Phytol hydrolysis during chlorophyll breakdown in tomato leaves is specifically catalyzed by pheophytinase, while during fruit ripening other, so far unknown, hydrolases are active.
Abstract
Chlorophyll breakdown occurs in different green plant tissues (e.g. during leaf senescence and in ripening fruits). For different plant species, the PHEOPHORBIDE A OXYGENASE (PAO)/phyllobilin pathway has been described to be the major chlorophyll catabolic pathway. In this pathway, pheophorbide (i.e. magnesium- and phytol-free chlorophyll) occurs as a core intermediate. Most of the enzymes involved in the PAO/phyllobilin pathway are known; however, the mechanism of dephytylation remains uncertain. During Arabidopsis (Arabidopsis thaliana) leaf senescence, phytol hydrolysis is catalyzed by PHEOPHYTINASE (PPH), which is specific for pheophytin (i.e. magnesium-free chlorophyll). By contrast, in fruits of different Citrus spp., chlorophyllase, hydrolyzing phytol from chlorophyll, was shown to be active. Here, we enlighten the process of chlorophyll breakdown in tomato (Solanum lycopersicum), both in leaves and fruits. We demonstrate the activity of the PAO/phyllobilin pathway and identify tomato PPH (SlPPH), which, like its Arabidopsis ortholog, was specifically active on pheophytin. SlPPH localized to chloroplasts and was transcriptionally up-regulated during leaf senescence and fruit ripening. SlPPH-silencing tomato lines were impaired in chlorophyll breakdown and accumulated pheophytin during leaf senescence. However, although pheophytin transiently accumulated in ripening fruits of SlPPH-silencing lines, ultimately these fruits were able to degrade chlorophyll like the wild type. We conclude that PPH is the core phytol-hydrolytic enzyme during leaf senescence in different plant species; however, fruit ripening involves other hydrolases, which are active in parallel to PPH or are the core hydrolases in fruits. These hydrolases remain unidentified, and we discuss the question of whether chlorophyllases might be involved.
Chlorophyll breakdown is an important physiological process in plants that occurs during different phases of plant development. Most obvious and eye-catching is the loss of green pigment color during autumnal leaf senescence in deciduous trees, but also the ripening phase of many fruits such as banana (Musa acuminata) and tomato (Solanum lycopersicum) includes massive degradation of chlorophyll.
For many years, chlorophyll degradation was considered a biological enigma (Hendry et al., 1987). Only the identification and structure determination of a first colorless nonfluorescent chlorophyll catabolite from senescing barley (Hordeum vulgare) as a (final) breakdown product (Kräutler et al., 1991) paved the way for the step-wise elucidation of a pathway of chlorophyll degradation (for review, see Hörtensteiner and Kräutler, 2011; Kräutler and Hörtensteiner, 2013; Christ and Hörtensteiner, 2014). This pathway leads to the ultimate degradation of chlorophyll to a group of colorless, linear tetrapyrroles, termed phyllobilins (Kräutler and Hörtensteiner, 2013).
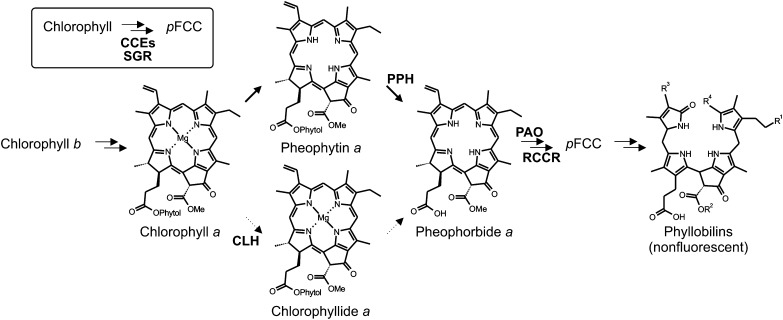
The pathway can be divided into two parts. Early reactions take place within senescing chloroplasts and result in the formation of a colorless primary fluorescent chlorophyll catabolite (pFCC; Fig. 1; Mühlecker et al., 1997). The reactions catalyzing the chlorophyll-to-pFCC conversion are commonly present in land plants (Hörtensteiner, 2013) and, therefore, represent the core part of the pathway. The second part of the chlorophyll degradation pathway is characterized by largely species-specific modifications at different peripheral positions within pFCC (indicated in Fig. 1 with R1–R4) and ultimate conversion to respective nonfluorescent phyllobilins that represent the end products of chlorophyll breakdown in most species and are stored in the vacuole (Kräutler and Hörtensteiner, 2013).
