The constitutive stress response induced by chloroplast dysfunction causes early differentiation via the activation of cell cycle inhibitors.
Abstract
The majority of research on cell cycle regulation is focused on the nuclear events that govern the replication and segregation of the genome between the two daughter cells. However, eukaryotic cells contain several compartmentalized organelles with specialized functions, and coordination among these organelles is required for proper cell cycle progression, as evidenced by the isolation of several mutants in which both organelle function and overall plant development were affected. To investigate how chloroplast dysfunction affects the cell cycle, we analyzed the crumpled leaf (crl) mutant of Arabidopsis (Arabidopsis thaliana), which is deficient for a chloroplastic protein and displays particularly severe developmental defects. In the crl mutant, we reveal that cell cycle regulation is altered drastically and that meristematic cells prematurely enter differentiation, leading to reduced plant stature and early endoreduplication in the leaves. This response is due to the repression of several key cell cycle regulators as well as constitutive activation of stress-response genes, among them the cell cycle inhibitor SIAMESE-RELATED5. One unique feature of the crl mutant is that it produces aplastidic cells in several organs, including the root tip. By investigating the consequence of the absence of plastids on cell cycle progression, we showed that nuclear DNA replication occurs in aplastidic cells in the root tip, which opens future research prospects regarding the dialogue between plastids and the nucleus during cell cycle regulation in higher plants.
Cell cycle regulation is the driving force of plant growth and development. The plant cell cycle has been studied extensively, and considerable progress has been made in our understanding of the molecular mechanisms involved. Because most proteins involved in cell cycle regulation are evolutionarily conserved, studies in plants have benefited from the knowledge attained in yeast and animals. CYCLIN-DEPENDENT KINASE (CDK)-CYCLIN (CYC) complexes are core cell cycle regulators that phosphorylate a variety of substrates to permit the orderly progression through the cycle phases (Harashima et al., 2013). Plant genomes encode many cyclins and CDKs, which have the potential to form a large number of protein complexes (Inagaki and Umeda, 2011). Investigation of the cell cycle interactome has identified several functional complexes in plant cells (Van Leene et al., 2010); according to these studies, CDKA-CYCD complexes are required for the entry into the cell cycle, the onset of S-phase, and S-phase progression, whereas CDKB-CYCB complexes regulate the G2 and G2/M transition (Van Leene et al., 2011). The assigned roles of these putative complexes are based on detected protein-protein interactions and gene expression profiles as well as genetic evidence supporting the function of a number of cyclins and CDKs. The regulatory cascade that governs entry into the cell cycle is highly conserved in all eukaryotes and results in the activation of D-type cyclins in late G1 (Berckmans and De Veylder, 2009). CDKA-CYCD complexes target the RETINOBLASTOMA PROTEIN-RELATED (RBR) protein, which binds E2 promoter binding factor transcription factors and inhibits their activity. Phosphorylation of RBR induces its release from chromatin and allows the activation of the S-phase genes, such as those encoding subunits of the prereplication complex (CDC10 TARGET1 [CDT1], CELL DIVISION CYCLE6 [CDC6], and MINI-CHROMOSOME MAINTENANCE [MCMs]), and proteins required for DNA synthesis, such as PROLIFERATING CELLS NUCLEAR ANTIGEN1 (PCNA1) and PCNA2 (Berckmans and De Veylder, 2009; Costas et al., 2011). Later in the cell cycle, CDKs from the B family are required to promote mitosis (Nowack et al., 2012), and the degradation of the B-type cyclins associated with CDKB is essential for the progression into mitosis and subsequent cell division (Brukhin et al., 2005). One distinctive feature of plant development is the common occurrence in differentiating cells of endoreduplication (i.e. several rounds of DNA replication without mitosis; De Veylder et al., 2011): both cell proliferation and cell growth associated with endoreduplication contribute to the final size of the organs (Gonzalez et al., 2012). The parameters governing the switch from the proliferative cell cycle to the endocycle are not well understood, but recent results suggest that chloroplasts play an important role during this developmental transition (Andriankaja et al., 2012), revealing a previously unsuspected role of interactions between different cellular compartments in cell cycle regulation.
Although the molecular mechanisms controlling nuclear DNA replication and the identical repartitioning of the genome between the two daughter cells are well described, much less is known about the dialogue between cellular compartments during the cell cycle. Nevertheless, such communication is likely to be essential to the coordinated activity of all organelles during cell proliferation and differentiation. Indeed, mutations in several chloroplastic components affect developmental processes (for review, see Inaba and Ito-Inaba, 2010; Lepistö and Rintamäki, 2012). For example, mutants deficient for the nucleus-encoded plastid RNA polymerase SCABRA3 display aberrant leaf development and mesophyll cell differentiation, suggesting that the proliferation of mesophyll cells and chloroplast biogenesis are coordinated during leaf development (Hricová et al., 2006). Moreover, the cell cycle is an energy-demanding process, and in all organisms, including plants, entry into the cell cycle occurs only if sufficient energy is available (Riou-Khamlichi et al., 2000). Because chloroplasts are the site of photosynthesis, and the source of energy for plant cells, efficient chloroplast function is likely to influence cell cycle progression. Indeed, plastid-derived signals have been implicated in at least two aspects of cell cycle regulation. First, the transition from heterotrophy to autotrophy was recently shown to be required for the reactivation of the root meristem during germination via Glc signaling (Xiong et al., 2013). Furthermore, as stated above, chloroplast biogenesis has been suggested to be an important component of the transition between cell proliferation and endoreduplication during leaf development (Andriankaja et al., 2012): inhibition of chloroplast differentiation during leaf development results in a delay in endoreduplication onset and prolonged cell proliferation. Likewise, functional chloroplasts have been shown to be strictly required for the extra cellular division induced by the high-light response in the mesophyll (Tan et al., 2008).
In agreement with a role of chloroplast-derived signals in the control of cell cycle progression, organelle-dependent checkpoints have been identified in unicellular algae. In Cyanidioschizon merolae, the onset of nuclear DNA replication is dependent on chloroplast DNA replication through the production of magnesium-protoporphyrin IX, an intermediate of chlorophyll biosynthesis that directly interacts with an F-box protein and prevents the degradation of CYCA1, thereby promoting nuclear DNA replication (Kobayashi et al., 2011). Although this checkpoint has been suggested to be conserved in higher plants (Kobayashi et al., 2009), all evidence was obtained in cell suspension; thus, how this checkpoint operates at the whole-plant level remains to be explored.
Finally, various studies have demonstrated that alteration of chloroplast homeostasis activates retrograde signaling pathways and affects nuclear gene expression. For example, mutation of a protein likely involved in chloroplast translation in the soldat10 mutant results in the constitutive activation of stress-responsive genes and antagonizes the cell death response caused by singlet oxygen production (Meskauskiene et al., 2009). Similarly, the happy on norflurazon5 mutant, which is deficient in the chloroplastic protease subunit ClpR4, is more tolerant to various stresses due to the constitutive activation of stress-response genes (Saini et al., 2011). However, how such perturbations of chloroplast function and subsequent activation of the stress response can impinge on cell cycle regulation or plant development has been little investigated to date. Interestingly, a mutation in CRUMPLED LEAF (CRL) was recently reported to result in constitutive activation of stress-response genes that are normally activated by chloroplast-to-nucleus retrograde signaling (Šimková et al., 2012). The crl mutant presents unique defects in chloroplast biogenesis as well as cell division and plant development (Asano et al., 2004) and thus appeared ideally suited to investigate how chloroplast-dependent signaling pathways modulate cell cycle regulation. We have examined how cell proliferation is modified in Arabidopsis (Arabidopsis thaliana) crl and have analyzed the contribution of reduced carbon assimilation to the observed defects. Because this mutant has the propensity to form aplastidic cells in some tissues (Chen et al., 2009), we also determined how entry into the cell cycle is affected in these cells.
RESULTS
Cell Proliferation Is Reduced Drastically in the crl Mutant
The crl full loss-of-function mutant displays reduced growth and altered division planes (Asano et al., 2004). These phenotypes are likely due to profound alterations of various chloroplastic functions. Indeed, although the efficiency of PSII was not modified in the mutant (Asano et al., 2004; Supplemental Fig. S1A), total chlorophyll fluorescence was reduced (Supplemental Fig. S1A) and the accumulation of some amino acids in chloroplasts as well as in the shoot was altered (Supplemental Table S1), indicating that primary metabolism occurring in chloroplasts is impaired in the mutant. In addition, cells of the inflorescence are enlarged in the mutant compared with wild-type plants, suggesting that cell proliferation is inhibited in crl (Asano et al., 2004). To more thoroughly analyze the development defects of crl mutants, we measured the area and cell size of cotyledons and leaves of wild-type and crl plants 28 d after sowing. We observed a 5- to 6-fold reduction in the size of the fourth leaf and a 2-fold reduction in the size of cotyledons. Although we observed a significant reduction of the average cell area in the mutant in all organs, this 1.4-fold reduction was not sufficient to account for the small size of cotyledons or leaves (Table I), demonstrating that the reduced stature of the mutant is due to a reduction of cell division. Indeed, the average leaf cell number was reduced more than 4-fold in crl. Because chloroplast differentiation is impaired in the mutant, some of the observed growth defects may be due to reduced carbon assimilation via photosynthesis. However, the addition of Suc in the growth medium did not rescue the leaf and cotyledon size defects in the crl mutants (Table I), suggesting that this is not likely the case.
Table I. Leaf and cotyledon size and average cell area in wild-type and crl plants grown with or without Suc.
Values are averages ± sd. Cotyledons and fully developed leaves (leaf 4) of 28-d-old crl and wild-type plants were measured using ImageJ software. Average epidermal cell area (excluding stomata guard cells) was also measured. Because this value is highly variable, cell number per surface unit was also counted. Globally, a mild decrease in cell size was observed in the crl mutant, as evidenced by the reduced average cell area and increased cell number per surface unit compared with the wild type (Student’s t test, P < 0.05), but this reduction could not account for the 6-fold reduction in leaf size and the 3-fold reduction in cotyledon size.
| Plant | Leaf Area | Leaf Cell Area | Leaf Cell No. per Surface Unit (2,000 µm2) | Average Cell No. per Leaf | Cotyledon Area | Cotyledon Cell Area | Cotyledon Cell No. per Surface Unit (2,000 µm2) | Average Cell No. per Cotyledon |
|---|---|---|---|---|---|---|---|---|
| mm2 | µm2 | mm2 | µm2 | |||||
| Wild type | 47.9 ± 7.9 | 5,642 ± 2,848 | 341 ± 39 | 8,490 ± 154 | 0.93 ± 0.1 | 7,697 ± 3,518 | 190 ± 24 | 121 ± 28 |
| crl | 7.8 ± 1.4 | 4,128 ± 2,546 | 459 ± 92 | 1,790 ± 64 | 0.30 ± 0.1 | 5,437 ± 2,406 | 245 ± 31 | 55 ± 34 |
| Wild type + Suc | 40.2 ± 1.4 | 6,752 ± 3,280 | 279 ± 27 | 5,580 ± 170 | 0.90 ± 0.1 | 7,082 ± 4,238 | 244 ± 50 | 127 ± 23 |
| crl + Suc | 8.5 ± 1.8 | 4,903 ± 2,553 | 391 ± 103 | 1,662 ± 93 | 0.42 ± 0.1 | 4,957 ± 2,093 | 298 ± 21 | 84 ± 47 |
In addition to shoot development defects, the crl mutant displays a severe reduction in root length (Asano et al., 2004). To better analyze how organ growth and cell proliferation are modified in roots of the crl mutant, we measured root elongation in the crl mutant and wild-type siblings. Root growth was drastically reduced in the crl mutant compared with the wild-type control, and supplementing the growth medium with Suc did not restore this reduction (Fig. 1A). Furthermore, because 25% of mutant seedlings arrested during the time-course analysis (7 d after sowing or later), their root growth could not be determined (Fig. 1B). This proportion of arrested seedlings was significantly higher than the 7% observed among wild-type plants (χ2 = 5.48). However, when plants were grown on Suc, the proportion of arrested seedlings was significantly reduced in the crl mutant (χ2 = 4.88) but still remained higher than that observed for the wild type. These results indicate that the reduced photosynthetic capacity of the crl mutant affects root growth and negatively affects the activity of the root meristem, but in plants where meristem activity is retained, the reduction of root growth cannot be attributed to reduced sugar availability.
