SUMMARY
Naïve mouse embryonic stem cells (mESCs) and primed epiblast stem cells (mEpiSCs) represent successive snapshots of pluripotency during embryogenesis. Using transcriptomic and epigenomic mapping we show that a small fraction of transcripts are differentially expressed between mESCs and mEpiSCs and these genes show expected changes in chromatin at their promoters and enhancers. Unexpectedly, the cis-regulatory circuitry of genes that are expressed at identical levels between these cell states also differs dramatically. In mESCs, these genes are associated with dominant proximal enhancers and dormant distal enhancers, which we term seed enhancers. In mEpiSCs, the naïve-dominant enhancers are lost, and the seed enhancers take up primary transcriptional control. Seed enhancers have increased sequence conservation and show preferential usage in downstream somatic tissues, often expanding into super enhancers. We propose that seed enhancers ensure proper enhancer utilization and transcriptional fidelity as mammalian cells transition from naïve pluripotency to a somatic regulatory program.
INTRODUCTION
At the molecular level, pluripotency is under the control of a complex array of regulatory mechanisms that maintain chromatin in a state permissive to differentiation into each of the early somatic and germ cell lineages. Recent evidence shows that pluripotency is not a single entity and can be maintained in either a ‘naïve’ or ‘primed’ state (Brons et al., 2007; Chenoweth et al., 2010; Nichols and Smith, 2009; Tesar et al., 2007). Naïve pluripotent cells, typified by mouse embryonic stem cells (mESCs), represent the pre-implantation inner cell mass and are widely utilized for developmental genetics as they are capable of extensive contribution to chimeric animals upon reintroduction back into the blastocyst (Bradley et al., 1984; Evans and Kaufman, 1981; Martin, 1981). On the other hand, primed pluripotent cells, typified by mouse epiblast stem cells (mEpiSCs) and human embryonic stem cells (hESCs), represent the post-implantation epiblast, the next successive stage of pluripotency, which occurs immediately prior to differentiation into the three germ cell lineages at gastrulation (Brons et al., 2007; Najm et al., 2011; Tesar et al., 2007; Thomson et al., 1998). There is tremendous interest in understanding the differences between the naïve and primed pluripotent states as they provide a direct window into the epigenetic dynamics in placental mammals that function to maintain pluripotency while simultaneously preparing to transition to a somatic regulatory program.
Enhancer elements establish and maintain expression patterns that drive normal development and cell identity. In comparison to promoters, the chromatin state of enhancers is divergent across different cell types. Even genes expressed broadly across different cell types can show dramatic differences in enhancer usage (Kieffer-Kwon et al., 2013). Recent evidence suggests that large genomic domains containing clusters of active enhancers, variously referred to as “super enhancers”, “stretch enhancers”, or “multiple enhancer variants” are particularly cell type-specific, and are proposed to mediate transcription of genes that are important for controlling cell identity (Corradin et al., 2014; Hnisz et al., 2013; Loven et al., 2013; Parker et al., 2013; Whyte et al., 2013). These discoveries have largely been garnered from comparisons of regulatory landscapes of cell types derived from very different tissues and distinct stages of development. Here, we employ genomic approaches to directly characterize the regulatory landscapes of two closely related cell types, mESCs and mEpiSCs. These cell types represent successive snapshots of early development, share the core property of pluripotency, and largely share a common transcriptional program; however, their maintenance relies on distinct signaling pathways. Our analyses show that enhancer usage differs not only for genes that are differentially expressed between mESCs and mEpiSCs, but also those that are similarly expressed between the two cell types. While enhancers unique to mESCs are decommissioned following the transition to primed pluripotency, those unique to mEpiSCs, which we term seed enhancers, are present in naïve pluripotency, become active in primed pluripotency, and retain activity in somatic derivatives, often contributing to super enhancers.
RESULTS
Enhancer profiles distinguish mouse pluripotent states
To understand the differences in transcriptional regulation between mESCs and mEpiSCs we performed epigenomic and transcriptome profiling of these two pluripotent cell types using high-quality ChIP-seq and RNA-seq datasets (see Table S1 and Figure S1A). We focused our epigenomic analysis on cis-regulatory regions known to be marked by specific chromatin features: H3K4me1, associated with putative enhancer elements (Heintzman et al., 2009; Heintzman et al., 2007; Wang et al., 2008); H3K4me3, associated with transcription start sites (TSSs) (Heintzman et al., 2009; Heintzman et al., 2007; Wang et al., 2008); H3K27ac, enriched at active promoters and enhancers (Creyghton et al., 2010; Rada-Iglesias et al., 2011; Wang et al., 2008; Zentner et al., 2011); H3K27me3, generally associated with transcriptionally repressed regions of chromatin (Rada-Iglesias et al., 2011; Schwartz and Pirrotta, 2007; Zentner et al., 2011); and DNase-seq, indicative of open regions of chromatin (Song and Crawford, 2010).
