Abstract
Kosakonia sacchari sp. nov. is a new species within the new genus Kosakonia, which was included in the genus Enterobacter. K sacchari is a nitrogen-fixing bacterium named for its association with sugarcane (Saccharum officinarum L.). K sacchari bacteria are Gram-negative, aerobic, non-spore-forming, motile rods. Strain SP1T (=CGMCC1.12102T=LMG 26783T) is the type strain of the K sacchari sp. nov and is able to colonize and fix N2 in association with sugarcane plants, thus promoting plant growth. Here we summarize the features of strain SP1T and describe its complete genome sequence. The genome contains a single chromosome and no plasmids, 4,902,024 nucleotides with 53.7% GC content, 4,460 protein-coding genes and 105 RNA genes including 22 rRNA genes, 82 tRNA genes, and 1 ncRNA gene.
Key words : endophyte, Enterobacter, Kosakonia, nitrogen fixation, plant growth-promoting bacteria, sugarcane
Introduction
The genus Enterobacter belonging to the family Enterobacteriaceae is polyphyletic based on 16S rRNA gene sequence analysis [1-3]. Recently, eleven species belonging to the genus Enterobacter were transferred into the genus Cronobacter and three novel genera (Lelliottia, Pluralibacter, and Kosakonia) based on multilocus sequence analysis of protein-coding genes, rpoB (RNA polymerase β-subunit gene), gyrB (DNA gyrase subunit B gene), infB (initiation translation factor 2 gene), and atpD (ATP synthase β-subunit gene) [1]. Enterobacter cowanii, E. radicincitans, E. oryzae and E. arachidis were reclassified as Kosakonia cowanii, K. radicincitans, K. oryzae and K. arachidis, respectively [1]. Enterobacter sacchari is a new species named for nitrogen-fixing bacteria in association with sugarcane (Saccharum officinarum L.) [2,4] and has been reclassified as Kosakonia sacchari [3]. K sacchari is able to colonize sugarcane plants, fix N2 in association with sugarcane plants and promote plant growth [4]. K sacchari strain SP1T was isolated from a surface-sterilized stem of sugarcane cultivar GT11 grown in Nanning, Guangxi, China in 1994. It has now been designated the type strain of K sacchari sp. nov [2,3]. Here we present a summary of its features [2] and the complete genome sequence and annotation for K sacchari strain SP1T (=CGMCC1.12102T=LMG 26783T).
Organism information
Classification and general features
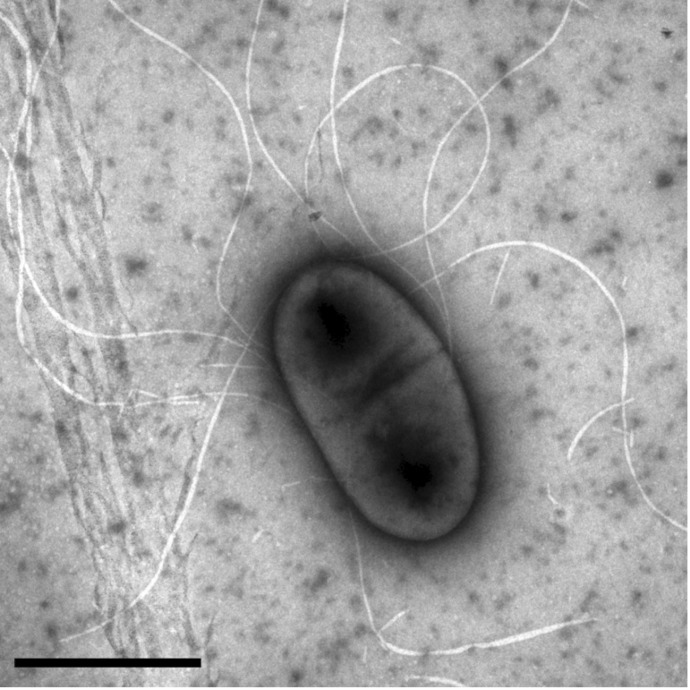
K sacchari type strain SP1T is a Gram-negative, non-spore-forming, motile rod with peritrichous flagella (Figure 1., Table 1.[2]). It grows aerobically but reduces N2 to NH3 at a low pO2. It is able to grow and fix N2 on media containing 10% (w/v) cane sugar or sucrose and forms circular, convex, smooth colonies with entire margins on solid media. It grows best around 30°C and pH 7.
