Summary
Posttranslational modifications of histones play fundamental roles in many biological functions. Specifically, histone H4-K20 methylation is critical in DNA synthesis and repair. However, little is known about how these functions are regulated by the upstream stimuli. Here, we identify a tyrosine phosphorylation site at Y72 of histone H4, which facilitates recruitment of histone methyltransferases (HMTases), SET8 and SUV4-20H, to enhance its K20 methylation, thereby promoting DNA synthesis and repair. Phosphorylation-defective histone H4 mutant is deficient in K20 methylation, leading to reduced DNA synthesis, delayed cell cycle progression, and decreased DNA repair ability. Disrupting the interaction between epidermal growth factor receptor (EGFR) and histone H4 by Y72 peptide significantly reduced tumor growth. Furthermore, EGFR expression clinically correlates with histone H4-Y72 phosphorylation, H4-K20 mono-methylation, and the Ki-67 proliferation marker. These findings uncover a mechanism by which EGFR transduces signal to chromatin to regulate DNA synthesis and repair.
Introduction
Cells compact a large amount of DNA by wrapping it in a nucleosome composed of a histone octamer of two of each of H2A, H2B, H3, and H4. Posttranslational modifications (PTMs) of histones such as acetylation, methylation, phosphorylation, ubiquitination, and ADP-ribosylation play important roles in chromatin dynamics and functions (Munshi et al., 2009). A number of PTMs of histone H4 have been identified, including acetylation at K5, K8, K12, and K16 and methylation at R3 and K20 in transcriptional regulation, and phosphorylation at S1 during mitosis (Peterson and Laniel, 2004). Lysine methylation on histone tails is functionally important in many biological events, such as transcriptional regulation, heterochromatin formation, and DNA synthesis and repair (Martin and Zhang, 2005). Of the five lysine residues in the N-terminal tail of histone H4 (K5, K8, K12, K16, and K20), K5 (Van Aller et al., 2012) and K20 (Pesavento et al., 2006) are methylated. K5 methylation has just been recently identified and is speculated to play a role in neoplastic process (Van Aller et al., 2012). H4-K20 methylation plays a key role in multiple functions, including DNA replication, mitosis, DNA repair, and chromatin condensation (Yang and Mizzen, 2009). K20 can be monomethylated by SET8 histone methytransferase (HMTase) (Fang et al., 2002) and dimethylated by SUV4-20H HMTase (Yang et al., 2008), which are responsible for DNA synthesis (Huen et al., 2008; Jorgensen et al., 2007) and DNA repair (Botuyan et al., 2006), respectively. Methylated lysine residues of histones recruit proteins containing chromodomain, tudor domain, or WD40-repeat domain in different biological processes (Martin and Zhang, 2005). Among these protein motifs, the tudor domain of 53BP1, a DNA damage checkpoint mediator (Fang et al., 2002), interacts directly with dimethylated H4-K20 in DNA repair processes (Botuyan et al., 2006). However, little is known about how these important functions are regulated by upstream signaling when these HMTases are recruited to histone H4 to methylate K20.
While the function of receptor tyrosine kinases (RTKs) as cell surface proteins have been well characterized, recent evidence indicates that RTKs, such as FGFR1 (Reilly and Maher, 2001; Stachowiak et al., 1997), c-Met (Gomes et al., 2008), VEGFR1 (Lee et al., 2007), and IGF1-R (Sehat et al., 2010), are also able to translocate to the nucleus. In fact, 11 out of 20 RTK subfamilies have been detected in the nucleus (Wang et al., 2013; Wang and Hung, 2012). In addition to the RTKs mentioned above, all of the epidermal growth factor receptor (EGFR) family members have been found in the nucleus (Wang and Hung, 2009) with the functions of EGFR in the nucleus being the most extensively studied (Dittmann et al., 2010; Han and Lo, 2012; Lo, 2010; Wang et al., 2013; Wang and Hung, 2012).
Nuclear EGFR (nEGFR) is involved in several biological functions, including DNA replication, DNA repair, transcriptional regulation, and resistance to therapy, through associations with various molecules (Chen et al., 2011; Huang et al., 2011; Huo et al., 2010; Li et al., 2009; Wang and Hung, 2009; Wheeler et al., 2010). Even though EGFR is known to recognize specific promoter sequences of its target genes (Lin et al., 2001; Lo et al., 2005), it does not actually have a DNA-binding domain. Instead, it was recently discovered that EGFR interacts with a DNA-binding protein, RNA helicase A, in the nucleus to regulate transcription (Huo et al., 2010). Interestingly, radiation has been shown to trigger EGFR translocation into nucleus, which leads to activation of DNA-dependent protein kinase (DNA-PK) in DNA repair process (Dittmann et al., 2010; Dittmann et al., 2005; Dittmann et al., 2008). More recently, nEGFR was shown to associate with DNA-PK to phosphorylate PNPase at S776 and inactivate its ribonuclease activity, which contributes to radioresistance of cancer cells (Yu et al., 2012). nEGFR also phosphorylates proliferating cell nuclear antigen (PCNA) at Y211 to maintain its stability and thus controls its functions in DNA replication and DNA repair (Wang et al., 2006). In addition, EGFR was shown to associate with histone H3 in a study using proteomic strategy to identify protein-protein interaction in the EGFR signaling pathway (Blagoev et al., 2003). Although this study did not reveal any specific functions of the EGFR/histone H3 interaction, it suggested possible interaction between nEGFR and histone proteins. The identified nuclear functions of EGFR and its potential interactions with the histone proteins raised an interesting question whether EGFR interacts with core histones and regulates their functions.
In the current study, we demonstrate that EGFR interacts with and phosphorylates histone H4 at Y72 (H4-Y72). Phosphorylated H4-Y72 then recruits SET8 and SUV4-20H through interaction with their non-SET domains and enables these HMTases to methylate H4-K20, thereby promoting DNA synthesis and repair. Both EGF and ionizing radiation (IR), which are known to enhance nuclear localization of EGFR, can induce this mechanism. Our results provide mechanistic insights into how the tyrosine kinase activity of EGFR in the nucleus regulates H4-K20 methylation and affects DNA synthesis and repair and open an avenue toward the understanding of how K20 methylation-mediated DNA synthesis and DNA repair may be regulated by upstream signaling pathway.
Results
EGFR interacts with and phosphorylates histone H4
To investigate the relationship between core histones and EGFR, EGFR-immunoprecipitated nuclear proteins that correspond to the sizes of the core histones (bands 1-4; Figure S1A) from the nucleus were subjected to electrospray ionization tandem mass (MS/MS) spectrometric analysis, and the results showed that the core histones, H2A, H2B, H3, and H4, associated with EGFR. The EGFR/histones association prompted us to perform an in vitro EGFR kinase assay to detect which histone is a potential substrate of EGFR. Histone H2B and H4 were phosphorylated by EGFR, with the tyrosine phosphorylation level of histone H4 being the strongest (Figure S1B). We then focused on EGFR/histone H4 and further validated their interaction by reciprocal immunoprecipitation in two different human cancer cell lines (Figure 1A). Their interaction occurred in the nucleus but not in the cytosol (Figure S1C). Upon EGF stimulation, the interaction between EGFR and histone H4 was enhanced, which was abolished by AG1478, an EGFR tyrosine kinase inhibitor (TKI; Figure 1B). We also observed similar phenomena under IR treatment (Figure 1C), indicating that the association between EGFR and histone H4 is EGF- and IR-inducible and requires the tyrosine kinase activity of EGFR. Isolation of Triton-resistant (chromatin-bound proteins) and Triton-extractable (chromatin-unbound proteins) fractions further suggested that the interaction between EGFR and histone H4 occurs in the chromatin-enriched fraction (Figure S1D). Confocal microscopy also showed that EGFR co-localized with histone H4 in the nucleus in both MDA-MB-468 (insets 2 and 4 vs. 1 and 3, Figure S1E) and A431 cells (insets 6 and 8 vs. 5 and 7, Figure S1E; See also 3D image in Figure S1F). Quantitative analysis indicated a 7- to 8-fold increase in EGF-stimulated nuclear co-localization of EGFR and histone H4 (Figure S1G). To visualize the in situ subcellular interaction between nEGFR and histone H4, Duolink proximity ligation assay was performed with results showing that both EGF and IR stimuli induced EGFR and histone H4 interaction in the nucleus in vivo (red spots, Figure 1D). Together, these results suggest that EGF- or IR-enhanced EGFR and histone H4 association occurs in the chromatin-enriched fraction.
