A peroxygenase controls floral transition and tolerance to oxidative stress.
Abstract
Contrasting with the wealth of information available on the multiple roles of jasmonates in plant development and defense, knowledge about the functions and the biosynthesis of hydroxylated oxylipins remains scarce. By expressing the caleosin RESPONSIVE TO DESSICATION20 (RD20) in Saccharomyces cerevisiae, we show that the recombinant protein possesses an unusual peroxygenase activity with restricted specificity toward hydroperoxides of unsaturated fatty acid. Accordingly, Arabidopsis (Arabidopsis thaliana) plants overexpressing RD20 accumulate the product 13-hydroxy-9,11,15-octadecatrienoic acid, a linolenate-derived hydroxide. These plants exhibit elevated levels of reactive oxygen species (ROS) associated with early gibberellin-dependent flowering and abscisic acid hypersensitivity at seed germination. These phenotypes are dependent on the presence of active RD20, since they are abolished in the rd20 null mutant and in lines overexpressing RD20, in which peroxygenase was inactivated by a point mutation of a catalytic histidine residue. RD20 also confers tolerance against stress induced by Paraquat, Rose Bengal, heavy metal, and the synthetic auxins 1-naphthaleneacetic acid and 2,4-dichlorophenoxyacetic acid. Under oxidative stress, 13-hydroxy-9,11,15-octadecatrienoic acid still accumulates in RD20-overexpressing lines, but this lipid oxidation is associated with reduced ROS levels, minor cell death, and delayed floral transition. A model is discussed where the interplay between fatty acid hydroxides generated by RD20 and ROS is counteracted by ethylene during development in unstressed environments.
Originally thought to be seed specific and confined to the surface of lipid droplets, caleosins were assumed to be structural proteins of these organelles (Chen et al., 1999; Naested et al., 2000; Purkrtova et al., 2008; Jiang and Tzen, 2010). Caleosins have now been endowed with new roles since they have also been detected in vegetative tissues, where they are associated with the endoplasmic reticulum (Hernandez-Pinzon et al., 2001), the vacuole (Carter et al., 2004), and the envelope of chloroplasts (Partridge and Murphy, 2009). In Arabidopsis (Arabidopsis thaliana), Arabidopsis thaliana Seed1 (ATS1; At4g26740; Nuccio and Thomas, 1999) appears to participate in the degradation of lipid reserves in seeds (Poxleitner et al., 2006), while the non-seed-specific Caleosin4 (CLO4; At1g70670; Naested et al., 2000) and RESPONSIVE TO DESSICATION20 (RD20; At2g33380; Yamaguchi-Shinozaki et al., 1992) are negative and positive regulators of abscisic acid (ABA) responses during germination and dehydration processes, respectively (Aubert et al., 2010; Kim et al., 2011). However, the molecular mechanisms by which distinct caleosins exert their functions remained unknown. The recent identification of ATS1 and CLO4 as peroxygenases (Hanano et al., 2006; Blée et al., 2012) has prompted the suggestion that caleosins might fulfill their physiological functions via their enzymatic production of oxylipins (Partridge and Murphy, 2009; Aubert et al., 2010; Kim et al., 2011). Among the primary oxylipins formed in vitro by peroxygenases are the unsaturated fatty acid hydroxides (FAOHs; Fig. 1A). Such oxylipins accumulate upon pathogen attack (Rustérucci et al., 1999; Göbel et al., 2002; Montillet et al., 2005), and they may play significant roles in plant-pathogen interactions. They could act directly through their antimicrobial properties (Prost et al., 2005) and/or they could be components of signaling pathways leading to defense responses. For example, 2-hydroxy-9,12,15-octadecatrienoic acid (2-HOT), when applied on both tobacco (Nicotiana tabacum) and Arabidopsis leaves, prevents necrosis due to bacterial infections (Ponce de León et al., 2002; Hamberg et al., 2003). 2-HOT also participates in the defense of Nicotiana attenuata against insect feeding (Gaquerel et al., 2012). In addition, 9-hydroxy-10,12,15-octadecatrienoic acid (9-HOT) was reported to antagonize the action of ethylene, triggering molecular events such as the accumulation of callose, the production of reactive oxygen species (ROS), and transcriptional changes for genes involved in plant defense (Hwang and Hwang, 2010; López et al., 2011). FAOHs also accumulate during oxidative stress, when their functions are still elusive (op den Camp et al., 2003; Montillet et al., 2004). Besides their roles in defensive responses, FAOHs are involved in signaling pathways controlling physiological processes such as the regulation of lateral root development (Vellosillo et al., 2007).
Figure 1.
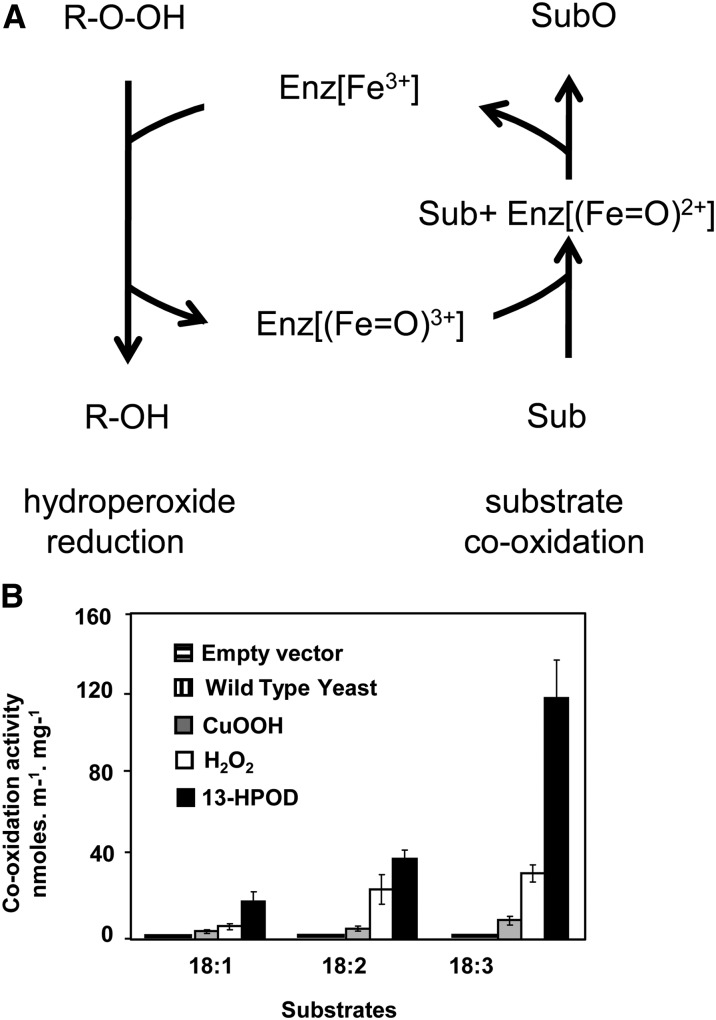
Characterization of recombinant RD20 activity in yeast. A, Mechanistic scheme for peroxygenase activity. Enz represents the peroxygenase, and Fe represents the heme of the protein; Sub and SubO represent the substrate and its oxidized form, respectively; R-O-OH and R-OH represent the hydroperoxide cosubstrate and its corresponding alcohol, respectively. B, Cooxidation of radiolabeled polyunsaturated fatty acids in the presence of cumene hydroperoxide (CuOOH), H2O2, or 13-HPOD. 14C-labeled substrates metabolized by microsomal fractions prepared from RD20-expressing yeast were separated by thin-layer chromatography and analyzed by radiodetection. 18:1, Oleic acid; 18:2, linoleic acid; 18:3, linolenic acid. Incubations of 14C-labeled substrates in the presence of microsomal fractions prepared from untransformed yeast and from yeast transformed with empty vector did not exhibit any activity.
The biosynthesis of FAOHs has only been partially elucidated. They most likely result from the reduction of fatty acid hydroperoxides (FAOOHs). FAOOHs can to be formed either chemically (e.g. in the presence of free radicals; Mosblech et al., 2009) or enzymatically by the oxygenation of C16 and C18 unsaturated fatty acids catalyzed by 9- and 13-lipoxygenases (9-LOXs and 13-LOXs; Feussner and Wasternack, 2002) or α-dioxygenases (α-DOXs; Hamberg et al., 1999). Thus, the biosynthesis of FAOOHS is well established, but the endogenous enzyme(s) reducing these hydroperoxides to FAOHs remain largely enigmatic. Peroxygenases were assumed to play such a role (Hamberg et al., 1999), because of their known ability to efficiently reduce many hydroperoxides in vitro (Ishimaru and Yamazaki, 1977; Blée and Schuber, 1990; Hamberg and Hamberg, 1990). Accordingly, a recent study reported the production of 2-HOT when recombinant α-DOX and the caleosin RD20 were incubated in vitro in the presence of linolenic acid. The resulting FAOH was postulated to be implicated in biotic stress responses as a phytoalexin in Arabidopsis (Shimada et al., 2014).
To ascertain the role of caleosin/peroxygenases in reducing endogenous FAOOHs and to test the implication of such an enzymatic activity in developmental and environmental responses, we have further studied RD20. This study describes the identification of RD20 as an unusual peroxygenase possessing substrate specificities restricted to polyunsaturated fatty acids and derivatives. This enzymatic activity is determinant for tolerance to oxidative stress conferred by RD20, resulting in reduced ROS accumulation and minor cell death. However, in an unstressed environment, overexpression of RD20 results in the accumulation of 13-hydroxy-9,11,15-octadecatrienoic acid (13-HOT), coinciding with enhanced ROS levels, early GA-dependent floral transition, and increased sensitivity to ABA. Based on our results, we discuss the role of ethylene in the generation and function of the products generated by the enzymatic activity of RD20 in the physiological responses during stress tolerance or escape. These studies allow a better understanding of the role of caleosin/peroxygenase in plant development and responses to abiotic stress.
RESULTS
Among all caleosin genes in Arabidopsis, RD20 is the most stress and hormone inducible (Supplemental Fig. S1). RD20 transcript levels respond to pathogen attack as well as to elicitors, hormones, and nutrient availability (Supplemental Fig. S1). Moreover, microarray data indicate that RD20 expression is also up-regulated in response to abiotic stresses such as wounding, drought, salt, cold, anoxia, osmotic stress, freezing recovery, and auxin-herbicide treatment (Supplemental Fig. S1). Consistent with these large-scale expression data, dedicated analyses also characterized RD20 as a general stress-inducible gene (Yamaguchi-Shinozaki et al., 1992; Takahashi et al., 2000; Partridge and Murphy, 2009). In addition, analysis of microarray databases using coexpression analysis (as described in Ehlting et al., 2008) shows that many genes with expression profiles similar to RD20 (Supplemental Fig. S2) are known to play roles in responses to environmental stress. These include homeobox-Leu zipper genes (Manavella et al., 2006; hormone data set), δ-1-pyrroline-5-carboxylate synthase involved in Pro biosynthesis (Yoshiba et al., 1995; hormone data set), remorins (Raffaele et al., 2007; stress data set), potassium transporter family members (Szczerba et al., 2009; stress data set), cold-responsive genes (cor15a; Baker et al., 1994; hormone data set), and the RD26 gene encoding a NAC (for NAM [no apical meristem]ATAF [Arabidopsis Transcription Factor], and CUC [cup-shaped cotyledon]) transcription factor involved in drought and ABA signaling (Fujita et al., 2004; hormone and pathogen data sets). Thus, these observed coexpression patterns provide independent support in favor of a key function of RD20 in stress responses. Moreover, microarray data, recently confirmed by GUS reporter gene analysis (Aubert et al., 2010), revealed an expression of RD20 in late stages of seed formation but also in stems, leaves, and flowers, suggesting multiple physiological roles for this caleosin in unstressed plants. The divergent roles of RD20 in development and stress responses might involve the enzymatic activity of this caleosin. However, the identity of RD20 as a peroxygenase is not definitively established. It does possess in its amino acid sequence essential features that typify such enzymes, such as an EF-hand calcium-binding motif and two strictly conserved His residues (Hanano et al., 2006). In addition, RD20 can form FAOHs in conjunction with α-DOX in vitro (Shimada et al., 2014), and a peroxygenase-like activity was detected in crude extracts of salt-stressed Arabidopsis, which was postulated to result from the induction of RD20 by salinity (Partridge and Murphy, 2009). Thus, further characterization of the oxidized compounds made in vivo by RD20 appears critical for a better understanding of its physiological function.
RD20 Is a Peroxygenase That Preferentially Accepts Lipids as Substrates
RD20 has been suggested to be a peroxygenase (Hanano et al., 2006; Shimada et al., 2014), and thus the enzymatic features of the recombinant protein expressed in yeast (Saccharomyces cerevisiae) were studied. Peroxygenases are typically able to catalyze cooxidations of diverse substrates in the presence of various hydroperoxides (Blée, 1998). In particular, all caleosin-type peroxygenases identified so far actively catalyze aniline hydroxylation in the presence of cumene hydroperoxide in vitro (Hanano et al., 2006; Blée et al., 2012). As a caleosin, RD20 was thus expected to catalyze such a reaction (Partridge and Murphy, 2009). Surprisingly, however, crude extracts from transformed yeast cells expressing RD20 were unable to hydroxylate aniline in the presence of cumene hydroperoxide. Attempts to optimize assay conditions by varying enzyme and substrate concentrations or by adding exogenous heme or calcium were unsuccessful. Nevertheless, analysis of public microarray data revealed that RD20 is closely coexpressed with several lipid metabolism- and signaling-related genes (Supplemental Fig. S2). These include the very-long-chain fatty acid-condensing enzyme CUT1, the lipoxygenase LOX2 (both in the organ data set), several putative lipid transfer proteins (in the stress and hormone data sets), a putative lipase, and a long-chain fatty acid CoA ligase (LACS8; in the hormone data set). This coexpression pattern and the ability of peroxygenases to epoxidize double bonds of unsaturated fatty acids (Blée and Schuber, 1990) prompted us to examine whether lipids were substrates for RD20. Such a hypothesis could still be consistent with the previously noted slight increase, and apparently cumene hydroperoxide-dependent, of RD20-mediated peroxygenase in salt-stressed leaves (Partridge and Murphy, 2009). For this, one should consider the possibility of RD20 having used endogenous FAOOHs present in the tested crude leaf extracts as cosubstrates to catalyze the hydroxylation of aniline rather than the exogenously added cumene hydroperoxide.
