During the past 54 million years of evolution, 94% of ancestral legume leucine-rich-repeat gene lineages have experienced deletions or expansions, while 6% have been maintained in a conservative manner.
Abstract
Proper utilization of plant disease resistance genes requires a good understanding of their short- and long-term evolution. Here we present a comprehensive study of the long-term evolutionary history of nucleotide-binding site (NBS)-leucine-rich repeat (LRR) genes within and beyond the legume family. The small group of NBS-LRR genes with an amino-terminal RESISTANCE TO POWDERY MILDEW8 (RPW8)-like domain (referred to as RNL) was first revealed as a basal clade sister to both coiled-coil-NBS-LRR (CNL) and Toll/Interleukin1 receptor-NBS-LRR (TNL) clades. Using Arabidopsis (Arabidopsis thaliana) as an outgroup, this study explicitly recovered 31 ancestral NBS lineages (two RNL, 21 CNL, and eight TNL) that had existed in the rosid common ancestor and 119 ancestral lineages (nine RNL, 55 CNL, and 55 TNL) that had diverged in the legume common ancestor. It was shown that, during their evolution in the past 54 million years, approximately 94% (112 of 119) of the ancestral legume NBS lineages experienced deletions or significant expansions, while seven original lineages were maintained in a conservative manner. The NBS gene duplication pattern was further examined. The local tandem duplications dominated NBS gene gains in the total number of genes (more than 75%), which was not surprising. However, it was interesting from our study that ectopic duplications had created many novel NBS gene loci in individual legume genomes, which occurred at a significant frequency of 8% to 20% in different legume lineages. Finally, by surveying the legume microRNAs that can potentially regulate NBS genes, we found that the microRNA-NBS gene interaction also exhibited a gain-and-loss pattern during the legume evolution.
To combat the constant challenges by pathogens, plants have evolved a sophisticated two-layered defense system, in which proteins encoded by disease RESISTANCE (R) genes serve to sense pathogen invasion signals and to elicit defense responses (Jones and Dangl, 2006; McDowell and Simon, 2006; Bent and Mackey, 2007). Over 140 R genes have been characterized from different flowering plants, which confer resistance to a large array of pathogens, including bacteria, fungi, oomycetes, viruses, and nematodes (Liu et al., 2007; Yang et al., 2013). Among these, about 80% belong to the NBS-LRR class, which encodes a central nucleotide-binding site (NBS) domain and a C-terminal leucine-rich repeat (LRR) domain. Based on whether their N termini are homologous to the Toll/Interleukin1 receptor (TIR), the angiosperm NBS-LRR genes are further divided into the TIR-NBS-LRR (TNL) subclass and the non-TIR-NBS-LRR (nTNL) subclass (Meyers et al., 1999; Bai et al., 2002; Cannon et al., 2002). The latter has been also called CC-NBS-LRR (CNL) subclass, since a coiled-coil (CC) structure is often detected at the N terminus (Meyers et al., 2003). Interestingly, a small group of nTNL genes have an N-terminal RPW8-like domain with a transmembrane region before the CC structure (Xiao et al., 2001). This group of RPW8-NBS-LRR (RNL) genes has been usually viewed as a specific lineage of CNLs (Bonardi et al., 2011; Collier et al., 2011); however, its real phylogenetic relationship with CNLs and TNLs requires further investigation.
NBS-LRR genes have been surveyed in many sequenced genomes of flowering plants, including four monocots: rice (Oryza sativa), maize (Zea mays), sorghum (Sorghum bicolor), and Brachypodium distachyon; one basal eudicot: Nelumbo nucifera; two asterid species: potato (Solanum tuberosum) and tomato (Solanum lycopersicum); and 14 rosids: Vitis vinifera, Populus trichocarpa, Ricinus communis, Medicago truncatula, soybean (Glycine max), Lotus japonicus, Cucumis sativus, Cucumis melo, Citrullus lanatus, Gossypium raimondii, Carica papaya, Arabidopsis (Arabidopsis thaliana), Arabidopsis lyrata, and Brassica rapa (Bai et al., 2002; Meyers et al., 2003; Monosi et al., 2004; Zhou et al., 2004; Yang et al., 2006, 2008b; Ameline-Torregrosa et al., 2008; Mun et al., 2009; Porter et al., 2009; Chen et al., 2010; Li et al., 2010a, 2010b; Guo et al., 2011; Zhang et al., 2011; Jupe et al., 2012; Lozano et al., 2012; Luo et al., 2012; Tan and Wu, 2012; Andolfo et al., 2013; Jia et al., 2013; Lin et al., 2013; Wan et al., 2013; Wei et al., 2013; Wu et al., 2014). Variable numbers (from dozens to hundreds) of NBS-LRR genes were reported from these genomes, making one wonder: how did these genes evolve so variably during flowering plant speciation?
Comparative genomic studies conducted in the available genome sequences of closely related species or subspecies revealed that a significant proportion of NBS-LRR genes are not shared. For example, 70 NBS-LRR genes between Arabidopsis and A. lyrata show the presence/absence of polymorphisms (Chen et al., 2010; Guo et al., 2011). Moreover, synteny analysis revealed that, among 363 NBS-LRR gene loci in indica (cv 93-11) and japonica (cv Nipponbare) rice, 124 loci exist in only one genome (Luo et al., 2012). Unequal crossover, homologous repair, and nonhomologous repair are the three ways that NBS-LRR gene deletions are caused in rice genomes (Luo et al., 2012).
In many surveyed genomes, the majority of NBS-LRR genes are found in a clustered organization (physically close to each other), with the rest exhibited as singletons. Many clusters are homogenous, with only duplicated members occupying the same phylogenetic lineage, whereas heterogenous clusters comprise members from distantly related clades (Meyers et al., 2003). Leister (2004) defined three types of NBS gene duplications: local tandem, ectopic, and segmental duplications. Although a general agreement on the widespread occurrence of local tandem duplications can be reached by various genome survey studies, the relative importance of ectopic and segmental duplications has been seldom investigated since the earliest surveys of the Arabidopsis genome (Richly et al., 2002; Baumgarten et al., 2003; Meyers et al., 2003; McDowell and Simon, 2006).
With more genomic data available in certain angiosperm families, NBS-LRR genes should be further investigated among phylogenetically distant species to fill the gaps in the understanding of their long-term evolutionary patterns. The legume family contains many economically important crop species, such as clover (Trifolium spp.), soybean, peanut (Arachis hypogaea), and common bean (Phaseolus vulgaris). Although these legumes are constantly threatened by various pathogens, only a few functional legume R genes have been characterized, and all of them belong to the NBS-LRR class (Ashfield et al., 2004; Hayes et al., 2004; Gao et al., 2005; Seo et al., 2006; Yang et al., 2008a; Meyer et al., 2009). Therefore, it would be interesting to investigate the NBS-LRR gene repertoire among different legume species. Here, we carried out a comprehensive analysis of NBS-LRR genes in four divergent legume genomes, M. truncatula, pigeon pea (Cajanus cajan), common bean, and soybean, which shared a common ancestor approximately 54 million years ago (MYA; Fig. 1; Lavin et al., 2005). Approximately 1,000 nTNL and 667 TNL subclass NBS-encoding genes were identified in our study. Their genomic distribution, organization modes, phylogenetic relationships, and syntenic patterns were examined to obtain insight into the long-term evolutionary patterns of NBS-LRR genes.
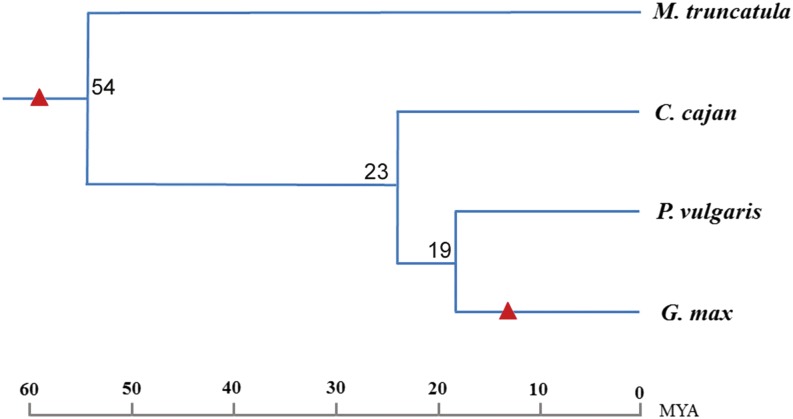
Figure 1.
The phylogenetic tree of four investigated legume species (M. truncatula, pigeon pea, common bean, and soybean). Two WGD events are indicated with triangles: one occurred approximately 59 MYA in the common ancestor of the four investigated legumes, and the other occurred approximately 13 MYA in the Glycine spp. lineage alone (Schmutz et al., 2010). The numbers at the branch nodes indicate divergence times (Lavin et al., 2005; Stefanovic et al., 2009). [See online article for color version of this figure.]
RESULTS
Identification and Classification of NBS-Encoding Genes in the Four Legume Genomes
A total of 1,662 NBS-encoding genes were identified from the four legume genomes: 571 from M. truncatula, 289 from pigeon pea, 337 from common bean, and 465 from soybean. A complete list of these 1,662 NBS genes, as well as their domain compositions, is shown in Supplemental Table S1.
In each legume genome, the identified NBS genes belonging to the nTNL subclass outnumbered those in the TNL subclass. The ratio of their numbers was calculated as 1.08 in M. truncatula, 1.58 in pigeon pea, 2.27 in common bean, and 1.61 in soybean (Table I). Within the nTNL subclass, a majority of the identified NBS genes were CNL genes and their close homologs, with only 17, 10, 10, and 18 RNL genes and their close homologs identified from the M. truncatula, pigeon pea, common bean, and soybean genomes, respectively. In each subclass, not all NBS genes had both an N-terminal domain (TIR, CC, or RPW8) and an LRR domain. For example, in M. truncatula, 44 NBSTIR-LRR genes lacked a detectable TIR domain and 49 TIR-NBS genes lacked an LRR domain. Some NBS genes lacked both the N-terminal and LRR domains (Table I). In addition to these structurally incomplete NBS genes, a total of 224 (39.2%), 145 (50.2%), 209 (62.0%), and 242 (52.0%) intact NBS genes were detected from the M. truncatula, pigeon pea, common bean, and soybean genomes, respectively (Table I). Notably, in the M. truncatula and soybean genomes, the identified NBSCC-LRR genes outnumbered the intact CNL genes (Table I). Reexamination of the identified NBSCC-LRR genes revealed that, in many of these genes, the N-terminal regions were comparable in length to the intact CNL genes; however, the CC structure was not identified, because a strict threshold (0.9) was used for the COILS program in this study (see “Materials and Methods”). A recent study (Wei et al., 2013) has shown that a less strict threshold would result in the identification of more CNL genes.