Figure 1. Histone H4 interacts with EGFR.

(A) Histone H4 and EGFR reciprocal immunoprecipitation (IP), followed by immunoblotting (IB) with the indicated antibody.
(B) Histone H4 IP/IB of serum-starved MDA-MB-468 cells treated with or without EGF and/or AG1478 for 30 min.
(C) Histone H4 IP/IB of serum-starved MDA-MB-468 cells treated with or without IR and/or AG1478 for 30 min.
(D) The in situ subcellular interaction between EGFR and histone H4 determined by Duolink proximity ligation assay in serum-starved MDA-MB-468 cells treated with or without EGF (20ng/ml) or post-irradiation (20 Gy) for 30 min (arrows indicated red spots). Blue color: DAPIstained nucleus.
(E) The in vitro translated myc-tagged FL-, ECD-, or ICD-EGFR (top) was individually incubated with purified recombinant histone H4 protein (middle) and immunoblotted to determine their interactions (bottom).
(F) HEK-293 cells were co-transfected with HA-histone H4 plasmid with myc-ECD-EGFR ormyc-ICD-EGFR plasmid for 24 h. Cell lysate was examined by reciprocal IP/IB with antibodies against myc and HA, respectively.
(G) Phosphotyrosine (p-Y) of histone H4 was examined by IP/IB with anti-p-Y or anti-histone H4 antibody, respectively, in 24 h-serum-starved cells treated with or without EGF and/orAG1478 for 30 min.
See also Figure S1.
To identify the specific domain of EGFR that associates with histone H4, we used an in vitro transcription/translation method (TNT system) to produce myc-tagged full-length EGFR (FL) and EGFR containing the extracellular domain (ECD) or the intracellular domain (ICD), and incubated them individually with recombinant histone H4. The results showed that EGFR interacts with histone H4 primarily through its ICD (Figure 1E), which was validated by reciprocal immunoprecipitation from lysate of HEK-293 cells co-transfected with plasmids harboring the ICD of EGFR and histone H4 (Figure 1F). The direct interaction between ICD of EGFR and histone H4 was validated by immunoprecipitation of purified recombinant EGFR-ICD protein with pure histone H4 protein (Figure S1H). Tyrosine phosphorylation of histone H4 was significantly enhanced in the presence of EGF but abolished by AG1478 in two cell lines (Figure 1G). Together, these results indicate that tyrosine phosphorylation of histone H4 is mediated by EGFR and the ICD of EGFR interacts with and phosphorylates histone H4.
EGFR mediates phosphorylation at Y72 of histone H4
To identify the tyrosine phosphorylation site of histone H4 in vivo, immunoprecipitated histone H4 from the nucleus of A431 cells was subjected to tandem mass (MS/MS) spectrometric analysis (Figure 2A). We reproducibly identified specific peptides containing phosphorylated tyrosine residue of histone H4 at Y72, indicating that this specific phosphorylation event occurs under physiological conditions. Sequence alignment of this region showed that this tyrosine residue is conserved across different species from Drosophila to human (inset, Figure 2A), implying that H4-Y72 phosphorylation may be an evolutionarily conserved event and is likely associated with important functions. We further performed in vitro kinase assay with purified recombinant histone H4 protein (His-H4Y72 or His-H4Y72F) and GST-EGFR kinase domain to demonstrate that EGFR directly phosphorylates histone H4 at Y72 but not the Y72F mutant (Figure S1I).
Figure 2. EGFR mediates tyrosine phosphorylation of H4-Y72.

(A) Mass (MS/MS) spectrometric analysis of A431 nuclear extract immunoprecipitated with anti-histone H4 antibody. Top, the flanking regions of histone H4-Y72 among different species. Dotted circle, phosphotyrosine-containing peptide fragments.
(B) Cells were serum-starved for 24 h and then treated with or without EGF and/or AG1478 for 30 min. Top, the level of histone H4-pY72 determined by IP/IB. Bottom, the endogenous levels of indicated proteins examined by IB.
(C) HEK-293 cells were co-transfected wild- type (WT) or Y72F mutant (Y72F) histone H4 plasmid with or without myc-EGFR plasmid and/or AG1478 or gefitinib for 24 h. Exogenous histone H4-Y72 phosphorylation was detected by IP/IB.
(D) MDA-MB-468 cells were infected with lentivirus containing shRNAs against luciferase (Luc) or EGFR (EGFR-1 or EGFR-2). Phosphorylated histone H4 at Y72 of each transfectant was examined.
See also Figure S2.
To investigate H4-Y72 phosphorylation in vivo, we generated an antibody against Y72-phosphorylated histone H4 (H4-pY72) and characterized its specificity. The H4-p-Y72 antibody recognized only the phospho (p-Y72)- but not the nonphospho (NP)-histone H4 peptide or other H4 phospho-peptides such as p-Y51 and p-Y88/p-Y98 (Figure S2A). The antibody immunoprecipitated only the wild-type HA-H4 but not the HA-Y72F mutant in the lysate from each stable transfectant (right panel, Figure S2B), and the signals were diminished by pretreatment of phosphatase (Figure S2C). Immunoprecipitation using our anti-histone H4-pY72 was efficient, as there was only a trace amount of tyrosine-phosphorylated H4 left in the unbound fractions (Figure S2D). We further performed immunoprecipitation using the antibody with denatured or native cell lysate and found that the anti-histone H4-pY72 antibody interacted mainly with histone H4 from native cell lysate (Figure S2E). We observed positive signals in EGF- or IR-treated cells but not in the EGFR knockdown cells from immunofluorescence staining using the anti-histone H4-pY72 antibody (Figure S2F). Taken together, these results indicate that the antibody recognizes primarily the native form of p-Y72 on histone H4. Using the H4-pY72 antibody, we showed that EGFR stimulated the H4-Y72 phosphorylation, which was inhibited by AG1478 in vivo in three different cell lines (Figure 2B). Expression of EGFR also enhanced the level of H4-Y72 phosphorylation in cells transfected with the wild-type histone H4 but not the Y72F mutant, and this phosphorylation can be abrogated by AG1478 or gefitinib, a clinically used TKI (Figure 2C). Consistently, knockdown of EGFR by two different short hairpin RNAs (shRNAs) also significantly reduced the H4-Y72 phosphorylation (Figure 2D). Unlike EGF-stimulated kinases, such as EGFR in which phosphorylation peaked at 1 h and then decreased at 4 h, EGF-stimulated phosphorylation at histone H4-Y72 was more stable and sustained for at least 4 h (Figure S2H). Collectively, these results demonstrate that EGFR is a tyrosine kinase for Y72 of histone H4 in vivo.
Y72 phosphorylation enhances histone H4 methylation at K20
To determine whether EGFR-mediated H4-Y72 phosphorylation might regulate other PTMs and modulate their physiological functions, we examined the effect of H4-Y72 phosphorylation on three well-known PTMs of histone H4 (methylation at K20 and acetylation at K5 and K8 of the histone H4 tail). The expression of EGFR significantly increased K20 methylation of wild-type histone H4 but not of the Y72F mutant (Figure S3A), suggesting that EGFR-mediated methylation is associated with H4-Y72 phosphorylation. We noticed that the level of H4-K5 acetylation was also increased in the presence of EGFR regardless of the H4-Y72 phosphorylation status, but neither EGFR nor H4-Y72 phosphorylation status affected H4-K8 acetylation (Figure S3A). These findings suggest that H4-Y72 phosphorylation is involved in regulation of H4-K20 methylation but is not associated with H4-K5 or -K8 acetylation.