Figure 1.
Transmission electron micrograph showing a negative-stained cell of the Kosakonia sacchari type strain SP1T [2]. The scale bar represents 1 μm.
Table 1. Classification and general features of Kosakonia sacchari type strain SP1T according to the MIGS recommendations.
| MIGS ID | Property | Term | Evidence code |
|---|---|---|---|
| Current classification | Domain Bacteria Phylum Proteobacteria Class Gammaproteobacteria Order Enterobacteriales Family Enterobacteriaceae Genus Kosakonia Species Kosakonia sacchari Type strain: SP1T |
TAS [16] TAS [17] TAS [18-20] TAS [21] TAS [22,23] TAS [1,3] TAS [2,3] TAS [2,3] |
|
| Gram stain | Negative | TAS [2] | |
| Cell shape | Rod | TAS [2] | |
| Motility | Motile | TAS [2] | |
| Sporulation | Non-sporulating | TAS [2] | |
| Temperature range | Mesophile | TAS [2] | |
| Optimum temperature | 28 – 32°C | TAS [2] | |
| Carbon source | Sucrose, glucose, fructose, galactose, maltose, mannitol, mannose, arabitol |
TAS [2] | |
| Energy source | Chemoorganotroph | TAS [2] | |
| MIGS-6 | Habitat | Soil, plants | IDA |
| MIGS-6.3 | Salinity | 0 – 4% NaCl | TAS [2] |
| MIGS-22 MIGS-23 |
Oxygen Isolation |
Aerobe Stem of sugarcane cultivar GT11 |
TAS [2] TAS [2] |
| MIGS-15 | Biotic relationship | Free-living, endophytic | IDA |
| MIGS-14 | Pathogenicity | Not reported | |
| MIGS-4 | Geographic location | Nanning, Guangxi, China | TAS [2] |
| MIGS-5 | Sample collection time | 1994 | TAS [2] |
| MIGS-4.1 MIGS-4.2 | Longitude Latitude |
108.33 22.84 |
NAS NAS |
| MIGS-4.3 | Depth | 0.1 – 0.5 m above the surface | IDA |
| MIGS-4.4 | Altitude | 76 m | NAS |
Evidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence).
Phylogenetic analysis of the 16S rRNA gene sequences from SP1T, the type strains of species of the genus Enterobacter and the type strains of type species of other genera in the family Enterobacteriaceae showed that SP1T formed a monophyletic group with the type strain of E. cloacae (the type species of the genus Enterobacter) [2]. However, phylogenetic analysis of the rpoB gene sequences showed that SP1T diverged from E. cloacae [2]. Here, phylogenetic analysis of the 16S rRNA gene sequences from SP1T, other type strains in the genus Kosakonia, and the type strain of E. cloacae showed that K sacchari formed a monophyletic group with K. radicincitans, K. oryzae, and K. arachidis and diverged from K. cowanii (the type species of the genus Kosakonia) and E. cloacae (Figure 2.).
Figure 2.
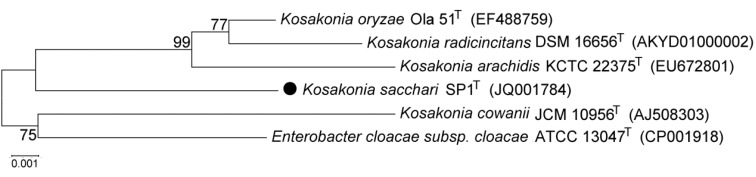
Phylogenetic tree based on 16S rRNA gene sequences of Kosakonia sacchari type strain SP1T (●), the type strains of other species in the genus Kosakonia, and the type strain of Enterobacter cloacae. The sequences were aligned with the CLUSTAL W program and were constructed with the neighbor-joining algorithm integrated in the MEGA 5.0 program [5]. The phylogenetic tree was tested with 1,000 bootstrap replicates. Bootstrap values are shown at the nodes. The GenBank accession numbers of the sequences are indicated in parentheses. The scale bar represents a 0.1% nucleotide sequence divergence.