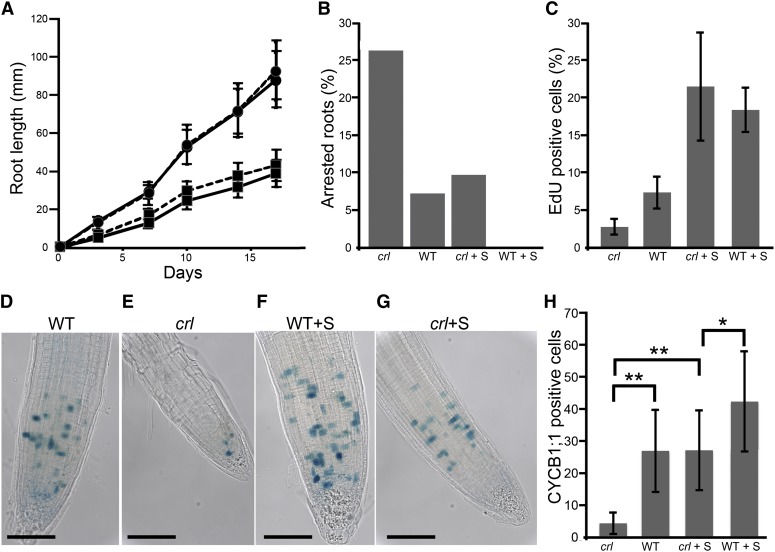
Figure 1.
Root growth and cell cycle progression are inhibited in the crl mutant. A, Time-course analysis of root length in crl mutants (squares) and wild-type siblings (circles) grown on MS medium (solid lines) or on MS medium supplemented with 1% (w/v) Suc (dashed lines). Values are averages ± sd (n > 40). B, Proportion of arrested roots in crl mutants and wild-type siblings (WT) grown on MS medium or on MS medium supplemented with Suc (+S). Identical results were obtained in two independent experiments. C, Proportion of EdU-positive cells in the root tip of crl mutants and wild-type siblings grown on MS medium or on MS medium supplemented with Suc. D to G, Histochemical staining of the GUS activity in crl (E and G) and wild-type siblings (D and F) harboring the CYCB1;1:DB-GUS construct. The number of stained cells reflects the number of cells at the G2/M transition. Plantlets were grown for 10 d on MS medium (D and E) or on MS medium supplemented with Suc (F and G). Bars = 100 µm. H, Quantification of G2/M cell number in the root tips of crl mutants and wild-type siblings grown in the presence or absence of Suc. Values are averages ± sd (n > 20). Asterisks indicate significant differences (Student t test: *P < 0.01, **P < 0.001). [See online article for color version of this figure.]
We next examined how cell cycle progression was affected in the mutant. To this end, DNA replication, G2/M cells, and mitoses were monitored in crl and wild-type siblings. To quantify DNA replication, we used the thymidine analog 5-ethynyl-2'-deoxyuridine (EdU) to label newly replicated DNA. In the crl mutant, the proportion of EdU-positive nuclei was reduced in the absence of Suc but identical to the wild type when Suc was added to the growth medium (Fig. 1C). To quantitate G2/M cells, we took advantage of the CYCB1;1:GUS construct (Colón-Carmona et al., 1999), which harbors the uidA gene (encoding GUS) downstream of a genomic CYCB1;1 fragment composed of the CYCB1 promoter and the beginning of the coding sequence, including the destruction box of CYCB1;1. In the CYCB1;1 reporter line, the GUS protein is targeted for degradation in M-phase, thereby allowing specific staining of G2/M cells (Colón-Carmona et al., 1999). The proportion of G2/M cells was strongly reduced in the crl mutant grown without Suc compared with wild-type plants, and this reduction was only partially rescued by the addition of Suc to the growth medium (Fig. 1, D–H). Finally, we counted the number of cells undergoing mitosis in the root tip of crl mutants and wild-type siblings grown with or without Suc. The number of mitotic events was reduced significantly in the crl mutant compared with wild-type plants and was not restored by Suc addition in the growth medium (Table II).
Table II. Number of mitotic events in the root tip of crl and wild-type siblings grown in the absence or presence of Suc.
Mitotic events were counted in the root tip of crl mutants and wild-type siblings grown with or without Suc after 4′,6-diamino-phenylindole staining of nuclei. Counting was performed on at least 15 roots for each condition. Student’s t test was calculated using the R software. n.s., Not significant.
| Plant | Average No. of Mitoses | sd | P (Student’s t Test, Comparison with the Wild Type Grown without Suc) |
|---|---|---|---|
| Wild type | 26.8 | 5.4 | |
| crl | 11 | 5.3 | <0.001 |
| Wild type + Suc | 21.6 | 3.6 | n.s. (>0.05) |
| crl + Suc | 12.2 | 4.6 | <0.001 |
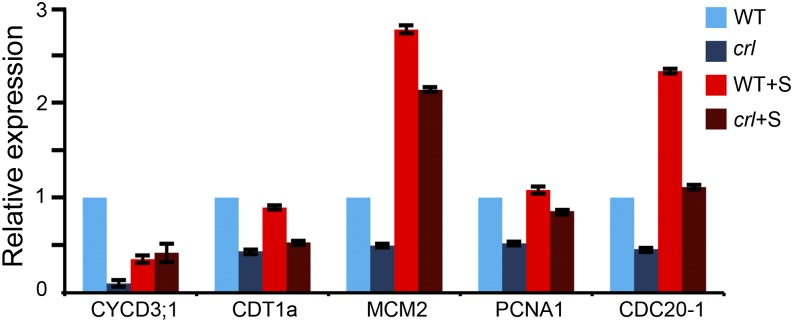
To gain insight into the molecular mechanisms underlying the impairment of cell cycle progression, we tested the expression of several cell cycle genes in 7-d-old mutant seedlings by quantitative reverse transcription (qRT)-PCR (Fig. 2). As markers for the different cell cycle phases, we monitored the expression of genes required for the G1/S transition and in early S-phase (CDT1 and MCM2), throughout S-phase (PCNA1), and in G2/M (CDC20-1) as well as in CYCD3;1, which is involved in the control of both the G1/S and G2/M transitions (Menges et al., 2002; Kevei et al., 2011). The expression of these genes was reduced at least 2-fold in the crl mutant compared with the wild type, with the expression of CYCD3;1 being reduced more than 10-fold. Adding Suc to the growth medium restored CYCD3;1 expression and partially restored the expression of MCM2 and PCNA1, but it was not sufficient to restore wild-type expression levels of the other tested cell cycle genes, suggesting that reduced carbon assimilation is not the only cause for the reduction of cell proliferation observed in the crl mutant. To confirm that this regulation occurred at the transcriptional level, we introduced a construct harboring the uidA gene downstream of the MCM3 promoter sequence (which drives the expression of a protein involved in the initiation of genome replication; Ni et al., 2009). The expression pattern of the MCM3 gene was unchanged in the crl mutant, but GUS staining was much weaker than in wild-type siblings (Supplemental Fig. S2, A–D), indicating that MCM3 promoter activity is reduced in the crl mutant, although this defect was partially restored by Suc. Together, these results indicate that cell cycle progression is to some extent impaired in the root tip of the crl mutant due to the reduced expression of cell cycle genes. Adding Suc to the growth medium restored EdU incorporation and the number of G2/M cells to a level close to that of the wild type, but it failed to restore wild-type mitosis and root growth.
Figure 2.
Cell cycle gene expression is globally reduced in the crl mutant. The expression of several cell cycle markers was analyzed in the crl mutant (dark blue bars) and wild-type siblings (WT; light blue bars) grown on MS medium or on MS medium supplemented with Suc (+S; dark red and light red bars, respectively). We monitored the expression of genes required for the G1/S transition and in early S-phase (CDT1 and MCM2), throughout S-phase (PCNA1), and in G2/M (CDC20-1) as well as in CYCD3;1, which is involved in the control of both the G1/S and G2/M transitions. The expression of all tested genes was drastically reduced in the mutant. Suc addition to the growth medium restored close to wild-type expression of CYCD3;1 and PCNA1 but failed to restore the expression of CDT1a and CDC20-1. Values are averages ± sd and were obtained in triplicate assays. The data presented here are representative of two biological replicates.
The Reduced Growth of crl Mutants Is Due to Premature Differentiation of the Cells, Which Cannot Be Overcome by CYCD3;1 Overexpression
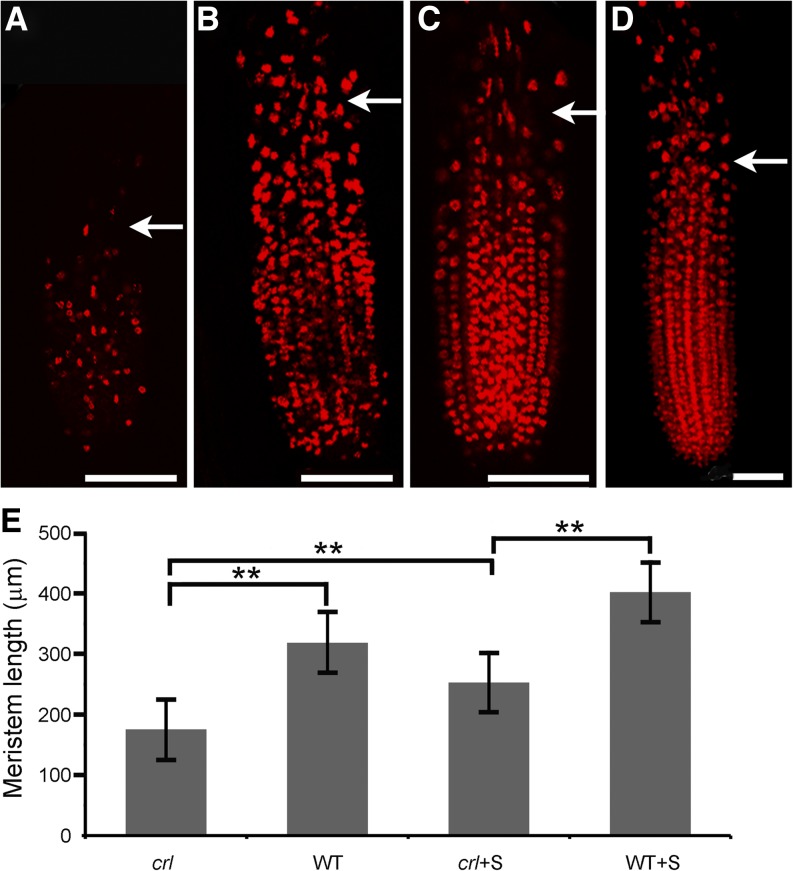
We next asked whether the reduction of root length could be due to defects in cell elongation. Root cell observations did not indicate a significant change in their size (data not shown), suggesting that other mechanisms are responsible for the dwarf phenotype of crl mutants. We reasoned that the reduced growth phenotype of the crl mutant could be due to a reduction of the meristem size and that even though the proportion of EdU-positive cells in the meristem may be similar in wild-type and crl mutant plants, the total number of meristematic cell may be reduced in the crl mutant, which would account for the reduction in the total number of G2/M and mitotic cells. To test this hypothesis, we performed EdU labeling and measured the length of the meristem in 10-d-old seedlings. EdU labeling was observed both in meristematic cells and in differentiating cells undergoing endoreduplication above the meristem (Fig. 3, A–D). The limit of the meristem was easily visualized because the distance between two nuclei was increased due to cell expansion. The meristem of crl mutants was shorter than that of their wild-type siblings, even on Suc-containing medium (Fig. 3E), indicating that the meristematic cells enter differentiation prematurely in the mutant.
