This study characterizes the cellular niche required for the expansion of human spermatogonial stem cells (SSCs), provides strong evidence supporting specific extracellular markers that define a testicular cell population highly enriched for human SSCs, and characterizes a novel testicular multipotent stromal cell population essential for SSC expansion. The results of this study are very important to understanding how human SSCs can be grown and expanded successfully in vitro for future therapeutic applications.
Keywords: Cancer, Cell surface markers, Clinical translation, Mesenchymal stem cells, Spermatogonial stem cells
Abstract
Prepubertal boys treated with high-dose chemotherapy do not have an established means of fertility preservation because no established in vitro technique exists to expand and mature purified spermatogonial stem cells (SSCs) to functional sperm in humans. In this study, we define and characterize the unique testicular cellular niche required for SSC expansion using testicular tissues from men with normal spermatogenesis. Highly purified SSCs and testicular somatic cells were isolated by fluorescence-activated cell sorting using SSEA-4 and THY1 as markers of SSCs and somatic cells. Cells were cultured on various established niches to assess their role in SSC expansion in a defined somatic cellular niche. Of all the niches examined, cells in the SSEA-4 population exclusively bound to adult testicular stromal cells, established colonies, and expanded. Further characterization of these testicular stromal cells revealed distinct mesenchymal markers and the ability to undergo differentiation along the mesenchymal lineage, supporting a testicular multipotent stromal cell origin. In vitro human SSC expansion requires a unique niche provided exclusively by testicular multipotent stromal cells with mesenchymal properties. These findings provide an important foundation for developing methods of inducing SSC growth and maturation in prepubertal testicular tissue, essential to enabling fertility preservation for these boys.
Introduction
Approximately 7,500 boys are diagnosed with cancers annually in the United States [1]. Although patients have seen significant survival improvements with treatments [2], many suffer from permanent sterilization [3, 4]. For postpubertal boys and adults, fertility preservation with cryopreserved semen is highly successful [5–7]. Unfortunately, this option is not feasible for prepubertal boys.
Two approaches may potentially help these infertile patients become fathers after cancer treatment. Testicular tissue taken prior to chemotherapy could be differentiated into mature sperm. This approach, combined with in vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI), has been successful in a neonatal mouse model [8]. Alternatively, autologous spermatogonial stem cell (SSC) transplant has been shown to restore spermatogenesis, leading to healthy offspring [9–11] in many nonprimate models for more than 15 years [11–19] and, most recently, in primates [9]. However, neither approach has been attempted in humans.
Despite advances in fertility treatment that have led to the routine use of IVF/ICSI for men with ejaculated or surgically obtained sperm concentrations approaching zero, these techniques are not possible for prepubertal boys because they do not produce sperm. However, their testicles do contain SSCs [20–22] with the potential to expand and differentiate into mature sperm. Developing a technique to expand SSCs for autologous transplantation or to differentiate SSCs into mature sperm would be of tremendous value. Current primary challenges include the identification, isolation, and expansion of highly purified SSCs primarily because of a gap of knowledge in identifying human SSCs based on extracellular marker expression and understanding the cellular environment (“niche”) necessary for SSC growth and differentiation [23–27].
Expansion of purified mouse SSCs using unique membrane protein markers such as THY1, GFRα1, GPR125, and CD49f has been reported [23, 28–30]. Allogeneic transplantation of in vitro expanded mouse SSCs into germ cell-depleted testes resulted in restoration of fertility [31]. Recent studies propose that human SSCs may be identified based on expression of THY1, CD49f, EpCAM, and SSEA-4 [32–35]. Of these markers, only SSEA-4 is highly expressed in embryonic stem cells and in both human fetal and prepubertal SSCs, suggesting that it may be a good marker of adult human SSCs [20, 36]. Although transplantation of human testicular cells into germ cell-depleted mouse testes resulted in either limited colonization of human cells or cells expressing germ cell markers, spermatogenesis was not detected, presumably because of interspecies differences [33–35, 37, 38]. Interestingly, mouse and human SSCs have been reported to have the capability of converting into testis-derived pluripotent stem cells during in vitro culture [33, 34, 39–43]. However, in contrast to studies in mice, recent studies suggest that the human testis-derived pluripotent stem cells derived from in vitro culture of putative human SSCs are actually cells of mesenchymal rather than germ cell origin. Filling this gap in our understanding of human SSC biology is critical [24–27].
Because of the limited availability of human tissues, the lack of in vitro or in vivo xenograft models capable of supporting human spermatogenesis, and the significant ethical and logistical challenges associated with human germ cell research, current data on the identification, isolation, and expansion of human SSCs are mixed and highly controversial. To shed light on this controversy and lay the groundwork for a new therapy for young male patients facing sterilizing treatments, a detailed characterization of SSCs and the required somatic niche capable of supporting SSC expansion is required [44].
In this study, we characterize the cellular niche required for the expansion of human SSCs, provide strong evidence supporting specific extracellular markers that define a testicular cell population highly enriched for human SSCs, and characterize a novel testicular multipotent stromal cell (TMSC) population essential for SSC expansion. The results of this study are very important to understanding how human SSCs can be grown and expanded successfully in vitro for future therapeutic applications.
Materials and Methods
Subjects
Thirteen adult subjects with normal spermatogenesis were enrolled in the institutional review board-approved University of California San Francisco LIFE and Human SSC studies.
Fluorescence-Activated Cell Sorting
Biopsied tissues and digested cells were incubated with the following antibodies: anti-SSEA-4 fluorescein isothiocyanate (FITC), anti-THY1 allophycocyanin, anti-CD45 Pacific Blue, anti-CD105 FITC, and anti-CD73 phosphatidylethanolamine (PE) in 1% bovine serum albumin for 30 minutes at 37°C (all from BD Pharmingen, San Jose, CA, http://www.bdbiosciences.com).
Confocal Microscopy
Images were captured using a Leica SP5 AOBS confocal microscope and a Leica DMI 4000B fluorescent microscope (Leica Microsystems Inc., Bannockburn, IL, http://www.leica.com) and analyzed using ImageJ v1.6.
Molecular Analyses
Subpopulations of testicular cells were sorted directly into RNA buffer. Quantitative polymerase chain reaction (qPCR) amplification was carried out using FastStart Universal SYBR Green Master Mix with ROX (Roche, Mannheim, Germany, http://www.roche.com) using the 7500 PCR system (Applied Biosystems, Foster City, CA, http://www.appliedbiosystems.com) (supplemental online Table 7).
RNA-seq data were generated from fluorescence-activated cell sorting (FACS)-sorted human testicular THY1+ cells and SSEA-4+ cells obtained from three healthy individuals. Briefly, THY1+ and SSEA-4+ cells were sorted directly into RNA buffer. Purified RNA was analyzed for quality using chip-based capillary electrophoresis (Bioanalyzer; Agilent Technologies, Palo Alto, CA, http://www.agilent.com), and quantity and purity were determined with a NanoDrop spectrometer. RNA-seq libraries were prepared with an ovation RNA-seq system v2 kit (NuGEN, San Carlos, CA, http://www.nugen.com/nugen). In this method, the mRNA is reverse transcribed to synthesize the first-strand cDNA using a combination of random hexamers and a poly-T chimeric primer. The RNA template is then partially degraded by heating, and the second strand cDNA is synthesized using DNA polymerase. The double-stranded DNA is then amplified using single primer isothermal amplification (SPIA) [45, 46]. SPIA is a linear cDNA amplification process in which RNase H degrades RNA in DNA/RNA heteroduplex at the 5′-end of the double-stranded DNA, after which the SPIA primer binds to the cDNA and the polymerase starts replication at the 3′-end of the primer by displacement of the existing forward strand. Random hexamers are then used to amplify the second-strand cDNA linearly. Finally, libraries from the SPIA-amplified cDNA were made using the Ultralow DR library kit (NuGEN). The RNA-seq libraries were analyzed by Bioanalyzer and quantified by qPCR (KAPA, Wilmington, MA, http://www.kapabiosystems.com). High-throughput sequencing was done using a HiSEquation 2500 (Illumina Inc., San Diego, CA, http://www.illumina.com).
