Appropriate complex formation at the plasma membrane is indispensable for a negative regulator of immune signaling in rice.
Abstract
Oryza sativa Pto-interacting protein 1a (OsPti1a), an ortholog of tomato (Solanum lycopersicum) SlPti1, functions as a negative regulator of innate immunity in rice (Oryza sativa). In ospti1a mutants, the activation of immune responses, including hypersensitive response-like cell death, is caused by loss of the OsPti1a protein; however, it is as yet unclear how OsPti1a suppresses immune responses. Here, we report that OsPti1a localizes to detergent-resistant membrane fractions of the plasma membrane through lipid modification of the protein’s amino terminus, which is highly conserved among Pti1 orthologs in several plant species. Importantly, mislocalization of OsPti1a after deletion of its amino terminus reduced its ability to complement the mutant phenotypes, including hypersensitive response-like cell death. Furthermore, complex formation of OsPti1a depends on its amino terminus-mediated membrane localization. Liquid chromatography-tandem mass spectrometry analysis of OsPti1a complex-interacting proteins identified several defense-related proteins. Collectively, these findings indicate that appropriate complex formation by OsPti1a at the plasma membrane is required for the negative regulation of plant immune responses in rice.
Plants are continuously exposed to attack by microorganisms under natural conditions. To protect themselves against pathogen challenges, plants have evolved two different layers of defense. The first layer is microbe-associated molecular pattern (MAMP)-triggered immunity (MTI). MAMPs are recognized by plant cells through plasma membrane receptors (pattern recognition receptors; Nürnberger and Kemmerling, 2006); however, compatible pathogens are able to evade the MTI response through the action of pathogen-encoded effectors that can suppress this line of defense. In turn, plants have evolved a second layer of plant defense through the recognition of specific effectors, called effector-triggered immunity (ETI), that is mediated by plant resistance proteins (Jones and Dangl, 2006). ETI is a more rapid and powerful response than MTI and is typically accompanied by rapid programmed cell death known as the hypersensitive response (Coll et al., 2011). Plant immune responses include ion fluxes across plasma membranes, oxidative bursts, activation of mitogen-activated protein kinase cascades, and transcriptional reprogramming of defense genes. Most of these responses are observed in both MTI and ETI, suggesting that plants employ common signaling machinery in response to these different stimuli (Navarro et al., 2004; Zipfel et al., 2006; Qi et al., 2011); however, the downstream signaling components after the initial recognition of pathogens by plant receptors are still not fully characterized.
Previously, we reported that knockout mutations of the Oryza sativa Pto-interacting protein 1a (OsPti1a) gene, which encodes a Ser/Thr protein kinase, activate a series of defense responses in rice (Oryza sativa) accompanied by hypersensitive response-like lesion formation over the entire leaf surface in the absence of pathogen challenge (Takahashi et al., 2007). We also reported that overexpression of OsPti1a results in enhanced susceptibility against compatible pathogens and reduces resistance against an incompatible race of rice blast fungus (Takahashi et al., 2007). These results suggest that OsPti1a functions as a negative regulator of both MTI and ETI in rice (Takahashi et al., 2007). Furthermore, suppression of OsRAR1 (required for mildew resistance locus A12 [Mla12] resistance), a rice homolog of Aradopsis thaliana (At) RAR1 and Hordeum vulgare (Hv) RAR1 (Shirasu et al., 1999; Muskett et al., 2002), in ospti1a mutants abolished unregulated lesion formation, the expression of pathogenesis-related genes, and enhanced basal resistance, suggesting that constitutive activation of defense responses in the ospti1a mutant is attributed to OsRAR1-dependent signaling. Additionally, OsPti1a-mediated basal defense is regulated by the phosphorylation of a conserved Thr through the upstream kinases rice oxidative signal-inducible1 (OsOxi1) and rice phosphoinositide-dependent protein kinase1 (OsPdk1; Matsui et al., 2010a, 2010b); however, it is still not clear how OsPti1a negatively regulates signaling to activate defense responses.
Pti1 proteins are highly conserved in many plant species, including tomato (Solanum lycopersicum), soybean (Glycine max), maize (Zea mays), rice, and Arabidopsis (Arabidopsis thaliana; Herrmann et al., 2006). Phylogenetic analysis revealed that Pti1 family proteins can be divided into three subgroups among angiosperms (I–III). Each subfamily has a highly conserved N-terminal domain with a specific consensus sequence. In these domains, myristoylation and/or palmitoylation sites were predicted. ZmPti1a in maize was reported to be localized to the plasma membrane, owing to its N-terminal myristoylation and/or S-acylation (Herrmann et al., 2006). Palmitoylation and myristoylation, the addition of fatty acids to specific sites on proteins, are well-known posttranslational modifications required for membrane binding or for complex formation (Hemsley and Grierson, 2008). These posttranslational modifications have important roles for a wide range of cellular processes in plants, such as salt tolerance, plant viability, and plant immunity (Ishitani et al., 2000; Kim et al., 2005; Pierre et al., 2007; Takemoto et al., 2012). The fact that the N-terminal domains of Pti1 homologs are highly conserved suggests that lipid modification of these domains could be related to Pti1 function; however, the biological significance of these modifications is still unknown.
In this article, we demonstrate that OsPti1a localizes to the plasma membrane and that N-terminal conserved palmitoylation sites are necessary for this localization. Deletion of the N terminus of OsPti1a leads to the aberrant activation of immune responses. We also demonstrate that at the plasma membrane, OsPti1a forms a protein complex and that the N terminus is indispensable for wild-type complex formation. We identified several defense-related proteins as candidates of the OsPti1a complex at the plasma membrane. These results indicate that appropriate complex formation at the plasma membrane is indispensable for OsPti1a function as a negative regulator of immune signaling in rice.
RESULTS
OsPti1a Is Restricted to the Plasma Membrane
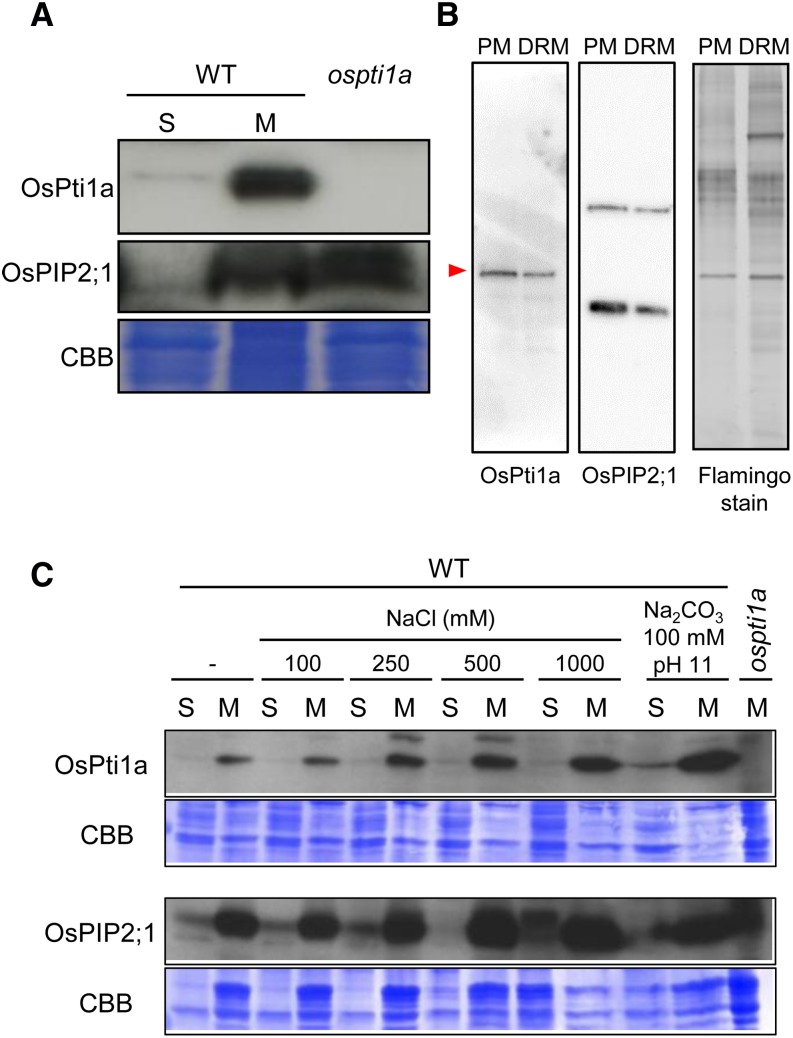
To elucidate the biological and physiological roles of OsPti1a in vivo, we investigated the subcellular localization of OsPti1a protein by cellular fractionation of rice suspension culture cells. An OsPti1a-specific antibody detected a strong signal in microsomal fractions compared with that in the soluble fractions (Fig. 1A). Although the microsomal fraction consists largely of microsomes, the microsomal fraction also frequently contains free ribosomes and fragments of the plasma membrane, the Golgi apparatus, mitochondria, and other subcellular structures (Boyes et al., 1998). To characterize OsPti1a localization in greater detail, we conducted an additional two-phase separation procedure that revealed that OsPti1a mainly localizes to the inner plasma membrane and exists in detergent-resistant membrane (DRM) fractions (Fig. 1B). Peripheral proteins are released from membranes under high ionic strength conditions or a pH change. To investigate how OsPti1a binds to the plasma membrane, we fractionated suspension-cultured cells after treatment with high salt (1 m NaCl) and high pH (Na2CO3, pH 11); however, these treatments did not remove the OsPti1a protein from the microsomal fraction (Fig. 1C). These data suggest that OsPti1a binds strongly to the plasma membrane and localizes preferentially in the DRM fractions.
