files were obtained from NCBI. The corresponding GenBank accession numbers are displayed in parentheses. A total of 515 conserved proteins were identified using cluster algorithm tool MCL. The 515 amino acid sequences were aligned using Clustal W [15] and the NJ tree was built using MEGA 5.05 [16] with boot strap value of 1,000.
Genome sequencing information
Genome project history
S. stibiiresistens SB22T was sequenced by Majorbio Bio-pharm Technology Co., Ltd, Shanghai, China. The draft genome sequence was deposited in NCBI with contigs less than 200 bp cut off. The GenBank accession number is AVFL00000000. A summary of the genome sequencing project information is shown in Table 2.
Table 2. Genome sequencing project information of S. stibiiresistens SB22T.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | High-quality draft |
| MIGS-28 | Libraries used | Illumina Paired-End library (300 bp insert size) |
| MIGS-29 | Sequencing platform | Illumina Hiseq2000 |
| MIGS-31.2 | Sequencing coverage | 184.5 × |
| MIGS-30 | Assemblers | SOAPdenovo v1.05 |
| MIGS-32 | Gene calling method | GeneMarkS+ |
| GenBank date of release | February 23, 2014 | |
| NCBI project ID | AVFL00000000 | |
| MIGS-13 | Source material identifier | SB22T |
| Project relevance | Genome comparison |
Growth condition and DNA isolation
S. stibiiresistens SB22T was grown aerobically in R2A medium at 28°C for 2 days. It can also grow on LB medium under the same conditions. The strain was apricot-colored after incubated 72 h at 28°C on R2A agar. The DNA was isolated using the QiAamp kit according to the manufacturer’s instruction (Qiagen, German).
Genome sequencing and assembly
The Illumina Hiseq2000 technology with Paired-End (PE) library strategy was used to determine the sequence of S. stibiiresistens SB22T. A total of 7,588,874 x 2 high quality reads totaling 1,454,191,294 bp data with an average coverage 184.5 x was generated. Illumina sequencing data was assembled with SOAPdenovo, version 1.05 (http://soap.genomics.org.cn/). The initial draft assembly contained 7,879,677 bp in 257 contigs. Then the draft genome sequence was deposited to the NCBI with contigs less than 200 bp nucleotides cut off.
Genome annotation
The draft genome sequence was deposited to NCBI and was annotated though the Prokaryotic Genome Annotation Pipeline (PGAP), using the Best-placed reference protein set and the gene caller GeneMarkS+. Signal peptides and transmembrane helices were predicted by SignalP [17] and SOSUI [18], respectively. The WebMGA-server [19] was used to identify the Clusters of Orthologs Groups (COG).
Genome properties
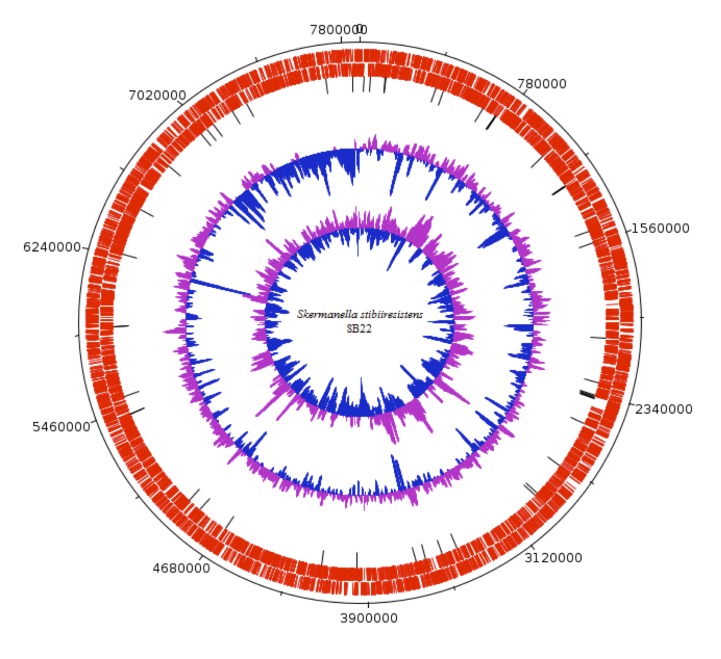
The final whole genome of S. stibiiresistens SB22T was 7,868,338 bp long in 190 contigs (with PEGs) with an average GC content of 65.88%. Of the total 7,378 predicted genes, 7,269 were protein-coding genes and 63 were RNA genes. A total of 5,176 protein-coding genes (71%) were assigned with putative functions with the remaining was annotated as hypothetical proteins. The property and the statistics of this genome are summarized in Table 3. We reordered the contigs using MAUVE, version 2.3 [20] with the complete genome sequence of Rhodospirillum centenum …( ) as a reference and a graphical circular map of S. stibiiresistens SB22T is shown in Figure 3. The distribution of genes into COGs functional categories is shown in Table 4.
