Abstract
Introduction
FASD is a leading cause of neurodevelopmental disability. Genetic factors can modify vulnerability to FASD, but these elements are poorly characterized.
Methods
We performed high-throughput transcriptional profiling to identify gene candidates that could potentially modify vulnerability to ethanol’s neurotoxicity. We interrogated a unique genetic resource, neuroprogenitor cells from two closely-related Gallus gallus lines having well-characterized robust or attenuated ethanol responses with respect to intracellular calcium mobilization and CaMKII / β-catenin-dependent apoptosis. Samples were not exposed to ethanol prior to analysis.
Results
We identified 363 differentially expressed genes in neuroprogenitors from these two lines. KEGG analysis revealed several gene clusters having significantly differential enrichment in gene expression. The largest and most significant cluster comprised ribosomal proteins (38 genes, p = 1.85 × 10−47). Other significantly enriched gene clusters included metabolism (25 genes, p = 0.0098), oxidative phosphorylation (18 genes, p = 1.10 × 10−11), spliceosome (13 genes, p = 7.02 × 10−8) and protein processing in the endoplasmic reticulum (9 genes, p = 0.0011). Inspection of GO-terms identified 24 genes involved in the calcium/β-catenin signals that mediate ethanol's neurotoxicity in this model, including β-catenin itself and both calmodulin isoforms.
Conclusions
Four of the identified pathways with altered transcript abundance mediate the flow of cellular information from RNA to protein. Importantly, ribosome biogenesis also senses nucleolar stress and regulates p53-mediated apoptosis in neural crest. Human ribosomopathies produce craniofacial malformations and eleven known ribosomopathy genes were differentially expressed in this model of neural crest apoptosis. Rapid changes in ribosome expression are consistently observed in ethanol-treated mouse embryo neural folds, a model that is developmentally similar to ours. The recurring identification of ribosome biogenesis suggests it is a candidate modifier of ethanol vulnerability. These results highlight this approach’s efficacy to formulate new, mechanistic hypotheses regarding ethanol’s developmental damage.
Keywords: Fetal alcohol spectrum disorders, transcriptome profiling, neural crest, apoptosis, ribosome biogenesis, Diamond-Blackfan anemia, oxidative phosphorylation, Gallus gallus
Introduction
Prenatal alcohol exposure (PAE) is a leading cause of neurodevelopmental disability (Mattson et al., 2013). Some affected individuals display distinctive craniofacial anomalies that are caused, in part, by the apoptotic depletion of neural crest progenitors during early development (Cartwright et al. 1998; Dunty et al. 2001). Individual vulnerability to Fetal Alcohol Spectrum Disorders (FASD) varies appreciably (Warren and Li, 2005) despite apparently equivalent patterns and intakes of alcohol. Environmental factors affecting phenotypic outcomes include maternal nutrition and age, stressors, polydrug abuse, and socioeconomic status (Carter et al. 2007; Leonardson and Loudenburg, 2003; May et al. 2011).
Genetic factors may also affect FASD outcomes. Monozygotic twins have greater concordance for FASD diagnosis than do dizygotic twins (Streissguth and Dehaene, 1993). Some of this discordance originates from differences in alcohol clearance rates. For example, polymorphisms in alcohol dehydrogenases, including ADH1B*2 and ADH1B*3, affect ethanol oxidation rates and blood-alcohol concentrations, and thus affect fetal ethanol exposure and FASD risk (Das et al., 2004; McCarver et al., 1997; Viljoen et al., 2001). Animal studies confirm that genetic factors modify individual sensitivity to ethanol’s teratogenesis including the face, brain, heart, and skeleton (Boehm et al. 1997; Chen et al. 2000: Debelak et al. 2000; Downing et al. 2012; Swartz et al. 2013; Gilliam and Kotch 1996; Goodlett et al. 1989; Su et al., 2001). This phenotypic heritability persists even when factors such as ethanol metabolism and exposure timing are equivalent, implying the existence of additional polymorphisms that modify ethanol vulnerability.
We have reported previously that embryos from chicken strains can differ significantly in their sensitivity to ethanol’s developmental damage (Debelak, et al. 2000; Su et al., 2001). In the well-characterized ethanol-sensitive line, Hy-Line W98S, ethanol (EC50 = 52 mM) elicits an intracellular calcium transient that originates from a G protein-coupled receptor and phosphoinoside signaling. This calcium signal mediates the CaMKII-dependent destabilization of transcriptionally active β-catenin, a trophic factor for neural crest progenitors (Flentke et al. 2011, 2013; Garic et al. 2011; Garic-Stankovic et al. 2005; Debelak-Kragtorp et al. 2003). In contrast, ethanol-mediated events including calcium mobilization, apoptosis, and facial dysmorphology are significantly attenuated in ethanol-resistant strains such as DeKalb Black and Hy-Line W98D despite equivalent ethanol exposure (Debelak, et al. 2000; Su et al., 2001). Because this ethanol-stimulated intracellular calcium transient mediates apoptosis in multiple species and diverse cell lineages including early mouse embryos, mouse cerebellar granule cells, zebrafish neural crest, and human cytotrophoblast lines (Kilburn et al. 2006; Kouzoukas et al. 2013; Flentke et al., 2014; Bolnick et al., 2014), a deeper understanding of this mechanism will more broadly inform studies of ethanol’s developmental toxicity.
RNA-Seq is a high-throughput sequencing technology that digitally records transcript abundance. Because this approach is genetically comprehensive, it eliminates several confounding analytical complications associated with hybridization-based technologies such as cross-hybridization and indirect quantification of signal intensity (Wang et al. 2009). Here, we perform RNA-Seq on neuroprogenitors of the W98S and W98D lines, which are closely-related but known previously to differ in their sensitivity to ethanol-induced calcium transients and apoptosis. We hypothesize that these cell populations’ de novo characteristics will provide insights into gene clusters and polymorphisms that potentially modulate vulnerability to FASD.
Methods
Genetic Stocks
Two closely-related commercial lines of the W98 White Leghorn chicken were obtained from Hy-Line International (West Des Moines, IA) from their facilities at Spencer, IA (W98S) and Dallas Center, IA (W98D), respectively. Both lines were derived from sib-grandparent stocks and were separated by at least ten generations (R. Muetzel, pers. comm.). W98D originated from W98S by continued selection for improved egg production; additional genetic and selection details are proprietary to Hy-Line. We noted that W98D also had reduced aggression.
Calcium Imaging
Ratiometric imaging to quantify intracellular calcium release in response to ethanol challenge was performed in embryos having 4 somites (stage 8−) as described (Garic et al. 2011). Data were analyzed using non-linear regression analysis (SigmaStat). We evaluated at least 6 embryos per dose and group.
