Abstract
Epigenetic modifications constitute the next frontier in tumor biology research. Post-translation modification of histones dynamically influences gene expression independent of alterations to the DNA sequence. These mechanisms are often mediated by histone linkers or by proteins associated with the recruitment of DNA-binding proteins, HDAC I and II interacting proteins and transcriptional activators, coactivators or corepressors. Early evidence suggested that histones and their modifiers are involved in sophisticated processes that modulate tumor behavior and cellular phenotype. In this review, we discuss how recent discoveries about chromatin modifications, particularly histone acetylation, are shaping our knowledge of cell biology and our understanding of the molecular circuitry governing tumor progression and consider whether recent insights may extend to novel therapeutic approaches. Furthermore, we discuss the latest oncogenomic findings in Head and Neck Squamous Cell Carcinoma (HNSCC) from studies using Next Generation Sequencing (NGS) technology and highlight the impact of mutations identified in histones and their modifiers.
Keywords: H3, H4, Oral cancer, Whole genome sequencing, Genetic code, Histone modifications, Acetylation, Chromatin compaction
Introduction
With the completion of the human genome it becomes evident the upcoming opportunities to explore and identify new genome-scale mechanisms involved in gene expression. The field of epigenetics presents a unique opportunity to understand the molecular mechanisms underlying the modulation of gene expression in the absence of alterations to the DNA sequence. Epigenetics is involved in the control of normal tissue homeostasis through maintenance of tissue-specific genetic signatures. However, deregulated environmental cues can corroborate to cellular transformation by altering local signaling circuitry, resulting in dysfunctional epigenetic equilibrium. By interacting with environmental cues, tumors undergo genetic deviations resulting in changes on cancer behavior, cellular phenotype, increase aggressiveness or induction of tumor dormancy. Therefore, it is important to understand the crosstalk between environmental signals and tumor behavior in epigenetics.
Oncogenomics, which uses high-throughput technologies to screen cancer-associated genes, is a novel strategy for tackling the complex interaction between DNA mutations and epigenetic driven deviations. DNA mutations originate from changes in the nucleotide sequence mainly derived from insertions, deletions, point mutations and chromosomal translocations that influence the normal balance between cellular proliferation and death. In an effort to identify new biomarkers and develop novel cancer therapeutics, multiple mega oncogenomic projects were initiated. These included the Cancer Genome Project at the Wellcome Trust Sanger Institute and the Cancer Genome Anatomy Project (CGAP), a National Cancer Institute initiative aimed at determining the genetic expression of normal, precancerous, and cancerous cells.
In the last several years, oncogenomics applications have extended from identifying genes associated with cancer to analyzing transcript abundance in tumors (Transcriptome) [1,2], DNA methylation (Methylome) and histone acetylation (Acetylome) [3,4]. The transcriptome is highly influenced by histone modifications that are primarily driven by acetylation and methylation. Histone modifications directly influence chromatin DNA packaging, resulting in a decondensed structure and enhanced interactions with transcription factors. This process, referred as the histone code, confers a high degree of cellular plasticity in both normal cells, such as the neuron synapse and lymphocytes [5,6], and tumor cells of the ovary and head and neck [7–9]. We highlight below how histones and its modifications are involved in stem cell biology and in the molecular circuitry governing tumor progression. We also present and discuss the involvement of histone acetyltransferase, histone deacetylases and its inhibitors as a therapeutic strategy used in various cancers. Further, we explore recent findings on head and neck cancer mutations of histones and histone modifiers identified using next-generation sequencing.
The Chromatin: Epigenetic Switches of the Genome
The genome is organized and packed into the nucleus through interactions with core histone proteins. Differential DNA folding regulates access to regulatory elements, resulting in alterations of the cellular response to environmental cues. DNA folding modifies cellular phenotypes by inducing cell type-specific chromatin organization [10]. We recently reported on functional chromatin folding in head and neck squamous cell carcinoma (HNSCC) cell lines during tumor response to environmental cues that dynamically modulates tumor behavior and cellular phenotype [9].
The ability of the chromatin to modulate cellular behavior is associated with how tightly DNA is spooled around H2A, H2B, H3 and H4 core histones [11]. Together, histones and DNA form nucleosomes, the fundamental units of chromatin. Each nucleosome is composed of 146bp of DNA coiled in sequence around a core of eight histone proteins [12].
Histones, the most abundant proteins associated with DNA, were highly conserved during evolution and are associated with the regulation of nuclear gene expression in several tissue types. Histones maintain contact with DNA through their flexible globular domain and amino acid residue regions (lysine, serine and arginine) that protrude from the nucleosome (histone “tail” domain). The majority of posttranslational modifications in histones, such as lysine acetylation, serine phosphorylation, lysine methylation, arginine methylation, and lysine ubiquitination, occur at the residue region [13–17]. The pattern of histone modifications determines chromatin status (euchromatin or heterochromatin) [18], the accessibility of DNA to nuclear factors and ultimately transcription. Euchromatin is relatively uncondensed and represents loci being actively transcribed. In contrast, heterochromatin is condensed and represents areas of the genome that are silenced. Several studies show that alterations in chromatin structure due to histone modifications correlate with gene expression, the cell cycle, DNA replication and damage, DNA repair and chromosome stability [19,20].
Dynamic changes in chromatin structure organization are mainly driven by histone acetylation and deacetylation, mechanisms responsible for important regulatory functions of gene expression. These processes are controlled by histone acetyltransferases (HAT) and histone deacetylases (HDACs), enzymes that are targeted to specific amino acid positions. In histone acetylation, HATs catalyze the addition of an acetyl group to the lysine residue in the tails of histones resulting in neutralization of the positive charge in the histone tail. The new acetyl group reduces the histone-DNA interaction, resulting in open or active chromatin that allows binding of transcription factors to promoters of target genes. Therefore, increased levels of histone acetylation are generally associated with increased transcriptional activity [21]. Acetyl groups are removed in histone deacetylation, resulting in increased histone-DNA interaction and chromatin condensation that blocks transcription by preventing the binding of transcription factors and regulatory complexes [21–23].
HATs and HDACs act in a coordinated manner to influence gene expression in different tissues. These enzymes induce conformational changes in histone-DNA interactions that alter the accessibility of transcription factors to DNA and are crucial epigenetic changes for the development and differentiation of many normal cell types. However, altered HAT and HDAC activity has also been observed in several types of cancer and may be promising therapeutic targets for these diseases [13,21,24].
Epigenetic regulation of gene expression is primarily controlled by histone acetylation and methylation, histone modifications, and changes in nucleosome conformation [20]. There is growing evidence that epigenetics is involved in gene regulation during mammalian development (reviewed in [25]). These processes are observed during the maintenance of embryonic stem cells (ES) in the early stages of embryogenesis. Polycomb group (PcG)-protein repressive system is an example of epigenetic control of gene expression. PcG form a multiprotein complex involved in chromatin organization and capable of epigenetically induce modifications on its structure thereby influencing gene transcription. In ES cells, PcG repressive system suppresses developmental pathways, thereby maintaining ES cell pluripotency and plasticity during embryonic development [26]. Furthermore, Boyer et al. identified several PcG target genes in ES cells bound by OCT4, SOX2 and NANOG, three transcription factors associated with pluripotency [27]. Similarly, the inability of the PcG protein repressive complex to suppress developmental genes in ES cells results in spontaneous cellular differentiation [26]. In accordance with the PcG-protein repressive complex, the effect of HDAC on chromatin organization regulates the maintenance of stem cell pluripotency in coordination with numerous signaling pathways [28,29]. In contrast, chromatin condensation in tumors is associated with chemoresistance [30–33] through the reactivation of stem cell-like transcription programs [34]. In addition to the known functions of the PcG-protein repressive complex, recent reports have shown that the histone variant H2A.Z plays a similar role in early development. H2A.Z assists with the dissociation of H2A–H2B dimers, resulting in modifications to the nucleosome surface that promotes transcription factor binding [35]. H2A.Z induces changes in chromatin configuration allowing access to transcription factors and cofactors that activate and repress regions in embryonic stem cells leading to self-renewal and differentiation [36,37]. This mechanism involves the depletion of nucleosomes at the Transcription Start Sites (TSSs) where gene transcription begins. Recently, Schones et al. found that nucleosomes containing H2A.Z are particularly susceptible to depletion [38]. Further, Li et al. observed by high-resolution nucleosome mapping and chromatin immunoprecipitation sequencing (ChIP-Seq) that co-occupancy of the transcriptional activator FoxA2 and H2A.Z results in nucleosome depletion during ES cell differentiation, coinciding with gene activation [37].
Histone Acetyltransferase (HAT) and Histone Deacetylases (HDAC)
The equilibrium between histone acetylation and deacetylation in the promoter region of chromatin is essential for the regulation of gene expression and the establishment of a transcriptionally competent chromatin state [39–42]. The antagonistic effects of HDACs and HATs tightly control the balance of steady-state acetylation. While HDACs promote transcriptional repression by removing acetyl groups from lysine, HATs increase transcriptional activity through the addition of acetyl groups. Besides these electrostatic changes, HATs and HDACs also play roles in gene activation or silencing through the recruitment of co-activators and co-repressors of transcription.
