Abstract
Background
Sweet potato chlorotic stunt virus (family Closteroviridae, genus Crinivirus) features a large bipartite, single-stranded, positive-sense RNA genome. To date, only three complete genomic sequences of SPCSV can be accessed through GenBank. SPCSV was first detected from China in 2011, only partial genomic sequences have been determined in the country. No report on the complete genomic sequence and genome structure of Chinese SPCSV isolates or the genetic relation between isolates from China and other countries is available.
Methodology/Principal Findings
The complete genomic sequences of five isolates from different areas in China were characterized. This study is the first to report the complete genome sequences of SPCSV from whitefly vectors. Genome structure analysis showed that isolates of WA and EA strains from China have the same coding protein as isolates Can181-9 and m2-47, respectively. Twenty cp genes and four RNA1 partial segments were sequenced and analyzed, and the nucleotide identities of complete genomic, cp, and RNA1 partial sequences were determined. Results indicated high conservation among strains and significant differences between WA and EA strains. Genetic analysis demonstrated that, except for isolates from Guangdong Province, SPCSVs from other areas belong to the WA strain. Genome organization analysis showed that the isolates in this study lack the p22 gene.
Conclusions/Significance
We presented the complete genome sequences of SPCSV in China. Comparison of nucleotide identities and genome structures between these isolates and previously reported isolates showed slight differences. The nucleotide identities of different SPCSV isolates showed high conservation among strains and significant differences between strains. All nine isolates in this study lacked p22 gene. WA strains were more extensively distributed than EA strains in China. These data provide important insights into the molecular variation and genomic structure of SPCSV in China as well as genetic relationships among isolates from China and other countries.
Introduction
Sweet potato (Ipomoea batatas) is the third most important root crop after potato and cassava [1], [2]. China is currently the largest producer of sweet potato; the country cultivates the crop over an average of 4.1 million hectares of planting area, which accounts for 48.29% of the worldwide total [3], [4]. Over 30 viruses are known to infect sweet potato [5]. Sweet potato chlorotic stunt virus (SPCSV) is the most devastating virus affecting sweet potato. SPCSV, which was previously known as sweet potato sunken vein virus, belongs to genus Crinivirus of family Closteroviridae [6]–[8]. SPCSV was first reported in the 1970s [9]. It is phloem-limited and transmitted in a semi-persistent manner by whitefly. The virus has the second largest genome and contains a single-stranded bipartite positive-sense RNA genome [10], [11]. SPCSV is often found in co-infection with Sweet potato feathery mottle virus (SPFMV), a member of the genus Potyvirus that causes a synergistic disease called sweet potato virus disease (SPVD); SPVD is the main viral constraint affecting sweet potatoes worldwide [12]–[14]. Plants with SPVD exhibit severe symptoms, such as leaf strapping, vein clearing, chlorosis, stunting, leaf distortion, and even death, and the disease causes yield losses ranging from 70% to 100% [15]–[18]. Molecular studies have shown that co-infection of SPCSV enhances SPFMV RNA viral titers by at least 600-fold, whereas SPCSV titers remain equal or are reduced compared with single infection [12], [13], [17], [19]. Besides SPFMV, several other viruses belonging to the genera Potyvirus, Carlavirus, Cucumovirus, Ipovovirus, and Cavemovirus can result in synergistic diseases and severely affect sweet potato yield upon co-infection with SPCSV [18], [20].
SPCSV is distributed worldwide and has been detected in all sweet potato production areas except those in the Pacific region [3], [5], [21], [22]. Based on serological studies, SPCSV can be divided into two distantly related strains: the East African (EA) strain and the West African (WA) strain [23], [24]. A similar subdivision into two genetic strains was revealed by phylogenetic analysis of the coat protein (cp) and heat shock protein 70 homolog (hsp70h) gene sequences [3], [25]–[27] as well as analysis of the RNA1 sequences [28]. WA strains are more widely distributed than EA strains [27], [29]–[31]. The complete genomic sequence of SPCSV was first determined by Kreuze [11]. As SPCSV has a low titer in sweet potato plants and always co-infects sweet potatoes with other viruses in field [22], [32]–[34], separation and purification of SPCSV is difficult to perform and determination of the SPCSV genomic sequence is greatly constrained. To date, only three SPCSV genomic sequences can be accessed through GenBank, including two complete sequences of the SPCSV EA strain and one sequence of the SPCSV WA strain [11], [29], [35]. Partial sequence analysis shows that not all EA strain isolates include the p22 open reading frame (ORF) at the 3′ end of RNA1; other isolates may lack a 767 nt region of RNA1 that includes the p22 gene [29]. The p22 gene has only been found in isolates from Uganda [28], [29], [36]. Regardless of the presence of p22, isolates of SPCSV act synergistically with SPFMV in sweet potato plants and significantly enhance SPFMV titers. However, co-infection of SPFMV with SPCSV isolates containing p22 causes more severe symptoms in the indicator plant Ipomoea setosa than co-infection of SPFMV with SPCSV isolates lacking p22 [28].
Recent studies have focused on the synergism of SPCSV with other unrelated viruses; the incidence, distribution, and effects of SPVD on sweet potato yield; and management approaches to SPVD [18], [33], [37]. Limited information is available on the genetic variability of SPCSV, and this information is based on analysis of the nucleotide sequences of hsp70h and cp genes on RNA2 and p7, p22, and RNase3 genes on RNA1 [3], [25]–[27], [36].
In China, SPCSV was first detected in 2011 [38], and SPVD was first reported in China in 2012 [39]. Subsequent to its discovery, the virus developed and caused a serious epidemic from 2012 to 2013 [27]. Despite the importance of the virus, however, knowledge of the molecular characterization and genetic diversity of SPCSV in China, which is an important sweet potato production area, remains limited. Moreover, the presence or absence of the p22 gene in China isolates is unclear.
The current study presents the complete genome sequences of five SPCSV EA and WA isolates obtained from different areas in China. This study is the first to report the complete genome sequences of SPCSV from whitefly vectors, and we provide a simplified and convenient method for cloning SPCSV genomes. The genomic structures of the five isolates were analyzed and compared with three isolate genomes obtained from GenBank. Nucleotide and amino acid identities, including the molecular variance of different isolates, were also compared and a phylogenetic tree was constructed. To confirm the genetic variability of the cp gene of WA strains and investigate whether or not the p22 gene is present in China, we also described the cp gene sequences of 20 other isolates and 4 genome segments of RNA1 flanking the p22 insertion region. Genetic variability and phylogenetic relationships among these isolates were further analyzed.
Results
Complete nucleotide sequence analyses of SPCSV from China and comparison with other isolates
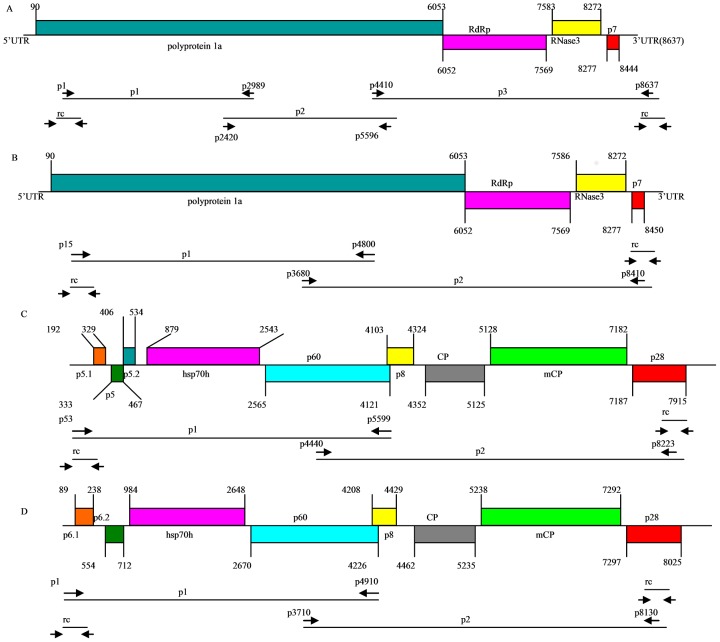
The complete genomic sequences of five isolates of SPCSV belonging to WA and EA strains from different areas in China were characterized by RT-PCR using viruliferous whitefly as materials. The 5′ and 3′ UTRs of virus genome segments RNA1 and RNA2 were also determined by rapid amplification of cDNA ends (RACE). The cloning strategy and a schematic representation of the SPCSV genome organization are shown in Figure 1. The complete genome nucleotide sequences of the five SPCSV isolates were deposited in GenBank under accession numbers KC146840–KC146843 and KC888961–KC888966 (Table 1). Table 2 shows the primer sequences and their corresponding positions in the SPCSV genome.
Figure 1. Schematic diagram of genomic organization and genome cloning strategy of SPCSV.
Solid lines represent RNA genomes and boxes represent ORFs. Putative protein products are indicated. p1, p2, and p3 represent RT-PCR-generated sequences using specific primers. rc represents the 5- and 3-terminal clones generated by 5′ and 3′ RACE, respectively. Primer sequences are shown in Table 2. (A) Schematic diagram of the genomic organization and genome cloning strategy of the SPCSV WA strain RNA1 segment. (B) Schematic diagram of the genomic organization and genome cloning strategy of the SPCSV EA strain RNA1 segment. (C) Schematic diagram of the genomic organization and genome cloning strategy of the SPCSV WA strain RNA2 segment. (D) Schematic diagram of the genomic organization and genome cloning strategy of the SPCSV EA strain RNA2 segment.