Apoptosis Quantitation
Cell death at 16–18 somites (stage 12/13), in response to ethanol challenge at 4 somites, was quantified as described using acridine orange, which detects apoptosis in this model (Garic et al. 2011). We enumerated the number of labeled neural progenitors within rhombomere 4, which normally lacks appreciable cell death, in >10 embryos per treatment. Data were analyzed using one-way ANOVA for within-line comparison and t-test for between-line comparisons (SigmaStat). A p value <0.05 was considered statistically significant.
Embryo manipulation
The neural folds anterior to somite pair two were dissected from embryos at stage 8+ to 9+ (5–8 somites). Each sample for sequencing contained 23 individual neural folds and all samples were developmentally-matched to contain 5 heads at stage 8+, 5 heads at stage 9−, 9 heads at stage 9, and 4 heads at stage 9+. Thus all sequencing data sets were identical with respect to somite number and developmental stage. These stages have equivalent ethanol responses (Cartwright and Smith, 1995; Debelak and Smith, 2000). Neural folds were not exposed to ethanol.
cDNA preparation and sequencing
RNA was extracted using a RNAqueous-4PCR Kit (Ambion/Applied Biosystems, Carlsbad, CA). Total RNA concentration and purity was determined using a NanoDrop-1000 spectrophotometer (ThermoFisher Scientific, Wilmington DE). RNA-Seq analysis was performed at the University of Wisconsin-Madison Biotechnology Center. RNA concentration and purity were reaffirmed using the Qubit 1.0 NanoDrop spectrophotometer (Life Technologies), and RNA integrity was assessed with an Agilent 2100 BioAnalyzer and RNA 6000 Nano kit (Agilent Technologies, Santa Clara, CA). cDNA libraries were constructed using the RNA Sequencing Sample Preparation kit (Illumina Inc., San Diego, CA). mRNA was purified from approximately 8 µg total purified RNA with poly-T oligo-attached magnetic beads. Double-stranded cDNA was synthesized using SuperScript II (Invitrogen, Carlsbad, CA) with random hexamer priming, purified with QIAquick PCR purification columns (Qiagen, Valencia, CA), ligated to Illumina adapters, and size-selected using electrophoresis to approximately 300bp. This fraction of size-enriched DNA was amplified in a Linker-Mediated PCR reaction (LM-PCR) and then column-purified (Zymo Research, Irvine, CA). Library quality was assessed using a DNA High Sensitivity series chip assay and a Qubit dsDNA HS assay on an Agilent 2100 Bioanalyzer and Qubit fluorometer (Invitrogen) respectively. Cluster generation was performed using Illumina’s Single Read Cluster Generation Kit and the Illumina cBot cluster amplification system. Paired-end reads (75 bp) were generated on an Illumina Genome Analyzer IIx with each sample occupying an independent lane on the same flow cell. For each line, we sequenced two biological replicates and each sequencing reaction represented an independent isolation of 23 stage-matched headfolds.
Data Analysis
Illumina’s Genome Analyzer Pipeline Software v1.8 was used to transform the primary imaging output into fastq files containing sequences of nucleic acids associated with quality scores. All RNA-Seq sequence reads were filtered through four trimming procedures that removed: (1) reads failing to meet or exceed Illumina’s default chastity threshold of 0.6 over the first 25 cycles; (2) reads containing more than two contiguous or interspersed ambiguous nucleotides; (3) reads with quality scores less than 20 (Phred scale; 1 error per 100 nucleotides); and (4) reads that were shorter than the expected 75bp.
The RNA-Seq module in CLC Genomics Workbench 5.5 (CLC Bio, Cambridge, MD) was used to map the filtered paired-end reads to 17,934 protein-coding genes (known or predicted) in Galgal3, assembly 2.1 (May 2006), of the Gallus gallus genome (Ensembl Release 70, January 2013). Reads were mapped to the reference genome, individually requiring at least 90% of each read to exhibit 80% or greater alignment similarity. Only reads that mapped uniquely to the reference were included in subsequent analyses. The number of read counts requires normalization to remove technical biases intrinsic to the RNA-Seq methodology, including the length of the RNA target and sequencing depth. Most often, Reads Per Kilobase per Million mapped reads (RPKM) is used (Mortazavi, et al., 2008). However, RPKM introduces significant bias in per-gene variances (Oshlack and Wakefield, 2009), and this bias is exceptional, especially for low-abundance transcripts, prompting the recommendation to abandon RPKM normalization altogether (Dillies, et al. 2013). Instead of RPKM, we used DE-Seq (v1.10.1; Anders and Huber, 2010) to normalize raw counts of mappings. In contrast to RPKM, the procedures in DE-Seq employ normalization factors within the statistical model used for identification of differential gene expression, rather than on the raw data themselves. Briefly, the geometric mean of read counts for each gene in each condition is used as a reference count value. To obtain the sequence depth of each gene relative to its reference, the observed read count for each gene in each condition is divided by its reference value. The median value is taken to represent the relative sequencing depth of each RNA-seq library. This method is robust to biases observed in real RNA-seq datasets (Dilles, et al. 2013).
In RNA-Seq data, the observed dispersion is expected to decrease as read count increases. Thus, empirical estimates of dispersion less than model-fitted values underestimate the true dispersion, the magnitude of which is dependent on the number of replicates performed (Anders and Huber, 2010). To be conservative, the DE-Seq procedure replaces empirical estimates of dispersion with the model-fitted value. However, it retains all empirical values equal to and larger than the fitted line values, even if the dispersion is overestimated in the latter case. This procedure dictates that as fewer read counts are observed (i.e., more dispersion) a larger difference between normalized counts between RNA-Seq libraries is required to achieve statistically significant differential expression. All p-values were adjusted with the Benjamin-Hochberg multiple testing correction to control the false discovery rate (FDR). Data were evaluated as mean normalized counts vs. dispersion, and as mean normalized counts vs. log2-fold change, plotting both empirical dispersion and fitted values. Genes were deemed differentially expressed if the adjusted level of significance was below 0.10. Genes identified as differentially expressed (DE) (p < 0.1) were submitted to DAVID (Huang et al., 2009a, 2009b) for GO term analysis. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (Kanehisa and Goto, 2000) was performed in DAVID using the above parameters.
