Summary
In eukaryotes, homologous recombination proteins such as RAD51 and RAD52 play crucial roles in DNA repair and genome stability. Human RAD52 is a member of a large single-strand annealing protein (SSAP) family [1] and stimulates Rad51-dependent recombination [2, 3]. In prokaryotes and phages, it has been difficult to establish the presence of RAD52 homologs with conserved sequences. Putative SSAPs were recently found in several phages that infect strains of Lactococcus lactis [4]. One of these SSAPs was identified as Sak and was found in the virulent L. lactis phage ul36, which belongs to the Siphoviridae family [4, 5]. In this study, we show that Sak is homologous to the N terminus of human RAD52. Purified Sak binds single-stranded DNA (ssDNA) preferentially over double-stranded DNA (dsDNA) and promotes the renaturation of long complementary ssDNAs. Sak also binds RecA and stimulates homologous recombination reactions. Mutations shown to modulate RAD52 DNA binding [6] affect Sak similarly. Remarkably, electron-microscopic reconstruction of Sak reveals an undecameric (11) subunit ring, similar to the crystal structure of the N-terminal fragment of human RAD52 [7, 8]. For the first time, we propose a viral homolog of RAD52 at the amino acid, phylogenic, functional, and structural levels.
Results and Discussion
The aim of this study was to investigate a possible link between Rad52 and Sak proteins. First, by using Pfam, we detected significant domains present in the protein sequence of Sak of lactococcal phage ul36 (252 amino acids) and the protein sequence of RAD52 (418 amino acids). Then, we aligned a eukaryote-only seed and reran this new Pfam domain on Sak. We got E values ranging from 0.01 to 0.028 (Table S1 available online). Comparison of the observed secondary structure of the human RAD52 with the predicted secondary structure of the viral Sak revealed a significant overlap between the N-terminal domains of the two proteins (Figures 1B and 1C, Figure S1). On the basis of the Pfam-motif alignment, we found 17% (26/152) identity and 32% (48/152) similarity between the Sak protein of phage ul36 and human RAD52, as well as 23% (35/152) identity and 36% similarity (55/152) with Saccharomyces cerevisiae Rad52. To further investigate this possible link between eukaryotic RAD52 and Sak, we generated a phylogenetic tree with a subset of the sequences associated to the Pfam domain PF04098 (Table S2). We observed that Sak is phylogenetically close to bacteria, a finding that suggests that the Rad52 domain is wide-spread in many other phages and bacteria (Figure 1A). The low eukaryote-only Pfam-domain E values, the agreement of the secondary structures, the identity and similarity of Sak to both human and yeast Rad52, the evolutionary tree linking Sak to the eukaryotes, and also the PSI-BLAST link described by [1] altogether suggest that Sak is a viral homolog of the eukaryotic RAD52.
Figure 1. Sak Is a Viral Ancestor of RAD52.
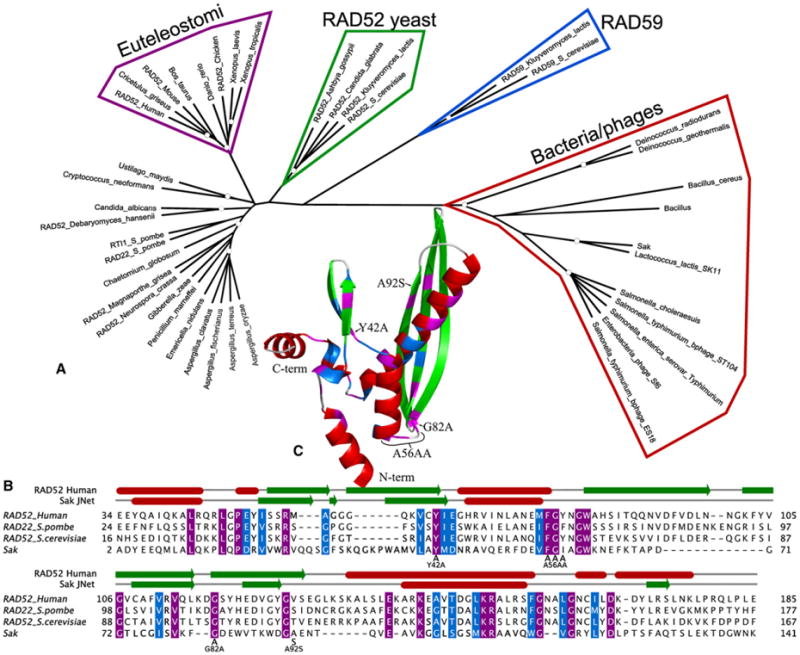
(A) Phylogenetic tree of the Rad52_Rad22 Pfam domain in which white dots represent 80% support.