Figure 1.
Structural outline of the PAO/phyllobilin pathway of chlorophyll breakdown showing the chemical constitutions of chlorophyll a and of selected chlorophyll catabolites that are relevant for this work. R1 to R4 indicate sites of modifications that are found in nonfluorescent phyllobilins of different plant species (Kräutler and Hörtensteiner, 2013). Relevant reactions (PPH, CLH, PAO, and RCCR) are indicated. Note that dephytylation by PPH was shown to be the major reaction of pheophorbide a formation during leaf senescence in Arabidopsis (Schelbert et al., 2009). The inset indicates that conversion of chlorophyll to pFCC requires the concerted action of different CCEs and of SGR.
To date, a total of four steps are known to be required for the conversion of chlorophyll a to pFCC. Except for the activity that is responsible for magnesium dechelation, genes encoding these catalytic activities have been identified in Arabidopsis (Arabidopsis thaliana) and other species. Since all except one of the phyllobilins that have been characterized structurally are derived from chlorophyll a (Hörtensteiner and Kräutler, 2011), the reductive part of the chlorophyll cycle that converts chlorophyll b into chlorophyll a has been considered an integral part of senescence-related chlorophyll breakdown (Tanaka et al., 2011).
The magnesium- and phytol-free intermediate of chlorophyll a, pheophorbide a, is a genuine breakdown product of chlorophyll (Langmeier et al., 1993). However, the means of pheophorbide formation during leaf senescence was (and still is) controversial, because the order of reactions—that is, dechelation versus dephytylation—was unclear (Amir-Shapira et al., 1987), although the favored hypothesis was that dephytylation by CHLOROPHYLLASE (CLH) would precede magnesium dechelation (Tanaka and Tanaka, 2006). We recently showed that the two CLHs of Arabidopsis are dispensable for leaf senescence (Schenk et al., 2007). Instead, we and others identified a novel esterase, PHEOPHYTINASE (PPH), which specifically dephytylates pheophytin, but not chlorophyll, and is required for chlorophyll breakdown in Arabidopsis and rice (Oryza sativa; Morita et al., 2009; Schelbert et al., 2009; Ren et al., 2010). Thus, PPH-deficient mutants exhibit a stay-green phenotype, which is characterized by a high retention of chlorophyll together with the accumulation of significant amounts of pheophytin during leaf senescence. This indicates that dechelation precedes dephytylation, at least during leaf senescence. By contrast, CLHs have been implicated in the postharvest senescence of broccoli (Brassica oleracea var italica) and citrus (Citrus spp.) fruit ripening (Jacob-Wilk et al., 1999; Azoulay Shemer et al., 2008; Chen et al., 2008; see below). Pheophorbide a, the last chlorin-type intermediate of chlorophyll breakdown, is oxygenolytically opened by PHEOPHORBIDE A OXYGENASE (PAO) to yield a red chlorophyll catabolite, which is further reduced to pFCC by RED CHLOROPHYLL CATABOLITE REDUCTASE (RCCR; Rodoni et al., 1997). PAO is responsible for the open tetrapyrrolic backbone of the phyllobilins. For this reason, the pathway described above is now termed the PAO/phyllobilin pathway of chlorophyll breakdown (Kräutler and Hörtensteiner, 2013).
Recently, it was shown that the chloroplast-localized chlorophyll catabolic enzymes (CCEs) physically interact at the thylakoid membrane, most likely to allow metabolic channeling of the breakdown intermediates upstream of pFCC that are potentially phototoxic (Sakuraba et al., 2012). STAY-GREEN (SGR), a chloroplast-localized protein (Hörtensteiner, 2009), is critical for these interactions; nonyellowing1-1, an Arabidopsis SGR mutant (Ren et al., 2007), is defective in CCE protein interaction (Sakuraba et al., 2012). This indicates that, rather being biochemically active itself, SGR may function as a scaffold protein to recruit CCEs for protein complex formation during chlorophyll breakdown. As a consequence, mutants that are deficient in SGR exhibit a stay-green phenotype (Barry, 2009; Hörtensteiner, 2009). In addition, SGR (negatively) regulates carotenoid biosynthesis during tomato fruit ripening (Luo et al., 2013) and (positively) regulates root nodule senescence in Medicago truncatula (Zhou et al., 2011), implying that SGR has diverse functions that are not restricted to chlorophyll degradation.