Results
Differential Ethanol Responses of W98 lines
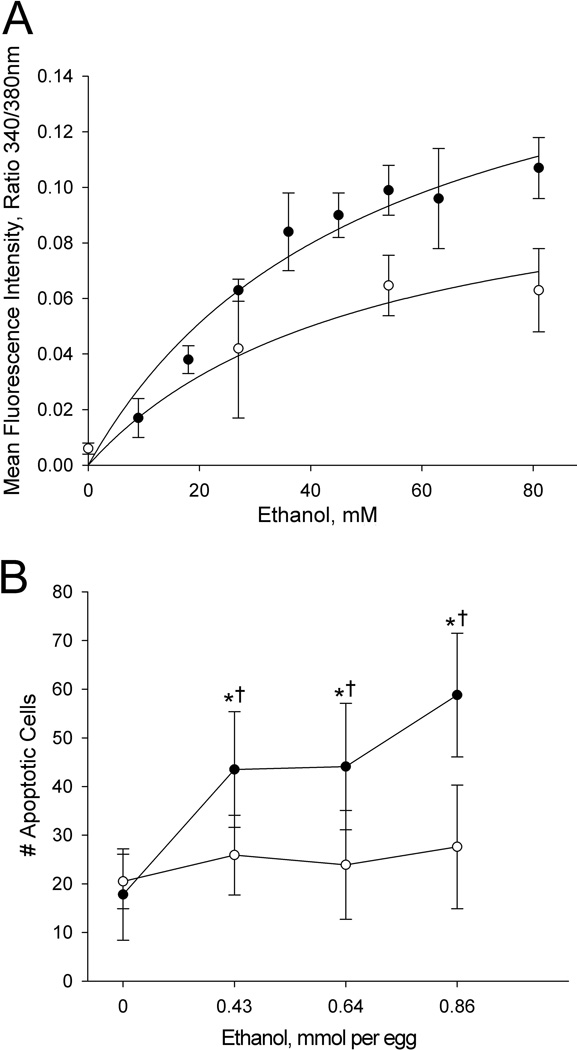
Neuroprogenitors from the well-characterized chick line W98S are sensitive to ethanol's neurotoxicity (Debelak and Smith, 2000; Debelak-Kragtorp et al., 2003; Garic et al., 2011; Garic-Stankovic et al., 2005). They initiate a robust and dose-dependent intracellular calcium transient within seconds of ethanol exposure (Figure 1A). This is followed by significantly increased apoptosis within neural crest and neural progenitors (Figure 1B), and we have shown elsewhere that the calcium transient initiates this apoptosis (Debelak-Kragtorp et al., 2003; Garic et al., 2005). In contrast, we discovered significantly blunted ethanol responses within equivalently-staged neuroprogenitors from a closely-related W98 line provided by the vendor's Dallas Center, IA, facility (hereafter referred to as W98D). The calcium transients within W98D neuroprogenitors averaged 60% to 67% of W98S responses at equivalent ethanol concentrations (Figure 1A). The differential response was not due to a shifted dose-response curve, because higher ethanol concentrations did not further increase the calcium levels in W98D. These same ethanol levels also failed to cause appreciable apoptosis within W98D neuroprogenitors, and the number of dead cells did not differ from that in saline-treated controls (Figure 1B). Their lack of cell death was not due to differences in apoptosis timing or developmental progression, and cell death levels remained low during the 22hr interval between ethanol challenge and cell death assessment, and for several hours thereafter (data not shown). Non-linear regression analysis of the calcium responses, using global curve fitting, revealed that the data best modeled an assumption that the two responses shared a similar Kd and had significantly different maxima (p<0.005). The apparent Kd for the ethanol-calcium dose-response was 55 mM for W98S and 51 mM for W98D; these values did not significantly differ and were similar to that previously calculated for W98S (52 mM, Garic-Stankovic et al., 2005). The similarity of the respective Kd values suggested that, at first approximation, there was no appreciable difference in ethanol affinity between the two lines, but that the lines differed in how the ethanol signal was transduced within these cells.
Figure 1. Differential ethanol sensitivity of W98S and W98D neural progenitors.
For both panels (●) is W98S and (○) is W98D. (A) Elevation of intracellular calcium within neural progenitors following ethanol challenge at the indicated concentrations as quantified using ratiometric imaging for Fura-2.Values are mean ± SD for 3–8 embryos per treatment. Data are plotted using non-linear regression analysis and global curve fitting to evaluate each line's response maxima and relative binding affinity for the dose-response. (B) The numbers of acridine orange-labeled cells within rhombomere 4 were enumerated within HH12/13 hindbrain following treatment at HH8 with the indicated ethanol doses. * indicates values significantly different from 0 mmol ethanol within that line at p< 0.001 using one-way ANOVA. †indicates values significantly different between lines at the same ethanol exposure at p< 0.001 using t-test. N = 9–15 embryos per treatment group.
Differential Gene Expression in W98S/D
To screen for gene candidates having differential expression and potentially contributing to ethanol sensitivity, we performed RNA-Seq on neuroprogenitors isolated from W98S and W98D. The number of trimmed reads obtained from 75 bp paired-end nucleotide sequencing was 67.5 (W98S) and 71.8 (W98D) million. The cumulative distribution of read alignments across transcript targets was nearly identical in each dataset. Of those sequences that mapped to an annotated region, 89.81% mapped uniquely to an annotated region in the Gallus gallus genome (Ensembl release 70). The number of non-specific mappings was very low and averaged 0.73% and 1.92%, respectively.
