Abstract
Background
A subset of patients with chronic myeloid leukemia (CML) can sustain a complete molecular response after discontinuing imatinib mesylate (IM). We focused on microRNAs (miRNAs), with the aim of finding a molecular biomarker to discriminate which patients can safely and successfully discontinue IM use.
Methods
To identify miRNAs that showed altered expression in patients who had discontinued IM (STOP-IM group), we first screened miRNA expression of peripheral blood mononuclear cells by using a TaqMan miRNA array on samples from five unselected patients from the STOP-IM group, seven CML patients receiving IM (IM group), and five healthy volunteers. We then performed miRNA quantification in 49 CML patients with deep molecular response. Mann–Whitney U and chi-square tests were used to determine statistical significance for comparisons between the control (healthy volunteers) and test groups (STOP-IM and IM groups). Multiple groups were compared by one-way analysis of variance.
Results
Downregulation of miR-148b was noted in patients in the STOP-IM group and in a subset of the IM group. We then subdivided the IM patients into two groups: one with downregulated miR-148b expression (IM-1; less than the cut-off value) and the other without downregulated miR-148b expression (IM-2; greater than the cut-off value). The number of patients who had a sustained stable molecular response was significantly lower in IM-2 group. This group also had a significantly lower percentage of natural killer cells.
Conclusion
Downregulated miR-148 may contribute to immune surveillance in STOP-IM patients and may therefore have potential as additive information in managing CML patients undergoing treatment with IM.
Keywords: chronic myeloid leukemia, imatinib mesylate, discontinuation, miR-148b
Background
The treatment of chronic myeloid leukemia (CML) has been greatly impacted by the development of imatinib mesylate (IM), the first specific inhibitor of BCR-ABL1.1 In newly diagnosed patients with chronic-phase CML, IM induces a complete hematological response in more than 90% of cases and a complete cytogenetic response in more than 80% of cases,1 and IM has been established as the standard therapy for CML. Recent reports have demonstrated that a subset of CML patients with a complete molecular response (currently referred to as undetectable minimal residual disease, UMRD) can sustain this state after IM discontinuation (STOP-IM).2–4 We and others have postulated that immunological surveillance may play an important role in these patients.5–8 More recently, Ng et al demonstrated that East Asian CML patients with a common intronic deletion polymorphism in the gene coding BIM (at intron 2) had an inferior response to tyrosine kinase inhibitors compared to those without the genetic variation.9,10 Taken together, immunological diversity as well as genetic background should be considered in discussions of the efficacy and safety of IM cessation in CML patients. Currently there is no definitive classifier to discriminate CML patients with UMRD who could safely stop IM and those who should not.
MicroRNAs (miRNAs), which are noncoding single-stranded RNAs of 21–25 nucleotides, have recently been implicated in the regulation of cellular processes such as development, differentiation, apoptosis, proliferation, and hematopoiesis. MiRNAs have also been implicated in the development of human cancers, either as tumor suppressors or as oncogenes.11,12 Approximately 100 miRNAs are known to be expressed in cells of the immune system and to play key roles in the development of innate and acquired immunity,13,14 therefore, aberrant expression of miRNAs that target the immune system may be related to the immunological background of cancer patients. In CML cells, abnormal expression of several miRNAs (miR-15a, miR-16, miR-31, miR-142, miR-155, miR-181, miR-221, miR-564, let7a, and the polycistronic miR-17–92 cluster) has been described.15–18
Although the miRNA expression profile in CML cells has been extensively studied, little is known about miRNA expression in CML patients with UMRD whose CML cells cannot be detected. We therefore determined the miRNA profiles of peripheral blood mononuclear cells (PBMCs) in CML patients with UMRD who were receiving IM or had discontinued IM, with the goal of clarifying whether altered miRNA expression is linked to pathophysiology in CML patients who safely discontinued IM. Here, we provide a distinct miRNA expression profile of circulating PBMCs in CML patients with successful IM cessation. We additionally describe a subset of CML patients receiving IM that showed similar miRNA expression profiles.
Materials and methods
Patients
Forty-nine consecutive patients with CML who had UMRD were enrolled in this study. In 16 patients who had discontinued IM and had sustained UMRD for more than 6 months (STOP-IM group), samples were collected when IM was stopped. In contrast, 33 patients who were receiving IM and had UMRD at the time of sample collection were categorized as the IM group. Fifteen healthy volunteers served as controls. CML patients before treatment and those who did not respond to IM were not included in this study. There was no significant difference in sex or age between the CML groups and the healthy volunteers. UMRD was defined as the absence of a BCR-ABL transcript as determined by nested reverse transcription-polymerase chain reaction (RT-PCR) or a highly sensitive transcription-mediated amplification (TMA) method. These PCR methods can detect at least a 4-log reduction in the BCR-ABL transcript (international scale <0.01%). The study was approved by the institutional review board of Tokyo Medical University (no 930, approved 24 June 2008). Written informed consent was obtained from all participants prior to specimen collection, in accordance with the Declaration of Helsinki.
RNA isolation from peripheral blood
PBMCs were obtained from 10 mL of whole blood by the Ficoll density gradient method (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). Cell pellets were quickly frozen at −80°C until use. Total RNA was isolated from cell pellets with a mirVana PARIS kit (Ambion, Austin, TX, USA), as reported previously.19 Briefly, 600 μL of cell lysate was mixed with an equal volume of denaturing solution, and the subsequent phenol extraction and cartridge filtration were carried out according to the manufacturer’s instructions.