Alterations in histone locus-specific post-translational modifications are proposed for describing a code, which defines the state transcriptional potential of a cell referred as the code of histones. A global comprehension of this code is necessary for dissecting the role of histones in physiology and pathology [43]. Initial steps towards this goal have been made with the characterization of the histone deacetylase-1 enzyme (HDAC1) as a key regulator of embryonic stem cells proliferation and repression of cell cycle inhibitors in mice indicating its requirement during embryonic development [44]. Impairment of HAT activity or overexpression of HDAC by aberrant gene expression is associated with the process of carcinogenesis and tumor progression [13,21,45,46]. Because histone modifications do not alter the DNA sequence, new agents have been developed that reverse this modification, resulting in reactivation of tumor suppressor genes and reestablishment of a signaling balance to maintain cellular homeostasis. Notably, inhibitors of histone deacetylases (HDACi) are a new group of anticancer agents that have shown promising results [47].
Histone acetyltransferase (HAT) and its Inhibitors
The HATs are divided into the following five distinct families based on sequence divergence in the HAT domain: (i) HAT1, (ii) Gcn5/PCAF, (iii) MYST, (iv). CBP/p300, and (v). Rtt109. HAT activities involve protein complexes and co-activators of transcription factors [48]. Various biological processes are associated with HAT function, including cell cycle progression, DNA damage repair, dosage compensation and hormone signaling [24].
The HAT family possesses diverse catalytic strategies for acetyl transfer, reflecting its long evolutionary time. Several HAT substrate peptides, including histone H3 (centered around Lysine 14) and histone H4 (centered around Lysine 8), have been described [24]. HATs have substrates beyond histones, such as non-histones proteins and transcription factors. Certain substrates are transcriptional cofactors for a range of cellular oncoproteins, such as MYB, JUN, FOS, RUNX, BRCA1, p53, and pRB, and viral oncoproteins, including E1A, E6, and SV40 large T [13, 24, 49, 50]. Of note, p53 was the first functional non-histone substrate identified to be acetylated by HATs [51] reviewed in [52]. P53 is acetylated by CBP/p300 at multiple lysine residues (positions 370, 372, 373, 381, and 382) in the C-terminal regulatory domain, which result in increased DNA binding activity and acetylation-induced conformational changes, and protein stabilization [53–55].
Translocation, amplification, overexpression or mutation of HAT genes are found in a variety of human pathologies, including cancer, inflammatory lung disease, viral infection, diabetes, fungal infection, and drug addiction [48,56–58]. The CBP/p300 HAT genes are considered functional tumor suppressors because they reside in regions frequently lost in tumors, and certain cancers exhibit mutations that abrogate p300 activity [59–61]. For example, the tumor suppressor function of CBP/p300 was confirmed in animal models with ablated Cbp [62,63]. Similarly, translocation partners, including MLL, MOZ, and MORF [64–66], disrupt CBP/p300 in leukemia. These findings established a new paradigm for transcriptional regulation and tumorigenesis by showing that epigenetic regulation of genes collaborates with genetic alterations in cancer development [18,51, 58,67].
Clinical trial assessment of HAT inhibitors is in the early stages, especially compared to the multitude of trials testing kinase inhibitors. There is also interest in natural compounds, such as anacardic acid and garcinol, that inhibit hPCAF and hp300, respectively. Other natural compounds like Curcumin, Epigallocatechin-3-gallate (EGCG) and plumbagin have been shown to selectively inhibit CBP/p300. New HAT inhibitors have been developed that display submicromolar inhibition activity and selectively inhibit Gcn5/PCAF and CBP/p300 [24,58,68]. There is great interest in developing additional HAT inhibitors with improved specificity and anti-tumor activity.
Histone deacetylases (HDACs) and HDAC inhibitors
HDACs are cellular enzymes expressed in many tissues that play important roles in differentiation, proliferation, and cancer. HDACs are primarily involved in the acetylation network, and several HDACs have been described in humans. Based on similarities, HDACs are divided into four functional classes based on sequence homology to their corresponding transcriptional regulator of yeast counterparts (Saccharomyces cerevisiae): 1) class I (HDAC1, HDAC2, HDAC3 and HDAC8); 2) class II is subdivided into class IIa (HDAC4, HDAC5, HDAC7, and HDAC9) and class IIb (HDAC6 and HDAC10); 3) class III (SIRT1 to SIRT7); and 4) class IV (HDAC11 and related enzymes) [13,39,69,70]. These enzymes are part of multi-protein complexes, and their activity depends on other factors, such as mSin3, N-CoR and SMART transcriptional co-repressors. Changes in these factors can indirectly interfere with HDAC activities [21].
HDACs also have numerous non-histone target proteins comprised of transcriptional regulators (Rb, DEK, HMGI(Y)/HMGA1 and others), transcription factors (p53, c-Myc, BCL-6, E2F1 and others), DNA repair enzymes (Ku70, TDG, NEIL2 and others), nuclear import regulators (Rch1 and importin-α7), chaperone proteins (HSP90), structural proteins (α-tubulin), viral proteins (E1A and others) and inflammatory mediators (HMGB1) [71]. These proteins are involved in cellular proliferation, differentiation and cell death, and their functions can be intrinsically modulated by protein acetylation [23,71].
Class I HDACs are ubiquitously expressed in all tissue types [72,73]. Of particular interest for this review, HDAC1 and HDAC2 are involved in proliferation, regulation of the cell cycle and apoptosis. Posttranslational modifications of HDAC1 and HDAC2 alter their enzymatic activity, subcellular localization, stability, and interaction with other binding sites in response to different stimuli. In addition, deregulation of HDAC1 and HDAC2 is associated with the onset of many diseases in humans, including cancer [73]. HDAC1 suppresses activity of the p21WAF/SIP1 (cyclin-dependent kinase inhibitor) pathway by deacetylating lysine 4 residues on histone H3 [73,74]. Acetylation of HDAC1 dramatically reduces its enzymatic and repressive functions in vitro and in vivo, and overexpression of HDAC1 has been observed in many epithelial malignant tumors, such as hepatocellular [75], colorectal, gastric [76], pancreatic [77], and prostate [32] carcinomas. In contrast, depletion of HDAC1 results in perturbation of the cell cycle, loss of mitotic cells and increased cell death in several human cancer cell lines [78]. HDAC2 is overexpressed during the polyp stage of familial adenomatous polyposis colon cancer [79]. High levels of HDAC2 are also found in cervical dysplasia and invasive carcinomas [77]. Increased expression of HDAC1, HDAC2 and HDAC3 is associated with advanced stage prostate, gastric and colorectal cancers [76].
SIRT1, a member of the class III HDACs, acts as an important regulatory route capable of influence the apoptotic machinery. SIRT1 represses DNA damage-induced apoptosis through deacetylation of p53 [80]. SIRT1 also interferes with activation of apoptosis by deacetylating FOXO3a and/or E2F1 to inhibit their transcriptional activity [81,82]. Along with the control over the apoptotic machinery, SIRT1 promotes tumorigenesis by inhibiting cellular senescence, impairing cellular differentiation and promoting cellular growth associated with angiogenesis. SIRT1 is found highly expressed in many cancers, including acute myeloid leukemia, prostate carcinoma, and colon carcinoma, among others [83]. Further, SIRT1 is known to induce tumor resistance to chemotherapy suggesting its role as an oncogene (review in [87]). In contrast, SIRT1 is also considered a tumor suppressor gene due to its role in genomic stability, in limiting the replicative life span, in regulate cellular response to genotoxic stress and in the suppression of intestinal tumorigenesis [84–86]. Together, contradictory reports on SIRT1 function strongly suggests distinct roles depending on the involved signaling machinery.
HDACs inhibitors (HDACi) constitute a novel and growing family of potent anticancer agents that activate or repress 4–12% of genes by promoting histone acetylation, relaxing DNA that is coiled around core histones and increasing access of DNA to transcription factors [48,88–92]. In cancer cells, HDACi impair survival pathways resulting in cellular differentiation mediated by cell cycle arrest, block angiogenesis, induce apoptosis by stimulating the death receptor and intrinsic apoptotic pathway, and prevent tumor cells from evading the immune system by enhancing host immunosurveillance antigenicity (reviewed in [88,92,93]). Similar to SIRT1, the mechanisms involved in HDACi function are complex, differ between tumor cell types, and depend on the drug type and exposure time [92,94]. In vitro studies reveal the effects of HDACi are different for each type of cell lineage. However, common among cell types, is the activation of p53 and p21WAF1 and downregulation of cyclins and cell cycle activity following HDACi administration [95].
HDACi vary structurally and are classified in four groups, including hydroxamic acid, benzamides, cyclic peptides and aliphatic acids [90,92,96]. The potential of HDACi to exert anti-cancer effects has been examined in several clinical trials; however, only two HDACi agents have been approved by the Food and Drug Administration to date. SAHA (Vorinostat) and Romidepsin (Istodax) are approved for use in cutaneous T cell lymphoma (CTCL) [96–99]. Current clinical trials are examining HDACi in hematologic malignancies and solid tumors where preliminary results indicate a weaker antitumoral activity of HDACi in solid tumors compared to hematological malignancies [92,96,100]. However, many clinical trials involving solid tumors are comprised of patients with advanced disease and with relapsed and metastatic sarcomas (Table 1) (http://www.cancer.gov/clinicaltrials).
Table 1.