To test our hypothesis, RD20-dependent degradation of 14C-labeled C18 fatty acids with one to three double bonds in the presence of cumene hydroperoxide, hydrogen peroxide (H2O2), or 13-hydroperoxy-9,11-octadecadienoic acid (13-HPOD) was followed by thin-layer chromatography coupled to radiodetection. The most active cosubstrate was 13-HPOD followed by H2O2, whereas cumene hydroperoxide only poorly promoted epoxidation of the unsaturated fatty acids (Fig. 1B). Among the substrates tested, linolenic acid, a triple unsaturated fatty acid, was most actively epoxidized by RD20, whereas the single unsaturated oleic acid was hardly metabolized regardless of the hydroperoxide used. As negative controls, neither extract from wild-type Wa6 yeast strain nor yeast transformed with an empty vector displayed any catalytic activity (Fig. 1B). To further characterize the peroxygenase identity of RD20, we tested whether it was able to catalyze the reduction of FAOOHs followed by oxidation of the resulting alcohol (i.e. cooxidation of FAOHs) in the absence of other oxidizable substrates (Fig. 1A). Such an activity was described previously for a soybean (Glycine max) peroxygenase and ATS1 (Blée et al., 1993; Hanano et al., 2006). To avoid the possibility of contaminant reductase activity present in yeast, recombinant RD20 fused to a C-terminal FLAG tag was purified to apparent homogeneity using affinity chromatography. SDS-PAGE analysis showed a single band, validated as RD20/FLAG by western blotting using a FLAG antibody (Fig. 2A). The purified fraction did not perform oxidation of substrates, like aniline, thiobenzamide, or unsaturated fatty acids, with cumene hydroperoxide. This confirmed that cumene hydroperoxide is a very poor substrate for RD20 and showed that the lack of aniline hydroxylation with crude extracts or microsomes from transformed yeasts was not due to the presence of inhibitors in yeast.
Figure 2.
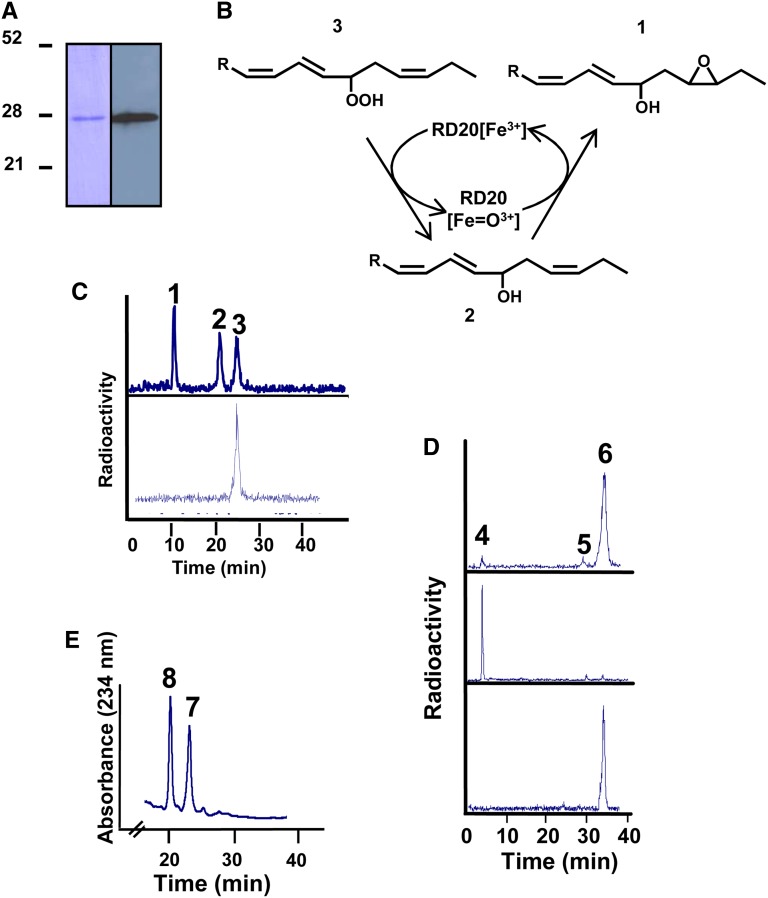
Characterization of purified recombinant RD20. A, Analysis by SDS-PAGE of a purified fraction of the recombinant protein (left) and western-blot analysis of this fraction using a monoclonal anti-FLAG antibody produced in mouse (right). B, Metabolism scheme showing the products expected from the peroxygenase-catalyzed transformation of FAOOH. C, Radio-HPLC analysis of the metabolization of [1-14C]13-HPOT by purified RD20. Peaks are as follows: 1, 15,16-epoxy-13-hydroxy-9,11-octadecenoic acid; 2, 13-HOT; 3, residual 13-HPOT. The bottom scan shows the absence of 13-HPOT transformation by boiled purified RD20 after 2 h of incubation at 27°C. D, Radio-HPLC analysis of the metabolization of [1-14C]13-HPOD by purified RD20. Top, products of [1-14C]13-HPOD formed by purified RD20 after 2 h of incubation at 27°C. Peaks are as follows: 4, trihydroxy-octadecanoates; 5, 13-hydroxy-9,11-octadecadienoic acid; 6, 13-HPOD. Middle, accumulation of trihydroxy-octadecanoates after overnight incubation of [14C]13-HPOD with purified RD20. Bottom, no epoxy or hydroxy derivatives were formed when [1-14C]13-HPOD was incubated overnight in the presence of a boiled purified RD20. E, Reduction of 9-FAOOH into its corresponding alcohol by purified RD20. Peaks are as follows: 7, 9-HPOT; 8, 9-HOT.
To test whether RD20 is able to catalyze the intramolecular transfer of an oxygen atom in FAOOHs (Blée et al., 1993; Fig. 2B), we incubated purified recombinant RD20 with [1-14C]13-HPOT (for 13-hydroperoxy-9,11,15-octadecatrienoic acid) or [1-14C]13-HPOD. The reaction products were analyzed by radio-HPLC. After 2 h of incubation, about 70% of 13-HPOT was converted into two more polar compounds (Fig. 2C, top). Peak 3 represented the residual substrate. Peak 2 (elution time, 21 min) was identified as 13-HOT by coelution with an authentic standard, and its structure was confirmed by mass spectrometry. Peak 1 (elution time, 10.5 min) absorbed at 234 nm, similar to 13-HOT, indicating that it still contained a Z,E-conjugated double bond system. Analysis by gas chromatography-mass spectrometry (GC-MS) of the trimethylsilyl ether derivative of its methyl ester featured a molecular ion at mass-to-charge ratio 396, indicative of an additional oxygen atom, compared with 13-HOT. Representative fragments were found identical to those observed with 15,16-epoxy-13-hydroxy-octadeca-9,11-dienoic acid (Supplemental Fig. S3; Blée et al., 1993). In contrast to 13-HPOT, 13-HPOD was only slowly metabolized by RD20 under identical incubation conditions (Fig. 2D, top). Peak 5 (elution time, 29.2 min) was 13-hydroxy-octadeca-9,11-dienoic acid (13-HOD), the methyl ester-trimethylsilyl ether derivative of this compound cochromatographed with an authentic standard, and the mass spectra of these two compounds were identical. A mixture of 9,10,13- and 9,12,13-trihydroxy-octadecanoates, identified by GC-MS (Hamberg, 1991), rapidly eluted as peak 4 (elution time, 3.7 min). These trihydroxy derivatives (probably resulting from the chemical opening of unstable epoxy alcohols) accumulated only after prolonged incubation times (i.e. overnight; Fig. 2D, middle). In negative control experiments using boiled purified RD20, 13-HPOT and 13-HPOD were not metabolized when incubated either for 2 h or overnight (Fig. 2, C and D, bottom). Thus, like ATS1, RD20 is capable, in the absence of any other oxidizable substrate, of catalyzing the reduction of FAOOHs with the concomitant formation of their corresponding epoxy alcohol. Of note, RD20 was also able to reduce the 9-hydroperoxide derivative of linolenic acid (Fig. 2E). Taken together, these experiments indicate that the apparent lack of the canonical peroxygenase activity of RD20 might result from its preferential use of lipophilic molecules (unsaturated fatty acids and their hydroperoxide derivatives) as substrates.
Molecular Characterization of RD20-Overexpressing and Transfer DNA Insertion Lines
To explore the function of RD20 in development and stress, we used a loss- and a gain-of-function approach. A single Arabidopsis transfer DNA (T-DNA) insertion line was available in public libraries at the onset of this project. The T-DNA was inserted into the second exon 572 bp downstream of the start codon. No RD20 transcripts were detectable in the rd20 null mutant compared with its wild-type Wassilewskija (Ws) background. Introduction of a 35S:RD20 overexpression construct into the rd20 mutant restored some RD20 expression (Fig. 3A). We also generated Arabidopsis Columbia-0 (Col-0) plants to overexpress RD20 under the control of the cauliflower mosaic virus 35S promoter. In parallel, void plasmid was transformed into wild-type Col-0 Arabidopsis and used as a control. The two RD20-overexpressing lines used, RD20-OE1 and RD20-OE2, showed 20- to 26-fold increases in RD20 transcript levels as assessed by quantitative reverse transcription (qRT)-PCR (Fig. 3B).
Figure 3.
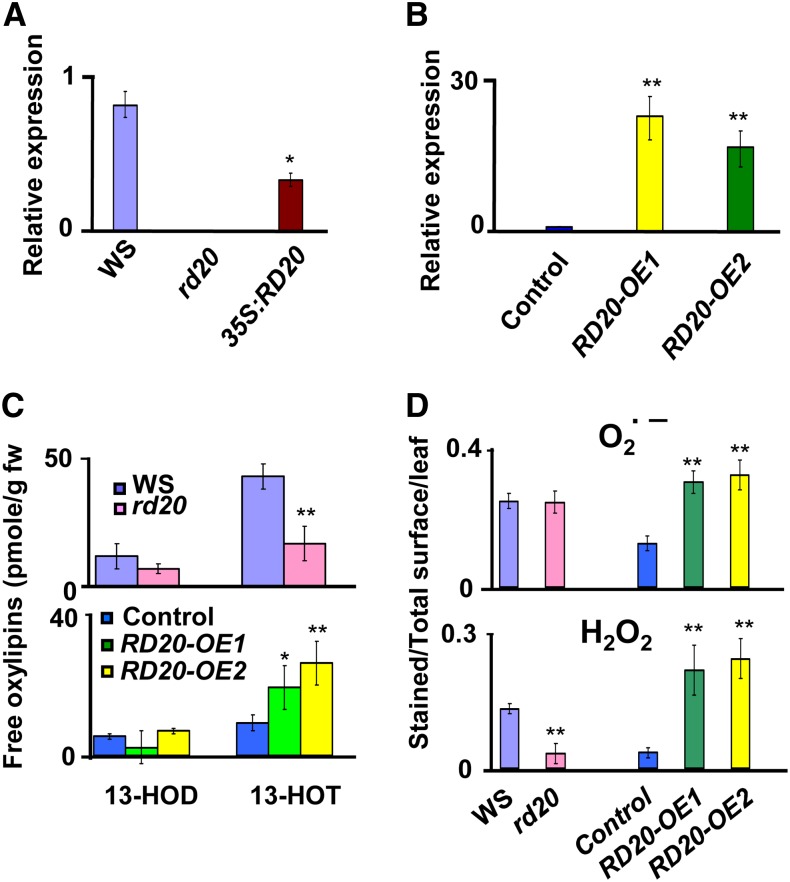
Characterization of RD20 transgenic plants. A and B, qRT-PCR analysis of RD20 gene expression in 3-week-old leaves. A, RD20 was expressed in the rd20 background, yielding the 35S::RD20 line. B, The control contains an empty vector used for the overexpression of RD20. Data represent means and se of three replicates obtained from three different biological samples. C, Oxylipin profiling. Free FAOHs present in leaves of the different lines were analyzed by HPLC. All lines were harvested at the bolting stage of RD20-overexpressing plants. Data represent means and se of two replicates from 20 different biological samples. fw, Fresh weight. D, ROS status of 3-week-old rosette leaves. Superoxide radical (O2∙−) levels were determined after staining leaves with NBT (top), whereas H2O2 contents were measured by staining leaves with DAB (bottom). Ratios of stained surface versus total surface were calculated with ImageJ. For all parts, results concerning transgenic lines versus their respective controls were considered statistically different as indicated: **P < 0.01, *P < 0.05 (n ≥ 30 for each line; Student’s t test analysis).
To assess whether RD20 accepts FAOOHs as substrates not only in vitro but also in planta, the FAOH content of the different RD20-transformed lines was analyzed by HPLC. Alcohols derived from 13-FAOOHs were the most abundant free oxylipins formed in untreated leaves of all lines studied. While 13-HOD levels were not significantly affected in any mutant, knock out of RD20 led to a 50% reduction of free 13-HOT compared with Ws wild-plant plants (Fig. 3C, top). The opposite metabolic phenotype is apparent in RD20-OE2 and RD20-OE1 lines, which contain 2- to 3-fold higher free 13-HOT levels, respectively, than their vector-only counterparts (Fig. 3C, bottom). These results were consistent with a functional expression of RD20 in the transgenic plants, able to reduce 13-HPOT, the main endogenous fatty acid hydroperoxide formed in green leaves.
