Abstract
The WNK1 (WNK lysine deficient protein kinase 1) protein is a serine/threonine protein kinase with emerging roles in cancer. WNK1 causes hypertension and hyperkalemia when overexpressed and cardiovascular defects when ablated in mice. In this study, the role of Wnk1 in angiogenesis was explored using the zebrafish model. There are two zebrafish wnk1 isoforms, wnk1a and wnk1b, and both contain all the functional domains found in the human WNK1 protein. Both isoforms are expressed in the embryo at the initiation of angiogenesis and in the posterior cardinal vein (PCV), similar to fms-related tyrosine kinase 4 (flt4). Using morpholino antisense oligonucleotides against wnk1a and wnk1b, we observed that wnk1 morphants have defects in angiogenesis in the head and trunk, similar to flk1/vegfr2 morphants. Furthermore, both wnk1a and wnk1b mRNA can partially rescue the defects in vascular formation caused by flk1/vegfr2 knockdown. Mutation of the kinase domain or the Akt/PI3K phosphorylation site within wnk1 destroys this rescue capability. The rescue experiments provide evidence that wnk1 is a downstream target for Vegfr2 (vascular endothelial growth factor receptor-2) and Akt/PI3K signaling and thereby affects angiogenesis in zebrafish embryos. Furthermore, we found that knockdown of vascular endothelial growth factor receptor-2 (flk1/vegfr2) or vascular endothelial growth factor receptor-3 (flt4/vegfr3) results in a decrease in wnk1a expression, as assessed by in situ hybridization and q-RT-PCR analysis. Thus, the Vegf/Vegfr signaling pathway controls angiogenesis in zebrafish via Akt kinase-mediated phosphorylation and activation of Wnk1 as well as transcriptional regulation of wnk1 expression.
Introduction
The development of the vascular system occurs via vasculogenesis and angiogenesis. Vasculogenesis refers to the de novo formation of vessels [1]; in angiogenesis, new blood vessels form by remodeling and extending old ones [2]. The most important molecules governing angiogenesis are the VEGF (vascular endothelial growth factor) family members and their receptors [3]. There are three different VEGF receptors, VEGFR1 (FLT1), VEGFR2 (KDR/FLK1) and VEGFR3 (FLT4). VEGFR2 mediates the majority of the downstream angiogenic effects of VEGF. These angiogenic effects include changes in microvascular permeability and endothelial cell proliferation, invasion, migration and survival. Upon activation by the binding of VEGF, the VEGFR2 tyrosine kinase phosphorylates downstream kinases, such as the phosphoinositide-dependent protein kinase (PI3 kinase), which then phosphorylates and activates the protein kinase Akt/PKB1. Multiple Akt/PKB substrates have been discovered, and WNK1 is a novel Akt/PKB substrate in insulin-stimulated 3T3-L1 adipocytes [4].
WNK1 (WNK lysine deficient protein kinase 1) protein is a novel mammalian serine/threonine protein kinase that lacks the invariant catalytic lysine found in subdomain II of all MAPKs, which is crucial for binding ATP and instead contains a catalytic lysine at position 233 in subdomain I [5]. Wnk1 cDNA was identified in a screen for MAPK family members in the mouse brain [5]. WNK1 was found to be overexpressed in invasive colorectal cell lines [6]. WNK1 activates ERK5 by phosphorylating MEKK2/3, which is upstream of ERK5 in human embryonic kidney 293 (HEK293) cells [7]. Knocking down Wnk1 in the C17.2 mouse neural progenitor cell line resulted in decreased Erk5 activity, which reduced cell proliferation, migration and differentiation [8]. Other reports indicate that the WNK1 protein has a protein kinase AKT/protein kinase B (PKB) phosphorylation consensus sequence, and it has been shown that AKT kinase phosphorylates threonine 60 (Thr60) of WNK1 [9]. It is possible that during angiogenesis, VEGF/VEGFR2 phosphorylates and activates PI3 kinase, which then phosphorylates and activates AKT kinase, which might then phosphorylate WNK1.
WNK kinases contain an autophosphorylation domain, and serine 382 in the activation loop was shown to be required for autophosphorylation. The autoinhibitory domain, which is conserved in all four WNKs, suppresses the activity of the WNK1 kinase domain, and the two key residues required for the function of the autoinhibitory domain have been identified [10].
Overexpression of WNK1 causes hypertension and hyperkalemia in humans by altering renal Na+ and K+ transport [11]. WNK1 activates the downstream protein kinases STE20/SPS1-related proline-alanine-rich protein kinase (SPAK) and oxidative stress responsive 1 (OSR1) through phosphorylation of the t-loop in the catalytic domain [12], [13], [14], [15]. Activated SPAK and OSR1 associate and then phosphorylate and activate other ion co-transporters, including Na+/K+/2Cl− co-transporter 1 (NKCC1) [16], [17]. NKCC1 is a ubiquitous ion transporter [18] that controls cell volume and maintains osmostasis through absorption of Na+, K+ and Cl− ions [19], [20], suggesting that under hyperosmotic conditions, WNK1 can regulate the activity of NKCC1 through SPAK and OSR1 [21].
In the past, all research on WNK1 has focused on its function in cancer cell proliferation, differentiation, migration and apoptosis. In studies of somatic cells, WNK1 involvement in renal Na+ and K+ transport is also well known. However, the physiological function of WNK1 outside the kidney remains unclear. Using gene disruption and rescue in mice, Xie et al. found that Wnk1 function is required for embryonic angiogenesis and cardiac development, with Wnk1 deletion affecting artery-vein specification [22]. The mechanism by which Wnk1 affects angiogenesis, however, remains largely unknown.
The zebrafish has emerged as a powerful vertebrate model system for development [23], [24], organogenesis [25], [26], [27], vasculogenesis [1], neurogenesis [28], [29] and carcinogenesis [30], [31]. Zebrafish and other vertebrates have highly conserved genomic sequences; thus, zebrafish can be used to analyze the developmental process of embryo formation as well as human disease pathology [1], [32]. Moreover, a range of forward and reverse genetic methods, including antisense morpholino oligonucleotide (MO)-based knockdown and Tol2 transgenesis, have been developed for functional analysis of genes in zebrafish [33], [34]. Green fluorescent protein (GFP) under the control of the fli1 or flk1 regulatory region is specifically expressed in angioblasts [35], [36]. The existence of transgenic lines expressing vessel-specific GFP also facilitates the study of vasculogenesis and angiogenesis in zebrafish.
Previous studies have shown that Wnk1 homozygous mutant mice die at E13 and have significant defects in angiogenesis, whereas Wnk1 heterozygous mice and wild-type mice do not differ significantly in hypotension, embryogenesis or angiogenesis [22], [37]. To further understand the role of Wnk1 in angiogenesis, we used the vessel-specific transgenic zebrafish line Tg(fli1:EGFP). We found that knockdown of wnk1 by MOs led to defects in the angiogenic sprouting of intersegmental vessels (ISVs). Unlike Wnk1 mutant mice, wnk1 zebrafish morphants survive. Analysis of these morphants has furthered our understanding of the role Wnk1 plays during angiogenesis and helped to identify the signaling pathway through which Wnk1 functions.
Materials and Methods
Zebrafish husbandry
Zebrafish embryos, larvae, and adult fish were maintained in the Zebrafish Core Facility at NTHU-NHRI (ZeTH) according to established protocols and methods [38]. Animal use protocols for zebrafish were approved by the Institutional Animal Care Use Committee (IACUC) of the National Health Research Institutes. The animal protocol number is NHRI-IACUC-096037-A.
Resources for sequence information
The zebrafish genomic sequence of WNK1 (NW_001878589) was obtained from the National Center for Biotechnology Information (NCBI). The human WNK1 protein sequence (gi|2711660|NP_061852.1) was obtained from the NCBI databank and used in a tBlastn search of the zebrafish RefSeq RNA database to identify zebrafish Wnk1 homologs.
RNA extraction, RT-PCR and quantitative polymerase chain reaction (q-RT-PCR)
Total RNA was extracted from twenty embryos at different embryonic stages with a NucleoSpin RNA II kit (MACHEREY-NAGEL). Complement DNA was synthesized from 900 ng of RNA using MutiScribe Reverse Transcriptase with the following program: 25°C-10 min → 48°C-30 min → 95°C-5 min → 12°C-indefinite. After the RT reaction, we diluted the cDNA 20-fold and performed the q-RT-PCR as described below.
Quantitative PCR studies were performed on selected clones using gene-specific primers. The rate of PCR amplification in one population vs. another (after normalizing against housekeeping genes) reflects differences in the expression of the gene in the two populations. We used GFP DNA as our standard to calculate the number of molecules after a given cycle number. Each q-RT-PCR Ct (cycle number) can be converted into number of molecules according to the standard curve and then converted into molecules per embryo by dividing by the number of embryos used.
DNA constructs: cloning and vectors
The wnk1a and wnk1b cDNA was generated by PCR amplification. The primers used to amplify the wnk1a and wnk1b cDNA are listed below.
wnk1a -F-NotI: 5′-ATATGCGGCCGC ATGGTCAAGTTCCTTTCCCC-3′ (bold letters indicate a NotI site, and italic letters denote the translation initiation site).
wnk1a-OUT-R: 5′-ACCCATTCGTGCCTCTATCA-3′.
wnk1b- F-NotI: 5′-ATATGCGGCCGCCTGGAAAGATGTCATCGGAAA-3′ (bold letters indicate a NotI site, and italic letters denote the translation initiation site).
wnk1b- OUT-R: 5′-CGTGGCATATTTGTGAGCAT-3′.
The PCR product was cloned into a cloning vector using the T&A cloning kit (Yeastern Biotech, catalog # YC001) and TaKaRa DNA ligation kit (TAKARA BIO INC, catalog #6024).
Rat Wnk1(1–449) was generated by PCR amplification from rat Wnk1(1–491)/pCMV5-Myc (kindly provided by Dr. Huang C. L., who obtained from Dr. Cobb’s lab). The primers used to amplify the rat Wnk1(1–449) are listed below.
Rat-EcoRI-Wnk1(1–449)-F: 5′-ATCGGAATTC ATGTCTGACGGCACCGCAGA-3′.(bold letters indicate an EcoRI site, and italic letters denote the translation initiation site).
Rat-XbaI-Wnk1(1–449)-R: 5′- ATCGTCTAGAAATTGCTACTTTGTCAAAAC-3′ (bold letters indicate an XbaI site).
Rat-Wnk1-seq-F: 5′-CCTAGTGTACCCGCAGTGGT-3′.
Rat-Wnk1-seq-R1: 5′-ACCACTGCGGGTACACTAGG-3′.