The PAO/phyllobilin pathway has largely been elucidated through investigations that focused on leaf senescence. Nevertheless, chlorophyll breakdown during fruit ripening was considered to be identical to the mechanism occurring during leaf senescence (Hörtensteiner and Kräutler, 2011). Deficiency of SGR, as for example in the tomato green flesh (gf) and the red pepper (Capsicum annuum) chlorophyll retainer mutants, causes a stay-green phenotype of these mutants in leaves and fruits (Barry et al., 2008; Borovsky and Paran, 2008), indicating that SGR is required for chlorophyll breakdown in both tissues. Similarly, PAO and RCCR were found to be active in chromoplast membranes isolated from tomato and red pepper fruits (Moser and Matile, 1997; Akhtar et al., 1999), and recently, different fluorescent and nonfluorescent phyllobilins were shown to occur in ripening apple (Malus domestica), pear (Pyrus communis), and banana (Kräutler, 2008; Moser et al., 2009). Finally, SGR and PAO have been identified in a recent proteome analysis of tomato chromoplasts (Barsan et al., 2010). In summary, these data indicate that the pathways of chlorophyll breakdown during fruit ripening and leaf senescence are identical. Yet, the identification of PPH as the major dephytylating enzyme of leaf senescence (Schelbert et al., 2009) challenges this view, because, contrary to the situation in leaves, CLH was shown to be involved during ethylene-induced ripening of citrus fruits (Jacob-Wilk et al., 1999; Harpaz-Saad et al., 2007; Azoulay Shemer et al., 2008).
The aim of this work, therefore, was to investigate whether PPH, besides its requirement for leaf senescence, is also involved in chlorophyll breakdown during fruit ripening. Using tomato as a model, we show that the PAO/phyllobilin pathway is active both during fruit ripening and leaf senescence, because genes encoding CCEs and SGR are transcriptionally up-regulated in both ripening fruits and senescing leaves. However, lines silenced in tomato PPH (SlPPH) were specifically deficient in leaf senescence-related chlorophyll breakdown, while the involvement of PPH in fruit ripening-related breakdown seems to be less important. Although our data show a transient delay of chlorophyll breakdown in the absence of PPH, SlPPH-silencing fruits ultimately degrade chlorophyll like the wild type. Pheophytin-specific phytol hydrolysis was reduced in chromoplasts of SlPPH-silencing lines, but substantial enzyme activity remained in these lines, which leads us to speculate that other hydrolases are important (in addition to PPH). The identity of these activities remains elusive.
RESULTS
The PAO/Phyllobilin Pathway Is Active during Chlorophyll Degradation in Tomato Leaves and Fruits
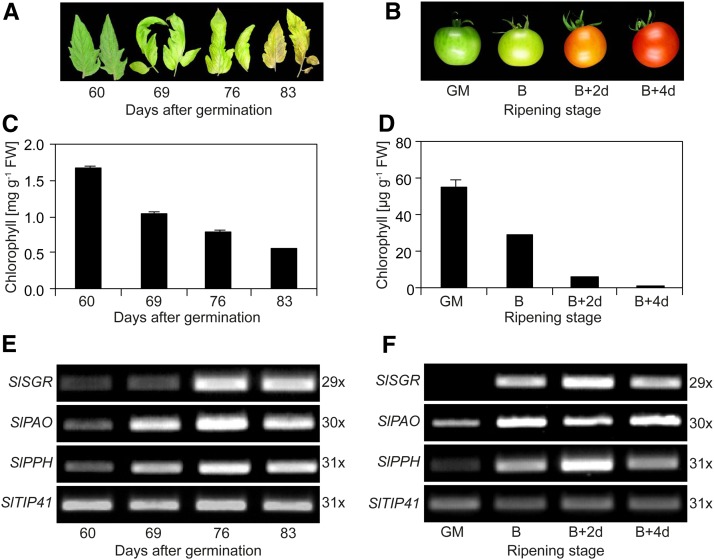
To enlighten whether the PAO/phyllobilin pathway is responsible for the loss of chlorophyll in tomato, CCE gene expression was analyzed during leaf senescence and fruit ripening. Yellowing was observed during the progression of natural senescence of tomato leaves starting at 60 d after germination (Fig. 2A), and within 23 d, the content of chlorophyll a and b decreased to around 30% of the initial amount (Fig. 2C). As shown in Figure 2, B and D, the chlorophyll content of tomato fruits at the breaker stage was reduced within 4 d of ripening, and red and yellow pigments, mainly carotenoids (Egea et al., 2010), became visible. Gene expression levels of SlSGR and SlPAO, as analyzed by semiquantitative reverse transcription (RT)-PCR, increased during both leaf senescence and fruit ripening (Fig. 2, E and F). These results confirmed published quantitative PCR (qPCR) data on CCE gene expression (Lira et al., 2014) and indicated that the PAO/phyllobilin pathway is activated during chlorophyll breakdown in tomato and that chlorophyll is degraded in a similar manner in tomato leaves and fruits. Nevertheless, it remained to be demonstrated whether the core phytol hydrolytic enzyme during chlorophyll degradation is PPH, as demonstrated in Arabidopsis leaves (Schelbert et al., 2009).
