Polymorphisms in the leucine-rich-repeat domains of two closely related soybean genes allow them to distinguish between two pathogen effectors detected by a single disease resistance gene in Arabidopsis.
Abstract
In Arabidopsis (Arabidopsis thaliana), the Pseudomonas syringae effector proteins AvrB and AvrRpm1 are both detected by the RESISTANCE TO PSEUDOMONAS MACULICOLA1 (RPM1) disease resistance (R) protein. By contrast, soybean (Glycine max) can distinguish between these effectors, with AvrB and AvrRpm1 being detected by the Resistance to Pseudomonas glycinea 1b (Rpg1b) and Rpg1r R proteins, respectively. We have been using these genes to investigate the evolution of R gene specificity and have previously identified RPM1 and Rpg1b. Here, we report the cloning of Rpg1r, which, like RPM1 and Rpg1b, encodes a coiled-coil (CC)-nucleotide-binding (NB)-leucine-rich repeat (LRR) protein. As previously found for Rpg1b, we determined that Rpg1r is not orthologous with RPM1, indicating that the ability to detect both AvrB and AvrRpm1 evolved independently in soybean and Arabidopsis. The tightly linked soybean Rpg1b and Rpg1r genes share a close evolutionary relationship, with Rpg1b containing a recombination event that combined a NB domain closely related to Rpg1r with CC and LRR domains from a more distantly related CC-NB-LRR gene. Using structural modeling, we mapped polymorphisms between Rpg1b and Rpg1r onto the predicted tertiary structure of Rpg1b, which revealed highly polymorphic surfaces within both the CC and LRR domains. Assessment of chimeras between Rpg1b and Rpg1r using a transient expression system revealed that AvrB versus AvrRpm1 specificity is determined by the C-terminal portion of the LRR domain. The P. syringae effector AvrRpt2, which targets RPM1 INTERACTOR4 (RIN4) proteins in both Arabidopsis and soybean, partially blocked recognition of both AvrB and AvrRpm1 in soybean, suggesting that both Rpg1b and Rpg1r may detect these effectors via modification of a RIN4 homolog.
Effector triggered immunity in plants involves highly specific recognition events in which plant resistance (R) proteins detect pathogen effector proteins directly or, alternatively, the modifications that they induce on host proteins (Bonardi et al., 2012). The largest group of R proteins belongs to the nucleotide-binding (NB)-leucine-rich repeat (LRR) family (McHale et al., 2006). The NB-LRR family can be further subdivided based on N-terminal domains into the Toll-Interleukin and R protein (TIR) class and non-TIR-NB-LRR class (McHale et al., 2006). The latter most often contain a coiled-coil (CC) domain at the N terminus. The contributions of the TIR, CC, and LRR domains to R protein specificity, and how new specificities evolve, remain important questions.
There are relatively few NB-LRR R proteins characterized to date that are thought to detect pathogen effectors directly; these include Pi-ta from rice (Oryza sativa), L and M variants from flax (Linum usitatissimum), and RESISTANCE TO RALSTONIA SOLANACEARUM1 and RESISTANCE TO PERONOSPORA PARASITICA1 (RPP1) from Arabidopsis (Arabidopsis thaliana; Jia et al., 2000; Deslandes et al., 2003; Dodds et al., 2006; Ueda et al., 2006; Catanzariti et al., 2010; Krasileva et al., 2010). In at least some of these examples, the R genes are found in clusters of NB-LRR paralogs in which multiple recognition specificities are represented (Ellis et al., 1995; Botella et al., 1998) or belong to allelic series (Ellis et al., 1995), arrangements that may promote evolution of recognition specificity via recombination between alleles and paralogs. Interestingly, sequence comparisons and domain swaps involving alleles at the L locus implicate both the LRR and TIR regions as determinants of recognition specificity (Ellis et al., 1999; Luck et al., 2000). Subsequently, domain swaps involving paralogs clustered at the barley (Hordeum vulgare) MILDEW A (MLA) and potato (Solanum tuberosum) Resistance to Potato Virus X (Rx)/Globodera pallida (Gpa) loci have provided additional support for the LRR domain playing a key role in conferring recognition specificity (Ellis et al., 1999; Luck et al., 2000; Shen et al., 2003; Rairdan and Moffett, 2006).
Several R proteins are known to detect the presence of pathogen effectors indirectly by monitoring the activity of pathogen effectors within the plant cell. For example, the Arabidopsis RESISTANCE TO PSEUDOMONAS MACULICOLA1 (RPM1) and RESISTANCE TO PSEUDOMONAS SYRINGAE2 (RPS2) R proteins detect modification of the effector target RPM1 INTERACTOR4 (RIN4), while the Arabidopsis RPS5 protein detects modification of the effector target AvrPphB SUSCEPTIBLE1 (Mackey et al., 2002, 2003; Axtell and Staskawicz, 2003; Shao et al., 2003). At least for the well-studied examples in Arabidopsis, R proteins that employ indirect recognition mechanisms are encoded by NB-LRR genes that are not members of large clusters, or allelic series, with variants encoding distinct recognition specificities. Correlated with this genomic structure, such loci are typically relatively stable, with RPM1 and RPS5 existing as presence/absence polymorphisms that have been maintained over long evolutionary periods (Stahl et al., 1999; Tian et al., 2002). Both functional and nonfunctional alleles of RPS2 have been isolated, but only a single recognition specificity has been detected at this locus, despite sequence polymorphisms between alleles (Caicedo et al., 1999).
Most likely, specificity for this class of R proteins is determined by a combination of the ability to associate with the host protein targeted by the effector and the ability to detect effector-induced modification of this target. Consistent with this hypothesis, it has been shown that the CC domains from at least some R proteins interact with the host proteins they are monitoring, even in the absence of pathogen effectors, in a prerecognition complex (Mackey et al., 2002; Ade et al., 2007). Hence, evolution of recognition specificity in R proteins that employ indirect recognition mechanisms may involve evolution of both the N-terminal CC and LRR domains.
To better understand the evolution and function of R proteins that detect pathogen effectors indirectly, we have been studying two soybean (Glycine max) R genes, with known recognition specificities, that are members of a complex NB-LRR cluster. The R genes involved, Resistance to Pseudomonas glycinea 1b (Rpg1b) and Rpg1r, mediate detection of the Pseudomonas syringae effector proteins AvrB and AvrRpm1, respectively (Staskawicz et al., 1984; Ashfield et al., 1995). We have previously cloned Rpg1b, which is a CC-NB-LRR (CNL) gene that maps to a cluster of R genes effective against a diverse range of pathogens (Ashfield et al., 1998, 2004). Rpg1r is present in the same cluster and maps 0.56 centiMorgans from Rpg1b (Ashfield et al., 1995); however, the evolutionary relationship shared by the two R genes is not known. The cluster is associated with numerous NB-LRR genes, of both the CC and TIR subgroups, spread over more than a megabase of soybean chromosome 13 (Peñuela et al., 2002; Hayes et al., 2004; Innes et al., 2008; Ashfield et al., 2012; Wen et al., 2013). The NB-LRR family in this region is evolving rapidly, with duplications/deletions of paralogs, recombination, and positive selection all playing a role (Ashfield et al., 2012).
While soybean can distinguish between AvrB and AvrRpm1, both effectors are detected by a single R protein, RPM1, in Arabidopsis (Bisgrove et al., 1994; Grant et al., 1995). It is known that RPM1 recognizes the effector proteins indirectly by detecting effector-dependent phosphorylation of a second Arabidopsis protein, RIN4 (Mackey et al., 2002; Chung et al., 2011; Liu et al., 2011). The available evidence suggests that a related strategy is employed by soybean, at least for the Rpg1b protein, despite the AvrB recognition specificity having evolved independently in these plant species (Ashfield et al., 2004; Selote and Kachroo, 2010; Selote et al., 2013). Soybean contains four RIN4 homologs (Chen et al., 2010), three of which interact physically with Rpg1b, with two required for full resistance conferred by this R gene (Selote and Kachroo, 2010; Selote et al., 2013). It is not known whether RIN4 homologs are required for Rpg1r function.
Here, we report the map-based cloning of the soybean Rpg1r gene. Comparison of the Rpg1r protein to Rpg1b, combined with structural modeling, revealed highly polymorphic surfaces in the CC and LRR domains. Transient expression of chimeric Rpg1 proteins demonstrated that specificity for AvrB versus AvrRpm1 is determined by the C-terminal LRR region. Finally, we provide evidence that Rpg1r, like Rpg1b, detects its corresponding pathogen effector indirectly, most likely by monitoring a RIN4 homolog, indicating convergent evolution of recognition mechanisms in separate plant families.
RESULTS
Genetic and Physical Mapping of Rpg1r
We have previously scored a small recombinant inbred line (RIL) population, derived from a cross between cv Flambeau (rpg1b Rpg1r) and Merit (Rpg1b rpg1r), for the segregation of both Rpg1b and Rpg1r. Based on these data, we determined that Rpg1b and Rpg1r are not allelic but are tightly linked, with Rpg1r mapping 0.56 ± 0.77 centiMorgans from Rpg1b (Ashfield et al., 1995). Rpg1b was subsequently mapped to soybean chromosome 13 (formally, molecular linkage group F), flanked by the microsatellite markers HSP176 and Sct33 (Ashfield et al., 1998), allowing the inference of a closely linked position for Rpg1r.
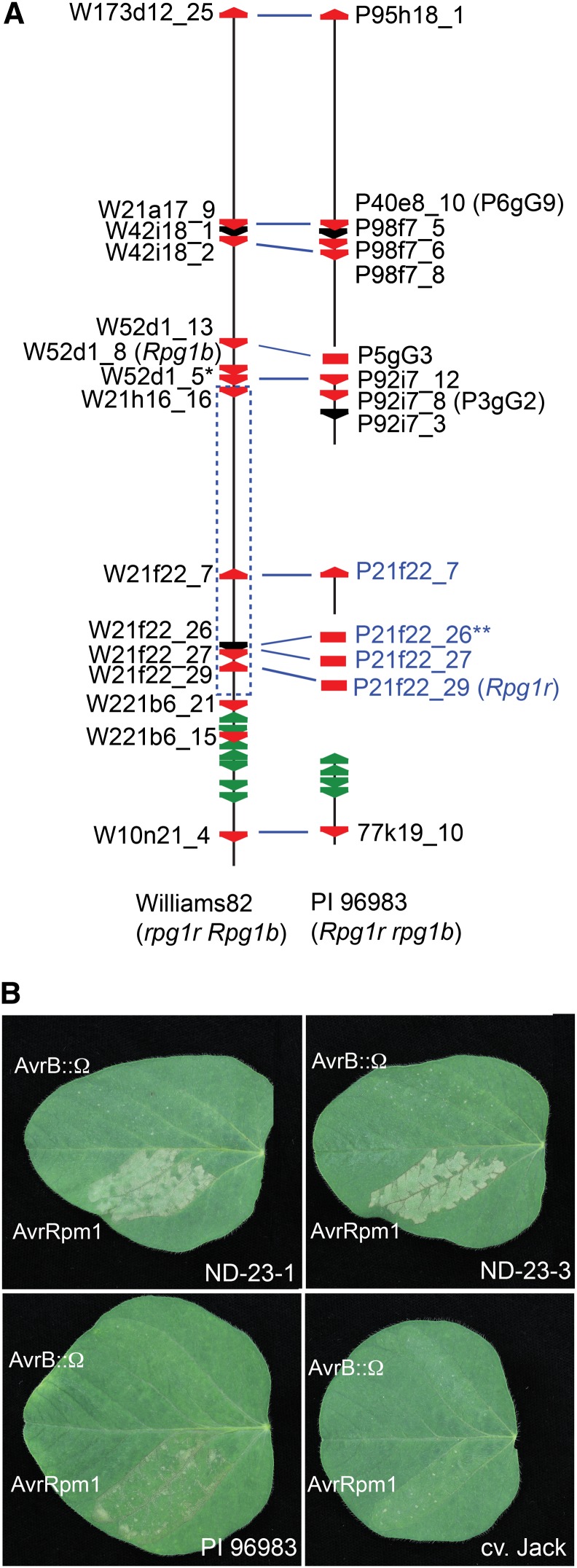
To further delimit the genetic interval that contains Rpg1r, we scored informative recombinants from the cv Flambeau × Merit RIL population with PCR-based markers corresponding to the bacterial artificial chromosome (BAC) contigs generated during the cloning of Rpg1b (Ashfield et al., 2003, 2004). In this manner, Rpg1r was delimited to an interval flanked by the markers EP-52d1r (for End Probe of BAC clone 52D1) and EP-221b6r (Supplemental Fig. S1). We have previously sequenced most of this interval in cv Williams 82 (Innes et al., 2008) using a BAC-based strategy, with one gap being filled from the soybean whole genome sequence (WGS; Schmutz et al., 2010). The genetically defined interval corresponds to a physical distance of approximately 291.5 kb in the cv Williams 82 sequence assembled from the BAC contigs. Interestingly, this region contains four intact CNL genes and one pseudogene belonging to the NB-LRR family (Fig. 1; Supplemental Fig. S1). In the soybean WGS, which is also derived from cv Williams 82, the genetically defined interval extends from gene Glyma13g26000 to gene Glyma13g26300 (chromosome 13, position 29,225,278 to 29,524,087 in the Glyma1 assembly of the soybean WGS), a physical distance of approximately 298.8 kb. While the BAC-derived sequence and WGS are mostly colinear, small indels are apparent when the sequences are aligned, thus explaining the minor difference in lengths (data not shown). Comparison of the predicted CNL gene models from the BAC-derived and WGS-derived sequences revealed sequence polymorphisms and/or indels in two of the four intact CNL genes, likely reflecting sequencing errors (Supplemental Fig. S2). When available, BAC-derived sequence and gene models were used for subsequent analyses.
Figure 1.