Table 3. Nucleotide content and gene count level in genome of S. stibiiresistens SB22T.
| Attribute | Value | % of Totala |
|---|---|---|
| Genome size (bp) | 7,868,388 bp | 100 |
| Number of contigs | 190 | |
| Contig N50 | 214,710 bp | |
| Total genes | 7,378 | 100 |
| Protein-coding genes | 7,269 | 98.52 |
| Pseudo genes | 46 | 0.62 |
| RNA genes | 63 | 0.86 |
| Frame shifted genes | 13 | |
| DNA coding region (bp) | 6,849,861 | 87.06 |
| Protein-coding genes with function prediction | 5,176 | 71.21 |
| Protein-coding genes assigned to COGs | 5,877 | 80.85 |
| Protein-coding genes with transmembrane helices | 1,673 | 23.02 |
| Protein-coding genes with signal peptides | 523 | 7.19 |
| CRISPR repeats | 2 |
a The total is based on either the size of the genome in base pairs or the total number of protein coding genes in the annotated genome.
Figure 3.
A graphical circular map of S. stibiiresistens SB22T. From outside to the center: Genes on forward strand, Genes on reverse strand, tRNA genes, GC content and GC skew.
Table 4. Number of protein-coding genes associated with the 25 general COG functional categories in S. stibiiresistens SB22T genome.
| Code | Value | %agea | COG Category |
|---|---|---|---|
| J | 188 | 2.59 | Translation, ribosomal structure and biogenesis |
| A | 1 | 0.01 | RNA processing and modification |
| K | 478 | 6.58 | Transcription |
| L | 240 | 3.30 | Replication, recombination and repair |
| B | 6 | 0.08 | Chromatin structure and dynamics |
| D | 50 | 0.69 | Cell cycle control, cell division, chromosome partitioning |
| Y | 0 | 0.00 | Nuclear structure |
| V | 73 | 1.00 | Defense mechanisms |
| T | 651 | 8.96 | Signal transduction mechanisms |
| M | 389 | 5.35 | Cell wall/membrane/envelope biogenesis |
| N | 147 | 2.02 | Cell motility |
| Z | 0 | 0.00 | Cytoskeleton |
| W | 0 | 0.00 | Extracellular structures |
| U | 76 | 1.05 | Intracellular trafficking, secretion, and vesicular transport |
| O | 195 | 2.68 | Posttranslational modification, protein turnover, chaperones |
| C | 405 | 5.57 | Energy production and conversion |
| G | 508 | 6.99 | Carbohydrate transport and metabolism |
| E | 678 | 9.33 | Amino acid transport and metabolism |
| F | 101 | 1.39 | Nucleotide transport and metabolism |
| H | 230 | 3.16 | Coenzyme transport and metabolism |
| I | 267 | 3.67 | Lipid transport and metabolism |
| P | 383 | 5.27 | Inorganic ion transport and metabolism |
| Q | 226 | 3.11 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 787 | 10.83 | General function prediction only |
| S | 573 | 7.88 | Function unknown |
| - | 617 | 8.49 | Not in COGS |
a The total is based on the total number of protein coding genes in the annotated genome.
