A Ca2+-dependent kinase is identified that phosphorylates and inhibits inward potassium channels expressed in guard cells, the overexpression of which results in a reduction of stomatal aperture.
Abstract
Ca2+-dependent protein kinases (CPKs) form a large family of 34 genes in Arabidopsis (Arabidopsis thaliana). Based on their dependence on Ca2+, CPKs can be sorted into three types: strictly Ca2+-dependent CPKs, Ca2+-stimulated CPKs (with a significant basal activity in the absence of Ca2+), and essentially calcium-insensitive CPKs. Here, we report on the third type of CPK, CPK13, which is expressed in guard cells but whose role is still unknown. We confirm the expression of CPK13 in Arabidopsis guard cells, and we show that its overexpression inhibits light-induced stomatal opening. We combine several approaches to identify a guard cell-expressed target. We provide evidence that CPK13 (1) specifically phosphorylates peptide arrays featuring Arabidopsis K+ Channel KAT2 and KAT1 polypeptides, (2) inhibits KAT2 and/or KAT1 when expressed in Xenopus laevis oocytes, and (3) closely interacts in plant cells with KAT2 channels (Förster resonance energy transfer-fluorescence lifetime imaging microscopy). We propose that CPK13 reduces stomatal aperture through its inhibition of the guard cell-expressed KAT2 and KAT1 channels.
Stomata are microscopic organs at the leaf surface, each made of two so-called guard cells forming a pore. Opening or closing these pores is the way through which plants control their gas exchanges with the atmosphere (i.e. carbon dioxide uptake to feed the photosynthetic process and transpirational loss of water vapor). Stomatal movements result from osmotically driven fluxes of water, which follow massive exchanges of solutes, including K+ ions, between the guard cells and the surrounding tissues (Hetherington, 2001; Nilson and Assmann, 2007).
Both Ca2+-dependent and Ca2+-independent signaling pathways are known to control stomatal movements (MacRobbie, 1993, 1998; Blatt, 2000; Webb et al., 2001; Mustilli et al., 2002; Israelsson et al., 2006; Marten et al., 2007; Laanemets et al., 2013). In particular, Ca2+ signals have been reported to promote stomatal closure through the inhibition of inward K+ channels and the activation of anion channels (Blatt, 1991, 1992, 2000; Thiel et al., 1992; Grabov and Blatt, 1999; Schroeder et al., 2001; Hetherington and Brownlee, 2004; Mori et al., 2006; Marten et al., 2007; Geiger et al., 2010; Brandt et al., 2012; Scherzer et al., 2012). However, little is known about the molecular identity of the links between Ca2+ events and Shaker K+ channel activity. Several kinases and phosphatases are believed to be involved in both the Ca2+-dependent and Ca2+-independent signaling pathways. Plants express two large kinase families whose activity is related to Ca2+ signaling. Firstly, CBL-interacting protein kinases (CIPKs; 25 genes in Arabidopsis [Arabidopsis thaliana]) are indirectly controlled by their interaction with a set of calcium sensors, the calcineurin B-like proteins (CBLs; 10 genes in Arabidopsis). This complex forms a fascinating network of potential Ca2+ signaling decoders (Luan, 2009; Weinl and Kudla, 2009), which have been addressed in numerous reports (Xu et al., 2006; Hu et al., 2009; Batistic et al., 2010; Held et al., 2011; Chen et al., 2013). In particular, some CBL-CIPK pairs have been shown to regulate Shaker channels such as Arabidopsis K+ Transporter1 (AKT1; Xu et al., 2006; Lan et al., 2011) or AKT2 (Held et al., 2011). Second, Ca2+-dependent protein kinases (CPKs) form an even larger family (34 genes in Arabidopsis) of proteins combining a kinase domain with the ability to bind Ca2+, thanks to the so-called EF hands (Harmon et al., 2000; Harper et al., 2004). CPKs, which, interestingly, are not found in animal cells, exhibit different calcium dependencies (Boudsocq et al., 2012). With respect to this, three types of CPKs can be considered: strictly Ca2+-dependent CPKs, Ca2+-stimulated CPKs (with a significant basal activity in the absence of Ca2+), and essentially Ca2+-insensitive CPKs (however, structurally close to kinases of groups 1 and 2).
Pioneering work by Luan et al. (1993) demonstrated in Vicia faba guard cells that inward K+ channels were regulated by some Ca2+-dependent kinases. Then, such a Ca2+-dependent kinase was purified from guard cell protoplasts of V. faba and shown to actually phosphorylate the in vitro-translated KAT1 protein, a Shaker channel subunit natively expressed in Arabidopsis guard cells (Li et al., 1998). KAT1 regulation by CPK was shown by the inhibition of KAT1 currents after the coexpression of KAT1 and CDPK from soybean (Glycine max) in oocytes (Berkowitz et al., 2000). Since then, several cpk mutant lines of Arabidopsis have been shown to be impaired in stomatal movements, for example cpk10 (Ca2+ insensitive), cpk4/cpk11 (Ca2+ dependent), and cpk3/cpk6/cpk23 (Ca2+ dependent; Mori et al., 2006; Geiger et al., 2010; Munemasa et al., 2011; Hubbard et al., 2012).
Of the nine genes encoding voltage-dependent K+ channels (Shaker) in Arabidopsis (Véry and Sentenac, 2002, 2003; Lebaudy et al., 2007; Hedrich, 2012), six are expressed in guard cells and play a role in stomatal movements: the Gated Outwardly-Rectifying K+ (GORK) gene, encoding an outward K+ channel subunit, and the AKT1, AKT2, Arabidopsis K+ Rectifying Channel1 (AtKC1), KAT1, and KAT2 genes, encoding inward K+ channel subunits (Pilot et al., 2001; Szyroki et al., 2001; Hosy et al., 2003; Pandey et al., 2007; Lebaudy et al., 2008a). Shaker channels result from the assembly of four subunits, and it has been shown that inward subunits tend to heterotetramerize, thus potentially widening the functional and regulatory scope of inward K+ conductance in guard cells (Xicluna et al., 2007; Jeanguenin et al., 2008; Lebaudy et al., 2008a, 2010). Inhibition of inward K+ channels has been shown to reduce stomatal opening (Liu et al., 2000; Kwak et al., 2001). This has grounded a strategy for disrupting inward K+ channel conductance in guard cells by expressing a nonfunctional KAT2 subunit (dominant negative mutation) in a kat2 knockout Arabidopsis line. The resulting Arabidopsis lines, named kincless, have no functional inward K+ channels and exhibit delayed stomatal opening (Lebaudy et al., 2008b) with, in the long term, a biomass reduction compared with the Arabidopsis wild-type line.
Among the CPKs presumably expressed in Arabidopsis guard cells (Leonhardt et al., 2004), we looked for CPK13, which belongs to the atypical Ca2+-insensitive type of CPKs (Kanchiswamy et al., 2010; Boudsocq et al., 2012; Liese and Romeis, 2013) and whose role remains unknown in stomatal movements. Here, we confirm first that CPK13 kinase activity is independent of Ca2+ and show that CPK13 expression is predominant in Arabidopsis guard cells using CPK13-GUS lines. We then report that overexpression of CPK13 in Arabidopsis induces a dramatic default in stomatal aperture. Based on the previously reported kincless phenotype (Lebaudy et al., 2008b), we propose that CPK13 could reduce the activity of inward K+ channels in guard cells, particularly that of KAT2. We confirm this hypothesis by voltage-clamp experiments and show an inhibition of KAT2 and KAT1 activity by CPK13 (but not that of AKT2). In addition, we present peptide array phosphorylation assays showing that CPK13 targets, with some specificity, several KAT2 and KAT1 polypeptides. Finally, we demonstrate that KAT2 and CPK13 interact in planta using Förster resonance energy transfer (FRET)-fluorescence lifetime imaging microscopy (FLIM).
RESULTS AND DISCUSSION
Kinase Activity of CPK13 Is Independent of Ca2+
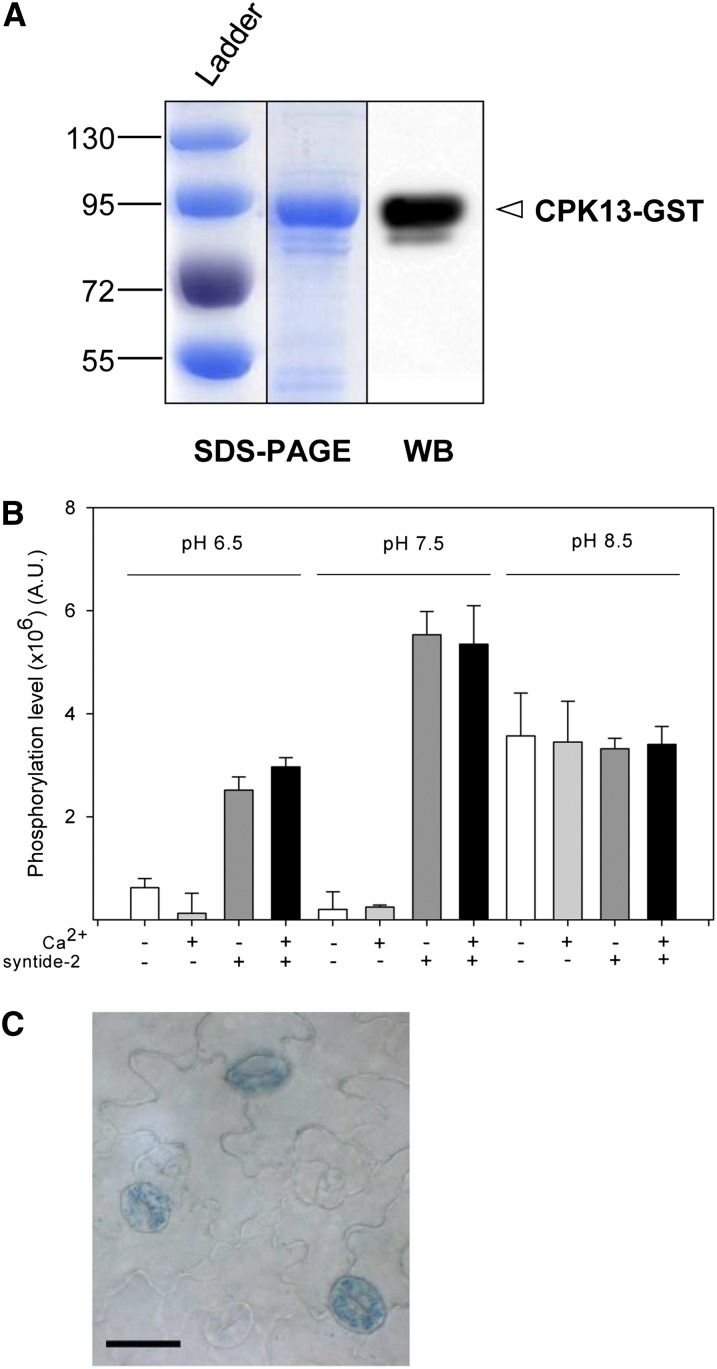
Coomassie Blue staining of an SDS-PAGE gel of purified glutathione S-transferase (GST)-tagged recombinant CPK13 shows a major band (Fig. 1A, left). Western blot with an anti-GST antibody resulted in a staining of this band that confirms the presence of the CPK13-GST fusion protein (Fig. 1A, right). Kinase activity of this GST-tagged CPK13 was determined in a set of buffers prepared with the aim of evaluating (1) the Ca2+ dependence of CPK13 kinase activity (buffers contained either 10 µm free or no Ca2+), (2) the transphosphorylation/autophosphorylation balance (buffers contained or not syntide-2 as a target phosphorylable peptide), and (3) the effect of pH (pH was set at 6.5, 7.5, or 8.5). The kinase activity of CPK13 was confirmed to be independent of Ca2+ (Fig. 1B), as reported previously (Kanchiswamy et al., 2010; Boudsocq et al., 2012; Liese and Romeis, 2013). The pH value had different effects on transphosphorylation and autophosphorylation resulting from the CPK13 kinase activity. Autophosphorylation (syntide-2 absent from the buffer) was very weak at pH 6.5 and 7.5 but increased dramatically at pH 8.5. As a consequence, the CPK13 kinase activity, which was poorly dependent on pH in the presence of syntide-2 (with, however, an optimum at the physiological pH 7.5), consisted essentially of transphosphorylation at pH 6.5 and 7.5 but was mostly autophosphorylation at pH 8.5 (Fig. 1B).