SSC Culture
Subpopulations of testicular cells were FACS-sorted and individually cultured in supplemented StemPro-34 (Invitrogen, Carlsbad, CA, http://www.invitrogen.com) [31, 34] with the following modifications: 1% penicillin-streptomycin, 1% insulin-transferrin-selenium (ITS) solution (Mediatech Inc., Manassas, VA, http://www.cellgro.com), recombinant human epidermal growth factor (20 ng/ml) (R&D Systems Inc., Minneapolis, MN, http://www.rndsystems.com), recombinant human glial cell line-derived neurotrophic factor (GDNF) (10 ng/ml) (R&D Systems), recombinant human leukemia inhibitory factor (LIF) (10 ng/ml) (Chemicon, Temecula, CA, http://www.chemicon.com) and 1% knockout serum replacement (KOSR). The medium was changed every 72 hours. Cells were cultured at 37°C in a humidified incubator with 5% CO2 and 20% O2 without stromal cells on either Matrigel (BD Biosciences, San Diego, CA, http://www.bdbiosciences.com)-coated dishes or 0.2% gelatin-coated dishes at a density of 50,000 cells per cm2 and on either mouse embryonic fibroblasts (MEFs), human fetal placental fibroblasts, human fetal testicular fibroblasts, or human adult testicular THY1+ cells at a density of 25,000 cells per cm2. Human fetal placental and testicular fibroblasts at 22 weeks of gestation were generated from discarded fetal tissues donated for research.
Results
Characterization of SSCs and TMSCs
Testicular THY1+ and SSEA-4+ cells represent distinct cell populations
Although controversial, both THY1 and SSEA-4 have been reported as putative markers of adult human testicular cells enriched for SSCs [20, 23, 33, 35]. However, although recent studies demonstrate that both THY1 and SSEA-4 are expressed in fetal gonocytes, only SSEA-4 continued to be expressed in prepubertal SSCs [20]. Thus, confocal microscopy was performed to detect the presence and precise location of cells expressing THY1, SSEA-4, and VASA. Two distinct populations of germ cells were observed by relative expression of VASA (Fig. 1A). VASA dim cells, located at the basement membrane, correlate anatomically with the known location of SSCs. In contrast, VASA bright cells, located predominantly away from the basement membrane toward the lumen, are consistent with differentiating spermatogonia, spermatocytes, and spermatids. SSEA-4 expression was primarily detected in VASA dim cells on the basement membrane, suggesting that it is a specific marker for SSCs (Fig. 1A). Additionally, evidence of meiosis, assessed by SYCP3 expression, was exclusively detected in VASA bright cells, suggesting that the SSEA-4/VASA dim population contains adult human SSCs (Fig. 1B). In contrast, THY1 expression was detected primarily on fibroblasts and myoid cells of the lamina propria with limited expression on peritubular cells (Fig. 1C). Additionally, the cell population located between the VASA dim and bright cells exclusively expressed both THY1 and WT1, consistent with Sertoli cells (Fig. 1C). All THY1+ cells expressed high levels of vimentin and did not express VASA, suggesting that they are likely of somatic and mesenchymal origin (Fig. 1D).
Figure 1.
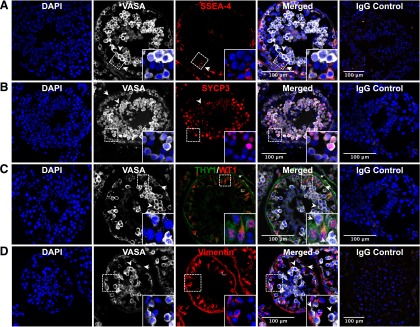
Confocal microscopy of seminiferous tubules. (A): VASA and SSEA-4 expression. Two populations of cells, VASA dim (arrow) and bright cells (arrowhead), were observed with VASA dim cells located exclusively on the basement membrane. Only VASA dim cells expressed SSEA-4. (B): VASA and SYCP3 expression. Only VASA bright cells (arrowhead) expressed SYCP3, whereas VASA dim cells (arrow) did not. (C): VASA, THY1, and WT1 expression. Sertoli cells reside in the region between VASA dim (arrow) and VASA bright (arrowhead). Nuclear WT1 expression (∗), specific to Sertoli cells, was readily detected in THY1+/VASA− cells. THY1 expression was also detected in cells making up the lamina propria. (D): VASA and vimentin expression. Vimentin expression is detected only in Sertoli cells and cells in the lamina propria. Abbreviation: DAPI, 4′,6-diamidino-2-phenylindole.
Seminiferous tubules were digested, and testicular cells were dual stained with THY1 and SSEA-4 antibodies for further characterization of the THY1+ and SSEA-4+ populations by FACS. Only live, CD45− testicular cells were gated for analyses with SYTOX/Pacific (CD45) Blue exclusion. Three distinct populations of testicular cells based on THY1 and SSEA-4 expression (THY1+, SSEA-4+, and THY1−/SSEA-4− populations) were observed (Fig. 2A). There were no cells that coexpressed both THY1 and SSEA-4 simultaneously. When THY1+, SSEA-4+, and THY1−/SSEA-4− populations were analyzed separately by backgating to examine their unique cellular characteristics, each demonstrated distinct forward and side scatter values, providing further confirmation that these three populations possessed different cellular physical properties (Fig. 2A). Immunofluorescent analyses of the sorted THY1+, SSEA-4+, and THY1−/SSEA-4− populations demonstrated the lack of SSEA-4, THY1, and either THY1/SSEA-4 expression in these populations, respectively.
Figure 2.
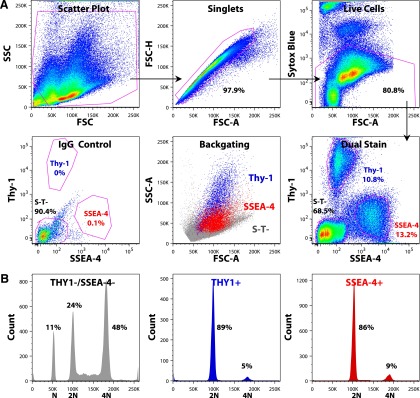
Fluorescence-activated cell sorting characterization of testicular THY1+ and SSEA-4+ cells. (A): Only single CD45− live cells were gated for analyses. Three distinct nonoverlapping populations (THY1+, SSEA-4+, and THY1−/SSEA-4−) were observed based on THY1 and SSEA-4 expression. Additionally, THY1+, SSEA-4+, and THY1−/SSEA-4− cells exhibited unique physical properties when they were gated individually for forward and side scatter analyses. (B): DNA content analyses of subpopulation of testicular cells. THY1−/SSEA-4− cells contain predominantly haploid (N) and tetraploid (4N) cells, whereas THY1+ and SSEA-4+ cells are predominantly 2N with small population of 4N. Abbreviations: FSC, forward scatter; SSC, side scatter.