Through analysis of the RNA-seq and ChIP-seq data, we determined the number of transcripts differentially expressed between mESCs and mEpiSCs, as well as the number of promoters and enhancers that showed chromatin state differences between the two cell types (Figure 1A). The transcriptomes of mESCs and mEpiSCs were remarkably similar to one another (R2 = 0.83; n = 4 biological replicates per cell type), with less than 6% (852 out of 15, 198) of expressed transcripts (FPKM > 0.25) showing a significant difference in abundance between mESCs and mEpiSCs. Among this list were transcripts of genes known to be mESC-specific, including Esrrb, Zfp42, Dppa3, and Klf4, as well as those known to be mEpiSC-specific, including Fgf5, Cer1, and Lefty1 (Figure 1B; see also Figure 1D and Table S2). Global promoter states were also largely similar, with over 73% of 10,560 active (H3K27ac+) promoters overlapping between the two cell types (Figure 1A and 1B). Even promoters of differentially expressed genes (referred to as mESC-enriched and mEpiSC-enriched) showed a similar state of DNase hypersensitive open chromatin (Figure 1D, 1E, and 1F). In stark contrast, chromatin states at the 22,156 H3K4me1+ H3K27ac+ enhancer loci were much more distinctive, showing only 27% overlap (Figure 1A and 1C). These data suggest that enhancer landscape may play a dominant role in defining differences between pluripotent cell types on a molecular level.
Figure 1. Enhancer profiles distinguish mouse pluripotent states.
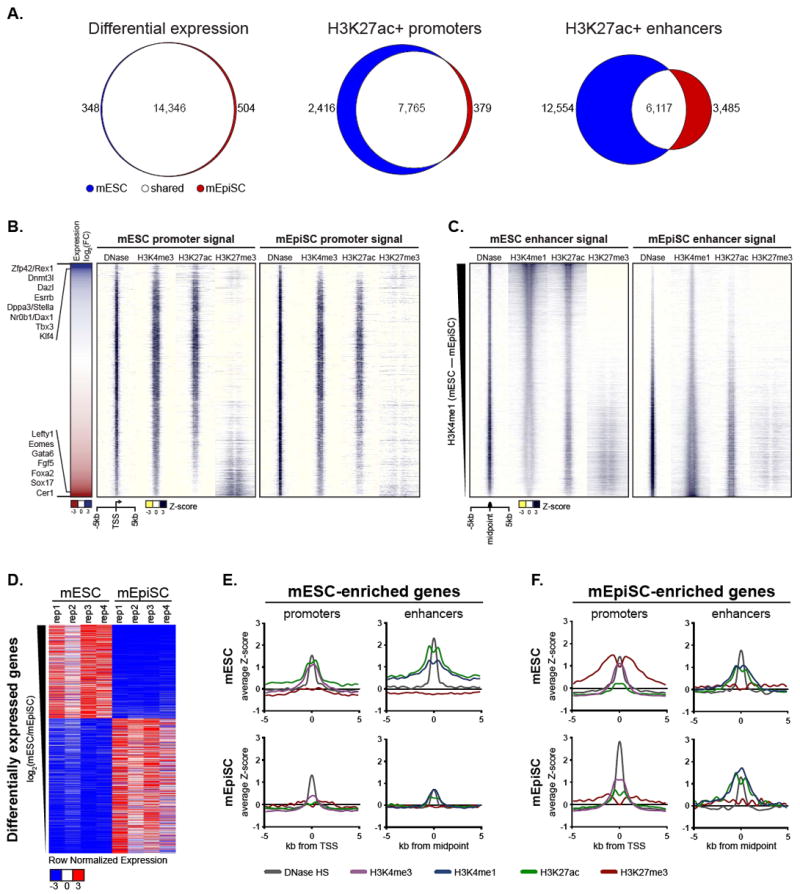
(A) Venn diagrams showing the number and overlap of expressed transcripts (left), H3K27ac+ promoters of expressed genes (center), and H3K27ac+ enhancers (right) detected in mESCs and mEpiSCs (B) Heatmap of expression differences between mESC and mEpiSC (log2 transformed, average of mESC replicates / average of mEpiSC replicates) ranked from high to low (left). Known mESC- and mEpiSC-enriched genes are listed to the left. Windowed chromatin heatmaps comparing DNase HS, H3K4me3, H3K27ac and H3K27me3 profiles ± 5kb of promoters in mESC and mEpiSC ranked in the same order as expression data (right). (C) Windowed heatmaps contrasting DNase HS, H3K4me1, H3K27ac, and H3K27me3 signal ± 5kb from the midpoint of DNase-centered putative enhancers identified in mESC or mEpiSC. Enhancers are ranked from most to least mESC-specific (compared to mEpiSCs) based on H3K4me1 peak intensities. (D) Row normalized expression of transcripts differentially expressed (also denoted as enriched) between mESC and mEpiSC, ranked as in B. (E) Aggregate plots depicting average ChIP-seq and DNase-seq signals at promoters and enhancers of mESC-enriched genes in mESCs and mEpiSCs (F) Same as E, but for mEpiSC-enriched genes. See also Figure S1 and Tables S1 and S2.