Figure 3.
Root meristem length is reduced in the crl mutant. A to D, Confocal images of EdU-labeled root tips of crl mutants (A and C) and wild-type siblings (B and D) grown on MS medium (A and B) or on MS medium supplemented with Suc (C and D). Bars = 75 µm. Arrows indicate the limit of the root meristem. E, Quantification of meristem length in crl and wild-type siblings (WT) grown on MS medium or MS medium supplemented with Suc (+S). Values are averages ± sd (n > 10). Asterisks indicate statistically significant differences (**P < 0.001).
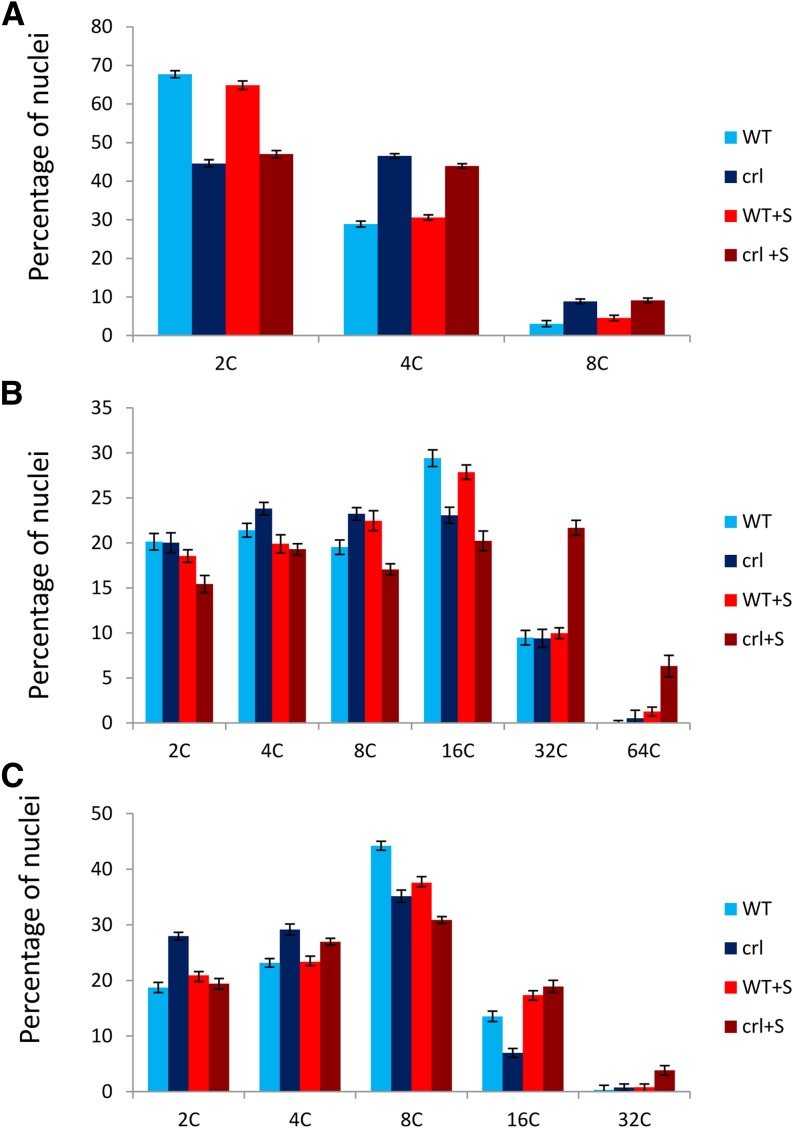
To determine if premature cell differentiation also occurred in aerial tissues, we measured the nuclear DNA content in the smallest collectible developed leaf of 21-d-old crl and wild-type plants. The crl mutant leaves displayed a higher proportion of 4C nuclei than in wild-type leaves of the same rank and size. Furthermore, endoreduplication was observed prematurely in crl mutants compared with wild-type plants, as evidenced by the presence of the 8C peak uniquely in the crl mutant (Fig. 4A).
Figure 4.
Endoreduplication is increased in the crl mutant. Flow cytometry analysis of DNA content distribution in nuclei extracted from crl mutants and wild-type siblings (WT) was performed on developing leaves (A), cotyledons (B), and the first two leaves (C) of 28-d-old plantlets grown in vitro in the presence or absence of Suc (+S). To analyze developing leaves, we identified the smallest leaf visible on the plant (leaf n) and collected leaf n − 2 for both the crl mutant and wild-type siblings. Values are averages ± sd obtained from three biological replicates.
To determine if extra rounds of endoreduplication followed this early differentiation, we measured endoreduplication in additional organs. In the cotyledons and first two leaves of 21-d-old seedlings, no difference was found in the DNA content of nuclei extracted from crl and wild-type plants (Fig. 4, B and C). However, when Suc was added to the growth medium, endoreduplication was increased slightly in the older leaves of crl mutants, as evidenced by the presence of the 32C peak that is almost undetectable in wild-type plants, and even more so in cotyledons, where nuclei with a 64C DNA content were observed almost uniquely in the crl mutant (Fig. 4, B and C).
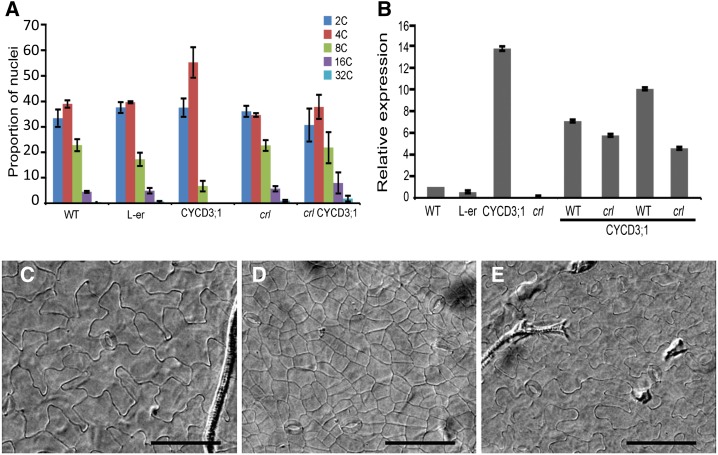
Overexpression of CYCD3;1 has been shown to block endoreduplication and maintain cells in an undifferentiated state (Riou-Khamlichi et al., 1999). Conversely, cycd3;1-3 triple mutants show enhanced endoreduplication (Dewitte et al., 2007). Since CYCD3;1 is down-regulated in crl, we tested whether CYCD3;1 overexpression (CYCD3OE) could reduce endoreduplication in the crl mutant. Surprisingly, the crl mutation completely abolished the G2 arrest observed in the CYCD3OE lines (Fig. 5A), even though the CYCD3;1 construct was highly expressed in the crl CYCD3OE lines (Fig. 5B). Furthermore, crl CYCD3OE plants did not show restored growth compared with crl (Supplemental Fig. S3), and the drastic reduction of cell size caused by the excessive proliferation in CYCD3OE was suppressed in the crl background (Fig. 5, C–E). These results indicated that early differentiation and endoreduplication in the crl mutant is not due to reduced CYCD3;1 expression and that the G2 block induced by CYCD3;1 overexpression is overcome in the crl mutant, leading to endoreduplication.
Figure 5.
The crl mutation overcomes the G2 block induced by CYCD3;1 overexpression and restores the normal differentiation of epidermal leaf cells. A, Flow cytometry analysis of DNA content in whole rosettes. The crl mutant was compared with wild-type siblings (WT), CYCD3;1OE (CYCD3;1), Landsberg erecta (L-er; because CYCD3;1OE is in the Landsberg erecta background), and crl and wild-type siblings obtained in the progeny of a cross between crl hemizygous mutants of CYCD3;1OE. F1 plants were sorted based on the presence of the phenotype associated with CYCD3;1 overexpression, and F2 plants were used for this analysis. Values are averages ± sd obtained on at least three independent plants. CYCD3;1OE crl plants did not display an increased proportion of 4C cells, which is at variance with wild-type CYCD3;1OE siblings from the same cross. B, qRT-PCR analysis of CYCD3;1 expression in the plants analyzed by flow cytometry. crl mutants and wild-type siblings accumulated more CYCD3;1 mRNA than the wild type, indicating that the absence of G2 arrest observed in CYCD3;1OE crl plants is not due to silencing of the construct. CYCD3;1 expression was monitored in crl mutants and wild-type siblings from two independent F2 populations. The data presented here are representative of three independent experiments. C to E, Microscopic images of the abaxial face of cleared leaves from the wild type (C), CYCD3;1OE (D), and crlCYCD3;1OE (E). Bars = 100 µm.
A Large Set of Defense Response Genes Is Constitutively Activated in crl Mutants
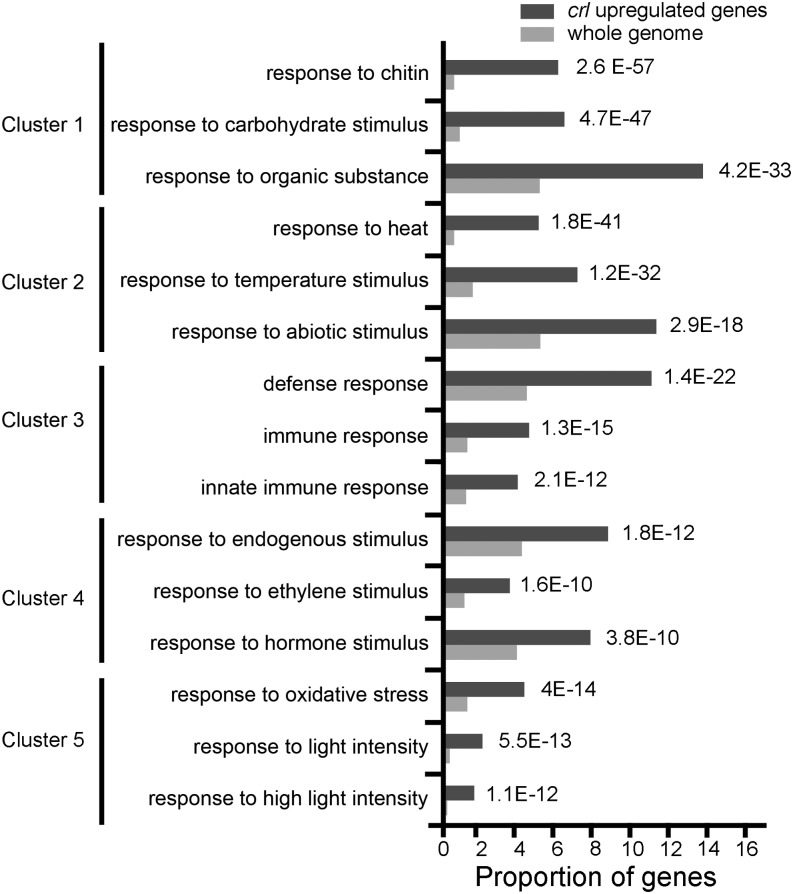
To gain further insight into the molecular basis underlying this altered cell cycle regulation, we performed a genome-wide transcriptome analysis on 7-d-old crl seedlings. A total of 1,199 and 689 genes were found to be significantly up-regulated and down-regulated, respectively (Supplemental Table S2). These results were confirmed by qRT-PCR for a set of up- and down-regulated genes (Supplemental Fig. S4). To identify the types of gene networks deregulated in the crl mutant, we performed a functional annotation clustering analysis of the up-regulated genes using the online software DAVID (http://david.abcc.ncifcrf.gov/summary.jsp), which finds enriched Gene Ontology (GO) terms and groups the genes belonging to enriched categories in clusters based on their expression patterns deduced from publicly available data. This type of analysis allows a finer visualization of the transcriptome results by associating in the same cluster GO terms corresponding to identical or overlapping sets of genes. In the crl mutant, the most significantly enriched clusters corresponded to biotic (clusters 1, 3, and 4) and abiotic (clusters 2 and 5) stress responses (Fig. 6), with enrichment in stress-related GO terms such as responses to chitin and high light. We next examined how the expression of the genes belonging to each GO category responded upon a variety of treatments and conditions represented in the Genevestigator software (Zimmermann et al., 2004). Interestingly, genes from the most significantly enriched categories were induced by pathogen infection or flagellin treatment and drought. The most similar expression profiles found in the Genevestigator database for genes corresponding to the GO term Response to Chitin are represented in Supplemental Figure S5A, but similar results were obtained with all clusters. In addition, we found that genes in these enriched categories also were expressed highly in the fluorescent (flu) mutant, which produces elevated levels of reactive singlet oxygen in chloroplasts due to a defect in chlorophyll biosynthesis (op den Camp et al., 2003). Indeed, among the list of 184 genes found to be up-regulated in the flu mutant (Laloi et al., 2007), 148 genes (80%) were also up-regulated in the crl mutant, indicating that the two mutants have highly similar transcriptomes.