Figure 1.
CPK13 expression pattern and biochemical assays. A, Coomassie Blue staining of an SDS-PAGE gel (left) and western-blot autoradiography (WB; right) showing a major band corresponding to the CPK13-GST fusion protein purified from the Rosetta E. coli strain (see “Materials and Methods”). B, In vitro phosphorylation activity of recombinant CPK13 kinase. Kinase activity (arbitrary units [A.U.]) of CPK13 (10 ng µL−1) in 15 min was measured either in the presence of 10 µm free Ca2+ or in the absence of Ca2+ and in the presence of 25 µm syntide-2 as a control target peptide (assay of both autophosphorylation and transphosphorylation) or in the absence of syntide-2 (assay of sole autophosphorylation). Buffers of three pH values were used: 6.5, 7.5, and 8.5. Data are shown as means ± se (n = 3). C, Spatial expression pattern of CPK13 in Arabidopsis leaf (partial view of an adaxial surface) is shown in guard cells, using the GUS gene as a reporter of CPK13 promoter activity. Bar = 20 µm. [See online article for color version of this figure.]
Similar experiments were carried out with recombinant CPK4, a typical CPK also expressed in guard cells and reported to regulate the stomatal response to abscisic acid (ABA; Zhu et al., 2007). The purification of recombinant CPK4 and determination of CPK4 kinase activity in the same conditions as CPK13 are shown in Supplemental Figure S1, A and B, respectively. At variance with CPK13, CPK4 showed little autophosphorylation (at any Ca2+ or pH condition tested) and strong Ca2+ and pH dependence for its transphosphorylation activity (here of syntide-2), which showed an optimum at pH 7.5 and was very weak in the absence of Ca2+ (Supplemental Fig. S1B).
Changes in cytosolic pH participate in the control of stomatal movements. Cytosolic alkalization occurs upon and even precedes ABA-, external Ca2+-, and/or methyl jasmonate-induced stomatal closure (Suhita et al., 2004; Islam et al., 2010), whereas cytosolic acidification would participate in promoting auxin-induced stomatal opening (Irving et al., 1992). At pH 6.5, transphosphorylation was still detected only for CPK13, whereas CPK4 did not show any kinase activity (Fig. 1B; Supplemental Fig. S1B). In contrast, at pH 8.5, a Ca2+-dependent transphosphorylation was detected only for CPK4, whereas CPK13 showed solely a Ca2+-independent autophosphorylation. Taken together, these results suggest that the two kinases are involved in two different signaling pathways, involving a cytosolic pH decrease for CPK13 and the reverse for CPK4.
CPK13 Is Expressed in Guard Cells
Based on transcriptomic data, it has been reported previously that CPK13 is expressed in guard cells (Leonhardt et al., 2004; Zhao et al., 2008). Therefore, we assessed the activity of the CPK13 promoter using the GUS reporter gene and found it in guard cells (Fig. 1C). Accordingly, subsequent work was aimed at identifying an eventual role of CPK13 in stomatal movements.
Overexpression of CPK13 Significantly Reduces Light-Induced Stomatal Opening
In preliminary experiments, the knockout line cpk13−, which has been reported to display lower defensin gene expression after wounding (Kanchiswamy et al., 2010), showed no measurable stomatal phenotype, and it was decided rather to study CPK13-overexpressing lines. Several such lines have been generated by transformation with the pGWB523-CPK13 construct. Among the homozygous lines from the T2 generation, two independent lines, denoted OE13#3 and OE13#4, were selected, and their stomatal movements under light or after ABA application were compared with wild-type plants.
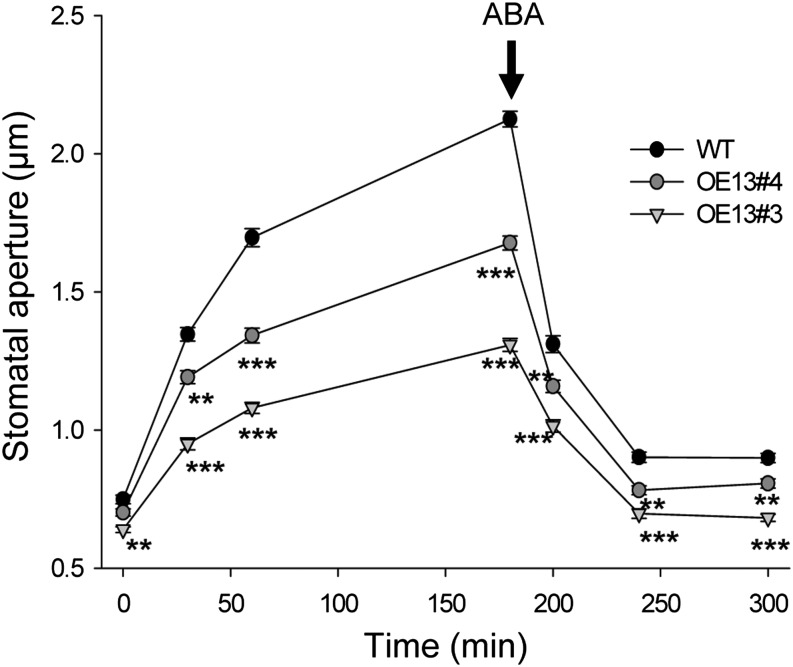
After a 12-h-long night, the wild type and the two OE13#3 and OE13#4 lines displayed similar stomatal aperture measured on epidermal peels (time 0 in Fig. 2). Then, light-induced stomatal opening was measured on the same epidermal peels at 30, 60, and 180 min. The OE13#3 and OE13#4 lines showed significant reductions in stomatal aperture (Fig. 2). At 180 min under light, stomatal closure in the presence of 10 µm ABA was tested. Stomatal closure induced by ABA was not affected in both mutant lines, but stomatal aperture remained significantly reduced (under light and ABA) in these mutants compared with the wild type (Fig. 2). This additive effect of ABA and CPK13 suggested that this kinase acts through a pathway independent of the hormone.
Figure 2.
Light- and ABA-triggered stomatal movements on epidermal strips peeled from wild-type and CPK13-overexpressing plants. Two CPK13-overexpressing lines (denoted OE13#3 and OE13#4) were compared with the Arabidopsis wild-type line (Columbia [WT]). After a 12-h-long period of darkness, light was switched on at time 0. Average stomatal aperture (35 or more stomata per epidermal strip) was measured at 0, 30, 60, and 180 min. At this time, stomatal closure was triggered by adding the plant stress hormone ABA (10 µm; black arrow) and average stomatal aperture (35 or more stomata per epidermal strip) was measured at 200, 240, and 300 min. Data are means ± se from 250 or more stomata from six independent plants by genotype. **P ≤ 0.01 and ***P ≤ 0.001 by Student’s t test.
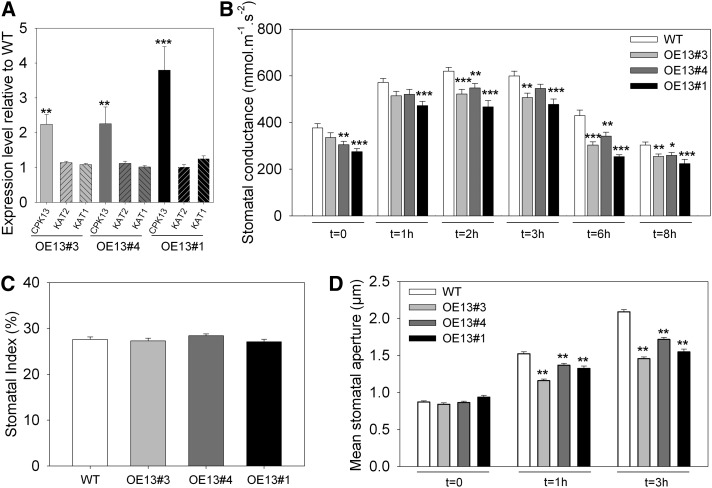
In another series of three independent runs, the strong impact of CPK13 overexpression on stomatal conductance and stomatal opening upon light onset was checked, with a third CPK13-overexpressing line incorporated in the experiments. This third mutant line, denoted OE13#1, was from the T1 generation and heterozygous for the pGWB523-CPK13 construct insertion. Compared with the wild type, the overexpression level of the CPK13 gene in the three lines OE13#3, OE13#4, and OE13#1 was found to be in the range of 2- to 4-fold (Fig. 3A).
Figure 3.