Rat-Wnk1-seq-R2: 5′-TAGCCATCTCAAGCATGCAC-3′.
The PCR product was digested with EcoRI and XbaI (NEW ENGLAND Biolabs Inc., catalog #R0101L and #R0145L), purified with a MinElute PCR Purification Kit (QIAGEN, catalog #28006), and subsequently cloned into an EcoRI- and XbaI-linearized pCS2+ vector using the T&A cloning kit (Yeastern Biotech, catalog # YC001) and TaKaRa DNA ligation kit (TAKARA BIO INC, catalog #6024).
Microinjection
Microinjections were carried out as previously described [38]. For the MO experiments, we injected 2.5–15 ng of MO were microinjected into each one-cell-stage embryo. For the RNA injections, 150–600 pg of mRNA was microinjected into each one cell-stage embryo.
Site-directed mutagenesis
We utilized the QuikChange II Site-Directed Mutagenesis Kit from Stratagene to generate the kinase dead and Akt phosphorylation site mutations in wnk1. wnk1 cDNA in the yT&A vector (50 ng) was used as the template DNA in each reaction. Following amplification (95°C for 5 minutes, 18 cycles of 95°C for 30 seconds, 70°C for 30 seconds, and 68°C for 7 minutes 30 seconds), the product was treated with 1 µl of the DpnI (10 U/µl) restriction enzyme to digest the parental methylated and hemimethylated dsDNA. The nicked vector DNA containing the desired mutations was then transformed into XL-1Blue super competent cells for nick repair and amplification.
The oligonucleotide sequences used to generate the mutations are given below. The underlined bases denote the mutation sites for site-directed mutagenesis.
Akt phosphorylation site mutant:
wnk1a 35 point mutation-F: 5′-CCGTCGTCGCCACCACGCCATGGATCGAGAACTGC-3′.
wnk1a 35 point mutation-R: 5′-GCAGTTCTCGATCCATGGCGTGGTGGCGACGACGG-3′.
Kinase-dead mutant:
wnk1a 206 point mutation-F: 5′-TAGGACGTGGCTCTTTTTGTACGGTCTACAAGGGACTG-3′.
wnk1a 206 point mutation-R: 5′-CAGTCCCTTGTAGACCGTACAAAAAGAGCCACGTCCTA-3′.
Morpholinos used to knockdown gene expression
Morpholino oligonucleotides (MOs) were custom-made by Gene Tools (Philomath, OR, USA) and injected at doses that generated distinct gene knockdown effects while yielding the highest proportion of living embryos with the knockdown phenotype. The MO sequences and target sites are given below.
wnk1a ATG MO: 5′-ACTTGACCATCTTGTCGTTGAGATT-3′
(Against wnk1a translation start codon, –15 to +10.)
wnk1a 5MM MO: 5′-ACTTCACGATCTTCTCCTTGACATT-3′
(Against wnk1a translation start codon, –15 to +10, but containing the five underlined mismatches.)
wnk1a Up MO: 5′-TCCACCAAGTGGAGCGTGAAGTTAG-3′
(Against wnk1a upstream of translation start codon, –52 to –28.)
wnk1b Up MO: 5′-TGCGTAAATTTCCTGCTCTTGCTT-3′
(Against wnk1b upstream of translation start codon, –100 to –77.)
wnk1b ATG MO: 5′-TGGGATTTTCCGATGACATCTTTCC-3′
(Against wnk1b translation start codon, –6 to +19.)
flk1 MO: 5′-GTCTGTTAAAATAACGTCCCGAATG-3′
(Against flk1 upstream of translation start codon, –28 to –4.)
flt4 MO: 5′-CTCTTCATTTCCAGGTTTCAAGTCC-3′
(Against flt4 translation start codon, –17 to +8.)
pi3kc2α: 5′-TATGTGGGCCATGGTGTCAGCTCT-3′
(Against pi3kc2α translation start codon, –12 to +12.)
Lyophilized MOs were resuspended to a final concentration of 10 mM in sterilized ddH2O, and the solution was heated for 5 minutes at 65°C to dissociate aggregates of the powder. The MO solution was divided into 10-µl aliquots. Before use, each aliquot was heated to 65°C for 5 to 10 minutes and then cooled to room temperature.
In vitro transcription of mRNA
We used the mMESSAGE mMACHINE T7 Ultra kit (Applied Biosystems, catalog # AM1345) for in vitro transcription of wild-type wnk1a and wnk1b and the two wnk1a mutants. The synthesized mRNAs were checked by agarose gel electrophoresis with RNA Millennium Markers (Applied Biosystems catalog # AM7150) for integrity. The concentration of in vitro transcribed mRNA was determined using a NanoDrop ND-1000 UV-Vis Spectrophotometer. Rat Wnk1(1–449) mRNA was generated from a pCS2+ vector construct by in vitro transcription using the mMESSAGE mMACHINE SP6 kit (Applied Biosystems, catalog # AM1340).
Whole-mount in situ hybridization (WMISH)
Whole-mount in situ hybridization was carried out as previously described [38].
To generate probes for in situ hybridization, we first amplified wnk1a, wnk1b, flt4, vegfc, and etv2 cDNA by PCR. The primers used to generate the DNA templates are listed in the supporting data (Data S1). We used the MEGAscript T7 kit (Ambion, catalog # AM1333) for in vitro transcription of digoxigenin (DIG)-labeled and fluorescein-labeled probes. Zebrafish embryos were collected and fixed in 4% paraformaldehyde at the indicated time points. To optimize hybridization, 5% dextran sulfate was added to the hybridization buffer for the DIG-labeled probes [39]. After post-hybridization washes to remove excess probe, the embryos were blocked with 1% blocking reagent (Roche Molecular Biochemicals, Mannheim, Germany) for 1 hour and then incubated overnight with pre-absorbed alkaline phosphatase (AP)-conjugated anti-DIG antibody (Roche Molecular Biochemical; 1∶2000 dilution) at 4 °C. Transcripts were visualized by AP-based NBT/BCIP staining under identical conditions and staining times.
To confirm the localization of wnk1a and wnk1b in the PCV of zebrafish embryo, double in situ hybridization was performed using fluorescein-labeled wnk1a or wnk1b probe mixed with DIG-labeled flt4 probe and detected with an AP-conjugated anti-fluorescein antibody followed by an AP-conjugated anti-DIG antibody. The wnk1a and wnk1b transcripts were visualized by AP-based NBT/BCIP, and the flt4 and vegfc transcripts were visualized by AP-based fast red (Roche Molecular Biochemicals, Mannheim, Germany) staining. Therefore, wnk1a and wnk1b expression appears blue while flt4 expression is labeled in red. In between visualization of the fluorescein- and DIG-labeled probes, the AP enzymatic reaction was inactivated by incubating embryos in 100 mM glycine-hydrochloride (pH2.2) for 30 minutes.
Construction of wnk1a-GFP and wnk1b-GFP for testing morpholino knockdown efficiency and specificity
To test the knockdown efficiency of wnk1a and wnk1b MOs, 263-bp and 286-bp fragments of wnk1a and wnk1b mRNA containing the MO targeting sites were amplified from zebrafish cDNA by PCR. For the wnk1a-GFP construct, the primers used were wnk1a-EcoRI (5′-AATAGAATTCTTCCACTTGGTTTAAAGCGG-3′, bold letters indicate an EcoRI site) and wnk1a-BamHI (5′-AATAGGATCCGCCTTCAGCAGTTCTCGATC-3′, bold letters indicate a BamHI site). For the wnk1b-GFP construct, the primers used were wnk1b-EcoRI (5′-AATAGAATTCAACTCTGTGGTTCACGTGAG-3′, bold letters indicate an EcoRI site) and wnk1b-BamHI (5′- AATAGGATCCTGGCGTCGCTTTCTGAC-3′, bold letters indicate a BamHI site). The PCR products were cloned into the pEGFP-N1 plasmid. Linearized wnk1a-GFP and wnk1b-GFP plasmids (200 pg) were microinjected into one-cell-stage embryos along with 10 ng of various wnk1a and wnk1b MOs as shown in Fig. S1.
Fluorescent microscopy
Embryos were removed from their chorions with watchmaker forceps. For live imaging, the embryos were anesthetized with 0.168 mg/ml tricaine (Sigma, catalog # A-5040) and placed on a 1% agarose plate. For imaging of fixed tissues, the embryos were incubated in a 4% formaldehyde-PBS (1x PBS) solution overnight at 4°C, washed with PBST (1x PBS, 0.1% Tween 20) and then mounted in 1% PBS-low melt agarose (Zymeset). Embryos of different stages were collected for GFP visualization and photography. All imaging was performed on a Zeiss SteREO Discovery.V8 fluorescent microscope equipped with a Zeiss AxioCam MRc CCD camera.
Results
Identification of zebrafish wnk1 and cloning of full-length wnk1 cDNA
To identify the zebrafish wnk1 orthologue, the human WNK1 protein sequence was used to search the translated zebrafish RefSeq RNA database using tBLASTn. Four sequences, XM_684564.5 (wnk1), XM_002666846.2 (wnk1), XM_003201205.1 (wnk3-like), and XM_680072.5 (wnk4-like), were found to be homologous to human WNK1.
We designed primers based on the first two homologs and cloned wnk1 from zebrafish using RT-PCR. The zebrafish wnk1a cDNA is 4794 bp long, which is slightly different from the previously reported NCBI GenBank sequence XM_678215.3 and very similar to XM_002666846.2. The zebrafish wnk1b cDNA is 5730 bp long and similar to XM_684564.5 with large internal deletions.