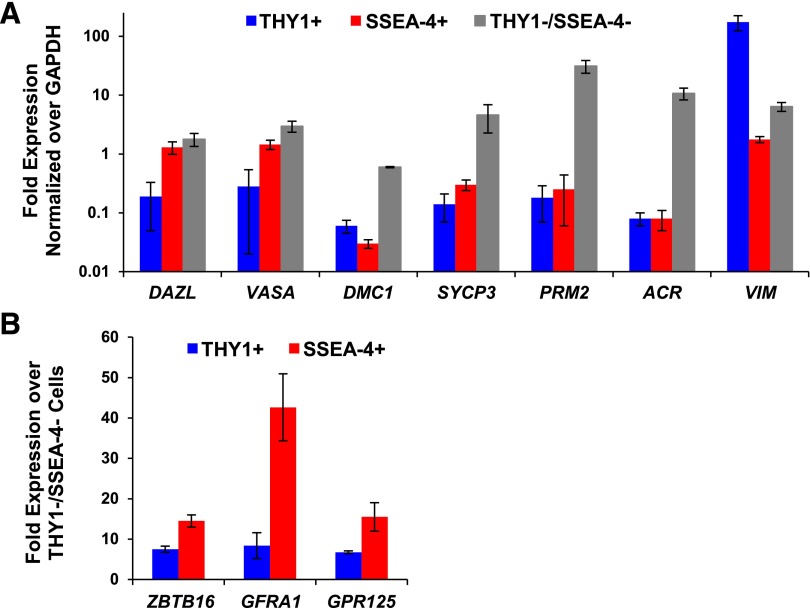
Sorted THY1+, SSEA-4+, and THY1−/SSEA-4− cells were subjected to DNA content and mRNA analyses. Significant amounts of haploid (N) and tetraploid (4N) cells, 11% and 48%, respectively, were identified exclusively in the THY1−/SSEA-4− population, suggesting that only the THY1−/SSEA-4− population contained differentiating germ cells (Fig. 2B). In addition to the lack of haploid cells, both THY1+ and SSEA-4+ populations contained a very small population of 4N cells, 5% and 9%, respectively, suggesting that they are mainly quiescent in normal homeostatic state (Fig. 2B). Although expression of DAZL and VASA were detected in the SSEA-4+ and THY1−/SSEA-4− cell populations, they were barely detectable in the THY1+ cells (Fig. 3A). Instead, THY1+ cells were found to express high levels of VIM, >98- and 27-fold more than SSEA-4+ and THY1−/SSEA-4− cells, respectively, suggesting a mesenchymal origin (Fig. 3A). Although both SSEA-4+ and THY1−/SSEA-4− populations expressed germ cell markers (DAZL and VASA), only the THY1−/SSEA-4− population contained haploid cells (Fig. 2B) and expressed high levels of both meiotic (DMC1, SYCP3) and spermatid markers (PRM2, ACR), demonstrating that SSEA-4+ population contains primitive spermatogonia that have not yet entered meiosis (Figs. 1A, 1B, 2B, 3A). As expected, very low levels of known mouse SSC markers, ZBTB16, GFRα1, and GPR125 were detected in the THY1−/SSEA-4− population. Although both THY1+ and SSEA-4+ populations expressed ZBTB16, GFRα1, and GPR125, the expression was significantly higher in the SSEA-4+ population (Fig. 3B), assessed by qPCR, and confirmed with FACS.
Figure 3.
Molecular characterization of testicular THY1+ and SSEA-4+ cells. (A): THY1+ cells expressed high levels of VIM but lack DAZL, VASA, DMC1, SYCP3, PRM2, and ACR. SSEA-4+ cells expressed high levels of DAZL and VASA with minimal expression of VIM and meiotic or spermatid markers. THY1−/SSEA-4− cells expressed high levels DAZL, VASA, DMC1, SYCP3, PRM2, and ACR. Expression levels between groups were significantly different for all genes examined. p < .05 by analysis of variance. (B): Significant differential expression of ZBTB16, GFRα-1, and GPR125 between THY1+ and SSEA-4+ populations. p < .05 by Student’s t test. Abbreviation: GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Characterization of the Niche Required for SSC Expansion
Testicular THY1+ Cells Are Critical for Successful SSC Expansion
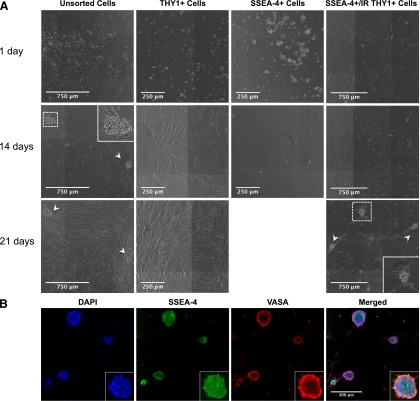
Unsorted, sorted THY1+, and sorted SSEA-4+ cells were subjected to in vitro expansion and monitored with time-lapse photography (supplemental online Videos 1–4). Unsorted testicular cells cultured on either uncoated or coated plates revealed two populations. The first adhered to the plates and exhibited fibroblast-like morphology within 48 hours. The second population of small round cells bound to these fibroblast-like adherent cells shortly after 48 hours, divided, and formed colonies after 2 weeks of culture (Fig. 4A). However, colonies began to disappear after 3 weeks of culture because the adherent cells became confluent (supplemental online Video 1). Although ∼98% of these in vitro expanded unsorted testicular cells expressed THY1, evaluated by FACS, after 3 weeks of culture, neither SSEA-4 nor VASA expression was detected by FACS, microscopy, or qPCR. Cell passage after 2 weeks of culture did not rescue expansion of SSC colonies because the adherent cells quickly grew to confluence, suggesting a preferential selection of THY1+ cells in this culture system.
Figure 4.
Human SSC colonies establishment. (A): Unsorted testicular cells formed colonies but disappeared after 21 days (arrowheads). THY1+ cells quickly bound to the culture dish and exhibited fibroblast like morphology without forming colonies. SSEA-4+ cells bound together in duplets or short chains but did not adhere to culture dish or form colonies. When SSEA-4+ cells were plated in culture dish with irradiated THY1+ cells, they bound to THY1+ cells and form colonies (arrowheads). (B): Confocal microscopy of SSC colonies for SSEA-4 and VASA expression. Expanded SSC colonies continue to coexpress both SSEA-4 and VASA. Abbreviation: DAPI, 4′,6-diamidino-2-phenylindole.
When plated on culture dishes uncoated or coated with either Matrigel or gelatin, THY1+ cells adhered to all plates within 24 hours, exhibited fibroblast morphology shortly after, and continued to expand without signs of quiescence (>20 passages) (Fig. 4A; supplemental online Video 2). Although DAZL and VASA were never detected by qPCR or confocal microscopy, this population continued to express high levels of THY1 and vimentin, assessed by immunofluorescent analyses. In contrast, SSEA-4+ (Fig. 4A) and THY1−/SSEA-4− cells did not adhere or form colonies when cultured on uncoated or coated plates, failed to expand, and died within 2 weeks of culture. Furthermore, immunofluorescent analyses did not detect any evidence of THY1 and vimentin expression in these two populations.
To overcome the rapid expansion of THY1+ cells in this system, sorted THY1+ cells were expanded and subjected to γ-irradiation to render them mitotically inactive. Sorted SSEA-4+ cells were then cocultured on the irradiated adherent THY1+ cells. SSEA-4+ cells bound to these adherent cells within 24 hours, formed SSC colonies (∼50 cells per colony) within 2 weeks, and continued to expand (Fig. 4A; supplemental online Video 3). The percentage of SSC colonies formed to SSEA-4+ cells plated ranged between 0.02% and 0.1% with an 8–12-fold increase in colony number and cell number (50–100 cells per colony) after each subsequent passage. These expanded colonies continued to express SSEA-4 and VASA with serial passaging (Fig. 4B; supplemental online Video 4). In contrast, THY1−/SSEA-4− cells failed to establish colonies when plated on irradiated THY1+ cells. Additionally, THY1+, SSEA-4+, and THY1−/SSEA-4− cells failed to establish colonies when cultured in the presence of MEFs, human placental, or fetal testicular stroma. Thus, adult testicular THY1+ cells uniquely provide the essential niche required for SSC expansion. Using this novel system, SSC colonies were successfully identified, isolated, passaged, and expanded in vitro.
Testicular THY1+ Cells Demonstrated Mesenchymal Properties
In addition to their lack of germ cell properties, sorted THY1+ cells immediately adhered to plastic and exhibited fibroblastic morphology, suggesting a mesenchymal origin. To investigate whether the THY1+ population exhibited mesenchymal characteristics, THY1+ cells were analyzed for coexpression of CD73 and CD105. 92% of THY1+ cells coexpressed both CD73 and CD105 (Fig. 5A). Of note, CD45+ cells were excluded from analyses in the SYTOX Blue gating using anti-CD45 conjugated with Pacific Blue. When THY1+ cells were expanded in vitro, they continued to coexpress both CD73 and CD105 (Fig. 5B).
Figure 5.