Exogenous application of 9-HOT was recently reported to trigger the formation of superoxide radicals in Arabidopsis leaves (López et al., 2011). To investigate whether such ROS accumulate when FAOH was produced endogenously by RD20, superoxide radical content was determined using the nitroblue tetrazolium (NBT) dye assay. No statistical difference could be observed between superoxide radical amounts in rd20 mutant and Ws plants (Fig. 3D, top). In contrast, about a 2-fold increased level of superoxide radical was found in the lines overexpressing RD20 compared with their controls (Fig. 3D, top). It thus appears that 13-LOX-derived oxylipins endogenously produced by RD20 trigger the accumulation of superoxide radical species, similar to what is observed with exogenously applied 9-HOT. Next, we investigated whether transgenic lines accumulate H2O2, the most chemically stable ROS. Considering the hypothesis that, in analogy with the recombinant protein in vitro (see above), RD20 would also be able to reduce H2O2 in planta, reduced levels of this hydroperoxide were expected in plants overexpressing RD20. However, this was not observed when H2O2 content was estimated with the 3,3′-diaminobenzidine (DAB) staining test. Instead, such plants accumulated 5 to 6 times more H2O2 than their respective controls (Fig. 3D, bottom). Conversely, rd20 leaves contained about 4-fold less H2O2 than the Ws wild type (Fig. 3D, bottom). Together, these results indicate that under unstressed conditions, RD20 leads to ROS accumulation.
Alteration of RD20 Expression Affects the Transition to Flowering
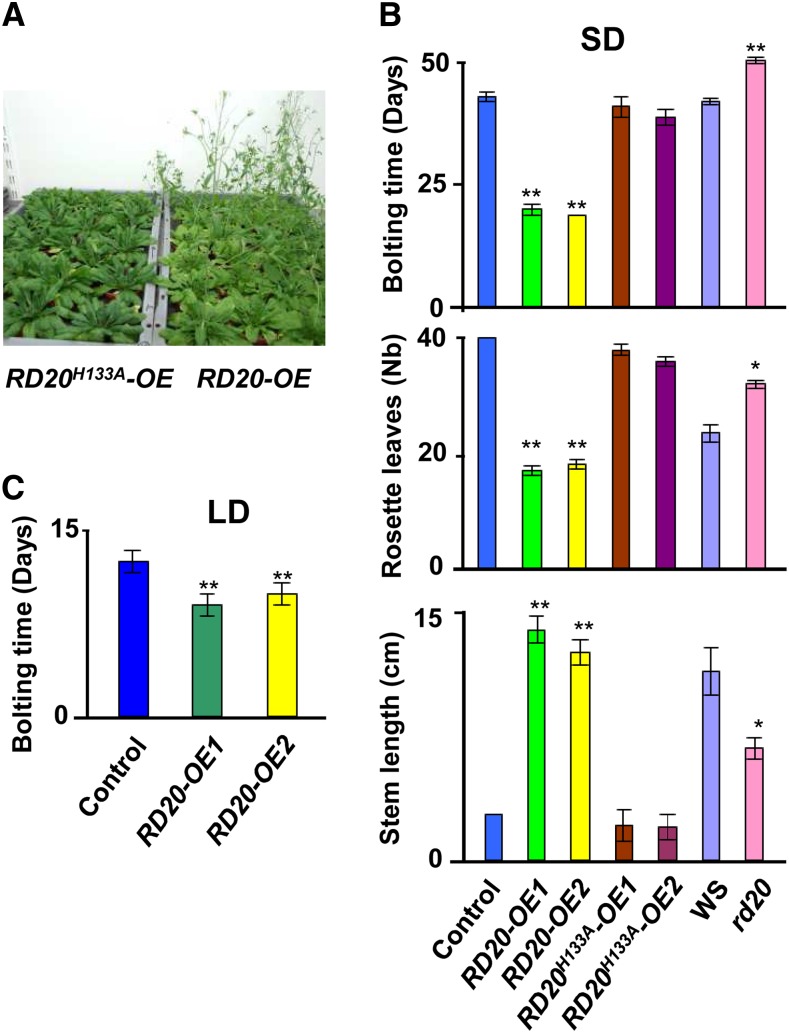
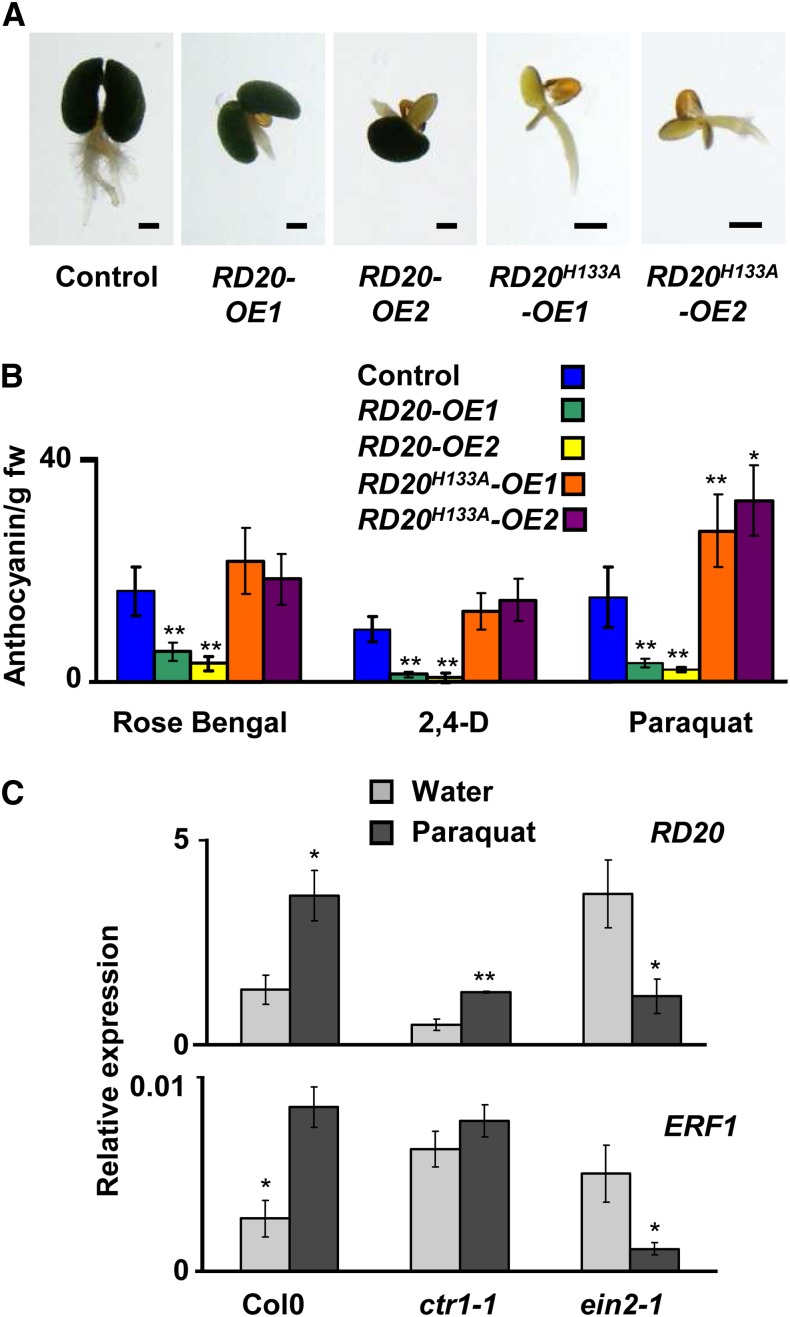
The most striking visual phenotypes of RD20 transgenic lines were changes in the timing of the floral transition. Both RD20-OE lines were in bloom about 3 weeks before control plants under short-day conditions. In contrast, the rd20 mutant showed delayed flowering compared with Ws wild-type plants. To test whether this phenotype is due to altered peroxygenase activity or due to altered RD20 levels per se, we generated transgenic Arabidopsis plants overexpressing catalytically inactive RD20. For this, we mutated His-133 to Ala. This His residue is strictly conserved in peroxygenases (Supplemental Fig. S4A) and is crucial for peroxygenase activity (Hanano et al., 2006). This substitution is assumed to prevent the cooxidation step, hence leading to an accumulation of the reaction intermediates (Fig. 1A) and ultimately to the inactivation of RD20 (Blée et al., 1993; Hanano et al., 2006). Accordingly, the recombinant RD20H133A was unable to catalyze epoxy derivative formation but still accumulated some 13-HOT when incubated with 13-HPOT (Supplemental Fig. S4B). We selected two lines, RD20H133A-OE1 and RD20H133A-OE2, with transgene expression levels similar to or higher than those found in plants overexpressing the wild-type enzyme (Fig. 3B). No difference in the accumulation of oxylipins was observed in these two lines in comparison with the wild type (Supplemental Fig. S4C). Importantly, these two transgenic lines displayed no apparent alterations in growth, morphology, or flowering time compared with the wild type. This shows that increased amounts of inactive RD20 are not sufficient to influence flowering time, indicating that, instead, increased oxylipin generation in the RD20-OE lines was required to accelerate the floral transition under nonstress conditions. These visual observations (Fig. 4A) were confirmed by quantifying bolting times, number of rosette leaves, and length of floral stems (Fig. 4B). In addition, phenotypes were preserved when plants were grown in phytochambers located at different places (Strasbourg, Bordeaux [both in France], and Damascus, Syria), indicating that they did not result from subtle differences in tap water or soil composition or local growth room conditions (e.g. light intensity/composition or temperature fluctuations).
Figure 4.
RD20 is involved in the floral transition. A, Peroxygenase activity was required for RD20 effects on flowering. Lines overexpressing enzymatically active (right) or inactive (left) RD20 were grown under short-day conditions for 4 weeks, when the photograph was taken. B, Modifications of RD20 expression or RD20 activity affect the floral transition of the mutant lines. Flowering changes were determined by measuring the bolting times (top), the number of rosette leaves at the bolting time of RD20-overexpressing lines (middle), and the length of the stems of 5-week-old plants (bottom). All lines were grown side by side under short-day conditions (SD). Data are means of two different biological samples (n = 35 per experiment per genotype). C, The RD20 accelerating effect on the floral transition is dependent on the photoperiod. Compared with controls, when grown under long-day conditions (LD), the bolting time is reduced in lines overexpressing RD20 (n = 28). The experiment was repeated twice. In all parts, differences between transgenic lines versus their respective controls were significant as indicated: **P < 0.01, *P < 0.05.
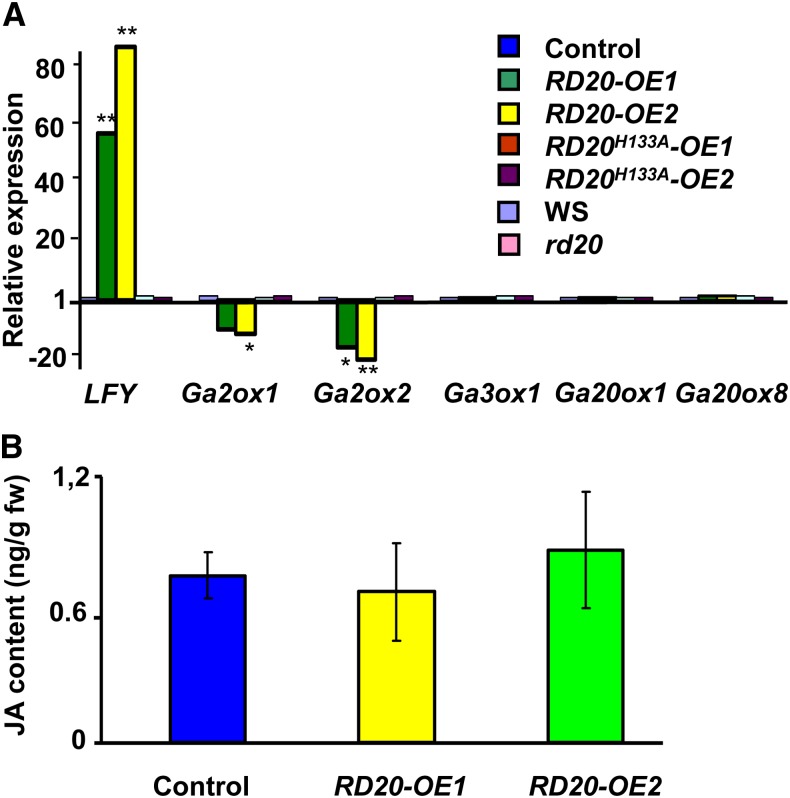
GAs are well-known regulators of the floral transition, and notably, this hormone class has a greater effect on flowering time under short-day than under long-day conditions in Arabidopsis (Wilson et al., 1992). The early-flowering phenotype conferred by RD20 overexpression was reduced from 3 weeks under short-day conditions (Fig. 4B, top) to only 4 d under long-day conditions (Fig. 4C). This immediately suggested a GA-mediated effect of RD20 on flowering. Among other mechanisms, GA promotes flowering through transcriptional activation of the floral meristem identity gene LEAFY (LFY; Wilson et al., 1992). LFY expression was up-regulated in both RD20-OE1 and RD20-OE2 lines (Fig. 5A). To investigate whether the RD20-dependent control of flowering time involves the induction of GA biosynthesis or turnover, we analyzed the expression of some of the genes involved in GA metabolism. While no induction of GA biosynthetic genes (i.e. GA3ox1, GA20ox1, and GA20ox8) was apparent in any of the transgenic lines, a down-regulation of the GA-deactivating genes GA2ox1 and GA2ox2 in plants overexpressing RD20 was indicative of a possible GA accumulation in these transgenic lines (Fig. 5A). These results were overall consistent with a GA signaling-dependent control of the floral transition in plants overexpressing RD20. Because the GA signaling pathway is antagonized by jasmonates (JAs; Yang et al., 2012), we next examined the possibility that alteration of JA level might lead to early flowering of the lines overexpressing RD20. Actually, both RD20 and allene oxide synthase, the first enzyme of the JA biosynthetic pathway, use 13-HOT as a substrate and thus might compete for it. According to such a scenario, a decrease of JA accumulation and not the production of oxylipins by caleosin would be responsible for the early-flowering phenotype of RD20-overexpressing lines. However, analysis of JA contents did not reveal any significant difference in hormone accumulation in lines overexpressing RD20 when compared with their controls (Fig. 5B), suggesting that the early flowering of the transgenic lines was unlikely driven by a competition between the JA and RD20 pathways.