Further phenotyping of CPK13-overexpressing plants. A, Expression levels of CPK13, KAT2, and KAT1 genes (normalized with the TUB4 housekeeping gene) relative to wild-type plants (WT) in three independent CPK13-overexpressing lines (OE13#3, OE13#4, and OE13#1). Data (means ± se, n ≥ 15) result from qRT-PCR performed on cDNA extracts from 15-d-old seedlings. B, Stomatal conductance of wild-type and CPK13-overexpressing plants (OE13#3, OE13#4, and OE13#1) over an 8-h-long illuminated period following a 12-h-long darkness period. Data are means ± se (n ≥ 6 plants in three independent experiments). Each individual value represents the leaf conductance assayed with a porometer at time 0 (light onset) and 1, 2, 3, 6, and 8 h after onset (same leaf of a given plant). All measurements were done as blind. C, Stomatal index measured as described by Royer (2001) for the wild type and OE13#3, OE13#4, and OE13#1 lines from epidermal peels featuring approximately 1,000 stomata. Data are means ± se. All measurements were done as blind. D, Mean stomatal apertures measured on epidermal strips peeled from wild-type and CPK13-overexpressing plants. Light-triggered stomatal opening assays were performed on the same plant genotypes as in A (the wild type, OE13#3, OE13#4, and OE13#1). Light onset was at time 0, and average stomatal aperture (400 or more stomata from six or more plants by genotype) were measured at 0, 1, and 3 h. All measurements were done as blind. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001 by Student’s t test.
In standard culture conditions, none of these three OE13 lines displayed a bulk phenotype. Their stomatal conductance (assayed on intact plants), however, was found to be significantly reduced (Fig. 3B).
It was checked whether this reduction in stomatal conductance (measured at the leaf level on intact plants) originated from a reduced stomatal index in the mutant lines compared with the wild type. Here, the three CPK13-overexpressing lines and the wild type displayed similar stomatal indices (Fig. 3C), thus suggesting that a default of stomatal aperture could explain the reduction in stomatal conductance of the OE13 lines (Fig. 3B).
Therefore, the stomatal phenotype was further characterized by measuring the stomatal pore width on epidermal peels. After a 12-h-long night, the wild type and the three overexpressing mutant lines displayed similar stomatal aperture (t = 0 in Fig. 3D). Then, light-induced stomatal opening of the four genotypes was checked at 1 and 3 h by measuring the average stomatal aperture on epidermal peels of these plants. After both 1 and 3 h of light, OE13#3, OE13#4, and OE13#1 plants showed significantly less open stomata than wild-type plants (Fig. 3D). In particular, when wild-type stomatal pores widened by an average of 1.22 µm between 0 and 3 h, OE13#3, OE13#4, and OE13#1 pores widened only on average 0.61, 0.85, and 0.65 µm, which makes reductions of 50%, 30%, and 47%, respectively, of the opening movement in the mutant lines compared with the wild type.
In summary, overexpressing CPK13 induces a default of stomatal aperture that is reminiscent of the previously reported phenotype of the kincless mutant line. This line, which is impaired in the expression of functional inward K+ channel KAT2 subunits (Lebaudy et al., 2008b), displays little, if any, K+ inward currents in guard cells. The expression level of KAT2 (and also that of its twin gene KAT1) was unchanged, compared with wild-type plants, in OE13#3, OE13#4, and OE13#1 plants (Fig. 3A). We then tested KAT2 as a putative target of CPK13.
CPK13 and KAT2 Colocalize and Interact in Planta
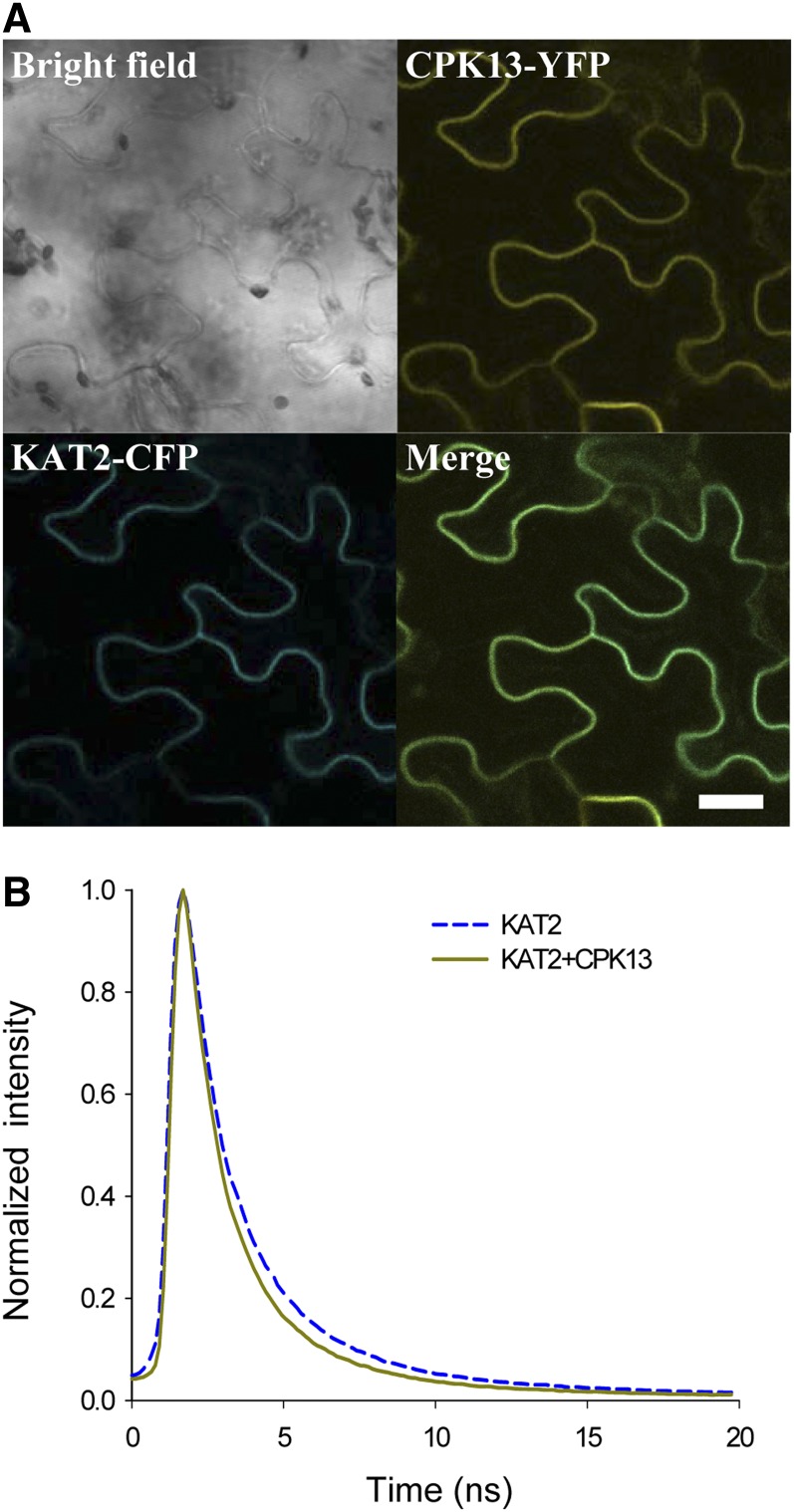
In order to determine if these two proteins interact in vivo, constructs allowing the expression of CPK13 fused to yellow fluorescent protein (CPK13-YFP) and of KAT2 fused to cyan fluorescent protein (KAT2-CFP) were obtained. Subcellular colocalization of the two tagged proteins was observed in Nicotiana benthamiana leaf epidermal cells expressing transiently the two constructs: KAT2-CFP and CPK13-YFP colocalized to the plasma membrane (Fig. 4A).
Figure 4.
Colocalization and interaction of KAT2 and CPK13 in plant cells. A, Confocal fluorescence microscopy of N. benthamiana epidermal leaf cells coexpressing KAT2-CFP and CPK13-YFP. Bar = 20 µm. B, FLIM of CFP in N. benthamiana leaf cells expressing KAT2-CFP only (blue dashed curve) and coexpressing KAT2-CFP and CPK13-YFP (dark yellow curve).
To provide evidence of interaction between these proteins in vivo, FRET experiments were then performed in N. benthamiana leaf epidermal cells. FRET was measured using a FLIM approach that allowed us to measure the fluorescence lifetime of CFP at the cell membrane in different conditions (Fig. 4B): the lifetime of CFP decreased significantly from 2.19 ± 0.05 ns (n = 115) when KAT2-CFP was expressed alone to 1.86 ± 0.04 ns (n = 146) when KAT2-CFP and CPK13-YFP were coexpressed (Table I). This decrease of CFP lifetime shows that FRET occurred between CFP and YFP when both KAT2-CFP and CPK13-YFP were present at the cell membrane. FRET efficiency is 15.07% (Table I) and strongly supports the physical interaction between KAT2 and CPK13 proteins in planta.
Table I. FLIM values of KAT2-CFP.
CFP fluorescent lifetime values are given for N. benthamiana plants that were transfected with pEARLEYGATE_102-KAT2 alone or cotransfected with pEARLEYGATE_101-CPK13 and pEARLEYGATE_102-KAT2. Fluorescence lifetime measurements are in nanoseconds (ns); n represents the total number of measurements from five independent experiments.
| Sample | Fluorescence Lifetime | Change in CFP Fluorescence Lifetime | n |
|---|---|---|---|
| ns | % | ||
| KAT2-CFP | 2.19 ± 0.05 | – | 115 |
| KAT2-CFP + CPK13-YFP | 1.86 ± 0.04 | −15.07 | 146 |
These results, which show that CPK13 and KAT2 interact in planta, suggest that CPK13 could phosphorylate KAT2.