We next used a computational approach called PreSTIGEouse to assign enhancers to mESC-enriched and mEpiSC-enriched genes (see Methods). Enhancers of mESC-enriched genes contained all signature features of active elements in mESCs (H3K4me1, H3K27ac, and DNase hypersensitivity), and were decommissioned in mEpiSCs (Figure 1E). Globally, enhancers of mEpiSC-enriched genes are marked by an active enhancer signature in both cell types (Figure 1F). Half of enhancers of mEpiSC-enriched genes lack marks of active enhancer elements in mESCs, while half are marked by an active enhancer signature in both. These data suggest that the enhancer landscape of mESCs is primed to activate mEpiSC-enriched genes at the successive developmental stage. However, fewer than 8% of enhancers that differ between mESCs and mEpiSCs could be accounted for by differences at mESC- and mEpiSC-enriched genes.
Pluripotency-enriched genes show dramatic enhancer differences between pluripotent states
To further explore the global enhancer differences between mESCs and mEpiSCs, we asked whether genes expressed at similar levels between mESCs and mEpiSCs, including many pluripotency-related factors, are regulated by distinct enhancers. It is known that the Pou5f1, also known as Oct3/4, locus is controlled by distinct enhancers in the pre-implantation inner cell mass versus the post-implantation epiblast in vivo as well as in mESCs versus mEpiSCs in vitro (Tesar et al., 2007; Yeom et al., 1996). However, it was not known whether this represents a global regulatory phenomenon. To test this hypothesis, we first defined a set of “pluripotency-enriched” genes using RNA-seq datasets from mESC, mEpiSCs, and a panel of 18 developmental and adult mouse cell and tissue types. We set stringent metrics to ensure that genes within this class are expressed at similar levels in mESCs and mEpiSCs (p>0.05 and <2-fold change between mESCs and mEpiSCs) as well as showing selectivity versus the panel of 18 different mouse tissues (Figure 2A and Table S2). As expected, this pluripotency-enriched gene class contained gene ontology terms consistent with pluripotent phenotypes (Figure S1B) and the genes share a nearly identical active chromatin state at their promoters (Figure 2B). Consistent with their role in regulating key cell identity genes, 25% of these pluripotency-enriched genes were associated with super enhancers in either of the two cell types (Figure S1C). However, the locations of these super enhancers in the two cell types were largely non-overlapping, indicating a distinct control mechanism even for these similarly expressed pluripotency-enriched genes.
Figure 2. Pluripotency-enriched genes show dramatic enhancer differences between pluripotent states.
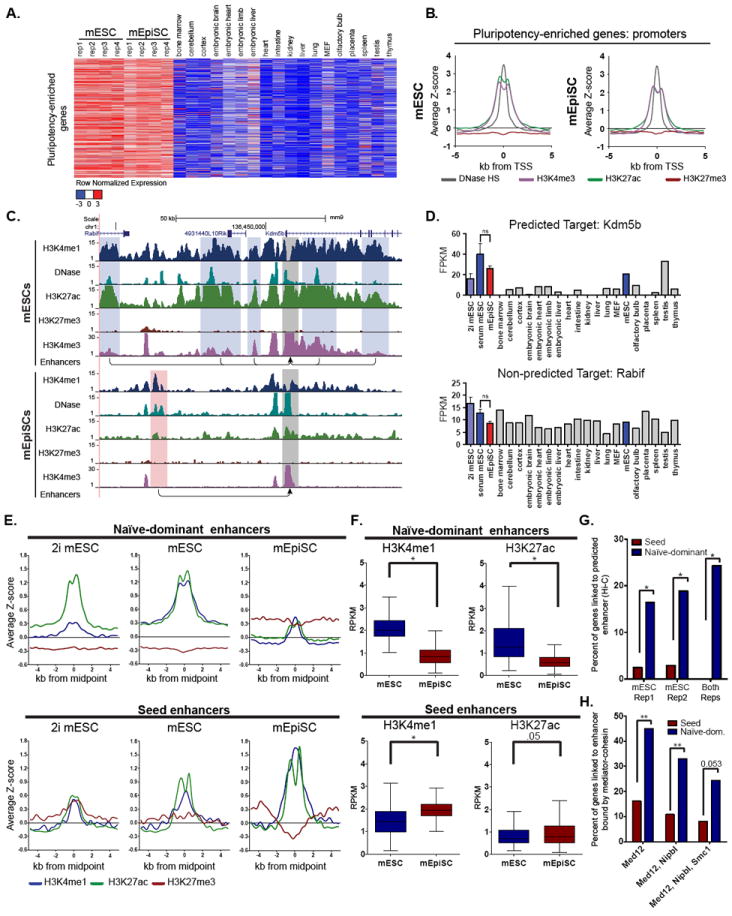
(A) Heatmap depicting row normalized expression of pluripotency-enriched genes. These genes are not differentially expressed (p-value>0.05), have <2-fold change between mESCs and mEpiSCs, and are relatively specific to mESCs and mEpiSCs compared in downstream tissues (see Methods). (B) Aggregate plots of DNase hypersensitivity and histone marks at the promoters of pluripotency-enriched genes (depicted in (A)), in mESC (left) and mEpiSC (right). (C) UCSC Browser image depicting the Kdm5b locus and the naïve-dominant enhancers (highlighted in blue) predicted to regulate its expression in mESC and the seed enhancers (highlighted in red) predicted to regulate its expression in mEpiSCs using the PreSTIGEouse methodology (see Methods). Grey boxes identify the Kdm5b promoter. The Rabif gene was not predicted to be regulated by these