Abstract
BACKGROUND
Expanded GGGGCC hexanucleotide repeats, ranging from hundreds to thousands in number, located in the noncoding region of the chromosome 9 open reading frame 72 (C9orf72) gene represent the most common genetic abnormality for familial and sporadic amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (abbreviated as C9ALS). Currently, three pathological mechanisms, such as haplo insufficiency of C9orf72, formation of nuclear RNA foci composed of sense and antisense repeats, and accumulation of unconventionally transcribed dipeptide-repeat (DPR) proteins, are proposed for C9ALS. However, at present, the central mechanism underlying neurodegeneration in C9ALS remains largely unknown.
METHODS
By using three distinct pathway analysis tools of bioinformatics, we studied molecular networks involved in C9ALS pathology by focusing on C9orf72 omics datasets, such as proteome of C9orf72 repeat RNA-binding proteins, transcriptome of induced pluripotent stem cells (iPSC)-derived motor neurons of patients with C9ALS, and transcriptome of purified motor neurons of patients with C9ALS.
RESULTS
We found that C9orf72 repeat RNA-binding proteins play a crucial role in the regulation of post-transcriptional RNA processing. The expression of a wide range of extracellular matrix proteins and matrix metalloproteinases was reduced in iPSC-derived motor neurons of patients with C9ALS. The regulation of RNA processing and cytoskeletal dynamics is disturbed in motor neurons of patients with C9ALS in vivo.
CONCLUSIONS
Bioinformatics data mining approach suggests a logical hypothesis that C9orf72 repeat expansions that deregulate post-transcriptional RNA processing disturb the homeostasis of cytoskeletal dynamics and remodeling of extracellular matrix, leading to degeneration of stress-vulnerable neurons in the brain and spinal cord of patients with C9ALS.
Keywords: bioinformatics, amyotrophic lateral sclerosis, C9orf72, frontotemporal dementia, KeyMolnet, molecular network, omics
Introduction
Recent evidence indicates that expanded GGGGCC hexanucleotide repeats, ranging from hundreds to thousands in number, located in the noncoding region of the chromosome 9 open reading frame 72 (C9orf72) gene represent the most common genetic abnormality for familial and sporadic amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD).1,2 Both constitute an overlapping continuum of a multisystem disorder affecting the central nervous system (CNS), abbreviated as C9ALS. C9orf72 is an evolutionarily conserved protein expressed most abundantly in neurons in the CNS, distantly related to the differentially expressed in normal and neoplastic cells (DENN) family of GDP–GTP exchange factors that activate Rab GTPases.3,4 Although the precise biological function of C9orf72 remains mostly unknown, C9orf72 regulates Rab GTPase-mediated endosome trafficking.5 The genetic mutations of C9orf72 are inherited in an autosomal dominant manner with incomplete penetrance. The patients with C9orf72 repeat expansions exhibit a clinical phenotype characterized by the earlier disease onset with bulbar involvement, cognitive and behavioral impairment, psychosis, and parkinsonism.6,7 The brains of patients with C9ALS not only show the TAR DNA-binding protein-43 (TDP-43) pathology but also exhibit an accumulation of TDP-43-negative p62-positive neuronal cytoplasmic and nuclear inclusions in the cerebellar granular cell layer and the dentate gyrus of the hippocampus.8
Currently, three pathological mechanisms are proposed for C9ALS.9,10 First, noncoding C9orf72 repeat expansions cause transcriptional silencing of the C9orf72 gene by epigenetic mechanisms involving hypermethylation of CpG islands and histone trimethylation, leading to haploinsufficiency causing loss of normal function of C9orf72.11,12 Expanded hexanucleotide repeats constitute thermodynamically stable, multimolecular G-quadruplex structures that directly interfere with transcription.13 Actually, C9orf72 protein levels are greatly reduced in brain tissues of patients with C9ALS.14 Furthermore, deletion of the C9orf72 ortholog causes degeneration of motor neurons in zebrafish and Caenorhabditis elegans, suggesting that loss of normal function of C9orf72 is detrimental for motor neuron survival.15,16 Second, messenger RNAs coding for expanded C9orf72 repeats, transcribed from sense and antisense strands of the mutated allele, are concentrated on nuclear RNA foci that sequester a panel of RNA-binding proteins, leading to aberrant mRNA splicing and processing of the genes pivotal for neuronal function.17,18 Third, bidirectional transcripts of expanded C9orf72 repeats are unconventionally transcribed in all the three reading frames into aggregation-prone dipeptide-repeat (DPR) proteins via repeat-associated non-ATG (RAN) translation.19,20 DPR proteins, accumulated in TDP-43-negative p62-positive neuronal cytoplasmic and nuclear inclusions in the early stage before the onset of TDP-43 pathology, could affect neuronal survival. However, at present, the central molecular mechanism underlying neurodegeneration in patients with C9ALS remains unknown.
Owing to the recent advance in microarray and next-generation sequencing technologies after the completion of the Human Genome Project, the global analysis of genome, transcriptome, proteome, and metabolome, collectively termed omics, helps us to characterize the genome-wide molecular basis of diseases and to identify disease-specific molecular signatures and biomarkers. Because omics approach produces high-throughput experimental data at one time, it is often difficult to extract biologically meaningful implications from these datasets. Recent progress in bioinformatics and systems biology enables us to illustrate the cell-wide map of complex molecular interactions with the aid of the literature-based knowledge base of molecular pathways.21 The logically arranged molecular networks construct the whole system characterized by robustness that maintains the proper function of the system in the face of genetic and environmental perturbations.22 In the scale-free molecular network, targeted disruption of limited numbers of critical components designated hubs, on which the biologically important molecular connections concentrate, could disturb the whole cellular function by destabilizing the network. Therefore, the integration of omics data derived from disease-affected cells and tissues with underlying molecular networks provides the highly efficient and rational approach for characterizing disease-relevant pathways to build up the most reasonable working hypothesis for disease mechanisms.