Figure 5.
Overexpression of RD20 modified the expression of the transcription factor LFY but not JA contents. A, Quantitative PCR analysis of gene expression involved in signaling (LFY), biosynthesis (Ga3ox1, GA20ox1, and Ga20ox8), and metabolization (Ga2ox1 and Ga2ox2) of GAs. Plants were harvested at the bolting time of RD20-overexpressing lines. Data represent means and se of three replicates. B, Analysis by liquid chromatography-mass spectrometry of JA contents of lines overexpressing RD20 and control plants. Data are means ± se of triplicate assays. Transgenic plants did not significantly differ from control plants when analyzed by Student’s t test, with P = 0.66 for RD20-OE1 versus control and P = 0.57 for RD20-OE2 versus control. fw, Fresh weight.
RD20 Affects ABA-Dependent Germination and Seed Dormancy
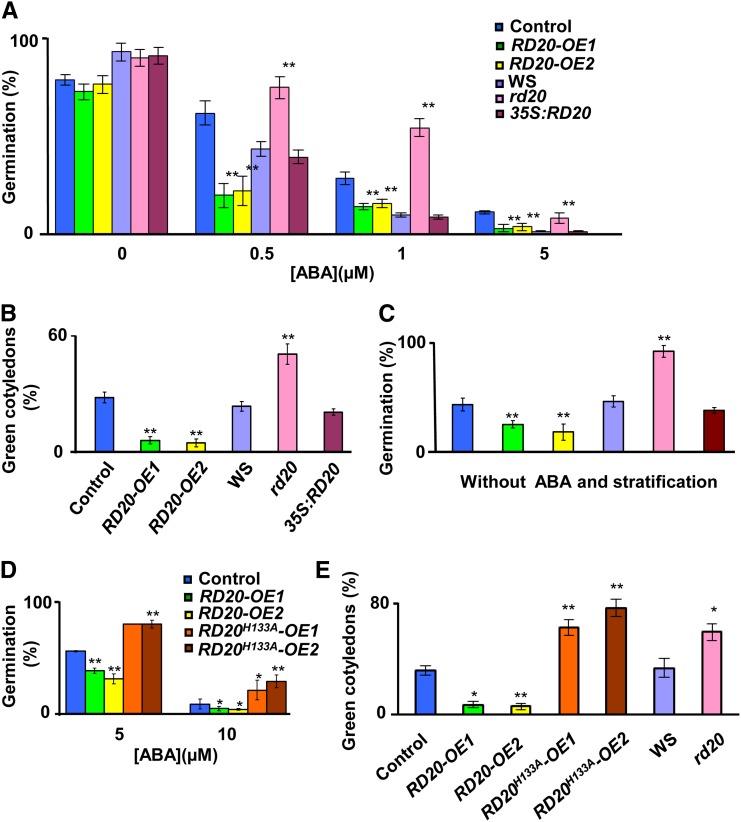
GA not only controls the floral transition but also acts as an ABA antagonist during germination (Debeaujon and Koornneef, 2000; Kucera et al., 2005). To investigate whether alterations in RD20 expression also impacted the sensitivity to exogenous ABA, we studied seed germination and postgermination growth of the different RD20 transgenic lines in the presence of increasing concentrations of ABA. As shown in Figure 6A, fewer RD20-OE1 and RD20-OE2 seeds germinated than wild-type seeds in an ABA dose-dependent manner. In contrast, rd20 seeds germinated at higher frequency and faster than did wild-type Ws seeds when treated with ABA (Fig. 6A; Supplemental Fig. S5A). Noteworthy, without the addition of ABA, the germination of rd20 seeds was slightly delayed compared with control seeds (Supplemental Fig. S5B). The insensitivity to ABA was abolished in the mutant when complemented with a 35S:RD20 transgene construct (Fig. 6A; Supplemental Fig. S5A). Similar results were observed when considering seedling growth. While ABA negatively affected the postemergence growth of RD20-overexpressing seedlings, more rd20 mutants showed expanded and green cotyledons after 6 d of light in the presence of 1 µm ABA compared with the wild type and the complemented line (Fig. 6B). Together, complementary loss- and gain-of-function studies clearly showed that ABA sensitivity is affected in plants with altered RD20 expression. It should be noted that, when assayed without any treatment, freshly harvested seeds of RD20-OE1 and RD20-OE2 showed a marked decrease of germination rate when compared with control seeds. In contrast, freshly harvested seeds of the rd20 mutant germinated at higher frequency than did wild-type Ws seeds (Fig. 6C). However, after a cold treatment (stratification) to break dormancy, rd20 and RD20-OE seeds displayed full germination indistinguishable from their respective wild-type controls (Fig. 6A). These results suggest that RD20 may impact seed dormancy by enhancing sensitivity to endogenous ABA.
Figure 6.
Alteration of RD20 expression affects ABA sensitivity. A, Germination of seeds of RD20 transgenic lines and their respective controls was scored as emergence of the radicle 4 d after stratification under various concentrations of ABA. B, Postemergence growth was estimated by the percentage of green cotyledons developed in the presence of 1 µm ABA 6 d after stratification. C, Germination of fresh seeds (without stratification) was also accessed in the absence of exogenous ABA. Data are means ± se for three different samples with n > 30 per line per replicate. D and E, Sensitivity to ABA conferred by RD20 depends on the catalytic activity of the encoded enzyme. The germination of seeds containing an empty vector or overexpressing active or catalytically impaired enzyme was scored in the presence of 5 and 10 µm ABA (D). Green cotyledons that developed in the presence of 5 µm ABA were counted 4 d after stratification (E). Data are means ± se of three different biological samples (n > 30 per line per sample). In all parts, differences between transgenic plants versus their respective controls were significant as indicated: *P < 0.05, **P < 0.01.
Are FAOHs Active Molecules during Germination?
To investigate whether peroxygenase activity was necessary for the altered ABA sensitivity of RD20 transgenic lines, the seed germination and dormancy of RD20H133A-OE1 and RD20H133A-OE2 lines were studied. Mutation of the RD20 active site His-133 (see above) rendered the resulting seeds resistant to ABA, as observed for the insertion mutant lines (Fig. 6D). This effect was particularly obvious when cotyledon expansion and greening were considered after 6 d of growth in the presence of 1 µm ABA. While less than 10% of the seedlings overexpressing active RD20 turned green, up to 80% of the seeds overexpressing the catalytically impaired enzyme developed green and expanded cotyledons. The ABA-dependent inhibition of greening in RD20H133A-OE lines was similar to the results obtained with knock-out mutant lines cultivated under the same conditions (Fig. 6E). Thus, the altered sensitivity to ABA of RD20 transgenic lines was conferred by the enzymatic activity of the product of this gene.
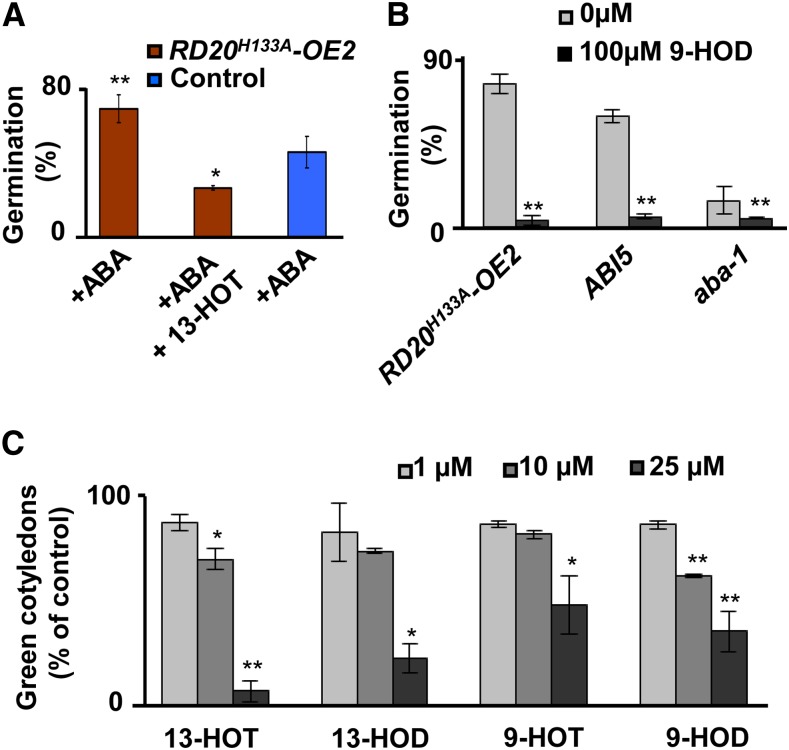
To identify what product resulting from RD20 catalysis was involved, we examined whether the ABA insensitivity of RD20H133A-OE seeds resulted from the action of FAOHs or from additional unidentified oxygenated compounds produced during the cooxidation reactions (Fig. 1A). To differentiate between these two possibilities, we attempted to chemically complement the deficiency of RD20 catalytic activity by supplying exogenous FAOHs to RD20H133A-OE seeds. As shown in Figure 7A, addition of 13-HOT fully reversed the ABA insensitivity of these seeds. In the absence of external ABA, the four hydroxylated derivatives of linoleic or linolenic acid, 13-HOD, 9-HOT, 13-HOT, and 9-HOD, impaired the development of green and fully expanded cotyledons of RD20H133A-OE2 plants (Fig. 7C). Inhibition of germination by FAOHs was observed not only for the RD20H133A-OE2 line but also for other ABA-insensitive lines, such as abi-5, which is devoid of a basic Leu zipper transcription factor required for ABA responses in seeds (Fig. 7B). Consistently, seed germination of the ABA-deficient aba1 mutant (devoid of zeaxanthin epoxidase, an enzyme involved in ABA biosynthesis) was also further delayed in the presence of 9-HOD (Fig. 7B). Together, these results strongly suggest a major role of the RD20-catalyzed formation of FAOHs in ABA-mediated signaling during seed germination and postgermination growth.
Figure 7.
FAOHs control ABA sensitivity and germination. A, Primary products of RD20 catalytic activity abolish ABA insensitivity. The insensitivity to 25 µm ABA of lines overexpressing inactive RD20 was abolished by 50 µm 13-HOT. B, Germination of ABA-insensitive ABI5 and the aba1 mutant was inhibited in the presence of 100 µm 9-HOD. C, Postemergence growth was inhibited in the presence of FAOHs. Data are means ± se of three different biological samples, with n = 25 per line per sample. In all parts, differences were significant as indicated: *P < 0.05, **P < 0.01 (by Student’s t test).