TaqMan low-density array screening
The miRNA expression profiles were determined with a TaqMan Array Human microRNA Card A (Thermo Fisher Scientific, Waltham, MA, USA). Ninety nanograms of total RNA was used as an input in each RT reaction. The RT reaction and pre-amplification step was set up according to the manufacturer’s recommendations. miRNAs were reverse transcribed with the Megaplex Primer Pools (Human Pools A version 2.1; Thermo Fisher Scientific). RT reaction products from the PBMC samples were further amplified with Megaplex PreAmp primers (Primers A version 2.1; Thermo Fisher Scientific). Quantitative RT-PCR was performed on an Applied Biosystems 7900HT thermocycler according to the manufacturer’s recommended program. With the use of SDS 2.4 software and DataAssist (Thermo Fisher Scientific), the expression of miRNA was calculated based on cycle threshold (Ct) values normalized by those of RNU6B. Data analysis was done using GeneSifter® software (Perkin Elmer, Waltham, MA, USA). P-values of less than 0.05 were considered to indicate a statistically significant difference, and the Benjamini–Hochberg algorithm was used for estimation of false discovery rates, as we have reported previously.19
Real-time quantitative RT-PCR
We determined the amounts of the individual miRNAs by real-time quantitative RT-PCR with a TaqMan MicroRNA Assay (Thermo Fisher Scientific) and the following miRNA-specific stem-loop primers: hsa-miR-148b (000471), hsa-miR-326 (000542), hsa-let7b (000378), and RNU6B (001093). Subsequently, quantitative real-time PCR was performed with an ABI Prism 7000 sequence detection system (Thermo Fisher Scientific). The reaction was initiated by incubation at 95°C for 2 minutes, followed by 50 cycles of 95°C for 15 seconds and then 60°C for 1 minute. All reactions were run in duplicate. Mean (Ct) values for all miRNAs were quantified with sequence detection system software (SDS version 1.02; Thermo Fisher Scientific). All miRNA expression was normalized to RNU6B expression, yielding a −ΔCt value, as reported elsewhere.20 The −ΔΔCt value was then calculated by subtracting the −ΔCt value of the normal sample from the respective −ΔCt values of patient samples. Expression of all miRNAs was normalized by using the 2−ΔΔCt method.
Flow cytometry
Freshly obtained PBMCs were separated using a Ficoll density gradient (GE Healthcare Bio-Sciences) and resuspended in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal calf serum. Immunophenotyping was performed on a FACSCanto II (BD Biosciences, San Jose, CA, USA) flow cytometer using CellQuest software (BD Biosciences) with a 5-color flow cytometry panel including antibodies against the following cell surface antigens and effector molecules: CCR7, CD3, CD8, CD45RO, and CD56 (eBioscience, San Diego, CA, USA, and BioLegend, San Diego, CA, USA). Cells were analyzed with the FACSCanto II (BD Biosciences) flow cytometer using CellQuest software.8
Statistical analysis and pathway analysis
Data are expressed as means ± standard deviation (SD). Mann–Whitney U and chi-square tests were used to determine statistical significance for comparisons between the control and test groups. Multiple groups were compared by one-way analysis of variance. GraphPad Prism software (version 5c for Macintosh; GraphPad Software Inc., La Jolla, CA, USA) was used for statistical analyses. Results were considered statistically significant when P<0.05.
Following identification of differentially expressing miRNAs, the predicted targets for these altered miRNAs were subjected to microRNA Target Filter in Ingenuity Pathways Analysis (IPA) software (Ingenuity System, Redwood City, CA, USA).
Results
The miRNA expression profiling by Taqman miRNA array
To identify candidate miRNAs that showed altered expression in patients in the STOP-IM group, we first screened miRNA expression using a TaqMan miRNA array (Thermo Fisher Scientific) on five unselected patients from the STOP-IM group, seven from the IM group, and five healthy volunteers. Among these three groups, we observed differential expression of 22 miRNAs as identified by using GeneSifter software (Figure 1; Gene Expression Omnibus [GEO] accession no GSE47652). Although 22 miRNAs were selected based on fold change, the expression profile was highly variable among individuals. We therefore selected three of these miRNAs (let-7b, miR-148b, and miR-326) for further validation on the basis of their averaged fold change in expression (log2 variation >0.6), significant P-value, quality control value, and their predicted target genes (Table S1).
Figure 1.

miRNA profiling by TaqMan (Thermo Fisher Scientific, Waltham, MA, USA) miRNA array.
Notes: A distinct expression pattern was found in the STOP-IM group (middle lane). Using SDS (version 2.4) and DataAssist software (Thermo Fisher), we calculated the expression of miRNAs based on their Ct values normalized by the Ct value of RNU6B. Data were analyzed with GeneSifter software (Perkin Elmer, Waltham, MA, USA). P-values of <0.05 were considered to indicate a statistically significant difference, and the Benjamini–Hochberg algorithm was used for estimation of false discovery rates.
Abbreviations: miRNA, microRNA; Ct, cycle threshold; IM, imatinib mesylate; STOP-IM, patients who had discontinued IM and had sustained UMRD for more than 6 months; UMRD, undetectable minimal residual disease.