(B) Multiple alignments of Sak, human RAD52, Rad22, and yeast Rad52 in which amino acids with purple background represent identity and amino acids with blue background represent similarity. On top of the alignment, the human RAD52 secondary structure as it is found in the PDB file 1KN0 is represented, along with a prediction of the Sak secondary structure from JNet. Red boxes represent helices and green arrows represent strands. The different mutations introduced in Sak are indicated.
(C) Identity and similarity displayed on human RAD52 structure (1KN0) with the same color code as the alignment and secondary structure of (B). The different mutations introduced in Sak are indicated.
Next, we determined whether Sak possessed functional and structural properties similar to RAD52. Wild-type Sak fused to a 10-histidine tag was purified to homogeneity from E. coli BL21(DE3) RP (Figure 2A). The native molecular weight of Sak was determined by gel filtration analysis. The bulk of protein was eluted in high-molecular-weight fractions, suggesting that Sak forms complexes containing a variable number of molecules (Figure 2B). It is now well recognized that RAD52 binds single-stranded DNA (ssDNA) preferentially over double-stranded DNA (dsDNA) [9]. This difference in affinity is critical for strand-annealing reactions because pairing occurs between ssDNAs. We used electrophoretic mobility shift assays to investigate the ssDNA- and dsDNA-binding properties of Sak. The Sak protein of lactococcal phage ul36 bound preferentially to ssDNA over dsDNA (Figure 2C). Quantification of the DNA-protein complexes at 0.25 μM Sak indicated that 14.3% of the input ssDNA was stably bound compared with 0.9% for dsDNA (Figure 2C, lanes c and j). When we incubated Sak with two 5′-32P-end-labeled complementary ssDNA of 400 bases in length, we observed the formation of duplex DNA products, consistent with a function in single-strand annealing (Figure 2D). At 0.25 μM of Sak, single-strand annealing was increased to a level of 37% (Figure 2D, lane e). Excess of Sak proteins (0.5 μM) was found to inhibit the reaction, perhaps as a consequence of nonspecific aggregation leading to nonproductive complex formation or the mechanism requiring ssDNA-Sak complexes and naked ssDNA. DNA-binding studies in these conditions revealed that Sak bound only to ssDNA, and the dsDNA product was released after annealing (Figure 2E, lanes d and e). Taken altogether, these biochemical results clearly showed that Sak of lactococcal phage ul36 belongs to the single-strand annealing protein (SSAP) family.
Figure 2. Purification and Properties of Lactococcal Phage Sak Proteins.

(A)SDS-PAGE of the purified wild-type Sakprotein. Lane a shows Mark12 molecular-weight markers, and lane b shows purified wild-type Sak (2.5 μg).
(B) Native molecular mass of purified His10-Sak as determined by gel filtration through Superdex 200. The top part shows size standards, and the bottom part shows western blotting of Sak with a His antibody.
(C) DNA-binding properties of Sak. DNA-binding reactions contained 100-mer ssDNA or 100 bp dsDNA. Reactions contained buffer alone without protein (lanes a and h) or the indicated concentrations of Sak protein (0.1–2 μM, lanes b–g and i–n) with ssDNA or dsDNA.
(D) Single-strand annealing by Sak protein. Lane a shows purified 400 bp duplex DNA. Reactions contained denatured 400 bp duplex DNA in buffer without protein (lane b) or Sak wild-type (0.05–2.5 μM, lanes c–j).