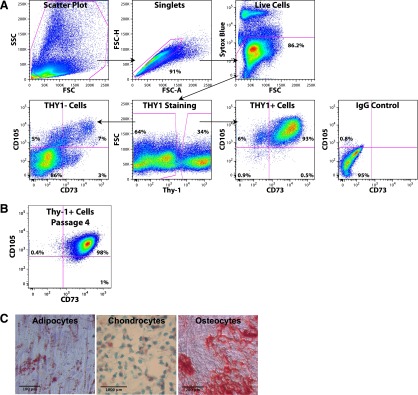
Testicular THY1+ cells exhibited mesenchymal properties. (A): Fluorescence-activated cell sorting analyses of testicular cells for THY1/CD73/CD105 expression. Only single CD45− live cells were gated for analyses. Cells were first evaluated for THY1 expression. THY1+ and THY1− cells were than gated for CD73 and CD105 expression. >92% of THY1+ cells coexpressed both CD73 and CD105, whereas only ∼7% of THY1− cells expressed both CD73 and CD105. (B): In vitro expanded THY1+ cells continue to express both CD73 and CD105. Shown here is passage 4. (C): Sorted THY1+ cells were subjected to differentiation along the mesenchymal lineage. Adipocytes, chondrocytes, and osteocytes were detected by histologic staining. Scale bars = 100 μm. Abbreviations: FSC, forward scatter; SSC, side scatter.
Upon differentiation, sorted THY1+/CD73+/CD105+ cells gave rise to adipocytes, chondrocytes, and osteocytes, further confirming the presence of mesenchymal properties within this population (Fig. 5C). Similarly, whereas VIM was highly expressed in the THY1+/CD73+/CD105+ population, neither DAZL nor VASA was detected in this population.
Comprehensive Molecular Characterization of the Testicular THY1+ and SSEA-4+ Cells by Next Generation Sequencing
Testicular SSEA-4+ Cells Expressed Genes Previously Identified as Enriched in Human and Mouse SSCs
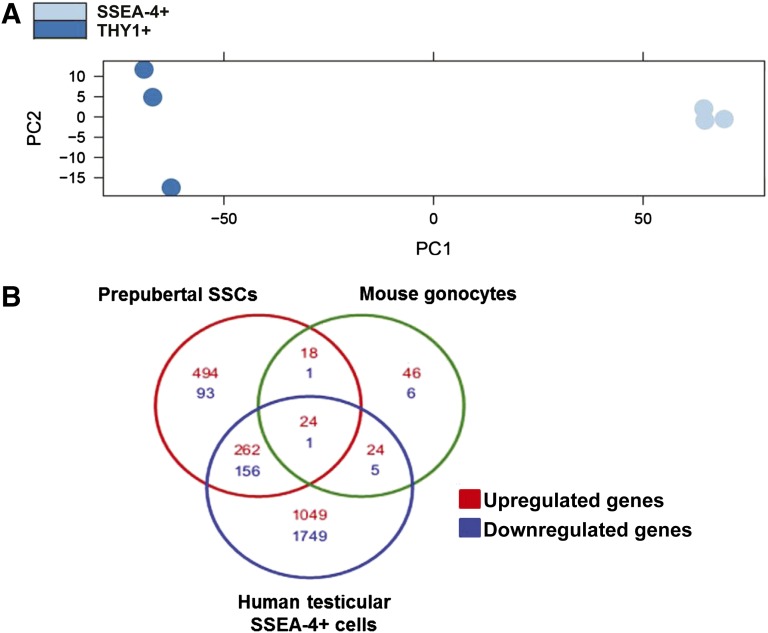
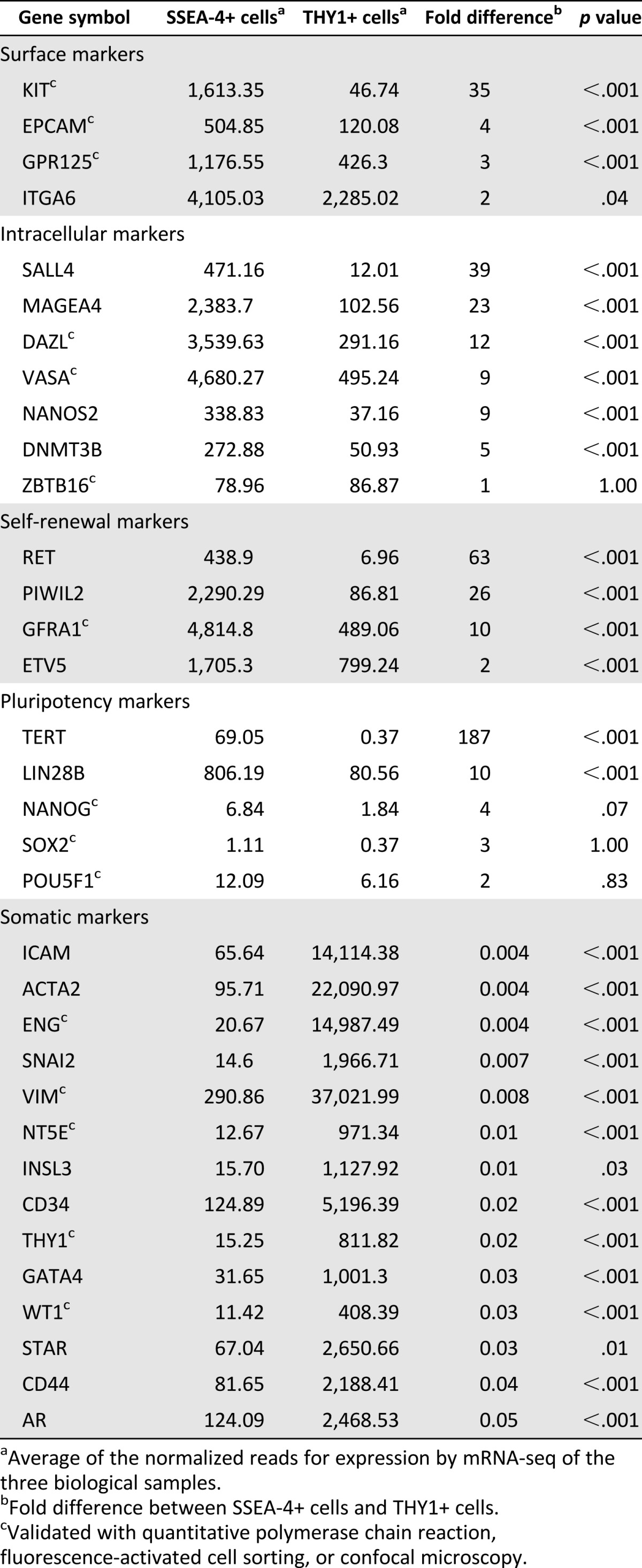
FACS-sorted THY1+ and SSEA-4+ cells were collected from three patients and subjected to mRNA sequencing. On average, there were 13,568,327 and 8,822,058 total reads from the THY1+ and SSEA-4+ populations, respectively. The principal component analysis is shown in Figure 6A. As demonstrated, the THY1+ and SSEA-4+ populations are significantly distinct from each other. Although the SSEA-4+ population clusters tightly, the THY1+ population shows more variability, reflecting this population’s innate heterogeneity. When log2 ± twofold change with a p value of <0.05 was used as the cutoff to define significant differential gene expression, there were 1,359 known upregulated and 1,911 downregulated genes in the SSEA-4+ population in comparison with the THY1+ population. Specifically, 29 genes were upregulated, and 232 were downregulated >100-fold (supplemental online Tables 1, 2). Genes previously reported to be enriched in human and mouse SSCs were examined. Table 1 demonstrates the enriched expression of known SSC genes in testicular SSEA-4+ cells. When further evaluated by FACS, EPCAM, GPR125, and ITGA6/CD49f were found to express in both THY1+ and SSEA-4+ populations (Table 1). However, qPCR confirmed the higher expression of GPR125 in the SSEA-4+ population (Fig. 3B). Although KIT was differentially expressed in the SSEA-4+ population, C-Kit was not detected by confocal microscopy or FACS. Known intracellular markers of SSCs were highly enriched in the SSEA-4+ cells. Although ZBTB16 was not found to be enriched with mRNA-seq, qPCR data demonstrated that it was significantly enriched (1.9-fold) in the SSEA-4+ population (Fig. 3B). Additionally, known genes (RET, GFRα1, and ETV5) in the GDNF-mediated SSC self-renewal pathway in rodent were also highly enriched in SSEA-4+ cells (Table 1; Fig. 3B) [47]. Furthermore, known pluripotency markers such as TERT and LIN28B were highly expressed in the SSEA-4+ cells, further suggesting that this population contains human SSCs. As expected and confirmed with qPCR, NANOG, SOX2, and POU5F1 were not expressed in any significant amount in either populations. In contrast, known somatic genes were highly expressed in the THY1+ population (Table 1). The diverse families of somatic genes expressed in the THY1+ cells confirmed that this is a heterogeneous population. Overall, the testicular SSEA-4+ and THY1+ mRNA transcriptome profiles confirmed that the SSEA-4+ population contains primitive spermatogonia in contrast to the profile of the THY1+ population that favors a somatic origin.