Stress-Induced RD20 Protects Arabidopsis against Oxidative Damage
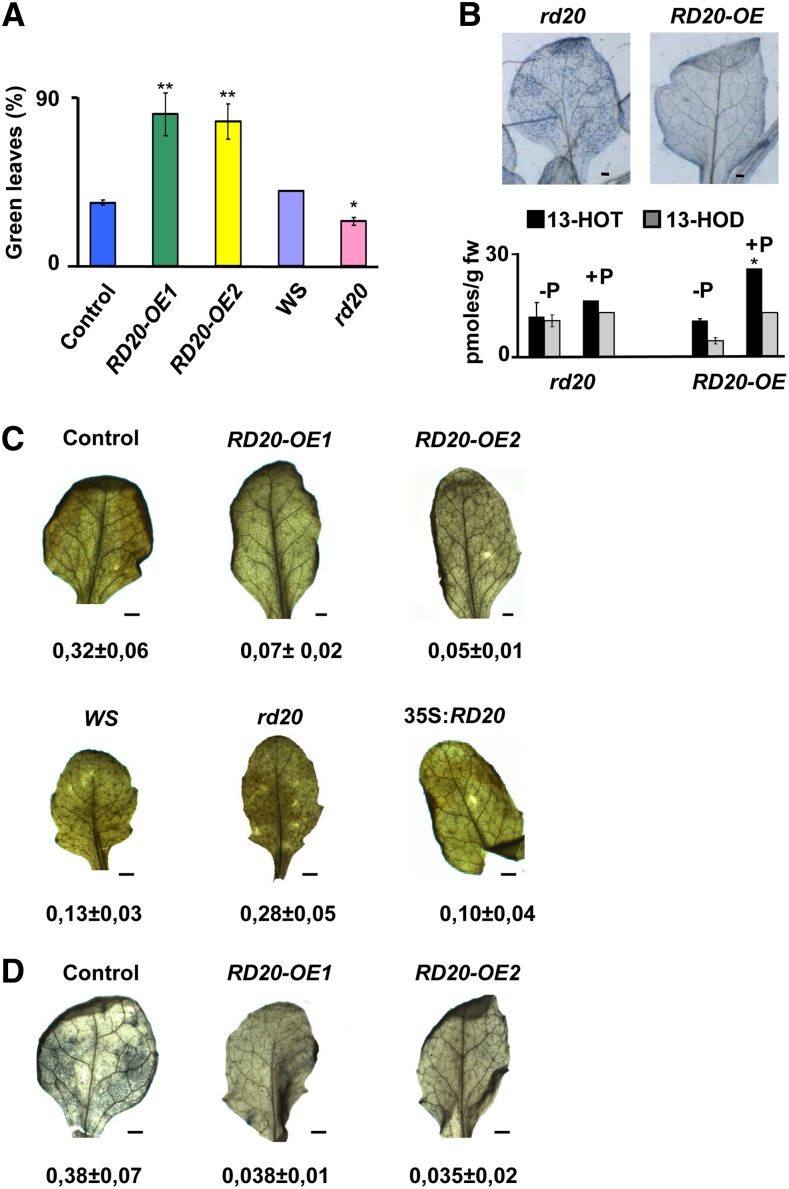
The strong stress response of RD20 expression suggests a role for the caleosin/peroxygenase RD20 in conditions known to generate oxidative damage (Supplemental Fig. S1). We first investigated the impact of the herbicide Paraquat on the development of the different transgenic lines. Paraquat is a generator of superoxide radical that strongly activates the expression of RD20 (Ramel et al., 2012). Leaf bleaching is the primary visual effect of Paraquat phytotoxicity, and less than 30% of the total leaves of Col-0 control plants remained green after 7 d of growth under continuous light in the presence of 0.5 µm Paraquat (Fig. 8A). In sharp contrast, up to 75% and 78% of leaves on RD20-OE1 and RD20-OE2 lines, respectively, remained green (Fig. 8A). Conversely, plants devoid of RD20 were more sensitive to Paraquat than Ws control plants (Fig. 8A). Leaf bleaching was observed about 1 week after germination in the presence of Paraquat, whereas cell death was apparent already 24 h after spraying the herbicide on mature leaves. As shown in Figure 8B (top), Paraquat-treated rd20 leaves exhibited much higher degrees of cell death, determined by Trypan Blue staining, than those from RD20-overexpressing lines. Decreased cell death was associated with an enhanced accumulation of 13-FAOHs in RD20-OE2 leaves (Fig. 8B, bottom). In contrast, Paraquat minimally affected FAOH levels in the null mutant, suggesting that Paraquat-induced production of these oxylipins is RD20 dependent (Fig. 8B, bottom). Additional oxylipin profiling of rd20 and RD20-OE plants revealed the induction of 9-ketodiene and 9-ketotriene derivatives (i.e. compounds that result from further transformations of FAOOHs by LOXs), suggesting that Paraquat likely has induced 9-LOX abundance/activity (Supplemental Fig. S6). 12-Hydroxy-9,12,15-octadecatrienoic acid and 16-hydroxy-9,12,15-octadecatrienoic acid levels were also minimally affected by Paraquat treatment (Supplemental Fig. S6). This suggested that 13-FAOHs likely resulted from the catalytic activity of 13-LOX/RD20. Since cell death is linked to enhanced generation of ROS in plants (Gechev et al., 2006), we examined whether RD20 may confer protection against cell death by lowering the accumulation of ROS. We first evaluated H2O2 levels in the transgenic plants treated with Paraquat. Quantitative analysis of whole-leaf staining with DAB revealed that the rd20 mutant accumulates significantly more H2O2 compared with Ws wild-type plants (Fig. 8C). Vice versa, RD20-OE lines contained 5- to 7-fold lower amounts of H2O2 compared with their controls (Fig. 8C). Likewise, superoxide radical content assessed by NBT staining decreased 10-fold in Paraquat-stressed RD20-OE leaves compared with controls (Fig. 8D). Collectively, these data are consistent with a protective function of RD20 against oxidative damage by lowering ROS accumulation.
Figure 8.
RD20 confers stress tolerance. A, RD20 protects leaves from Paraquat toxicity. Green leaves from RD20-overexpressing lines, the rd20 mutant, and their respective controls were counted after 1 week of growth under continuous light in the presence of 0.5 µm Paraquat. The experiment was repeated twice. Data-averaged values are means ± se (n = 60 per genotype; differences were significant at *P < 0.05 and **P < 0.01 when analyzed by Student’s t test). B, RD20 protects against cell death. Cell death was visualized by staining leaves of RD20-overexpressing lines and the rd20 mutant with Trypan Blue (top). Oxylipins were measured from two independent experiments (bottom); data are means ± se and differ between with or without Paraquat (P) at *P < 0.05 (Student’s t test analysis). fw, Fresh weight. C and D, RD20 decreases the accumulation of ROS. H2O2 (C) and superoxide radical (D) contents of leaves of transgenic lines and control plants were measured by DAB and NBT staining, respectively. ROS accumulation was estimated by the ratio of stained surface versus total leaf surface using ImageJ (n = 30). Differences between transgenic lines and their respective controls are highly significant with P < 0.01 by Student’s t test. Bars = 1 mm.
To examine whether peroxygenase activity was necessary to realize the protective role of RD20, the two lines overexpressing inactive and active RD20 as well control plants were grown in the presence of the synthetic auxin 1-naphthaleneacetic acid (NAA). Control plants developed green cotyledons and several adventitious roots, characteristic of NAA action (Fig. 9A). Both RD20-OE lines also developed green cotyledons but lacked additional roots, indicating that the presence of enzymatically active RD20 protected Arabidopsis against NAA effects (Fig. 9A). In sharp contrast, the H133A mutation in RD20-overexpressing lines (see above) led to the development of small white/yellowish seedlings that resulted from the deleterious effects of NAA acting as an herbicide (Fig. 9A). This suggested that abolishing peroxygenase activity in RD20-overexpressing plants impaired the protective function of RD20. Anthocyanin levels have recently been used as valuable markers to assess plant stress responses to environmental conditions (Izumi et al., 2009). Analysis of the red pigment content of the four lines overexpressing either active or inactive RD20, grown in the presence of Paraquat, Rose Bengal, and 2,4-dichlorophenoxyacetic acid, suggested reduced stress levels in RD20-OE lines compared with controls (Fig. 9B). In contrast, overexpression of enzymatically inactive RD20 led to higher degrees of stress in RD20H133A-OE plants (Fig. 9B). Taken together, these results underline that peroxygenase activity is required for RD20-mediated protection against oxidative stress in planta.
Figure 9.
Peroxygenase activity is a prerequisite for the protective function of RD20. A, Seedlings of control lines and lines overexpressing active or catalytically impaired enzyme were grown on NAA. Bars = 1 mm. B, The degree of stress of such lines was estimated by measuring their anthocyanin contents after 1 week of growth in the presence of ROS inducers (n = 10 in four independent experiments; **P < 0.01 and *P < 0.05 when analyzed by Student’s t test). fw, Fresh weight. C, qRT-PCR was used to access the expression of RD20 and ERF1 in leaves of wild-type plants and ctr1-1 and ein2-1 ethylene mutants treated with Paraquat. The results shown represent means ± se of at least four biological replicates (*P ≤ 0.05 and **P < 0.01 when analyzed by the Student-Newman-Keuls test).
RD20 Is Positively Regulated by Ethylene during Stress
Oxylipins such 9-HOT may exert protective roles through antagonistic action with ethylene. While the ethylene-overproducing eto1 mutant fails to survive under oxidative stress (López et al., 2011), the constitutive ethylene response mutant ctr1 is resistant to high salinity (Achard et al., 2006), and the ethylene-insensitive ein2 mutant is vulnerable to many stresses, including Paraquat-induced oxygen radicals (Alonso et al., 1999; Silverman et al., 2005). Thus, there is an apparent discrepancy between ethylene-overproducing and ethylene response mutants. Because eto1 failed to enhance RD20 expression under oxidative stress (López et al., 2011), we hypothesized that the opposing stress tolerance phenotypes of ethylene response are due to the presence of RD20 that generates FAOHs such as 9-HOT. To test this hypothesis, we analyzed by qRT-PCR the expression of RD20 in ctr1-1 and ein2-1 mutants after treatment with 5 µm Paraquat or water. In addition, the expression of the ethylene response factor ERF1 was analyzed, since a recent report highlighted its critical function during stress responses (Cheng et al., 2013). When sprayed with water, ctr1 accumulated RD20 transcripts to significantly lower levels as compared with Col-0, while increased levels of RD20 transcripts were found in ein2 (Fig. 9C). When sprayed with Paraquat, a significant induction of both RD20 and ERF1 expression was observed in Col-0 seedlings compared with those sprayed with water (Fig. 9C). In contrast, the expression of both genes in the ein2 mutant was largely reduced after treatment with Paraquat as compared with unstressed conditions (Fig. 9C). In ctr1 mutants, oxidative stress minimally affected EFR1 expression but significantly induced RD20 expression when compared with water treatment (Fig. 9C). Such data suggest that the induction of RD20 expression is positively regulated by Paraquat in ethylene perception mutants.
Copper-Induced Oxidative Stress Alters the Floral Transition in RD20 Transgenic Lines
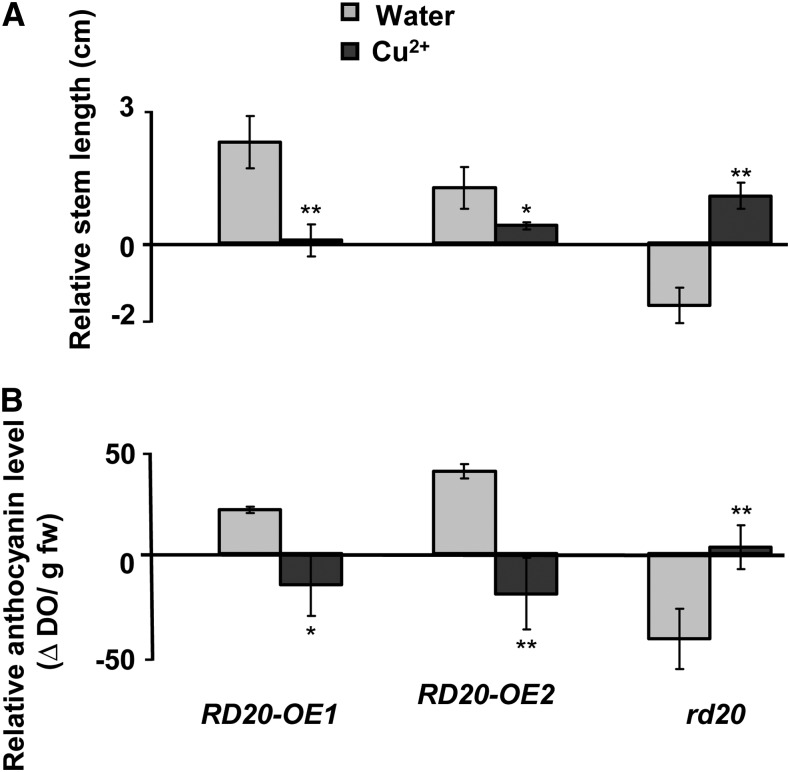
Our results indicated that the enzymatic activity of RD20 was involved in the floral transition and protection against oxidative stress. To investigate the interplay between these two roles, we examined the growth of transgenic plants in the presence of high copper concentrations known to trigger oxidative stress. Whole plants displayed reduced development under these conditions. However, independently of the photoperiod duration, inflorescence stems of stressed RD20-OE1 and RD20-OE2 lines decreased in length by 53% and 47%, respectively, compared with controls, indicating that their growth was delayed by stress (Fig. 10A). The opposite effect was apparent in rd20 mutants, where copper treatment led to longer inflorescence stems compared with the wild type, suggesting an earlier flowering of this mutant line (Fig. 10A). Noteworthy, rd20 mutants contained less anthocyanins than the wild-type plants in the absence of Cu2+. In contrast, the anthocyanin content of rd20 leaves in the presence of the metal increased up to those observed in wild-type plants (Fig. 10B). These results suggested that loss of RD20 increased sensitivity to copper and that rd20 may escape stress by entering the floral stage early. In contrast, in the absence of copper, more anthocyanins were present in the leaves of RD20-OE1 and RD20-OE2 lines than in those of the controls (Fig. 10B), whereas in the presence of copper, both RD20-OE1 and RD20-OE2 lines exhibited significantly lower anthocyanin amounts than those of their controls. Together, these data suggested that RD20-overexpressing lines were less affected by copper than their controls, extending vegetative growth to favor defense mechanisms over early induction of reproduction.
Figure 10.
Copper affects the flowering and stress tolerance of RD20-overexpressing and knock-out lines compared with their respective controls. A, Growth was estimated by recording the relative stem length of the transgenic lines versus their respective controls after 7 weeks of growth in the presence of 485 µg Cu2+ g−1 soil. B, The degree of stress of such lines was estimated by measuring their relative anthocyanin contents versus their respective controls. All experiments were repeated three times; data are means ± se. Treatment with copper was statistically different from water-treated controls when analyzed by Student’s t test (*P < 0.05, **P < 0.01). fw, Fresh weight.
DISCUSSION
RD20 encodes a peroxygenase that uses FAOOHs as substrates; thus, at least one member of the small family of caleosins is able to reduce this type of endogenous oxylipins. Beyond or instead of being structural proteins of lipid droplets, caleosins are endowed with distinctive peroxygenase activities, allowing them to produce various oxygenated compounds (Hanano et al., 2006; Blée et al., 2012; this article). Our data show that the peroxygenase activity of RD20 is necessary for the diverse biological functions implied by our reverse genetics analyses and highlight the long neglected importance of these enzymes in plant development and stress response.