Table 1. Name, strain assignment, geographic origin, segment length, reference, and GenBank accession number of all isolates or samples used in this study.
| Isolate names | accession numbers | Serotype | Segments | Segment length (bp) | Geographical origin | Reference |
| Jiangsu-2011 | KC146840 | WA | RNA1 | 8637 | Jiangsu | This study |
| Jiangsu-2011 | KC146841 | WA | RNA2 | 8107 | Jiangsu | This study |
| Guangdong-2011 | KC146842 | EA | RNA1 | 8622 | Guangdong | This study |
| Guangdong-2011 | KC146843 | EA | RNA2 | 8217 | Guangdong | This study |
| Sichuan-12-8 | KC888964 | WA | RNA1 | 8637 | Sichuan | This study |
| Sichuan-12-8 | KC888961 | WA | RNA2 | 8107 | Sichuan | This study |
| Sichuan-12-12 | KC888965 | WA | RNA1 | 8637 | Sichuan | This study |
| Sichuan-12-12 | KC888962 | WA | RNA2 | 8107 | Sichuan | This study |
| Chongqing-12-8 | KC888966 | WA | RNA1 | 8637 | Chongqing | This study |
| Chongqing-12-8 | KC888963 | WA | RNA2 | 8107 | Chongqing | This study |
| m2-47 | HQ291259 | EA | RNA1 | 8637 | Peru | Cuellar et al. 2011 |
| m2-47 | HQ291260 | EA | RNA2 | 8219 | Peru | Cuellar et al. 2011 |
| Uganda | AJ428554 | EA | RNA1 | 9407 | Uganda | Kreuze et al. 2002 |
| Uganda | AJ428555 | EA | RNA2 | 8223 | Uganda | Kreuze et al. 2002 |
| Can181-9 | FJ807784 | WA | RNA1 | 8637 | Spain | Trenado et al. 2012 |
| Can181-9 | FJ807785 | WA | RNA2 | 8108 | Spain | Trenado et al. 2012 |
| Jiangsu-11-19 | KC243096 | WA | RNA1 | 1356 | Jiangsu | This study |
| Anhui-11-2 | KC243097 | WA | RNA1 | 1356 | Anhui | This study |
| Chongqing-11-8 | KC243098 | WA | RNA1 | 1356 | Chongqing | This study |
| Zhejiang-11-4 | KC243099 | EA | RNA1 | 1356 | Zhejiang | This study |
| Sichuan-11-28 | KC243079 | WA | RNA2 | 774 | Sichuan | This study |
| Jiangxi-11-3 | KC243080 | WA | RNA2 | 774 | Jiangxi | This study |
| Guangxi-11-5 | KC243081 | WA | RNA2 | 774 | Guangxi | This study |
| Guangxi-11-7 | KC243082 | WA | RNA2 | 774 | Guangxi | This study |
| Hebei-11-5 | KC243083 | WA | RNA2 | 774 | Hebei | This study |
| Shanxi-11-6 | KC243084 | WA | RNA2 | 774 | Shanxi | This study |
| Shandong-11-7 | KC243085 | WA | RNA2 | 774 | Shandong | This study |
| Shaanxi-11-1 | KC243086 | WA | RNA2 | 774 | Shaanxi | This study |
| Jiangsu-11-17 | KC243087 | WA | RNA2 | 774 | Jiangsu | This study |
| Jiangsu-11-19 | KC243088 | WA | RNA2 | 774 | Jiangsu | This study |
| Anhui-11-2 | KC243089 | WA | RNA2 | 774 | Anhui | This study |
| Hunan-11-4 | KC243090 | WA | RNA2 | 774 | Hunan | This study |
| Chongqing-11-3 | KC243091 | WA | RNA2 | 774 | Chongqing | This study |
| Henan-11-28 | KC243092 | WA | RNA2 | 774 | Henan | This study |
| Guangdong-11-9 | KC243093 | WA | RNA2 | 774 | Guangdong | This study |
| Zhejiang-11-3 | KC243094 | WA | RNA2 | 774 | Zhejiang | This study |
| Zhejiang-11-4 | KC243095 | WA | RNA2 | 774 | Zhejiang | This study |
| Guangdong-2009 | HM773432 | EA | RNA2 | 774 | Guangdong | Qiao et al. 2011 |
| Sichuan-10-77 | KC146844 | WA | RNA2 | 774 | Sichuan | This study |
Table 2. Polymerase chain reaction primers and PCR conditions used to amplify the SPCSV genome, CP gene, and RNA1 partial sequences.
| Primer name | Sequence (5′ to 3′) | Nucleotide position | Size of amplificons (bp) | polarity | PCR conditions |
| WA-RNA1-1 | GAAATACTTCCAGCTATCCAAATTTGGTG | 1–29 | 2989 | Sense | 94°C, 3 minutes, 1 cycle; 94°C, 30 seconds, 55°C, 30 seconds, |
| WA-RNA1-2989 | ATACGTCTCTCTCCAACGACAAC | 2967–2989 | Antisense | 72°C, 3 minutes, 30 cycles; 72°C, 10 minutes, 1 cycle | |
| WA-RNA1-2420 | AGAGAACAACTTCATTTCTACTCAATTGT | 2421–2449 | 3205 | Sense | 94°C, 3 minutes, 1 cycle; 94°C, 30 seconds, 55°C, 30 seconds, |
| WA-RNA1-5596 | GATAAATAAGTTTACCTGTATTGTCGGTC | 5597–5625 | Antisense | 72°C, 3.5 minutes, 30 cycles; 72°C, 10 minutes, 1 cycle | |
| WA-RNA1-4410 | GAGCATCAACTGTGGACGGCGAACCAAGC | 4411–4439 | 4227 | Sense | 94°C, 3 minutes, 1 cycle; 94°C, 30 seconds, 55°C, 30 seconds, |
| WA-RNA1-8637 | AACCTAGTTATTTAAATACTAGGTTTTCC | 8609–8637 | Antisense | 72°C, 4.5 minutes, 30 cycles; 72°C, 10 minutes, 1 cycle | |
| WA-RNA2-53 | CATTGGTTGTCGTCATGACTCGCAT | 53–77 | 5574 | Sense | 94°C, 3 minutes, 1 cycle; 94°C, 30 seconds, 55°C, 30 seconds, |
| WA-RNA2-5599 | CCAACTTACCAGATTTCGAGAACTGTAC | 5599–5626 | Antisense | 72°C, 6 minutes, 30 cycles; 72°C, 10 minutes, 1 cycle | |
| WA-RNA2-4440 | ATGCGTCTCGTCGTGACGTCCAGACTG | 4440–4466 | 3668 | Sense | 94°C, 3 minutes, 1 cycle; 94°C, 30 seconds, 55°C, 30 seconds, |
| WA-RNA2-8223 | GGCCTAGTTATTTAAATACTAGGTTTTCC | 8079–8107 | Antisense | 72°C, 4 minutes, 30 cycles; 72°C, 10 minutes, 1 cycle | |
| EA-RNA1-15 | TATCCAAATTTGGTGTGTTCTGCAG | 15–39 | 4815 | Sense | 94°C, 3 minutes, 1 cycle; 94°C, 30 seconds, 55°C, 30 seconds, |
| EA-RNA1-4800 | TTCCAATTGTGATAGATAAAGATCTACC | 4802–4829 | Antisense | 72°C, 5 minutes, 30 cycles; 72°C, 10 minutes, 1 cycle | |
| EA-RNA1-3680 | GGTGAGTAAGAAGGATAAATTGATCTCG | 3680–3707 | 4754 | Sense | 94°C, 3 minutes, 1 cycle; 94°C, 30 seconds, 55°C, 30 seconds, |
| EA-RNA1-8410 | TTCTAATACTCAAAAGGCAATATACAAAC | 8405–8433 | Antisense | 72°C, 5 minutes, 30 cycles; 72°C, 10 minutes, 1 cycle | |
| EA-RNA2-1 | GAAATACTACCCAGGTTTTTCCATGAGT | 1–28 | 4933 | Sense | 94°C, 3 minutes, 1 cycle; 94°C, 30 seconds, 55°C, 30 seconds, |
| EA-RNA2-4910 | GCATGTGATTGATGAAACTATGAGTTC | 4907–4933 | Antisense | 72°C, 5 minutes, 30 cycles; 72°C, 10 minutes, 1 cycle | |
| EA-RNA2-3710 | TTACCTAGGGACGTTGACGAATTGGT | 3705–3730 | 4452 | Sense | 94°C, 3 minutes, 1 cycle; 94°C, 30 seconds, 55°C, 30 seconds, |
| EA-RNA2-8130 | TTCATACACACACTCTAAATAGAAATACG | 8128–8156 | Antisense | 72°C, 4.5 minutes, 30 cycles; 72°C, 10 minutes, 1 cycle | |
| RdRp-F | CAANACNAANGAATTGAACAT | 7176–7196 | 1356 or 2081 | Sense | 94°C, 3 minutes, 1 cycle; 94°C, 30 seconds, 53°C, 30 seconds, |
| SVV-R3 | TTTTTGAGNTTTTANAATACACAC | 8508–8531 | Antisense | 72°C, 2 minutes, 30 cycles; 72°C, 10 minutes, 1 cycle | |
| CP-F | ATGGCTGATAGCACTAAAGTCGA | 4352–4374 | 774 | Sense | 94°C, 3 minutes, 1 cycle; 94°C, 30 seconds, 58°C, 30 seconds, |
| CP-R | TCAACAGTGAAGACCTGTTCCAG | 5103–5125 | Antisense | 72°C, 45 seconds, 30 cycles; 72°C, 10 minutes, 1 cycle | |
| Hsp70h-F | AGTGGTGAYGTAATAGTCGGTGG | 1008–1030 | 365 | Sense | 94°C, 3 minutes, 1 cycle; 94°C, 30 seconds, 58°C, 30 seconds, |
| Hsp70h-R | GCTAACGATTCACADACAGACTTCA | 1348–1372 | Antisense | 72°C, 30 seconds, 30 cycles; 72°C, 10 minutes, 1 cycle |
The nucleotide sequences of the Guangdong-2011 isolate genome RNA1 and RNA2 comprised 8622 and 8217 bp, respectively. The genome segment RNA1 of Guangdong-2011 isolate had an 89 nt 5′-UTR, a 172 nt 3′-UTR, and four ORFs: nucleotides 90–6053 (p227), 6052–7569 (RdRp), 7586–8272 (RNase3), and 8277–8450 (p7). The genome segment RNA2 had an 88 nt 5′-UTR, a 192 nt 3′-UTR, and contained eight ORFs: nucleotides 89–238 (p6.1), 554–712 (p6.2), 984–2648 (hsp70h), 2670–4226 (p60), 4208–4429 (p8), 4462–5235 (major cp), 5238–7292 (minor cp), and 7297–8025 (p28). The nucleotide sequences of the Jiangsu-2011, Sichuan-12-8, Sichuan-12-12, and Chongqing-12-8 isolate genome RNA1 and RNA2 comprised 8637 and 8107 bp, respectively. The genomic segment RNA1 of these isolates had an 89 nt 5′-UTR, a 193 nt 3′-UTR, and contained four ORFs: nucleotides 90–6053 (1a), 6052–7569 (RdRp), 7583–8272 (RNase3), and 8277–8444 (p7). The genomic segment RNA2 of these isolates had a 191 nt 5′-UTR, a 192 nt 3′-UTR, and contained nine ORFs: nucleotides 192–329 (p5.2), 333–467 (p5), 406–534 (p5.1), 879–2543 (hsp70h), 2565–4121 (p60), 4103–4324 (p8), 4352–5125 (major cp), 5128–7182 (minor cp), and 7187–7915 (p28). Comparisons of the genomic structures of RNA1 and RNA2 between this study and the reported isolates are shown in Tables 3 and 4.