CPK13 Phosphorylates KAT2 in Vitro
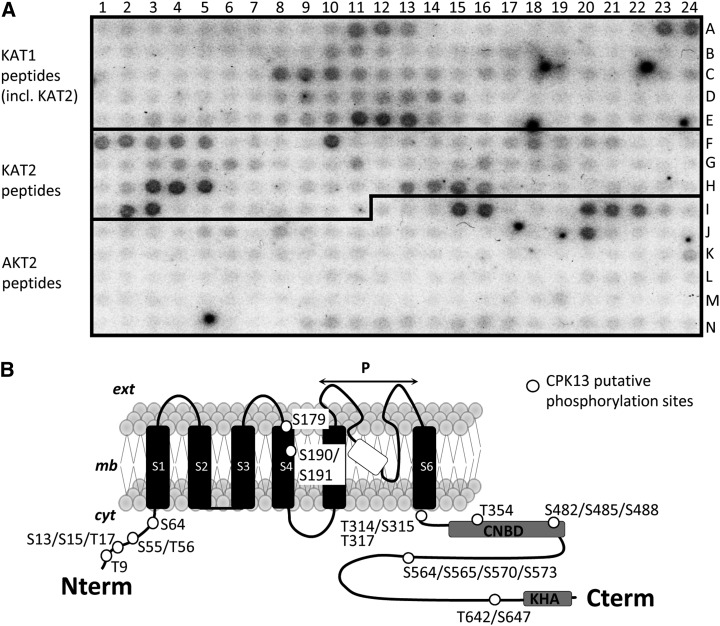
The phosphorylation by CPK13 of putative target sites in KAT2 (Ser and Thr residues) was tested using peptide arrays featuring 384 peptides in duplicate, including 52 KAT1-, 52 KAT2-, and 60 AKT2-derived wild-type peptides. KAT1 peptides were included because of the strong homology/identity with KAT2 and the in vivo heteromerization of these two K+ channel subunits (Lebaudy et al., 2008a). AKT2 peptides were included because AKT2 is expressed in guard cells and is also able to form heteromeric channels with KAT2 (Xicluna et al., 2007). A representative autoradiograph of one of these peptide arrays is shown (Fig. 5A) after incubation with recombinant CPK13 (Fig. 1A) in the presence of [γ-32P]ATP and Ca2+. Stained spots in Figure 5A represent phosphorylated peptides. The peptide sequence of KAT1 is highly homologous to that of KAT2. On the peptide array built for this study, 11 of the KAT1-labeled peptides in Figure 5A also belong to KAT2. The 41 KAT2-labeled peptides in Figure 5A are only those from KAT2 that are different from KAT1. The wild-type channel-derived peptides (from KAT1, KAT2, and AKT2) featured by the peptide arrays are listed in Supplemental Table S1. A quantitative analysis of the staining of all spots was made using ImageJ (Supplemental Fig. S2). In the first instance, a peptide was considered as significantly phosphorylated when its labeling was significantly above a threshold (dashed line in Supplemental Fig. S2). At least 11 KAT2 polypeptides were found to be phosphorylated by recombinant CPK13, showing that KAT2 is actually a target of this kinase (Supplemental Fig. S2A). These 11 polypeptides correspond to 16 Ser and seven Thr residues (white spots on the channel structure in Fig. 5B); both the N-terminal and C-terminal regions of KAT2 bear several of these targeted sites. The distribution of phosphorylated spots in KAT1 (nine phosphorylated peptides in Supplemental Fig. S2B) over the sequence of this polypeptide is rather similar to that in KAT2. In contrast, only four AKT2 peptides (comprising four Ser residues and one Thr residue) were phosphorylated by CPK13, mostly in the N-terminal region of AKT2 (Supplemental Fig. S2C). The other guard cell-expressed kinase, CPK4, of which the purification and dependence on Ca2+ and pH are shown in Supplemental Figure S1, was tested in parallel phosphorylation assays of the peptide arrays. CPK4 is poorly able to phosphorylate any of the three studied Shaker subunits (only one peptide for KAT2 and three peptides for KAT1; Supplemental Fig. S3). Therefore, in guard cells, CPK13 would be rather specific in its ability to target inward K+ channels incorporating KAT2 subunits.
Figure 5.
In vitro phosphorylation of peptide arrays by CPK13. A, Representative autoradiography of KAT1, KAT2, and AKT2 peptide arrays after phosphorylation by recombinant CPK13. Peptide arrays contain 52 peptides (15–18 amino acid each) corresponding to the wild-type KAT1 sequence (including 11 in common with KAT2), 41 peptides corresponding to the wild-type KAT2 sequence (different from KAT1), and 60 peptides corresponding to the wild-type AKT2 sequence. The list of these peptides is given in Supplemental Table S1. A number of mutant peptides are spotted between some wild-type peptides (not listed in Supplemental Table S1). Black spots correspond to phosphorylated peptides. B, Schematic drawing of the secondary structure of the KAT2 Shaker channel subunit, highlighting the putative sites phosphorylated by CPK13 (white dots) as deduced from the on-chip assay of kinase activity shown in A. ext, cyt, and mb stand for extracellular, cytoplasmic side, and membrane; S1 to S6, P, CNBD, and KHA stand for transmembrane segments 1 to 6, pore domain, putative cyclic nucleotide-binding domain, and domain rich in hydrophobic and acidic residues, respectively.
As CPK13 overexpression reduces stomatal aperture (Figs. 2 and 3) and targets both protein-protein interaction (Fig. 4) and phosphorylation (Fig. 5; Supplemental Fig. S2), the simplest hypothesis is that CPK13 inhibits the K+ channel activity of KAT2.
CPK13 Inhibits KAT2 Inward Currents in Xenopus laevis Oocytes
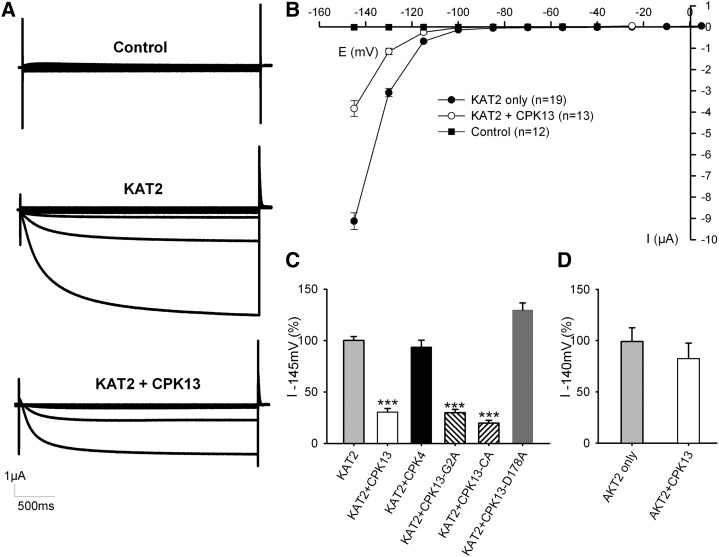
A standard voltage-clamp protocol was used to assess KAT2 channel activity when expressed alone or coexpressed with CPK13 in X. laevis oocytes. Membrane hyperpolarization activated the inward-rectifying KAT2 currents, which, in the same conditions, were dramatically reduced if CPK13 was coexpressed with KAT2 (Fig. 6A). Average current-voltage plots (Fig. 6B) showed that this effect is unlikely to be due to an impact of the kinase on channel voltage-dependent gating (no shift of the activation threshold) but rather to fewer channels being able to participate in the membrane conductance. This effect, in the 60% range, was somehow specific for the KAT2-CPK13 pair, as CPK4, a typical CPK expressed in guard cells, was unable to significantly change KAT2 currents (Fig. 6C). This KAT2-CPK13 specificity is also reinforced by the fact that the current of another hyperpolarization-activated K+ channel, AKT2 (also expressed in guard cells), is essentially unchanged upon coexpression with CPK13 (Fig. 6D).
Figure 6.
Inhibition of KAT2 channels by CPK13 in X. laevis oocytes. A, Representative current recordings obtained in noninjected oocytes (control; top) or in oocytes injected with KAT2 cRNA (KAT2; middle) or with KAT2 + CPK13 cRNAs (KAT2 + CPK13; bottom) using TEVC. In all recordings, 3-s voltage steps to values ranging between +5 and −145 mV in −15-mV decrements were applied from a −40-mV holding potential. B, Average current-voltage relationships from recordings obtained in the same conditions as in A. Data are means ± se for noninjected oocytes (black squares; n = 12) and oocytes injected with either KAT2 cRNA (black circles; n = 19) or KAT2 + CPK13 cRNAs (white circles; n = 13). C, Comparative effects on KAT2 of CPK13, CPK4, CPK13-G2A (nonmyristoylated mutant CPK13), CPK13-CA (constitutively active mutant CPK13; see text), and CPK13-D178A (mutant CPK13 devoid of kinase activity). The bar chart represents the current recorded at −145 mV in oocytes expressing KAT2 with a kinase as a percentage of current recorded at −145 mV in oocytes expressing KAT2 only within the same experiment. Data are means ± se from two or three independent experiments with n = 53 for KAT2, n = 40 for KAT2 + CPK13, n = 8 for KAT2 + CPK4, n = 30 for KAT2 + CPK13-G2A, n = 18 for KAT2 + CPK13-CA, and n = 20 for KAT2 + CPK13-D178A. ***P ≤ 0.001 by Student’s t test. D, Effect of CPK13 on AKT2. The bar chart represents the current recorded at −140 mV for oocytes expressing AKT2 only (n = 10) or AKT2 + CPK13 (n = 8). Data are means ± se.
Similar inhibition of KAT1 currents (up to 70%) was observed upon coexpression of this other guard cell Shaker channel subunit with CPK13 in X. laevis oocytes (Supplemental Fig. S4). KAT1 and KAT2 subunits make heteromeric channels in Arabidopsis guard cells (Lebaudy et al., 2008a, 2010). Therefore, it was also determined that strong inhibition of KAT1-KAT2 currents is observed in oocytes when CPK13 is coexpressed with these two Shaker channel subunits (Supplemental Fig. S5).
Like some other CPKs, CPK13 bears a myristoylation site (Gly residue in position 2) that is responsible for protein anchoring to the plasma membrane (Martín and Busconi, 2000; Benetka et al., 2008; Lu and Hrabak, 2013). The mutant G2A (with Gly in position 2 substituted by Ala), which cannot be myristoylated, was reported to be located at the cytosol and the nucleus of plant cells (Benetka et al., 2008; Lu and Hrabak, 2013). CPK13-G2A was coexpressed with KAT2 in oocytes. This G2A mutant CPK13 was as efficacious as wild-type CPK13 in inhibiting KAT2, suggesting that anchoring to the plasma membrane is not necessary for the kinase to target the channel to the plasma membrane (Fig. 6C). To confirm that CPK13 activity does not require Ca2+ to regulate KAT2 activity, the calmodulin and autoinhibitor domains (Harper et al., 1994, 2004; Cheng et al., 2002) of CPK13 were deleted, as described (Boudsocq et al., 2010; Dubiella et al., 2013), to generate a constitutively active mutant of CPK13 (denoted CPK13-CA). The coexpression of CPK13-CA with KAT2 was still able to inhibit KAT2 currents in oocytes (Fig. 6C). To confirm that CPK13 kinase activity is required for KAT2 inhibition, an inactive CPK13-D178A was generated. Substituting the Asp by an Ala in the conserved RDLKPEN motif (also found in other CPKs; Supplemental Fig. S6) was reported to abolish the Ser/Thr kinase activity of the yeast enzyme TPK1 (Gibbs and Zoller, 1991). This Asp-to-Ala point mutation has been subsequently reported to also abolish the Ser/Thr kinase activity of several CPKs (see references in Supplemental Fig. S6), including in particular CPK23 (Geiger et al., 2010), which is a Ca2+-independent CPK like CPK13. When coexpressed with KAT2 in oocytes, CPK13-D178A failed to inhibit the potassium current (Fig. 6C), indicating a kinase activity-dependent effect of CPK13. After expression of CPK13-D178A, the KAT2 current was even greater than in the control condition, suggesting a possible protective effect of this inactive kinase against some endogenous kinases of the X. laevis oocyte.