Figure 1.
OsPti1a localizes with the microsomal fraction. A, Distribution of OsPti1a protein. Total protein derived from wild-type (WT) rice suspension-cultured cells was fractionated into soluble (S) and microsomal (M) samples by ultracentrifugation. The lysates were run on SDS-PAGE and subjected to immunoblot analysis with anti-OsPti1a antibody. Total protein of ospti1a mutants was used as a negative control. OsPIP2;1 indicates control integral membrane protein. B, Immunoblot analysis using an anti-OsPti1a antibody to probe the plasma membrane (PM) fraction and the DRM fraction proteins. The red arrowhead indicates the band of OsPti1a protein. OsPIP2;1 is the control integral membrane protein. Flamingo-stained SDS-PAGE of the two-phase partitioned plasma membrane and purified DRM fractions is shown at right. C, Top gels are immunoblots of soluble and microsomal fractions derived from wild-type total microsomal fraction after treatment with the indicated buffer. The microsomal fraction of the ospti1a mutant is a negative control. Bottom gels show Coomassie Brilliant Blue (CBB) staining of membrane proteins after immunoblotting.
The N-Terminal Domain of OsPti1a Is Required for Plasma Membrane Localization
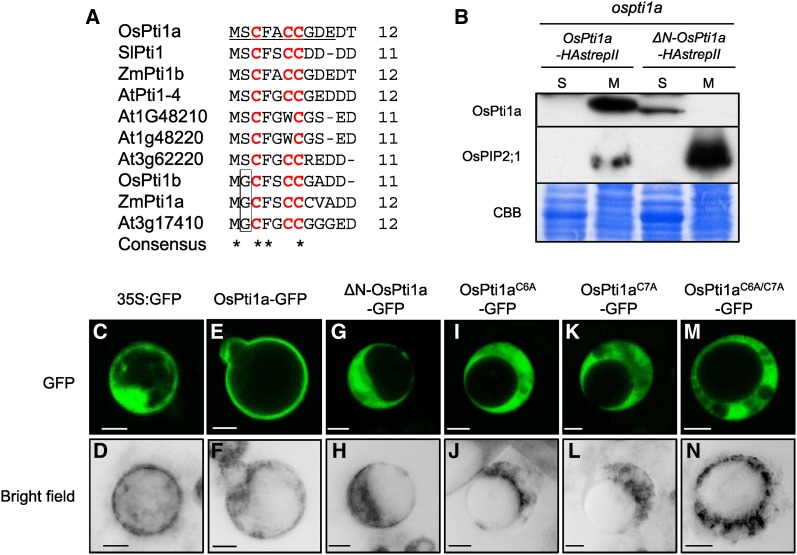
Since OsPti1a does not contain a transmembrane domain, membrane localization probably depends on interaction with other membrane-associated protein(s) or by anchoring directly to the membrane through a posttranslational modification. Phylogenetic analysis revealed that OsPti1a belongs to the group II Pti1 family that has potential lipid modification sites within the N-terminal region (Fig. 2A; Herrmann et al., 2006). To test if the conserved N-terminal domain is required for localization, we produced transgenic suspension-cultured cells expressing OsPti1a whose 10 N-terminal amino acids were deleted (ΔN-OsPti1a) in the ospti1a mutant background. To confirm whether tagged OsPti1a is functional, we produced transgenic ospti1a homozygous mutant plants expressing N-terminal or C-terminal hemagglutinin-strepII (HAstrepII)-tagged OsPti1a. Although transgenic plants expressing C-terminal HAstrepII-tagged OsPti1a were phenotypically similar to wild-type plants (Supplemental Fig. S1A), indicating that C-terminally tagged OsPti1a (OsPti1a-HAstrepII) is functional, the expression of N-terminally tagged OsPti1a (HAstrepII-OsPti1a) did not complement the ospti1a mutant phenotype, including dwarfism and aberrant lesion formation (Supplemental Fig. S1B). This result suggests that the N terminus is important for OsPti1a function. Therefore, we decided to employ the C-terminally tagged OsPti1a in further analyses. Figure 2B shows that OsPti1a-HAstrepII was mostly detected in the microsomal fraction like the endogenous OsPti1a protein. On the other hand, the ΔN-OsPti1a-HAstrepII protein was detected in the soluble fraction and not in the microsomal fractions. Similar results were obtained using Agrobacterium tumefaciens-mediated transient expression in Nicotiana benthamiana (Supplemental Fig. S2A). These results indicate that the N-terminal domain of OsPti1a is essential for localization to the membrane.
Figure 2.
The N-terminal domain of OsPti1a is essential for protein localization in the microsomal fraction. A, Alignment of N-terminal amino acid sequences of OsPti1a homologs that belong to the group II Pti1 family. Underlined letters indicate amino acids of OsPti1a that were deleted for the construction of ΔN-OsPti1a. Red letters indicate putative palmitoylation sites in Pti1 family proteins. The box delimits putative myristorylation sites. B, ΔN-OsPti1a-HAstrepII protein is localized in the cytosolic fraction. Immunoblots show soluble (S) and microsomal (M) fractions derived from cultured cells expressing OsPti1a-HAstrepII or ΔN-OsPti1a-HAstrepII in the ospti1a mutant background. Cellular fractionation was confirmed by immunoblotting using anti-OsPIP2;1 antibody as a control for integral membrane proteins. CBB, Coomassie Brilliant Blue. C to N, Free GFP (C and D), OsPti1aWT-GFP (E and F), ΔN-OsPti1a-GFP (G and H), OsPti1aC6A-GFP (I and J), OsPti1aC7A-GFP (K and L), and OsPti1aC6A/C7A-GFP (M and N) were transiently expressed under the control of the cauliflower mosaic virus (CaMV) 35S promoter in rice protoplasts. The top row shows the green fluorescence of GFP-tagged proteins visualized with a confocal laser scanning microscope, and the bottom row shows bright-field images. Bars = 10 µm.