Like typical members in the genera Enterobacter and Kosakonia, K sacchari SP1T utilizes L-alanine, D-cellobiose, citrate, D-fructose, D-galactose, D-glucose, glycerol, maltose, D-mannitol and D-mannose [2,6,7]. K sacchari differentiates from E. cloacae by utilization of D-arabitol and L-fucose, differentiates from K. radicincitans by utilization of putrescine, D-arabitol, L-fucose and α-methyl-D-glucoside, and differentiates from K. oryzae by utilization of putrescine, D-arabitol and L-rhamnose [2].
Genome sequencing information
Genome project history
K sacchari SP1T was selected for sequencing because it is the type strain of K sacchari, and on the basis of its scientific interest as an endophyte that has the potential to promote the growth of agriculturally important crops by nitrogen fixation [8]. Its 16S rRNA gene sequence is deposited in GenBank under the accession number JQ001784. Its genome sequence is deposited in GenBank under the accession number CP007215.2. A summary of the genome sequencing project information and its association with MIGS version 2.0 compliance is shown in Table 2.
Table 2. Genome sequencing project information for Kosakonia sacchari type strain SP1T.
| MIGS ID | Property | Term |
|---|---|---|
| MIGS-31 | Finishing quality | Finished |
| MIGS-28 | Libraries used | Pacbio 4 – 10 Kb library |
| MIGS-29 | Sequencing platforms | PacBio RS II |
| MIGS-31.2 | Fold coverage | 63 × |
| MIGS-30 | Assemblers | HGAP in smrtanalysis-2.1.1 |
| MIGS-32 | Gene calling method | GeneMarkS+ |
| Genome Database release | Genbank | |
| Genbank ID | CP007215.2 | |
| Genbank Date of Release | May 23, 2014 | |
| Project relevance | Taxonomy, biotechnology |
Growth conditions and DNA isolation
K sacchari SP1T was grown in liquid Luria-Bertani (LB) medium at 30°C to early stationary phase. The genome DNA was extracted from the cells by using a TIANamp bacterial DNA kit (Tiangen Biotech, Beijing, China). DNA quality and quantity were determined with a Nanodrop spectrometer (Thermo Scientific, Wilmington, USA).
Genome sequencing and assembly
The genome DNA of K sacchari strain SP1T was first constructed into a 500-bp-insert library and sequenced by an Illumina HiSeq 2000 sequencing system. A draft genome of 4,945,084 nucleotides containing 239 contigs was obtained and deposited at DDBJ/EMBL/GenBank under the accession no. AMSC00000000 [8]. However, 84,628 nucleotides (203 short contigs) of the draft genome were accidently contaminated by sequences from eukaryotic organisms. Therefore, the genome of SP1T was resequenced at the Duke University Genome Sequencing & Analysis Core Resource using the Pacific Biosciences’ Single Molecule, Real-Time (SMRT) sequencing technology (http://www.pacificbiosciences.com/). A 4 – 10 Kb insert library was constructed. Sequencing was run on a single SMRT Cell. The sequencing data were assembled using the Hierarchical Genome Assembly Process (HGAP) with smrtanalysis-2.1.1. The final assembly of the chromosome produced 63-fold coverage of the genome.
Genome annotation
Automated genome annotation was completed using the NCBI Prokaryotic Genome Annotation Pipeline. Product description annotations were obtained using searches against the KEGG, InterPro, and COG databases. Genes with signal peptides were predicted using SignalP [9]. Genes with transmembrane helices were predicted using TMHMM [10]. Genes for tRNA were found by tRNAScanSE [11]. Ribosomal RNAs were found by using BLASTN vs. ribosomal RNA databases, and 5S rRNA hits were further refined using Cmsearch (http://manpages.ubuntu.com/manpages/raring/man1/cmsearch.1.html). Two hundred twenty seven disrupted genes were replaced by the complete gene sequences obtained from the first Illumina HiSeq 2000 sequencing.