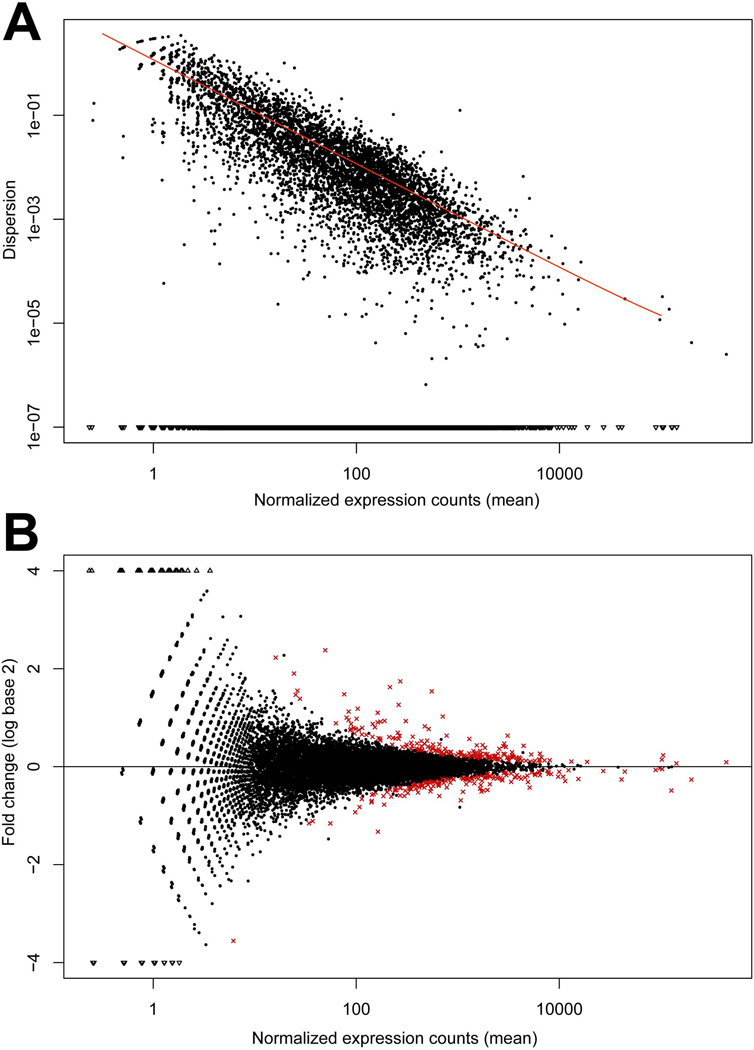
The magnitude of dispersion is directly related to the biological variation present in the pooled DNA sample population and is represented by plotting normalized expression vs. magnitude of dispersion. For these data, as read count increased, the dispersion decreased as represented by the fitted line in Figure 2A, indicating that the variation between the replicate runs was minimal. Analysis of these datasets using a FDR = 0.10 and Benjamin-Hochberg correction revealed 363 differentially expressed genes between the W98S/D lines (Figure 2B; Supplemental Table 1). Of these, 52.9% (192 genes) had significantly decreased transcript levels in W98S, and 47.1% (171 genes) were significantly increased. Following this analysis, Galgal4 became available in Ensembl (e71; Apr 2013). However, because Galgal4 has not been integrated into any additional software applications (KEGG, DAVID, STRING, GO term), the analyses presented herein utilize Galgal3 (Ensembl release70). Gene names were updated with Galgal4 annotation when applicable.
Figure 2. Distribution plots for differentially expressed transcripts in neural folds of ethanol-sensitive (W98S) and ethanol-resistant (W98D) lines in the absence of ethanol treatment.
(A) Normalized dispersion plot showing variance in Read Counts across the W98S and W98D RNA-Seq libraries. Black dots indicate the empirical dispersion. As expected, dispersion decreases as read count increases, as indicated by the fitted line (red). (B). Comparison of transcriptome abundance between W98S and W98D neural progenitors including neural crest. Fold-changes (mean expression W98D – mean expression W98S) are presented as log2. Genes with significantly different expression are shown in red.
Biotype analysis identified at least one transcript mapping to five of ten identified biotypes for each line (Supplemental Figure 1). This included transcripts for protein-coding genes, pseudogenes, mitochondrial RNA, miscellaneous RNA and snoRNA. Transcripts encoding ribosomal and transfer RNAs were not isolated. This further endorsed that the RNAs under analysis were high quality.
Gene Ontogeny (GO) and KEGG Analysis
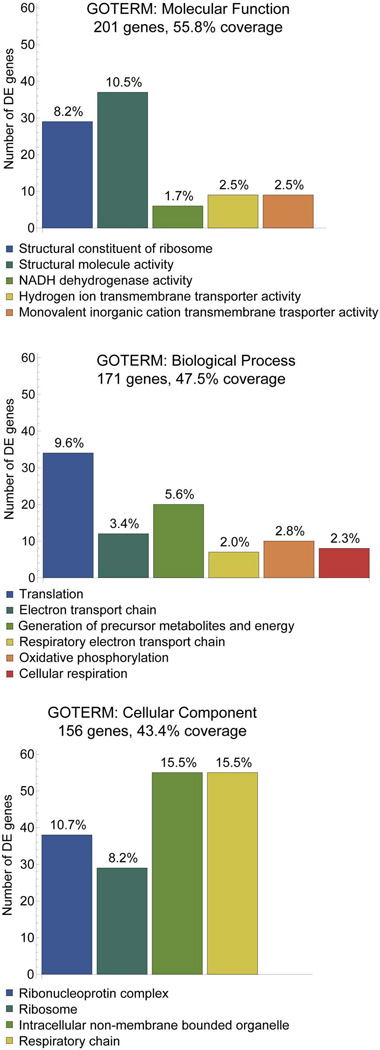
For the 363 differentially expressed genes identified, we performed GO-term and KEGG-pathway enrichment analysis with DAVID on 17,934 genes (Galgal3, Ensembl release 70) with at least one transcript expressed in all samples combined. The three most significantly enriched GO terms were Molecular Function (201 genes, 55.8% coverage), Biological Process (171 genes, 47.5% coverage), and Cellular Component (156 genes, 43.4% coverage). Within these sub-ontogenies, the most abundant members were associated with ribosomes, translation, metabolic pathways, oxidative phosphorylation, and energy generation (Figure 3).
Figure 3. Gene ontogeny modules showing significant differential enrichment in ethanol-sensitive versus ethanol-resistant neural crest and neural progenitors at p≤0.05.
The top enrichments of GOTERMS involved Molecular Function (A), Biological Processes (B), and Cellular Components (C). For each category, the number of transcripts represented out of the 363 DE genes is indicated. For each column the percentage of transcripts within each GOTERM is indicated.
KEGG analysis indicated that 33.9% of the 363 genes were associated with one or more biologically characterized pathways at level of significance p≤0.05 (Table 1). Of these ten pathways, those containing the greatest number of genes were Ribosomal (N=38 genes, p = 1.85 × 10−47), Metabolism (N=25, p=0.0098), and Oxidative Phosphorylation (N=18, p = 1.10 × 10−11); enrichment of Glycolysis / Gluconeogenesis approached significance (N=18 genes, p=0.053). Other pathways that had significant gene enrichment included Spliceosome (N=13, p = 7.02 × 10−8), Protein Processing in the Endoplasmic Reticulum (N=9, p=0.0011), and Cardiac Muscle Contraction (N=10, p = 5.71 × 10−9).
Table 1.