Figure 6.
Stress-related GO terms are enriched among up-regulated genes in the crl mutant. Analysis was performed using the functional annotation clustering analysis tool using the DAVID online software (http://david.abcc.ncifcrf.gov/summary.jsp). This analysis finds enriched GO terms and groups the genes belonging to enriched categories in clusters based on their expression patterns. Dark gray bars represent the proportion of genes corresponding to each GO term among up-regulated genes, whereas light gray bars represent the proportion of genes corresponding to these GO terms in the whole Arabidopsis genome. All these terms were significantly enriched (P values are indicated on the right of the graph).
A similar analysis performed on down-regulated genes resulted in fewer significantly overrepresented GO terms, with the most significantly enriched terms being related to hormone response, and more specifically to auxin response (Supplemental Table S2). Consistently, about 50% of these genes were induced by auxin application in publicly available microarray databases (Supplemental Fig. S5B). Together, these results suggest that many of the growth defects observed in crl mutants likely are due to the constitutive activation of stress responses, consistent with the identification of a crl allele in a genetic screen for constitutive activators of the stress-related ATPase associated with diverse activities (AAA-ATPase; At3g28580; Šimková et al., 2012). One puzzling observation was that this AAA-ATPase gene was not among the up-regulated genes found in our transcriptome analysis. However, our subsequent qRT-PCR analysis showed that this gene was up-regulated in crl mutants (Supplemental Fig. S3).
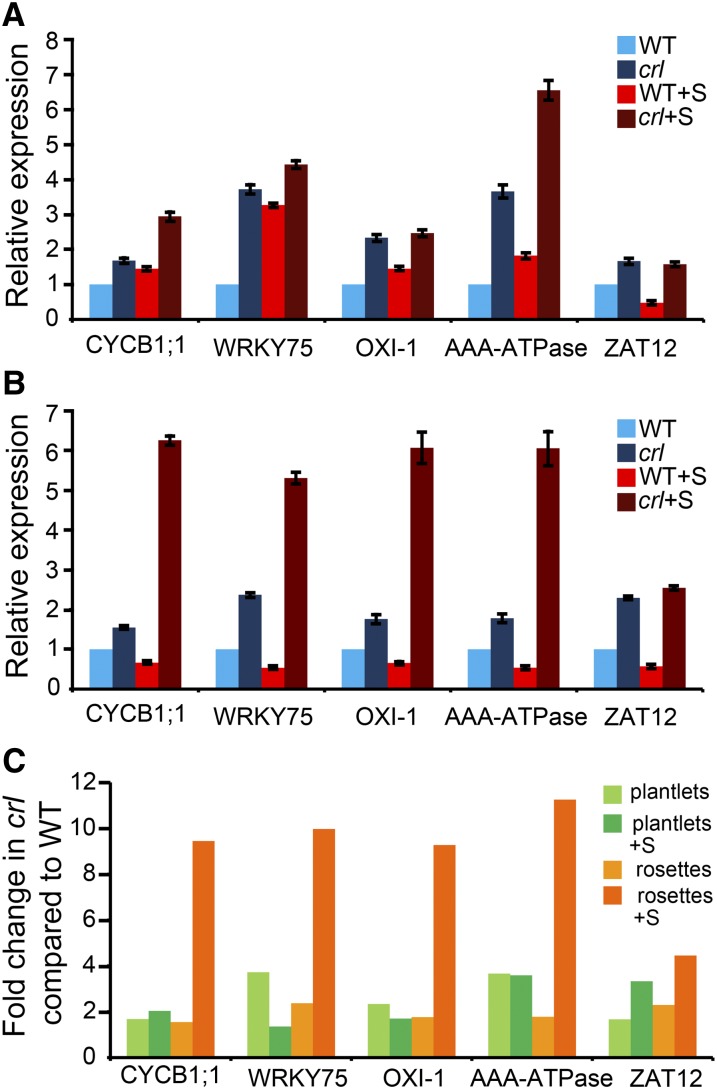
Because increased endoreduplication was observed in plants grown on Suc-containing medium, it is possible that the addition of Suc to the growth medium enhanced the constitutive stress response. However, with the exception of the AAA-ATPase gene, Suc addition did not significantly alter the expression of a tested subset of up-regulated stress genes in seedlings (Fig. 7, A and C). By contrast, up-regulated genes in crl were hyperinduced in older plants (Fig. 7, B and C), and this increase was even more striking when plants were grown on Suc-containing medium.
Figure 7.
The stress response is enhanced by age and Suc availability in the crl mutant. A, qRT-PCR analysis of the expression of a set of stress-related genes in 10-d-old plantlets of crl and wild-type siblings (WT) grown with (+S) and without Suc. B, qRT-PCR analysis of the expression of the same set of genes in 28-d-old rosettes of crl and wild-type siblings grown with and without Suc. C, Fold change in the expression of the same set of genes in the crl mutant compared with the wild type under the same conditions calculated from the data shown in A and B: plantlets grown without or with Suc (light and dark green bars) and rosettes without or with Suc (light and dark orange bars). All tested genes were hyperinduced in rosettes in the presence of Suc. For A and B, values are averages ± sd obtained from three technical replicates. Similar results were obtained for at least two biological replicates.
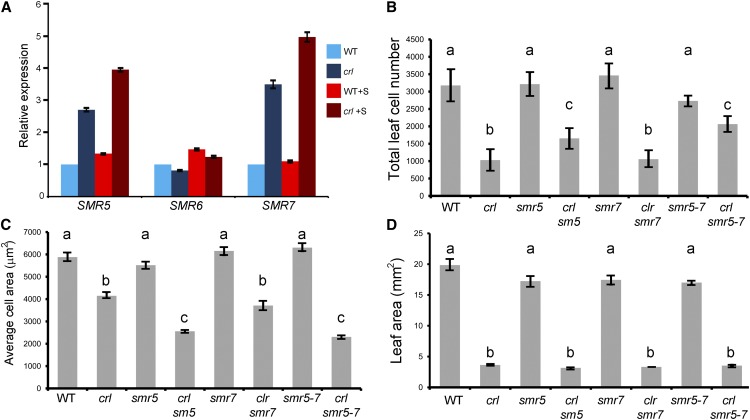
Enhanced Expression of the Cell Cycle Inhibitor SIAMESE-RELATED5 Contributes to the Inhibition of Cell Proliferation in crl
We next examined the transcriptomic data to identify core cell cycle regulators that could be responsible for the early exit from cell proliferation and the enhanced endoreduplication observed in crl mutants. Among a list of 199 core cell cycle genes, only three were up-regulated in our transcriptomic set: CYCB1;1, SIAMESE-RELATED5 (SMR5), and CKL2 CASEIN KINASE 1-LIKE PROTEIN2. Induction of CYCB1;1 was unexpected, based on the results obtained in root meristems using the CYCB1;1:GUS reporter construct (Fig. 1). However, aerial parts (cotyledons and young leaves) represent most of the biomass of seedlings, suggesting that CYCB1;1 is differentially regulated in the root and the shoot of the crl mutant. Consistently, quantitative PCR analysis of CYCB1;1 expression in the root of crl mutants grown on Suc-containing medium showed that the expression of CYCB1;1 is not induced in roots (Supplemental Fig. S6). However, we did not observe a reduction of its expression, suggesting that posttranscriptional regulation accounts for the discrepancy between GUS protein accumulation and accumulation of the CYCB1;1 transcript. Induction of CYCB1;1 in response to DNA stress has been reported in several mutants (Cools and De Veylder, 2009) and has been suggested to prevent the loss of all division-competent cells by blocking endoreduplication. SMR5 encodes a CDK-CYC inhibitor that is induced by several stress conditions, including drought, heat, high light, and salt, all of which lead to oxidative stress in plant cells (Peres et al., 2007). Recently, SMR5 was involved in the oxidative stress cell cycle arrest together with SMR7 (Yi et al., 2014). qRT-PCR analysis revealed that the expression of SMR5 and SMR7 but not of SMR6 was enhanced in the crl mutant (Fig. 8A), suggesting that the reduced cell proliferation observed in crl could be due at least in part to the inhibition of CDK-CYC complexes. Furthermore, induction of SMR5 and SMR7 upon oxidative stress has been shown to depend on the SUPPRESSOR OF GAMMA RESPONSE1 (SOG1) transcription factor (Yi et al., 2014), and we found that among the 30 genes that are induced by γ-irradiation in a SOG1-dependent manner described by Yoshiyama et al. (2009), 16 were induced in the crl mutant according to microarray results (Supplemental Table S3) and the 14 others were all found to be induced, particularly in the presence of Suc, by qRT-PCR analysis (Supplemental Fig. S7), providing evidence for cross talk between chloroplast dysfunction and the activation of the SOG1 pathway. To determine whether this up-regulation of putative SOG1 target genes is a common response to several types of chloroplast-deficient mutants, we searched data from publicly available microarray results. We first examined the expression level of these genes in the genomes uncoupled4-1 (gun4-1) mutant, which is deficient for chlorophyll biosynthesis and plants expressing artificial microRNAs (amiR-White) targeting GUN4 (Schwab et al., 2006). As shown in Supplemental Table S4, only two of these genes were up-regulated in gun4, and five were down-regulated. More of these genes were misregulated in amiR-White lines, consistent with the observation that their phenotype is much more severe than that of gun4-1. However, half of the misregulated genes were up-regulated and half were down-regulated, indicating that the stress response activated by the crl mutation is not activated by defects in chlorophyll biosynthesis. The same comparison was performed with the transcriptome of lincomycin-treated plantlets (Koussevitzky et al., 2007). Almost half of the examined genes were up-regulated and none were down-regulated, indicating that inhibition of chloroplast transcription activates a similar stress response to the crl mutation, possibly due to defects in the repair of the photosynthetic machinery leading to oxidative stress.