Table I. Number of identified NBS-encoding genes in the four legume genomes.
| Domain Composition | Abbreviation | M. truncatula | Pigeon Pea | Common Bean | Soybean |
|---|---|---|---|---|---|
| TNL subclass | |||||
| TNL and homologs | 275 (48.1%) | 112 (38.7%) | 103 (30.5%) | 178 (38.3%) | |
| TIR-NBS-LRR | TNL | 121 | 78 | 76 | 124 |
| TIR-NBS | TN | 49 | 6 | 13 | 24 |
| NBSTIR-LRR | NTIRL | 44 | 15 | 12 | 17 |
| NBSTIR only | NTIR | 45 | 11 | 0 | 12 |
| Others | 16 | 2 | 2 | 1 | |
| nTNL subclass | |||||
| CNL and homologs | 279 (48.9%) | 167 (57.8%) | 224 (66.5%) | 269 (57.8%) | |
| CC-NBS-LRR | CNL | 94 | 63 | 128 | 109 |
| CC-NBS | CN | 16 | 7 | 9 | 8 |
| NBSCC-LRR | NCCL | 95 | 53 | 84 | 120 |
| NBSCC only | NCC | 63 | 43 | 2 | 30 |
| Others | 11 | 1 | 1 | 2 | |
| RNL and homologs | 17 (3%) | 10 (3.5%) | 10 (3%) | 18 (3.9%) | |
| RPW8-NBS-LRR | RNL | 8 | 4 | 5 | 9 |
| RPW8-NBS | RN | 0 | 0 | 0 | 1 |
| NBSRPW8-LRR | NRPW8L | 6 | 5 | 4 | 8 |
| NBSRPW8 only | NRPW8 | 3 | 1 | 1 | 0 |
| Others | 0 | 0 | 0 | 0 | |
| Ratio of nTNLs to TNLs | 1.08 | 1.58 | 2.27 | 1.61 | |
| Total No. | 571 | 289 | 337 | 465 |
Distribution and Organization of NBS-Encoding Genes in Legume Genomes
The NBS-encoding genes are distributed unevenly among legume chromosomes (Supplemental Fig. S1). For example, among the eight chromosomes of M. truncatula, chromosome 3 has a maximum number of NBS genes (141; 24.7%) and chromosome 1 has the least number (17; 3%). On a given chromosome, NBS genes are often distributed close to each other. In a previous study, a window size of 250 kb was used to scan all identified NBS genes in the M. truncatula genome and to show that 83.6% of NBS genes are present in clusters of at least two genes (Ameline-Torregrosa et al., 2008). When the same criterion was applied to the completed data of M. truncatula in this study (see “Materials and Methods”), it was found that 479 (88.2%) chromosome-anchored NBS genes were organized into 72 clusters, with each cluster containing two to 23 genes. Meanwhile, 64 NBS genes appeared as singletons in M. truncatula (Table II). Similarly, for all anchored NBS genes in the pigeon pea, common bean, and soybean genomes, 29, 37, and 71 clustered loci and 40, 53, and 116 singleton loci were identified, respectively (Table II). NBS genes in clustered loci were much more abundant than NBS genes in singleton loci, and their ratio ranged from 2.45 to 7.48 among the four legume genomes (Table II). On average, a cluster in M. truncatula (6.65) or common bean (7.35) contained more NBS genes than a cluster in pigeon pea (3.38) or soybean (4.83). The largest cluster was found in common bean, which contains a maximum of 51 NBS genes (Table II). The information on the locus assignment of NBS genes in each legume genome is provided in Supplemental Table S1.
Table II. Organization of NBS-encoding genes in the four legume genomes.
| Genes and Loci | M. truncatula | Pigeon Pea | Common Bean | Soybean |
|---|---|---|---|---|
| No. of chromosome-anchored NBS loci and genes | 136 (543) | 69 (138) | 90 (325) | 187 (459) |
| No. of singleton loci (No. of NBS genes) | 64 (64) | 40 (40) | 53 (53) | 116 (116) |
| No. of clustered loci (No. of NBS genes) | 72 (479) | 29 (98) | 37 (272) | 71 (343) |
| Clustered NBS genes/singleton NBS genes | 7.48 | 2.45 | 5.13 | 2.96 |
| Average (median) No. of NBS genes in clusters | 6.65 (5) | 3.38 (3) | 7.35 (4) | 4.83 (3) |
| No. of clusters with 10 or more NBS genes | 18 | 0 | 7 | 6 |
| No. of NBS genes in the largest cluster | 23 (locus 41) | 9 (locus 28) | 51 (locus 90) | 25 (locus 156) |
Reconstructing the NBS Gene Phylogenies
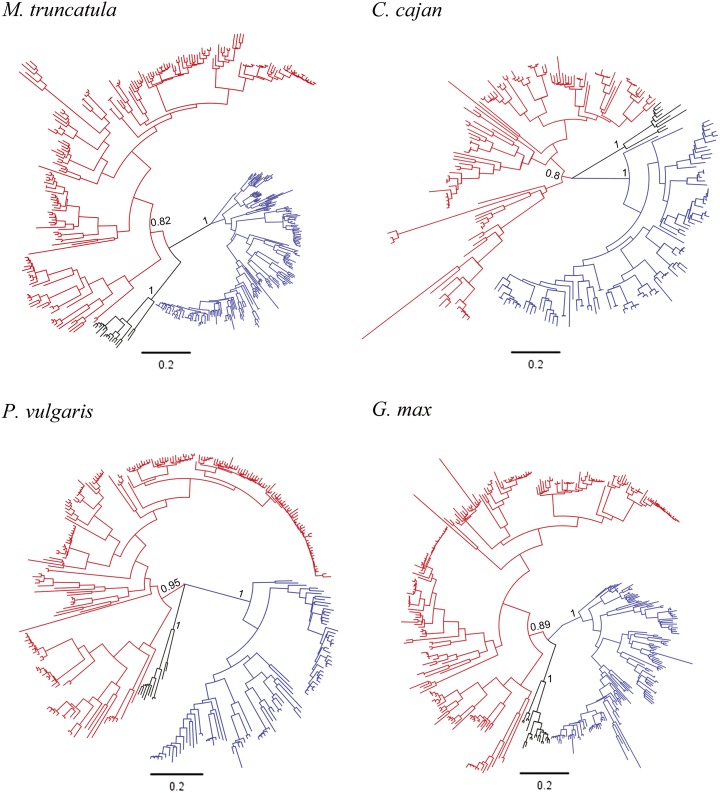
Using the conserved NBS domain sequences in each legume genome, the identified TNL, CNL, and RNL genes and their close homologs were aligned together. After removing short sequences with large deletions, 399, 226, 304, and 378 NBS sequences were finally used to reconstruct NBS gene phylogenies for M. truncatula, pigeon pea, common bean, and soybean, respectively. As shown in Figure 2, the four obtained phylogenies are similar in their overall topology, with TNL, CNL, and RNL genes and their close homologs forming three independent monophyletic clades. It is noteworthy that the RNL clade classified into the nTNL subclass is not embedded into, but sister to, the CNL clade.
Figure 2.
Phylogenetic relationships of NBS-encoding genes in each of the four legume genomes. The TNL clade (blue), CNL clade (red), and RNL clade (black) are shown. The scale bars represent numbers of nucleotide substitutions per site. Support values (SH-aLRT values) for basal nodes are indicated.
To investigate the evolutionary changes of NBS genes within the legume family, and also to explore the variation of main NBS gene lineages beyond the legume family, we reconstructed two phylogenetic trees based on conserved NBS domain sequences, one for the nTNL subclass (including 783 legume NBS sequences and 59 Arabidopsis sequences; Fig. 3; Supplemental Fig. S2) and the other for the TNL subclass (including 524 legume sequences and 99 Arabidopsis sequences; Fig. 4; Supplemental Fig. S3). Both phylogenies were then reconciled with a real species tree to restore the NBS gene duplication and loss events that occurred at different stages of evolution (Supplemental Figs. S4 and S5).
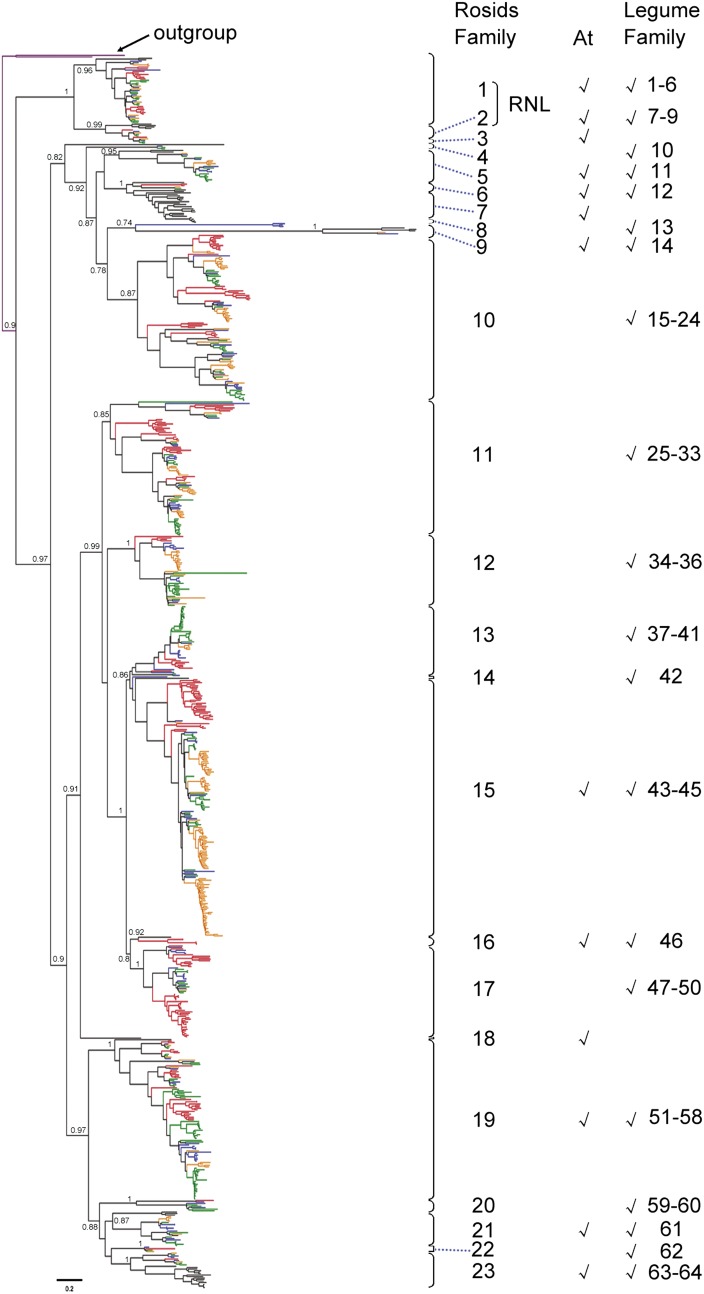
Figure 3.
Phylogenetic tree of nTNL subclass NBS genes based on conserved NBS domain sequences. A total of 842 nTNL subclass NBS sequences were used: 202 sequences from M. truncatula (shown in red), 131 from pigeon pea (blue), 216 from common bean (orange), 234 from soybean (green), and 59 from Arabidopsis (At; black). Two TNL sequences from Arabidopsis (purple) were used as an outgroup. The reconstructed nTNL phylogeny is divided into 23 nTNL-rosid NBS gene families and 64 nTNL-legume NBS gene families (see “Materials and Methods”). The presence of Arabidopsis or legume sequences in nTNL-rosid families is indicated with check marks. The scale bar represents the number of nucleotide substitutions per site. The support values (SH-aLRT values) for basal nodes are indicated.