Quantification of individual miRNA by real-time quantitative RT-PCR
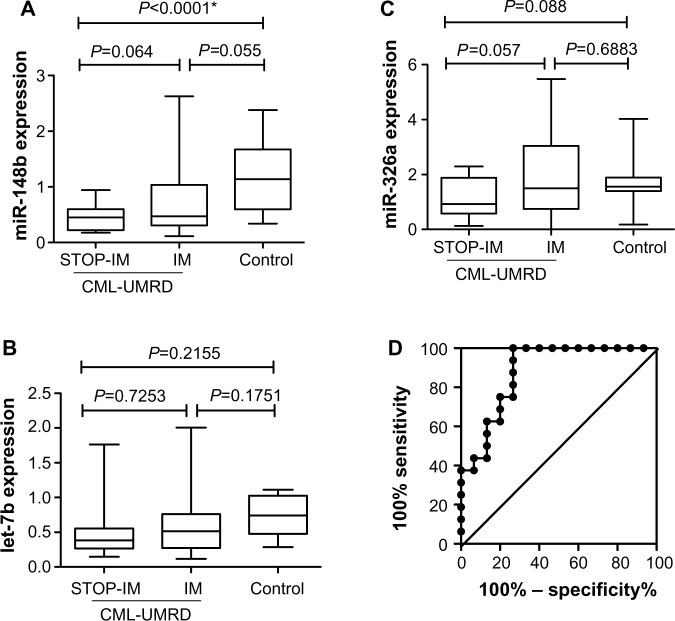
We then validated the miRNA array results by real-time quantitative RT-PCR using samples from 16 patients of the STOP-IM group, 33 of the IM group, and 15 healthy volunteers. Validation of miR-148b, miR-362, and let-7b revealed that expression of miR-148b was significantly lower in the STOP-IM group than in the healthy volunteers (Figure 2A). The miR-148b expression tended to be lower in the STOP-IM group than in the IM group, although the difference was not statistically significant. We observed no statistically significant difference in the expression of let-7b between the STOP-IM and IM groups (Figure 2B). Despite miR-362 being upregulated in the STOP-IM group based on miRNA array screening, the validation analysis using a large number of samples revealed that miR-326 tended to be downregulated in the STOP-IM group in comparison to the IM group and the control group. We observed no statistically significant difference in the expression of miR-326 between each group (Figure 2C).
Figure 2.
Expression of miR-148b (A), let-7b (B), and miR-326b (C) in the STOP-IM, IM, and control groups. (D) Cut-off level for miR-148b expression between the STOP-IM and control groups.
Notes: The asterisk indicates a statistically significant difference. The area under the ROC curve was 0.8792 with 75% sensitivity (95% CI, 47.62%–90%) and 80% specificity (95% CI, 51.91%–95%). The cut-off value was thus determined to be 0.5880, which was used in the following analyses of miR-148b expression.
Abbreviations: IM, imatinib mesylate; ROC, receiver operating characteristic; CI, confidence interval; CML, chronic myeloid leukemia; STOP-IM, patients who had discontinued IM and had sustained UMRD for more than 6 months; UMRD, undetectable minimal residual disease.
Clinical and biological relevance of miR-148b expression
Based on the statistical significant results from quantifying individual miRNAs, we particularly focused on miR-148b. To estimate a cut-off value for miR-148b expression in mononuclear cells for distinguishing between the STOP-IM and healthy control groups, we performed receiver operating characteristic (ROC) curve analysis (Figure 2D). The area under the ROC curve was 0.8792 with 75% sensitivity (95% confidence interval [CI], 47.62%–90%) and 80% specificity (95% CI, 51.91%–95%). The cut-off value was thus determined to be 0.5880, which was used in subsequent analyses of miR-148b expression. We next subdivided the IM patients into two groups: one with downregulated miR-148b expression (IM-1; less than the cut-off value) and the other without downregulated miR-148b expression (IM-2; greater than the cut-off value) (Table 1).
Table 1.
Clinico-hematological, immunological, and molecular characteristics of patients with and without downregulation of miR-148b expression
| Downregulation of miR-148b expression
|
STOP-IM
|
IM-1
|
IM-2
|
P-value |
|---|---|---|---|---|
| Yes
|
Yes
|
No
|
||
| Number of patients | 16 | 18 | 15 | |
| Age (years) | 57.5±11.6 | 60.4±9.1 | 54.5±15.4 | 0.404 |
| Sex (male/female) | 7/9 | 12/6 | 11/4 | 0.201§ |
| Sokal score | 0.74±0.22 | 0.70±0.09 | 0.73±0.24 | 0.849 |
| Total imatinib dose (grams) |
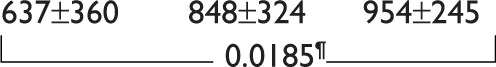
|
0.0579 | ||
| Duration of imatinib therapy (months) | 67.4±34.9 | 75.8±29.3 | 88.9±76.0 | 0.2264 |
| Months to achieve UMRD from diagnosis | 59.3±68.0 | 48.2±39.8 | 67.5±44.9 | 0.5473 |
| Stable MR* (yes/no) |

|
0.0454§ | ||
| Prior IFN-α (yes/no) | 4/12 | 3/15 | 5/10 | 0.3888§ |
| NK cells (%) |
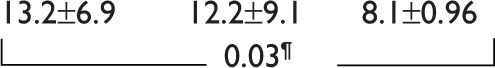
|
0.141 | ||
| CCR7−CR45RO− CD8+ cells (%) | 8.4±3.9 | 10.5±5.4 | 7.6±6.1 | 0.1733 |
| BIM deletion (yes/no) | 1/10 | 2/16 | 1/13 | 0.9288§ |
Notes:
If BCR-ABL1 remained undetectable for a continuous period of 24 months of imatinib treatment, the response was termed stable molecular response (MR). Patients who received IM and did not have a sustained UMRD but maintained a major molecular response for more than 24 months (fluctuating UMRD) were not referred as “stable MR”. P values are calculated by one-way ANOVA unless otherwise indicated
Student’s t-test
chi-square test.
