Abstract
As a strategy to identify gene expression changes affected by human polynucleotide phosphorylase (hPNPaseold-35), we performed gene expression analysis of HeLa cells in which hPNPaseold-35 was overexpressed. The observed changes were then compared to those of HO-1 melanoma cells in which hPNPaseold-35 was stably knocked down. Through this analysis, 90 transcripts, which positively or negatively correlated with hPNPaseold-35 expression, were identified. The majority of these genes were associated with cell communication, cell cycle and chromosomal organization gene ontology categories. For a number of these genes, the positive or negative correlations with hPNPaseold-35 expression were consistent with transcriptional data extracted from the TCGA (The Cancer Genome Atlas) expression datasets for colon adenocarcinoma (COAD), skin cutaneous melanoma (SKCM), ovarian serous cyst adenocarcinoma (OV), and prostate adenocarcinoma (PRAD). Further analysis comparing the gene expression changes between Ad.hPNPaseold-35 infected HO-1 melanoma cells and HeLa cells overexpressing hPNPaseold-35 under the control of a doxycycline-inducible promoter, revealed global changes in genes involved in cell cycle and mitosis. Overall, this study provides further evidence that hPNPaseold-35 is associated with global changes in cell cycle-associated genes and identifies potential gene targets for future investigation.
Introduction
Human polynucleotide phosphorylase, an evolutionarily conserved 3′-5′ exoribonuclease [Deutscher MP, 1993; Andrade et al., 2009; Ibrahim et al., 2008; Leszczyniecka et al., 2004], has been implicated in numerous cellular functions [Sarkar et al., 2006a; Das et al., 2011; Sokhi et al., 2013a] since its discovery as an upregulated transcript in terminally differentiated melanoma cells and senescent fibroblasts [Leszczyniecka et al., 2002]. Some of the major findings relating to this interesting enzymatic protein revolve around its ability to cause growth suppression of cancer cells as a result of c-myc mRNA degradation [Sarkar et al., 2003], and more recently due to its post-transcriptional regulation of miR-221 [Das et al., 2010]. hPNPaseold-35 is also implicated in the generation of reactive oxygen species leading to chronic inflammation, suggesting a probable role for this enzyme in senescence-associated degenerative diseases [Sarkar et al., 2004; Sarkar et al., 2006a; Sarkar et al., 2006b]. In addition, hPNPaseold-35 plays a major role in the maintenance of cellular homeostasis, import of RNA into the mitochondria, mediating mtRNA processing, while also being part of a complex responsible for mtRNA decay [Chen et al., 2006; Wang et al., 2010; Wang et al., 2011; Nagaike et al., 2005; Slomovic et al., 2008; Wang et al., 2009; Szczesny et al., 2010; Sokhi et al., 2013a]. Although these findings are a major step forward in delineating the functional roles this enzyme plays in cellular physiology, there is still limited understanding at the molecular level regarding the specific RNA species the enzyme targets for degradation, given the role of hPNPaseold-35 in RNA processing. To this end, our prior work showed that manipulation of hPNPaseold-35 expression in melanoma cells, i.e., its depletion or overexpression, caused genome wide alterations in numerous genes and pathways, with some of the most significant changes associated with mitochondrial function, cholesterol biosynthesis, cell cycle and cellular growth and proliferation [Sokhi et al., 2013b]. Apart from the identification of global cellular patterns of gene expression changes resulting from manipulation of hPNPaseold-35, we were able to validate certain known cellular targets of hPNPaseold-35 function (e.g., mitochondria) and identify novel potential targets of hPNPaseold-35, which could pave the way for future therapeutic opportunities pertaining to hPNPaseold-35 related disease states. Thus, these studies produced the most extensive assessment of hPNPaseold-35 regulated gene expression changes to date. However, since these studies were performed in a single cell system (i.e., melanoma), we were interested to evaluate whether the changes observed in this scenario were specific to the cell type under analysis or if they were a more global cell type-independent phenomena, which could provide even more valuable and concrete biological insights into the workings of hPNPaseold-35.
In order to interrogate the above premise and unravel additional RNA molecules under hPNPaseold-35 regulatory control at a more global level, we performed genome-wide expression analysis of human cancer cell lines with gained or lost hPNPaseold-35 functionality. This was accomplished through evaluation of genome-wide expression changes (via microarray analysis) following ectopic hPNPaseold-35 overexpression in HeLa cells. The resulting transcriptional changes were then compared with those previously reported when hPNPaseold-35 was either overexpressed or knocked down in melanoma cells [Sokhi et al., 2013b], to gain insight into the normal biological pathways influenced by hPNPaseold-35 and identify cell type independent global regulatory patterns of hPNPaseold-35 induced gene expression changes and their future implications. Specifically, this extensive analysis revealed that (i) hPNPaseold-35 primarily regulates cell cycle progression events as evidenced by the global deregulation of genes and pathways associated with the same, (ii) the cell cycle associated genes reported as potentially regulated by hPNPaseold-35 in this study provide extremely valuable information regarding the networks it regulates and hold immense promise as future candidates, as they were identified in two separate microarray analyses performed in two histologically unrelated cancer cell lines (HO-1 and HeLa) using different overexpression approaches, (iii) genes reported as potentially directly or indirectly regulated by hPNPaseold-35 commonly identified both in this study and our previous one in melanoma could be valid hPNPaseold-35 targets, (iv) the tissue correlations of some of the hPNPaseold-35 target genes identified through TCGA correlation studies exponentially increases the validity of pursuing some of these genes as possibly regulated by hPNPaseold-35 in certain cancers. In other words, the exploration of the TCGA database allowed us to verify if the cell line-based findings are also reflected in cancer tissue samples. This is a very important observation as it underscores the cell type-independent relevance of the genes identified, and (v) there is a minimal role of the c-myc transcription factor in the regulation of the gene networks identified, suggesting that the changes are not a secondary consequence of alteration in c-myc expression.
Overall, this study represents a targeted genomic approach towards global evaluation of the cellular processes possibly associated with hPNPaseold-35 functionality and opens up new biological avenues to investigate. We refer to these global common changes occurring as a consequence of altering hPNPaseold-35 expression as the “hPNPaseold-35 reactome”, now setting the stage for critically evaluating new potential direct and indirect targets of this evolutionary conserved enzyme.
Materials and methods
Establishing a conditional hPNPaseold-35 over-expressing cell line
The full-length cDNA of hPNPaseold-35 with a C-terminal HA tag was cloned into a modified Tet-On pTRIPZ vector (Open Biosystems; obtained from Dr. Kristoffer Valerie) to permit inducible expression of the gene in the presence of doxycycline. HeLa-Tet-On cell lines were established by transfecting HeLa cells with the pTRIPZ -hPNPaseold-35 plasmid (sequence was verified before use) and stable clones were selected with the help of puromycin (2 μg/ml). All cell lines were grown in Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen Life Technologies) supplemented with 10% fetal bovine serum (FBS; Sigma) and 5% penicillin/streptomycin (Gibco). All transfections were performed using Lipofectamine® 2000 Transfection Reagent (Life Technologies) according to the manufacturer’s protocol. All cell lines were maintained in a 5% CO2 95% O2 humidified incubator at 37°C. The melanoma cell lines used in this study have been extensively described in our previous work and verified as mycoplasma free [Sokhi et al., 2013b].
Total RNA isolation for microarray analyses
Total RNA from the hPNPaseold-35 knockdown cell line (HO-1 melanoma cells stably expressing shRNA for hPNPaseold-35 and scrambled control shRNA) [Sokhi et al., 2013b] and the hPNPaseold-35 conditionally over-expressing cell line (HeLa cells stably transfected with pTRIPZ-hPNPaseold-35 induced with doxycycline for 24 hrs and uninduced cells as control) was isolated from cell lysates in TRIZOL reagent (InvitrogenTM Life Technologies, Carlsbad, CA). Cell lysates were subjected to an automated extraction method using the MagMAX™-96 for Microarrays Total RNA Isolation Kit (Ambion/InvitrogenTM Life Technologies, Carlsbad, CA) on the MagMAX™ Express Magnetic Particle Processor (Applied Biosystems/InvitrogenTM Life Technologies, Carlsbad, CA).
