A selection approach is described that uses lentivirus vectors to express green fluorescent protein under the promoter region of FOXA2, which gives rise to dopamine neurons during embryogenesis. Our results suggest that FOXA2 is not sufficient to induce a dopaminergic fate in this system but that a combined approach of promoter activation and lentivirus vector technology can be used to select a defined cell population from a variety of human pluripotent stem cell lines.
Keywords: Cell selection, Dopamine, Neurogenesis, Floor plate, FACS, Promoter
Abstract
Human embryonic and induced pluripotent stem cells are potential cell sources for regenerative approaches in Parkinson disease. Inductive differentiation protocols can generate midbrain dopamine neurons but result in heterogeneous cell mixtures. Therefore, selection strategies are necessary to obtain uniform dopamine cell populations. Here, we developed a selection approach using lentivirus vectors to express green fluorescent protein under the promoter region of FOXA2, a transcription factor that is expressed in the floor plate domain that gives rise to dopamine neurons during embryogenesis. We first validated the specificity of the vectors in human cell lines against a promoterless construct. We then selected FOXA2-positive neural progenitors from several human pluripotent stem cell lines, which demonstrated a gene expression profile typical for the ventral domain of the midbrain and floor plate, but failed to enrich for dopamine neurons. To investigate whether this was due to the selection approach, we overexpressed FOXA2 in neural progenitors derived from human pluripotent stem cell lines. FOXA2 forced expression resulted in an increased expression of floor plate but not mature neuronal markers. Furthermore, selection of the FOXA2 overexpressing fraction also failed to enrich for dopamine neurons. Collectively, our results suggest that FOXA2 is not sufficient to induce a dopaminergic fate in this system. On the other hand, our study demonstrates that a combined approach of promoter activation and lentivirus vector technology can be used as a versatile tool for the selection of a defined cell population from a variety of human pluripotent stem cell lines.
Introduction
Parkinson disease is a common neurodegenerative disorder characterized by selective degeneration of the midbrain dopamine (DA) neurons of the substantia nigra (SN) that project to the dorsal striatum and regulate motor programs. Cell replacement to restore nigro-striatal DA levels has been successfully achieved using fetal midbrain transplants in patients [1–4] with remarkable long-term benefit [5]. Notwithstanding, both ethical and practical issues limit the clinical application of fetal cells. Additionally, fetal grafts are heterogeneous, and the presence of non-DA cells in the transplants has been related to motor complications such as graft-induced dyskinesias [6, 7]. Upon striatal transplantation, DA neurons derived from pluripotent or reprogrammed cells can restore DA levels, modulate DA storage and synaptic release, and correct motor deficits in animal models [8–10]. However, the successful application of pluripotent stem cells (PSCs) in regenerative medicine relies on our ability to implement efficient differentiation paradigms to obtain the desired cells at high purity and exclude contaminant populations, which can pose a risk or diminish efficacy. Optimized inductive protocols combining morphogens and small molecules and improved culture conditions have been developed to better reproduce the in vivo conditions during DA neurogenesis [11–13]. However, inductive protocols habitually produce a mixed variety of cell types, and selection procedures have been assayed to enrich for defined phenotypes [14–16]. Regarding DA neurons, most selection approaches have been based on transgenic PSC lines that express fluorescent markers controlled by tissue specific transcription factors or on cells stably transformed to overexpress key DA factors [17–21]. These approaches are useful, but their application is limited to the transgenic cell line. An alternative would be to use cell-surface markers. Nevertheless, despite their indisputable interest, there is limited knowledge about surface markers for human DA neurons, in addition to generic neuronal codes that have been used to exclude nonneuronal cells [22, 23]. Here, we devised a strategy based on lentiviral-mediated expression of a reporter gene under the control of the FOXA2 (Forkhead box A2, HNF3β, or TCF-3B) specific promoter. Our approach could provide the necessary flexibility to select target cells derived from any PSC line, which would be convenient for the expanding number of available human induced pluripotent stem cells (iPSCs).
During early embryogenesis, FOXA2 is transiently expressed in midline structures such as the node and notochord and, importantly for this study, at the base of the folding neural plate, where it is required for floor plate development [24, 25]. FOXA2 is induced by Sonic hedgehog (SHH), and FOXA2-positive neuroepithelial cells in the floor plate reciprocally express high levels of SHH [26–28]. FOXA2 plays both SHH-dependent and independent roles in floor plate specification [29]. At the midbrain level, both FOXA2 and FOXA1 have been implicated in the generation of DA neurons of the SN and ventral tegmental area [30–34], as well as other (non-DA) neurons that contribute to the red nucleus and the oculomotor complex [35, 36]. Given the apparent role of FOXA2 in floor plate specification, in midbrain neurogenesis, and in particular in DA neurogenesis [30, 37], we chose the FOXA2 promoter to drive the expression of green fluorescent protein (GFP) in a lentiviral construct, as a flexible system to select floor plate and ventral midbrain progenitors derived from any human PSC line.
Materials and Methods
Human ES Cell Propagation
Human embryonic stem cells (hESCs) lines: H9 (WA-09, passages 51–80), ES3 (CMR[B], passages 37–54), and ES5 (CMR[B], passages 27–55) and a human iPS cell line derived from a healthy individual in our laboratory, iPSC C1 (BP-143) (supplemental online Fig. 1) (passages 7–36), were maintained on irradiated human foreskin fibroblasts (HFF1, SCRC-1041; American Type Culture Collection [ATCC], Manassas, VA, http://www.atcc.org) in human embryonic stem (hES) medium composed of knockout Dulbecco’s modified Eagle’s medium (DMEM) (10829-018; Invitrogen, Carlsbad, CA, http://www.invitrogen.com), supplemented with 2 mM l- GlutaMAX (35050038; Invitrogen), 50 μM β-mercaptoethanol (31350-010; Invitrogen), 1× nonessential amino acids (M7145; Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com), 20% knockout serum (KSR; Invitrogen), 50 U/ml penicillin, 50 μg/ml streptomycin, and 10 ng/ml of fibroblast growth factor (FGF) 2 (100-18B; PeproTech).
In Vitro DA Differentiation
DA differentiation of hES cells followed, with some modifications, the protocol described by Perrier et al. [38]. For neural induction, we combined the dual SMAD inhibition protocol [13] with other small molecules [12] and recombinant morphogens as follows. Two days after plating undifferentiated cells on MS5 stromal cells, hESC medium was supplemented with 500 ng/ml of recombinant human noggin (120-10C; PeproTech, Rocky Hill, NJ, http://www.peprotech.com) or 100 nM of 4-[6-(4-piperazin-1-yl phenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline hydrochloride (LDN193189 (130-096-226; Miltenyi Biotec, Bergisch Gladbach, Germany, http://www.miltenyibiotec.com) until day in vitro (DIV) 12. 4-[4-(1,3-Benzodioxol-5-yl)-5-pyridin-2-yl-1H-imidazol-2-yl]benzamide hydrate (SB)-431542 (1614; Tocris Bioscience, Bristol, U.K., http://www.tocris.com) at 10 μM, was supplemented from DIV2 to DIV6. Recombinant mouse SHH (C25II-N, 464-SH; R&D Systems Inc., Minneapolis, MN, http://www.rndsystems.com) at 250 ng/ml or the SHH agonists purmorphamine (2 μM, 130-095-560; Miltenyi Biotech) or N-methyl-N′-(3-pyridinylbenzyl)-N′-(3-chlorobenzo[b]thiophene-2-carbonyl)-1,4-diaminocyclohexane (SAG) (0.5 μM, 566660; Millipore, Billerica, MA, http://www.millipore.com) were applied from DIV2 to DIV12 in addition to 6-[[2-[[4-(2,4-dichlorophenyl)-5-(5-methyl-1H-imidazol-2-yl)-2-pyrimidinyl]amino]ethyl]amino]-3-pyridinecarbonitrile (CHIR)-99021 (0.5 μM, 13122; Cayman Chemicals, Ann Arbor, MI, http://www.caymanchem.com). From DIV12 to DIV14, hES medium was gradually shifted to N2 medium (DMEM/F-12 with 1× N2 supplement (17502048; Invitrogen). At this time, the SAG concentration was reduced to 100 nM. On days 12–14, neural rosettes were manually picked and passaged onto plates coated with 0.1% gelatin (GL, G1393; Sigma-Aldrich), 15 μg/ml polyornithine, and 1 μg/ml fibronectin and expanded at high cell densities (>300,000/cm2) in BASF medium (N2 medium supplemented with 20 ng/ml brain-derived neurotrophic factor [BDNF], 200 μM ascorbic acid (AA), 100 nM SAG, 100 ng/ml FGF8, and 10 ng/ml FGF2). Subsequent passages of neural progenitors were done using Accutase solution (SCR005; Millipore). For terminal differentiation, BASF was replaced with neurobasal medium containing 1× B27 supplement (17504-044; Invitrogen), 2 mM l-glutamine (G7513; Sigma-Aldrich), 20 ng/ml BDNF, 20 ng/ml glial cell-derived neurotrophic factor, 200 μM AA, 0.5 mM dibutyryl cAMP, and 1 ng/ml transforming growth factor βIII.