Palmitoylation Sites Cys-6 and Cys-7 Are Necessary for OsPti1a Localization to the Plasma Membrane
Alignment of the N-terminal sequences of group II Pti1 orthologs indicated that OsPti1a possesses conserved putative palmitoylation sites at Cys-3, Cys-6, and Cys-7, as predicted by the CSS-Palm program (Ren et al., 2008; Fig. 2A). Since Cys-3 had a low prediction score compared with the other two Cys residues using CSS-Palm version 4.0, we focused on Cys-6 and Cys-7. To test whether palmitoylation of OsPti1a is required for membrane binding, we analyzed the effect of amino acid substitutions in the N-terminal sequences of OsPti1a using agroinfiltration of N. benthamiana (Supplemental Fig. S2, B and C). OsPti1aC6A and OsPti1aC7A mutant proteins mainly accumulated in the soluble fraction, although OsPti1aWT protein was detected in the microsomal fraction. Furthermore, accumulation of the OsPti1aC6A/C7A mutated protein was observed completely in the soluble fraction. This result indicates that residues Cys-6 and Cys-7 are required for OsPti1a membrane binding. To confirm the importance of Cys-6 and Cys-7 of OsPti1a for binding to rice membranes, OsPti1a variants fused to GFP were expressed in rice protoplasts (Fig. 2, C–N). As expected, OsPti1a-GFP clearly localized to the membrane, whereas ΔN-OsPti1a-GFP, OsPti1aC6A-GFP, OsPti1aC7A-GFP, and OsPti1aC6A/C7A-GFP were not membrane localized and were observed in the cytosol. These results indicate that OsPti1a localization to the plasma membrane, which contains DRM, is dependent on the putative palmitoylation of N-terminal Cys-6 and Cys-7.
The N-Terminal Domain of OsPti1a Has a Critical Role in Suppressing the Lesion-Mimic Phenotype of ospti1a
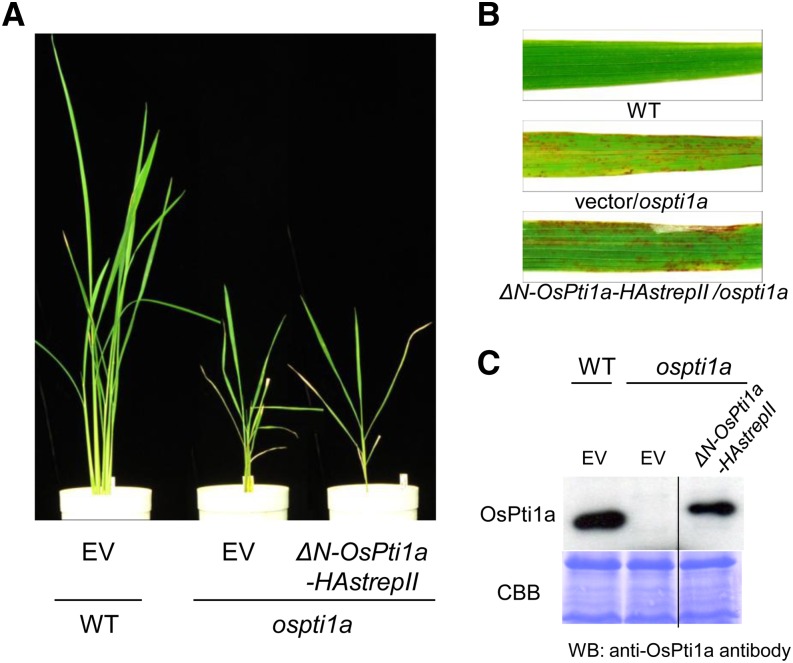
To investigate the biological function of membrane localization mediated by the N-terminal domain of OsPti1a, we generated transgenic plants expressing ΔN-OsPti1a-HAstrepII in the ospti1a mutant background. As expected, ΔN-OsPti1a-HAstrepII transgenic plants did not complement lesion formation or the dwarf phenotype of ospti1a mutants in T0 transgenic plants (Fig. 3, A and B), despite accumulation of the OsPti1a protein (Fig. 3C). Consistent results were observed in at least six independent transgenic lines. These results indicated that the N-terminal domain of OsPti1a is essential for OsPti1a function in negatively regulating defense signaling in rice. Taken together, we conclude that the localization of OsPti1a at the rice plasma membrane through its N-terminal domain is indispensable for OsPti1a function in immunity signal regulation.
Figure 3.
Expression of ΔN-OsPti1a-HAstrepII does not complement the mutant phenotypes of ospti1a mutant plants. A, Phenotypes of 8-week-old transgenic ospti1a plants transformed with either ΔN-OsPti1a-HAstrepII or an empty vector (EV). B, Lesion phenotypes of transgenic plants. Plants were photographed 8 weeks after sowing. C, Accumulation of ΔN-OsPti1a protein was detected by immunoblots using the anti-OsPti1a antibody. The bottom gel shows Coomassie Brilliant Blue (CBB) staining of the membrane after immunoblotting. WT, Wild type. WB, western blotting.
OsPti1a Forms a Protein Complex at the Plasma Membrane
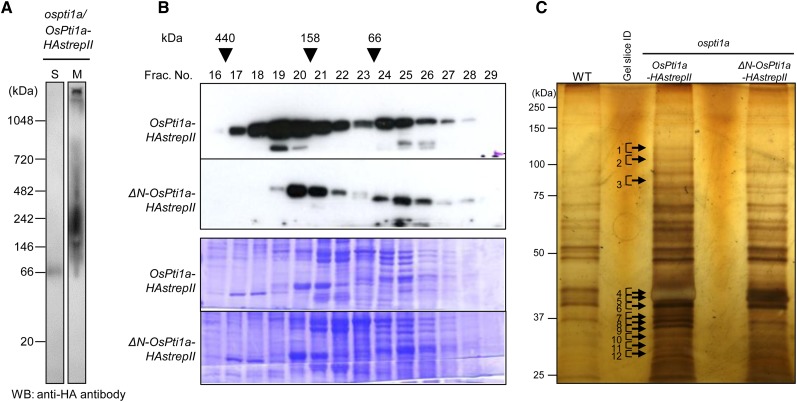
DRMs are known to be involved in various signaling processes. In Arabidopsis, several membrane-associated proteins important for plant immunity (e.g. Receptor-like kinases and plasma membrane-H+-ATPases) have been reported to be localized in DRMs (Liu et al., 2009b; Keinath et al., 2010). The fact that OsPti1a could be purified with the DRM fraction raised the hypothesis that OsPti1a potentially regulates defense signaling through interaction with such proteins in DRMs. To inspect the possible association of OsPti1a with other signaling components at the plasma membrane, we examined whether OsPti1a forms protein complexes using blue native-PAGE. As shown in Figure 4A, the OsPti1a-HAstrepII protein was detected in the microsomal fraction of the OsPti1a-HAstrepII-overexpressing cells by the anti-hemagglutinin (anti-HA) antibody at a wide range of electrophoretic mobilities ranging from 146 to over 720 kD. In contrast, the OsPti1a-HAstrepII protein was mainly detected as a monomer in the soluble fraction. These results suggest that OsPti1a forms protein complexes at the plasma membrane. When the anti-OsPti1a antibody that was raised against the N-terminal sequence (residues 11–24) of OsPti1a was used for the detection, no signal was observed (data not shown). This result may indicate that the OsPti1a N-terminal region is masked by components of the OsPti1a complexes under native conditions (Takahashi et al., 2007). To confirm whether membrane localization is required for complex formation, we investigated OsPti1a protein complex formation by size-exclusion chromatography using transgenic cultured cells expressing OsPti1a-HAstrepII or ΔN-OsPti1a-HAstrepII in the ospti1a background. The majority of OsPti1a protein was found to be broadly distributed from about 440 kD to less than 66 kD (fractions 17–28) in ospti1a/OsPti1a-HAstrepII cells, with the signal peak present in fraction 19 (Fig. 4B). On the other hand, signals of ΔN-OsPti1a shifted to a lower size (fractions 19–28) with the signal peak detected in fraction 20, presumably because ΔN-OsPti1a forms an incomplete protein complex (or complexes) nonidentical to the complex formed by intact OsPti1a. These results indicate that membrane localization of OsPti1a is necessary for endogenous, wild-type OsPti1a complex formation.