Abbreviations: IM, imatinib mesylate; UMRD, undetectable minimal residual disease; ANOVA, analysis of variance; IFN, interferon; NK, natural killer; STOP-IM, patients who had discontinued IM and had sustained UMRD for more than 6 months.
Comparison of the STOP-IM, IM-1, and IM-2 groups revealed no significant differences in sex, age, Sokal score, duration of IM therapy, months to achieve UMRD from diagnosis, or prior interferon (IFN)-α therapy (Table 1). The total IM dose was significantly higher (P=0.0289) in the IM-2 group (893±294 g) than in the STOP-IM group (636±369 g), while there was no statistically significant difference between the IM-1 group and STOP-IM group. Similarly, the number of patients who sustained stable molecular response was significantly lower (P=0.0232) in the IM-2 group than in the STOP-IM group. Notably, the percentage of natural killer (NK) cells was significantly lower (P=0.03) in the IM-2 group than in the STOP-IM group. We could not find any statistical significance of BIM deletion among the 3 groups.
Network analysis of miRNA-148b
ROC analysis of the miR-148b suggests that the miRNA signature in PBMCs may predict whether IM discontinuation is appropriate. Therefore, using in silico network analysis we assessed whether predicted targets of miR-148b signature could provide insights into analogous changes enriched in the PBMC signaling pathways in CML-UMRD patients. Network analysis on predicted targets of miRNA-148 was carried out using IPA (Ingenuity System), which provides an independent method of assessing the role of predicted miR-148b targets. The top network has ten nodal molecules that are predicted targets of miR-148b like DNMT3B, NR1I2, and TLR3 (Figure 3).
Figure 3.
Network analysis of miR-148b by Ingenuity Pathways Analysis.
Notes: The miR-148b-3p is located in the center. Solid lines indicate a direct relation, and broken lines indicates an indirect relation.
Discussion
One of the major questions in research on STOP-IM centers is the genetic and epigenetic markers for discriminating patients who can safely stop IM. Several empirical clinical studies have shown that durable stable UMRD before discontinuation, prior IFN therapy, and early deep molecular response are important to avoid molecular relapse.2,7,21,22 In this context, we particularly focused on miRNA expression profiling of PBMCs in CML patients who could safely stop IM. Since all patients in this study exhibited deep molecular response, we did not do miRNA profiling of CML cells; therefore, we could compare CML patients with or without IM and healthy volunteers. We found a distinct miRNA profile that was specific for the STOP-IM group, and we concluded that the miR-148b expression level might be a possible diagnostic marker for safely stopping IM.
miR-148a, miR-148b, and miR-152 are the three members of the miR148/152 family.23 Aberrant expression of the miR148/152 family has been reported in tumor as well as non-tumor disease. The present study was performed using non-tumor PBMCs; target genes regulated by miR-148b might be different from those in tumor tissue. For this reason, we tried to identify possible targets through use of the IPA software (Ingenuity System). There were several candidate targets for miR-148b as shown in Figure 3, such as cholecystokinin B receptor (CCBR), DNA methyltransferase 1 (DNMT1), DNMT3, nuclear receptor subfamily 1, group 1, member 2 (NR112), peroxisome proliferator-activated receptor alpha (PPARA), ribosomal protein S6 kinase, and polypeptide 5 (RPS6KA). Notably, DNMT1 and DNMT3 were among the candidates. The CpG island in the first intron of Foxp3 DNA is demethylated in naturally occurring human and murine T-regulatory (Treg) cells, and the methylation status is linked with Foxp3 expression.24 A recent report by Wang et al demonstrated Foxp3+ Treg cells require Dnmt1 expression to prevent development of lethal autoimmunity.25 Therefore, we could not rule out the possibility that down-regulation of miR-148b induces DNMT1 expression and promotes Treg suppression regulated by Foxp3 expression. In the present study, we were not able to analyze Treg function; however, Rea et al recently reported that NK cells rather than Treg cells are associated with clinical outcome after IM discontinuation.26 Since imatinib itself is known to inhibit CD4+CD25+ Treg activity,27 regulation of Treg after IM discontinuation might be partly compensated by down-regulation of miR-148b in a subset of STOP-IM patients.