H4-K20 can be mono- and di-methylated to regulate DNA replication (Huen et al., 2008; Jorgensen et al., 2007) and DNA repair (Botuyan et al., 2006), respectively. Thus, we further asked how H4-Y72 phosphorylation affects H4-K20 methylation and its associated biological consequences. The levels of both H4-K20 mono- and di-methylation were increased under EGF stimulation but reduced by AG1478 or gefitinib treatment in MDA-MB-468 cells (Figure 3A). Inhibition of H4-K20 methylation by AG1478 was also evident in another cell line (Figure S3B). As shown in Figure S2I, about 35% of histone H4 was phosphorylated at Y72 upon EGF stimulation. The amount of histone H4 left in unbound fractions (non-phosphorylated histone H4-Y72) was 92, 47, and 89% of total input histone H4 in control, EGF-stimulated, and EGF plus AG1478-treated cells, respectively. Around 30∼40% of total K20-methylated H4 was not phosphorylated at Y72 in the unbound fraction. Similarly, co-transfection of EGFR and wild-type histone H4 also enhanced the levels of H4-K20 methylations in HEK-293 cells but suppressed by AG1478. However, only a trace amount of H4-K20 methylation of H4-Y72F was detected by co-transfection of EGFR and the H4-Y72F mutant (Figure 3B), suggesting that EGFR-increased K20 methylation is mediated through EGFR-histone H4-Y72 phosphorylation.
Figure 3. Phosphorylation at Y72 of histone H4 enhances its methylation at K20.

(A) The expressions of indicated proteins from 4-day-serum-starved MDA-MB-468 cells treated with or without EGF and/or AG1478 for or gefitinib 30 min.
(B) HEK-293 cells were co-transfected with HA-histone H4 or -H4Y72F plasmid with or without myc-EGFR plasmid and/or AG1478. The exogenous HA-histone H4 or HA-H4Y72F was immunoprecipitated with anti-HA antibody, followed by IB with specific antibodies against methylated histone H4 at K20.
(C) HEK-293 cells were co-transfected with HA-tagged histone H4 plasmid with or without ΔNLS- or wild-type-EGFR plasmid. Nuclear and cytosolic of the indicated proteins are shown below.
(D) SET8 or SUV4-20H HMTase was immunoprecipitated from the nuclear extract of MDA-MB-468 cells with specific antibody. An increasing amount of immunoprecipitated SET8 or SUV4-20H nuclear extract (50, 100, or 200 μg) was incubated with recombinant histone H4 or H4Y72F protein in the HMTase reaction buffer. Histone H4 K20 methylation and input were detected by IB.
(E) The recombinant wild-type and Y72F mutant histone H4 proteins were treated with or without EGFR kinase, followed by HMTase assay with immunoprecipitated SET8 or SUV4-20H from nuclear extract (50 μg). The histone H4-K20me1, H4-K20me2, and input of the histone were also examined.
(F) The same sequential in vitro EGFR kinase and HMTase assay as in (E) was performed with or without EGF and/or AG1478.
See also Figure S3.
To ensure that the Y72F mutant still retains its functions to associate with chromatin and other histone core proteins, we established MDA-MB-468 stable transfectants expressing HA-tagged wild-type or Y72F mutant histone H4 (HA-H4). The stable transfectants expressed equal amounts of exogenous HA-H4 (Figure S3C). The Y72F mutant did not affect its binding ability to chromatin as evident from similar levels of HA-H4 WT and Y72F mutant in the Triton-resistant chromatin bound fraction (Figure S3D); it also did not alter its interaction with other core histones in the chromatin (H3, H2A, and H2B; Figure S3E). Both EGF- and IR-stimulated histone H4-Y72 phosphorylation and subsequent K20 methylation in H4-Y72F stable transfectant were less than that in mock- or wild-type H4 transfectant (Figures S3F and S3G).
We expressed an EGFR mutant lacking the nuclear localization sequence (NLS), which is defective in nuclear translocation but still retains its kinase activity (Hsu and Hung, 2007; Hsu et al., 2009). This mutant could not be detected in the nuclear fraction, consistent with the previous reports (ΔNLS, Figure 3C), and hence was unable to increase H4-Y72 phosphorylation and H4-K20 methylation (top, Figure 3C), indicating that H4-Y72 phosphorylation-mediated H4-K20 methylation indeed depends on the presence of EGFR in the nucleus. Together, the results suggest that nEGFR-mediated Y72 phosphorylation of histone H4 enhances both its monomethylation and dimethylation at K20 but does not affect its ability to associate with other histone core proteins or its binding to chromatin.
Next, we asked whether Y72 phosphorylation of histone H4 affects its methylation by SET8 and SUV4-20H in vitro. Using an in vitro HMTase activity assay, we showed that the wild-type histone H4 but not the Y72F mutant was mono- and di-methylated at K20 by immunoprecipitated SET8 and SUV4-20H, respectively, in a dose-dependent manner (Figure 3D). To further establish the enhancement of H4-K20 methylation by H4-Y72 phosphorylation, sequential in vitro EGFR kinase and HMTase activity assays were performed (Figure 3E), which showed that H4-K20 methylation was enhanced by pretreating histone H4 with EGFR kinase (lane 4 vs. 3, Figure 3E) whereas AG1478 diminished the in vitro EGFR kinase-enhanced HMTase activity toward H4-K20 mono- and di-methylation (Figure 3F). However, pretreatment of EGFR kinase had no effect on the H4-Y72F mutant (lane 8 vs. 7, Figure 3E). Thus, the results support that EGFR-mediated H4-Y72 phosphorylation enhances histone H4 to serve as a substrate for SET8 and SUV4-20H HMTases.
Y72 phosphorylation of histone H4 enhances recruitment of SET8 and SUV4-20H
Next, we asked whether H4-Y72 phosphorylation affects its interactions with SET8 and SUV4-20H. Under EGF stimulation, the association between histone H4 and SUV4-20H or SET8 was significantly increased but abolished by AG1478 (Figures S4A and 4A). However, SUV39H, a H3-K9 methylase, did not interact with histone H4 even after EGFR stimulation (Figure 4A). In addition, wild-type histone H4 but not Y72F mutant associated with SUV4-20H and SET8 (Figure 4B), suggesting that Y72 phosphorylation of histone H4 may be critical for it to recruit SET and SUV4-20H. To further address whether SUV4-20H and SET8 specifically recognize the phospho-Y72 residue, we used 13-amino-acid peptides derived from histone H4 that contain either p-Y72 or non-phospho-Y72 (NP) in the center of the peptides as bait to determine the interaction between Y72 of histone H4 and SUV4-20H and SET8. As evident from the dot blot analysis, both SET8 and SUV4-20H from cell lysates interacted only with the p-Y72 peptide (Figure S4B). In addition, these two purified HMTases (Figure 4C) also recognized the p-Y72 but not the NP peptide (Figure 4D), and their interactions can be disrupted by the pY72 but not the NP peptide in a competition assay (Figure 4E). The pull-down assay using the pY72 peptide recognized only GST-SET8 and GST-SUV4-20H2 but not GST-SUV39H1 (Figure 4F). To determine which region of these two HMTases interacts with p-Y72 peptide, we first constructed SET8 and SUV4-20H with or without the SET domain that contains the K20 HMTase active site. We then transfected the full- length (FL), SET-containing (S) domain, or non-SET-containing (NS) domain of these two HMTases into HEK293 cells (Figure 4G). The results showed that the p-Y72 peptide mainly associated with the NS region of SET8 and SUV4-20H2 (Figure 4H), indicating that the NS region, which does not have a well-defined function, interacts with p-Y72 of histone H4. The results suggest that EGF/EGFR-mediated Y72 phosphorylation of histone H4 enhances its interaction with the SET8 and SUV4-20H methyltransferases through the NS domain to bring the SET domain closer to K20 for methylation.
Figure 4. Phosphorylation at Y72 of histone H4 enhances its interaction with SET8 and SUV4-20H.

(A) Serum-starved MDA-MB-468 cells were treated with or without 20 ng/ml EGF and/or AG1478 (10 μM) for 30 min. Triton-resistant extract was isolated and immunoprecipitated with anti-histone H4 antibody to determine its association with SET8, SUV4-20H, or SUV39H1HMTases by IB.
(B) Histone H4 in nuclear fractions from indicated stable transfectants with or without AG1478 (10 μM) was immunoprecipitated to determine its association with SET8 or SUV4-20H(top). Endogenous levels of indicated proteins in the nuclear fraction (bottom).
(C) Purified GST-SET8 and -SUV4-20H2 proteins were stained with Coomassie blue (top) orimmunoblotted with specific antibodies (bottom).