Figure 4.
OsPti1a forms a protein complex in the membrane fraction. A, Blue native-PAGE was performed using soluble (S) and microsomal (M) fractions derived from cultured rice cells expressing OsPti1a-HAstrepII. OsPti1a protein was detected using an anti-HA antibody. WB, Western blotting. B, Gel filtration fractions of protein extracts from rice cultured cells expressing OsPti1a-HAstrepII or ΔN-OsPti1a-HAstrepII were subjected to immunoblot analyses (top gel) using an anti-HA antibody. The fraction numbers and molecular masses (kD) are indicated at the top. The membrane after immunoblotting was stained with CBB. C, Biochemical purification and identification of OsPti1a-associated factors. OsPti1a protein complexes were affinity purified. The gels were silver stained for protein visualization. The arrows indicate OsPti1a-specific bands in ospti1a/OsPti1a-HAstrepII cultured cells. WT, Wild type.
Since OsPti1a is involved in MTI signaling, changes in the OsPti1a complex may have an important role in regulating MTI responses. To investigate whether the size of the OsPti1a complex changes in response to MAMP treatment, we carried out gel filtration analyses after treatment with chitin (Supplemental Fig. S3); however, the size of the complex was not observed to change in response to chitin treatment even after 180 min. This result suggests that OsPti1a forms a stable complex that is attached to the membrane fraction and functions as a negative regulator by interaction with complex-associated proteins.
Membrane-Localized OsPti1a Forms Complexes with Rice Homologs of Reported Plant Immune Regulators
In order to understand the significance of complex formation at the plasma membrane for plant immune system regulation, we characterized the components in the membrane-localized OsPti1a complexes. OsPti1a-HAstrepII and ΔN-OsPti1a-HAstrepII were affinity purified from transgenic cell lines, and associated proteins were identified by liquid chromatography (LC)-tandem mass spectrometry (MS/MS) analysis. Untransformed O. sativa cv Nipponbare cell suspension cultures (wild type) were used as a negative control. As shown in Figure 4C, we were able to detect protein bands that were specifically associated with OsPti1a-HAstrepII but not with ΔN-OsPti1a-HAstrepII and analyzed these bands to identify proteins involved in the membrane-localized OsPti1a complexes. Using this approach, we identified 39 proteins that potentially form complexes with OsPti1a at the plasma membrane (Table I). Importantly, many of these proteins, such as OSA2 (for Oryza sativa PM H+-ATPase isoform2), OSA7, vacuolar ATP synthase subunit d, and Phytochrome-Associated Phosphatase Type 2C (PAPP2C), are suggested to be membrane-localized proteins from analyses of homologous proteins in other plant species (Schumacher, 2006; Fujiwara et al., 2009; Liu et al., 2009b; Wang et al., 2012). Moreover, Arabidopsis homologs of OSA7, jacalin domain protein, and PAPP2C have been reported to play important roles in plant immunity (Liu et al., 2009a, 2009b; Wang et al., 2012). These results suggest that plasma membrane localization of OsPti1a is important for complex formation with certain proteins to regulate plant immunity.
Table I. Proteins that copurified with OsPti1a.
Proteins with at least two unique peptides were regarded as confident identifications.
| No. of Identified Unique Peptides |
||||||
|---|---|---|---|---|---|---|
| Gel Slice Identifier | Accession No. | Protein Function | Mass | Wild Type | OsPti1a | ΔN-OsPti1a |
| kD | ||||||
| 1 | LOC_Os04g56160.1 | OSA2 | 105 | 0 | 6 | 0 |
| LOC_Os07g09340.1 | OSA7 | 105 | 0 | 6 | 0 | |
| LOC_Os11g07470.1 | Expressed protein | 127 | 0 | 7 | 6 | |
| 2 | LOC_Os04g56160.1 | OSA2 | 105 | 0 | 2 | 0 |
| LOC_Os05g35460.1 | Patellin protein | 64 | 0 | 3 | 0 | |
| LOC_Os01g08560.1 | DnaK family protein | 93 | 0 | 5 | 2 | |
| LOC_Os01g13430.1 | Importin-α reexporter | 108 | 0 | 3 | 3 | |
| LOC_Os10g10244.1 | Alanyl-tRNA synthetase | 109 | 0 | 6 | 3 | |
| 3 | LOC_Os01g61920.1 | Core histone H2A/H2B/H3/H4 domain-containing protein | 11 | 0 | 3 | 0 |
| LOC_Os01g64640.1 | Histone H3 | 15 | 0 | 2 | 0 | |
| LOC_Os02g36974.1 | 14-3-3 protein | 30 | 0 | 2 | 0 | |
| LOC_Os03g50290.1 | 14-3-3 protein | 29 | 0 | 2 | 0 | |
| LOC_Os08g33370.1 | 14-3-3 protein | 29 | 0 | 3 | 0 | |
| LOC_Os08g37490.1 | 14-3-3 protein | 29 | 0 | 3 | 0 | |
| 4 | LOC_Os01g40470.1 | Vacuolar ATP synthase subunit d | 41 | 0 | 2 | 0 |
| LOC_Os03g63410.1 | Elongation factor Tu | 48 | 0 | 2 | 0 | |
| LOC_Os08g01760.1 | Dehydrogenase | 45 | 0 | 3 | 0 | |
| LOC_Os09g07830.1 | Acetyl-CoA acetyltransferase | 41 | 0 | 2 | 0 | |
| LOC_Os09g04670.1 | Differentiation and greening protein, chloroplast precursor | 43 | 0 | 2 | 0 | |
| LOC_Os03g08440.1 | Ribosomal protein S2 | 33 | 0 | 3 | 2 | |
| 5 | LOC_Os01g21320.1 | NAD-dependent epimerase/dehydratase family domain-containing protein | 46 | 0 | 2 | 0 |
| LOC_Os01g21560.1 | Esterase/lipase/thioesterase family protein | 36 | 0 | 2 | 0 | |
| LOC_Os01g71200.1 | RNA recognition motif-containing protein | 49 | 0 | 2 | 0 | |
| LOC_Os03g16980.1 | NAD-dependent epimerase/dehydratase family domain-containing protein | 39 | 0 | 2 | 0 | |
| LOC_Os09g33810.1 | Ankyrin repeat domain-containing protein | 38 | 0 | 2 | 0 | |
| LOC_Os12g33958.1 | NADH-ubiquinone oxidoreductase 49-kD subunit | 45 | 0 | 2 | 0 | |
| LOC_Os03g08440.1 | Ribosomal protein S2 | 33 | 0 | 2 | 2 | |
| LOC_Os02g14110.1 | Aminotransferase, classes I and II, domain-containing protein | 48 | 0 | 0 | 3 | |
| 6 | LOC_Os03g11440.1 | Protein transport protein Secretion61 subunit α | 52 | 0 | 4 | 2 |
| LOC_Os01g02880.1 | Fru-bisP aldolase isozyme | 42 | 0 | 0 | 4 | |
| 7 | LOC_Os08g02120.1 | Kinase, phosphofructokinase B family | 36 | 0 | 2 | 0 |
| LOC_Os05g02520.1 | Cupin domain-containing protein | 38 | 0 | 6 | 5 | |
| 9 | LOC_Os01g24710.1 | Jacalin-like lectin domain-containing protein | 15 | 0 | 2 | 0 |
| LOC_Os01g36090.1 | DNA-damage-repair/toleration protein DRT102 | 33 | 0 | 3 | 0 | |
| LOC_Os02g56180.1 | Dehydrogenase | 34 | 0 | 2 | 0 | |
| 10 | LOC_Os04g37904.1 | Arabidopsis PAPP2C homolog | 31 | 0 | 3 | 0 |
| LOC_Os02g36974.1 | 14-3-3 protein | 30 | 0 | 2 | 2 | |
| LOC_Os03g50290.1 | 14-3-3 protein | 29 | 0 | 2 | 2 | |
| LOC_Os04g40730.1 | Oxidoreductase, short-chain dehydrogenase/reductase | 35 | 0 | 3 | 2 | |
| LOC_Os11g47550.1 | Glycosyl hydrolase | 32 | 0 | 2 | 3 | |
| 11 | LOC_Os04g55040.1 | Vacuolar ATP synthase subunit d1 | 29 | 0 | 2 | 0 |
| LOC_Os07g47510.1 | Stress-related protein | 27 | 0 | 2 | 0 | |
| LOC_Os03g50290.1 | 14-3-3 protein | 29 | 0 | 5 | 4 | |
| 12 | LOC_Os03g21460.1 | Metallo-β-lactamase family protein | 29 | 0 | 2 | 0 |
DISCUSSION
The N-Terminal Domain of OsPti1a Is Required for Plasma Membrane Localization
