A high-resolution spatiotemporal transcriptome atlas of maize seed uncovers the genetic control of embryo and endosperm development.
Abstract
Maize (Zea mays) is an excellent cereal model for research on seed development because of its relatively large size for both embryo and endosperm. Despite the importance of seed in agriculture, the genome-wide transcriptome pattern throughout seed development has not been well characterized. Using high-throughput RNA sequencing, we developed a spatiotemporal transcriptome atlas of B73 maize seed development based on 53 samples from fertilization to maturity for embryo, endosperm, and whole seed tissues. A total of 26,105 genes were found to be involved in programming seed development, including 1,614 transcription factors. Global comparisons of gene expression highlighted the fundamental transcriptomic reprogramming and the phases of development. Coexpression analysis provided further insight into the dynamic reprogramming of the transcriptome by revealing functional transitions during maturation. Combined with the published nonseed high-throughput RNA sequencing data, we identified 91 transcription factors and 1,167 other seed-specific genes, which should help elucidate key mechanisms and regulatory networks that underlie seed development. In addition, correlation of gene expression with the pattern of DNA methylation revealed that hypomethylation of the gene body region should be an important factor for the expressional activation of seed-specific genes, especially for extremely highly expressed genes such as zeins. This study provides a valuable resource for understanding the genetic control of seed development of monocotyledon plants.
Maize (Zea mays) is one of the most important crops and provides resources for food, feed, and biofuel (Godfray et al., 2010). It has also been used as a model system to study diverse biological phenomena, such as transposons, heterosis, imprinting, and genetic diversity (Bennetzen and Hake, 2009). The seed is a key organ of maize that consists of the embryo, endosperm, and seed coat. Maize seed development initiates from a double fertilization event in which two pollen sperm fuse with the egg and central cells of the female gametophyte to produce the progenitors of the embryo and endosperm, respectively (Dumas and Mogensen, 1993; Chaudhury et al., 2001). The mature embryo inherits the genetic information for the next plant generation (Scanlon and Takacs, 2009), whereas the endosperm, which is storage tissue for the embryo, persists throughout seed development and functions as the site of starch and protein synthesis (Sabelli and Larkins, 2009). Elucidation of the genetic regulatory mechanisms involved in maize seed development will facilitate the design of strategies to improve yield and quality, and provide insight that is applicable to other monocotyledon plants.
A key means to explore the mechanisms of seed development is to identify gene activities and functions. Genetic studies have uncovered a number of genes that play major roles in governing embryogenesis and accumulation of endosperm storage compounds, such as Viviparous1, KNOTTED1, Indeterminate gametophyte1, Shrunken1 (Sh1), Opaque2 (O2), and Defective kernel1 (Chourey and Nelson, 1976; McCarty et al., 1991; Smith et al., 1995; Vicente-Carbajosa et al., 1997; Lid et al., 2002; Evans, 2007). Furthermore, the activity of some genes has also been extensively studied. Typical examples are zein genes that encode primary storage proteins in endosperm. Woo et al. (2001) examined zein gene expression and showed that they were the most highly expressed genes in endosperm based on EST data, whereas their dynamic expression patterns were revealed in a later study (Feng et al., 2009). Nevertheless, information on the global gene expression network throughout seed development is still very limited.
The transcriptome is the overall set of transcripts, which varies based on cell or tissue type, developmental stage, and physiological condition. Analysis of transcriptome dynamics aids in implying the function of unannotated genes, identifying genes that act as critical network hubs, and interpreting the cellular processes associated with development. In Arabidopsis (Arabidopsis thaliana), the genes expressed in developing seed and its subregions at several development stages have been analyzed with Affymetrix GeneChips (Le et al., 2010; Belmonte et al., 2013). In maize, microarray-based atlases of global transcription have provided insight into the programs controlling development of different organ systems (Sekhon et al., 2011). Compared with microarray, high-throughput RNA sequencing (RNA-seq) is a powerful tool to comprehensively investigate the transcriptome at a much lower cost, but with higher sensitivity and accuracy (Wang et al., 2009b). Several studies have taken advantage of the RNA-seq strategy to interpret the dynamic reprogramming of the transcriptome during leaf, shoot apical meristem, and embryonic leaf development in maize (Li et al., 2010; Takacs et al., 2012; Liu et al., 2013).
To date, only two studies have focused on identifying important regulators and processes required for embryo, endosperm, and/or whole seed development in maize based on a genome-wide transcriptional profile produced by RNA-seq (Liu et al., 2008; Teoh et al., 2013). However, these studies were limited by the low number of samples used and they did not provide an extensive, global view of transcriptome dynamics over the majority of seed development stages. Here, we present a comprehensive transcriptome study of maize embryo, endosperm, and whole seed tissue from fertilization to maturity using RNA-seq, which serves as a valuable resource for analyzing gene function on a global scale and elucidating the developmental processes of maize seed.
RESULTS
Generation and Analysis of the RNA-seq Data Set
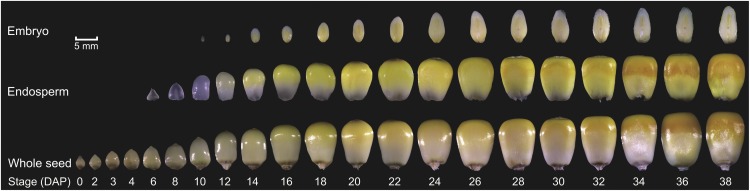
To systematically investigate the dynamics of the maize seed transcriptome over development, we generated RNA-seq libraries of B73 seed tissues from different developmental stages, including 15 embryo, 17 endosperm, and 21 whole seed samples (Fig. 1). Utilizing paired-end Illumina sequencing technology, we generated around 1.9 billion high-quality reads, 80.2% of which could be uniquely mapped to the B73 reference genome (Schnable et al., 2009; Supplemental Table S1). The genic distribution of reads was 66.6% exonic, 25.6% splice junction, and 2.6% intronic, leaving about 5% from unannotated genomic regions, demonstrating that most of the detected genes have been annotated. Uniquely mapped reads were used to estimate normalized transcription level as reads per kilobase per million (RPKM). To reduce the influence of transcription noise, genes from the B73 filtered gene set (FGS) were included for analysis only if their RPKM values were ≥ 1. Considering that our purpose was not to identify minor differential expression of genes between two time points of development, but to provide an atlas of gene expression profile across tissues using time series biological samples, we only randomly selected 12 samples to have a biological replicate (Supplemental Fig. S1) to assess our data quality. Comparisons of biological replicates showed that their expression values were highly correlated (average R2 = 0.96). For the samples with biological replicates, we took the average RPKM as the expression quantity. To further evaluate the quality of our expression data, we compared the transcript abundance patterns of a number of selected genes with previously measured expression profiles (Supplemental Fig. S2). For example, LEAFY COTYLEDON1 (LEC1), which functions in embryogenesis, was mainly expressed in the early stage of the embryo (Lotan et al., 1998). Globulin2 (Glb2) had high expression late in embryogenesis, in accordance with its function as an important storage protein in the embryo (Kriz, 1989). Similarly, O2, a transcription factor (TF) that regulates zein synthesis (Vicente-Carbajosa et al., 1997), and Fertilization independent endosperm1 (Fie1), a repressor of endosperm development in the absence of fertilization (Danilevskaya et al., 2003), were almost exclusively expressed in the endosperm. Expression patterns of these selected genes were identified exclusively in their known tissue of activity, indicating that the embryo and endosperm samples were processed well.
Figure 1.
Overview of the time series maize seed samples used for RNA-seq analysis. The photographs show the changes in maize embryo, endosperm, and whole seed during development. The 53 samples shown here were used to generate RNA-seq libraries. Bar = 5 mm.