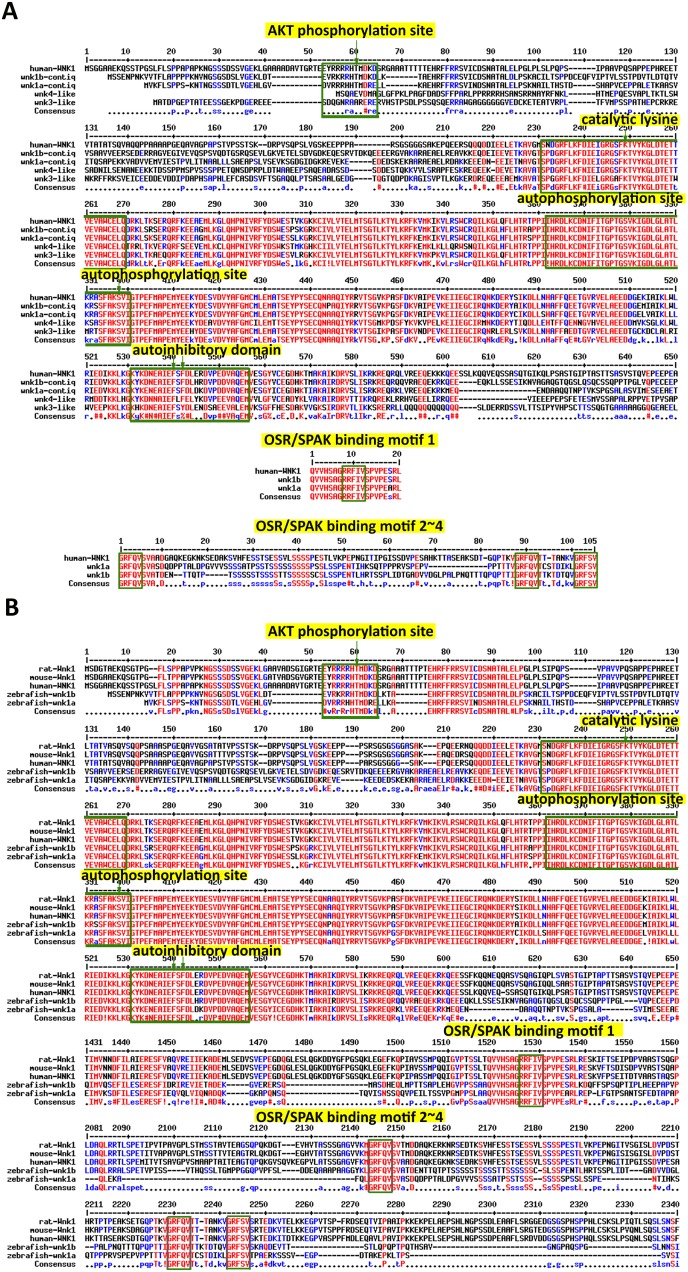
The translated amino acid sequences of the four zebrafish wnks were compared with the human WNK1 protein sequence (Fig. 1A). As in human WNK1, a glycine in the glycine string of the MAPK motif is replaced by lysine in all four Wnk1 homologs in zebrafish. Additionally, all four zebrafish Wnk proteins contain an auto-inhibitory domain and a serine residue that is susceptible to auto-phosphorylation (Fig. 1A).
Figure 1. Sequence comparison between human WNK1 and zebrafish Wnk proteins.
(A) Alignment of four zebrafish Wnks with the human WNK1 protein sequence. The WNK1 signature domain is highlighted. The AKT phosphorylation site at threonine 60 (arrow) in WNK1 is specific to Wnk1a and Wnk1b. The catalytic lysine (arrow) is located in a highly conserved region among all Wnk1s. The autophosphorylation site and the autoinhibitory domain are conserved between all four Wnk1s. The four OSR/SPAK binding motifs found in human WNK1 are specific to Wnk1a and Wnk1b. (B) Alignment of WNK1 protein sequences from mouse, rat, human and zebrafish.
Among the four mammalian WNK family members, only WNK1 has an Akt phosphorylation site and the four RFXV (arginine-phenylalanine-any amino acid-valine) motifs required for binding to oxidative stress responsive 1 (OSR1) and STE20/SPS1-related proline-alanine-rich protein kinase (SPAK). Akt can phosphorylate WNK1, and OSR1 and SPAK are endogenous substrates for WNK1. Of the four zebrafish Wnks, only Wnk1a and Wnk1b have the Akt phosphorylation site and four OSR1/SPAK binding motifs (Fig. 1A). Based on these unique signatures, we confirmed that zebrafish wnk1a and wnk1b are mammalian WNK1 homologs.
The WNK1 amino acid sequences from rat, mouse, human and zebrafish were also compared (Fig. 1B). In all examined species, the Wnk1 protein exhibited strong similarity in the AKT phosphorylation site, the catalytic domain, the autophosphorylation domain, the autoinhibitory domain and four OSR/SPAK binding domains. Previous work on Wnk1 has identified important residues for each of the domains [10]; these residues are also conserved between species. The rat Wnk1(1–449) truncation has been proven to be constitutively active using an in vitro kinase assay [10].
Spatial and temporal expression of wnk1 in zebrafish development
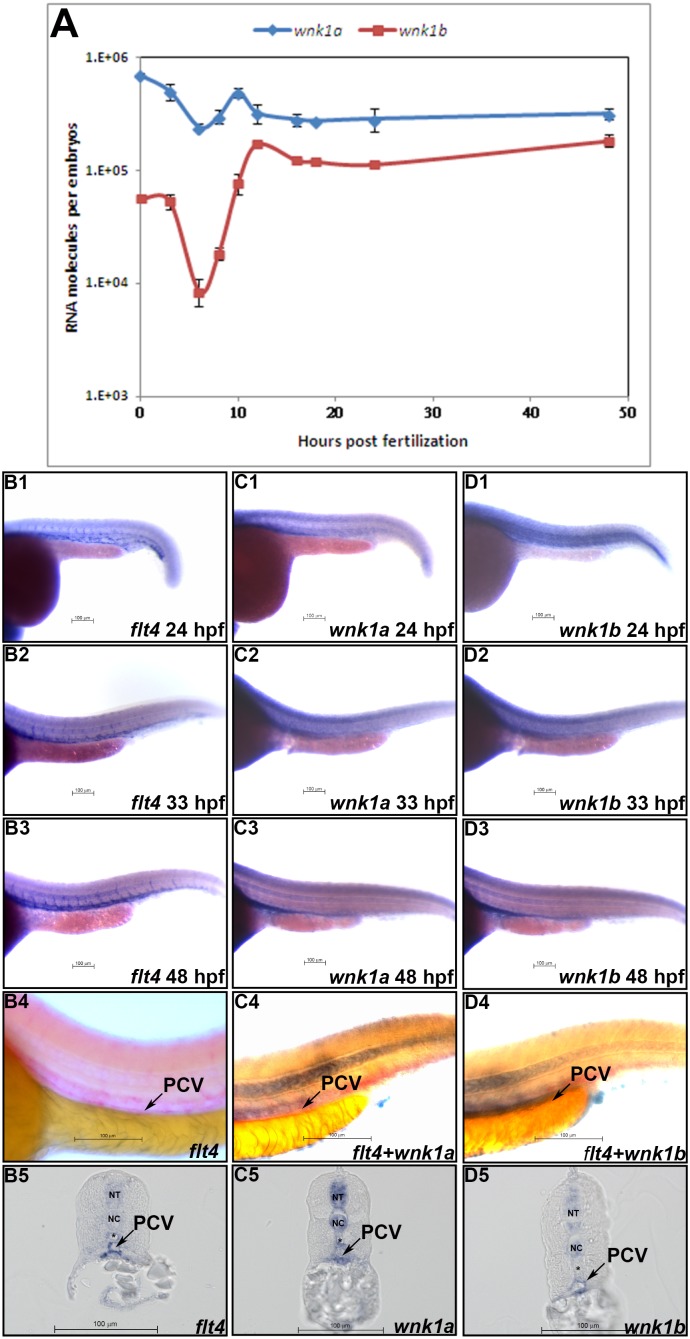
To understand the role of wnk1 in embryogenesis, we examined the temporal expression profile of wnk1 by quantitative reverse transcription polymerase chain reaction (q-RT-PCR) (Fig. 2A). wnk1a and wnk1b mRNAs are maternally deposited, and zygotic expression of both begins as early as 10 hpf (hours post fertilization) and remains high at all stages examined. The expression of flk1/vegfr2 begins as early as the 5 to 9 somite stage (12 to 15 hpf) in the intermediate cell mass of the mesoderm [40] and the 14 to 19 somite stage (16 to 19 hpf) in the head and vessel endothelial cells [41]. Additional studies have reported that flk1/vegfr2 is expressed in the intersegmental vessels at 24 hpf [42] when the single circulatory loop in zebrafish is established [43], [44]. Overall, the temporal expression of wnk1 is consistent with its role in angiogenesis. We did not detect significant expression of wnk3 or wnk4 in the zebrafish embryo prior to 48 hpf (data not shown).
Figure 2. Spatial and temporal expression patterns of wnk1a and wnk1b.
(A) wnk1a and wnk1b mRNA expression profiles as determined by q-RT-PCR. At least three replicates were performed, and the average number of molecules was calculated using a standard curve from a q-RT-PCR assay. The standard deviations are shown in the graph. The red and blue lines indicate the wnk1a and wnk1b expression profiles, respectively. (B∼D) Whole-mount in situ hybridization for flt4 (B) wnk1a (C), and wnk1b (D) was performed at the indicated time points. flt4 (B1∼B3), wnk1a (C1∼C3), and wnk1b (D1∼D3) mRNA expression in the tail is shown at 24, 33 and 48 hpf. Double in situ hybridization for flt4 alone (B4), wnk1a+flt4 (C4), and wnk1b+flt4 (D4) shows the co-localization of wnk1a and wnk1b with flt4 in the PCV. Expression of flt4 (B5), wnk1a (C5) and wnk1b (D5) is seen in the PCV (arrow) in sections from stained embryos. wnk1a and wnk1b are also expressed in the neural tube (NT) and notochord (NC). The dorsal aorta (*) is negative for wnk1a and wnk1b expression. Scale bar: 100 µm.
q-RT-PCR was performed using cDNA from whole embryos, including non-vascular tissues. We next examined wnk1a and wnk1b expression in blood vessels by whole-mount in situ hybridization. At 3 hpf, maternal wnk1a and wnk1b mRNA are ubiquitously expressed in embryos (Fig. S2-A1, B1). At 6 and 8 hpf, expression of both wnk1a and wnk1b transcripts is decreased (Fig. S2-A2, A3, B2, B3). At 10 and 12 hpf, zygotic wnk1a and wnk1b are expressed ubiquitously (Fig. S2-A4, A5, B4, B5). At 18 hpf, wnk1a is strongly expressed in head and somite tissues (Fig. S2-A6), whereas wnk1b is also expressed in the notochord (Fig. S2-B6). Similarly, from 24 to 48 hpf, wnk1a is detected in the head, neural tube, somite, and PCV (Fig. S2-A7∼A12), whereas wnk1b is detected in all those tissues plus the notochord. Similar to wnk1a, wnk1b is expressed in the PCV after 33 hpf (Fig. S2-B7∼B12). We used flt4, which is known to be expressed in the posterior cardinal vein (PCV), as a marker and found that both wnk1a and wnk1b are expressed in the PCV from 24 to 48 hpf (Fig. 2A1∼A3, B1∼B3 and C1∼C3).