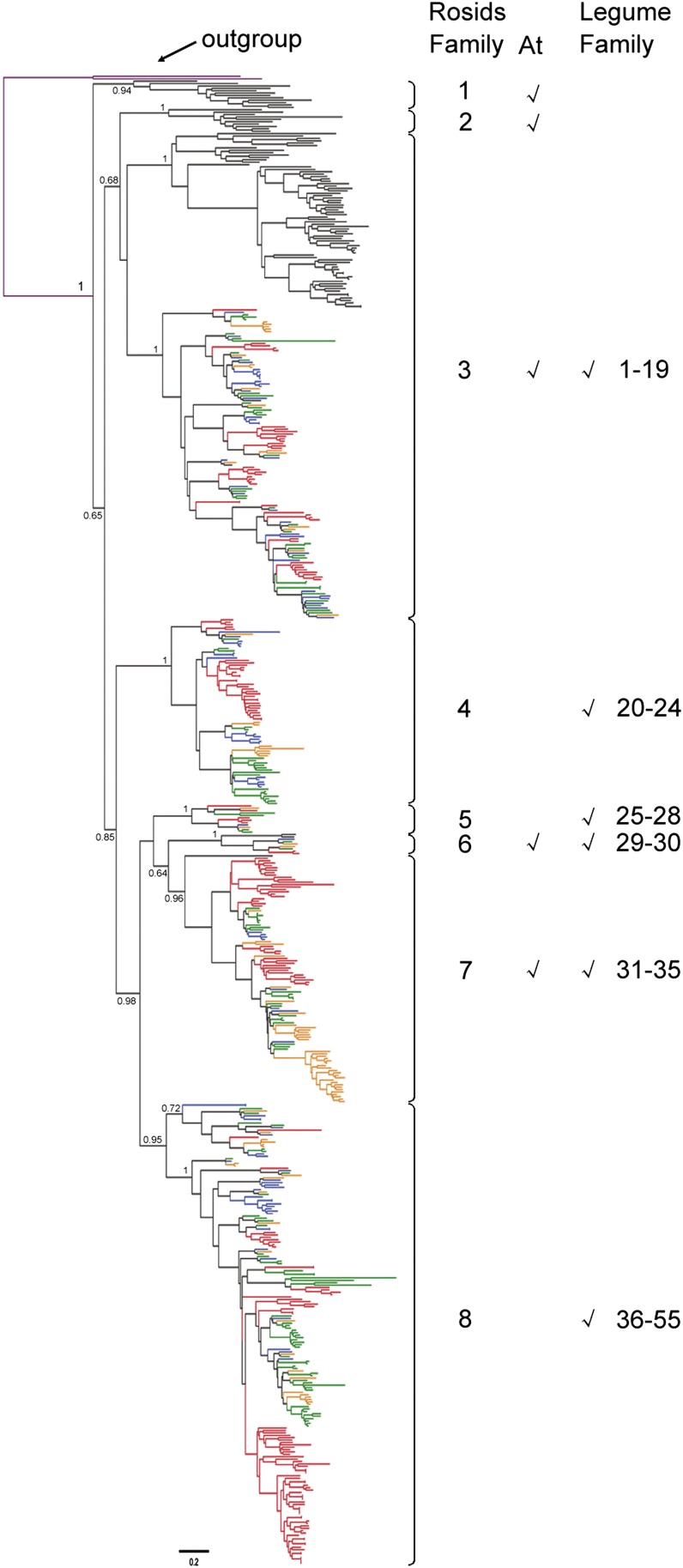
Figure 4.
Phylogenetic tree of TNL subclass NBS genes based on conserved NBS domain sequences. A total of 623 nTNL subclass NBS sequences were used: 197 sequences from M. truncatula (shown in red), 95 from pigeon pea (blue), 88 from common bean (orange), 144 from soybean (green), and 99 from Arabidopsis (At; black). Two nTNL sequences from Arabidopsis (purple) were used as an outgroup. The reconstructed TNL phylogeny is divided into eight TNL-rosid NBS gene families and 55 TNL-legume NBS gene families (see “Materials and Methods”). The presence of Arabidopsis or legume sequences in nTNL-rosid families is indicated with check marks. The scale bar represents the number of nucleotide substitutions per site. The support values (SH-aLRT values) for basal nodes are indicated.
It was revealed that some NBS gene lineages had originated before the divergence of legumes (rosid I clade; Fabaceae) and Arabidopsis (rosid II clade; Brassicaceae). For convenience, NBS genes derived from each of these lineages were defined as a rosid NBS gene family (see “Materials and Methods”). On the reconstructed nTNL phylogeny, 23 rosid NBS gene families were defined, with the first two belonging to an RNL clade and the other 21 forming a CNL clade sister to the RNL clade (Fig. 3; Supplemental Table S2). Notably, not all nTNL-rosid families contain NBS sequences from both Arabidopsis and legumes. NBS sequences were lost from Arabidopsis in families 4, 8, 10 to 14, 17, 20, and 22, while legume NBS sequences were missing in nTNL-rosid families 3, 7, and 18. Overall, in 10 out of 23 nTNL-rosid families, both Arabidopsis and legumes kept NBS gene sequences inherited from their common ancestor. Eight monophyletic rosid NBS gene families were recovered on the reconstructed TNL phylogeny (Fig. 4; Supplemental Table S3). Legume NBS genes were missing from TNL-rosid families 1 and 2, and the Arabidopsis genome had no NBS genes belonging to TNL-rosid families 4, 5, and 8. Therefore, NBS genes are maintained in both Arabidopsis and legumes only in three out of eight TNL-rosid families (families 3, 6, and 7).
Even shared by both rosid lineages, the NBS gene maintenance pattern could be highly different. In TNL-rosid family 7, only one sequence is maintained in Arabidopsis, and it is sister to 103 legume sequences (Fig. 4; Supplemental Table S3). Also in nTNL-rosid families 15 and 19, one Arabidopsis sequence is found sister to 199 and 135 legume sequences, respectively (Supplemental Table S2). Conversely, in nTNL-rosid family 23, 16 sequences were found in Arabidopsis, while only one to three sequences were maintained in four legume species (Fig. 3; Supplemental Table S2).
Differential NBS Gene Losses and Frequent Duplications during Legume Evolution
Many rosid NBS gene lineages were seen to further diverge in the common ancestor of legumes. For example, the main NBS gene lineage of nTNL-rosid family 1 further diverged into six monophyletic sublineages, with each sublineage defined as a legume NBS gene family (see “Materials and Methods”; Supplemental Figs. S2 and S4). In total, 119 legume NBS gene families were defined on the nTNL and TNL phylogenies (Supplemental Figs. S4 and S5).
Among these families, NBS genes were often deleted from one or more legume species, with only 17 out of 64 nTNL families and 10 out of 55 TNL families containing NBS genes from all four legume species (Supplemental Tables S2 and S3). Meanwhile, in approximately 45% of families, NBS genes had expanded their numbers to five or more copies in at least one legume (Supplemental Tables S2 and S3). An exaggerated example comes from nTNL-legume family 45. Up to 106 common bean NBS genes accumulated in this sublineage, representing 45.3% of all identified nTNL genes in this species, while M. truncatula, pigeon pea, and soybean duplicated two, 17, and 34 NBS genes, respectively.
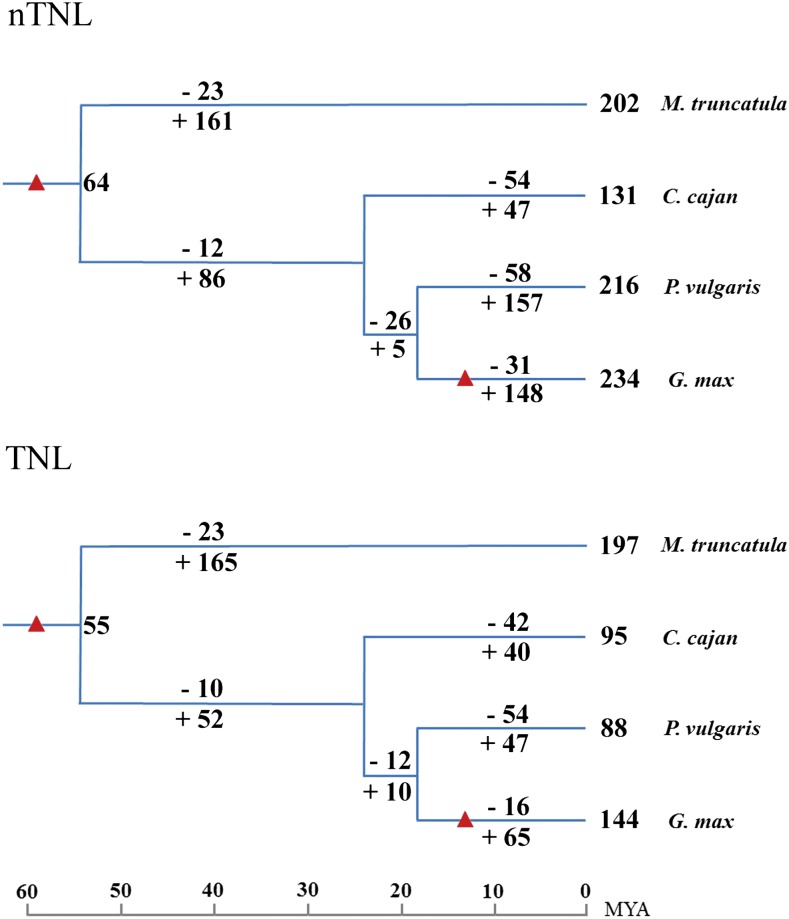
Figure 5 shows the number variations of nTNL and TNL subclass NBS genes at different stages of legume evolution according to the reconstructed phylogenies seen in Supplemental Figures S4 and S5. Since the divergence, the Medicago spp. lineage lost a total of 46 nTNL and TNL genes (23 each) and duplicated 161 nTNL and 165 TNL genes. Moreover, in the lineage leading to the common ancestor of pigeon pea, common bean, and soybean, 12 ancestral nTNL genes and 10 TNL genes were lost, while 86 nTNL and 52 TNL genes were duplicated (Fig. 5). Moreover, on the two terminal branches leading to common bean and soybean, NBS gene losses in the soybean branch appeared to have occurred less frequently than in the common bean branch for both nTNL (−31 versus −58) and TNL (−16 versus −54) subclasses. On the other hand, both branches accumulated more nTNL genes than TNL genes (157 versus 47 and 148 versus 65).
Figure 5.
Number variations of nTNL and TNL subclass NBS genes at different stages of legume evolution. Differential gene losses and gains are indicated by numbers with − or + on each branch. WGD events are indicated with triangles as in Figure 1. [See online article for color version of this figure.]
Despite the frequent deletions and duplications occurring on the majority of NBS gene families during the past 54 million years of legume evolution, there are also seven not-so-active legume NBS gene lineages (nTNL families 51, 52, 62, and 63 and TNL families 7, 13, and 43), in which the NBS genes were maintained rather conservatively among legume species (Supplemental Tables S2 and S3). For example, in both nTNL-legume family 62 and TNL-legume family 43, only one NBS sequence was kept in each legume species. Similarly, in nTNL-legume families 51, 52, and 63 and TNL-legume families 7 and 13, the NBS genes from the four surveyed legumes are all maintained at very low copy numbers (no more than three copies).
NBS Gene Duplication Types and Their Relative Roles in Legume NBS Gene Expansion
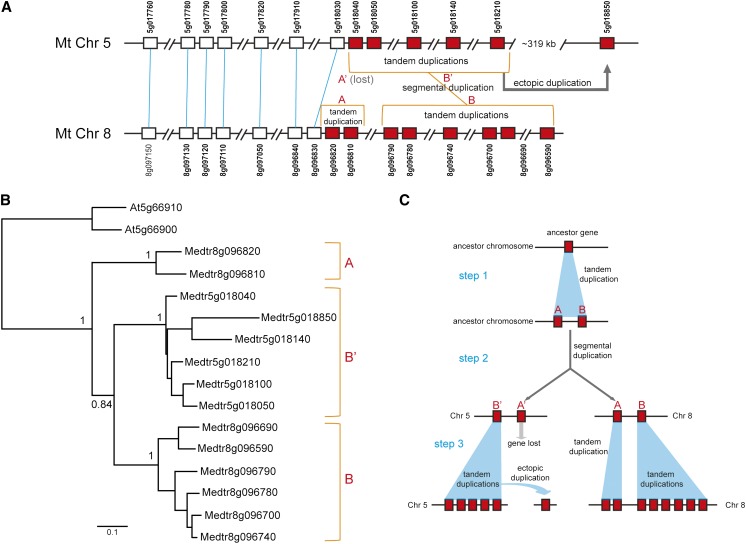
Leister (2004) defined three types of NBS gene duplications: local tandem, ectopic, and segmental duplications. In this study, based on the reconstructed TNL and nTNL phylogenies, we further examined the genomic locations of closely related NBS genes to explore the relative importance of these three duplication types during legume NBS gene expansion (Supplemental Figs. S4 and S5). In general, local tandem duplications produce closely related NBS genes that are also located together. However, both ectopic and segmental duplications produce closely related NBS genes that are located distantly on a chromosome or on different chromosomes. The syntenic relationship of many neighboring genes should be examined to determine whether the duplication is a large-scale event (segmental duplication) or a small-scale event (ectopic duplication). Figure 6 shows an example of how these three different duplication mechanisms occurred in the Medicago spp. genome. Fourteen Medicago spp. NBS genes that belong to nTNL-rosid NBS gene family 1 (Supplemental Table S2) were located on chromosome 5 (loci 68 and 69) and chromosome 8 (locus 134; Supplemental Table S1). Within-genome synteny analysis detected collinearity between these two regions (Fig. 6A). Phylogenetic analysis clearly showed that these 14 sequences form three monophyletic groups (Fig. 6B): group A (8g096810 and 8g096820), group B (8g096590, 8g096690, 8g096700, 8g096740, 8g096780, and 8g096790), and group B′ (5g018040, 5g018050, 5g018100, 5g018140, 5g018210, and g018850). Groups B and B′ are sister to each other and together form a clade sister to group A. Moreover, within group B′, the gene 5g018850 (locus 69) is located approximately 319 kb away from the other five sequences (locus 68). Based on these results, we can reasonably replay the evolutionary steps leading to these NBS genes (Fig. 6C): an ancient NBS gene became two copies (genes A and B), probably through a local tandem duplication; a segmental duplication event occurred to form four genes (A, B, A′, and B′), and multiple local tandem duplications made genes A, B, and B′ become two, six, and five copies, respectively. Furthermore, during the evolutionary process, a gene loss event occurred on gene A′ and an ectopic duplication event resulted in the gene 5g018850. Supplemental Figure S2 shows that steps 1 and 2 should have occurred before the Medicago spp. lineage separated from the other legumes, while step 3 occurred after the Medicago spp. lineage branched. In step 3, four, one, and five local tandem duplications likely took place in groups B′, A, and B, respectively, while one ectopic duplication occurred in group B′.