enhancers. (D) Expression levels (mean ± SD) of pluripotency-enriched transcript Kdm5b (top), and non-pluripotency-enriched transcript Rabif (bottom) in indicated cell types. (E) Aggregate plots of enhancers associated with the pluripotency-enriched genes. Naïve-dominant enhancers are predicted to regulate expression of pluripotency-enriched genes in the mESC state, but not in the mEpiSC state. Seed enhancers are predicted to regulate pluripotency-enriched genes in mEpiSCs, but not in mESCs. mESCs grown in standard conditions and 2i conditions are shown. (F) Boxplots depicting differences in the levels of enhancer histone marks between the two cell types as measured by RPKM (reads per kilobase per million mapped reads) at naïve-dominant enhancers (top) and seed enhancers (bottom) (paired sample Wilcoxon signed rank test, *p-value<0.0001). (G) Percentage of pluripotency-enriched genes associated with naïve-dominant enhancers and seed enhancers in mESC Hi-C datasets (Fisher’s Exact Test, *p-value<0.002). (H) Percentage of pluripotency-enriched genes for which both the gene target and an associated enhancer are bound by subunits of the mediator-cohesin complex (Med12, Nipbl, Smc1) (Fischer’s Exact Test, **p-value<0.0001). See also Figures S2 and S3.
We assigned enhancer elements to each pluripotency-enriched gene using a computational approach called PreSTIGEouse (see Methods). Of the 602 pluripotency-enriched genes, 97% showed evidence of differential enhancer usage between the two pluripotent cell types. An example is shown in Figure 2C. Here, the five enhancer elements predicted in mESCs to regulate Kdm5b, an exemplar pluripotency-enriched gene, are highlighted in blue (Figure 2C and 2D). All five enhancers are located in open chromatin and contain high levels of H3K4me1 and H3K27ac (Figure 2C). In mEpiSCs these five enhancers are virtually devoid of H3K4me1 and H3K27ac, and a different enhancer (highlighted in red) is predicted to regulate Kdm5b.
We next selected all enhancers that, like those associated with Kdm5b in mESCs, were predicted to regulate pluripotency-enriched genes exclusively in mESCs. These enhancers, which we call “naive-dominant”, generally contained high levels of H3K4me1 and H3K27ac in mESCs relative to mEpiSCs, where enhancer-histone signals were low or near background levels (Figure 2E, 2F, and S2A). Thus, most naïve-dominant enhancers are largely inactivated, or “lost” upon transition of mESCs to the mEpiSC state. We next selected “primed-dominant” enhancers, or those predicted to regulate the pluripotency-enriched genes exclusively in mEpiSCs. As expected, these enhancers contained high levels of the active enhancer marks H3K4me1 and H3K27ac in mEpiSCs (Figure 2E, 2F, and S2B). However, unexpectedly, these same enhancers were enriched for H3K4me1 and H3K27ac in mESCs, albeit at lower levels than in mEpiSCs (Figure 2E, 2F, and S2B). These enhancers were not assigned to gene targets in mESCs due to the relatively low levels of the associated enhancer histone marks. As a result of their apparent switch from dormancy in the naïve state to active transcriptional regulation in the primed state, we refer to these as seed enhancers. We compared seed enhancers to a class of “shared enhancers”, i.e., enhancers predicted to regulate pluripotency-enriched genes in both cell types. Despite their relatively low signal intensity in mESCs, seed enhancers contain the highest levels of H3K4me1 and H3K27ac amongst all mEpiSC enhancers of pluripotency-enriched genes (Figure S2C) suggesting the importance of the seed enhancer in the active regulation of the pluripotency-enriched genes.