To confirm the localization of wnk1a and wnk1b in the PCV of zebrafish embryos, double in situ hybridization with antisense wnk1a or wnklb riboprobes and a flt4 riboprobe was performed. The flt4 transcript was detected with an AP-conjugated anti-DIG antibody and visualized with AP-Fast Red (Fig. 2A4) while wnk1a and wnk1b were detected with an AP-conjugated anti-fluorescein antibody. Our data showed that wnk1a and wnk1b colocalized with flt4 in the PCV. We also observed wnk1a and wnk1b expression in the notochord (Fig. 2B4, C4). Embryos hybridized with the sense probes for wnk1a and wnk1b have no staining, indicating that the staining observed with the antisense probe is specific (Fig. S2-A13 and B13). Thus, colocalization studies using double in situ hybridization to detect both wnk1a or wnk1b and the PCV marker flt4 in the same embryo indicate that wnk1a and wnk1b are expressed in vascular structures specific to the PCV.
We also examined sections of embryos following whole-mount in situ hybridization for flt4, wnk1a or wnk1b. We found flt4 expression in the PCV whereas wnk1a and wnk1b expression was observed in the PCV, the neural tube (NT) and the notochord (NC) (Fig. 2A5, B5 and C5). All of these results suggest that wnk1a and wnk1b are indeed expressed in the vascular structures of the PCV.
Quantitative analysis of the effect of wnk1 knockdown on angiogenesis
To investigate the functional role of wnk1 in zebrafish embryos, we used the Tg(fli1:EGFP) strain, which allows immediate and direct in situ monitoring of vessel formation. Translation of endogenous wnk1 was suppressed in Tg(fli1:EGFP) by targeting wnk1a and wnk1b with specific antisense morpholino oligonucleotides (MOs). Morpholino specificity was verified by testing their ability to suppress the expression of a fusion protein that contained the targeted wnk1 sequence (Fig. S1). The primer and nucleotide sequence for the wnk1a-GFP and wnk1b-GFP constructs are provided in the supporting data (Data S1). Figure S1 shows the target sites for the MOs, the number of injected embryos, and the quantification of GFP expression in the morphants. Morpholinos that target the 5′UTRs of wnk1a and wnk1b also efficiently inhibit the expression of the corresponding GFP fusion construct (Fig. S1-A4, B4). In contrast, the control MO does not suppress the expression of the GFP fusion protein (Fig. S1-A5, B5). Furthermore, wnk1b Up MOs do not inhibit wnk1a-GFP (Fig. S1-A6) and wnk1a MOs do not suppress wnk1b-GFP (Fig. S1-B6, B7). Our results demonstrate that wnk1a MOs specifically inhibit wnk1a-GFP expression and wnk1b MOs specifically inhibit wnk1b-GFP expression.
To determine the role that wnk1 plays in angiogenesis, we designed and tested MOs against wnk1a, wnk1b and other genes required for angiogenesis. ISVs sprout and elongate dorsally from the DA and the PCV and connect to the DLAV between 29 and 36 hpf [45]. We analyzed embryos at 33 hpf, when the ISVs are forming. Because ISV development begins in the anterior trunk and proceeds posteriorly, we calculated the lengths of the ISVs from the middle of the yolk to the yolk extension. ISVs were observed in age-matched embryos and categorized as having extended over 100%, 75%, 50%, 25% or 0% of the distance from the DA (or PCV) to the DLAV (Fig. S3-A). Due to variations in the effects of morpholino injection from embryo to embryo, we examined many embryos in each group. The phenotype and motility of the embryos were classified as described in Fig. S3-B. The number of embryos analyzed (n) and the means, standard deviation and significance from three independent experiments are indicated in the figures.
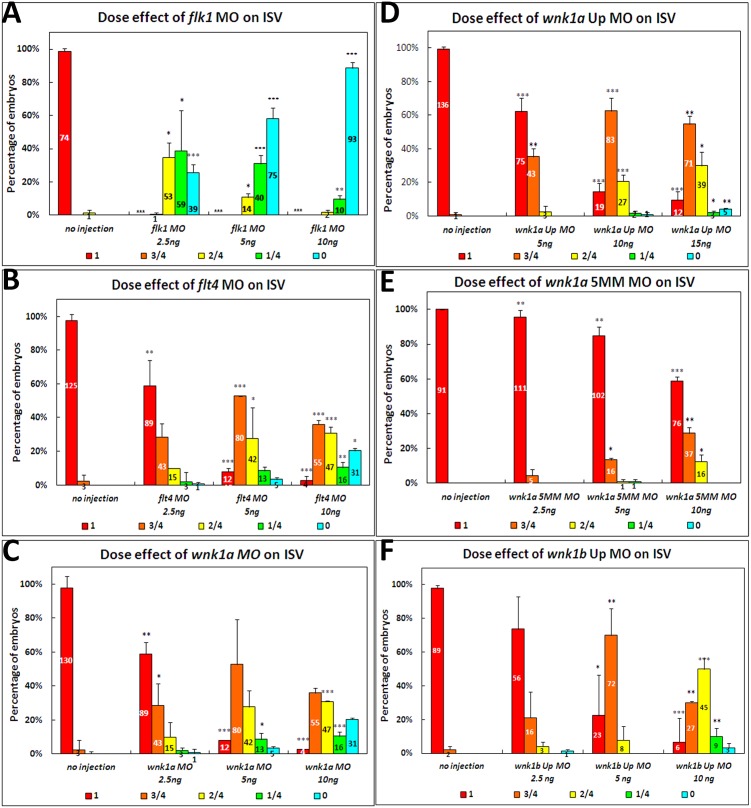
In mice, VEGFR2 (KDR/FLK1) is the primary VEGFA receptor in the developing endothelial lineage and it is essential for endothelial differentiation during embryonic vasculogenesis as well as angiogenesis [46]. VEGFR2 also plays important roles in physiological as well as pathological postnatal angiogenesis [47]. In zebrafish, VEGFR2 is essential for angiogenesis only, and knockdown of vegfr2/flk1 using morpholinos causes angiogenesis defects as shown by the shortening of ISVs (Fig. 3A). The inhibition of ISVs by flk1 MO knockdown is dose-dependent.
Figure 3. Statistical analysis of the length of intersegmental vessels in flk1, flt4, wnk1a, wnk1a 5MM, wnk1a upstream and wnk1b upstream morpholino-injected embryos.
The effects of (A) flk1 MO, (B) flt4 MO, (C) wnk1a MO, (D) wnk1a 5 base mismatch MO, (E) wnk1a upstream MO, and (F) wnk1b upstream MO on the length of intersegmental vessels. Morpholino injection has a dose-dependent effect on intersegmental vessel formation and growth. All experiments were performed at least three times, and the average ISV length was calculated along with the standard deviation, which is labeled on each bar. Red indicates that the ISVs grew to full length, orange indicates that the ISVs were 75% of the normal length, yellow indicates 50%, green indicates 25%, and light blue indicates that no ISVs were observed in the embryos. The differences between treatments were assessed using a two-tailed Student’s t-test. Significant differences between the morphants and controls are indicated (*, P<0.05; **, P<0.01; and ***, P<0.001).
VEGFR3 (Flt4) is predominantly expressed in embryonic blood endothelium, lymphatic endothelial cells, monocytes and macrophages [48]. VEGFR3 is important in lymphangiogenesis, but recent studies have shown that VEGFR3 also functions during sprouting angiogenesis [49]. In mouse, blocking VEGFR3 suppresses angiogenic sprouting and reduces the retina vessel density [50]. We found that flt4 MOs inhibit the formation of ISVs, although to a lesser extent than flk1 MOs (Fig. 3B).
Compared with vegfr2 and vegfr3 morphants, wnk1a morphants display a similar inhibition of ISV growth (Fig. 3C), suggesting that wnk1a is involved in angiogenesis in the vessel during sprouting or elongation. A 5-base mismatch (5MM) control morpholino has no inhibitory activity, with ISV lengths in the morphants comparable to those in wild-type embryos (Fig. 3D). A second morpholino targeted to the 5′-untranslated region of wnk1a causes similar inhibition (Fig. 3E). We also designed a translation-inhibiting MO and an upstream MO for wnk1b knockdown and found that the wnk1b upstream MO significantly inhibits ISV growth (Fig. 3F).
Zebrafish wnk1 knockdown results in defective angiogenesis but not vasculogenesis
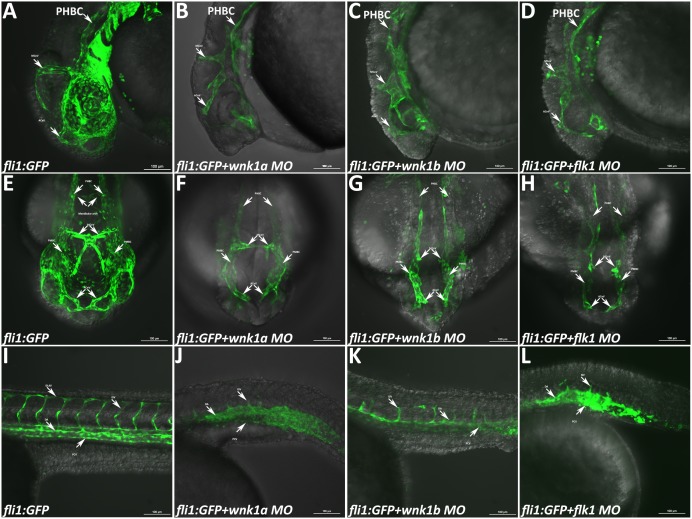
Representative age/somite matched images of uninjected Tg(fl1i:EGFP) embryos as well as wnk1a, wnk1b, and flk1 morphants are shown in Figure 4. Since wnk1a and wnk1b are expressed strongly in the head as well as the trunk, we examined the embryos from a lateral head view (Fig. 4A–D), a frontal head view (Fig. 4E–H), and a lateral trunk view (Fig. 4I–L). Knockdown of either wnk1a or wnk1b causes significant defects in angiogenesis in both the head and trunk vessels. The vessel formation defects caused by knockdown of wnk1a or wnk1b in zebrafish are similar to the defects observed in Wnk1 mutant mice, which include atresic branches of the internal carotid artery and primary head veins and disorganization of the intersomitic vessels and their branches [22]. Knockdown of either wnk1a or wnk1b in zebrafish inhibits the density and formation of vessels in the head region and body (Fig. 4). Notably, only the formation of small vessels in the head, such as the caudal division of the internal carotid artery (CaDI), the primitive mesencephalic artery (PMsA) and the partial optic artery (OA), are inhibited. Major vessel structures, such as the vessels of the mandibular arch, the anterior (rostral) cerebral vein (ACeV), the cranial division of the internal carotid artery (CrDI), the middle cerebral vein (MCeV), the primordial hindbrain channel (PHBC), the primitive internal carotid artery (PICA), and the primordial midbrain channel (PMBC), are not affected by wnk1a MO (Fig. 4B, F, J) or wnk1b MO (Fig. 4C, G, K); this result is similar to that seen in flk1 morphants (Fig. 4D, H, L).