Table 3. Comparison of genomic structures of RNA1.
| Can181-9 | Chongqing-12-8 | Jiangsu-2011 | Sichuan-12-8 | Sichuan-12-12 | Guangdong-2011 | m2-47 | Uganda | |
| 5′UTR | 1–89 | 1–89 | 1–89 | 1–89 | 1–89 | 1–89 | 1–89 | 1–89 |
| polyprotein 1a | 90–6053 | 90–6053 | 90–6053 | 90–6053 | 90–6053 | 90–6053 | 90–6053 | 90–6053 |
| RdRP | 6052–7569 | 6052–7569 | 6052–7569 | 6052–7569 | 6052–7569 | 6052–7569 | 6004–7569 | 6052–7569 |
| RNase3 | 7583–8272 | 7583–8272 | 7583–8272 | 7583–8272 | 7583–8272 | 7586–8272 | 7586–8272 | 7586–8272 |
| p7 | 8277–8444 | 8277–8444 | 8277–8444 | 8277–8444 | 8277–8444 | 8277–8450 | 8277–8450 | 8277–8450 |
| p22 | 8606–9181 | |||||||
| 3′UTR | 8445–8637 | 8445–8637 | 8445–8637 | 8445–8637 | 8445–8637 | 8451–8622 | 8451–8637 | 9182–9407 |
Table 4. Comparison of genomic structures of RNA2.
| Can181-9 | Chongqing-12-8 | Jiangsu-2011 | Sichuan-12-8 | Sichuan-12-12 | Guangdong-2011 | m2-47 | Uganda | |
| 5′UTR | 1–191 | 1–191 | 1–191 | 1–191 | 1–191 | 1–88 | 1–90 | 1–90 |
| p5.2/p6.1 | 192–329 | 192–329 | 192–329 | 192–329 | 192–329 | 89–238 | 91–240 | 91–240 |
| p5 | 333–467 | 333–467 | 333–467 | 333–467 | 333–467 | |||
| p5.1/p6.2 | 406–534 | 406–534 | 406–534 | 406–534 | 406–534 | 554–712 | 556–714 | |
| hsp70h | 880–2544 | 879–2543 | 879–2543 | 879–2543 | 879–2543 | 984–2648 | 986–2650 | 987–2651 |
| p60 | 2566–4122 | 2565–4121 | 2565–4121 | 2565–4121 | 2565–4121 | 2670–4226 | 2672–4228 | 2673–4229 |
| p8 | 4104–4325 | 4103–4324 | 4103–4324 | 4103–4324 | 4103–4324 | 4208–4429 | 4210–4431 | 4211–4432 |
| CP | 4353–5126 | 4352–5125 | 4352–5125 | 4352–5125 | 4352–5125 | 4462–5235 | 4464–5237 | 4465–5241 |
| mCP | 5129–7183 | 5128–7182 | 5128–7182 | 5128–7182 | 5128–7182 | 5238–7292 | 5240–7294 | 5244–7298 |
| p28 | 7188–7916 | 7187–7915 | 7187–7915 | 7187–7915 | 7187–7915 | 7297–8025 | 7299–8027 | 7303–8031 |
| 3′UTR | 7917–8108 | 7916–8107 | 7916–8107 | 7916–8107 | 7916–8107 | 8026–8217 | 8028–8217 | 8032–8223 |
Multiple sequence comparisons showed that the RNA1 sequence of the Guangdong-2011 isolate was similar to that of the SPCSV EA strain. RNA1 and RNA2 genome segments of the Guangdong-2011 isolate were 99.27% and 99.68% identical to that of the EA m2-47 isolate and 82.44% and 69.99% identical to that of the WA Can181-9 isolate, respectively. The Jiangsu-2011, Sichuan-12-8, Sichuan-12-12, and Chongqing-12-8 isolates showed a closer genetic relationship to the WA Can181-9 isolate than to the EA m2-47 isolate. The nucleotide sequence identities of RNA1 and RNA2 segments respectively ranged from 98.90% to 99.26% and from 98.80% to 99.17% when these four isolates were compared with the Can181-9 isolate of the WA strain. By contrast, the nucleotide sequence identities of RNA1 and RNA2 segments respectively ranged from 82.33% to 82.41% and from 69.80% to 69.98% when these four isolates were compared with the m2-47 isolate of the EA strain.
The pairwise percent identity of complete genomic sequences of the five isolates in China and three isolates retrieved from GenBank was calculated in multiple alignment (Table 5). Results showed that the RNA1 and RNA2 segment nt sequence identities of all isolates ranged from 81.5% to 99.6% and from 70.9% to 99.7%, respectively. The nt sequence identities in the WA strain were 98.8%–99.6% for RNA1 and 98.8%–99.7% for RNA2. The nt sequence identities in EA strains were 97.5%–99.4% for RNA1 and 98.3%–99.7% for RNA2. The nt identities between WA and EA strains were 81.5%–82.7% for RNA1 and 70.9%–71.2% for RNA2. This study revealed that the complete genomic sequences of the same strain group display a high degree of conservation. Compared with the reported sequences, the Chinese isolates showed high conservation and low molecular variation in their genomic sequences.
Table 5. Analysis of nucleotide sequence identities (%) of RNA1 (lower diagonal) and RNA2 (upper diagonal) segments of SPCSV isolates in this study and those retrieved from the database.
| Isolates | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| 1. Can181-9 | 99.0 | 98.8 | 99.2 | 99.2 | 71.0 | 70.9 | 71.0 | |
| 2. Chongqing-12-8 | 98.9 | 99.1 | 99.1 | 99.1 | 71.0 | 70.9 | 71.0 | |
| 3. Jiangsu-2011 | 98.9 | 99.6 | 98.8 | 98.8 | 71.1 | 71.0 | 71.2 | |
| 4. Sichuan-12-8 | 99.3 | 98.8 | 98.8 | 99.7 | 71.0 | 70.9 | 71.1 | |
| 5. Sichuan-12-12 | 99.3 | 98.8 | 98.8 | 99.6 | 71.0 | 70.9 | 71.0 | |
| 6. Guangdong-2011 | 82.6 | 82.5 | 82.5 | 82.5 | 82.6 | 99.7 | 98.3 | |
| 7. m2-47 | 82.7 | 82.6 | 82.5 | 82.6 | 82.6 | 99.4 | 98.4 | |
| 8. Uganda | 81.6 | 81.5 | 81.5 | 81.6 | 81.6 | 97.5 | 97.6 |
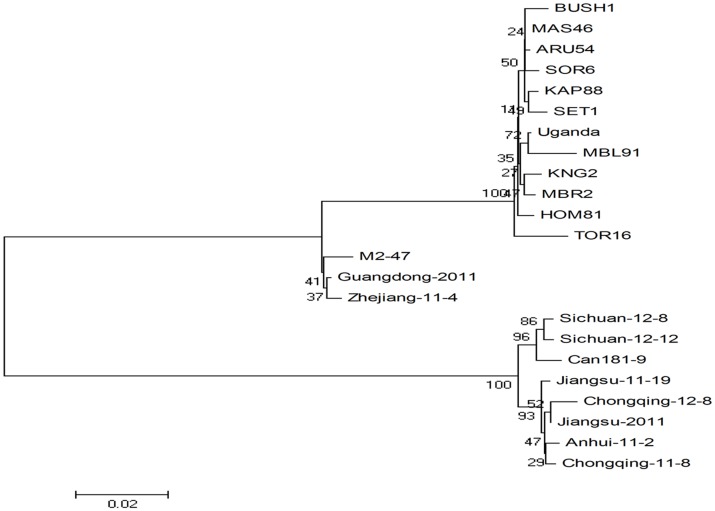
MEGA 4.0 was used to construct a phylogenetic tree of the five complete genomic sequences determined in this study and three isolates obtained from GenBank (Figure 2). Results indicated that Jiangsu-2011, Sichuan-12-8, Sichuan-12-12, and Chongqing-12-8 belong to the same branch as the Can181-9 isolate while Guangdong-2011 belongs to the same branch as the m2-47 and Uganda isolates.
Figure 2. Phylogenetic analysis of genomic RNA1 (A) and RNA2 (B) segments of five isolates determined in China and three isolates retrieved from GenBank.
Neighbor-joining trees were constructed via the maximum composite likelihood substitution model using MEGA (version 4.0). Numbers at branches represent bootstrap values of 1000 replicates. The scale bar shows the number of nucleotide substitutions per site.
Comparison of the length and nucleotide identity of the 5′ and 3′ UTRs of RNA1 and RNA2
Analysis of RNA1 5′ UTRs indicated 89 nt in Guangdong-2011, m2-47, Uganda (EA strain), Jiangsu-2011, Sichuan-12-8, Sichuan-12-12, Chongqing-12-8 and Can181-9 (WA strain), a nucleotide identity of over 98% between different isolates in the WA and EA strains, and a nucleotide identity of 85.4%–87.6% between WA and EA strains (Table 6).
Table 6. Nucleotide (lower diagonal) and amino acid (upper diagonal) (%) sequence identities of UTRs, polyprotein 1a (p227), RdRp, RNase3, p7, hsp70h, p60, p8, CP, mCP, and p28 among SPCSV isolates.