KEGG Pathway Enrichments in W98S vs. W98D.
| KEGG Identity | # Genes | Significance | Genes |
|---|---|---|---|
| 3010 Ribosomal | 38 | 1.85 × 10−47 | GGA.41946, RPL5, RPL6, RPL7, RPL10A, RPL11, RPL12, RPL13, RPL14, RPL17L, RPL17L, RPL18A, RPL21, RPL23, RPL23A, RPL24, RPL26, RPL27, RPL29, RPL31, RPL35, RPL35A, RPL37, RPL37A, RPL39, RPL38, RPLP1, RPS2, RPS3A, RPS4, RPS6, RPS8, RPS10, RPS11, RPS12, RPS15, RPS17, RPS23 |
| 190 Oxidative Phosphorylation | 18 | 1.10 × 10−11 | ALG8, ATP5A1, ATP5G1, ATP5H, ATP6, ATXN3, COX8A, gga-mir-3527, MT-CO1, MT-CO3, MT-CYB, MT-ND2, MT-ND3, MT-ND4, ND4L, ND6, NDUFA4, UQCR10 |
| 4260 Cardiac Muscle | 10 | 5.71 × 10−9 | ACTC1, ATP1B3, COX8A, gga-mir-3527, MT-CO1, MT-CO3, MT-CYB, TNNT2, TPM1, UQCR10 |
| 3040 Spliceosome | 13 | 7.02 × 10−8 | HNRPK, HNRNPA3, HSPA8, RBM17, SF3A1, SF3B4, SNRNP40, SNRPD3, SNRPB, SNRPF, SRSF3, TRA2A, WBP11 |
| 4141 Protein Processing In ER | 9 | 0.00109 | ATXN3, HSP90AB1, HSP90B1, HSPA8, PDIA4, SEC61A1, SEC61G, SSR2, UBE2E1 |
| 4530 Epithelial Tight Junctions | 6 | 0.00309 | ACTB, CGN, CLDN1, CTNNB1, EXOC4, MYH9 |
| 4114 Oocyte meiosis | 6 | 0.00585 | CALM, CALM2, CDC2, YWHAE, YWHAQ, YWHAZ |
| 3013 RNA Transport | 7 | 0.00676 | CHSAP18, EIF4G2, NUP160, PABPC1, SUMO2, TACC3, XPO5 |
| 1100 Metabolism | 25 | 0.00983 | ALG8, ATP5G1, ATP5H, AK2, ATP5A1, ATP6, ATXN3, B3GNT2, COX8A, DNMT3A, ENO1, FUK, gga-mir-3527, MT-CO1, MT-CO3, MT-CYB, MT-ND2, MT-ND3, MT-ND4, ND4L, ND6, NDUFA4, PRPS1, ST3GAL4, UQCR10 |
| 4110 Cell cycle | 6 | 0.0140 | CDC2, MCM5, STT3A, YWHAE, YWHAQ, YWHAZ |
| 10 Glycolysis / Gluconeogenesis | 18 | 0.0530 | ATXN3, CALM, CALM2, ENO1, HSP90AB1, LDHA, LGMN, NDUFA4, PRDX1, PTPRF, RPL35, RPS3A, RPS8, STT3A, TRA2A, PDHA1 YWHAZ |
| 4145 Phagosome | 5 | 0.0600 | ACTB, CTSL2, E1C520_CHICK, SEC61A1, SEC61G |
Because the Gallus gallus genome is incompletely annotated as compared with species such as human and mouse, we visually screened the KEGG descriptors of these 363 genes to identify those genes having known contributions to the calcium-β-catenin apoptosis pathway that mediates these cells' ethanol response. This revealed genes associated with β-catenin/Wnt signaling (N=18), neural crest development (N=21), calcium signaling (N=9), and apoptosis (N=14; Table 2). Noteworthy among these, transcripts encoding both calmodulin isoforms, which we have shown convert ethanol’s calcium transient into a pro-apoptosis signal in these cells (Garic-Stankovic et al. 2005; Garic et al. 2011), were reduced in ethanol-sensitive W98S (CALM 0.899-fold, p=0.017; CALM2 0.847-fold, p=0.060). W98S also had lower levels of the β-catenin antagonist Dapper-2 (0.810-fold, p=0.002) and reduced expression of beta-catenin itself (0.917-fold, p=0.0059), whose repression by ethanol mediates the apoptosis (Flentke et al., 2011). Other transcripts with notable differential expression included ApoB (11.76-fold, p=0.085), ROMO1 (0.30-fold, p=2.52 × 10−29), somatostatin (0.34-fold, p=0.089), and multiple novel genes, one of which was not detected in W98D (ENSGALG00000023495, p=2.94 × 10−7).
Table 2.
Expanded DE Categories
| Pathway | W98S/W98D | Gene Names |
|---|---|---|
| Wnt/β-Catenin Signals (18) | Decreased | CALM, CALM2, CTNNB1, CDH2, DACT2, PTPRF, GPC4, NRP2, PTN, SUMO2, TAX1BP3, XIRP1, YWHAZ |
| Increased | GPR177, CHPKC1, SFRP2, SHISA2, TCF7L2 | |
| Calcium Signaling (9) | Decreased | CALM, CALM2, CDH2, CLSTN1, TNNT2 |
| Increased | CDH11, NEDD4L, SLC24A6, TMEM38A, | |
| Apoptosis (14) | Decreased | AK2, ANP32E, C1QBP, ERC1, GHITM, HDGF, HTRA2, ROMO1, SUMO2, YWHAZ |
| Increased | F1NGP2 (Clusterin), CYT_Chick, SERBP1, TCTP, VDAC2 | |
| Alcohol/Addiction (5) | Decreased | CALM, CALM2, DSTN, PEBP1 |
| Increased | AUTS2 | |
| Neural Crest (21) | Decreased | CTNNB1, CALM, GPC4, KIRREL, PHB2, PIGY, RPL24, YWHAE |
| Increased | CDH11, CHSAP18, CNIH4, FBLN1, GPR177, PBX1A, PES1, PIGG, SETBP1, SFRP2, TSPAN3, WWP2, ZIC1 |
Discussion
Insufficient attention has focused on identifying genes that may affect vulnerability to ethanol’s developmental damage, apart from nucleotide polymorphisms that modify maternal-fetal ethanol metabolism. Our comparison of neural crest and neural progenitors from embryos having differential ethanol sensitivities has identified 363 genes that are differentially expressed in these genetically-related lines. Importantly, these expression-level differences between W98S/D were identified in the absence of ethanol challenge, and thus ethanol itself does not factor into these results. The W98S/D lines were selected for characteristics independent of ethanol response and, in the absence of genetic lineage groups, many of their gene differences are unlikely to affect ethanol responses. However, several of these significantly altered genes encode proteins that directly participate in or modify the calcium-CaMKII-β-catenin pathway that governs the ethanol-induced apoptosis studied here (Garic et al. 2011; Flentke et al. 2011, 2013). Thus, they are good candidates to shape cellular responses to ethanol. Our most important finding is that several of the most significantly altered gene clusters identified here were independently identified in screens designed to detail early transcriptional changes in response to ethanol challenge (Green et al. 2007; Downing et al. 2012). Moreover, those latter studies used embryonic models that were developmentally very similar to our own, and compared gene expression within ethanol-treated neural folds isolated from mouse strains having high (C57Bl/6J) or low (C57Bl/6N, DBA/2J) susceptibility to ethanol teratogenesis. Those studies identified the same KEGG enrichments include ribosomes, mRNA splicing, tight junctions, and glycolysis / gluconeogenesis. The consistent detection of these clusters in all three studies suggests a model where changes in one or more of these clusters may have primed naïve neuroprogenitors for ethanol sensitivity or resistance. Our identification of calcium/Wnt components, as well as previously-recognized gene clusters, underscores the potential of this unbiased analysis to formulate novel hypotheses regarding the mechanisms that underlie ethanol’s developmental damage.