To establish a logically supported hypothesis accounting for the pathological role of C9orf72, we characterized molecular networks involved in C9ALS pathology by using three distinct pathway analysis tools of bioinformatics. We selected three C9orf72 omics datasets ideal for molecular network analysis, such as proteome of C9orf72 hexanucleotide repeat RNA-binding proteins, transcriptome of induced pluripotent stem cells (iPSC)-derived motor neurons of patients with C9ALS and transcriptome of purified motor neurons of patients with C9ALS.
Methods
RNA pull-down assay dataset of C9orf72 hexanucleotide repeat RNA-binding proteins
We studied two comprehensive datasets of C9orf72 hexanucleotide repeat RNA-binding proteins in human cells identified by recent studies.13,23 One is the dataset that comprised 288 interactors for biotinylated (GGGGCC)4 RNAs in G-quadruplex and hairpin conformations in HEK293T cells labeled by stable isotope labeling amino acids in cell culture (SIALC).13 They were identified by RNA pull-down assay, followed by mass spectrometry. The other is the dataset containing 103 interactors for biotinylated (GGGGCC)5 RNA in human cerebellum homogenates and SH-SY5Y extracts.23 They were identified by UV crosslinking and RNA pull-down assay, followed by mass spectrometry. We combined both datasets, and searched the presence of RNA recognition motif termed RRM (SM00360) on target proteins by Simple Modular Architecture Research Tool (SMART) v7 (smart.embl.de) and prion-like domain (PrLD)-containing proteins by referring to the established dataset.24
RNA-Seq dataset of iPSC-derived motor neurons of patients with C9ALS
We retrieved a RNA deep sequencing (RNA-Seq) dataset from Gene Expression Omnibus under the accession number GSE52202 (SRP032798). It contains eight transcriptome data derived from iPSC-derived choline acetyltransferase-positive motor neurons established from four normal controls (Group 1: 00i, 03i, 14i, and 83i) and four patients with C9ALS (Group 2: 28i, 29i, 30i, and 52i).25 The patients numbered 28i, 29i, and 52i expressed ~800 repeats, while the patient 30i showed ~70 repeats. Transcriptome profiling was performed by single end-sequencing on HiSeq 2000 (Illumina). After removing poly-A tails and low-quality reads from the original data, we mapped short reads on the human genome reference sequence hg19 by TopHat2.0.9 (ccb.jhu.edu/software/tophat/index.shtml), and identified differentially expressed genes that satisfy the significance expressed as q-value (FDR-adjusted P value) < 0.05 by Cufflinks2.1.1 (cufflinks.cbcb.umd.edu). The processed data were visualized on CummeRbund (compbio.mit.edu/cummerbund).
Exon array dataset of purified motor neurons of patients with C9ALS
We retrieved an exon array dataset of postmortem frozen tissues from the recent publication.26 It contains 12 transcriptome data derived from cervical cord motor neurons purely isolated by laser capture microdissection (LCM). They are derived from six normal controls, three patients with sporadic ALS, and three patients with C9ALS. Transcriptome profiling was performed on Human Exon 1.0 ST Array (Affymetrix), followed by gene- and exon-level analysis using the Partek Genomics Suite (www.partek.com/pgs). We extracted the set of genes and exons that satisfy the significance of P < 0.01 and fold change either >1.5 or <−1.5 from the original dataset comprising 742 genes and 6449 exons differentially expressed between total number of patients with ALS (n = 6) and controls (n = 6).26
Molecular network analysis
To identify biologically relevant molecular networks and pathways, we utilized three distinct pathway analysis tools of bioinformatics endowed with the comprehensive knowledge base. They include Kyoto Encyclopedia of Genes and Genomes (KEGG) (www.kegg.jp), Ingenuity Pathways Analysis (IPA) (Ingenuity Systems; www.ingenuity.com), and KeyMolnet (KeyMolnet Data; www.km-data.jp/keymolnet). First, we imported Entrez Gene IDs of corresponding genes into the Functional Annotation tool of Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (david.abcc.ncifcrf.gov).27 DAVID automatically identifies KEGG pathways, and Gene Ontology (GO) categories composed of the genes enriched in the given set with statistical significance evaluated by the modified Fisher’s exact test with Bonferroni multiple comparison test. KEGG includes manually curated reference pathways that cover a wide range of metabolic, genetic, environmental, and cellular processes, and human diseases.28 Currently, KEGG contains 302,965 distinct pathways generated from 463 reference pathways.
IPA is a knowledge base that contains approximately 3,000,000 biological and chemical interactions and functional annotations with definite scientific evidence. By uploading the list of Gene IDs, the network-generation algorithm identifies focused genes integrated in a global molecular network. IPA calculates the score P value that reflects the statistical significance of association between the genes and the networks by the Fisher’s exact test.
KeyMolnet contains knowledge-based contents on 164,000 relationships among human genes and proteins, small molecules, diseases, pathways and drugs.21 They include the core contents collected from selected review articles with the highest reliability. By importing the list of Gene ID, KeyMolnet automatically provides corresponding molecules as nodes on the network. The neighboring network-search algorithm selected one or more molecules as starting points to generate the network of all kinds of molecular interactions around starting molecules, including direct activation/inactivation, transcriptional activation/repression, and the complex formation within one path from starting points. The generated network was compared side by side with 501 human canonical pathways of the KeyMolnet library. The algorithm counting the number of overlapping molecular relations between the extracted network and the canonical pathway makes it possible to identify the canonical pathway showing the most significant contribution to the extracted network.21
Results
Molecular network of C9orf72 hexanucleotide repeat RNA-binding proteins
First, we extracted the set of 353 human C9orf72 hexanucleotide repeat RNA-binding proteins from two proteome datasets (Supplementary Table 1). They included 38 proteins overlapped between them. SMART revealed that 65 proteins (18.4%) have one or more RRM domains, among them 17 proteins have PrLD (Table 1). Importantly, the list contains a wide variety of pre-mRNA splicing factors, along with FUS, EWSR1, HNRNPA1, HNRNPA2B1, and TAF15, all having RRM and PrLD, whose genetic mutations are causative of ALS.29–32
Table 1.