Figure 2.
The PAO/phyllobilin pathway is active during chlorophyll degradation in tomato leaves and fruits. A, Phenotypic appearance of the first true leaves from wild-type tomato during natural senescence starting from 60 d after germination. B, Phenotypes of fruits during ripening. GM, Green mature; B, breaker. C and D, Quantification of total chlorophyll during natural leaf senescence (C) and fruit ripening (D). Total leaves and fruit exocarp and mesocarp tissues at the indicated times were used for chlorophyll quantification. Data represent means of three technical replicates ± sd. FW, Fresh weight. E and F, Analysis of gene expression during natural leaf senescence (E) and fruit ripening (F). SlTIP41 was used as a control (Expósito-Rodríguez et al., 2008). Expression was analyzed with the number of PCR cycles as indicated. PCR products were separated on agarose gels and visualized with ethidium bromide.
SlPPH Is Expressed in Tomato and Localizes to Chloroplasts
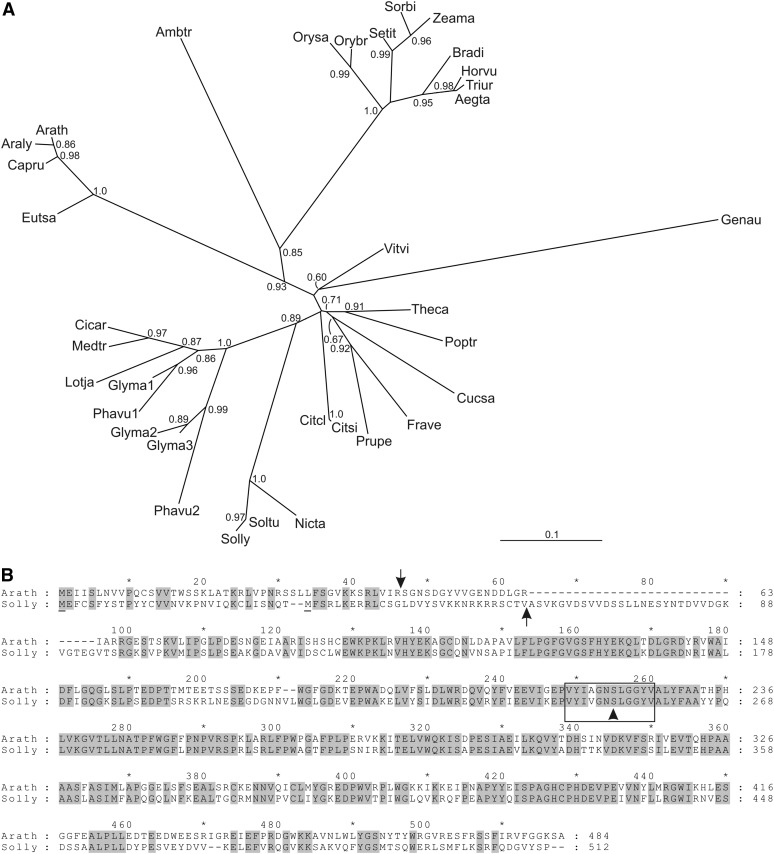
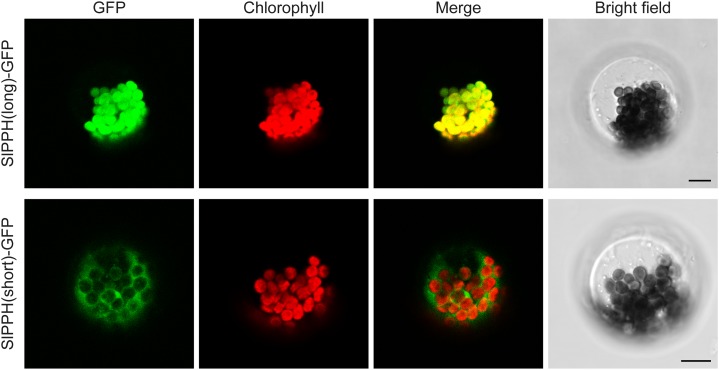
BLASTP searches (Altschul et al., 1997) for PPH protein homologs in tomato identified SlPPH (Solyc01g088090). Highly homologous PPH proteins were present in all sequenced plant genomes as single proteins, except for soybean (Glycine max) and common bean (Phaseolus vulgaris), with three and two PPHs, respectively (Fig. 3A). PPHs of species within different plant families, including Fabaceae, Brassicaceae, Solanaceae, and Gramineae, clustered into separate clades. Overall protein sequence identity within families was between 65% and 96%, and even the most divergent PPH from Genlisea aurea was more than 58% identical to the other protein sequences. An alignment of SlPPH and AtPPH, which exhibits 62.8% sequence identity, is shown in Figure 3B. The conserved PPH domain (Schelbert et al., 2009) with its proposed active-site Ser residue (boxed in Fig. 3B) was present in all PPH proteins included in the phylogenetic tree of Figure 3A. Expression of SlPPH, as analyzed by semiquantitative RT-PCR, increased with the onset of leaf senescence and fruit ripening and correlated with the transcript levels of SlPAO and SlSGR (Fig. 2, E and F). From these results, we concluded that SlPPH is involved in chlorophyll breakdown and likely acts as the phytol hydrolytic enzyme in leaves and fruits. In order to analyze the subcellular localization of SlPPH, which based on its proposed function was expected to localize to plastids, we constructed C-terminal GFP fusions (SlPPH-GFP). The sequence of the predicted SlPPH complementary DNA (cDNA) contained two possible in-frame start codons (underlined Met residues in Fig. 