Cloning of the soybean Rpg1r gene. A, Distribution of NB-LRR genes across an approximately 1-Mb interval encompassing the Rpg1r locus in cv Williams 82 and PI 96983. The vertical black lines represent genomic sequence from soybean cv Williams 82 (rpg1r Rpg1b) and PI 96983 (Rpg1r rpg1b) as described by Innes et al. (2008). The region corresponds to the interval from Glyma13g25440 to Glyma13g26530 in the soybean WGS (Schmutz et al., 2010). Breaks in the lines indicate lack of available sequence. The red and green polygons correspond to CNL and TIR-NB-LRR family members, respectively. Black polygons represent CNL pseudogenes or paralogs that extend beyond the available sequence. Gene names printed in black correspond to CNL genes identified in the available genomic sequence, while those printed in blue correspond to CNL genes amplified from PI 96983 total genomic DNA. The blue dashed rectangle indicates the genetically defined region containing Rpg1r (see Supplemental Fig. S1). The blue lines linking CNL genes in the cv Williams 82 and PI 96983 sequences indicate probable alleles. A single asterisk indicates W52d1_5 has the atypical structure CNL:CNL. Double asterisks indicate gene 21f22_26 is a pseudogene in both cv Williams 82 and PI 96983. B, Functional complementation of Rpg1r recognition specificity in soybean stable transformants. Primary leaves from accession PI 96983 (Rpg1r), untransformed cv Jack (rpg1r), and cv Jack transformants expressing an Rpg1r transgene were injected with PsgR4 expressing avrRpm1 or a nonfunctional avrB allele disrupted with an Ω sequence (avrB::Ω). The responses in two independent transformants (ND-23-1 and ND-23-3) are shown. Photographs were taken approximately 24 h after injection.
Because Rpg1b belongs to the CNL family, we reasoned that Rpg1r would as well and thus focused our attention on CNL genes in the genetic interval containing Rpg1r. As cv Williams 82 does not express Rpg1r, putative alleles for these genes were isolated from the Rpg1r-expressing soybean accession PI 96983. This was accomplished using two strategies. Firstly, one gene (P92i7_8; with the P designating a CNL from PI 96983) was obtained from a previously reported contig of PI 96983 BACs that extends into the genetically defined region that contains Rpg1r (Fig. 1; Supplemental Fig. S1; Innes et al., 2008). Secondly, an additional three intact genes (P21f22_7, P21f22_27, and P21f22_29), and one pseudogene (P21f22_26), were amplified from PI 96983 genomic DNA using primers that flank CNLs in the target region of the cv Williams 82 sequence (Fig. 1A; Supplemental Fig. S1). Alignment of the amino acid sequences of the three intact CNLs amplified from PI 96983 with the corresponding cv Williams 82 CNLs used for primer design revealed that while P21f22_7 and P21f22_27 are very similar to their presumed cv Williams 82 alleles, the P21f22_29/W21f22_29 pair are more divergent, consistent with differing recognition specificities in the two soybean genotypes (Supplemental Fig. S3). Interestingly, one of the PI 96983 CNLs (P92i7_8) from the Rpg1r target region has been previously reported as a candidate to encode the Resistance to Soybean Mosaic Virus1 gene, a resistance gene effective against certain strains of soybean mosaic virus (P92i7_8 is referred to as 3gG2 in previous studies; Hayes et al., 2004; Wen et al., 2013).
Functional Complementation Confirms that Rpg1r Is Encoded by a CNL Type R Protein
Given that the pathogen effectors detected by Rpg1b and Rpg1r are recognized by a single R protein in Arabidopsis, we hypothesized that these two soybean R genes may guard the same (or related) plant protein(s) and that this might be reflected in a close evolutionary relationship, with one gene derived from the other. We therefore assessed the phylogenetic relationship of the Rpg1r candidate CNL genes to the previously cloned Rpg1b using a phylogenetic tree based on the NB domain (hereafter referred to as the NB-ARC domain for NB domain found in Apoptotic Protease Activating Factor1 (Apaf1), R genes, and Cell Death Protein4; van der Biezen and Jones, 1998) of all CNL genes in the 1-Mb Rpg1b region found in both cv Williams 82 (Rpg1b genotype) and PI 96983 (Rpg1r genotype; Supplemental Fig. S4). Only a single cv Williams 82 CNL gene (W21f22_29; with the W designating a CNL from cv Williams 82) located within the genetically defined region containing Rpg1r was found to be in the strongly supported clade containing Rpg1b. Based on this observation, further analysis was focused on the PI 96983 CNL (P21f22_29) that was amplified using primers that flank this gene.
To test for Rpg1r function, we transiently coexpressed this CNL gene (P21f22_29) with AvrRpm1 in leaves of common bean (Phaseolus vulgaris) using an Agrobacterium tumefaciens delivery system. Encouragingly, coexpression of P21f22_29 with AvrRpm1 resulted in a weak hypersensitive response (HR). To unambiguously confirm the identity of Rpg1r, we then transformed a soybean cultivar (cv Jack) that lacks Rpg1r function with a genomic clone of P21f22_29 that included the presumed promoter and transcriptional terminator regions and tested for gain in the ability to respond to AvrRpm1. Seeds from three independent transformants were grown and screened for the ability to respond with an HR after inoculation with P. syringae expressing AvrRpm1. Progeny from two of the independent transformants segregated for the ability to mount an HR in response to AvrRpm1 (Fig. 1B). Segregation ratios obtained using PCR with transfer DNA (T-DNA)-specific primers were consistent with both these transformed lines containing multiple insertions. All T2 progeny found to lack an insertion failed to respond to AvrRpm1, consistent with recognition of the effector being dependent on the presence of the T-DNA and indicating that CNL P21f22_29 represents a functional Rpg1r allele.
Structure of the Rpg1r Gene
The Rpg1r allele from PI 96983 (P21f22_29) encodes a CNL protein 1,212 amino acids in length (Fig. 2). The COILs software predicts two regions within the CC domain that may participate in CC structures (predictions supported with probabilities of 88% and 94%, respectively; Fig. 2A). However, additional residues between the first two heptad repeats, and the presence of hydrophilic residues at expected hydrophobic positions, argue against the second region participating in a CC, and it is not marked in Figure 2. Also present is the EDVID motif (present as EDLLD in Rpg1r), a motif previously implicated in intradomain interactions (Rairdan et al., 2008; Fig. 2A).
Figure 2.
Structural domains of the Rpg1r protein. A, The CC domain. The predicted CC is printed in orange, with the hydrophobic residues at the a and d positions in the heptad repeats underlined. The conserved EDVID motif (Rairdan et al., 2008) is printed in blue. B, The NB domain. Shown is the region corresponding to the known structure of the human Apaf1 NB-ARC1-ARC2 region. Previously described conserved motifs (van der Biezen and Jones, 1998; Meyers et al., 1999) are printed in blue in the following order: P-loop (kinase 1a), kinase 2, GLPL, RNBS-D, and MHD. An unusual substitution with a nonacidic residue within the kinase 2 motif is printed in red. C, LRR domain. Regions predicted to form α helices and β strands are highlighted in green and blue, respectively. Residues matching the consensus for intracellular LRRs are printed in red. D, Consensus for intracellular LRRs (Jones and Jones, 1997), where x can be any residue and L can be substituted with I, VM, or F.
The NB-ARC domain contains the expected P-loop (kinase 1a), kinase 2, RNBS-D, GLPL, and MHD motifs described previously for proteins of this class (Fig. 2B; McHale et al., 2006; van Ooijen et al., 2008). Interestingly, the kinase 2 motif has the sequence LLVLDN, with an Asn in place of the second acidic residue present in most CNL proteins. It is thought that this acidic residue may be important for the catalytic activity (nucleotide hydrolysis) of the domain (Tameling et al., 2006).
Analysis of the LRR domain primary sequence, together with the predicted secondary structure for the region, indicates that the domain contains 24 canonical repeat units at least partially matching the consensus [LxxLxxLxLxxN/C/Tx(x) LxxIPxx] for intracellular LRR domains (Fig. 2C; Jones and Jones, 1997)). The repeats show considerable divergence from the consensus outside the core LxxLxLxxN/C/T region associated with the repeating β-strand and adjacent loops. Strikingly, a Cys is present in 13 of the 24 repeats at the N/C/T position in the consensus.
Rpg1r Belongs to the Same Subclade of Arabidopsis CNLs as Rpg1b But Is Not Orthologous to RPM1
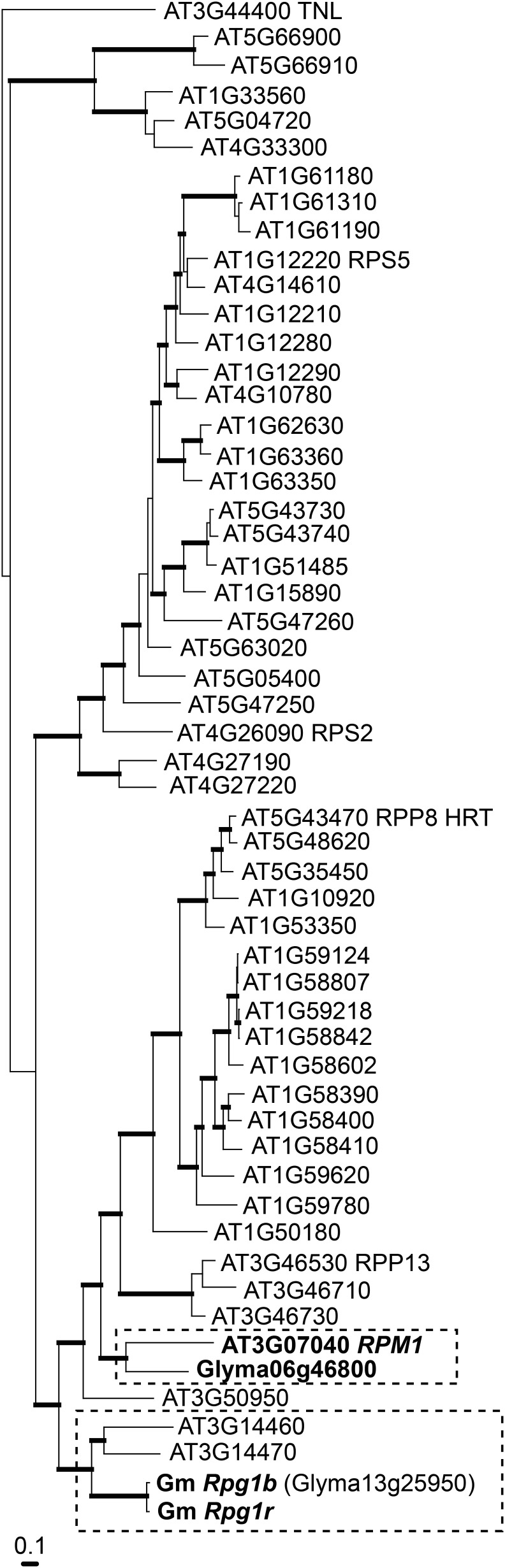
To assess the evolutionary relationship of Rpg1r to the Arabidopsis RPM1 gene, we constructed a phylogenetic tree that included the NB-ARC domains from the entire set of intact Arabidopsis CNLs, together with the soybean Rpg1b and Rpg1r genes. The NB-ARC domain was used, as its conserved structure facilitates alignment of distantly related CNLs and it has historically been used to define NB-LRR clades and because we have previously observed that this domain is less prone to interparalog recombination events that can confound phylogenetic analyses (Ashfield et al., 2012). Significantly, as we previously observed for Rpg1b, Rpg1r and RPM1 belong to distinct clades, consistent with the AvrRpm1 recognition specificity having evolved independently in soybean and Arabidopsis (Fig. 3). The two Rpg1 genes are closely related and are found in a well-supported clade containing two Arabidopsis CNLs of unknown function, AT3G14460 and AT3G14470. It should be noted that while Rpg1r and Rpg1b, together with one additional sequence described below, are the only soybean genes included in the tree, the Rpg1 genes are members of a larger group of closely related CNLs in the soybean genome. The majority of these closely related paralogs are located in the 1-Mb interval centered on the Rpg1b gene (shown in Fig. 1) and in segmental duplications of this region (Ashfield et al., 2012).
Figure 3.
The soybean Rpg1r and Rpg1b genes are closely related, but neither is orthologous to the RPM1 gene from Arabidopsis. A Bayesian phylogenetic tree based on an amino acid alignment of the NB-ARC domains from the soybean Rpg1r and Rpg1b genes, the complete set of Arabidopsis CNLs, and the soybean CNL (Glyma06g46800) most closely related to RPM1 (based on a BLAST search of the soybean proteome). Glyma13g25950.2 represents the Rpg1b locus in the soybean WGS (Schmutz et al., 2010) but contains a small internal deletion relative to the known BAC-derived sequence. Sequence AT3G44400 belongs to the TIR-NB-LRR subclass of NB-LRR genes and was used as an outgroup. Branches supported with posterior probabilities greater than 90% are printed in bold.
Also included in the tree was the top BLAST hit, Glyma06g46800, from a search of the whole soybean genome proteome using the Arabidopsis RPM1 NB-ARC domain sequence as the query. RPM1 forms a well-supported clade with Glyma06g46800, indicating that, at least with respect to the NB-ARC domain, RPM1 is more closely related to this soybean gene than to any other Arabidopsis CNL. As with the Rpg1 genes, Glyma06g46800 has a number of close relatives in the soybean genome (Zhang et al., 2011), revealing that this subfamily of CNLs has either expanded in soybean or contracted in the Arabidopsis lineage. No known AvrB- or AvrRpm1-specific R genes map close to Glyma06g46800 in the soybean genome, consistent with it having a recognition specificity different from RPM1. However, it is interesting to note that Glyma06g46800 and the Rpg1b/Rpg1r genes are located in homeologous segments of the soybean genome (i.e. regions duplicated by polyploidy), consistent with their lineages having been present in the same ancestral cluster (Chen et al., 2010).