The set of 65 C9orf72 hexanucleoride repeat RNA-binding proteins with RRM motifs.
| ENTREZ GENE ID | GENE SYMBOL | GENE NAME | PrLD |
|---|---|---|---|
| 10189 | ALYREF | Aly/REF export factor | |
| 1153 | CIRBP | cold inducible RNA binding protein | |
| 11052 | CPSF6 | cleavage and polyadenylation specific factor 6, 68kDa | |
| 79869 | CPSF7 | cleavage and polyadenylation specific factor 7, 59kDa | |
| 26528 | DAZAP1 | DAZ associated protein 1 | yes |
| 8662 | EIF3B | eukaryotic translation initiation factor 3, subunit B | |
| 8666 | EIF3G | eukaryotic translation initiation factor 3, subunit G | |
| 1975 | EIF4B | eukaryotic translation initiation factor 4B | |
| 7458 | EIF4H | eukaryotic translation initiation factor 4H | |
| 2130 | EWSR1 | Ewing sarcoma breakpoint region 1 | yes |
| 2521 | FUS | fusion (involved in t(12;16) in malignant liposarcoma) | yes |
| 10146 | G3BP1 | GTPase activating protein (SH3 domain) binding protein 1 | |
| 9908 | G3BP2 | GTPase activating protein (SH3 domain) binding protein 2 | |
| 2926 | GRSF1 | G-rich RNA sequence binding factor 1 | |
| 10949 | HNRNPA0 | heterogeneous nuclear ribonucleoprotein A0 | yes |
| 3178 | HNRNPA1 | heterogeneous nuclear ribonucleoprotein A1 | yes |
| 3181 | HNRNPA2B1 | heterogeneous nuclear ribonucleoprotein A2/B1 | yes |
| 220988 | HNRNPA3 | heterogeneous nuclear ribonucleoprotein A3 | yes |
| 3182 | HNRNPAB | heterogeneous nuclear ribonucleoprotein A/B | yes |
| 3183 | HNRNPC | heterogeneous nuclear ribonucleoprotein C (C1/C2) | |
| 3184 | HNRNPD | heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) | yes |
| 3185 | HNRNPF | heterogeneous nuclear ribonucleoprotein F | |
| 3187 | HNRNPH1 | heterogeneous nuclear ribonucleoprotein H1 (H) | yes |
| 3188 | HNRNPH2 | heterogeneous nuclear ribonucleoprotein H2 (H’) | yes |
| 3189 | HNRNPH3 | heterogeneous nuclear ribonucleoprotein H3 (2H9) | yes |
| 3191 | HNRNPL | heterogeneous nuclear ribonucleoprotein L | |
| 4670 | HNRNPM | heterogeneous nuclear ribonucleoprotein M | |
| 10236 | HNRNPR | heterogeneous nuclear ribonucleoprotein R | |
| 9987 | HNRPDL | heterogeneous nuclear ribonucleoprotein D-like | yes |
| 10642 | IGF2BP1 | insulin-like growth factor 2 mRNA binding protein 1 | |
| 9782 | MATR3 | matrin 3 | |
| 4691 | NCL | nucleolin | |
| 7936 | NELFE | negative elongation factor complex member E | |
| 4841 | NONO | non-POU domain containing, octamer-binding | |
| 26986 | PABPC1 | poly(A) binding protein, cytoplasmic 1 | |
| 8761 | PABPC4 | poly(A) binding protein, cytoplasmic 4 (inducible form) | |
| 84271 | POLDIP3 | polymerase (DNA-directed), delta interacting protein 3 | |
| 55269 | PSPC1 | paraspeckle component 1 | yes |
| 125950 | RAVER1 | ribonucleoprotein, PTB-binding 1 | |
| 8241 | RBM10 | RNA binding motif protein 10 | |
| 10432 | RBM14 | RNA binding motif protein 14 | yes |
| 84991 | RBM17 | RNA binding motif protein 17 | |
| 55696 | RBM22 | RNA binding motif protein 22 | |
| 5935 | RBM3 | RNA binding motif (RNP1, RRM) protein 3 | |
| 9584 | RBM39 | RNA binding motif protein 39 | |
| 9939 | RBM8a | RNA binding motif protein 8A | |
| 27316 | RBMX | RNA binding motif protein, X-linked | |
| 6294 | SAFB | scaffold attachment factor B | |
| 57466 | SCAF4 | SR-related CTD-associated factor 4 | |
| 6421 | SFPQ | splicing factor proline/glutamine-rich (polypyrimidine tract binding protein associated) | yes |
| 6431 | SFRS6 | serine/arginine-rich splicing factor 6 | |
| 81892 | SLIRP | SRA stem-loop interacting RNA binding protein | |
| 6426 | SRSF1 | serine/arginine-rich splicing factor 1 | |
| 10772 | SRSF10 | serine/arginine-rich splicing factor 10 | |
| 6427 | SRSF2 | serine/arginine-rich splicing factor 2 | |
| 6428 | SRSF3 | serine/arginine-rich splicing factor 3 | |
| 6430 | SRSF5 | serine/arginine-rich splicing factor 5 | |
| 6432 | SRSF7 | serine/arginine-rich splicing factor 7 | |
| 8683 | SRSF9 | serine/arginine-rich splicing factor 9 | |
| 6741 | SSB | Sjogren syndrome antigen B (autoantigen La) | |
| 10492 | SYNCRIP | synaptotagmin binding, cytoplasmic RNA interacting protein | |
| 8148 | TAF15 | TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa | yes |
| 7307 | U2AF1 | U2 small nuclear RNA auxiliary factor 1 | |
| 113 3 8 | U2AF2 | U2 small nuclear RNA auxiliary factor 2 |
Notes: The set of 353 C9orf72 hexanucleotide repeat RNA-binding proteins are collected from two published datasets.13,23 Among them, the genes encoding 65 proteins with RRM motifs are listed with Entrez Gene ID, Gene Symbol, Gene Name, and the presence of prion-like domains (PrLD). The entire list is shown in Supplementary Table 1.