Figure 6.
A typical example of Medicago spp. NBS gene evolution showing three different duplication types: local tandem duplication, ectopic duplication, and segmental duplication. A, Collinearity shared by Medicago spp. NBS gene loci 68 and 69 on chromosome 5 and locus 134 on chromosome 8. NBS genes are indicated by red boxes, non-NBS genes are indicated by white boxes, with syntenic genes linked by lines. B, Phylogenetic analysis revealed three monophyletic NBS gene groups (A, B, and B′). Two relevant Arabidopsis sequences were used as an outgroup, and all major nodes were supported with high confidence. C, A postulated evolutionary history of these NBS genes: steps 1 and 2 occurred before the Medicago spp. lineage separated from the other legumes, and step 3 occurred mainly in the Medicago spp. lineage.
We applied a similar reasoning process to all other lineages on the reconstructed TNL and nTNL trees. The newly duplicated NBS genes on the terminal branches leading to M. truncatula, common bean, and soybean were evaluated to investigate the relative roles of the three duplication types. As a result (Table III), approximately 80% of all NBS genes in the Medicago spp. lineage were due to local tandem duplications, whereas the rest (approximately 20%) were likely caused by ectopic duplications. No reliable collinearity evidence was found in Medicago spp. to support the formation of NBS genes through lineage-specific segmental duplications. As a matter of fact, the segmental duplication event in Figure 6 occurred before Medicago spp. diverged; therefore, it is not counted (see “Materials and Methods”). In the Phaseolus spp. lineage (after it diverged from the Glycine spp. lineage), 134 (85%) nTNL and 43 (91%) TNL subclass genes were duplicated locally, while 15 (10%) nTNL and two (4%) TNL subclass genes were duplicated ectopically to distant locations (Table III). We also detected one pair of syntenic NBS genes (5g016500 and 6g019300) in the common bean genome. These syntenic genes likely arose by segmental duplication, and the calculated synonymous substitution rate between these two genes in syntenic blocks was only 0.095. In contrast, in the soybean genome, up to 21 pairs of syntenic NBS genes were detected, suggesting that at least 21 NBS genes were born through segmental duplication. The calculated synonymous substitution rate among these 21 pairs of NBS genes ranged from 0.044 to 0.370, with a mean value of 0.158. Furthermore, 111 (75%) nTNL and 52 (80%) TNL genes evolved via local tandem duplications in the soybean genome, while 20 (14%) nTNL and 5 (8%) TNL NBS genes arose ectopically (Table III).
Table III. Relative contributions of three duplication types in producing new NBS genes during the evolution of three legume species.
| Different Types of Duplication | M. truncatula | Common Bean | Soybean | |||
|---|---|---|---|---|---|---|
| nTNL | TNL | nTNL | TNL | nTNL | TNL | |
| Total No. of new duplicated genes | 161 | 165 | 157 | 47 | 148 | 65 |
| Local tandem duplication | 128 (80%) | 130 (79%) | 134 (85%) | 43 (91%) | 111 (75%) | 52 (80%) |
| Ectopic duplication | 33 (20%) | 35 (21%) | 15 (10%) | 2 (4%) | 20 (14%) | 5 (8%) |
| Segmental duplication | 0 | 0 | 0 | 1 (2%) | 14 (9%) | 7 (11%) |
| Unanchored genes | 0 | 0 | 8 | 1 | 3 | 1 |
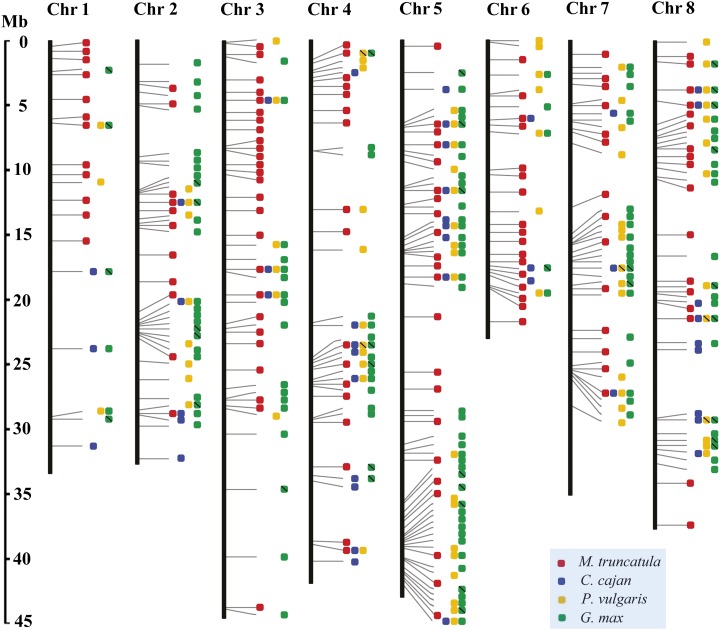
An Integrated Map of NBS Gene Loci for the Four Legume Genomes
A total of 136, 69, 90, and 187 NBS gene loci were assigned to the M. truncatula, pigeon pea, common bean, and soybean genomes, respectively (Table II; Supplemental Table S1). To explore the conservation pattern of these loci across genomes, synteny analyses were performed by mapping syntenic NBS gene loci from different legume genomes onto the M. truncatula chromosomes (see “Materials and Methods”). As a result (Fig. 7; Supplemental Table S4), a total of 298 integrated NBS gene loci were identified. Among them, 225 loci were actually species specific. Either new NBS gene loci were rapidly produced in individual species or common NBS gene loci were lost convergently from multiple species. The other 73 integrated NBS gene loci were maintained in syntenic regions of at least two legume genomes. Among them, only 13 were shared by all four legumes (Fig. 7; Supplemental Table S4). Further examination of these 13 conserved loci revealed that they could be TNL (five cases), CNL (six cases), or RNL (two cases) loci. The previously identified seven not-so-active legume NBS families (nTNL families 51, 52, 62, and 63 and TNL families 7, 13, and 43) were all covered in the 13 conserved loci.
Figure 7.
An integrated map of NBS gene loci showing synteny relationships among four legume genomes. The eight chromosomes of M. truncatula are used as background to map NBS loci from different legume genomes to their syntenic positions. The NBS loci are indicated by squares in different colors: red, M. truncatula; blue, pigeon pea; yellow, common bean; and green, soybean. If two or more loci are mapped to one syntenic position, then a slash is added to the square.
NBS Gene Evolution within nTNL Family 45: A Close Examination of the Evolutionary History of the Rpg1-b/Rsv1 Locus
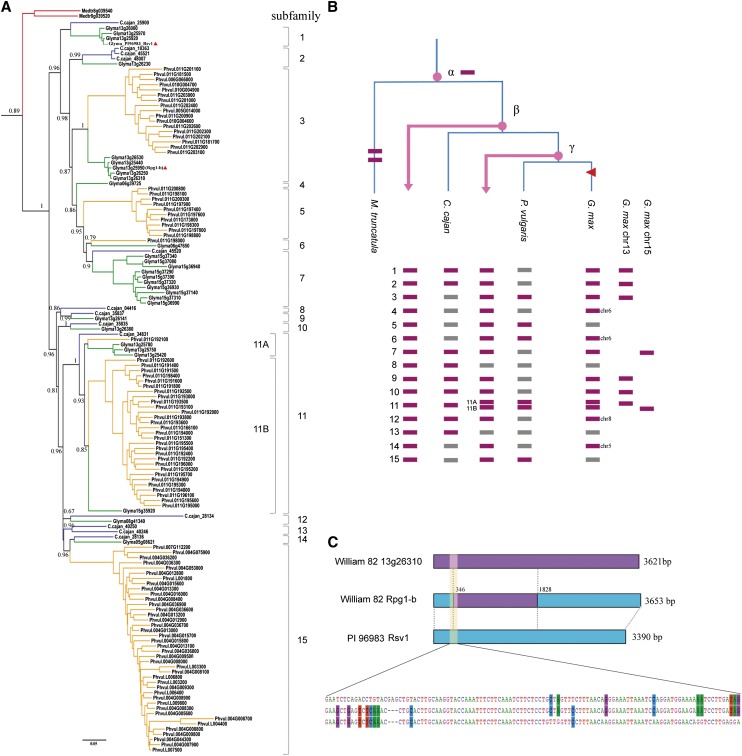
As mentioned previously, among all the defined legume NBS gene families, nTNL family 45 is the largest one, containing a total of 159 NBS genes from four legumes (Supplemental Table S2). This family also contains two known functional R genes from soybean: Rpg1-b, conferring resistance to the bacterial blight pathogen Pseudomonas syringae pv glycinea, and Rsv1, conferring resistance to Soybean mosaic virus (Ashfield et al., 2004; Hayes et al., 2004). We examined this lineage closely to explore how NBS genes became so expanded in this family and to understand the evolutionary relationship between Rpg1-b and Rsv1.
Our data showed that NBS gene expansion in nTNL family 45 mainly occurred after the Medicago spp. lineage separated out. In the common ancestor of pigeon pea, common bean, and soybean, at least 15 copies of NBS genes had been duplicated (Fig. 8). For convenience, we defined genes derived from these copies as subfamilies. According to the phylogeny, pigeon pea later lost subfamilies 3 to 6, 14, and 15, while common bean and soybean together lost subfamilies 8 and 13. Before common bean and soybean diverged, subfamily 11 had duplicated into subfamilies 11A and 11B. Common bean later lost subfamilies 1, 2, 4, 7, 9, 10, 12, and 14, whereas soybean lost only subfamilies 5 and 15 (Fig. 8B).
Figure 8.
An evolutionary history of the legume nTNL family containing known functional genes Rpg1-b and Rsv1. A, The reconstructed phylogeny of NBS genes belonging to nTNL-legume family 45 with the soybean Rsv1 gene incorporated. A total of 15 subfamilies that had evolved in the common ancestor of pigeon pea, common bean, and soybean are labeled. Soybean Rpg1-b and Rsv1 are indicated by red triangles. B, NBS gene subfamilies were differentially lost among later-evolving legume lineages. Common ancestors (α, β, and γ) are indicated by pink dots. The presence/absence of 15 subfamilies in pigeon pea, common bean, soybean, and common ancestor β and γ are indicated with purple/gray bars. C, Graph showing the recombination event that occurred on soybean Rpg1-b. Major parental sequence (PI96983 Rsv1) and minor parental sequence (13g26310) are shown in light blue and purple, respectively. Also, the sequence recombination boundary before the NBS domain is shown with a 111-bp-long alignment.