| Can181-9 | Chongqing-12-8 | Jiangsu-2011 | Sichuan-12-8 | Sichuan-12-12 | Guangdong-2011 | m2-47 | Uganda | |
| RNA1-5′UTR | ||||||||
| Can181-9 | ||||||||
| Chongqing-12-8 | 98.9 | |||||||
| Jiangsu-2011 | 97.8 | 98.9 | ||||||
| Sichuan-12-8 | 98.9 | 100 | 98.9 | |||||
| Sichuan-12-12 | 98.9 | 100 | 98.9 | 100 | ||||
| Guangdong-2011 | 85.4 | 86.5 | 87.6 | 86.5 | 86.5 | |||
| m2-47 | 85.4 | 86.5 | 87.6 | 86.5 | 86.5 | 100 | ||
| Uganda | 85.4 | 86.5 | 87.6 | 86.5 | 86.5 | 98.9 | 98.9 | |
| polyprotein 1a | Can181-9 | Chongqing-12-8 | Jiangsu-2011 | Sichuan-12-8 | Sichuan-12-12 | Guangdong-2011 | m2-47 | Uganda |
| Can181-9 | 98.8 | 98.7 | 99.3 | 99.5 | 87.2 | 87.4 | 87.5 | |
| Chongqing-12-8 | 99.0 | 99.2 | 98.7 | 98.9 | 87.3 | 87.5 | 87.6 | |
| Jiangsu-2011 | 98.9 | 99.6 | 98.6 | 98.8 | 87.4 | 87.6 | 87.6 | |
| Sichuan-12-8 | 99.3 | 98.8 | 98.7 | 99.5 | 87.2 | 87.4 | 87.5 | |
| Sichuan-12-12 | 99.3 | 98.8 | 98.7 | 99.5 | 87.3 | 87.5 | 87.6 | |
| Guangdong-2011 | 81.2 | 81.2 | 81.0 | 81.2 | 81.2 | 98.7 | 97.6 | |
| m2-47 | 81.3 | 81.3 | 81.1 | 81.2 | 81.3 | 99.4 | 98.0 | |
| Uganda | 81.3 | 81.2 | 81.1 | 81.2 | 81.3 | 98.3 | 98.6 | |
| RdRp | Can181-9 | Chongqing-12-8 | Jiangsu-2011 | Sichuan-12-8 | Sichuan-12-12 | Guangdong-2011 | m2-47 | Uganda |
| Can181-9 | 99.2 | 99.6 | 99.4 | 99.2 | 96.2 | 96.4 | 96.6 | |
| Chongqing-12-8 | 99.1 | 99.6 | 99.0 | 98.8 | 96.0 | 96.2 | 96.4 | |
| Jiangsu-2011 | 99.3 | 99.8 | 99.4 | 99.2 | 96.2 | 96.4 | 96.6 | |
| Sichuan-12-8 | 99.4 | 99.1 | 99.3 | 99.4 | 96.0 | 96.2 | 96.4 | |
| Sichuan-12-12 | 99.4 | 99.1 | 99.3 | 99.7 | 95.8 | 96.0 | 96.2 | |
| Guangdong-2011 | 87.6 | 87.4 | 87.5 | 87.4 | 87.4 | 99.2 | 99.4 | |
| m2-47 | 87.6 | 87.4 | 87.5 | 87.4 | 87.4 | 99.5 | 99.8 | |
| Uganda | 87.7 | 87.5 | 87.5 | 87.4 | 87.4 | 99.4 | 99.6 | |
| RNase3 | Can181-9 | Chongqing-12-8 | Jiangsu-2011 | Sichuan-12-8 | Sichuan-12-12 | Guangdong-2011 | m2-47 | Uganda |
| Can181-9 | 97.4 | 97.8 | 98.3 | 97.8 | 80.7 | 79.8 | 81.1 | |
| Chongqing-12-8 | 98.3 | 99.6 | 97.8 | 97.4 | 80.7 | 79.8 | 81.1 | |
| Jiangsu-2011 | 98.8 | 99.4 | 98.3 | 97.8 | 81.1 | 80.3 | 81.6 | |
| Sichuan-12-8 | 98.8 | 98.6 | 99.1 | 98.7 | 81.1 | 80.3 | 81.6 | |
| Sichuan-12-12 | 98.7 | 98.4 | 99.0 | 99.6 | 80.7 | 79.8 | 81.1 | |
| Guangdong-2011 | 83.0 | 82.8 | 83.4 | 83.3 | 83.1 | 99.1 | 99.1 | |
| m2-47 | 82.8 | 82.7 | 83.3 | 83.1 | 83.0 | 99.4 | 98.2 | |
| Uganda | 83.1 | 83.0 | 83.6 | 83.4 | 83.3 | 99.1 | 98.7 | |
| p7 | Can181-9 | Chongqing-12-8 | Jiangsu-2011 | Sichuan-12-8 | Sichuan-12-12 | Guangdong-2011 | m2-47 | Uganda |
| Can181-9 | 98.2 | 98.2 | 98.2 | 100 | 63.6 | 63.6 | 61.8 | |
| Chongqing-12-8 | 98.8 | 100 | 96.4 | 98.2 | 63.6 | 63.6 | 61.8 | |
| Jiangsu-2011 | 98.8 | 100 | 96.4 | 98.2 | 63.6 | 63.6 | 61.8 | |
| Sichuan-12-8 | 99.4 | 98.2 | 98.2 | 98.2 | 61.8 | 61.8 | 60.0 | |
| Sichuan-12-12 | 100 | 98.8 | 98.8 | 99.4 | 63.6 | 63.6 | 61.8 | |
| Guangdong-2011 | 78.0 | 76.8 | 76.8 | 77.4 | 78.0 | 100 | 98.2 | |
| m2-47 | 78.0 | 76.8 | 76.8 | 77.4 | 78.0 | 100 | 98.2 | |
| Uganda | 76.2 | 75.0 | 75.0 | 75.6 | 76.2 | 98.3 | 98.3 | |
| RNA1-3′UTR | Can181-9 | Chongqing-12-8 | Jiangsu-2011 | Sichuan-12-8 | Sichuan-12-12 | Guangdong-2011 | m2-47 | Uganda |
| Can181-9 | ||||||||
| Chongqing-12-8 | 99.0 | |||||||
| Jiangsu-2011 | 100 | 99.0 | ||||||
| Sichuan-12-8 | 99.5 | 98.4 | 99.5 | |||||
| Sichuan-12-12 | 100 | 99.0 | 100 | 99.5 | ||||
| Guangdong-2011 | 90.7 | 90.1 | 90.7 | 90.1 | 90.7 | |||
| m2-47 | 90.9 | 90.4 | 90.9 | 90.4 | 90.9 | 100 | ||
| Uganda | 80.3 | 80.3 | 80.3 | 79.8 | 80.3 | 80.2 | 81.8 | |
| RNA2-5′UTR | Can181-9 | Chongqing-12-8 | Jiangsu-2011 | Sichuan-12-8 | Sichuan-12-12 | Guangdong-2011 | m2-47 | Uganda |
| Can181-9 | ||||||||
| Chongqing-12-8 | 100 | |||||||
| Jiangsu-2011 | 100 | 100 | ||||||
| Sichuan-12-8 | 99.5 | 99.5 | 99.5 | |||||
| Sichuan-12-12 | 99.5 | 99.5 | 99.5 | 100 | ||||
| Guangdong-2011 | 96.6 | 96.6 | 96.6 | 96.6 | 96.6 | |||
| m2-47 | 93.2 | 93.2 | 93.2 | 93.2 | 93.2 | 96.6 | ||
| Uganda | 95.5 | 95.5 | 95.5 | 95.5 | 95.5 | 98.9 | 97.8 | |
| Hsp70h | Can181-9 | Chongqing-12-8 | Jiangsu-2011 | Sichuan-12-8 | Sichuan-12-12 | Guangdong-2011 | m2-47 | Uganda |
| Can181-9 | 99.1 | 98.9 | 99.1 | 99.3 | 90.4 | 90.4 | 90.6 | |
| Chongqing-12-8 | 99.0 | 99.1 | 98.6 | 98.7 | 90.1 | 90.1 | 90.3 | |
| Jiangsu-2011 | 98.9 | 99.5 | 98.4 | 98.6 | 90.1 | 90.1 | 90.3 | |
| Sichuan-12-8 | 99.4 | 98.8 | 98.9 | 99.1 | 90.1 | 90.1 | 90.3 | |
| Sichuan-12-12 | 99.4 | 98.8 | 98.9 | 99.6 | 90.1 | 90.1 | 90.3 | |
| Guangdong-2011 | 76.9 | 76.5 | 76.6 | 76.9 | 76.9 | 100 | 99.8 | |
| m2-47 | 77.0 | 76.5 | 76.7 | 76.9 | 77.0 | 99.9 | 99.8 | |
| Uganda | 76.9 | 76.6 | 76.8 | 76.9 | 76.9 | 98.7 | 98.9 | |
| p60 | Can181-9 | Chongqing-12-8 | Jiangsu-2011 | Sichuan-12-8 | Sichuan-12-12 | Guangdong-2011 | m2-47 | Uganda |
| Can181-9 | 97.5 | 97.3 | 97.5 | 97.5 | 78.8 | 78.8 | 79.5 | |
| Chongqing-12-8 | 98.7 | 99.4 | 98.5 | 98.5 | 79.7 | 79.7 | 80.5 | |
| Jiangsu-2011 | 98.3 | 99.4 | 98.6 | 98.6 | 79.3 | 79.3 | 80.1 | |
| Sichuan-12-8 | 99.0 | 98.7 | 99.0 | 99.2 | 79.5 | 79.5 | 80.3 | |
| Sichuan-12-12 | 98.9 | 98.7 | 98.9 | 99.7 | 79.5 | 79.5 | 80.3 | |
| Guangdong-2011 | 72.4 | 72.7 | 72.4 | 72.4 | 72.4 | 99.4 | 98.6 | |
| m2-47 | 72.5 | 72.8 | 72.5 | 72.5 | 72.4 | 99.8 | 98.8 | |
| Uganda | 72.7 | 73.0 | 72.7 | 72.7 | 72.6 | 98.8 | 98.9 | |
| p8 | Can181-9 | Chongqing-12-8 | Jiangsu-2011 | Sichuan-12-8 | Sichuan-12-12 | Guangdong-2011 | m2-47 | Uganda |
| Can181-9 | 98.6 | 95.9 | 97.3 | 98.6 | 68.5 | 68.5 | 68.5 | |
| Chongqing-12-8 | 98.2 | 97.3 | 98.6 | 100 | 69.9 | 69.9 | 69.9 | |
| Jiangsu-2011 | 98.2 | 98.2 | 95.9 | 97.3 | 68.5 | 68.5 | 68.5 | |
| Sichuan-12-8 | 98.6 | 98.6 | 98.6 | 98.6 | 69.9 | 69.9 | 69.9 | |
| Sichuan-12-12 | 98.6 | 98.6 | 98.6 | 99.1 | 69.9 | 69.9 | 69.9 | |
| Guangdong-2011 | 72.1 | 72.5 | 71.6 | 72.1 | 72.1 | 100 | 93.2 | |
| m2-47 | 72.1 | 72.5 | 71.6 | 72.1 | 72.1 | 100 | 93.2 | |
| Uganda | 73.0 | 73.4 | 72.5 | 73.0 | 73.0 | 95.5 | 95.5 | |
| CP | Can181-9 | Chongqing-12-8 | Jiangsu-2011 | Sichuan-12-8 | Sichuan-12-12 | Guangdong-2011 | m2-47 | Uganda |
| Can181-9 | 99.2 | 99.2 | 99.6 | 98.4 | 79.8 | 79.4 | 77.8 | |
| Chongqing-12-8 | 99.5 | 98.4 | 99.6 | 98.4 | 79.4 | 79.0 | 77.4 | |
| Jiangsu-2011 | 99.4 | 98.8 | 98.8 | 97.7 | 80.2 | 79.8 | 78.2 | |
| Sichuan-12-8 | 99.6 | 99.6 | 99.0 | 98.8 | 79.8 | 79.4 | 77.8 | |
| Sichuan-12-12 | 99.4 | 99.4 | 98.7 | 99.5 | 78.6 | 78.2 | 76.7 | |
| Guangdong-2011 | 73.3 | 73.3 | 74.0 | 73.3 | 72.9 | 99.2 | 95.7 | |
| m2-47 | 73.3 | 73.3 | 74.0 | 73.3 | 72.9 | 99.7 | 96.5 | |
| Uganda | 73.2 | 73.2 | 73.8 | 73.2 | 72.8 | 98.6 | 98.8 | |
| mCP | Can181-9 | Chongqing-12-8 | Jiangsu-2011 | Sichuan-12-8 | Sichuan-12-12 | Guangdong-2011 | m2-47 | Uganda |
| Can181-9 | 98.4 | 98.5 | 98.7 | 99.0 | 61.5 | 61.3 | 61.4 | |
| Chongqing-12-8 | 99.2 | 98.1 | 98.8 | 99.1 | 61.4 | 61.1 | 61.3 | |
| Jiangsu-2011 | 98.9 | 98.5 | 98.4 | 98.7 | 62.3 | 62.0 | 62.1 | |
| Sichuan-12-8 | 99.4 | 99.5 | 98.7 | 99.4 | 61.5 | 61.3 | 61.4 | |
| Sichuan-12-12 | 99.5 | 99.6 | 98.8 | 99.8 | 61.7 | 61.4 | 61.5 | |
| Guangdong-2011 | 63.5 | 63.2 | 63.6 | 63.3 | 63.4 | 99.1 | 98.7 | |
| m2-47 | 63.2 | 62.9 | 63.3 | 63.0 | 63.1 | 99.7 | 98.1 | |
| Uganda | 63.4 | 63.1 | 63.6 | 63.1 | 63.2 | 98.4 | 98.2 | |
| p28 | Can181-9 | Chongqing-12-8 | Jiangsu-2011 | Sichuan-12-8 | Sichuan-12-12 | Guangdong-2011 | m2-47 | Uganda |
| Can181-9 | 98.9 | 98.3 | 97.9 | 97.9 | 85.1 | 85.1 | 85.5 | |
| Chongqing-12-8 | 98.9 | 98.8 | 99.2 | 99.2 | 86.0 | 86.0 | 86.4 | |
| Jiangsu-2011 | 98.9 | 98.6 | 98.8 | 98.8 | 86.0 | 86.0 | 86.4 | |
| Sichuan-12-8 | 99.0 | 99.6 | 98.8 | 99.2 | 86.0 | 86.0 | 86.4 | |
| Sichuan-12-12 | 99.0 | 99.6 | 98.8 | 99.7 | 86.0 | 86.0 | 86.4 | |
| Guangdong-2011 | 77.4 | 77.6 | 77.9 | 77.5 | 77.5 | 99.2 | 97.9 | |
| m2-47 | 77.4 | 77.6 | 77.9 | 77.5 | 77.5 | 99.7 | 98.8 | |
| Uganda | 77.9 | 78.2 | 78.5 | 78.1 | 78.1 | 97.9 | 98.2 | |
| RNA2-3′UTR | Can181-9 | Chongqing-12-8 | Jiangsu-2011 | Sichuan-12-8 | Sichuan-12-12 | Guangdong-2011 | m2-47 | Uganda |
| Can181-9 | ||||||||
| Chongqing-12-8 | 99.5 | |||||||
| Jiangsu-2011 | 100 | 99.5 | ||||||
| Sichuan-12-8 | 99.5 | 100 | 99.5 | |||||
| Sichuan-12-12 | 99.5 | 100 | 99.5 | 100 | ||||
| Guangdong-2011 | 79.7 | 79.7 | 79.7 | 79.7 | 79.7 | |||
| m2-47 | 79.7 | 79.7 | 79.7 | 79.7 | 79.7 | 99.0 | ||
| Uganda | 79.7 | 79.7 | 79.7 | 79.7 | 79.7 | 99.0 | 100 |
Analysis of RNA1 3′ UTRs showed that this UTR, being 193 nt long (Table 3), is conserved between different isolates in WA strains and that its nucleotide identity is 98.4%–100% (Table 6). Isolates in the EA strain differed from each other significantly: the length of the 3′ UTR of isolate Uganda was 226 nt, that of isolate m2-47 was 187 nt long, and that of isolate Guangdong-2011 was 172 nt long (Table 3). The nt identity between Guangdong-2011 and m2-47 isolates was 100%, that between Uganda and m2-47 was 81.8%, and that between Guangdong-2011 and Uganda was 80.2% (Table 6).