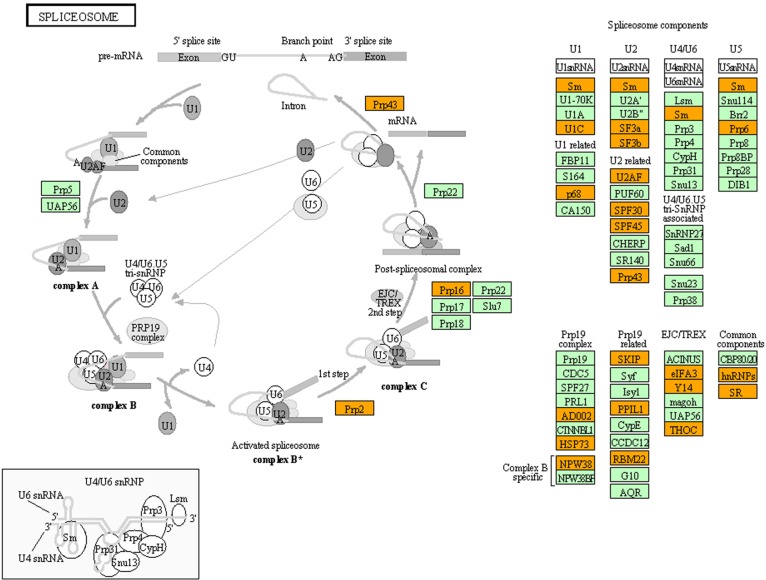
DAVID showed that 144 of 353 proteins (40.8%) belonged to the GO category termed as “ribonucleoprotein complex” (GO:0031012; P = 9.74E−46 corrected by Bonferroni). Molecular network analysis of 353 proteins by KEGG indicated a significant relationship with the pathways termed as “Ribosome” (hsa03010; P = 4.20E−81 corrected by Bonferroni) and “Spliceosome” (hsa03040; P = 2.36E−33 corrected by Bonferroni) (Fig. 1). IPA identified a significant relationship of these with functional networks defined by “RNA Post-Transcriptional Modification, Infectious Disease, Organismal Injury and Abnormalities” (P = 1.00E−128) (Supplementary Fig. 1) and “Gene Expression, Protein Synthesis, RNA Post-Transcriptional Modification” (P = 1.00E−102). KeyMolnet extracted the complex network comprising 1097 molecules and 1399 molecular relations (Supplementary Fig. 2). They showed a significant relationship with “HSP90 signaling pathway” (P = 3.85E−145), “Spliceosome assembly” (P = 2.23E−97), and “Intermediate filament signaling pathway” (P = 6.02E−88). Thus, three distinct pathway analysis tools commonly suggest that C9orf72 hexanucleotide repeat RNA-binding proteins play a pivotal role in the regulation of post-transcriptional RNA processing, particularly of sliceosome assembly.
Figure 1.
KEGG molecular network of C9orf72 hexanucleotide repeat RNA-binding proteins. When Entrez Gene IDs of 353 C9orf72 hexanucleotide repeat RNA-binding proteins were imported into the Functional Annotation tool of DAVID, it identified “Spliceosome” (hsa03040) as the second-rank relevant KEGG pathway. Orange nodes represent C9orf72 hexanucleotide repeat RNA-binding proteins.
Molecular network of differentially expressed genes in iPSC-derived motor neurons of patients with C9ALS and controls
Next, we reanalyzed a RNA-Seq dataset numbered GSE52202 and identified the set of 282 differentially expressed genes in iPSC-derived motor neurons of patients with C9ALS and normal controls (q value < 0.05) (Supplementary Table 2). C9orf72 mRNA expression levels were not significantly different in these cells between patients with C9ALS and controls (log2_fold change = 0.368586, q value = 0.999956). Unexpectedly, we did not find any significant differences in splicing patterns between patients with C9ALS and controls, in part attributable to the lower coverage of single-end RNA-Seq data. Among 282 differentially expressed genes, 11 were upregulated and 271 were down-regulated in patients with C9ALS. Thus, the great majority of them were underexpressed in patients with C9ALS. The set of upregulated genes include CBLN4, CBLN2, CBLN1, DPP6, and SLITRK2 as reported previously,25 supporting the validity of our analysis.
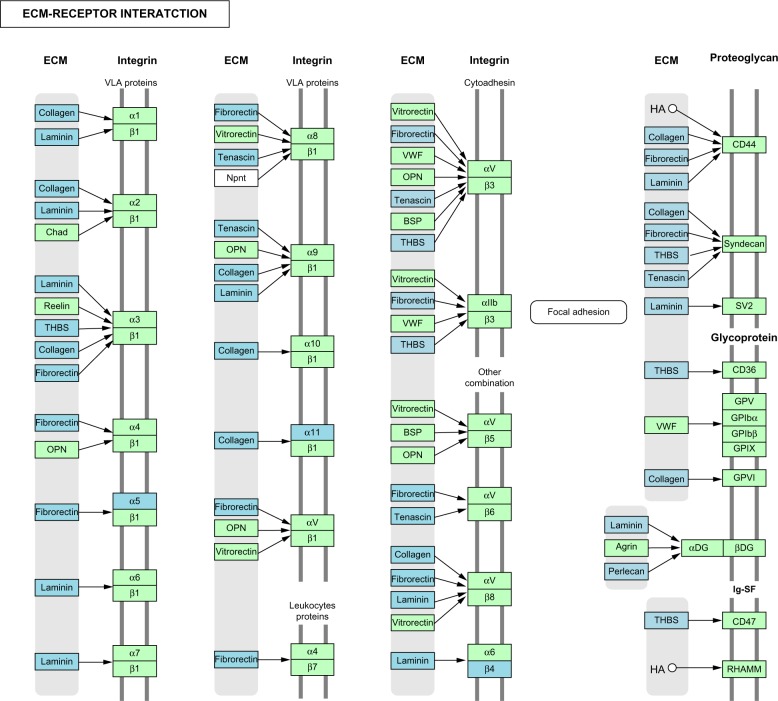
DAVID showed that 66 of the 282 genes (23.4%) belonged to the GO category termed as “extracellular matrix” (GO:0031012; P = 9.74E−46 corrected by Bonferroni). Molecular network analysis of 282 genes by KEGG indicated a significant relationship with the pathways defined as “ECM-receptor interaction” (hsa04512; P = 1.95E−14 corrected by Bonferroni) (Fig. 2), “Focal adhesion” (hsa04510; P = 6.23E−12), and “TGF-beta signaling pathway” (hsa04350; P = 0.0156). Consistent with the results of KEGG, IPA identified a significant relationship of these with functional network defined by “Connective Tissue Disorders, Dermatological Diseases and Conditions, Organismal Injury and Abnormalities” (P = 1.00E−76) where ECM proteins are enriched substantially (Supplementary Fig. 3). KeyMolnet extracted the complex network composed of 1291 molecules and 1735 molecular relations (Supplementary Fig. 4). They showed a significant relationship with “MMP signaling pathway” (P = 1.26E−232), “Transcriptional regulation by Ets-1/2” (P = 8.31E−164), and “Calpain signaling pathway” (P = 1.29E−122). Thus, three distinct tools commonly suggest that the expression of extracellular matrix proteins and matrix metalloproteinases is reduced in iPSC-derived motor neurons of patients with C9ALS.