Ribosomal Pathways
The most compelling finding from this unbiased search for candidate genes is that four of the significantly represented KEGG pathways and 18.6% of the genes participate in the flow of cellular information from RNA to protein, including networks of the spliceosome, RNA transport, the ribosome, and protein processing in the endoplasmic reticulum. Of these, the cluster showing the largest and most significant enrichment included 38 genes that participate in ribosome biosynthesis including 27 large and 11 small ribosomal subunit proteins, as well as two additional participants in ribosome biogenesis, pescadillo (PES-1, 1.198-fold) and NSA2 (1.340-fold). Two separate interrogations of mouse neural folds, 3–4hr post-ethanol treatment, have each found significant decreases in ribosome transcripts (Downing et al. 2012; Green et al. 2007), as have studies of ethanol-treated neuronal cultures (Rahman and Miles, 2001; Gutala et al., 2004). Our own preliminary findings have similarly identified ribosome biogenesis as the most significantly altered gene cluster in these chick neuroprogenitors 6hr post-ethanol challenge (ME Berres, A Garic, SM Smith, unpublished). Thus, differential ribosomal protein expression has repeatedly emerged from multiple comparisons of neuroprogenitors having differential ethanol sensitivity. This suggests that ribosomal activity may be an integral component of cellular ethanol responses and is a strong candidate to modulate ethanol vulnerability.
How might ribosomal proteins shape cellular ethanol responses within neural crest progenitors? Ribosomal components not only mediate protein translation but also regulate cell proliferation and differentiation. Because the energy demands of ribosome biogenesis are high, proliferating cells monitor ribosome biogenesis as a key sensor of nucleolar stress, wherein a subset of ribosomal proteins directly interact with and silence the protein MDM2/HDM2, an E3 ubiquitin ligase that targets p53 for degradation. MDM2 silencing by these unchaparoned ribosomal proteins causes p53 accumulation and p53-mediated apoptosis (Fumagalli and Thomas, 2011; Kruse and Gu, 2009). Human ribosomopathies, such as Treacher-Collins syndrome and Diamond-Blackfan anemia, can feature macrocytic anemia, short stature, thumb and cardiac defects, and, importantly, facial deficits with parallels to FASD including epicanthal folds, flat nasal bridge, hypertelorism, and cleft lip/palate (Narla and Ebert, 2010; Teng et al. 2013). In neural crest, impaired ribosome biogenesis initiates their p53-mediated apoptosis and decreases their proliferation, producing cranioskeletal hypoplasia (Trainor, 2010). This mechanism strongly parallels ethanol’s effects on neural crest as detailed in animal models of FASD (Debelak et al. 2003; Dunty et al. 2001). It may be noteworthy that five of the ribosomal proteins that regulate MDM2/p53 interactions had significantly differential expression in W98S/D neural crest progenitors including RPL5 (1.244-fold), RPL11 (0.886-fold), RPL12 (0.748-fold), RPL23 (0.870-fold) and RPS15 (1.169-fold; Table 1) (Narla and Ebert, 2010). Additionally, nine of these altered ribosomal proteins are causative in the craniofacial ribosomopathy Diamond-Blackfan anemia and include RPL5 (1.244-fold), RPL11 (0.886-fold), RPL26 (0.888-fold), RPL27 (0.831-fold), RPL35A (0.859-fold), RPL36 (0.492-fold), RPS10 (0.932-fold), RPLS15 (1.167-fold), and RPS17 (0.765-fold) (Narla and Ebert, 2010; Teng et al. 2013). While adult W98S and W98D birds lacked obvious craniofacial defects, in human ribosomopathies haploinsufficiency often does not produce overt facial changes (Trainor, 2010). Whether the expression-level changes described here contribute to the divergent ethanol responses of these neural crest populations is currently unknown. For the W98S/D model, it is possible that modest differences in ribosome expression might affect free ribosomal protein abundance and thereby shift the ethanol threshold that activates the apoptotic p53-ribosome biosensor within these cells. Given its critical role in neural crest development and its persistent association with ethanol responsiveness, ribosome biogenesis may be an important candidate modifier of ethanol vulnerability and it merits follow-up study.
β-Catenin/Wnt Signaling Pathway
A limitation of our GO term and KEGG pathway analysis is the incomplete annotation of Gallus gallus databases, which may preclude identification of additional candidate pathways. Because the molecular events in this ethanol exposure model are well understood, our expanded analysis identified multiple genes with known relevance to neural crest development and these signaling events. Importantly, multiple participants in this calcium/Wnt cell signaling pathway had differential expression between these lines. Ethanol-sensitive W98S had significantly lower expression of calmodulin 1 and 2, which is well-documented to detect ethanol’s calcium signal and convert it into a lasting pro-apoptotic signal by activating the calmodulin-dependent kinase CaMKII (Debelak-Kragtorp et al. 2003; Garic et al. 2011; Flentke et al. 2011, 2013). The CaMKII-mediated destabilization of β-catenin is necessary and sufficient to cause this ethanol-mediated cell death (Flentke et al. 2011, 2013), and thus it is especially noteworthy that several proteins that modify Wnt signaling, including β-catenin itself, emerged from this analysis. Differential expression in this pathway could shape cellular responses by shifting the critical threshold of ethanol necessary to reduce β-catenin’s transcriptional activity and thereby trigger apoptosis. These genes are thus obvious candidate modifiers of cellular vulnerability to ethanol. Wnt/β-catenin can also cross-communicate with p53/ribosome biosynthesis in neural crest (Bugner et al., 2011), adding additional complexity to these cells’ ethanol responses.
Energy Metabolism and Oxidative Phosphorylation
Three of the ten enriched pathways involved cellular energy generation and included Oxidative Phosphorylation, Glycolysis / Gluconeogenesis, and Metabolism Pathways. Multiple components of Complex I / NADH dehydrogenase, cytochrome c oxidase, and ATP synthase were significantly increased in W98S, and several components of cytochrome c reductase and cytochrome c oxidase were reduced. Whether these changes would alter flux through oxidative phosphorylation is unknown because the W98S line no longer exists. Energy generation is rate-limiting with respect to embryo growth and efficient nutrient conversion is valued commercially. Oxidative flux also affects cellular responses to stressors and thus could shape cellular sensitivity to ethanol. Metabolic stress is also a significant activator of the ribosome/p53 biosensor (Fumagelli and Thomas, 2011; Kruse and Gu, 2009). It is likely that differences in these energetic pathways resulted from selective pressures to improve growth and production characteristics in these White Leghorn layers. Endorsing this is our additional identification of genes mediating growth and energetics including somatostatin (0.341-fold), ApoB (11.76-fold), and NDUFA4 (0.499-fold). Those same commercial concerns may have also shaped differential expression within the oocyte meiosis cluster (6 genes, p=0.0058) and egg white proteins (B–G, 2.232-fold; HEP21, 2.164-fold) to affect layer potential and egg yield.