Gene Expression Microarrays
Gene expression profiles were ascertained using GeneChip® Human Genome U133A 2.0 (HG-U133A 2.0) arrays (Affymetrix, Santa Clara, CA) as previously described [Dumur et al., 2009]. Every chip was scanned at a high resolution, with pixelations ranging from 2.5 μm down to 0.51 μm, by the Affymetrix GeneChip® Scanner 3000 according to the GeneChip® Expression Analysis Technical Manual procedures (Affymetrix, Santa Clara, CA). After scanning, the raw intensities for every probe were stored in electronic files (in .DAT and .CEL formats) by the GeneChip® Operating Software (GCOS) (Affymetrix, Santa Clara, CA). The overall quality of each array was assessed by monitoring the 3′/5′ ratios for a housekeeping gene (GAPDH) and the percentage of “Present” genes (%P); where arrays exhibiting GAPDH 3′/5′ < 3.0 and %P > 40% were considered good quality arrays [Dumur et al., 2009]. All experiments were done in biological triplicates. The microarray data generated for this study are available online at the Gene Expression Omnibus repository under the accession numbers GSE46884 and GSE53604.
Statistical Analyses
The Robust Multiarray Average method (RMA) was used for normalization and obtaining probe set expression summaries for the gene expression assays. These values were then used to identify genes significantly altered among the different conditions (i.e., hPNPaseold-35 up- and down-regulation), and analyzed for significance across replicate experiments with the help of TM4-MeV (MultiExperiment Viewer) analysis software by using a permutation method performed with the Significance Analysis of Microarray (SAM) program from Stanford University [Stanford, CA] [Saeed et al., 2003]. Once the program reported the list of ranked genes whose expression changed in the opposite direction in the two datasets, the “delta value” was adjusted to a stringent false discovery rate (FDR) of 5%. Cluster analysis was done using the Cluster and TreeView programs 4. Genes reported by SAM were analyzed by hierarchical clustering with average linkage grouping.
In order to identify transcripts whose expression negatively or positively correlated with the expression of c-myc, we employed a Pearson correlation coefficient of 0.01 and statistical significance of the identified genes was set at p-value cutoff of 0.05 using standard t-test (Table S1).
Enrichment analysis
The Protein ANalysis THrough Evolutionary Relationships (PANTHER) resource was used to identify Gene Ontology (GO) categories and generate pie charts for the hPNPaseold-35 responsive genes (www.pantherdb.org) [Mi et al., 2013a; Mi et al., 2013b; Hitomi et al., 2008]. PANTHER can categorize genes based on their molecular functions, biological processes and protein classes with the help of available literature and evolutionary relationships.
Functional annotation cluster analysis was done using the Database for Annotation, Visualization and Integrated Discovery (DAVID) for the Gene ontology-based functional enrichment of the gene lists, using their respective gene symbols [Huang et al., 2007; Babenko et al., 2012; Hanaoka et al., 2012]. This kind of analysis allows us to group functionally related genes together, which in turn provides a global view of altered biological networks.
The MetaCore™ (GeneGo Inc. USA) bioinformatics software was used in order to gain a deeper understanding of the plausible molecular networks or biological pathways the hPNPaseold-35 responsive genes might be involved in [Hellman et al., 2010; Malaney et al., 2013]. The build network tool assigns pathway significance based upon the number of genes represented within a pathway and the direction of change.
The comparison analysis tool in the Ingenuity Pathway Analysis (IPA, Ingenuity® Systems, http://www.ingenuity.com) software was used to compare the common biological processes and canonical pathways between the doxycycline-inducible HeLa hPNPaseold-35 overexpression system and the Ad.hPNPaseold-35 infected HO-1 cells.
Analysis of TCGA Genome-wide Expression Datasets
The processed, Illumina Hi Seq 2000-generated TCGA (The Cancer Genome Atlas) genome-wide expression datasets for colon adenocarcinoma (COAD), skin cutaneous melanoma (SKCM), OV (ovarian serous cystadenocarcinoma) and PRAD (prostate adenocarcinoma) were obtained through the UCSC Cancer Genomics Browser (https://genome-cancer.ucsc.edu/) [Goldman et al., 2013]. The sample breakdown for each dataset is as follows: COAD (193 primary, tumors, 18 solid tissue normals), SKCM (241 metastatic samples, 43 primary tumors), OV (262 primary tumors, 4 recurrent tumors), PRAD (176 primary tumors, 44 solid tissue normals). In addition, the UCSC pan cancer (PANCAN) dataset [Cline et al., 2013], which resulted from merging (and normalization) of all of the 22 TCGA genome wide expression (Illumina Hi Seq 2000) cancer cohorts (including COAD, OV, PRAD, and SKCM) with a combined total of 6040 samples (4982 primary tumors, 271 metastatic, 27 recurrent tumors, 173 peripheral blood, 587 solid tissue normals), was downloaded from the same website and subsequently analyzed. The primary objective of the analyses was to examine the possible transcriptional correlations between hPNPaseold-35 (PNPT1) and genes identified through the cell line-based experiments. Through the use of the Gene-E program (Broad Institute, Cambridge, MA, http://www.broadinstitute.org/cancer/software/GENE-E/index.html) and the JMP 10 Pro statistical software (SAS, Cary, NC), the correlations between hPNPaseold-35 expression levels and those of other genes of interest were examined by calculation of Pearson correlation coefficients (Table S2), as well as generation of relevant heat maps and Cartesian plots.
cDNA synthesis and quantitative real-time RT-PCR (qRT-PCR)
Total RNA was harvested using the RNeasy purification kit (Qiagen). The quality and concentrations of isolated RNA samples were assessed using the NanoDrop 2000 (Thermo Scientific). 2 μg of RNA was used in a total volume of 20 μl to synthesize cDNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) according to the manufacturer’s instructions. Real-time quantitative PCR was conducted using the ViiA™ 7 Real-Time PCR System (Applied Biosystems) and performed in a total volume of 20 μl that contained the TaqMan Gene Expression Master Mix (Applied Biosystems), 1 μl of the cDNA template generated and the target-specific TaqMan Gene expression assays (Applied Biosystems) according to following cycle parameters: 95°C for 10 minutes followed by 40 cycles at 95°C for 15 seconds and at 60°C for 1 minute. Each sample was run in triplicate using three biological replicates and normalized to the housekeeping gene GAPDH used as an internal control in each case. The ΔΔCt method was used for comparing relative fold expression differences in the genes of interest between different test samples. Statistical significance (*P<0.05, **P<0.01, ***P<0.001) was determined using two-tailed student’s t-test and one-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparison test wherever required.