Figure 2.
KEGG molecular network of differentially expressed genes in iPSC-derived motor neurons of C9ALS patients and controls. When Entrez Gene IDs of 282 differentially expressed genes in iPSC-derived motor neurons of C9ALS patients and controls were imported into the Functional Annotation tool of DAVID, it identified “ECM-receptor interaction” (hsa04512) as the most relevant KEGG pathway. Light blue nodes indicate downregulated genes in C9ALS.
Molecular network of differentially expressed genes and exons in purified motor neurons of patients with C9ALS and controls
Finally, we studied an exon array data-set and identified the set of 353 genes and 3579 exons differentially expressed in purified motor neurons between patients with C9ALS and normal controls in the setting of the significance that satisfies P < 0.01 and fold change either >1.5 or <−1.5 (Supplementary Table 3 for genes and Supplementary Table 4 for exons). Among 353 differentially expressed genes, 134 were upregulated, whereas 219 were downregulated in patients with C9ALS. Among 3579 exons, 1574 were upregulated, while 2005 were downregulated in patients with C9ALS. Thus, downregulated genes and exons outnumbered upregulated classes in patients with C9ALS. Importantly, the list includes C9orf72 as one of the downregulated genes and exons, in addition to TARDBP (TDP-43) as one of the upregulated genes and exons in patients with C9ALS.
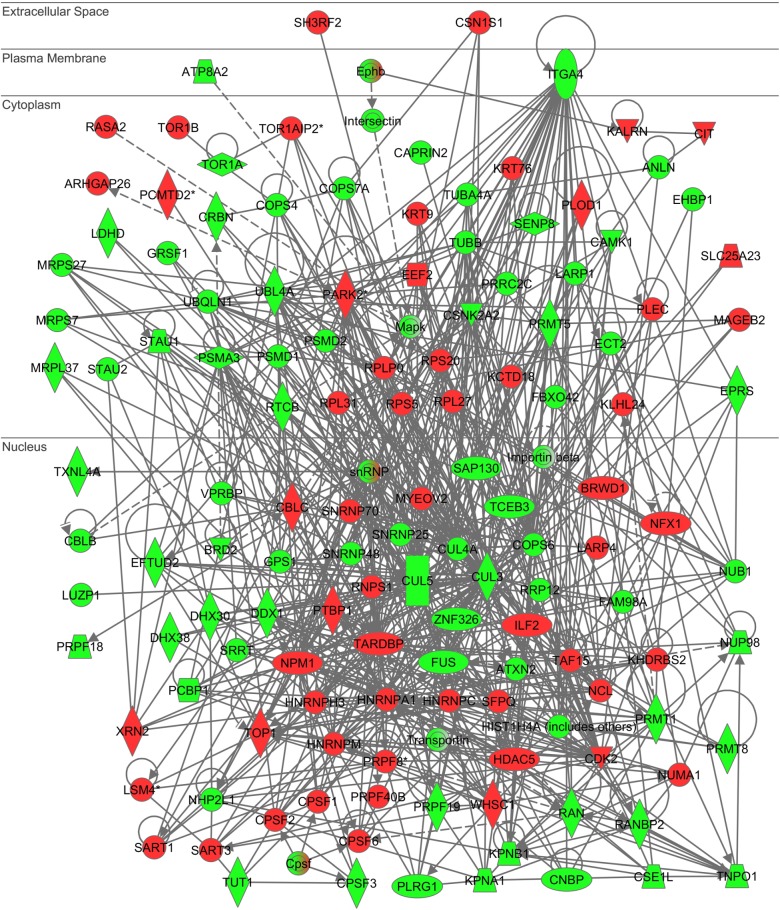
DAVID showed that 23 of the 353 differentially expressed genes belonged to the GO category termed as “RNA splicing” (GO:0008380; P = 6.45E−6 corrected by Bonferroni) and 460 of 3579 genes corresponding to differentially expressed exons belonged to the GO category termed as “nucleotide binding” (GO:0000166; P = 1.59E−11 corrected by Bonferroni). Molecular network analysis by KEGG indicated a discernible relationship with the pathways defined as “Spliceosome” (hsa03040; P = 0.0042, uncorrected) for differentially expressed genes and defined as “Regulation of actin cytoskeleton” (hsa04810; P = 0.0116 corrected by Bonferroni) for differentially expressed exons. IPA identified the most significant relationship with functional networks defined by “RNA Post-Transcriptional Modification, RNA Damage and Repair, Protein Synthesis” (P = 1.00E−93) for differentially expressed genes (Supplementary Fig. 5) and “RNA Post-Transcriptional Modification, Amino Acid Metabolism, Post-Translational Modification” (P = 1.00E−134) for differentially expressed exons (Fig. 3). KeyMolnet extracted the highly complex network composed of 1673 molecules and 2183 molecular relations for differentially expressed genes, exhibiting the most significant relationship with “Calpain signaling pathway” (P = 1.58E−141) (data not shown). KeyMolnet identified the extremely complex network composed of 4525 molecules and 8799 molecular relations for differentially expressed exons, showing the most significant relationship with “Kinesin family signaling pathway” (P = 1.82E−157) (data not shown). Thus, all these results suggest that the regulation of post-transcriptional RNA processing process, along with cytoskeletal dynamics and intracellular molecular transport, is disturbed in motor neurons of patients with C9ALS in vivo.