N-terminally tagged OsPti1a did not complement lesion formation in ospti1a mutants (Supplemental Fig. S1). Indeed, OsPti1a protein with deletions of only 10 amino acids in the N terminus lost the ability to complement the knockout mutant phenotype. These results indicate that the N-terminal region of the OsPti1a protein is essential for suppressing defense activation in rice. In this article, we showed that the N terminus of the OsPti1a protein has two potential palmitoylation sites (Fig. 2A). OsPti1aC6A and OsPti1aC7A mutants clearly showed that these palmitoylation sites strongly affect OsPti1a localization, leading to membrane detachment (Fig. 2, C–N; Supplemental Fig. S2C). Palmitoylation is important for the regulation of not only subcellular localization but also signal transduction (Asai et al., 2013; Hemsley et al., 2013). In Pti1 family proteins, Cys-6 and Cys-7 are highly conserved in monocots and dicots (Fig. 2A), indicating that these palmitoylation sites possibly play a critical role in controlling Pti1-mediated signaling in plants. Despite being highly conserved in both monocots and dicots, Pti1 proteins may have different downstream signaling functions in these different plant groups. Unfortunately, we are not able to explain how Pti1 family proteins in dicots are regulated by palmitoylation, because no data have yet been obtained from dicot pti1 loss-of-function mutants; however, our results suggest that Pti1 family proteins, which have conserved N-terminal amino acid sequences, are probably modified by palmitoylation and can function in the suppression of immunity in rice plants.
Treatment with chitin, a major MAMP, did not change the size of the OsPti1a complex (Supplemental Fig. S3), suggesting that this complex is stable during MTI. Therefore, if the ability of OsPti1a to suppress MTI depends on OsPti1a complex formation, it is possible that the downstream signal is conveyed through protein interactions between this stable complex and temporarily interacting proteins at the plasma membrane. Alternatively, the OsPti1a complex may change the permeability of the plasma membrane or the activity of other membrane-associated complexes in response to MAMPs. In order to resolve this question, further experiments investigating whether the OsPti1a complex is required to induce or suppress MTI must be conducted.
In the ETI response, there are no data about the dynamics of the OsPti1a complex. Previously, we hypothesized that OsPti1a is a guardee or decoy, a potential target of pathogen effectors in rice plants (Takahashi et al., 2007; Matsui et al., 2010b). If OsPti1a is not able to form a complex in the membrane correctly, then defense responses are activated. Direct interacting targets of OsPti1a might be used to monitor OsPti1a to activate defense responses.
In this study, we have identified proteins that potentially form complexes with membrane-localized functional OsPti1a (Fig. 4C; Table I; Supplemental Tables S1 and S2). We detected several proteins that were in common between OsPti1a-HAstrepII and ΔN-OsPti1a-HAstrepII cultured cells but were not localized at the plasma membrane (Supplemental Table S1). This result is consistent with the hypothesis that ΔN-OsPti1a is able to form complexes in the cytosolic fraction that are not able to localize at the plasma membrane. Among the identified proteins that form complexes with OsPti1a protein, several proteins were predicted to have a role in plant immunity. For instance, the Arabidopsis homolog of OSA7, Arabidopsis thaliana PM H+-ATPase isoform2 (AHA2), plays an important role in stomatal defense (Liu et al., 2009b). AHA2 is also known to interact with the Arabidopsis Resistance to Pseudomonas syringae pv maculicola1 (RPM1)-interacting protein (RIN4) protein, a guardee for two independent resistance proteins, RPM1 and Resistance to P. syringae2 (RPS2; Belkhadir et al., 2004). Furthermore, the rice homolog of jacalin domain protein, a RIN4-associated protein in Arabidopsis, was also identified from the OsPti1a-HAstrepII cultured cells (Liu et al., 2009a). These intriguing results raise the possibility that OsPti1a forms a complex in the plasma membrane analogous to the Arabidopsis RIN4 complex. Additionally, PAPP2C is known to interact with Resistance to powdery mildew8 (RPW8), which confers mildew resistance in Arabidopsis (Wang et al., 2012). Transgenic rice plants expressing PAPP2C RNA interference constructs show the stunted and cell death phenotypes (Wang et al., 2012). The morphological phenotype of the ospti1a mutant might be associated with PAPP2C, regulating both plant growth and defense mechanisms in rice. These identified proteins may contribute to the activation of the immune system through OsPti1a complexes in rice plants. Future work will investigate whether the molecular function of OsPti1a-associated protein contributes to the regulation of plant immunity during pathogen infection.
DRM Localization
OsPti1a is localized in the membrane fraction, especially in DRM fractions that are resistant to treatment with nonionic detergents (Fig. 1B). From animal and yeast studies, DRMs are thought to be involved in regulating signal transduction pathways such as endocytosis and exocytosis, protein secretion, apoptosis, and the actin cytoskeleton (Simons and Toomre, 2000; Parton and Richards, 2003; Helms and Zurzolo, 2004). Recent studies have revealed that DRMs in the plasma membrane are involved in signal transduction for plant innate immunity in Arabidopsis and rice (Fujiwara et al., 2009; Keinath et al., 2010). In rice, OsRac1 (for Rac/Rho of plant [Rop]-type GTPase1), a plant-specific Rac/Rop small GTPase, and its immune complex have been well characterized (Kawasaki et al., 1999; Ono et al., 2001; Suharsono et al., 2002; Fujiwara et al., 2006, 2009; Kawano et al., 2010). In this case, the OsRac1 complex is localized in DRMs and functions as a molecular switch for defense responses. Possibly, OsPti1a regulates immune signaling through OsRac1 activity. To test this hypothesis, we analyzed the interaction between OsPti1a and OsRac1 by a coimmunoprecipitation experiment; however, OsPti1a did not interact with OsRac1 (data not shown). In addition, a dominant-negative form of OsRac1 (OsRac1DN) did not affect the ospti1a mutant phenotype (data not shown). These results indicate that OsPti1a and OsRac1 probably form two separate complexes in DRMs and function independently. On the other hand, OsPti1a and OsRac1 both have a relationship with OsRAR1 in activating defense responses against pathogens in rice. Therefore, it is possible that downstream components of OsPti1a may be shared with OsRac1 through OsRAR1-mediated signaling.
Taken together, our results indicate that the membrane localization of OsPti1a through its N-terminal lipid modification is crucial for the regulation of immune signaling by OsPti1a. Additionally, we revealed that OsPti1a forms a protein complex at the membrane and that the N-terminal region of OsPti1a is required for wild-type complex formation. These findings indicate that OsPti1a functions as a negative regulator of immune signaling dependent on complex formation in the plasma membrane.