Protein isolation and Western blot analysis
Cells were harvested by centrifugation. The pellets were washed in PBS and subsequently lysed in ice-cold 1X cell lysis buffer (Cell Signaling) supplemented with PhosSTOP Phosphatase Inhibitor Cocktail Tablets and complete Mini Protease Inhibitor Cocktail Tablets (Roche), followed by centrifugation at 13,000 rpm for 15 minutes at 4°C. The supernatant or whole cell lysate was collected in a fresh tube, and protein concentration was measured using the Bio-Rad Protein Assay Dye Reagent Concentrate (BIO-RAD). Thirty μg of total cell lysate was mixed with SDS sample buffer and heated for 5 minutes at 95°C. The proteins were separated in 8–10% SDS-PAGE gels and transferred onto nitrocellulose membranes, and blocked using 5% non-fat milk supplemented with 1% bovine serum albumin (BSA) in TBS-T for 1 hour. After washing three times with TBS-T for 10 minutes each, the membranes were incubated with primary antibodies overnight at 4°C. The primary antibodies used were anti-HA (mouse, 1:1000), anti-hPNPaseold-35 (chicken; 1:5000), anti-EF1α (mouse, 1:1000). After ~24 h, the membranes were washed as before, and incubated with the relevant horseradish-peroxidase conjugated secondary antibodies for 1 h at room temperature. The membranes were washed three times with TBS-T for 10 minutes each. This was followed with protein detection through the use of ECL Western Blotting detection reagent (GE Healthcare Life Sciences) and exposure to X-ray film.
Results
Identification of hPNPaseold-35 responsive genes
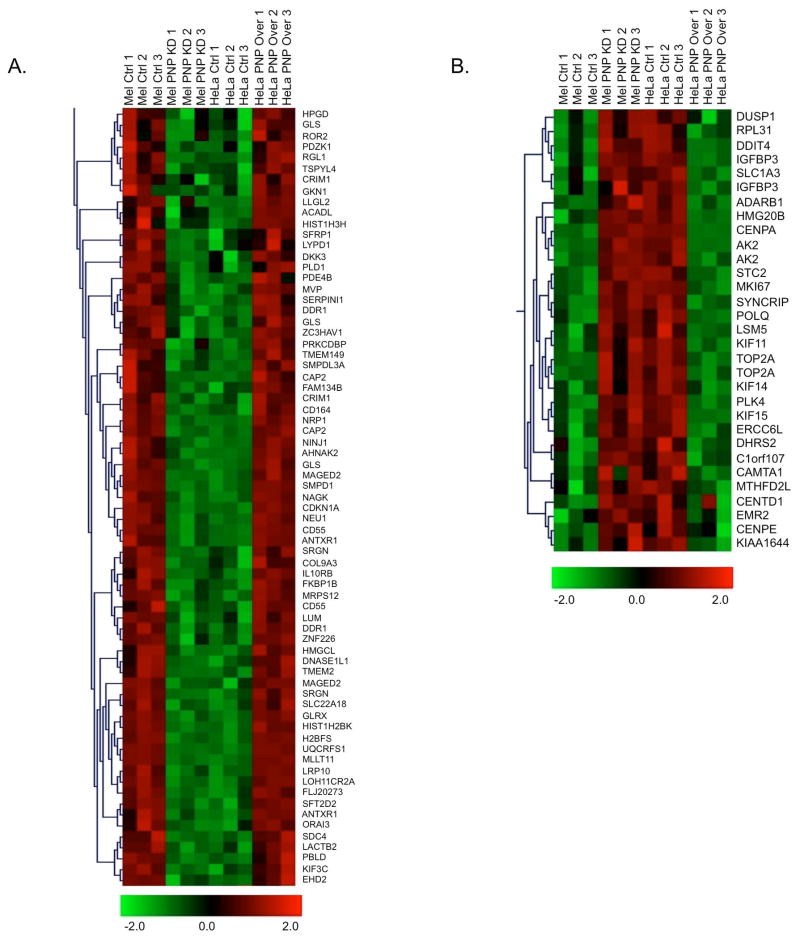
To explore the effects of hPNPaseold-35 on the global regulation of gene expression, HeLa cells were stably transfected with a doxycycline-inducible hPNPaseold-35 expression vector (to attain physiologically relevant levels of hPNPaseold-35 overexpression) (Figure S1), while HO-1 melanoma cells were knocked down for hPNPaseold-35 expression through the use of shRNA vector [Sokhi et al., 2013b]. The cells were then profiled using cDNA microarray analyses. Unsupervised hierarchical cluster analyses resulted in the six HeLa and six HO-1 array experiments clustering within their respective classes (Figure S2), indicating that the microarray results were robust and able to differentiate the two cell lines. A comparative analysis between the two datasets (using the TM4-MeV analysis software) led to the identification of 62 transcripts whose expression was significantly down regulated upon hPNPaseold-35 over-expression (in HeLa cells) (Figure 1A). On the other hand, 28 transcripts were significantly upregulated upon hPNPaseold-35 repression (in HO-1 cells) (Figure 1B). Some genes are represented on the microarray by multiple probe sets and thus were identified more than once (as evident in Figure 1). Apart from providing an additional internal control, this finding also increases confidence in the identification of certain genes as being responsive to hPNPaseold-35 levels.
Figure 1. Comparison of hPNPaseold-35-induced changes in gene expression in HO-1 and HeLa cells.
Hierarchical clustering of hPNPaseold-35 positively (A) and negatively (B) correlating genes as identified by microarray analysis. Each column represents a single biological replicate and each row corresponds to a gene, with red and green representing high and low expression levels, respectively.
Functional categorization of hPNPaseold-35 responsive genes
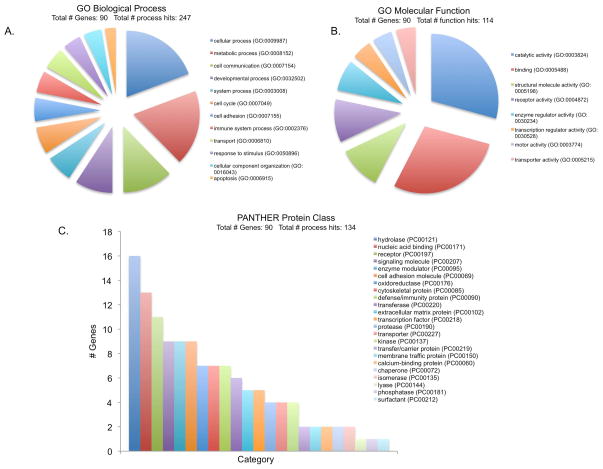
The 90 hPNPaseold-35 responsive genes (Figure 1B) were categorized according to Gene Ontology (GO) biological processes, molecular functions and protein classes, through the PANTHER database and its tools. The 12 GO biological processes describing at least five of the 90 hPNPaseold-35 responsive genes are shown in Figure 2A. Of the 12, the cellular and metabolic processes included the highest percentages of genes (18.5% and 17.6 %, respectively). The cellular processes subcategories included cell communication (36.4%), cell cycle (19.5%) and cell adhesion (16.9%). Metabolic processes were sub-classified mostly as primary metabolic processes (86%), which in turn comprised of nucleic acid (38.3%), protein (34%), lipid (14.9%), amino acid (8.5%) and carbohydrate (4.3%) metabolisms. The molecular functions attributed to the hPNPaseold-35 responsive genes were catalytic activity (29.2%, mainly hydrolases, oxidoreductases and transferases) and binding (28.3%, mainly protein and nucleic acid binding) (Figure 2B). When subcategorized according to protein functions, the hPNPaseold-35 responsive genes were hydrolases (12.4%), nucleic acid binding proteins (10%) and receptors (8.5%) (Figure 2C).
Figure 2. Gene Ontology (GO) terms for PNPase responsive genes.
GO analysis corresponding to Biological process (A), Molecular function (B) and PANTHER Protein class (C) represented as pie charts generated by PANTHER classification system (http://www.pantherdb.org/).