In soybean, a total of 34 NBS genes belong to nTNL family 45. Among them, 28 are distributed on chromosomes 13 and 15, and their surrounding genes share syntenic relationships, suggesting that they resulted from the recent whole-genome duplication (WGD) event (Innes et al., 2008). Interestingly, NBS gene subfamilies were inherited differentially between these two homologous loci, with six on chromosome 13 and another two on chromosome 15 (Fig. 8B). This pattern was also reported in a recent study (Ashfield et al., 2012). On the other hand, five subfamilies were retained in the common bean genome. The 106 common bean NBS sequences belonging to nTNL-legume family 45 are mainly clustered on chromosomes 11 (56 sequences) and 4 (35 sequences). Geffroy et al. (2009) paid particular attention to sequences on chromosome 4, because some gene members could confer resistance to the fungus Colletotrichum lindemuthianum. The NBS sequences on chromosome 4 are derived from sequences on chromosome 11 through an ancient ectopic recombination event that occurred after the divergence of the Medicago spp. lineage (Geffroy et al., 2009) and before the divergence of Phaseolus and Glycine spp. (David et al., 2009). Our phylogeny corroborates this hypothesis and further dates the ectopic recombination event back to the common ancestor of pigeon pea, common bean, and soybean (Fig. 8).
On the reconstructed phylogeny, soybean Rpg1-b (Glyma13g25950) grouped with Glyma13g26250, Glyma13g26310, Glyma13g26530, and Glyma13g25440, which all belong to subfamily 3. Instead, soybean Rsv1 (isolated from cv PI96983) was close to Glyma13g25920, Glyma13g25970, and Glyma13g26000 in subfamily 1. Notably, Rpg1-b (Glyma13g25950) is physically close to NBS genes in subfamily 1 but phylogenetically close to subfamily 3 genes. Recombination analyses of these soybean sequences revealed that Rpg1-b had acquired an approximately 1.5-kb fragment from a close homolog, Glyma13g26310 (98% identity), through recombination (Fig. 8C).
Legume NBS Genes That Are Potentially Regulated by MicroRNAs
Previously, 263 and 219 families of microRNAs were identified from the M. truncatula and soybean genomes, respectively (Zhai et al., 2011; Kozomara and Griffiths-Jones, 2014). In this study, we identified all NBS genes from the two legume genomes and screened all those microRNAs to find potential regulators of NBS genes.
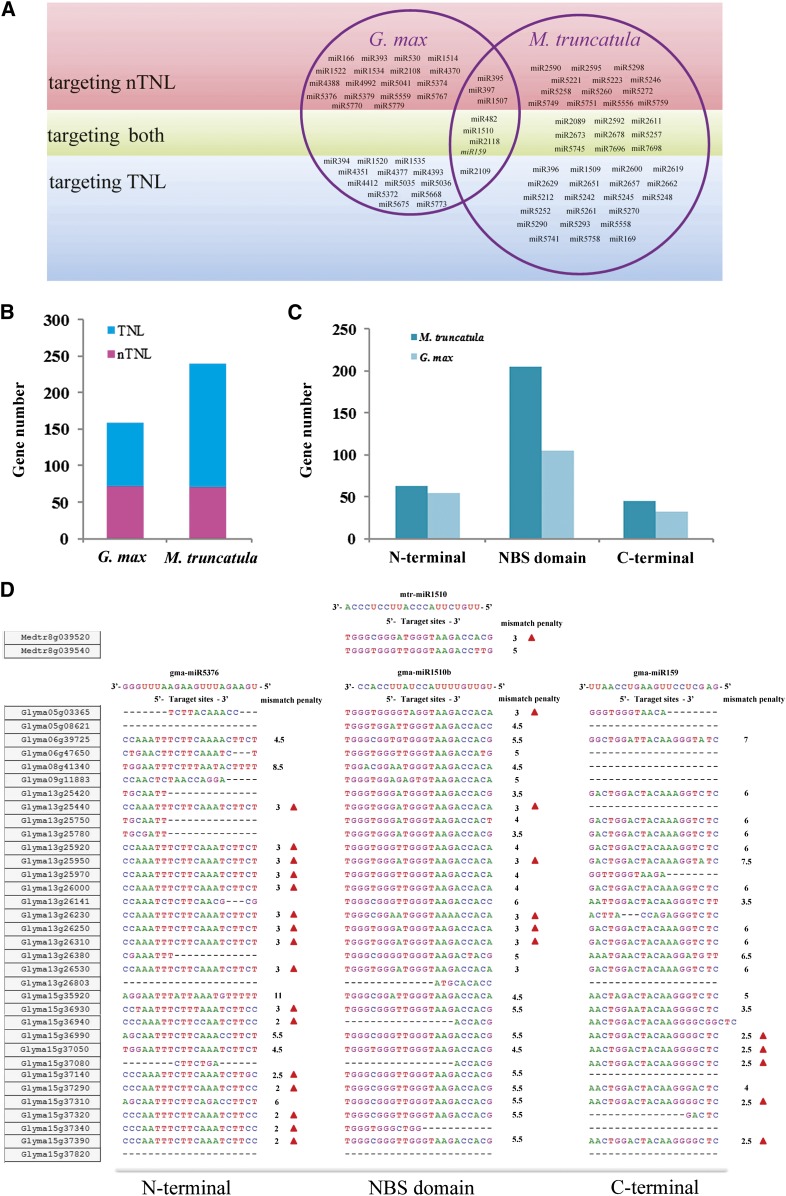
As a result, 51 and 39 families of microRNAs were predicted to regulate NBS genes in M. truncatula and soybean, respectively (Supplemental Table S5). Among them, eight are shared by both legumes and the other 74 seem to regulate NBS genes solely in one genome (Fig. 9A). Interestingly, some microRNA families showed a pattern to recognize multiple NBS genes from successively diverged legume NBS gene lineages (Supplemental Table S5). For example, in Medicago spp., miR1507 was predicted to regulate NBS genes from nTNL families 29, 30, 31, 32, 35, 38, and 43 (note that Medicago spp. had lost NBS genes in nTNL families 33, 39, 41, and 42); miR5261 could recognize NBS genes from TNL families 31, 32, 33, and 34. Such a continuous recognition pattern lends some support to the idea that the regulation relationships had been established before those legume lineages diverged. Some other microRNA families, however, were predicted to recognize only a few closely related NBS genes within a legume NBS gene family, suggesting that the regulation relationships were established during legume evolution. Some examples fitting this pattern include Medicago spp. miR1509, miR5252, miR5290, and miR5749 and Glycine spp. miR4393, miR4412, miR5374, miR5376, and miR5559. Moreover, for 45 microRNA families, each was predicted to recognize only one NBS gene, possibly suggesting newly established regulations.
Figure 9.
Predicted regulation of microRNAs on NBS genes in M. truncatula and soybean. A, Summary of microRNA families predicted to target NBS genes in M. truncatula and soybean. The prediction process was performed by using the psRNATarget server with default settings, in which both the mismatch penalty (expectation value) and target accessibility (allowed maximum energy to unpair the target site) were taken into consideration. B, Numbers of TNL and nTNL subclass NBS genes that were predicted to be targeted by microRNAs in M. truncatula and soybean. C, Localization of microRNA target sites at different domains of NBS genes. D, An example showing the predicted NBS gene regulation pattern within nTNL-legume family 45 by three major microRNAs (miR5376, miR1510, and miR159). Certain NBS gene members were predicted to be regulated by the three microRNAs (shown with red triangles), and they all meet two conditions: the calculated mismatch penalty (expectation value) is no more than 3, and the estimated target accessibility value does not exceed 25 kcal mol−1. Nucleotide mutations, insertions, and deletions at the potential targeting sites often could be detected on genes that were not predicted to be targeted by a given microRNA. All missing nucleotides are indicated with dashes.
A total of 240 and 159 NBS genes were predicted to be regulated by microRNAs in M. truncatula and soybean, respectively. These NBS genes are distributed in 35 nTNL-legume NBS gene families and 46 TNL-legume NBS gene families (Supplemental Table S5). In each legume, the predicted number of TNL genes targeted by microRNAs outnumbered the nTNL genes (Fig. 9B). Furthermore, the targeting sites of microRNAs were mainly located within the NBS domain, although, they could also be located at N-terminal or C-terminal regions (Fig. 9C). Among the 81 legume NBS gene families containing microRNA-targeted members, 12 families (nTNL families 12, 30, 43, 45, 50, and 58 and TNL families 9, 11, 32, 46, 49, and 55) were detected in both legumes, and often some common microRNAs were involved (Supplemental Table S5).
For instance, we again consider nTNL family 45, wherein we aligned 36 M. truncatula and G. max sequences and examined the recognition sites of the three major regulating microRNAs (miR5376, miR1510, and miR159). Our results showed that the shared miR1510 could regulate one out of two M. truncatula genes and six out of 34 G. max genes in this lineage. This further supported the idea that some microRNAs had originated before the divergence of legumes and were maintained in different legume genomes to regulate NBS genes of the same lineages. Furthermore, miR159 and miR5376 were predicted to regulate five and 16 genes, respectively. Together, these three microRNAs could potentially regulate 21 out of 34 G. max NBS genes in this family. Accumulated nucleotide mutations, deletions, and insertions had occurred in many NBS gene members of this lineage, resulting in the failure of recognition by relevant microRNAs (Fig. 9D).
DISCUSSION
Since the pioneering studies in the rice and Arabidopsis genomes (Bai et al., 2002; Meyers et al., 2003), studies have been conducted to survey NBS-LRR genes in other angiosperm genomes, especially in rosid families and other economically important species. The availability of many angiosperm genome sequences provides the opportunity to study long-term evolutionary patterns of NBS-LRR genes. In a recent study, Luo et al. (2012) investigated the genomes of four Poaceae species and revealed a remarkable presence/absence pattern of NBS genes between different grass species, suggesting the occurrence of frequent deletions and translocations of NBS genes in the grass family. However, since monocot genomes contain only the nTNL subclass of NBS genes, the case in the TNL subclass is not inferred.
Besides Poaceae, multiple genome sequences are also available for Fabaceae (rosid I) and Brassicaceae (rosid II). Previous surveys of individual genomes of these two families had revealed that both nTNL and TNL subclass NBS genes are present (Meyers et al., 2003; Ameline-Torregrosa et al., 2008). Therefore, these two families are ideal for studying long-term evolutionary patterns of both nTNL and TNL subclass NBS genes. In this study, we mainly focused on Fabaceae. NBS-LRR genes were isolated from four divergent legume genomes and were further analyzed together with Arabidopsis NBS sequences, which provided a deep view into the history of NBS gene evolution.
The RNL Group Is Not Phylogenetically Embedded in the CNL Clade But Forms Its Own Clade
Within the nTNL subclass, there is a small but special group of NBS-LRR genes that possess an RPW8 domain at their N-terminal region (Xiao et al., 2001). This RNL group is often regarded as a special lineage of CNL genes in the literature and is designated as CCR-NB-LRR (Bonardi et al., 2011; Collier et al., 2011). However, its real phylogenetic relationship with true CNL genes remains unclear. In this study, by using all isolated NBS genes in each legume genome, we reconstructed phylogenies to explore the relationships among RNL, CNL, and TNL genes. Among the four obtained topologies (Fig. 2), RNL, CNL, and TNL genes form independent and monophyletic clades. Therefore, in legumes, the RNL clade does not appear to be a lineage of the CNL clade but sister to the CNL clade, if both RNL and CNL are still viewed as nTNL subclass NBS genes.