Rpg1r and Rpg1b Share Closely Related NB-ARC Domains as a Consequence of Sequence Exchange during Their Recent Evolution
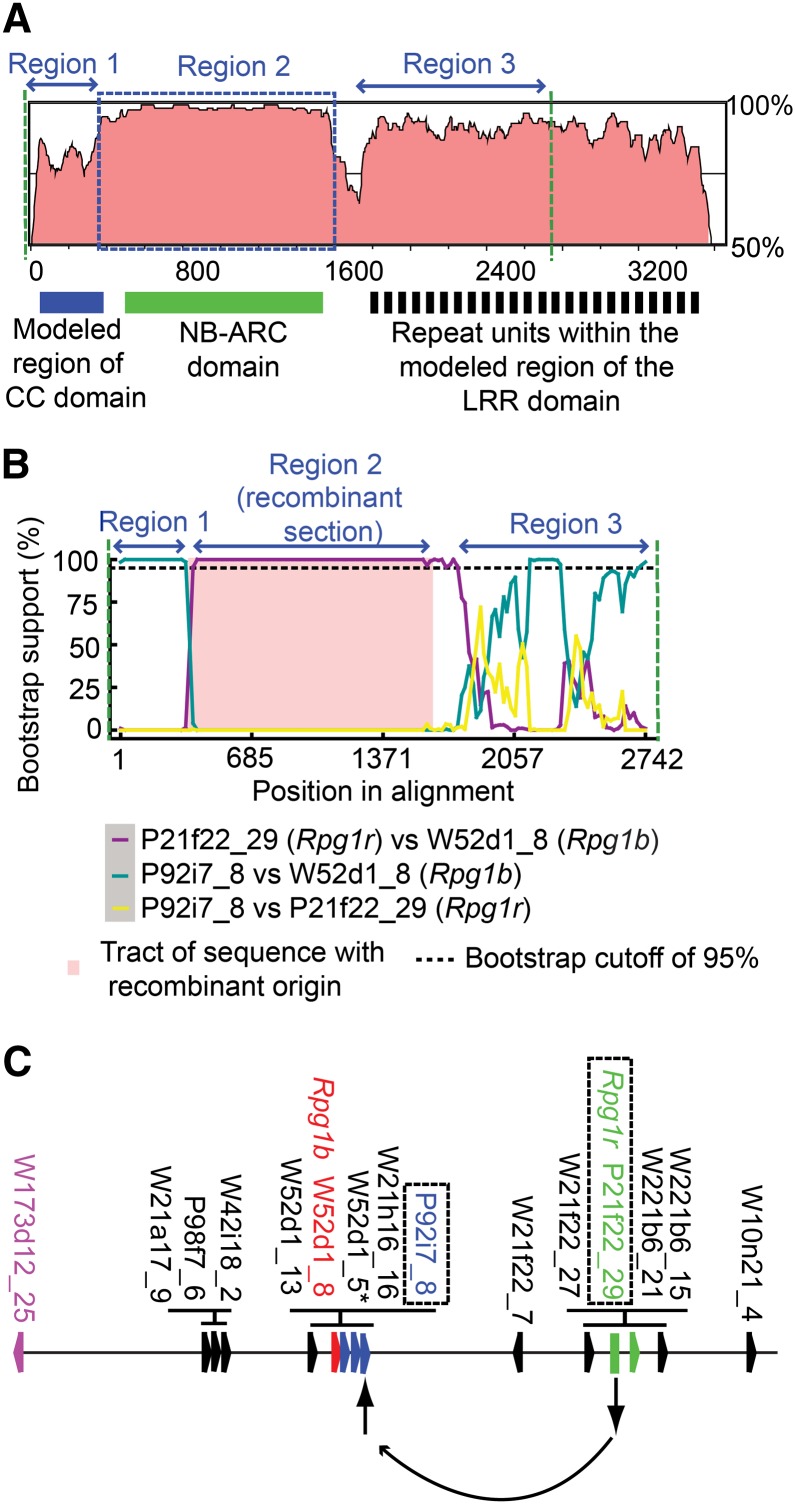
The cloning of Rpg1r allowed a comparison of its amino acid sequence to those from both Rpg1b and the Arabidopsis RPM1 protein. An alignment of the sequence of Rpg1r with the previously cloned Rpg1b revealed striking differences in the percentage nucleotide identity observed in different domains (Fig. 4A; Supplemental Fig. S5). Rpg1b and Rpg1r are most similar over a stretch that includes the C-terminal portion of the CC and the complete NB-ARC domain, with the N-terminal half of the CC domain and the LRR domain being less similar (Fig. 4A). Such a pattern could be explained by different strengths of selection acting on different domains or by sequence exchange between the ancestral lineages. To distinguish between these possibilities, we used the Recombination Detection Program (RDP) to analyze an alignment of Rpg1b, Rpg1r, and a single allele from each of the other CNL loci distributed across the 1-Mb interval in cv Williams 82 and accession PI 96983 that contains the Rpg1 loci. For the purposes of this analysis, we define recombination as any exchange of DNA sequence between paralogs or alleles, including gene conversion, and thus do not attempt to address whether flanking markers have undergone reciprocal exchanges. To reduce the chance of artifacts resulting from poor alignment, the C-terminal part of the LRR domain was excluded from the analysis, as not all the sequences could be aligned accurately in this region.
Figure 4.
As a consequence of recombination, Rpg1r and Rpg1b share closely related NB-ARC domains but more divergent CC and LRR regions. A, Comparison of the nucleotide sequences of Rpg1r and Rpg1b. The Rpg1 sequences were compared using the VISTA program and a 100-nucleotide sliding window. The y axis shows the percentage nucleotide identity between the two genes, and the x axis shows the position in the Rpg1b open reading frame. The interval between the two dashed green lines corresponds to the region subsequently analyzed for evidence of recombination (as described in B). A region found to have a strong signal for recombination is indicated with a blue dashed box (corresponds to the pink shaded region in B). B, Evidence for a recombinant origin for the Rpg1b NB-ARC domain. Shown is output from the Bootscan program. The recombinant region in Rpg1b (region 2) completely encompasses the NB-ARC domain and is indicated by pink shading. C, Location of recombinant Rpg1b, and the predicted parental sequences, within the 1-Mb interval that contains the Rpg1 genes. The locations of CNL paralogs are indicated with colored polygons arranged on the horizontal line. The image shown represents a hypothetical ancestral chromosome that includes all the unique CNL loci represented in both cv Williams 82 and PI 986983. Rpg1b is shown in red, and a second paralog (W173d12_25) predicted to carry the same recombination event is shown in magenta. The two parental sequences, P92i7_8 and P21f22_29 (Rpg1r), predicted to have recombined to generate the ancestral Rpg1b locus are printed in blue and green, respectively.
The RDP analysis indicated that the Rpg1b NB-ARC domain does have a recombinant origin. The initial RDP screen identified three possible recombination events within the analyzed region of Rpg1b, with the most strongly supported event encompassing the C-terminal portion of the CC domain and the entire NB-ARC (Region 2 in Fig. 4, A and B). This event was supported by all five methodologies deployed in the RDP screen (P < 0.001 threshold). For example, using the Bootscan method, which assesses changes in the phylogenetic relationship between the paralogs using a sliding-window approach, Rpg1b groups with CNL P92i7_8 over the N-terminal portion of the CC domain (Fig. 4B, Region 1) but with Rpg1r (P21f22_29) over the remainder of the CC domain and the entire NB-ARC domain (Fig. 4B, Region 2). Across the LRR domain, Rpg1b groups with P92i7_8 for only a short segment and not at all with Rpg1r (Fig. 4B, Region 3), suggesting that additional recombination events have occurred in this region. Interestingly, RDP identified a second CNL in the cluster, W173d12_25, as potentially sharing the same recombination event as Rpg1b, implying duplication and translocation of the ancestral Rpg1b locus subsequent to the recombination event discussed above (Fig. 4C).
As a consequence of the event described above, the recombinant and nonrecombinant segments of Rpg1b cluster with different subsets of CNL paralogs in neighbor-joining trees (Supplemental Fig. S6). It should be noted that, at least in the trees corresponding to Regions 1 and 2, the two potential parental sequences identified by RDP (Rpg1r and P92i7_8) belong to distinct clades that each contain several closely related paralogs (colored green and blue, respectively, in Fig. 4C and Supplemental Fig. S6). It is possible that these related CNLs, or others not present in the soybean genotypes sampled, could be the actual parental sequences.
To determine if different strengths of selection acting across the Rpg1 genes might also be contributing to the observed pattern of sequence similarity in the Rpg1b/Rpg1r alignment (Fig. 4A), the ratio of non-silent to silent substitutions (dN/dS) was calculated from the alignment for the three regions described in Figure 4 (Supplemental Table S1). In fact, the recombinant region in Rpg1b (Region 2 in Fig. 4) was under almost neutral selection (dN/dS = 1.0853), while both the flanking regions (Regions 1 and 3 in Fig. 4) had dN/dS values less than 1 (0.4871 and 0.8494, respectively), consistent with purifying selection. It is therefore clear that the island of high sequence similarity observed in the alignment of Rpg1b and Rpg1r is not the result of strong purifying selection acting on this region but is instead the consequence of recombination. In summary, during the evolution of the Rpg1b gene, recombination between CNLs in the cluster has combined an Rpg1r-like NB-ARC with the N-terminal section of a CC domain closely related to the other CNLs immediately adjacent to the present-day Rpg1b (Fig. 4C).
Dating the Recombination Event in Rpg1b Suggests a Recent Origin for This R Gene
To estimate the date of the recombination event spanning the NB-ARC region of Rpg1b, we used the dS value for this section of the Rpg1b/Rpg1r alignment (Region 2 in Fig. 4; Supplemental Table S1) and a mutation rate (Ks) estimate of 5.2 × 10–9 per year (Pfeil et al., 2005). This calculation gives an estimated age for the recombination event of 1.46 million years ago. Although it is a formal possibility that the capacity to recognize AvrB predates this recombination event, the rapid rate of duplication and deletion of NB-LRR genes in this region (Ashfield et al., 2012) supports a recent origin of AvrB specificity. Consistent with this view, we have not observed AvrB recognition capacity in common bean (Chen et al., 2010), which diverged from the Glycine spp. lineage approximately 19 million years ago (Lavin et al., 2005).
Comparison of the RPM1 Protein with Rpg1b and Rpg1r Reveals Only Limited Sequence Conservation between the Arabidopsis and Soybean CNLs Specific for AvrB and AvrRpm1
Although the ability to detect AvrB and AvrRpm1 appears to have evolved independently in the Arabidopsis and soybean lineages, it is possible that R proteins specific for these effectors will have conserved features. To investigate this possibility, Rpg1r was aligned with both the Rpg1b and RPM1 amino acid sequences. This analysis revealed only limited sequence conservation between the soybean R proteins and RPM1 outside the EDVID motif in the CC domain and conserved motifs of the NB-ARC domain (Supplemental Fig. S7). While the alignment across the LRR domain is highly fragmented, reflecting the presence of 24 canonical repeats in both the soybean Rpg1 proteins versus only 15 in RPM1 (Grant et al., 1995; Ashfield et al., 2004), the first four repeats align without insertions or deletions, raising the possibility of a conserved function for this region. Based on functional analyses of the Arabidopsis RPS5 CNL, this is likely associated with keeping the R proteins in an off state in the absence of elicitation (Qi et al., 2012).
Plotting Polymorphic Positions on Predicted Tertiary Structures for the Rpg1b CC and LRR Domains Reveals Highly Polymorphic Surfaces
As described above, a comparison of the Rpg1r and Rpg1b sequences revealed that both the CC and LRR domains are highly polymorphic, suggesting a role for one, or both, of these domains in conferring the ability to distinguish between AvrB and AvrRpm1. We hypothesized that polymorphic residues involved in recognition specificity would be located on solvent-exposed surfaces of Rpg1b. We therefore mapped the polymorphic positions in the Rpg1r/Rpg1b amino acid alignment onto models of the tertiary structures of the Rpg1b CC and LRR domains.
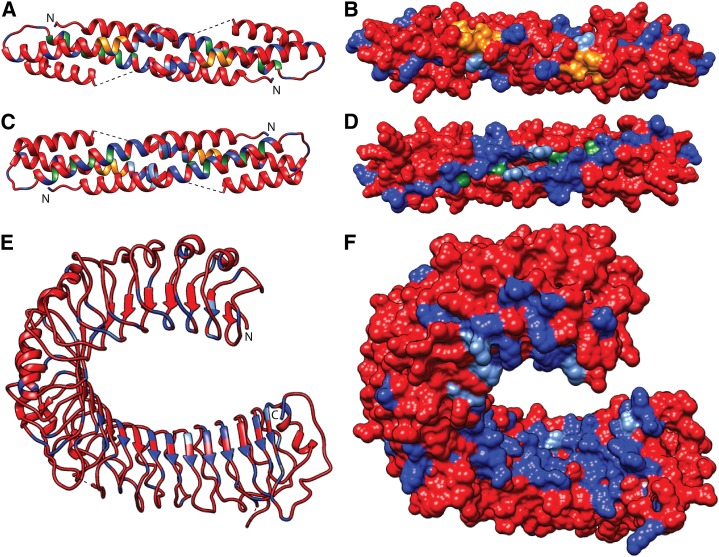
We have previously described modeling of the Rpg1b CC domain tertiary structure using the known MLA10 CC structure as the template (Ashfield et al., 2012). The MLA10 domain is known to form a homodimer with a conserved sequence motif (EDVID), previously shown to interact with the NB-ARC domain, being exposed on one surface of the dimer (Maekawa et al., 2011), and we propose that Rpg1b will form a similar structure. A model for the Rpg1b CC homodimer, generated using the Phyre2 modeling service, is shown in Figure 5, A to D. Based on the MLA structure, we predict that the α1 helices of the two Rpg1b monomers will form a CC structure. This hypothesis is supported by analysis of the Rpg1b sequence using the COILs program, which predicts that the region from Q38 to K61 in Rpg1b will participate in such a structure (P > 98%). Hydrophobic residues predicted to reside at the key a and d positions in the CC heptad repeats are shown in green in the modeled structure (Fig. 5, A–D). These include residues L41, L44, and I51, which correspond, in the alignment of the Rpg1b and MLA10 sequences, to three residues in MLA10 (I32, L36, and M43, respectively) shown previously to be required for dimerization in yeast (Saccharomyces cerevisiae) two-hybrid assays (Maekawa et al., 2011). Intriguingly, when residue positions that differ between the Rpg1b and Rpg1r proteins were mapped onto the Rpg1b homodimer model, an uneven distribution was observed. While the surface that contains the conserved EDVID motifs (EDILD in Rpg1b) contains relatively few nonconservative substitutions (residues of the EDVID motif printed in orange and polymorphic residues in blue; Fig. 5, A and B; Supplemental Movies S1 and S2), a highly polymorphic surface was observed on the opposite side of the dimer (Fig. 5, C and D). This highly polymorphic surface is generated by a region of the Rpg1b model that corresponds to the α1 helix in the MLA10 structure, a region previously shown to interact with WRKY domain-containing transcription factors (Shen et al., 2007).