Summary of Main HDACi Drugs and Agents in Clinical Trials.
| Class | Compound | HDAC target | Phase | Company | Condition tested in Clinical Trials |
|---|---|---|---|---|---|
| Hydroxamic acid | Zolinza, vorinostat (SAHA) | Classes I and II | FDA | Merck | Approved for CTCL, phase I/ II for gastric cancer, NSCLC, thyroid cancer and advanced cancers |
| Panobinostat (LBH589) | Classes I and II | III | Novartis | Phase III trial for CTCL and phase II for CML, MM, NHL, HL, BC, chordoma, prostate and others advanced solid tumors | |
| Belinostat (PXD101) | Classes I and II | II | TopoTarget | Relapsed ovarian cancer, TCL, thymoma, thymic carcinoma, MM, NSCLC, brain tumors | |
| Givinostat or Gavinostat (ITF2357) | Classes I and II | II | Italfarmaco | Relapsed leukemias and MM | |
| Trichostatin (TSA) | Classes I and II | N/A | N/A | N/A | |
| Abexinostat (PCI-24781) | Classes I and II | II | Pharmacyclics | BCL, sarcoma | |
| LAQ-824 | Classes I and II | I | Novartis | Solid and hematologic malignancies | |
| Pracinostat (SB939) | Classes I, II and SIRT 1 | II | MEI Pharma | Prostate cancer, solid tumors, hematologic malignancies | |
| JNJ-26481585 | HDAC1, HDAC6 | I | Johnson & Johnson | Leukemia, CTCL and solid tumors | |
| CUDC-101 | Classes I and II | I | Curis | Head and neck, gastric, BC, liver cancer, MM, NSCLC | |
| CHR-3996 | Class I | I | Chroma Therapeutics | Solid tumors | |
| CHR-2845 | Class I | I | Chroma Therapeutics | Hematological diseases or lymphoid malignancies | |
| Quisinostat (JNJ 26481585) | Class I | II | MedKoo bioscience | CTCL, MM | |
| CG200745 | Pan-HDAC | I | CrystalGenomics, Inc, | Solid tumors | |
| Rocilinostat (ACY-1215) | HDAC6 | I and II | MedKoo bioscience | MM | |
| Benzamides | Entinostat (MS-275 SNDX275) | HDAC1,HDAC2,HDAC3 | II | Schering | HL, lung cancer, ALL, BC, melanoma and advanced solid tumors |
| Mocetinostat (MGCD0103) | HDAC1 and HDAC2 | II | Methylgene | AML | |
| Tacedinaline (CI-994) | HDAC1 and HDAC2 | II and III | Parke-Davis Pharmaceutical | MM, pancreatic cancer, phase III for Lung Cancer | |
| AR-42 | Pan-HDAC | I | Arno Therapeutics | MM, CLL or lymphoma | |
| 4SC-202 | HDAC1, HDAC2, HDAC3 | I | MedKoo bioscience | Hematological malignancies | |
| Resminostat (4SC-201) | Classes I and II | I and II | MedKoo bioscience | Hepatocellular carcinoma, colorectal cancer and HL | |
| Cyclic Peptides | Romidepsin/Istodax (FK228 and FR901228) | HDAC1 and HDAC2 | FDA | Celgene | Aproved for CTCL |
| Aliphatic Acids | Valproic Acid | Classes I and IIa | II and III | Abbot | Phase III trials for cervical cancer, ovarian cancer and phase II for sarcomas, BC, malignant melanoma, AML, brain tumors and advanced cancers |
| Phenylbutyrate (PB) | Classes I and IIa | I and II | Targon Corporation | Colorectal cancer, malignant gliomas, solid tumors, leukemias, lymphomas | |
| Butyrate | Classes I and IIa | I, II and III | Lymphoproliferative disorders, prostate cancer or other solid tumors, phase III for ovarian cancer |
ALL: acute lymphoblastic leukemia; AML: Acute Myeloid leukemia; BC: Breast Cancer; BCL: B-Cell Lymphoma; CLL: Chronic Lymphocytic Leukemia; CML: Chronic Myeloid Leukemia; CTCL: Cutaneous T Cell Lymphoma; HL: Hodgkin Lymphoma; MM: Multiple Myeloma; N/A: not available; NHL: Non-Hodgkin Lymphoma; NSCLC: Non-Small- Cell Lung Carcinoma; TCLT: T Cell Lymphoma.
The most promising HDACi results involve its synergistic or additive effects with other therapeutic regimens, including radiotherapy, chemotherapy, proteasome inhibitors, death receptor agonists and kinase inhibitors [92,96,100,101]. This is evidenced in HNSCC, where in vitro administration of HDACi exhibits good anticancer properties; however, HDACi shows only limited clinical utility as a single agent [99,102–105]. HDACi is only effective against HNSCC when administered in combination with radiotherapy [106]. For example, Romidepsin shows preclinical activity as a radiosensitizer [101,107].
Although promising, the potential use of HDACi as a new treatment for HNSCC remains poorly explored. Kim et al. [108] examined the pharmacological efficiency of combining the PS-341proteasome inhibitor, also known as Bortezomib, with Trichostatin A, an HDAC inhibitor. They found the drugs synergistically induce apoptosis in HNSCC by a mechanism that preserves PS-341-induced endoplasmic reticulum stress. Erlich et al. analyzed the antitumor effects of AKT and PI3K-mTOR inhibitors delivered as monotherapies, or in combination with HDACi, in models of HNSCC. As a single agent, Vorinostat (HDACi) induces selective cytotoxicity in squamous cell carcinoma, an effect enhanced by PI3K inhibitors that cause persistent AKT inhibition and generation of reactive oxygen species [91]. Further, the use of Panobinostat, a non-selective HDACi resulted in a dose-dependent hyperacetylation of histone H3 and inhibition of tumor proliferation and viability when administered alone. Further, the cytotoxicity effect observed upon administration of Panobinostat was continuously enhanced by co-treatment with PI3K inhibitors. Clinically, the administration of PI3K inhibitors enhanced the cancer cell specific cytotoxicity induced by Panobinostat. There are currently only four registered clinical trials testing HDACi and others therapeutic modalities in HNSCC. These studies involve Valproic acid and platinum-based chemoradiation in advanced HNSCC, Vorinostat in the treatment of advanced staged oropharyngeal squamous cell carcinoma, Capecitabine and Vorinostat in patients with recurrent and/or metastatic HNSCC and CUDC-101 plus cisplatin and radiation in HNSCC (Table 2) (http://www.cancer.gov/clinicaltrials).
Table 2.
Clinical trials involving HDACi and head and neck squamous cell carcinoma.
| Intervention Drugs |
Condition | Sponsor | Patients enrolled |
Phase | Period | Clinical trial number |
|---|---|---|---|---|---|---|
| Vorinostat (SAHA), Cisplatin, Radiation | Stage III or IV of SCC of the oropharynx | Ohio University Comprehensive Cancer Center | 38 | I | 2010 2013 |
NCT01064921 |
| Capecitabine (Xeloda), Vorinostat (SAHA) | Recurrent and/or metastatic head and neck cancer, Nasopharyngeal carcinoma, stage III and IV HNSSC and verrucous carcinoma |
National (NCI) Cancer Institute | 1445 | II | 2010 2013 |
NCT01267240 |
| CUDC-101 Cisplatin, Radiation | Head and neck cancer | Curis, Inc. | 22 | I | 2011 2013 |
NCT01384799 |
| Valproic acid, Patinum and Radiation | Locally advanced head and neck SCC (oral cavity and oropharynx) | Intituto do Cancer do Estado de São Paulo | 40 | II | 2012 2014 |
NCT01695122 |
SCC: squamous cell carcinoma HNSCC: head and neck squamous cell carcinoma
Although the use of HDACi has generated great enthusiasm, adverse side effects and resistance have been observed in clinical trials of different cancer types. The main side effects are related to hematologic complications, but nausea, asthenia/fatigue, infections, vomiting, anorexia, diarrhea and pyrexia have been noted. Along with the side effects, the development of tumor resistance to HDACi have been noted although the involved mechanism is far from understood. Additional studies are needed to better understand the mechanisms underlying HDACi action, to explore various combination therapies and to identify which types of cancer will better respond to HDACi treatment [71,100].
The Histone Code of Chromatin Acetylation
Posttranslational chromatin modifications occur through changes to histones. Histones can be modified by acetylation, methylation, and ubiquitination of lysines, methylation of arginines, phosphorylation of serines, sumoylation and ubiquitination, among other recently identified covalent histone modifications [109]. The histone code hypothesizes that histone modifications dynamically influence transcription by recruiting a variety of proteins that activate transcription. Among the numerous histone modifications, we are particularly interested in the effects of chromatin acetylation on tumor behavior. Core histones are composed of a histone-fold domain and an amino-terminal tail domain. Histone acetylation is mediated through interactions of the amino-terminal domain with regulatory proteins that reduce the affinity of the histone to DNA [110]. Therefore, binding efficiency of transcription factors is directly associated with histone mediated changes in DNA configuration induced by acetylation [111,112].
We are just beginning to elucidate the effects of chromatin acetylation on cellular function. Initial observations by Meegee et al. [113] demonstrated the requirement of lysines to be acetylated in the histone 4 tail domain for cell cycle progression and genomic stability. Further, the acetylation status of histone 4 lysine 16 is implicated in numerous physiological functions across several organisms. In humans, loss of monoacetylation of histone 4 lysine 16 serve as an epigenetic marker for leukemia, breast, lung and colon cancer cells [114] and may be a key event associated with epigenetic silencing of tumor suppressor genes (TSGs) [115]. Interestingly, other histone 4 acetylation residues, such as lysine 5, 8, and 12, are not found in tumors, suggesting that suppression of histone 4 lysine 16 acetylation may be important for cancer prevention. Global patterning of histone acetylation is also observed during the development of the nervous system. Cho et al. demonstrated dynamic histone acetylation expression during neuronal differentiation, as evidenced by increased expression of histone 3 and histone 4 in the cortical plate from rat cerebral cortex (E17) and chick spinal cord (E7) and increased histone 3 in adult mouse dentate gyrus [116]. In the cardiac development system, the HATs p300 and CBP are coactivators that promote the physiological and pathological growth of cardiac myocytes. Similarly, lungs and small intestines require p300 expression during normal development [117]. Collectively, these data suggest a role for acetylated histones in mitosis and differentiation during tissue development and maintenance. In fact, the identification of 3,600 lysine acetylation sites using quantitative mass spectrometry (MS)-based proteomics in 1,750 human proteins indicates that protein modifications mediated by acetylation is far more common than anticipated [118,119]. Although epigenetic modifications result in changes on gene expression independent from alterations on the DNA sequence, tumor epigenome may be influenced by heritable alterations on histones and histones associated transcriptional regulators. Indeed, next-generation sequencing of HNSCC has unveiled several of these mutations suggesting the differential modulation of epigenetic among normal and cancer cells and with that the existence of a cancer histone code. However, the functional and biological impact of these mutations in HNSCC is largely unknown.