Functional annotation clustering by DAVID resulted in the identification of 32 clusters of related genes, with the highest enrichment score being 1.91. The most significantly enriched gene clusters along with their GO categories are represented in Figure S3. These cluster of genes are associated with chromosomal organization (score=1.91, 18 genes: HIST1H2BK, HIST1H3H, H2BFS, CENPA, TSPYL4, CENPE, TOP2A, HMG20B, ERCC6L, SYNCRIP, KIF14, MKI67, ZNF226, DNASE1L1, POLQ, KIF15, CDKN1A, PLK4), signaling molecules (score=1.67, 32 genes: AK2, GLRX, UQCRFS1, TMEM2, TMEM149, SLC1A3, SDC4, GKN1, LYPD1, NEU1, KIAA1644, ANTXR1, SERPINI1, CRIM1, CD164, EMR2, ROR2, LRP10, DNASE1L1, CD55, IL10RB, SMPDL3A, COL9A3, STC2, LUM, SRGN, SMPD1, NRP1, DDR1, DKK3, SFRP1, IGFBP3), and cell cycle (score=1.37, 15 genes: CENPE, KIF11, KIF15, KIF3C, KIF14, ERCC6L, MKI67, CENPA, CDKN1A, PLK4, TOP2A, LLGL2, HMG20B, DUSP1, AK2).
MetaCore™ (GeneGo Inc. USA) analysis of the hPNPaseold-35 responsive genes resulted in the identification a number of gene interaction networks, two of which are represented in Figure S4. Network A includes 14 genes (CENPA, CENPE, TOP2A, PLK4, ERCC6L, DDIT4, SFRP1, CRIM1, ZC3HAV1, MVP, DKK3, FAM134B, LOH11CR2A, and MAGED2) from the list of hPNPaseold-35 responsive genes. p53 seems to be a critical player in this signaling cascade. Network B includes 11 (CENPA, STC2, IGFBP3, DUSP1, ZC3HAV1, PDZK1, SDC4, NRP1, CDKN1A, RGL1, TOP2A) of the hPNPaseold-35 responsive genes, with STAT1 and p21 appearing as major regulatory proteins in this pathway.
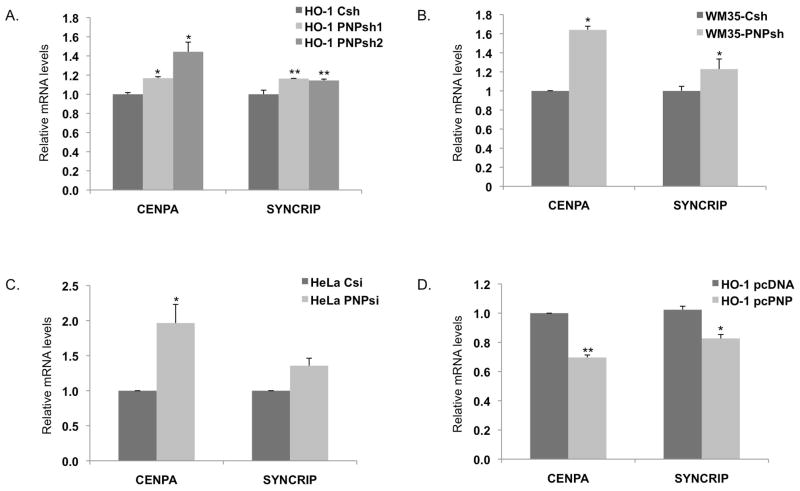
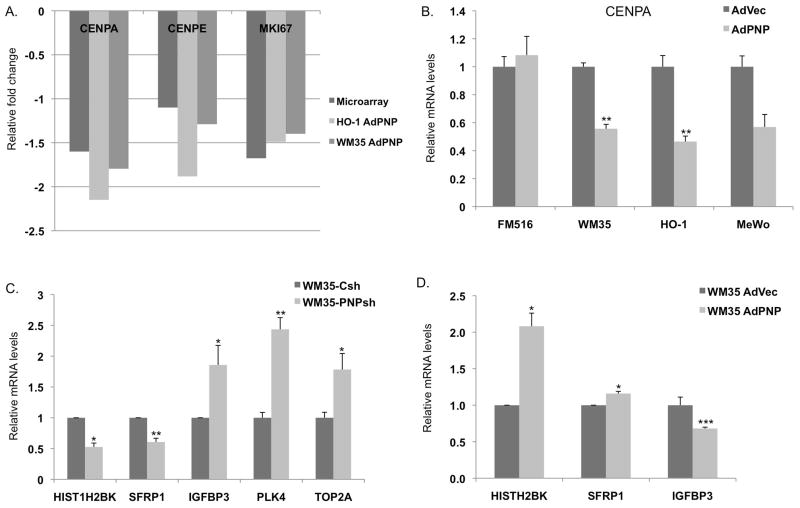
The expression levels of representative genes in the two networks such as CENPA (Centromere protein A) and CENPE (Centromere protein E) were validated using qRT-PCR. As predicted, both genes were down regulated in response to hPNPaseold-35 overexpression (Figures 3 and 4). The hPNPaseold-35 stable knockdown in HO-1 and WM35 melanoma cells resulted in a ~1.2 fold increase in SYNCRIP (Synaptotagmin binding, cytoplasmic RNA interacting protein) and a ~1.2–1.6 fold increase in CENPA transcript levels as determined by qRT-PCR (Figure 3A–B). Transient knockdown of hPNPaseold-35 in HeLa cells also resulted in a 1.4 and ~2 fold elevation of SYNCRIP and CENPA RNA levels respectively (Figure 3C), proving that these gene dysregulations are neither cell line specific effect, nor an indirect effect due to stable knockdown. Plasmid mediated overexpression of hPNPaseold-35 in HO-1 melanoma cells resulted in a 20% and 30% reduction in SYNCRIP and CENPA RNA levels respectively (Figure 3D). Figure 4A shows a graphical representation of the fold decrease in the RNA levels of CENPA, CENPE and MKI67 following adenoviral mediated overexpression of hPNPaseold-35 in HO-1 and WM35 melanoma cells, in comparison to their respective fold-changes as obtained in our microarray experiments. Adenoviral mediated overexpression of hPNPaseold-35 in three different melanoma cell lines (HO-1, WM35 and MeWo) all resulted in ~40% reduction in CENPA levels (Figure 4B). Additional genes validated by qRT-PCR in WM35 melanoma cells following hPNPaseold-35 knockdown or overexpression are shown in Figures 4C and D, respectively, and were as follows: HIST1H2BK (histone cluster 1, H2bk), SFRP1 (Secreted frizzled-related protein 1), IGFBP3 (Insulin-like growth factor-binding protein 3), PLK4 (polo-like kinase 4) and TOP2A (Topoisomerase (DNA) II Alpha (170kD)).
Figure 3. Real-time PCR validation of genes identified by microarray analysis.
(A) hPNPaseold-35–stable knockdown HO-1cells, (B) hPNPaseold-35–stable knockdown WM35 cells, (C) HeLa cells transiently transfected with siRNA against hPNPaseold-35 and (D) HO-1 cells transfected with pcDNA3.1-hPNPaseold-35. Error bars represent mean ± S.E. of three independent experiments done in triplicate. Statistical significance was determined by a one-way analysis of variance followed by Dunnett’s multiple comparison test and a two-tailed student’s t-test, *P<0.05, **P<0.01, ***P<0.001.
Figure 4. Real-time PCR validation of genes identified by microarray analysis following Ad.hPNPaseold-35 –mediated overexpression.
Error bars represent mean ± S.E. of three (4A, B, C) or two (4D) independent experiments done in triplicate. Statistical significance was determined by two-tailed student’s t-test, *P<0.05, **P<0.01.