Figure 5.
Modeling of the Rpg1b protein structure, and comparison to Rpg1r, reveals highly polymorphic surfaces on both the LRR and CC domains. A to D, Model of the proposed Rpg1b CC domain homodimer showing the location of polymorphic residues with respect to Rpg1r. The residues conserved between the Rpg1 proteins are shown in red, and those that differ shown in blue (conservative and nonconservative substitutions are shown in light and dark blue, respectively). Residues that reside at the a and d positions in predicted CC heptad repeats are shown in green. Residues that are part of the conserved EDVID sequence motifs are shown in orange. A and B, Surface containing the conserved EDVID sequence motifs. C and D, Surface on the opposite side of the structure as the EDVID motifs (i.e. model rotated through 180°). E and F, Model of the Rpg1b LRR domain showing the location of polymorphic residues with respect to Rpg1r. Residue coloring as for the CC domain.
Next, we generated a model for the Rpg1b LRR domain tertiary structure using the modeler software and the known Toll-Like Receptor3 (TLR3) structure as the template (Fig. 5, E and F). The TLR3 LRR ectodomain forms a horseshoe structure characteristic of other LRR proteins for which crystal structures are known, with a series of β strands forming a solvent-exposed β sheet across the concave surface. Regular β strands are also predicted in the Rpg1b LRR sequence using the PSIPHED secondary structure prediction server, and as expected, the majority of these are positioned across the concave surface of the model (Fig. 5E). As with the CC domain, mapping of residues that differ between the two Rpg1 proteins onto the Rpg1b LRR model revealed an uneven distribution. Clustering of polymorphisms was particularly striking across the concave surface of the structure, a region known to interact with ligands for at least some LRR proteins (Fig. 5, E and F; Supplemental Movies S3 and S4). Interestingly, most of the polymorphic positions are located within the C-terminal portion of the concave surface, where they form a highly polymorphic pocket with respect to Rpg1r.
AvrB versus AvrRpm1 Specificity Is Determined by the C-Terminal Portions of the LRR Domains of Rpg1b and Rpg1r
To test the predictions made by our structural modeling, we developed a transient expression system that enabled us to reconstitute the Rpg1b and Rpg1r signaling pathways in Nicotiana glutinosa (R. Kessens, T. Ashfield, S.H. Kim, and R.W. Innes, unpublished data). We employed N. glutinosa rather than the more commonly used Nicotiana benthamiana because transient expression of AvrB and AvrRpm1 in the latter induces rapid cell death, indicating that N. benthamiana contains R genes that recognize these effectors. To establish the efficacy of N. glutinosa, we coexpressed Rpg1rfused to super Yellow Fluorescent Protein (sYFP) and AvrRpm1 using A. tumefaciens-mediated transient expression under control of a dexamethasone-inducible promoter. Coexpression of the two genes together induced visible tissue collapse, which was not observed when either one was expressed alone (Fig. 6A). Importantly, coexpression of Rpg1r-sYFP with AvrB did not induce tissue collapse, demonstrating that Rpg1r specifically recognizes AvrRpm1. Repeating the experiment using untagged Rpg1r revealed significant R gene autoactivity, independent of the presence of AvrRpm1, indicating that the YFP tag attenuates the signaling activity of Rpg1r, which, in the context of Rpg1r overexpression, was useful. Analogous experiments with Rpg1b, however, revealed that sYFP nearly eliminated Rpg1b signaling ability (data not shown).
Figure 6.
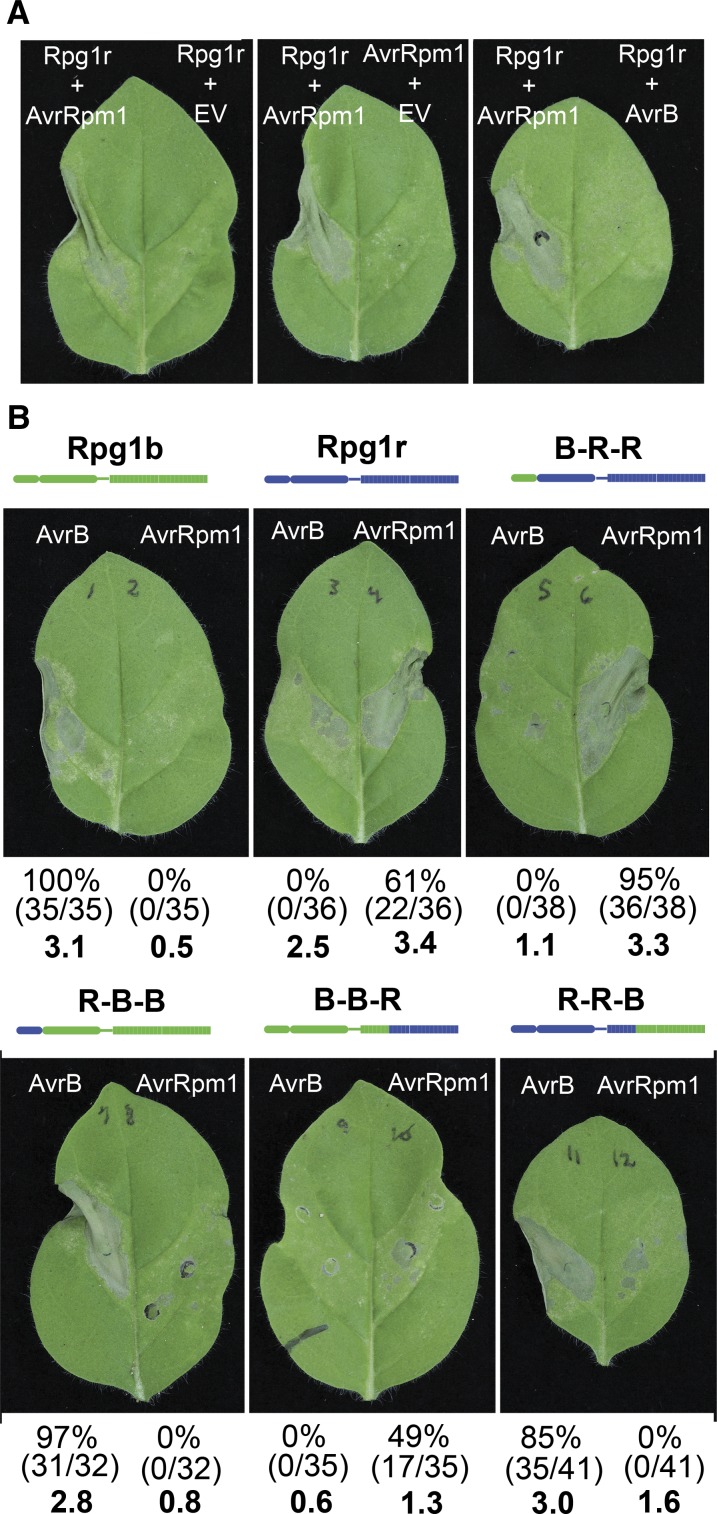
The LRR domains of Rpg1b and Rpg1r determine AvrB versus AvrRpm1 specificity. A, Rpg1r specificity is retained when transiently expressed in N. glutinosa. Agroinfiltrations were used to transiently express combinations of Rpg1r-YFP, AvrRpm1, AvrB, or empty vector (EV). All transgenes were under the control of a dexamethasone-inducible promoter. Photographs were taken approximately 24 h after gene induction. B, Expression of Rpg1b-Rpg1r chimeras reveals that the LRR domain determines effector specificity. The indicated chimeras were coexpressed with GmRIN4b and AvrB or AvrRpm1. The percentages given below each leaf indicate the percentage of leaves showing a stronger response to AvrB than AvrRpm1 (left value) and the percentage of leaves showing a stronger response to AvrRpm1 than AvrB (right value). The actual number of leaves used to calculate the percentages are shown in parentheses (number of leaves showing stronger response/total number of leaves injected). The values for leaves in which the responses to both effectors were similar are not shown. The numbers in bold indicate average HR scores. Photographs were taken approximately 48 h after dexamethasone application.
To circumvent the dual problems of attenuation of Rpg1b signaling by sYFP, and Rpg1r autoactivity when highly overexpressed, we switched to a vector in which the Rpg1 genes were expressed without an epitope tag and in which expression was driven by a Cauliflower mosaic virus 35S promoter to avoid the rapid accumulation resulting from an inducible promoter. Using this system, coexpression of Rpg1b and AvrB induced a strong tissue collapse, but only when we also coexpressed any one of the four soybean RIN4 genes (Fig. 6B; (R. Kessens, T. Ashfield, S.H. Kim, and R.W. Innes, unpublished data)). No such requirement for a GmRIN4 was noted for Rpg1r in this transient expression system (R. Kessens, T. Ashfield, S.H. Kim, and R.W. Innes, unpublished data). Untagged Rpg1b did not induce tissue collapse when coexpressed with AvrRpm1 and GmRIN4b (Fig. 6B); thus, the specificity of Rpg1r and Rpg1b are maintained in this heterologous system. Untagged Rpg1r induced some tissue collapse when expressed by itself or with AvrB and GmRIN4b (Fig. 6B), but this collapse was consistently weaker than that observed when coexpressed with AvrRpm1. We thus adopted a scoring system in which the responses to AvrB and AvrRpm1 were always compared within a single leaf and assessed for which induced the stronger collapse. Figure 6B summarizes data from three independent experiments in which 32 to 41 leaves in total were injected for each construct. For Rpg1r, 22 of 36 leaves responded more strongly to AvrRpm1, and none responded more strongly to AvrB (14 leaves gave a similar response to both effectors, with levels of collapse similar to that observed for Rpg1r alone).
Having established the efficacy of the N. glutinosa transient system, we tested the predictions of our structural modeling by assessing the effector specificity of two sets of Rpg1b-Rpg1r chimeras. In the first set of chimeras, we swapped the N-terminal regions containing the CC domain with the junction located just N terminal to the P-loop. We refer to these chimeras as B-R-R (indicating CC domain from Rpg1b and NB-LRR domains from Rpg1r) and R-B-B (CC domain from Rpg1r and NB-LRR domains from Rpg1b). In the second set, we swapped the C-terminal portion of the LRR domain starting at LRR repeat number 8 (R-R-B and B-B-R). We made the junction at this position because the majority of the polymorphisms between Rpg1b and Rpg1r were localized distal to this point. As predicted by the structural modeling, swapping the LRR domains (starting at repeat 8) was sufficient to swap recognition specificity, with R-R-B recognizing AvrB and B-B-R recognizing AvrRpm1, albeit weakly (Fig. 6B). These findings indicate that the CC domain is not required for differential recognition of AvrB and AvrRpm1. Consistent with this conclusion, swapping just the CC domains did not alter specificity (Fig. 6B). In summary, we conclude that the ability to distinguish between AvrB and AvrRpm1 is controlled by polymorphisms located within the C-terminal 17 LRR repeats of Rpg1b and Rpg1r.
Soybean Contains Four RIN4 Homologs and All Can Be Cleaved by AvrRpt2
RPM1-dependent recognition of AvrB and AvrRpm1 in Arabidopsis requires effector-dependent phosphorylation of a second plant protein, RIN4 (Mackey et al., 2002; Chung et al., 2011; Liu et al., 2011; Baltrus et al., 2012). RIN4 is also targeted and cleaved at two sites (AvrRpt2 Cleavage Site1 [RCS1] and RCS2) by a third P. syringae effector, AvrRpt2, which is a Cys protease (Axtell et al., 2003; Mackey et al., 2003; Chisholm et al., 2005). One consequence of this activity is that AvrRpt2 blocks recognition of AvrB and AvrRpm1 in Arabidopsis (Kim et al., 2005) and has the potential to be a useful tool for assessing the role of RIN4 homologs, or additional targets of AvrRpt2, in effector recognition in other species.