Figure 8.
Expression of SMR5 and SMR7 is induced in the crl mutant, and loss of SMR5 partially restores cell proliferation. A, Expression of SMR5, SMR6, and SMR7 was monitored by qRT-PCR in crl and wild-type siblings (WT) grown without Suc (dark and light blue bars) or in the presence of Suc (dark and light red bars). Values are averages ± sd obtained from three technical replicates. Similar results were obtained for at least two biological replicates. B, Average epidermal cell number in the first two leaves of crl, crl smr5, crl smr7, and crl smr5 smr7 mutants. Values are averages ± sd. Measurements were made on at least 100 cells and at least six leaves for each genotype. C, Average epidermal cell area in the first two leaves of crl, crl smr5, crl smr7, and crl smr5 smr7 mutants. Values are averages ± sd. Measurements were made on at least 100 cells. D, Average leaf area in the first two leaves of crl, crl smr5, crl smr7, and crl smr5 smr7 mutants. Values are averages ± sd. Measurements were made on at least 10 leaves for each genotype. For B to D, different letters above the bars indicate significantly different values (Student’s t test, P < 0.05).
To determine whether the activation of SMR5 and SMR7 is responsible for the inhibition of cell division observed in the crl mutant, we generated crl smr5, crl smr7, and crl smr5 smr7 mutants. As shown of Figure 8B, loss of SMR5, but not SMR7, induced a 1.5-fold increase in the total leaf cell number in crl, indicating that the induction of SMR5 contributes to the reduction of cell division. However, this increase in cell division was not sufficient to compensate the 4-fold decrease in leaf cell number compared with the wild type. Consistently, early endoreduplication was still observed in young leaves of crl smr5 and crl smr5 smr7 mutants (data not shown). In addition, leaf cell area was reduced in crl smr5 and crl smr5 smr7 mutants compared with crl (Fig. 8C); the partial restoration of leaf cell number, therefore, was not correlated with an increase in total leaf area (Fig. 8D). Together, these results indicate that SMR5 induction contributes to the premature cell cycle exit and growth of cells in the crl mutant.
Nuclear DNA Replication Still Occurs in Aplastidic Cells in the Root Tip of the crl Mutant
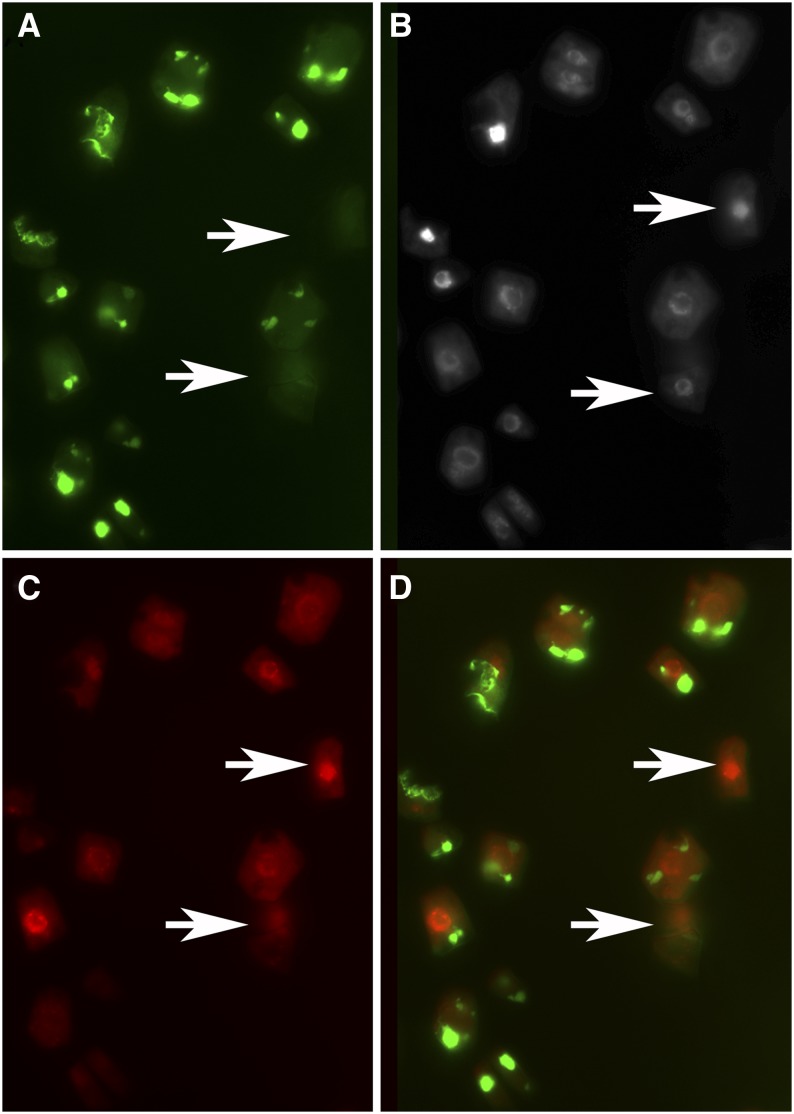
Taken together, our results suggest that alterations in chloroplast function influence cell cycle progression to inhibit proliferation and promote cell differentiation and endoreduplication. Another open question is whether some cell cycle checkpoints help to prevent the formation (or proliferation) of cells lacking plastids in plant tissues. Indeed, plastid number is divided by two during mitosis, and their number is maintained by binary fission of preexisting organelles. In unicellular algae containing only one or a few chloroplasts, chloroplast division occurs just before or during mitosis, and replication of organelle DNA appears to be a prerequisite for entry into the cell cycle (Kobayashi et al., 2009). In addition to its reduced growth, abnormal chloroplast biogenesis, and aberrant chloroplast division, the crl mutant also contains plastid-devoid cells (aplastidic cells) in various tissues and organs (Chen et al., 2009). We took advantage of this original phenotype to test whether this organelle checkpoint is conserved in higher plants and whether cell cycle progression is blocked in aplastidic cells. To this end, we performed EdU incorporation in crl mutants expressing a plastid-targeted GFP to easily monitor the locations of plastids, using an optimized protocol to avoid the destruction of GFP fluorescence during EdU revelation (S. Brown, personal communication). EdU incorporation was detected in aplastidic cells (Fig. 9). Furthermore, the percentage of EdU-positive cells after 10 h of incorporation was not significantly different between plastid-devoid cells and normal cells (30% versus 38%, respectively; Table III). This result differs from the mechanism described in the red alga C. merolae, where organelle DNA replication is required for entry into S-phase.
Figure 9.
Aplastidic cells of the crl mutant can replicate their nuclear DNA. DNA replication was monitored by EdU incorporation in 10-d-old plantlets. Plantlets expressing the TPFtsZ::YFP construct (Chen et al., 2009) were incubated in 0.5× MS medium supplemented with EdU for 10 h. After EdU revelation using a procedure that does not alter GFP fluorescence and Hoechst counterstaining, root tips were squashed and observed using an epifluorescence microscope. Aplastidic cells were identified based on the absence of GFP fluorescence. A, GFP fluorescence. B, Hoechst fluorescence labeling all nuclei. C, AlexaFluor 647 fluorescence labeling EdU-positive cells. D, Merged image. Arrows point to aplastidic cells with EdU-positive nuclei.
Table III. Proportion of EdU-positive cells among aplastidic cells in the crl mutant.
After EdU staining and Hoechst counterstaining, root tips of crl mutants expressing a plastid-targeted yellow fluorescent protein were squashed, and EdU-positive cells were counted among normal and aplastidic cells (identified based on the absence of yellow fluorescent protein). n.s., Not significant.
| Experiment | Proportion of EdU-Positive Cells among Normal Cells | Proportion of EdU-Positive Cells among Aplastidic Cells | χ2 |
|---|---|---|---|
| 1 | 0.53 (n = 905) | 0.36 (n = 29) | 3.12 (n.s.) |
| 2 | 0.38 (n = 934) | 0.30 (n = 66) | 1.56 (n.s.) |
DISCUSSION
Due to their sessile lifestyle, plants are continuously exposed to adverse environmental conditions and, thus, need to adapt their growth behavior by modulating the rate of cell proliferation and differentiation. How environmental cues contribute to regulate cell cycle progression is not well understood, but GAs, via the activity of DELLA proteins, light, and sugars have all been reported to impact core cell cycle regulators (for review, see Komaki and Sugimoto, 2012). Chloroplasts are a hub of metabolic processes, and their function is extremely sensitive to changes in external conditions. Therefore, they are uniquely placed to act as environmental sensors of stress and coordinate nucleus-encoded adaptive stress responses (Chan et al., 2010). Indeed, several studies have highlighted the role of chloroplast-derived signals in plant acclimation to environmental changes (Barajas-López et al., 2013). Nevertheless, little progress has been made to understand the role of chloroplast-derived signals during cell cycle progression. To unravel the role of chloroplast-derived retrograde signals in the cell cycle of higher plants, we exploited the crl mutant, which lacks the CRL protein in the chloroplast outer envelope and displays pleiotropic growth and developmental defects (Asano et al., 2004) and was reported to show constitutive activation of some stress-response genes due to chloroplast dysfunction (Šimková et al., 2012). We found that the reduced stature of the crl mutant was caused primarily by a reduction in cell proliferation. In the absence of Suc, both the G1/S and G2/M transitions were inhibited in the mutant. Surprisingly, although the crl mutant’s dwarf phenotype could be expected to result from poor carbon assimilation, adding Suc to the growth medium only mildly improved these defects: DNA replication was increased on Suc-containing medium, as was the number of G2/M cells, but the number of mitotic events remained unchanged, indicating an inhibition of the G2/M transition. Nevertheless, adding Suc to the growth medium reduced the proportion of crl plants that displayed root growth arrest after the transition to photoautotrophy, consistent with the result that Glc signaling is required for the reactivation of the root meristem during germination (Xiong et al., 2013). Hence, it is likely that meristem arrest in crl seedlings is due at least in part to limited carbon availability. However, in seedlings that did not display development arrest, adding Suc had no effect on root growth or leaf size, indicating that other factors account for the phenotype of the crl mutant. Consistent with this result, providing an external carbon source to the mutants restored the expression of some cell cycle regulators, but not all of them, and did not fully restore meristem size in the crl mutant. In the presence of Suc, EdU incorporation and the number of G2/M cells were increased in the meristem but the frequency of mitosis remained low, suggesting that the length of the G2 phase was prolonged in the mutant. Because chloroplasts are not only the site of photosynthesis but also the site of the biosynthesis of numerous metabolites, reduced production of other molecules is likely to contribute to the observed reduction of cell proliferation.