To test whether the chromatin signal at seed enhancers in mESCs could be an artifact caused by metastable heterogeneity within mESC cultures, we tested if the seed enhancers were present in mESCs grown under defined “2i” culture conditions which are thought to promote a more homogeneous population of naïve cells (Ying et al., 2008). We find that, similar to mESCs grown in standard conditions, nearly all seed enhancer loci are marked by H3K4me1 in mESCs grown in 2i conditions and that half of these sites are also marked with H3K27ac, while the other half are marked with H3K27me3 (Figure 2E and S2B). Additionally, the levels of Pecam1 expression can distinguish mESCs in culture that are more naïve-like from cells that are more epiblast-like (Furusawa et al., 2004; Hayashi et al., 2008). We used FACS to separate mESCs into Pecam1-high and Pecam1-low populations (Figure S2D), followed by immediate fixation and ChIP-seq. The seed enhancers were found to be present at nearly identical levels in both populations (Figure S2E), further supporting that the presence of seed enhancers in mESCs is not the result of a contaminating epiblast-like population. Using publically available chromatin interaction maps generated through Hi-C experiments, we validated that compared to naïve-dominant enhancers seed enhancers rarely physically interact with the promoters of pluripotency-enriched genes in mESCs (Figure 2G). Additionally, compared to naïve-dominant enhancers, seed enhancers are infrequently occupied by components of the mediator-cohesin complex (Med12, Nipbl and Smc1) in mESCs, which are known to physically link enhancers with their target promoters (Figure 2H) (Kagey et al., 2010).
Seed enhancers show increased sequence conservation and are generally located farther from the transcription start sites of genes they control than naïve-dominant enhancers (Figure S2F, G and H). Motif analysis of the two classes revealed that many of the transcription factors enriched at seed enhancers overlap with those found at naïve-dominant enhancers (Table S3). Interestingly naïve-dominant enhancers exclusively are enriched for both known naïve-specific factors, such as Esrrb and Tcfcp2l1 and pluripotency factors, such as Oct4, Nanog and Sox2. Additionally, naïve-dominant enhancers are enriched for motifs of Smad2/3 and Smad4, key downstream mediators of the Activin/Nodal pathway required for mEpiSC maintenance (Brons et al., 2007; Nomura and Li, 1998; Tesar et al., 2007; Weinstein et al., 1998). Collectively, these results suggest a mechanism by which Activin/Nodal signaling may be required for repression of naïve-dominant enhancers during the transition to primed pluripotency.
We asked if the control of pluripotency-enriched genes by seed enhancers was a mouse specific molecular feature or if it extended more broadly to other mammalian pluripotent cells such as hESCs. Although embryo-derived hESCs exist in a primed pluripotent state similar to mEpiSCs, recent work has shown the ability to convert hESCs into a naïve-like state using extrinsic factors (Chan et al., 2013; Gafni et al., 2013; Ware et al., 2014). We took advantage of available enhancer-histone modification ChIP-seq data from this model system to test if the dynamic enhancer changes observed in mESCs and mEpiSCs are recapitulated in human pluripotent stem cells. To do this, a set of human pluripotency-enriched genes were selected and assigned to enhancers using methods similar to those used in mouse. Similar to our observations in mouse cells, these human cells contained robust naïve-dominant enhancers that lack chromatin markers of enhancer activity in the primed state, as well as “seed”-like enhancers with more robust activity in the primed state than the naïve cell state (Figure S3A-I). These data suggest that an early developmental transition from a naïve pluripotency-reinforcing epigenomic state to a primed somatic differentiation-capable state may be a general phenomenon of mammalian development.
Seed enhancers are utilized in downstream tissues where they expand into multi-enhancer clusters
Collectively, our findings suggest that the global transcriptional control of pluripotency genes is quite distinct between the naïve and primed phases of pluripotency. While genes have been shown to be controlled by distinct enhancers in completely different cell types (Kieffer-Kwon et al., 2013), we found it puzzling as to why genes expressed at virtually identical levels in successive cell stages would undergo enhancer switching. This question prompted us to investigate additional features of seed enhancers. Given that seed enhancers show greater sequence conservation than typical enhancers we asked if they might play a role at later stages of development. Using available H3K27ac ChIP-seq data from 15 different mouse embryonic and adult tissues, we found that 21% of seed enhancers were significantly enriched for H3K27ac in at least one tissue (Figure 3A and 3B). The rate of seed enhancer usage in downstream tissues was nearly double that of naïve-dominant enhancers, shared enhancers, and a control set of mouse embryonic fibroblast enhancers (indicative of the background rate of enhancer usage in multiple tissues) (Figure 3C). This trend held true across all fifteen tissues (Figure 3D).
Figure 3. Seed enhancers are utilized in downstream tissues where they expand into multi-enhancer clusters.