Profiles of metabolic networks and pathways
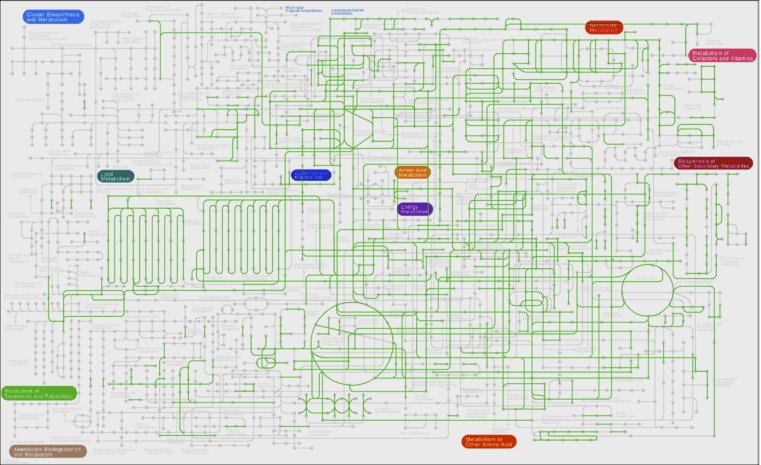
The Kyoto Encyclopedia of Genes and Genomes (KEGG) [21] was used to reconstruct the pathways of S. stibiiresistens SB22T [Figure 4]. The metabolic pathways suggest that strain SB22T possess necessary encoding genes for central carbohydrate metabolism, such as glycolysis, the TCA cycle and the pentose phosphate pathway, to support basic growth. But it has relatively few lipid metabolism pathways. Only one integrated pathway that could synthesize tetracosanoyl-CoA from acyl-CoA is found in cytoplasm. This result indicates that strain SB22T might have certain limitations in some lipid utilization.
Figure 4.
Metabolic pathways of S. stibiiresistens SB22T as predicted using KEGG. Green lines indicate pathways that are possessed by this strain.
Genes involved in carbon fixation
Genus Skermanella belongs to family Rhodospirillaceae, but Skermanella species cannot fix carbon as Rhodospirillum centenum [22] and Azospirillum amazonense [23] do. Genomic analysis of S. stibiiresistens SB22T, shows that the RuBisCO encoding genes cbbL1S1 [24,25] are not present, which is in agreement with the strain’s inability to fix carbon.
Genes involved in nitrogen metabolism
Strain S. stibiiresistens SB22T is closely related to some species of genera Azospirillum and Rhodospirillum [6]. Genus Rhodospirillum is described as a photosynthetic non-sulfur purple bacterium that favors growth in an anoxygenic, photosynthetic, nitrogen-fixing environment [26]. Some aerobic nitrogen fixing strains of Azospirillum have significant effects on crop plants [27]. But genus Skermanella is unable to fix nitrogen under microaerophilic conditions [3,4,6]. Even though nitrogenase genes nifDKH are present in the genome of S. stibiiresistens SB22T, we found that the nitrogenase delta subunit gene anfG is absent.