KAT2 inhibition by CPK13 might result from an alteration of the channel targeting to the plasma membrane (as reported for the channel/kinase couple AKT2/CIPK6; Held et al., 2011) or from an inhibition of KAT2 channels at the cell membrane level, or eventually from a combination of both these effects.
The structural bases for targeting of the KAT2 channel have recently been addressed in detail (Nieves-Cordones et al., 2014), although on an isoform lacking the first 11 amino acids (MLKRKHLNTRP) found in that described by Pilot et al. (2001) and used herein. The domain linking the sixth (and last) membrane-spanning segment to the large cytosolic C-terminal region of KAT2 was shown to control the targeting. In particular, by substituting Ser-382 by a Pro (found at the corresponding position in the endoplasmic reticulum [ER]-retained AtKC1 Shaker polypeptide), Nieves-Cordones et al. (2014) obtained a retention in the ER of the mutant channel, this assessed both by subcellular localization of GFP-tagged KAT2 and the absence of measurable KAT2 current. As substituting this Ser by an Ala had no impact on KAT2 targeting and KAT2 current level, Nieves-Cordones et al. (2014) concluded that phosphorylation of this Ser is not necessary for KAT2 targeting to the membrane. However, it could be that phosphorylation of this residue alters KAT2 targeting to the plasma membrane and promotes some retention in the ER. In the KAT2 isoform studied here, this Ser residue is at position 393 in the amino acid sequence and at spot G20 on the peptide array described in Supplemental Table S1 (position 382 and spot C21 for KAT1). Based on peptide array phosphorylation data (Supplemental Fig. S2), CPK13 seems unable to phosphorylate this Ser residue. This and the fact that KAT2-CFP and CPK13-YFP expressed in N. benthamiana epidermal cells colocalize to the plasma membrane (Fig. 4) do not support a retention of KAT2 in the ER due to its interaction with CPK13. Therefore, we rather hypothesize that phosphorylation by CPK13 inhibits the K+ channel activity of KAT2.
CPK13 phosphorylates at least 11 oligopeptides derived from KAT2 (including two common peptides, A11 and B11, with KAT1; Fig. 5; Supplemental Fig S2; Supplemental Table S1), while it targets only four of those derived from AKT2, a CPK13-insensitive channel. Inhibition of KAT2 would then result from phosphorylation of this channel subunit by CPK13, putatively in the large C-terminal cytoplasmic region. Among the common phosphorylated peptides of KAT2 and KAT1, only the peptide B11 is targeted by both CPK13 (Fig. 5) and CPK4 (Supplemental Fig. S3), suggesting that phosphorylation of these residues might not be sufficient to reduce KAT2 and KAT1 activities. Ongoing experiments currently aim at identifying in the KAT2 (and/or KAT1) sequence the active phosphorylation site or the several sites responsible for the inhibitory effect of CPK13. Until now, none of the KAT2 single mutants tested in oocytes lost inhibition by CPK13. Thus, a channel inhibition effect could require multiple phosphorylations at different sites. Unraveling the molecular mechanism of phosphorylation-dependent inhibition of KAT2 (KAT1) channels by CPK13 will certainly require much further work.
CONCLUSION
Transcriptomic data revealed that ABA signal transduction has no effect on CPK13 expression (Leonhardt et al., 2004), suggesting that CPK13 is not involved in ABA signaling in guard cells. The rather additive effects of ABA and CPK13 on stomatal closure (Fig. 2) further suggest that this kinase would act through a pathway independent of the phytohormone. The CPK13-overexpressing mutants OE13#3 and OE13#4 show no default of ABA-triggered stomatal closure. Independent regulatory pathways for stomatal aperture and closure have been reported previously by Assmann’s group (Wang et al., 2001). They showed that inhibition of stomatal opening by ABA is impaired in the knockout mutant G protein alpha subunit-1, whereas stomatal closure is still maintained with ABA. Our results obtained with the CPK13-overexpressing lines show that stomatal opening is altered but that ABA is still able to promote stomatal closure. Thus, CPK13 would act in an ABA-independent manner through an inhibition of KAT2 (and KAT1) channels. Of course, like most kinases, CPK13 might have several other targets that could participate in the CPK13-induced reduction in stomatal aperture.
The other guard cell-expressed CPK studied here, CPK4, has been shown to participate in some Ca2+-dependent transduction pathway of the ABA signal and to promote stomatal closure (Zhu et al., 2007), Interestingly, CPK4 has no effect on KAT2 inward K+ channels (Fig. 4C) and is unable to phosphorylate inward K+ channel-derived peptides except a few of them (in a likely unspecific way). Then, reducing the activity of inward channels (like KAT1 and KAT2) would be appropriate for limiting stomatal opening but not enough for promoting stomatal closure. It is considered that activation, with some dependence on Ca2+, of anion channels is a key and early step of the cascade resulting in ABA-triggered stomatal closure (Scherzer et al., 2012). CPK4 may eventually combine its action with that of other guard cell-expressed kinases, like OPEN STOMATA1 (Acharya et al., 2013; Imes et al., 2013) and CPK3/CPK6/CPK21/CPK23 (Mori et al., 2006; Brandt et al., 2012), which have been reported as promoting stomatal closure through their action on anion channels.
Here, the noncanonical CPK13 is shown to be a new negative regulator of stomatal aperture, through the inhibition of KAT2 (and KAT1) channel activity. In our hands, a knockout line, cpk13- (Kanchiswamy et al., 2010), showed the same stomatal pore aperture as wild-type plants. This suggests that the basal level of CPK13 activity is not efficient in inhibiting KAT2 and that some up-regulation of this activity, as in our CPK13-overexpressing lines, is required for a stomatal phenotype to be observable. Such an up-regulation may result from some posttranslational regulation of CPK13 and/or from an increase in CPK13 expression.
As regards the posttranslational regulation of CPK13, we solely observed that cytosolic acidification seems to shift the essentially Ca2+- and pH-insensitive kinase activity of CPK13 from autophosphorylation to transphosphorylation (Fig. 1B). As auxin-induced stomatal opening involves some cytosolic acidification (Irving et al., 1992), CPK13 may eventually behave as a negative regulator of this pathway.
On the other hand, one can assume that in planta regulation of CPK13 activity could result from a tight control of the transcriptional level of CPK13. Rather limited overexpression levels of CPK13 were sufficient to result in clear stomatal effects of CPK13, and an efficient physiological control of CPK13 transcript accumulation is suggested by the low amount of these transcripts found in our overexpressing lines despite the use of a strong promoter.
Physiological control of the CPK13 transcription level would not be mediated by ABA (Leonhardt et al., 2004). Data mining for likely candidates, using Arabidopsis eFP Browser (http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi), pinpointed two stresses among those tested for their impact on the Arabidopsis transcriptome: cold and oxidative stresses (Kilian et al., 2007) that appeared to favor the overexpression of CPK13. For both of these stresses, the integrative plant response is known to involve stomatal aperture control (McAinsh et al., 1996; Wilkinson et al., 2001; Kim et al., 2013), which, as suggested by the data presented here, could imply CPK13.
MATERIALS AND METHODS
Cloning of Shaker Channel Subunits (KAT1, KAT2, and AKT2) and of CPKs (CPK4 and CPK13)
Arabidopsis (Arabidopsis thaliana) KAT1, KAT2, AKT2, CPK4, and CPK13 full-length complementary DNAs (cDNAs) were amplified by PCR using the primers listed in Supplemental Table S2. PCR fragments with and without the stop codon were amplified with reverse primers RV-STOP and RV-END (for C-terminal fusion), respectively. The truncated constitutively active CPK13-CA was amplified with CPK13-CA-FW and CPK13-CA-RV primers (Supplemental Table S2). All PCR products were introduced into a Gateway pDONR entry vector (pDONR207) by recombination as described by the manufacturer (BP cloning; Invitrogen). The recombinant pDONR-based constructs were checked (by sequencing) before further transfer of target cDNA (by LR Gateway recombination) into one of the four following destination Gateway vectors: (1) pGEMGWC, to obtain subsequently (in vitro transcription) copy RNA (cRNA) transcripts for heterologous expression in oocytes; (2) pDEST15, for recombinant protein production; (3) pGWB523 (Nakagawa et al., 2007), to obtain transgenic overexpressing plants; and (4) pEarleyGate (Earley et al., 2006), for subcellular localization and in planta interaction (FRET-FLIM) studies.
Mutagenesis
Point mutations (G2A and D178A) were generated with the Stratagene QuickChange Site-Directed Mutagenesis Kit. Briefly, the plasmid pGEMGWC-CPK13 was amplified with the primers FW and RV harboring the mutation (for primer sequences, see Supplemental Table S2). The amplified plasmids were digested by DpnI to cut only the original plasmid (that does not contain the mutation). Bacteria were transformed with the mutated plasmid. Each point mutation was checked and confirmed by sequencing.