In total, we detected 26,105 genes expressed in at least 1 of the 53 samples (Supplemental Data Set S1). The distribution of these genes is revealed by a Venn diagram (Fig. 2A), which shows that 20,360 genes were common among all three tissue types. The number of genes detected in endosperm tissue during the developmental stages was lower and much more variable compared with embryo or whole seed tissue, and a greater number of genes were expressed in the tissue during the early and late phases. Several thousand fewer expressed genes were detected 14 d after pollination (DAP) in the endosperm compared with 6 or 8 DAP (Fig. 2B; Supplemental Data Set S2). In addition, the median expression level in the embryo was roughly 2-fold greater than that of the endosperm from 10 to 30 DAP (Fig. 2C). Of the 1,506 genes unique to the whole seed samples, 451 were present in RNA-seq data of 14 and/or 25 DAP pericarp tissue (Morohashi et al., 2012). Considering that the maternal tissue is the vast majority of the content of the early seed (Márton et al., 2005; Pennington et al., 2008), we inferred that most of these genes might be expressed exclusively in maternal tissue such as the pericarp or nucellus. Moreover, 1,062 genes in the embryo and endosperm with low expression were not detected in whole seed samples (Fig. 2C).
Figure 2.
Analysis of global gene expression among different samples. A, Venn diagram of the 26,105 genes detected among embryo, endosperm, and whole seed. B, Number of genes expressed in each of the samples. C, Comparison of expression levels of genes detected in embryo and endosperm tissues.
Division of Development Phases by Global Gene Expression Patterns
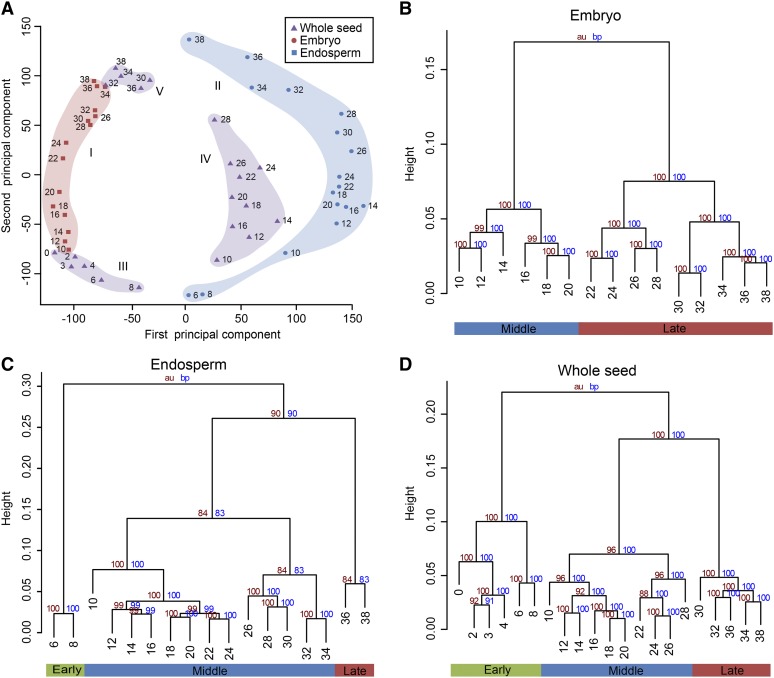
To gain insight into the relationships among the different transcriptomes, we performed principal component analysis (PCA) on the complete data set, which can graphically display the transcriptional signatures and developmental similarity. The first component (40.7% variance explained) separated samples based on tissue identity and clearly distinguished embryo from endosperm samples, with whole seed samples located in between (Fig. 3A). The second component (24.4% variance explained) discriminated early, middle, and late stages of development for all three tissues (Fig. 3A). The wider area occupied by endosperm samples than embryo demonstrates stronger transcriptome reprogramming in developing endosperm, which is mainly attributable to drastic changes in the early and late stages. Moreover, whole seed samples of 0 to 8 DAP and 30 to 38 DAP clustered closely to the embryo, but 10 to 28 DAP samples were close to the endosperm.
Figure 3.
Global transcriptome relationships among different stages and tissues. A, PCA of the RNA-seq data for the 53 seed samples shows five distinct groups: I for embryo (light red), II for endosperm (light blue), and III to V for early (III), middle (IV), and late (V) whole seed (light purple). B to D, Cluster dendrogram showing global transcriptome relationships among time series samples of embryo (B), endosperm (C), and whole seed (D). The y axis measures the degree of variance (see the “Materials and Methods”). The bottom row indicates the developmental phases according to the cluster dendrogram of the time series data. au, Approximately unbiased.
Cluster analysis of the time series data for the tissues grouped samples well along the axis of developmental time (Fig. 3, B–D). Embryo samples from 10 to 20 DAP and 22 to 38 DAP were the primary clusters, which correspond to morphogenesis and maturation phases of development (Fig. 3B). This is consistent with the embryo undergoing active DNA synthesis, cell division, and differentiation, and then switching to synthesis of storage reserve and desiccation (Vernoud et al., 2005). Expression differences in endosperm samples resulted in three primary clusters, which correspond to early, middle, and late phases of development (Fig. 3C). The earliest time point (6 and 8 DAP) is an active period of cell division and cell elongation that terminates at about 20 to 25 DAP (Duvick, 1961). The samples of 10 to 24 DAP formed one subgroup and 26 to 34 DAP formed another subgroup, suggesting that they mark the period forming the main cell types and maturation of endosperm, respectively (Fig. 3C). The two subgroups form a larger cluster for active accumulation of storage compounds during 10 to 34 DAP. The distinct cluster of 36 and 38 DAP is in accordance with the end of storage compound accumulation in the endosperm and the activation of biological processes involved in dormancy and dehydration. In whole seed tissue, a primary cluster was formed from the earliest time points with 0 to 4 DAP and 6 to 8 DAP samples as subgroups, separating the nucellus degradation as well as endosperm syncytial and cellularization phases from the rapid expansion of the endosperm and development of embryonic tissues (Fig. 3D). After 10 DAP, the embryo and endosperm dominate the formation of seed, as shown in Figure 1 and morphological observation (Pennington et al., 2008). As effected by both embryo and endosperm, 10 to 28 DAP whole seed samples clustered together and 30 to 38 DAP formed another group (Fig. 3D). These results confirm that the expression data successfully captured the characteristic seed development phases and should therefore contain valuable insights about corresponding changes in the transcriptome.
Integration of Gene Activity and Cellular Function across Development Phases
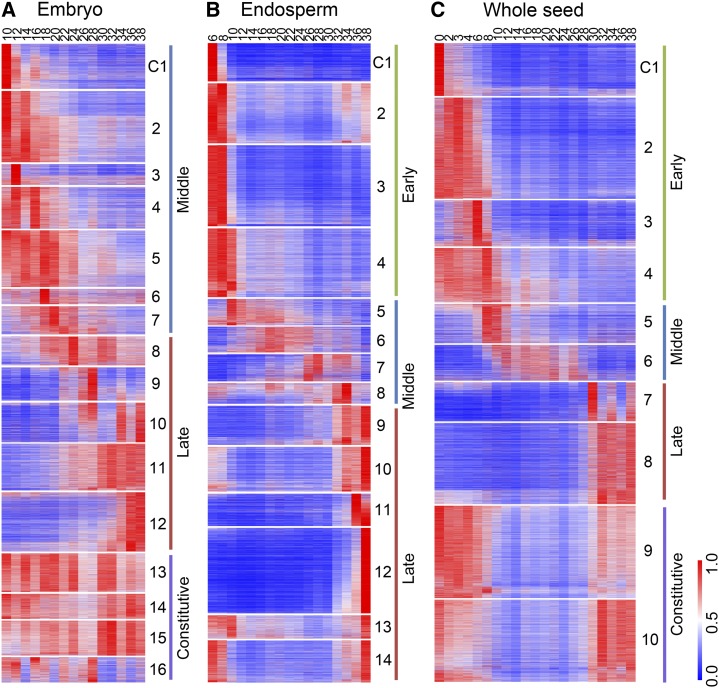
The PCA and hierarchical clustering analysis graphically display the relationship among different samples, but do not indicate the detailed cellular function. Using the k-means clustering algorithm, we classified the detected genes into 16, 14, and 10 coexpression modules for embryo, endosperm, and whole seed, respectively, each of which contains genes that harbor similar expression patterns (Fig. 4). We then used MapMan annotation to assign genes to functional categories (Supplemental Fig. S3). Thus, we can aggregate genes over continuous time points and obtain a view of functional transitions along seed development.