Viral Vectors
Viruses were produced in a dedicated core facility in Inbiomed. Cloning was performed using standard molecular biological techniques, and all constructs were confirmed by sequence analysis. The self-inactivating lentiviral vector pRRLSIN.cPPT.PGK-GFP.WPRE [39] was kindly provided by Dr. D. Trono (University of Geneva, Geneva, Switzerland). To drive GFP from the human FOXA2 promoter, the phosphoglycerate kinase gene promoter was excised using XhoI and AgeI and replaced with two different sequences of the FOXA2 promoter previously amplified by PCR. One construct contained a 400-base pair (bp) fragment located upstream of exon 1 (pFOXA2.GFP). A second construct contained 1,600 bp of the promoter region upstream of the ATG start codon in exon 2 and included the untranslated exon 1 and intron 1 sequences (pFOXA2.5′.GFP). Replication incompetent, vesicular stomatitis virus G-coated lentiviral particles were packaged in 293T cells (CRL-11268; ATCC). Viral supernatants were concentrated by the ViraBind Lentivirus Concentration and Purification Kit (VPK-090; Bionova Cientifica S.L., Madrid, Spain, http://www.bionova.es) or by ultracentrifugation (Optima L-100 XP Ultracentrifuge; Beckman Coulter, Fullerton, CA, http://www.beckmancoulter.com) and thereafter titrated with the Lenti-X qRT-PCR Titration Kit (631235; Clontech, Mountain View, CA, http://www.clontech.com) and/or by flow cytometry (FACSCalibur; Becton, Dickinson and Company, Franklin Lakes, NJ, http://www.bd.com). The vector pRRL.CMV.GFP was previously developed in the platform and was always produced in parallel with similar titers. The retroviral construct pMXs-FoxaPTVppEGFP was kindly provided by Dr. Alessandra Giorgetti. A control vector lacking a promoter sequence was generated as follows: the FOXA2 promoter was excised from the vector pRRLSIN.cPPT.pFOXA2.GFP.WPRE using XhoI and AgeI, and the linearized plasmid was subsequently purified. The large (Klenow) fragment of the DNA polymerase I was used to fill in the 5′ overhangs to form blunt ends, which were then religated. Lentiviral vectors were produced as described above.
Transduction Experiments
ATCC cell lines (HepG2-HB8065, P19; HEK-293T CRL-3216, P22) were transduced with the pFOXA2.GFP or pFOXA2.5′.GFP vectors. Attached cells were infected with viral supernatant at a multiplicity of infection (MOI) of 10 with 8 μg/ml hexadimethrine bromide (Polybrene) overnight (ON). Three days later, cells were trypsinized and analyzed for GFP expression by flow cytometry. Untransduced cells from each line were used as negative controls for GFP expression, and other cells were fixed to examine endogenous FOXA2 expression by immunofluorescence (IF).
Neural precursors derived from hESCs were transduced around DIV28 of differentiation, as stated in the results and figure legends for individual experiments. Transductions were done in attachment or in suspension using MOIs of ∼8–10 and 2 μg/ml of Polybrene. Concentrated viral supernatants were diluted in BASF medium and added to the cells ON. The next day the virus was removed, and cells were expanded in fresh medium.
Fluorescence-Activated Cell Sorting
Human neural progenitors were sorted for GFP following optimized fluorescence-activated cell sorting (FACS) conditions [22] with some modifications. Briefly, cells were incubated with 5 μM of the Rock inhibitor Y-27632 (1614; Tocris Bioscience) 30 minutes before harvesting, gently dissociated into a single-cell suspension with Accutase, resuspended directly in sorting buffer, and filtered through cell strainer caps (40 μm).
Cell sorting was performed in a FACSAria III cell sorter with the FACSDiva software (BD Biosciences, San Diego, CA, http://www.bdbiosciences.com). The population of interest was identified by forward- and side-scatter gating, using the viability markers, 7-aminoactinomycin D (A9400; Sigma-Aldrich) or To-Pro 3 (E3341F; Thermo Fisher Scientific, Waltham, MA, http://www.thermofisher.com). Using a 488-nm laser for excitation, eGFP was determined according to fluorescence intensity with a biparametric analysis in the fluorescein isothiocyanate and phycoerythrin channels (490-nm long-pass and 510/20-nm band-pass filters). Nontransduced cells at the same stage of differentiation were used as the eGFP-negative control. Cells were recovered in the corresponding expansion or differentiation medium supplemented with 10 ng/ml of FGF2 and 50% fetal bovine serum. The purity of all sorted eGFP fractions was determined by reanalysis using FACS. Cells were sorted using a purity mask excluding doublets and conflicts. A 100-μm nozzle, sheath pressure of 20–25 psi, and an acquisition rate of 1,000–2,000 events per second were used as the standard conditions [21, 40]. Further flow cytometric analysis was performed using FlowJo software (Tree Star, Ashland, OR, http://www.treestar.com). Cells to be further analyzed in vitro after FACS for eGFP expression were replated in droplets at high densities (>300,000/cm2) onto polyornithine/fibronectin-coated coverslips or on irradiated primary striatal rat astrocytes (R-CpAs-522; Lonza, Walkersville, MD, http://www.lonza.com) in expansion or differentiation medium supplemented with 10 ng/ml of FGF2.
Inmunofluorescence and Confocal Analysis
Cells and tissues were analyzed by IF staining as previously described [8] and examined using an LSM510 Meta confocal microscope equipped with ultraviolet, argon, and helium-neon lasers (Carl Zeiss, Jena, Germany, http://www.zeiss.com).
Briefly, cells on coverslips were fixed with 4% paraformaldehyde in 1× phosphate-buffered saline (PBS) for 20 minutes at room temperature (RT). Cells were then incubated in blocking buffer consisting of 10% normal donkey serum (017000121-10ml; Jackson Immunoresearch), 0.1% Triton X-100 in 1× PBS for 1 hour at RT, followed by incubation overnight with the primary antibodies at 4°C. Primary antibodies are listed in supplemental online Table 1. After washing in 1× PBS with 10% normal donkey serum, the coverslips were incubated in fluorescence-labeled Alexa Fluor secondary antibodies (at 1:500 dilution) for 1 hour at RT, washed, and counterstained with Hoechst 33258 (861405; Sigma-Aldrich, diluted 1/1,000 in H2O). The coverslips were finally washed in H2O and mounted onto slides with Vectashield mounting medium H-1000, Vector Laboratories (Burlingame, CA, https://www.vectorlabs.com/). On selected coverslips, the primary antibody was omitted to verify specificity of staining.
For identification of signal colocalization within a cell, optical thickness was kept to a minimum, and orthogonal reconstructions were obtained from optical stacks. Quantitative analysis was performed on randomly selected visual fields from at least two independent experiments. In each field, images were acquired at ×40 magnification, and cells were counted using ImageJ 1.42q (NIH, Bethesda, MD, http://rsb.info.nih.gov/ij, cell counter plugin). On average, 16 visual fields were acquired per coverslip, and a total of 1,000–5,000 cells were counted per experiment in a blinded manner by two investigators.