The Next Generation of DNA Sequencing in HNSCC
Recent technological advances in the field of genome sequencing culminated in the development of Next-Generation Sequencing (NGS). This new technology has substantially improved discovery of single-base changes and the identification of larger structural variants characterized by insertions, deletions, translocations and viral insertions. Several sequencing platforms in use are characterized by amplification of single strands of fragmented DNA followed by sequencing [120].
The work of Agrawal et al. and Stransky et al. comprise the first HNSCC tumor sequencing studies using NGS technology and have provided a novel perspective for this complex disease. This wealth of data derives from 106 sequenced tumors from the head and neck anatomical area, including the oral cavity, oropharynx, hypopharynx, larynx, and sinonasal cavity. Stransky et al. have identified mutations of well-known genes, including TP53, which is present in 62% of sequenced tumors, and the CDKN2A gene, which occurs in 12% of tumors. In addition to TP53 and CDKN2A, more novel gene mutations were also identified. For example, nonsense, missense and splice site mutations in Notch1 were found in 11% of 74 tumor samples analyzed [121]. Using NGS in 32 head and neck samples, Agrawal et al. found NOTCH1 highly mutated in HNSCC, and approximately 40% of the mutations result in loss of function [122]. Both NGS studies found an inherited genomic difference between HPV-positive and HPV-negative head and neck tumors, with HPV-positive tumors containing less than half the number of mutations evident in HPV-negative tumors [121,122]. This suggests that HPV-positive HNSCC is genetically distinct from HPV-negative HNSCCs, a notion previously implied by their differential responses to chemotherapy and radiotherapy [123–125].
Novel NGS findings on HNSCC histones Mutations and its Modifiers
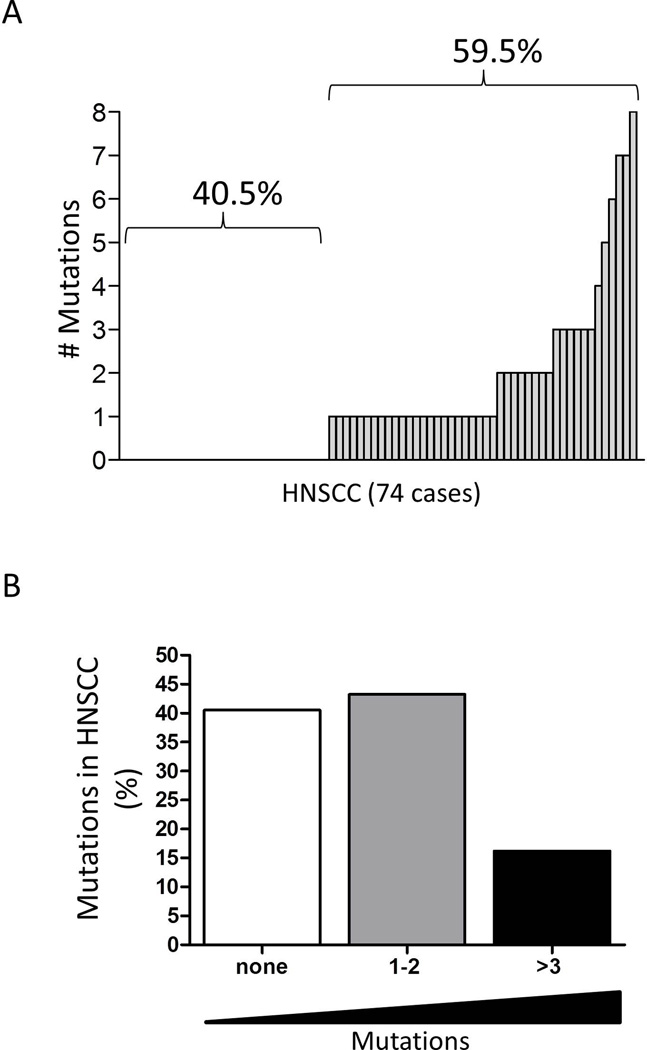
Within the thousands of genes found mutated in HNSCC, NGS have also identified mutations in histones and histones modifiers (Table 3). Of the 74 human head and neck tumor samples sequenced by Stransky et al. 44 tumors (59.5%) have mutated histones and/or transcriptional coactivators (Figure 1A). NGS analysis also revealed that 12 HNSCC have three or more mutations, and as many as eight mutations were found in a single tumor; 32 tumors harbor one or two mutations in histones and histone associated transcriptional regulators (Figure 1B). The most prevalent mutations are associated with genes encoding the H2A, H2B, H3, and H4 histone families. Similar mutations have been reported in the H3F3A and HIST1H3B genes in pediatric diffuse glioma [126,127], in HIST1H4I in clear cell renal cell carcinoma [128], and in HIST1H4E in human colon and rectal cancer [129]. Interestingly, mutations were also found in HIST1H1B, HIST1H1C, HIST1H1D and HIST1H1E linker histones. Linker histones are characterized by the absence of the histone fold domain and play a key role in organizing maximal nucleosome compaction within the chromatin [130,131] and chromatin binding affinity [132,133], among other functions. Notably, depletion of histone H1 induces aberrant mitotic figures [134], a common histological finding in tumor cells.
Table 3.
Histones and histones modifiers found mutated in HNSCC (NGS)
| Gene Name |
Gene Alias | UniProt | Protein Name | Protein Alias | cDNA Change | Mutation type | Protein Change |
|---|---|---|---|---|---|---|---|
| BCOR | KIAA1575 | Q6W2J9 | BCL-6 corepressor | BCoR | c.5486G>T | Nonsense | p.E1732 |
| BCORL1 | Q5H9F3 | BCoR-L1 | BCoR-like protein 1 | c.3989G>A | Missense | p.R1292Q | |
| CREBBP | CBP | Q92793 | CREB-binding protein | --- | c.4541G>A c.4765G>T c.1711C>T c.3053C>T c.964G>C |
Missense Missense Nonsense Missense Missense |
p.R1446H p.D1521Y p.Q503 p.T950M p.A254P |
| EHMT1 | UHMTASE1, GLP, KIAA1876, KMT1D | Q9H9B1 | EHMT1 | Eu-HMTase1, GLP, GLP1, H3-K9-HMTase 5. | c.1157G>A c.168G>C |
Missense Missense |
p.D374N p.G44A |
| EHMT2 | BAT8, C6orf30, G9A, KMT1C, NG36 | Q96KQ7 | EHMT2 | Euchromatic histone-lysine N-methyltransferase 2, HLA-B-associated transcript 8, H3-K9-HMTase 3 | c.1155C>T | Missense | p.S384L |
| EP300 | P300 | Q09472 | p300 HAT | E1A-associated protein p300 | c.4549G>T c.4590G>A c.4154G>T |
Missense Missense Missense |
p.C1385F p.D1399N p.C1385F |
| H2BFWT | --- | Q7Z2G1 | Histone H2B type W-T | H2B histone family member W testis-specific | c.451C>T | Missense | p.R143C |
| H3F3A | H3.3A, H3F3, H3.3B | P84243 | H3.3 | --- | c.122C>T | Missense | p.R3C |
| H3F3C | --- | Q6NXT2 | H3.3C | H3.5 | c.439A>G c.172A>G |
Missense Missense |
p.K122E p.T33A |
| HDAC3 | --- | O15379 | HD3 | RPD3-2, SMAP45 | c.968G>T | Missense | p.R301L |
| HDAC4 | KIAA0288 | P56524 | HD4 | --- | c.3452G>T | Missense | p.G887V |
| HDAC9 | HDAC7, HDAC7B, HDRP, KIAA0744, MITR | Q9UKV0 | HD9 | HD7, HD7b, Histone deacetylase-related protein, MEF2-interacting transcription repressor MITR | c.2643C>A | Missense | p.H868N |
| HIST1H1B | H1F5 | P16401 | H1.5 | H1a, H1b, H1s-3 | c.727G>C | Missense | p.K225N |
| HIST1H1E | H1F4 | P10412 | H1.4 | H1b, H1s-4 | c.146G>A | Missense | p.G29D |
| HIST1H2AJ | H2AFE | Q99878 | H2A type 1-J | H2A/e | c.89G>A c.56C>T |
Missense Missense |
p.R30Q p.S19F |
| HIST1H2BF | HIST1H2BC, HIST1H2BE, HIST1H2BG, HIST1H2BI | P62807 | H2B type 1-C/E/F/G/I | H2B/a, H2B/g, H2B/h, H2B/k, H2B/l | c.229G>A c.152C>G |
Missense Missense |
p.E77K p.P51A |
| HIST1H2BH | H2BFJ | Q93079 | H2B type 1-H | H2B/j | c.9T>G c.67C>A |
Missense Missense |
p.D3E p.Q23K |
| HIST1H3A | HIST1H3B, HIST1H3C, HIST1H3D, HIST1H3E, HIST1H3F, HIST1H3G, HIST1H3H, HIST1H3I, HIST1H3J | P68431 | H3.1 | H3/a, H3/b, H3/c, H3/d, H3/f, H3/h, H3/I, H3/j, H3/k, H3/l | c.125A>G c.90G>A c.120A>T c.103A>T |
Missense Missense Missense Missense |
p.Y42C p.R27Q p.K37M p.T33S |
| HIST1H4B | HIST1H4A, HIST1H4C, HIST1H4D, HIST1H4E, HIST1H4F, HIST1H4H, HIST1H4I, HIST1H4J, HIST1H4K, HIST1H4L, HIST2H4A, HIST2H4B, HIST4H4 | P62805 | H4 | --- | c.202C>T c.77A>T c.162G>C c.11G>A c.262G>T c.118C>G c.39A>T |
Missense Missense Missense Missense Missense Missense |
p.R68W p.N26I p.E54D p.R4K p.V88F p.R40G p.K13I |
| HIST2H2AB | --- | Q8IUE6 | H2A type 2-B | --- | c.133G>A | Missense | p.G45R |
| HIST2H2BE | H2BFQ | Q16778 | H2B type 2-E | H2B-GL105, H2B/q | c.247G>A | Missense | p.D69N |
| KDM3A | JHDM2A, JMJD1, JMJD1A, KIAA0742, TSGA | Q9Y4C1 | Lysine-specific demethyla, se 3A | Jumonji domain-containing protein 1A | c.3698C>G c.4178C>A |
Missense Nonsense |
p.S1124C p.S1284 |
| KDM4A | JHDM3A, JMJD2, JMJD2A, KIAA0677 | O75164 | Lysine-specific demethylase 4A | Jumonji domain-containing protein 2A | c.174C>T c.503G>A c.2150G>T |
Missense Missense Missense |
p.S3F p.E113K p.A662S |
| KDM4B | JHDM3B, JMJD2B, KIAA0876 | O94953 | Lysine-specific demethylase 4B | Jumonji domain-containing protein 2B | c.1418T>C | Missense | p.S398P |
| KDM5A | JARID1A, RBBP2, RBP2 | P29375 | Lysine-specific demethylase 5A | Jumonji/ARID domain-containing protein 1A, RBBP-2 | c.5287G>C c.2708G>A |