Figure 4.
Coexpression modules. A to C, Expression patterns of coexpression modules of embryo (A), endosperm (B), and whole seed (C), ordered according to the sample time points of their peak expression. For each gene, the RPKM value normalized by the maximum value of all RPKM values of the gene over all time points is shown.
According to the cluster analysis results, most modules of the embryo can be divided into middle (10–20 DAP) and late (22–38 DAP) stages (Fig. 4A). The middle stage, best represented by modules C1 to C7, is typified by the overrepresentation of glycolysis, tricarboxylic acid cycle, mitochondrial electron transport, redox, RNA regulation, DNA and protein synthesis, cell organization, and division-related genes. This is consistent with the high requirement of energy during embryo formation. The late stage represented by C8 to C12 exhibited up-regulation of the cell wall, hormone metabolism (ethylene and jasmonate), stress, storage proteins, and transport-related genes, which coincides with the maturation of the embryo. The modules C13 to C16 included genes that were broadly expressed across the time points sampled and were related to hormone metabolism (brassinosteroid), cold stress, RNA processing and regulation, amino acid activation, and protein targeting.
All of the 14 coexpression modules of endosperm can be roughly divided into early (6–8 DAP), middle (10–34 DAP), and late (36–38 DAP) stages (Fig. 4B). The early stage (represented by modules C1 to C4) is exemplified by high expression of hormone metabolism (gibberellin), cell wall, cell organization and cycle, amino acid metabolism, DNA, and protein synthesis–related genes, which is consistent with differentiation, mitosis, and endoreduplication. Genes in the tricarboxylic acid cycle and mitochondrial electron transport are also overrepresented and related to energy demands at that time. The middle stage (best represented by C5 to C8, in which different modules have distinct profiles) is the active storage accumulation phase and exhibits high expression of carbohydrate metabolism genes, as expected. Increased expression of protein degradation-related genes around 26 to 34 DAP in C7 and C8 coincides with the process of endosperm maturation. Genes involved in protein degradation, secondary metabolism, oxidative pentose phosphate, receptor kinase signaling, and transport were up-regulated in the late stage in modules C9 to C14 during the concluding phase of endosperm maturation.
Ten DAP and later time points of whole seed samples reflect the additive combination of embryo and endosperm expression. Genes that are active early in development (0–8 DAP) in clusters C1 to C4 are expected to be related to maternal tissue, which is the bulk of the seed at that time. A group of genes are highly expressed in C1 at 0 DAP, but their expression drops rapidly by 2 DAP, suggesting that they have functional roles that precede pollination. This group includes photosynthesis light reaction members and some TFs involved in RNA regulation. Genes related to cell wall and protein degradation, signaling, nucleotide metabolism, DNA synthesis, cell organization, and mitochondrial electron transport are overrepresented in C2 to C4, which has increased expression after pollination, in accordance with the degradation of nucellus tissue and development of embryonic tissues. The expression patterns and functional categories of the 1,506 genes detected only in whole seed samples are shown in Supplemental Figure S4. Because these genes tend to be expressed at high levels mainly before 8 DAP, they are presumed to have functions in early seed development.
Together, these data show that the transition of major biochemical processes along the developmental time axis of the seed is produced partly by highly coordinated transcript dynamics.
TF Expression during Seed Development
Of the 2,297 identified maize TFs (Zhang et al., 2011), 1,614 (70%) are included in our analysis (Supplemental Data Set S3), which accounts for 6.18% of the total number of genes detected in seed tissue. The number of TFs detected in the different samples is shown in Supplemental Figure S5A. Their proportion to the total genes expressed in each tissue time point was always greater in embryo than endosperm samples (Supplemental Fig. S5B). Shannon entropy has been used to determine the specificity of gene expression, with lower values indicating a more time-specific profile (Makarevitch et al., 2013). The Shannon entropy of TFs was significantly lower than all other genes in both embryo (P = 2.3 × 10−10) and endosperm (P < 1.5 × 10−3), indicating that TFs tended to be expressed more time specifically than other genes (Supplemental Fig. S5, C and D).
The number of TFs from each family used in the seed development program, along with the proportion of members present in the coexpression modules relative to the total members of the family expressed in the tissue, is shown in Supplemental Figure S6. Enrichment of these TF families in the coexpression modules based on observed numbers was evaluated with Fisher’s exact test. Significant TF family enrichment was identified for specific coexpression modules. For example, 12 auxin-response factor TFs (38.7%) were expressed in embryo module C2 during morphogenesis and one-half of the detected members of the WRKY family were active in endosperm module C12 late in endosperm development. The WRKY family has been reported to be mainly involved in the physiological programs of pathogen defense and senescence (Eulgem et al., 2000; Pandey and Somssich, 2009). Twenty-one MIKC family TFs (56.8%) were present in whole seed module C2, implying an important role in regulating genes involved in response to fertilization. The developmental specificity of the detected TFs makes them excellent candidates for reverse genetics approaches to investigate their role in grain production.
Tissue-Specific Genes of Seed
Identification of uncharacterized tissue-specific genes can help to explain their function and understand the underlying control of tissue or organ identity. To generate a comprehensive catalog of seed-specific genes, results from this study were compared with 25 published nonseed RNA-seq data sets (Jia et al., 2009; Wang et al., 2009a; Li et al., 2010; Davidson et al., 2011; Bolduc et al., 2012), including root, shoot, shoot apical meristem, leaf, cob, tassel, and immature ear (Supplemental Table S2). In total, we identified 1,258 seed-specific genes, including 91 TFs from a variety of families (Supplemental Data Set S4). To gain further insight into the spatial expression trend in the developing seed, we divided these genes into four groups: embryo specific, endosperm specific, expressed in both embryo and endosperm, and other as only expressed in whole seed (Table I). The dynamic expression patterns of these genes reflect their roles in corresponding development stages (Supplemental Fig. S7). The largest numbers of seed-specific genes were observed in the endosperm, consistent with a study in maize using microarrays (Sekhon et al., 2011), perhaps reflecting the specific function of endosperm.
Table I. Total number of detected seed-specific genes and TFs.
Data are presented as n.
| Tissue Type | Specific Genes | Specific TFs |
|---|---|---|
| Embryo | 249 | 23 |
| Endosperm | 742 | 59 |
| Embryo and endosperm | 219 | 6 |
| Othera | 48 | 3 |
| Total | 1,258 | 91 |
These genes only detected expression in whole seeds.