(E) Sak binds ssDNA preferentially and releases dsDNA after ssDNA annealing. Reactions were performedasin(D),but rather than being deproteinized, the samples were fixed by glutaraldehyde. Lane a shows purified 400 bp duplex DNA. Reactions contained denatured 400 bp duplex DNA and no protein (lane b) or Sak wild-type (0.05–1.5 μM, lanes c–h).
In solution, full-length RAD52 protein forms homo-oligomers of seven or more subunits [10]. Crystal structures of the truncated mutants hRAD52(1–212) and hRAD52(1–209) also revealed undecameric ring structures [7, 8]. It is proposed that the RAD52 ring binds DNA breaks via interactions with the terminal base, leading to the formation of a precisely organized ssDNA-RAD52 complex in which the DNA lies on an exposed surface of the protein ring. Hence, the protein-DNA arrangement may facilitate the DNA-DNA interactions necessary for RAD52-mediated annealing of complementary DNA strands [11]. Therefore, we investigated whether Sak could also form ring structures. Remarkably, Sak (252 aa) formed ring structures in the absence of DNA (Figure 3A, top left), indicating that Sak has the ability to self-associate and possesses a similar architecture to RAD52. Consistent with a specific interaction with ssDNA in gel retardation assays, wild-type Sak selectively bound circular ssDNA (Figure 3A). Also apparent were filaments consisting of stacked rings (Figure 3B). Irregular regions in structure could also be distinguished. These experiments established that Sak forms toroidal structures and is a single-strand annealing protein.
Figure 3. The Biochemical Properties of Sak Are Similar to RAD52.
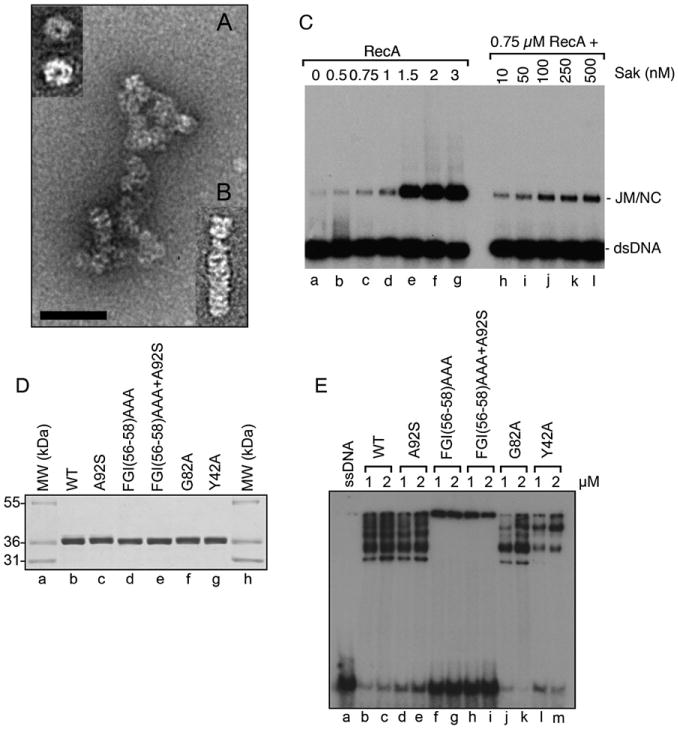
(A and B) Electron-microscopic visualization of complexes formed by Sak ([A] and [B], 5 μM) with single-stranded DNA (5 μM). The insert (top left) indicates an enlargement of a typical Sak wild-type ring. The magnification bar represents 50 nm.
(C) Stimulation of RecA strand exchange by Sak. Lane a shows no protein, lanes b–g show titration of RecA (0.5– 3 uM), and lanes h–l show reactions with RecA (0.75 μM) and the indicated concentrations of Sak (10–500 nM). Joint molecules and nicked circles (NC) are indicated.
(D) SDS-PAGE of purified Sak and mutated derivatives. Lanes a and h show Mark12 molecular-weight markers. Lane b shows purified wild-type Sak, lane c shows Sak A92S, lane d shows Sak FGI(56–58)AAA, lane e shows Sak FGI(56–58) AAA+A92S, lane f shows Sak G82A, and lane g shows Sak Y42A.