Reverse Transcription and Quantitative Real-Time Polymerase Chain Reaction
Total RNA from plated cells at different stages of the differentiation protocol was prepared using TRIzol reagent (15596-026; Invitrogen) followed by terminal purification with RNeasy Mini Kit (74104; Qiagen, Hilden, Germany, http://www.qiagen.com). Total RNA from sorted samples included TRIzol treatment followed by terminal purification with RNAqueous Micro Kit (AM1931; Ambion, Austin, TX, http://www.ambion.com). For quantitative real-time polymerase chain reaction (qPCR), up to 2 μg of total RNA was transcribed into cDNA using the High Capacity cDNA Reverse Transcription kit with RNAse Inhibitor (4374966; Applied Biosystems, Foster City, CA, http://www.appliedbiosystems.com) and random hexamer primers. Quantitative analysis of gene expression was done in the StepOne Plus Real-Time PCR System (Applied Biosystems). Primers are listed in supplemental online Table 2. Amplifications were performed in 20 μl containing 50 nM of each primer, 0.5× Power SYBR Green PCR Master Mix (436870; Applied Biosystems), and 10 ng of cDNA. Forty PCR cycles were performed with a temperature profile of 95°C for 30 seconds and annealing temperature of 60°C for 1 minute. The dissociation curve of each PCR product was determined to test the specificity of the fluorescent signals. RNA from mouse and human tissue was isolated and reverse transcribed, and standard curves with serial 10-fold cDNA dilutions were built to validate primers sets. The fluorescent signals from specific qPCR products were normalized against that of a housekeeping gene (glyceraldehyde 3-phosphate dehydrogenase [GAPDH] or eukaryotic translation elongation factor 1 alpha 1 [EEF1A1]), and the relative expression was calculated by the ΔΔCt method [41]. Two to three replicates were done for each sample. Primers for detection of reporter and viral constructs were previously published [42].
Western Blot
Protein extracts were prepared from the phenol:ethanol phase left over after RNA extraction with TRIzol according to the supplier’s guidelines (Life Technologies, Rockville, MD, http://www.lifetech.com). Ten percent SDS-PAGE and immunoblotting were carried out as previously described [43]. Primary antibodies are listed in supplemental online Table 1. Visualization of horseradish peroxidase-labeled proteins was performed using enzyme-linked chemifluorescence (Thermo Fisher Scientific) and quantified using ImageJ software.
Results
Expression of FOXA2 in Human Neurons Derived From Pluripotent Stem Cells
FOXA2 has been proposed to be a master regulator involved in the specification, maturation, and maintenance of midbrain DA neurons [30, 32, 44, 45]. We first confirmed the expression of FOXA2 in tyrosine hydroxylase (TH)pos cells during development in the mouse embryo (supplemental online Fig. 2A; supplemental online data), and in agreement with previous studies [8, 30, 45], we found that FOXA2 expression was maintained in THpos neurons in the adult rodent [8] (supplemental online Fig. 2B) and nonhuman primate SN (supplemental online Fig. 2C). In primary ventral midbrain cultures, FOXA2 was initially expressed in both THpos and THneg neurons but became progressively restricted to THpos cells (supplemental online Fig. 2D–2E), and at DIV20, 80% of FOXA2pos cells were THpos.
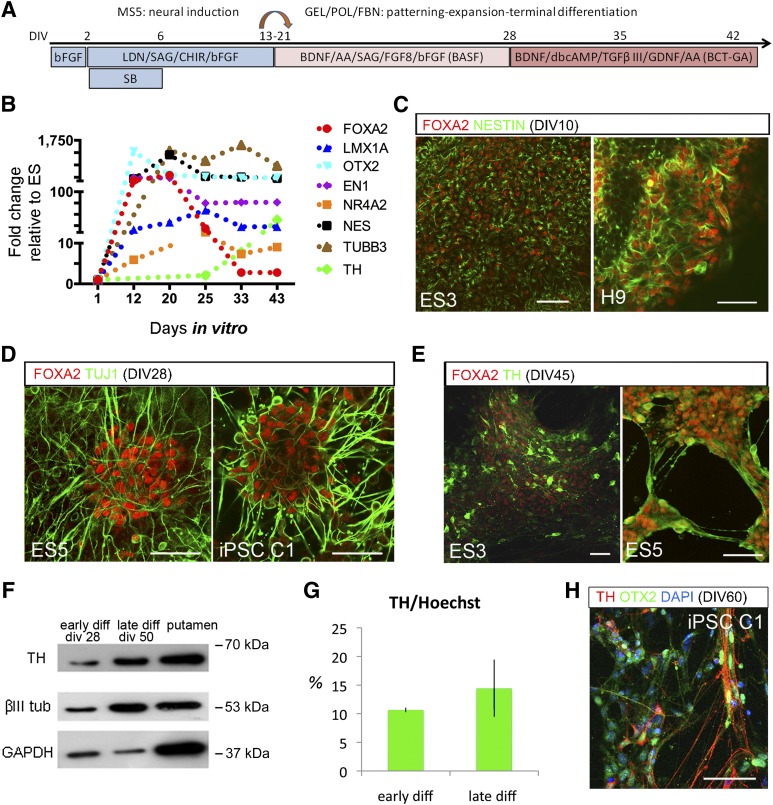
We next examined FOXA2 expression in human neurogenesis from PSCs. To differentiate human PSCs toward a midbrain floor plate phenotype, we followed an inductive protocol combining dual SMAD inhibition (Fig. 1A) [13] and glycogen synthase kinase3 β inhibition [12]. Despite the inherent transcriptional variability between human PSCs lines, there was an early and sustained expression of ventral and floor plate transcription factors, OTX2, FOXA2/1, EN1/2, and LMX1A (Fig. 1B). In parallel, there was a strong induction of nestin followed by the neuron-specific βIII tubulin and, later on, by TH. Transcripts for OCT4 were undetectable after in vitro day 10, and other pluripotency genes and mesoderm and endoderm markers were below detection threshold (data not shown). Using IF, we corroborated the expression of FOXA2 in nestinpos progenitors, in βIII tubulin (TuJ1)pos neuroblasts, and, upon differentiation, in THpos neurons (Fig. 1C–1E). βIII tubulin and TH protein content, measured by Western blot, increased in late differentiation stages (Fig. 1F). The average percentage of THpos cells was 10.6% ± 0.41% (n = 4) (Fig. 1G) at 4 weeks (early differentiation) and ∼15% at later stages (>6 weeks) (Fig. 1G). THpos cells expressed OTX2 and other midbrain regional transcription factors (Fig. 1H and supplemental online Fig. 1F).
Figure 1.
FOXA2 expression in midbrain dopamine neurons derived from human pluripotent cells. (A): DA differentiation paradigm for neural induction of human PSCs lines (human embryonic stem cells: H9, ES3, ES5, and iPSC C1; see supplemental online Fig. 1). (B): Transcriptional profile by quantitative real-time polymerase chain reaction shows early induction of FOXA2 and the region-specific transcription factors En1 and OTX2. (C–E): Expression of FOXA2 in neural progenitors and neurons was corroborated by immunofluorescence in all PSCs lines by coexpression with Nestin, TuJ1, and TH at corresponding differentiation stages. (F): βIII tubulin and TH protein content increased in the cultures upon differentiation, shown in a representative Western Blot. (G): Quantification of the percentage of THpos cells in cultures at early (4 weeks) and late (>6 weeks) differentiation stages. (H): THpos cells also expressed other ventral markers like OTX2 (see also supplemental online Fig. 1) Scale bars = 100 μm. Abbreviations: AA, ascorbic acid; BDNF, brain-derived neurotrophic factor; bFGF, basic fibroblast growth factor; CHIR, 6-[[2-[[4-(2,4-dichlorophenyl)-5-(5-methyl-1H-imidazol-2-yl)-2-pyrimidinyl]amino]ethyl]amino]-3-pyridinecarbonitrile; DAPI, 4[prime],6-diamidino-2-phenylindole; diff, differentiation; div or DIV, day in vitro; FBN, fibronectin; FGF, fibroblast growth factor; GDNF, glial cell-derived neurotrophic factor; GEL, gelatin; iPSC, induced pluripotent stem cell; LDN, 4-[6-(4-Piperazin-1-ylphenyl)pyrazolo[1,5-a]pyrimidin-3-yl]quinoline hydrochloride; POL, polyornithine; SB, 4-[4-(1,3-benzodioxol-5-yl)-5-pyridin-2-yl-1H-imidazol-2-yl]benzamide hydrate; SAG, N-methyl-N′-(3-pyridinylbenzyl)-N′-(3-chlorobenzo[b]thiophene-2-carbonyl)-1,4-diaminocyclohexane; TGF, transforming growth factor; TH, tyrosine hydroxylase; tub, tubulin.