CPK13 Promoter Activity Using the GUS Reporter Gene
The promoter region of CPK13 (2291 bp upstream of the start codon) was cloned into pDONR207 using the Gateway method as described above. The promoter was amplified from genomic DNA with the primers pCPK13-FW and pCPK13-RV (for primer sequences, see Supplemental Table S2). The promoter sequence was checked by sequencing and then transferred into the vector pGWB533 (Nakagawa et al., 2007) by LR recombination for the GUS gene reporter study. The final plasmid was introduced in Agrobacterium tumefaciens (GV3101) by electroporation to transform Arabidopsis with the floral dip method (Clough and Bent, 1998). T2 homozygous transgenic plants were isolated with hygromycin (50 µg mL−1) in one-half-strength Murashige and Skoog medium supplemented with Suc (1%, w/v). To analyze the GUS expression pattern, 4-week-old plant leaves were incubated for 45 min in pre-fix solution (50 mm NaPO4, 1.5% [v/v] formaldehyde, and 0.05% [v/v] Triton X-100, pH 7) and then overnight at 37°C in GUS-fix solution (50 mm NaPO4, 0.5 mm ferricyanide, 0.5 mm ferrocyanide, 0.05% [v/v] Triton X100, and 1 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, pH 7), then washed in successive baths of 50%, 70%, 90%, and 100% (v/v) ethanol. Observations were made using a Color-View II camera (Olympus soft imaging system).
Production and Purification of Recombinant Proteins
Rosetta Escherichia coli strains were transformed with pDEST15-CPK13 or pDEST15-CPK4 plasmids that allow the production of recombinant CPK13 and CPK4 proteins upon induction by isopropyl β-d-1-thiogalactopyranoside. Bacteria were grown in 200 mL of Löwenstein-Jensen TB medium supplemented with ampicillin (50 µg mL−1) and chloramphenicol (30 µg mL−1). At 0.7 to 0.8 optical density at 600 nm, protein production was induced using 0.8 m isopropyl β-d-1-thiogalactopyranoside. After 12 h of culture at 20°C, bacteria were centrifuged. The bacterial pellet was then resuspended in phosphate-buffered saline (pH 7.3) with 0.5 mg mL−1 lysozyme (Roth) and antiprotease cocktail (Roche), and cells were lysed by sonication. After centrifugation at 12,000g at 4°C for 12 min, the bacterial lysate was incubated with 500 µL of Glutathione Sepharose 4B (GE Healthcare) at 4°C for 2.5 h. Beads were washed four times with phosphate-buffered saline buffer, and recombinant GST-CPKs were eluted with 20 mm reduced glutathione (Sigma) in 50 mm Tris-HCl (pH 8) following the manufacturer’s protocol (GE Healthcare).
In Vitro Kinase Assays
Kinase activities were monitored following the manufacturer’s procedure (Promega; Kinase-Glo Luminescent Kinase Assays). Recombinant CPK (500 ng) was incubated at 25°C with or without syntide-2 (25 µm) as a substrate in 50 µL of kinase buffer containing 25 µm ATP, 10 mm MgCl2, 2 mm dithiothreitol, 4 mm EGTA, and 50 mm HEPES. The pH of the buffer was adjusted to 6.5, 7.5, or 8.5 with NaOH. The total amounts of CaCl2 supplied in each buffer at different pH levels to get 10 µm free Ca2+ concentrations were estimated with the software Maxchelator (http://www.stanford.edu/~cpatton/downloads.htm). Incubations were realized on 96-well plates. The phosphorylation reaction was stopped after 2 and 15 min for CPK4 and CPK13, respectively, by adding 50 µL of Kinase-Glo reagent. Luminescent signals correlated to the ATP amount remaining in the solution were measured with a microplate reader (Xenuis; Safas). For each condition at different pH levels and with or without Ca2+, initial values (T0) corresponding to 25 µm ATP were estimated and subtracted from experiment values assayed after kinase reactions to obtain the consumed amount of ATP in each condition.
Obtaining Transgenic Plants Overexpressing CPK13
The pGWB523-CPK13 construct was introduced in A. tumefaciens (GV3101::pMP90) with CPK13-FW and CPK13-RV-END primers (Supplemental Table S2) and then used to transform the Arabidopsis Columbia-0 ecotype by the floral dip method (Clough and Bent, 1998). Transgenic plants overexpressing CPK13 were selected on hygromycin (50 µg mL−1) and checked by quantitative real-time (qRT)-PCR. Two homozygous CPK13-overexpressing lines (OE13#3 and OE13#4) of T2 plants and one heterozygous CPK13-overexpressing line (OE13#1) of T1 plants were used in our experiments.
qRT-PCR
Fifteen seedlings of each line (15 d old) were frozen in liquid N2 in 2-mL tubes containing one steel bead (2.5 mm diameter). Tissues were disrupted during 1 min at 30 s−1 in a Retsch mixer mill MM301 homogenizer (Retsch). RNA was extracted using the RNeasy-Plus mini kit (Qiagen) following the manufacturer’s procedure. Total RNA was quantified with nanodrop (ND 1000; Thermo). One microgram of total RNA was treated with DNase (RQ1 RNase free; Promega) before use as a template for cDNA synthesis with the GoScript Reverse Transcription System (Promega). qRT-PCR analysis was carried out with the Light-Cycler 480 (Roche) using Syber Green I master mix (Roche). The Light-Cycler experimental run protocol was as follows: denaturation (95°C for 5 min) followed by amplification and quantification repeated 45 times (95°C for 10 s, 60°C for 10 s, and 72°C for 10 s). Results were analyzed using the comparative critical threshold method (Pfaffl, 2001). TUB4 (for tubulin β-chain4) was used as a control gene. Primers used for qRT-PCR are listed in Supplemental Table S2.
Porometer Measurement
The stomatal conductance of 4-week-old plants was recorded with a porometer (AP4 Leaf Porometer; Delta-T Devices). Wild-type, OE13#3, OE13#4, and OE13#1 plants were grown in a chamber at 22°C with 70% relative humidity in short-day conditions (200 µE m−2 s−1 light for 8 h d−1). Before measurements (all made as blind), the porometer was calibrated in the growth chamber, and stomatal conductances were recorded at T0 and at 1, 2, 3, 6, and 8 h after light onset. Three to four mature leaves were measured on six plants per genotype. Data are expressed as means ± se (n = 22 leaves by genotype).
Stomatal Index
Stomatal index measurements (all made as blind) were realized from epidermal strips from abaxial leaf faces of 4-week-old plants (Hosy et al., 2003). Cell counts were taken from three different leaves of each genotype. Images of 10 different areas from each epidermal strip were captured with the light microscope at low magnification. The number of stomata and epidermal cells per square millimeter were counted using ImageJ freeware (Schneider et al., 2012) to calculate the stomatal index according to Royer (2001).
Stomatal Aperture Measurements
Epidermal strips were peeled from abaxial leaf faces of 4-week-old plants as described (Hosy et al., 2003) and incubated in 30 mm KCl and 10 mm MES-KOH, pH 6. For stomatal aperture, epidermal strips were kept in darkness to ensure stomatal closure. These strips were then exposed to white light (300 µE m−2 s−1), and stomatal apertures were measured (all measurements made as blind) at different times as indicated in Figures 2 and 3D (time 0 being the time of light onset). In a first set of experiments (Fig. 2), stomatal pore width values were measured with an optical microscope (Optiphot; Nikon) and a digitizing table (Houston Instrument), and data are expressed as means of at least 250 values from six plants for each of the three studied genotypes (the wild type, OE13#3, and OE13#4). To induce stomatal closure at 180 min under light, 10 µm ABA was added in the same buffer (30 mm KCl and 10 mm MES-KOH, pH 6). In a second set of experiments (Fig. 3D), stomatal pore width values were measured with ImageJ from photographs of epidermal strips taken at the indicated times (0, 1, and 3 h). The number of stomata measured at the different times was more than 650 from 10 wild-type plants, more than 600 from nine OE13#3 plants, more than 660 from nine OE13#4 plants, and more than 400 from six OE13#1 plants (during three independent experiments; all measurements done as blind). Data are expressed as means ± se.
Expression in Xenopus laevis Oocytes and Electrophysiological Recordings
Oocyte preparation, injection, and two-electrode voltage-clamp (TEVC) measurements were performed as described (Véry et al., 1995). In vitro transcriptions were performed using the mMESSAGE mMACHINE T7 Ultra kit (Ambion). X. laevis oocytes were either injected with 20 ng of KAT2 (or KAT1, KAT1 + KAT2, or AKT2) cRNA alone or coinjected with 20 ng of KAT2 (or KAT1, KAT1 + KAT2, or AKT2) cRNA and 20 ng of CPK13 (or CPK4) cRNA. Before recordings, oocytes were incubated for 48 h at 20°C in a solution containing 96 mm NaCl, 2 mm KCl, 1.8 mm CaCl2, 2 mm MgCl2, 2.5 mm sodium pyruvate, 5 mm HEPES-NaOH (pH 7.5), and 50 µg mL−1 gentamycin. Whole-cell currents were recorded using the TEVC technique with oocytes bathed in 10 mm KCl, 90 mm NaCl, 1 mm MgCl2, 1 mm CaCl2, and 10 mm HEPES-NaOH (pH 7.5). Three-second-long voltage steps to values ranging between +5 and −145 mV (recordings for KAT2) or between +10 and −140 mV (recordings for KAT1, KAT1 + KAT2, or AKT2) in −15-mV decrements were applied from a −40-mV holding potential.
Colocalization and Interaction (FRET-FLIM) Studies in Plant Tissues
Nicotiana benthamania leaves were transformed by agroinfiltration using A. tumefaciens (GV3101::pMP90) harboring different pEarleyGate recombinant plasmids (Earley et al., 2006): p101-CPK13 and p102-KAT2 were used to yield the expression of CPK13-YFP and KAT2-CFP, respectively. Coexpression of both CPK13-YFP and KAT2-CFP was obtained by coinfiltration with A. tumefaciens harboring p101-CPK13 and p102-KAT2. Leaves of 4- to 5-week-old N. benthamiana plants were infiltrated by bacterial suspensions (optical density at 600 nm = 0.5) in a buffer containing 10 mm MES-KOH (pH 5.8), 10 mm MgCl2, and 200 µm acetosyringone. Expression and localization of the proteins were checked 2 d after infiltration using a confocal microscope (LSM SP2 AOBS; Leica Microsystems) equipped with a 40× water-immersion objective and an argon ion laser for excitation at 458 and 514 nm. The emission range was set to 470 to 500 nm and 535 to 590 nm for CFP and YFP, respectively.
FLIM was performed using a Streak-FLIM system (Streakscope C4334; Hamamatsu Photonics) coupled to a high-sensitivity CCD camera (model C8800-53C; Hamamatsu). The light source (wavelength of 439 nm) was a pulsed diode laser working at 2 MHz (Hamamatsu). All images were acquired with a 60× oil-immersion objective (Plan Apo 1.4 numerical aperture; Infra-Red) mounted on an inverted microscope (Eclipse TE2000E; Nikon) coupled to the FLIM system through a band-pass filter for CFP. Fluorescence-decay images were analyzed with homemade software to calculate fluorescence lifetimes by fitting fluorescence decay curves with a biexponential function using the Levenberg-Marquardt nonlinear least-squares algorithm. For each condition, at least 20 measurements were made. Comparisons between control (KAT2-CFP) and coexpressed (KAT2-CFP + CPK13-YFP) assays were performed with Student’s t test. Results are expressed as means ± se of data from five independent biological experiments.