In addition to DNMT1, we noted that miR-148b is indirectly linked to toll-like receptor 3 (TLR3) in the network presented by IPA (Ingenuity System). Liu et al recently reported that miRNA-148 and miRNA-152 fine-tune the regulation of the innate immune response and the antigen-presenting capacity of dendritic cells, and this fine-tuning may contribute to immune homeostasis and immune regulation.28 The expression of miR-148a, 148b, and miR152 was upregulated in mouse dendritic cells upon maturation, and activation was induced by Tlr3, Tlr4, and Tlr9 agonists. In the clinical context, we subdivided CML patients based on miR-148b expression and found that CML patients in the STOP-IM group and the IM-2 group showed different immunological features. Additionally, the percentage of NK cells was significantly lower in the IM-2 group. Although we did not intend to focus on immunological surveillance in STOP-IM patients at the beginning of this miRNA study, the present data may support previous reports regarding a possible role of NK cells in STOP-IM patients.8,10,26
In line with this, CML patients in the IM-1 group could stop IM, and miR-148 expression in PBMCs might be an additive biomarker for IM cessation. The correlation between miR-148b levels and treatment-free remission is not clear at this moment (Figure S1A); therefore, follow-up study is needed to clarify the clinical significance of miR-148b. We only had two patients who relapsed shortly after IM discontinuation (Figure S1B), and all STOP-IM patients enrolled in the current study had sustained UMRD for more than 12 months. Consequently, we could not determine whether miR-148b levels are correlated with relapse after stopping IM. Since the number of patients in the present study was rather small, an analysis of a larger number of patients in an independent cohort may clarify this point.
Furthermore, it is a matter for discussion whether PBMCs are suitable for miRNA analysis in clinical specimens. Analysis of the lymphocyte fractions (eg, NK cells and T cells) would be useful, but using fractionated samples from clinical specimens is difficult in practice. For this reason, we used circulating lymphocytes rather than purified NK or T cells; however, we could not completely rule out the possibility that the difference in miRNA expression was due to differences in cellular composition. Another limitation in the current study is that the miRNA profile was not from cancer cells; therefore, we could not perform a functional analysis of the target genes. Unfortunately, we could not perform biological experiments to demonstrate a possible association between downregulation of miR-148b and the immune surveillance in CML. We alternatively applied an IPA (Ingenuity System) bioinformatics tool to estimate the network between miR-148b and its target genes (Figure 3). Although our bioinformatics analysis is mainly based on previous knowledge base, it may help to understand the biological relevance of miR-148b in the current study. Finally, we could not find any relationship between BIM deletion and the level of miR-148b expression. This point should be clarified in a larger cohort in the near future.
Conclusion
Our observation of downregulated expression of miR-148b supports the idea that miRNA may contribute to immune surveillance in STOP-IM patients and may therefore have potential as an additive information marker in managing CML patients being treated with IM. Whether miR-148b may be used as a biomarker among other factors (eg, NK cells) to predict disease relapse after IM discontinuation needs to be investigated.
Supplementary materials
(A) miR-148b expression and treatment-free survival (TFS) in STOP-IM patients. Two patients who relapsed shortly after IM discontinuation are shown in open circles. (B) Sequential analysis of miR-148b expression in two patients who relapsed shortly after IM discontinuation.
Note: Patients who had discontinued IM and had sustained UMRD for more than 6 months (STOP-IM group). In contrast, patients who were receiving IM and had UMRD at the time of sample collection were categorized as the IM group.
Abbreviation: IM, imatinib mesylate; UMRD, undetectable minimal residual disease.