Our results indicate that the growth reduction observed in the crl mutant is due to inhibition of both the G1/S and G2/M transitions and an early exit of the cells from the proliferative state to enter differentiation. Surprisingly, we did not detect reduced expression of cell cycle genes in our transcriptomic analysis. However, it is possible that the low expression level of cell cycle genes limits the ability to detect a reduction in their accumulation using microarray analysis, as suggested by the observation that changes in cell cycle gene expression were only observed by qRT-PCR (Fig. 2A). In addition, it is likely that these modifications of cell cycle regulation are also due to the constitutive activation of stress responses in the crl mutant. Indeed, a second crl mutant allele was isolated recently in a genetic screen aimed at identifying constitutive activators of the stress-related AAA-ATPase gene (Šimková et al., 2012). Consistent with this finding, our transcriptome analysis of the crl mutant showed that most up-regulated genes were involved in the stress response. Furthermore, we observed a significant overlap between the transcriptomes of the crl mutant and the flu mutant, in which singlet oxygen production results in oxidative stress (Laloi et al., 2007), consistent with the hypothesis that, in crl mutants, perturbation of the chloroplast homeostasis interferes with chloroplast-to-nucleus signaling and results in constitutive activation of defense responses (Šimková et al., 2012). How this chloroplast signal is transmitted to the nucleus and how it interferes with cell cycle regulation remains to be fully determined. It has been shown that plastid signals can rewire light signaling by regulating the expression of particular genes (Ruckle et al., 2012). Indeed, inhibition of chloroplast function by lincomycin treatment abolishes the light-dependent activation of cell cycle genes such as CDKB2;2 and MCM5 in seedlings (Ruckle et al., 2012). We postulate that a similar mechanism could be at work in crl mutants, leading to the reduced expression of key cell cycle regulators such as CDT1 or CDC20-1.
The reduction of cell proliferation and the early onset of cell differentiation in the crl mutant could also be due to an enhanced production of cell cycle inhibitors. Indeed, both SMR5 and SMR7 are strongly induced in the crl mutant, and overexpression of these two factors stimulates endoreduplication (Yi et al., 2014). This overaccumulation of SMR5 and SMR7 in crl may account for the observation that the crl mutation overcomes the G2 block induced by CYCD3;1 overexpression, since SMRs can target CDKA-CYCD complexes (Peres et al., 2007). Very recently, Yi et al. (2014) showed that SMR5 and SMR7 can inhibit cell proliferation in response to oxidative stress: they reported that oxidative stress promotes the phosphorylation of the SOG1 transcription factor by the ataxia-telangiectasia mutated kinase and that SOG1 directly binds the promoter of SMR5 and SMR7. This signaling pathway has been described for its role in the DNA stress response (Yoshiyama et al., 2009, 2013). Here, we show that loss of SMR5 partially restores cell proliferation in the crl mutant, although the total number of leaf cells remains lower than in the wild type. Together, our results suggest that in the leaves of crl mutants, alteration of chloroplast homeostasis activates the SOG1-dependent checkpoint, resulting in the induction of SMR5 and early cell differentiation. The observation that loss of SMR7 did not have a similar effect suggests that other members of the SMR family may participate in the inhibition of cell division in crl. In addition, the observed up-regulation of CYCB1;1 may also account for the reduction of cell proliferation by inducing a G2 arrest of the cell cycle. The signaling pathway leading from chloroplast dysfunction to the activation of CYCB1;1 and SMR gene expression remains to be investigated. Indeed, SOG1 can be activated by oxidative stress, but we could not detect any production of reactive oxygen species in the crl mutant using 3,3'-diaminobenzidine staining, possibly because this method is not sensitive enough. However, the transcriptome of the crl mutant is very similar to that of the flu mutant, in which the production of singlet oxygen is increased, suggesting that the chloroplastic defects in crl mimic oxidative stress without leading to enhanced reactive oxygen species accumulation or induce low levels of reactive oxygen species accumulation. A wealth of retrograde signaling pathways have been reported to alter nuclear gene expression in response to chloroplast-derived signals (Chi et al., 2013), and several reports have shown that altering the basal functions of plastids, such as translation or proteolysis, results in activation of the stress response (Coll et al., 2009; Meskauskiene et al., 2009; Saini et al., 2011). However, how the signal is transmitted from the chloroplast to the nucleus remains elusive. Although the response to singlet oxygen has been shown to be mediated by EXECUTER (EX) proteins (Lee et al., 2007), the activation of the AAA-ATPase in crl is independent of EX1 and EX2 (Šimková et al., 2012), indicating that other signaling pathways are at work in the mutant. Possible candidates that could play a role in activation of the stress response in crl include heat shock transcription factors such as HSFA1D, which has been shown to mediate the plant response to excess light and changes in the redox state of the electron transfer chain (Jung et al., 2013).
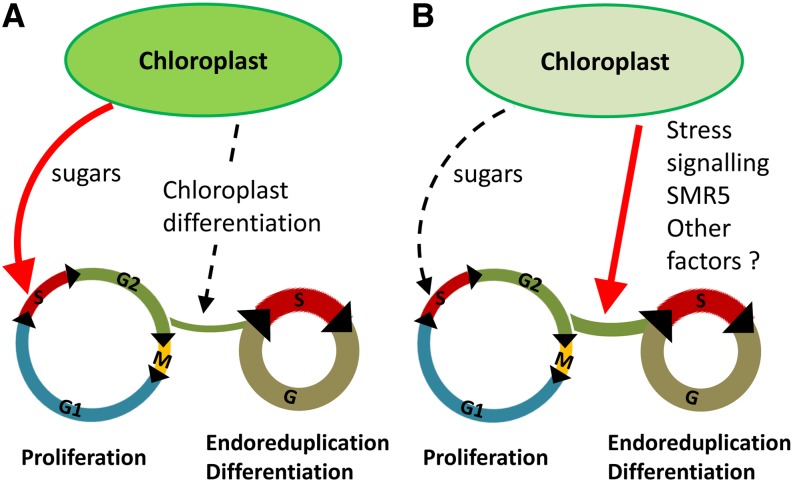
The results reported here combined with previous work from other groups reveal the complexity of the chloroplast-nucleus dialogue in the control of cell proliferation. Indeed, in addition to the expected role for photoassimilates as positive regulators of cell proliferation, normal chloroplast differentiation seems to stimulate cell differentiation in leaves (Andriankaja et al., 2012; Fig. 10A). However, in the crl mutant, alteration of chloroplast function results in reduced cell proliferation due to the low availability of energy sources, but the main effect of chloroplast dysfunction is the constitutive activation of stress signaling that promotes early cell differentiation via the activation of SMR5. Nevertheless, the lack of full complementation suggests that other pathways also negatively affect cell cycle progression in the crl mutant. Furthermore, we observed a compensation mechanism between cell division and cell growth that maintained leaf size in crl. Since sugar addition to the growth medium did not restore leaf area in crl smr5 mutants compared with crl (data not shown), we conclude that the lack of other factors that remain to be identified accounts for the reduced stature of crl mutants (Fig. 10B).
Figure 10.
Effect of chloroplast function on cell cycle regulation. A, In wild-type plants, sugars produced by photosynthesis are required to sustain cell proliferation both in roots and shoots. In addition, Andriankaja et al. (2012) have shown that chloroplast differentiation positively regulates differentiation in developing leaves. B, In the crl mutants, reduced photosynthesis results in an inhibition of cell proliferation via the down-regulation of core cell cycle regulators. In parallel, constitutive activation of the stress response via the induction of SMR5 and other factors promotes early differentiation, which cannot be overcome by external supply of sugars.
Finally, the mechanisms that down-regulate cell proliferation in the root may differ from the ones we described for the shoot. GA signaling may play a role in this process, since DELLA proteins can control cell proliferation in Arabidopsis (Achard et al., 2009) and are involved in the stress response (Claeys et al., 2012). Interestingly, DELLA proteins inhibit cell proliferation under drought stress by promoting the degradation of CYCB1;1 via the anaphase-promoting complex/cyclosome (Claeys et al., 2012). Such a mechanism would account for our results, since the down-regulation of CYCB1;1 in the crl roots appears to involve posttranscriptional regulation. Alternatively, inhibition of root growth in crl could be due to altered auxin signaling, as suggested by the down-regulation of several auxin-responsive genes we observed in the mutant. Indeed, redox and auxin signaling are tightly connected (Tognetti et al., 2012), and a more oxidized environment seems to lead to a reduced auxin response and to inhibit cell proliferation in the root meristem (De Tullio et al., 2010). In addition, auxin is produced mainly in young actively growing leaf primordia, which are particularly severely affected in the crl mutant.
To summarize, our results suggest that altered chloroplast function in the crl mutant results in two distinct and partially antagonistic responses (Fig. 10). First, the reduction of carbon assimilation results in the reduced expression of cell cycle regulators such as CYCD3;1 and impairs both the reactivation of the cell cycle in the root meristem during early development and the global maintenance of meristem activity, resulting in development arrest. This phenotype is likely shared by many chloroplast-deficient mutants with reduced photosynthesis, resulting in growth retardation. Indeed, analysis of publicly available microarray data revealed that many core cell cycle genes, and particularly genes involved in the control of S-phase onset or progression, are down-regulated in GUN4-deficient plants (Supplemental Table S5). By contrast, expression of most of these genes was not affected by lincomycin treatment. In addition, deficiencies in other aspects of chloroplast metabolism likely also affect cell proliferation. Another feature of the crl mutant is that activation of the stress responses due to the perturbation of chloroplast homeostasis results in an increase in the length of the G2 phase, early cell differentiation, and enhanced endoreduplication in the plants that continue to develop, partly due to the expression of SMR5. The full induction of this response is counteracted by the low availability of energy in crl cells; as such, adding Suc to the growth medium results in further endoreduplication and hyperinduction of stress-response genes. This response is at least partly mimicked by lincomycin treatment but not by the inhibition of chlorophyll biosynthesis. These results highlight the complexity of signaling pathways connecting chloroplasts to the nucleus and regulating cell proliferation. In addition to the elucidation of the signaling pathways activated in the crl mutant, analysis of other chloroplast-deficient mutants will be necessary to obtain a full picture of how chloroplast-derived signals can affect cell cycle progression.
Finally, the crl mutant offered a unique tool with which to investigate the existence of a chloroplast-dependent cell cycle checkpoint in whole plants rather than in cellular suspensions, because the crl mutant produces aplastidic cells in various tissues. Using EdU incorporation, we showed that aplastidic cells enter the S-phase of the cell cycle at the same frequency as normal plastid-containing cells. Although we cannot rule out the possibility that some of the aplastidic EdU-positive cells originate from the division of a cell that still contained one plastid at the onset of S-phase, this possibility would not account for the unchanged frequency of EdU-positive cells in aplastidic cells. In addition, because the 10-h EdU labeling time used is shorter than the 18-h length of an entire cell cycle (West et al., 2004), the proportion of labeled cells arising from the division of a labeled cell would be expected to be too low to account for the unchanged frequency. This observation is surprising, since organelle DNA replication (and hence the presence of organelles) is strictly required for entry into the S-phase of the cell cycle in the unicellular alga C. merolae and in tobacco (Nicotiana tabacum) BY-2 cells (Kobayashi et al., 2009, 2011). Several hypotheses may account for this apparent discrepancy. First, the chloroplast-dependent checkpoint may be at work in the aerial part of the plant rather than in roots: even though BY-2 cells are nonphotosynthetic and grow in the dark, they may activate some cellular responses that do not operate in the root meristem. Second, although several reports suggest that chloroplast-derived signals act cell autonomously (Kim et al., 2005; Tan et al., 2008), plastid-devoid cells may replicate their DNA, because magnesium-protoporphyrin IX diffuses from neighboring cells in which plastid DNA replication occurs normally. Finally, it is possible that in some tissues of higher plants, this cell cycle checkpoint does not exist, or that it operates later in the cell cycle, for example during the G2 phase.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds were surface sterilized by treatment with bayrochlore for 20 min and washed and imbibed in sterile water for 2 to 4 d at 4°C to obtain homogenous germination. Seeds were sown on commercially available 0.5× Murashige and Skoog (MS) medium (Basal Salt Mixture M0221; Duchefa) supplemented or not with 1% (w/v) Suc solidified with 0.8% (w/v) agar (Phyto-Agar HP696; Kalys) and grown in a long-day (16 h of light, 8 h of dark, 21°C) growth chamber. After 2 weeks, the plants were transferred to soil in a glasshouse or in a growth chamber under short-day conditions (8 h of light at 20°C, 16 h of dark at 18°C) for 2 weeks before being transferred to long-day conditions.