Design and Validation of Lentiviral Vectors
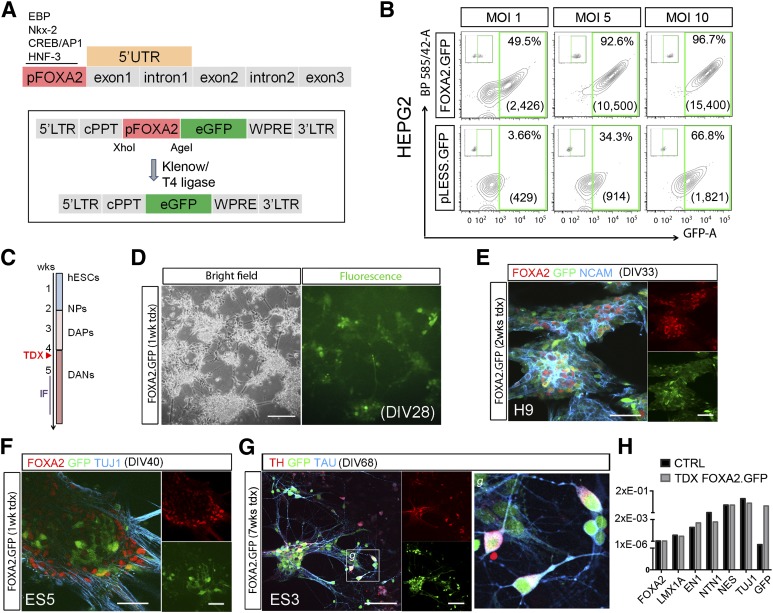
To drive gene expression from the FOXA2 promoter, we used a cloned 5′ fragment that contains the 400-bp putative promoter region [46] and generated lentiviral vectors expressing the GFP cDNA [39] controlled by this promoter sequence, pFOXA2.GFP (Fig. 2A). To validate the specificity of the FOXA2 promoter sequence, we excised this region from the pFOXA2.GFP construct and generated a promoterless vector (pLess.GFP) (Fig. 2A). We first tested the functionality and specificity of the vectors in HepG2 cells, a human hepatocarcinoma cell line that expresses high levels of FOXA2 [47], and HEK-293T cells, as a negative control (Fig. 2B and supplemental online Fig. 3A). We transduced HepG2 cells with either pFOXA2.GFP or the pLess.GFP vector at different MOIs and measured GFP expression 1 week later by flow cytometry. At an MOI of 1, less than 4% of cells transduced with the promotor-less virus were GFPpos, in contrast to 50% GFPpos cells in the pFOXA2.GFP transductions (Fig. 2B). At higher MOIs (5 and 10), we observed an increase in the fraction of GFPpos cells with the promotor-less virus, although the mean fluorescence intensity remained low (8%–12% of pFOXA2.GFP). GFPpos cells were observed in pFOXA2.GFP-transduced HEK-293T cells (22%; supplemental online Fig. 3A), but importantly, the mean fluorescence intensity was very low (<10% of HepG2). This can be explained by a low expression of FOXA2 in these cells that is detectable at the protein level in the cytoplasm by IF and Western blot (WB) (supplemental online Fig. 3B, 3C). The GFP signal was very low in HEK-293T cells transduced with the promoterless virus (<5% at MOI 10; supplemental online Fig. 3A), further supporting the idea that the signal observed in these cells with the pFOXA2.GFP is specific and corresponds to the low level of transcript present in these cells (supplemental online Fig. 3D). Collectively, these data demonstrated that GFP expression from the pFOXA2.GFP construct is specific, even if at high MOIs there can be some ectopic reporter gene expression. Transduction with this vector did not modify the levels of FOXA2 RNA transcript (supplemental online Fig. 3E). Inclusion of the 5′-untranslated region comprising exon 1 and intron 1, in another vector, pFOXA2.5′.GFP, increased the percentage and intensity (more than twofold) of GFP signal in HepG2 cells but also in the 293T cells (supplemental online Fig. 4), making this vector unsuitable to select FOXA2pos cells.
Figure 2.
Validation of lentiviral vector. (A): Representation of the human FOXA2 gene with the promoter region (in red) and the 5′-untranslated region (in orange) used to regulate the expression of GFP (see also supplemental online Fig. 4) and later removed to generate a promoterless vector (pLESS.GFP). (B): Quantification of GFPpos cells (%) and mean fluorescence intensity by flow cytometry, 3 days after transduction of HepG2 cells, a human cell line with robust constitutive expression of FOXA2. Excision of the promoter sequence from the vector demonstrates the specificity of the construct, at low MOI; at high MOIs, it showed leakage with increase in the percentage of GFPpos cells, although the mean intensity was always very low. (C, D): Transduction of human neural progenitors was carried out at approximately DIV28 with sustained expression of GFP in the neural cultures. Shown are representative microphotographs of live human embryonic stem cell-derived neurons 1 week after lentiviral transduction. (E, F): IF and confocal analysis confirmed that GFP expression was restricted to FOXA2pos cells and was colocalized with neuronal markers like NCAM and βIII tubulin. (G): GFPpos neurons expressed Tau and TH and displayed mature features with axonal localization of Tau. Inset (g) shows a higher magnification of the boxed area in (G). (H): Transduction with this vector did not modify the endogenous levels of FOXA2 transcript or the expression of other relevant genes by quantitative real-time polymerase chain reaction, 1 week post-transduction. Scale bars = 50 μm in (E) and (F) and 100 μm in (D) and (G). Abbreviations: cPPT, central polypurine tract; CTRL, control; DANs, dopamine neurons; DAPs, dopamine progenitors; DIV, day in vitro; eGFP, enhanced green fluorescent protein; GFP, green fluorescent protein; hESC, human embryonic stem cell; IF, immunofluorescence; LTR, long terminal repeat; MOI, multiplicity of infection; NCAM, neural cell adhesion molecule; NPs, neural progenitors; tdx, transduction; UTR, untranslated region; wk, week; WPRE, Woodchuck hepatitis virus Postranscriptional Regulatory Element.
We next examined the expression in PSCs-derived neural precursors transduced at DIV28–DIV34 with the pFOXA2.GFP lentiviral vector (Fig. 2C, 2D) or with a CMV.GFP control virus, in which GFP expression was ubiquitously and constitutively expressed, to control for transduction efficiency (∼60%) (data not shown). pFOXA2.GFP transduced cells expressed GFP, FOXA2, and the neuronal markers neural cell adhesion molecule (NCAM) and βIII tubulin (Fig. 2E, 2F), and Tau and TH at later stages (Fig. 2G). Transduction with this vector did not alter the level of expression of FOXA2 as determined by qPCR (Fig. 2H). These results demonstrated that the pFOXA2.GFP reporter construct recapitulated FOXA2 expression as in neural development and labeled stem cell-derived neuronal cells.