In Vitro Phosphorylation of Peptide Arrays
From the primary sequence of different target polypeptides, including KAT1, KAT2, and AKT2, we designed peptide arrays bearing 15 to 18 amino acid peptides. These peptides were synthesized and spotted (covalent link to a glass slide) by Proteomic Solutions. Whole sequences of KAT1 (GenBank/EBI accession no. U25088), KAT2 (GenBank/EBI accession no. AJ288900), and AKT2 (GenBank/EBI accession no. U40154) were covered through 52, 52, and 60 wild-type peptides, respectively (Supplemental Table S1), of the 384 peptides spotted in duplicate on each of these peptide arrays. In vitro phosphorylation of peptide arrays was performed using 5 µg of recombinant CPK13 or CPK4 in 5 mL of a kinase buffer (10 mm MgCl2, 2 mm dithiothreitol, 4 mm EGTA, and 50 mm HEPES-NaOH, pH 7.4) supplemented with 4.86 mm CaCl2. A concentration of 830 µm free Ca2+ was estimated with Maxchelator software (http://www.stanford.edu/~cpatton/downloads.htm). Phosphorylation reaction was started by adding 5 µCi of [γ-32P]ATP and lasted 1 h at 25°C upon slow agitation. Peptide arrays were then washed five times using 5 mL of washing solution (150 mm NaCl, 5 mm EDTA, 0.05% [v/v] Triton X-100, and 50 mm HEPES-NaOH, pH 7.4). Radiolabeling of phosphorylated peptides was revealed by autoradiography. Image analyses (quantification of phosphorylation) were done in ImageJ freeware (Schneider et al., 2012).
Accession numbers of sequences relevant for this article are as follows: At3g51850 (CPK13), At4g09570 (CPK4), At2g26650 (KAT1), At4g18290 (KAT2), At4g22200 (AKT2), and At5g44340 (TUB4).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Biochemical assays on recombinant CPK4.
Supplemental Figure S2. Quantification of on-chip phosphorylation by CPK13 of KAT2-, KAT1-, and AKT2-derived synthetic peptides.
Supplemental Figure S3. Quantification of on-chip phosphorylation by CPK4 of KAT2-, KAT1-, and AKT2-derived synthetic peptides.
Supplemental Figure S4. Inhibition of inward K+ currents by CPK13 in X. laevis oocytes expressing the KAT1 Shaker channel subunit.
Supplemental Figure S5. Inhibition of inward K+ currents by CPK13 in X. laevis oocytes expressing both the KAT1 and KAT2 Shaker channel subunits.
Supplemental Figure S6. Alignment of CPKs from Arabidopsis with TPK1 from yeast reveals a conserved active site.
Supplemental Table S1. Sequences of 153 wild-type peptides of KAT1, KAT2, and AKT2 featured by the peptide arrays used in this work.
Supplemental Table S2. List of primers used for cloning and qRT-PCR.
Acknowledgments
We thank Drs. Benoît Lacombe and Gabriel Krouk for helpful discussions of this work, Hugues Baudot for technical assistance in growing plants. Dr. Marie Boudsocq for advice and comments on CPKs, Dr. Tsuyoshi Nakagawa for the gift of pGWB vectors, and Dr. Isabel A. Lefevre for helpful comments on the article.
Glossary
- FRET
Förster resonance energy transfer
- FLIM
fluorescence lifetime imaging microscopy
- ABA
abscisic acid
- ER
endoplasmic reticulum
- cDNA
complementary DNA
- cRNA
copy RNA
- qRT
quantitative real-time
- TEVC
two-electrode voltage-clamp
Footnotes
This work was supported by the Agropolis Foundation (grant no. AA0803–022), by the European Commission Framework Programme 7 (grant no. 268393), and by the Institut National de la Recherche Agronomique and the Centre National de la Recherche Scientifique (to E.R.).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
References
- Acharya BR, Jeon BW, Zhang W, Assmann SM. (2013) Open Stomata 1 (OST1) is limiting in abscisic acid responses of Arabidopsis guard cells. New Phytol 200: 1049–1063 [DOI] [PubMed] [Google Scholar]
- Batistic O, Waadt R, Steinhorst L, Held K, Kudla J. (2010) CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J 61: 211–222 [DOI] [PubMed] [Google Scholar]
- Benetka W, Mehlmer N, Maurer-Stroh S, Sammer M, Koranda M, Neumüller R, Betschinger J, Knoblich JA, Teige M, Eisenhaber F. (2008) Experimental testing of predicted myristoylation targets involved in asymmetric cell division and calcium-dependent signalling. Cell Cycle 7: 3709–3719 [DOI] [PubMed] [Google Scholar]
- Berkowitz G, Zhang X, Mercie R, Leng Q, Lawton M. (2000) Co-expression of calcium-dependent protein kinase with the inward rectified guard cell K+ channel KAT1 alters current parameters in Xenopus laevis oocytes. Plant Cell Physiol 41: 785–790 [DOI] [PubMed] [Google Scholar]
- Blatt MR. (1991) Ion channel gating in plants: physiological implications and integration for stomatal function. J Membr Biol 124: 95–112 [DOI] [PubMed] [Google Scholar]
- Blatt MR. (1992) K+ channels of stomatal guard cells: characteristics of the inward rectifier and its control by pH. J Gen Physiol 99: 615–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatt MR. (2000) Ca2+ signalling and control of guard-cell volume in stomatal movements. Curr Opin Plant Biol 3: 196–204 [PubMed] [Google Scholar]
- Boudsocq M, Droillard MJ, Regad L, Laurière C. (2012) Characterization of Arabidopsis calcium-dependent protein kinases: activated or not by calcium? Biochem J 447: 291–299 [DOI] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, Bush J, Cheng SH, Sheen J. (2010) Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464: 418–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt B, Brodsky DE, Xue S, Negi J, Iba K, Kangasjärvi J, Ghassemian M, Stephan AB, Hu H, Schroeder JI. (2012) Reconstitution of abscisic acid activation of SLAC1 anion channel by CPK6 and OST1 kinases and branched ABI1 PP2C phosphatase action. Proc Natl Acad Sci USA 109: 10593–10598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wang QQ, Zhou L, Ren F, Li DD, Li XB. (2013) Arabidopsis CBL-interacting protein kinase (CIPK6) is involved in plant response to salt/osmotic stress and ABA. Mol Biol Rep 40: 4759–4767 [DOI] [PubMed] [Google Scholar]
- Cheng SH, Willmann MR, Chen HC, Sheen J. (2002) Calcium signaling through protein kinases: the Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol 129: 469–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, Romeis T. (2013) Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci USA 110: 8744–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629 [DOI] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Marten I, Ache P, Matschi S, Liese A, Wellmann C, Al-Rasheid KA, Grill E, et al. (2010) Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci USA 107: 8023–8028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs CS, Zoller MJ. (1991) Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J Biol Chem 266: 8923–8931 [PubMed] [Google Scholar]
- Grabov A, Blatt MR. (1999) A steep dependence of inward-rectifying potassium channels on cytosolic free calcium concentration increase evoked by hyperpolarization in guard cells. Plant Physiol 119: 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon AC, Gribskov M, Harper JF. (2000) CDPKs: a kinase for every Ca2+ signal? Trends Plant Sci 5: 154–159 [DOI] [PubMed] [Google Scholar]
- Harper JF, Breton G, Harmon A. (2004) Decoding Ca2+ signals through plant protein kinases. Annu Rev Plant Biol 55: 263–288 [DOI] [PubMed] [Google Scholar]
- Harper JF, Huang JF, Lloyd SJ. (1994) Genetic identification of an autoinhibitor in CDPK, a protein kinase with a calmodulin-like domain. Biochemistry 33: 7267–7277 [DOI] [PubMed] [Google Scholar]
- Hedrich R. (2012) Ion channels in plants. Physiol Rev 92: 1777–1811 [DOI] [PubMed] [Google Scholar]
- Held K, Pascaud F, Eckert C, Gajdanowicz P, Hashimoto K, Corratgé-Faillie C, Offenborn JN, Lacombe B, Dreyer I, Thibaud JB, et al. (2011) Calcium-dependent modulation and plasma membrane targeting of the AKT2 potassium channel by the CBL4/CIPK6 calcium sensor/protein kinase complex. Cell Res 21: 1116–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington AM. (2001) Guard cell signaling. Cell 107: 711–714 [DOI] [PubMed] [Google Scholar]
- Hetherington AM, Brownlee C. (2004) The generation of Ca2+ signals in plants. Annu Rev Plant Biol 55: 401–427 [DOI] [PubMed] [Google Scholar]
- Hosy E, Vavasseur A, Mouline K, Dreyer I, Gaymard F, Porée F, Boucherez J, Lebaudy A, Bouchez D, Very AA, et al. (2003) The Arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. Proc Natl Acad Sci USA 100: 5549–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu HC, Wang YY, Tsay YF. (2009) AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J 57: 264–278 [DOI] [PubMed] [Google Scholar]
- Hubbard KE, Siegel RS, Valerio G, Brandt B, Schroeder JI. (2012) Abscisic acid and CO2 signalling via calcium sensitivity priming in guard cells, new CDPK mutant phenotypes and a method for improved resolution of stomatal stimulus-response analyses. Ann Bot (Lond) 109: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imes D, Mumm P, Böhm J, Al-Rasheid KA, Marten I, Geiger D, Hedrich R. (2013) Open stomata 1 (OST1) kinase controls R-type anion channel QUAC1 in Arabidopsis guard cells. Plant J 74: 372–382 [DOI] [PubMed] [Google Scholar]
- Irving HR, Gehring CA, Parish RW. (1992) Changes in cytosolic pH and calcium of guard cells precede stomatal movements. Proc Natl Acad Sci USA 89: 1790–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MM, Hossain MA, Jannat R, Munemasa S, Nakamura Y, Mori IC, Murata Y. (2010) Cytosolic alkalization and cytosolic calcium oscillation in Arabidopsis guard cells response to ABA and MeJA. Plant Cell Physiol 51: 1721–1730 [DOI] [PubMed] [Google Scholar]
- Israelsson M, Siegel RS, Young J, Hashimoto M, Iba K, Schroeder JI. (2006) Guard cell ABA and CO2 signaling network updates and Ca2+ sensor priming hypothesis. Curr Opin Plant Biol 9: 654–663 [DOI] [PubMed] [Google Scholar]