The landmark work done by Meyers et al. (2003) showed that RNL genes form a monophyletic group (CNL-A group), which is located at a very basal, but not the most basal, position on the reconstructed nTNL phylogeny. Instead, two sequences (At4g19050 and At5g45510) having an NBS-LRR structure are located at the most basal position. However, the branch node received a support value below 50%, suggesting that these two sequences probably occupied a wrong position. We further rebuilt the NBS gene phylogenies for five Brassicaceae genomes either separately or jointly for lucidity. It turned out that both At4g19050 and At5g45510 belong other places in the CNL clade, whereas the RNL clade locates at the most basal position of the nTNL tree (Y.-M. Zhang, Q. Wang, Y.-Y. Hang, J.-Y. Xue, Z.-Q. Shao, B. Wang, and J.-Q. Chen, unpublished data). Likewise, in two recent studies, NBS genes identified from the M. truncatula and potato genomes were used to build their phylogenies (Ameline-Torregrosa et al., 2008; Jupe et al., 2012). Our results corroborate the hypothesis that the RNL genes in both cases form a monophyletic group and are located at the basal position of the nTNL part (Fig. 2). Therefore, the RNL clade is distinguishable from both the CNL clade and the TNL clade, not only functionally but also phylogenetically.
Thirty-One Ancient Rosid NBS Gene Lineages Gave Birth to All Extant NBS Genes in the Legume and Arabidopsis Genomes
In this study, we reconstructed two NBS gene phylogenies, one for the nTNL subclass (Fig. 3; Supplemental Fig. S2) and the other for the TNL subclass (Fig. 4; Supplemental Fig. S3). These two phylogenies provided invaluable information on the long-term evolutionary pattern of NBS genes.
The two phylogenies explicitly recovered a total of 23 nTNL (two RNL and 21 CNL) and eight TNL NBS gene lineages that were present in the common ancestor of legumes and Arabidopsis (Figs. 3 and 4). These lineages were then differentially inherited. Up to 20 nTNL and six TNL lineages are present in legume genomes today, while only 13 nTNL and five TNL lineages are present in the Arabidopsis genome. Therefore, both legumes and Arabidopsis had lost some ancient NBS gene lineages since they diverged approximately 108 MYA (Magallón and Sanderson, 2005). We also surveyed four other genomes in Brassicaceae and identified the same 13 nTNL and five TNL lineages (Y.-M. Zhang, Q. Wang, Y.-Y. Hang, J.-Y. Xue, Z.-Q. Shao, B. Wang, and J.-Q. Chen, unpublished data). Overall, only 10 nTNL and three TNL lineages are retained by both legumes and Arabidopsis. In three such lineages, one Arabidopsis gene was present (At3g14460 in nTNL rosid family 15, At3g07040 in nTNL rosid family 19, and At5g36930 in TNL rosid family 7), and each is sister to abundant legume NBS genes. This observation presents two intriguing scenarios: (1) the rosid common ancestor contained one copy, and the Arabidopsis lineage maintained this single copy for approximately 108 million years while the legume-inherited copy had become sufficiently diversified via duplications; and (2) the Arabidopsis lineage inherited the ancient copy, and it also diverged in the past, but today only one copy is left, either by chance or through selection. The former scenario would suggest that some NBS-LRR genes (either TNL or CNL) could be preserved for an extremely long period of time, and they probably have conserved functions (e.g. recognizing conserved pathogen effectors or guarding conserved host proteins targeted by pathogen effectors). The second scenario is also interesting, since it suggests that NBS genes can experience an evolutionary cycle from a single copy to multiple copies and then back to a single copy. Further investigations revealed that the three Arabidopsis genes were also present as single copies in some other Brassicaceae genomes, which make the former scenario tangible. In fact, one of the three genes, At3g07040, also known as the functional R gene RESISTANCE TO PSEUDOMONAS SYRINGAE1 (RPM1), can confer resistance to Pseudomonas syringae by guarding the target protein RPM1-INTERACTING PROTEIN4 RIN4; (Mackey et al., 2002).
A Majority of Legume NBS Gene Lineages Experienced Deletions or Expansions, While Seven Lineages Were Maintained Conservatively
Furthermore, according to Supplemental Figures S4 and S5, the 20 nTNL lineages and six TNL lineages inherited by legumes had further diverged into 64 nTNL and 55 TNL-legume NBS genes in the legume ancestor. After 54 million years of evolution, 42, 47, 37, and 38 original nTNL NBS gene lineages and 32, 34, 27, and 37 original TNL NBS gene lineages were retained in the M. truncatula, pigeon pea, common bean, and soybean genomes, respectively. Thus, 32% to 46% of the original 119 legume NBS gene lineages were lost from the four legume genomes, and the pattern was largely species specific (Supplemental Tables S2 and S3). Differential loss of NBS genes was also observed in comparing two Arabidopsis and two rice genomes (Chen et al., 2010; Guo et al., 2011; Luo et al., 2012). Collectively, these results suggest that the differential loss of original copies is indeed a main evolutionary feature for NBS gene evolution in angiosperms. Although a significant proportion of original NBS gene lineages was lost, the four legume genomes today still possess a few hundred NBS genes, indicating that gene duplications must have occurred in those retained NBS gene lineages. Among 97 out of 119 original legume NBS gene lineages, NBS gene duplication had occurred at least once in a legume species (Supplemental Tables S2 and S3). In seven cases (nTNL-legume families 16, 31, 45 and 58 and TNL-legume families 3, 38, and 55), NBS genes were duplicated in all four surveyed legumes. Collectively, these duplications gained a total number of 787 nTNL NBS genes and 518 TNL NBS genes in the four legumes during the past 54 million years. Therefore, independent gene duplication is another major feature of NBS gene evolution in legumes.
Although gene losses and duplications had occurred in many legume NBS gene families, we also detected some not-so-active lineages, such as nTNL-legume families 51, 52, 62, and 63 and TNL-legume families 7, 13, and 43 (Supplemental Tables S2 and S3; Supplemental Figures S4 and S5). Among these families, NBS gene numbers were maintained at one to three copies in each legume. Synteny analysis further confirmed that NBS genes in these seven families are all located at syntenic regions shared by the four legumes (Fig. 7). Therefore, these NBS genes, as well as their surrounding genes, probably have been maintained conservatively for approximately 54 million years.
Ectopic Duplication Occurred at a Significant Frequency and Created Many Novel NBS Gene Loci in Legume Genomes
In this study, we also aimed to evaluate the relative importance of three different duplication types responsible for new NBS genes in legume evolution. The case of the pigeon pea lineage was not considered, since many NBS genes in that genome were not anchored to the chromosome yet. Moreover, it is difficult to determine which type of duplications had occurred in the ancient lineages, since the locations of the duplicated genes were not known. Therefore, for the sake of accuracy, we only counted NBS genes that were duplicated on the terminal branches leading to M. truncatula, common bean, and soybean.
We found that, in legumes (Table III), more than three-quarters of the newly expanded NBS genes were produced by local tandem duplications. Furthermore, ectopic duplications contributed approximately 20% new NBS genes in the Medicago spp. lineage; however, in the common bean and soybean lineages, this contribution ratio was lower, especially for the TNL subclass. These results provided new evidence to support the so-called rapid rearrangement model, which acknowledges the importance of ectopic duplications (Leister, 2004; McDowell and Simon, 2006). However, an alternative model argues that many ectopically duplicated NBS genes are caused by segmental duplications followed by rapid deletions/arrangements of surrounding genes (Baumgarten et al., 2003). This possibility should be taken into consideration, as it might explain the higher ratio of ectopic duplications in Medicago spp. lineage (diverged longer) than in Phaseolus and Glycine spp. lineages in our study.
After reexamining all identified ectopic duplication events in Medicago and Phaseolus spp. lineages, we found that, in about 30% of cases, the duplicated NBS gene located on another chromosome was phylogenetically close to a recently duplicated member in the original NBS gene cluster, suggesting that the duplication event must have occurred recently. For example, nTNL-legume family 43 had 28 M. truncatula NBS genes (Supplemental Table S2). Among these, 27 genes were physically close to each other and were located on chromosome 3, with the only other gene, 7g020800, located on chromosome 7. Gene 7g020800 was found to be phylogenetically close to gene 3g030870, which was embedded in the middle of the family phylogeny. It is highly unlikely that this was a result of segmental duplications for two reasons. First, no other closely related NBS genes were found around 7g020800. This is strange, since the duplicated segment should contain multiple copies of NBS genes, as indicated by previously diverged lineages on the family tree (Supplemental Fig. S2). Second, collinearity was not detected around genes 3g030870 and 7g020800. This is also strange, since the collinearity should have been detected in surrounding genes if this were a recent event. Moreover, a recent study has shown that in a duplicated segment, NBS genes, instead of the surrounding non-NBS genes, are more likely to be deleted first (Innes et al., 2008).
Overall, our data in legumes support the following NBS gene duplication pattern: first, local tandem duplications constantly occur and create many homogenous clustered loci that were seen in each legume genome; second, ectopic duplications also occur at a certain frequency (8%–20%), which can take an NBS gene from a homogenous cluster to a distant location. This explains why the majority of 298 integrated NBS gene loci (Fig. 7) in legumes are species specific. Similarly, homogenous clusters could become heterogenous if distantly related NBS genes moved in through ectopic duplication. Finally, when a WGD event does not occur, only a few NBS genes are produced via segmental duplications.
Frequent Gain and Loss of MicroRNA Regulation in NBS Gene Evolution
Plants maintaining a large number of NBS genes have potential fitness costs (Tian et al., 2003; Orgil et al., 2007). Thus, to balance the resistance benefits and fitness costs, plants have recruited microRNAs to regulate NBS gene expression (Zhai et al., 2011; Shivaprasad et al., 2012; Fei et al., 2013; Källman et al., 2013). In this study, we screened out 82 microRNA families that can potentially target a total of 399 NBS genes in the M. truncatula and soybean genomes.
Among the identified microRNAs, many microRNA families were predicted to recognize only one NBS gene or a few closely related NBS genes, suggesting that these interactions were formed only recently. Some other microRNA families, however, could target multiple NBS genes derived from successively diverged legume families. Such observations strongly support the idea that certain microRNA-NBS gene regulation relationships must have been established in the common ancestor of legumes. The fact that some shared microRNAs could regulate the same lineage of NBS genes in both the M. truncatula and soybean genomes gave this idea further support. Furthermore, we detected microRNA families that can target both nTNL and TNL genes from diverse legume NBS families. Some of these microRNAs have ancient origins and can target NBS genes in other plants (Zhai et al., 2011; Shivaprasad et al., 2012). Considering the fact that their targets are distributed across phylogenetically distant legume NBS families of both nTNLs and TNLs, one explanation is that the regulation had been established before the separation of different NBS subclasses. However, the possibility that NBS genes from different families or subclasses may independently recruit microRNAs to regulate their expression cannot be ruled out, because some key motifs of the NBS domain targeted by microRNAs are highly conserved among NBS subclasses.
NBS genes have higher substitution rates than housekeeping genes. Thus, a microRNA may not maintain its recognition ability when its target motif is changed by nucleotide substitution or insertion/deletion. Additionally, mutations outside of the target sequence may also disrupt the recognition, as they would affect the secondary structure of this region and hinder the accessibility of microRNAs (Kertesz et al., 2007). Indeed, frequent losses of microRNA regulation due to the rapid evolution of NBS genes were seen in nTNL-legume family 45 (Fig. 9D). On the other hand, some NBS genes that escaped regulation by ancient microRNAs could be targeted by other recently evolved microRNAs, suggesting that plants are consistently recruiting new microRNAs to regulate NBS genes. Overall, the above observations indicate that the microRNA regulation of NBS genes shows a frequent gain-and-loss pattern.
In summary, we found that RNL is an ancient NBS gene clade that is phylogenetically independent of the TNL and CNL clades. Legume NBS genes have experienced long-term evolution with incessant gene losses and independent gene duplications. Meanwhile, some conservatively maintained NBS gene families are also present, which may have functions to recognize conserved pathogen effectors or to guard conserved host proteins targeted by pathogen effectors. This study also revealed that ectopic duplications play a significant role in NBS gene duplication, which created many novel NBS gene loci in individual legume genomes. Furthermore, our study revealed that microRNAs are involved in NBS gene regulation and that they exhibit a gain-and-loss pattern during legume evolution.