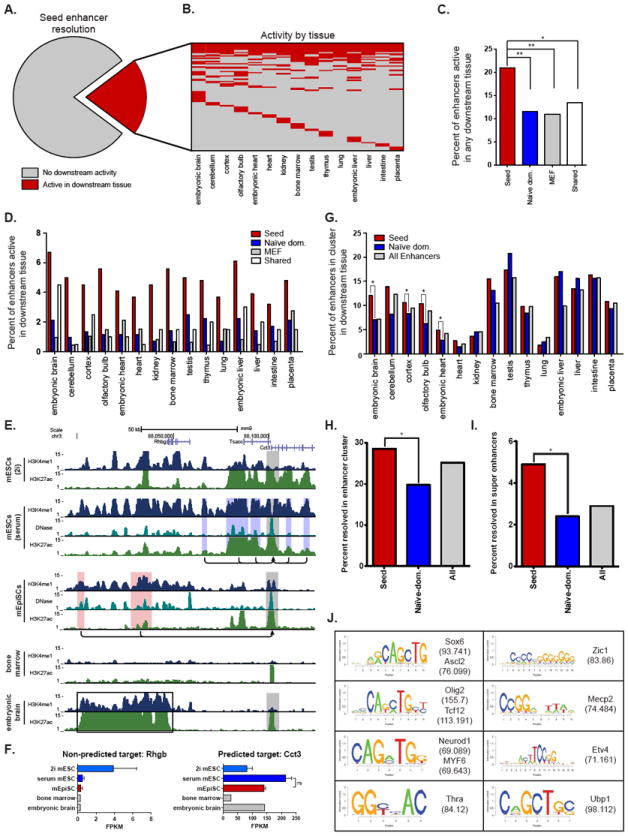
(A) Pie chart depicting the fraction of seed enhancers that show a chromatin state indicative of enhancer activity (H3K27ac+) in downstream tissues (red). (B) Heatmap displaying the downstream tissues in which each seed enhancer is active. Each row represents a seed enhancer. Red denotes that the enhancer displays H3K27ac signal intensity above threshold in the given cell type. (C) Percentage of seed enhancers (red) active in at least one downstream tissue compared to naïve-dominant enhancers (blue), MEF enhancers (grey), and enhancers shared between mESC and mEpiSC (white) (Fisher’s Exact Test, *p-value<0.03, **<0.0001). MEF datasets here provide an approximate measure of the background level of enhancer utilization. (D) Percent of seed enhancers active in each downstream tissue (red) compared to naïve-dominant (blue), MEF (grey) and enhancers common to mESC and mEpiSC (shared; white). (E) Genome browser image depicting the Cct3 gene and the enhancers predicted to regulate its transcription in mESCs and mEpiSCs. Grey boxes demarcate active promoters, while blue boxes identify naïve-dominant enhancers. The red boxes highlight two seed enhancers, which become components of a super enhancer in embryonic brain (black box), but not bone marrow. (F) Expression (mean ± SD) of Cct3 (right) is high in pluripotent cells and embryonic brain, but low in bone marrow. None of the enhancers in this region are predicted to target neighboring gene Rhbg (left). (G) Percent of enhancers located in a cluster of enhancers (defined as 4 or more active enhancer elements within a 100-kb window) in a downstream tissue. Seed enhancers (red) are significantly more likely to occur in enhancer clusters than naïve-dominant enhancers (blue) in embryonic brain, cortex, olfactory bulb, and embryonic heart (Fisher’s Exact Test, *p-value<0.003). The background rate for all mESC and mEpiSC enhancers (grey) is included for comparison. (H) Percentage of seed enhancers (red), naïve-dominant enhancers (blue) and all enhancers (grey) within a region that becomes an enhancer cluster (defined as in G) in one of the four neural tissues in the panel (embryonic brain, cortex, olfactory bulb, and cerebellum; Fisher’s Exact Test, *p-value<0.003). (I) As in H, but for regions that are super enhancers in neural tissues. (J) Motifs enriched amongst seed enhancers that are active in downstream tissues. See also Table S3.
Upon visual inspection of the seed enhancers in the downstream tissues, we noticed that several were contained within large domains of chromatin broadly marked with H3K4me1. For example, the Cct3 gene is a pluripotency-enriched gene regulated by two seed enhancers in mEpiSCs (Figure 3E) that is also expressed in embryonic brain (Figure 3F). Both seed enhancers become components of a large enhancer domain in embryonic brain. Domains like these are likely comprised of multiple individual enhancer elements, and are reminiscent of super and stretch enhancers. To test the significance of these observations, we determined the number of seed enhancers that lie within an enhancer cluster (defined as 4 or more enhancers within 100-kb) in a downstream tissue (Figure 3G). Seed enhancers were more likely than naïve-dominant enhancers to lie within enhancer clusters in four different tissues: embryonic brain, cortex, olfactory bulb and embryonic heart (Figure 3G). By comparison, naïve-dominant enhancers were not significantly enriched in enhancer clusters of any downstream tissue tested.
Given the propensity for seed enhancers to lie within neural-related enhancer clusters, we identified how often a seed enhancer gives rise to an enhancer cluster in at least one of the four neural tissues in our panel (embryonic brain, cortex, olfactory-bulb and cerebellum) and found that 29% of all seed enhancers fall within an enhancer cluster in one of these neural tissues (Figure 3H). We found the same enrichment for seed enhancers in regions that become super enhancers in these four neural tissues (Figure 3I). Interestingly, given the tendency for seed enhancers to contribute to super enhancers or enhancer clusters in neural tissues, seed enhancers that resolve in any downstream tissue are enriched for motifs associated with neural lineage transcription factors, including Ascl2, Sox6, Olig2, and Neurod1 (Figure 3J and Table S3).