Missense Missense |
p.G1642R p.R782Q |
| KDM5B | JARID1B, PLU1, RBBP2H1 | Q9UGL1 | Lysine-specific demethylase 5B | CT31, Histone demethylase JARID1B, Jumonji/ARID domain-containing protein 1B, PLU-1, RBP2-H1 | c.1741A>T c.2368G>A |
Missense Missense |
p.Y542F p.C751Y |
| KDM5C | DXS1272E, JARID1C, SMCX, XE169 | P41229 | Lysine-specific demethylase 5C | Histone demethylase JARID1C, Jumonji/ARID domain-containing protein 1C, SmcX, protein Xe169 | c.1539G>C | Missense | p.D336H |
| MYST1 | KAT8, MOF | Q9H7Z6 | MYST-1 | Lysine acetyltransferase 8, MOZ, YBF2/SAS3, SAS2 and TIP60 protein1, hMOF | c.829C>G | Missense | p.L271V |
| MYST2 | KAT7, HBO1, HBOa | O95251 | MYST-2 | KAT7, Histone acetyltransferase binding to ORC1 Lysine acetyltransferase 7, MOZ, YBF2/SAS3, SAS2 and TIP60 protein 2 | c.179C>T c.1016_1017delTT |
Missense Frame_Shift_Del |
p.S18F p.L297fs |
| MYST3 | MOZ, KAT6A, RUNXBP2, ZNF220 | Q92794 | MYST-3 | KAT6A, MOZ, YBF2/SAS3, SAS2 and TIP60 protein 3 | c.3999G>T | Missense | p.W1152L |
| NAT10 | ALP, KIAA1709 | Q9H0A0 | N-acetyltransferase 10 | c.1598G>A | Missense | p.A452T | |
| PHF21A | BHC80, KIAA1696 | Q96BD5 | PHD finger protein 21A | BHC80a BRAF35-HDAC complex protein BHC80 | c.2553C>G c.2491C>T |
Missense Missense |
p.I643M p.R623C |
| RCOR1 | KIAA0071, RCOR | Q9UKL0 | REST corepressor 1 | CoREST | c.e3_splice_site | Splice_Site_SNP | |
| RCOR2 | --- | Q8IZ40 | REST corepressor 2 | --- | c.731A>T | Nonsense | p.K115 |
| RCOR3 | KIAA1343 | Q9P2K3 | REST corepressor 3 | --- | c.559A>T | Missense | p.M122L |
| TNP2 | --- | Q05952 | TP-2 | --- | c.206C>A | Missense | p.S55R |
Data represents HNSCC mutations by Stransky N. et al, and Agrawal et al supplemented by UniProt database (http://www.uniprot.org/), and COSMIC (http://cancer.sanger.ac.uk/cancergenome/project/cosmic/). Only missense and nonsense mutations along with frame shift deletions and splice site SNP of histones and histones associated modifiers were included. Synonymous mutations were excluded.
Figure 1. HNSCC mutations in histones and histone associated transcription regulators.
(A) Bar graph of 74 sequenced HNSCC tumor samples [121] containing mutations in epigenetic associated genes that encode histones and their linkers, proteins associated with the recruitment of DNA-binding proteins, HDAC I and II interacting proteins, corepressor proteins and transcriptional activators and coactivators. (B) Bar graph depicting the percentage of mutations found in HNSCC samples (n=74). (Graph generated from the analysis of the HNSCC NGS supporting data published by Stransky et. al. 2011.
The Histone Lysine Demethylases (KDMs) are the second major family of genes mutated in HNSCC. KDMs, a new class of epigenetic enzymes, remove histone marks associated with gene expression and suppression. KDMs play a major role in prostate cancer by suppressing or promoting tumorigenesis [135].
NGS of HNSCC identified additional interesting gene mutations involved in transcription regulation. BCOR, and the BCORL1 encoded corepressor proteins are two examples of transcription regulators capable of selectively inhibit gene expression by interacting with the sequence specific DNA-binding protein BCL6. BCOR interacts with class I and II HDACs and may play an important role in repression. Both BCOR and BCORL1 are found in bone sarcomas and in acute myeloid leukemia [136,137]. The RCOR1 gene also found mutated in HNSCC encodes a corepressor protein that binds to the repressor element-1 silencing transcription factor (REST), but little is known about its involvement in tumor progression.
Transcriptional activators and coactivators are also mutated in HNSCC. Stransky’s NGS identified mutations in CREBBP and EP300, genes that encode the well-known CREB-binding protein and histone acetyltransferase p300 (p300 HAT) transcriptional activators. CREB-binding protein and histone acetyltransferase p300 coactivators have inherent histone acetyltransferase activity and stabilize proteins at the transcription complex. Mutations in CREBBP and EP300 are associated with the development of Rubinstein-Taybi syndrome and are involved in the development of acute myeloid leukemia. EP300 is mutated in several epithelial tumors, including gastric, breast, cervical, colorectal, pancreatic, and gastric carcinomas [59,60,138–140]. The MYST family of histone acetyltransferases is composed of five human HATs: Tip60 (KAT5), MOZ (MYST3), MORF (MYST4), HBO1 (MYST2) and MOF (MYST1). MYST1, MYST2, and MYST3 are mutated in HNSCC and were recently linked to cancer (reviewed in [141]). A putative role for MYST1 is in modulation of ATM and acetylation of multiple residues of histone 4 [142]. MYST2 is also involved in acetylation of histone 4, but it plays a larger role in DNA replication and is a common site for retroviral integration [143]. MYST2 occurs in a complex with the ING4 and ING5 tumor suppressors, suggesting it plays a suppressive role in carcinogenesis [144]. Similar to MYST2, MYST3 forms a complex with ING5, suggesting that mutations in MYST3 alter the ability of ING5 to regulate p53 function [145]. Lastly, mutation in the EHMT1 histone methyltransferase suggests that transcriptional repression is a potential mechanism involved in tumor progression. The protein encoded by EHMT1 is responsible for directly methylating lysine 9 of histone H3 and is responsible for Kleefstra syndrome (9q34 deletion syndrome) [146].
Conclusions
Increased knowledge of the biology of cancer is reflected in steady improvements in the overall survival rate of various cancers and the continuous reduction in cancer related death in the United States since the 1990s. The latest report on clinical advances in cancer therapy from the American Society of Clinical Oncology (ASCO) highlights the use of multitargeted drugs as the most efficient approach to manage different types of cancer. Emerging therapeutic strategies are using different drugs to target multiple components of common molecular pathways and have shown a positive impact on overall survival in patients with Gastrointestinal Stromal Tumors (GIST), metastatic colorectal carcinomas, neuroblastoma, Anaplastic Large-Cell Lymphoma (ALCL), medullary thyroid carcinoma, breast cancer, and metastatic sarcomas (http://www.cancerprogress. net). Unfortunately, the overall survival rate of HNSCC, one of the most common malignancies worldwide [147], has only marginally improved in the last 50 years despite significant efforts in identify new prognostic markers and understand the molecular signatures involved in tumor progression [148]. Until recently, the molecular circuitry associated with tumor invasion, metastasis and the development of chemoresistance was attributed to genetic instability driven by the accumulation of mutations. Recently, a better understanding of stem cells and developmental biology, in addition to technological advances in the field of full genome sequencing, are providing strong evidence that epigenetics equally contributes to cancer development.