RD20 Is Involved in Regulating Plant Development
We show here that changes in either RD20 expression or RD20 catalytic activity lead to alterations of GA-controlled floral transition, seed dormancy, and sensitivity to exogenous ABA. From these observations, a question is immediately raised: how could the catalytic activity of RD20 bring about such pleiotropic effects? A possible answer is that the reductase activity of RD20 might alter the accumulation of JA. When RD20 is overexpressed, 13-HPOT, which is a precursor of JA, might be channeled away from the JA biosynthetic pathway to be converted into 13-HOT, thus leading to reduced JA levels. JA has been reported to antagonize the GA signaling pathway (Yang et al., 2012) and GA biosynthesis by inhibiting the accumulation of Ga20ox and Ga3ox transcripts (Heinrich et al., 2013). If this occurs in lines overexpressing RD20, reduced JA accumulation could result in early flowering of these lines. However, we did not detect any alteration in JA content in the leaves of transgenic plants, in accordance with the absence of an accumulation of the transcripts of the biosynthetic genes GA20ox and GA3ox. Thus, the production of oxylipins and not the decrease in JA accumulation seems to be the origin of phenotypes that result from the overexpression of RD20 Another possible explanation is that RD20-generated oxylipins positively influence GA signaling. Such an assumption would be consistent with the delay of flowering of the rd20 mutant as well as its slower seed germination compared with wild-type plants. However, this hypothesis could hardly explain the rapid break of dormancy of the rd20 mutant seeds or their insensitivity to exogenous ABA during germination. Alternatively, oxylipins formed by RD20 catalysis might antagonize ethylene, as recently demonstrated for 9-HOT (López et al., 2011). This oxylipin in particular has been found to trigger ROS accumulation. Therefore, a hypothetic antagonism between ethylene and FAOHs generated by RD20 would lead to ROS accumulation in RD20-overexpressing lines and, inversely, to a reduced quantity of ROS in rd20. That is what we observed when superoxide radical and H2O2 levels were estimated. In agreement with the proposed antagonism between ethylene and oxylipins is the accumulation of RD20 transcripts in the ethylene-insensitive mutant ein2. Consistently, this contrasts with the low expression of the caleosin gene in the ethylene constitutive response mutant ctr1 (Fig. 9C).
Furthermore, this hypothesis predicts that the rd20 mutant, impaired in the production of oxylipins, would be more sensitive to ethylene perception and/or signaling. Ethylene has been reported to delay flowering by inhibiting GA signaling (Achard et al., 2007). In agreement, rd20 mutants flowered late compared with controls, similar to plants treated with ethylene. In sharp contrast, the RD20-OE1 and RD20-OE2 lines, by generating high amounts of oxylipins, would be expected to be less sensitive to ethylene. Indeed, these mutants flowered earlier than their control counterparts. Ethylene is also known as a negative regulator of the ABA response during seed germination (Beaudoin et al., 2000; Ghassemian et al., 2000). In agreement with an antagonism between ethylene and oxylipins, lines overexpressing RD20 showed an increased responsiveness to ABA similar to ethylene-insensitive mutants. Both fresh seeds of RD20-overexpressing and ein2 lines germinated later than their respective controls. Similarly, the germination of seeds of RD20-OE1 and RD20-OE2 lines was hypersensitive to exogenous ABA, as was that of ein2 seeds. Together, all these results support the hypothesis that oxylipins generated by RD20 interact with ethylene to promote GA-dependent flowering, ABA sensitivity, and ROS accumulation. They also suggest that, among the oxylipins produced by RD20, FAOHs are the active molecules. 13-HOT appears to be a major factor in the regulation of GA-dependent flowering. The absence of other FAOHs in oxylipin profiling, such as 12-hydroxy-9,12,15-octadecatrienoic acid or 16-hydroxy-9,12,15-octadecatrienoic acid, which would be characteristic of a chemical lipid oxidation pathway, suggests that active 13-HOT is of enzymatic origin. In accordance, an increase of chloroplastic 13-LOX activity has been shown to occur at the developmental transition to flowering (Ye et al., 2000; Bañuelos et al., 2008). Moreover, 13-LOX stimulation was also associated with ROS increases, mainly due to a decline in ascorbate peroxidase activity acting as an enzymatic H2O2 scavenger (Ye et al., 2000; Chai et al., 2012).
RD20 Confers Stress Tolerance
Our results are in agreement with an antagonism between FAOHs and ethylene during development, leading to an accumulation of ROS. However, ROS levels were strongly reduced in Paraquat-treated plants overexpressing RD20 compared with controls, suggesting that an antagonism between FAOH and ethylene is unlikely to be relevant during defense responses. In support of this conclusion, lines overexpressing RD20, presumably less sensitive to ethylene in the scenario of an FAOH and ethylene antagonism, are remarkably resistant to Paraquat, while the ethylene-insensitive ein2 mutant was susceptible to the herbicide (Alonso et al., 1999). Actually, tolerance to Paraquat appears to be linked to the presence of RD20 rather than to ethylene perception, as shown by the analysis of the accumulation of RD20 transcripts (Fig. 9C). This observation, however, is not in agreement with a recent report showing that 9-HOT counteracts ethylene responses under oxidative stress (López et al., 2011). The latter conclusion was mainly supported by the inability of the ethylene-overproducing mutant eto1 to survive Rose Bengal treatment. Since ethylene was shown to induce defense genes, including RD20, through the activation of EFR1 in response to harmful environmental conditions (Cheng et al., 2013), one possible explanation resides in the apparent lack of stimulation of ERF1 expression in eto1 by superoxide radical (López et al., 2011), which might impair RD20 expression and subsequent defense responses. Alternatively, we cannot exclude that ethylene biosynthesis and not its perception is determinant for its interplay with FAOHs.
Both the 13- and 9-LOX pathways were induced by Paraquat. Sequential up-regulation of these pathways was also reported under high-light conditions and after severe cadmium treatment (Montillet et al., 2004). Among the oxylipins formed by these pathways, 13-LOX-derived FAOHs (in particular 13-HOT) accumulate after Paraquat treatment. Similarly, 13-HOT has been found to accumulate during the initial stress responses to the release of singlet oxygen. This oxylipin was postulated to act as a second messenger in ROS signaling (op den Camp et al., 2003). A putative role of caleosin in ROS signaling is further supported by the recent report of the binding of a wheat (Triticum aestivum) RD20 homolog to G-proteins (Khalil et al., 2011) that are involved in signaling mediated by ROS (Joo et al., 2005). The inability of the rd20 mutant to enhance the production of 13-LOX-derived FAOHs rendered this mutant sensitive to Paraquat. Similarly, the absence of the 9-LOX pathway in the lox1 lox5 mutant made this mutant particularly vulnerable to Rose Bengal (López et al., 2011). Thus, it seems that tolerance to oxidative stress requires the production of oxylipins derived from both the 13- and 9-LOX pathways. Additionally, α-dioxygenases might contribute to FAOOH accumulation and stress tolerance. In support of this hypothesis is the induction of α-DOX1 by salt stress (Tirajoh et al., 2005; Aung, 2009) and the high sensitivity to Paraquat and salt exhibited by α-dox1 mutants (Ponce de León et al., 2002; Aung, 2009). Of note, roots of these mutants were reported to accumulate H2O2 under salt treatment (Aung, 2009). Thus, at a given time, the composition of the FAOOH mixture would depend on the induction of lipid peroxidation and/or on the expression/activity of the different biosynthetic enzymes that vary based on the nature of the stress (e.g. photooxidation, heavy metals, or salt) and on the fatty acid composition of the organ examined. In this context, RD20 plays a determinant role by generating FAOHs that affect the oxidative status. From our data and the points discussed here, we propose a model according to which RD20 produces oxylipins that alleviate stress through tolerance mechanisms that include ROS reduction and, thereby, allow the delay of flowering. In consequence, the rd20 mutant, depleted in FAOHs, is more vulnerable to stress and in response activates the reproductive stage early to escape stress. Our results clearly highlight the importance of the peroxygenase catalytic activity of RD20 in the regulation of ROS accumulation during the floral transition and stress tolerance and open new perspectives for caleosin function.
MATERIALS AND METHODS
The Arabidopsis (Arabidopsis thaliana) ecotypes Col-0 and Ws were used for this study. Generally, seeds were sown on a standard soil compost mixture, and seedlings were grown individually in growth chambers under white fluorescent lamps (60 µmol m−2 s−1. The temperature was 22°C during the day (12 h) and 19°C during the night (12 h). To study the effect of the photoperiod on flowering, young seedlings (about 1 cm in height) were placed in growth chambers with similar controlled temperature and humidity under either short (8 h) or long (16 h) light conditions. Copper toxicity studies were performed with transgenic and control lines that were transferred as young seedlings (about 1 cm in height) into soil containing pentahydrated CuSO4 (485 µg g−1 soil). To study the effect of oxidative stress, seeds were sterilized using the vapor-phase sterilization protocol. To that end, seeds were placed in open 1.5-mL tubes under a bell jar containing a beaker with 20 mL of bleach (sodium hypochlorite solution). Two milliliters of fuming hydrochloric acid (37% v/v) was added to the bleach, and seeds were sterilized for 4 h. Sterilized seeds were sown on 2.2 g L−1 Murashige and Skoog basal medium (Sigma-Aldrich) with 7% (w/v) agar and 15 g L−1 Suc at pH 5.7 supplemented with 0.5 µm Paraquat, 20 µm Rose Bengal, or 0.1 µm 2,4-dichlorophenoxyacetic acid. After 2 d of stratification at 4°C, plants were grown under continuous light, and 7-d-old leaves were analyzed for anthocyanin contents. Alternatively, plants were grown at 22°C during the day (16 h) under 70 to 90 µmol m−2 s−1 light and 20°C at night (8 h) without the presence of superoxide inducers. Two-week-old seedlings were then sprayed with water or Paraquat solutions (5 µm in water). At 12 h after this treatment, green leaves were counted, and after an additional 12 h, the leaves were harvested and immediately frozen in liquid nitrogen for quantitative PCR analysis. ROS status, number of green leaves, and cell death were analyzed on 7-d-old plants after 24 h of treatment with 5 µm Paraquat or water. Importantly, to ensure the reproducibility of our results, we compared plants sown and grown side by side under identical environmental conditions. To test germination, seeds were rapidly washed with ethanol containing 0.01% Tween 20 (v/v) and then surface sterilized with 40% (v/v) bleach and washed five times with distilled water. Sterile seeds were plated on freshly prepared Murashige and Skoog medium supplemented with 1.5% (w/v) Suc and ABA, or FAOHs, as needed. FAOHs were purchased from Cayman Chemical (SPI-BIO), and their purity was controlled by HPLC. Freshly made plates were stratified in darkness for 2 to 4 d at 4°C and then transferred to a tissue culture room at 23°C under 4,000 lx light intensity with a 16-h-light period. Seeds were considered as germinated when radicles completely penetrated the seed coats. Postemergence growth was estimated by counting green cotyledons. Of note, the transgenic plants and their respective controls underwent two consecutive reproductive developmental cycles in a similar environment before their seeds were collected. Seeds were stored under identical conditions for at least 1 month before the experiments. In contrast, seeds used to study the break of dormancy were used immediately after harvesting and analyzed for germination without the stratification step.
Generation of RD20-Overexpressing Plants
To obtain overexpressing plants, RD20 full-length complementary DNA (cDNA) was PCR amplified (primers 1 and 2; Supplemental Table S1) and transferred to the binary vector pB7WG2 using Gateway technology procedures (Invitrogen, Life Technologies). Plasmids containing RD20 or the empty vector were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. Transgenic Arabidopsis plants were generated by the floral dip method (Clough and Bent, 1998) and screened to homozygosity with the herbicide phosphinothricin (Basta; 0.3 g L−1). To generate 35S:RD20H133A lines, the residue His-133 was mutated to Val using the QuickChange site-directed mutagenesis kit of Stratagene (Agilent Technologies) with the sense mutations primer called H133 in Supplemental Table S1 (the modified codon is underlined, and the nucleotide changed is indicated in boldface). The mutated gene was transferred to the vector pB7WG2, and the resulting plasmid was used to transform A. tumefaciens and wild-type Col-0 Arabidopsis.
T-DNA Insertion Mutant and Complementation
rd20 was discovered in the Arabidopsis thaliana Integrated Database, and seeds were obtained from the Arabidopsis thaliana Resource Centre for Genomics (line DSA78, T-DNA_LB.FLAG.237F07; http://www.ijpb.versailles.inra.fr). Primers 3, 4, and 5 (Supplemental Table S1) were used to screen for homozygous mutants. For complementation of the rd20 mutant, we used Gateway technology procedures to transfer the RD20 gene from the vector pB7WG2 to the binary pH2GW7 vector to gain hygromycin resistance. The latter vector was used to transform Ws wild-type Arabidopsis via A. tumefaciens transformation and floral dipping.
Protein Expression and Purification
Full-length RD20 cDNA was PCR amplified using primers 6 and 7 (Supplemental Table S1). The amplified product after sequencing verification was transferred to the yeast constitutive vector pVT102U (Vernet et al., 1987) in the BamHI and XbaI sites. The expression of the recombinant FLAG-tagged RD20 in Saccharomyces cerevisiae Wa6 was conducted as described for other peroxygenases (Hanano et al., 2006), and its purification on anti-FLAG M2 affinity gel was mostly according to the manufacturer’s procedures (Sigma-Aldrich), except that each step was realized in the presence of detergent (0.1% CHAPS). Western-blot analysis was performed as reported previously (Hanano et al., 2006) by using a monoclonal anti-FLAG M2 antibody produced in mouse (Sigma-Aldrich).