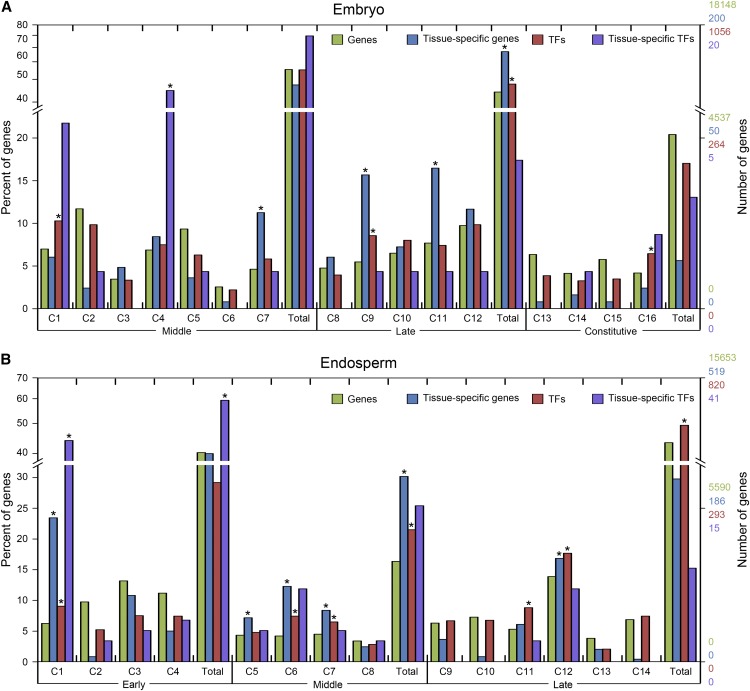
We compared the distribution of tissue-specific genes and TFs in embryo and endosperm coexpression modules to identify important phases in the underlying transcription network. Coexpression modules with an enrichment of tissue-specific genes or TFs may provide insight about uncharacterized genes and preparation for subsequent developmental processes. A feature of gene activity is shown in Figure 5. Fisher’s exact test (P < 0.05) was used to determine modules with significant enrichment of tissue-specific genes and TFs in the embryo and endosperm. In the embryo, TFs and tissue-specific genes were significantly enriched in the late phase, suggesting a specific process during maturation. In the endosperm, we observed that TFs and tissue-specific genes were overrepresented in the middle phase, which conforms to the role of endosperm in storage compound accumulation and to specific progress at this phase.
Figure 5.
Distribution and enrichment of genes, tissue-specific genes, TFs, and tissue-specific TFs in coexpression modules of embryo and endosperm. A and B, Bars indicate the percentage of all detected genes (green), tissue-specific genes (blue), TFs (red), and tissue-specific TFs (purple) observed in a coexpression module (C) or in the development phase (Total) relative to the total number of each group detected across samples for embryo (A) and endosperm (B). The number of genes represented by the percentage is shown on the right y axis. Enrichment for tissue-specific genes and TFs was evaluated with Fisher’s exact test based on the number of genes observed in each coexpression module, whereas enrichment for tissue-specific TFs was evaluated based on the number of TFs observed in each coexpression module. Asterisks represent significant enrichment at a false discovery rate ≤ 0.05.
To gain further insight into the functional significance of tissue-specific genes, overrepresented gene ontology (GO) terms were examined using the WEGO online tool (Ye et al., 2006; Supplemental Fig. S8). All overrepresented GO terms were observed for the middle embryo development phase, including the biosynthetic process, cellular metabolic process, macromolecule, and nitrogen compound metabolic process. Similarly, overrepresented GO terms for the endosperm were mostly observed in the early phase, including the macromolecule, nitrogen compound metabolic process, DNA binding, and transcription regulator. Genes involved in the nutrient reservoir class were enriched in the middle phase. The oxidoreductase class was overrepresented in late phase, and is known to be involved in maturation (Zhu and Scandalios, 1994).
The Expression of Zein Genes in Endosperm
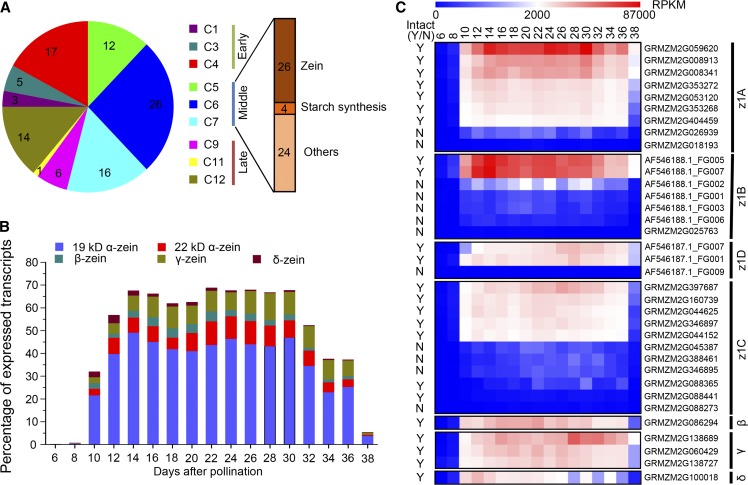
Zeins are the most important storage proteins in maize endosperm and are an important factor in seed quality. According to Xu and Messing (2008), there are 41 α, 1 β, 3 γ, and 2 δ zein genes. In order to explore their expression pattern, we first confirmed the gene models by mapping publicly available full-length complementary DNAs of zein subfamily genes to B73 bacterial artificial chromosomes, and then mapped these back to the reference genome. Because some of these zein genes were not assembled in the current B73 reference genome or were only annotated in the working gene set, we confirmed a final set of 35 zein genes in the FGS of the B73 annotation, including 30 α, 1 β, 3 γ, and 1 δ zein genes (Supplemental Table S3). About three-quarters (26) of these were in the list of the 100 most highly expressed genes in the endosperm, based on mean expression across all endosperm samples (Supplemental Table S4). The distribution of these most highly expressed genes clearly showed that the 26 zeins and 4 starch synthesis genes were actively expressed in the middle phase of endosperm development, characteristic of storage compound accumulation (Fig. 6A).
Figure 6.
Analysis of highly expressed genes in the endosperm. A, The distribution of the 100 most highly expressed genes in the endosperm ordered by mean expression in different modules. B, The dynamic transcript levels of different zein gene family members in the endosperm as reflected by their percentage among all detected gene transcript levels. C, Heat map showing RPKM values of 35 zein genes in the different development stages of the endosperm. +, Having intact coding regions; −, with premature_stop; N, no; Y, yes.
Previous research has shown that the zein genes constitute approximately 40% to 50% of the total transcripts in the endosperm (Marks et al., 1985; Woo et al., 2001), but these results are based on EST data from a single tissue or pooled tissues and only a few zein genes were assessed. Thus, we reevaluated the transcriptomic contribution of zein genes across endosperm development using our RNA-seq data, which is able to overcome the high structural similarity among them, especially in the α family (Xu and Messing, 2008). Zein genes stably accounted for about 65% of transcripts from 10 to 34 DAP, with 19-kD α zeins (approximately 42%), 22-kD α zeins (approximately 8%), and γ zeins (approximately 10%) representing the most abundant transcripts (Fig. 6B). The expression of different members within a given family was highly variable (Fig. 6C). Three genes contributed over 76% expression of the entire α zeins A (z1A) family, and five genes accounted for about 98% expression of the z1C family. Expression of z1B and z1D families was almost entirely derived from two genes (Supplemental Table S3). All of the highly expressed genes appear to have intact coding regions as reported by Feng et al. (2009). However, we also detected expression of genes with a premature stop codon, and the expression level of these truncated zein genes is actually much higher than other nonzein genes (Supplemental Fig. S9).