Comparison of RNA2 5′UTRs showed that the WA strain length is 191 nt (Table 4) and that the nucleotide identity between different isolates is 99.5%–100% (Table 6). The nucleotide identity between the three isolates in the EA strain was 96.6%–98.9% (Table 6), the length of isolate Guangdong-2011 was 88 nt, and the lengths of the two other isolates were both 90 nt. Despite differences in the 5′ UTR lengths among WA and EA strains, the nucleotide sequences of these strains were rather conserved; nt identities between the eight isolates were 93.2%–100% (Table 6).
Comparison of RNA2 3′ UTRs suggested that the length of both WA and EA strains is 192 nt (Table 4) and that the nt identities between different isolates in the WA and EA strains exceed 99%. By contrast, the nt identity between WA and EA strains was 79.7%, which is rather low (Table 6).
Comparison of the 5′ and 3′ UTRs of RNA1 and RNA2 indicated that the nt identity of 5′UTRs between RNA1 and RNA2 is low (around 40%) whereas that of 3′UTRs of RNA1 and RNA2 is high (around 80%). The nt identity of 3′ UTRs of RNA1 and RNA2 of isolate Uganda was about 99%.
Genome structure analysis and nucleotide/amino acid sequence comparison of proteins
Analysis of genome structures suggested that the RNA1 segment contained four ORFs: 1a (p227), RdRp, RNase3, and p7. The genome positions of these ORFs were listed in Table 5. Between protein p7 and the 3′UTR, the Uganda isolate contains a p22 ORF (located at 8606–9181 nt of the genome); none of the seven other isolates had the p22 gene. Nucleotide and amino acid identities between these proteins are listed in Table 6.
Analysis of the nucleotide and amino acid identity of protein 1a (p227), RdRp, RNase3, and p7 encoded by eight isolates in RNA1 showed that the nucleotide identities of encoding proteins between the eight isolates are as follows: protein 1a (81.0%–99.6%), RdRp (87.4%–99.8%), RNase3 (82.7%–99.6%) and p7(75.0%–100%). The amino acid identities of encoding proteins between the eight isolates are as follows: protein 1a (87.2%–99.5%), RdRp (95.8%–99.8%), RNase3 (79.8%–99.6%), and p7 (60.0%–100%). The nt/aa identities between different isolates in the WA strain are as follows: protein 1a (98.7%–99.6%/98.6%–99.5%), RdRp (99.1%–99.8%/98.8%–99.6%), RNase3 (98.3%–99.6%/97.4%–99.6%), and p7 (98.2%–100%/96.4%–100%). The nt/aa identities between different isolates in the EA strain are as follows: protein 1a (98.3%–99.4%/97.6%–98.7%), RdRp (99.4%–99.6%/99.2%–99.8%), RNase3 (98.7%–99.4%/98.2%–99.1%), and p7 (98.3%–100%/98.2%–100%). The nt/aa identities between WA and EA strains are as follows: protein 1a (81.0%–81.3%/87.2%–87.6%), RdRp (87.4%–87.6%/95.8%–96.6%), RNase3 (82.7%–83.6%/79.8%–81.6%), and p7 (75.0%–78.0%/60.0%–63.6%). Analysis of RdRp protein sequences demonstrated that the 5′ end of isolate m2-47 is 16 aa longer than that of the other isolates. RNase3 sequence analysis showed that the ORF for RNase3 is 684 nt (228 aa) in all EA strain isolates but 687 nt (229 aa) in WA strain isolates. The 5′ end of the RNase3 ORF in EA strains was 1 aa longer than that of WA strains. P7 sequence analysis showed that the ORF for p7 is 171 nt (57 aa) in all EA strain isolates but 165 nt (55 aa) in WA strain isolates. The 3′ end of the p7 ORF in EA strains was 2 aa longer than that of WA strains.
Analysis of genomic structures indicated that the segment RNA2 contains at least six ORFs: Hsp70h, p60, p8, CP, mCP, and p28. The locations of these ORFs in the genome are listed in Table 4. The nucleotide and amino acid identities between these proteins are listed in Table 6.
Isolates Guangdong-2011, m2-47, and Uganda of the EA strain contained an ORF for p6 (named p6.1 in this study) in the upstream region of hsp70h, and the nt/aa identity of the p6.1 protein between these three isolates was 100%. Between p6.1 and hsp70h, similar to isolate m2-47, the Guangdong-2011 isolate contained one ORF for the p6 protein (named p6.2 in this study) more than isolate Uganda; the nt and aa identities of this protein were 98.7% and 96.2%, respectively. Similar to Can181-9, isolates Jiangsu-2011, Sichuan-12-8, Sichuan-12-12, and Chongqing-12-8 of the WA strain contained three ORFs in the upstream region of hsp70h, namely p5.2, p5, and p5.1. The nt and aa identities of the five isolates are as follows: p5.2 (97.8%–100% and 100%), p5 (97.0%–100% and 90.9%–100%), and p5.1 (95.3%–100% and 90.5%–100%).
Analysis showed that the nucleotide identities of the encoded proteins between the eight isolates are as follows: Hsp70h (76.5%–99.9%), p60 (72.4%–99.8%), p8 (72.1%–100%), CP (72.8%–99.7%), mCP (62.9%–99.8%), and p28 (77.4%–99.7%). The aa identities of these proteins are as follows: Hsp70h (90.1%–100%), p60 (78.8%–99.4%), p8 (68.5%–100%), CP (76.7%–99.6%), mCP (61.1%–99.4%), and p28 (85.1%–99.2%). The nt/aa identities between different isolates of WA strain are as follows: Hsp70h (98.8%–99.6%/98.4%–99.3%), p60 (98.3%–99.7%/97.3%–99.4%), p8 (98.2%–99.1%/95.9%–100%), CP (98.7%–99.6%/97.7%–99.6%), mCP (98.5%–99.8%/98.1%–99.4%), and p28 (98.6%–99.7%/97.9%–99.2%). The nt/aa identities between different isolates within the EA strain are as follows: Hsp70h (98.7%–99.9%/99.8%–100%), p60 (98.8%–99.8%/98.6%–99.4%), p8 (95.5%–100%/93.2%–100%), CP (98.6%–99.7%/95.7%–99.2%), mCP (98.2%–99.7%/98.1%–99.1%), and p28 (97.9%–99.7%/97.9%–99.2%). The nt/aa identities between the WA and EA strains are as follows: Hsp70h (76.5%–77.0%/90.1%–90.6%), p60 (72.4%–73.0%/78.8%–80.5%), p8 (72.1%–73.4%/68.5%–69.9%), CP (72.8%–74.0%/76.7%–80.2%), mCP (62.9%–63.6%/61.1%–62.3%), and p28 (77.4%–78.5%/85.1%–86.4%). CP protein sequence analysis also showed that isolate Uganda has one methionine residue more than the seven other isolates in locus 231.
Absence of the p22 gene in RNA1
Genome structure analysis showed that the p22 gene was not present in any of the aforementioned isolates from China. To investigate whether or not p22 is present in other Chinese regions, the primers RdRp-F and SVV-R3 were used to amplify the region from the RdRp gene to the 3′-UTR of RNA1; these primers were designed according to the reference [28]. Sweet potato samples infected by SPCSV were collected from Jiangsu, Zhejiang, Anhui, and Chongqing, China. Using RT-PCR, the partial RNA1 sequences from four different samples were obtained (relevant data, such as name, strain assignment, geographic origin, segment length, reference, and GenBank accession numbers, are listed in Table 1). Sequence analysis indicated that Zhejiang-11-4 shares nucleotide identities of 99.6% with Guangdong-2011 and 84.4% with isolate Jiangsu-2011. Isolates Jiangsu-11-19, Anhui-11-2, and Chongqing-11-8 shared nucleotide identities of 84.4% with isolate Guangdong-2011 and 99.4%–99.6% with isolate Jiangsu-2011. Overall, genome organization analysis showed that p22 is missing in all nine isolates from China.
