Arabidopsis proteins resolve intermediary structures of DNA recombination and repair by symmetrically oriented incisions and provide mechanistic insight in processing of nicked structures.
Abstract
Holliday junctions (HJs) are physical links between homologous DNA molecules that arise as central intermediary structures during homologous recombination and repair in meiotic and somatic cells. It is necessary for these structures to be resolved to ensure correct chromosome segregation and other functions. In eukaryotes, including plants, homologs of a gene called XPG-like endonuclease1 (GEN1) have been identified that process HJs in a manner analogous to the HJ resolvases of phages, archaea, and bacteria. Here, we report that Arabidopsis (Arabidopsis thaliana), a eukaryotic organism, has two functional GEN1 homologs instead of one. Like all known eukaryotic resolvases, AtGEN1 and Arabidopsis single-strand DNA endonuclease1 both belong to class IV of the Rad2/XPG family of nucleases. Their resolvase activity shares the characteristics of the Escherichia coli radiation and UV sensitive C paradigm for resolvases, which involves resolving HJs by symmetrically oriented incisions in two opposing strands. This leads to ligatable products without the need for further processing. The observation that the sequence context influences the cleavage by the enzymes can be interpreted as a hint for the existence of sequence specificity. The two Arabidopsis paralogs differ in their preferred sequences. The precise cleavage positions observed for the resolution of mobile nicked HJs suggest that these cleavage positions are determined by both the substrate structure and the sequence context at the junction point.
To counter the effects of endogenous and exogenous factors that threaten the genome integrity, efficient mechanisms have evolved to ensure the faithful transmission of genetic information (Tuteja et al., 2001). Double-strand breaks, induced by conditions such as ionizing radiation or replication fork (RF) stalling, are among the most deleterious lesions (Jackson and Bartek, 2009). To protect the genome from consequences of these lesions, the cells have ancient double-strand break repair mechanisms, including the homologous recombination (HR) pathway. The HR mechanism is also of great importance in the intentional genetic recombination during sexual reproduction. A key intermediate in HR is the so-called Holliday junction (HJ), a structure that was first suggested in the context of a gene conversion model in fungi (Holliday, 1964) and later shown to arise in somatic and meiotic cells (Szostak et al., 1983; Schwacha and Kleckner, 1995; Cromie et al., 2006; Bzymek et al., 2010).
HJs are structures consisting of four DNA strands of two homologous DNA helices (e.g. homologous chromosomes or sister chromatids). They arise through invasion of one single strand from each of two helices into the other double strand. This results in two continuous strands (one per helix) and two strands that cross from one helix into the other. Schematics often depict the HJs with a parallel orientation of the helices, in which the crossing strands cross each other as was originally postulated (Holliday, 1964). However, HJs based on oligonucleotides have been shown to adopt an antiparallel conformation (for review, see Lilley, 2000). In this configuration, the junction resembles the letter H in a lateral view, and the crossing strands actually perform U turns. The crossing strands represent physical links between the two DNA strands involved. If a RF is restored by HR-mediated repair during mitosis, the resulting HJ usually involves the two sister chromatids of one chromosome (Li and Heyer, 2008). In meiosis, the physical links in the shape of HJs arise because of meiotic crossover between homologous chromosomes. In either case, these links must be resolved to ensure unperturbed cell survival.
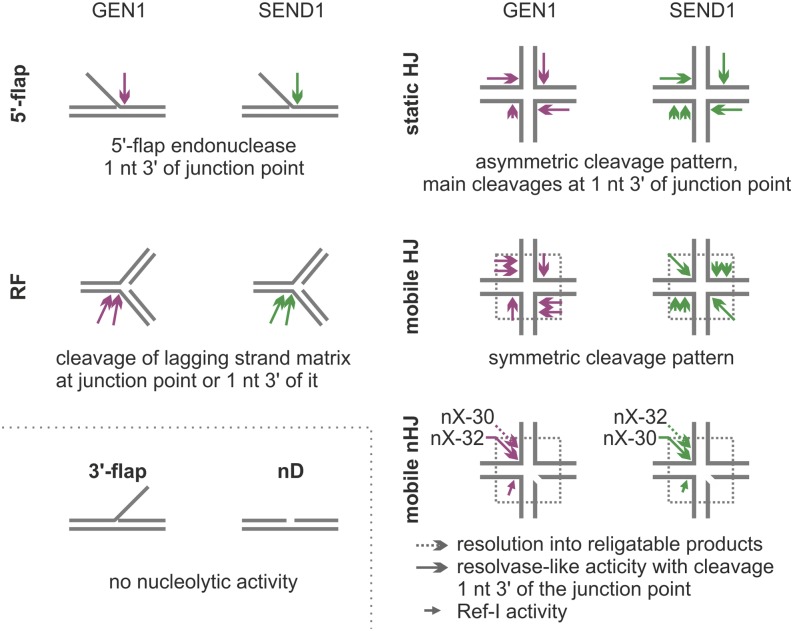
The importance of resolving the HJs for the survival of cells and organisms is highlighted by the phenotypes described for mutants defective for the known pathways of HJ resolution. One of these pathways is the resolution by canonical HJ resolvases, enzymes that cleave the two opposing strands of a HJ in perfectly symmetric positions relative to the junction point, which results in readily ligatable nicked duplex (nD) products (Svendsen and Harper, 2010). This property distinguishes the canonical HJ resolvases from the noncanonical resolvases (see below).
The main resolvase of Escherichia coli is radiation and UV sensitive C (RuvC), which is part of the E. coli resolvasome (RuvABC complex; Otsuji et al., 1974; Sharples et al., 1990, 1999). In this complex, a HJ is sandwiched between two RuvA tetramers (Panyutin and Hsieh, 1994). Two RuvB complexes form ATP-dependent motors of branch migration, with two opposing helical arms of the junction threaded through their central openings. For the resolution of the HJ, one RuvA tetramer is replaced by a RuvC homodimer. This homodimer positions two active sites at the center of the junction that are poised to cleave the junction point if a preferred consensus sequence of the form 5′-(A/T)TT↓(G/C)-3′ is encountered. The requirement for this correct sequence is quite strict; even a single base change can lead to a drastic reduction of the cleavage efficiency (Shah et al., 1994). Isolated EcRuvC is also active in vitro and binds only HJ structures with high specificity. This binding is independent of the sequence context, but the cleavage depends on the specific sequence (Iwasaki et al., 1991; Benson and West, 1994; Dunderdale et al., 1994). The exact cleavage position has been determined to be either one nucleotide 3′ or 5′ from the junction or at the junction point (Bennett and West, 1996; Shida et al., 1996; Osman et al., 2009). The well-characterized EcRuvC is often referred to as a paradigm of canonical HJ resolution.
Eukaryotes have evolved a more complex interplay of different HJ resolution pathways (Schwartz and Heyer, 2011; Zakharyevich et al., 2012). A defined complex, consisting of a recombination deficiency Q (RecQ) helicase (AtRECQ4A in Arabidopsis [Arabidopsis thaliana], Bloom syndrome protein in human, and Slow growth suppression1 (Sgs1) in yeast [Saccharomyces cerevisiae]), a type IA topoisomerase (DNA topoisomerase 3-alpha [TOP3A] in Arabidopsis, HsTOPOIIIα in human, and ScTop3 in yeast), and the structural protein RecQ-mediated genome instability1 (AtRMI1 in Arabidopsis, HsRMI1 in human, and ScRmi1 in yeast; RTR complex), mediates the so-called dissolution pathway. The crossing points of a double HJ are brought together by branch migration catalyzed by the helicase followed by decatenation catalyzed by the topoisomerase (Wu and Hickson, 2003; Hartung et al., 2007a, 2008; Mankouri and Hickson, 2007; Yang et al., 2010). In addition to the catalytic activities, a functional RTR complex also requires structural functions based on protein-protein interactions, for which RMI1 plays an essential role (Mullen et al., 2005; Chen and Brill, 2007; Bonnet et al., 2013; Schröpfer et al., 2014). Dissolution leads to noncross-over products and therefore, is a major mechanism in somatic yeast cells (Gangloff et al., 1994; Ira et al., 2003; Matos et al., 2011). In Arabidopsis, the loss of RTR component function leads to elevated rates of HR as well as sensitivity to UV light and methylmethane sulfonate (MMS; Bagherieh-Najjar et al., 2005; Hartung et al., 2007a; Bonnet et al., 2013). Mutants of AtRMI1 and AtTOP3A exhibit severe and unique meiotic phenotypes (Chelysheva et al., 2008; Hartung et al., 2008). This meiosis I arrest is dependent on HR, but the exact nature of the recombination intermediates that are involved remains unclear (Li et al., 2004; Hartung et al., 2007b; Knoll et al., 2014).
Dissolution acts in parallel with a second pathway mediated by the structure-specific endonuclease MMS and UV-sensitive protein81 (MUS81) as shown by the fact that the additional mutation of ScSgs1/AtRECQ4A leads to synthetic lethality (Mullen et al., 2001; Hartung et al., 2006; Mannuss et al., 2010). Single mutants of MUS81 in yeast, human, Drosophila melanogaster, and Arabidopsis are sensitive to DNA-damaging agents that perturb RFs and show reduced HR after induction of double-strand breaks (Boddy et al., 2001; Hanada et al., 2006; Hartung et al., 2006). The MUS81 homologs form heterodimers with the noncatalytic subunit essential meiotic endonuclease1 (EME1; ScMms4 in S. cerevisiae). SpMus81-Eme1 was, to our knowledge, the first nuclear endonuclease reported to be capable of resolving HJs (Boddy et al., 2001). The Arabidopsis complexes can be formed with the two different subunits: AtEME1A or AtEME1B (Geuting et al., 2009). AtMUS81-EME1A/B, like the fission yeast ortholog, preferentially cleaves nicked Holliday junctions (nHJs) and 3′-flaps but also shows weaker activity on intact HJs in vitro (Boddy et al., 2001; Osman et al., 2003; Geuting et al., 2009; Schwartz and Heyer, 2011). MUS81 homologs are key players in meiotic cross-over generation (Osman et al., 2003; Berchowitz et al., 2007; Higgins et al., 2008). Although cross-over formation is solely dependent on SpMus81 in fission yeast, this function was shown to be shared with ScYen1 in budding yeast (Osman et al., 2003; Blanco et al., 2010; Ho et al., 2010; Tay and Wu, 2010). Tightly regulated by cell division cycle5-dependent hyperphosphorylation at the end of prophase I, the main activity of ScMus81-Mms4 is timed to coordinate with the formation of chiasmata and HJs that link the homologous chromosomes. This role in meiosis I is shown by the failure of chromosome segregation at the end of meiosis I in ScMus81 mutants (Matos et al., 2011). Interestingly, the chromosomes could be segregated at the end of meiosis II because of the presence of ScYen1. In contrast to canonical HJ resolvases, the hallmark of the MUS81-EME1 cleavage mechanism is the asymmetry of the second incision relative to either a first incision or a preexisting nick. This difference classifies MUS81-EME1 as a noncanonical resolvase. Its products need additional processing by gap-filling or flap-cleaving enzymes to allow religation (Boddy et al., 2001; Geuting et al., 2009).
In very recent studies, HsMUS81-EME1 was found to constitute an essential canonical HJ resolvase with HsSLX1-SLX4 (SLX for synthetic lethal of unknown function), in which a first incision is made by HsSLX1-SLX4 followed by the enhanced action of the HsMUS81-EME1 subunits on the resulting nHJ (Garner et al., 2013; Wyatt et al., 2013). HsSLX1-SLX4 had previously been described as a canonical resolvase, albeit producing only a low level of symmetrically cut ligatable products (Fekairi et al., 2009).