It has been reported in rice (Oryza sativa) that CpG and CHG hypomethylation is a major mechanism for the activation of genes in the endosperm (Zemach et al., 2010). To obtain a global view of the relationship between DNA methylation in gene body regions and the expression of genes in maize endosperm, we compared the RNA-seq data with recently published high-throughput bisulfite sequencing data on the DNA methylation levels of B73 shoot, 12 DAP embryo, and endosperm in CpG and CHG contexts (Zhang et al., 2014). We found that for endosperm-specific genes, there is a significant enrichment of differentially methylated regions (DMRs). These DMRs are hypomethylated in the endosperm compared with shoot and embryo tissue, regardless of CpG or CHG context (Supplemental Table S5). Of the zein genes, which were all expressed specifically in the endosperm, 31 harbored a hypomethylation state in the CHG context, 22 harbored a hypomethylation state in the CpG context, and 21 harbored a hypomethylation state in both CpG and CHG contexts (Supplemental Table S3). Three zein genes without associated DMRs seem to be an exception. Nevertheless, among these three, the 16-kD γ zein (GRMZM2G060429) and 15-kD β zein (GRMZM2G086294) were still hypomethylated, whereas a z1B zein (GRMZM2G025763) was hypermethylated in all three tissues studied. Thus, we predict that gene body hypomethylation should have an important effect on activating the expression of genes in the endosperm, including zein genes.
DISCUSSION
We constructed a high-resolution transcriptome atlas of maize seed development, using time series samples of embryo, endosperm, and whole seed from fertilization to maturity. Our results show that at least 26,105 genes, including 1,614 TFs, are required to program maize seed development, which involves about 65.7% of the B73 FGS genes. Global comparisons of gene expression highlighted the fundamental transcriptomic reprogramming and well-demonstrated development phases present in embryo, endosperm, and whole seed. Coexpression analysis illustrates the dynamic reprogramming of the transcriptome during maturation and reveals the functional transitions that take place during seed development. Combined with published nonseed RNA-seq data, we identified 1,258 seed development specific genes, including 91 TFs, which should facilitate the identification of functional processes and regulatory networks that are important for programming seed development. Although very early embryo and endosperm tissues were not available because of the difficulty in manual sample collection, our data serve as a valuable resource for the in-depth understanding of the dynamics of gene expression throughout maize seed development.
Expression Patterns Imply Gene Function and Inform Cellular Processes
The development of seed is highly complex and requires a vast interaction of genes and cooperation of processes. Confirming the phases of different processes has an important significance in expounding seed development and guiding the selection of tissue development stages for different purposes. Many seed development studies were previously performed from the perspective of morphology, storage accumulation, and gene expression (Kiesselbach, 1949; Woo et al., 2001; Lee et al., 2002; Liu et al., 2008), but the deep profiling in this study fills in many missing details.
Although embryo and endosperm samples were well distinguished, common variance in expression was observed over the stages of development by PCA (Fig. 3A) and expression of genes present in both embryo and endosperm tended to be positively correlated (Supplemental Fig. S10), indicating that large sets of genes change coordinately in the embryo and endosperm over development. It is reasonable to assume that there are some regulators shared between the embryo and endosperm, so that the two relatively independent seed tissues can proceed and complete the development process at the same time. In Arabidopsis, a global comparison of mRNA populations also suggested that substantial coordination of biological processes occurs in embryo and endosperm regions (Belmonte et al., 2013). However, the regulators responsible for coordinating the embryo and endosperm development remain to be determined.
Starch constitutes most of the biomass of the mature modern maize hybrid and is the main yield component being improved by breeding. Starch biosynthesis requires the coordinated activities of several enzymes, for which different isozymes have different effects (Jeon et al., 2010). The major form of Suc synthase and granule-bound starch synthase (SS) in the endosperm is encoded by Sh1 and Waxy1, respectively (Carlson and Chourey, 1996; Vrinten and Nakamura, 2000). These can be revealed by the expression of Sh1 and Waxy1 and they account for almost all of the transcripts of Suc synthase and granule-bound SS, respectively (Supplemental Fig. S11). Our results are also consistent with earlier studies that three of the eight soluble SS paralogs (Ss1, SSIIa, and Dull1), two of the four starch branching enzyme (SBE) paralogs (Sbe1 and SBEIIb), and two of the four starch debranching enzyme paralogs (Sugary1 and Pullulanase-type starch debranching enzyme1) play a major role in starch synthesis (Yun and Matheson, 1993; Beatty et al., 1999; Cao et al., 1999; Ball and Morell, 2003; Zhang et al., 2004; Fujita et al., 2006). Except for Pullulanase-type starch debranching enzyme1, all of these main genes are members of two similar endosperm coexpression modules (C5 and C6), as well as Brittle2 (Bt2) and Shrunken2, two key enzymes in starch synthesis (Ballicora et al., 2004). Genes functioning in the same pathway tend to appear in the same or similar expression modules, indicating that the coexpression data provide excellent inferences for uncharacterized genes through guilt by association. For example, the maize orthologs of Floury endosperm2 and Floury endosperm4 in rice belong to module C6 and C5 of the endosperm, respectively, which indicates that they may function in starch accumulation similar to rice (Kang et al., 2005; She et al., 2010).
Roles of Seed-Specific Genes
About 4.8% (1,258) of the total genes detected during seed development were identified as seed-specific genes, which is more than the number reported by Sekhon et al. (2011), owing to the RNA-seq technology and more extensive sampling in our study. In rice, Wang et al. (2010) predicted 589 seed-specific genes, 411 of which can be identified in the Nipponbare reference genome. According to the MaizeGDB, 155 of these genes have orthologs in maize, nearly one-half of which (68) were found to have seed-specific ortholog genes in maize. This result suggests high conservation between rice and maize in whether a gene is specifically expressed in the seed (P < 2.2 × 10−16), even though different methods of calculating gene expression and different criteria were used to determine the tissue specificity.
One critical question concerns the functions that these specific genes play in seed development. Previous genetic studies with seed defective mutants have demonstrated the functional significance of certain seed-specific genes. For example, Viviparous1 and LEC1 are important for embryogenesis and maturation (McCarty et al., 1991; Lotan et al., 1998). Other examples include O2, Shrunken2, Bt1, and Bt2, which are involved in storage compound accumulation (Russell et al., 1993; Vicente-Carbajosa et al., 1997; Shannon et al., 1998); Fie1 and Maternally expressed gene1, which are two endosperm imprinting genes (Gutiérrez-Marcos et al., 2004; Raissig et al., 2011); Glb1 and Glb2, which are two embryo storage protein genes (Kriz, 1989); and Floury2, which is a zein gene in the endosperm (Fontes et al., 1991). Such well-studied genes represent a small proportion of the 1,258 identified seed-specific genes, but this study provides information on when all of these genes are actively expressed (Supplemental Fig. S7). The timing of seed-specific gene expression in development combined with predictions of gene function based on their annotation (Supplemental Fig. S8) is very useful for accelerating understanding of their role.
Potential Mechanisms in Endosperm for Extremely High Expression of Zein Genes
Of the 100 most highly expressed genes in the endosperm, 54 genes fall into expression modules in the middle phase of endosperm development. Among these highly expressed genes, the zeins collectively account for about 65% of the total endosperm transcripts, consistent with previous observations (Marks et al., 1985; Woo et al., 2001). Therefore, there must be some mechanisms in the endosperm that allow the rapid and drastic increase in the expression level of particular genes. One possible mechanism is to increase the metabolic capacity of the endosperm by endoreduplication with a large-scale increase in the amount of transcriptional templates to support high transcription rates (Sugimoto-Shirasu and Roberts, 2003). Based on our data, although storage accumulation is the longest phase of endosperm development, only 16.3% (3,654) of genes were actively expressed during this stage and a large number of genes are actually down-regulated (Figs. 2C and 4B). Thus, we propose that there is an active mechanism in the endosperm to limit a number of metabolic processes to save resources for the synthesis and accumulation of storage compounds in this special period. In addition, the excessive overdraft to perform such a specific function of storage accumulation over such a long time for seed filling may be a reason that the starchy endosperm cells undergo programmed cell death prior to desiccation and quiescence (Young and Gallie, 2000). By contrast, there is little accumulation of zein proteins in aleurone cells (Reyes et al., 2011), consistent with the fact that the aleurone cells do not undergo programmed cell death until germination (Fath et al., 2000).