TCGA correlations of hPNPaseold-35 responsive genes
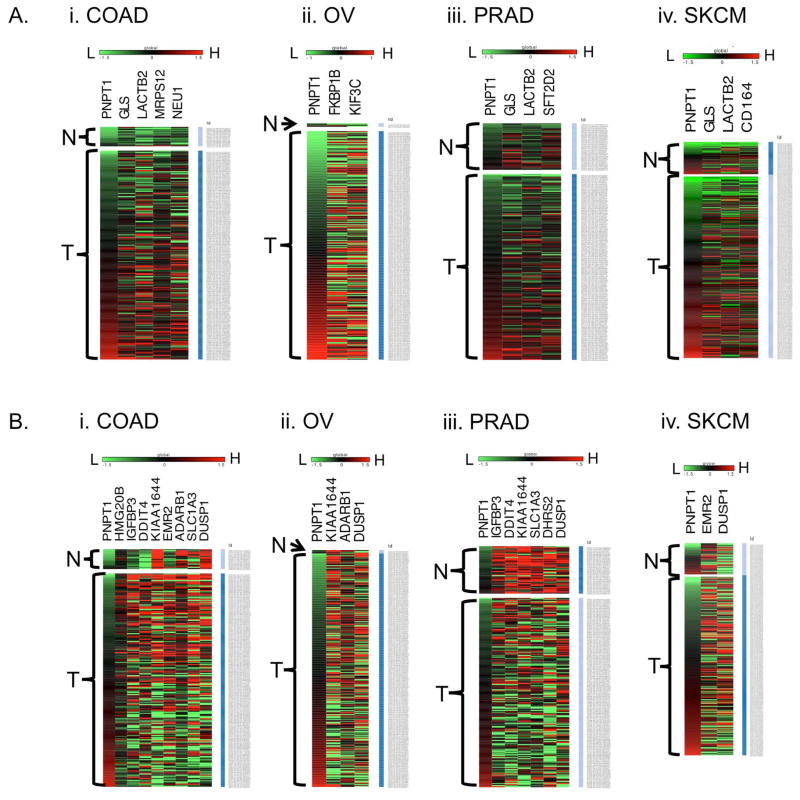
The Cancer genome atlas (TCGA) database is a very comprehensive repository of integrated genomic (including genome-wide expression) data of various adult cancer types. In order to evaluate if our observations are also reflected in human cancer samples, we examined the correlation between hPNPaseold-35 expression and the hPNPaseold-35 responsive genes we identified, in TCGA datasets. The genes identified as positively or negatively correlating with hPNPaseold-35 expression in different cancer types are represented in the heat maps shown in Figures 5A and B respectively. Prior to heatmap generation, each gene expression value (at log, base 2 scale) for every sample was transformed by subtracting the average expression across all samples in a given dataset. As shown in Figure 5, the samples in the TCGA OV, COAD, SKCM, and PRAD cohorts were arranged in order of increasing hPNPaseold-35 expression level, going from green to red. One gene that stood out is DUSP1. As the hPNPaseold-35 expression of OV, COAD, SKCM, and PRAD samples increased, DUSP1 level decreased (also indicated by negative Pearson correlation coefficients). The negative correlation between hPNPaseold-35 and DUSP1 was clearly observed in a larger PANCAN dataset (See the Cartesian plot, Figure S5). The Pearson correlation coefficient (r) between hPNPaseold-35 and DUSP1 registered at −0.362. This r-value was 221st lowest (99th percentile) among the 20500 genes whose expression was related to hPNPaseold-35. As the heat maps indicate, opposite expression trends (compared to hPNPaseold-35) were observed for the genes KI1644 (OV, COAD, PRAD), ADARB1 (OV, COAD), HMG20B (COAD), IGFBP3 (COAD, PRAD), DDIT4 (COAD, PRAD), EMR2 (COAD, SKCM), SLC1A3 (COAD, PRAD), and DHRS2 (PRAD). In contrast, the expression levels of the genes GLS (COAD, SKCM, PRAD), LACTB2 (COAD, SKCM, PRAD), MRPS12 (COAD), NEU1 (COAD), CD164 (SKCM), FKBP1B (OV), KIF3C (OV), and SFT2D2 (PRAD) exhibited positive correlation with hPNPaseold-35.
Figure 5. Heat maps representing correlations of hPNPaseold-35 responsive genes with hPNPaseold-35 in different tumor types identified using TCGA datasets.
Shown are the hPNPaseold-35 positively (A) and negatively (B) correlating genes in (i) colon adenocarcinoma (COAD), (ii) ovarian serous cyst adenocarcinoma (OV), (iii) prostate adenocarcinoma (PRAD), and (iv) skin cutaneous melanoma (SKCM), respectively. The samples are subdivided into solid normal (N) and primary tumor (T) groups, and arranged (top to bottom) in order of increasing (low=green to high=red) expression of PNPT1.
Identification of Myc-correlating genes
c-myc mRNA was previously shown to be regulated by hPNPaseold-35 at the post-transcriptional level [Sarkar et al., 2003; Sarkar et al., 2005]. Since c-myc is an essential cell cycle regulator and also a transcription factor implicated in the regulation of expression of numerous genes, we were interested to identify if genes differentially regulated by hPNPaseold-35 following its gain or loss of function were in any way under c-myc transcriptional control. Thus, in order to dissect the potential role of myc in the regulation of hPNPaseold-35 responsive genes, we identified genes in our two datasets (hPNPaseold-35 knockdown and overexpression) whose expression correlated with c-myc expression. Thirty-one genes were identified with expression negatively correlating with c-myc and 29 with expression positively correlating with c-myc (Table S1). We further grouped the 60 c-myc correlating genes into biological categories using the functional annotation cluster analysis tool in DAVID [Huang et al., 2007; Babenko et al., 2012; Hanaoka et al., 2012], which resulted in the identification of 26 gene enrichment clusters with the highest enrichment score being 2.02. Some of the functional categories identified represented in Figure S6 were apoptosis, DNA repair, cytoskeleton and mRNA processing. Eleven of c-myc correlating genes identified were also among the hPNPaseold-35 regulated genes we report here. Of these 11 genes, the expression of DHRS2, LSM5, STC2, TOP2A positively correlated with c-myc while IL10RB, LOH11CR2A, MLLT11, MRPS12, NEU1, SMPD1, UQCRFS1 negatively correlated with c-myc.
hPNPaseold-35 overexpression in HO-1 and HeLa cells causes global changes in cell cycle-associated genes
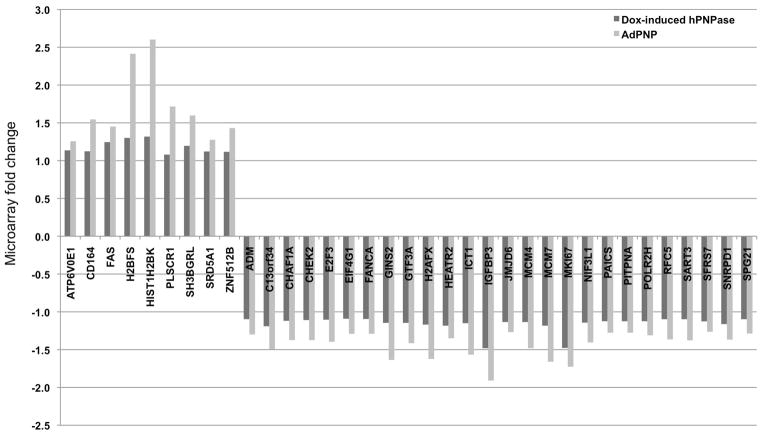
The online tool VENNY [Oliveros JC, 2007] was able to identify 105 (Table S3) differentially regulated genes (FDR<5%) common between HeLa cells in which hPNPaseold-35 was overexpressed through a doxycycline-inducible system, and Ad.hPNPaseold-35 infected HO-1 cells [Sokhi et al., 2013b]. Some of these genes are represented in Figure 6. Overall, there were 31 upregulated and 74 downregulated overlapping transcripts in both studies. In order to classify these genes into biological categories, we employed the ToppGene suite of web applications (Table 1), which was able to identify cell cycle, mitosis and DNA metabolism-associated functions as the most significantly overrepresented categories. We then compared the top biological functions and canonical pathways associated with all the differentially regulated transcripts in both systems (Figure S7) to further understand the global implications of hPNPaseold-35 overexpression through IPA (Ingenuity Pathway Analysis). This analysis identified hPNPaseold-35–associated biological processes and canonical pathways as mostly related to cell cycle.