Figure 6.
mRNA profiles of testicular THY1+ and SSEA-4+ cells. (A): PCA analysis of fluorescence-activated cell sorting-sorted THY1+ and SSEA-4+ cell transcriptomes obtained by mRNA sequencing. (B): Venn diagram depicts overlapping differentially expressed genes between human testicular SSEA-4+/THY1+ cells, human prepubertal SSCs/somatic cells, and mouse gonocytes/somatic cells at log2 ± twofold change with an adjusted p value of <0.05. Human prepubertal SSCs and mouse SSCs profiles were previously published by Wu et al. [22]. Abbreviation: SSCs, spermatogonial stem cells.
Table 1.
Expression of genes in human testicular SSEA-4+ and THY1+ cells previously identified as enriched in either human prepubertal or mouse SSCs or somatic cells
Testicular SSEA-4+ Cells Expressed Genes Previously Identified as Enriched in Human Prepubertal SSCs and Mouse Gonocytes
Because the results thus far support the hypothesis that testicular SSEA-4+ population contains adult human SSCs, it is important to compare this population with known pure populations of human or mouse SSCs. Hence, the transcriptome of the testicular SSEA-4+ cells reported here was compared with previously published human prepubertal SSC transcriptome because current information on adult human SSCs is limited and controversial. When log2 ± twofold-change with a p value of <0.05 was used to define significant differential gene expression, there were 798 genes that were upregulated and 251 that were downregulated in the human prepubertal SSCs in comparison with their respected somatic population with the top 50 up- and downregulated genes shown in supplemental online Table 3. In contrast, 112 genes were upregulated and 13 were downregulated in the mouse gonocytes in comparison with their somatic populations (supplemental online Table 4). There were 24 common upregulated genes found between the three groups (Fig. 6B; supplemental online Table 5). Mouse gonocytes shared 35% (44 of 125) of their differentially expressed genes with human prepubertal SSCs (Fig. 6B). In contrast, human prepubertal SSCs and mouse gonocytes shared 42% (443 of 1,049) and 43% (54 of 125) of their differentially expressed genes, respectively, with SSEA-4+ cells (Fig. 6B). Alternatively, when only the top 50 upregulated genes from the human prepubertal SSCs and mouse gonocytes were evaluated, 68% (34 of 50) and 54% (27 of 50) of the differentially expressed genes, respectively, were in common with differentially upregulated genes seen in human testicular SSEA-4+ cells. Thus, the similarity between the transcriptome profiles of human testicular SSEA-4+ cells, human prepubertal SSCs, and mouse gonocytes further demonstrates that human adult SSCs are within the testicular SSEA-4+ population.
Discussion
Developing the ability to isolate SSCs and understanding the testicular niche optimal for SSC growth and development are required to develop effective therapeutic options for pediatric cancer patients facing sterilizing treatments. Using confocal microscopy, FACS-sorted subpopulations of testicular cells, time-lapse photography, comprehensive mRNA sequencing, and a novel in vitro culture system, this study identifies a subpopulation of adult human testicular cells highly enriched for SSCs and the support cells critical to their growth based on distinct extracellular markers SSEA-4 and THY1, respectively (supplemental online Table 6). These insights provide valuable information for the development of future treatments to preserve and restore fertility.
Both testicular THY1+ and SSEA-4+ cells have been reported to contain SSCs based on mouse and human studies [23, 33, 35]. Specifically, transplantation of both enriched human testicular THY1+ [33] and SSEA-4+ [35] cells into mouse testes resulted in germ cell colony formation. These findings are controversial, however, because in vitro culture of human unsorted testicular cells and enriched THY1+ cells with the current system did not select for long-term SSC expansion [24–27]. Rather, recent human studies suggest that the in vitro systems selected for cells of mesenchymal origin [24–27]. Our data demonstrated that SSC colonies disappeared as THY1+ cells expanded in culture over time. Using confocal microscopy, we demonstrated that THY1+ cells were predominantly located in lamina propria with some expression on Sertoli cells. Sorted THY1+ cells did not express germ cell markers as evaluated by microscopy (VASA) or qPCR (DAZL and VASA), suggesting a somatic origin. In contrast, previous studies demonstrated that human THY1+ populations contain SSCs after mouse xenotransplantation [33]. It is possible that this observation was the result of germ cell contamination, given that an enriched rather than sorted cell population was analyzed. When sorted testicular THY1+ cells were used to establish mRNA profile by RNA sequencing, the THY1+ transcriptome was consistent with the many somatic cell types making up the seminiferous tubules. Additionally, the transcriptome profiles of the THY1+ and SSEA-4+ populations were quite distinct, as shown in the principal component analysis (PCA) (Fig. 6A). Further functional evidence that supports sorted testicular THY1+ cells as the population containing TMSCs includes rapid binding to plastic; expression of consensus mesenchymal markers (CD73 and 105); the ability to differentiate into adipocytes, chondrocytes, and osteocytes; and the absence of CD45 expression. Given the anatomic location of the THY1+ cells within the seminiferous tubules, the lack of germ cell markers, high expression of vimentin, and the ability to differentiate into all mesenchymal lineages, the TMSC (THY1+) population, demonstrated here, is of somatic origin and not germ cells.
SSEA-4 is a marker of undifferentiated pluripotent human embryonic stem cells, cleavage to blastocyst stage embryo, human fetal SSCs, and prepubertal SSCs [20, 36]. Anatomically, SSEA-4+ cells are located predominantly at the basement membrane, suggesting that they are SSCs. This is in contrast to previous studies that demonstrated limited coexpression of THY1 and SSEA-4 on subpopulations of human testicular cells using conventional microscopy [35]. However, these earlier studies did not use multicolor FACS or confocal microscopy analyses. In our studies, multicolor FACS, confocal microscopy, and transcriptomes on sorted cells confirmed that THY1+ and SSEA-4+ cells were two distinct populations with different physical, cellular, and molecular profiles. When the tubules were stained for VASA, VASA bright (SSEA-4−) cells contained haploid cells and located toward the lumen, whereas the VASA dim (SSEA-4+) cells were found at the basement membrane. This finding is consistent with previous human studies demonstrating that a high level of VASA expression was associated with maturing germ cells [48, 49]. In contrast to the absence of markers associated with meiosis (DMC1 and SYCP3) or differentiating spermatids (PRM2 and ACR), SSEA-4+ cells expressed high levels of putative SSC markers (ZBTB16, GFRα1, SALL4, MAGEA4, and GPR125), and pluripotency genes (TERT and LIN28B), consistent with primitive spermatogonia containing SSCs. Lower levels of ZBTB16, GFRα1, and GPR125 expression were also detected in THY1+ cells in comparison with SSEA-4+ cells in these studies. This is similar to previous human and mouse studies in which ZBTB16, GFRα1, and GPR125 are markers of SSCs, but low-level expression was also detected in testicular somatic cells [22]. Of note, the genes (GFRα1, RET, and ETV5) involved in the GDNF-mediated SSC self-renewal were also highly expressed in the SSEA-4+ cells [47]. These results support previous studies demonstrating germ cell colonization in mouse xenograft model following transplantation of primary SSEA-4+ sorted cells [35]. Interestingly, recent studies demonstrate that human bone marrow-derived mesenchymal stem cells (BmMSCs) also express SSEA-4 but lack CD45 expression [50]. Although CD45+ cells were excluded from any FACS analyses, we cannot eliminate the possibility of BmMSC contamination. Perhaps the small number of BmMSCs contributed to a very low level of VIM expression detected in our SSEA-4+ population. However, sorted SSEA-4+ cells failed to survive in vitro in the absence of other cell types, suggesting that this possible contamination is very low at best.