In addition to the mechanisms described above, an activity resembling that of EcRuvC had long been known to be present in mammalian cell-free extracts. In 2008, the group of Steven C. West succeeded in identifying, to their knowledge, the first nuclear proteins analogous to the EcRuvC paradigm: ScYen1 and Homo sapiens XPG-like endonuclease1 (HsGEN1; Ip et al., 2008). These proteins are members of the large and well-characterized Rad2/XPG family of nucleases. The Rad2/XPG family consists of the Xeroderma pigmentosum group G-complementing protein (XPG) endonucleases of the nucleotide excision repair (class I), the flap endonuclease1 (FEN1) replication-associated flap endonucleases (class II), the exodeoxyribonuclease1 (EXO1) exonucleases of recombination and repair (class III), and class IV (containing the [putative] eukaryotic HJ resolvases). This last class was introduced after the identification of the rice (Oryza sativa) single-strand DNA endonuclease1 (OsSEND-1) based on sequence homology. The class IV members show a domain composition homologous to FEN1 and EXO1, with no spacer region between their N-terminal XPG (XPG-N) and internal XPG (XPG-I) domains, whereas the primary structure of these domains is more similar to the sequence of the nuclease domain of XPG (Furukawa et al., 2003).
Although all Rad2/XPG homologs share a common cleavage mechanism as observed for the typical 5′-flap substrate (Tsutakawa et al., 2011; Tsutakawa and Tainer, 2012), the striking evolutionary difference between classes I, II, and III on the one hand and the HJ resolvases (class IV) on the other hand is the ability of class IV members to form homodimers in vitro at their preferred substrate, the HJs (Rass et al., 2010). The homodimer configuration ensures the presence of two active sites positioned on the opposing strands of the HJ, which is necessary for resolution. The mode of eukaryotic HJ resolution is largely similar to the bacterial paradigm: (1) cleavage occurs one nucleotide in the 3′ direction of a static junction point (equivalent to the main cleavage site on 5′-flaps), (2) the incisions occur with almost perfect point symmetry, (3) the incisions result in readily ligatable nDs, and (4) certain sites within a migratable HJ core are preferred, providing evidence for a (yet to be determined) sequence specificity (Ip et al., 2008; Bailly et al., 2010; Rass et al., 2010; Yang et al., 2012).
In the absence of MUS81-EME1/Mms4, the proteins HsGEN1, ScYen1, and CeGEN-1 have been shown to play a role in response to replication-associated perturbations, such as MMS- and UV-induced DNA damage (Bailly et al., 2010; Blanco et al., 2010; Tay and Wu, 2010; Gao et al., 2012; Muñoz-Galván et al., 2012). It is also likely that these proteins provide a backup mechanism in mitosis and meiosis, ensuring proper chromosome segregation after a failure of other mechanisms, including MUS81-EME1/Mms4 (Blanco et al., 2010; Matos et al., 2011).
Although canonical HJ resolvases in animals and fungi are a current topic of great interest, very little is known about these proteins in plants. In rice, two members of the Rad2/XPG class IV have been described: OsSEND-1 (the founding member) and OsGEN-like (OsGEN-L). OsSEND-1 was shown to digest single-stranded circular DNA, and its expression is induced on MMS-induced genotoxic stress, whereas OsGEN-L is implicated in late spore development (Furukawa et al., 2003; Moritoh et al., 2005). Both studies (Furukawa et al., 2003; Moritoh et al., 2005) proposed putative homologs in other plants, and the gene locus At1g01880 of Arabidopsis, coding for the protein AtGEN1, is considered the ortholog of HsGEN1 and ScYen1 (Ip et al., 2008). However, currently, only OsGEN-L has been further investigated and described to possess in vitro properties similar to both Rad2/XPG nucleases and EcRuvC. This protein shows a well-defined 5′-flap activity as well as a poorly characterized ability, similar to that of EcRuvC, to resolve mobile HJs (Yang et al., 2012).
Thus, of two members of Rad2/XPG class IV of plants, only one member has so far been analyzed with respect to a possible HJ resolvase activity. However, Arabidopsis expression data show that both proteins are expressed in plants and do not reveal marked differences (Laubinger et al., 2008). In this study, the goal was, therefore, to characterize the in vitro activities of not only AtGEN1 but also, AtSEND1, focusing on the idea that Arabidopsis and (seed) plants in general might encode not one but actually two HJ resolvases with functional homology to EcRuvC.
RESULTS
GEN1 and SEND1 Homologs Are Conserved among Seed Plants
Searches in the Arabidopsis genome for sequence homologs of the previously characterized human and yeast resolvases resulted in two hits corresponding to gene loci At1g01880 and At3g48900, which share 34% amino acid similarity in their conserved N-terminal regions. Their domain structure and sequence similarity place them in the Rad2/XPG family of nucleases, subclass IV, as previously postulated based on partial protein sequences (Furukawa et al., 2003; Moritoh et al., 2005). Sequence similarities to OsGEN-L and OsSEND-1 enabled us to assign AtGEN1 to At1g01880 and AtSEND1 to At3g48900.
AtGEN1 and AtSEND1 code for proteins of 599 and 600 amino acids, respectively, with the typical organization of all class IV members: XPG-N and XPG-I separated by a short spacer and followed by a partially overlapping helix-hairpin-helix domain (Fig. 1A). The sequence of the N-terminal half of the proteins is conserved between animals, fungi, and plants, whereas the C-terminal half shows little to no sequence similarity, except among plant paralogs (Supplemental Information S1). Both AtGEN1 and AtSEND1 share between approximately 18% and 28% identity on the amino acid level with their homologs from Caenorhabditis elegans, D. melanogaster, S. cerevisiae, and human. No sequence conservation could be found to noneukaryotic HJ resolvases (data not shown).
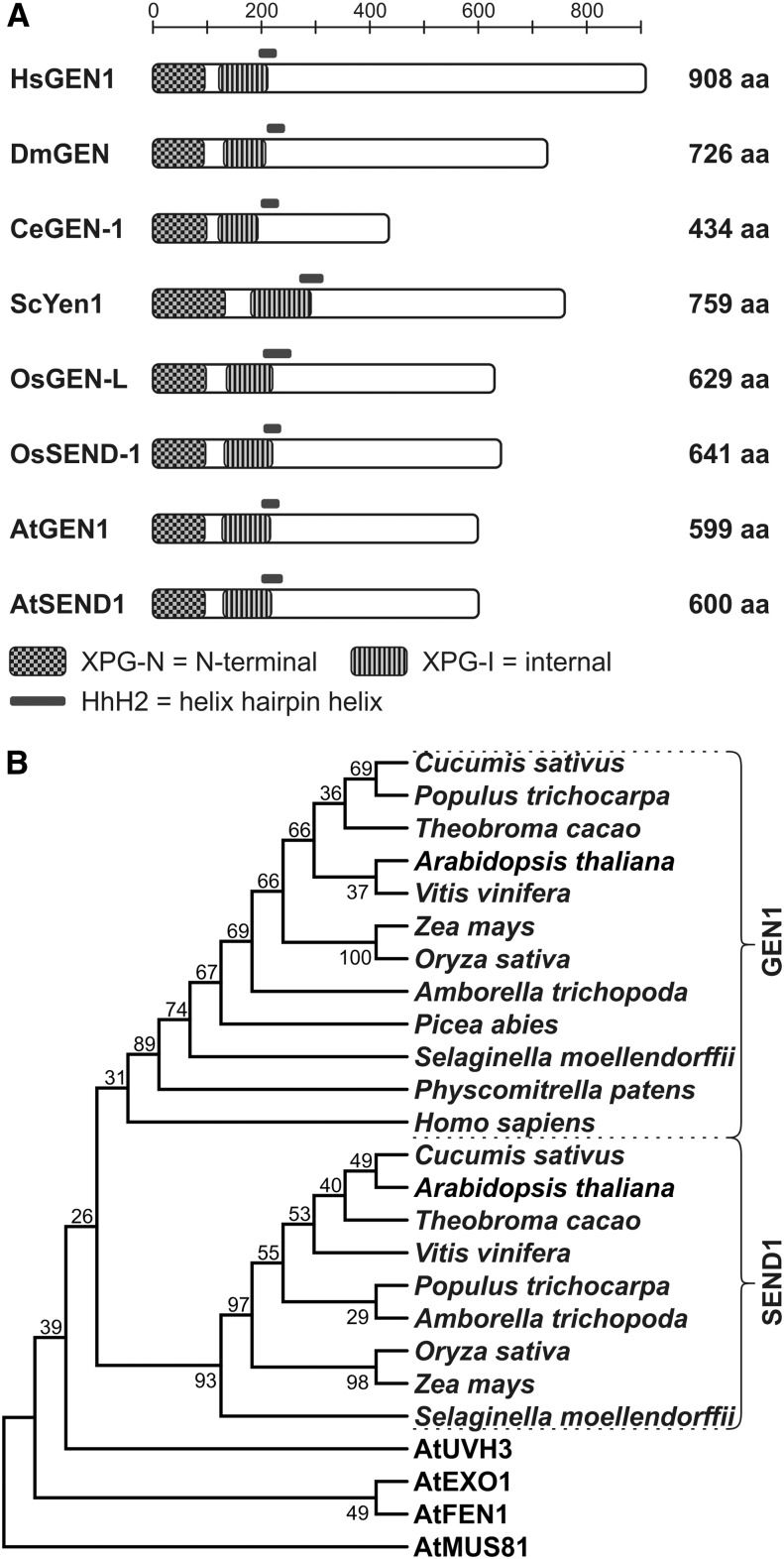
Figure 1.
Eukaryotic HJ resolvases. A, Domain structure of subclass IV of the Rad2/XPG superfamily. All members share a common N-terminal domain organization, whereas the C-terminal half contains no defined functional domains. XPG-N and XPG-I domains constitute the nuclease domain and mediate metal ion coordination by conserved amino acids. The helix-hairpin-helix2 domain is implicated in binding of double-stranded DNA. B, The evolutionary history was inferred using the maximum parsimony method. Tree 1 of the two most parsimonious trees (length = 2,351) is shown. AtMUS81 was defined as the outgroup. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The maximum parsimony tree was obtained using the Subtree-Pruning-Regrafting algorithm with search level 1, in which the initial trees were obtained by the random addition of sequences (10 replicates). aa, Amino acid.
To determine whether the presence of two Rad2/XPG class IV members (hence, two putative canonical HJ resolvases) is a common feature among plants, we investigated the conservation of GEN1 and SEND1 in several plant genomes. The corresponding phylogenetics of selected species are depicted in Figure 1B. Although animals and fungi have one resolvase (GEN1/Yen1), a second resolvase gene (SEND1) was found in most plants with a higher organization than moss (Physcomitrella patens). The plant orthologs share an average identity of 37% to 59%, whereas paralogs share an average identity of 22% to 28%, dependent on the evolutionary distances among the species. GEN1 and SEND1 form two separate clades in the tree. In Picea abies, as a representative of conifers, we were only able to confirm the presence of one homolog. A similar situation was found in Malus domestica, Prunus persica, and Fragaria vesca. It is not clear whether these organisms have eliminated one paralog or if the absence is because of the quality of the sequencing data, which is the case in Carica papaya. By BLAST analysis, we verified that the genome of P. patens contains only one gene, which is located in the GEN1 clade. Thus, a gene duplication event before the development of seed plants might have led to the evolution of two paralogous genes, which might both encode functional HJ resolvases.
To investigate the possible functions of both paralogs, we cloned the respective complementary DNAs (cDNAs), and sequencing confirmed that the gene models provided by The Arabidopsis Information Resource are correct. The overexpressed recombinant full-length AtGEN1 and AtSEND1 were purified using C-terminal affinity tags (Supplemental Fig. S1) that left the N terminus unaltered. The latter criterion proved to be required, because initial constructs with an N-terminal His tag were inactive (data not shown), whereas the proteins used in this study exhibit a robust nuclease activity on various substrates. Proteins featuring an amino acid exchange in their nuclease domain (D75A for AtGEN1 and D76A for AtSEND1) showed no such activities and were used as controls for purification quality (Supplemental Fig. S2).