Figure 3.
IPA molecular network of differentially expressed exons in purified motor neurons of C9ALS patients and controls. When Entrez Gene IDs of the genes encoding 3579 differentially expressed exons in LCM-isolated purified motor neurons of C9ALS patients and controls were imported into the core analysis tool of IPA, it identified “RNA Post-Transcriptional Modification, Amino Acid Metabolism, Post-Translational Modification” as the most relevant functional network. Red nodes indicate genes encoding upregulated exons, while green nodes represent genes encoding downregulated exons in C9ALS.
Discussion
By using three distinct pathway analysis tools of bioinformatics, we studied molecular networks involved in C9ALS pathology by focusing on three different C9orf72 omics data-sets. They include proteome of C9orf72 hexanucleotide repeat RNA-binding proteins providing the most valuable biochemical information on molecular basis of C9ALS, transcriptome of iPSC-derived motor neurons of patients with C9ALS serving as the most representative cell culture model, and transcriptome of purified motor neurons of patients with C9ALS acting as the most clinically relevant in vivo source.
Molecular network analysis indicated that the proteome of human C9orf72 hexanucleotide repeat RNA-binding proteins plays an active role in the function of ribosome, spliceosome, and RNA post-transcriptional modification. Importantly, it is enriched in the category of RNA-binding proteins having RRM and PrLD. They have the capacity to self-assemble through PrLD, characterized by cluster of uncharged polar amino acids and glycine, resulting in generation of self-propagating amyloids.32,33 Previous studies showed that C9orf72 hexanucleotide repeats interact with HNRNPA1, HNRNPA2B1, HNRNPA3, HNRNPH, PURA, SRSF1, and ADARB2.18,34–37 In this study, the proteome included all these except for ADARB2. Notably, the proteome list contains FUS, EWSR1, HNRNPA1 HNRNPA2B1, and TAF15, all of which have RRM and PLD and are causative of ALS and/or FTD.29–32 Furthermore, a recent study of familial ALS patients identified missense mutations in the MATR3 gene encoding an RNA- and DNA-binding nuclear matrix protein interacting with TDP-43 and DEAH (Asp–Glu–Ala–His) box helicase 9 (DHX9).38 We identified both MARN3 and DHX9 as C9orf72 repeat RNA-binding proteins. Poly- A-binding protein, cytoplasmic 1 (PABPC1) having four RRM domains shuttles between the nucleus and cytoplasm, binds to the 3′-poly-A tail of mRNAs, and promotes ribosome recruitment and translation initiation. PABPC1 protein accumulates in inclusions of ALS spinal cord motor neurons.39 Enhanced phosphorylation of eIF2α (EIF2S1) serves as a marker for stress granules and an indicator for translational suppression in a Drosophila model of TDP-43-associated neurodegeneration.39 We identified both PABPC1 and EIF2S1 as C9orf72 repeat RNA-binding proteins.
A previous study showed that the amount of collagen proteins is greatly reduced in the cervical spinal cord of patients with ALS.40 Supporting this, we identified down-regulation of various collagen genes in iPSC-derived C9ALS motor neurons. They include COL1A2, COL2A1, COL4A1, COL4A2, COL4A5, COL5A1, COL5A2, COL5A3, COL6A2, COL6A3, COL8A2, COL11A1, COL12A1, COL13A1, COL14A1, COL15A1, and COL16A1, along with reduction of BGN, DCN, LUM, PLOD1, and P4H2 involved in synthesis, assembly, and crosslinking of collagen fibrils. Importantly, the levels of integrin (ITGB4, ITGA5, ITGA11), laminin (LAMB1, LAMA2, LAMC1), tenacin (TNC), fibronectin (FN1), fibulin (FBN1, FBN2), thrombospondin (THBS1), and proteoglycan (HSPG2, GPC3, GPC4, CSPG4), all of which play a role in cell-to-cell and cell-to-matrix interactions essential for neuronal migration and axonal guidance during development, synaptic plasticity, and neuronal regeneration are also decreased in patients with C9ALS. Furthermore, the genes involved in regulation of actin cytoskeleton dynamics (ACTA1, ACTA2, ACTC1, ACTG2, FLNB, GSN, and TAGLN) are downregulated in iPSC-derived motor neurons of patients with C9ALS. Previous studies showed that matrix metalloproteinases, such as MMP9 and MMP2, are overexpressed in the spinal cord of ALS and SOD1G93 A transgenic mice.41,42 In contrast, we found that the levels of MMP2, MMP11, and MMP14, along with ADAMTS1 and ADAMTS9, are decreased in iPSC-derived motor neurons of patients with C9ALS, suggesting that remodeling of extracellular matrix proteins is downregulated in patients with C9ALS. C9ALS patient-derived iPSC expresses intranuclear RNA foci containing hexanucleotide expansions that potentially affect processing of various RNAs by sequestering RNA-binding proteins.25,34 Importantly, aberrantly spliced genes in spinal cord motor neurons of sporadic ALS patients are enriched in the category of cell–matrix adhesion.43 We also found downregulation of the genes regulated by Ets transcription factors and those involved in calpain signaling in iPSC-derived motor neurons of patients with C9ALS. However, they are seemingly contradictory to the findings that Ets-2 immunoreactivity is enhanced in astrocytes in the spinal cord of patients with ALS and activation of calpain promotes the carboxyl-terminal cleavage of TDP-43, enhancing mislocalization of TDP-43 in affected neurons.44,45
Finally, we identified the set of 353 genes and 3579 exons differentially expressed in LCM-isolated purified motor neurons of patients with C9ALS and controls. Importantly, they constitute the molecular networks showing the most significant relationship with RNA post-transcriptional modification, consistent with the network of C9orf72 repeat RNA-binding proteins. These results suggest that a battery of genes encoding post-transcriptional RNA processing machinery components are aberrantly expressed and/or spliced in C9ALS motor neurons in vivo due to formation of nuclear RNA foci that sequester principal RNA-binding proteins. This constitutes a vicious circle for the proper RNA metabolism. A previous study found that a pre-mRNA splicing factor SRSF2 sequestered in RNA foci acts as a key molecule in aberrant regulation of splicing.23
Conclusions
By using three distinct pathway analysis tools, we studied molecular networks involved in C9ALS pathology by focusing on C9orf72 omics datasets. We found that C9orf72 repeat RNA-binding proteins play a crucial role in the regulation of post-transcriptional RNA processing. The expression of a wide range of extracellular matrix proteins and matrix metalloproteinases is reduced in iPSC-derived motor neurons of patients with C9ALS. The regulation of RNA processing and cytoskeletal dynamics is disturbed in motor neurons of patients with C9ALS in vivo. Taken all together, bioinformatics data mining approach suggests a logical hypothesis that C9orf72 repeat expansions that deregulate post-transcriptional RNA processing disturb the homeostasis of cytoskeletal dynamics and remodeling of extracellular matrix, leading to degeneration of stress-vulnerable neurons in the brain and spinal cord of patients with C9ALS/FTD.