The significant similarity in differentially expressed genes in human testicular SSEA-4+/THY1+ in comparison with human prepubertal SSCs/somatic cells and mouse gonocytes/somatic cells further solidify that human adult SSCs reside with the SSEA-4+ population [22]. Interestingly, of the 24 common upregulated genes between the 3 groups, 16 genes (ASF1B, ASPM, BUB1, CASC5, CENPA, CENPF, CENPO, EXO1, HELLS, KIF11, KNTC1, MCM8, RAD51AP1, RAD54B, STAG3, and TOP2A) are involved cell cycle, DNA replication, meiosis, and DNA repair regulations, as demonstrated in the interactive pathway analysis (supplemental online Fig. 1). Of the 24 common genes, 3 (DAZL, PIWIL4, and BNC1) are intricately involved with germ cell maintenance and fertility [51–53]. When the top 50 differentially upregulated genes from human prepubertal SSCs were compared with human testicular SSEA-4+ cells, 19 of the 34 commonly upregulated genes (STK31, MAGEA4, TPTE, VASA, TKTL1, MAEL, ELAVL2, LIN28B, TEX15, SNAP91, CENPE, SLC25A31, DPPA2, FGFR3, DPPA4, DAZL, CASC5, TOP2A, and CENPF) are also involved in pluripotency, cell cycle, meiosis, DNA repair, and germ cell regulations [54–63]. Of the 16 nonoverlapping genes, SOHLH2 and POLB were significantly upregulated in human SSEA-4+ population but at <log2 twofold change. Of the 14 remaining nonoverlapping genes, only 1 (CD109) has been well characterized [64]. CD109 is well characterized in the hematopoietic system, and its expression is likely due to contamination with the prepubertal SSC isolation because the SSCs were manually isolated [22]. In contrast, when the top 50 upregulated genes in the mouse gonocytes were compared with human testicular SSEA-4+ cells, 20 of the 27 commonly upregulated genes (EPCAM, BNC1, CENPF, CDCA7L, DAZL, TOP2A, STAG3, MCM8, BUB1, RAD54B, KNTC1, EXO1, CASC5, PIWIL4, CDCA5, SALL4, RAD51, ERCC6L, TPX2, and CDCA2) are involved in the pluripotency, cell cycle, meiosis, DNA repair, and germ cell regulations [65, 66]. Thus, these findings highlight the evolutionary conserved genes in SSCs that are essential for reproduction.
When sorted THY1+, SSEA-4+, and THY1−/SSEA-4− cells were cultured separately on either uncoated or coated plates, only THY1+ cells adhered and grew in a monolayer fashion regardless of surface substrate. In contrast to the lack of germ cell expression, THY1+ cells continued to express THY1+/CD73+/CD105+ and vimentin after more than 20 passages, further confirming a mesenchymal origin. When unsorted testicular cells were cultured, germ cells quickly bound to the adherent cells, formed colonies within 2 weeks of culture, and continued to express SSEA-4 and VASA. However, the adherent cells quickly outgrew and subsequently inhibited the SSC colonies growth by 4 weeks of culture. To delineate the relationship between SSC growth and its required niche, sorted SSEA-4+ cells were cultured in the presence of irradiated MEFs, human placental fibroblasts, human fetal testicular fibroblasts, and sorted adult testicular THY1+ cells. SSEA-4+ cells bound exclusively to testicular THY1+ cells within 48 hours and established colony formations. Under these conditions, SSC colonies continued to expand in size and underwent many subsequent passages without loss of SSEA-4 and VASA expression. This observation is consistent with recent studies that have demonstrated the importance of testicular stromal cells as a source of essential growth factors required by SSCs [67]. Previous studies demonstrated the presence of both SSCs and testis-derived pluripotent stem cells when unsorted testicular cells were cultured in vivo [34]. Thus far, we have not observed this phenomenon with either unsorted testicular cells or sorted testicular SSEA-4+ cells. All observed colonies in our studies expressed SSEA-4 and VASA. Although the percentage of SSC colonies formed per sorted SSEA-4+ cells (25,000 cells per experiment) appeared to be low (0.02%–0.1%) using our system, the cell number required for engraftment of SSEA-4+ colonies using a mouse model was more than 300,000 cells per transplant [35]. This suggests that the SSEA-4+ population is enriched for SSCs and that only a small portion of the population can form colonies and repopulate in vivo. The number of germ cells needed for engraftment in autologous nonhuman primate transplant model was ∼100 × 106 cells [9]. Thus, the low initial concentration of SSCs in testicular tissue highlights the need for in vitro expansion of SSCs prior to developing clinically viable SSC transplantation techniques in the future. Although MEFs were found to support in vitro mouse SSC expansion [31], this was not the case with human SSCs, suggesting basic differences between human and mouse SSCs. These data provide strong evidence that SSEA-4 is a specific marker expressed in primitive SSCs, whereas the somatic THY1+ cells are TMSCs that play an instrumental role in providing the appropriate niche required for SSC expansion.
A weakness of the study includes the lack of SSEA-4+ cell xenotransplantation, before and after in vitro expansion [11]. Although allotransplantation of mouse SSCs into germ cell-depleted testes is an ideal in vivo assay to evaluate for the ability of mouse SSCs to rescue spermatogenesis, it is suboptimal for human SSCs. Presumably because of interspecies differences, xenograft of human SSCs into mouse testes resulted in only colonization of human cells without differentiation. Furthermore, recent studies demonstrate that allogeneic mesenchymal stem cell (MSC) xenotransplantation also resulted in formation of germ cell colonies, highlighting this model’s lack of specificity, especially in the absence of in vivo differentiation [68]. Thus, there is no current ideal in vivo assay to evaluate human SSC activity.
These data have many significant strengths. Using a combination of multicolor FACS, confocal microscopy analyses, molecular profiling, and cell culture with sorted subpopulations of testicular cells monitored with time-lapse photography, we were able to definitively identify and differentiate the stromal and SSC compartments within the human testicular niche. Of critical importance was the finding that specific interactions between adult testicular THY1+ cells and SSCs are essential for in vitro human SSC expansion. Furthermore, these data likely explain the controversy regarding MSC expansion with the traditional in vitro culture system using testicular cells.
Conclusion
Identifying and growing SSC have important implications for patients. To date, prepubertal males facing sterilizing chemotherapy do not have a proven means of protecting their fertility. In a small number of centers around the world, testicular biopsies are performed prior to chemotherapy in the hopes that new fertility restoring treatments will become available. Development of methods to expand purified SSCs exponentially, free of malignant cell contamination, for future autologous transplant or to differentiate prepubertal SSC to mature sperm has enormous therapeutic potential. We have characterized the essential steps of isolating and expanding highly purified human SSCs using a defined somatic niche provided by the TMSCs. This is a critical step forward in developing strategies of prepubertal SSC expansion and autologous SSC therapy for fertility preservation treatments.
Supplementary Material
Acknowledgments
We acknowledge Dr. Grace Wei for her assistance with time-lapse photography. Funding for the study design and conduct of the study and the collection, management, analysis, and interpretation of the data was provided in part by an American Society of Reproductive Medicine New Investigator Award and through institutional and departmental funding.
Author Contributions
J.F.S. and N.D.T.: conception and design, administrative and financial support, provision of study materials or patients, data analysis and interpretation, manuscript writing, final approval of manuscript; P.Y. and E.A.: data analysis and interpretation, collection and assembly of data, manuscript writing, final approval of manuscript; S.C.: provision of study materials or patients, data analysis and interpretation, manuscript writing, final approval of manuscript; A.P., A.M.Z., M.R., and P.C.K.: data analysis and interpretation, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
M.R. has an uncompensated consultancy with Ovascience.
References
- 1.Howlader N, Noone A, Krapcho M et al. SEER Cancer Statistics Review, 1975–2008. Available at http://seer.cancer.gov/csr/1975_2008/, 2011. Accessed February 1, 2014.