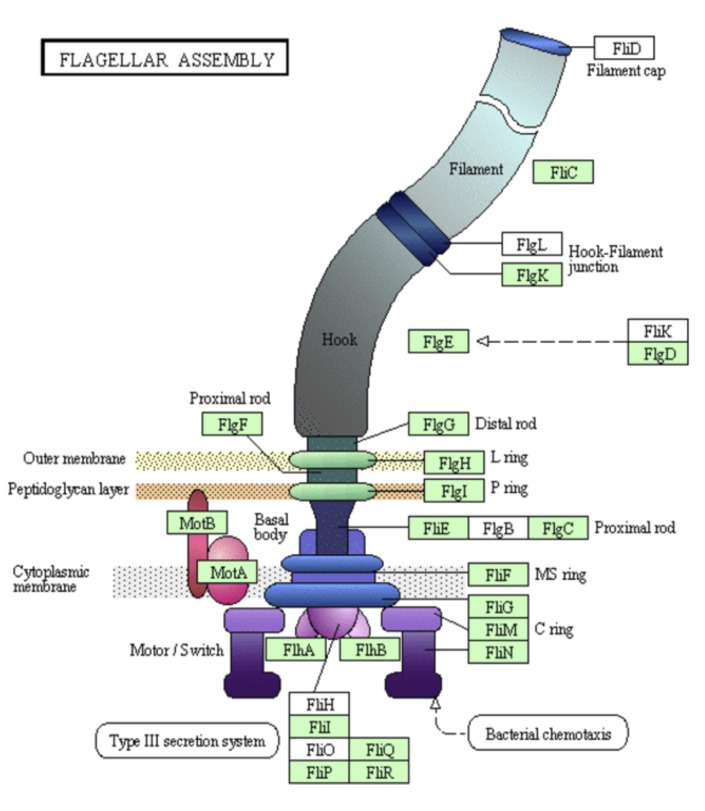
Flagella and chemotaxis
Many flagella genes were identified in S. stibiiresistens SB22T genome. A KEGG map demonstrates that this strain possesses a series of genes belonging to the families flg, fli and flh in flagellar assembly (Figure 5). These genes enable strain SB22T to move to a more suitable environment. In the same region of the genome as the flagella genes, we also identified genes encoding the central signal transduction pathway for chemotaxis (che), such as the conserved cheAWYBR genes and a group of transmembrane chemoreceptors (known as MCPs or methyl-accepting proteins), which are present in nearly all motile bacteria [23]. The MCPs may sense environment signals and transfer them to the signal transduction histidine kinase CheA, whose activity is positively regulated by CheW. CheA in turn phosphorylates a response regulator CheYVB, which controls the rotational direction of the flagella motor [28].
Figure 5.
The KEGG flagellar assembly map. Green labels represent the flagella proteins that are encoded on the S. stibiiresistens SB22T genome.
Acknowledgement
This work was supported by the National Natural Science Foundation of China (31170106).
References
- 1.Skerman VBD, Sly LI. Williamson M-L. Conglomeromonas largomobilis gen. nov., sp. nov., a sodium-sensitive, mixed-flagellated organism from fresh waters. Int J Syst Bacteriol 1983; 33:300-308;. 10.1099/00207713-33-2-300 [DOI] [Google Scholar]
- 2.Ben Dekhil S, Cahill M, Stackebrandt E, Sly LI. Transfer of Conglomeromonas largomobilis subsp. largomobilis to the genus Azospirillum as Azospirillum largomobile comb. nov., and elevation of Conglomeromonas largomobilis subsp. parooensis to the new type species of Conglomeromonas, Conglomeromonas parooensis sp. nov. Syst Appl Microbiol 1997; 20:72-77;. 10.1016/S0723-2020(97)80050-1 [DOI] [Google Scholar]
- 3.Sly LI, Stackebrandt E. Description of Skermanella parooensis gen. nov., sp. nov. to accommodate Conglomeromonas largomobilis subsp. parooensis following the transfer of Conglomeromonas largomobilis subsp. largomobilis to the genus Azospirillum. Int J Syst Bacteriol 1999; 49:541-544;. 10.1099/00207713-49-2-541 [DOI] [Google Scholar]
- 4.Weon HY, Kim BY, Hong SB, Joa JH, Nam SS, Lee KH. Kwon S-W. Skermanella aerolata sp. nov., isolated from air, and emended description of the genus Skermanella. Int J Syst Evol Microbiol 2007; 57:1539-1542; , 10.1099/ijs.0.64676-0 [DOI] [PubMed] [Google Scholar]
- 5.An H, Zhang L, Tang Y, Luo X, Sun T, Li Y, Wang Y, Dai J, Fang C. Skermanella xinjiangensis sp. nov., isolated from the desert of Xinjiang, China. Int J Syst Evol Microbiol 2009; 59:1531-1534; , 10.1099/ijs.0.003350-0 [DOI] [PubMed] [Google Scholar]
- 6.Luo G, Shi Z, Wang H, Wang G. Skermanella stibiiresistens sp. nov., a highly antimony-resistant bacterium isolated from coal-mining soil, and emended description of the genus Skermanella. Int J Syst Evol Microbiol 2012; 62:1271-1276; , 10.1099/ijs.0.033746-0 [DOI] [PubMed] [Google Scholar]
- 7.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nat Biotechnol 2008; 26:541-547; , 10.1038/nbt1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci USA 1990; 87:4576-4579; , 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1. [Google Scholar]