Understanding the link between cancer mutations and epigenetic modulation of transcription will allow design of new therapeutic strategies that may improve the overall survival rate of HNSCC and other cancers. Advances in oncogenomics have also identified mutations in epigenetic associated genes that encode histones and their linkers, proteins associated with the recruitment of DNA-binding proteins, HDAC I and II interacting proteins, corepressor proteins and transcriptional activators and coactivators. Such mutations can dynamically confer tumor advantage over the transcriptional control of pro-survival pathways supporting tumor growth. The complexity of these interactions is evident in the differential effects of HDACi in in vitro models compared to the unpredictable outcomes observed in the clinic. Important questions remain to be answered including how mutations in histones and its modifiers influence tumor response to chemotherapeutic agents; how epigenetic signals presenting in the tumor microenvironment modulates tumor behavior; and how mutated epigenetic modifiers are capable of deregulate the balance between active and repressed transcriptional regions of the chromatin. Thus, understanding which gene mutations drive histone modifications of enhancer and promoter regions may help us understand tumor susceptibility to HAT and HDAC inhibitors and guide the development new combination therapies.
Acknowledgement
This work was funded by the National Institutes of Health (NIH/NCI) P50- CA97248 (University of Michigan Head and Neck SPORE). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions
MDM and RMC have contributed to the design, organization and writing of the review article.
References
- 1.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 2.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 3.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 4.Schafer S, Jung M. Chromatin modifications as targets for new anticancer drugs. Arch Pharm (Weinheim) 2005;338:347–357. doi: 10.1002/ardp.200500984. [DOI] [PubMed] [Google Scholar]
- 5.Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baxter J, Sauer S, Peters A, John R, Williams R, et al. Histone hypomethylation is an indicator of epigenetic plasticity in quiescent lymphocytes. EMBO J. 2004;23:4462–4472. doi: 10.1038/sj.emboj.7600414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bapat SA, Jin V, Berry N, Balch C, Sharma N, et al. Multivalent epigenetic marks confer microenvironment-responsive epigenetic plasticity to ovarian cancer cells. Epigenetics. 2010;5:716–729. doi: 10.4161/epi.5.8.13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berry NB, Bapat SA. Ovarian cancer plasticity and epigenomics in the acquisition of a stem-like phenotype. J Ovarian Res. 2008;1:8. doi: 10.1186/1757-2215-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giudice FS, Pinto DS, Jr, Nor JE, Squarize CH, Castilho RM. Inhibition of histone deacetylase impacts cancer stem cells and induces epithelial-mesenchyme transition of head and neck cancer. PLoS One. 2013;8:e58672. doi: 10.1371/journal.pone.0058672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 11.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 12.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 13.Gallinari P, Di Marco S, Jones P, Pallaoro M, Steinkuhler C. HDACs, histone deacetylation and gene transcription: from molecular biology to cancer therapeutics. Cell Res. 2007;17:195–211. doi: 10.1038/sj.cr.7310149. [DOI] [PubMed] [Google Scholar]
- 14.Rando OJ. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr Opin Genet Dev. 2012;22:148–155. doi: 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 16.Song SH, Han SW, Bang YJ. Epigenetic-based therapies in cancer: progress to date. Drugs. 2011;71:2391–2403. doi: 10.2165/11596690-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 17.Spotswood HT, Turner BM. An increasingly complex code. J Clin Invest. 2002;110:577–582. doi: 10.1172/JCI16547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lund AH, van Lohuizen M. Epigenetics and cancer. Genes Dev. 2004;18:2315–2335. doi: 10.1101/gad.1232504. [DOI] [PubMed] [Google Scholar]
- 19.Wolffe AP, Guschin D. Review: chromatin structural features and targets that regulate transcription. J Struct Biol. 2000;129:102–122. doi: 10.1006/jsbi.2000.4217. [DOI] [PubMed] [Google Scholar]
- 20.Blancafort P, Jin J, Frye S. Writing and rewriting the epigenetic code of cancer cells: from engineered proteins to small molecules. Mol Pharmacol. 2013;83:563–576. doi: 10.1124/mol.112.080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ropero S, Esteller M. The role of histone deacetylases (HDACs) in human cancer. Mol Oncol. 2007;1:19–25. doi: 10.1016/j.molonc.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan O, La Thangue NB. HDAC inhibitors in cancer biology: emerging mechanisms and clinical applications. Immunol Cell Biol. 2012;90:85–94. doi: 10.1038/icb.2011.100. [DOI] [PubMed] [Google Scholar]
- 23.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Yuan H, Marmorstein R. Histone acetyltransferases: Rising ancient counterparts to protein kinases. Biopolymers. 2013;99:98–111. doi: 10.1002/bip.22128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 26.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 27.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Li L, Pandey R, Byun JS, Gardner K, et al. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell. 2012;11:163–178. doi: 10.1016/j.stem.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun G, Fu C, Shen C, Shi Y. Histone deacetylases in neural stem cells and induced pluripotent stem cells. J Biomed Biotechnol. 2011;2011:835968. doi: 10.1155/2011/835968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 31.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Halkidou K, Gaughan L, Cook S, Leung HY, Neal DE, et al. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate. 2004;59:177–189. doi: 10.1002/pros.20022. [DOI] [PubMed] [Google Scholar]
- 33.Song J, Noh JH, Lee JH, Eun JW, Ahn YM, et al. Increased expression of histone deacetylase 2 is found in human gastric cancer. APMIS. 2005;113:264–268. doi: 10.1111/j.1600-0463.2005.apm_04.x. [DOI] [PubMed] [Google Scholar]
- 34.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 35.Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu G, Cui K, Northrup D, Liu C, Wang C, et al. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2013;12:180–192. doi: 10.1016/j.stem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, et al. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell. 2012;151:1608–1616. doi: 10.1016/j.cell.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 40.Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, et al. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- 41.Gorisch SM, Wachsmuth M, Toth KF, Lichter P, Rippe K. Histone acetylation increases chromatin accessibility. J Cell Sci. 2005;118:5825–5834. doi: 10.1242/jcs.02689. [DOI] [PubMed] [Google Scholar]
- 42.Kouraklis G, Theocharis S. Histone deacetylase inhibitors and anticancer therapy. Curr Med Chem Anticancer Agents. 2002;2:477–484. doi: 10.2174/1568011023353921. [DOI] [PubMed] [Google Scholar]
- 43.Turner BM. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- 44.Lagger G, O'Carroll D, Rembold M, Khier H, Tischler J, et al. Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lehrmann H, Pritchard LL, Harel-Bellan A. Histone acetyltransferases and deacetylases in the control of cell proliferation and differentiation. Adv Cancer Res. 2002;86:41–65. doi: 10.1016/s0065-230x(02)86002-x. [DOI] [PubMed] [Google Scholar]
- 46.Mellor J. Dynamic nucleosomes and gene transcription. Trends Genet. 2006;22:320–329. doi: 10.1016/j.tig.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 47.Richon VM, O'Brien JP. Histone deacetylase inhibitors: a new class of potential therapeutic agents for cancer treatment. Clin Cancer Res. 2002;8:662–664. [PubMed] [Google Scholar]
- 48.Acharya MR, Sparreboom A, Venitz J, Figg WD. Rational development of histone deacetylase inhibitors as anticancer agents: a review. Mol Pharmacol. 2005;68:917–932. doi: 10.1124/mol.105.014167. [DOI] [PubMed] [Google Scholar]
- 49.Caron C, Col E, Khochbin S. The viral control of cellular acetylation signaling. Bioessays. 2003;25:58–65. doi: 10.1002/bies.10202. [DOI] [PubMed] [Google Scholar]
- 50.Iyer NG, Ozdag H, Caldas C. p300/CBP and cancer. Oncogene. 2004;23:4225–4231. doi: 10.1038/sj.onc.1207118. [DOI] [PubMed] [Google Scholar]
- 51.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 52.Brooks CL, Gu W. The impact of acetylation and deacetylation on the p53 pathway. Protein Cell. 2011;2:456–462. doi: 10.1007/s13238-011-1063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 54.Luo J, Nikolaev AY, Imai S, Chen D, Su F, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 55.Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, et al. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci U S A. 2004;101:2259–2264. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang F, Marshall CB, Ikura M. Transcriptional/epigenetic regulator CBP/p300 in tumorigenesis: structural and functional versatility in target recognition. Cell Mol Life Sci. 2013 doi: 10.1007/s00018-012-1254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ichim G, Mola M, Finkbeiner MG, Cros MP, Herceg Z, et al. The histone acetyltransferase component TRRAP is targeted for destruction during the cell cycle. Oncogene. 2013 doi: 10.1038/onc.2012.570. [DOI] [PubMed] [Google Scholar]