Figure 6.
Genes differentially regulated in response to hPNPaseold-35 overexpression as identified by microarray analysis in HO-1 melanoma and HeLa cells.
Table 1.
Functional annotation of genes differentially regulated in response to hPNPaseold-35 overexpression by ToppGene suite.
| Functional category | Source | p-value | no. of genes |
|---|---|---|---|
| RNA binding | GO: Molecular Function | 1.90E-02 | 16/857 |
| Cell Cycle phase | GO: Biological Process | 5.81E-11 | 30/864 |
| Mitotic Cell Cycle | GO: Biological Process | 1.31E-08 | 26/785 |
| DNA metabolic process | GO: Biological Process | 9.19E-07 | 25/879 |
| Chromosome | GO: Cellular Component | 3.89E-08 | 21/641 |
| Nucleoplasm | GO: Cellular Component | 6.32E-06 | 28/1483 |
| Genes involved in cell cycle, Mitotic pathway | MSigDB | 2.61E-04 | 14/306 |
Discussion
This study focuses on furthering our understanding of the global gene dysregulation patterns resulting from hPNPaseold-35 gain- or loss-of-function. We compared the gene expression changes resulting from hPNPaseold-35 knockdown and overexpression in two different cells lines and were able to identify significantly dysregulated genes common between the two. In the current analysis, we identified 62 hPNPaseold-35 -induced and 28 hPNPaseold-35 -repressed genes. Gene ontology analysis revealed that a number of these hPNPaseold-35 -induced and -repressed transcripts are involved in cell cycle, chromosomal organization and cellular signaling. Among the genes whose expression were significantly elevated upon hPNPaseold-35 overexpression were KIF3C [Harper et al., 1993] and CDKN1A [Toh et al., 2011], two genes known to be upregulated during growth arrest. On the other hand, genes involved in cellular growth and proliferation such as CENPA, CENPE, PLK4, TO2PA, and POLQ were found to be downregulated after hPNPaseold-35 overexpression. Additional bioinformatics analysis indicated that these hPNPaseold-35 -associated genes belong to two networks related to mitosis/cell cycle and regulation by p21 (a known regulator of growth arrest and cellular senescence). We also compared the gene expression profiles of two different cell lines in which hPNPaseold-35 were overexpressed using two different mechanisms: a doxycycline-inducible expression vector and an adenoviral vector. We not only observed 105 overlapping genes between both systems, but interestingly almost all these genes were associated with cell cycle, mitosis and chromosomal functions as identified by gene ontology categories. Apart from the overlapping genes, comparative pathway analysis of both the hPNPaseold-35 overexpression datasets revealed that the most significantly affected pathways were related to cellular growth and proliferation. Overall, these results suggest that hPNPaseold-35 may have some cell cycle- and cell proliferation-associated functionalities. Nonetheless, future studies are needed in order to gain more mechanistic insights of these possible hPNPaseold-35 regulatory functions.
Interestingly, some of the hPNPaseold-35-associated genes identified in this study have been reported in our previous study [Sokhi et al., 2013b]. These include two genes with roles in cellular proliferation: MKI67 (a cellular proliferation marker) and CENPE (Centromere protein E), both of which negatively correlate with hPNPaseold-35 expression. This study also identified another centomeric protein (CENPA, Centromere protein A), whose transcription level negatively correlates with that of hPNPaseold-35. CENPA is reported to be a prognostic marker for lung adenocarcinoma [Toh et al., 2011] and ER-positive breast cancer [McGovern et al., 2012].
The transcriptional relationships between hPNPaseold-35 and some of the genes identified in this study are also evident in genome-wide expression data for four adult cancer types included in the TCGA project. For instance, TCGA data analysis shows the negative correlation between hPNPaseold-35 and: DUSP1 (implicated in promotion of angiogenesis and metastasis of non-small-cell-lung cancer [Moncho-Amor et al., 2011]); ADARB1 (reported to be upregulated in numerous carcinomas [Flanagan et al., 2009; Shaikhibrahim et al., 2013; Valles et al., 2012); and EMR2 (associated with poor prognosis in glioblastoma [Rutkowski et al., 2011]). These data point to the possibility that hPNPaseold-35 has a regulatory role in cancer progression. Nevertheless, additional experiments are needed to prove this hypothesis.
It is also possible that the genes we identified to be hPNPaseold-35-associated may not be directly regulated by the protein. For instance, it is possible that what hPNPaseold-35 regulates (by degradation) are the miRNAs, which in turn control the expression of such genes. Another factor we cannot discount is the previously demonstrated hPNPaseold-35 regulation of c-myc (through exoribonucleolytic degradation), which as a transcription factor regulates the expression of numerous genes involved in cellular processes including but not restricted to apoptosis, cell cycle and metabolism. Hence, it is plausible that the observed negative correlation between hPNPaseold-35 expression and those of the numerous genes reported here might just be a consequence of hPNPaseold-35–driven degradation of c-myc. For one thing, our expression array data indicated that most of the c-myc expression correlations were with those of other genes associated with apoptosis, cell cycle, mRNA processing and DNA repair. However, after mining the Myc target gene database (http://www.myccancergene.org), we found that none of the 11 c-myc-correlated/hPNPaseold-35-responsive genes are under known direct regulation by c-myc.
In summary, this current report (along with our previous one [Sokhi et al., 2013b]) suggests that hPNPaseold-35 is associated with the regulation of cell cycle- and cell proliferation-associated genes. Our bioinformatics analysis points to hPNPaseold-35 overexpression tipping the balance towards growth arrest, perhaps due to the resulting downregulation of cellular proliferation-related genes such as E2F3, MCM4, MCM7 and EIF4G1, and the upregulation of pro-apoptotic transcripts like FAS. In any case, follow-up experiments are needed to clarify the observations reported here and determine if the genes whose expression is regulated by hPNPaseold-35 are primary (direct) or secondary (associated) targets of this enzyme. Based on the modification of expression of overlapping (common) and distinct sets of genes, and likely common and unique sets of miRNAs [Das et al., 2010], in different cell types by hPNPaseold-35 we are referring to these global changes in cells as their “hPNPaseold-35 reactome”. Molecularly and biochemically defining these changes and their roles in cellular physiology will be challenging, but clearly a worthwhile and informative endeavor.
Supplementary Material
Figure S1. Doxycycline induced overexpression of hPNPaseold-35 post 24 hours at the mRNA (A) and protein levels (B) in HeLa cells. Error bars represent mean ± S.E. of three independent experiments done in triplicate. Statistical significance determined by two-tailed student’s t-test, *P<0.05, **P<0.01.
Figure S2. Unsupervised hierarchical cluster analysis. Unsupervised cluster dendrogram using centered correlation and average linkage, based on 22,227 probe sets for the 12-array dataset.
Figure S3. Functional annotation of genes significantly regulated by hPNPaseold-35 represented as enrichment clusters generated by the DAVID analysis software. (A) Cluster of 18 genes which were grouped together based on their involvement in chromosomal organization and related functions, (B) Cluster of 32 signaling molecules and cellular receptors, (C) Cluster of 15 genes grouped together based on their link to cell cycle-associated functions. Green - corresponding gene-term association positively reported in DAVID; black - corresponding gene-term association not reported yet in DAVID.
Figure S4. Pathway analysis of hPNPaseold-35 responsive genes. (A) Network 1 is associated with mitosis and involves 14 of our reported hPNPaseold-35 responsive genes. (B) Network 2 is associated with p21 signaling and involves 11 of our reported hPNPaseold-35 responsive genes. Representative genes such as CENPA and CENPE are validated by qRT-PCR, which are downregulated in response to hPNPaseold-35 overexpression.