- Jeanguenin L, Lebaudy A, Xicluna J, Alcon C, Hosy E, Duby G, Michard E, Lacombe B, Dreyer I, Thibaud JB. (2008) Heteromerization of Arabidopsis Kv channel alpha-subunits: data and prospects. Plant Signal Behav 3: 622–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchiswamy CN, Takahashi H, Quadro S, Maffei ME, Bossi S, Bertea C, Zebelo SA, Muroi A, Ishihama N, Yoshioka H, et al. (2010) Regulation of Arabidopsis defense responses against Spodoptera littoralis by CPK-mediated calcium signaling. BMC Plant Biol 10: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kim HS, Oh JM, Luan S, Carlson JE, Ahn SJ. (2013) Cold stress causes rapid but differential changes in properties of plasma membrane H+-ATPase of camelina and rapeseed. J Plant Physiol 170: 828–837 [DOI] [PubMed] [Google Scholar]
- Kwak JM, Murata Y, Baizabal-Aguirre VM, Merrill J, Wang M, Kemper A, Hawke SD, Tallman G, Schroeder JI. (2001) Dominant negative guard cell K+ channel mutants reduce inward-rectifying K+ currents and light-induced stomatal opening in Arabidopsis. Plant Physiol 127: 473–485 [PMC free article] [PubMed] [Google Scholar]
- Laanemets K, Brandt B, Li J, Merilo E, Wang YF, Keshwani MM, Taylor SS, Kollist H, Schroeder JI. (2013) Calcium-dependent and -independent stomatal signaling network and compensatory feedback control of stomatal opening via Ca2+ sensitivity priming. Plant Physiol 163: 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan WZ, Lee SC, Che YF, Jiang YQ, Luan S. (2011) Mechanistic analysis of AKT1 regulation by the CBL-CIPK-PP2CA interactions. Mol Plant 4: 527–536 [DOI] [PubMed] [Google Scholar]
- Lebaudy A, Hosy E, Simonneau T, Sentenac H, Thibaud JB, Dreyer I. (2008a) Heteromeric K+ channels in plants. Plant J 54: 1076–1082 [DOI] [PubMed] [Google Scholar]
- Lebaudy A, Pascaud F, Véry AA, Alcon C, Dreyer I, Thibaud JB, Lacombe B. (2010) Preferential KAT1-KAT2 heteromerization determines inward K+ current properties in Arabidopsis guard cells. J Biol Chem 285: 6265–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebaudy A, Vavasseur A, Hosy E, Dreyer I, Leonhardt N, Thibaud JB, Véry AA, Simonneau T, Sentenac H. (2008b) Plant adaptation to fluctuating environment and biomass production are strongly dependent on guard cell potassium channels. Proc Natl Acad Sci USA 105: 5271–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebaudy A, Véry AA, Sentenac H. (2007) K+ channel activity in plants: genes, regulations and functions. FEBS Lett 581: 2357–2366 [DOI] [PubMed] [Google Scholar]
- Leonhardt N, Kwak JM, Robert N, Waner D, Leonhardt G, Schroeder JI. (2004) Microarray expression analyses of Arabidopsis guard cells and isolation of a recessive abscisic acid hypersensitive protein phosphatase 2C mutant. Plant Cell 16: 596–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Lee YR, Assmann SM. (1998) Guard cells possess a calcium-dependent protein kinase that phosphorylates the KAT1 potassium channel. Plant Physiol 116: 785–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liese A, Romeis T. (2013) Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim Biophys Acta 1833: 1582–1589 [DOI] [PubMed] [Google Scholar]
- Liu K, Fu H, Bei Q, Luan S. (2000) Inward potassium channel in guard cells as a target for polyamine regulation of stomatal movements. Plant Physiol 124: 1315–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu SX, Hrabak EM. (2013) The myristoylated amino-terminus of an Arabidopsis calcium-dependent protein kinase mediates plasma membrane localization. Plant Mol Biol 82: 267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S. (2009) The CBL-CIPK network in plant calcium signaling. Trends Plant Sci 14: 37–42 [DOI] [PubMed] [Google Scholar]
- Luan S, Li W, Rusnak F, Assmann SM, Schreiber SL. (1993) Immunosuppressants implicate protein phosphatase regulation of K+ channels in guard cells. Proc Natl Acad Sci USA 90: 2202–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRobbie EA. (1993) Ca2+ and cell signalling in guard cells. Semin Cell Biol 4: 113–122 [DOI] [PubMed] [Google Scholar]
- MacRobbie EA. (1998) Signal transduction and ion channels in guard cells. Philos Trans R Soc Lond B Biol Sci 353: 1475–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marten H, Konrad KR, Dietrich P, Roelfsema MR, Hedrich R. (2007) Ca2+-dependent and -independent abscisic acid activation of plasma membrane anion channels in guard cells of Nicotiana tabacum. Plant Physiol 143: 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín ML, Busconi L. (2000) Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitoylation. Plant J 24: 429–435 [DOI] [PubMed] [Google Scholar]
- McAinsh MR, Clayton H, Mansfield TA, Hetherington AM. (1996) Changes in stomatal behavior and guard cell cytosolic free calcium in response to oxidative stress. Plant Physiol 111: 1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori IC, Murata Y, Yang Y, Munemasa S, Wang YF, Andreoli S, Tiriac H, Alonso JM, Harper JF, Ecker JR, et al. (2006) CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol 4: e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemasa S, Hossain MA, Nakamura Y, Mori IC, Murata Y. (2011) The Arabidopsis calcium-dependent protein kinase, CPK6, functions as a positive regulator of methyl jasmonate signaling in guard cells. Plant Physiol 155: 553–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J. (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. (2007) Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Nieves-Cordones M, Chavanieu A, Jeanguenin L, Alcon C, Szponarski W, Estaran S, Chérel I, Zimmermann S, Sentenac H, Gaillard I. (2014) Distinct amino acids in the C-linker domain of the Arabidopsis K+ channel KAT2 determine its subcellular localization and activity at the plasma membrane. Plant Physiol 164: 1415–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilson SE, Assmann SM. (2007) The control of transpiration: insights from Arabidopsis. Plant Physiol 143: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Zhang W, Assmann SM. (2007) Roles of ion channels and transporters in guard cell signal transduction. FEBS Lett 581: 2325–2336 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilot G, Lacombe B, Gaymard F, Chérel I, Boucherez J, Thibaud JB, Sentenac H. (2001) Guard cell inward K+ channel activity in Arabidopsis involves expression of the twin channel subunits KAT1 and KAT2. J Biol Chem 276: 3215–3221 [DOI] [PubMed] [Google Scholar]
- Royer DL. (2001) Stomatal density and stomatal index as indicators of paleoatmospheric CO2 concentration. Rev Palaeobot Palynol 114: 1–28 [DOI] [PubMed] [Google Scholar]
- Scherzer S, Maierhofer T, Al-Rasheid KA, Geiger D, Hedrich R. (2012) Multiple calcium-dependent kinases modulate ABA-activated guard cell anion channels. Mol Plant 5: 1409–1412 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder JI, Allen GJ, Hugouvieux V, Kwak JM, Waner D. (2001) Guard cell signal transduction. Annu Rev Plant Physiol Plant Mol Biol 52: 627–658 [DOI] [PubMed] [Google Scholar]
- Suhita D, Raghavendra AS, Kwak JM, Vavasseur A. (2004) Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate- and abscisic acid-induced stomatal closure. Plant Physiol 134: 1536–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyroki A, Ivashikina N, Dietrich P, Roelfsema MR, Ache P, Reintanz B, Deeken R, Godde M, Felle H, Steinmeyer R, et al. (2001) KAT1 is not essential for stomatal opening. Proc Natl Acad Sci USA 98: 2917–2921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel G, MacRobbie EA, Blatt MR. (1992) Membrane transport in stomatal guard cells: the importance of voltage control. J Membr Biol 126: 1–18 [DOI] [PubMed] [Google Scholar]
- Véry AA, Gaymard F, Bosseux C, Sentenac H, Thibaud JB. (1995) Expression of a cloned plant K+ channel in Xenopus oocytes: analysis of macroscopic currents. Plant J 7: 321–332 [DOI] [PubMed] [Google Scholar]
- Véry AA, Sentenac H. (2002) Cation channels in the Arabidopsis plasma membrane. Trends Plant Sci 7: 168–175 [DOI] [PubMed] [Google Scholar]
- Véry AA, Sentenac H. (2003) Molecular mechanisms and regulation of K+ transport in higher plants. Annu Rev Plant Biol 54: 575–603 [DOI] [PubMed] [Google Scholar]
- Wang XQ, Ullah H, Jones AM, Assmann SM. (2001) G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science 292: 2070–2072 [DOI] [PubMed] [Google Scholar]
- Webb AA, Larman MG, Montgomery LT, Taylor JE, Hetherington AM. (2001) The role of calcium in ABA-induced gene expression and stomatal movements. Plant J 26: 351–362 [DOI] [PubMed] [Google Scholar]
- Weinl S, Kudla J. (2009) The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol 184: 517–528 [DOI] [PubMed] [Google Scholar]
- Wilkinson S, Clephan AL, Davies WJ. (2001) Rapid low temperature-induced stomatal closure occurs in cold-tolerant Commelina communis leaves but not in cold-sensitive tobacco leaves, via a mechanism that involves apoplastic calcium but not abscisic acid. Plant Physiol 126: 1566–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xicluna J, Lacombe B, Dreyer I, Alcon C, Jeanguenin L, Sentenac H, Thibaud JB, Chérel I. (2007) Increased functional diversity of plant K+ channels by preferential heteromerization of the shaker-like subunits AKT2 and KAT2. J Biol Chem 282: 486–494 [DOI] [PubMed] [Google Scholar]
- Xu J, Li HD, Chen LQ, Wang Y, Liu LL, He L, Wu WH. (2006) A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125: 1347–1360 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zhang W, Stanley BA, Assmann SM. (2008) Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell 20: 3210–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu SY, Yu XC, Wang XJ, Zhao R, Li Y, Fan RC, Shang Y, Du SY, Wang XF, Wu FQ, et al. (2007) Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]