Structure-Specific Cleavage of Flapped DNAs
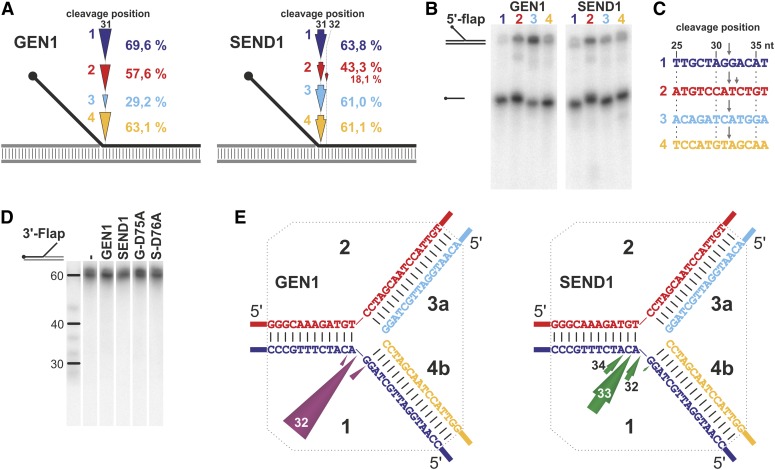
Because AtGEN1 and AtSEND1 are members of the Rad2/XPG family, we tested their activity on 5′-flaps, the shared substrate of all described nucleases within this superfamily. Both plant homologs showed a robust activity with a specific cleavage site in the 5′ overhang strand exactly one nucleotide 3′ of the branch point (Fig. 2A; Supplemental Fig. S3), the identical location to the preferred cleavage site of the FEN1, XPG, and GEN1 homologs. Testing of four different 5′-flaps, each featuring one of the four strands of the X0-HJ (see below) as the flap strand, revealed that each of the Arabidopsis proteins exhibits a weaker activity toward one of the four substrates (Fig. 2, A and B; Supplemental Fig. S3). For AtGEN1, the weaker activity was observed toward the 5′-flap3, whereas AtSEND1 shows a slightly reduced activity on 5′-flap2 and an additional cleavage site two nucleotides 3′ of the branch point (Supplemental Fig. S3). This argues for an influence of the sequence around the branch point (Fig. 2C) on the activities of AtGEN1 and AtSEND1, with different sequence preferences for the two paralogs.
Figure 2.
Structure-specific cleavage of flapped DNA structures. A, Incision mapping at four static 5′-flaps (1–4; indicated by the respective colors) with different sequence compositions. The dots mark the labeled 5′ end of the cleaved flapped strand. The length of the arrowheads and percentages give the 31-nucleotide product as the portion of the total DNA content. B, AtGEN1 and AtSEND1 cleave the four 5′-flaps with different efficiencies: AtGEN1 is less active with 5′-flap3, and AtSEND1 is less active with 5′-flap2 (native gel electrophoresis). C, Sequence context of the flapped strands around the junction point. The main and secondary cleavage sites of AtGEN1 and AtSEND1 are indicated by arrows. D, AtGEN1 and AtSEND1 do not cleave 3′-flaps as shown by denaturing gel electrophoresis of the 5′-labeled flapped strand. E, Cleavage of a RF (RF2) by AtGEN1 and AtSEND1 occurs exclusively in the matrix strand of lagging strand synthesis and preferentially around the branch point of the junction. The cleavage positions, which are equivalent to the lengths of the cleaved oligonucleotides measured from the 5′ end, are indicated at or in the arrowheads. The dotted lines demarcate the migratable homologous core of the RF. nt, Nucleotide.
A 3′-flap proved not to be a suitable substrate for either AtGEN1 or AtSEND1 (Fig. 2D). However, a model RF, which can be considered as a flapped structure with a double-stranded flap, was well recognized by both enzymes. Incision mapping showed that AtGEN1 preferentially cleaves strand 1 exactly at the branch point, whereas AtSEND1 preferentially cleaves one nucleotide in the 3′ direction of the branch point (Fig. 2E). AtSEND1 shows a less pronounced preference for this main cleavage site as indicated by the fact that the two neighboring positions are also cleaved in approximately 40% of cases. Cleavage in strand 2 was not detected with either AtGEN1 or AtSEND1 (data not shown), indicating a preference for cleavage of the lagging strand matrix as described for HsGEN1 and OsGEN-L (Rass et al., 2010; Yang et al., 2012).
GEN1 and SEND1 Cleave HJs near the Junction Point
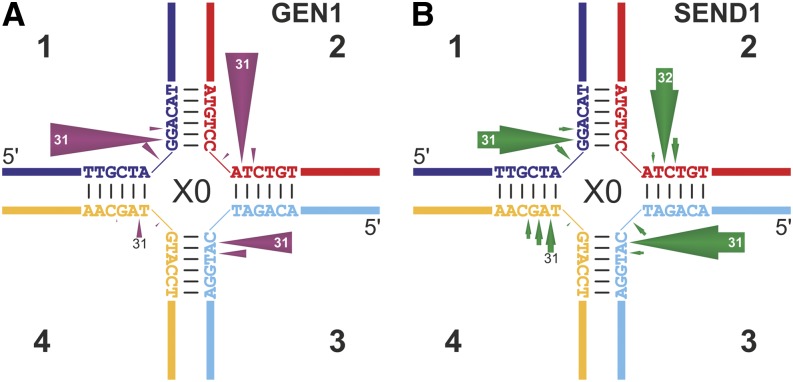
With respect to their putative roles as HJ resolvases, we tested whether AtGEN1 and AtSEND1 cleave HJs. A common substrate for the characterization of HJ-processing enzymes is a static HJ like the X0-HJ, because it is characterized by a fixed junction point caused by the heterologous sequences of four contributing oligonucleotides (X0 because of 0 bp homology around the junction point). These static HJs are used for the determination of the cleavage position, although the overall outcome of the reaction may differ compared with the natural situation of migratable HJs with homologous sequences (see below). The main incisions by AtGEN1 and AtSEND1 are introduced exactly one nucleotide 3′ of the fixed junction point in seven of eight cases (Fig. 3; Supplemental Fig. S4). The only exception is the incision by AtSEND1 in strand 2, which is displaced to the position two nucleotides 3′ of the junction point (Fig. 3B; Supplemental Fig. S4). Notably, this same strand produces the flapped arm of 5′-flap2, which shows the additional cleavage site described above. These data suggest that AtGEN1 and AtSEND1 preferentially cleave the sugar-phosphate backbone one nucleotide 3′ of the junction point, as is the case with several HJ resolvases (Ip et al., 2008; Osman et al., 2009; Bailly et al., 2010; Yang et al., 2012).
Figure 3.
Schematic summary of incision mapping at the static HJ X0 with AtGEN1 (A) and AtSEND1 (B). Cleavage events (Supplemental Fig. S4) were quantified, and the relative frequencies are represented by the lengths of the arrows. The cleavage positions, which are equivalent to the lengths of the cleaved oligonucleotide measured from the 5′ end, are indicated at or in the arrowheads. Cleavage products that represent less than 5% of the cleavage events are not shown.
An additional similarity to previously characterized HJ resolvases, like EcRuvC and OsGEN-L (the most closely related of the characterized resolvase), is the apparent asymmetric distribution of activity on the four strands of a static X0-HJ (Osman et al., 2009; Yang et al., 2012). Both Arabidopsis proteins favor strands 1 to 3 over strand 4. This asymmetry is especially pronounced for AtGEN1 (Fig. 3A) but also apparent with AtSEND1 (Fig. 3B). A similar pattern has been observed with OsGEN-L (Yang et al., 2012) and is, in case of EcRuvC, the result of the well-characterized sequence specificity of the bacterial HJ resolvase (Osman et al., 2009). This observation suggests the potential for sequence preference of the plant resolvases, which will be further evaluated.
Symmetrical Cleavage Action as Resolvases Leads to Ligatable Products
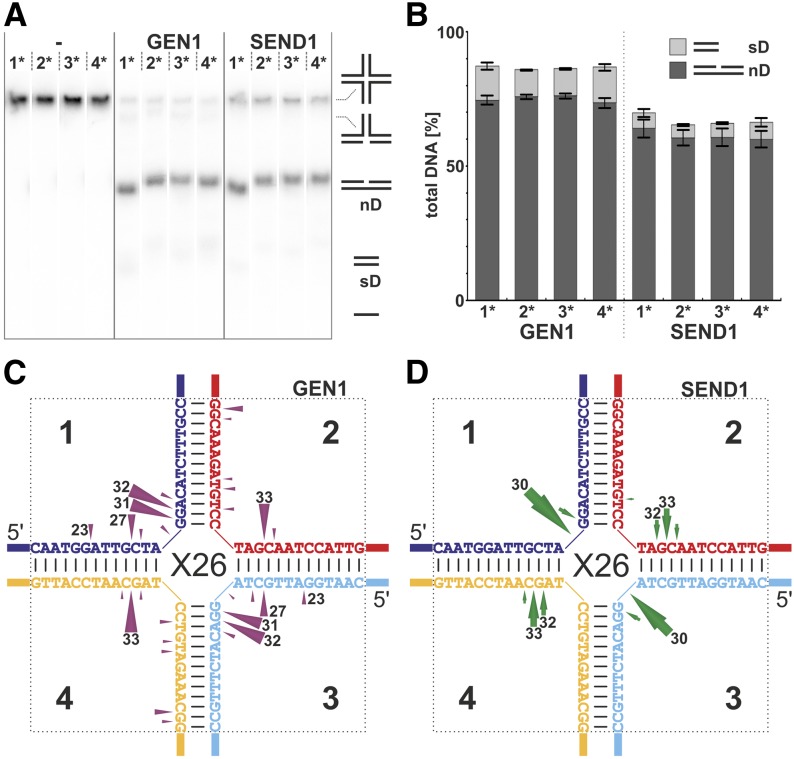
The spatial structure of a static HJ, such as the X0, may resemble that of a naturally occurring HJ, but the main difference is the heterologous sequence that fixates the junction point. We, therefore, tested another HJ substrate, the X26, which comprises a homologous core of 26 bp that mimics the natural character of a migratable junction. AtGEN1 and AtSEND1 were both able to resolve the X26. The resulting major product migrates at the position of nD DNA in native gel electrophoresis (Fig. 4A), whereas a minor product is a faster migrating species equivalent to a short duplex (sD; Fig. 4B). Analysis of the product lengths for each strand of the junction reveals cleavage patterns almost perfectly symmetrical with respect to the junction point (Supplemental Fig. S5). These results are summarized in Figure 4, C and D. AtGEN1 introduces multiple incisions in both cleavage axes throughout the homologous core, with three pronounced cleavage sites at 32, 31, and 27 nucleotides in strands 1 and 3 and a single main cleavage position at 33 nucleotides in strands 2 and 4 (Fig. 4C; Supplemental Fig. S5). Because the HJs were labeled at their 5′ ends, the sizes of the cleavage products (as determined by sequencing gel analysis) are indicative of the cleavage positions. A cleavage product of 32 nucleotides means that the enzyme cut the sugar-phosphate backbone between nucleotides 32 and 33, as is visualized by the arrowheads in the schematic drawings in Figure 4, C and D and named here a cleavage site at position 32. In contrast, the cleavage pattern of AtSEND1 is more concentrated around the center of the X26, with only one main position (30 nucleotides) accompanied by a single secondary position (31 nucleotides) in strands 1 and 3 and two main incisions (33 and 32 nucleotides) in strands 2 and 4 (Fig. 4D; Supplemental Fig. S5).
Figure 4.
Symmetric cleavage pattern at a HJ providing a migratable, homologous core (HJ X26). Four versions of the X26 were used as substrates (differing in which strand was labeled; numbers with asterisks). A, Native gel electrophoresis of the reaction products of AtGEN1 and AtSEND1 compared with buffer controls (−). With the help of marker structures, the reaction products were identified. B, Quantification of the results of independent experiments as shown in A. C and D, Schematic summary of incision mapping at the mobile HJ X26 with AtGEN1 (C) and AtSEND1 (D). The cleavage events (Supplemental Fig. S5) were quantified, and the relative frequencies are represented by the lengths of the arrows. The cleavage positions, which are equivalent to the lengths of the cleaved oligonucleotide measured from the 5′ end, are indicated at the arrowheads. Cleavage products that represent less than 5% of the cleavage events are not shown. The lengths of the main products are indicated.