Figure S5. Cartesian plot showing negative correlation between hPNPaseold-35 and DUSP1 as observed in the TCGA PANCAN dataset.
Figure S6. Cluster analysis of functional enrichments for c-Myc correlating genes. (A) Cluster of 13 genes involved in apoptosis, (B) Cluster of 16 genes involved in cytoskeleton associated functions, (C) Cluster of 13 genes involved in DNA repair, (D) Cluster of 10 genes involved in mRNA processing. Green - corresponding gene-term association positively reported in DAVID; black - corresponding gene-term association not reported yet in DAVID.
Figure S7. Biological functions (A) and canonical Pathways (B) identified by IPA corresponding to genes differentially regulated in response to hPNPaseold-35 overexpression. HeLa pnp = dox-inducible hPNPaseold-35 overexpression in HeLa cells, mel adpnp = Ad.hPNPaseold-35-mediated overexpression in melanoma cells.
Table S1. List of c-myc correlating genes.
Table S2. Pearson correlation of hPNPaseold-35 responsive genes identified through TCGA database.
Table S3. List of genes differentially upregulated (A) and downregulated (B) in response to hPNPaseold-35 overexpression as identified by microarray analysis in HO-1 melanoma and HeLa cells.
Acknowledgments
The present study was supported in part by NIH grants R01 CA097318 and the National Foundation for Cancer Research to P. B. Fisher; NIH grant R01 CA138540 and the James S. McDonnell Foundation to D. Sarkar. D. Sarkar is a Harrison Scholar in Cancer Research and a Blick Scholar. P.B. Fisher holds the Thelma Newmeyer Corman Chair in Cancer Research in the VCU Massey Cancer Center.
References
- Andrade J, Pobre V, Silva I, Domingues S, Arraiano C. The role of 3′-5′ exoribonucleases in RNA degradation. Prog Mol Biol Transl Sci. 2009;85:187–229. doi: 10.1016/S0079-6603(08)00805-2. [DOI] [PubMed] [Google Scholar]
- Babenko O, Golubov A, Ilnytskyy Y, Kovalchuk I, Metz GA. Genomic and epigenomic responses to chronic stress involve miRNA-mediated programming. PLoS One. 2012;7(1):e29441. doi: 10.1371/journal.pone.0029441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HW, Rainey RN, Balatoni CE, Dawson DW, Troke JJ, Wasiak S, Hong JS, McBride HM, Koehler CM, Teitell MA, French SW. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol Cell Biol. 2006;26(22):8475–8487. doi: 10.1128/MCB.01002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline MS, Craft B, Swatloski T, Goldman M, Ma S, Haussler D, Zhu J. Exploring TCGA Pan-Cancer Data at the UCSC Cancer Genomics Browser. Sci Rep. 2013;3:2652. doi: 10.1038/srep02652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SK, Sokhi UK, Bhutia SK, Azab B, Su ZZ, Sarkar D, Fisher PB. Human polynucleotide phosphorylase selectively and preferentially degrades microRNA-221 in human melanoma cells. Proc Natl Acad Sci USA. 2010;107(26):11948–11953. doi: 10.1073/pnas.0914143107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SK, Bhutia SK, Sokhi UK, Dash R, Azab B, Sarkar D, Fisher PB. Human polynucleotide phosphorylase (hPNPase(old-35)): An evolutionary conserved gene with an expanding repertoire of RNA degradation functions. Oncogene. 2011;30(15):1733–1743. doi: 10.1038/onc.2010.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher MP. Ribonuclease multiplicity, diversity, and complexity. J Biol Chem. 1993;268:13011–13014. [PubMed] [Google Scholar]
- Dumur CI, Ladd AC, Wright HV, Penberthy LT, Wilkinson DS, Powers CN, Garrett CT, DiNardo LJ. Genes involved in radiation therapy response in head and neck cancers. Laryngoscope. 2009;119:91–101. doi: 10.1002/lary.20005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JM, Funes JM, Henderson S, Wild L, Carey N, Boshoff C. Genomics screen in transformed stem cells reveals RNASEH2A, PPAP2C, and ADARB1 as putative anticancer drug targets. Mol Cancer Ther. 2009;8(1):249–60. doi: 10.1158/1535-7163.MCT-08-0636. [DOI] [PubMed] [Google Scholar]
- Goldman M, Craft B, Swatloski T, Ellrott K, Cline M, Diekhans M, Ma S, Wilks C, Stuart J, Haussler D, Zhu J. The UCSC Cancer Genomics Browser: update 2013. Nucleic Acids Res. 2013;41:D949–54. doi: 10.1093/nar/gks1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka M, Ito M, Droma Y, Ushiki A, Kitaguchi Y, Yasuo M, Kubo K. Comparison of gene expression profiling between lung fibrotic and emphysematous tissues sampled frompatients with combined pulmonary fibrosis and emphysema. Fibrogenesis Tissue Repair. 2012;5(1):17. doi: 10.1186/1755-1536-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75(4):805–16. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Hellman U, Mörner S, Engström-Laurent A, Samuel JL, Waldenström A. Temporal correlation between transcriptional changes and increased synthesis of hyaluronan in experimentalcardiac hypertrophy. Genomics. 2010;96(2):73–81. doi: 10.1016/j.ygeno.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J. Identification of a molecular signaling network that regulates a cellular necrotic cell deathpathway. Cell. 2008;135(7):1311–23. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8(9):R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim H, Wilusz J, Wilusz C. RNA recognition by 3′-to-5′ exonucleases: The substrate perspective. Biochimica Et Biophysica Acta. 2008;1779(4):256–265. doi: 10.1016/j.bbagrm.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leszczyniecka M, Kang DC, Sarkar D, Su ZZ, Holmes M, Valerie K, Fisher PB. Identification and cloning of human polynucleotide phosphorylase, hPNPaseold-35, in the context of terminal differentiation and cellular senescence. Proc Natl Acad Sci USA. 2002;99(26):16636–16641. doi: 10.1073/pnas.252643699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leszczyniecka M, DeSalle R, Kang DC, Fisher PB. The origin of polynucleotide phosphorylase domains. Mol Phylogenet Evol. 2004;31(1):123–130. doi: 10.1016/j.ympev.2003.07.012. [DOI] [PubMed] [Google Scholar]
- Malaney P, Pathak RR, Xue B, Uversky VN, Davé V. Intrinsic Disorder in PTEN and its Interactome Confers Structural Plasticity and Functional Versatility. Sci Rep. 2013;3:2035. doi: 10.1038/srep02035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGovern SL, Qi Y, Pusztai L, Symmans WF, Buchholz TA. Centromere protein-A, an essential centromere protein, is a prognostic marker for relapse in estrogen receptor-positive breast cancer. Breast Cancer Res. 2012;14(3):R72. doi: 10.1186/bcr3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013a;8(8):1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013b;41:D377–86. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncho-Amor V, Ibañez de Cáceres I, Bandres E, Martínez-Poveda B, Orgaz JL, Sánchez-Pérez I, Zazo S, Rovira A, Albanell J, Jiménez B, Rojo F, Belda-Iniesta C, García-Foncillas J, Perona R. DUSP1/MKP1 promotes angiogenesis, invasion and metastasis in non-small-cell lung cancer. Oncogene. 2011;30(6):668–78. doi: 10.1038/onc.2010.449. [DOI] [PubMed] [Google Scholar]
- Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J Biol Chem. 2005;280(20):19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- Oliveros JC. VENNY. An interactive tool for comparing lists with Venn Diagrams. 2007 http://bioinfogp.cnb.csic.es/tools/venny/index.html.