Despite the importance of zein protein accumulation in the endosperm, the mechanisms by which these genes are regulated are still not clear. Previous studies have shown that DNA methylation status may be an important factor. For example, a recent study demonstrated that the promoter regions of zein genes have different methylation patterns with differences between tissues (Miclaus et al., 2011). The low methylation level in these promoters is not a result of O2 regulation (Locatelli et al., 2009). The gene bodies of storage protein genes were also found to be undermethylated in the endosperm compared with other tissues (Bianchi and Viotti, 1988). Our study extends the analysis to gene bodies of all of the zein genes and shows that the high expression of zein genes in the endosperm is highly correlated with DNA hypomethylation in the gene body, which is consistent with a recent study in rice (Zemach et al., 2010). Therefore, gene body hypomethylation seems to be a common mechanism for activating the expression of endosperm-specific genes.
MATERIALS AND METHODS
Plant Material and Sequencing
The maize (Zea mays) inbred line B73 was grown in the field in the summer of 2012 in Beijing, China. Ears were self-pollinated on the same day. All samples of embryo, endosperm, and whole seed were collected by manual dissection, frozen immediately in liquid nitrogen, and stored at −80°C before processing. Each sample was obtained from at least three plants.
Total RNA was extracted using TRIzol reagent (Invitrogen). RNA-seq libraries were prepared according the manufacturer’s protocol of the Illumina Standard mRNA-seq library preparation kit (Illumina) and were sequenced to generate 101-nucleotide paired-end reads on a Illumina HiSeq platform.
Read Mapping and Gene Expression Analysis
The raw reads were aligned to the B73 reference genome (RefGen_v2) using Tophat 2.0.6 (http://ccb.jhu.edu/software/tophat/index.shtml; Trapnell et al., 2009) with maximum intron length set to 30 kb, with default settings for other parameters. We calculated the number of uniquely mapped reads for each gene model in the B73 FGS by parsing the alignment output files from Tophat, and then normalized the resulting read counts by RPKM to measure the gene expression level. We calculated the Pearson correlation coefficient between biological replicates with R software using genes expressed in at least one of the samples. Maize reference genome sequence data and the FGS 5b were downloaded at http://ensembl.gramene.org/Zea_mays/Info/Index.
PCA and Hierarchical Clustering
To facilitate graphical interpretation of relatedness among 53 different seed samples, we reduced the dimensional expression data to two dimensions by PCA using the prcomp function in R software with default settings (R Development Core Team, 2012). Hierarchical clustering was performed by the pvclust package with default settings using Pearson’s correlation distance (Suzuki and Shimodaira, 2006). The transformed and normalized gene expression values with log2 (RPKM + 1) were used for the analysis of PCA and hierarchical clustering.
Gene Coexpression Analysis
We conducted the coexpression analysis for embryo, endosperm, and whole seed, respectively, using MeV, which was downloaded from http://www.tm4.org/mev.html. Before running the MeV for a tissue, we calculated the relative expression values of the genes by dividing their expression level at different time points with their maximum observed RPKM. The relative expression levels were used as input for MeV and clustered using the k-means method and Pearson correlation coefficients among the genes. The figure of merit (Yeung et al., 2001) was used to determine the optimal cluster number. Functional category enrichment for each cluster was evaluated with the MapMan annotation (Thimm et al., 2004). Before conducting the MapMan annotation, the longest protein of each FGS gene was chosen as a representative protein and the Mercator was run with default settings. Overrepresentation and underrepresentation of functional categories in a cluster were determined with Fisher’s exact test. Resulting P values were adjusted to Q values by the Benjamini-Hochberg correction, and a false discovery rate of 5% was applied.
Identification of Seed-Specific Gene Expression
To identify genes that are preferentially expressed in seed tissues, we downloaded 25 nonseed RNA-seq data sets (Supplemental Table S2) from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/) and calculated the gene expression level. We then used the Z score value to determine whether a gene expressed specificity in seed. The expression across all of the samples was normalized using log2 (RPKM + 0.01). For a given gene, we calculated Z scores of the gene in different seed samples compared with the nonseed sample data set using the normalized expression level. The gene was determined to be seed-specifically expressed if it had a Z score above 3 in at least one of the seed samples.
We then divided the identified seed-specific genes into different parts (Table I). We found 45 genes that were not expressed in any of the embryo or endosperm samples, but were present in whole seed samples and were thus classified as other. The remaining seed-specific genes were further determined if they were specifically expressed in the embryo or endosperm. The Z score value was used to identify seed-specific genes. The difference is that the raw RPKM value was used instead of the log2 transformed value. A gene was determined to be embryo specific if it had a Z score above 3 in at least one of the embryo samples compared with the endosperm samples. Similarly, a gene was determined to be endosperm specific if it had a Z score above 3 in at least one of the endosperm samples compared with the embryo samples. In addition, genes were considered to have expression in both the embryo and endosperm if they could not be classified as embryo or endosperm specific.
Identification of DMRs
The DMRs among embryo (12 DAP), endosperm (12 DAP), and shoot tissue were identified as described in our previous study (Zhang et al., 2014) with some modifications. Briefly, high-throughput bisulfite sequencing reads were first processed by SolexaQA software (Cox et al., 2010) to filter out low-quality bases (<13). Next, processed reads were aligned to the B73 reference genome (RefGen_v2) using Bismark (Krueger and Andrews, 2011). Only uniquely mapped reads were used to determine the methylation level. When identifying DMRs, we applied a sliding-window approach to overcome the limit of sequencing depth with a window size of 200 bp and a slide step of 100 bp across the whole genome. Windows that contained more than five CGs/CHGs and were supported with at least five reads were kept. Fisher’s exact test was used to weight the significance of DMRs, and resulting P values were adjusted to Q values. Regions with a Q value ≤ 0.01 and methylation level differences of more than 30% between two samples were determined to be DMRs, and overlapping DMRs within 200 bp were merged. For the CG context, the hypermethylated sample was also required to have methylation levels of more than 40% to be considered a DMR.
Sequence data from this article can be found in the National Center for Biotechnology Information Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) under accession number SRP037559.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Correlation between biological replicates.
Supplemental Figure S2. Expression profiles of LEC1, Glb2, Fie1, and O2 in the 53 samples.
Supplemental Figure S3. MapMan functional categories enriched in different coexpression modules of embryo, endosperm, and whole seed.
Supplemental Figure S4. Expression patterns and MapMan functional categories of the 1,506 genes only detected expression in the whole seed samples.
Supplemental Figure S5. Analysis of TF expression across samples.
Supplemental Figure S6. TF distribution in the embryo, endosperm, and whole seed coexpression modules.
Supplemental Figure S7. Expression patterns of seed-specific genes in all seed and nonseed samples.
Supplemental Figure S8. WEGO annotation of embryo- and endosperm-specific genes.
Supplemental Figure S9. Comparison of expression levels of different gene types in endosperm.
Supplemental Figure S10. Distribution of coexpression between embryo and endosperm samples.
Supplemental Figure S11. Genes involved in the starch biosynthesis pathway.
Supplemental Table S1. Summary of RNA-seq read mapping results.
Supplemental Table S2. List of published nonseed RNA-seq data used to find seed-specific genes.
Supplemental Table S3. Summary of zein genes detected in B73.
Supplemental Table S4. List of the 100 most highly expressed genes in endosperm shown in Figure 6A.
Supplemental Table S5. Summary of DMRs among embryo, endosperm, and shoot samples.
Supplemental Data Set S1. Expression level of FGS genes in different samples.