In summary, this work identifies candidate genes that potentially modify cellular vulnerability to ethanol, including genes that were directly mapped to the apoptotic pathway that mediates ethanol’s neurotoxicity in these lines. The recurring identification of ribosome biogenesis in several FASD models highlights this pathway as a potential influence upon individual vulnerability to ethanol’s developmental damage. Thus, unbiased transcriptome comparative analysis in a non-treatment paradigm offers new insights into the potential mechanisms underlying ethanol’s neurotoxicity.
Supplementary Material
Supplemental Figure 1. Distribution of RNA Biotypes identified in W98S (left) and W98D (right) neural crest and neural progenitors at HH8+ to HH9+.
Supplemental Table 1. List of gene transcripts showing significantly differential transcript abundance in W98S versus W98D neural crest and neural progenitors.
Acknowledgements
We thank Ron Muetzel at Hy-Line International for providing the W98S/D lines. Supported by NIH Award R37-AA11085 and an ARRA supplement to S.M.S.
References
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm SL, 2nd, Lundahl KR, Caldwell J, Gilliam DM. Ethanol teratogenesis in the C57BL/6J, DBA/2J, and A/J inbred mouse strains. Alcohol. 1997;14:389–395. doi: 10.1016/s0741-8329(97)87950-5. [DOI] [PubMed] [Google Scholar]
- Bolnick JM, Karana R, Chiang PJ, Kilburn BA, Romero R, Diamond MP, Smith SM, Armant DR. Apoptosis of alcohol-exposed human placental cytotrophoblast cells is downstream of intracellular calcium signaling. Alcohol Clin Exp Res. 2014 doi: 10.1111/acer.12417. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugner V1, Tecza A, Gessert S, Kühl M. Peter Pan functions independently of its role in ribosome biogenesis during early eye and craniofacial cartilage development in Xenopus laevis. Development. 2011;138:2369–2378. doi: 10.1242/dev.060160. [DOI] [PubMed] [Google Scholar]
- Carter RC, Jacobson SW, Molteno CD, Jacobson JL. Fetal alcohol exposure, iron-deficiency anemia, and infant growth. Pediatrics. 2007;120:559–567. doi: 10.1542/peds.2007-0151. [DOI] [PubMed] [Google Scholar]
- Cartwright MM, Smith SM. Stage dependent effects of ethanol on cranial neural crest cell development: Partial basis for the phenotypic variations observed in Fetal Alcohol Syndrome. Alcohol Clin Exp Res. 1995;19:1454–1462. doi: 10.1111/j.1530-0277.1995.tb01007.x. [DOI] [PubMed] [Google Scholar]
- Cartwright MM, Tessmer LA, Smith SM. Ethanol-induced neural crest apoptosis is coincident with their endogenous death but is mechanistically distinct. Alcohol Clin Exp Res. 1998;22:142–149. [PubMed] [Google Scholar]
- Chen SY, Periasamy A, Yang B, Herman B, Jacobson K, Sulik KK. Differential sensitivity of mouse neural crest cells to ethanol-induced toxicity. Alcohol. 2000;20:75–81. doi: 10.1016/s0741-8329(99)00058-0. [DOI] [PubMed] [Google Scholar]
- Das UG, Cronk CE, Martier SS, Simpson PM, McCarver DG. Alcohol dehydrogenase 2*3 affects alterations in offspring facial morphology associated with maternal ethanol intake in pregnancy. Alcohol Clin Exp Res. 2004;28:1598–1606. doi: 10.1097/01.alc.0000141816.14776.97. [DOI] [PubMed] [Google Scholar]
- Debelak KA, Smith SM. Avian genetic background modulates the neural crest apoptosis induced by ethanol exposure. Alcohol Clin Exp Res. 2000;24:307–314. [PubMed] [Google Scholar]
- Debelak-Kragtorp KA, Armant DR, Smith SM. Ethanol-induced cephalic apoptosis requires phospholipase C-dependent intracellular calcium signaling. Alcohol Clin Exp Res. 2003;27:515–523. doi: 10.1097/01.ALC.0000056615.34253.A8. [DOI] [PubMed] [Google Scholar]
- Dillies MA, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N, Keime C, Marot G, Castel D, Estelle J, Guernec G, Jagla B, Jouneau L, Laloë D, Le Gall C, Schaëffer B, Le Crom S, Guedj M, Jaffrézic F French StatOmique Consortium. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 2013;14:671–683. doi: 10.1093/bib/bbs046. [DOI] [PubMed] [Google Scholar]
- Downing C, Balderrama-Durbin C, Kimball A, Biers J, Wright H, Gilliam D, Johnson TE. Quantitative trait locus mapping for ethanol teratogenesis in BXD recombinant inbred mice. Alcohol Clin Exp Res. 2012;36:1340–1354. doi: 10.1111/j.1530-0277.2012.01754.x. [DOI] [PubMed] [Google Scholar]
- Dunty WC, Jr, Chen SY, Zucker RM, Dehart DB, Sulik KK. Selective vulnerability of embryonic cell populations to ethanol-induced apoptosis: implications for alcohol-related birth defects and neurodevelopmental disorder. Alcohol Clin Exp Res. 2001;36:1340–1354. [PubMed] [Google Scholar]
- Flentke GR, Garic A, Amberger E, Hernandez M, Smith SM. Calcium-mediated repression of β-catenin and its transcriptional signaling mediates neural crest cell death in an avian model of fetal alcohol syndrome. Birth Defects Res A Clin Mol Teratol. 2011;91:591–602. doi: 10.1002/bdra.20833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flentke GR, Garic A, Hernandez M, Smith SM. CaMKII represses transcriptionally-active β-catenin to mediate acute ethanol neurodegeneration and can phosphorylate β-catenin. J Neurochem. 2013;128:523–535. doi: 10.1111/jnc.12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flentke GR, Klingler RH, Tanguay RL, Carvan MJ, 3rd, Smith SM. An evolutionarily-conserved mechanism of calcium-dependent neurotoxicity in a zebrafish model of FASD. [2014 Feb 11];Alcohol Clin Exp Res. 2014 doi: 10.1111/acer.12360. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Ivy Yu HM, Maruyama T, Mirando AJ, Hsu W. Gpr177/mouse Wntless is essential for Wnt-mediated craniofacial and brain development. Dev Dyn. 2011;240:365–371. doi: 10.1002/dvdy.22541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S, Thomas G. The role of p53 in ribosomopathies. Sem Hematol. 2011;48:97–105. doi: 10.1053/j.seminhematol.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Garic A, Flentke GR, Amberger E, Hernandez M, Smith SM. CaMKII activation is a novel effector of alcohol's neurotoxicity in neural crest stem/progenitor cells. J Neurochem. 2011;118:646–657. doi: 10.1111/j.1471-4159.2011.07273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garic-Stankovic A, Hernandez MR, Chiang PJ, Debelak-Kragtorp KA, Flentke GR, Armant DR, Smith SM. Ethanol triggers neural crest apoptosis through the selective activation of a pertussis toxin-sensitive G protein and a phospholipase Cβ-dependent Ca2+ transient. Alcohol Clin Exp Res. 2005;29:1237–1246. doi: 10.1097/01.alc.0000172460.05756.d9. [DOI] [PubMed] [Google Scholar]