- Rutkowski MJ, Sughrue ME, Kane AJ, Kim JM, Bloch O, Parsa AT. Epidermal growth factor module-containing mucin-like receptor 2 is a newly identified adhesion G protein-coupled receptor associated with poor overall survival and an invasive phenotype in glioblastoma. J Neurooncol. 2011;105(2):165–71. doi: 10.1007/s11060-011-0576-7. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V, Quackenbush J. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Leszczyniecka M, Kang DC, Lebedeva IV, Valerie K, Dhar S, Pandita TK, Fisher PB. Down-regulation of myc as a potential target for growth arrest induced by human polynucleotide phosphorylase (hPNPaseold-35) in human melanoma cells. J Biol Chem. 2003;278(27):24542–24551. doi: 10.1074/jbc.M302421200. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Lebedeva IV, Emdad L, Kang DC, Baldwin AS, Fisher PB. Human polynucleotide phosphorylase (hPNPaseold-35): A potential link between aging and inflammation. Cancer Res. 2004;64(20):7473–7478. doi: 10.1158/0008-5472.CAN-04-1772. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Park ES, Emdad L, Randolph A, Valerie K, Fisher PB. Defining the domains of human polynucleotide phosphorylase (hPNPaseOLD-35) mediating cellular senescence. Mol Cell Biol. 2005;25(16):7333–7343. doi: 10.1128/MCB.25.16.7333-7343.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar D, Fisher PB. Polynucleotide phosphorylase: An evolutionary conserved gene with an expanding repertoire of functions. Pharmacol Ther. 2006a;112(1):243–263. doi: 10.1016/j.pharmthera.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Fisher PB. Human polynucleotide phosphorylase (hPNPase old-35): An RNA degradation enzyme with pleiotrophic biological effects. Cell Cycle. 2006b;5(10):1080–1084. doi: 10.4161/cc.5.10.2741. [DOI] [PubMed] [Google Scholar]
- Sarkar D, Fisher PB. Molecular mechanisms of aging-associated inflammation. Cancer Lett. 2006c;236(1):13–23. doi: 10.1016/j.canlet.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Shaikhibrahim Z, Lindstrot A, Ochsenfahrt J, Fuchs K, Wernert N. Epigenetics-related genes in prostate cancer: expression profile in prostate cancer tissues, androgen-sensitive and -insensitive cell lines. Int J Mol Med. 2013;31(1):21–5. doi: 10.3892/ijmm.2012.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slomovic S, Schuster G. Stable PNPase RNAi silencing: Its effect on the processing and adenylation of human mitochondrial RNA. RNA. 2008;14(2):310–323. doi: 10.1261/rna.697308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokhi UK, Das SK, Dasgupta S, Emdad L, Shiang R, DeSalle R, Sarkar D, Fisher PB. Human polynucleotide phosphorylase (hPNPaseold-35): should I eat you or not- that is the question? Adv Cancer Res. 2013a;19:161–90. doi: 10.1016/B978-0-12-407190-2.00005-8. [DOI] [PubMed] [Google Scholar]
- Sokhi UK, Bacolod MD, Dasgupta S, Emdad L, Das SK, Dumur CI, Miles MF, Sarkar D, Fisher PB. Identification of genes potentially regulated by human polynucleotide phosphorylase (hPNPaseold-35) using melanoma as a model. PloS ONE. 2013b;8(10):e76284. doi: 10.1371/journal.pone.0076284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny R, Borowski L, Brzezniak L, Dmochowska A, Gewartowski K, Bartnik E, Stepien PP. Human mitochondrial RNA turnover caught in flagranti: Involvement of hSuv3p helicase in RNA surveillance. Nucleic Acids Res. 2010;38(1):279–298. doi: 10.1093/nar/gkp903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh SH, Prathipati P, Motakis E, Kwoh CK, Yenamandra SP, Kuznetsov VA. A robust tool for discriminative analysis and feature selection in paired samples impacts the identification of the genes essential for reprogramming lung tissue to adenocarcinoma. BMC Genomics. 2011;12(Suppl 3):S24. doi: 10.1186/1471-2164-12-S3-S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valles I, Pajares MJ, Segura V, Guruceaga E, Gomez-Roman J, Blanco D, Tamura A, Montuenga LM, Pio R. Identification of novel deregulated RNA metabolism-related genes in non-small cell lung cancer. PLoS One. 2012;7(8):e42086. doi: 10.1371/journal.pone.0042086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Shu Z, Lieser S, Chen P, Lee W. Human mitochondrial SUV3 and polynucleotide phosphorylase form a 330-kDa heteropentamer to cooperatively degrade double-stranded RNA with a 3′-to-5′ directionality. J Biol Chem. 2009;284(31):20812–20821. doi: 10.1074/jbc.M109.009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Chen HW, Oktay Y, Zhang J, Allen E, Smith GM, Fan KC, Hong JS, French SW, McCaffery JM, Lightowlers RN, Morse HC, 3rd, Koehler CM, Teitell MA. PNPASE regulates RNA import into mitochondria. Cell. 2010;142(3):456–467. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Shimada E, Koehler CM, Teitell MA. PNPASE and RNA trafficking into mitochondria. Biochim Biophys Acta. 2012;1819(9–10):998–1007. doi: 10.1016/j.bbagrm.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Doxycycline induced overexpression of hPNPaseold-35 post 24 hours at the mRNA (A) and protein levels (B) in HeLa cells. Error bars represent mean ± S.E. of three independent experiments done in triplicate. Statistical significance determined by two-tailed student’s t-test, *P<0.05, **P<0.01.
Figure S2. Unsupervised hierarchical cluster analysis. Unsupervised cluster dendrogram using centered correlation and average linkage, based on 22,227 probe sets for the 12-array dataset.
Figure S3. Functional annotation of genes significantly regulated by hPNPaseold-35 represented as enrichment clusters generated by the DAVID analysis software. (A) Cluster of 18 genes which were grouped together based on their involvement in chromosomal organization and related functions, (B) Cluster of 32 signaling molecules and cellular receptors, (C) Cluster of 15 genes grouped together based on their link to cell cycle-associated functions. Green - corresponding gene-term association positively reported in DAVID; black - corresponding gene-term association not reported yet in DAVID.
Figure S4. Pathway analysis of hPNPaseold-35 responsive genes. (A) Network 1 is associated with mitosis and involves 14 of our reported hPNPaseold-35 responsive genes. (B) Network 2 is associated with p21 signaling and involves 11 of our reported hPNPaseold-35 responsive genes. Representative genes such as CENPA and CENPE are validated by qRT-PCR, which are downregulated in response to hPNPaseold-35 overexpression.
Figure S5. Cartesian plot showing negative correlation between hPNPaseold-35 and DUSP1 as observed in the TCGA PANCAN dataset.
Figure S6. Cluster analysis of functional enrichments for c-Myc correlating genes. (A) Cluster of 13 genes involved in apoptosis, (B) Cluster of 16 genes involved in cytoskeleton associated functions, (C) Cluster of 13 genes involved in DNA repair, (D) Cluster of 10 genes involved in mRNA processing. Green - corresponding gene-term association positively reported in DAVID; black - corresponding gene-term association not reported yet in DAVID.
Figure S7. Biological functions (A) and canonical Pathways (B) identified by IPA corresponding to genes differentially regulated in response to hPNPaseold-35 overexpression. HeLa pnp = dox-inducible hPNPaseold-35 overexpression in HeLa cells, mel adpnp = Ad.hPNPaseold-35-mediated overexpression in melanoma cells.
Table S1. List of c-myc correlating genes.
Table S2. Pearson correlation of hPNPaseold-35 responsive genes identified through TCGA database.
Table S3. List of genes differentially upregulated (A) and downregulated (B) in response to hPNPaseold-35 overexpression as identified by microarray analysis in HO-1 melanoma and HeLa cells.