Supplementary Material
Supplementary Figure 1. IPA molecular network of C9orf72 hexanucleotide repeat RNA-binding proteins.
Supplementary Figure 2. KeyMolnet molecular network of C9orf72 hexanucleotide repeat RNA-binding proteins.
Supplementary Figure 3. IPA molecular network of differentially expressed genes in iPSC-derived motor neurons of C9ALS patients and controls.
Supplementary Figure 4. KeyMolnet molecular network of differentially expressed genes in iPSC-derived motor neurons of C9ALS patients and controls.
Supplementary Figure 5. IPA molecular network of differentially expressed genes in purified motor neurons of C9ALS patients and controls.
Supplementary Table 1. The set of 353 C9orf72 hexanucleoride repeat RNA-binding proteins.
Supplementary Table 2. The set of 282 differentially expressed genes in iPSC-derived motor neurons established from C9ALS patients and controls.
Supplementary Table 3. The set of 353 differentially expressed genes in LCM-dissected motor neurons of C9ALS patients and controls.
Supplementary Table 4. The set of 3,579 differentially expressed exons in LCM-dissected motor neurons of C9ALS patients and controls.
Footnotes
Author Contributions
JS designed data analysis and drafted the manuscript. YY, SK, MT, NA and YK performed molecular network analysis. All authors have read and approved the final manuscript.
ACADEMIC EDITOR: Alexander Rotenberg, Editor in Chief
FUNDING: This study was supported by the JSPS KAKENHI (C22500322 and C25430054), the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan; and the grant from the National Center for Geriatrics and Gerontology (NCGC26-20).
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
This paper was subject to independent, expert peer review by a minimum of two blind peer reviewers. All editorial decisions were made by the independent academic editor. All authors have provided signed confirmation of their compliance with ethical and legal obligations including (but not limited to) use of any copyrighted material, compliance with ICMJE authorship and competing interests disclosure guidelines and, where applicable, compliance with legal and ethical guidelines on human and animal research participants.
REFERENCES
- 1.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki N, Maroof AM, Merkle FT, et al. The mouse C9ORF72 ortholog is enriched in neurons known to degenerate in ALS and FTD. Nat Neurosci. 2013;16(12):1725–7. doi: 10.1038/nn.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics. 2013;29(4):499–503. doi: 10.1093/bioinformatics/bts725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farg MA, Sundaramoorthy V, Sultana JM, et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet. 2014;23(13):3579–95. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cruts M, Gijselinck I, Van Langenhove T, van der Zee J, Van Broeckhoven C. Current insights into the C9orf72 repeat expansion diseases of the FTLD/ALS spectrum. Trends Neurosci. 2013;36(8):450–9. doi: 10.1016/j.tins.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Cooper-Knock J, Shaw PJ, Kirby J. The widening spectrum of C9ORF72-related disease; genotype/phenotype correlations and potential modifiers of clinical phenotype. Acta Neuropathol. 2014;127(3):333–45. doi: 10.1007/s00401-014-1251-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Sarraj S, King A, Troakes C, et al. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011;122(6):691–702. doi: 10.1007/s00401-011-0911-2. [DOI] [PubMed] [Google Scholar]
- 9.Stepto A, Gallo JM, Shaw CE, Hirth F. Modelling C9ORF72 hexanucleotide repeat expansion in amyotrophic lateral sclerosis and frontotemporal dementia. Acta Neuropathol. 2014;127(3):377–89. doi: 10.1007/s00401-013-1235-1. [DOI] [PubMed] [Google Scholar]
- 10.Gendron TF, Belzil VV, Zhang YJ, Petrucelli L. Mechanisms of toxicity in C9FTLD/ALS. Acta Neuropathol. 2014;127(3):359–76. doi: 10.1007/s00401-013-1237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belzil VV, Bauer PO, Prudencio M, et al. Reduced C9orf72 gene expression in c9FTD/ALS is caused by histone trimethylation, an epigenetic event detectable in blood. Acta Neuropathol. 2013;126(6):895–905. doi: 10.1007/s00401-013-1199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xi Z, Zinman L, Moreno D, et al. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am J Hum Genet. 2013;92(6):981–9. doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haeusler AR, Donnelly CJ, Periz G, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507(7491):195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waite AJ, Bäumer D, East S, et al. Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging. 2014;35(7):e5–1779. doi: 10.1016/j.neurobiolaging.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ciura S, Lattante S, Le Ber I, et al. Loss of function of C9orf72 causes motor deficits in a zebrafish model of amyotrophic lateral sclerosis. Ann Neurol. 2013;74(2):180–7. doi: 10.1002/ana.23946. [DOI] [PubMed] [Google Scholar]
- 16.Therrien M, Rouleau GA, Dion PA, Parker JA. Deletion of C9ORF72 results in motor neuron degeneration and stress sensitivity in C elegans. PLoS One. 2013;8(12):e83450. doi: 10.1371/journal.pone.0083450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagier-Tourenne C, Baughn M, Rigo F, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci USA. 2013;110(47):E4530–9. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee YB, Chen HJ, Peres JN, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5(5):1178–86. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ash PE, Bieniek KF, Gendron TF, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77(4):639–46. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mori K, Weng SM, Arzberger T, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339(6125):1335–8. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 21.Satoh J. Bioinformatics approach to identifying molecular biomarkers and networks in multiple sclerosis. Clin Exp Neuroimmunol. 2010;1(3):127–40. [Google Scholar]