- 10.List Editor Validation List No. 107. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 2006; 56:1-6; , 10.1099/ijs.0.64188-0 [DOI] [PubMed] [Google Scholar]
- 11.Garrity GM, Bell JA, Lilburn T. Class I. Alphaproteobacteria class. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part C, Springer, New York, 2005, p. 1. [Google Scholar]
- 12.Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Bacteriol 1980; 30:225-420;. 10.1099/00207713-30-1-225 [DOI] [PubMed] [Google Scholar]
- 13.Pfennig N, Trüper HG. Higher taxa of the phototrophic bacteria. Int J Syst Bacteriol 1971; 21:17-18;. 10.1099/00207713-21-1-17 [DOI] [Google Scholar]
- 14.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000; 25:25-29; , 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 1994; 22:4673-4680 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731-2739; , 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 2011; 8:785-786; , 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- 18.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 1998; 14:378-379; , 10.1093/bioinformatics/14.4.378 [DOI] [PubMed] [Google Scholar]
- 19.Wu S, Zhu Z, Fu L, Niu B, Li W. WebMGA: a Customizable Web Server for Fast Metagenomic Sequence Analysis. BMC Genomics 2011; 12:444; , 10.1186/1471-2164-12-444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 2004; 14:1394-1403 10.1101/gr.2289704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 2014; 42:D199-D205; , 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu YK, Marden J, Han M, Swingley W, Mastrian S, Chowdhury S, Hao J, Helmy T, Kim S. A Kurdoglu A, J Matthies H, Rollo D, Stothard P, E Blankenship R, Bauer C, W Touchman J. Metabolic flexibility revealed in the genome of the cyst-forming α-1 proteobacterium Rhodospirillum centenum. BMC Genomics 2010; 11:325; , 10.1186/1471-2164-11-325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sant'Anna HF, Almeida LG, Cecagno R, Reolon AL, Siqueira FM, Machado MR, Vasconcelos AT, Schrank IS. Genomic insights into the versatility of the plant growth-promoting bacterium Azospirillum amazonense. BMC Genomics 2011; 12:409; , 10.1186/1471-2164-12-409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Badger MR, Bek EJ. Multiple Rubisco forms in Proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J Exp Bot 2007; 59:1525-1541; , 10.1093/jxb/erm297 [DOI] [PubMed] [Google Scholar]
- 25.Tabita FR. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a different perspective. Photosynth Res 1999; 60:1-28;. 10.1023/A:1006211417981 [DOI] [Google Scholar]
- 26.Favinger J, Stadtwald R, Gest H. Rhodospirillum centenum, sp. nov., a thermotolerant cyst-forming anoxygenic photosynthetic bacterium. Antonie van Leeuwenhoek 1989; 55:291-296; , 10.1007/BF00393857 [DOI] [PubMed] [Google Scholar]
- 27.Steenhoudt O, Vanderleyden J. Azospirillum, a free-living nitrogen-fixing bacterium closely associated with grasses: genetic, biochemical and ecological aspects. FEMS Microbiol Rev 2000; 24:487-506; , 10.1111/j.1574-6976.2000.tb00552.x [DOI] [PubMed] [Google Scholar]
- 28.Falke JJ, Bass RB, Butler SL, Chervitz SA, Danielson MA. The two-component signaling pathway of bacterial chemotaxis: a molecular view of signal transduction by receptors, kinases, and adaptation enzymes. Annu Rev Cell Dev Biol 1997; 13:457-512; , 10.1146/annurev.cellbio.13.1.457 [DOI] [PMC free article] [PubMed] [Google Scholar]