Figure 4. Phenotype of Tg(fli1:GFP) embryos injected with various morpholinos and imaged with a confocal microscope.
(A–D) Lateral views of the heads of uninjected control embryos and wnk1a, wnk1b and flk1 morphants at 33 hpf. (E–H) Frontal views of the heads of uninjected control embryos and wnk1a, wnk1b and flk1 morphants at 33 hpf. (I–L) Lateral views of the trunk in uninjected control embryos and wnk1a, wnk1b and flk1 morphants at 33 hpf. Important vessels are indicated with arrows and labeled, with the full name given in the text. Scale bar: 100 µm.
In the trunk, we found that growth of the intersegmental vessels (ISVs) is inhibited in wnk1 morphants and that formation of the dorsal longitudinal anastomotic vessel (DLAV) is also affected. However, blood vessels formed by the vasculogenesis process, including the dorsal aorta (DA), the caudal artery (CA), the caudal vein (CV) and the posterior cardinal vein (PCV), are not affected (Fig. 4I–L). The phenotype of zebrafish wnk1 morphants is consistent with that of wnk1 knockout mice, with effects on angiogenesis but not vasculogenesis [22].
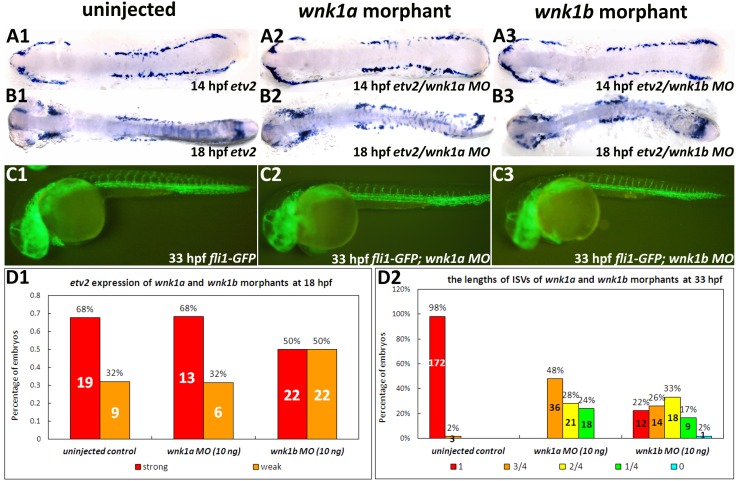
Expression of the vasculogenesis marker etv2 is unaffected in wnk1 morphants
To further verify that wnk1 functions in angiogenesis rather than vasculogenesis, we examined the expression of the vasculogenesis marker etv2 in somite matched control, wnk1a and wnk1b morphants. The embryonic expression of etv2 is restricted to the earliest precursors of vascular endothelial cells. Morpholino knockdown of etv2 results in the absence of vasculogenesis [51]. wnk1a and wnk1b morpholinos were injected into fli1:GFP embryos, and the effect on ISVs was assayed. We found that there is no difference in etv2 expression between the wnk1a or wnk1b morphants and control embryos at 14 hpf (Fig. 5A1– A3) and 18 hpf (Fig. 5B1– B3), whereas the development of ISVs is significantly affected (Fig. 5C1–C3). The quantified results for etv2 expression at 18 hpf and the length of ISVs at 33 hpf were shown in Figure 5D1 and D2 respectively.
Figure 5. Effect of wnk1a or wnk1b knockdown on vasculogenesis and angiogenesis in Tg(fli1:GFP) embryos.
(A, B) Whole mount in situ hybridization for etv2 at 14 and 18 hpf in uninjected control embryos (A1, B1), wnk1a morphants (A2, B2), and wnk1b morphants (A3, B3). Flat mounts of de-yolked embryos were prepared. (C) GFP fluorescence was used to assay ISV formation in uninjected control embryos (C1), wnk1a morphants (C2), and wnk1b morphants (C3) at 33 hpf. (D1) etv2 expression in uninjected control, wnk1a and wnk1b morphants as a percentage of embryos exhibit strong or expression, (D2) ISV length in wnk1a and wnk1b morphants.
Knockdown of endothelial-specific pi3kc2α inhibits the growth of intersegmental vessels
Previous studies have demonstrated that VEGF/VEGFR signaling regulates endothelial cell survival and growth through the activation of the PI3K/Akt pathway, and endothelial cell permeability through the activation of endothelial NO synthase (eNOS) [52], [53], [54]. The WNK1 protein contains an Akt phosphorylation motif; therefore, we suspect that Wnk1 participates in angiogenesis after being phosphorylated and activated by Akt kinase downstream of Vegf/Vegfr signaling.
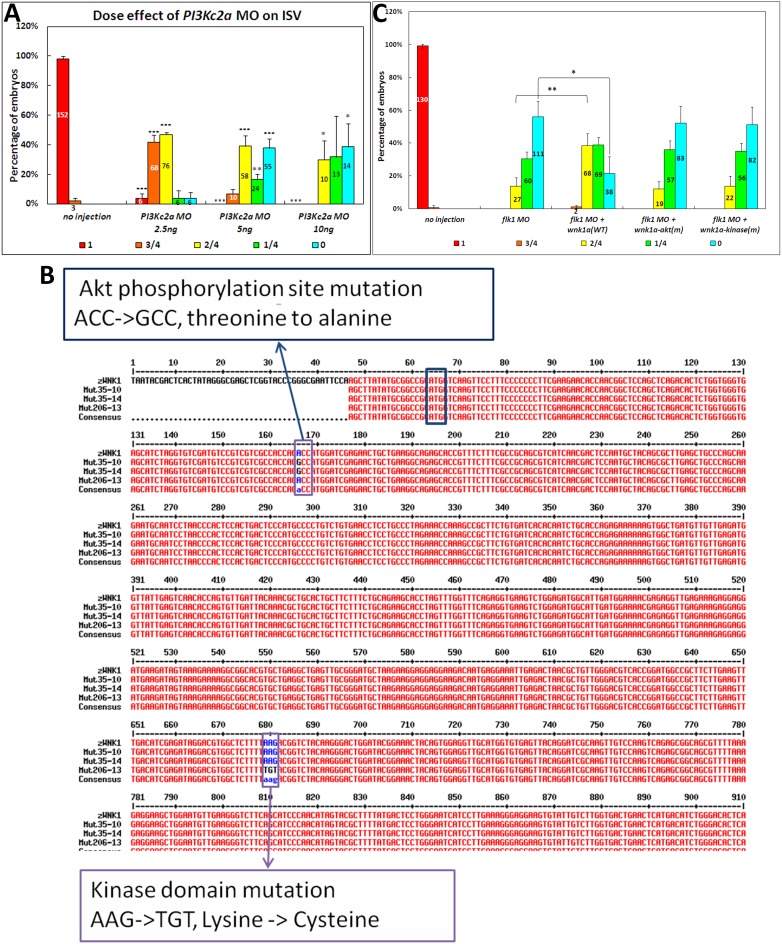
To further analyze whether Wnk1 acts through the Vegf/Vegfr2-PI3K/Akt pathway in endothelial cell activation, we wanted to inhibit PI3K, which falls between Vegfr2 and Wnk1 in the proposed signaling cascade, to check the integrity of this pathway in angiogenesis. To date, there are eight known PI3K isoforms with varying structural features and lipid substrate preferences; they are divided into three classes (class I, class II and class III), and these isoforms are involved in metabolic control, immunity, angiogenesis and cardiovascular homeostasis [55]. From the literature, we know that endothelial cells only express PI3KC2α [56]. From an NCBI GenBank search, we found that the zebrafish gene phosphoinositide-3-kinase, class 2, alpha polypeptide (pik3c2a) is similar to the human PI3KC2α homologs. The pi3kc2α mRNA expression pattern from ZFIN (Bernard Thisse’s group, Expression of the zebrafish genome during embryogenesis. ZFIN Direct Data Submission (http://zfin.org/cgi-bin/webdriver?MIval=aa-ZDB_home.apg)) showed that pi3kc2α is expressed in axial vasculature at 24 hpf (Fig. S4), providing support for a role for pi3kc2α in angiogenesis.
We then designed a MO to knockdown pik3c2a. The pi3kc2α morphant exhibits growth inhibition in the ISVs similar to that observed in the vegfr2, wnk1a and wnk1b morphants (Fig. 6A), suggesting that Pi3kc2α and Wnk1 may be part of the same pathway. We hypothesized that upon Vegf binding to Vegfr, Pi3kc2α is activated, which phosphorylates and activates Wnk1, which in turn activates downstream targets. Additionally, we found that injection of 10 ng of pi3kc2α MO into embryos significantly increases mortality relative to controls, suggesting that Pi3kc2α might participate in other physiological functions, possibly through other signaling pathways. The number of embryos examined and the phenotypic and mortality analyses of pi3kc2a morphants versus other morphants are provided in Fig. S5.
Figure 6. Effect of knockdown of the PI3K ortholog on ISVs and the rescue effects of wild-type wnk1a, wnk1a containing an Akt phosphorylation site mutation and kinase-deficient wnk1a on flk1 morphants.
(A) The effects of pi3kc2α MO on the length of intersegmental vessels. (B) Two Thr35 mutations and one Lys206 mutation were generated using site-directed mutagenesis. The sequence of wild-type wnk1a aligned with Thr35 and Lys206 mutants, which are Akt phosphorylation site and kinase domain mutants, respectively. (C) Co-injection of wnk1a mRNA rescues flk1 morphants. Quantitative analysis of the length of the ISVs in flk1 morphants co-injected with various wnk1a mRNAs. Experiments were performed at least three times, and the number of embryos analyzed is shown in the bar graph. Red indicates that the ISVs grew to full length, orange indicates that the ISVs were 75% of the normal length, yellow indicates 50%, green indicates 25%, and light blue indicates no ISVs were observed in the embryos. The differences between treatments were assessed using a two-tailed Student’s t-test. Significant differences between the morphants and controls are indicated (*, P<0.05; **, P<0.01; and ***, P<0.001).
Partial Rescue of the angiogenesis defect in vegfr2 morphants by wild-type, but not Akt-phosphorylation site mutant or kinase-dead mutant, wnk1a mRNA
Based on the results presented in Figure 3 and 6A, we hypothesized that the binding of Vegf to Vegfr2 activates Wnk1a through phosphorylation by the PI3K-Akt kinase cascade. To test this theory, we used site-directed mutagenesis to generate two mutants of Wnk1a, one missing the Akt phosphorylation site and the other lacking kinase activity (Fig. 6B). We transcribed mRNA for wild-type wnk1a and the two mutants in vitro and then co-injected these with the vegfr2 MO for rescue experiments.