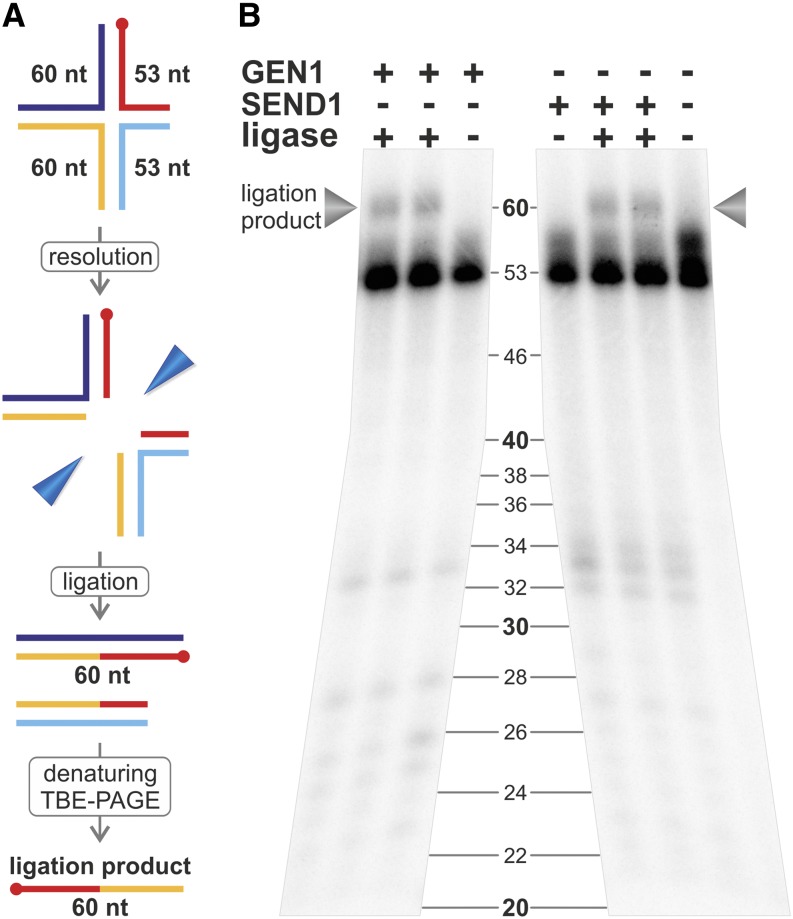
Because even a cleavage pattern with perfect symmetry alone does not constitute proof of HJ resolvase activity according to the EcRuvC paradigm, we tested whether the activities resulted in ligatable nD products as suggested by the native gels and the pattern symmetry. For that purpose, we performed a religation experiment as was recently performed for other eukaryotic HJ resolvases (Ip et al., 2008; Yang et al., 2012). The asymmetric X26-S features three 30 and one 23 bp long helical arms in its centered conformation. Radioactive labeling of strand 2S enables us to detect a ligation-specific product of 60 nucleotides (Fig. 5). For both AtGEN1 and AtSEND1, this ligation product is detected in reactions containing the T4 DNA ligase. This ligation product can only arise because of symmetrical resolution of the HJ, proving that the symmetrical cleavage patterns of AtGEN1 and AtSEND1 do not arise because of statistically distributed nicking events. Instead, both enzymes are able to act as canonical HJ resolvases.
Figure 5.
Ligation of cleavage products. A, Schematic illustration of the ligation assay principle. The asymmetric X26-S HJ containing a label (red dot on red oligonucleotide) was used. Symmetric cleavage followed by ligation yields a longer labeled oligonucleotide of 60 nucleotides (red and yellow segments). B, Resolution products of AtGEN1 and AtSEND1 are ligatable, which is shown by a denaturing sequencing gel after performing the experiments according to the scheme in A. nt, Nucleotide.
Resolution Takes Place within the Life Time of an Enzyme-Substrate Complex
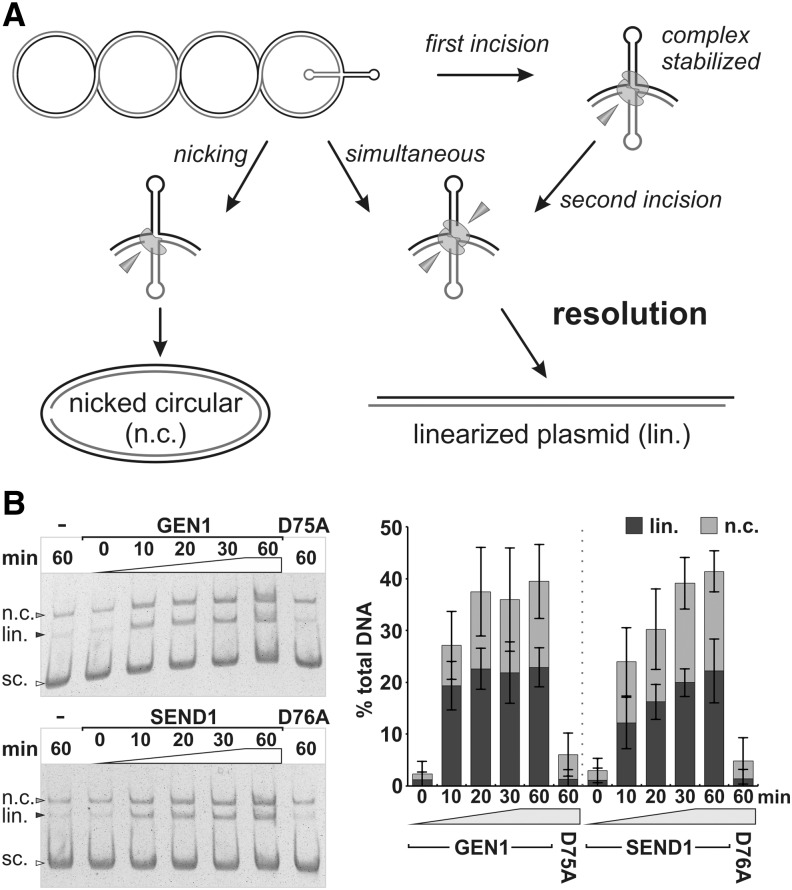
The action of a canonical HJ resolvase is also characterized by a rapid succession of the two incisions. They may either take place simultaneously or as two sequential events in which the complex of HJ and resolvase remains intact, thus stabilizing the junction structure. The assay system described in Figure 6A using a plasmid-based cruciform structure as a HJ substrate allows us to distinguish between nicking and resolution (Lilley and Markham, 1983; Rass et al., 2010). In contrast to the substrate described for the analysis of human GEN1, our plasmid, called pIR9, also included a portion of the sequence of the homologous core of the X26 as part of the inserted inverted repeat (see “Materials and Methods”).
Figure 6.
Resolvase and nicking activities of AtGEN1 and AtSEND1 on plasmid pIR9. A, The plasmid pIR9 features an inverted repeat, which forms a HJ stabilized by negative supercoiling. A single incision leads to relaxation and reabsorption of the HJ if the junction is not stabilized by the DNA-protein complex. The resulting nicked circular (n.c.) plasmid, therefore, is a nicking product. Resolution into linear (lin.) products through two opposing incisions can happen either simultaneously or in a successive manner in which the HJ structure is maintained by the bound resolvase dimer. B, Separation of the nicking and resolution products from the uncleaved supercoiled (sc.) plasmid. For quantification, the percentage of the products was corrected for the background of nicked circular and linear DNA in the buffer reaction (−). The fraction of supercoiled plasmid resistant to EcoRI digestion was determined and set to 100%, representing the available portion of plasmids extruding the cruciform structure.
In the reactions using the negative controls AtGEN1 D75A and AtSEND1 D75A, no significant cleavage was observed, indicative of the fact that the cleavage observed in the reactions with AtGEN1 and AtSEND1 are catalyzed by the proteins of interest (Fig. 6B; Supplemental Fig. S2). Both Arabidopsis resolvases show a similar activity, with AtGEN1 reaching a plateau after 20 min, whereas AtSEND1 takes 60 min (Fig. 6B). The linearized plasmid, the product of the enzymes’ resolvase activities, constitutes up to 70% of the total products for AtGEN1 (at 10 min) and roughly one-half of all products for AtSEND1. The remaining products were nicked circular plasmids, which run the slowest in the gel. These products indicate the presence of a nicking activity for both enzymes.
Cleavage Pattern at nHJs Is Condensed
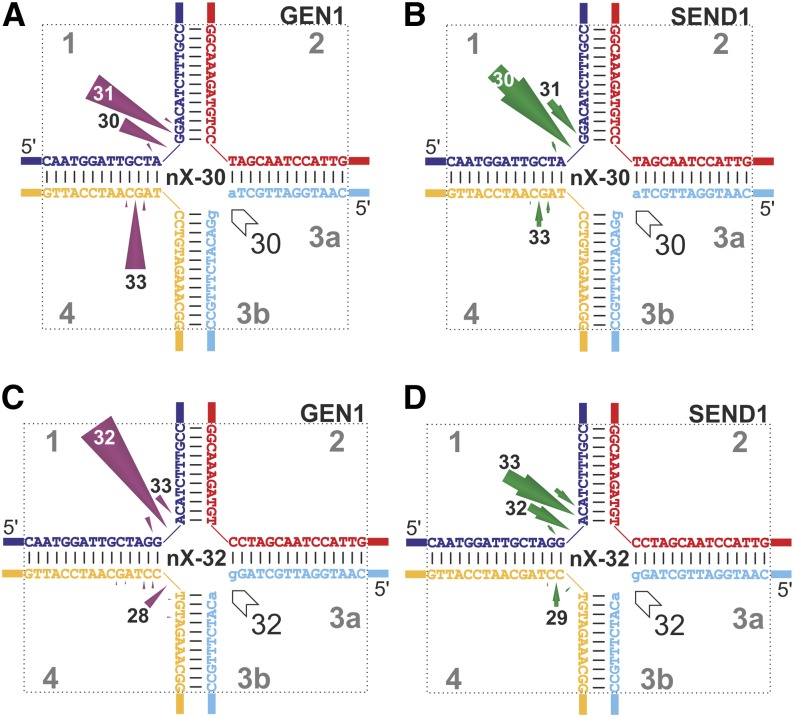
HJ resolvases are known to act on nHJs, which may play a role in vivo as an early recombination intermediate. To date, all in vitro experiments using nHJs have been carried out with static junctions, which fix the nick at a specific position. Usually, the nick has been positioned at the structurally preferred cleavage site. In this study, we tested two nHJs with homologous cores based on the migratable X26, thus providing the enzymes with a more natural imitation of an nHJ. The nHJs in Figure 7 are shown such that that the discontinuity is placed in the junction center, because Pöhler et al. (1994) proposed that this was the manner in which the junction would migrate. As Figure 7 summarizes, the main cleavage activities can be detected in strand 1 opposite of the discontinuity (nick). The pattern of cleavage of strand 1 of the nHJs by AtGEN1 is further condensed compared with the cleavage of the intact X26 (compare Fig. 4C with Fig. 7, A and C). Because AtSEND1 already cleaved strand 1 of the X26 mainly at position 30, a prominent condensation cannot be detected (compare Fig. 4D with Fig. 7, B and D). Interestingly, in the nX-32, the incisions by AtSEND1 are displaced to positions 32 and 33 (Fig. 7D). Taken together, for both AtSEND1 and AtGEN1, the main incisions in strand 1 are either directly opposite the nick in the center of the junction (position 30 in the nX-30 and position 32 in the nX-32) or one nucleotide 3′ of the junction center (position 31 in the nX-30 and position 33 in the nX-32).
Figure 7.
nHJs are primarily cut in the strand opposite the nick but also in the strand that hybridizes with the 5′ end of the nick. Results of incision mapping of two different nHJs treated with AtGEN1 or AtSEND1 are shown. A, AtGEN1 at nX-30. B, AtSEND1 at nX-30. C, AtGEN1 at nX-32. D, AtSEND1 at nX-32. The cleavage events were quantified, and the relative frequencies are represented by the lengths of the arrows. Cleavage products that represent less than 5% of the cleavage events are not shown. The cleavage positions equivalent to the lengths of the main products are indicated (in nucleotides) in or at the arrowheads.
Although no cleavage can be detected in strand 2, incisions are made in strand 4. The extent of cleavage of strand 4 was, in general, less than cleavage in strand 1, but it differed depending on the enzyme and the respective nHJ. However, this detection of cleavage in strand 4 without corresponding incisions in strand 2 is not consistent with a resolution-like activity of the enzymes. Therefore, we endeavored to unravel the source of this cleavage pattern.