DISCUSSION
Here we compared the transcriptomes and epigenomic landscapes of naïve mESCs and primed mEpiSCs. We report two novel and unexpected observations. First, pluripotency-enriched genes shared between mESCs and mEpiSCs are regulated by distinct enhancer elements in each cell type. Thus, the known differential enhancer regulation of the Oct3/4 locus between naïve and primed states may represent a well-studied example of a more general mechanism of enhancer switching at other pluripotency genes. Second, we show that enhancers actively regulating pluripotency-enriched genes in mEpiSCs often exist as dormant seed enhancers in mESCs. The seed enhancers are not in physical contact with the promoters of the pluripotency-enriched genes in mESCs, but then appear to take up primary transcriptional control in the sequential mEpiSC state. Seed enhancers appear to be epigenetically and functionally distinct from previously described “poised” and “latent” enhancers. Poised enhancers are co-marked with H3K4me1 and H3K27me3 in mESCs, whereas many seed enhancers lack H3K27me3 in mESCs (Rada-Iglesias et al., 2011; Zentner et al., 2011). Additionally, poised enhancers are not active in mESCs, nor do they regulate genes that are transcriptionally active in pluripotent cells. In contrast to seed enhancers that take over control of already expressed genes, poised enhancers regulate genes that become active upon differentiation. Latent enhancers altogether lack the signature enhancer-histone marks, and acquire them only upon response to cell stimulation (Ostuni et al., 2013). By comparison, seed enhancers are present but not engaged in mESCs, and then persist through the mEpiSC state into more terminally differentiated cell types.
This raises the questions why seed enhancers exist and what is their precise function? While further studies are clearly required, one possibility is that seed enhancers function in mESCs to ensure transcriptional robustness of the associated target gene. This hypothesis has been proposed for shadow enhancers in Drosophila melanogaster (Hong et al., 2008; Perry et al., 2010). However, if seed enhancers ensure proper gene expression, we might expect them to be in physical contact with their target gene promoters in mESCs, which was clearly not the case. A more attractive hypothesis is that seed enhancers act as “placeholders” in the naive mESC state to ensure that the proper enhancer is utilized in the primed pluripotent state and in subsequent stages of development. It will be particularly interesting to test whether seed enhancers are a phenomenon specific to the unique pre- to post-implantation transition of placental mammals or something more general within developmental and stem cell hierarchies.
EXPERIMENTAL PROCEDURES
Cell culture
Pluripotent stem cells were cultured as previously described (Chenoweth and Tesar, 2010; Najm et al., 2011; Tesar et al., 2007). For details see Supplemental Experimental Procedures.
Sequencing experiments
ChIP and DNase sequencing experiments were performed as previously described (Schmidt et al., 2009; Song et al., 2011). RNA sequencing libraries were prepared using the Illumina TruSeq RNA Sample Preparation Kit according to the manufacturer’s protocol. For details see Supplemental Experimental Procedures.
Datasets
Datasets used in this study are summarized in Table S1 (Buecker et al., 2014; Encode Project Consortium, 2011; Gafni et al., 2013; Kagey et al., 2010; Marks et al., 2012; Selvaraj et al., 2013; Shen et al., 2012; Wamstad et al., 2012). mEpiSC ChIP-seq, mEpiSC DNase-seq, and mESC and mEpiSC RNA-seq datasets generated for this publication are available on the NCBI Gene Expression Omnibus website in GEO Series GSE57409.
Predicting gene targets of enhancers with PreSTIGEouse
The PreSTIGE (Predicting Specific Tissue Interactions of Genes and Enhancers) bioinformatics algorithm was utilized to associate enhancer/gene pairs (Corradin et al., 2014). PreSTIGE predicts enhancer-gene pairs based on genomic proximity (<100 kb) and shared specificity of enhancer H3K4me1 signal and gene expression, as compared to a panel of 12 tissues. PreSTIGE was initially developed to interrogate human enhancer-gene pairs and was adapted for application to mouse (PreSTIGEouse). Predictions are available at http://genetics.case.edu/prestige/. For details see Supplemental Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Enhancer switching is a general mechanism of pluripotent gene regulation
Seed enhancers are dormant in naïve cells but take up control in primed cells
Seed enhancers have increased sequence conservation relative to typical enhancers
Seed enhancers give rise to super enhancers in downstream somatic lineages
Acknowledgments
This research was supported by funding from the NIH (R01HD056369) and (R01CA160356), the New York Stem Cell Foundation, the Mt. Sinai Health Care Foundation, and the Genomics and Cytometry core facilities of the Case Comprehensive Cancer Center (P30CA43703), CWRU Cellular and Molecular Biology training grant T32GM008056 (O.C.). P.J.T. is New York Stem Cell Foundation Robertson Investigator. We are grateful to Thomas LaFramboise, Cindy Bartels, Simone Edelheit and Neil Molyneaux for technical assistance, James Morrow and Tyler Miller for discussion, and Joanna Wysocka and Jacob Hanna for sharing data.