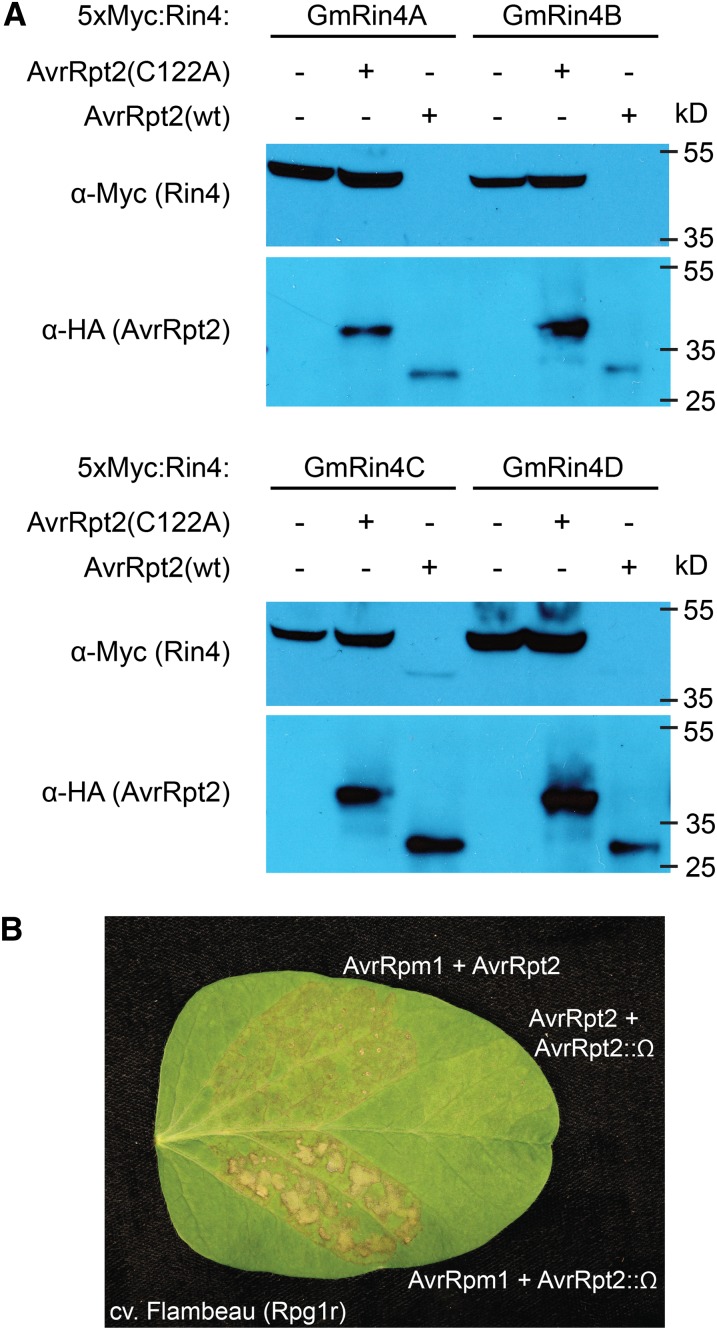
Soybean expresses four RIN4 homologs (GmRIN4s), and all four contain at least the C-terminal RCS2, with no more than a single conservative substitution (GmRIN4a–GmRIN4d; Chen et al., 2010; Supplemental Fig. S8). To test whether AvrRpt2 can cleave one or more of the GmRIN4s, each was transiently coexpressed in N. benthamiana with either AvrRpt2 or a proteolytically inactive variant of the effector (C122A; Axtell et al., 2003). To facilitate detection, each GmRIN4 was fused to an N-terminal 5×-Myc epitope tag and overexpressed using a dexamethasone-inducible expression vector. When expressed alone, or coexpressed with C122A, all four GmRIN4s accumulated strongly (Fig. 7A). However, coexpression with a functional AvrRpt2 transgene resulted in the complete, or near complete, disappearance of all four GmRIN4s (Fig. 7A). In some experiments, a cleavage product was detectable for GmRIN4c and/or GmRIN4d (Fig. 7A). Examination of the alignment of the GmRIN4s (Supplemental Fig. S8) reveals substitutions in the RCS1 cleavage site for both GmRIN4c and GmRIN4d that could inhibit cleavage and thus enable accumulation of a product cleaved at only RCS2.
Figure 7.
AvrRpt2 cleaves all four soybean RIN4 homologs and suppresses Rpg1r-mediated HR. A, Cleavage of GmRIN4s by AvrRpt2. Soybean 5× Myc-tagged RIN4 homologs (GmRIN4a, GmRIN4b, GmRIN4c, and GmRIN4d) were transiently coexpressed with or without HA-tagged AvrRpt2 (wild type [WT]), or a protease-inactive variant (C122A), in N. benthamiana, and total protein was extracted and immunoblotted with the indicated antibodies. B, Rpg1r-mediated recognition of AvrRpm1 in soybean is partially blocked by AvrRpt2. PsgR4 expressing AvrRpm1 was coinfiltrated into soybean leaves (cv Flambeau) expressing Rpg1r together with PsgR4 strains expressing either a functional (AvrRpt2), or nonfunctional (AvrRpt2::Ω), avrRpt2 allele. Infiltration of a mixture of the PsgR4 (avrRpt2) and PsgR4 (avrRpt2::Ω) strains was used to confirm that AvrRpt2 failed to elicit a visible HR in cv Flambeau. Photographs were taken approximately 48 h after infiltration.
AvrRpm1 Recognition in Soybean Is Partially Blocked by AvrRpt2
We have previously shown that Rpg1b-dependent detection of AvrB in soybean is inhibited by the presence of AvrRpt2 and have hypothesized that this was caused by cleavage of soybean RIN4 homologs (Ashfield et al., 2004). The cleavage results described above support this hypothesis. If Rpg1r function is also dependent on RIN4 homologs, AvrRpm1 detection in soybean should also be inhibited by AvrRpt2.
To test this prediction, mixtures of P. syringae strains were injected into leaves of an Rpg1r-expressing soybean cultivar (cv Flambeau). Coinjection of a strain expressing avrRpm1 with a strain expressing a disrupted avrRpt2 gene induced a typical HR by 24 to 48 h post injection (Fig. 7B). By contrast, coinjection of a strain expressing avrRpm1 with a strain expressing a functional avrRpt2 gene induced a weaker HR that was slower to develop, indicating that AvrRpt2 partially suppresses AvrRpm1 recognition (Fig. 7B). Coinjection of strains expressing functional AvrRpt2 and disrupted AvrRpt2 genes resulted in no tissue collapse, demonstrating that AvrRpt2 does not elicit a visible HR in cv Flambeau.
The above observations indicate that Rpg1r function is at least partially dependent on a target(s) of the AvrRpt2 protease, supporting the hypothesis that recognition of AvrRpm1 in soybean requires at least one soybean RIN4 homolog. The lack of such a requirement in the N. glutinosa transient system is likely explained by the presence of endogenous RIN4 homologs in N. glutinosa (R. Kessens, T. Ashfield, S.H. Kim, and R.W. Innes, unpublished data).
DISCUSSION
While the Arabidopsis RPM1 R gene can mediate detection of both the AvrB and AvrRpm1 pathogen effector proteins (Bisgrove et al., 1994), in soybean, two tightly linked R genes can distinguish between them (Ashfield et al., 1995). The soybean Rpg1b gene can only mediate detection of AvrB, while the Rpg1r gene can only mediate detection of AvrRpm1. This was an intriguing observation because it demonstrated that, at least in soybean, the two effectors likely have distinct activities or targets and yet are detected by a single R protein in Arabidopsis. Cloning RPM1, and the two soybean R genes, has been an important goal because it has the potential to provide insights into the evolution of specificity, persistence over time, and mode of activity of R genes that detect pathogen effectors indirectly. RPM1 and Rpg1b have been cloned previously (Grant et al., 1995; Ashfield et al., 2004). Here, we describe the map-based cloning of the soybean Rpg1r gene, allowing a detailed comparison of the three functionally related R genes.
Like the previously cloned Rpg1b gene, Rpg1r belongs to the CNL family of R genes. Overall, the two genes display high overall sequence similarity, sharing 86% amino acid identity. However, as discussed below, the level of sequence identity varies dramatically between domains. Interestingly, while the Rpg1r protein contains the domains and motifs typical of CNL R proteins, its NB-ARC domain contains an unusual substitution. Specifically, the second Asp residue of the kinase 2 (Walker B) domain (consensus hhhhDD for R proteins, where h is usually a hydrophobic residue; Takken et al., 2006) has been replaced with an Asn. An acidic residue is highly conserved, but not invariant, in this position in both plant R proteins and other members of the STAND class of the P-loop nucleotide triphosphatase (NTPase) family (Leipe et al., 2004). This residue is proposed to play a key role in the NTPase activity of the domain in at least some classes of P-loop NTPases, priming water for nucleophilic attack of the NTP (Muneyuki et al., 2000; Geourjon et al., 2001). At least some R proteins have known NTPase activity, and it has been proposed that this activity is required for returning the protein to an inactive state after elicitation (Tameling et al., 2002; Takken and Tameling, 2009). Consistent with this hypothesis, a D283E substitution in the tomato (Solanum lycopersicum) I-2 protein affected its ATPase activity, but not ATP binding, and leads to autoactivity (Tameling et al., 2006). The observation that Rpg1r can function with an Asn in this location suggests either that ATPase activity is not required for its successful operation or, in this case, the domain retains its enzymatic activity despite the absence of the second conserved Asp.
Given that both AvrB and AvrRpm1 are detected by the RPM1 protein in Arabidopsis (Bisgrove et al., 1994; Grant et al., 1995), it was possible that one or both of the soybean Rpg1 genes would be an ortholog of RPM1 (i.e. an ancestral R gene with the ability to respond to AvrB and/or AvrRpm1 that has survived since before the divergence of the soybean and Arabidopsis lineages). We have previously reported that Rpg1b and RPM1 are not orthologs (Ashfield et al., 2004), and here we confirm phylogenetically that Rpg1r is not orthologous with RPM1 either. We therefore conclude that the ability to recognize both AvrB and AvrRpm1 evolved independently in Arabidopsis and soybean, consistent with the general observation that NB-LRR genes experience a high rate of turnover through species-specific duplications and deletions (Cannon et al., 2002; McHale et al., 2006).
Curiously, the soybean CNLs with NB-ARC domains phylogenetically most closely related to RPM1 are found in a region homeologous to the Rpg1 cluster; that is, the two regions were generated by duplication of an ancestral cluster as a consequence of polyploidization (Chen et al., 2010). This suggests that in the common ancestor of soybean and Arabidopsis, the ancestral Rpg1 region contained CNL lineages closely related to both RPM1 and the Rpg1 genes, and these lineages were subsequently parsed between the two duplicated regions in soybean. This raises the possibility that while the functional AvrB- and AvrRpm1-specific R genes evolved independently in Arabidopsis and soybean, some property of the ancestral cluster predisposed the subsequent evolution of R genes able to detect these pathogen effectors. As has been discussed previously, such a property might be the ability of at least some of the CNLs present in the cluster to monitor RIN4-like proteins for pathogen-induced modifications (Chen et al., 2010).
The hypothesis that the CNL cluster in which the Rpg1 genes reside is predisposed to evolve AvrB/AvrRpm1-specific R genes is further supported by the observation that an R gene (Resistance to Pseudomonas syringae effector AvrRpm1 number1) with the ability to detect AvrRpm1 is also found in common bean in a region orthologous to the Rpg1b/Rpg1r R gene cluster in soybean (Chen et al., 2010). While it is possible that the Rpg1r gene has survived for at least the approximately 19 million years since the divergence of the Glycine and Phaseolus spp. lineages, the available evidence from mapping CNL subfamilies relative to the AvrRpm1-specific R gene in Phaseolus spp. argues against this (Chen et al., 2010). The alternative explanation is that the AvrRpm1-specific genes evolved independently in soybean and common bean. Only the cloning of the AvrRpm1-specific gene in Phaseolus spp. will allow the relationship between the functionally analogous genes to be unambiguously resolved. In contrast to the situation observed for AvrRpm1, no common bean cultivars with the ability to respond to AvrB have been identified (Chen et al., 2010). Interestingly, the ability to respond to AvrB and/or AvrRpm1 has been observed in N. benthamiana (Chung et al., 2011), lettuce (Lactuca sativa), and pepper (Capsicum annuum; Wroblewski et al., 2009), suggesting that these recognition specificities have evolved independently in multiple plant lineages. Whether the AvrB/AvrRpm1-specific R genes found in the N. benthamiana and lettuce reside in Rpg1-orthologous regions is not known.
It should be noted that while functional R genes typically do not persist long enough to be found in different plant families, the available evidence indicates that the downstream signaling components are far more conserved. For example, the MLA1 R gene from the monocot barley has been shown to function in the dicot Arabidopsis, demonstrating that the downstream components of the MLA1-triggered pathway have been conserved since before the monocot/dicot split (Maekawa et al., 2012). This observation has practical implications because it raises the possibility of cloned R genes effective against economically important pathogens being transferred transgenically to distantly related crop species.
Many of the sites that differ between Rpg1b and Rpg1r were found to localize to the C-terminal portion of the LRR domain in a region predicted to form a concave surface of a horseshoe-like structure (Fig. 5). This is consistent with previous observations that this surface binds ligands for at least some LRR proteins (Kobe and Deisenhofer, 1995). We thus predict that this surface on Rpg1b and Rpg1r mediates detection of AvrB- and AvrRpm1-mediated modifications of soybean RIN4 homolog(s), respectively. In support of this model, transient expression of chimeric Rpg1b-Rpg1r proteins confirmed that AvrB versus AvrRpm1 specificity is controlled by the C-terminal 17 LRR repeats (Fig. 6). The C-terminal half of the LRR domain has recently been implicated in determining the specificity of the CNL N and L proteins of Nicotiana sylvestris and Capsicum spp., which mediate detection of tobamovirus coat proteins (Sekine et al., 2012). Similarly, structural modeling of the RPP1 protein of Arabidopsis, a TIR-NB-LRR that likely directly binds its corresponding effector, Arabidopsis thaliana Recognised1, identified the C-terminal LRR region as containing hotspots of polymorphism that correlate with effector specificity (Steinbrenner et al., 2012). Like Rpg1b and Rpg1r, these polymorphisms localize to the concave surface of the LRR horseshoe.