(E) DNA-binding properties of Sak on ssDNA. Reactions contained buffer alone without protein (lane a) or the indicated concentrations of Sak (lanes b and c), Sak A92S (lanes d and e), Sak FGI(56–58)AAA (lanes f and g), Sak FGI(56–58) AAA+A92S (lanes h and i), Sak G82A (lanes j and k), and Sak Y42A (lanes l and m).
Another key feature of RAD52 homologs is that they stimulate homologous recombination reactions during DNA double-strand break repair. In order for double-strand breaks to be repaired accurately during homologous recombination, the broken DNA ends must undergo resection followed by the binding of RAD51, an ortholog of Escherichia coli RecA [12]. RAD51 polymerizes as helical nucleoprotein filaments that subsequently search for DNA homology, leading to the formation of DNA joint molecules. Nucleation of RAD51 on ssDNA is a limiting step and is stimulated by protein-protein interactions with RAD52 [2, 3, 13]. We investigated whether we could find the same stimulatory effect with RecA and Sak. First, immunoprecipitations were conducted with E. coli cell extracts expressing L. lactis RecA and His-tagged Sak. Sak antibodies pulled down a stable complex between L. lactis RecA and Sak (Figure S2A, lane d). Affinity pull-downs also revealed a complex between E. coli RecA and Sak (Figure S2B). Next, we performed strand-exchange assays [14] and determined whether the addition of Sak increased strand-exchange activity of the RecA protein. When limiting concentrations of RecA were used (0.75 μM), only 1.8% of displaced ssDNA was produced (Figure 3C, lane c). However, when RecA was supplemented with Sak at concentrations of 10–500 nM, strand exchange occurred at a higher level. Sak alone did not promote strand exchange (data not shown). Quantification of the products at 0.75 μM RecA and 250 nM Sak revealed a 2.3-fold increase in activity compared to RecA alone. Therefore, Sak, like RAD52 protein, can stimulate homologous recombination mediated by the recombinases with which it can normally interact.
Via alanine-scanning mutagenesis of the N-terminal domain of human RAD52, several residues likely to be in direct contact with ssDNA were previously identified [6]. For example, RAD52 mutants in residues FGY (amino acids 79–81) did not show detectable DNA binding, whereas mutation Y65A reduced binding by4-fold without disrupting the oligomeric ring. Therefore, we analyzed whether these corresponding mutations in Sak (Sak mutants FGI(56–58)AAA and Y42A, respectively), mutation A92S (which confers resistance to the antiviral protein AbiK [4]), and mutation G82A in a conserved glycine (Figures 1B, 1C, and 3D) could also affect DNA binding and oligomeric-ring formation. The results are summarized in Table S3.
When Sak or Sak A92S DNA binding was analyzed on lower-percentage Tris-glycine gel, DNA-protein complexes were observed as a ladder (Figure 3E, lanes b and c, lanes d and e). Both proteins behaved similarly, except that the number of stacked rings was decreased in Sak A92S (Table S3). Sak FGI(56–58)AAA with or without the A92S mutation aggregated DNA to a high extent because no complexes could enter the gel (Figure 3E, lanes f–i). The FGI(56–58)AAA mutation disrupted the oligomeric nature of the protein as judged by electron microscopy (Figure 4A) and gel filtration analysis (Figure S3). The corresponding FGY(79–81)AAA RAD52 mutant also displayed similar features because it is completely inhibited for DNA binding [6]. This mutant has not been observed by electron microscopy, but in the crystal structures of the RAD52 1–212 and 1–209 proteins, F79 and Y81 are positioned at the bottom of the DNA-binding groove near the subunit interface [7, 8]. In light of our data, which show that these residues are important for ring formation, we can suggest that DNA-binding defects for these mutants may result from disruption of the oligomeric state of the protein. Consistent with these observations, corresponding residues F94 and Y96 in S. cerevisiae Rad52 have been shown to be important for efficient γ-radiation repair or mitotic recombination [15]. At 250 nM protein, Sak DNA binding was decreased by 2.3-fold when mutation G82A was introduced and the number of stacks were greatly reduced (Figure 3E, lanes j and k, and Figure 4A). To clarify this process and find out whether the formation of stacks could affect DNA binding, we mutated Y42A of Sak, corresponding to Y65A in human RAD52. This residue protrudes from the N-terminal surface of RAD52 and does not disrupt the oligomeric ring but decreases DNA binding by 4-fold [6]. Interestingly, although the oligomeric nature of the ring was preserved, Sak stacked rings were not observed in the Y42A mutant (Figure 4A), and an 8.4-fold decrease in DNA binding was observed at 250 nM (Figure 3E, lanes l and m and data not shown). These results suggest that Sak stacked rings contribute to DNA binding and that similar residues in human RAD52 and phage Sak proteins mediate protein-DNA interactions.