Resolvase Activity Is Cryptic at nHJs
nHJs with homologous cores and therefore, the potential to migrate are clearly more natural models than static nHJs. Furthermore, it is clear that testing two similar substrates leads to more globally valid data. We, therefore, chose to test two nHJs, nX-30 and nX-32, based on the previously obtained data. We have already shown that AtGEN1 and AtSEND1 differ in their cleavage efficiency toward several substrates. The efficiency depends on the precise sequence used, possibly because of different sequence specificities. Because the X26 is cut at position 30 of strand 3 only by AtSEND1, testing only the nX-30 with the nick at this position would have given an incomplete and biased view. The choice of the position of the discontinuities in strand 3 was based on the cleavage patterns of AtGEN1 and AtSEND1 at the X26, as is illustrated in Supplemental Figure S6. Briefly, the nX-30 is an X26 featuring a nick between nucleotides 30 and 31 in strand 3 and might represent a favorable substrate for AtSEND1. The discontinuity of nX-32 is located between nucleotides 32 and 33 and provides a substrate that might be favored for AtGEN1.
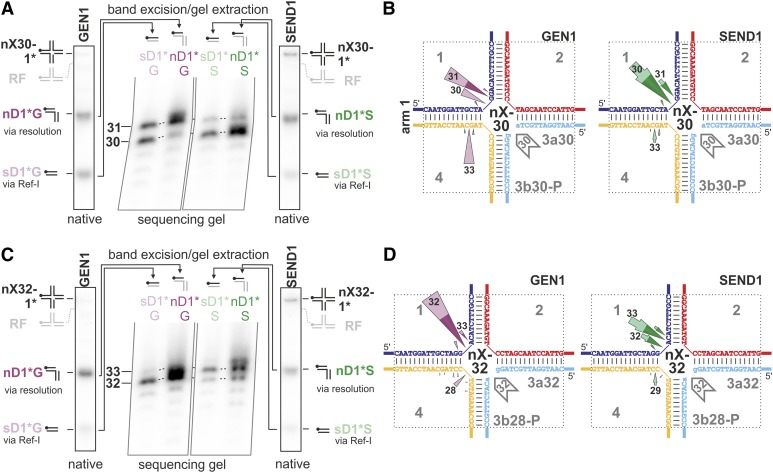
As stated above, both enzymes make incisions in strand 4 but not strand 2 of the two tested nHJs, which is inconsistent with a resolvase-like activity. The activity of AtGEN1 on strand 4 is considerably more pronounced for the nX-30 substrate compared with the nX-32. The cleavage activity of AtSEND1 in strand 4 is detectable but low with both substrates. We analyzed the outcome of cleavage by AtGEN1 and AtSEND1 on the nHJs by native gel electrophoresis (Supplemental Fig. S7). The observed patterns of the native product structures for the two enzymes and the two different nHJs differ in quantity but are markedly similar in quality. Specifically, when the substrates are labeled on strand 1 or 4, an sD and a long duplex that most likely corresponds to the nD product are present. In contrast, when the nHJs are labeled on strand 2 or 3, only nD products can be detected. The identity of the sD in lanes 4* (indicating that the nHJs are labeled at strand 4) can be easily explained by the activities of both AtGEN1 and AtSEND1 on strand 4 of the nHJs: the observed incisions in strand 4 of the nHJs (Fig. 7) in the presence of a discontinuity in strand 3 lead to the separation of a duplex region composed of the oligonucleotide 3b and the 5′-terminal region of oligonucleotide 4. This is equivalent to the separation of arm 4 from the nHJ and would leave a RF-like structure. Indeed, minor amounts of such a RF-like structure are visible for AtGEN1 with the nX-30 (Supplemental Fig. S7). The considerably smaller amount of this RF structure compared with the amount of arm 4 suggests that the RF structure is only an intermediate, which is further processed. We, therefore, named this activity Ref-I for the involved RF intermediate. Because strand 2 of the nHJs is not cleaved (Fig. 7), the processing of the RF-like structure most likely involves cleavage of strand 1. This would also be consistent with the exclusive cleavage of a RF in the lagging strand matrix as shown above. Indeed, a significant amount of an sD product, corresponding to arm 1 of the nHJ, is present when the nHJs are labeled on strand 1 (sD1*; Fig. 8). Separately prepared nD structures were not processed by either AtGEN1 or AtSEND1 (data not shown), supporting the idea that sD1* products can only arise from processing of a RF intermediate.
Figure 8.
nHJs are processed through both a resolution-like pathway and a pathway involving a RF intermediate (Ref-I pathway). In the resolution-like pathway, the position of incision in the strand opposing the nick is dependent on the sequence context at the junction point. A, The nHJ nX-30 was labeled on strand 1, and native gel electrophoresis after incubation with AtGEN1 or AtSEND1 reveals more products than the nD structure expected by a solely canonical HJ resolvase. The sD1* product is the result of a first incision in strand 4, yielding a RF intermediate, and then a second in strand 1 (see text). The nD1* and sD1* products were analyzed separately on sequencing gels to identify the exact cleavage positions for the two different pathways. Supporting data regarding the identity and processing of the RF intermediates are presented in Supplemental Figures S7 and S8. B, Assignment of the cleavage events for the nX-30 to the two different pathways. The portions of the arrowheads ascribed to resolvase-like (dark colors) and Ref-I (light colors) pathways are based on the quantifications shown in Figure 7 and Supplemental Figure S8, H to K. The cleavage positions, which are equivalent to the lengths of the 5′ cleavage products, are indicated in or at the colored arrowheads. The relative frequencies are represented by the lengths of the arrows. Cleavage products that represent less than 5% of the cleavage events are not shown. C is like A but with the nX-32. D, Assignment of the cleavage events for the nX-32 to the two different pathways, like in B.
In addition to this Ref-I activity, we can assume that both enzymes possess a resolution-like activity, because we can detect an nD product composed of arms 1 and 4 (nDs in lanes 1* or 4* and nD1* in Fig. 8; scheme of native product structures in Supplemental Fig. S7). The proposed composition of this duplex is as described above and in Figure 8, because strand 2 is not incised (Fig. 7). Incisions by AtGEN1 and AtSEND1 only occur in strands 1 and 4. Because incisions in strand 4 lead into the Ref-I pathway that separates arm 1 from arm 4, the presence of unseparated arms 1 and 4 (nDs in lane 1* or 4*) indicates exclusive cleavage in strand 1 and thus, represents a resolvase-like activity.
Both activities, resolvase-like and Ref-I, lead to cleavage of strand 1 and were previously analyzed together. Therefore, we wanted to know if specific cleavage positions in strand 1 can be attributed to one activity or the other. Because the sD1* product arises only by the Ref-I activity and the nD1* product is exclusively the product of the resolution-like activity, we separated the two species by native gel electrophoreses, isolated the DNA, and analyzed the respective cleavage positions and their relative quantities on a sequencing gel (Fig. 8, A and C). Interestingly, no prominent differences are visible in the cleavage patterns of AtSEND1 for nD1* and sD1* (Fig. 8, A and C). This indicates that both the Ref-I and the resolvase-like activities have the same cleavage patterns. The same is true for AtGEN1 with the nX-32 (Fig. 8C). However, for AtGEN1 with the nX-30, the cleavage event that is directly opposite the discontinuity (position 30) occurs almost exclusively through the Ref-I pathway, whereas the incision that occurs one nucleotide 3′ of the junction center is the result of both Ref-I and resolvase-like activities (Fig. 8A).
To gain additional insights into the contributions of the two activities, we also constructed the different RFs that we expect to be intermediates of the Ref-I pathway and analyzed the quantity and position of the cleavage events (Supplemental Fig. S8). The data obtained are in good accordance with the cleavage positions and estimated quantities determined after gel extraction of the sD1* bands (Fig. 8, A and C). With the data presented above, we were able to define the contributions of the two pathways for the enzyme-derived products of the nHJs: the main resolution-like pathway and the secondary Ref-I pathway (Fig. 8, B and D). Considering only the resolution-like pathway, it is obvious that the main incision of AtGEN1 in strand 1 of the nX-30 is one nucleotide 3′ of the junction point (Fig. 8B). The incision one nucleotide 3′ of the junction point is reminiscent of the behavior toward the static X0-HJ (Fig. 3). In contrast, in the case of the nX-32, AtGEN1 primarily incises strand 1 directly opposite the discontinuity in strand 3 (Fig. 8D), which would correspond to the cut of a canonical resolvase and lead to ligatable products. AtSEND1, however, shows two main cleavage sites, both directly opposite the centered nick and dislocated one nucleotide 3′ of the junction point. Thus, the differences detected for the two different nHJs are not as strong for AtSEND1 as they are for AtGEN1. Still, the situation described for AtGEN1 is inversely mirrored by AtSEND1: the nX-30 is preferentially cut into ligatable products by an incision directly opposite the discontinuity (Fig. 8B), whereas the main incision in the nX-32 is one nucleotide 3′ of the junction point (Fig. 8D).
Incorporating the results of the analysis of both the nX-30 and the nX-32 paints the picture. If, on the one hand, the discontinuity leading to a nHJ is located at a preferred incision site for the Arabidopsis resolvase, the enzyme will preferentially cut directly opposite the nick, which should lead to ligatable products. If, on the other hand, the discontinuity leading to a nHJ is at a position that is not a preferred incision site, the enzyme will preferentially cut one nucleotide 3′ of the junction point, as was observed with static HJs for the Arabidopsis resolvases and resolvases from other organisms (Ip et al., 2008; Osman et al., 2009; Bailly et al., 2010; Yang et al., 2012).
DISCUSSION
The ability to resolve HJs is conserved throughout all living organisms, although resolution represents analogous rather than homologous evolution (West, 2009). The underlying principle is universal: resolution involves two diametrically opposed incisions that separate the two DNA helices into easily ligatable double strands.
Plants Possess Two Paralogs of Canonical HJ Resolvases
OsGEN-L, the homolog of HsGEN1 and AtGEN1, was the first functional HJ resolvase to be described in plants (Yang et al., 2012). Additional members of the same family include OsSEND-1 and its Arabidopsis ortholog AtSEND1 (Furukawa et al., 2003; Moritoh et al., 2005). In this study, we have shown that AtGEN1, AtSEND1, and their respective orthologs are conserved throughout the spermatophyta and in lycophytes. In contrast, the more ancient mosses carry only one gene coding for a canonical resolvase homologous to AtGEN1 and AtSEND1. This leads us to postulate that a gene duplication took place in the early development of higher plants after separation from the mosses.
Our biochemical data also support the concept that AtGEN1 and AtSEND1 are, indeed, functional enzymes of paralogous origin. Both gene products are canonical HJ resolvases that show strong similarities to both the eukaryotic Rad2/XPG family members and the EcRuvC paradigm of resolution. They also possess additional properties that might represent a gain-of-function variant compared with other eukaryotes (see below). This makes Arabidopsis the first organism, to our knowledge, to be shown to have two functional paralogous resolvases.
The Resolvases of Arabidopsis Show Broadened Substrate Specificities
In contrast to EcRuvC, HsGEN1, ScYen1, and CeGEN-1, both Arabidopsis resolvases are active on a broad range of substrates with no marked preference for HJs (Fig. 9; Benson and West, 1994; Ip et al., 2008; Bailly et al., 2010). This is somewhat reminiscent of the broad substrate specificity observed for the phage resolvases T7 endo I and T4 endo VII (Dickie et al., 1987; Benson and West, 1994). These different substrate specificities are summarized in Supplemental Table S1. The broad substrate specificity is also similar to the ability of HsSLX1-SLX4 to act on these structures, although this human protein recognizes 3′-flaps (Wyatt et al., 2013), which are not substrates for AtGEN1 and AtSEND1. The 5′-flap activities of AtGEN1 and AtSEND1 seem identical to that of HsGEN1 as well as HsFEN1, further supporting the postulated universal cleavage mechanism for the superfamily (Tsutakawa et al., 2011).
Figure 9.