- 58.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 59.Muraoka M, Konishi M, Kikuchi-Yanoshita R, Tanaka K, Shitara N, et al. p300 gene alterations in colorectal and gastric carcinomas. Oncogene. 1996;12:1565–1569. [PubMed] [Google Scholar]
- 60.Gayther SA, Batley SJ, Linger L, Bannister A, Thorpe K, et al. Mutations truncating the EP300 acetylase in human cancers. Nat Genet. 2000;24:300–303. doi: 10.1038/73536. [DOI] [PubMed] [Google Scholar]
- 61.Ozdag H, Batley SJ, Forsti A, Iyer NG, Daigo Y, et al. Mutation analysis of CBP and PCAF reveals rare inactivating mutations in cancer cell lines but not in primary tumours. Br J Cancer. 2002;87:1162–1165. doi: 10.1038/sj.bjc.6600554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kung AL, Rebel VI, Bronson RT, Ch'ng LE, Sieff CA, et al. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 2000;14:272–277. [PMC free article] [PubMed] [Google Scholar]
- 63.Kang-Decker N, Tong C, Boussouar F, Baker DJ, Xu W, et al. Loss of CBP causes T cell lymphomagenesis in synergy with p27Kip1 insufficiency. Cancer Cell. 2004;5:177–189. doi: 10.1016/s1535-6108(04)00022-4. [DOI] [PubMed] [Google Scholar]
- 64.Borrow J, Stanton VP, Jr, Andresen JM, Becher R, Behm FG, et al. The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nat Genet. 1996;14:33–41. doi: 10.1038/ng0996-33. [DOI] [PubMed] [Google Scholar]
- 65.Sobulo OM, Borrow J, Tomek R, Reshmi S, Harden A, et al. MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t(11;16)(q23;p13.3) Proc Natl Acad Sci U S A. 1997;94:8732–8737. doi: 10.1073/pnas.94.16.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Panagopoulos I, Fioretos T, Isaksson M, Samuelsson U, Billstrom R, et al. Fusion of the MORF and CBP genes in acute myeloid leukemia with the t(10;16)(q22;p13) Hum Mol Genet. 2001;10:395–404. doi: 10.1093/hmg/10.4.395. [DOI] [PubMed] [Google Scholar]
- 67.Liu X, Wang D, Zhao Y, Tu B, Zheng Z, et al. Methyltransferase Set7/9 regulates p53 activity by interacting with Sirtuin 1 (SIRT1) Proc Natl Acad Sci U S A. 2011;108:1925–1930. doi: 10.1073/pnas.1019619108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Santer FR, Hoschele PP, Oh SJ, Erb HH, Bouchal J, et al. Inhibition of the acetyltransferases p300 and CBP reveals a targetable function for p300 in the survival and invasion pathways of prostate cancer cell lines. Mol Cancer Ther. 2011;10:1644–1655. doi: 10.1158/1535-7163.MCT-11-0182. [DOI] [PubMed] [Google Scholar]
- 69.Nathan D, Sterner DE, Berger SL. Histone modifications: Now summoning sumoylation. Proc Natl Acad Sci U S A. 2003;100:13118–13120. doi: 10.1073/pnas.2436173100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc Natl Acad Sci U S A. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26:5541–5552. doi: 10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 72.Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: What are the cancer relevant targets? Cancer Lett. 2009;277:8–21. doi: 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- 73.Segre CV, Chiocca S. Regulating the regulators: the post-translational code of class I HDAC1 and HDAC2. J Biomed Biotechnol. 2011;2011:690848. doi: 10.1155/2011/690848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zupkovitz G, Grausenburger R, Brunmeir R, Senese S, Tischler J, et al. The cyclin-dependent kinase inhibitor p21 is a crucial target for histone deacetylase 1 as a regulator of cellular proliferation. Mol Cell Biol. 2010;30:1171–1181. doi: 10.1128/MCB.01500-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rikimaru T, Taketomi A, Yamashita Y, Shirabe K, Hamatsu T, et al. Clinical significance of histone deacetylase 1 expression in patients with hepatocellular carcinoma. Oncology. 2007;72:69–74. doi: 10.1159/000111106. [DOI] [PubMed] [Google Scholar]
- 76.Weichert W, Roske A, Gekeler V, Beckers T, Stephan C, et al. Histone deacetylases 1, 2 and 3 are highly expressed in prostate cancer and HDAC2 expression is associated with shorter PSA relapse time after radical prostatectomy. Br J Cancer. 2008;98:604–610. doi: 10.1038/sj.bjc.6604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weichert W. HDAC expression and clinical prognosis in human malignancies. Cancer Lett. 2009;280:168–176. doi: 10.1016/j.canlet.2008.10.047. [DOI] [PubMed] [Google Scholar]
- 78.Senese S, Zaragoza K, Minardi S, Muradore I, Ronzoni S, et al. Role for histone deacetylase 1 in human tumor cell proliferation. Mol Cell Biol. 2007;27:4784–4795. doi: 10.1128/MCB.00494-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mariadason JM. Making sense of HDAC2 mutations in colon cancer. Gastroenterology. 2008;135:1457–1459. doi: 10.1053/j.gastro.2008.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Olmos Y, Brosens JJ, Lam EW. Interplay between SIRT proteins and tumour suppressor transcription factors in chemotherapeutic resistance of cancer. Drug Resist Updat. 2011;14:35–44. doi: 10.1016/j.drup.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 82.Frazzi R, Valli R, Tamagnini I, Casali B, Latruffe N, et al. Resveratrol-mediated apoptosis of hodgkin lymphoma cells involves SIRT1 inhibition and FOXO3a hyperacetylation. Int J Cancer. 2013;132:1013–1021. doi: 10.1002/ijc.27748. [DOI] [PubMed] [Google Scholar]
- 83.Kriegl L, Vieth M, Kirchner T, Menssen A. Up-regulation of c-MYC and SIRT1 expression correlates with malignant transformation in the serrated route to colorectal cancer. Oncotarget. 2012;3:1182–1193. doi: 10.18632/oncotarget.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Firestein R, Blander G, Michan S, Oberdoerffer P, Ogino S, et al. The SIRT1 deacetylase suppresses intestinal tumorigenesis and colon cancer growth. PLoS One. 2008;3:e2020. doi: 10.1371/journal.pone.0002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kabra N, Li Z, Chen L, Li B, Zhang X, et al. SirT1 is an inhibitor of proliferation and tumor formation in colon cancer. J Biol Chem. 2009;284:18210–18217. doi: 10.1074/jbc.M109.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cho IR, Koh SS, Malilas W, Srisuttee R, Moon J, et al. SIRT1 inhibits proliferation of pancreatic cancer cells expressing pancreatic adenocarcinoma up-regulated factor (PAUF), a novel oncogene, by suppression of beta-catenin. Biochem Biophys Res Commun. 2012;423:270–275. doi: 10.1016/j.bbrc.2012.05.107. [DOI] [PubMed] [Google Scholar]
- 87.Bosch-Presegue L, Vaquero A. The dual role of sirtuins in cancer. Genes Cancer. 2011;2:648–662. doi: 10.1177/1947601911417862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Monneret C. Histone deacetylase inhibitors. Eur J Med Chem. 2005;40:1–13. doi: 10.1016/j.ejmech.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 89.Baradari V, Huether A, Hopfner M, Schuppan D, Scherubl H. Antiproliferative and proapoptotic effects of histone deacetylase inhibitors on gastrointestinal neuroendocrine tumor cells. Endocr Relat Cancer. 2006;13:1237–1250. doi: 10.1677/erc.1.01249. [DOI] [PubMed] [Google Scholar]
- 90.Lawlor ER, Thiele CJ. Epigenetic changes in pediatric solid tumors: promising new targets. Clin Cancer Res. 2012;18:2768–2779. doi: 10.1158/1078-0432.CCR-11-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Erlich RB, Kherrouche Z, Rickwood D, Endo-Munoz L, Cameron S, et al. Preclinical evaluation of dual PI3K-mTOR inhibitors and histone deacetylase inhibitors in head and neck squamous cell carcinoma. Br J Cancer. 2012;106:107–115. doi: 10.1038/bjc.2011.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cherenkov VG. Use of key clinical symptoms in oncology for activating the educational process. Vopr Onkol. 1988;34:92–97. [PubMed] [Google Scholar]
- 93.Licciardi PV, Karagiannis TC. Regulation of immune responses by histone deacetylase inhibitors. ISRN Hematol. 2012;2012:690901. doi: 10.5402/2012/690901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bi G, Jiang G. The molecular mechanism of HDAC inhibitors in anticancer effects. Cell Mol Immunol. 2006;3:285–290. [PubMed] [Google Scholar]
- 95.Heider U, Kaiser M, Sterz J, Zavrski I, Jakob C, et al. Histone deacetylase inhibitors reduce VEGF production and induce growth suppression and apoptosis in human mantle cell lymphoma. Eur J Haematol. 2006;76:42–50. doi: 10.1111/j.1600-0609.2005.00546.x. [DOI] [PubMed] [Google Scholar]
- 96.Blumenschein G., Jr Sorafenib in lung cancer: clinical developments and future directions. J Thorac Oncol. 2008;3:S124–S127. doi: 10.1097/JTO.0b013e318174e085. [DOI] [PubMed] [Google Scholar]
- 97.Grant S, Easley C, Kirkpatrick P. Vorinostat. Nat Rev Drug Discov. 2007;6:21–22. doi: 10.1038/nrd2227. [DOI] [PubMed] [Google Scholar]
- 98.StatBite: FDA oncology drug product approvals in 2009. J Natl Cancer Inst. 2010;102:219. doi: 10.1093/jnci/djq030. [DOI] [PubMed] [Google Scholar]
- 99.Piekarz RL, Frye R, Turner M, Wright JJ, Allen SL, et al. Phase II multi-institutional trial of the histone deacetylase inhibitor romidepsin as monotherapy for patients with cutaneous T-cell lymphoma. J Clin Oncol. 2009;27:5410–5417. doi: 10.1200/JCO.2008.21.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qiu T, Zhou L, Zhu W, Wang T, Wang J, et al. Effects of treatment with histone deacetylase inhibitors in solid tumors: a review based on 30 clinical trials. Future Oncol. 2013;9:255–269. doi: 10.2217/fon.12.173. [DOI] [PubMed] [Google Scholar]