(D) Different amounts of histone H4-Y72 peptides (IRDAVT-Y-TEHAKR) containing non-phospho- (NP) or phospho-residue at Y72 (p-Y72) were dotted onto the PVDF membrane, stained with Ponceau S solution (bottom), and then incubated with purified GST-SET8 or GST-SUV4-20H2 protein. Binding of SET8 or SUV4-20H to the coated H4 peptides was determined by IB (top).
(E) Histone H4-pY72 peptide was dotted onto membrane, incubated with GST-SET8 or GST-SUV4-20H2 protein with or without H4-Y72 peptide (NP or p-Y72) followed by IB.
(F) Histone H4-pY72 peptide was conjugated on agarose beads and then applied to pull down against GST-SET8, GST-SUV4-20H2, and GST-SUV39H1 recombinant proteins.
(G) HEK-293 cells were transfected with full- length (FL), SET-containing (S), or non-SET-containing (NS) region of SET8 or SUV4-20H2 for 24 h. Expression of each fragment was examined by IB against V5.
(H) Cell lysate from each transfectant was incubated with the H4-pY72 peptide-coated membrane followed by IB against V5.
See also Figure S4.
H4-Y72 phosphorylation promotes DNA synthesis
SET8 is known to monomethylate H4-K20 during DNA synthesis and is required for DNA replication (Huen et al., 2008). Thus, to determine whether H4-Y72 phosphorylation affects DNA synthesis, cells were pulse-labeled with BrdU and the newly synthesized DNA associated with histone H4 (wild-type or Y72F mutant) was detected by chromatin immunoprecipitation to measure the relative amounts of immunoprecipitated BrdU-labeled DNA. As shown in Figure 5A, EGF treatment increased the amounts of newly synthesized DNA in wild-type histone H4 but not in mutant Y72F or K20R. Addition of AG1478 attenuated EGF-stimulated DNA synthesis in the wild-type histone H4. It should be mentioned that the K20R mutant behaved similarly to the Y72F mutant, having lower ability of DNA synthesis, and this is in line with the impairment of K20 methylation in the Y72F mutant. These results support that H4-Y72 phosphorylation promotes DNA synthesis.
Figure 5. Histone H4-Y72 phosphorylation is involved in DNA synthesis.

(A) Stable transfectants were serum starvated for 24 h, then were treated with or without EGF and/or AG1478, and pulse-labeled with 100 μM BrdU for 12 h. Histone H4-associated newly synthesized DNA was determined. Top right, expression of HA-histone H4 in each stable transfectant.
(B) Stable transfectants were pulse-labeled with 100 μM BrdU for 4 h. Subsequently, the DNA contents and the incorporated BrdU in each transfectant were determined by flow cytometry. The percentage of cells distributed in each phase of cell cycle is shown.
(C) Stable transfectants were treated with nocodazole and/or AG1478 for 24 h. The distributions of cells in G1, S, and G2/M phases of cell cycle were analyzed by flow cytometer (Figure S5A). The bar plot is the G1 (%) in each treatment.
(D) Stable transfectants were synchronized at G1 phase and then released to normal culture medium for indicated time intervals. The distributions of cells in each stage of cell cycle were analyzed by flow cytometry (Figure S5B). Bar graph shows quantitative results.
(E) Relative number of cells of each stable transfectant was determined at indicated time intervals. Error bars, mean ± SD.
See also Figure S5.
To assess the speed of DNA synthesis, cells were pulse-labeled with BrdU and analyzed by flow cytometry according to a previous report (Jorgensen et al., 2007). In BrdU incorporation experiments, the BrdU signal from non-labeled cells was used as a cut-off line. Cells in the S phase distributed above the line were in replicating S phase and labeled as S (R); cells distributed below the line were in non-replicating S phase and labeled as S (NR). Remarkably, 30-40% of mock and wild-type histone H4 stable transfectants were in the replicating S phase, whereas only around 10% of the H4-Y72F and H4-K20R transfectants were in the replicating S phase. Consistent with the lower percentage of cells in the replicating S phase, we observed a higher percentage (around 10%) of H4-Y72F and H4K20R stable transfectants in the non-replicating S phase than in the mock and wild-type histone H4 transfectants (around 5%) (Figure 5B). Together, these results suggest that DNA synthesis is slower in the H4-Y72F mutant, delaying the replicating S phase as a result of defective K20 methylation.
In support of this notion, we measured the effect of phosphorylated H4-Y72 on cell cycle progression using a previously established protocol (Jorgensen et al., 2007). Stable transfectants expressing wild-type or Y72F mutant histone H4 were treated with a mitotic spindle inhibitor, nocodazole, to inhibit mitosis and prevent cells from progressing to the next G1 phase. Under this condition, if the cell cycle progresses faster, more cells will transition from G1 to S and stop at the G2/M phase; therefore, the number of cells in the G1 phase will be reduced, and vice versa. Treatment of the histone H4-Y72F transfectant with nocodazole resulted in 20% of cells in the G1 phase compared with 6∼9% of cells in the G1 phase for the mock and wild-type transfectants, indicating that Y72F mutant reduced cell cycle progression (Figures 5C and S5A). In contrast, treatment with AG1478 abolished the differences we observed between the mock, wild-type H4, and Y72F transfectants, with all three having about 18∼20% of cells in the G1 phase (Figures 5C and S5A). Cells were synchronized at G1 phase by double thymidine block and released to normal culture medium for different time intervals to progress cell cycle. As shown in Figures 5D and S5B, the decrease in G1 phase and increase in S phase in mock and wild-type H4 transfectants were more than that in H4-Y72F transfectants, particularly at the at the 4 h time point, indicating that H4-Y72F transfectants entered S phase more slowly than mock or wild-type transfectants. Moreover, EGF stimulated histone H4-Y72 phosphorylation, K20 methylation (Figure S5C), and DNA synthesis (Figure S5D) in a dose-dependent manner. Finally,cell proliferation assay revealed that H4-Y72F transfectant grew slower than mock or wild-type H4 transfectant. Regression analysis of the growth curve of each transfectant indicated that cell doubling time was 26, 26, and 31 h in mock, wild-type H4, and H4-Y72F mutant transfectants, respectively (Figure 5E). Collectively, these results suggest that the Y72F mutant impairs DNA synthesis and delays S phase progression and cell growth as a result of defective K20 methylation.
H4-Y72 phosphorylation regulates DNA double-stranded break repair in response to ionizing radiation
Dimethylation at K20 of histone H4 is involved in DNA double-stranded break (DSB) repair (Botuyan et al., 2006; Sanders et al., 2004). To determine whether H4-Y72 phosphorylation affects this function, we determined the recovery of IR-induced DNA damage by observing the level of phospho-histone H2AX at S139 (γ-H2AX), which is indicative of IR-induced DNA damage. At 1 h after irradiation, γ-H2AX was induced, and within 8 h of recovery, the increased level of γ-H2AX was gradually reduced to the basal level in wild-type histone H4 cells; however, the reduction rate of γ-H2AX was slower in the Y72F mutant (Figure 6A). Consistently, the intensity of γ-H2AX in immunofluorescence staining, which peaked at 1 h after irradiation, was reduced by 50% in cells expressing wild-type histone H4 but only by 20% in those expressing the Y72F mutant within a 2-h recovery period (Figures 6B and S6A), suggesting that the Y72F mutant reduced its DNA repair activity.
Figure 6. Histone H4-Y72 phosphorylation is involved in DNA DSB repair.