Supplemental Data Set S2. Number of genes and TFs detected in different seed samples.
Supplemental Data Set S3. List of TFs expressed in seed samples.
Supplemental Data Set S4. Summary of different types of seed-specific genes shown in Table I.
Glossary
- RNA-seq
RNA sequencing
- RPKM
reads per kilobase per million
- FGS
filtered gene set
- TF
transcription factor
- DAP
d after pollination
- PCA
principal component analysis
- GO
gene ontology
- DMR
differentially methylated region
- SS
starch synthase
- SBE
starch branching enzyme
Footnotes
This work was supported by the National High Technology Research and Development Program of China (863 Project, grant no. 2012AA10A305 to J.L.) and the National Natural Science Foundation of China (grant no. 31225020 to J.L.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Ball SG, Morell MK. (2003) From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol 54: 207–233 [DOI] [PubMed] [Google Scholar]
- Ballicora MA, Iglesias AA, Preiss J. (2004) ADP-glucose pyrophosphorylase: a regulatory enzyme for plant starch synthesis. Photosynth Res 79: 1–24 [DOI] [PubMed] [Google Scholar]
- Beatty MK, Rahman A, Cao H, Woodman W, Lee M, Myers AM, James MG. (1999) Purification and molecular genetic characterization of ZPU1, a pullulanase-type starch-debranching enzyme from maize. Plant Physiol 119: 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, et al. (2013) Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci USA 110: E435–E444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Hake SC. (2009) Handbook of Maize: Genetics and Genomics. Springer, New York [Google Scholar]
- Bianchi MW, Viotti A. (1988) DNA methylation and tissue-specific transcription of the storage protein genes of maize. Plant Mol Biol 11: 203–214 [DOI] [PubMed] [Google Scholar]
- Bolduc N, Yilmaz A, Mejia-Guerra MK, Morohashi K, O’Connor D, Grotewold E, Hake S. (2012) Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev 26: 1685–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Imparl-Radosevich J, Guan H, Keeling PL, James MG, Myers AM. (1999) Identification of the soluble starch synthase activities of maize endosperm. Plant Physiol 120: 205–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson SJ, Chourey PS. (1996) Evidence for plasma membrane-associated forms of sucrose synthase in maize. Mol Gen Genet 252: 303–310 [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Koltunow A, Payne T, Luo M, Tucker MR, Dennis ES, Peacock WJ. (2001) Control of early seed development. Annu Rev Cell Dev Biol 17: 677–699 [DOI] [PubMed] [Google Scholar]
- Chourey PS, Nelson OE. (1976) The enzymatic deficiency conditioned by the shrunken-1 mutations in maize. Biochem Genet 14: 1041–1055 [DOI] [PubMed] [Google Scholar]
- Cox MP, Peterson DA, Biggs PJ. (2010) SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics 11: 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danilevskaya ON, Hermon P, Hantke S, Muszynski MG, Kollipara K, Ananiev EV. (2003) Duplicated fie genes in maize: expression pattern and imprinting suggest distinct functions. Plant Cell 15: 425–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RM, Hansey CN, Gowda M, Childs KL, Lin H, Vaillancourt B, Sekhon RS, de Leon N, Kaeppler SM, Jiang N, et al. (2011) Utility of RNA sequencing for analysis of maize reproductive transcriptomes. Plant Genome 4: 191–203 [Google Scholar]
- Dumas C, Mogensen HL. (1993) Gametes and fertilization: maize as a model system for experimental embryogenesis in flowering plants. Plant Cell 5: 1337–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick DN. (1961) Protein granules of maize endosperm cells. Cereal Chem 38: 4–385 [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE. (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Evans MM. (2007) The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo Sac and leaf development. Plant Cell 19: 46–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath A, Bethke P, Lonsdale J, Meza-Romero R, Jones R. (2000) Programmed cell death in cereal aleurone. Plant Mol Biol 44: 255–266 [DOI] [PubMed] [Google Scholar]
- Feng L, Zhu J, Wang G, Tang Y, Chen H, Jin W, Wang F, Mei B, Xu Z, Song R. (2009) Expressional profiling study revealed unique expressional patterns and dramatic expressional divergence of maize α-zein super gene family. Plant Mol Biol 69: 649–659 [DOI] [PubMed] [Google Scholar]
- Fontes EB, Shank BB, Wrobel RL, Moose SP, OBrian GR, Wurtzel ET, Boston RS. (1991) Characterization of an immunoglobulin binding protein homolog in the maize floury-2 endosperm mutant. Plant Cell 3: 483–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Asakura N, Ohdan T, Miyao A, Hirochika H, Nakamura Y. (2006) Function and characterization of starch synthase I using mutants in rice. Plant Physiol 140: 1070–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfray HCJ, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C. (2010) Food security: the challenge of feeding 9 billion people. Science 327: 812–818 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Marcos JF, Costa LM, Biderre-Petit C, Khbaya B, O’Sullivan DM, Wormald M, Perez P, Dickinson HG. (2004) maternally expressed gene1 Is a novel maize endosperm transfer cell-specific gene with a maternal parent-of-origin pattern of expression. Plant Cell 16: 1288–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon JS, Ryoo N, Hahn TR, Walia H, Nakamura Y. (2010) Starch biosynthesis in cereal endosperm. Plant Physiol Biochem 48: 383–392 [DOI] [PubMed] [Google Scholar]
- Jia Y, Lisch DR, Ohtsu K, Scanlon MJ, Nettleton D, Schnable PS. (2009) Loss of RNA-dependent RNA polymerase 2 (RDR2) function causes widespread and unexpected changes in the expression of transposons, genes, and 24-nt small RNAs. PLoS Genet 5: e1000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HG, Park S, Matsuoka M, An G. (2005) White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J 42: 901–911 [DOI] [PubMed] [Google Scholar]
- Kiesselbach TA. (1949) The structure and reproduction of corn. In Research Bulletin No. 161. University of Nebraska Agricultural Experiment Station, Lincoln, NE, pp 61–84 [Google Scholar]
- Kriz AL. (1989) Characterization of embryo globulins encoded by the maize Glb genes. Biochem Genet 27: 239–251 [DOI] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Williams ME, Tingey SV, Rafalski AJ. (2002) DNA array profiling of gene expression changes during maize embryo development. Funct Integr Genomics 2: 13–27 [DOI] [PubMed] [Google Scholar]
- Li P, Ponnala L, Gandotra N, Wang L, Si Y, Tausta SL, Kebrom TH, Provart N, Patel R, Myers CR, et al. (2010) The developmental dynamics of the maize leaf transcriptome. Nat Genet 42: 1060–1067 [DOI] [PubMed] [Google Scholar]
- Lid SE, Gruis D, Jung R, Lorentzen JA, Ananiev E, Chamberlin M, Niu X, Meeley R, Nichols S, Olsen OA. (2002) The defective kernel 1 (dek1) gene required for aleurone cell development in the endosperm of maize grains encodes a membrane protein of the calpain gene superfamily. Proc Natl Acad Sci USA 99: 5460–5465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu WY, Chang YM, Chen SC, Lu CH, Wu YH, Lu MY, Chen DR, Shih AC, Sheue CR, Huang HC, et al. (2013) Anatomical and transcriptional dynamics of maize embryonic leaves during seed germination. Proc Natl Acad Sci USA 110: 3979–3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Fu J, Gu D, Liu W, Liu T, Peng Y, Wang J, Wang G. (2008) Genome-wide analysis of gene expression profiles during the kernel development of maize (Zea mays L.). Genomics 91: 378–387 [DOI] [PubMed] [Google Scholar]
- Locatelli S, Piatti P, Motto M, Rossi V. (2009) Chromatin and DNA modifications in the Opaque2-mediated regulation of gene transcription during maize endosperm development. Plant Cell 21: 1410–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Yee KM, West MA, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. (1998) Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell 93: 1195–1205 [DOI] [PubMed] [Google Scholar]