MATERIALS AND METHODS
Identification of NBS-Encoding Genes from Four Legume Genomes
The genome sequences and gene models of Medicago truncatula (Assembly version 3.5 and Annotation version 3.0; Young et al., 2011), soybean (Glycine max; Assembly version 1.0 and Annotation version 1.1; Schmutz et al., 2010), and common bean (Phaseolus vulgaris; Assembly version 1.0 and Annotation version 1.0) were obtained from the Phytozome database (http://www.phytozome.org/; Schmutz et al., 2014). The genome sequence and gene model for pigeon pea (Cajanus cajan; Assembly version 5.0 and Annotation version 5.0; Varshney et al., 2012) were downloaded from the Web site of the International Initiative for Pigeon Pea Genomics (http://www.icrisat.org/gt-bt/iipg/Genome_Manuscript.html).
To identify NBS-encoding genes from the four legumes, both hidden Markov model (HMM) and BLAST searches were performed. All of the protein sequences in each legume genome were first searched against the HMM profile of the NB-ARC domain (Pfam no. PF00931) in hmmer3.0 (http://hmmer.org) using default parameter settings. The amino acid sequence of the NB-ARC domain was then used to run a BLASTp search against all protein sequences in each genome. The threshold expectation value was set to 1.0 as in a previous study (Li et al., 2010a). All hits obtained using HMM or BLAST searches were then merged together, and the redundant hits were removed. The remaining sequences were further subjected to the online Pfam analysis (http://pfam.sanger.ac.uk/) to verify whether they indeed possessed the NBS domain, with the E-value setting to 10−4. Finally, the identified NBS domain-encoding genes were further examined to see whether they encode TIR, RPW8, CC, or LRR domains using the Pfam analysis, SMART protein motif analyses (http://smart.embl-heidelberg.de/), MEME (for Multiple Expectation Maximization for Motif Elicitation; Bailey et al., 2006), and also COILS (Lupas et al., 1991).
Classification of Identified NBS-Encoding Genes in Each Legume Genome
NBS genes containing both the N-terminal domain (TIR, CC, or RPW8) and the LRR domain are deemed as intact NBS genes (TNL, CNL, or RNL), while NBS genes lacking the LRR domain (TN, CN, or RN), lacking the N-terminal domain (NL), and lacking both domains (N only) are regarded as incomplete or truncated NBS genes (Meyers et al., 2003; Plocik et al., 2004; Mun et al., 2009). In each legume genome, all of the identified NBS genes were first classified into eight categories: TNL, CNL, RNL, TN, CN, RN, NL, and N only. Furthermore, for genes in the NL and N only categories, BLASTn searches were performed to examine whether they were homologous to TNL, CNL, or RNL genes. Finally, genes with TNL, TN, NTIRL, and NTIR domain structures were all classified into the TNL subclass; genes with CNL, CN, NCCL, and NCC structures and genes with RNL, RN, NRPW8L, and NRPW8 structures were all classified into the nTNL subclass.
Chromosomal Distribution of NBS-Encoding Genes and Cluster Assignment
In each legume genome, for all identified NBS-encoding genes, their chromosomal locations were determined by retrieving relevant information from the downloaded annotation files. Thereafter, we examined the occurrence numbers of TNL and nTNL subclass NBS genes on different chromosomes. The criterion used previously for Medicago spp. was followed for cluster assignment (Ameline-Torregrosa et al., 2008): if two neighboring NBS genes are separated by no more than 250 kb on a chromosome, then these two genes are regarded as gene members of the cluster. Based on this criterion, the NBS genes in each legume genome were assigned to a number of singleton loci and clustered loci, which were ordered along chromosomes.
Sequence Alignment and Phylogenetic Analyses
Reconstructing the NBS Gene Phylogeny for Each Legume
In each legume genome, the NBS domain sequences of all identified NBS genes were aligned in MEGA 5.0 by using the ClustalW program with default settings (Tamura et al., 2011). The obtained alignments were then subjected to visional inspections and manual adjustments to improve their quality. Short sequences containing large deletions were removed, since these NBS sequences could often prove to be problematic in lateral phylogenetic analyses. A maximal likelihood method was then adopted to reconstruct the NBS gene phylogeny in each legume, with the reliability of internal nodes evaluated by calculating the Shimodaira-Hasegawa approximate likelihood ratio test (SH-aLRT) values (Guindon et al., 2010). This new evaluating method has been shown to provide a compelling alternative to standard bootstrap methods, offering not only a speed advantage but also excellent levels of accuracy and power, especially for large data sets (Anisimova et al., 2011).
Reconstructing the TNL and nTNL Gene Phylogenies for Four Legumes and Arabidopsis
The NBS domain sequences of TNL subclass and nTNL (CNL and RNL) subclass genes identified from the four legumes as well as from the Arabidopsis (Arabidopsis thaliana) genome (Meyers et al., 2003) were aligned separately. We used the same procedures as described above to reconstruct the TNL and nTNL phylogenies. Furthermore, to restore the NBS gene duplication and loss events that occurred during legume evolution, the reconstructed TNL and nTNL trees were compared with the real species tree lineage by lineage, using the Notung software (Stolzer et al., 2012). If a lineage of NBS genes is monophyletic and it originated in the common ancestor of legumes, then this lineage is defined as a legume NBS gene family. If a monophyletic lineage of NBS genes originated in the common ancestor of legumes and Arabidopsis, then this lineage is defined as a rosid NBS gene family, since it is shared by both Fabaceae (rosid I clade) and Brassicaceae (rosid II clade). Accordingly, various numbers of rosid NBS gene families and legume NBS gene families were defined on both TNL and nTNL trees.
For those short NBS sequences that were precluded in phylogenetic analyses, BLASTn searches against all identified NBS genes were conducted to define their potential positions on the tree by finding their closest relatives.
Synteny Analyses within and across Legume Genomes
MCScanX, a package developed by the Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/), was adopted to perform synteny analysis among legume genomes (Wang et al., 2012; Lee et al., 2013). This package can efficiently identify syntenic blocks within a legume genome or between different legume genomes through BLASTp searches. The presence of NBS genes on various syntenic blocks was then examined for two purposes: (1) to identify NBS gene pairs resulting from segmental duplications within a genome, especially in soybean, which underwent a recent WGD event; and (2) to explore the conservation pattern of NBS gene loci among four legume genomes. By mapping the NBS gene loci identified from other genomes to syntenic regions on M. truncatula chromosomes, an integrated map of NBS gene loci was finally constructed.
NBS Gene Duplications and Cluster Composition Analysis
There are three types of NBS gene duplications: local tandem duplication, ectopic duplication, and segmental duplication (for definitions, see Leister, 2004). The closely related NBS genes were checked lineage by lineage on the reconstructed nTNL and TNL phylogenies. Their chromosomal locations coupled with their within-genome syntenic relationships (especially for soybean) helped estimate the number of duplicated genes resulting from each kind of duplication. In order to make the estimation accurate, we only considered the duplications occurring on terminal branches leading to three legume species: M. truncatula, common bean, and soybean. This is because the ancient tandem, ectopic, and segmental duplications are often hard to distinguish due to the accumulated chromosomal activities (splits, fusions, and rearrangements). Furthermore, the pigeon pea lineage was not examined, since many NBS genes in that genome are not anchored to chromosomes yet. For identified syntenic NBS gene pairs due to segmental duplications, synonymous substitution rate values were calculated using MEGA 5.0 (Tamura et al., 2011). Moreover, all clustered NBS gene loci were analyzed for their compositions. Clusters containing only gene members belonging to one legume NBS gene family on the reconstructed nTNL or TNL phylogeny are regarded as homogenous; clusters containing gene members from multiple legume NBS gene families are regarded as heterogenous. Among the heterogenous clusters, if both TNL subclass and nTNL subclass NBS genes were contained, then such clusters are regarded as mixed clusters.
Searching the NBS Genes That Can Be Potentially Targeted by MicroRNAs
The mature microRNA sequences of M. truncatula and soybean were retrieved from miRBase (version 20A; http://www.mirbase.org/; Kozomara and Griffiths-Jones, 2014). Then, these microRNA sequences, as well as all the NBS gene sequences identified by this study from the two legume genomes, were submitted to the online server psRNATarget (for Plant Small RNA Target Analysis Server; http://plantgrn.noble.org/psRNATarget/; Dai and Zhao, 2011) to search for NBS genes that could be potentially targeted by certain families of microRNAs. psRNATarget searches target genes of microRNAs based on both complementarity scoring analysis and target-site accessibility evaluation by calculating the unpaired energy required to open a secondary structure around a small RNA’s target site on mRNA. The penalty score for noncanonical Watson-Crick pairings is 1.0, except for guanine:uracil (G:U) wobble pairings (0.5 instead). Each insertion or deletion receives a penalty of 2.0. Notably, any mismatch other than guanine:uracil (G:U) wobble at positions 2 to 7 at the 5′ end is further penalized 0.5 point. By testing predicted results with experimentally verified target genes for 10 known Arabidopsis microRNAs, the authors of psRNATarget found that if the mismatch penalty score was set to 3, all validated target genes could be predicted. However, if the mismatch penalty score was set to 2, about 17% of validated targets were missed by the prediction. Thus, this study followed the default settings (penalty score of 3) of psRNATarget to maximally cover the potential targets of microRNAs among legume NBS genes.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The chromosomal distribution of identified NBS-encoding genes in four legume genomes.
Supplemental Figure S2. Phylogenetic tree of nTNL subclass NBS genes based on conserved NBS domain sequences: a complete version showing all sequence names and branch support values.
Supplemental Figure S3. Phylogenetic tree of TNL subclass NBS genes based on conserved NBS domain sequences: a complete version showing all sequence names and branch support values.
Supplemental Figure S4. Reconciled nTNL tree based on the real legume species phylogeny, with various loss events restored.
Supplemental Figure S5. Reconciled TNL tree based on the real legume species phylogeny, with various loss events restored.
Supplemental Table S1. A list of identified NBS-encoding genes in four legume genomes, with domain analysis and cluster analysis results shown.
Supplemental Table S2. Gene number variations among 23 nTNL-rosid NBS gene families and 64 nTNL-legume NBS gene families.
Supplemental Table S3. Gene number variations among 8 TNL-rosid NBS gene families and 55 TNL-legume NBS gene families.
Supplemental Table S4. A list of integrated NBS loci showing synteny relationships among four legume genomes.
Supplemental Table S5. A list of legume NBS genes that are targeted by microRNAs.