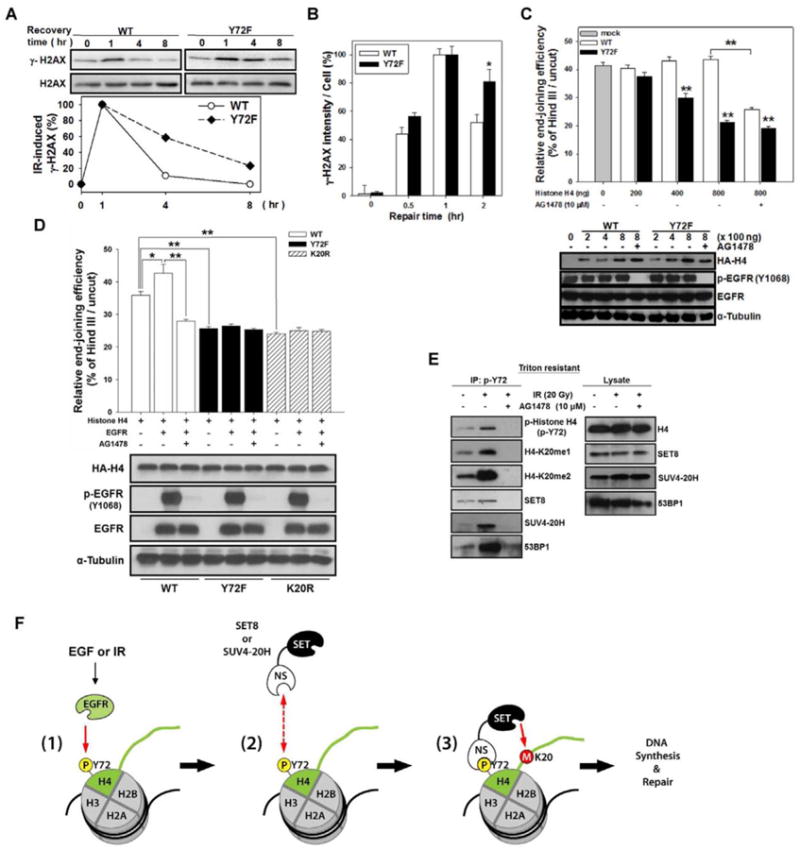
(A) Top, each stable transfectant was irradiated with 20 Gy and incubated at 37 °C to recover for the indicated time. Expression of γ-H2AX was examined by IB. Bottom, relative quantity of γ-H2AX was normalized to H2AX 1 h after irradiation.
(B) The quantitation of γ-H2AX in Figure S6A.
(C) HEK-293 cells were co-transfected with circular (cir-pGL3) or linearized (lin-pGL3) luciferase plasmid with myc-EGFR and increased amounts of wild-type (WT) or Y72F mutant histone H4 plasmid for 48 h with or without AG1478. The relative end-joining efficiency was determined by the percentage of luciferase activity from cells with lin-pGL3 over that with cir-pGL3. Error bars, mean ± SD. Bottom, the expression of transfected plasmids as indicated.
(D) A similar experiment as described in (C) was also performed using another histone H4K20R mutant plasmid (800 ng) with or without EGFR and/or AG1478.
(E) Serum-starved MDA-MB-468 cells were treated with or without irradiation and/or AG1478. After 1 h, Triton-resistant fraction was immunoprecipitated against histone H4-p-Y72 followed by IB with the indicated antibodies. Right, endogenous level of indicated proteins.
(F) A proposed model of nEGFR-mediated H4-Y72 phosphorylation in the regulation of H4-K20 methylation.
See also Figure S6.
To further validate that H4-Y72 phosphorylation affects DNA repair, we examined the end-joining capacity by transfecting HEK-293 cells with linearized luciferase plasmid, which is inactive unless it is re-circularized by end-joining DNA repair. In the presence of wild-type histone H4, luciferase activity remained the same whereas expression of the Y72F mutant significantly reduced luciferase activity in a dose-dependent manner (up to 40% of inhibition; Figure 6C), indicating that the mutant impaired end-joining activity. AG1478 also decreased end-joining activity in cell expressing wild-type histone H4 but had no effect on the Y72F mutant. Consistently, overexpression of EGFR significantly increased end-joining ability in cells with wild-type histone H4, which was reduced by AG1478. However, EGFR expression had no effect on the histone H4 Y72F or K20R mutant (Figure 6D), suggesting that EGFR-enhanced end-joining activity is modulated by Y72 phosphorylation and K20 methylation.
Dimethylated H4-K20 participates in DNA repair by recruiting 53BP1 to the damaged site (Botuyan et al., 2006), and IR enhances nuclear translocation of EGFR in the activation of DNA repair (Dittmann et al., 2010; Dittmann et al., 2005; Dittmann et al., 2008). To determine whether H4-Y72 phosphorylation, which enhances H4-K20 methylation, also affects the recruitment of 53BP1 in DNA repair, we analyzed H4-Y72 phosphorylation after IR and examined its interaction with 53BP1. After IR, H4-Y72 phosphorylation was increased in the chromatin-enriched Triton-resistant fraction but reduced in the presence AG1478 (Figure 6E). Co-immunoprecipitation also showed that H4-Y72 phosphorylation increased H4-K20 methylation (me1/me2) to recruit SET8 and SUV4-20H and subsequently enhanced the levels of 53BP1 in the chromatin-enriched Triton-resistant fraction upon IR (Figure 6E). A dose-dependent response to IR was observed for histone H4-Y72 phosphorylation, K20 methylation, and association of 53BP1 in DNA repair (Figure S6B).
The functional link between nEGFR and IR-induced DNA repair has been reported in which EGFR-induced PI3K/AKT activation and subsequent activation of DNA-PK regulates radiosensitivity (Qu et al., 2013; Toulany et al., 2006). We further tested whether PI3K/AKT is involved in EGFR-mediated histone H4-Y72 phosphorylation upon IR. To this end, we examined the effects of PI3K/AKT inhibitors on histone H4-Y72 phosphorylation and K20 methylation in IR-induced DNA repair. As shown in Figure S6C, addition of PI3K/AKT inhibitors (LY294002, API2, and AKT inhibitor X) did not affect the IR-stimulated histone H4-Y72 phosphorylation and K20 methylation, suggesting that EGFR-induced histone H4-Y72 phosphorylation and K20 methylation are not mediated by activation of PI3K/AKT pathway. All together, these results suggest that IR increased the levels of nEGFR for H4-Y72 phosphorylation to recruit SET8 and SUV4-20H for H4-K20 methylation and subsequent binding by 53BP1 for DNA repair. These findings provide a plausible mechanism linking the two well known DNA replication and DNA repair events, namely EGFR and histone H4-K20 and should open a new avenue toward the understanding of how growth factor receptors may regulate these two events through the histone H4-Y72 phosphorylation/K20 methylation cascade.
In summary, we report a link between nEGFR and histone H4 function. As depicted in Figure 6F, nEGFR first phosphorylates histone H4 at Y72 (1). Then, the non-SET domain of SET8 and SUV4-20H binds to p-Y72 (2) and brings the SET domain in close proximity to K20 for methylation (3). Methylation at K20 leads to increased DNA synthesis and DNA repair.
Disruption of the interaction between EGFR and histone H4 by Y72 peptide demonstrates tumor suppressive activity in vivo
To determine whether disruption of the EGFR-histone H4 binding interface affects tumor growth, we first evaluated the effect of several tyrosine peptides on the EGFR-histone H4 interaction and found that the Y72 peptide more potently disrupted their interaction than other phosphotyrosine peptides (Figure S7A) or scrambled control (Figure 7A). We then tested the anti-tumor effect of Y72 peptide in an MDA-MB-468 breast cancer xenograft tumor model. Mice treated with Y72 peptide had significantly reduced tumor size (Figures 7B and S7B) and weight (Figure 7C) compared with those treated with PBS or scrambled peptide. The levels of histone H4-pY72 were also reduced in the tumor mass from mice treated with the Y72 peptide (Figure 7D). Together, these results further validate a biologically important role of EGFR-mediated histone H4 Y72 phosphorylation and demonstrate that blocking the EGFR-histone H4 interaction has therapeutic potential.
Figure 7. Y72 peptide demonstrates tumor suppressive activity in vivo, and histone H4-pY72 and K20me1 correlate with EGFR expression in clinical samples of human breast cancer.

(A) MDA-MB-468 cell lysates were subjected to IP with anti-EGFR antibody in the presence or absence of Y72 or scrambled peptide (200 μM) followed by IB against histone H4 and EGFR.
(B) The tumor volume of mice treated with PBS or the indicated peptides. Error bars, mean ± SD.
(C) Bar graph shows the quantative tumor weight in each group. Error bars, mean ± SD.
(D) Protein lysates from tumors in (C) were subjected to Western blot analysis with the indicated antibodies. To detect histone H4-pY72, lysates were subjected to IP first with anti-histone H4-pY72 antibody followed by IB with anti-histone H4 antibody.
(E) Correlations between expression of EGFR and histone H4-pY72, histone H4-K20me1, or Ki-67 in surgical specimens of breast cancer were analyzed by Pearson Chi-Square test. ¶M=membrane, N= nucleus, C= cytoplasm.