Enzymatic Activity of RD20
Aniline hydroxylation in the presence of a hydroperoxide cosubstrate produces N-phenylhydroxylamine that is spontaneously transformed into nitrobenzene. The absorbance of nitrobenzene was followed at 310 nm in a spectrometric cuvette containing yeast extract or microsomes expressing RD20 or RD20 purified fraction in 0.1 m KH2PO4 (pH 7.4) containing 1 mm aniline. The reaction was initiated by adding 1 mm cumene hydroperoxide (total volume, 1 mL). Epoxidase activities supported by peroxygenase were assayed as described before (Blée and Durst, 1987; Blée and Schuber, 1989, 1990). Hydroperoxide reductase activity of purified RD20 was measured using radiolabeled FAOOHs. [1-14C]13-HPOD and [1-14C]13-HPOT were synthesized enzymatically from [1-14C]linoleic acid (50 Ci mol−1) and [1-14C]linolenic acid (50 Ci mol−1; PerkinElmer Life Sciences) in the presence of soybean (Glycine max) lipoxygenase (Sigma-Aldrich). Potato (Solanum tuberosum) lipoxygenase (Cayman Chemical, SPI-BIO) was used to obtain 9-HPOT from linolenic acid as described (Galliard and Phillips, 1971). [1-14C]FAOOHs were incubated with purified RD20 (5 µg of protein) in 500 µL of 0.1 m sodium acetate (pH 5.5) for 2 h or overnight at 27°C. The reaction was stopped by adding two drops of 4 n HCl, and the products were extracted by 3× 2 mL of a mixture of dichloromethane:ether (1:1, v/v). The organic phase was evaporated under argon, and the residue was dissolved into 25 µL of acetonitrile. Metabolites of FAOOHs were separated by HPLC on a Lichrospher 100 RP-18 (5 µm) column (Interchim) using a mixture of acetonitrile:water:acetic acid (50:50:0.1, v/v/v) as solvent (0.5 mL min−1). Radioactivity was analyzed with a 500 TR Radiomatic-Flo-one detector (Packard Instrument, PerkinElmer). Identification of the metabolites of FAOOHs was performed using a GC-MS spectrometer (Agilent 5973N) with an ionizing energy of 70 eV. The sample was injected directly in splitless mode (injector temperature of 250°C) into a DB-5-coated fused column (30 m, 0.25 mm i.d.) with a temperature program of 6°C min−1 from 60°C to 100°C followed by 3°C min−1 from 100°C to 300°C (mass-to-charge ratio = 50–700).
RNA Analysis
Total RNA was isolated from pooled leaf tissues using the Nucleospin RNA plant kit (Macherey-Nagel). cDNA was synthesized from total RNA using SuperScript III (Invitrogen, Life Technologies) with random hexamer primers according to the manufacturer’s instructions. qRT-PCR plates were prepared with a Biomek 3000 (Beckman Coulter) and run on a Light Cycler 480 II (Roche). Each reaction was prepared using 2 µL of cDNA, 5 µL of Light Cycler 480 SYBR Green I Master (Roche), and 250 nm of forward and reverse primers in a total volume of 10 µL. The amplification profile consisted of 95°C for 10 min and 40 cycles (95°C denaturation for 10 s, annealing at 60°C for 15 s, and extension at 72°C for 15 s), followed by melting curve analysis from 55°C to 95°C to check the specificity of transcripts. All reactions were performed in triplicate. Primers used for all the quantitative PCRs performed in this study are listed in Supplemental Table S2. Vacuolar protein Monensin Sensitivity1 (At2g28390), TAP42 Interacting Protein1-like (At4g34270), and the uncharacterized conserved protein UCP022280 (At4g26410) were taken as reference genes to normalize the expression of genes of interest.
Anthocyanin Determination
The pigments were extracted and their content in leaves was estimated by spectrophotometry essentially as described (Rabino and Mancinelli, 1986). Plant tissues (about 9 mg) in 300 µL of acidic methanol (containing 1% [w/v] HCl) were rapidly frozen in liquid nitrogen and ground (2× 30 s) in the presence of two steel balls and left for 48 h at 4°C in the dark to extract the pigments. The absorbance of the methanol layer was measured on a Shimadzu MPS-2000 spectrophotometer at wavelengths of 530 and 657 nm. The formula A530 − 0.25 × A657 was used to compensate for the contribution of chlorophyll and its degradation products to the absorption at 530 nm.
Oxylipin Profiling
For analysis of free oxylipins, 0.8 to 2.1 g of frozen plant material was extracted as described previously (Göbel et al., 2002) with some modifications. After adding 20 mL of extraction medium (3:2 [v/v] n-hexane:2-propanol with 0.0025% [w/v] butylated hydroxytoluene) containing 13-hydroperoxy-6,9,11-octadecatrienoic acid, plant material was immediately homogenized with an Ultra Turrax homogenizer under streaming argon on ice for 30 s. The extract was shaken for 10 min and centrifuged at 3,200g at 4°C for 10 min. The upper phase was collected, and a 6.7% (w/v) solution of potassium sulfate was added to a volume of 32.5 mL. After vigorous shaking and centrifugation at 3,200g at 4°C for 10 min, the upper hexane-rich layer was subsequently dried under streaming nitrogen. The remaining lipids were redissolved in methanol:water:acetic acid (75:25:0.1, v/v/v). Further analysis was carried out on an Agilent 1100 HPLC system coupled to a diode array detector. At first, oxylipins were purified by reverse-phase HPLC on an ET250/2 Nucleosil 120-5 C18 column (2.1 × 250 mm, 5 µm particle size; Macherey-Nagel), with a solvent system of methanol:water:acetic acid (85:15:0.1, v/v/v) and a flow rate of 0.18 mL min−1. For the detection of FAOHs and FAOOHs, A234 indicating the conjugated diene system was recorded. For the quantification of FAOOHs and FAOHs, straight-phase HPLC was carried out on a Zorbax Rx-SIL column (2.1 × 150 mm, 5 µm particle size; Agilent) with a solvent system of n-hexane:2-propanol:trifluoroacetic acid (100:1:0.02, v/v/v) and a flow rate of 0.2 mL min−1. Free oxylipins were quantified using 13-hydroperoxy-6,9,11-octadecatrienoic acid as an internal standard to determine the recovery of FAOHs and FAOOHs. Calibration curves (five-point measurements) for 13-HOD and 13-HOT were established.
Characterization and Quantification of JA
Characterization and quantification of JA were performed by comparing retention times and mass spectrometry and tandem mass spectrometry transition analyses using an ultra-performance liquid chromatograph coupled to a tandem mass spectrometer. All analyses were performed using a Waters Quattro Premier XE equipped with an electrospray ionization source and coupled to an Acquity ultra-performance liquid chromatography system (Waters). Chromatographic separation was achieved using the Acquity UPLC BEH C18 column (100 × 2.1 mm, 1.7 µm; Waters) coupled to the Acquity UPLC BEH C18 precolumn (2.1 × 5 mm, 1.7 µm; Waters). The mobile phase consisted of a mixture of water (99.9%; acidified with 0.1% formic acid [A]) and methanol (99.9%; acidified with 0.1% formic acid [B]). The following gradient was used: 95% solvent A and 5% solvent B (1 min), linear gradient of 95% A to 100% B (10 min), 100% B was maintained during 2 min, and then during 2 min the gradient was set to the initial condition. The total run time was 15 min. The column was operated at 35°C with a flow rate of 0.35 mL min−1 (sample injection volume of 3 µL). Nitrogen generated from pressurized air in an N2G nitrogen generator (Mistral) was used as the drying and nebulizing gas. The nebulizer gas flow was set to approximately 50 L h−1 and the desolvation gas flow to 900 L h−1. The interface temperature was set at 400°C and the source temperature at 135°C. The capillary voltage was set at 3.2 kV and the cone voltage at 25 V. The ionization mode (positive and negative) was adjusted by using commercially available standard molecules. Low-mass and high-mass resolution were 13 for both mass analyzers, ion energies 1 and 2 were 0.5 V, entrance and exit potentials were 2 and 1 V, and detector (multiplier) gain was 650 V. Collision-induced dissociation of protonated or deprotonated parent ions was accomplished with a collision energy of 10 V. Daughter scan monitoring permitted us to identify for JA the transition from the parent ion. The combination of chromatographic retention time, parent mass, and unique fragment ion analysis was used to selectively monitor JA (209 > 59). JA quantification was obtained by injecting different concentrations (ng mL−1 to µg mL−1) of commercially available standard phytohormone. The peak area of each peak obtained after ultra-performance liquid chromatography-tandem mass spectrometry analysis was used for the establishment of calibration curves. Data acquisition and analysis were performed with the MassLynx software (version 4.1) running under Windows XP Professional on a Pentium personal computer.
ROS Content and Cell Death Estimations
Detection of H2O2 by DAB staining and superoxide radical by NBT staining were performed by standard methods as described (Thordal-Christensen et al., 1997). Images were obtained with a Leica macrofluo Z16 APO equipped with a Leica DFC camera (Leica Microsystems). Quantification of the staining was performed with ImageJ (Schneider et al., 2012) in arbitrary units. Leaves were stained with Trypan Blue to visualize cell death, according to Koch and Slusarenko (1990).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression analysis of the RD20 caleosin/peroxygenase gene in Arabidopsis.
Supplemental Figure S2. Coexpression analysis of RD20.
Supplemental Figure S3. Mass spectrum of the methyl trimethylsilyl ester of 15,16-epoxy,13-hydroxy-9,11-octadecenoic acid.
Supplemental Figure S4. Inhibition of peroxygenase activity by mutation of the catalytic residue His-133.
Supplemental Figure S5. Deletion of RD20-modified seed germination and ABA-dependent germination.
Supplemental Figure S6. Free oxylipin analysis of rd20 and RD20-OE leaves sprayed with water or with 5 µm parathion.
Supplemental Table S1. Primers used for the generation of transgenic plants and expression in yeast.
Supplemental Table S2. Primers used for reverse transcription-PCR analysis.
Acknowledgments
We thank the following colleagues from the Institut de Biologie Moléculaire des Plantes for support: Martine Flenet for molecular cloning; Vincent Compagnon for primary reverse transcription-PCR analysis; Martha Ramel, Michel Kerneis, and Sébastien Staerck for plant production; Jérôme Mutterer and Mathieu Erhardt for microscopy; Alain Rahier for GC-MS technique; and Patrick Achard for stimulating discussions. We thank Sabine Freitag from the Albrecht-von-Haller Institute for technical help with oxylipin analysis and also Francis Schuber from School of Pharmacy, University of Strasbourg, Illkirch, France for critical reading of the article.
Glossary
- FAOH
fatty acid hydroxide
- 2-HOT
2-hydroxy-9,12,15-octadecatrienoic acid
- 9-HOT
9-hydroxy-10,12,15-octadecatrienoic acid
- ABA
abscisic acid
- ROS
reactive oxygen species
- FAOOH
fatty acid hydroperoxide
- 13-HOT
13-hydroxy-9,11,15-octadecatrienoic acid
- 13-HPOD
13-hydroperoxy-9,11-octadecadienoic acid
- H2O2
hydrogen peroxide
- GC-MS
gas chromatography-mass spectrometry
- 13-HOD
13-hydroxy-octadeca-9,11-dienoic acid
- T-DNA
transfer DNA
- Ws
Wassilewskija
- Col-0
Columbia-0
- qRT
quantitative reverse transcription
- NBT
nitroblue tetrazolium
- DAB
3,3′-diaminobenzidine
- JA
jasmonate
- NAA
1-naphthaleneacetic acid
- cDNA
complementary DNA
Footnotes
This work was supported by the Centre National de la Recherche Scientifique and the French Ministry for Research.
The online version of this article contains Web-only data.