Selection of Human PSC-Derived FOXA2pos Cells Based on Endogenous Expression Using Lenti pFOXA2.GFP
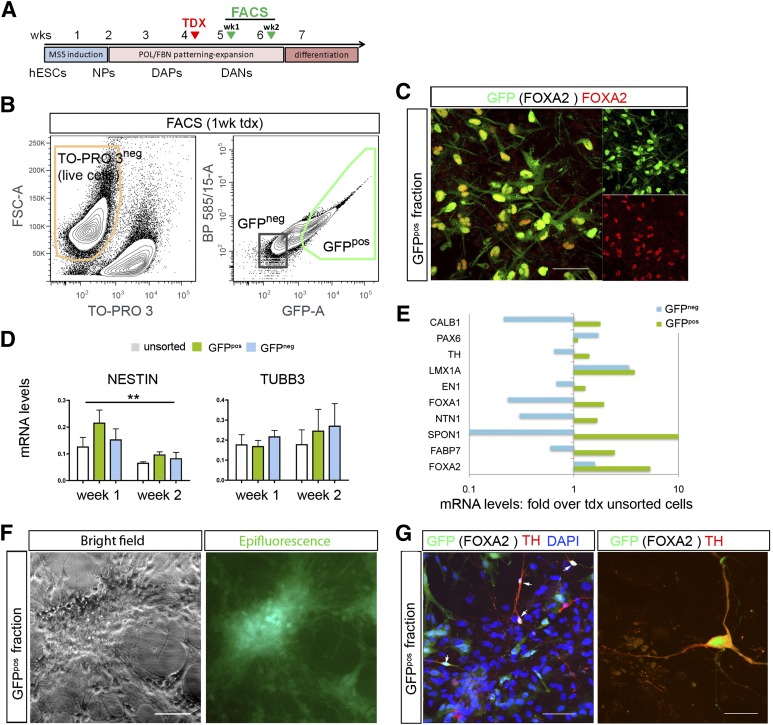
Stem cell-derived cultures were sorted at different time points during differentiation (from 4 days to 6 weeks post-transduction, n = 17 independent experiments) (Fig. 3A). The detailed gating procedures followed to exclude debris, dead cells, and doublets are provided in supplemental online Figure 5. The percentage of GFPpos cells in all experiments was remarkably stable, with an average of 27.3 ± 2.7% (Fig. 3B). Because transduction efficiency was ∼60%, this would roughly correspond to ∼45% FOXA2 cells in the culture at the time of transduction. There was good colocalization of GFP and FOXA2 after sorting (Fig. 3C), further corroborating the specificity of the vector. Most experiments (n = 11) were performed either at 1 week or at 2 weeks post-transduction. At 2 weeks, there was a clear trend toward a decrease in nestin and an increase in βIII tubulin mRNA content in all the fractions, reflecting a progressive maturation of the cells at these stages. The change in nestin content was significant (F = 10.667; p < .005), with no significant differences between the fractions (Fig. 3D). Next we analyzed the relative expression of several floor plate and DA genes in the sorted populations (Fig. 3E; values are plotted in a log scale for clarity). Taking together all experiments, there was a significant difference in the expression of these markers between the GFPpos fraction and the unsorted cells (two-tailed Wilcoxon signed rank test, p < .0002), whereas the GFPneg population was not different from the unsorted cells (p = .087). In addition to the enrichment for FOXA2 (5.4-fold), in the GFPpos fraction, there was a relative enrichment for LMX1A (3.8-fold), FOXA1 (∼2-fold), FABP7 (also known as BLBP) (2.5-fold), and f-spondin (SPON1) (19-fold) consistent with floor plate fates. On the other hand, there was little effect on TH (1.4-fold) and EN1 (1.3-fold). Replated cells from the positive fraction were later analyzed by IF. Survival was improved by plating the cells on irradiated astrocytes [21] (Fig. 3F), but only a few GFPpos cells corresponded to THpos neurons (Fig. 3G).
Figure 3.
Selection of human dopamine progenitors transduced with the lentiviral vector pFOXA2.GFP. (A): Schematic panel showing the protocol of human pluripotent stem cell differentiation with cells transduced at week 4 and selected by FACS 1 or 2 weeks later. (B): Representative gating strategy for isolation of viable GFPpos cells (shown is a selection of H9-derived cells, 1 week after transduction) (see supplemental online Fig. 5 for methodological details and postsort analysis). (C): Postsorting immunofluorescence (IF) confirmed a good overlap of GFP and FOXA2 (∼90%). (D): Analysis of nestin and βIII tubulin at 1 and 2 weeks in the unsorted, GFPpos, and GFPneg fractions by quantitative real-time polymerase chain reaction immediately after FACS. Shown are pooled data from 11 independent experiments. There was a significant decrease in nestin in the 2 weeks (p < .01). (E): Transcriptional profiles for floor-plate, DA, neural, and neuronal genes in the FACS cellular fractions showed enrichment for midbrain floor plate transcripts in the GFPpos fraction; data are shown here as fold change relative to unsorted cells for the corresponding transduction experiments, in a log scale for clarity. (F): Isolated GFPpos fractions could be maintained in vitro when plated at high cell densities in coculture with rat striatal astrocytes. (G): Analysis by IF of the GFPpos cells plated on astrocytes (GFP negative nuclei) showed few THpos cells that were GFPpos (arrows) at 2 weeks postsorting. Scale bars = 50 μm in (C), (F), and left panel in (G) and 20 μm for right panel in (G). Abbreviations: DANs, dopamine neurons; DAPs, dopamine progenitors; DAPI, 4[prime],6-diamidino-2-phenylindole; FACS, fluorescence-activated cell sorting; FSC, forward scatter; GFP, green fluorescent protein; hESC, human embryonic stem cell; NPs, neural progenitors; POL, polyornithine; tdx or TDX, transduction; TH, tyrosine hydroxylase; wk, week.
Collectively, these results showed that the pFOXA2.GFP vector can be used to select human ES-derived neural progenitors and enrich for floor plate cells. However, the selection of FoxA2pos cells did not enrich for DA progenitors or postmitotic DA neurons after differentiation.
Overexpression of FOXA2 to Promote DA Neurogenesis
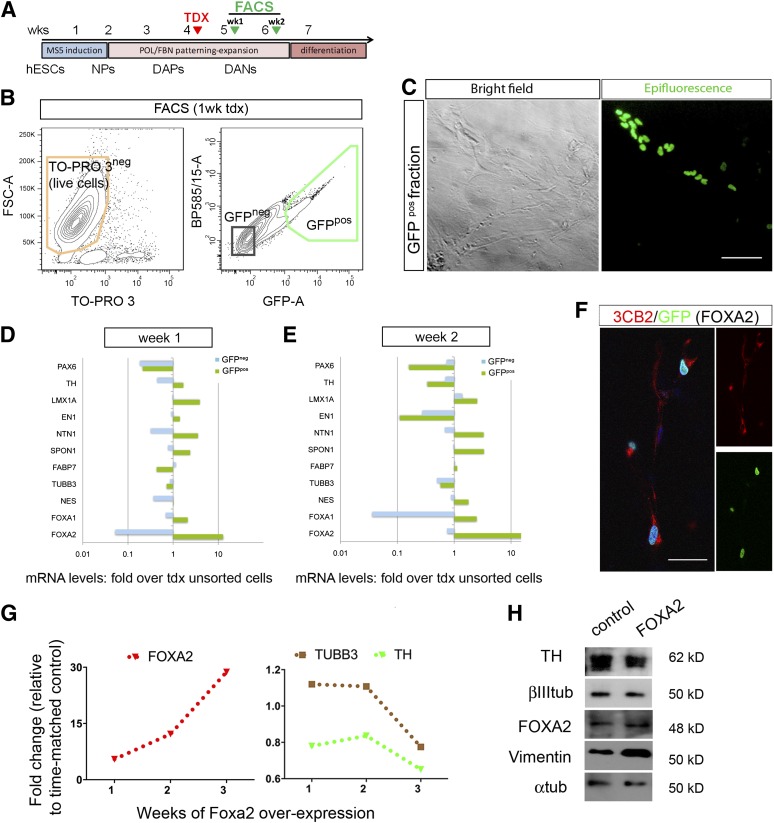
To exclude technical reasons for the lack of DA enrichment using the putative promoter of FOXA2, we used a retroviral vector to overexpress FOXA2 in the cells at similar differentiation times (Fig. 4A; supplemental online Fig. 6A). Retroviral transduction was less efficient; we obtained an average of 6.7 ± 0.34% of GFPpos cells in the sorting experiments (n = 6) (Fig. 4B, 4C). Despite the low number of transduced cells by IF, we found a significant increase in the number of FOXA2pos cells: 27.8% at 3 weeks compared with 7% FoxA2pos cells in untransduced cultures at the same time of differentiation (supplemental online Fig. 6B and data not shown). We sorted the cells at 1 or 2 weeks after transduction. Multivariate analyses with FOXA2 as covariate (F(1,1)= 509; p < .05) showed a significant effect of the sorting (F(2,1)= 746; p < .05). Gene expression profiles of the selected cell fractions showed that FOXA2 expression was increased in the GFPpos cell fraction, by 12-fold at 1 week and 40-fold at 2 weeks. There was also an increase of typical floor plate transcripts similar to what was found using the FOXA2 promoter construct (Fig. 3E). At 2 weeks there was an increase in netrin (3.3-fold), SPON1 (3.3-fold), FOXA1 (2.5-fold), and LMX1A (2.5-fold) in the positive fraction, and a relative decrease in TH, TUBB3, PAX6, and EN1 in the sorted fractions (Fig. 4E). At 2 weeks, IF showed few neurons in the positive fraction, and most GFP cells colocalized with the radial glial marker 3CB2 (Fig. 4F). Indeed, in transduced unsorted cultures there was a relative decrease in TH and TUBB3 at 3 weeks compared with untransduced controls (Fig. 4G). A similar trend was observed in the protein content by WB analysis with a relative increase in vimentin (3CB2; Figure 4H).