(F) IHC staining of human breast tumor tissue sections for EGFR and histone H4-K20me1. The plot represents the quantitative results of histone H4-K20me1 in the tissue sections with different EGFR status. EGFR N+, nuclear EGFR positive; EGFR C+, cytosolic EGFR positive; EGFR-, EGFR negative. Error bars, mean ± SD.
See also Figure S7.
EGFR expression correlates with histone H4-Y72 phosphorylation and K20 methylation in human tumor samples
To determine the clinical relevance of this pathway, we examined the levels of EGFR, histone H4-pY72, histone H4-K20me1, and Ki-67 in human breast tumor samples using immunohistochemical (IHC) staining. We first verified the specificity of our anti-histone H4-pY72 antibody for IHC staining by peptide competition. The IHC staining of histone H4-pY72 was reduced only by phospho-histone H4-pY72 peptide but not non-phospho-histone H4-Y72 (NP) and phospho-histone H4-pY51 peptide (Figure S2G), indicating that our anti-histone H4-pY72 antibody is specific to phosphorylated Y72 of histone H4. Multiple-variable analysis of the results from IHC staining revealed that EGFR expression was significantly correlated with H4-pY72 (p = 0.049), H4-K20me1 (p = 0.011), and proliferation marker Ki-67 (p = 0.002) (Figure 7E; also see Figure S7C for representative images of IHC staining). In addition, the level of H4-K20me1 was higher in cells expressing nuclear EGFR than in cells with cytosolic EGFR (p = 0.0598). However, there was no detectable difference in H4-K20me1 between the cells with cytosolic EGFR and without EGFR (p = 0.5329) (Figure 7F), suggesting the potential role of nuclear EGFR in regulation of histone H4-K20 methylation in breast cancer. Taken together, these findings provide a significant clinical relevance between EGFR, and histone H4-Y72 phosphorylation and K20 methylation in human breast cancer.
Discussion
Aside from their prominent role in cell surface signal transduction, EGFR family members also have non-canonical functions, such as transcriptional regulation, DNA synthesis, and DNA repair in the nucleus (Dittmann et al., 2008). However, the molecular mechanisms underlying these nuclear functions are not completely understood. The present study identifies a mechanism by which nEGFR regulates DNA synthesis and repair. We showed that nEGFR phosphorylates histone H4 at Y72 which facilitates the recruitment of SET8 and SUV4-20H HMTases for H4-K20 monomethylation mono- and di-methylation to regulate DNA synthesis and DNA repair (Figure 6F).
It is commonly known that catalytically inactive enzymes associate with their substrates more avidly when they are unable to modify their substrates. While this is generally true for enzyme-substrate interactions, there are many exceptions when substrates are proteins. For example, the interaction between Grb10 and insulin receptor and insulin-like growth factor receptor depends on the receptor's tyrosine kinase activity. Mutation of the tyrosine residues by Y1150F and Y1151F within the insulin receptor's activation loop dramatically reduced the interaction. (He et al., 1998). Activation of focal adhesion kinase by Y397 phosphorylation is also required for its association with phosphatidylinositol 3-kinase (Chen et al., 1996). In addition, the binding of ATR to fragile DNAs also requires its kinase activity (Wang et al., 2010b). Similarly, we found that EGF- or IR-induced association of EGFR and histone H4 requires the tyrosine kinase activity of EGFR (Figures 1B and 1C).
Methylation at histone H4-K20 is a sequential and dynamic process during cell cycle progression or DNA repair. In general, SET8-mediated histone H4-K20 mono-methylation is required for G1/S transition and S phase (Huen et al., 2008; Jorgensen et al., 2007); SUV4-20H-mediated histone H4-K20 di-methylation is involved in the recruitment of 53BP1 for DNA repair (Botuyan et al., 2006). However, it has been reported that SUV4-20H-mediated di- and/or tri-methylation of histone H4-K20 also play important roles for recruitment of the origin recognition complex (ORC) to the replication origins (Beck et al., 2012); de novo histone H4-K20 mono-methylation by SET8 is also involved for 53BP1 recruitment in DNA repair (Oda et al., 2010). Thus, SET8 and SUV4-20H may dynamically interact with histone H4 in both DNA replication and DNA repair.
SH2 and PTB are common phosphotyrosine binding domains. Although we could not predict a distinct SH2 or PTB domain on the non-SET regions of SET8 and SUV4-20H, sequence alignment of these two non-SET regions of SET8 and SUV4-20H revealed 7 consensus arginine (R) residues (Figure S4C). Arginine residues on SH2 domain are known to have particularly prominent roles as key contacts with the phosphotyrosines (Yaffe, 2002), suggesting that these consensus R residues on the non-SET regions of SET8 and SUV4-20H may be key residues for phosphotyrosine binding. Further study would be required to demonstrate this.
There is recent evidence showing that tyrosine phosphorylation of histones affects their functions. WSTF, a transcription factor, has intrinsic tyrosine kinase activity toward Y142 of histone H2AX to maintain S139 phosphorylation and IR-induced foci formation, which is crucial for regulation of the DNA damage response (Xiao et al., 2009). Rad53-associated Y99 phosphorylation of histone H3 is also critical for efficient ubiquitination and degradation in the regulation of histone levels (Singh et al., 2009). JAK2 phosphorylates Y41 of histone H3 and prevents HP1α binding to chromatin (Dawson et al., 2009). WEE1 phosphorylates Y37 of histone H2B to suppress mRNA synthesis of replication-dependent core histone genes (Mahajan et al., 2012). Specifically, our study supports the concept that H4-Y72 phosphorylation can regulate H4-K20 methylation to affect histone-associated functions and raises interesting questions about the functional role of nuclear tyrosine kinases.
It is worthwhile to mention that both nEGFR and H4-K5 acetylation have been shown to be involved in transcriptional regulation (Fukuda et al., 2006; Huo et al., 2010; Peterson and Laniel, 2004; Wang and Hung, 2009). We demonstrated that EGFR stimulates H4-K5 acetylation, but this may not be regulated through H4-Y72 phosphorylation as the Y72F mutant still responded to EGFR-enhanced H4-K5 acetylation (Figure S3A). Further systematic study would be needed to address how EGFR upregulates H4-K5 acetylation and whether this contributes to nEGFR-mediated transcriptional regulation.
Although it is conceptually difficult to accept that an integral membrane-embedded RTK can escape from the lipid bilayer and enter the nucleus, recent studies have provided a comprehensive model of integral membrane-embedded EGFR trafficking to the nucleus in a vesicle via COPI-mediated retrograde trafficking (Wang et al., 2012; Wang et al., 2010a; Wang et al., 2010c), which enables EGFR to stay in its membrane-embedded form throughout the entire trafficking pathway from the cell surface to the nucleus (Wang and Hung, 2012; Wang et al., 2010b). The current study further provides a comprehensive molecular mechanism governing the function of nEGFR, namely, the regulation of DNA synthesis and DNA repair through nEGFR-mediated H4-Y72 phosphorylation to enhance recruitment of SET8 and SUV4-20H for methylation at K20 of histone H4. These findings together with recent studies that have unraveled a nuclear EGFR translocation trafficking pathway, providing compelling evidence for more investigations into the potentially important roles of a vast number of cell surface receptors in the nucleus.
Experimental Procedures
Duolink Proximity Ligation Assay
The in situ proximity ligation assay was performed using the Duolink II fluorescence kit (Olink Bioscience) according to the instruction. In brief, cells were seeded on cover slides the day before the experiment. After treatment, cells were fixed, permeabilized, blocked, and then incubated with primary antibodies against EGFR and histone H4 at 4°C overnight. After washing, the oligonucleotides (Minus and Plus) conjugated secondary antibodies were incubated for another hour at 37°C. Subsequently, cells were washed and incubated with ligation solution for 30 min at 37°C. Finally, the ligated nucleotide circles were amplified by polymerase via rolling-circle amplification (RCA), and the RCA products were visualized by hybridization with fluorescence labeled oligonucleotides. The visualized fluorescence spots represent the clusters of protein-protein interactions.