MATERIALS AND METHODS
Plant Materials
Rice (Oryza sativa cv Nipponbare) was used as the wild-type plant, and the ospti1a mutant was described by Takahashi et al. (2007). Rice cells were suspension cultured at 25°C in liquid N6D medium (30 g L−1 sucrose, 0.3 g L−1 casamino acids, Chu [N6] Medium Salt Mixture [Wako Pure Chemical Industries], 2.878 g L−1 proline, 2 mg L−1 glycine, N6-vitamins and 2 mg L−1 2,4-dichlorophenoxyacetic acid (2,4-d), pH = 5.8) and subcultured in fresh medium every 7 d under constitutive light conditions. Cells at 5 d after subculture were used for experiments. Nicotiana benthamiana was grown at 25°C under constitutive light conditions.
Plasmid Construction
OsPti1a constructs and mutant variants were constructed as follows. Amplification and site-directed mutagenesis were performed using the primers listed in Supplemental Table S3. The cDNA fragments of HAstrepII-OsPti1a or OsPti1a-HAstrepII were generated by PCR and cloned into pCR4 TOPO (Invitrogen). The HAstrepII-tagged protein can be detected by both an anti-HA antibody and an anti-strep-tagII antibody. After sequence confirmation, these fragments were introduced into pPZP2Ha3(−) by restriction enzyme-based cloning (Fuse et al., 2001). The ΔN-OsPti1a-HAstrepII fragment was amplified by PCR using specific primers that added restriction enzyme sites and was cloned into pENTR/D-TOPO (Invitrogen). The ΔN-OsPti1a-HAstrepII fragment was introduced into pPZP2Ha3(−). Rice transformation was carried out as described previously (Takahashi et al., 2007). C-terminal synthetic GFP-fused OsPti1a was generated by PCR and was cloned into pENTR/D-TOPO. This plasmid was used as a template to generate ospti1a mutant variants. The ospti1a mutant variants were cloned into pENTR/D-TOPO. After sequence confirmation, these constructs were introduced into pEl2Ω-MCS that was modified for Gateway cloning (Mitsuhara et al., 1996). Gene expression of OsPti1a-GFP and OsPti1a variants was controlled under CaMV 35S promoter.
Subcellular Fractionation Analysis of OsPti1a
To determine the membrane localization of OsPti1a, rice suspension-cultured cells and leaves of N. benthamiana were collected and homogenized in membrane isolation buffer (50 mm Tris-HCl [pH 7.5], 0.25 m sorbitol, 2 mm EDTA, 5 mm ascorbic acid, and Complete Protease Inhibitor Cocktail tablets [Roche]). Samples were ground to a powder using liquid nitrogen and resuspended in membrane isolation buffer. Cell debris was removed by centrifugation at 10,000g at 4°C for 10 min. The supernatant was ultracentrifuged at 100,000g at 4°C for 60 min using a Beckman TLA100.3 rotor. The resulting supernatants were used as the soluble fractions, and the pellets were used as the microsomal fractions in experiments.
To characterize the membrane association of OsPti1a, microsomal fractions were diluted (10-fold) into membrane isolation buffer or membrane isolation buffer supplemented with 250 mm NaCl, 500 mm NaCl, 1 m NaCl, or 100 mm Na2CO3 (pH 11.5). Samples were incubated for 30 min on ice followed by centrifugation (Beckman TLA100.3; 100,000g, 30 min, and 4°C). Pellet fractions were washed with membrane isolation buffer, and protein equivalents of supernatant and pellet fractions were analyzed by immunoblotting using the anti-OsPti1a antibody (Takahashi et al., 2007) or anti-PIP2;1 (for aquaporin Plasma membrane intrinsic protein2;1) antibody (Cosmo Bio). Prior to immunoblotting, the nitrocellulose membrane was analyzed by CBB staining to confirm protein recovery and equal loading. Purification of DRMs was as described in detail by Fujiwara et al. (2009).
Rice suspension cell cultures were harvested 4 d after subculture and ground in homogenization buffer (50 mm MOPS/KOH [pH 7.6], 5 mm EGTA, 5 mm EDTA, 0.5 m d-sorbitol, 2 mm phenylmethylsulfonyl fluoride, 2.5 mm dithiothreitol, and Complete Protease Inhibitor Cocktail tablets [Roche]). After centrifugation at 13,000g for 15 min, the supernatant was centrifuged at 100,000g for 1 h at 4°C. The resulting pellet was subjected to a polyethylene glycol-dextran (6.4%, w/w) aqueous two-phase partitioning system for plasma membrane purification. The plasma membrane fractions were suspended in TED buffer (50 mm Tris-HCl [pH 7.4], 3 mm EDTA, and 1 mm dithiothreitol), Triton X-100 was added to a detergent:plasma membrane protein ratio of 15:1, and the mixture was incubated for 30 min on ice. After incubation, the sample was diluted with a Suc solution in TED buffer to a final concentration of 52% (w/w) Suc, overlaid with 40%, 35%, and 5% (w/w) Suc in TED buffer, and centrifuged for 16 h at 150,000g in a SW40Ti rotor (Beckman). DRM fractions were recovered above the 5% and 35% interfaces, diluted five times with TED buffer, and centrifuged for 1 h at 200,000g. Final pellets were suspended in SDS-PAGE sample buffer containing 1% (v/v) N-octyl glucoside for SDS-PAGE.
Immunoblotting
Immunoblot analysis was performed as described previously (Matsui et al., 2010b). To detect OsPti1a protein in the plasma membrane and DRM fractions by immunoblot analysis, sample proteins were separated by SDS-PAGE and transferred onto an Immobilon-P membrane (Millipore). The membrane was blocked for 1 h in phosphate-buffered saline (137 mm NaCl, 8.1 mm Na2HPO4, 2.68 mm KCl, and 1.47 mm KH2PO4) containing 5% (w/v) skim milk and incubated for 2 h with anti-Pti1a antibody (1:2,000 dilution; Takahashi et al., 2007). After washing with phosphate-buffered saline containing 0.1% (v/v) Tween 20, the membranes were incubated for 1.5 h with anti-rabbit IgG conjugated to horseradish peroxidase (1:10,000 dilution; GE Healthcare). Chemical enhancement was performed using ECL PLUS Western Blotting Detection Reagents (GE Healthcare). The enhanced signals were detected with an LAS-3000mini luminescent image analyzer (Fujifilm).
Transient Expression Assay Using Rice Protoplasts
Methods for protoplast isolation from rice cultured cells were essentially as described previously (Kyozuka and Shimamoto 1991). Plasmid transformation was performed using the polyethylene glycol method (Sheen, 2002 unpublished data http://genetics.mgh.harvard.edu/sheenweb/, Chen et al., 2006). Protoplasts isolated from rice cell line Oc suspension-cultured cells were adjusted to 2.5 × 106 cells mL−1. Plasmid DNAs (5 μg of DNA of each construct) were mixed with 100-μL aliquots of suspended protoplasts in each transformation experiment. The cells were observed between 12 and 16 h after transformation at 30°C. Confocal microscopy was performed using a TCS SP5 instrument (Leica). Fluorescence was excited with an argon laser at 488 nm and detected at wavelengths of 500 to 520 nm. Images were processed and arranged using LAS AF software (Leica) and Adobe Photoshop CC software.
Agroinfiltration of N. benthamiana
Agroinfiltration of N. benthamiana was performed as described previously (Kobayashi et al., 2007) with modifications. Binary vector constructs were made by inserting DNA fragments of OsPti1aWT, ΔN-OsPti1a, OsPti1aC6A, OsPti1aC7A, and OsPti1aC6A/C7A into the binary vector pEl2Ω-MCS-3xHA (Mitsuhara et al., 1996). Gene expression of OsPti1a and its variants was controlled under the control of the CaMV 35S promoter in pEl2Ω-MCS-3xHA. Transgenic lines of Rhizobium radiobacter (Agrobacterium tumefaciens) strain EHA105 carrying the binary constructs were used to infiltrate 5-week-old N. benthamiana leaves. Three days after infiltration, samples were frozen in liquid nitrogen until their use in experiments.