For root growth assays, plants were grown vertically. Plates were scanned at 2- or 3-d intervals, and root length was measured using ImageJ software (http://rsb.info.nih.gov/ij/).
RNA Extraction and qRT-PCR
Total RNA were extracted from seedlings or leaves with the Nucleospin RNA II kit (Macherey-Nagel) according to the manufacturer’s instructions. First-strand complementary DNA was synthesized from 2 µg of total RNA using Improm-II reverse transcriptase (A3802; Promega) according to the manufacturer’s instructions. One-twenty-fifth of the synthesized complementary DNA was mixed with 100 nm each primer and LightCycler 480 Sybr Green I master mix (Roche Applied Science) for quantitative PCR analysis. Products were amplified and fluorescent signals acquired with the LightCycler 480 detection system. The specificity of amplification products was determined by melting curves. Ubiquitin10 and Protein Phosphatase 2A subunit A3 were used as internal controls for signal normalization. Exor4 relative quantification software (Roche Applied Science) automatically calculates the relative expression levels of the selected genes with algorithms based on the delta delta cycle threshold method. Data were from duplicates of at least two biological replicates. qRT-PCR analysis was performed three time for each biological replicate. The sequences of primers used in this study are provided in Supplemental Table S6.
EdU Incorporation Assay
Ten-day-old seedlings were incubated in 0.5× MS liquid medium (Basal Salt Mixture M0221; Duchefa) with or without 1% (w/v) Suc supplemented with 10 µm EdU (Life Technologies) for 10 h on a six-well plate with shaking in a long-day (16 h of light, 8 h of dark, 21°C) growth chamber. Plantlets were infiltrated with the fixative 3.7% (w/v) paraformaldehyde (PFA) in 1× phosphate-buffered saline (PBS), pH 7.4, under vacuum during 1 h at room temperature and washed twice with PBS-bovine serum albumin (BSA; 1× PBS, pH 7.4, and 3% [w/v] BSA). Plantlets were then permeabilized in PBS-Triton (1× 0.5% [w/v] Triton-X100, pH 7.4) for 20 min at room temperature. Plantlets were washed twice in PBS-BSA prior to the Click-iT reaction. The Click-iT reaction mix for EdU visualization was prepared according to the manufacturer’s instructions (Click-iT EdU AlexaFluor 647 Imaging Kit; Life Technologies), and the reaction was performed in the dark for 30 min at room temperature. Nuclei were counterstained with Hoechst (supplied in the Click-iT kit and diluted 2,000-fold in 1× PBS). Roots were mounted on slides in water under coverslips and squashed with a cone. Observations were done with an epifluorescence microscope (SVII; Zeiss), and images were captured with a color CCD camera (Power HAD; Sony).
For root meristem size measurement, 10-d-old plantlets were used for 10-h EdU incorporation. After revelation, roots were mounted in water and directly imaged with a TCS-SP2 upright microscope (Leica Microsystems) with 543-nm excitation, 488/543/633-nm beam-splitter filter, and 610- to 680-nm (red channel) detection windows. Transmitted light was also collected. All images were acquired with similar gain adjustments. Meristem size measurement was performed with the ImageJ 1.28u software.
For GFP and EdU simultaneous observations, the procedure was modified as follows for some steps, according to M. Bourge, C. Fort, M.-N. Soler, B. Satiat-Jeunemaître, and S.C. Brown (unpublished data): plantlets were fixated in PFA and kept in PFA at 4°C for 1 week, we did not perrmeabilize cells with Triton to avoid GFP destruction, and only half of the CuSO4 quantity was used in the Click-iT reaction mix. After Hoescht staining, the last 1× PBS, pH 7.4, wash solution was replaced with 40 mm EDTA, pH 5.8, and was incubated in the dark overnight with shaking at room temperature. A last 1× PBS, pH 7.4, wash was necessary the day after, and plantlets could be kept in PBS at 4°C until the observations. Observations were done with an epifluorescence microscope (SVII; Zeiss), and images were captured with a color CCD camera (Power HAD; Sony).
Histochemical Staining of GUS Activity
After 15 min of fixation in 100% cold acetone, GUS activity was revealed as described previously (Ni et al., 2009). After 1 h at 37°C, samples were washed in 70% (v/v) ethanol, fixed with PFA during 20 min under vacuum, and then cleared using chloral hydrate solution overnight at room temperature (8 g of chloral hydrate [Sigma], 2 mL of 50% glycerol [w/v], and 1 mL of water). Images were captured on a macroscope (AZ100; Nikon) with a Nikon RI1 video camera.
Light and Fluorescence Microscopy
For cell size measurement, cotyledons and leaves of identical ranks of 3-weeks-old plantlets were fixed in ethanol:acetic acid (3:1) and washed in 70% (v/v) ethanol during 20 min at room temperature. Plantlets were subsequently cleared with chloral hydrate overnight. The day after, samples were mounted on slides in water under coverslips, and differential interference contrast macroscopy (AZ100; Nikon) was used to capture images with a Nikon RI1 video camera. Cell area measurement was performed with the ImageJ 1.28u software. The area of at least 60 cells of the abaxial epidermis located between 25% and 75% of the distance between the tip and the base of the leaf, halfway between the midrib and leaf margin, was measured on at least six leaves. The total number of cells per leaf was estimated by dividing the leaf area by the average cell area.
For mitotic index, 7-d-old plantlets were fixed in PFA, washed in PBS, and stained with Hoescht during 30 min. Mitosis figures were counted with an epifluorescence microscope.
Flow Cytometry
For flow cytometric nuclei analysis, tissues were chopped with a razor blade in 1 mL of 45 Galbraith buffer supplemented with 1% (w/v) polyvinylpyrrolidone 10,000, 5 mm metabisulfite, and 5 mg mL−1 RNase from a stock solution at 50 units mg−1.
Propidium iodide was added to the filtered supernatants at 50 µg mL−1. Endoreplication levels of 5,000 to 10,000 stained nuclei were determined using a Cyflow SL flow cytometer (Partec) with 532-nm solid-state laser (100 mW) excitation and emission collected after a 590-nm long-pass filter.
Transcriptome Studies
Microarray analysis was carried out at the Unité de Recherche en Génomique Végétale using the CATMAv6.2 array based on Roche-NimbleGen technology. A single high-density CATMAv6.2 microarray slide contains 12 chambers, each containing 219,684 primers representing all the Arabidopsis genes: 37,309 probes corresponding to The Arabidopsis Information Resource 8 annotation (including 476 probes of mitochondrial and chloroplast genes) plus 1,796 probes corresponding to EuGene software predictions. Moreover, it included 5,328 probes corresponding to repeat elements, 1,322 probes for microRNA, 329 probes for other RNAs (ribosomal RNA, tRNA, small nuclear RNA, and small nucleolar RNA), and finally several controls. Each long primer is triplicate in each chamber for robust analysis and in both strands. Two independent biological replicates were produced. For each biological repetition and each point, RNA samples were obtained by pooling RNAs from more than 100 7-d-old plantlets grown on 0.5× MS medium. Total RNA was extracted as described above. For each comparison, one technical replicate with fluorochrome reversal was performed for each biological replicate (i.e. four hybridizations per comparison). The labeling of copy RNAs with Cy3-dUTP or Cy5-dUTP (Perkin-Elmer-NEN Life Science Products) and the hybridization to slides were performed as described (Lurin et al., 2004). Two-micrometer scanning was performed with the InnoScan900 scanner (Innopsys), and raw data were extracted using Mapix software (Innopsys).
Statistical Analysis of Microarray Data
Experiments were designed with the statistics group of the Unité de Recherche en Génomique Végétale. For each array, the raw data comprised the logarithm of median feature pixel intensity at wavelengths 635 nm (red) and 532 nm (green). For each array, a global intensity-dependent normalization using the loess procedure (Yang et al., 2002) was performed to correct the dye bias. The differential analysis is based on the log ratios averaging over the duplicate probes and over the technical replicates. Hence, the numbers of available data for each gene equals the numbers of biological replicates and are used to calculate the moderated Student’s t test (Smyth, 2004).
Under the null hypothesis, no evidence that the specific variances vary between probes is highlighted by Limma; consequently, the moderated t-statistic is assumed to follow a standard normal distribution. To control the false discovery rate, adjusted P values found using the optimized false discovery rate approach of Storey and Tibshirani (2003) are calculated. We considered as being differentially expressed the probes with an adjusted P ≤ 0.05.
Analysis was done with the R software. The function SqueezeVar of the library Limma was used to smooth the specific variances by computing empirical Bayes posterior means. The library kerfdr was used to calculate the adjusted P values.
Data Deposition
Microarray data from this article were deposited at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession no. GSE48465) and at CATdb (http://urgv.evry.inra.fr/CATdb/ [Gagnot et al., 2008]; project AU13-04_CDT1bis) according to the Minimum Information about a Microarray Experiment standards.
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CDC20-1 (AT4G33270), CDT1a (AT2G31270), CYCB1;1 (AT4G37490), CYCD3;1 (AT4G34160), MCM3 (AT5G46280), SMR5 (AT1G07500), SMR6 (AT5G40460), SMR7 (AT3G27630), MCM2 (AT1G44900), PCNA2 (AT2G29570), ZAT12 (AT5G59820), OXI-1 (AT3G25250), CAT2 (AT4G35090), WRKY75 (AT5G13080), AAA-ATPase (AT3G28580), ARF10 (AT2G28350), and ARF11 (AT1G19220).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. PSII fluorescence is reduced in the crl mutant.
Supplemental Figure S2. Expression of the MCM3 gene is reduced at the transcriptional level in the crl mutant.
Supplemental Figure S3. Overexpression of CYCD3;1 does not rescue growth defects in crl.
Supplemental Figure S4. Validation of microarray data by qRT-PCR.
Supplemental Figure S5. Expression profile of differentially regulated genes in the crl mutant.
Supplemental Figure S6. CYCB1;1 is not up-regulated in the root of crl mutants.
Supplemental Figure S7. Stress-response genes regulated by the SOG1 transcription factor are up-regulated in the crl mutant.
Supplemental Table S1. Amino acid accumulation is modified in the shoot and isolated chloroplast of crl mutants.
Supplemental Table S2. List of differentially expressed genes in the crl mutant.
Supplemental Table S3. Several SOG1 target genes are induced in the crl mutant.
Supplemental Table S4. Expression levels of putative SOG1 target genes in response to chloroplast dysfunction.
Supplemental Table S5. Expression levels of core cell cycle genes in response to chloroplast dysfunction.
Supplemental Table S6. Primers used for quantitative PCR analysis.
Acknowledgments
We thank Allison Mallory for critical reading of the article, Francis-André Wollman for welcoming us into the laboratory for chlorophyll fluorescence analysis, and Spencer Brown for helpful discussions and advice for flow cytometry and simultaneous observation of GFP and EdU.
Glossary
- qRT
quantitative reverse transcription
- GO
Gene Ontology
- MS
Murashige and Skoog
- PBS
phosphate-buffered saline
- BSA
bovine serum albumin
Footnotes
This work was supported by the Agence Nationale de la Recherche of France (grant no. ANR 2010 JCJC1207 01) and the Interuniversity Attraction Poles Program (grant no. IUAP P7/29 MARS), initiated by the Belgian Science Policy Office, as well as by the facilities and expertise of the Imagif Cell Biology Unit of the Gif Campus, which is supported by the Infrastructures en Biologie Sante et Agronomie, the French National Research Agency, under Investments for the Future programs France-BioImaging Infrastructure (grant no. ANR–10–INSB–04–01), Saclay Plant Sciences (grant no. ANR–10–LABX–0040–SPS), and the Conseil Général de l’Essonne.