Table S1.
Fold changes, P-values, and quality control values of 22 genes differentially expressed among 3 groups (GEO accession number GSE47652).
| ID | ANOVA | Mean 1 | Mean 2 | Mean 3 | SEM 1 | SEM 2 | SEM 3 | Quality 1 | Quality 2 | Quality 3 |
|---|---|---|---|---|---|---|---|---|---|---|
| hsa-miR-96-4373372 | 0.027347 | 0.055553 | 0.310587 | 0.205137 | 0.017893 | 0.081936 | 0.063318 | 0 | 0 | 0 |
| hsa-miR-338-3p-4395363 | 0.010837 | 0.725438 | 1.7903 | 0.98898 | 0.178231 | 0.194392 | 0.524248 | 0 | 0.2 | 0.142857 |
| hsa-miR-362-3p-4395228 | 0.009886 | 1.7976 | 4.79113 | 1.02369 | 0.588755 | 0.763221 | 0.200925 | 0 | 0.6 | 0.142857 |
| hsa-miR-326-4373050 | 0.01577 | 0.697104 | 1.6751 | 0.306825 | 0.384774 | 0.295916 | 0.130009 | 0 | 0.2 | 0 |
| hsa-miR-323-3p-4395338 | 0.019971 | 1.58278 | 0.694484 | 2.70994 | 0.238736 | 0.118801 | 1.0002 | 0 | 0 | 0.142857 |
| hsa-miR-210-4373089 | 0.036942 | 99.0538 | 41.6222 | 151.605 | 43.0505 | 9.48987 | 32.3182 | 1 | 1 | 1 |
| hsa-miR-329-4373191 | 0.044299 | 0.227578 | 0.112847 | 0.263752 | 0.074624 | 0.011992 | 0.048371 | 0 | 0 | 0 |
| hsa-miR-454-4395434 | 0.0394 | 732.624 | 287.192 | 738.77 | 190.599 | 62.7271 | 154.128 | 1 | 1 | 1 |
| hsa-let-7b-4395446 | 0.040507 | 924.122 | 304.309 | 1128.06 | 333.482 | 84.3708 | 265.746 | 1 | 1 | 1 |
| hsa-miR-148b-4373129 | 0.016993 | 70.1809 | 19.1018 | 122.726 | 24.9349 | 5.41085 | 27.5564 | 1 | 1 | 1 |
| hsa-miR-449a-4373207 | 0.04547 | 0.268017 | 0.073807 | 0.406233 | 0.101705 | 0.022575 | 0.116278 | 0 | 0 | 0 |
| hsa-miR-487b-4378102 | 0.014862 | 5.60136 | 1.50581 | 6.74418 | 0.9649 | 0.323029 | 4.30608 | 0.6 | 0.4 | 0.142857 |
| hsa-let-7c-4373167 | 0.042081 | 6.04992 | 1.27731 | 8.23423 | 2.83505 | 0.284943 | 2.29054 | 0.4 | 0 | 0.428571 |
| hsa-miR-127-3p-4373147 | 0.000099 | 16.1789 | 7.28123 | 13.2454 | 0.098157 | 0.826206 | 4.04516 | 1 | 0.6 | 0.714286 |
| hsa-miR-625-4395542 | 0.021171 | 27.7682 | 5.90378 | 30.5204 | 10.5496 | 1.2195 | 6.98524 | 1 | 0.8 | 1 |
| hsa-miR-107-4373154 | 0.013937 | 12.7226 | 4.78675 | 8.64006 | 1.88017 | 0.770132 | 2.13526 | 1 | 0.6 | 0.571429 |
| hsa-miR-551b-4380945 | 0.02058 | 0.349719 | 0.118071 | 0.293659 | 0.059812 | 0.036671 | 0.063495 | 0 | 0 | 0 |
| hsa-miR-495-4381078 | 0.024363 | 19.448 | 7.30647 | 12.057 | 3.20282 | 0.834396 | 4.05008 | 1 | 0.6 | 0.571429 |
| hsa-miR-539-4378103 | 0.043747 | 5.21212 | 1.31043 | 3.7235 | 1.19105 | 0.313569 | 2.09332 | 0.6 | 0.4 | 0.142857 |
| hsa-miR-23a-4373074 | 0.004595 | 100.709 | 31.6829 | 17.4238 | 14.1188 | 14.0881 | 9.88996 | 1 | 0.6 | 0.428571 |
| hsa-miR-503-4373228 | 0.007081 | 1.17271 | 0.064389 | 2.96314 | 0.460532 | 0.025522 | 0.643415 | 0 | 0 | 0.142857 |
| hsa-miR-18a-4395533 | 0.006033 | 80.6828 | 4.76815 | 85.372 | 29.6422 | 0.762338 | 17.4761 | 1 | 0.6 | 1 |
Abbreviations: ANOVA, analysis of variance ID, identification document; SEM, standard error of measurement; GEO, Gene Expression Omnibus.
Acknowledgments
This work was supported by the Private University Strategic Research Based Support Project: Epigenetics Research Project Aimed at General Cancer Cure Using Epigenetic Targets from MEXT (Ministry of Education, Culture, Sports, Science and Technology), Tokyo, Japan.
Authors’ contributions
Junko H Ohyashiki designed and performed the research; Kazushige Ohtsuki performed PCR analysis; Seiichiro Katagiri and Kazuma Ohyashiki contributed essential reagents or tools; Izuru Mizoguchi, Takayuki Yoshimoto performed the FACS analysis; Tomohiro Umezu performed the bioinformatic analysis; Junko H Ohyashiki wrote the paper; and Kazuma Ohyashiki performed critical reading. All authors contributed toward data analysis, drafting, and revising the manuscript.
Disclosure
Kazuma Ohyashiki received research support from Novartis and BMS. Junko H Ohyashiki received research support from Novartis. This work was supported by the Private University Strategic Research Based Support Project: Epigenetics Research Project Aimed at General Cancer Cure Using Epigenetic Targets from MEXT (Ministry of Education, Culture, Sports, Science and Technology), Tokyo, Japan. The authors report no other conflicts of interest in this work.