In summary, we conclude that the AvrB- and AvrRpm1-specific R genes in Arabidopsis and soybean evolved independently but probably from CNL lineages originally present in the same ancestral R gene cluster. Despite both effectors being detected by a single R gene (RPM1) in Arabidopsis, the AvrB- and AvrRpm1-specific genes in soybean contain numerous polymorphisms in both the CC and LRR domains. The work presented here indicates that the LRR domain is the primary specificity determinant, which raises the question of whether the polymorphisms in the CC domain are functionally important. We are currently investigating whether the CC domain polymorphisms impact the affinity of Rpg1b and Rpg1r for the four soybean RIN4 homologs. This information should provide insights into the apparent propensity of CNL paralogs present in the Rpg1, and homologous, clusters to develop into AvrB- and AvrRpm1-specific R genes.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Seed for soybean (Glycine max) accession PI 96983 (Rpg1r rpg1b) was obtained from M.A. Saghai-Maroof (Department of Crop and Soil Environmental Sciences, Virginia Tech). Seed for cv Williams 82 (rpg1r Rpg1b) was ordered from the U.S. Department of Agriculture Soybean Germplasm Collection via the National Plant Germplasm System Web portal (http://www.ars-grin.gov/npgs). The soybean ‘Flambeau’ (Rpg1r rpg1b) × ‘Merit’ (rpg1r Rpg1b) RIL population has been described previously (Ashfield et al., 1995). Soybean plants were grown in clay pots containing a compost:Metro-Mix 360 (Sun Gro Horticulture):vermiculite:perlite (4:2:1:1) mix supplemented with osmocote slow-release fertilizer. Pathogenicity tests were conducted 2 to 3 weeks after planting with the growth chamber set for 23°C and a 16-h photoperiod.
Nicotiana glutinosa accession 241768 was obtained from the U.S. Department of Agriculture National Plant Germplasm System Nicotiana Collection at North Carolina State University. Both it and Nicotiana benthamiana were grown in a 3:1 mixture of Metro-Mix 360 potting soil (Sun Gro Horticulture) and expanded perlite in a growth room at 22°C. N. glutinosa plants were germinated under long-day conditions (16-h light/8-h dark) for 12 d to facilitate seedling establishment and then transferred to short-day conditions (9-h light/15-h dark cycle; 150 µE m–2 s–1) for 2 to 3 more weeks before transformation. The initial growth period under long-day conditions was omitted for N. benthamiana.
Genetic and Physical Mapping
Primers for PCR-based genetic markers used for the genetic fine mapping of Rpg1r in the cv Flambeau × Merit RIL population, as shown in Supplemental Figure S1, are listed in Supplemental Table S2 and the markers are described in Supplemental Table S3. The PCR was conducted as described by Ashfield et al. (1998). The scoring of the RIL population for response to Pseudomonas syringae expressing AvrRpm1, and the identification of lines containing informative recombination events in the region containing the Rpg1 genes, has been described previously (Ashfield et al., 1995, 2003).
The generation of the BAC-based sequence centered on the Rpg1b locus for both the cv Williams 82 and PI 96983 soybean genotypes and the identification of CNL genes within these sequences were described by Innes et al. (2008). The WGS (Glyma1 assembly) for soybean ‘Williams 82’ (Schmutz et al., 2010) and the associated gene set (v1.1 gene set) were accessed from the Phytozome Web portal (http://www.phytozome.net).
Amplifying Rpg1r Candidate CNLs from PI 96983 Genomic DNA
The primers and PCR annealing temperatures used to amplify CNLs from soybean accession PI 96983 (Rpg1r) genomic DNA, using locus-specific primers corresponding to noncoding sequences that flank CNLs in cv Williams 82 (rpg1r), are given in Supplemental Table S4. Primer sequences are listed in Supplemental Table S2.
PI 96983 genomic DNA was isolated as described by Ashfield et al. (2003) or using the Qiagen DNeasy Plant Mini Kit; in both cases, small (<1 cm in length), only partially expanded trifoliate leaves were used as the source tissue. PCR was accomplished using the TripleMaster polymerase (Eppendorf) according to the manufacture’s instructions. PCR products were isolated from agarose gel slices using the QIAquick gel extraction kit (Qiagen). The purified DNA fragments were subsequently sequenced using the ABI PRISM BigDye v3.0 Terminator cycle sequencing kit (PE Applied Biosystems) according to the manufacture’s instructions, except that reaction volumes were reduced to 10 μL, consisting of 1 μL of BigDye terminator ready reaction mix, 3 μL of MgCl2 (5 mm), 2 μL of primer (2 µm), 2 μL of template DNA, and 2 μL of water.
Complementation of Rpg1r Function in Soybean Stable Transformants
The approximately 5-kb genomic region containing the Rpg1r (CNL P21f22_29) coding region and flanking sequences was PCR amplified from soybean accession PI 96983 (Rpg1r) genomic DNA using locus-specific primers as described above (details in Supplemental Table S4). The amplified fragment was initially cloned into the pGEM-T Easy vector (Promega), then excised as a NotI fragment, and subsequently ligated into NotI-digested pGREENII 229 (http://www.pgreen.ac.uk; Hellens et al., 2000) to generate the plasmid pTAI-30.9. Soybean ‘Jack’ (Rpg1b rpg1r) was transformed with pTAI-30.9 by the Plant Transformation Core Facility, University of Missouri.
The presence of the pTAI-30.9 T-DNA in the primary transformants and T2 progeny was monitored using PCR and the T-DNA-specific primers TA129 and TA130 (Supplemental Table S2).
Construction of Rpg1b/Rpg1r Chimeras
Overlap PCR was used to generate Rpg1r/Rpg1b chimeric genes in which either the CC domains, or C-terminal portions of the LRRs, were swapped. The splice sites are indicated in Supplemental Figure S5. The Rpg1r and Rpg1b CC domains were amplified using primer pairs attB1-Rpg1r-F + CC/NB-R and attB1-Rpg1b-F + CC/NB-R, respectively, while the Rpg1r and Rpg1b NB-ARC-LRR regions were amplified using primer pairs CC/NB-F + attB2-Rpg1r-R and CC/NB-F + attB2-Rpg1b-R, respectively (Supplemental Table S2). The PCR products were gel purified, the appropriate combinations were mixed, and the chimeric Rpg1 coding sequences were amplified using flanking primers.
The Rpg1r/Rpg1b chimeric genes in which the C-terminal portion of the LRR domains were swapped were generated in a similar manner. In this instance, the Rpg1r and Rpg1b CC-NB-ARC-N-terminal LRR regions were amplified using primer pairs attB1-Rpg1r-F + LRR-R and attB1-Rpg1b-F + LRR-R, respectively. The Rpg1r and Rpg1b C-terminal LRR regions were amplified using primer pairs LRR-F + attB2-Rpg1r-R and LRR-F + attB2-Rpg1b-R, respectively.
The amplified chimeric sequences were subsequently cloned in the pEG100 destination vector (Earley et al., 2006) using Gateway (Invitrogen) technology to permit in planta expression under control of the 35S promoter.
Transient Expression Assays in N. glutinosa
For transient expression in planta, Rpg1r-sYFP, Rpg1b-sYFP, untagged AvrB, and untagged AvrRpm1 were cloned into a Gateway-compatible derivative of pTA7002, which places the transgene under control of a dexamethasone-inducible promoter (Aoyama and Chua, 1997). Untagged Rpg1b, Rpg1r, and the untagged chimeric Rpg1 constructs were cloned in the pEG100 vector (Earley et al., 2006) to allow expression by the 35S promoter. The 35S-GmRIN4B construct has been described previously (Selote et al., 2013).
The R gene constructs, GmRIN4b, and the AvrB/AvrRpm1 effectors were transiently expressed in N. glutinosa by Agrobacterium tumefaciens-mediated transformation. A. tumefaciens strain GV3101 (pMP90) was used for all constructs except GmRIN4b, in which case LBA4404 was used. A. tumefaciens strains were grown overnight (at least 16 h) at 30°C in Luria-Bertani media with 50 µg mL–1 of kanamycin and 50 µg mL–1 of gentamycin for the GV3101 strains and 100 μg mL–1 spectinomycin and 50 µg mL–1 streptinomycin for LBA4404 harboring the GmRIN4b construct. After overnight culture, bacterial cells were pelleted by centrifugation, washed with 10 mm MgCl2, resuspended in 10 mm MgCl2 containing 100 µm acetosyringone, and incubated at room temperature for at least 2 h. When assessing the wild-type Rpg1 genes with C-terminal sYFP tags, inoculum was prepared by mixing the indicated strains so that each transgene-containing strain had an individual optical density at 600 nm (OD600) of 0.1. When assessing the untagged Rpg1 genes or chimeras, the density of the strains harboring the R genes and GmRIN4b were reduced to an OD600 of 0.05. The inoculum was injected into 3.5- to 4.5-week-old N. glutinosa plants with a needleless syringe, nicking the leaf surface with a razor blade prior to injection to facilitate fluid entry when necessary. Older plants were avoided, as they transformed less reliably. Transgene expression was induced approximately 40 h post injection by spraying the plants with 50 µm dexamethasone and 0.02% (v/v) L77 surfactant (Momentive Performance Materials, Inc.). The leaves were scored for tissue collapse approximately 24 to 48 h later, and leaves were removed for photography shortly after. Responses were scored as follows: 0, no visible response; 1, white speckling typically associated with a nonspecific response to the A. tumefaciens; 2, chlorosis or tissue discoloration; 3, partial collapse of the injected panel; and 4, complete collapse of the injected panel.
HR Tests and Testing for Suppression of Rpg1r Function by AvrRpt2
The response of soybean plants to P. syringae expressing AvrRpm1 was assessed using HR hand inoculation tests as described by Ashfield et al. (1995). In brief, P. syringae pv glycinea race4 (PsgR4; Ashfield et al., 1995) was injected into the apoplast of soybean leaves at an OD600 of either 0.1 or 0.2 using a 1-mL disposable syringe. Rpg1r-depependent recognition of AvrRpm1 was associated with brown discoloration of the leaf veins, and typically tissue collapse, within the injected panels. Injected leaves were scored 24 to 48 h after injection.
Suppression of AvrRpm1 recognition by AvrRpt2 was demonstrated using mixed inoculations in which the component PsgR4 strains were coinfiltrated at equal densities with each strain at an OD600 of 0.1. The PsgR4 (AvrRpm1), PsgR4 (AvrRpt2), and PsgR4 (AvrRpt2::Ω) strains have been described previously (Whalen et al., 1991; Ashfield et al., 1995). The nonfunctional AvrRpt2::Ω allele was generated by insertion of the Ω fragment into the AvrRpt2 clone pLH12 to generate plasmid pLH12Ω (Whalen et al., 1991). HR responses were scored 48 h after injection.
Sequence Analyses
Sequence alignments were generated either using pairwise BLAST (Altschul et al., 1990) or ClustalX (Larkin et al., 2007). Sequence conservation in Clustal alignments was highlighted using the BoxShade 3.21 server (http://www.ch.embnet.org/software/BOX_form.html).
Variation in conservation of nucleotide identity between Rpg1b and Rpg1r across their coding sequences was assessed using the VISTA server (http://genome.lbl.gov/vista/index.shtml; Frazer et al., 2004) with the mVISTA and LAGAN (Brudno et al., 2003) settings.
The COILs server (http://www.ch.embnet.org/software/COILS_form.html; Lupas et al., 1991) was used to screen the Rpg1b and Rpg1r CC domains for regions likely to participate in CC structures. The MTIDK matrix was used together with a 2.5-fold weighting for positions a and d in the heptad repeats.
Phylogenetic Analyses
The phylogenetic tree presented in Figure 3 is based on an amino acid alignment of the NB-ARC domains (VGMGG to MHDLL in Rpg1b) from the complete set of intact Arabidopsis (Arabidopsis thaliana) CNLs together with three soybean genes. The soybean CNLs are the cv Williams 82 allele of Rpg1b (GenBank accession no. AY452685), the PI 96983 allele of Rpg1r (P21f22_29), and the top hit in a BLASTp search (using the BLAST tool available at http://www.phytozome.org) of the soybean whole-genome proteome (Schmutz et al., 2010) using the NB-ARC domain sequence from the Arabidopsis RPM1 gene (AT3G07040). The Arabidopsis TIR-NB-LRR gene AT3G44400 was also included in the analysis as an outgroup. The Arabidopsis CNL sequences are available from The Arabidopsis Information Resource Web site (http://www.Arabidopsis.org). The soybean sequences were added to a previously described alignment of Arabidopsis CNL genes (Meyers et al., 2003) using Clustal (Larkin et al., 2007) and the BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) interface. The Bayesian tree was then generated using the MrBayes 3.1.2 software (Ronquist and Huelsenbeck, 2003) and the following parameters: two parallel runs with four chains in each; model jumping between fixed-rate amino acid models; 1 million generations; sampling every 100 generations; and first 25% of trees discarded before the consensus tree calculated. The runs were considered to have converged when the average sd of split frequencies between the two runs had reduced to less than 0.01.
The neighbor-joining tree shown in Supplemental Figure S4 is based on an alignment of the NB-ARC domains of the soybean CNL genes from the 1-Mb interval centered on the Rpg1b gene in cv Williams 82 and the corresponding region in accession PI 96983 (Ashfield et al., 2012). The tree was generated using ClustalX 2.1 (Larkin et al., 2007), and clades were assessed using 1,000 bootstrap repeats. Trees were visualized using TreeView X (Page, 1996).