- 2.Ferrara F, Schiffer CA. Acute myeloid leukaemia in adults. Lancet. 2013;381:484–495. doi: 10.1016/S0140-6736(12)61727-9. [DOI] [PubMed] [Google Scholar]
- 3.Borgmann-Staudt A, Rendtorff R, Reinmuth S, et al. Fertility after allogeneic haematopoietic stem cell transplantation in childhood and adolescence. Bone Marrow Transplant. 2012;47:271–276. doi: 10.1038/bmt.2011.78. [DOI] [PubMed] [Google Scholar]
- 4.Salooja N, Szydlo RM, Socie G, et al. Pregnancy outcomes after peripheral blood or bone marrow transplantation: A retrospective survey. Lancet. 2001;358:271–276. doi: 10.1016/s0140-6736(01)05482-4. [DOI] [PubMed] [Google Scholar]
- 5.Kliesch S, Behre HM, Jürgens H, et al. Cryopreservation of semen from adolescent patients with malignancies. Med Pediatr Oncol. 1996;26:20–27. doi: 10.1002/(SICI)1096-911X(199601)26:1<20::AID-MPO3>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 6.Keene DJ, Sajjad Y, Makin G, et al. Sperm banking in the United Kingdom is feasible in patients 13 years old or older with cancer. J Urol. 2012;188:594–597. doi: 10.1016/j.juro.2012.04.023. [DOI] [PubMed] [Google Scholar]
- 7.van Casteren NJ, van Santbrink EJ, van Inzen W, et al. Use rate and assisted reproduction technologies outcome of cryopreserved semen from 629 cancer patients. Fertil Steril. 2008;90:2245–2250. doi: 10.1016/j.fertnstert.2007.10.055. [DOI] [PubMed] [Google Scholar]
- 8.Sato T, Katagiri K, Gohbara A, et al. In vitro production of functional sperm in cultured neonatal mouse testes. Nature. 2011;471:504–507. doi: 10.1038/nature09850. [DOI] [PubMed] [Google Scholar]
- 9.Hermann BP, Sukhwani M, Winkler F, et al. Spermatogonial stem cell transplantation into rhesus testes regenerates spermatogenesis producing functional sperm. Cell Stem Cell. 2012;11:715–726. doi: 10.1016/j.stem.2012.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shinohara T, Inoue K, Ogonuki N, et al. Birth of offspring following transplantation of cryopreserved immature testicular pieces and in-vitro microinsemination. Hum Reprod. 2002;17:3039–3045. doi: 10.1093/humrep/17.12.3039. [DOI] [PubMed] [Google Scholar]
- 11.Brinster RL, Avarbock MR. Germline transmission of donor haplotype following spermatogonial transplantation. Proc Natl Acad Sci USA. 1994;91:11303–11307. doi: 10.1073/pnas.91.24.11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proc Natl Acad Sci USA. 1994;91:11298–11302. doi: 10.1073/pnas.91.24.11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlatt S, Rosiepen G, Weinbauer GF, et al. Germ cell transfer into rat, bovine, monkey and human testes. Hum Reprod. 1999;14:144–150. doi: 10.1093/humrep/14.1.144. [DOI] [PubMed] [Google Scholar]
- 14.Honaramooz A, Behboodi E, Blash S, et al. Germ cell transplantation in goats. Mol Reprod Dev. 2003;64:422–428. doi: 10.1002/mrd.10205. [DOI] [PubMed] [Google Scholar]
- 15.Honaramooz A, Megee SO, Dobrinski I. Germ cell transplantation in pigs. Biol Reprod. 2002;66:21–28. doi: 10.1095/biolreprod66.1.21. [DOI] [PubMed] [Google Scholar]
- 16.Izadyar F, Den Ouden K, Stout TA, et al. Autologous and homologous transplantation of bovine spermatogonial stem cells. Reproduction. 2003;126:765–774. [PubMed] [Google Scholar]
- 17.Mikkola M, Sironen A, Kopp C, et al. Transplantation of normal boar testicular cells resulted in complete focal spermatogenesis in a boar affected by the immotile short-tail sperm defect Reprod Domest Anim 2006;41:124–128 [DOI] [PubMed] [Google Scholar]
- 18.Herrid M, Olejnik J, Jackson M, et al. Irradiation enhances the efficiency of testicular germ cell transplantation in sheep. Biol Reprod. 2009;81:898–905. doi: 10.1095/biolreprod.109.078279. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y, Turner D, Nelson J, et al. Production of donor-derived sperm after spermatogonial stem cell transplantation in the dog. Reproduction. 2008;136:823–831. doi: 10.1530/REP-08-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altman E, Smith JF, Yango P et al. Identification and characterization of human fetal, prepubertal, and adult spermatogonial stem cells. Paper presented at: American Society of Reproductive Medicine Annual Meeting; October 12–17, 2013; Boston, MA. [Google Scholar]
- 21.Altman E, Smith JF, Yango P et al. Progressive initiation of germ cell meiosis in the human testes occurs during early second trimester. Paper presented at: American Society of Reproductive Medicine Annual Meeting; October 12–17, 2013; Boston, MA. [Google Scholar]
- 22.Wu X, Schmidt JA, Avarbock MR, et al. Prepubertal human spermatogonia and mouse gonocytes share conserved gene expression of germline stem cell regulatory molecules. Proc Natl Acad Sci USA. 2009;106:21672–21677. doi: 10.1073/pnas.0912432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dym M, Kokkinaki M, He Z. Spermatogonial stem cells: Mouse and human comparisons. Birth Defects Res C Embryo Today. 2009;87:27–34. doi: 10.1002/bdrc.20141. [DOI] [PubMed] [Google Scholar]
- 24.Ko K, Araúzo-Bravo MJ, Tapia N, et al. Human adult germline stem cells in question Nature 2010;465:E1–E3 [DOI] [PubMed] [Google Scholar]
- 25.Ko K, Reinhardt P, Tapia N, et al. Brief report: Evaluating the potential of putative pluripotent cells derived from human testis. Stem Cells. 2011;29:1304–1309. doi: 10.1002/stem.671. [DOI] [PubMed] [Google Scholar]
- 26.Tapia N, Araúzo-Bravo MJ, Ko K, et al. Concise review: Challenging the pluripotency of human testis-derived ESC-like cells. Stem Cells. 2011;29:1165–1169. doi: 10.1002/stem.669. [DOI] [PubMed] [Google Scholar]
- 27.Chikhovskaya JV, Jonker MJ, Meissner A, et al. Human testis-derived embryonic stem cell-like cells are not pluripotent, but possess potential of mesenchymal progenitors. Hum Reprod. 2012;27:210–221. doi: 10.1093/humrep/der383. [DOI] [PubMed] [Google Scholar]
- 28.Seandel M, James D, Shmelkov SV, et al. Generation of functional multipotent adult stem cells from GPR125+ germline progenitors. Nature. 2007;449:346–350. doi: 10.1038/nature06129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebata KT, Zhang X, Nagano MC. Expression patterns of cell-surface molecules on male germ line stem cells during postnatal mouse development. Mol Reprod Dev. 2005;72:171–181. doi: 10.1002/mrd.20324. [DOI] [PubMed] [Google Scholar]
- 30.Grisanti L, Falciatori I, Grasso M, et al. Identification of spermatogonial stem cell subsets by morphological analysis and prospective isolation. Stem Cells. 2009;27:3043–3052. doi: 10.1002/stem.206. [DOI] [PubMed] [Google Scholar]
- 31.Kanatsu-Shinohara M, Ogonuki N, Inoue K, et al. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod. 2003;69:612–616. doi: 10.1095/biolreprod.103.017012. [DOI] [PubMed] [Google Scholar]
- 32.Dovey SL, Valli H, Hermann BP, et al. Eliminating malignant contamination from therapeutic human spermatogonial stem cells. J Clin Invest. 2013;123:1833–1843. doi: 10.1172/JCI65822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conrad S, Renninger M, Hennenlotter J, et al. Generation of pluripotent stem cells from adult human testis. Nature. 2008;456:344–349. doi: 10.1038/nature07404. [DOI] [PubMed] [Google Scholar]
- 34.Sadri-Ardekani H, Mizrak SC, van Daalen SK, et al. Propagation of human spermatogonial stem cells in vitro. JAMA. 2009;302:2127–2134. doi: 10.1001/jama.2009.1689. [DOI] [PubMed] [Google Scholar]
- 35.Izadyar F, Wong J, Maki C, et al. Identification and characterization of repopulating spermatogonial stem cells from the adult human testis. Hum Reprod. 2011;26:1296–1306. doi: 10.1093/humrep/der026. [DOI] [PubMed] [Google Scholar]