Blue Native-PAGE
Blue native-PAGE was performed as described by the manufacturer’s protocol (Invitrogen) using the fractionated samples from transgenic calli expressing 35S:OsPti1a-HAstrepII in the ospti1a background. Immunoblot analysis was performed using anti-HA antibody (1:5,000 dilution; Covance).
Size-Exclusion Chromatography
Rice suspension-cultured cells (1 g) were homogenized in liquid nitrogen with 4 mL of extraction buffer (50 mm Tris [pH 7.5], 2 mm EDTA, 150 mm NaCl, 5 mm MgCl2, 10% glycerol, 0.8% [w/v] Triton X-100, and 1× Complete Protease Inhibitor Cocktail tablets [Roche]) for 20 min at 4°C. The extracts were centrifuged at 10,000g for 20 min at 4°C, and the supernatant was filtered through a 0.22-μm filter (Millipore). The filtrate was applied to a Superdex 200 HR column (GE Healthcare) attached to an FPLC system (LCC501 Plus; GE Healthcare) using extraction buffer without protease inhibitor cocktail as the running buffer. Fractions (0.5 mL each) were collected, and 400-μL aliquots were concentrated by acetone precipitation. The precipitates were resuspended in 50 μL of SDS-PAGE sample buffer and heated for 15 min at 60°C. A total of 15 μL of each sample was subjected to SDS-PAGE and immunoblot analysis.
Coaffinity Purification and Nano-LC-MS/MS Analysis
The method of protein extraction was the same as that used for size-exclusion chromatography described above. For coaffinity purification, the total protein extract was incubated with 100 µL of Strep-Tactin beads (IBA) at 4°C for 1 h with gentle rocking. The matrix was washed three times with the extraction buffer. Bound proteins were eluted with 400 µL of extraction buffer containing 2.5 mm biotin. After that, samples were concentrated by acetone precipitation and centrifuged at 15,000g at 4°C for 20 min. The precipitates were resuspended in 30 μL of SDS-PAGE sample buffer and heated for 15 min at 60°C. Each sample (15 μL) was subjected to SDS-PAGE, and proteins were detected by silver staining.
In-gel digestions were performed as described previously (Shevchenko et al., 2006). Digested peptides in the gel pieces were recovered by adding 5% formic acid/acetonitrile, desalted using StageTips with C18 disk membranes (EMPORE; 3M; Rappsilber et al., 2003), dried in a vacuum evaporator, and dissolved in 9 μL of 5% acetonitrile containing 0.1% trifluoroacetic acid. An LTQ-Orbitrap XL (Thermo Fisher Scientific) coupled with a Dionex Ultimate3000 pump and an HTC-PAL autosampler (CTC Analytics) were used for nano-LC-MS/MS analyses. A self-pulled needle (150 mm length × 100 μm i.d., 6-μm opening) packed with ReproSil C18 resin (3 μm; Dr. Maisch GmbH) was used as an analytical column with a stone-arch frit (Ishihama et al., 2002). A spray voltage of 2,400 V was applied. The injection volume was 6 μL, and the flow rate was 500 nL min−1. The mobile phase consisted of 0.5% acetic acid (A) and 0.5% acetic acid and 80% acetonitrile (B). A two-step linear gradient of 0% to 40% B in 30 min, 40% to 100% B in 5 min, and 100% B for 10 min was employed. The mass spectrometry (MS) scan range was mass-to-charge ratio 300 to 1,400. The top 10 precursor ions were selected in the MS scan by Orbitrap at 100,000 resolution and for subsequent MS/MS scans by ion trap in the automated gain control mode, where automated gain control values of 5.00e + 05 and 1.00e + 04 were set for full MS and MS/MS, respectively. The normalized collision-induced dissociation was set to 35. A lock mass function was used for the LTQ-Orbitrap XL to obtain constant mass accuracy during gradient analysis (Olsen et al., 2005). Selected sequenced ions were dynamically excluded for 60 s after sequencing.
Mass Navigator version 1.3 (Mitsui Knowledge Industry) with default parameters for LTQ-Orbitrap XL was used to create peak lists on the basis of the recorded fragmentation spectra. The mass-to-charge ratio values of the isotope peaks were converted to the corresponding monoisotopic peaks when the isotope peaks were selected as the precursor ions. To improve the quality of the MS/MS spectra, Mass Navigator discarded all peaks with absolute intensities of less than 10 and with peak intensities of less than 0.1% of the most intense peak in the MS/MS spectra (Ravichandran et al., 2009). Peptides and proteins were identified by means of automated database searching using Mascot version 2.3.02 (Matrix Science) in the Michigan State University Rice Genome Annotation Project Database (ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_7.0/all.dir/) with a precursor mass tolerance of 3 ppm, a fragment ion mass tolerance of 0.8 D, and strict trypsin specificity (Olsen et al., 2004), allowing for up to two missed cleavages. Carbamidomethylation of Cys was set as a fixed modification, and oxidation of Met was allowed as a variable modification. Proteins with at least two unique peptides were regarded as confident identifications.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number OsPti1a (AK104870).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Complementation test of HAstrepII-OsPti1a or OsPti1a-HAstrepII in the ospti1a mutant background.
Supplemental Figure S2. Transient expression analysis of OsPti1a mutants in N. benthamiana.
Supplemental Figure S3. The size of the OsPti1a complex does not change in response to chitin treatment.
Supplemental Table S1. Total unique peptides in nano-LC-MS/MS analysis.
Supplemental Table S2. Peptide report in this assay.
Supplemental Table S3. PCR primers used in this study.
Acknowledgments
We thank Mayuko Harada-Yamazaki for technical assistance and Pamela Gan, both at the RIKEN Center for Sustainable Resource Science, for critically reading the article.