Figure 4.
Selection of human neural progenitors that overexpress FOXA2. (A): Cells were differentiated, transduced, and selected as in the promoter experiments shown in Fig. 3. (B): GFPpos cells overexpressing FOXA2 were selected by FACS 1 or 2 weeks after transduction. The image shows a representative gating strategy for isolation of viable GFPpos cells 1 week after transduction (exclusion of doublets is not shown for clarity; see Materials and Methods and supplemental online Fig. 5). (C): Representative microphotographs of neural progenitors transduced at DIV33. (D, E): Quantitative real-time polymerase chain reaction (qPCR) transcriptional profiles for midbrain and floor plate genes are shown for cultures sorted at 1 and 2 weeks. Fold increase over unsorted cells revealed a remarkable increase in the GFPpos fraction for FOXA2 as expected and for other midbrain floor plate markers, in particular at 2 weeks with decreases in TH and PAX6 among others. Data are shown as fold change over unsorted controls in a log scale for clarity. (F): Most GFPpos cells coexpressed the radial glial marker 3CB2, shown at 10 days post sorting. (G): qPCR analysis showed a persistent increase of FOXA2 and a concomitant decline in TH and TUBB3 in transduced unsorted cells compared with untransduced control cultures. (H): A similar trend was observed at the protein level in unsorted cultures at 2 weeks post-transduction, with an increase in vimentin by Western blot. Scale bars = 100 μm in (C) and 50 μm in (F). Abbreviations: DANs, dopamine neurons; DAPs, dopamine progenitors; FACS, fluorescence-activated cell sorting; FSC, forward scatter; GFP, green fluorescent protein; hESC, human embryonic stem cell; NPs, neural progenitors; POL, polyornithine; tdx or TDX, transduction; TH, tyrosine hydroxylase; tub, tubulin; wk, week.
Finally, to rule out the possibility of a delayed effect of FOXA2 forced expression on DA neurogenesis, we followed another set of transduced (unsorted) cells for up to 8 weeks (supplemental online Fig. 6). The results of these experiments both at the RNA and protein levels (supplemental online Fig. 6B, 6C) failed to demonstrate any effect on DA neurogenesis (or indeed on neurogenesis at all, because there was no evidence of enrichment for serotonergic, red nucleus, or cranial motor neurons) with persistent expression of radial glia and floor plate markers in these cultures.
Taken together, these overexpression experiments showed that FOXA2 is not sufficient to enhance DA neurogenesis from human PSCs in this system.
Discussion
Cellular heterogeneity is a problem for the development of cellular therapies because it decreases safety, efficacy, and efficiency. Homogeneous populations are also desirable for in vitro disease modeling to facilitate the identification of disease phenotypes [48]. Here, we developed a selection approach using lentivirus vectors to express GFP under the promoter region of FOXA2, a transcription factor that is expressed in the floor plate domain that gives rise to DA neurons during embryogenesis. Previous reports have shown that FOXA2 is involved in the specification, maturation, and maintenance of DA neurons [30, 37, 45, 49, 50]. Deletion of FOXA1/2 in postmitotic DA neurons results in loss of phenotype, at least in part through reduced binding of NURR1 to the promoter region of TH [45]. Two other independent studies have also shown that FOXA2 functions cooperatively with LMX1a to promote DA neuron development [49, 50]. Moreover, characterization of the FOXA2 cis-regulatory network by chromatin immunoprecipitation has shown its ability to regulate the transcription of most determinants of ventral midbrain progenitors contributing to the DA neuronal fate, including LMX1A, LMX1B, MSX1, and FERD3L, while repressing components of the SHH signaling pathway like Patched-1, GLI 1-3, NKX2.2, and NKX2.9 [32]. Thus, it appears indisputable that FOXA2 plays a permissive role in patterning of the midbrain floor plate and that FOXA2 floor plate cells give rise to DA progenitors. However, our data strongly suggest that FOXA2 is not sufficient for the production of human midbrain DA neurons.
The profile that we observed in our selected cultures is compatible with the midbrain floor plate. The floor plate extends from the preoptic area caudally into the midbrain, hindbrain, and spinal cord and has different properties along the anteroposterior axis [36]. Anterior floor plate cells express SIX6 and NOV, which were not enriched in our paradigm (data not shown), most probably because we used FGF8 in our inductive protocol. It has been shown that anterior floor plate cells derived from hES cells can be caudalized by the addition of retinoic acid, WNT1, or FGF8 [51]. f-Spondin is an extracellular matrix protein involved in axon guidance that, like netrin-1, is secreted by the midbrain floor plate. FOXA2 also plays a role in the regulation of axonal guidance across the floor plate [32]. The regional midbrain identity of the floor plate cells that we selected for, using the FOXA2 constructs, should have the capacity to generate neuroblasts and give rise to DA neurons [30, 37, 52–54]. However, we failed to enrich for DA neurons using both the endogenous promoter and the forced expression approaches. One explanation is that FOXA2 can play a repressor role on ventro-lateral midbrain genes [32] that may be important for the progression toward postmitotic phenotypes. This can also explain the relatively low efficiency of our protocol, compared with other protocols that are similar but use a high dose of SHH for shorter periods [10, 11]. In addition, lineage-tracing studies [55] suggest that SHH signaling is not directly involved in DA development and specification, which would be under the control of the canonical WNT pathway [56]. The neuroepithelial midline cells express FOXA2 and SHH that are reciprocally induced. Floor plate cells are responsive to SHH, i.e., GLI2a induces GLI1 in response to SHH and is inhibited when the cells start expressing SHH themselves. Although it has been proposed that the two pathways, WNT and SHH, cooperate at different stages of specification and differentiation [45, 49], it is plausible that we overestimated the role of SHH-FOXA2 in DA neurogenesis. WNT1 is expressed in a restricted ventral domain in the midbrain floor plate that is sufficient to antagonize SHH, allowing neurogenesis, and specifically the production of DA neurons. On the other hand, FOXA2 expression is robust in adult SN neurons and collaborates with NURR1 to regulate the expression of TH and AADC at the promoter level [45]. Thus, FOXA2 may be useful for selection at later stages and could be used to retrospectively identify specific cell-surface markers for enrichment without genetic manipulation. A recent study [57] used early selection of cells expressing CORIN, a floor plate surface marker, toward enrichment in FOXA2/LMX1A DA progenitors from human iPSC. Altogether, our selection experiments, using both endogenous and forced FOXA2 expression, did not increase the proportion of DA neurons derived from human PSC lines. These results suggest that FOXA2 is not sufficient to induce the DA fate, in line with the hypothesis that DA neurogenesis is under the direct control of canonical WNT and not SHH signaling.