References
- 1.Druker BJ, Guilhot F, O’Brien SG, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355(23):2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 2.Branford S, Yeung DT, Ross DM, et al. Early molecular response and female sex strongly predict stable undetectable BCR-ABL1, the criteria for imatinib discontinuation in patients with CML. Blood. 2013;121(19):3818–3824. doi: 10.1182/blood-2012-10-462291. [DOI] [PubMed] [Google Scholar]
- 3.Mahon FX, Rea D, Guilhot J, et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 2010;11(11):1029–1035. doi: 10.1016/S1470-2045(10)70233-3. [DOI] [PubMed] [Google Scholar]
- 4.Ross DM, Branford S, Seymour JF, et al. Safety and efficacy of imatinib cessation for CML patients with stable undetectable minimal residual disease: results from the TWISTER Study. Blood. 2013;122(4):515–522. doi: 10.1182/blood-2013-02-483750. [DOI] [PubMed] [Google Scholar]
- 5.Melo JV, Ross DM. Minimal residual disease and discontinuation of therapy in chronic myeloid leukemia: can we aim at a cure? Hematology Am Soc Hematol Educ Program Book. 2011:136–142. doi: 10.1182/asheducation-2011.1.136. [DOI] [PubMed] [Google Scholar]
- 6.Ohyashiki K, Katagiri S, Tauchi T, et al. Increased natural killer cells and decreased CD3(+)CD8(+)CD62L(+) T cells in CML patients who sustained complete molecular remission after discontinuation of imatinib. Br J Haematol. 2012;157(2):254–256. doi: 10.1111/j.1365-2141.2011.08939.x. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi N, Kyo T, Maeda Y, et al. Discontinuation of imatinib in Japanese patients with chronic myeloid leukemia. Haematologica. 2012;97(6):903–906. doi: 10.3324/haematol.2011.056853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mizoguchi I, Yoshimoto T, Katagiri S, et al. Sustained upregulation of effector natural killer cells in chronic myeloid leukemia after discontinuation of imatinib. Cancer Sci. 2013;104(9):1146–1153. doi: 10.1111/cas.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ng KP, Hillmer AM, Chuah CT, et al. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18(4):521–528. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- 10.Katagiri S, Umezu T, Ohyashiki JH, Ohyashiki K. The BCL2L11 (BIM) deletion polymorphism is a possible criterion for discontinuation of imatinib in chronic myeloid leukaemia patients. Br J Haematol. 2013;160(2):269–271. doi: 10.1111/bjh.12111. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.He L, Thomson JM, Hemann MT, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nature Immunol. 2008;9(8):839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 14.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10(2):111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 15.Yu Y, Yang L, Zhao M, et al. Targeting microRNA-30a-mediated autophagy enhances imatinib activity against human chronic myeloid leukemia cells. Leukemia. 2012;26(8):1752–1760. doi: 10.1038/leu.2012.65. [DOI] [PubMed] [Google Scholar]
- 16.Venturini L, Battmer K, Castoldi M, et al. Expression of the miR-17–92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood. 2007;109(10):4399–4405. doi: 10.1182/blood-2006-09-045104. [DOI] [PubMed] [Google Scholar]
- 17.San Jose-Eneriz E, Roman-Gomez J, Jimenez-Velasco A, et al. MicroRNA expression profiling in Imatinib-resistant Chronic Myeloid Leukemia patients without clinically significant ABL1-mutations. Mol Cancer. 2009;8:69. doi: 10.1186/1476-4598-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rokah OH, Granot G, Ovcharenko A, et al. Downregulation of miR-31, miR-155, and miR-564 in chronic myeloid leukemia cells. PLoS One. 2012;7(4):e35501. doi: 10.1371/journal.pone.0035501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Umezu T, Ohyashiki K, Ohyashiki JH. Detection method for quantifying global DNA methylation by fluorescence correlation spectroscopy. Anal Biochem. 2011;415(2):145–150. doi: 10.1016/j.ab.2011.04.035. [DOI] [PubMed] [Google Scholar]
- 20.Ohyashiki K, Umezu T, Yoshizawa S, et al. Clinical impact of down-regulated plasma miR-92a levels in non-Hodgkin’s lymphoma. PLoS One. 2011;6(2):e16408. doi: 10.1371/journal.pone.0016408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Etienne G, Dulucq S, Nicolini FE, et al. Achieving deeper molecular response is associated with a better clinical outcome in chronic myeloid leukemia patients on imatinib front-line therapy. Haematologica. 2014;99(3):458–464. doi: 10.3324/haematol.2013.095158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rousselot P, Charbonnier A, Cony-Makhoul P, et al. Loss of major molecular response as a trigger for restarting tyrosine kinase inhibitor therapy in patients with chronic-phase chronic myelogenous leukemia who have stopped imatinib after durable undetectable disease. J Clin Oncol. 2014;32(5):424–430. doi: 10.1200/JCO.2012.48.5797. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y, Song YX, Wang ZN. The microRNA-148/152 family: multi-faceted players. Mol Cancer. 2013;12:43. doi: 10.1186/1476-4598-12-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204(7):1543–1551. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Liu Y, Beier UH, et al. Foxp3+ T-regulatory cells require DNA methyltransferase 1 expression to prevent development of lethal autoimmunity. Blood. 20132;121(18):3631–3639. doi: 10.1182/blood-2012-08-451765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rea D, Dulphy N, Henry G, et al. Low Natural Killer (NK) Cell counts and functionality are associated with molecular relapse after imatinib discontinuation in patients (pts) with chronic phase (CP)-chronic myeloid leukemia (CML) with undetectable BCR-ABL transcripts for at least 2 years: preliminary results rrom immunostim, on behalf of STIM investigators. Blood. 2013;122(21) Abstract #856. [Google Scholar]
- 27.Larmonier N, Janikashvili N, LaCasse CJ, et al. Imatinib mesylate inhibits CD4+ CD25+ regulatory T cell activity and enhances active immunotherapy against BCR-ABL- tumors. J Immunol. 2008;181(10):6955–6963. doi: 10.4049/jimmunol.181.10.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Zhan Z, Xu L, et al. MicroRNA-148/152 impair innate response and antigen presentation of TLR-triggered dendritic cells by targeting CaMKIIalpha. J Immunol. 2010;185(12):7244–7251. doi: 10.4049/jimmunol.1001573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) miR-148b expression and treatment-free survival (TFS) in STOP-IM patients. Two patients who relapsed shortly after IM discontinuation are shown in open circles. (B) Sequential analysis of miR-148b expression in two patients who relapsed shortly after IM discontinuation.