Summary of tested substrate types and detected activities. All types of oligonucleotide-based substrates used in the in vitro experiments are shown, and the corresponding activities of AtGEN1 (violet) and AtSEND1 (green) are presented in a simplified manner. The length of the arrows is a rough approximation of the activities of the enzymes on all tested substrate subtypes (e.g. RF1–RF3) to present a more general picture than shown in the detailed data presentation before. The schematic of the mobile nHJs is to be read as follows: AtGEN1 cleaves strand 1 of the nX-32 exactly opposite the discontinuity in strand 3 (continuous arrows), leading to religatable products, whereas for the nX-30, the inherent preference of AtGEN1 to cleave at a certain site in relation to the junction point leads to an incision one nucleotide 3′ of the junction point (dashed arrows). For AtSEND1, the situation is inverted. Cleavage of strand 4 of the nHJs (small arrows) leads to a RF intermediate, which is processed as shown for the RF substrate type. Note that the Ref-I activity can also be observed at the static X0 and to a limited degree, the intact X26. nt, Nucleotide.
Possible Sequence Specificities
Sequence specificity, present to various degrees, is a common feature of nucleases (for example, the type II restriction enzymes and the bacterial resolvases, such as EcRuvC; Shah et al., 1994; Kovall and Matthews, 1999). Furthermore, it is known that the three-dimensional structure in which a HJ is bound by a resolvase may exert some influence on the cleavage decision. Free HJs in solution exist as an equilibrium of different three-dimensional structures, with the two opposite forms of the stacked X structure as extremes and the open planar form as the central intermediate (for review, see Lilley, 2000). Which of the two possible stacked X conformations a HJ adopts depends on the sequence surrounding the junction point (Altona, 1996; Lilley, 2000). Therefore, the selectivity for a tertiary structure of the junction might be confused with sequence specificity. For example, the specificity of AtGEN1 and AtSEND1 for certain positions within the homologous core of the migratable X26 could stem from a preference for certain junction conformations. However, for most characterized HJ resolvases, the three-dimensional conformation of the junction is altered upon binding to the enzyme. The resulting structures often feature unstacked helical arms and resemble the open planar conformation to various degrees (Bennett and West, 1995; Déclais and Lilley, 2000; Ceschini et al., 2001; Biertümpfel et al., 2007). Therefore, the structural influence of the sequence composition on the cleavage preference could be diminished.
We found that AtGEN1 and AtSEND1 showed distinct preferences using four 5′-flap substrates with different sequence compositions. A similar behavior has not been described for HsGEN1, ScYen1, OsGEN-L, or to our knowledge, FEN1. However, using different substrates, the existence of sequence specificities can be assumed (Ip et al., 2008; Bailly et al., 2010; Yang et al., 2012). Marked preferences for cleavage of certain strands of a static HJ by EcRuvC were proposed to stem from the enzyme’s well-characterized sequence specificity (Osman et al., 2009) and also observed for AtGEN1 and AtSEND1. Furthermore, the presence of distinct cleavage patterns for AtGEN1 and AtSEND1 within the mobile core of the X26 might be because of structural or sequence specificities (see above). Many canonical HJ resolvases, including T7 Endo I, EcRuvC, and the eukaryotic resolvases, have been shown to act with sequence specificity on oligonucleotide-based HJs or α-structures (Picksley et al., 1990; Shah et al., 1994; Ip et al., 2008; Bailly et al., 2010; Yang et al., 2012).
Whereas the specific consensus sequences for EcRuvC and in part, the phage enzymes are known, additional investigation using more substrates with different sequences will be needed in case of the eukaryotic HJ resolvases.
Resolution of nHJs Reveals Common Resolvase-Like Characteristics of AtGEN1 and AtSEND1
Despite the masking effect of the Ref-I activity, we were able to discern an inherent property of HJ resolvases in the activity of AtGEN1 and AtSEND1 toward nHJs: the structure-dependent cleavage one nucleotide 3′ of the junction point. Although such an activity would not lead to ligatable products without additional processing, this function retains the resolvase character in cleaving the strand opposite the nick and the structure-specific position. We, therefore, designated this activity as resolvase like.
It has been proposed that a mobile nHJ would adapt a conformation with the nick exactly at the junction point as depicted in Figure 7 (Pöhler et al., 1994). If the Arabidopsis resolvases act on a nHJ in which the junction point is centered at the nick but the sequence differs from their putative sequence preference, both enzymes favor incisions at the position displaced by one nucleotide 3′ from the nick and hence, the junction point. This position, one nucleotide 3′ of the junction point, is exactly the cleavage position that is defined by the structure of the junction, as was observed for the activities of AtGEN1, AtSEND1, EcRuvC, and the other eukaryotic resolvases on 5′-flaps and static HJs (Ip et al., 2008; Osman et al., 2009; Bailly et al., 2010; Yang et al., 2012). Interestingly, AtSEND1 also shows a somewhat broadened cleavage pattern if its sequence preference is not met, which is the case with the nX-32, strand 4 of the static X0, and 5′-flap2. However, AtSEND1 may resolve a nHJ more often canonically than AtGEN1, even if the sequence around the junction point deviates from its putative preference. Mus81-Eme1 from Schizosaccharomyces pombe is known to depend on the exact fitting of nucleotides into the active site, facilitated by junction flexibility, for efficient second-strand cleavage (Chang et al., 2008; Osman et al., 2009). Applied to AtSEND1, this property might explain how AtSEND1 achieves the higher canonical resolution rate with the nHJ: because the nick enhances the flexibility of the junction, one could imagine that this flexibility is used to align the bases directly adjoining to the opposite site of the nick into the active site, such as if it is searching for a better cleavage site. In summary, the processes that occur to resolve nHJs are not a matter of simple cleavage but a complex interplay of at least structure and sequence specificity, if the latter is a property of the respective enzyme.
AtGEN1 and AtSEND1 as Members of the HJ Processing Toolkit
The unfaithful resolution into nonligatable products or through the Ref-I mechanism by AtGEN1 and AtSEND1 in vitro might be considered detrimental for DNA repair and cell survival in vivo. The first possibility would necessitate additional processing, and the second case would reintroduce a double-strand break. Because an intact mobile HJ is predominantly resolved in a canonical manner by AtGEN1 and AtSEND1, it is unclear whether nHJs are physiologically relevant substrates. If they are, the need for processing to ensure ligatability might pose a relatively minor problem, because nHJs occur early during repair processes (Schwartz and Heyer, 2011). In contrast, the activity of AtGEN1 and AtSEND1 on intact mobile HJs is in good agreement with findings from yeast. ScYen1 was shown to act late during meiosis and mitosis, thus serving as a backup mechanism if previous repair pathways, such as dissolution or ScMus81-Mms4-mediated cleavage, should fail to do the job (Matos et al., 2011).
The products from the action of AtGEN1 and AtSEND1 on the static HJs and nHJs observed in this study are similar to the pattern seen for OsGEN-L (Yang et al., 2012). We, therefore, think that it is likely that OsGEN-L acts on HJs in a manner similar to that of AtGEN1 and AtSEND1, including the presence of the alternative Ref-I activity. Another example of a similar activity is HsSLX1-SLX4. During preparation of this article, a functional interaction of HsMUS81-EME1 and HsSLX1-SLX4, which together form a heterotetrameric bona fide HJ resolvase, was described (Wyatt et al., 2013). Despite earlier findings showing that HsSLX1-SLX4 can act on its own as a canonical HJ resolvase, these studies describe the heterodimer HsSLX1-SLX4 as a nicking enzyme (Fekairi et al., 2009; Wyatt et al., 2013). This is based on a low yield of ligatable products compared with HsGEN1 and the substantial quantity of nicking products assayed using a plasmid-based HJ similar to the one used in this study.
In comparison, both AtGEN1 (up to 70%) and AtSEND1 (50%) show substantially higher percentages of resolution products in the plasmid assay than the 20% seen with HsSLX1-SLX4. However, they possess more nicking activity than HsGEN1, which acts almost exclusively as a resolvase (Rass et al., 2010). Another very recent work showed that HsSLX1-SLX4 defines one of three parallel pathways of HJ processing: the other two pathways are resolution by HsGEN1 and dissolution (Garner et al., 2013). Strikingly, although AtGEN1 and AtSEND1 are orthologs of HsGEN1, several rounds of BLAST analyses with SLX4 from S. cerevisiae and the human ortholog (also known as HsBTBD12) failed to identify a putative SLX4 ortholog in Arabidopsis (M. Bauknecht and D. Kobbe, unpublished data). If Arabidopsis possesses an SLX4 homolog, it is most likely heavily altered and therefore, might be unlikely to fulfill the same functions as in other organisms. Thus, Arabidopsis might be missing the important scaffold protein, which is known in humans to interact with HsMUS81, HsSLX1, and HsXPF-ERCC1 and enhance their activities (Fekairi et al., 2009; Muñoz et al., 2009; Svendsen et al., 2009; Andersen et al., 2011).
Our data define AtGEN1 and AtSEND1 as enzymes with in vitro properties intermediate between HsGEN1 and HsSLX1-SLX4 with respect to resolution and their alternative activity. Thus, AtGEN1 and AtSEND1 provide examples of duplicated proteins that have been conserved in plant evolution, which have some properties similar to their mammalian counterparts but also differ in some aspects from those and each other (Hartung et al., 2007a; Geuting et al., 2009; Knoll and Puchta, 2011; Schröpfer et al., 2014). Although the basic principles of pathways in DNA repair and recombination are conserved, plants feature differences in the detailed organization of the pathways.
CONCLUSION
We suggest that, in contrast to mosses and animals like humans, which have one GEN1 homolog, seed plants have two functional, canonical HJ resolvases, which we have shown here for Arabidopsis in vitro. The two evolutionarily related paralogs have similar biochemical properties (e.g. with respect to their relatively broad substrate spectra and processing mechanisms), but they differ slightly in their possible sequence preferences. This allowed us to unravel a common inherent mechanism in processing of nHJs by the two paralogs AtGEN1 and AtSEND1. Because the two paralogs are evolutionarily conserved, it is likely that they both play important, possibly nonredundant roles in the nucleic acid metabolism of the cell.
MATERIALS AND METHODS
Bioinformatics Analyses
BLAST analyses were conducted with the database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). Multiple sequence alignments were performed using the multiple sequence comparison by log expectation algorithm at the European Molecular Biology Laboratory-European Bioinformatics Institute (http://www.ebi.ac.uk). MEGA 6.0 (Tamura et al., 2013) was used to calculate the cladogram. A more detailed description of tools, databases, and parameters used is provided online (Supplemental Methods S1).
Cloning of AtGEN1 and AtSEND1
All primers and oligonucleotides used for cloning are listed in Supplemental Table S2. Total RNA was extracted from 2-week-old Arabidopsis (Arabidopsis thaliana) ecotype Columbia seedlings and reverse transcribed into cDNA. For sequencing, the cDNA was amplified using primers 1 + 2 (AtGEN1) and 3 + 4 (AtSEND1) and subcloned into pGEM-T Easy (Promega Corporation). Restriction sites for BamHI and KpnI were then attached to the full-length open reading frames by PCR primers 5 + 6 (AtGEN1) and 7 + 8 (AtSEND1). The coding sequences for the nuclease-deficient proteins AtGEN1 D75A and AtSEND1 D76A were created by overlap extension PCR using the pGEM-T Easy clones as templates and primers 5, 6, 9, and 10 and 7, 8, 11, and 12, resulting in an exchange from Asp to Ala at positions 75 (AtGEN1) and 76 (AtSEND1), respectively.
For the expression constructs, the vector pETDuet1 (EMD Millipore) was modified (oligonucleotides 13–18; Supplemental Table S2; Supplemental Methods S2) to yield recombinant proteins featuring an unchanged/native N terminus and a C terminus with a PreScission cleavage site + His tag + StrepII tag (final additional C-terminal amino acid sequence: GTLEVLFQGPTGHHHHHHLESGSTSAWSHPQFEK).