- 36.Henderson JK, Draper JS, Baillie HS, et al. Preimplantation human embryos and embryonic stem cells show comparable expression of stage-specific embryonic antigens. Stem Cells. 2002;20:329–337. doi: 10.1634/stemcells.20-4-329. [DOI] [PubMed] [Google Scholar]
- 37.Sadri-Ardekani H, Akhondi MA, van der Veen F, et al. In vitro propagation of human prepubertal spermatogonial stem cells. JAMA. 2011;305:2416–2418. doi: 10.1001/jama.2011.791. [DOI] [PubMed] [Google Scholar]
- 38.Geens M, Goossens E, De Block G, et al. Autologous spermatogonial stem cell transplantation in man: Current obstacles for a future clinical application. Hum Reprod Update. 2008;14:121–130. doi: 10.1093/humupd/dmm047. [DOI] [PubMed] [Google Scholar]
- 39.Guan K, Nayernia K, Maier LS, et al. Pluripotency of spermatogonial stem cells from adult mouse testis. Nature. 2006;440:1199–1203. doi: 10.1038/nature04697. [DOI] [PubMed] [Google Scholar]
- 40.Kanatsu-Shinohara M, Lee J, Inoue K, et al. Pluripotency of a single spermatogonial stem cell in mice. Biol Reprod. 2008;78:681–687. doi: 10.1095/biolreprod.107.066068. [DOI] [PubMed] [Google Scholar]
- 41.Kossack N, Meneses J, Shefi S, et al. Isolation and characterization of pluripotent human spermatogonial stem cell-derived cells. Stem Cells. 2009;27:138–149. doi: 10.1634/stemcells.2008-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Golestaneh N, Kokkinaki M, Pant D, et al. Pluripotent stem cells derived from adult human testes. Stem Cells Dev. 2009;18:1115–1126. doi: 10.1089/scd.2008.0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizrak SC, Chikhovskaya JV, Sadri-Ardekani H, et al. Embryonic stem cell-like cells derived from adult human testis. Hum Reprod. 2010;25:158–167. doi: 10.1093/humrep/dep354. [DOI] [PubMed] [Google Scholar]
- 44.Oatley JM, Brinster RL. The germline stem cell niche unit in mammalian testes. Physiol Rev. 2012;92:577–595. doi: 10.1152/physrev.00025.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurn N, Chen P, Heath JD, et al. Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clin Chem. 2005;51:1973–1981. doi: 10.1373/clinchem.2005.053694. [DOI] [PubMed] [Google Scholar]
- 46.Dafforn A, Chen P, Deng G, et al. Linear mRNA amplification from as little as 5 ng total RNA for global gene expression analysis. Biotechniques. 2004;37:854–857. doi: 10.2144/04375PF01. [DOI] [PubMed] [Google Scholar]
- 47.Oatley JM, Brinster RL. Regulation of spermatogonial stem cell self-renewal in mammals. Annu Rev Cell Dev Biol. 2008;24:263–286. doi: 10.1146/annurev.cellbio.24.110707.175355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gkountela S, Li Z, Vincent JJ, et al. The ontogeny of cKIT+ human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat Cell Biol. 2013;15:113–122. doi: 10.1038/ncb2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson RA, Fulton N, Cowan G, et al. Conserved and divergent patterns of expression of DAZL, VASA and OCT4 in the germ cells of the human fetal ovary and testis. BMC Dev Biol. 2007;7:136. doi: 10.1186/1471-213X-7-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosu-Myles M, McCully J, Fair J, et al. The globoseries glycosphingolipid SSEA-4 is a marker of bone marrow-derived clonal multipotent stromal cells in vitro and in vivo. Stem Cells Dev. 2013;22:1387–1397. doi: 10.1089/scd.2012.0547. [DOI] [PubMed] [Google Scholar]
- 51.Ruggiu M, Speed R, Taggart M, et al. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature. 1997;389:73–77. doi: 10.1038/37987. [DOI] [PubMed] [Google Scholar]
- 52.Sasaki T, Shiohama A, Minoshima S, et al. Identification of eight members of the Argonaute family in the human genome. Genomics. 2003;82:323–330. doi: 10.1016/s0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X, Chou W, Haig-Ladewig L, et al. BNC1 is required for maintaining mouse spermatogenesis. Genesis. 2012;50:517–524. doi: 10.1002/dvg.22014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Kopylow K, Kirchhoff C, Jezek D, et al. Screening for biomarkers of spermatogonia within the human testis: A whole genome approach. Hum Reprod. 2010;25:1104–1112. doi: 10.1093/humrep/deq053. [DOI] [PubMed] [Google Scholar]
- 55.von Kopylow K, Staege H, Schulze W, et al. Fibroblast growth factor receptor 3 is highly expressed in rarely dividing human type A spermatogonia. Histochem Cell Biol. 2012;138:759–772. doi: 10.1007/s00418-012-0991-7. [DOI] [PubMed] [Google Scholar]
- 56.Brower JV, Lim CH, Jorgensen M, et al. Adenine nucleotide translocase 4 deficiency leads to early meiotic arrest of murine male germ cells. Reproduction. 2009;138:463–470. doi: 10.1530/REP-09-0201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maldonado-Saldivia J, van den Bergen J, Krouskos M, et al. Dppa2 and Dppa4 are closely linked SAP motif genes restricted to pluripotent cells and the germ line. Stem Cells. 2007;25:19–28. doi: 10.1634/stemcells.2006-0269. [DOI] [PubMed] [Google Scholar]
- 58.Olesen C, Nyeng P, Kalisz M, et al. Global gene expression analysis in fetal mouse ovaries with and without meiosis and comparison of selected genes with meiosis in the testis. Cell Tissue Res. 2007;328:207–221. doi: 10.1007/s00441-006-0205-5. [DOI] [PubMed] [Google Scholar]
- 59.Chi P, San Filippo J, Sehorn MG, et al. Bipartite stimulatory action of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes Dev. 2007;21:1747–1757. doi: 10.1101/gad.1563007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tapparel C, Reymond A, Girardet C, et al. The TPTE gene family: Cellular expression, subcellular localization and alternative splicing. Gene. 2003;323:189–199. doi: 10.1016/j.gene.2003.09.038. [DOI] [PubMed] [Google Scholar]
- 61.Soper SF, van der Heijden GW, Hardiman TC, et al. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell. 2008;15:285–297. doi: 10.1016/j.devcel.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wiszniak SE, Dredge BK, Jensen KB. HuB (elavl2) mRNA is restricted to the germ cells by post-transcriptional mechanisms including stabilisation of the message by DAZL. PLoS One. 2011;6:e20773. doi: 10.1371/journal.pone.0020773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang F, Eckardt S, Leu NA, et al. Mouse TEX15 is essential for DNA double-strand break repair and chromosomal synapsis during male meiosis. J Cell Biol. 2008;180:673–679. doi: 10.1083/jcb.200709057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murray LJ, Bruno E, Uchida N, et al. CD109 is expressed on a subpopulation of CD34+ cells enriched in hematopoietic stem and progenitor cells. Exp Hematol. 1999;27:1282–1294. doi: 10.1016/s0301-472x(99)00071-5. [DOI] [PubMed] [Google Scholar]
- 65.Gassei K, Orwig KE. SALL4 expression in gonocytes and spermatogonial clones of postnatal mouse testes. PLoS One. 2013;8:e53976. doi: 10.1371/journal.pone.0053976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carofiglio F, Inagaki A, de Vries S, et al. SPO11-independent DNA repair foci and their role in meiotic silencing. PLoS Genet. 2013;9:e1003538. doi: 10.1371/journal.pgen.1003538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cheng CY, Wong EW, Yan HH, et al. Regulation of spermatogenesis in the microenvironment of the seminiferous epithelium: New insights and advances. Mol Cell Endocrinol. 2010;315:49–56. doi: 10.1016/j.mce.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cakici C, Buyrukcu B, Duruksu G, et al. Recovery of fertility in azoospermia rats after injection of adipose-tissue-derived mesenchymal stem cells: The sperm generation Biomed Res Int 2013;2013:529589. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.