Note: Patients who had discontinued IM and had sustained UMRD for more than 6 months (STOP-IM group). In contrast, patients who were receiving IM and had UMRD at the time of sample collection were categorized as the IM group.
Abbreviation: IM, imatinib mesylate; UMRD, undetectable minimal residual disease.
Table S1.
Fold changes, P-values, and quality control values of 22 genes differentially expressed among 3 groups (GEO accession number GSE47652).
| ID | ANOVA | Mean 1 | Mean 2 | Mean 3 | SEM 1 | SEM 2 | SEM 3 | Quality 1 | Quality 2 | Quality 3 |
|---|---|---|---|---|---|---|---|---|---|---|
| hsa-miR-96-4373372 | 0.027347 | 0.055553 | 0.310587 | 0.205137 | 0.017893 | 0.081936 | 0.063318 | 0 | 0 | 0 |
| hsa-miR-338-3p-4395363 | 0.010837 | 0.725438 | 1.7903 | 0.98898 | 0.178231 | 0.194392 | 0.524248 | 0 | 0.2 | 0.142857 |
| hsa-miR-362-3p-4395228 | 0.009886 | 1.7976 | 4.79113 | 1.02369 | 0.588755 | 0.763221 | 0.200925 | 0 | 0.6 | 0.142857 |
| hsa-miR-326-4373050 | 0.01577 | 0.697104 | 1.6751 | 0.306825 | 0.384774 | 0.295916 | 0.130009 | 0 | 0.2 | 0 |
| hsa-miR-323-3p-4395338 | 0.019971 | 1.58278 | 0.694484 | 2.70994 | 0.238736 | 0.118801 | 1.0002 | 0 | 0 | 0.142857 |
| hsa-miR-210-4373089 | 0.036942 | 99.0538 | 41.6222 | 151.605 | 43.0505 | 9.48987 | 32.3182 | 1 | 1 | 1 |
| hsa-miR-329-4373191 | 0.044299 | 0.227578 | 0.112847 | 0.263752 | 0.074624 | 0.011992 | 0.048371 | 0 | 0 | 0 |
| hsa-miR-454-4395434 | 0.0394 | 732.624 | 287.192 | 738.77 | 190.599 | 62.7271 | 154.128 | 1 | 1 | 1 |
| hsa-let-7b-4395446 | 0.040507 | 924.122 | 304.309 | 1128.06 | 333.482 | 84.3708 | 265.746 | 1 | 1 | 1 |
| hsa-miR-148b-4373129 | 0.016993 | 70.1809 | 19.1018 | 122.726 | 24.9349 | 5.41085 | 27.5564 | 1 | 1 | 1 |
| hsa-miR-449a-4373207 | 0.04547 | 0.268017 | 0.073807 | 0.406233 | 0.101705 | 0.022575 | 0.116278 | 0 | 0 | 0 |
| hsa-miR-487b-4378102 | 0.014862 | 5.60136 | 1.50581 | 6.74418 | 0.9649 | 0.323029 | 4.30608 | 0.6 | 0.4 | 0.142857 |
| hsa-let-7c-4373167 | 0.042081 | 6.04992 | 1.27731 | 8.23423 | 2.83505 | 0.284943 | 2.29054 | 0.4 | 0 | 0.428571 |
| hsa-miR-127-3p-4373147 | 0.000099 | 16.1789 | 7.28123 | 13.2454 | 0.098157 | 0.826206 | 4.04516 | 1 | 0.6 | 0.714286 |
| hsa-miR-625-4395542 | 0.021171 | 27.7682 | 5.90378 | 30.5204 | 10.5496 | 1.2195 | 6.98524 | 1 | 0.8 | 1 |
| hsa-miR-107-4373154 | 0.013937 | 12.7226 | 4.78675 | 8.64006 | 1.88017 | 0.770132 | 2.13526 | 1 | 0.6 | 0.571429 |
| hsa-miR-551b-4380945 | 0.02058 | 0.349719 | 0.118071 | 0.293659 | 0.059812 | 0.036671 | 0.063495 | 0 | 0 | 0 |
| hsa-miR-495-4381078 | 0.024363 | 19.448 | 7.30647 | 12.057 | 3.20282 | 0.834396 | 4.05008 | 1 | 0.6 | 0.571429 |
| hsa-miR-539-4378103 | 0.043747 | 5.21212 | 1.31043 | 3.7235 | 1.19105 | 0.313569 | 2.09332 | 0.6 | 0.4 | 0.142857 |
| hsa-miR-23a-4373074 | 0.004595 | 100.709 | 31.6829 | 17.4238 | 14.1188 | 14.0881 | 9.88996 | 1 | 0.6 | 0.428571 |
| hsa-miR-503-4373228 | 0.007081 | 1.17271 | 0.064389 | 2.96314 | 0.460532 | 0.025522 | 0.643415 | 0 | 0 | 0.142857 |
| hsa-miR-18a-4395533 | 0.006033 | 80.6828 | 4.76815 | 85.372 | 29.6422 | 0.762338 | 17.4761 | 1 | 0.6 | 1 |
Abbreviations: ANOVA, analysis of variance ID, identification document; SEM, standard error of measurement; GEO, Gene Expression Omnibus.