Overexpression and Purification
Heterologous protein expression was performed in Escherichia coli strain ER2566 (NEB) liquid cultures cotransformed with the vector pTf16 (Takara Bio Inc.). Chaperone expression was induced with 0.5 mg mL−1 of l-arabinose at the inoculation start. After the cultures reached an absorbance at 600 nm of 0.7 after growth at 28°C and 200 rpm, they were cooled to 16°C for 30 min. The expression of the target proteins was then induced with 0.2 mm isopropyl-β-d-thiogalactopyranoside for 3 h at 16°C and 200 rpm. The cells were harvested by centrifugation.
All purification steps were performed at 4°C. The pellets from 2.2 L of liquid culture were thawed and resuspended in 75 mL of buffer A (100 mm Tris-HCl, 500 mm NaCl, 1 mm dithiothreitol [DTT], and 0.1% [v/v] Tween20, pH 8.0). The cells were lysed by incubation with 0.1 mg mL−1 of lysozyme for 30 min on ice followed by sonication. The lysate was centrifuged for 30 min at 40,000g, and the supernatant was filtered using a glass fiber/polyester membrane and supplemented with 1 µg mL−1 of Avidin. This clear lysate was loaded at a flow rate of approximately 1 mL min−1 on self-packed 6-mL streptactin (Strep-Tactin Superflow; IBA) gravity flow columns equilibrated with buffer A. After washing with 41.4 mL of buffer A, the proteins were eluted in three steps: 1 × 2.34 mL, 1 × 9 mL, and 1 × 3 mL of buffer B (buffer A with 3 mm desthiobiotin). The second fraction, containing most of the protein, was supplemented with 20 mm imidazole and loaded on a HisTrap FF crude column (GE Healthcare) at a flow rate of 0.25 mL min−1. All of the following steps were conducted at a flow rate of 1 mL min−1. An initial washing step with 45 mL of buffer C (100 mm Tris-HCl, 500 mm NaCl, 20 mm imidazole, and 5% [v/v] glycerol, pH 8.0) + 0.1% (v/v) Triton X-100 was followed by another wash with 15 mL of buffer C. The proteins were eluted with buffer D (100 mm Tris-HCl, 500 mm NaCl, 400 mm imidazole, and 5% [v/v] glycerol, pH 8.0), the peak fractions were pooled, and their buffer was exchanged for buffer A using PD10 columns (GE Healthcare) according to the manufacturer’s instructions. The final fractions were supplemented with 50% (v/v) glycerol and stored in aliquots at −80°C.
The purified proteins were identified by colloidal Coomassie Brilliant Blue-stained (Neuhoff et al., 1988) SDS-PAGE gels and western blot. Quantification was performed with the ImageJ software (http://rsb.info.nih.gov/ij/) using bovine serum albumin (Bio-Rad) as a standard.
DNA Substrates
Most substrates were based on oligonucleotides (Supplemental Tables S3 and S4). The HJ substrates X0, X26, and X26-S as well as the 3′-flap and 5′-flap4 were composed of the same oligonucleotides as described elsewhere (Ip et al., 2008), but a different protocol was used. For the nX-30, nX-32, 3′-flap, and 5′\x{2011}flaps, one 5′ 32P-labeled oligonucleotide was annealed with a 4-fold molar excess of unlabeled oligonucleotides in 1× buffer of 70 mm Tris-HCl, 10 mm MgCl2, and 5 mm DTT (pH 7.6) by heating to 95°C for 5 min followed by cooling to room temperature (RT). In case of X0, X26, X26-S, and the RFs, oligonucleotides constituting strands 1 and 4 were first annealed in an equimolar ratio by heating to 95°C for 5 min followed by cooling to RT. Similarly, strands 2 and 3 were annealed in a separate reaction. In a second step, the previously annealed splayed arm structures were combined and incubated at 37°C for 30 min followed by another 30 min at RT. All substrates (except the X26-S) were purified by 10% [w/v] native Tris-borate/EDTA (TBE) -PAGE and electroelution into Tris-borate/MgCl2 buffer (44.5 mm Tris-Base, 44.5 mm boric acid, and 5 mm MgCl2) using D-Tube Dialyzers (Merck). All HJs (except the X26-S) were labeled on one oligonucleotide for each HJ, resulting in four differently labeled substrates per HJ.
To create pIR9, pAT153 (MoBiTec) was linearized by EcoRI digestion, and the single-strand overhangs were converted to blunt ends using mung bean nuclease (NEB). After religation, a plasmid without the EcoRI site was obtained and verified by sequencing. The inverted repeat was created by self-annealing of oligonucleotide 19 (Supplemental Table S2) and inserted into the BamHI site of the vector just described. Oligonucleotide 19 was designed with overhangs corresponding to a BamHI digest, sequences of the homologous core of the X26 HJ (underlined in Supplemental Table S2) and the inverted repeat sequence (italics in Supplemental Table S2), and central EcoRI site (bold in Supplemental Table S2) used for pIRbke8 (Lilley and Markham, 1983). The formation of the HJ was favored by incubation at 37°C in HJ extrusion buffer (50 mm Tris-HCl, 50 mm NaCl, 0.1 mm EDTA, pH 7.5) before performing the assays.
Endonuclease Assays
The standard reactions (20 µL) contained 1.5 nm 32P-labeled substrate and 30 nm protein in 1× reaction buffer (25 mm HEPES-KOH, 5 mm Tris-HCl, 50 mm NaCl, 2 mm MgCl2, 1.05 mm DTT, 0.1 mg mL−1 of bovine serum albumin, 0.005% [v/v] Tween20, and 5% [v/v] glycerol, pH 8.0). An additional 50 mm NaCl (final concentration of 100 mm) was added to samples containing AtSEND1. Unless otherwise indicated, the reactions were incubated at 37°C for 30 min and terminated by the addition of 10 µL of native stopping solution (50 mm EDTA, 0.6% [v/v] SDS, 20% [v/v] glycerol, 0.1% [v/v] xylenxyanole FF, 0.1% [v/v] bromophenol blue, and 0.95 of mg mL−1 proteinase K) or 20 µL of denaturing stopping solution (89 mm Tris-HCl, 7 m urea, 12% [v/v] Ficoll, 0.8% [v/v] SDS, 0.02% [v/v] xylenxyanole FF, 0.01% [v/v] bromophenol blue, and 1 mg mL−1 of proteinase K, pH 8.0) followed by additional incubation at 37°C for 15 min.
For the ligation experiments, T4 ligase (Fermentas) was added to a final concentration of 40 units µL−1, and the incubation was continued for 30 min at RT. The reactions were stopped by the addition of an equal volume of denaturing stopping solution and incubated for another 15 min at 37°C.
The 32P-labeled reaction products were separated by 12% (w/v) native TBE-PAGE, 20% (w/v) denaturing TBE-PAGE (containing 7 m urea), or sequencing gels with 7 m urea and 13% (v/v) TBE-PAGE followed by autoradiography. Quantification was carried out using a CR 35 Bio for phosphorimaging and the Advanced Image Data Analyzer software (raytest).
For assays with plasmid pIR9, the reactions containing 4.5 nm plasmid and 90 nm enzyme were incubated under the same conditions as for the oligonucleotide-based substrates. Aliquots were taken at each time point. The reaction products were separated on vertical 1% (w/v) Tris-acetate EDTA agarose gels run under native conditions and visualized using GelStar (Lonza Group Ltd) staining. To determine the proportion of supercoiled plasmids extruding the cruciform structure, the samples were digested with EcoRI under the respective assay conditions. The fraction of plasmid containing the HJ structure was defined as 100% of the total substrate DNA.
The Arabidopsis Genome Initiative locus identifiers for AtGEN1 and AtSEND1 are At1g01880 and At3g48900, respectively.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Quantification of the purified recombinant proteins.
Supplemental Figure S2. Structures of the recombinant proteins and negative controls.
Supplemental Figure S3. Cleavage mapping at static 5′-flaps reveals a single main cleavage site.
Supplemental Figure S4. Cleavage mapping at the static HJ X0.
Supplemental Figure S5. Cleavage mapping at the mobile HJ X26.
Supplemental Figure S6. Rational for the design of nX-30 and nX-32.
Supplemental Figure S7. Cleavage pattern at two different nHJs analyzed by native gel electrophoresis reveals both a canonical resolution activity and an activity involving a RF intermediate (Ref-I).
Supplemental Figure S8. Cleavage pattern with different RF-like structures sustains the hypothesis of a RF intermediate in the Ref-I pathway that exists beside the classical resolvase-like pathway in the resolution of an nHJ.
Supplemental Table S1. Examples of characterized canonical HJ resolvases.
Supplemental Table S2. PCR primers and oligonucleotides used in cloning.
Supplemental Table S3. Oligonucleotide combinations used for model substrates in the in vitro assays.
Supplemental Table S4. Oligonucleotide sequences used for model substrates in the in vitro assays.
Supplemental Methods S1. Bioinformatics analysis.
Supplemental Methods S2. Cloning of expression constructs.
Supplemental Information S1. Alignment used for the calculation of the phylogenetic tree shown in Figure 1.
Acknowledgments
We thank Jasmin Jansen, Julia Gebhardt, and Tobias Sprissler for contributions to the cloning of expression vectors and pIR9; Dominik Graf for preliminary experiments with AtGEN1 and AtSEND1; Maria Männle for technical support; Alexander Knoll for discussions on phylogeny; and Manfred Focke and Holger Puchta for general discussions and critical reading of this article.