Footnotes
AUTHOR CONTRIBUTIONS
DCF, OC, PCS, and PJT designed the study, analyzed and interpreted all data, and wrote the manuscript. DCF, GEZ, JGC, RDM, and PJT cultured cells and performed all ChIP-seq and RNA-seq experiments. LS and GEC performed DNase-seq experiments. OC, AS, and PCS developed and implemented the PreSTIGEouse software package. All authors edited and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
All authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bradley A, Evans M, Kaufman MH, Robertson E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature. 1984;309:255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Buecker C, Srinivasan R, Wu Z, Calo E, Acampora D, Faial T, Simeone A, Tan M, Swigut T, Wysocka J. Reorganization of enhancer patterns in transition from naïve to primed pluripotency. Cell stem cell. 2014;14 doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YS, Goke J, Ng JH, Lu X, Gonzales KA, Tan CP, Tng WQ, Hong ZZ, Lim YS, Ng HH. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell stem cell. 2013;13:663–675. doi: 10.1016/j.stem.2013.11.015. [DOI] [PubMed] [Google Scholar]
- Chenoweth JG, McKay RD, Tesar PJ. Epiblast stem cells contribute new insight into pluripotency and gastrulation. Dev Growth Differ. 2010;52:293–301. doi: 10.1111/j.1440-169X.2010.01171.x. [DOI] [PubMed] [Google Scholar]
- Chenoweth JG, Tesar PJ. Isolation and Maintenance of Mouse Epiblast Stem Cells. Methods Mol Biol. 2010;636:25–44. doi: 10.1007/978-1-60761-691-7_2. [DOI] [PubMed] [Google Scholar]
- Corradin O, Saiakhova A, Akhtar-Zaidi B, Myeroff L, Willis J, Cowper-Sallari R, Lupien M, Markowitz S, Scacheri PC. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome research. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encode Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS biology. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Furusawa T, Ohkoshi K, Honda C, Takahashi S, Tokunaga T. Embryonic stem cells expressing both platelet endothelial cell adhesion molecule-1 and stage-specific embryonic antigen-1 differentiate predominantly into epiblast cells in a chimeric embryo. Biology of reproduction. 2004;70:1452–1457. doi: 10.1095/biolreprod.103.024190. [DOI] [PubMed] [Google Scholar]
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature. 2013;504:282–286. doi: 10.1038/nature12745. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Lopes SM, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell stem cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genetics. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon KR, Tang Z, Mathe E, Qian J, Sung MH, Li G, Resch W, Baek S, Pruett N, Grontved L, et al. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks H, Kalkan T, Menafra R, Denissov S, Jones K, Hofemeister H, Nichols J, Kranz A, Stewart AF, Smith A, et al. The transcriptional and epigenomic foundations of ground state pluripotency. Cell. 2012;149:590–604. doi: 10.1016/j.cell.2012.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm FJ, Chenoweth JG, Anderson PD, Nadeau JH, Redline RW, McKay RD, Tesar PJ. Isolation of epiblast stem cells from preimplantation mouse embryos. Cell stem cell. 2011;8:318–325. doi: 10.1016/j.stem.2011.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell stem cell. 2009;4:487–492. doi: 10.1016/j.stem.2009.05.015. [DOI] [PubMed] [Google Scholar]
- Nomura M, Li E. Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature. 1998;393:786–790. doi: 10.1038/31693. [DOI] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, Program NCS, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Current biology : CB. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48:240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nature reviews Genetics. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Selvaraj S, J RD, Bansal V, Ren B. Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nature biotechnology. 2013;31:1111–1118. doi: 10.1038/nbt.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harbor protocols. 2010;2010 doi: 10.1101/pdb.prot5384. pdb prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome research. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh T-Y, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nature Genetics. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware CB, Nelson AM, Mecham B, Hesson J, Zhou W, Jonlin EC, Jimenez-Caliani AJ, Deng X, Cavanaugh C, Cook S, et al. Derivation of naive human embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4484–4489. doi: 10.1073/pnas.1319738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein M, Yang X, Li C, Xu X, Gotay J, Deng CX. Failure of egg cylinder elongation and mesoderm induction in mouse embryos lacking the tumor suppressor smad2. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:9378–9383. doi: 10.1073/pnas.95.16.9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom YI, Fuhrmann G, Ovitt CE, Brehm A, Ohbo K, Gross M, Hubner K, Scholer HR. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome research. 2011;21:1273–1283. doi: 10.1101/gr.122382.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