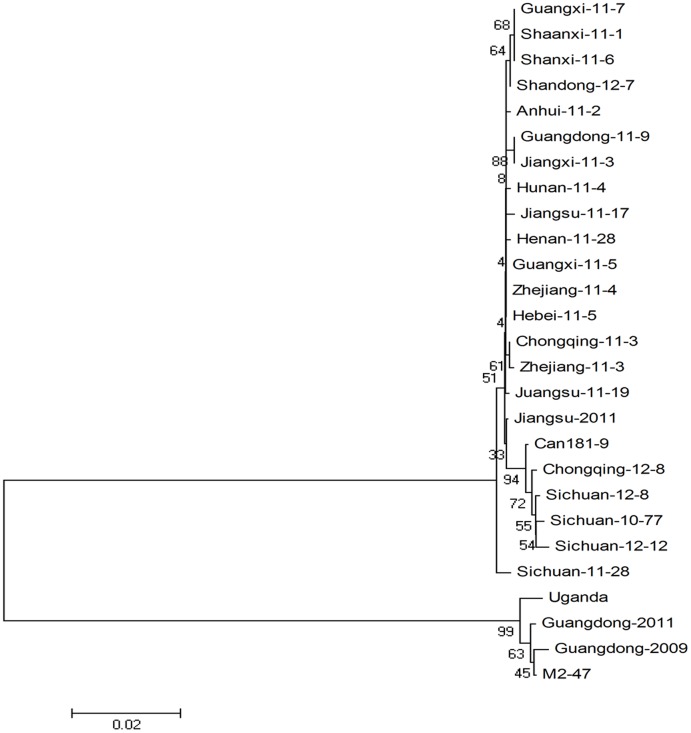
The nt sequences of 9 SPCSV isolates characterized in this study and 14 isolates deposited in GenBank were subjected to phylogenetic analysis, including the previously characterized WA strain Can181-9 isolate from Spain, EA strain m2-47 isolate from Peru, and 12 EA strain isolates from Uganda. Phylogenetic analysis (Figure 3) of partial genomic sequences of RNA1 was used to assign isolates of SPCSV from China to the two relatively distantly related strains EA and WA. Isolates Zhejiang-11-4 and Guangdong-2011 in this study belonged to the EA strain. Isolates Jiangsu-11-19, Anhui-11-2, Chongqing-11-8, Jiangsu-2011, Sichuan-12-8, Sichuan-12-12, and Chongqing-12-8 belonged to the WA strain.
Figure 3. Phylogenetic tree based on partial sequences of RNA1.
Phylogenetic tree illustrating relationships between isolates obtained from China and representative isolates of sweet potato chlorotic stunt virus (SPCSV) deposited in GenBank. Neighbor-joining trees were constructed via the maximum composite likelihood substitution model using MEGA (version 4.0). Numbers at branches represent bootstrap values of 1000 replicates. The scale bar shows the number of nucleotide substitutions per site.
Molecular variability and phylogenetic analysis of the cp gene in WA strains
To study the genetic variation of SPCSV in different Chinese regions, we used the nucleotide sequence of the cp gene of the SPCSV WA strain in GenBank (GenBank accession number FJ807785) to design primers named CP-F and CP-R to amplify the cp gene of SPCSV WA isolates from different regions. A total of 20 cp gene sequences from 14 Chinese provinces or cities were obtained (name, strain assignment, geographic origin, segment length, reference, and GenBank accession numbers are listed in Table 1). Sequencing results suggested that the cp gene sequence length of 20 isolates is 774 bp, as expected.
Sequence alignment and phylogenetic analysis using the neighbor-joining method were performed with MEGA software (version 4.0). Nucleotide and amino acid sequence identity analysis of 25 isolates acquired in this research showed that, except for isolate Guangdong-2011 (EA strain), SPCSV isolates belonging to the WA strain exhibit 98.4%–100% nt sequence identity and 97.7%–100% deduced aa sequence identity. These findings demonstrate that isolates of the SPCSV WA strain from China show nearly identical. A phylogenetic tree was constructed (Figure 4) based on CP sequences to illustrate the probable genetic relationships between the 25 isolates and Guangdong-2009, m2-47, Uganda, and Can181-9. Results showed that Chinese SPCSV isolates could be divided into two branches. Except for those obtained from the Guangdong region, SPCSV isolates from other Chinese regions belonged to the WA strain. This finding indicates that at least two different strains may be observed among Chinese SPCSV isolates and that the WA strain is more prevalent than the EA strain.
Figure 4. Phylogenetic tree based on CP gene sequences.
Phylogenetic tree illustrating relationships between the CP genes of isolates obtained from China and representative isolates of sweet potato chlorotic stunt virus (SPCSV) deposited in GenBank. Neighbor-joining trees were constructed via the maximum composite likelihood substitution model using MEGA (version 4.0). The bar in this figure represents 0.02 Kimura nucleotide unites. Numbers at branches represent bootstrap values of 1000 replicates.
Analysis of the phylogenetic tree based on partial RNA1 sequences showed that Zhejiang-11-4 from Zhejiang Province belongs to the EA strain. However, when the tree was constructed using the CP gene, the isolate appeared to belong to the WA strain. This finding suggests that Zhejiang-11-4 occurs through co-infection by WA and EA.
This study reveals that nucleotide and amino acid identities are highly conserved between different isolates of Chinese strains and that the WA strain is distributed more extensively than the EA strain.
Discussion
The complete genomic sequences of five different SPCSV isolates were acquired from main sweet potato production areas in China by RT-PCR and 5′ and 3′-RACE using virus-transmitting vector whitefly as a material. Guangdong-2011, which was isolated from Guangdong Province, belongs to the EA strain, whereas Jiangsu-2011, Sichuan-12-8, Sichuan-12-8, and Chongqing-12-8, which were isolated from Jiangsu, Sichuan, and Chongqing, belong to the WA strain. To the best of our knowledge, this study is the first to report the complete genomic sequences of Chinese SPCSV isolates as well as the complete SPCSV genomic sequence acquired from whitefly.
Factors such as low titers of SPCSV, heterogeneous distribution of the virus in sweet potato, and phenol and polysaccharide enrichment in sweet potato leaves [40], [41] significantly decrease the quality of extracted RNA from sweet potato leaves and the amplification efficiency of conventional RT-PCR. SPCSV usually co-infects sweet potato with other viruses in the field; thus SPCSV virions are difficult to separate and purify. These factors severely restrict SPCSV genome sequencing [22], [32]–[34]. Only three SPCSV complete genomic sequences can be accessed via GenBank today, and only one complete sequence of SPCSV WA strain is available [11], [29], [35]. In the present study, the SPCSV titer in whitefly was found to be significantly higher than that in sweet potato and I. setosa. This feature of the virus-transmitting vector whitefly is advantageous for acquiring the complete SPCSV genomic sequence. In addition, using viruliferous whitefly as an experimental material can preclude the interference of other viruses, phenols, and polysaccharides in sweet potato leaves. Our research provides a simplified and convenient method for cloning SPCSV genomic sequences.
The complete genomic sequences of the five isolates from different Chinese areas were used to analyze molecular variations and phylogenesis compared with the genomic sequences of three isolates obtained from GenBank. Analysis of the nucleotide identity of complete genomic sequences indicated that the nt identities in SPCSV strain are highly conserved and significant differences were found between WA and EA strains. Analyses of nt and aa identities based on the cp gene and partial RNA1 sequences are consistent with aforementioned results. High conservation was observed between different isolates in the same strain, but significant differences were found between WA and EA strains, which are inferred to be caused by the short time since SPCSV was transmitted into China, so the virus shows only limited genetic variability. This result is consistent with previous reports [3], [27], [28], [36]. Tugume [36] analyzed the genetic variability of p7, RNase3, and p22 genes of different SPCSV isolates infecting sweet potato and wild species in Uganda and suggested that the three genes show only limited genetic variability among strains. The EA and WA strain isolates showed nt (aa) sequence identities of 83.8%–84.3% (81.6%–82.5%) for RNase3 and 76.2%–79.8% (60.7%–66.7%) for p7 [36]. The nt and deduced aa sequence identities of RNase3, p7 and hsp70h of EA strain isolates Ug, Tug2, Unj2, Mis1, m2-47, and WA strain isolate Is were analyzed. The nt and aa sequences of RNase3 and hsp70h in the EA strain isolates were nearly identical but the nt and aa of p7 showed more variation [28]. We previously analyzed the genetic variability of partial hsp70h gene of different SPCSV isolates in China, and our results demonstrated that the hsp70 gene sequences of the same strain group display a high degree of conservation and that strain group WA has a wider geographic distribution in China than the EA strain [27].
The genome organization of SPCSV shares many similarities with other criniviruses [10]. However, the genome of SPCSV possesses unique features particularly concerning the gene content of RNA1. Downstream from the ORF for the replicase, RNA1 contains ORFs for Class 1 RNase III enzyme, a putative hydrophobic protein (p7), and a 22 kDa protein that shows no significant similarity to known proteins from any organism [11], [28], [42]. The 3′-proximal part of RNA1 constitutes an interesting genomic region for study owing to its unique gene functions. RNase3 contains a single endoribonuclease domain and a dsRNA-binding domain [43], [44]. RNase3 inhibits posttranscriptional gene silencing and exerts a key function in the development of severe diseases in sweet potato plants co-infected with other viruses [42], [43]. The p22 gene was identified as a SPCSV RNA silencing suppressor and it could suppress silencing induced by dsRNA [42]. Similar activities were exhibited by homologs of p22 encoded by other members of the family Closteroviridae. Cañizares identified Tomato chlorosis virus (ToCV) RNA1-encoded p22 protein as an effective silencing suppressor by using an agrobacterium co-infiltration assay. ToCV p22 suppressed local RNA silencing induced either by sense RNA or dsRNA very efficiently but did not interfere with short or long-distance systemic spread of silencing [45]. Data showed that the Beet yellow virus p21, Beet yellow stunt virus p22, Citrus tristeza virus p20, and Grapevine leafroll-associated virus-2 p24, which are homologs of p22, are silencing suppressors [46]–[48]. Recent studies have revealed that many SPCSV isolates lacking p22 still synergize with unrelated viruses [18], [28], [43], which indicates that p22 is dispensable for synergy between SPCSV and other viruses. While p22 is a pathogenicity enhancer of SPCSV, co-infection of SPFMV with SPCSV isolates containing p22 causes more severe symptoms than co-infection with SPCSV isolates lacking p22 in the indicator plant I. setosa [28]. The p22 gene is present only in Ugandan isolates of SPCSV, and SPCSV isolates from other areas do not contain the p22 gene [36]. The partial RNA1 sequences of nine isolates from different Chinese regions were determined in this study, and no isolate (neither EA nor WA strains) containing the p22 gene was observed in our research.