Glossary
- cDNA
complementary DNA
- DTT
dithiothreitol
- HJ
Holliday junction
- HR
homologous recombination
- MMS
methylmethane sulfonate
- nD
nicked duplex
- nHJ
nicked Holliday junction
- RF
replication fork
- RT
room temperature
- sD
short duplex
- TBE
Tris-borate/EDTA
Footnotes
This work was supported by the German Research Foundation (grant no. KO 4049/1–1 to D.K.), and the Young Investigator Group (grant no. YIG 9–109 to D.K.) received financial support from the Concept for the Future of Karlsruhe Institute of Technology within the framework of the German Excellence Initiative.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Altona C. (1996) Classification of nucleic acid junctions. J Mol Biol 263: 568–581 [DOI] [PubMed] [Google Scholar]
- Andersen SL, Kuo HK, Savukoski D, Brodsky MH, Sekelsky J. (2011) Three structure-selective endonucleases are essential in the absence of BLM helicase in Drosophila. PLoS Genet 7: e1002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagherieh-Najjar MB, de Vries OM, Hille J, Dijkwel PP. (2005) Arabidopsis RecQI4A suppresses homologous recombination and modulates DNA damage responses. Plant J 43: 789–798 [DOI] [PubMed] [Google Scholar]
- Bailly AP, Freeman A, Hall J, Déclais AC, Alpi A, Lilley DMJ, Ahmed S, Gartner A. (2010) The Caenorhabditis elegans homolog of Gen1/Yen1 resolvases links DNA damage signaling to DNA double-strand break repair. PLoS Genet 6: e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett RJ, West SC. (1995) Structural analysis of the RuvC-Holliday junction complex reveals an unfolded junction. J Mol Biol 252: 213–226 [DOI] [PubMed] [Google Scholar]
- Bennett RJ, West SC. (1996) Resolution of Holliday junctions in genetic recombination: RuvC protein nicks DNA at the point of strand exchange. Proc Natl Acad Sci USA 93: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson FE, West SC. (1994) Substrate specificity of the Escherichia coli RuvC protein. Resolution of three- and four-stranded recombination intermediates. J Biol Chem 269: 5195–5201 [PubMed] [Google Scholar]
- Berchowitz LE, Francis KE, Bey AL, Copenhaver GP. (2007) The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet 3: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biertümpfel C, Yang W, Suck D. (2007) Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature 449: 616–620 [DOI] [PubMed] [Google Scholar]
- Blanco MG, Matos J, Rass U, Ip SC, West SC. (2010) Functional overlap between the structure-specific nucleases Yen1 and Mus81-Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair (Amst) 9: 394–402 [DOI] [PubMed] [Google Scholar]
- Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, III, Russell P. (2001) Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107: 537–548 [DOI] [PubMed] [Google Scholar]
- Bonnet S, Knoll A, Hartung F, Puchta H. (2013) Different functions for the domains of the Arabidopsis thaliana RMI1 protein in DNA cross-link repair, somatic and meiotic recombination. Nucleic Acids Res 41: 9349–9360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. (2010) Double Holliday junctions are intermediates of DNA break repair. Nature 464: 937–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceschini S, Keeley A, McAlister MS, Oram M, Phelan J, Pearl LH, Tsaneva IR, Barrett TE. (2001) Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2. EMBO J 20: 6601–6611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JH, Kim JJ, Choi JM, Lee JH, Cho Y. (2008) Crystal structure of the Mus81-Eme1 complex. Genes Dev 22: 1093–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelysheva L, Vezon D, Belcram K, Gendrot G, Grelon M. (2008) The Arabidopsis BLAP75/Rmi1 homologue plays crucial roles in meiotic double-strand break repair. PLoS Genet 4: e1000309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CF, Brill SJ. (2007) Binding and activation of DNA topoisomerase III by the Rmi1 subunit. J Biol Chem 282: 28971–28979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. (2006) Single Holliday junctions are intermediates of meiotic recombination. Cell 127: 1167–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déclais AC, Lilley DM. (2000) Extensive central disruption of a four-way junction on binding CCE1 resolving enzyme. J Mol Biol 296: 421–433 [DOI] [PubMed] [Google Scholar]
- Dickie P, McFadden G, Morgan AR. (1987) The site-specific cleavage of synthetic Holliday junction analogs and related branched DNA structures by bacteriophage T7 endonuclease I. J Biol Chem 262: 14826–14836 [PubMed] [Google Scholar]
- Dunderdale HJ, Sharples GJ, Lloyd RG, West SC. (1994) Cloning, overexpression, purification, and characterization of the Escherichia coli RuvC Holliday junction resolvase. J Biol Chem 269: 5187–5194 [PubMed] [Google Scholar]
- Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, III, Russell P, et al. (2009) Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell 138: 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T, Kimura S, Ishibashi T, Mori Y, Hashimoto J, Sakaguchi K. (2003) OsSEND-1: a new RAD2 nuclease family member in higher plants. Plant Mol Biol 51: 59–70 [DOI] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R. (1994) The yeast type I topoisomerase Top3 interacts with Sgsl, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol 14: 8391–8398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Rendtlew Danielsen J, Wei LZ, Zhou DP, Xu Q, Li MM, Wang ZQ, Tong WM, Yang YG. (2012) A novel role of human holliday junction resolvase GEN1 in the maintenance of centrosome integrity. PLoS ONE 7: e49687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner E, Kim Y, Lach FP, Kottemann MC, Smogorzewska A. (2013) Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced Holliday junctions. Cell Reports 5: 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuting V, Kobbe D, Hartung F, Dürr J, Focke M, Puchta H. (2009) Two distinct MUS81-EME1 complexes from Arabidopsis process Holliday junctions. Plant Physiol 150: 1062–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. (2006) The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J 25: 4921–4932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Suer S, Bergmann T, Puchta H. (2006) The role of AtMUS81 in DNA repair and its genetic interaction with the helicase AtRecQ4A. Nucleic Acids Res 34: 4438–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Suer S, Knoll A, Wurz-Wildersinn R, Puchta H. (2008) Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana. PLoS Genet 4: e1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Suer S, Puchta H. (2007a) Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 18836–18841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Wurz-Wildersinn R, Fuchs J, Schubert I, Suer S, Puchta H. (2007b) The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. Plant Cell 19: 3090–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins JD, Buckling EF, Franklin FC, Jones GH. (2008) Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J 54: 152–162 [DOI] [PubMed] [Google Scholar]
- Ho CK, Mazón G, Lam AF, Symington LS. (2010) Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol Cell 40: 988–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. (1964) A mechanism for gene conversion in fungi. Genet Res 5: 282–304 [DOI] [PubMed] [Google Scholar]
- Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. (2008) Identification of Holliday junction resolvases from humans and yeast. Nature 456: 357–361 [DOI] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE. (2003) Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115: 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H, Takahagi M, Shiba T, Nakata A, Shinagawa H. (1991) Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure. EMBO J 10: 4381–4389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. (2009) The DNA-damage response in human biology and disease. Nature 461: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll A, Puchta H. (2011) The role of DNA helicases and their interaction partners in genome stability and meiotic recombination in plants. J Exp Bot 62: 1565–1579 [DOI] [PubMed] [Google Scholar]
- Knoll A, Schröpfer S, Puchta H. (2014) The RTR complex as caretaker of genome stability and its unique meiotic function in plants. Front Plant Sci 5: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall RA, Matthews BW. (1999) Type II restriction endonucleases: structural, functional and evolutionary relationships. Curr Opin Chem Biol 3: 578–583 [DOI] [PubMed] [Google Scholar]
- Laubinger S, Zeller G, Henz SR, Sachsenberg T, Widmer CK, Naouar N, Vuylsteke M, Schölkopf B, Rätsch G, Weigel D. (2008) At-TAX: a whole genome tiling array resource for developmental expression analysis and transcript identification in Arabidopsis thaliana. Genome Biol 9: R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Chen C, Markmann-Mulisch U, Timofejeva L, Schmelzer E, Ma H, Reiss B. (2004) The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc Natl Acad Sci USA 101: 10596–10601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Heyer WD. (2008) Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 18: 99–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley DMJ. (2000) Structures of helical junctions in nucleic acids. Q Rev Biophys 33: 109–159 [DOI] [PubMed] [Google Scholar]
- Lilley DMJ, Markham AF. (1983) Dynamics of cruciform extrusion in supercoiled DNA: use of a synthetic inverted repeat to study conformational populations. EMBO J 2: 527–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri HW, Hickson ID. (2007) The RecQ helicase-topoisomerase III-Rmi1 complex: a DNA structure-specific ‘dissolvasome’? Trends Biochem Sci 32: 538–546 [DOI] [PubMed] [Google Scholar]
- Mannuss A, Dukowic-Schulze S, Suer S, Hartung F, Pacher M, Puchta H. (2010) RAD5A, RECQ4A, and MUS81 have specific functions in homologous recombination and define different pathways of DNA repair in Arabidopsis thaliana. Plant Cell 22: 3318–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J, Blanco MG, Maslen S, Skehel JM, West SC. (2011) Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147: 158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritoh S, Miki D, Akiyama M, Kawahara M, Izawa T, Maki H, Shimamoto K. (2005) RNAi-mediated silencing of OsGEN-L (OsGEN-like), a new member of the RAD2/XPG nuclease family, causes male sterility by defect of microspore development in rice. Plant Cell Physiol 46: 699–715 [DOI] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. (2001) Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157: 103–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ. (2005) Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol Cell Biol 25: 4476–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz IM, Hain K, Déclais AC, Gardiner M, Toh GW, Sanchez-Pulido L, Heuckmann JM, Toth R, Macartney T, Eppink B, et al. (2009) Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell 35: 116–127 [DOI] [PubMed] [Google Scholar]
- Muñoz-Galván S, Tous C, Blanco MG, Schwartz EK, Ehmsen KT, West SC, Heyer WD, Aguilera A. (2012) Distinct roles of Mus81, Yen1, Slx1-Slx4, and Rad1 nucleases in the repair of replication-born double-strand breaks by sister chromatid exchange. Mol Cell Biol 32: 1592–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhoff V, Arold N, Taube D, Ehrhardt W. (1988) Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis 9: 255–262 [DOI] [PubMed] [Google Scholar]
- Osman F, Dixon J, Doe CL, Whitby MC. (2003) Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol Cell 12: 761–774 [DOI] [PubMed] [Google Scholar]
- Osman F, Gaskell L, Whitby MC. (2009) Efficient second strand cleavage during Holliday junction resolution by RuvC requires both increased junction flexibility and an exposed 5′ phosphate. PLoS ONE 4: e5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuji N, Iyehara H, Hideshima Y. (1974) Isolation and characterization of an Escherichia coli ruv mutant which forms nonseptate filaments after low doses of ultraviolet light irradiation. J Bacteriol 117: 337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panyutin IG, Hsieh P. (1994) The kinetics of spontaneous DNA branch migration. Proc Natl Acad Sci USA 91: 2021–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picksley SM, Parsons CA, Kemper B, West SC. (1990) Cleavage specificity of bacteriophage T4 endonuclease VII and bacteriophage T7 endonuclease I on synthetic branch migratable Holliday junctions. J Mol Biol 212: 723–735 [DOI] [PubMed] [Google Scholar]
- Pöhler JR, Duckett DR, Lilley DMJ. (1994) Structure of four-way DNA junctions containing a nick in one strand. J Mol Biol 238: 62–74 [DOI] [PubMed] [Google Scholar]
- Rass U, Compton SA, Matos J, Singleton MR, Ip SC, Blanco MG, Griffith JD, West SC. (2010) Mechanism of Holliday junction resolution by the human GEN1 protein. Genes Dev 24: 1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröpfer S, Kobbe D, Hartung F, Knoll A, Puchta H. (2014) Defining the roles of the N-terminal region and the helicase activity of RECQ4A in DNA repair and homologous recombination in Arabidopsis. Nucleic Acids Res 42: 1684–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. (1995) Identification of double Holliday junctions as intermediates in meiotic recombination. Cell 83: 783–791 [DOI] [PubMed] [Google Scholar]
- Schwartz EK, Heyer WD. (2011) Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma 120: 109–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah R, Bennett RJ, West SC. (1994) Genetic recombination in E. coli: RuvC protein cleaves Holliday junctions at resolution hotspots in vitro. Cell 79: 853–864 [DOI] [PubMed] [Google Scholar]
- Sharples GJ, Benson FE, Illing GT, Lloyd RG. (1990) Molecular and functional analysis of the ruv region of Escherichia coli K-12 reveals three genes involved in DNA repair and recombination. Mol Gen Genet 221: 219–226 [DOI] [PubMed] [Google Scholar]
- Sharples GJ, Ingleston SM, Lloyd RG. (1999) Holliday junction processing in bacteria: insights from the evolutionary conservation of RuvABC, RecG, and RusA. J Bacteriol 181: 5543–5550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shida T, Iwasaki H, Saito A, Kyogoku Y, Shinagawa H. (1996) Analysis of substrate specificity of the RuvC holliday junction resolvase with synthetic Holliday junctions. J Biol Chem 271: 26105–26109 [DOI] [PubMed] [Google Scholar]
- Svendsen JM, Harper JW. (2010) GEN1/Yen1 and the SLX4 complex: solutions to the problem of Holliday junction resolution. Genes Dev 24: 521–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, Elledge SJ, Harper JW. (2009) Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell 138: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. (1983) The double-strand-break repair model for recombination. Cell 33: 25–35 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay YD, Wu L. (2010) Overlapping roles for Yen1 and Mus81 in cellular Holliday junction processing. J Biol Chem 285: 11427–11432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, et al. (2011) Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell 145: 198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Tainer JA. (2012) Double strand binding-single strand incision mechanism for human flap endonuclease: implications for the superfamily. Mech Ageing Dev 133: 195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N, Singh MB, Misra MK, Bhalla PL, Tuteja R. (2001) Molecular mechanisms of DNA damage and repair: progress in plants. Crit Rev Biochem Mol Biol 36: 337–397 [DOI] [PubMed] [Google Scholar]
- West SC. (2009) The search for a human Holliday junction resolvase. Biochem Soc Trans 37: 519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID. (2003) The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874 [DOI] [PubMed] [Google Scholar]
- Wyatt HD, Sarbajna S, Matos J, West SC. (2013) Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol Cell 52: 234–247 [DOI] [PubMed] [Google Scholar]
- Yang J, Bachrati CZ, Ou J, Hickson ID, Brown GW. (2010) Human topoisomerase IIIalpha is a single-stranded DNA decatenase that is stimulated by BLM and RMI1. J Biol Chem 285: 21426–21436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ishino S, Yamagami T, Kumamaru T, Satoh H, Ishino Y. (2012) The OsGEN-L protein from Oryza sativa possesses Holliday junction resolvase activity as well as 5′-flap endonuclease activity. J Biochem 151: 317–327 [DOI] [PubMed] [Google Scholar]
- Zakharyevich K, Tang S, Ma Y, Hunter N. (2012) Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149: 334–347 [DOI] [PMC free article] [PubMed] [Google Scholar]