- 101.Thurn KT, Thomas S, Moore A, Munster PN. Rational therapeutic combinations with histone deacetylase inhibitors for the treatment of cancer. Future Oncol. 2011;7:263–283. doi: 10.2217/fon.11.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bansal SK. The recommended measure. Indian Pediatr. 1988;25:1226. [PubMed] [Google Scholar]
- 103.Murakami Y, Hotei H, Tsumura H, Kohmo N, Nakai S, et al. A case of right adrenal myelolipoma diagnosed preoperatively and review of literature in Japan. Nihon Geka Gakkai Zasshi. 1988;89:464–469. [PubMed] [Google Scholar]
- 104.Gillenwater AM, Zhong M, Lotan R. Histone deacetylase inhibitor suberoylanilide hydroxamic acid induces apoptosis through both mitochondrial and Fas (Cd95) signaling in head and neck squamous carcinoma cells. Mol Cancer Ther. 2007;6:2967–2975. doi: 10.1158/1535-7163.MCT-04-0344. [DOI] [PubMed] [Google Scholar]
- 105.Gan CP, Hamid S, Hor SY, Zain RB, Ismail SM, et al. Valproic acid: growth inhibition of head and neck cancer by induction of terminal differentiation and senescence. Head Neck. 2012;34:344–353. doi: 10.1002/hed.21734. [DOI] [PubMed] [Google Scholar]
- 106.Kano M, Yamada S, Hoshino I, Murakami K, Akutsu Y, et al. Effects of carbon-ion radiotherapy combined with a novel histone deacetylase inhibitor, cyclic hydroxamic-acid-containing peptide 31 in human esophageal squamous cell carcinoma. Anticancer Res. 2009;29:4433–4438. [PubMed] [Google Scholar]
- 107.Groselj B, Sharma NL, Hamdy FC, Kerr M, Kiltie AE. Histone deacetylase inhibitors as radiosensitisers: effects on DNA damage signalling and repair. Br J Cancer. 2013;108:748–754. doi: 10.1038/bjc.2013.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kim J, Guan J, Chang I, Chen X, Han D, et al. PS-341 and histone deacetylase inhibitor synergistically induce apoptosis in head and neck squamous cell carcinoma cells. Mol Cancer Ther. 2010;9:1977–1984. doi: 10.1158/1535-7163.MCT-10-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tan M, Luo H, Lee S, Jin F, Yang JS, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hong L, Schroth GP, Matthews HR, Yau P, Bradbury EM. Studies of the DNA binding properties of histone H4 amino terminus. Thermal denaturation studies reveal that acetylation markedly reduces the binding constant of the H4 "tail" to DNA. J Biol Chem. 1993;268:305–314. [PubMed] [Google Scholar]
- 111.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 112.Vettese-Dadey M, Grant PA, Hebbes TR, Crane-Robinson C, Allis CD, et al. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 113.Megee PC, Morgan BA, Smith MM. Histone H4 and the maintenance of genome integrity. Genes Dev. 1995;9:1716–1727. doi: 10.1101/gad.9.14.1716. [DOI] [PubMed] [Google Scholar]
- 114.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 115.Kapoor-Vazirani P, Kagey JD, Powell DR, Vertino PM. Role of hMOF-dependent histone H4 lysine 16 acetylation in the maintenance of TMS1/ASC gene activity. Cancer Res. 2008;68:6810–6821. doi: 10.1158/0008-5472.CAN-08-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cho B, Kim HJ, Kim H, Sun W. Changes in the Histone Acetylation Patterns during the Development of the Nervous System. Exp Neurobiol. 2011;20:81–84. doi: 10.5607/en.2011.20.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shikama N, Lutz W, Kretzschmar R, Sauter N, Roth JF, et al. Essential function of p300 acetyltransferase activity in heart, lung and small intestine formation. EMBO J. 2003;22:5175–5185. doi: 10.1093/emboj/cdg502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 119.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 120.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 121.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Psyrri A, Gouveris P, Vermorken JB. Human papillomavirus-related head and neck tumors: clinical and research implication. Curr Opin Oncol. 2009;21:201–205. doi: 10.1097/cco.0b013e328329ab64. [DOI] [PubMed] [Google Scholar]
- 124.Eriksen JG, Lassen P, Overgaard J. Do all patients with head and neck cancer benefit from radiotherapy and concurrent cetuximab? Lancet Oncol. 2010;11:312–313. doi: 10.1016/S1470-2045(10)70035-8. [DOI] [PubMed] [Google Scholar]
- 125.Lassen P. The role of Human papillomavirus in head and neck cancer and the impact on radiotherapy outcome. Radiother Oncol. 2010;95:371–380. doi: 10.1016/j.radonc.2010.04.022. [DOI] [PubMed] [Google Scholar]
- 126.Wu X, Northcott PA, Dubuc A, Dupuy AJ, Shih DJ, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature. 2012;482:529–533. doi: 10.1038/nature10825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 128.Guo G, Gui Y, Gao S, Tang A, Hu X, et al. Frequent mutations of genes encoding ubiquitin-mediated proteolysis pathway components in clear cell renal cell carcinoma. Nat Genet. 2012;44:17–19. doi: 10.1038/ng.1014. [DOI] [PubMed] [Google Scholar]
- 129.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Huynh VA, Robinson PJ, Rhodes D. A method for the in vitro reconstitution of a defined "30 nm" chromatin fibre containing stoichiometric amounts of the linker histone. J Mol Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 131.Butler PJ, Thomas JO. Changes in chromatin folding in solution. J Mol Biol. 1980;140:505–529. doi: 10.1016/0022-2836(80)90268-5. [DOI] [PubMed] [Google Scholar]
- 132.Talasz H, Sapojnikova N, Helliger W, Lindner H, Puschendorf B. In vitro binding of H1 histone subtypes to nucleosomal organized mouse mammary tumor virus long terminal repeat promotor. J Biol Chem. 1998;273:32236–32243. doi: 10.1074/jbc.273.48.32236. [DOI] [PubMed] [Google Scholar]
- 133.Orrego M, Ponte I, Roque A, Buschati N, Mora X, et al. Differential affinity of mammalian histone H1 somatic subtypes for DNA and chromatin. BMC Biol. 2007;5:22. doi: 10.1186/1741-7007-5-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Maresca TJ, Freedman BS, Heald R. Histone H1 is essential for mitotic chromosome architecture and segregation in Xenopus laevis egg extracts. J Cell Biol. 2005;169:859–869. doi: 10.1083/jcb.200503031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Crea F, Sun L, Mai A, Chiang YT, Farrar WL, et al. The emerging role of histone lysine demethylases in prostate cancer. Mol Cancer. 2012;11:52. doi: 10.1186/1476-4598-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pierron G, Tirode F, Lucchesi C, Reynaud S, Ballet S, et al. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat Genet. 2012;44:461–466. doi: 10.1038/ng.1107. [DOI] [PubMed] [Google Scholar]
- 137.Tiacci E, Grossmann V, Martelli MP, Kohlmann A, Haferlach T, et al. The corepressors BCOR and BCORL1: two novel players in acute myeloid leukemia. Haematologica. 2012;97:3–5. doi: 10.3324/haematol.2011.057901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ionov Y, Matsui S, Cowell JK. A role for p300/CREB binding protein genes in promoting cancer progression in colon cancer cell lines with microsatellite instability. Proc Natl Acad Sci U S A. 2004;101:1273–1278. doi: 10.1073/pnas.0307276101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bourguignon LY, Xia W, Wong G. Hyaluronan-mediated CD44 interaction with p300 and SIRT1 regulates beta-catenin signaling and NFkappaB-specific transcription activity leading to MDR1 and Bcl-xL gene expression and chemoresistance in breast tumor cells. J Biol Chem. 2009;284:2657–2671. doi: 10.1074/jbc.M806708200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ohshima T, Suganuma T, Ikeda M. A novel mutation lacking the bromodomain of the transcriptional coactivator p300 in the SiHa cervical carcinoma cell line. Biochem Biophys Res Commun. 2001;281:569–575. doi: 10.1006/bbrc.2001.4389. [DOI] [PubMed] [Google Scholar]
- 141.Avvakumov N, Cote J. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene. 2007;26:5395–5407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- 142.Vonlaufen N, Naguleswaran A, Coppens I, Sullivan WJ., Jr MYST family lysine acetyltransferase facilitates ataxia telangiectasia mutated (ATM) kinase-mediated DNA damage response in Toxoplasma gondii. J Biol Chem. 2010;285:11154–11161. doi: 10.1074/jbc.M109.066134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Suzuki T, Shen H, Akagi K, Morse HC, Malley JD, et al. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002;32:166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 144.Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 145.Shiseki M, Nagashima M, Pedeux RM, Kitahama-Shiseki M, Miura K, et al. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 2003;63:2373–2378. [PubMed] [Google Scholar]
- 146.Kleefstra T, Smidt M, Banning MJ, Oudakker AR, Van Esch H, et al. Disruption of the gene Euchromatin Histone Methyl Transferase1 (Eu-HMTase1) is associated with the 9q34 subtelomeric deletion syndrome. J Med Genet. 2005;42:299–306. doi: 10.1136/jmg.2004.028464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 148.Molinolo AA, Amornphimoltham P, Squarize CH, Castilho RM, Patel V, et al. Dysregulated molecular networks in head and neck carcinogenesis. Oral Oncol. 2009;45:324–334. doi: 10.1016/j.oraloncology.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]