A novel virus isolate related to viruses in genus Crinivirus carrying predicted ORFs for proteins homologous to the RNase3 and p7 of SPCSV was detected and designated as KML33b. The sequences of KML33b were highly divergent from SPCSV isolates and showed <60% sequence identities for both nt and aa [36]. According to the species demarcation criteria from the Ninth Report of the International Committee on Taxonomy of Viruses (ICTV), the criteria for demarcating species in the genus Crinivirus are: (a) genome structure and organization (number and relative location of the ORFs) and (b) amino acid sequences of relevant gene products (polymerase, CP, Hsp70h) differing by more than 25% [49]. In the present study, comparison of RNA1 and RNA2 between WA and EA strains showed that the nucleotide identities exhibit significant differences between strains. Genomic structure analysis suggested that the quantity of proteins encoded by the RNA2 segment between SPCSV WA and EA strains differ. Before hsp70h, WA encodes three but EA encodes one or two hypothetical proteins. Comparison of the 3′ and 5′ UTRs of the genome shows that the lengths of RNA1 3′UTR and RNA2 5′UTR are different between WA and EA strains. The nucleotide identity of the RNA1 3′UTR and RNA2 5′UTR between WA and EA strains is low. Although the RNA1 5′ and RNA2 3′UTRs of the WA and EA strains showed the same length, their nucleotide identity was low. Comparison of aa identities suggested that proteins with an aa identity <75% between the WA and EA strains include p7 in RNA1, p8, and mCP in RNA2. Tairo previously suggested that EA and WA of SPCSV may belong to different species in the genus Crinivirus [3]. As mentioned above, the low sequence identity and differences in genomic structures between the EA and WA strains of SPCSV support this proposal. However, as the biological and serological relationships between the strains remain incompletely understood, a systematic assessment of these differences should be given the top priority for future research.
Materials and Methods
Ethics statement
Samples were collected from private land with the owner's permission. No specific permissions were required for sampling from any other location. Field studies did not involve endangered or protected species.
Collection of virus isolates
From 2010 to 2012, five sweet potato vine cuttings were collected from the main sweet potato-producing areas (Jiangsu, Guangdong, Sichuan Provinces, and Chongqing City) of China (Table 1) and grown in an insect-proof greenhouse. These cuttings were proven to be infected with SPCSV by nitrocellulose membrane (NCM)-ELISA and RT-PCR. The SPCSV-infected sweet potato was placed in an insect cage to feed the whiteflies. Non-viruliferous whiteflies were separately fed in SPCSV-infected sweet potato plants; here, whiteflies that had been fed for more than 3 d were considered viruliferous. Adult viruliferous whiteflies were collected and quick-frozen in liquid nitrogen, and then stored at −70°C to amplify complete genomic sequence.
From 2010 to 2012, a total of 20 vine cuttings were collected from the main sweet potato-producing areas in 14 provinces of China (Table 1). I. setosa was inoculated by side-grafting with infected sweet potato scions and grown in an insect-proof greenhouse (temperature, 25–30°C; relative humidity, 70%) under natural daylight. I. setosa-infecting SPCSV were used to clone the cp gene and partial RNA1 sequences.
Design and synthesis of primers used in this study
The primers used in this study are shown in Table 2. Primers for amplification of the RNA1 and RNA2 regions of WA strains were designed according to the sequences of SPCSV and deposited in GenBank (GenBank accession numbers FJ807784 and FJ807785). The primers were named WA-RNA1-P1, WA-RNA1-P2989, WA-RNA1-P2420, WA-RNA1-P5596, WA-RNA1-P4410, WA-RNA1-P8637, WA-RNA2-P53, WA-RNA2-P5599, WA-RNA2-P4440, and WA-RNA2-P8223. Primers for amplification of the RNA1 and RNA2 regions of EA strains were designed according to the sequences of SPCSV and deposited in GenBank (GenBank accession numbers HQ291259, HQ291260, AJ428554, and AJ428555). The primers were named EA-RNA1-P15, EA-RNA1-P4800, EA-RNA1-P3680, EA-RNA1-P8410, EA-RNA2-P1, EA-RNA2-P4910, EA-RNA2-P3710, and EA-RNA2-P8130. Primers for amplification of the cp gene were designed according to the sequences of SPCSV and deposited in GenBank (GenBank accession number FJ807785). These primers were named CP-F and CP-R. Primers for amplification of the 3′ genomic region of RNA1 were synthesized according to the reference [28] and named RdRp-F and SVV-R3.
Serological detection of SPCSV
SPCSV was detected using an NCM-ELISA kit from the International Potato Center (CIP). Briefly, 150 mg of leaf material was ground in a mortar with 1 mL of extraction buffer (1 M Tris-HCl containing 0.2% Na2SO3). The homogenate was transferred to a 1.5 mL Eppendorf tube and spun at 6000×g for 5 min. Aliquots of the supernatant (20 µL) were placed on membranes. The membranes were blocked with blocking solution at room temperature and then incubated for 60 min. After washing twice for 3 min each time, the membranes were incubated in virus-specific antibodies to the SPCSV coat protein (provided by CIP) at 4°C overnight. After washing twice for 3 min each time, the membranes were incubated in the conjugated anti-SPCSV antibody at room temperature for 1 h and then washed again twice for 3 min each time. The color reaction was developed using NBT/BCIP as the substrate. Color development was ceased by discarding the substrate solution and immersing the membranes in tap water. NCMs were washed in distilled water for 10 min.
RNA isolation
Total RNA was isolated from 100 mg of viruliferous whiteflies or I. setosa leaves as templates using the Total Plant RNA Extraction Miniprep System (Sangon, Shanghai, China). The amount and quality of the RNA were verified using agarose gel electrophoresis.
RT-PCR detection of SPCSV
RNA was used for reverse transcription using Moloney murine leukemia virus (M-MLV) reverse transcriptase (TaKaRa, Shiga, Japan) according to the manufacturer's instructions. Synthesized cDNA was amplified using Ex Taq DNA polymerase (TaKaRa). The partial sequence of hsp70h gene was amplified to confirm samples infected SPCSV. Degenerate primers for amplifying the hsp70h gene were designed according to the sequences of the SPCSV WA and EA strains and deposited in GenBank. These primers were respectively named Hsp70h-F and Hsp70h-R (Table 2).
Cloning and sequence analysis
RNA was used for reverse transcription using M-MLV reverse transcriptase (TaKaRa) according to the manufacturer's instructions. Synthesized cDNA was amplified using LA Taq DNA polymerase (TaKaRa). Genome RNA1 or RNA2 sequences of the WA strain were acquired by two or three overlapping RT-PCRs, whereas genome RNA1 or RNA2 sequences of the EA strain were obtained by two overlapping RT-PCRs. All PCR conditions in this study are shown in Table 2. Rapid amplification of cDNA ends (RACE) was used to determine the 5′ and 3′ ends of the viral genomic segments RNA1 and RNA2. 5′ and 3′ RACE was conducted by TaKaRa. The cp gene and the partial sequence of RNA1 obtained from the 3′-promixmal region of the RdRp gene to the middle of the 3′UTR were determined by cloning and sequencing obtained amplicons by standard RT-PCR.
The amplified products were purified from agarose gels using an AxyPrep DNA Gel Extraction Kit (Axygen, Hangzhou, China) and cloned into the PMD19-T vector (TaKaRa). Recombinant plasmids were transformed into Escherichia coli strain TG1 competent cells, purified using Plasmid Miniprep Kits (Bioteke, Beijing, China), and sequenced by Sangon Biotech Company (Sangon, Shanghai, China). Sequencing was conducted in both directions for each of the two independent amplicons of all isolates.
The sequences obtained were compared by BLAST search with the existing sequences in the NCBI database. Sequences were analyzed using DNAMAN Version 6.0. SPCSV Can181-9, m2-47 and Uganda isolates were used to explore the genome organization of the SPCSV WA and EA strains in China. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 4.0 [50]. Multiple alignments of protein-coding sequences were obtained using the default options in Clustal W [51]. Phylogenetic trees were constructed based on the aligned protein-coding sequences using the neighbor-joining method. The statistical significance of tree branching was tested by performing 1000 bootstrap replications.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
Financial support provided by the Earmarked Fund for China Agricultural Research System (CARS-11-B-07) and Independent Innovation Foundation of Henan Academy of Agricultural Sciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.FAOSTAT (2008) FAO Statistical Database. http://faostat.fao.org/.
- 2. Rännäli M, Czekaj V, Jones RAC, Fletcher JD, Davis RI, et al. (2008) Molecular genetic characterization of Sweet potato virus G (SPVG) isolates from areas of the Pacific Ocean and southern Africa. Plant Dis 92: 1313–1320 10.1094/PDIS-92-9-1313 [DOI] [PubMed] [Google Scholar]
- 3. Tairo F, Mukasa SB, Jones RAC, Kullaya A, Rubaihayo PR, et al. (2005) Unraveling the genetic diversity of the three main viruses involved in sweetpotato virus disease (SPVD), and its practical implications. Mol Plant Pathol 6: 199–211 10.1111/j.1364-3703.2005.00267.x [DOI] [PubMed] [Google Scholar]
- 4.Ma D, Li H, Tang J, Xie Y, Li Q, et al.. (2010) Current status and future prospects of development of sweet potato industry in China. Sweet potato in food and energy security: Proceedings of China Xuzhou 4th International Sweet potato Symposium & 4th China-Japan-Korea Sweet potato Workshop. Xuzhou, China, pp. 3–10.
- 5. Clark CA, Davis JA, Abad JA, Cuellar W, Fuentes S, et al. (2012) Sweet potato viruses: 15 years of progress on understanding and managing complex diseases. Plant Dis 96: 168–185 10.1094/PDIS-07-11-0550 [DOI] [PubMed] [Google Scholar]
- 6.Cohen J, Franck A, Vetten HJ, Lesemann DE, Loebenstein G (1992) Purification and properties of closterovirus-like particles associated with a whitefly-transmitted disease of sweet potato. Ann. Appl. Biol. 121: , 257–268. doi:10.1111/j.1744-7348.1992.tb03438.x. [Google Scholar]
- 7. Gibson RW, Mpembe I, Alicai T, Carey EE, Mwanga ROM, et al. (1998b) Symptoms, aetiology and serological analysis of sweet potato virus disease in Uganda. Plant Pathol 47: 95–102 10.1046/j.1365-3059.1998.00196.x [DOI] [Google Scholar]
- 8.Van Regenmortel MHV, Fauquet CM, Bishop DHL, Carstens EB, Estes MK, et al.. (2000) The Seventh Report of the International Committee on Taxonomy of Viruses. San Diego: Academic Press.
- 9. Schaefers GA, Terry ER (1976) Insect transmission of sweet potato disease agents in Nigeria. Phytopathology 66: 642–645 10.1094/Phyto-66-642 [DOI] [Google Scholar]
- 10. Dolja VV, Kreuze JF, Valkonen JPT (2006) Comparative and functional genomics of closteroviruses. Virus Res 117: 38–51 10.1016/j.virusres.2006.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kreuze JF, Savenkov E, Valkonen JPT (2002) Complete genome sequence and analyses of the subgenomic RNAs of Sweet potato chlorotic stunt virus reveal several new features for the genus Crinivirus. J Virol 76: 9260–9270 10.1128/JVI.76.18.9260-9270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karyeija RF, Kreuze JF, Gibson RW, Valkonen JPT (2000) Synergistic interactions of a potyvirus and a phloem limited crinivirus in sweetpotato plants. Virology 269: 26–36 10.1006/viro.1999.0169 [DOI] [PubMed] [Google Scholar]
- 13. Mukasa SB, Rubaihayo PR, Valkonen JPT (2006) Interactions between a crinivirus, an ipomovirus and a potyvirus in coinfected sweetpoato plants. Plant Pathol 55: 458–467 10.1111/j.1365-3059.2006.01350.x [DOI] [Google Scholar]
- 14. Untiveros M, Quispe D, Kreuze J (2010) Analysis of complete genomic sequences of isolates of the Sweet potato feathery mottle virus strains C and EA: molecular evidence for two distinct potyvirus species and two P1 protein domains. Arch Virol 155: 2059–2063 10.1007/s00705-010-0805-y [DOI] [PubMed] [Google Scholar]
- 15.Salazar LE, Fuentes S (2001) Current knowledge on major virus diseases of sweetpotatoes. Pages 14-19 in: Proc. Int. Workshop Sweetpotato Cultivar Decline study, Sept. 8–9, 2000, Miyakonojo, Japan.