Figure 4. Electron-Microscopic Visualization of Wild-Type Sak and Mutants.

(A) Electron micrographs of Lactococcus lactis Sak, Sak A92S, Sak FGI(56– 58)AAA, Sak FGI(56–58)AAA+A92S, Sak G82A, and Sak Y42A in the absence of DNA. The magnification bar represents 100 nm.
(B) An averaged “top view” of the wild-type Sak rings (∼155 angstroms) with no symmetry imposed (n = 1310), clearly showing the undecameric nature (top left). The scale bar represents 100 angstroms. For comparison, the top right shows a low-resolution projection of the Rad52 N-terminal domain crystal structure (∼130 angstroms; PDB ID 1KN0). The averaged (n = 230) Sak filament image (bottom) shows the 55-angstrom periodicity in the spacing between rings in these stacks.
Further similarity between Sak and RAD52 was evident when electron-microscopic images of Sak were used for analysis. A total of 1310 images of rings were collected from preparations that had been negatively stained with uranyl acetate. Strikingly, similar to hRAD52 1–209 [8] and hRAD52 1–212 [7], Sak protein formed an undecameric ring (Figure 4B). The inner dimensions of the Sak and human RAD52 1–212 rings were similar (∼30 angstroms and 25 angstroms [7], respectively), but the radius of the Sak ring (∼155 angstroms) was slightly larger than the RAD52 1–212 ring (120 angstroms [7]). A possible explanation is that the Sak C-terminal domain might extrude radially from the RAD52-like inner core. The undecameric structure of Sak is consistent with a bioinformatics comparison revealing that the first 166 amino acids of Sak are homologous to RAD52. It is not clear how Sak uses its toroidal shape to facilitate ssDNA annealing; it might function like RAD52 to present ssDNA appropriately for rapid annealing. SSAPs of bacteriophages, such as protein Beta of coliphage lambda and protein Erf of Salmonella phage P22, are usually involved in genome circularization after the entry of their linear DNA, in DNA repair of concatemers, and, in some cases, in DNA replication [4, 16, 17]. Whether the toroidal structure of Sak is important in these processes will be the subject of future studies.
The results presented here show that Sak is a viral homolog of RAD52 at the amino acid, phylogenic, functional, and structural levels and raise the possibility of a common origin. Our results suggest that proteins homologous to the N terminus of RAD52, such as RAD52 splice variants, but also RAD59, should form rings. Although yeast Rad52 plays key functions in DNA repair, lack of RAD52 leads only to mild phenotypes in mammalian cells. However, the transduction rate of recombinant HIV-1 was found to be 10-fold greater in Rad52−/− mouse embryonic stem cells compared to Rad52+/+ cells [18]. Because Sak is involved in viral infection and is likely to be an ancestor of RAD52, it might suggest that functions related to viral infection have been retained as RAD52 evolved. Collectively, our results suggest that key functions involved in DNA repair and viral infection in prokaryotes and eukaryotes are more related than previously appreciated.
Supplementary Material
Acknowledgments
We thank Louis-Charles Fortier for the cloning of WT Sak into pET16b. We are grateful to members of the Masson and Moineau laboratories for stimulating discussion. A.B. is a recipient of a NSERC Canada Graduate Scholarship. This research was supported by the National Sciences and Engineering Research Council (Strategic Grant to S.M. and J.Y.M.), the National Cancer Institute of Canada (017121 to J.Y.M.), the Swiss National Science Foundation (3100A0-116275 to A.S.), and the National Institutes of Health (GM03569 to E.H.E).