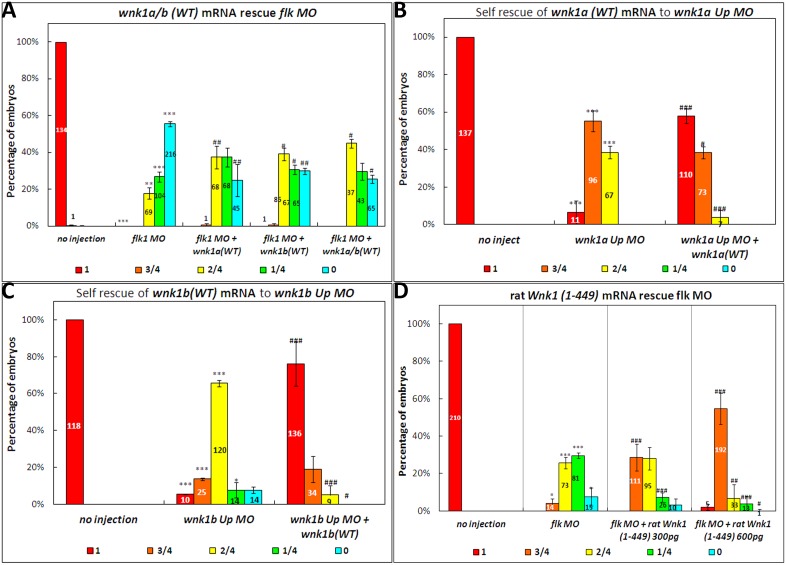
Figure 6C illustrates the result of the rescue experiments. Co-injection of vegfr2 MO with wild-type wnk1a RNA, but not wnk1a-akt(m) or wnk1a-kinase(m), leads to a statistically significant increase in angiogenesis compared with vegfr2 MO injection alone. Representative images of rescued morphants are provided in Figure S6. Notably, the rescue is incomplete: ISVs in the vegfr2 morphants rescued with wnk1a RNA are only half as long as in wildtype. It is possible that some Vegfr2 activity remains in morphant embryos, and hence wild-type Wnk1 can be activated and subsequently phosphorylate downstream targets to achieve rescue. Therefore, the rescue is partially due to the morpholino knockdown the Vegfr2.
Because there are two Wnk1 isoforms in zebrafish, we wondered if the ISVs could be completely rescued by providing both isoforms. Co-injection of both wnk1a and wnk1b mRNA with flk1 MO, however, is comparable to co-injection with wnk1a or wnk1b RNA alone (Fig. 7A). Wild-type wnk1a and wnk1b mRNA fully rescue the angiogenesis defects in wnk1a and wnk1b morphants, respectively (Fig. 7B and 7C). Thus, the partial rescue is not due to poor quality or low abundance of the mRNA.
Figure 7. Effect of wild-type wnk1a and wnk1b mRNA injection on flk1, wnk1a and wnk1b morphants.
(A) Co-injection of wnk1a, wnk1b or both wnk1a and wnk1b mRNA rescues flk1 morphants. (B) Co-injection of wnk1a mRNA rescues the ISV defect caused by the wnk1a upstream MO. (C) Co-injection of wnk1b mRNA rescues the ISV defect caused by the wnk1b upstream MO. (D) Injection of rat Wnk1(1–449) rescues flk1 morphants. Experiments were performed at least three times, and the number of embryos analyzed is shown in the bar graph. Red indicates that the ISVs grew to full length, orange indicates that the ISVs were 75% of the normal length, yellow indicates 50%, green indicates 25%, and light blue indicates no ISVs were observed in the embryos. The differences between treatments were assessed using a two-tailed Student’s t-test. Significant differences between the morphants and controls are indicated (*, P<0.05; **, P<0.01; and ***, P<0.001); significant differences between co-injection of RNA with morpholino and morpholino alone are also indicated (#, P<0.05; ##, P<0.01; and ###, P<0.001).
Because no biochemical assays have been performed to confirm the expected effects of truncation on zebrafish wnk1a and wnk1b, we used a constitutively active rat Wnk1 truncation, for which biochemical data are available, in rescue assays. Previous biochemical studies on rat Wnk1 have demonstrated that the Wnk1(1–449) truncation is constitutively active due to the lack of an autoinhibitory domain [10]. The truncated form of Wnk1 was expected to be better able to rescue flk1 morphants than wild-type Wnk1. We obtained rat Wnk1 cDNA from Dr. Cobb’s lab, subcloned the Wnk1(1–449) fragment and then generated mRNA for co-injection with flk1 MOs. We demonstrated that rat Wnk1(1–449) effectively rescues ISV formation in flk1 morphants in a dose-dependent manner (Fig. 7D).
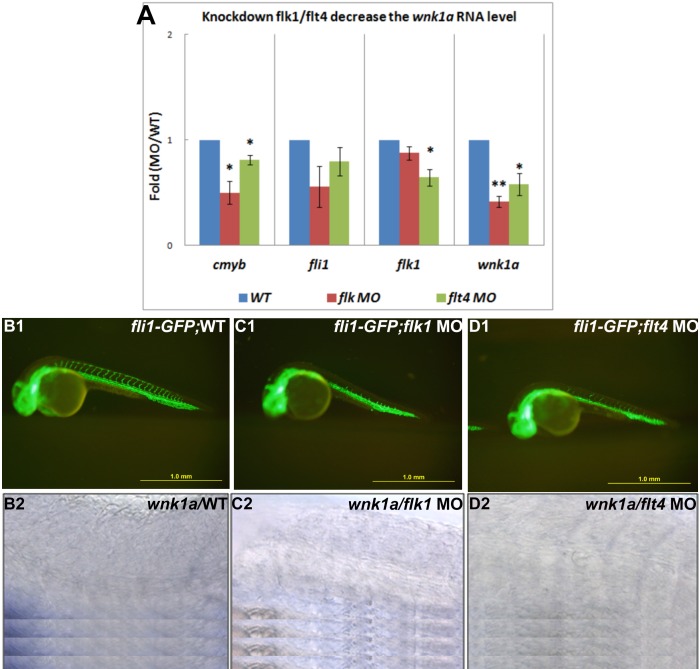
Knockdown of flk1 or flt4 decreases expression of wnk1 mRNA
Another possible mechanism to explain how VEGF signal transduction affects angiogenesis through wnk1 involves transcriptional regulation. VEGF signal transduction might also regulate wnk1 gene expression. The co-localization of flt4 (vegfr3) and wnk1 in the PCV strengthens this possibility. Furthermore, we have analyzed flk1 and flt4 morphants for wnk1 expression. q-RT-PCR analysis of wnk1a and other endothelial genes demonstrated a significant reduction in wnk1a RNA in flk1 and flt4 morphants relative to controls (Fig. 8A). Several genes for blood/vessel formation (cmyb, fli1 and flk1) are also down regulated in flk1 and flt4 morphants. ISVs are inhibited by flk1 and flt4 MO injection (Fig. 8B1, C1 and D1) and expression of wnk1a in the PCV is decreased in flk1 and flt4 morphants compared with uninjected controls (Fig. 8 B2, C2 and D2).
Figure 8. Injections of flk1 MO and flt4 MO decrease wnk1 mRNA expression.
(A) Relative fold-change comparisons between flk1 and flt4 morphants and uninjected controls. Comparison of mRNA expression levels at 33 hpf for important transcription factors, receptors, and wnk1. Blue, red, and green bars denote mRNA expression in wild-type embryos, flk1 morphants and flt4 morphants, respectively. The x-axis indicates the expressed genes, and the y-axis shows the fold differences between the morphants and the control. The differences between treatments were assessed using a two-tailed Student’s t-test. Significant differences between the morphants and the controls are indicated (*, P<0.05 and**, P<0.01). (B1, C1, D1) The ISVs are affected in morphants compared with wild-type embryos. (B2, C2 and D2) The expression of wnk1a is reduced in the PCV in flk1 and flt4 morphants. wnk1a mRNA was detected with in situ hybridization in wild-type embryos (B2), flk1 morphants (C2) and flt4 morphants (D2).
Discussion
In this study, we found that knockdown of vegfr2, vegfr3, wnk1 or pi3kc2a causes angiogenesis defects. From published works, we also know that Wnk1 can be phosphorylated by Akt kinase, and our zebrafish Wnk1a protein sequence contains an Akt phosphorylation site. Therefore, we propose that Wnk1a is downstream of the Vegfr2/Fkl1-PI3K-Akt signaling pathway in zebrafish. In wnk1a mRNA rescue experiments, we found that wild-type wnk1a, but not wnk1a with an Akt phosphorylation site mutation, partially rescues the angiogenesis defect of vegfr2 knockdown. The finding that the Akt site mutant fails to rescue the angiogenesis defect of flk1 morphants supports the hypothesis that binding of Vegf to Vegfr2 activates PI3K and Akt kinase, which then phosphorylate Wnk1 to regulate angiogenesis. In addition, we found that rat Wnk1(1–449), a constitutively active form that lacks the autoinhibitory domain, effectively rescues the flk1 morphant. One important question arising from these results, though, is why wild-type wnk1a RNA rescues flk1 morphants if the function of Vegfr2/Flk1 pathway is to phosphorylate and activate Wnk1.
To address this issue, we postulated that the Vegfr2 pathway might also regulate the expression of wnk1a. We performed real-time q-RT-PCR and in situ hybridization to investigate this possibility. Our q-RT-PCR results reveal that knockdown of either flk1 or flt4 causes a decrease in the wnk1a mRNA level. The reduction in wnk1a expression is modest, perhaps because wnk1a is broadly expressed in a variety of tissues, including some not affected by the relatively vascular-specific knockdown of flk1 or flt4. The in situ hybridization results also suggest that the expression of wnk1a in the PCV is decreased in flk1 and flt4 morphants. Together, the q-RT-PCR and in situ results support the notion that the expression of wnk1a is down regulated by knockdown of flk1 and flt4. This downregulation of wnk1a would explain why co-injection of wild-type wnk1a RNA rescues the angiogenesis defect of fkl1 morphants. Because morpholino knockdown is incomplete, residual Flk1 protein must remain in flk1 morphants. It is conceivable that overexpression of Wnk1a proteins (by mRNA injection) can partially overcome decreased phosphorylation of Wnk1a by the Flk1-PI3K-Akt cascade. How Vegfr2/Flk1 regulates wnk1a expression remains unknown and awaits future investigation.