Glossary
- MAMP
microbe-associated molecular pattern
- MTI
microbe-associated molecular pattern-triggered immunity
- ETI
effector-triggered immunity
- DRM
detergent-resistant membrane
- HA
hemagglutinin
- LC
liquid chromatography
- MS/MS
tandem mass spectrometry
- CaMV
cauliflower mosaic virus
- MS
mass spectrometry
Footnotes
This work was supported by the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation; grant no. PMI0005) and by Grants-in-Aid for Scientific Research (KAKENHI; grant no. 23780047 to A.T., grant nos. 23780048 and 25850034 to H.M., and grant nos. 24688007 and 26650106 to H.N.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Asai S, Ichikawa T, Nomura H, Kobayashi M, Kamiyoshihara Y, Mori H, Kadota Y, Zipfel C, Jones JD, Yoshioka H. (2013) The variable domain of a plant calcium-dependent protein kinase (CDPK) confers subcellular localization and substrate recognition for NADPH oxidase. J Biol Chem 288: 14332–14340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Nimchuk Z, Hubert DA, Mackey D, Dangl JL. (2004) Arabidopsis RIN4 negatively regulates disease resistance mediated by RPS2 and RPM1 downstream or independent of the NDR1 signal modulator and is not required for the virulence functions of bacterial type III effectors AvrRpt2 or AvrRpm1. Plant Cell 16: 2822–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Nam J, Dangl JL. (1998) The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA 95: 15849–15854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL. (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Coll NS, Epple P, Dangl JL. (2011) Programmed cell death in the plant immune system. Cell Death Differ 18: 1247–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Hamada S, Hiratsuka M, Fukao Y, Kawasaki T, Shimamoto K. (2009) Proteome analysis of detergent-resistant membranes (DRMs) associated with OsRac1-mediated innate immunity in rice. Plant Cell Physiol 50: 1191–1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Umemura K, Kawasaki T, Shimamoto K. (2006) Proteomics of Rac GTPase signaling reveals its predominant role in elicitor-induced defense response of cultured rice cells. Plant Physiol 140: 734–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse T, Sasaki T, Yano M. (2001) Ti-plasmid vectors useful for function analysis of rice genes. Plant Biotechnol 18: 219–222 [Google Scholar]
- Helms JB, Zurzolo C. (2004) Lipids as targeting signals: lipid rafts and intracellular trafficking. Traffic 5: 247–254 [DOI] [PubMed] [Google Scholar]
- Hemsley PA, Grierson CS. (2008) Multiple roles for protein palmitoylation in plants. Trends Plant Sci 13: 295–302 [DOI] [PubMed] [Google Scholar]
- Hemsley PA, Weimar T, Lilley KS, Dupree P, Grierson CS. (2013) A proteomic approach identifies many novel palmitoylated proteins in Arabidopsis. New Phytol 197: 805–814 [DOI] [PubMed] [Google Scholar]
- Herrmann MM, Pinto S, Kluth J, Wienand U, Lorbiecke R. (2006) The PTI1-like kinase ZmPti1a from maize (Zea mays L.) co-localizes with callose at the plasma membrane of pollen and facilitates a competitive advantage to the male gametophyte. BMC Plant Biol 6: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama Y, Rappsilber J, Andersen JS, Mann M. (2002) Microcolumns with self-assembled particle frits for proteomics. J Chromatogr A 979: 233–239 [DOI] [PubMed] [Google Scholar]
- Ishitani M, Liu J, Halfter U, Kim CS, Shi W, Zhu JK. (2000) SOS3 function in plant salt tolerance requires N-myristoylation and calcium binding. Plant Cell 12: 1667–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kawano Y, Akamatsu A, Hayashi K, Housen Y, Okuda J, Yao A, Nakashima A, Takahashi H, Yoshida H, Wong HL, et al. (2010) Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe 7: 362–375 [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Henmi K, Ono E, Hatakeyama S, Iwano M, Satoh H, Shimamoto K. (1999) The small GTP-binding protein rac is a regulator of cell death in plants. Proc Natl Acad Sci USA 96: 10922–10926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinath NF, Kierszniowska S, Lorek J, Bourdais G, Kessler SA, Shimosato-Asano H, Grossniklaus U, Schulze WX, Robatzek S, Panstruga R. (2010) PAMP (pathogen-associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. J Biol Chem 285: 39140–39149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL. (2005) The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc Natl Acad Sci USA 102: 6496–6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Ohura I, Kawakita K, Yokota N, Fujiwara M, Shimamoto K, Doke N, Yoshioka H. (2007) Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19: 1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka J, Shimamoto K (1991) Transformation and regeneration of rice protoplast. In K Lindsey, ed, Plant Tissue Culture Manual. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp B1, 1–16 [Google Scholar]
- Liu J, Elmore JM, Coaker G. (2009a) Investigating the functions of the RIN4 protein complex during plant innate immune responses. Plant Signal Behav 4: 1107–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, Coaker G. (2009b) RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol 7: e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H, Miyao A, Takahashi A, Hirochika H. (2010a) Pdk1 kinase regulates basal disease resistance through the OsOxi1-OsPti1a phosphorylation cascade in rice. Plant Cell Physiol 51: 2082–2091 [DOI] [PubMed] [Google Scholar]
- Matsui H, Yamazaki M, Kishi-Kaboshi M, Takahashi A, Hirochika H. (2010b) AGC kinase OsOxi1 positively regulates basal resistance through suppression of OsPti1a-mediated negative regulation. Plant Cell Physiol 51: 1731–1744 [DOI] [PubMed] [Google Scholar]
- Mitsuhara I, Ugaki M, Hirochika H, Ohshima M, Murakami T, Gotoh Y, Katayose Y, Nakamura S, Honkura R, Nishimiya S, et al. (1996) Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol 37: 49–59 [DOI] [PubMed] [Google Scholar]
- Muskett PR, Kahn K, Austin MJ, Moisan LJ, Sadanandom A, Shirasu K, Jones JD, Parker JE. (2002) Arabidopsis RAR1 exerts rate-limiting control of R gene-mediated defenses against multiple pathogens. Plant Cell 14: 979–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Zipfel C, Rowland O, Keller I, Robatzek S, Boller T, Jones JD. (2004) The transcriptional innate immune response to flg22: interplay and overlap with Avr gene-dependent defense responses and bacterial pathogenesis. Plant Physiol 135: 1113–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger T, Kemmerling B. (2006) Receptor protein kinases: pattern recognition receptors in plant immunity. Trends Plant Sci 11: 519–522 [DOI] [PubMed] [Google Scholar]
- Olsen JV, de Godoy LMF, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics 4: 2010–2021 [DOI] [PubMed] [Google Scholar]
- Olsen JV, Ong SE, Mann M. (2004) Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol Cell Proteomics 3: 608–614 [DOI] [PubMed] [Google Scholar]
- Ono E, Wong HL, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K. (2001) Essential role of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA 98: 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton RG, Richards AA. (2003) Lipid rafts and caveolae as portals for endocytosis: new insights and common mechanisms. Traffic 4: 724–738 [DOI] [PubMed] [Google Scholar]
- Pierre M, Traverso JA, Boisson B, Domenichini S, Bouchez D, Giglione C, Meinnel T. (2007) N-Myristoylation regulates the SnRK1 pathway in Arabidopsis. Plant Cell 19: 2804–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Tsuda K, Glazebrook J, Katagiri F. (2011) Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis. Mol Plant Pathol 12: 702–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem 75: 663–670 [DOI] [PubMed] [Google Scholar]
- Ravichandran A, Sugiyama N, Tomita M, Swarup S, Ishihama Y. (2009) Ser/Thr/Tyr phosphoproteome analysis of pathogenic and non-pathogenic Pseudomonas species. Proteomics 9: 2764–2775 [DOI] [PubMed] [Google Scholar]
- Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. (2008) CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng Des Sel 21: 639–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K. (2006) Endomembrane proton pumps: connecting membrane and vesicle transport. Curr Opin Plant Biol 9: 595–600 [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1: 2856–2860 [DOI] [PubMed] [Google Scholar]
- Shirasu K, Lahaye T, Tan MW, Zhou F, Azevedo C, Schulze-Lefert P. (1999) A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99: 355–366 [DOI] [PubMed] [Google Scholar]
- Simons K, Toomre D. (2000) Lipid rafts and signal transduction. Nat Rev Mol Cell Biol 1: 31–39 [DOI] [PubMed] [Google Scholar]
- Suharsono U, Fujisawa Y, Kawasaki T, Iwasaki Y, Satoh H, Shimamoto K. (2002) The heterotrimeric G protein alpha subunit acts upstream of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA 99: 13307–13312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Agrawal GK, Yamazaki M, Onosato K, Miyao A, Kawasaki T, Shimamoto K, Hirochika H. (2007) Rice Pti1a negatively regulates RAR1-dependent defense responses. Plant Cell 19: 2940–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D, Rafiqi M, Hurley U, Lawrence GJ, Bernoux M, Hardham AR, Ellis JG, Dodds PN, Jones DA. (2012) N-terminal motifs in some plant disease resistance proteins function in membrane attachment and contribute to disease resistance. Mol Plant Microbe Interact 25: 379–392 [DOI] [PubMed] [Google Scholar]
- Wang WM, Ma XF, Zhang Y, Luo MC, Wang GL, Bellizzi M, Xiong XY, Xiao SY. (2012) PAPP2C interacts with the atypical disease resistance protein RPW8.2 and negatively regulates salicylic acid-dependent defense responses in Arabidopsis. Mol Plant 5: 1125–1137 [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. (2006) Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]