Genome properties
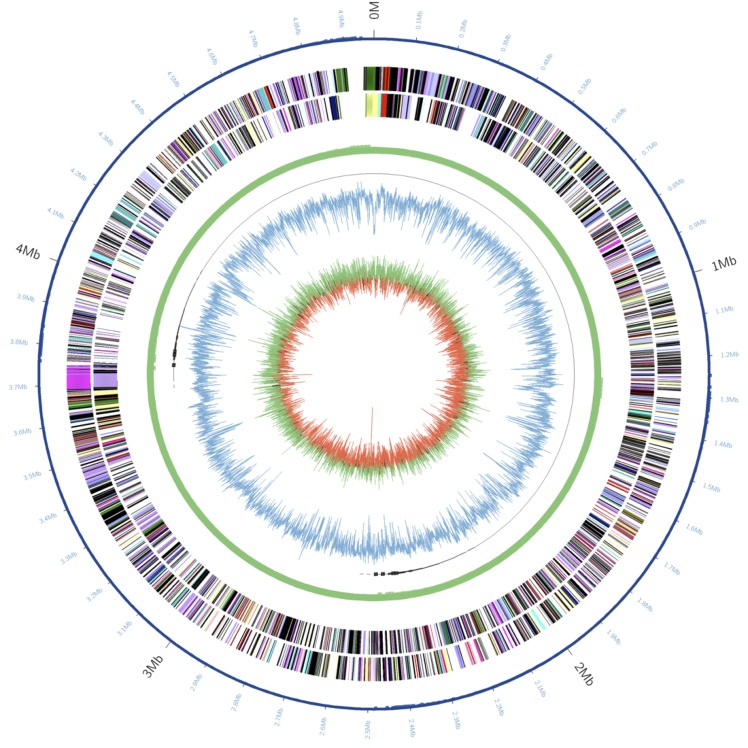
The genome of K sacchari SP1T contains a single chromosome of 4,902,024 nucleotides with 53.7% GC content and no plasmids (Table 3, Figure 3.). The genome contains 4,585 predicted genes, 4,460 protein-coding genes and 105 RNA genes including 22 rRNA genes, 82 tRNA genes and 1 ncRNA gene. A total of 3,752 genes (81.8%) were assigned a putative function. The remaining genes were annotated as hypothetical or unknown proteins (Table 3). The distribution of genes into COGs functional categories is presented in Table 4.
Table 3. Nucleotide content and gene count levels of the genome.
| Attribute | Value | % of total |
|---|---|---|
| Size (bp) | 4,902,024 | 100.00 |
| G+C content (bp) | 2,634,551 | 53.74 |
| Coding region (bp) | 4,281,189 | 87.34 |
| Total genes | 4,585 | 100.00 |
| RNA genes | 105 | 2.29 |
| Protein-coding genes | 4,460 | 97.27 |
| Pseudo genes | 20 | 0.44 |
| Genes assigned to COGs | 3,786 | 82.57 |
| Genes with signal peptides | 452 | 9.86 |
| Genes with transmembrane helices | 1096 | 23.90 |
Figure 3.
Graphical circular map of the chromosome of Kosakonia sacchari type strain SP1T. From outside to the center: Genes on forward strand (color by SEED subsystems [12]), Genes on reverse strand (color by SEED subsystems), genome structure (a circular chromosome with no gaps), GC content, GC skew.
Table 4. Number of genes associated with the 25 general COG functional categories.
| Code | Value | % of totala | Description |
|---|---|---|---|
| J | 193 | 4.33 | Translation |
| A | 2 | 0.04 | RNA processing and modification |
| K | 386 | 8.65 | Transcription |
| L | 170 | 3.81 | Replication, recombination and repair |
| B | 0 | 0.00 | Chromatin structure and dynamics |
| D | 38 | 0.85 | Cell cycle control, mitosis and meiosis |
| Y | 0 | 0.00 | Nuclear structure |
| V | 52 | 1.17 | Defense mechanisms |
| T | 269 | 6.03 | Signal transduction mechanisms |
| M | 251 | 5.63 | Cell wall/membrane biogenesis |
| N | 128 | 2.87 | Cell motility |
| Z | 0 | 0.00 | Cytoskeleton |
| W | 0 | 0.00 | Extracellular structures |
| U | 107 | 2.40 | Intracellular trafficking and secretion |
| O | 144 | 3.23 | Posttranslational modification, protein turnover, chaperones |
| C | 268 | 6.01 | Energy production and conversion |
| G | 394 | 8.83 | Carbohydrate transport and metabolism |
| E | 414 | 9.28 | Amino acid transport and metabolism |
| F | 90 | 2.02 | Nucleotide transport and metabolism |
| H | 186 | 4.17 | Coenzyme transport and metabolism |
| I | 117 | 2.62 | Lipid transport and metabolism |
| P | 265 | 5.94 | Inorganic ion transport and metabolism |
| Q | 82 | 1.84 | Secondary metabolites biosynthesis, transport and catabolism |
| R | 481 | 10.78 | General function prediction only |
| S | 382 | 8.57 | Function unknown |
| - | 674 | 15.11 | Not in COGs |
a) The total is based on the total number of protein coding genes in the annotated genome.