Angiogenesis comprises four main steps: selection of sprouting endothelial cells, sprout outgrowth and guidance, sprout fusion, and perfusion/maturation [49]. These processes are regulated by different growth factors, receptors and downstream signal molecules. In the wnk1a morphants, we observed that the number of endothelial cells participating in the ISVs and the intervals between ISVs are similar to those in wild-type embryos. Knockdown of wnk1 suppresses the growth of the ISV toward the DLAV. We also found that ISVs become straighter and finer in embryos overexpressing wnk1a mRNA than in wild-type embryos (Fig. S5). Therefore, we speculate that Wnk1a may be involved in sprout outgrowth, migration and elongation of the selected tip cells, rather than in the selection of sprouting endothelial cells or lateral inhibition among endothelial cells. We speculate that Wnk1a has no interaction with the Notch signaling pathway, which is involved in lateral inhibition.
The expression pattern of WNK1 is ubiquitous in human, mouse and zebrafish. In a previous study [22], it was found that Wnk1 mRNA is expressed in endothelial tissue such as the heart, the brachial arches, the dorsal aorta, pericytes and other manifestations of the cardiovascular system. Wnk1 is also expressed in the neural tube and the gut. From our in situ data, we found the zebrafish wnk1 is expressed in the neural tube, notochord, and the vascular structure-posterior cardinal vein (PCV). In Wnk1-null mice, Wnk1 mRNA was absent in all these tissues. Using a Cre recombinase system, endothelial-specific Wnk1 expression rescues angiogenesis in Wnk1-null mice. This result suggests that Wnk1 expression in tissues beyond the vascular endothelial cells is not necessary for angiogenesis. Although we have not shown that endothelial-specific wnk1 can rescue angiogenesis in zebrafish wnk1 null mutants, we believe the function of wnk1 in angiogenesis is highly conserved among vertebrates. Only endothelial-specific expression of wnk1 can rescue angiogenesis in the Wnk1 –/– background [22].
VEGF is an important factor in promoting angiogenesis. Compared with normal tissues, most tumors grow faster in vivo and often require additional blood to supply the necessary nutrients and oxygen; thus, many tumors secrete various angiogenic factors and induce vascular endothelial cells to promote proliferation and tumor expansion. In the healthy body, the role of VEGF is limited mainly to wound healing and menstruation. However, in the process of tumor growth and metastasis, VEGF and downstream genes play an indispensable role. Therefore, VEGF therapies that inhibit the formation of new blood vessels have become very important in cancer treatment. In future work, we will xenotransplant cancer cells into Tg(fli1:EGFP) zebrafish to observe angiogenesis occurring near the cancer cells in live animals. Using drugs that inhibit VEGF as well as the wnk1 morpholino antisense oligonucleotides, we will test the feasibility of using anti-wnk1 MOs to inhibit angiogenesis, tumor growth and metastasis.
Supporting Information
Morpholino specificity revealed by co-injections of wnk1a-GFP or wnk1b-GFP with various morpholinos. (A1, B1) Schematic of wnk1a-GFP and wnk1b-GFP constructs and the location of morpholino target sites. (A2, B2) Injection of wnk1a-GFP or wnk1b-GFP only. Co-injection of wnk1a-GFP or wnk1b-GFP with (A3, B3) wnk1a or wnk1b MOs targeted to the ATG, (A4, B4) wnk1a or wnk1b MOs that bind upstream of the translation start site, (A5, B5) scrambled control MOs, (B6) MOs targeted to the other isoform’s ATG, and (A6, A7) MOs targeted to the 5′ untranslated region of the other isoform.
(TIF)
Temporal expression patterns of wnk1a and wnk1b. Whole mount in situ hybridization to detect wnk1a (A1∼A12) and wnk1b (B1∼B12) mRNA expression was performed at the indicated time points. Whole mount in situ hybridization of sense probes for wnk1a (A13) and wnk1b (B13) at 48 hpf showed no signal. All pictures are lateral views. Scale bar: 100 µm.
(TIF)
Measurement of the length of ISVs and representative images of phenotypic classification. (A1) Illustration of the region used to measure the ISVs. (A2∼A6) ISVs categorized as having extended over 100%, 75%, 50%, 25% or 0% of the distance from the DA (or PCV) to the DLAV at 33 hpf. (B1∼B4) Phenotypes were characterized as normal, class 1, class 2 or class 3 at 24 hpf.
(TIF)
Whole mount in situ hybridization for pi3kc2a mRNA at the indicated time points. Images were obtained from ZFIN.
(TIF)
Phenotypic classification of 24 hpf flk1 morphants. flt4 morphant (A), pi3kc2a morphant (B), wnk1a morphant (C), wnk1a UP morphant (D) and wnk1a 5 MM morphant (F). For each morphant, there are two figures. The first figure shows the number of embryos analyzed, and the second figure shows the percentage of morphants displaying a phenotype.
(TIF)
Phenotype of Tg(fli1:GFP) embryos injected with various morpholinos and imaged with a florescence microscope. (A–E) Uninjected control embryos (A), flk1 morphants (B) and flk1 morphants co-injected with wnk1a (WT) mRNA (C), wnk1a-akt(m) mRNA (D), or wnk1a-kinase(m) mRNA (E) at 33 hpf. (F–H) Frontal views of the heads of uninjected control embryos (F), wnk1b morphants (G) or wnk1b morphants co-injected with wnk1b mRNA at 33 hpf. (I and J) wnk1a mRNA injected embryo (I) compared to wild-type control (J) at 29 hpf.
(TIF)
Detailed information regarding q-RT-PCR primers, in situ probe location, morpholino design, and wnk1 -GFP constructs.
(DOCX)
Acknowledgments
We would like to thank the Taiwan Zebrafish Core Facility at NTHU-NHRI for providing fish lines and resources; TZeTH is supported by a grant from the NSC (102-2321-B-400-018). We are thankful to B. Thisse for providing the in situ data for phosphoinositide-3-kinase, class 2, alpha polypeptide. Additional funding provided to C.H.Y. by the National Health Research Institute is gratefully acknowledged.
Funding Statement
This research was supported by grants from the NSC grant from Taiwan (99-3112-B-400-010, 101-2321-B-400-017 and 102-2321-B-400-016) to C.H.Y., and the NIH grant from USA (RO1DK59530) to C.L.H. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Baldessari D, Mione M (2008) How to create the vascular tree? (Latest) help from the zebrafish. Pharmacol Ther 118: 206–230. [DOI] [PubMed] [Google Scholar]
- 2. Risau W (1997) Mechanisms of angiogenesis. Nature 386: 671–674. [DOI] [PubMed] [Google Scholar]
- 3. Shibuya M (2008) Vascular endothelial growth factor-dependent and -independent regulation of angiogenesis. BMB Rep 41: 278–286. [DOI] [PubMed] [Google Scholar]
- 4. Jiang ZY, Zhou QL, Holik J, Patel S, Leszyk J, et al. (2005) Identification of WNK1 as a substrate of Akt/protein kinase B and a negative regulator of insulin-stimulated mitogenesis in 3T3-L1 cells. J Biol Chem 280: 21622–21628. [DOI] [PubMed] [Google Scholar]
- 5. Xu B, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, et al. (2000) WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem 275: 16795–16801. [DOI] [PubMed] [Google Scholar]
- 6. Verissimo F, Jordan P (2001) WNK kinases, a novel protein kinase subfamily in multi-cellular organisms. Oncogene 20: 5562–5569. [DOI] [PubMed] [Google Scholar]
- 7. Xu BE, Stippec S, Lenertz L, Lee BH, Zhang W, et al. (2004) WNK1 activates ERK5 by an MEKK2/3-dependent mechanism. J Biol Chem 279: 7826–7831. [DOI] [PubMed] [Google Scholar]
- 8. Sun X, Gao L, Yu RK, Zeng G (2006) Down-regulation of WNK1 protein kinase in neural progenitor cells suppresses cell proliferation and migration. J Neurochem 99: 1114–1121. [DOI] [PubMed] [Google Scholar]
- 9. Vitari AC, Deak M, Collins BJ, Morrice N, Prescott AR, et al. (2004) WNK1, the kinase mutated in an inherited high-blood-pressure syndrome, is a novel PKB (protein kinase B)/Akt substrate. Biochem J 378: 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu BE, Min X, Stippec S, Lee BH, Goldsmith EJ, et al. (2002) Regulation of WNK1 by an autoinhibitory domain and autophosphorylation. J Biol Chem 277: 48456–48462. [DOI] [PubMed] [Google Scholar]
- 11. Wilson FH, Disse-Nicodeme S, Choate KA, Ishikawa K, Nelson-Williams C, et al. (2001) Human hypertension caused by mutations in WNK kinases. Science 293: 1107–1112. [DOI] [PubMed] [Google Scholar]
- 12. Richardson C, Rafiqi FH, Karlsson HK, Moleleki N, Vandewalle A, et al. (2008) Activation of the thiazide-sensitive Na+-Cl− cotransporter by the WNK-regulated kinases SPAK and OSR1. J Cell Sci 121: 675–684. [DOI] [PubMed] [Google Scholar]
- 13. Zagorska A, Pozo-Guisado E, Boudeau J, Vitari AC, Rafiqi FH, et al. (2007) Regulation of activity and localization of the WNK1 protein kinase by hyperosmotic stress. J Cell Biol 176: 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Anselmo AN, Earnest S, Chen W, Juang YC, Kim SC, et al. (2006) WNK1 and OSR1 regulate the Na+, K+, 2Cl- cotransporter in HeLa cells. Proc Natl Acad Sci U S A 103: 10883–10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Moriguchi T, Urushiyama S, Hisamoto N, Iemura S, Uchida S, et al. (2005) WNK1 regulates phosphorylation of cation-chloride-coupled cotransporters via the STE20-related kinases, SPAK and OSR1. J Biol Chem 280: 42685–42693. [DOI] [PubMed] [Google Scholar]
- 16. Gagnon KB, England R, Delpire E (2006) Characterization of SPAK and OSR1, regulatory kinases of the Na-K-2Cl cotransporter. Mol Cell Biol 26: 689–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Richardson C, Alessi DR (2008) The regulation of salt transport and blood pressure by the WNK-SPAK/OSR1 signalling pathway. J Cell Sci 121: 3293–3304. [DOI] [PubMed] [Google Scholar]
- 18. Orlov SN, Gossard F, Pausova Z, Akimova OA, Tremblay J, et al. (2010) Decreased NKCC1 activity in erythrocytes from African Americans with hypertension and dyslipidemia. Am J Hypertens 23: 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flatman PW (2007) Cotransporters, WNKs and hypertension: important leads from the study of monogenetic disorders of blood pressure regulation. Clin Sci (Lond) 112: 203–216. [DOI] [PubMed] [Google Scholar]