Detection of Recombination
An alignment was generated using ClustalX for Rpg1b (W52d1_8), Rpg1r (P21f22_29), and the remaining intact CNLs from the 1-Mb interval in cv Williams 82 shown in Figure 1. Also included were two additional genes from the corresponding region in accession PI 96983 (P98f7_6 and P92i7_8) that lack alleles in the cv Williams 82 sequence. The alignment was then trimmed to remove the 3′ end of the LRR domain (leaving nucleotides 1–2,664 for Rpg1b), as this region was hard to align accurately. The alignment was then screened for evidence of recombination using the RDP v.4.9 (Martin et al., 2010) using the parameters described by Ashfield et al. (2012) and the following methods: RDP (Martin and Rybicki, 2000), GENECONV (Padidam et al., 1999), Bootscan (Martin et al., 2005), Chimaera (Posada and Crandall, 2001), and MAXCHI (Smith, 1992). Only events supported by two or more methods at a Bonferroni-corrected P value of 0.001 or better were considered. After manual assessment of the RDP output associated with the NB-ARC-encompassing recombination event detected in Rpg1b, the position of the N-terminal breakpoint was refined and the alignment then rescanned.
Calculating Ka and Ks Values and Estimating the Age of Recombinant Segments
The ratio of nonsynonymous to synonymous mutations (Ka/Ks) was calculated for three segments of the alignment of nonredundant CNL genes used in the RDP recombination analysis (segments shown in Figure 4, A and B). Region 1 refers to the interval between the start of the coding regions and the recombinant region detected in Rpg1b (W52d1_8). Region 2 refers to the part of alignment corresponding to the recombinant region in Rpg1b (ending with the first of the two possible breakpoints; see Supplemental Fig. S9). Region 3 refers to the region from the end of recombinant segment in Rpg1b to the end of the trimmed alignment (starting with the second of the two possible breakpoints; see Supplemental Fig. S9). Ka, Ks, and Ka/Ks values were calculated using the method described by Yang and Nielsen (2000) as implemented by the PAML software package (Yang, 2007).
The age of the recombinant section identified in Rpg1b was estimated using a substitution rate of 5.2 × 10–9 Ks per year (Pfeil et al., 2005) and the formula T = K/2r, where T is time, K is the Ks value for the Rpg1b (W52d1_8)/Rpg1r (P21f22_29) pairwise comparison for this region, and r = substitution rate.
Tertiary Structure Prediction and Plotting Polymorphic Positions
The Rpg1b CC domain monomer structure was modeled using the Phyre2 server with the normal mode modeling setting (http://www.sbg.bio.ic.ac.uk/phyre2/; Kelley and Sternberg, 2009). Phyre2 modeled 120 residues in Rpg1b (A13–K132) using the MLA10 CC structure (Protein Data Bank [PDB] code 3qfl; Maekawa et al., 2011) as the template. Before assembly of the Rpg1b homodimer, a 14-residue stretch (Q99–T112; associated with an insertion in Rpg1b and a gap in the MLA10 structure; shown in blue in Supplemental Fig. S10A) that could not be threaded onto known MLA10 structure was deleted. The Rpg1b dimer was assembled and visualized using the University of California, San Francisco (UCSF) Chimera package (v. 1.7; http://www.cgl.ucsf.edu/chimera/; Pettersen et al., 2004) and the biological unit information present in the MLA10 PDB structure file.
The Rpg1b LRR domain structure was modeled using the modeler server (Eswar et al., 2006) accessed remotely using the UCSF Chimera package (v 1.7; Pettersen et al., 2004). Default settings were used. The known structure of the Human TLR3 ectodomain (PDB no. 1ziw; Choe et al., 2005) was identified as a suitable template using the Phyre2 server (http://www.sbg.bio.ic.ac.uk/phyre2/; Kelley and Sternberg, 2009) and the structure file obtained from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (http://www.pdb.org; Berman et al., 2000). The Rpg1b LRR and TLR3 amino acid sequences were first aligned using ClustalX (Larkin et al., 2007) as implemented by the BioEdit sequence alignment editor (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). The alignment was then refined to improve the alignment of the β strands (secondary structure known in TLR3 and predicted in Rpg1b) and associated xxLxLxx sequence motifs (where L can be substituted by another hydrophobic residue), characteristic of the concave surfaces typical of LRR domains (see Supplemental Fig. S11 for alignment). TLR3 secondary structure information was sourced from the 1ziw PDB file. Rpg1b secondary structure was predicted using the PSIPRED protein structure prediction server (http://bioinf.cs.ucl.ac.uk/psipred/; Jones, 1999; Bryson et al., 2005). Secondary structure visualization and alignment editing was accomplished using the SBAL program (www.structuralchemistry.org/pcsb/; Wang et al., 2012). After modeling, two insertions present in Rpg1b (886Q-898C and 1059P-1075I; shown in blue in Supplemental Fig. S10C), likely representing segments of additional repeat units absent in the TLR3 template structure, were deleted from the model to generate the structure shown in Supplemental Figure S10D.
Sequence conservation between the Rpg1 proteins over the modeled regions was determined by aligning the Rpg1b and Rpg1r amino acid sequences using ClustalX (Larkin et al., 2007). Subsequently, a custom Perl script was used to substitute the b factor values in the Rpg1b CC and LRR model PDB files with values reflecting the substitution pattern. For this analysis, substitutions between the following amino acid pairs were considered to be conservative: FW, IL, IV, IM, LV, LM, VM, RK, RH, KH, GA, TS, DE, and NQ. The models were viewed, and images and movies were generated, using the UCSF Chimera package (v 1.7; Pettersen et al., 2004).
Cleavage of Soybean RIN4 Homologs by AvrRpt2
N-terminally Myc-tagged GmRIN4 expression constructs were generated using multisite Gateway (Invitrogen) technology with protocols provided by the manufacturer. Briefly, the soybean GmRIN4a (Glyma03g19920), GmRIN4b (Glyma16g12160), GmRIN4c (Glyma18g36000), and GmRIN4d (Glyma08g46400) complementary DNA sequences (see Supplemental Fig. S8 for an alignment) were amplified from EST clones for GmRIN4a to GmRIN4c and complementary DNA for GmRIN4d derived from soybean ‘Williams 82’ using primers that included Gateway attB sites. The gene-specific primers (minus the attB tails) are provided in Supplemental Table S2. BP cloning was used to introduce these PCR products into a modified Bluescript vector (Agilent Technologies) containing the appropriate attP sites (Qi et al., 2012). Subsequently, multisite LR cloning was used to combine the GmRIN4 sequences with 5× Myc N-terminal tags in a Gateway-compatible variant of the pTA7002 expression vector (Aoyama and Chua, 1997). All constructs were sequence verified. Cloning in pTA7002 allowed dexamethasone-inducible expression of the GmRIN4s in transformed plant tissue. AvrRpt2, and the proteolytically inactive variant C122A, were expressed under the control of the 35S promoter, and the constructs used have been described previously (Axtell et al., 2003).
A. tumefaciens GV3101 strains carrying the various dexamethasone-inducible GmRIN4 and AvrRpt2 constructs were grown, and inoculum prepared for transient transformation of N. benthamiana leaves, based on protocols described by Wroblewski et al. (2005). All strains were resuspended in water at an OD600 of 0.8. Suspensions for mixed inoculations were combined in a 1:1 ratio prior to infiltrating the leaves of 4-week-old N. benthamiana. For treatments where each GmRIN4 was expressed alone, A. tumefaciens suspensions were mixed in a 1:1 ratio with water prior to infiltration. Plants were sprayed with 50 µm dexamethasone (Sigma) 36 h following injection to induce expression. Samples were collected 3.5 h after dexamethasone treatment and flash frozen in liquid nitrogen. Protein extraction, SDS-PAGE, and immunoblotting were conducted as described by Qi et al. (2012).
Sequence data from this article can be found in the GenBank data library under accession numbers AY452685 (Rpg1b from cv Williams 82); KF958751 (Rpg1r from PI96983); AC151963 (sequence for BAC clone gmw1-21f22, from which the Williams 82 allele of Rpg1r was extracted, along with NB-LRR genes W21f22_7 and W21f22_27); KF958753 (P21F22_7); KF958752 (P21f22_27); X87851 (Arabidopsis RPM1); GU132851 (GmRIN4a); GU132855 (GmRIN4B); GU132852 (GmRIN4C); and GU132853 (GmRIN4D).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genetic and physical mapping of Rpg1r region.
Supplemental Figure S2. Comparison of corresponding CNL gene sequences from BAC-derived and soybean WGS-derived sequences.
Supplemental Figure S3. Comparison of cv Williams 82 and PI 96983 CNL alleles.
Supplemental Figure S4. Phylogenetic relationships of the CNL genes in cv Williams 82 (rpg1r Rpg1b) and PI 96983 (Rpg1r rpg1b) from the approximately 1-Mb region containing the Rpg1 genes.
Supplemental Figure S5. Alignment of Rpg1r and Rpg1b amino acid sequences.
Supplemental Figure S6. The Rpg1b CC and NB-ARC domains group with different paralogs from the Rpg1b region in phylogenetic analyses.
Supplemental Figure S7. Comparison of the Arabidopsis RPM1 and soybean Rpg1r and Rpg1b amino acid sequences.
Supplemental Figure S8. Alignment of the Arabidopsis RIN4 amino acid sequence with the four soybean homologs (GmRIN4a, GmRIN4b, GmRIN4c, and GmRIN4d).
Supplemental Figure S9. Identifying the location of the breakpoints that flank the recombination event in Rpg1b that encompasses the NB-ARC domain.
Supplemental Figure S10. Comparison of the Rpg1b CC and LRR domain-modeled structures with the templates on which the models are based.
Supplemental Figure S11. Secondary structure-guided alignment of the Rpg1b and TLR3 LRR domains using the SBAL aligner and manual editing.
Supplemental Table S1. dN, dS, and Ω (dN/dS) values calculated from an alignment between Rpg1b and Rpg1r.
Supplemental Table S2. Primer sequences used in this study.
Supplemental Table S3. Details of markers used for genetic mapping.
Supplemental Table S4. Details of amplification of CNLs from soybean accession PI 96983 (Rpg1r) using primers that flank CNLs from cv Williams82 (rpg1r).
Supplemental Movie S1. Model of the proposed Rpg1b CC domain dimer showing the location of polymorphic residues with respect to Rpg1r with the structure represented as a ribbon.
Supplemental Movie S2. Model of the proposed Rpg1b CC domain dimer showing the location of polymorphic residues with respect to Rpg1r with the structure surface shown.
Supplemental Movie S3. Model of the Rpg1b LRR domain showing the location of polymorphic residues with respect to Rpg1r with the structure represented as a ribbon.
Supplemental Movie S4. Model of the Rpg1b LRR domain showing the location of polymorphic residues with respect to Rpg1r with the structure surface shown.
Acknowledgments
We thank Michelle Metcalf and Brian Rutter for technical assistance, Blake Meyers for an alignment of the Arabidopsis CNL genes, and the U.S. Department of Agriculture Soybean Germplasm Collection for soybean seed.