Comparison with the genome of Enterobacter sp. strain R4-368
The chromosome of K sacchari SP1T shows the highest sequence similarities ranging from 69.5% to 100% to the chromosome of Enterobacter sp. strain R4-368, which is an endophytic nitrogen-fixing bacterium isolated from the biofuel plant Jatropha curcas [13]. The genome of the strain R4-368 comprises a single circular chromosome of 5,039,027 bp with 54.0% GC content (deposited in GenBank under the accession number CP005991) and one plasmid pENT01 of 116,007 bp with 52.8% GC content (deposited in GenBank under the accession number CP005992) [13].
The chromosome of K sacchari SP1T shares 4,105 genes (89.5%) with the chromosome of strain R4-368. The digital DNA-DNA hybridization values between the two chromosomes calculated by the online Genome-to-Genome Distance Calculator [14,15] (version 2.0; http://ggdc.dsmz.de) are 90.2%, 57.7%, and 86.6% under the distance Formula 1, 2 (recommended for dealing with incomplete genomes), and 3, respectively. The probabilities of same species for the two strains (DDH > 70%) assessed via logistic regression are 97.4%, 44.3%, and 98.8%, respectively. Likely, strain R4-368 belongs to the species K sacchari.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant 31171504 and 31200003), Guangxi Key Laboratory Construction Program, Guangxi Expert Special Fund Project, and BAGUI Scholar Program of Guangxi, China.
References
- 1.Brady C, Cleenwerck I, Venter S, Coutinho T, De Vos P. Taxonomic evaluation of the genus Enterobacter based on multilocus sequence analysis (MLSA): Proposal to reclassify E. nimipressuralis and E. amnigenus into Lelliottia gen. nov. as Lelliottia nimipressuralis comb. nov. and Lelliottia amnigena comb. nov., respectively, E. gergoviae and E. pyrinus into Pluralibacter gen. nov. as Pluralibacter gergoviae comb. nov. and Pluralibacter pyrinus comb. nov., respectively, E. cowanii, E. radicincitans, E. oryzae and E. arachidis into Kosakonia gen. nov. as Kosakonia cowanii comb. nov., Kosakonia radicincitans comb. nov., Kosakonia oryzae comb. nov. and Kosakonia arachidis comb. nov., respectively, and E. turicensis, E. helveticus and E. pulveris into Cronobacter as Cronobacter zurichensis nom. nov., Cronobacter helveticus comb. nov. and Cronobacter pulveris comb. nov., respectively, and emended description of the genera Enterobacter and Cronobacter. Syst Appl Microbiol 2013; 36:309-319 10.1016/j.syapm.2013.03.005 [DOI] [PubMed] [Google Scholar]
- 2.Zhu B, Zhou Q, Lin L, Hu C, Shen P, Yang L, An Q, Xie G, Li Y. Enterobacter sacchari sp. nov., a nitrogen-fixing bacterium associated with sugar cane (Saccharum officinarum L.). Int J Syst Evol Microbiol 2013; 63:2577-2582 10.1099/ijs.0.045500-0 [DOI] [PubMed] [Google Scholar]
- 3.Gu CT, Li CY, Yang LJ, Huo GC. Enterobacter xiangfangensis sp. nov., isolated from Chinese traditional sourdough and reclassification of Enterobacter sacchari Zhu et al. as Kosakonia sacchari comb. nov. Int J Syst Evol Microbiol 2014; in press [DOI] [PubMed] [Google Scholar]
- 4.Lin L, Li Z, Hu C, Zhang X, Chang S, Yang L, Li Y, An Q. Plant growth-promoting nitrogen-fixing Enterobacteria are in association with sugarcane plants growing in Guangxi, China. Microbes Environ 2012; 27:391-398 10.1264/jsme2.ME11275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011; 28:2731-2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grimont F. Grimont PAD. The genus Enterobacter. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds), The Prokaryotes Volume 6: Proteobacteria: Gamma Subclass, Springer, New York, 2006, pp. 197-214. [Google Scholar]