- Makarevitch I, Eichten SR, Briskine R, Waters AJ, Danilevskaya ON, Meeley RB, Myers CL, Vaughn MW, Springer NM. (2013) Genomic distribution of maize facultative heterochromatin marked by trimethylation of H3K27. Plant Cell 25: 780–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks MD, Lindell JS, Larkins BA. (1985) Quantitative analysis of the accumulation of Zein mRNA during maize endosperm development. J Biol Chem 260: 16445–16450 [PubMed] [Google Scholar]
- Márton ML, Cordts S, Broadhvest J, Dresselhaus T. (2005) Micropylar pollen tube guidance by egg apparatus 1 of maize. Science 307: 573–576 [DOI] [PubMed] [Google Scholar]
- McCarty DR, Hattori T, Carson CB, Vasil V, Lazar M, Vasil IK. (1991) The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell 66: 895–905 [DOI] [PubMed] [Google Scholar]
- Miclaus M, Xu JH, Messing J. (2011) Differential gene expression and epiregulation of alpha zein gene copies in maize haplotypes. PLoS Genet 7: e1002131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K, Casas MI, Falcone Ferreyra ML, Mejía-Guerra MK, Pourcel L, Yilmaz A, Feller A, Carvalho B, Emiliani J, Rodriguez E, et al. (2012) A genome-wide regulatory framework identifies maize pericarp color1 controlled genes. Plant Cell 24: 2745–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE. (2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington PD, Costa LM, Gutierrez-Marcos JF, Greenland AJ, Dickinson HG. (2008) When genomes collide: aberrant seed development following maize interploidy crosses. Ann Bot (Lond) 101: 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2012) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- Raissig MT, Baroux C, Grossniklaus U. (2011) Regulation and flexibility of genomic imprinting during seed development. Plant Cell 23: 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes FC, Chung T, Holding D, Jung R, Vierstra R, Otegui MS. (2011) Delivery of prolamins to the protein storage vacuole in maize aleurone cells. Plant Cell 23: 769–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DA, Deboer DL, Stark DM, Preiss J, Fromm ME. (1993) Plastid targeting of E. coli β-glucuronidase and ADP-glucose pyrophosphorylase in maize (Zea mays L.) cells. Plant Cell Rep 13: 24–27 [DOI] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA. (2009) The development of endosperm in grasses. Plant Physiol 149: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlon MJ, Takacs EM. (2009) Kernel biology. In Bennetzen J, Hake S, eds, Handbook of Maize: Its Biology. Springer, New York, pp 121–143 [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Sekhon RS, Lin H, Childs KL, Hansey CN, Buell CR, de Leon N, Kaeppler SM. (2011) Genome-wide atlas of transcription during maize development. Plant J 66: 553–563 [DOI] [PubMed] [Google Scholar]
- Shannon JC, Pien FM, Cao H, Liu KC. (1998) Brittle-1, an adenylate translocator, facilitates transfer of extraplastidial synthesized ADP—glucose into amyloplasts of maize endosperms. Plant Physiol 117: 1235–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She KC, Kusano H, Koizumi K, Yamakawa H, Hakata M, Imamura T, Fukuda M, Naito N, Tsurumaki Y, Yaeshima M, et al. (2010) A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell 22: 3280–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LG, Jackson D, Hake S. (1995) Expression of knotted1 marks shoot meristem formation during maize embryogenesis. Dev Genet 16: 344–348 [Google Scholar]
- Sugimoto-Shirasu K, Roberts K. (2003) “Big it up”: endoreduplication and cell-size control in plants. Curr Opin Plant Biol 6: 544–553 [DOI] [PubMed] [Google Scholar]
- Suzuki R, Shimodaira H. (2006) Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22: 1540–1542 [DOI] [PubMed] [Google Scholar]
- Takacs EM, Li J, Du C, Ponnala L, Janick-Buckner D, Yu J, Muehlbauer GJ, Schnable PS, Timmermans MC, Sun Q, et al. (2012) Ontogeny of the maize shoot apical meristem. Plant Cell 24: 3219–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh KT, Requesens DV, Devaiah SP, Johnson D, Huang X, Howard JA, Hood EE. (2013) Transcriptome analysis of embryo maturation in maize. BMC Plant Biol 13: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernoud V, Hajduch M, Khaled A, Depège N, Rogowsky MP. (2005) Maize embryogenesis. Maydica 50: 469–483 [Google Scholar]
- Vicente-Carbajosa J, Moose SP, Parsons RL, Schmidt RJ. (1997) A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc Natl Acad Sci USA 94: 7685–7690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrinten PL, Nakamura T. (2000) Wheat granule-bound starch synthase I and II are encoded by separate genes that are expressed in different tissues. Plant Physiol 122: 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xie W, Chen Y, Tang W, Yang J, Ye R, Liu L, Lin Y, Xu C, Xiao J, et al. (2010) A dynamic gene expression atlas covering the entire life cycle of rice. Plant J 61: 752–766 [DOI] [PubMed] [Google Scholar]
- Wang X, Elling AA, Li X, Li N, Peng Z, He G, Sun H, Qi Y, Liu XS, Deng XW. (2009a) Genome-wide and organ-specific landscapes of epigenetic modifications and their relationships to mRNA and small RNA transcriptomes in maize. Plant Cell 21: 1053–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. (2009b) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo YM, Hu DW, Larkins BA, Jung R. (2001) Genomics analysis of genes expressed in maize endosperm identifies novel seed proteins and clarifies patterns of zein gene expression. Plant Cell 13: 2297–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu JH, Messing J. (2008) Organization of the prolamin gene family provides insight into the evolution of the maize genome and gene duplications in grass species. Proc Natl Acad Sci USA 105: 14330–14335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, Wang J. (2006) WEGO: a web tool for plotting GO annotations. Nucleic Acids Res 34: W293–W297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung KY, Haynor DR, Ruzzo WL. (2001) Validating clustering for gene expression data. Bioinformatics 17: 309–318 [DOI] [PubMed] [Google Scholar]
- Young TE, Gallie DR. (2000) Programmed cell death during endosperm development. Plant Mol Biol 44: 283–301 [DOI] [PubMed] [Google Scholar]
- Yun SH, Matheson NK. (1993) Structures of the amylopectins of waxy, normal, amylose-extender, and wx:ae genotypes and of the phytoglycogen of maize. Carbohydr Res 243: 307–321 [DOI] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Silva P, Rodrigues JA, Dotson B, Brooks MD, Zilberman D. (2010) Local DNA hypomethylation activates genes in rice endosperm. Proc Natl Acad Sci USA 107: 18729–18734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Jin J, Tang L, Zhao Y, Gu X, Gao G, Luo J. (2011) PlantTFDB 2.0: update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res 39: D1114–D1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Xie S, Dong X, Zhao X, Zeng B, Chen J, Li H, Yang W, Zhao H, Wang G, et al. (2014) Genome-wide high resolution parental-specific DNA and histone methylation maps uncover patterns of imprinting regulation in maize. Genome Res 24: 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Colleoni C, Ratushna V, Sirghie-Colleoni M, James MG, Myers AM. (2004) Molecular characterization demonstrates that the Zea mays gene sugary2 codes for the starch synthase isoform SSIIa. Plant Mol Biol 54: 865–879 [DOI] [PubMed] [Google Scholar]
- Zhu D, Scandalios JG. (1994) Differential accumulation of manganese-superoxide dismutase transcripts in maize in response to abscisic acid and high osmoticum. Plant Physiol 106: 173–178 [DOI] [PMC free article] [PubMed] [Google Scholar]