- 20. Gamba G (2005) Molecular physiology and pathophysiology of electroneutral cation-chloride cotransporters. Physiol Rev 85: 423–493. [DOI] [PubMed] [Google Scholar]
- 21. Vitari AC, Deak M, Morrice NA, Alessi DR (2005) The WNK1 and WNK4 protein kinases that are mutated in Gordon's hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochem J 391: 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie J, Wu T, Xu K, Huang I, Cleaver O, et al.. (2009) Endothelial-specific expression of WNK1 kinase is essential for angiogenesis and heart development in mice. Am J Pathol: 1315–1321. [DOI] [PMC free article] [PubMed]
- 23. Driever W, Stemple D, Schier A, Solnica-Krezel L (1994) Zebrafish: genetic tools for studying vertebrate development. Trends Genet 10: 152–159. [DOI] [PubMed] [Google Scholar]
- 24. Lele Z, Krone PH (1996) The zebrafish as a model system in developmental, toxicological and transgenic research. Biotechnol Adv 14: 57–72. [DOI] [PubMed] [Google Scholar]
- 25. Drummond IA (2000) The zebrafish pronephros: a genetic system for studies of kidney development. Pediatr Nephrol 14: 428–435. [DOI] [PubMed] [Google Scholar]
- 26. Langenberg T, Brand M, Cooper MS (2003) Imaging brain development and organogenesis in zebrafish using immobilized embryonic explants. Dev Dyn 228: 464–474. [DOI] [PubMed] [Google Scholar]
- 27. Glickman NS, Yelon D (2002) Cardiac development in zebrafish: coordination of form and function. Semin Cell Dev Biol 13: 507–513. [DOI] [PubMed] [Google Scholar]
- 28. Avanesov A, Malicki J (2004) Approaches to study neurogenesis in the zebrafish retina. Methods Cell Biol 76: 333–384. [DOI] [PubMed] [Google Scholar]
- 29. Lam CS, Marz M, Strahle U (2009) gfap and nestin reporter lines reveal characteristics of neural progenitors in the adult zebrafish brain. Dev Dyn 238: 475–486. [DOI] [PubMed] [Google Scholar]
- 30. Goessling W, North TE, Zon LI (2007) Ultrasound biomicroscopy permits in vivo characterization of zebrafish liver tumors. Nat Methods 4: 551–553. [DOI] [PubMed] [Google Scholar]
- 31. Peterson SM, Freeman JL (2009) Cancer cytogenetics in the zebrafish. Zebrafish 6: 355–360. [DOI] [PubMed] [Google Scholar]
- 32. Amsterdam A, Hopkins N (2006) Mutagenesis strategies in zebrafish for identifying genes involved in development and disease. Trends Genet 22: 473–478. [DOI] [PubMed] [Google Scholar]
- 33. Malicki JJ, Pujic Z, Thisse C, Thisse B, Wei X (2002) Forward and reverse genetic approaches to the analysis of eye development in zebrafish. Vision Res 42: 527–533. [DOI] [PubMed] [Google Scholar]
- 34. Malicki J (2000) Harnessing the power of forward genetics–analysis of neuronal diversity and patterning in the zebrafish retina. Trends Neurosci 23: 531–541. [DOI] [PubMed] [Google Scholar]
- 35. Lawson ND, Weinstein BM (2002) In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol 248: 307–318. [DOI] [PubMed] [Google Scholar]
- 36. Jin SW, Beis D, Mitchell T, Chen JN, Stainier DY (2005) Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 132: 5199–5209. [DOI] [PubMed] [Google Scholar]
- 37. Zambrowicz BP, Abuin A, Ramirez-Solis R, Richter LJ, Piggott J, et al. (2003) Wnk1 kinase deficiency lowers blood pressure in mice: a gene-trap screen to identify potential targets for therapeutic intervention. Proc Natl Acad Sci U S A 100: 14109–14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tseng WF, Jang TH, Huang CB, Yuh CH (2011) An evolutionarily conserved kernel of gata5, gata6, otx2 and prdm1a operates in the formation of endoderm in zebrafish. Dev Biol 357: 541–557. [DOI] [PubMed] [Google Scholar]
- 39. Lauter G, Soll I, Hauptmann G (2011) Two-color fluorescent in situ hybridization in the embryonic zebrafish brain using differential detection systems. BMC Dev Biol 11: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tao S, Cai Y, Sampath K (2009) The Integrator subunits function in hematopoiesis by modulating Smad/BMP signaling. Development 136: 2757–2765. [DOI] [PubMed] [Google Scholar]
- 41. Siekmann AF, Standley C, Fogarty KE, Wolfe SA, Lawson ND (2009) Chemokine signaling guides regional patterning of the first embryonic artery. Genes Dev 23: 2272–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zeng XXI, Zheng XJ, Xiang Y, Cho HP, Jessen JR, et al. (2009) Phospholipase D1 is required for angiogenesis of intersegmental blood vessels in zebrafish. Developmental Biology 328: 363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nicoli S, De Sena G, Presta M (2009) Fibroblast growth factor 2-induced angiogenesis in zebrafish: the zebrafish yolk membrane (ZFYM) angiogenesis assay. J Cell Mol Med 13: 2061–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Habeck H, Odenthal J, Walderich B, Maischein H, Schulte-Merker S (2002) Analysis of a zebrafish VEGF receptor mutant reveals specific disruption of angiogenesis. Curr Biol 12: 1405–1412. [DOI] [PubMed] [Google Scholar]
- 45. Isogai S, Horiguchi M, Weinstein BM (2001) The vascular anatomy of the developing zebrafish: an atlas of embryonic and early larval development. Dev Biol 230: 278–301. [DOI] [PubMed] [Google Scholar]
- 46. Hirashima M (2009) Regulation of endothelial cell differentiation and arterial specification by VEGF and Notch signaling. Anat Sci Int 84: 95–101. [DOI] [PubMed] [Google Scholar]
- 47. Shibuya M (2006) Differential roles of vascular endothelial growth factor receptor-1 and receptor-2 in angiogenesis. J Biochem Mol Biol 39: 469–478. [DOI] [PubMed] [Google Scholar]
- 48. Jakobsson L, Bentley K, Gerhardt H (2009) VEGFRs and Notch: a dynamic collaboration in vascular patterning. Biochem Soc Trans 37: 1233–1236. [DOI] [PubMed] [Google Scholar]
- 49. Adams RH, Alitalo K (2007) Molecular regulation of angiogenesis and lymphangiogenesis. Nat Rev Mol Cell Biol 8: 464–478. [DOI] [PubMed] [Google Scholar]
- 50. Tammela T, Zarkada G, Wallgard E, Murtomaki A, Suchting S, et al. (2008) Blocking VEGFR-3 suppresses angiogenic sprouting and vascular network formation. Nature 454: 656–660. [DOI] [PubMed] [Google Scholar]
- 51. Sumanas S, Lin S (2006) Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol 4: e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dayanir V, Meyer RD, Lashkari K, Rahimi N (2001) Identification of tyrosine residues in vascular endothelial growth factor receptor-2/FLK-1 involved in activation of phosphatidylinositol 3-kinase and cell proliferation. J Biol Chem 276: 17686–17692. [DOI] [PubMed] [Google Scholar]
- 53. Fujio Y, Walsh K (1999) Akt mediates cytoprotection of endothelial cells by vascular endothelial growth factor in an anchorage-dependent manner. J Biol Chem 274: 16349–16354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Makinen T, Veikkola T, Mustjoki S, Karpanen T, Catimel B, et al. (2001) Isolated lymphatic endothelial cells transduce growth, survival and migratory signals via the VEGF-C/D receptor VEGFR-3. EMBO J 20: 4762–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B (2010) The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol 11: 329–341. [DOI] [PubMed] [Google Scholar]
- 56. El Sheikh SS, Domin J, Tomtitchong P, Abel P, Stamp G, et al. (2003) Topographical expression of class IA and class II phosphoinositide 3-kinase enzymes in normal human tissues is consistent with a role in differentiation. BMC Clin Pathol 3: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Morpholino specificity revealed by co-injections of wnk1a-GFP or wnk1b-GFP with various morpholinos. (A1, B1) Schematic of wnk1a-GFP and wnk1b-GFP constructs and the location of morpholino target sites. (A2, B2) Injection of wnk1a-GFP or wnk1b-GFP only. Co-injection of wnk1a-GFP or wnk1b-GFP with (A3, B3) wnk1a or wnk1b MOs targeted to the ATG, (A4, B4) wnk1a or wnk1b MOs that bind upstream of the translation start site, (A5, B5) scrambled control MOs, (B6) MOs targeted to the other isoform’s ATG, and (A6, A7) MOs targeted to the 5′ untranslated region of the other isoform.
(TIF)
Temporal expression patterns of wnk1a and wnk1b. Whole mount in situ hybridization to detect wnk1a (A1∼A12) and wnk1b (B1∼B12) mRNA expression was performed at the indicated time points. Whole mount in situ hybridization of sense probes for wnk1a (A13) and wnk1b (B13) at 48 hpf showed no signal. All pictures are lateral views. Scale bar: 100 µm.
(TIF)
Measurement of the length of ISVs and representative images of phenotypic classification. (A1) Illustration of the region used to measure the ISVs. (A2∼A6) ISVs categorized as having extended over 100%, 75%, 50%, 25% or 0% of the distance from the DA (or PCV) to the DLAV at 33 hpf. (B1∼B4) Phenotypes were characterized as normal, class 1, class 2 or class 3 at 24 hpf.
(TIF)
Whole mount in situ hybridization for pi3kc2a mRNA at the indicated time points. Images were obtained from ZFIN.
(TIF)
Phenotypic classification of 24 hpf flk1 morphants. flt4 morphant (A), pi3kc2a morphant (B), wnk1a morphant (C), wnk1a UP morphant (D) and wnk1a 5 MM morphant (F). For each morphant, there are two figures. The first figure shows the number of embryos analyzed, and the second figure shows the percentage of morphants displaying a phenotype.
(TIF)
Phenotype of Tg(fli1:GFP) embryos injected with various morpholinos and imaged with a florescence microscope. (A–E) Uninjected control embryos (A), flk1 morphants (B) and flk1 morphants co-injected with wnk1a (WT) mRNA (C), wnk1a-akt(m) mRNA (D), or wnk1a-kinase(m) mRNA (E) at 33 hpf. (F–H) Frontal views of the heads of uninjected control embryos (F), wnk1b morphants (G) or wnk1b morphants co-injected with wnk1b mRNA at 33 hpf. (I and J) wnk1a mRNA injected embryo (I) compared to wild-type control (J) at 29 hpf.
(TIF)
Detailed information regarding q-RT-PCR primers, in situ probe location, morpholino design, and wnk1 -GFP constructs.
(DOCX)