On a technical level, lentivirus promoter-based cell selection is a versatile tool applicable to a broad number of human PSCs lines and could be important not only for direct therapeutic applications but also for drug screening and disease models, to study cell autonomous and nonautonomous mechanisms. As our study shows, it is critical to verify appropriate expression from the promoter sequence in the specific cell context, taking into account ectopic expression of the reporter gene and possible requirements for tissue-specific enhancers. As for FACS sorting, survival of cells grown in attachment and with complex morphology such as neurons is poor. It is important to consider that some cell types (or cells at certain maturation stages) are more resilient, which could result in their relative enrichment in both positive and negative fractions. Likewise, more vulnerable postmitotic neurons may decrease in both fractions relative to the unsorted cells. Despite these limitations, we show that it is feasible to use this approach for selection of defined human populations derived from any PSC line. Finally, it may be worth considering selection strategies not only aiming to obtain a uniform homogeneous population but to establish a “structured” contextual heterogeneity in the culture (in lieu of the random heterogeneity generated in vitro) by using regional domain determinants. In this sense, a selection approach based on expression of an early multilineage regional determinant, such as FOXA2 for the ventral floor plate, could provide this kind of contextual regional heterogeneity to favor survival and functional maturation of the target cells.
Supplementary Material
Acknowledgments
Funding for this project was provided by the Harvard Stem Cell Institute, by the Spanish Government (Grant SAF2008-04615) and the Basque Government Department of Industry (Grant SAIOTEK PE10IB04) and Education (EC2011-47) to R.S.P., by the Swedish Medical Research Council (2011-2651) to E.H., and in part by the National Institute of Neurological Disorders and Stroke R21 NS067335 to K.C.S. J.C.A. was supported by a MINECO FPI fellowship (BES-2009-028305). M.G. was supported by a CONACYT fellowship (CVU 357631, Mexico). We thank Drs. Xonia Carvajal, Kausalia Vijayaragavan, and Marc Bosse for helpful discussion and Drs. Alessandra Giorgetti and Julio Castaño for the retroviral vectors and experimental advice. Several antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by the University of Iowa Department of Biology (Iowa City, IA). J.C.A. is currently affiliated with the Department of Neuroscience, Karolinska Institutet Stockholm, Sweden.
Author Contributions
J.C.A.: collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; A.B. and M.P.L.M.: collection and/or assembly of data, data analysis and interpretation, final approval of manuscript; J.v.A.: conception and design, manuscript writing, final approval of manuscript; A.S., N.V., A.A., M.G., and R.L.d.M.: collection and/or assembly of data, final approval of manuscript; E.H.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing, final approval of manuscript; K.-C.S.: conception and design, data analysis and interpretation, manuscript writing, final approval of manuscript, virus production; R.S.-P.: conception and design, data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Lindvall O, Björklund A. Cell therapeutics in Parkinson’s disease. Neurotherapeutics. 2011;8:539–548. doi: 10.1007/s13311-011-0069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brundin P, Barker RA, Parmar M. Neural grafting in Parkinson’s disease Problems and possibilities. Prog Brain Res. 2010;184:265–294. doi: 10.1016/S0079-6123(10)84014-2. [DOI] [PubMed] [Google Scholar]
- 3.Björklund A, Dunnett SB, Brundin P, et al. Neural transplantation for the treatment of Parkinson’s disease. Lancet Neurol. 2003;2:437–445. doi: 10.1016/s1474-4422(03)00442-3. [DOI] [PubMed] [Google Scholar]
- 4.Mendez I, Sanchez-Pernaute R, Cooper O, et al. Cell type analysis of functional fetal dopamine cell suspension transplants in the striatum and substantia nigra of patients with Parkinson’s disease. Brain. 2005;128:1498–1510. doi: 10.1093/brain/awh510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kefalopoulou Z, Politis M, Piccini P et al. Long-term clinical outcome of fetal cell transplantation for Parkinson disease: Two case reports. JAMA Neurol 2014;71:83–87. [DOI] [PMC free article] [PubMed]
- 6.Politis M, Wu K, Loane C, et al. Serotonergic neurons mediate dyskinesia side effects in Parkinson’s patients with neural transplants. Sci Transl Med. 2010;2:38ra46. doi: 10.1126/scitranslmed.3000976. [DOI] [PubMed] [Google Scholar]
- 7.Barker RA, Kuan WL. Graft-induced dyskinesias in Parkinson’s disease: What is it all about? Cell Stem Cell. 2010;7:148–149. doi: 10.1016/j.stem.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez-Pernaute R, Lee H, Patterson M, et al. Parthenogenetic dopamine neurons from primate embryonic stem cells restore function in experimental Parkinson’s disease. Brain. 2008;131:2127–2139. doi: 10.1093/brain/awn144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wernig M, Zhao JP, Pruszak J, et al. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson’s disease. Proc Natl Acad Sci USA. 2008;105:5856–5861. doi: 10.1073/pnas.0801677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kriks S, Shim JW, Piao J, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xi J, Liu Y, Liu H, et al. Specification of midbrain dopamine neurons from primate pluripotent stem cells. Stem Cells. 2012;30:1655–1663. doi: 10.1002/stem.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li W, Sun W, Zhang Y, et al. Rapid induction and long-term self-renewal of primitive neural precursors from human embryonic stem cells by small molecule inhibitors. Proc Natl Acad Sci USA. 2011;108:8299–8304. doi: 10.1073/pnas.1014041108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambers SM, Fasano CA, Papapetrou EP, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hedlund E, Perlmann T. Neuronal cell replacement in Parkinson’s disease. J Intern Med. 2009;266:358–371. doi: 10.1111/j.1365-2796.2009.02155.x. [DOI] [PubMed] [Google Scholar]
- 15.Sonntag KC, Simunovic F, Sanchez-Pernaute R. Stem cells and cell replacement therapy for Parkinson’s disease. J Neural Transm Suppl. 2009;73:287–299. doi: 10.1007/978-3-211-92660-4_24. [DOI] [PubMed] [Google Scholar]
- 16.Aguila JC, Hedlund E, Sanchez-Pernaute R.Cellular programming and reprogramming: Sculpting cell fate for the production of dopamine neurons for cell therapy. Stem Cells Int 2012;2012:412040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganat YM, Calder EL, Kriks S, et al. Identification of embryonic stem cell-derived midbrain dopaminergic neurons for engraftment. J Clin Invest. 2012;122:2928–2939. doi: 10.1172/JCI58767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung S, Moon JI, Leung A, et al. ES cell-derived renewable and functional midbrain dopaminergic progenitors. Proc Natl Acad Sci USA. 2011;108:9703–9708. doi: 10.1073/pnas.1016443108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung S, Shin BS, Hedlund E, et al. Genetic selection of sox1GFP-expressing neural precursors removes residual tumorigenic pluripotent stem cells and attenuates tumor formation after transplantation. J Neurochem. 2006;97:1467–1480. doi: 10.1111/j.1471-4159.2006.03841.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung S, Sonntag KC, Andersson T, et al. Genetic engineering of mouse embryonic stem cells by Nurr1 enhances differentiation and maturation into dopaminergic neurons. Eur J Neurosci. 2002;16:1829–1838. doi: 10.1046/j.1460-9568.2002.02255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hedlund E, Pruszak J, Lardaro T, et al. Embryonic stem cell-derived Pitx3-enhanced green fluorescent protein midbrain dopamine neurons survive enrichment by fluorescence-activated cell sorting and function in an animal model of Parkinson’s disease. Stem Cells. 2008;26:1526–1536. doi: 10.1634/stemcells.2007-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruszak J, Sonntag KC, Aung MH, et al. Markers and methods for cell sorting of human embryonic stem cell-derived neural cell populations. Stem Cells. 2007;25:2257–2268. doi: 10.1634/stemcells.2006-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan SH, Martin J, Elia J, et al. Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PLoS One. 2011;6:e17540. doi: 10.1371/journal.pone.0017540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasaki H, Hogan BL. HNF-3 beta as a regulator of floor plate development. Cell. 1994;76:103–115. doi: 10.1016/0092-8674(94)90176-7. [DOI] [PubMed] [Google Scholar]