- 22.Kitano H. A robustness-based approach to systems-oriented drug design. Nat Rev Drug Discov. 2007;6(3):202–10. doi: 10.1038/nrd2195. [DOI] [PubMed] [Google Scholar]
- 23.Cooper-Knock J, Walsh MJ, Higginbottom A, et al. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain. 2014;137(t7):2040–51. doi: 10.1093/brain/awu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Couthouis J, Hart MP, Shorter J, et al. A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci USA. 2011;108(52):20881–90. doi: 10.1073/pnas.1109434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sareen D, O’Rourke JG, Meera P, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5(208):208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Highley JR, Kirby J, Jansweijer JA, Webb PS, Hewamadduma CA, Heath PR, et al. Loss of nuclear TDP-43 in ALS causes altered expression of splicing machinery and widespread dysregulation of RNA splicing in motor neurons. Neuropathol Appl Neurobiol. 2014 Apr 18; doi: 10.1111/nan.12148. [Epub ahead of print] PMID: 24750229. [DOI] [PubMed] [Google Scholar]
- 27.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40(Database issue):D109–114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vance C, Rogelj B, Hortobágyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323(5918):1208–11. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Couthouis J, Hart MP, Erion R, et al. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21(13):2899–911. doi: 10.1093/hmg/dds116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ticozzi N, Vance C, Leclerc AL, et al. Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis. Am J Med Genet B Neuropsychiatr Genet. 2011;156B(3):285–90. doi: 10.1002/ajmg.b.31158. [DOI] [PubMed] [Google Scholar]
- 32.Kim HJ, Kim NC, Wang YD, et al. Mutations in prion-like domains in hnRN-PA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495(7442):467–73. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell. 2013;154(4):727–36. doi: 10.1016/j.cell.2013.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donnelly CJ, Zhang PW, Pham JT, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80(2):415–28. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mori K, Lammich S, Mackenzie IR, et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013;125(3):413–23. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- 36.Reddy K, Zamiri B, Stanley SY, Macgregor RB, Jr, Pearson CE. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem. 2013;288(14):9860–66. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Z, Poidevin M, Li X, et al. Expanded GGGGCC repeat RNA associated with amyotrophic lateral sclerosis and frontotemporal dementia causes neurodegeneration. Proc Natl Acad Sci USA. 2013;110(19):7778–83. doi: 10.1073/pnas.1219643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson JO, Pioro EP, Boehringer A, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci. 2014;17(5):664–6. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim HJ, Raphael AR, LaDow ES, et al. Therapeutic modulation of eIF2α phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat Genet. 2014;46(2):152–60. doi: 10.1038/ng.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ono S, Imai T, Munakata S, et al. Collagen abnormalities in the spinal cord from patients with amyotrophic lateral sclerosis. J Neurol Sci. 1998;160(2):140–47. doi: 10.1016/s0022-510x(98)00223-8. [DOI] [PubMed] [Google Scholar]
- 41.Lim GP, Backstrom JR, Cullen MJ, Miller CA, Atkinson RD, Tökés ZA. Matrix metalloproteinases in the neocortex and spinal cord of amyotrophic lateral sclerosis patients. J Neurochem. 1996;67(1):251–9. doi: 10.1046/j.1471-4159.1996.67010251.x. [DOI] [PubMed] [Google Scholar]
- 42.Fang L, Teuchert M, Huber-Abel F, et al. MMP-2 and MMP-9 are elevated in spinal cord and skin in a mouse model of ALS. J Neurol Sci. 2010;294:1–2. 51–6. doi: 10.1016/j.jns.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 43.Rabin SJ, Kim JM, Baughn M, et al. Sporadic ALS has compartment-specific aberrant exon splicing and altered cell-matrix adhesion biology. Hum Mol Genet. 2010;19(2):313–8. doi: 10.1093/hmg/ddp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee J, Kannagi M, Ferrante RJ, Kowall NW, Ryu H. Activation of Ets-2 by oxidative stress induces Bcl-xL expression and accounts for glial survival in amyotrophic lateral sclerosis. FASEB J. 2009;23(6):1739–49. doi: 10.1096/fj.08-121046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamashita T, Hideyama T, Hachiga K, et al. A role for calpain-dependent cleavage of TDP-43 in amyotrophic lateral sclerosis pathology. Nat Commun. 2012;3:1307. doi: 10.1038/ncomms2303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. IPA molecular network of C9orf72 hexanucleotide repeat RNA-binding proteins.
Supplementary Figure 2. KeyMolnet molecular network of C9orf72 hexanucleotide repeat RNA-binding proteins.
Supplementary Figure 3. IPA molecular network of differentially expressed genes in iPSC-derived motor neurons of C9ALS patients and controls.
Supplementary Figure 4. KeyMolnet molecular network of differentially expressed genes in iPSC-derived motor neurons of C9ALS patients and controls.
Supplementary Figure 5. IPA molecular network of differentially expressed genes in purified motor neurons of C9ALS patients and controls.
Supplementary Table 1. The set of 353 C9orf72 hexanucleoride repeat RNA-binding proteins.
Supplementary Table 2. The set of 282 differentially expressed genes in iPSC-derived motor neurons established from C9ALS patients and controls.
Supplementary Table 3. The set of 353 differentially expressed genes in LCM-dissected motor neurons of C9ALS patients and controls.
Supplementary Table 4. The set of 3,579 differentially expressed exons in LCM-dissected motor neurons of C9ALS patients and controls.