References
- Achard P, Baghour M, Chapple A, Hedden P, Van Der Straeten D, Genschik P, Moritz T, Harberd NP. (2007) The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc Natl Acad Sci USA 104: 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Hirayama T, Roman G, Nourizadeh S, Ecker JR. (1999) EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284: 2148–2152 [DOI] [PubMed] [Google Scholar]
- Aubert Y, Vile D, Pervent M, Aldon D, Ranty B, Simonneau T, Vavasseur A, Galaud JP. (2010) RD20, a stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol 51: 1975–1987 [DOI] [PubMed] [Google Scholar]
- Aung TST (2009) Regulation of α-dioxygenase expression and functional analysis in salt-stressed Arabidopsis thaliana. PhD thesis. Simon Fraser University, Burnaby, Canada [Google Scholar]
- Baker SS, Wilhelm KS, Thomashow MF. (1994) The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol 24: 701–713 [DOI] [PubMed] [Google Scholar]
- Bañuelos GR, Argumedo R, Patel K, Ng V, Zhou F, Vellanoweth RL. (2008) The developmental transition to flowering in Arabidopsis is associated with an increase in leaf chloroplastic lipoxygenase activity. Plant Sci 174: 366–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin N, Serizet C, Gosti F, Giraudat J. (2000) Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12: 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blée E. (1998) Phytooxylipins and plant defense reactions. Prog Lipid Res 37: 33–72 [DOI] [PubMed] [Google Scholar]
- Blée E, Durst F. (1987) Hydroperoxide-dependent sulfoxidation catalyzed by soybean microsomes. Arch Biochem Biophys 254: 43–52 [DOI] [PubMed] [Google Scholar]
- Blée E, Flenet M, Boachon B, Fauconnier ML. (2012) A non-canonical caleosin from Arabidopsis efficiently epoxidizes physiological unsaturated fatty acids with complete stereoselectivity. FEBS J 279: 3981–3995 [DOI] [PubMed] [Google Scholar]
- Blée E, Schuber F. (1989) Mechanism of S-oxidation reactions catalyzed by a soybean hydroperoxide-dependent oxygenase. Biochemistry 28: 4962–4967 [Google Scholar]
- Blée E, Schuber F. (1990) Efficient epoxidation of unsaturated fatty acids by a hydroperoxide-dependent oxygenase. J Biol Chem 265: 12887–12894 [PubMed] [Google Scholar]
- Blée E, Wilcox AL, Marnett LJ, Schuber F. (1993) Mechanism of reaction of fatty acid hydroperoxides with soybean peroxygenase. J Biol Chem 268: 1708–1715 [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV. (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16: 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai L, Wang JM, Fan ZY, Lio ZB, Wen GQ, Li XF, Yang Y. (2012) Regulation of the flowering time of Arabidopsis thaliana by thylakoid ascorbate peroxidase. Afr J Biotechnol 11: 7151–7157 [Google Scholar]
- Chen JCF, Tsai CC, Tzen JT. (1999) Cloning and secondary structure analysis of caleosin, a unique calcium-binding protein in oil bodies of plant seeds. Plant Cell Physiol 40: 1079–1086 [DOI] [PubMed] [Google Scholar]
- Cheng MC, Liao PM, Kuo WW, Lin TP. (2013) The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol 162: 1566–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Koornneef M. (2000) Gibberellin requirement for Arabidopsis thaliana seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol 122: 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J, Sauveplane V, Olry A, Ginglinger JF, Provart NJ, Werck-Reichhart D. (2008) An extensive (co-)expression analysis tool for the cytochrome P450 superfamily in Arabidopsis thaliana. BMC Plant Biol 8: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner I, Wasternack C. (2002) The lipoxygenase pathway. Annu Rev Plant Biol 53: 275–297 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Maruyama K, Seki M, Hiratsu K, Ohme-Takagi M, Tran LS, Yamaguchi-Shinozaki K, Shinozaki K. (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J 39: 863–876 [DOI] [PubMed] [Google Scholar]
- Galliard T, Phillips DR. (1971) Lipoxygenase from potato tubers: partial purification and properties of an enzyme that specifically oxygenates the 9-position of linoleic acid. Biochem J 124: 431–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaquerel E, Steppuhn A, Baldwin IT. (2012) Nicotiana attenuata α-DIOXYGENASE1 through its production of 2-hydroxylinolenic acid is required for intact plant defense expression against attack from Manduca sexta larvae. New Phytol 196: 574–585 [DOI] [PubMed] [Google Scholar]
- Gechev TS, Van Breusegem F, Stone JM, Denev I, Laloi C. (2006) Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. BioEssays 28: 1091–1101 [DOI] [PubMed] [Google Scholar]
- Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P. (2000) Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12: 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göbel C, Feussner I, Hamberg M, Rosahl S. (2002) Oxylipin profiling in pathogen-infected potato leaves. Biochim Biophys Acta 1584: 55–64 [DOI] [PubMed] [Google Scholar]
- Hamberg M. (1991) Regio- and stereochemical analysis of trihydroxyoctadecenoic acids derived from linoleic acid 9- and 13-hydroperoxides. Lipids 26: 407–415 [DOI] [PubMed] [Google Scholar]
- Hamberg M, Hamberg G. (1990) Hydroperoxide-dependent epoxidation of unsaturated fatty acids in the broad bean (Vicia faba L.). Arch Biochem Biophys 283: 409–416 [DOI] [PubMed] [Google Scholar]
- Hamberg M, Sanz A, Castresana C. (1999) Alpha-oxidation of fatty acids in higher plants: identification of a pathogen-inducible oxygenase (piox) as an alpha-dioxygenase and biosynthesis of 2-hydroperoxylinolenic acid. J Biol Chem 274: 24503–24513 [DOI] [PubMed] [Google Scholar]
- Hamberg M, Sanz A, Rodriguez MJ, Calvo AP, Castresana C. (2003) Activation of the fatty acid alpha-dioxygenase pathway during bacterial infection of tobacco leaves: formation of oxylipins protecting against cell death. J Biol Chem 278: 51796–51805 [DOI] [PubMed] [Google Scholar]
- Hanano A, Burcklen M, Flenet M, Ivancich A, Louwagie M, Garin J, Blée E. (2006) Plant seed peroxygenase is an original heme-oxygenase with an EF-hand calcium binding motif. J Biol Chem 281: 33140–33151 [DOI] [PubMed] [Google Scholar]
- Heinrich M, Hettenhausen C, Lange T, Wünsche H, Fang J, Baldwin IT, Wu J. (2013) High levels of jasmonic acid antagonize the biosynthesis of gibberellins and inhibit the growth of Nicotiana attenuata stems. Plant J 73: 591–606 [DOI] [PubMed] [Google Scholar]
- Hernandez-Pinzon I, Patel K, Murphy DJ. (2001) The Brassica napus calcium-binding protein caleosin has distinct endoplasmic reticulum- and lipid body-associated isoforms. Plant Physiol Biochem 39: 615–622 [Google Scholar]
- Hwang IS, Hwang BK. (2010) The pepper 9-lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens. Plant Physiol 152: 948–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru A, Yamazaki I. (1977) Hydroperoxide-dependent hydroxylation involving “H2O2-reducible hemoprotein” in microsomes of pea seeds: a new type enzyme acting on hydroperoxide and a physiological role of seed lipoxygenase. J Biol Chem 252: 6118–6124 [PubMed] [Google Scholar]
- Izumi C, Mori IC, Utsugi S, Tanakamaru S, Tani A, Enomoto T, Katsuhara M. (2009) Biomarkers of green roof vegetation: anthocyanin and chlorophyll as stress marker pigments for plant stresses of roof environments. J Environ Econ Manage 19: 21–27 [Google Scholar]
- Jiang PL, Tzen JTC. (2010) Caleosin serves as the major structural protein as efficient as oleosin on the surface of seed oil bodies. Plant Signal Behav 5: 447–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo JH, Wang S, Chen JG, Jones AM, Fedoroff NV. (2005) Different signaling and cell death roles of heterotrimeric G protein α and β subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell 17: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil HB, Wang Z, Wright JA, Ralevski A, Donayo AO, Gulick PJ. (2011) Heterotrimeric Gα subunit from wheat (Triticum aestivum), GA3, interacts with the calcium-binding protein, Clo3, and the phosphoinositide-specific phospholipase C, PI-PLC1. Plant Mol Biol 77: 145–158 [DOI] [PubMed] [Google Scholar]
- Kim YY, Jung KW, Yoo KS, Jeung JU, Shin JS. (2011) A stress-responsive caleosin-like protein, AtCLO4, acts as a negative regulator of ABA responses in Arabidopsis. Plant Cell Physiol 52: 874–884 [DOI] [PubMed] [Google Scholar]
- Koch E, Slusarenko A. (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucera B, Cohn MA, Leubner-Metzger G. (2005) Plant hormone interactions during seed dormancy release and germination. Seed Sci Res 15: 281–307 [Google Scholar]
- López MA, Vicente J, Kulasekaran S, Vellosillo T, Martínez M, Irigoyen ML, Cascón T, Bannenberg G, Hamberg M, Castresana C. (2011) Antagonistic role of 9-lipoxygenase-derived oxylipins and ethylene in the control of oxidative stress, lipid peroxidation and plant defence. Plant J 67: 447–458 [DOI] [PubMed] [Google Scholar]
- Manavella PA, Arce AL, Dezar CA, Bitton F, Renou JP, Crespi M, Chan RL. (2006) Cross-talk between ethylene and drought signalling pathways is mediated by the sunflower Hahb-4 transcription factor. Plant J 48: 125–137 [DOI] [PubMed] [Google Scholar]
- Montillet JL, Cacas JL, Garnier L, Montané MH, Douki T, Bessoule JJ, Polkowska-Kowalczyk L, Maciejewska U, Agnel JP, Vial A, et al. (2004) The upstream oxylipin profile of Arabidopsis thaliana: a tool to scan for oxidative stresses. Plant J 40: 439–451 [DOI] [PubMed] [Google Scholar]
- Montillet JL, Chamnongpol S, Rustérucci C, Dat J, van de Cotte B, Agnel JP, Battesti C, Inzé D, Van Breusegem F, Triantaphylidès C. (2005) Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol 138: 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosblech A, Feussner I, Heilmann I. (2009) Oxylipins: structurally diverse metabolites from fatty acid oxidation. Plant Physiol Biochem 47: 511–517 [DOI] [PubMed] [Google Scholar]
- Naested H, Frandsen GI, Jauh GY, Hernandez-Pinzon I, Nielsen HB, Murphy DJ, Rogers JC, Mundy J. (2000) Caleosins: Ca2+-binding proteins associated with lipid bodies. Plant Mol Biol 44: 463–476 [DOI] [PubMed] [Google Scholar]
- Nuccio ML, Thomas TL. (1999) ATS1 and ATS3: two novel embryo-specific genes in Arabidopsis thaliana. Plant Mol Biol 39: 1153–1163 [DOI] [PubMed] [Google Scholar]
- op den Camp RG, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Göbel C, Feussner I, et al. (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge M, Murphy DJ. (2009) Roles of a membrane-bound caleosin and putative peroxygenase in biotic and abiotic stress responses in Arabidopsis. Plant Physiol Biochem 47: 796–806 [DOI] [PubMed] [Google Scholar]
- Ponce de León I, Sanz A, Hamberg M, Castresana C. (2002) Involvement of the Arabidopsis α-DOX1 fatty acid dioxygenase in protection against oxidative stress and cell death. Plant J 29: 61–62 [DOI] [PubMed] [Google Scholar]
- Poxleitner M, Rogers SW, Lacey Samuels A, Browse J, Rogers JC. (2006) A role for caleosin in degradation of oil-body storage lipid during seed germination. Plant J 47: 917–933 [DOI] [PubMed] [Google Scholar]
- Prost I, Dhondt S, Rothe G, Vicente J, Rodriguez MJ, Kift N, Carbonne F, Griffiths G, Esquerré-Tugayé MT, Rosahl S, et al. (2005) Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol 139: 1902–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purkrtova Z, Le Bon C, Kralova B, Ropers MH, Anton M, Chardot T. (2008) Caleosin of Arabidopsis thaliana: effect of calcium on functional and structural properties. J Agric Food Chem 56: 11217–11224 [DOI] [PubMed] [Google Scholar]
- Rabino I, Mancinelli AL. (1986) Light, temperature, and anthocyanin production. Plant Physiol 81: 922–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Mongrand S, Gamas P, Niebel A, Ott T. (2007) Genome-wide annotation of remorins, a plant-specific protein family: evolutionary and functional perspectives. Plant Physiol 145: 593–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylidès C, Havaux M. (2012) Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA 109: 5535–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustérucci C, Montillet JL, Agnel JP, Battesti C, Alonso B, Knoll A, Bessoule JJ, Etienne P, Suty L, Blein JP, et al. (1999) Involvement of lipoxygenase-dependent production of fatty acid hydroperoxides in the development of the hypersensitive cell death induced by cryptogein on tobacco leaves. J Biol Chem 274: 36446–36455 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada TL, Takano Y, Shimada T, Fujiwara M, Fukao Y, Mori M, Okazaki Y, Saito K, Sasaki R, Aoki K, et al. (2014) Leaf oil body functions as a subcellular factory for the production of a phytoalexin in Arabidopsis. Plant Physiol 164: 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman FP, Petracek PD, Fledderman CM, Ju Z, Heiman DF, Warrior P. (2005) Salicylate activity. 1. Protection of plants from paraquat injury. J Agric Food Chem 53: 9764–9768 [DOI] [PubMed] [Google Scholar]
- Szczerba MW, Britto DT, Kronzucker HJ. (2009) K+ transport in plants: physiology and molecular biology. J Plant Physiol 166: 447–466 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Katagiri T, Yamaguchi-Shinozaki K, Shinozaki K. (2000) An Arabidopsis gene encoding a Ca2+-binding protein is induced by abscisic acid during dehydration. Plant Cell Physiol 41: 898–903 [DOI] [PubMed] [Google Scholar]
- Thordal-Christensen H, Zhang Z, Wei Y, Collonge DB. (1997) Subcellular localization of H2O2 in plants: H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 2: 1187–1194 [Google Scholar]
- Tirajoh A, Aung TS, McKay AB, Plant AL. (2005) Stress-responsive alpha-dioxygenase expression in tomato roots. J Exp Bot 56: 713–723 [DOI] [PubMed] [Google Scholar]
- Vellosillo T, Martínez M, López MA, Vicente J, Cascón T, Dolan L, Hamberg M, Castresana C. (2007) Oxylipins produced by the 9-lipoxygenase pathway in Arabidopsis regulate lateral root development and defense responses through a specific signaling cascade. Plant Cell 19: 831–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet T, Dignard D, Thomas DY. (1987) A family of yeast expression vectors containing the phage f1 intergenic region. Gene 52: 225–233 [DOI] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR. (1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Koizumi M, Urao S, Shinozaki K. (1992) Molecular cloning and characterization of 9 cDNAs for genes that are responsive to desiccation in Arabidopsis thaliana: sequence analysis of one cDNA that encodes a putative transmembrane channel protein. Plant Cell Physiol 33: 217–224 [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun TP, Li J, Deng XW, et al. (2012) Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proc Natl Acad Sci USA 109: E1192–E1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z, Rodriguez R, Tran A, Hoang H, de los Santos D, Brown S, Vellanoweth RL. (2000) The developmental transition to flowering represses ascorbate peroxidase activity and induces enzymatic lipid peroxidation in leaf tissue in Arabidopsis thaliana. Plant Sci 158: 115–127 [DOI] [PubMed] [Google Scholar]
- Yoshiba Y, Kiyosue T, Katagiri T, Ueda H, Mizoguchi T, Yamaguchi-Shinozaki K, Wada K, Harada Y, Shinozaki K. (1995) Correlation between the induction of a gene for delta 1-pyrroline-5-carboxylate synthetase and the accumulation of proline in Arabidopsis thaliana under osmotic stress. Plant J 7: 751–760 [DOI] [PubMed] [Google Scholar]