In vitro Histone Methyltransferase Assay
HMTases SET8 and SUV4-20H were immunoprecipitated from the nuclear fraction of MDA-MB-468 cells. The immunoprecipitated protein was incubated with 1 μg of recombinant histone wild-type or Y72F mutant histone H4 protein as substrate and S-adenosyl-L-methionine as the methyl donor in the reaction buffer (50 mM Tris-HCl at pH 8.5, 100 mM NaCl, 10 mM DTT) at 30 °C for 1 h. The reaction mixture was then resolved on 15% SDS-PAGE. The amount of methylated histone H4 was detected by Western blotting with specific antibodies.
Irradiation and DSB End-Joining Capacity Assay
To generate DNA damage, cells were irradiated with 100-kV photons (RX-650 Cabinet X-Radiator from Faxitron X-Ray) at a dose rate of 4 Gy/min. After irradiation, cells were incubated at 37°C for the indicated time and then harvested for further experiments. For the DSB end-joining capacity assay, approximately 2 × 105 cells were seeded on each well of a 24-well plate. Cells were transfected with 200 ng of Hind III-linearized or circular pGL3-CMV plasmid, 10 ng of pRL-TK-Luc, 200 ng of myc-EGFR, and indicated amount of histone H4 expression plasmids for 48 h to permit DSB repair and expression of luciferase. Luciferase activity was measured by a Dual-Luciferase Reporter System (Promega). Cells were ruptured in the passive lysis buffer for 15 min by shaking at room temperature. Five microliters of lysate were applied to dual luciferase assay reagent, and its luminescence was measured with a luminometer (Berthold). The Renilla luciferase activity from pRL-TK-Luc was used for normalizing the transfection efficiency. The DSB end-joining capacity was determined from the ratio of the normalized luciferase activity from cells transfected with lin-pGL3 and cir-pGL3 (Zhong et al., 2002).
Animal Studies
Tumor xenografts were established by orthotopic implantation of MDA-MB-468 cells (1×107 cells in a 1:1 mixture of PBS and Matrigel (BD Biosciences) in a total volume of 100 μl) into the mammary fat pad of each 6-8-week old female severe combined immunodeficient (SCID) mice. When the tumors were palpable, mice were randomly separated into 3 groups. The mice were administered PBS, scrambled peptide, or Y72 peptides (200 nmole/mouse) by intratumoral injection every three days. The tumor size (mm3) was measured at every three days by the following formula: V= 0.5 × length × diameter2 for 30 days. All animal experiments were approved by the Institutional Review Board of Animal Experiments, China Medical University Hospital.
Statistical Analysis
Data was analyzed by t-test and the IHC result from clinical tissues was calculated by Pearson Chi-Square test. *, p < 0.05; ** p < 0.01.
Supplementary Material
Highlights.
EGF- or IR-induced nEGFR interacts with and phosphorylates histone H4 at Y72
H4 p-Y72 enhances SET8 and SUV4-20H recruitment to facilitate K20 methylation
H4 Y72 phosphorylation promotes DNA synthesis and DNA double-strand break repair
Disrupting the EGFR-histone H4 interaction inhibits tumor growth in vivo
Acknowledgments
We thank the National RNAi Core Facility (Academia Sinica, Taipei, Taiwan) for providing the shRNAs. We appreciate Dr. Stephanie A. Miller (Department of Molecular and Cellular Oncology) and Dr. Tamara Locke (Scientific Publications at the MD Anderson) for their assistance in editing this manuscript and Jung-Mao Hsu for drawing the proposed model. This research is supported in part by the National Institutes of Health (Cancer Center Support Grant CA016672, CA109311, and CA099031), and The University of Texas MD Anderson–China Medical University and Hospital Sister Institution Fund, NSC99-2320-B-039-030-MY3, NSC101-2321-B-039-004, NHRI-EX102-10245BI (to Y.L.Y.), NHRI-EX98-9603BC (to L.Y.L.), DOH102-TD-C-111-005 (to L.Y.L. and M.C.H.), NSC3111-B-039 and NSC2623-13-039 (to M.C.H., Y.L.Y. and L.Y.L.), and NSC101-2311-B-039-001, CMU100-N2-09, CMU101-N2-04 (to R.H.C) from Taiwan.
Footnotes
Conflict of interest: The authors have no conflicts of interest to declare.
A complete description of the materials and methods is provided in Supplemental Experimental Procedures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beck DB, Burton A, Oda H, Ziegler-Birling C, Torres-Padilla ME, Reinberg D. The role of PR-Set7 in replication licensing depends on Suv4-20h. Genes Dev. 2012;26:2580–2589. doi: 10.1101/gad.195636.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat Biotechnol. 2003;21:315–318. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HC, Appeddu PA, Isoda H, Guan JL. Phosphorylation of tyrosine 397 in focal adhesion kinase is required for binding phosphatidylinositol 3-kinase. J Biol Chem. 1996;271:26329–26334. doi: 10.1074/jbc.271.42.26329. [DOI] [PubMed] [Google Scholar]
- Chen YJ, Huang WC, Wei YL, Hsu SC, Yuan P, Lin HY, Wistuba II, Lee JJ, Yen CJ, Su WC, et al. Elevated BCRP/ABCG2 expression confers acquired resistance to gefitinib in wild-type EGFR-expressing cells. PLoS One. 2011;6:e21428. doi: 10.1371/journal.pone.0021428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Bannister AJ, Gottgens B, Foster SD, Bartke T, Green AR, Kouzarides T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–822. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Kehlbach R, Rodemann HP. Nuclear EGFR shuttling induced by ionizing radiation is regulated by phosphorylation at residue Thr654. FEBS Lett. 2010;584:3878–3884. doi: 10.1016/j.febslet.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Raju U, Milas L, Chen DJ, Kehlbach R, Rodemann HP. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005;280:31182–31189. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- Dittmann K, Mayer C, Kehlbach R, Rodemann HP. Radiation-induced caveolin-1 associated EGFR internalization is linked with nuclear EGFR transport and activation of DNA-PK. Mol Cancer. 2008;7:69. doi: 10.1186/1476-4598-7-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Feng Q, Ketel CS, Wang H, Cao R, Xia L, Erdjument-Bromage H, Tempst P, Simon JA, Zhang Y. Purification and functional characterization of SET8, a nucleosomal histone H4-lysine 20-specific methyltransferase. Curr Biol. 2002;12:1086–1099. doi: 10.1016/s0960-9822(02)00924-7. [DOI] [PubMed] [Google Scholar]
- Fukuda H, Sano N, Muto S, Horikoshi M. Simple histone acetylation plays a complex role in the regulation of gene expression. Brief Funct Genomic Proteomic. 2006;5:190–208. doi: 10.1093/bfgp/ell032. [DOI] [PubMed] [Google Scholar]
- Gomes DA, Rodrigues MA, Leite MF, Gomez MV, Varnai P, Balla T, Bennett AM, Nathanson MH. c-Met must translocate to the nucleus to initiate calcium signals. J Biol Chem. 2008;283:4344–4351. doi: 10.1074/jbc.M706550200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Lo HW. Landscape of EGFR signaling network in human cancers: biology and therapeutic response in relation to receptor subcellular locations. Cancer Lett. 2012;318:124–134. doi: 10.1016/j.canlet.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Rose DW, Olefsky JM, Gustafson TA. Grb10 interacts differentially with the insulin receptor, insulin-like growth factor I receptor, and epidermal growth factor receptor via the Grb10 Src homology 2 (SH2) domain and a second novel domain located between the pleckstrin homology and SH2 domains. J Biol Chem. 1998;273:6860–6867. doi: 10.1074/jbc.273.12.6860. [DOI] [PubMed] [Google Scholar]
- Hsu SC, Hung MC. Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J Biol Chem. 2007;282:10432–10440. doi: 10.1074/jbc.M610014200. [DOI] [PubMed] [Google Scholar]
- Hsu SC, Miller SA, Wang Y, Hung MC. Nuclear EGFR is required for cisplatin resistance and DNA repair. Am J Transl Res. 2009;1:249–258. [PMC free article] [PubMed] [Google Scholar]