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, Beemster GT, Genschik P. (2009) Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol 19: 1188–1193 [DOI] [PubMed] [Google Scholar]
- Andriankaja M, Dhondt S, De Bodt S, Vanhaeren H, Coppens F, De Milde L, Mühlenbock P, Skirycz A, Gonzalez N, Beemster GT, et al. (2012) Exit from proliferation during leaf development in Arabidopsis thaliana: a not-so-gradual process. Dev Cell 22: 64–78 [DOI] [PubMed] [Google Scholar]
- Asano T, Yoshioka Y, Kurei S, Sakamoto W, Machida Y, Sodmergen (2004) A mutation of the CRUMPLED LEAF gene that encodes a protein localized in the outer envelope membrane of plastids affects the pattern of cell division, cell differentiation, and plastid division in Arabidopsis. Plant J 38: 448–459 [DOI] [PubMed] [Google Scholar]
- Barajas-López JdD, Blanco NE, Strand Å. (2013) Plastid-to-nucleus communication, signals controlling the running of the plant cell. Biochim Biophys Acta 1833: 425–437 [DOI] [PubMed] [Google Scholar]
- Berckmans B, De Veylder L. (2009) Transcriptional control of the cell cycle. Curr Opin Plant Biol 12: 599–605 [DOI] [PubMed] [Google Scholar]
- Brukhin V, Gheyselinck J, Gagliardini V, Genschik P, Grossniklaus U. (2005) The RPN1 subunit of the 26S proteasome in Arabidopsis is essential for embryogenesis. Plant Cell 17: 2723–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Crisp PA, Estavillo GM, Pogson BJ. (2010) Chloroplast-to-nucleus communication: current knowledge, experimental strategies and relationship to drought stress signaling. Plant Signal Behav 5: 1575–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Asano T, Fujiwara MT, Yoshida S, Machida Y, Yoshioka Y. (2009) Plant cells without detectable plastids are generated in the crumpled leaf mutant of Arabidopsis thaliana. Plant Cell Physiol 50: 956–969 [DOI] [PubMed] [Google Scholar]
- Chi W, Sun X, Zhang L. (2013) Intracellular signaling from plastid to nucleus. Annu Rev Plant Biol 64: 559–582 [DOI] [PubMed] [Google Scholar]
- Claeys H, Skirycz A, Maleux K, Inzé D. (2012) DELLA signaling mediates stress-induced cell differentiation in Arabidopsis leaves through modulation of anaphase-promoting complex/cyclosome activity. Plant Physiol 159: 739–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll NS, Danon A, Meurer J, Cho WK, Apel K. (2009) Characterization of soldat8, a suppressor of singlet oxygen-induced cell death in Arabidopsis seedlings. Plant Cell Physiol 50: 707–718 [DOI] [PubMed] [Google Scholar]
- Colón-Carmona A, You R, Haimovitch-Gal T, Doerner P. (1999) Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Cools T, De Veylder L. (2009) DNA stress checkpoint control and plant development. Curr Opin Plant Biol 12: 23–28 [DOI] [PubMed] [Google Scholar]
- Costas C, Sanchez MdL, Sequeira-Mendes J, Gutierrez C. (2011) Progress in understanding DNA replication control. Plant Sci 181: 203–209 [DOI] [PubMed] [Google Scholar]
- De Tullio MC, Jiang K, Feldman LJ. (2010) Redox regulation of root apical meristem organization: connecting root development to its environment. Plant Physiol Biochem 48: 328–336 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Larkin JC, Schnittger A. (2011) Molecular control and function of endoreplication in development and physiology. Trends Plant Sci 16: 624–634 [DOI] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V, Murray JA. (2007) Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci USA 104: 14537–14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnot S, Tamby JP, Martin-Magniette ML, Bitton F, Taconnat L, Balzergue S, Aubourg S, Renou JP, Lecharny A, Brunaud V. (2008) CATdb: a public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res 36: D986–D990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N, Vanhaeren H, Inzé D. (2012) Leaf size control: complex coordination of cell division and expansion. Trends Plant Sci 17: 332–340 [DOI] [PubMed] [Google Scholar]
- Harashima H, Dissmeyer N, Schnittger A. (2013) Cell cycle control across the eukaryotic kingdom. Trends Cell Biol 23: 345–356 [DOI] [PubMed] [Google Scholar]
- Hricová A, Quesada V, Micol JL. (2006) The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiol 141: 942–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T, Ito-Inaba Y. (2010) Versatile roles of plastids in plant growth and development. Plant Cell Physiol 51: 1847–1853 [DOI] [PubMed] [Google Scholar]
- Inagaki S, Umeda M. (2011) Cell-cycle control and plant development. Int Rev Cell Mol Biol 291: 227–261 [DOI] [PubMed] [Google Scholar]
- Jung HS, Crisp PA, Estavillo GM, Cole B, Hong F, Mockler TC, Pogson BJ, Chory J. (2013) Subset of heat-shock transcription factors required for the early response of Arabidopsis to excess light. Proc Natl Acad Sci USA 110: 14474–14479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevei Z, Baloban M, Da Ines O, Tiricz H, Kroll A, Regulski K, Mergaert P, Kondorosi E. (2011) Conserved CDC20 cell cycle functions are carried out by two of the five isoforms in Arabidopsis thaliana. PLoS ONE 6: e20618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim GT, Yano S, Kozuka T, Tsukaya H. (2005) Photomorphogenesis of leaves: shade-avoidance and differentiation of sun and shade leaves. Photochem Photobiol Sci 4: 770–774 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Imamura S, Hanaoka M, Tanaka K. (2011) A tetrapyrrole-regulated ubiquitin ligase controls algal nuclear DNA replication. Nat Cell Biol 13: 483–487 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kanesaki Y, Tanaka A, Kuroiwa H, Kuroiwa T, Tanaka K. (2009) Tetrapyrrole signal as a cell-cycle coordinator from organelle to nuclear DNA replication in plant cells. Proc Natl Acad Sci USA 106: 803–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki S, Sugimoto K. (2012) Control of the plant cell cycle by developmental and environmental cues. Plant Cell Physiol 53: 953–964 [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719 [PubMed] [Google Scholar]
- Laloi C, Stachowiak M, Pers-Kamczyc E, Warzych E, Murgia I, Apel K. (2007) Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 672–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Kim C, Landgraf F, Apel K. (2007) EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 10270–10275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepistö A, Rintamäki E. (2012) Coordination of plastid and light signaling pathways upon development of Arabidopsis leaves under various photoperiods. Mol Plant 5: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, Hennig L, Gruissem W, Murray JA. (2002) Cell cycle-regulated gene expression in Arabidopsis. J Biol Chem 277: 41987–42002 [DOI] [PubMed] [Google Scholar]
- Meskauskiene R, Würsch M, Laloi C, Vidi PA, Coll NS, Kessler F, Baruah A, Kim C, Apel K. (2009) A mutation in the Arabidopsis mTERF-related plastid protein SOLDAT10 activates retrograde signaling and suppresses (1)O(2)-induced cell death. Plant J 60: 399–410 [DOI] [PubMed] [Google Scholar]
- Ni DA, Sozzani R, Blanchet S, Domenichini S, Reuzeau C, Cella R, Bergounioux C, Raynaud C. (2009) The Arabidopsis MCM2 gene is essential to embryo development and its over-expression alters root meristem function. New Phytol 184: 311–322 [DOI] [PubMed] [Google Scholar]
- Nowack MK, Harashima H, Dissmeyer N, Zhao X, Bouyer D, Weimer AK, De Winter F, Yang F, Schnittger A. (2012) Genetic framework of cyclin-dependent kinase function in Arabidopsis. Dev Cell 22: 1030–1040 [DOI] [PubMed] [Google Scholar]
- op den Camp RG, Przybyla D, Ochsenbein C, Laloi C, Kim C, Danon A, Wagner D, Hideg E, Göbel C, Feussner I, et al. (2003) Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15: 2320–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres A, Churchman ML, Hariharan S, Himanen K, Verkest A, Vandepoele K, Magyar Z, Hatzfeld Y, Van Der Schueren E, Beemster GT, et al. (2007) Novel plant-specific cyclin-dependent kinase inhibitors induced by biotic and abiotic stresses. J Biol Chem 282: 25588–25596 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Huntley R, Jacqmard A, Murray JA. (1999) Cytokinin activation of Arabidopsis cell division through a D-type cyclin. Science 283: 1541–1544 [DOI] [PubMed] [Google Scholar]
- Riou-Khamlichi C, Menges M, Healy JM, Murray JA. (2000) Sugar control of the plant cell cycle: differential regulation of Arabidopsis D-type cyclin gene expression. Mol Cell Biol 20: 4513–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle ME, Burgoon LD, Lawrence LA, Sinkler CA, Larkin RM. (2012) Plastids are major regulators of light signaling in Arabidopsis. Plant Physiol 159: 366–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini G, Meskauskiene R, Pijacka W, Roszak P, Sjögren LL, Clarke AK, Straus M, Apel K. (2011) ‘happy on norflurazon’ (hon) mutations implicate perturbance of plastid homeostasis with activating stress acclimatization and changing nuclear gene expression in norflurazon-treated seedlings. Plant J 65: 690–702 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimková K, Kim C, Gacek K, Baruah A, Laloi C, Apel K. (2012) The chloroplast division mutant caa33 of Arabidopsis thaliana reveals the crucial impact of chloroplast homeostasis on stress acclimation and retrograde plastid-to-nucleus signaling. Plant J 69: 701–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. (2004) Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3: e3. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. (2003) Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan W, Bögre L, López-Juez E. (2008) Light fluence rate and chloroplasts are sources of signals controlling mesophyll cell morphogenesis and division. Cell Biol Int 32: 563–565 [DOI] [PubMed] [Google Scholar]
- Tognetti VB, Mühlenbock P, Van Breusegem F. (2012) Stress homeostasis: the redox and auxin perspective. Plant Cell Environ 35: 321–333 [DOI] [PubMed] [Google Scholar]
- Van Leene J, Boruc J, De Jaeger G, Russinova E, De Veylder L. (2011) A kaleidoscopic view of the Arabidopsis core cell cycle interactome. Trends Plant Sci 16: 141–150 [DOI] [PubMed] [Google Scholar]
- Van Leene J, Hollunder J, Eeckhout D, Persiau G, Van De Slijke E, Stals H, Van Isterdael G, Verkest A, Neirynck S, Buffel Y, et al. (2010) Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol Syst Biol 6: 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West G, Inzé D, Beemster GT. (2004) Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiol 135: 1050–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J. (2013) Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi D, Kamei CLA, Cools T, Vanderauwera S, Takahashi N, Okushima Y, Eekhout T, Yoshiyama KO, Larkin J, Van den Daele H, et al. (2014) The Arabidopsis SIAMESE-RELATED cyclin-dependent kinase inhibitors SMR5 and SMR7 regulate the DNA damage checkpoint in response to reactive oxygen species. Plant Cell 26: 296–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama K, Conklin PA, Huefner ND, Britt AB. (2009) Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc Natl Acad Sci USA 106: 12843–12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama KO, Kobayashi J, Ogita N, Ueda M, Kimura S, Maki H, Umeda M. (2013) ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep 14: 817–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]