- Gilliam DM, Kotch LE. Dose-related growth deficits in LS but not SS mice prenatally exposed to alcohol. Alcohol. 1996;13:47–51. doi: 10.1016/0741-8329(95)02010-1. [DOI] [PubMed] [Google Scholar]
- Goodlett CR, Gillam DM, Nichols JM, West JR. Genetic influences on brain growth restriction induced by developmental exposure to alcohol. Neurotoxicity. 1989;10:321–334. [PubMed] [Google Scholar]
- Green ML, Singh AV, Zhang Y, Nemeth KA, Sulik KK, Knudsen TB. Reprogramming of genetic networks during initiation of the Fetal Alcohol Syndrome. Dev Dyn. 2007;236:613–631. doi: 10.1002/dvdy.21048. [DOI] [PubMed] [Google Scholar]
- Gutala R, Wang J, Kadapakkam S, Hwang Y, Ticku M, Li MD. Microarray analysis of ethanol-treated cortical neurons reveals disruption of genes related to the ubiquitin-proteasome pathway and protein synthesis. Alcohol Clin Exp Res. 2004;28:1779–1788. doi: 10.1097/01.alc.0000148117.17707.b4. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009a;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009b;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburn BA, Chiang PJ, Wang J, Flentke GR, Smith SM, Armant DR. Rapid induction of apoptosis in gastrulating mouse embryos by ethanol and its prevention by HB-EGF. Alcohol Clin Exp Res. 2006;30:127–134. doi: 10.1111/j.1530-0277.2006.00008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzoukas DE, Li G, Takapoo M, Moninger T, Bhalla RC, Pantazis NJ. Intracellular calcium plays a critical role in the alcohol-mediated death of cerebellar granule neurons. J Neurochem. 2013;124:323–335. doi: 10.1111/jnc.12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:3907–3920. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardson GR, Loudenburg R. Risk factors for alcohol use during pregnancy in a multistate area. Neurotoxicol Teratol. 2003;25:651–658. doi: 10.1016/j.ntt.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Mattson SN, Roesch SC, Glass L, Deweese BN, Coles CD, Kable JA, May PA, Kalberg WO, Sowell ER, Adnams CM, Jones KL, Riley EP CIFASD. Further development of a neurobehavioral profile of fetal alcohol spectrum disorders. Alcohol Clin Exp Res. 2013;37:517–528. doi: 10.1111/j.1530-0277.2012.01952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May PA, Tabachnick BG, Gossage JP, Kalberg WO, Marais AS, Robinson LK, Manning M, Buckley D, Hoyme HE. Maternal risk factors predicting child physical characteristics and dysmorphology in fetal alcohol syndrome and partial fetal alcohol syndrome. Drug Alcohol Depend. 2011;119:18–27. doi: 10.1016/j.drugalcdep.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarver DG, Thomasson HR, Martier SS, Sokol RJ, Li T. Alcohol dehydrogenase-2*3 allele protects against alcohol-related birth defects among African Americans. J Pharmacol Exp Ther. 1997;283:1095–1101. [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshlack A, Wakefield MJ. Transcript length bias in RNA-seq data confounds systems biology. Biol Direct. 2009;4:14. doi: 10.1186/1745-6150-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman S, Miles MF. Identification of novel ethanol-sensitive genes by expression profiling. Pharmacol Ther. 2001;92:123–134. doi: 10.1016/s0163-7258(01)00163-2. [DOI] [PubMed] [Google Scholar]
- Streissguth AP, Dehaene P. Fetal alcohol syndrome in twins of alcoholic mothers: concordance of diagnosis and IQ. Am J Med Genet. 1993;47:857–861. doi: 10.1002/ajmg.1320470612. [DOI] [PubMed] [Google Scholar]
- Su B, Debelak KA, Tessmer LL, Cartwright MM, Smith SM. Genetic influences on craniofacial outcome in an avian model of prenatal alcohol exposure. Alcohol Clin Exp Res. 2001;25:60–69. [PubMed] [Google Scholar]
- Swartz ME, Wells MB, Griffin M, McCarthy N, Lovely CB, McGurk P, Rozacky J, Eberhart JK. A screen of zebrafish mutants identifies ethanol-sensitive genetic loci. Alcohol Clin Exp Res. 2014;38:694–703. doi: 10.1111/acer.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng T, Thomas G, Mercer CA. Growth control and ribosomopathies. Curr Opin Genet Dev. 2013;23:63–71. doi: 10.1016/j.gde.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Trainor PA. Craniofacial birth defects: the role of neural crest cells in the etiology and pathogenesis of Treacher Collins syndrome and the potential for prevention. Am J Med Genet Part A. 2010;152A:2984–2994. doi: 10.1002/ajmg.a.33454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viljoen DL, Carr LG, Foroud TM, Brooke L, Ramsay M, Li TK. Alcohol dehydrogenase-2*2 allele is associated with decreased prevalence of fetal alcohol syndrome in the mixed-ancestry population of the Western Cape Province, South Africa. Alcohol Clin Exp Res. 2001;25:1719–1722. [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren KR, Li TK. Genetic polymorphisms: impact on the risk of fetal alcohol spectrum disorders. Birth Defects Res Part A. 2005;73:195–203. doi: 10.1002/bdra.20125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Distribution of RNA Biotypes identified in W98S (left) and W98D (right) neural crest and neural progenitors at HH8+ to HH9+.
Supplemental Table 1. List of gene transcripts showing significantly differential transcript abundance in W98S versus W98D neural crest and neural progenitors.