- Huang WC, Chen YJ, Li LY, Wei YL, Hsu SC, Tsai SL, Chiu PC, Huang WP, Wang YN, Chen CH, et al. Nuclear translocation of epidermal growth factor receptor by Akt-dependent phosphorylation enhances breast cancer-resistant protein expression in gefitinib-resistant cells. J Biol Chem. 2011;286:20558–20568. doi: 10.1074/jbc.M111.240796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Sy SM, van Deursen JM, Chen J. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J Biol Chem. 2008;283:11073–11077. doi: 10.1074/jbc.C700242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo L, Wang YN, Xia W, Hsu SC, Lai CC, Li LY, Chang WC, Wang Y, Hsu MC, Yu YL, et al. RNA helicase A is a DNA-binding partner for EGFR-mediated transcriptional activation in the nucleus. Proc Natl Acad Sci U S A. 2010;107:16125–16130. doi: 10.1073/pnas.1000743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, Helleday T, Helin K, Sorensen CS. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol. 2007;179:1337–1345. doi: 10.1083/jcb.200706150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TH, Seng S, Sekine M, Hinton C, Fu Y, Avraham HK, Avraham S. Vascular endothelial growth factor mediates intracrine survival in human breast carcinoma cells through internally expressed VEGFR1/FLT1. PLoS Med. 2007;4:e186. doi: 10.1371/journal.pmed.0040186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Iida M, Dunn EF, Ghia AJ, Wheeler DL. Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene. 2009;28:3801–3813. doi: 10.1038/onc.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- Lo HW. Nuclear mode of the EGFR signaling network: biology, prognostic value, and therapeutic implications. Discov Med. 2010;10:44–51. [PMC free article] [PubMed] [Google Scholar]
- Lo HW, Hsu SC, Ali-Seyed M, Gunduz M, Xia W, Wei Y, Bartholomeusz G, Shih JY, Hung MC. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell. 2005;7:575–589. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- Mahajan K, Fang B, Koomen JM, Mahajan NP. H2B Tyr37 phosphorylation suppresses expression of replication-dependent core histone genes. Nat Struct Mol Biol. 2012;19:930–937. doi: 10.1038/nsmb.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Munshi A, Shafi G, Aliya N, Jyothy A. Histone modifications dictate specific biological readouts. J Genet Genomics. 2009;36:75–88. doi: 10.1016/S1673-8527(08)60094-6. [DOI] [PubMed] [Google Scholar]
- Oda H, Hubner MR, Beck DB, Vermeulen M, Hurwitz J, Spector DL, Reinberg D. Regulation of the histone H4 monomethylase PR-Set7 by CRL4(Cdt2)-mediated PCNA-dependent degradation during DNA damage. Mol Cell. 2010;40:364–376. doi: 10.1016/j.molcel.2010.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento JJ, Mizzen CA, Kelleher NL. Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: human histone H4. Anal Chem. 2006;78:4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Qu YY, Hu SL, Xu XY, Wang RZ, Yu HY, Xu JY, Chen L, Dong GL. Nimotuzumab enhances the radiosensitivity of cancer cells in vitro by inhibiting radiation-induced DNA damage repair. PLoS One. 2013;8:e70727. doi: 10.1371/journal.pone.0070727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly JF, Maher PA. Importin beta-mediated nuclear import of fibroblast growth factor receptor: role in cell proliferation. J Cell Biol. 2001;152:1307–1312. doi: 10.1083/jcb.152.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- Sehat B, Tofigh A, Lin Y, Trocme E, Liljedahl U, Lagergren J, Larsson O. SUMOylation mediates the nuclear translocation and signaling of the IGF-1 receptor. Sci Signal. 2010;3:ra10. doi: 10.1126/scisignal.2000628. [DOI] [PubMed] [Google Scholar]
- Singh RK, Kabbaj MH, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol. 2009;11:925–933. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stachowiak EK, Maher PA, Tucholski J, Mordechai E, Joy A, Moffett J, Coons S, Stachowiak MK. Nuclear accumulation of fibroblast growth factor receptors in human glial cells--association with cell proliferation. Oncogene. 1997;14:2201–2211. doi: 10.1038/sj.onc.1201057. [DOI] [PubMed] [Google Scholar]
- Toulany M, Kasten-Pisula U, Brammer I, Wang S, Chen J, Dittmann K, Baumann M, Dikomey E, Rodemann HP. Blockage of epidermal growth factor receptor-phosphatidylinositol 3-kinase-AKT signaling increases radiosensitivity of K-RAS mutated human tumor cells in vitro by affecting DNA repair. Clin Cancer Res. 2006;12:4119–4126. doi: 10.1158/1078-0432.CCR-05-2454. [DOI] [PubMed] [Google Scholar]
- Van Aller GS, Reynoird N, Barbash O, Huddleston M, Liu S, Zmoos AF, McDevitt P, Sinnamon R, Le B, Mas G, et al. Smyd3 regulates cancer cell phenotypes and catalyzes histone H4 lysine 5 methylation. Epigenetics. 2012;7:340–343. doi: 10.4161/epi.19506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SC, Hung MC. Nuclear translocation of the epidermal growth factor receptor family membrane tyrosine kinase receptors. Clin Cancer Res. 2009;15:6484–6489. doi: 10.1158/1078-0432.CCR-08-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SC, Nakajima Y, Yu YL, Xia W, Chen CT, Yang CC, McIntush EW, Li LY, Hawke DH, Kobayashi R, et al. Tyrosine phosphorylation controls PCNA function through protein stability. Nat Cell Biol. 2006;8:1359–1368. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- Wang YN, Hsu JL, Hung MC. Nuclear Functions and Trafficking of Receptor Tyrosine Kinases. In: Yarden Y, Tarcic G, editors. Vesicle Trafficking in Cancer. Springer; 2013. pp. 159–176. [Google Scholar]
- Wang YN, Hung MC. Nuclear functions and subcellular trafficking mechanisms of the epidermal growth factor receptor family. Cell Biosci. 2012;2:13. doi: 10.1186/2045-3701-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YN, Lee HH, Lee HJ, Du Y, Yamaguchi H, Hung MC. Membrane-bound trafficking regulates nuclear transport of integral epidermal growth factor receptor (EGFR) and ErbB-2. J Biol Chem. 2012;287:16869–16879. doi: 10.1074/jbc.M111.314799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YN, Wang H, Yamaguchi H, Lee HJ, Lee HH, Hung MC. COPI-mediated retrograde trafficking from the Golgi to the ER regulates EGFR nuclear transport. Biochem Biophys Res Commun. 2010a;399:498–504. doi: 10.1016/j.bbrc.2010.07.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YN, Yamaguchi H, Hsu JM, Hung MC. Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene. 2010b;29:3997–4006. doi: 10.1038/onc.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YN, Yamaguchi H, Huo L, Du Y, Lee HJ, Lee HH, Wang H, Hsu JM, Hung MC. The translocon Sec61beta localized in the inner nuclear membrane transports membrane-embedded EGF receptor to the nucleus. J Biol Chem. 2010c;285:38720–38729. doi: 10.1074/jbc.M110.158659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat Rev Clin Oncol. 2010;7:493–507. doi: 10.1038/nrclinonc.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, Ishibe-Murakami S, Wang B, Tempst P, Hofmann K, et al. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB. Phosphotyrosine-binding domains in signal transduction. Nat Rev Mol Cell Biol. 2002;3:177–186. doi: 10.1038/nrm759. [DOI] [PubMed] [Google Scholar]
- Yang H, Mizzen CA. The multiple facets of histone H4-lysine 20 methylation. Biochem Cell Biol. 2009;87:151–161. doi: 10.1139/O08-131. [DOI] [PubMed] [Google Scholar]
- Yang H, Pesavento JJ, Starnes TW, Cryderman DE, Wallrath LL, Kelleher NL, Mizzen CA. Preferential dimethylation of histone H4 lysine 20 by Suv4-20. J Biol Chem. 2008;283:12085–12092. doi: 10.1074/jbc.M707974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YL, Chou RH, Wu CH, Wang YN, Chang WJ, Tseng YJ, Chang WC, Lai CC, Lee HJ, Huo L, et al. Nuclear EGFR suppresses ribonuclease activity of polynucleotide phosphorylase through DNAPK-mediated phosphorylation at serine 776. J Biol Chem. 2012;287:31015–31026. doi: 10.1074/jbc.M112.358077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Chen CF, Chen PL, Lee WH. BRCA1 facilitates microhomology-mediated end joining of DNA double strand breaks. J Biol Chem. 2002;277:28641–28647. doi: 10.1074/jbc.M200748200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