- 25.Monaghan AP, Kaestner KH, Grau E, et al. Postimplantation expression patterns indicate a role for the mouse forkhead/HNF-3 alpha, beta and gamma genes in determination of the definitive endoderm, chordamesoderm and neuroectoderm. Development. 1993;119:567–578. doi: 10.1242/dev.119.3.567. [DOI] [PubMed] [Google Scholar]
- 26.Ding Q, Motoyama J, Gasca S, et al. Diminished Sonic hedgehog signaling and lack of floor plate differentiation in Gli2 mutant mice. Development. 1998;125:2533–2543. doi: 10.1242/dev.125.14.2533. [DOI] [PubMed] [Google Scholar]
- 27.Lee J, Platt KA, Censullo P, et al. Gli1 is a target of Sonic hedgehog that induces ventral neural tube development. Development. 1997;124:2537–2552. doi: 10.1242/dev.124.13.2537. [DOI] [PubMed] [Google Scholar]
- 28.Sasaki H, Hui C, Nakafuku M, et al. A binding site for Gli proteins is essential for HNF-3beta floor plate enhancer activity in transgenics and can respond to Shh in vitro. Development. 1997;124:1313–1322. doi: 10.1242/dev.124.7.1313. [DOI] [PubMed] [Google Scholar]
- 29.Bayly RD, Brown CY, Agarwala S. A novel role for FOXA2 and SHH in organizing midbrain signaling centers. Dev Biol. 2012;369:32–42. doi: 10.1016/j.ydbio.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kittappa R, Chang WW, Awatramani RB, et al. The foxa2 gene controls the birth and spontaneous degeneration of dopamine neurons in old age. PLoS Biol. 2007;5:e325. doi: 10.1371/journal.pbio.0050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye W, Shimamura K, Rubenstein JL, et al. FGF and Shh signals control dopaminergic and serotonergic cell fate in the anterior neural plate. Cell. 1998;93:755–766. doi: 10.1016/s0092-8674(00)81437-3. [DOI] [PubMed] [Google Scholar]
- 32.Metzakopian E, Lin W, Salmon-Divon M, et al. Genome-wide characterization of Foxa2 targets reveals upregulation of floor plate genes and repression of ventrolateral genes in midbrain dopaminergic progenitors. Development. 2012;139:2625–2634. doi: 10.1242/dev.081034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hynes M, Porter JA, Chiang C, et al. Induction of midbrain dopaminergic neurons by Sonic hedgehog. Neuron. 1995;15:35–44. doi: 10.1016/0896-6273(95)90062-4. [DOI] [PubMed] [Google Scholar]
- 34.Hynes M, Poulsen K, Tessier-Lavigne M, et al. Control of neuronal diversity by the floor plate: Contact-mediated induction of midbrain dopaminergic neurons. Cell. 1995;80:95–101. doi: 10.1016/0092-8674(95)90454-9. [DOI] [PubMed] [Google Scholar]
- 35.Joksimovic M, Patel M, Taketo MM, et al. Ectopic Wnt/beta-catenin signaling induces neurogenesis in the spinal cord and hindbrain floor plate. PLoS One. 2012;7:e30266. doi: 10.1371/journal.pone.0030266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Placzek M, Briscoe J. The floor plate: Multiple cells, multiple signals. Nat Rev Neurosci. 2005;6:230–240. doi: 10.1038/nrn1628. [DOI] [PubMed] [Google Scholar]
- 37.Ferri AL, Lin W, Mavromatakis YE, et al. Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development. 2007;134:2761–2769. doi: 10.1242/dev.000141. [DOI] [PubMed] [Google Scholar]
- 38.Perrier AL, Tabar V, Barberi T, et al. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci USA. 2004;101:12543–12548. doi: 10.1073/pnas.0404700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zufferey R, Dull T, Mandel RJ, et al. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hedlund E, Pruszak J, Ferree A, et al. Selection of embryonic stem cell-derived enhanced green fluorescent protein-positive dopamine neurons using the tyrosine hydroxylase promoter is confounded by reporter gene expression in immature cell populations. Stem Cells. 2007;25:1126–1135. doi: 10.1634/stemcells.2006-0540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Geraerts M, Willems S, Baekelandt V, et al. Comparison of lentiviral vector titration methods. BMC Biotechnol. 2006;6:34. doi: 10.1186/1472-6750-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez de Maturana R, Aguila JC, Sousa A, et al. Leucine-rich repeat kinase 2 modulates cyclooxygenase 2 and the inflammatory response in idiopathic and genetic Parkinson’s disease. Neurobiol Aging. 2014;35:1116–1124. doi: 10.1016/j.neurobiolaging.2013.11.018. [DOI] [PubMed] [Google Scholar]
- 44.Ang SL. Foxa1 and Foxa2 transcription factors regulate differentiation of midbrain dopaminergic neurons. Adv Exp Med Biol. 2009;651:58–65. doi: 10.1007/978-1-4419-0322-8_5. [DOI] [PubMed] [Google Scholar]
- 45.Stott SR, Metzakopian E, Lin W, et al. Foxa1 and foxa2 are required for the maintenance of dopaminergic properties in ventral midbrain neurons at late embryonic stages. J Neurosci. 2013;33:8022–8034. doi: 10.1523/JNEUROSCI.4774-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Navas MA, Vaisse C, Boger S, et al. The human HNF-3 genes: Cloning, partial sequence and mutation screening in patients with impaired glucose homeostasis. Hum Hered. 2000;50:370–381. doi: 10.1159/000022943. [DOI] [PubMed] [Google Scholar]
- 47.Unterman TG, Fareeduddin A, Harris MA, et al. Hepatocyte nuclear factor-3 (HNF-3) binds to the insulin response sequence in the IGF binding protein-1 (IGFBP-1) promoter and enhances promoter function. Biochem Biophys Res Commun. 1994;203:1835–1841. doi: 10.1006/bbrc.1994.2401. [DOI] [PubMed] [Google Scholar]
- 48.Merkle FT, Eggan K. Modeling human disease with pluripotent stem cells: From genome association to function. Cell Stem Cell. 2013;12:656–668. doi: 10.1016/j.stem.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 49.Chung S, Leung A, Han BS, et al. Wnt1-lmx1a forms a novel autoregulatory loop and controls midbrain dopaminergic differentiation synergistically with the SHH-FoxA2 pathway. Cell Stem Cell. 2009;5:646–658. doi: 10.1016/j.stem.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin W, Metzakopian E, Mavromatakis YE, et al. Foxa1 and Foxa2 function both upstream of and cooperatively with Lmx1a and Lmx1b in a feedforward loop promoting mesodiencephalic dopaminergic neuron development. Dev Biol. 2009;333:386–396. doi: 10.1016/j.ydbio.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 51.Fasano CA, Chambers SM, Lee G, et al. Efficient derivation of functional floor plate tissue from human embryonic stem cells. Cell Stem Cell. 2010;6:336–347. doi: 10.1016/j.stem.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Nissim-Eliraz E, Zisman S, Schatz O, et al. Nato3 integrates with the Shh-Foxa2 transcriptional network regulating the differentiation of midbrain dopaminergic neurons. J Mol Neurosci. 2013;51:13–27. doi: 10.1007/s12031-012-9939-6. [DOI] [PubMed] [Google Scholar]
- 53.Norton WH, Mangoli M, Lele Z, et al. Monorail/Foxa2 regulates floorplate differentiation and specification of oligodendrocytes, serotonergic raphé neurones and cranial motoneurones. Development. 2005;132:645–658. doi: 10.1242/dev.01611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ono Y, Nakatani T, Sakamoto Y, et al. Differences in neurogenic potential in floor plate cells along an anteroposterior location: Midbrain dopaminergic neurons originate from mesencephalic floor plate cells. Development. 2007;134:3213–3225. doi: 10.1242/dev.02879. [DOI] [PubMed] [Google Scholar]
- 55.Zervas M, Millet S, Ahn S, et al. Cell behaviors and genetic lineages of the mesencephalon and rhombomere 1. Neuron. 2004;43:345–357. doi: 10.1016/j.neuron.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 56.Joksimovic M, Yun BA, Kittappa R, et al. Wnt antagonism of Shh facilitates midbrain floor plate neurogenesis. Nat Neurosci. 2009;12:125–131. doi: 10.1038/nn.2243. [DOI] [PubMed] [Google Scholar]
- 57.Doi D, Samata B, Katsukawa M, et al. Isolation of human induced pluripotent stem cell-derived dopaminergic progenitors by cell sorting for successful transplantation. Stem Cell Rev. 2014;2:337–350. doi: 10.1016/j.stemcr.2014.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.