- 16. Njeru RW, Mburu MWK, Cheramgoi E, Gibson RG, Kiburi ZM, et al. (2004) Studies on the physiological effects of viruses on sweet potato yield in Kenya. Ann Appl Biol 145: 71–76 10.1111/j.1744-7348.2004.tb00360.x [DOI] [Google Scholar]
- 17. Kokkinos CD, Clark CA (2006) Interactions among Sweet potato chlorotic stunt virus and different potyviruses and potyvirus strains infecting sweetpotato in the United States. Plant Dis 90: 1347–1352 10.1094/PD-90-1347 [DOI] [PubMed] [Google Scholar]
- 18. Untiveros M, Fuentes S, Salazar LF (2007) Synergistic interaction of Sweet potato chlorotic stunt virus (Crinivirus) with carla-, cucumo-, ipomo-, and potyviruses infecting sweetpotato. Plant Dis 91: 669–676 10.1094/PDIS-91-6-0669 [DOI] [PubMed] [Google Scholar]
- 19. Untiveros M, Fuentes S, Kreuze J (2008) Molecular variability of sweet potato feathery mottle virus and other potyviruses infecting sweet potato in Peru. Arch Virol 153: 473–483 10.1007/s00705-007-0019-0 [DOI] [PubMed] [Google Scholar]
- 20. Cuellar WJ, De Souza J, Barrantes I, Fuentes S, Kreuze JF (2011b) Distinct cavemoviruses interact synergistically with Sweet potato chlorotic stunt virus (genus Crinivirus) in cultivated sweet potato. J Gen Virol 92: 1233–1243 10.1099/vir.0.029975-0 [DOI] [PubMed] [Google Scholar]
- 21.Loebenstein G, Thottappilly G, Fuentes S, Cohen J (2009) Virus and phytoplasma diseases. Pages 105–134 in; The Sweetpotato, Loebenstein G, Totthappilly G, eds. Springer Sciences Business Media BV, Dordrecht, The Netherlands. doi: 10.1007/978-1-4020-9475-0-8.
- 22. Mukasa SB, Rubaihayo PR, Valkonen JPT (2003) Incidence of viruses and viruslike diseases of sweetpotato in Uganda. Plant Dis. 87: 329–335 10.1094/PDIS.2003.87.4.329 [DOI] [PubMed] [Google Scholar]
- 23. Hoyer U, Maiss E, Jelkmann W, Lesemann DE, Vetten HJ (1996) Identification of the coat protein gene of a Sweet potato sunken vein closterovirus isolate from Kenya and evidence for a serological relationship among geographically diverse closterovirus isolates from sweetpotato. Phytopathology 86: 744–750 10.1094/Phyto-86-744 [DOI] [Google Scholar]
- 24.Vetten HJ, Hoyer U, Maiss E, Lesemann DE, Jelkmann W (1996) Serological detection and discrimination of geographically diverse isolates of sweet potato sunken vein closterovirus. Phytopathology 100 (Suppl, abstract no 891A). [Google Scholar]
- 25. Alicai T, Fenby NS, Gibson RW, Adipala E, Vetten HJ, et al. (1999) Occurrence of two serotypes of sweetpotato chlorotic stunt virus in East Africa and their associated differences in coat protein and Hsp70 homologue gene sequences. Plant Pathol 48: 718–726 10.1046/j.13653059.1999.00402.x [DOI] [Google Scholar]
- 26. Aritua V, Barg E, Adipala E, Gibson RW, Vetten HJ (2008) Further evidence for limited genetic diversity among East African isolates of Sweet potato chlorotic stunt virus . J Phytopathol 156: 181–189 doi:0.1111/j.1439-0434.2007.01338.x [Google Scholar]
- 27. Qin Y, Zhang Z, Qiao Q, Zhang D, Tian Y, et al. (2013) Molecular variability of sweet potato chlorotic stunt virus (SPCSV) and five potyviruses infecting sweet potato in China. Arch Virol 158: 491–495 10.1007/s00705-012-1503-8 [DOI] [PubMed] [Google Scholar]
- 28. Cuellar WJ, Tairo F, Kreuze JF, Valkonen JPT (2008) Analysis of gene content in Sweet potato chlorotic stunt virus RNA1 reveals the presence of p22 RNA silencing suppressor in only few isolates: implications to viral evolution and synergism. J Gen Virol 89: 573–582 10.1099/vir.0.83471-0 [DOI] [PubMed] [Google Scholar]
- 29. Cuellar WJ, Cruzado KR, Fuentes S, Untiveros M, Soto M, et al. (2011a) Sequence characterization of a Peruvian isolate of Sweet potato chlorotic stunt virus: further variability and a model for p22 acquisition. Virus Res 157: 111–115 10.1016/j.viruses.2011.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fenby NS, Foster GD, Gibson RW, Seal SE (2002) Partial sequence of HSP70 homologue gene shows diversity between West African and East African isolates of Sweet potato chlorotic stunt virus . Tropical Agric 79: 26–30. [Google Scholar]
- 31. Sivparsad BJ, Gubba A (2012) Molecular resolution of the genetic variability of major viruses infecting sweet potato (Ipomoea batatas L.) in the province of KwaZulu-Natal in the Republic of South Africa. Crop Protect 41: 49–56 10.1016/j.cropro.2012.04.020 [DOI] [Google Scholar]
- 32.Rukarwa RJ, Mashingaidze AB, Kyamanywa S, Mukasa SB (2010) Detection and elimination of sweetpotato viruses. African Crop Sci J 18 : pp. 223–233. doi:10.4314/acsj.v18i4.68651. [Google Scholar]
- 33. Valverde RA, Clark CA, Valkonen JPT (2007) Viruses and Virus Disease Complexes of Sweetpotato. Plant Viruses 1: 116–126. [Google Scholar]
- 34. Tairo F, Kullaya A, Valkonen JPT (2004) Incidence of Viruses Infecting Sweetpotato in Tanzania. Plant Dis 88: 916–920 10.1094/PDIS.2004.88.9.916 [DOI] [PubMed] [Google Scholar]
- 35.Trenado HP, Franco AO, Navas-Castillo J (2009) http://www.ncbi.nlm.nih.gov/nuccore/FJ807784 and http://www.ncbi.nlm.nih.gov/nuccore/FJ807785.
- 36. Tugume AK, Amayo R, Weinheimer I, Mukasa SB, Rubaihayo PR (2013) Genetic Variability and Evolutionary Implications of RNA Silencing Suppressor Genes in RNA1 of Sweet Potato Chlorotic Stunt Virus Isolates Infecting Sweetpotato and Related Wild Species. PLoS ONE 8: e81479 10.1371/journal.pone.0081479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gutiérrez DL, Fuentes S, Salazar LF (2003) Sweet potato Virus Disease (SPVD): Distribution, Incidence, and Effect on Sweetpotato Yield in Peru. Plant Dis 87: 297–302 10.1094/PDIS.2003.87.3.297 [DOI] [PubMed] [Google Scholar]
- 38. Qiao Q, Zhang ZC, Qin YH, Zhang DS, Tian YT, et al. (2011) First report of Sweet potato chlorotic stunt virus infecting sweet potato in China. Plant Dis 95: 356 10.1094/PDIS-09-10-0675 [DOI] [PubMed] [Google Scholar]
- 39. Zhang ZC, Qiao Q, Qin YH, Zhang DS, Tian YT, et al. (2012) First Evidence for occurrence of Sweet potato virus disease (SPVD) caused by dual infection of plants with Sweet Potato Feathery Mottle Virus and Sweet potato chlorotic stunt virus in China. Acta Phytopathologica Sinica 42: 10–16. [Google Scholar]
- 40. Cali BB, Moyer JW (1981) Purification, serology, and particle morphology of two russet crack strains of sweet potato feathery mottle virus. Phytopathology 71: 302–305 10.1094/Phyto-71-302 [DOI] [Google Scholar]
- 41. Moyer JW, Kennedy GG (1978) Purification and properties of sweet potato feathery mottle virus. Phytopathology 68: 998–1004 10.1094/Phyto-68-998 [DOI] [Google Scholar]
- 42. Kreuze JF, Savenkov EI, Cuellar W, Li X, Valkonen JPT (2005) Viral class 1 RNase III involved in suppression of RNA silencing. J Virol 79: 7227–7238 10.1128/JVI.79.11.7227-7238.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cuellar WJ, Kreuze JF, Rajamäki ML, Cruzado KR, Untiveros M, et al. (2009) Elimination of antiviral defense by viral RNase III. Proc Natl Acad Sci U S A 106: 10354–10358 10.1073/pnas.0806042106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinheimer I, Boonrod K, Moser M, Wassenegger M, Krczal G, et al.. (2013) Binding and processing of dsRNA molecules by the Class 1 RNase III protein encoded by sweet potato chlorotic stunt virus. J Gen Virol doi:10.1099/vir.0.058693-0. [DOI] [PubMed]
- 45. Cañizares MC, Navas-Castillo J, Moriones E (2008) Multiple suppressors of RNA silencing encoded by both genomic RNAs of the crinivirus, Tomato chlorosis virus. Virology 379: 168–174 10.1016/j.virol.2008.06.020 [DOI] [PubMed] [Google Scholar]
- 46. Chiba M, Reed JC, Prokhnevsky AI, Chapman EJ, Mawassi M, et al. (2006) Diverse suppressors of RNA silencing enhance agroinfection by a viral replicon. Virology 346: 7–14 10.1016/j.virol.2005.09.068 [DOI] [PubMed] [Google Scholar]
- 47. Lu R, Folimonov A, Shintaku M, Li WX, Falk BW, et al. (2004) Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proc Natl Acad Sci U S A 101: 15742–15747 10.1073/pnas.0404940101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reed JC, Kasschau KD, Prokhnevsky AI, Gopinath K, Pogue GP, et al. (2003) Suppressor of RNA silencing encoded by Beet yellows virus . Virology 306: 203–209 10.1006/S0042-6822(02)00051-X [DOI] [PubMed] [Google Scholar]
- 49.King A, Lefkowitz E, Adams MJ (2011) Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses: An Elsevier Title.
- 50. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- 51. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nuc Acids Res 22: 4673–4680 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.