Footnotes
Supplemental Data: Supplemental Data include Supplemental Experimental Procedures, three figures, and three tables and can be found with this article online at http://www.current-biology.com/cgi/content/full/18/15/1142/DC1/.
References
- 1.Iyer LM, Koonin EV, Aravind L. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics. 2002;3:8. doi: 10.1186/1471-2164-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.New JH, Sugiyama T, Zaitseva E, Kowalczykowski SC. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein-A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 3.Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 4.Bouchard JD, Moineau S. Lactococcal phage genes involved in sensitivity to AbiK and their relation to single-strand annealing proteins. J Bacteriol. 2004;186:3649–3652. doi: 10.1128/JB.186.11.3649-3652.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Labrie S, Moineau S. Complete genomic sequence of bacteriophage ul36: Demonstration of phage heterogeneity within the P335 quasi-species of lactococcal phages. Virology. 2002;296:308–320. doi: 10.1006/viro.2002.1401. [DOI] [PubMed] [Google Scholar]
- 6.Lloyd JA, McGrew DA, Knight KL. Identification of residues important for DNA binding in the full-length human Rad52 protein. J Mol Biol. 2005;345:239–249. doi: 10.1016/j.jmb.2004.10.065. [DOI] [PubMed] [Google Scholar]
- 7.Kagawa W, Kurumizaka H, Ishitani R, Fukai S, Nureki O, Shibata T, Yokoyama S. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol Cell. 2002;10:359–371. doi: 10.1016/s1097-2765(02)00587-7. [DOI] [PubMed] [Google Scholar]
- 8.Singleton MR, Wentzell LM, Liu Y, West SC, Wigley DB. Structure of the single-strand annealing domain of human RAD52 protein. Proc Natl Acad Sci USA. 2002;99:13492–13497. doi: 10.1073/pnas.212449899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Dyck E, Hajibagheri NMA, Stasiak A, West SC. Visualisation of human Rad52 protein and its complexes with hRad51 and DNA. J Mol Biol. 1998;284:1027–1038. doi: 10.1006/jmbi.1998.2203. [DOI] [PubMed] [Google Scholar]
- 10.Stasiak AZ, Larquet E, Stasiak A, Müller S, Engel A, Van Dyck E, West SC, Egelman EH. The human Rad52 protein exists as a heptameric ring. Curr Biol. 2000;10:337–340. doi: 10.1016/s0960-9822(00)00385-7. [DOI] [PubMed] [Google Scholar]
- 11.Parsons CA, Baumann P, Van Dyck E, West SC. Precise binding to single-stranded DNA termini by RAD52 protein. EMBO J. 2000;19:4175–4181. doi: 10.1093/emboj/19.15.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sung P, Klein H. Mechanism of homologous recombination: Mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 13.Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in genetic recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 14.Sauvageau S, Stasiak AZ, Banville I, Ploquin M, Stasiak A, Masson JY. Fission yeast rad51 and dmc1, two efficient DNA recombinases forming helical nucleoprotein filaments. Mol Cell Biol. 2005;25:4377–4387. doi: 10.1128/MCB.25.11.4377-4387.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mortensen UH, Erdeniz N, Feng Q, Rothstein R. A molecular genetic dissection of the evolutionarily conserved N terminus of yeast Rad52. Genetics. 2002;161:549–562. doi: 10.1093/genetics/161.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poteete AR, Sauer RT, Hendrix RW. Domain structure and quaternary organization of the bacteriophage P22 Erf protein. J Mol Biol. 1983;171:401–418. doi: 10.1016/0022-2836(83)90037-2. [DOI] [PubMed] [Google Scholar]
- 17.Passy SI, Yu X, Li Z, Radding CM, Egelman EH. Rings and filaments of b protein from bacteriophage I suggest a superfamily of recombination proteins. Proc Natl Acad Sci USA. 1999;96:4279–4284. doi: 10.1073/pnas.96.8.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau A, Kanaar R, Jackson SP, O'Connor MJ. Suppression of retroviral infection by the RAD52 DNA repair protein. EMBO J. 2004;23:3421–3429. doi: 10.1038/sj.emboj.7600348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