Glossary
- CC
coiled-coil
- NB
nucleotide-binding
- LRR
leucine-rich repeat
- RIL
recombinant inbred line
- BAC
bacterial artificial chromosome
- WGS
whole genome sequence
- CNL
coiled-coil-nucleotide-binding-leucine-rich repeat
- HR
hypersensitive response
- T-DNA
transfer DNA
- RDP
Recombination Detection Program
- OD600
optical density at 600 nm
- PsgR4
Pseudomonas syringae pv glycinea race4
- UCSF
University of California, San Francisco
Footnotes
This work was supported by the National Science Foundation (Plant Genome Research Program grant no. DBI–0321664 to R.W.I.), the National Institute of General Medical Sciences at the National Institutes of Health (grant nos. R01GM046451 and R01GM063761 to R.W.I.), and the Faculty Research Support Program of Indiana University.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription
References
- Ade J, DeYoung BJ, Golstein C, Innes RW. (2007) Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci USA 104: 2531–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Aoyama T, Chua NH. (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Ashfield T, Bocian A, Held D, Henk AD, Marek LF, Danesh D, Peñuela S, Meksem K, Lightfoot DA, Young ND, et al. (2003) Genetic and physical localization of the soybean Rpg1-b disease resistance gene reveals a complex locus containing several tightly linked families of NBS-LRR genes. Mol Plant Microbe Interact 16: 817–826 [DOI] [PubMed] [Google Scholar]
- Ashfield T, Danzer JR, Held D, Clayton K, Keim P, Saghai Maroof MA, Webb PM, Innes RW. (1998) Rpg1, a soybean gene effective against races of bacterial blight, maps to a cluster of previously identified disease resistance genes. Theor Appl Genet 96: 1013–1021 [Google Scholar]
- Ashfield T, Egan AN, Pfeil BE, Chen NW, Podicheti R, Ratnaparkhe MB, Ameline-Torregrosa C, Denny R, Cannon S, Doyle JJ, et al. (2012) Evolution of a complex disease resistance gene cluster in diploid Phaseolus and tetraploid Glycine. Plant Physiol 159: 336–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield T, Keen NT, Buzzell RI, Innes RW. (1995) Soybean resistance genes specific for different Pseudomonas syringae avirulence genes are allelic, or closely linked, at the RPG1 locus. Genetics 141: 1597–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfield T, Ong LE, Nobuta K, Schneider CM, Innes RW. (2004) Convergent evolution of disease resistance gene specificity in two flowering plant families. Plant Cell 16: 309–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Chisholm ST, Dahlbeck D, Staskawicz BJ. (2003) Genetic and molecular evidence that the Pseudomonas syringae type III effector protein AvrRpt2 is a cysteine protease. Mol Microbiol 49: 1537–1546 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. (2003) Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Baltrus DA, Nishimura MT, Dougherty KM, Biswas S, Mukhtar MS, Vicente J, Holub EB, Dangl JL. (2012) The molecular basis of host specialization in bean pathovars of Pseudomonas syringae. Mol Plant Microbe Interact 25: 877–888 [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. (2000) The Protein Data Bank. Nucleic Acids Res 28: 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove SR, Simonich MT, Smith NM, Sattler A, Innes RW. (1994) A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell 6: 927–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonardi V, Cherkis K, Nishimura MT, Dangl JL. (2012) A new eye on NLR proteins: focused on clarity or diffused by complexity? Curr Opin Immunol 24: 41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella MA, Parker JE, Frost LN, Bittner-Eddy PD, Beynon JL, Daniels MJ, Holub EB, Jones JD. (1998) Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10: 1847–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno M, Do CB, Cooper GM, Kim MF, Davydov E, Green ED, Sidow A, Batzoglou S. (2003) LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res 13: 721–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. (2005) Protein structure prediction servers at University College London. Nucleic Acids Res 33: W36–W38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo AL, Schaal BA, Kunkel BN. (1999) Diversity and molecular evolution of the RPS2 resistance gene in Arabidopsis thaliana. Proc Natl Acad Sci USA 96: 302–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Zhu H, Baumgarten AM, Spangler R, May G, Cook DR, Young ND. (2002) Diversity, distribution, and ancient taxonomic relationships within the TIR and non-TIR NBS-LRR resistance gene subfamilies. J Mol Evol 54: 548–562 [DOI] [PubMed] [Google Scholar]
- Catanzariti AM, Dodds PN, Ve T, Kobe B, Ellis JG, Staskawicz BJ. (2010) The AvrM effector from flax rust has a structured C-terminal domain and interacts directly with the M resistance protein. Mol Plant Microbe Interact 23: 49–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen NW, Sévignac M, Thareau V, Magdelenat G, David P, Ashfield T, Innes RW, Geffroy V. (2010) Specific resistances against Pseudomonas syringae effectors AvrB and AvrRpm1 have evolved differently in common bean (Phaseolus vulgaris), soybean (Glycine max), and Arabidopsis thaliana. New Phytol 187: 941–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Dahlbeck D, Krishnamurthy N, Day B, Sjolander K, Staskawicz BJ. (2005) Molecular characterization of proteolytic cleavage sites of the Pseudomonas syringae effector AvrRpt2. Proc Natl Acad Sci USA 102: 2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe J, Kelker MS, Wilson IA. (2005) Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science 309: 581–585 [DOI] [PubMed] [Google Scholar]
- Chung EH, da Cunha L, Wu AJ, Gao Z, Cherkis K, Afzal AJ, Mackey D, Dangl JL. (2011) Specific threonine phosphorylation of a host target by two unrelated type III effectors activates a host innate immune receptor in plants. Cell Host Microbe 9: 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA 100: 8024–8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CI, Ayliffe MA, Kobe B, Ellis JG. (2006) Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci USA 103: 8888–8893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Ellis JG, Lawrence GJ, Finnegan EJ, Anderson PA. (1995) Contrasting complexity of two rust resistance loci in flax. Proc Natl Acad Sci USA 92: 4185–4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JG, Lawrence GJ, Luck JE, Dodds PN. (1999) Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11: 495–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. (2007) Comparative protein structure modeling using Modeller. Curr Protoc Protein Sci http://dx.doi.org/10.1002/0471140864.ps0209s50 [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. (2004) VISTA: computational tools for comparative genomics. Nucleic Acids Res 32: W273–W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geourjon C, Orelle C, Steinfels E, Blanchet C, Deléage G, Di Pietro A, Jault JM. (2001) A common mechanism for ATP hydrolysis in ABC transporter and helicase superfamilies. Trends Biochem Sci 26: 539–544 [DOI] [PubMed] [Google Scholar]
- Grant MR, Godiard L, Straube E, Ashfield T, Lewald J, Sattler A, Innes RW, Dangl JL. (1995) Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269: 843–846 [DOI] [PubMed] [Google Scholar]
- Hayes AJ, Jeong SC, Gore MA, Yu YG, Buss GR, Tolin SA, Maroof MA. (2004) Recombination within a nucleotide-binding-site/leucine-rich-repeat gene cluster produces new variants conditioning resistance to soybean mosaic virus in soybeans. Genetics 166: 493–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Innes RW, Ameline-Torregrosa C, Ashfield T, Cannon E, Cannon SB, Chacko B, Chen NW, Couloux A, Dalwani A, Denny R, et al. (2008) Differential accumulation of retroelements and diversification of NB-LRR disease resistance genes in duplicated regions following polyploidy in the ancestor of soybean. Plant Physiol 148: 1740–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19: 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DA, Jones JDG. (1997) The role of leucine-rich repeat proteins in plant defences. Adv Bot Res 24: 89–167 [Google Scholar]
- Jones DT. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292: 195–202 [DOI] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4: 363–371 [DOI] [PubMed] [Google Scholar]
- Kim HS, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL. (2005) The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc Natl Acad Sci USA 102: 6496–6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. (1995) A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature 374: 183–186 [DOI] [PubMed] [Google Scholar]
- Krasileva KV, Dahlbeck D, Staskawicz BJ. (2010) Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell 22: 2444–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF. (2005) Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst Biol 54: 575–594 [DOI] [PubMed] [Google Scholar]
- Leipe DD, Koonin EV, Aravind L. (2004) STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J Mol Biol 343: 1–28 [DOI] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Lin ZJ, Coaker G. (2011) A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host Microbe 9: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck JE, Lawrence GJ, Dodds PN, Shepherd KW, Ellis JG. (2000) Regions outside of the leucine-rich repeats of flax rust resistance proteins play a role in specificity determination. Plant Cell 12: 1367–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. (1991) Predicting coiled coils from protein sequences. Science 252: 1162–1164 [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389 [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF, III, Wiig A, Dangl JL. (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Cheng W, Spiridon LN, Töller A, Lukasik E, Saijo Y, Liu P, Shen QH, Micluta MA, Somssich IE, et al. (2011) Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9: 187–199 [DOI] [PubMed] [Google Scholar]
- Maekawa T, Kracher B, Vernaldi S, Ver Loren van Themaat E, Schulze-Lefert P. (2012) Conservation of NLR-triggered immunity across plant lineages. Proc Natl Acad Sci USA 109: 20119–20123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D, Rybicki E. (2000) RDP: detection of recombination amongst aligned sequences. Bioinformatics 16: 562–563 [DOI] [PubMed] [Google Scholar]
- Martin DP, Lemey P, Lott M, Moulton V, Posada D, Lefeuvre P. (2010) RDP3: a flexible and fast computer program for analyzing recombination. Bioinformatics 26: 2462–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DP, Posada D, Crandall KA, Williamson C. (2005) A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retroviruses 21: 98–102 [DOI] [PubMed] [Google Scholar]
- McHale L, Tan X, Koehl P, Michelmore RW. (2006) Plant NBS-LRR proteins: adaptable guards. Genome Biol 7: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND. (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20: 317–332 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muneyuki E, Noji H, Amano T, Masaike T, Yoshida M. (2000) F0F1-ATP synthase: general structural features of ‘ATP-engine’ and a problem on free energy transduction. Biochim Biophys Acta 1458: 467–481 [DOI] [PubMed] [Google Scholar]
- Padidam M, Sawyer S, Fauquet CM. (1999) Possible emergence of new geminiviruses by frequent recombination. Virology 265: 218–225 [DOI] [PubMed] [Google Scholar]
- Page RD. (1996) TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12: 357–358 [DOI] [PubMed] [Google Scholar]
- Peñuela S, Danesh D, Young ND. (2002) Targeted isolation, sequence analysis, and physical mapping of non-TIR NBS-LRR genes in soybean. Theor Appl Genet 104: 261–272 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. (2004) UCSF Chimera: a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Pfeil BE, Schlueter JA, Shoemaker RC, Doyle JJ. (2005) Placing paleopolyploidy in relation to taxon divergence: a phylogenetic analysis in legumes using 39 gene families. Syst Biol 54: 441–454 [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. (2001) Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci USA 98: 13757–13762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi D, DeYoung BJ, Innes RW. (2012) Structure-function analysis of the coiled-coil and leucine-rich repeat domains of the RPS5 disease resistance protein. Plant Physiol 158: 1819–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan GJ, Collier SM, Sacco MA, Baldwin TT, Boettrich T, Moffett P. (2008) The coiled-coil and nucleotide binding domains of the Potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 20: 739–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairdan GJ, Moffett P. (2006) Distinct domains in the ARC region of the potato resistance protein Rx mediate LRR binding and inhibition of activation. Plant Cell 18: 2082–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Sekine KT, Tomita R, Takeuchi S, Atsumi G, Saitoh H, Mizumoto H, Kiba A, Yamaoka N, Nishiguchi M, Hikichi Y, et al. (2012) Functional differentiation in the leucine-rich repeat domains of closely related plant virus-resistance proteins that recognize common avr proteins. Mol Plant Microbe Interact 25: 1219–1229 [DOI] [PubMed] [Google Scholar]
- Selote D, Kachroo A. (2010) RPG1-B-derived resistance to AvrB-expressing Pseudomonas syringae requires RIN4-like proteins in soybean. Plant Physiol 153: 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selote D, Robin GP, Kachroo A. (2013) GmRIN4 protein family members function nonredundantly in soybean race-specific resistance against Pseudomonas syringae. New Phytol 197: 1225–1235 [DOI] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233 [DOI] [PubMed] [Google Scholar]
- Shen QH, Saijo Y, Mauch S, Biskup C, Bieri S, Keller B, Seki H, Ulker B, Somssich IE, Schulze-Lefert P. (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315: 1098–1103 [DOI] [PubMed] [Google Scholar]
- Shen QH, Zhou F, Bieri S, Haizel T, Shirasu K, Schulze-Lefert P. (2003) Recognition specificity and RAR1/SGT1 dependence in barley Mla disease resistance genes to the powdery mildew fungus. Plant Cell 15: 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM. (1992) Analyzing the mosaic structure of genes. J Mol Evol 34: 126–129 [DOI] [PubMed] [Google Scholar]
- Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. (1999) Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature 400: 667–671 [DOI] [PubMed] [Google Scholar]
- Staskawicz BJ, Dahlbeck D, Keen NT. (1984) Cloned avirulence gene of Pseudomonas syringae pv. glycinea determines race-specific incompatibility on Glycine max (L.) Merr. Proc Natl Acad Sci USA 81: 6024–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbrenner AD, Goritschnig S, Krasileva KV, Schreiber KJ, Staskawicz BJ. (2012) Effector recognition and activation of the Arabidopsis thaliana NLR innate immune receptors. Cold Spring Harb Symp Quant Biol 77: 249–257 [DOI] [PubMed] [Google Scholar]
- Takken FL, Albrecht M, Tameling WI. (2006) Resistance proteins: molecular switches of plant defence. Curr Opin Plant Biol 9: 383–390 [DOI] [PubMed] [Google Scholar]
- Takken FL, Tameling WI. (2009) To nibble at plant resistance proteins. Science 324: 744–746 [DOI] [PubMed] [Google Scholar]
- Tameling WI, Elzinga SD, Darmin PS, Vossen JH, Takken FL, Haring MA, Cornelissen BJ. (2002) The tomato R gene products I-2 and MI-1 are functional ATP binding proteins with ATPase activity. Plant Cell 14: 2929–2939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameling WI, Vossen JH, Albrecht M, Lengauer T, Berden JA, Haring MA, Cornelissen BJ, Takken FL. (2006) Mutations in the NB-ARC domain of I-2 that impair ATP hydrolysis cause autoactivation. Plant Physiol 140: 1233–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Araki H, Stahl E, Bergelson J, Kreitman M. (2002) Signature of balancing selection in Arabidopsis. Proc Natl Acad Sci USA 99: 11525–11530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Yamaguchi Y, Sano H. (2006) Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein N factor during the hypersensitive response in tobacco plants. Plant Mol Biol 61: 31–45 [DOI] [PubMed] [Google Scholar]
- van der Biezen EA, Jones JDG. (1998) The NB-ARC domain: a novel signaling motif shared by plant resistance gene products and regulators of cell death in animals. Curr Biol 8: R226–R227 [DOI] [PubMed] [Google Scholar]
- van Ooijen G, Mayr G, Kasiem MM, Albrecht M, Cornelissen BJ, Takken FL. (2008) Structure-function analysis of the NB-ARC domain of plant disease resistance proteins. J Exp Bot 59: 1383–1397 [DOI] [PubMed] [Google Scholar]
- Wang CK, Broder U, Weeratunga SK, Gasser RB, Loukas A, Hofmann A. (2012) SBAL: a practical tool to generate and edit structure-based amino acid sequence alignments. Bioinformatics 28: 1026–1027 [DOI] [PubMed] [Google Scholar]
- Wen RH, Khatabi B, Ashfield T, Saghai Maroof MA, Hajimorad MR. (2013) The HC-Pro and P3 cistrons of an avirulent soybean mosaic virus are recognized by different resistance genes at the complex Rsv1 locus. Mol Plant Microbe Interact 26: 203–215 [DOI] [PubMed] [Google Scholar]
- Whalen MC, Innes RW, Bent AF, Staskawicz BJ. (1991) Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3: 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski T, Caldwell KS, Piskurewicz U, Cavanaugh KA, Xu H, Kozik A, Ochoa O, McHale LK, Lahre K, Jelenska J, et al. (2009) Comparative large-scale analysis of interactions between several crop species and the effector repertoires from multiple pathovars of Pseudomonas and Ralstonia. Plant Physiol 150: 1733–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski T, Tomczak A, Michelmore R. (2005) Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol J 3: 259–273 [DOI] [PubMed] [Google Scholar]
- Yang Z. (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591 [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. (2000) Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol 17: 32–43 [DOI] [PubMed] [Google Scholar]
- Zhang X, Feng Y, Cheng H, Tian D, Yang S, Chen JQ. (2011) Relative evolutionary rates of NBS-encoding genes revealed by soybean segmental duplication. Mol Genet Genomics 285: 79–90 [DOI] [PubMed] [Google Scholar]