- 7.Hoffmann H, Stindl S, Stumpf A, Mehlen A, Monget D, Heesemann J, Schleifer KH, Roggenkamp A. Description of Enterobacter ludwigii sp. nov., a novel Enterobacter species of clinical relevance. Syst Appl Microbiol 2005; 8:206-212 10.1016/j.syapm.2004.12.009 [DOI] [PubMed] [Google Scholar]
- 8.Zhu B, Chen M, Lin L, Yang L, Li Y, An Q. Genome sequence of Enterobacter sp. strain SP1, an endophytic nitrogen-fixing bacterium isolated from sugarcane. [PubMed]. J Bacteriol 2012; 194:6963-6964 10.1128/JB.01933-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. [PubMed]. J Mol Biol 2004; 340:783-795 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]
- 10.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. [PubMed]. J Mol Biol 2001; 305:567-580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 11.Lowe TM, Eddy SR. t-RNAscan-SE: a program for improved detection of transfer RNA gene in genomic sequence. Nucleic Acids Res 1997; 25:955-964 10.1093/nar/25.5.0955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. [PubMed]. Nucleic Acids Res 2005; 33:5691-5702 10.1093/nar/gki866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madhaiyan M, Peng N, Ji L. Complete genome sequence of Enterobacter sp. strain R4-368, an endophytic N-fixing gammaproteobacterium isolated from surface-sterilized roots of Jatropha curcas L. [PubMed]. Genome Announc 2013; 1:e00544-e13 10.1128/genomeA.00544-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auch AF, Klenk HP, Göker M. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. [PubMed]. Stand Genomic Sci 2010; 2:142-148 10.4056/sigs.541628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auch AF, Von Jan M, Klenk HP, Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. [PubMed]. Stand Genomic Sci 2010; 2:117-134 10.4056/sigs.531120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. [PubMed]. Proc Natl Acad Sci USA 1990; 87:4576-4579 10.1073/pnas.87.12.4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garrity GM, Bell JA, Lilburn T. Phylum XIV. Proteobacteria phyl. nov. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Part B, Springer, New York, 2005, p. 1. [Google Scholar]
- 18.Garrity GM, Bell JA, Lilburn T. Class III. Gammaproteobacteria class. nov. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 2, Springer, New York, 2005, p. 1. [Google Scholar]
- 19.List Editor Validation of publication of new names and new combinations previously effectively published outside the IJSEM. List no. 106. Int J Syst Evol Microbiol 2005; 55:2235-2238 10.1099/ijs.0.64108-0 [DOI] [PubMed] [Google Scholar]
- 20.Williams KP. Kelly DP. Proposal for a new class within the phylum Proteobacteria, Acidithiobacillia classis nov., with the type order Acidithiobacillales, and emended description of the class Gammaproteobacteria. Int J Syst Evol Microbiol 2013; 63:2901-2906 10.1099/ijs.0.049270-0 [DOI] [PubMed] [Google Scholar]
- 21.Garrity GM, Holt JG. Taxonomic Outline of the Archaea and Bacteria. In: Garrity GM, Boone DR, Castenholz RW (eds), Bergey's Manual of Systematic Bacteriology, Second Edition, Volume 1, Springer, New York, 2001, p. 155-166. [Google Scholar]
- 22.Judicial Commission Conservation of the family name Enterobacteriaceae, of the name of the type genus, and designation of the type species OPINION NO. 15. Int Bull Bacteriol Nomencl Taxon 1958; 8:73-74 [Google Scholar]
- 23.Skerman VBD. McGowan V, Sneath PHA. Approved Lists of Bacterial Names. Int J Syst Bacteriol 1980; 30:225-420 10.1099/00207713-30-1-225 [DOI] [PubMed] [Google Scholar]