Glossary
- NBS
nucleotide-binding site
- LRR
leucine-rich repeat
- CC
coiled-coil
- TIR
Toll/Interleukin1 receptor
- TNL
TIR-NBS-LRR
- nTNL
non-TIR-NBS-LRR
- CNL
CC-NBS-LRR
- RNL
RPW8-NBS-LRR
- MYA
million years ago
- WGD
whole-genome duplication
- HMM
hidden Markov model
- SH-aLRT
Shimodaira-Hasegawa approximate likelihood ratio test
- psRNATarget
Plant Small RNA Target Analysis Server
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 30930008, 31000105, 31170210, 91231102, and 31300190), the National Key Project for Gene Transformation (grant no. 2009ZX08009–027B), the China Postdoctoral Science Foundation (grant nos. 2013M540435 and 2014T70503), the Postdoctoral Science Foundation of Jiangsu Province (grant no. 1302131C), and the Natural Science Foundation of Jiangsu Province (grant no. BK20130565).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Ameline-Torregrosa C, Wang BB, O’Bleness MS, Deshpande S, Zhu H, Roe B, Young ND, Cannon SB. (2008) Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol 146: 5–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andolfo G, Sanseverino W, Rombauts S, Van de Peer Y, Bradeen JM, Carputo D, Frusciante L, Ercolano MR. (2013) Overview of tomato (Solanum lycopersicum) candidate pathogen recognition genes reveals important Solanum R locus dynamics. New Phytol 197: 223–237 [DOI] [PubMed] [Google Scholar]
- Anisimova M, Gil M, Dufayard JF, Dessimoz C, Gascuel O. (2011) Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst Biol 60: 685–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield T, Egan AN, Pfeil BE, Chen NW, Podicheti R, Ratnaparkhe MB, Ameline-Torregrosa C, Denny R, Cannon S, Doyle JJ, et al. (2012) Evolution of a complex disease resistance gene cluster in diploid Phaseolus and tetraploid Glycine. Plant Physiol 159: 336–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW. (2004) Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell 16: 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Pennill LA, Ning J, Lee SW, Ramalingam J, Webb CA, Zhao B, Sun Q, Nelson JC, Leach JE, et al. (2002) Diversity in nucleotide binding site-leucine-rich repeat genes in cereals. Genome Res 12: 1871–1884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34: W369–W373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten A, Cannon S, Spangler R, May G. (2003) Genome-level evolution of resistance genes in Arabidopsis thaliana. Genetics 165: 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Mackey D. (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Bonardi V, Tang S, Stallmann A, Roberts M, Cherkis K, Dangl JL. (2011) Expanded functions for a family of plant intracellular immune receptors beyond specific recognition of pathogen effectors. Proc Natl Acad Sci USA 108: 16463–16468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Zhu H, Baumgarten AM, Spangler R, May G, Cook DR, Young ND. (2002) Diversity, distribution, and ancient taxonomic relationships within the TIR and non-TIR NBS-LRR resistance gene subfamilies. J Mol Evol 54: 548–562 [DOI] [PubMed] [Google Scholar]
- Chen Q, Han Z, Jiang H, Tian D, Yang S. (2010) Strong positive selection drives rapid diversification of R-genes in Arabidopsis relatives. J Mol Evol 70: 137–148 [DOI] [PubMed] [Google Scholar]
- Collier SM, Hamel LP, Moffett P. (2011) Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein. Mol Plant Microbe Interact 24: 918–931 [DOI] [PubMed] [Google Scholar]
- Dai XB, Zhao PX. (2011) psRNATarget: a plant small RNA target analysis server. Nucl Acids Res 39: W155–W159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David P, Chen NW, Pedrosa-Harand A, Thareau V, Sévignac M, Cannon SB, Debouck D, Langin T, Geffroy V. (2009) A nomadic subtelomeric disease resistance gene cluster in common bean. Plant Physiol 151: 1048–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Xia R, Meyers BC. (2013) Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 25: 2400–2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Narayanan NN, Ellison L, Bhattacharyya MK. (2005) Two classes of highly similar coiled coil-nucleotide binding-leucine rich repeat genes isolated from the Rps1-k locus encode Phytophthora resistance in soybean. Mol Plant Microbe Interact 18: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Geffroy V, Macadré C, David P, Pedrosa-Harand A, Sévignac M, Dauga C, Langin T. (2009) Molecular analysis of a large subtelomeric nucleotide-binding-site-leucine-rich-repeat family in two representative genotypes of the major gene pools of Phaseolus vulgaris. Genetics 181: 405–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321 [DOI] [PubMed] [Google Scholar]
- Guo YL, Fitz J, Schneeberger K, Ossowski S, Cao J, Weigel D. (2011) Genome-wide comparison of nucleotide-binding site-leucine-rich repeat-encoding genes in Arabidopsis. Plant Physiol 157: 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes AJ, Jeong SC, Gore MA, Yu YG, Buss GR, Tolin SA, Maroof MA. (2004) Recombination within a nucleotide-binding-site/leucine-rich-repeat gene cluster produces new variants conditioning resistance to soybean mosaic virus in soybeans. Genetics 166: 493–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes RW, Ameline-Torregrosa C, Ashfield T, Cannon E, Cannon SB, Chacko B, Chen NW, Couloux A, Dalwani A, Denny R, et al. (2008) Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean. Plant Physiol 148: 1740–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia RZ, Ming R, Zhu YJ. (2013) Genome-wide analysis of nucleotide-binding site (NBS) disease resistance (R) genes in sacred lotus (Nelumbo nucifera Gaertn.) reveals their transition role during early evolution of land plants. Tropical Plant Biology 6: 19 [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jupe F, Pritchard L, Etherington GJ, Mackenzie K, Cock PJ, Wright F, Sharma SK, Bolser D, Bryan GJ, Jones JD, et al. (2012) Identification and localisation of the NB-LRR gene family within the potato genome. BMC Genomics 13: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Källman T, Chen J, Gyllenstrand N, Lagercrantz U. (2013) A significant fraction of 21-nucleotide small RNA originates from phased degradation of resistance genes in several perennial species. Plant Physiol 162: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. (2007) The role of site accessibility in microRNA target recognition. Nat Genet 39: 1278–1284 [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF. (2005) Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol 54: 575–594 [DOI] [PubMed] [Google Scholar]
- Lee TH, Tang H, Wang X, Paterson AH. (2013) PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res 41: D1152–D1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. (2004) Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet 20: 116–122 [DOI] [PubMed] [Google Scholar]
- Li J, Ding J, Zhang W, Zhang Y, Tang P, Chen JQ, Tian D, Yang S. (2010a) Unique evolutionary pattern of numbers of gramineous NBS-LRR genes. Mol Genet Genomics 283: 427–438 [DOI] [PubMed] [Google Scholar]
- Li X, Cheng Y, Ma W, Zhao Y, Jiang H, Zhang M. (2010b) Identification and characterization of NBS-encoding disease resistance genes in Lotus japonicus. Plant Syst Evol 289: 10 [Google Scholar]
- Lin X, Zhang Y, Kuang H, Chen J. (2013) Frequent loss of lineages and deficient duplications accounted for low copy number of disease resistance genes in Cucurbitaceae. BMC Genomics 14: 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Liu X, Dai L, Wang G. (2007) Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. J Genet Genomics 34: 765–776 [DOI] [PubMed] [Google Scholar]
- Lozano R, Ponce O, Ramirez M, Mostajo N, Orjeda G. (2012) Genome-wide identification and mapping of NBS-encoding resistance genes in Solanum tuberosum group phureja. PLoS ONE 7: e34775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Zhang Y, Hu Q, Chen J, Li K, Lu C, Liu H, Wang W, Kuang H. (2012) Dynamic nucleotide-binding site and leucine-rich repeat-encoding genes in the grass family. Plant Physiol 159: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. (1991) Predicting coiled coils from protein sequences. Science 252: 1162–1164 [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF, III, Wiig A, Dangl JL. (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- Magallón SA, Sanderson MJ. (2005) Angiosperm divergence times: the effect of genes, codon positions, and time constraints. Evolution 59: 1653–1670 [DOI] [PubMed] [Google Scholar]
- McDowell JM, Simon SA. (2006) Recent insights into R gene evolution. Mol Plant Pathol 7: 437–448 [DOI] [PubMed] [Google Scholar]
- Meyer JD, Silva DC, Yang C, Pedley KF, Zhang C, van de Mortel M, Hill JH, Shoemaker RC, Abdelnoor RV, Whitham SA, et al. (2009) Identification and analyses of candidate genes for rpp4-mediated resistance to Asian soybean rust in soybean. Plant Physiol 150: 295–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND. (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20: 317–332 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monosi B, Wisser RJ, Pennill L, Hulbert SH. (2004) Full-genome analysis of resistance gene homologues in rice. Theor Appl Genet 109: 1434–1447 [DOI] [PubMed] [Google Scholar]
- Mun JH, Yu HJ, Park S, Park BS. (2009) Genome-wide identification of NBS-encoding resistance genes in Brassica rapa. Mol Genet Genomics 282: 617–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgil U, Araki H, Tangchaiburana S, Berkey R, Xiao S. (2007) Intraspecific genetic variations, fitness cost and benefit of RPW8, a disease resistance locus in Arabidopsis thaliana. Genetics 176: 2317–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plocik A, Layden J, Kesseli R. (2004) Comparative analysis of NBS domain sequences of NBS-LRR disease resistance genes from sunflower, lettuce, and chicory. Mol Phylogenet Evol 31: 153–163 [DOI] [PubMed] [Google Scholar]
- Porter BW, Paidi M, Ming R, Alam M, Nishijima WT, Zhu YJ. (2009) Genome-wide analysis of Carica papaya reveals a small NBS resistance gene family. Mol Genet Genomics 281: 609–626 [DOI] [PubMed] [Google Scholar]
- Richly E, Kurth J, Leister D. (2002) Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol Biol Evol 19: 76–84 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schmutz J, McClean PE, Mamidi S, Wu GA, Cannon SB, Grimwood J, Jenkins J, Shu S, Song Q, Chavarro C, et al. (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46: 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo YS, Rojas MR, Lee JY, Lee SW, Jeon JS, Ronald P, Lucas WJ, Gilbertson RL. (2006) A viral resistance gene from common bean functions across plant families and is up-regulated in a non-virus-specific manner. Proc Natl Acad Sci USA 103: 11856–11861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaprasad PV, Chen HM, Patel K, Bond DM, Santos BA, Baulcombe DC. (2012) A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24: 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic S, Pfeil BE, Palmer JD, Doyle JJ. (2009) Relationships among phaseoloid legumes based on sequences from eight chloroplast regions. Systematic Botany 34: 14 [Google Scholar]
- Stolzer M, Lai H, Xu M, Sathaye D, Vernot B, Durand D. (2012) Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics 28: i409–i415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S, Wu S. (2012) Genome wide analysis of nucleotide-binding site disease resistance genes in Brachypodium distachyon. Comp Funct Genomics 2012: 418208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J. (2003) Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 423: 74–77 [DOI] [PubMed] [Google Scholar]
- Varshney RK, Chen W, Li Y, Bharti AK, Saxena RK, Schlueter JA, Donoghue MT, Azam S, Fan G, Whaley AM, et al. (2012) Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol 30: 83–89 [DOI] [PubMed] [Google Scholar]
- Wan H, Yuan W, Bo K, Shen J, Pang X, Chen J. (2013) Genome-wide analysis of NBS-encoding disease resistance genes in Cucumis sativus and phylogenetic study of NBS-encoding genes in Cucurbitaceae crops. BMC Genomics 14: 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, et al. (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Li W, Sun X, Zhu S, Zhu J. (2013) Systematic analysis and comparison of nucleotide-binding site disease resistance genes in a diploid cotton Gossypium raimondii. PLoS ONE 8: e68435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Shao ZQ, Wu XZ, Wang Q, Wang B, Chen JQ, Hang YY, Xue JY. (2014) Loss/retention and evolution of NBS-encoding genes upon whole genome triplication of Brassica rapa. Gene 540: 54–61 [DOI] [PubMed] [Google Scholar]
- Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M, Turner JG. (2001) Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291: 118–120 [DOI] [PubMed] [Google Scholar]
- Yang S, Feng Z, Zhang X, Jiang K, Jin X, Hang Y, Chen JQ, Tian D. (2006) Genome-wide investigation on the genetic variations of rice disease resistance genes. Plant Mol Biol 62: 181–193 [DOI] [PubMed] [Google Scholar]
- Yang S, Gao M, Xu C, Gao J, Deshpande S, Lin S, Roe BA, Zhu H. (2008a) Alfalfa benefits from Medicago truncatula: the RCT1 gene from M. truncatula confers broad-spectrum resistance to anthracnose in alfalfa. Proc Natl Acad Sci USA 105: 12164–12169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Li J, Zhang X, Zhang Q, Huang J, Chen JQ, Hartl DL, Tian D. (2013) Rapidly evolving R genes in diverse grass species confer resistance to rice blast disease. Proc Natl Acad Sci USA 110: 18572–18577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Zhang X, Yue JX, Tian D, Chen JQ. (2008b) Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol Genet Genomics 280: 187–198 [DOI] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KF, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, Li Y, González AJ, Yan Z, Kitto SL, Grusak MA, et al. (2011) MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev 25: 2540–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Feng Y, Cheng H, Tian D, Yang S, Chen JQ. (2011) Relative evolutionary rates of NBS-encoding genes revealed by soybean segmental duplication. Mol Genet Genomics 285: 79–90 [DOI] [PubMed] [Google Scholar]
- Zhou T, Wang Y, Chen JQ, Araki H, Jing Z, Jiang K, Shen J, Tian D. (2004) Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol Genet Genomics 271: 402–415 [DOI] [PubMed] [Google Scholar]