3B); however, none of these encoded a PPH version that would contain an N-terminal chloroplast transit peptide according to the prediction by ChloroP (Emanuelsson et al., 1999). Therefore, both varieties, SlPPH(long) and SlPPH(short), were cloned. The fusion proteins were transiently expressed in senescing Arabidopsis mesophyll protoplasts and analyzed by confocal laser-scanning microscopy. As shown in Figure 4, the overlay of GFP fluorescence and chlorophyll autofluorescence indicated that the long SlPPH version localized to the chloroplast, while the GFP signal of the short version was detected in the cytosol. From these results, we conclude that SlPPH is indeed located in the chloroplast and that SlPPH(long) represents the full-length SlPPH version, with a likely 61-amino acid chloroplast transit peptide as predicted by ChloroP (Emanuelsson et al., 1999; Fig. 3B).
Figure 3.
Analysis of PPH proteins from different plant species. A, Maximum likelihood phylogenetic tree of PPH proteins from different higher plant species. Branch support values are based on 100 bootstrap replicates and are indicated when higher than 0.6. Aegta, Aegilops tauschii; Ambtr, Amborella trichopoda; Araly, Arabidopsis lyrata; Arath, Arabidopsis; Bradi, Brachypodium distachyon; Capru, Capsella rubella; Cicar, Cicer arietinum; Citcl, Citrus clementina; Citsi, Citrus sinensis; Cucsa, Cucumis sativus; Eutsa, Eutrema salsugineum; Frave, Fragaria vesca; Genau, Genlisea aurea; Glyma, soybean; Horvu, barley; Lotja, Lotus japonicus; Medtr, Medicago truncatula; Nicta, Nicotiana tabacum; Orybr, Oryza brachyantha; Orysa, rice; Phavu, common bean; Poptr, Populus trichocarpa; Prupe, Prunus persica; Setit, Setaria italica; Solly, tomato; Soltu, Solanum tuberosum; Sorbi, Sorghum bicolor; Theca, Theobroma cacao; Triur, Triticum urartu; Vitvi, Vitis vinifera; Zeama, Zea mays. B, Alignment of PPH proteins from Arabidopsis (Arath) and tomato (Solly). Two potential start Met residues are underlined. Cleavage sites of the chloroplast transit peptide sequences as predicted by ChloroP (Emanuelsson et al., 1999) are indicated with arrows. The PPH motif (Schelbert et al., 2009) containing the active-site Ser residue (arrowhead) is boxed. Identical amino acids are shaded in gray.
Figure 4.
Subcellular localization of SlPPH. Two SlPPH varieties, SlPPH(long) and SlPPH(short), were transiently expressed as GFP fusions in Arabidopsis protoplasts isolated from senescent leaves. GFP fluorescence (GFP) and chlorophyll autofluorescence (chlorophyll) were examined by confocal laser-scanning microscopy. Merged images show the overlay of GFP and autofluorescence. Bars = 10 µm.
SlPPH Is a Genuine PPH
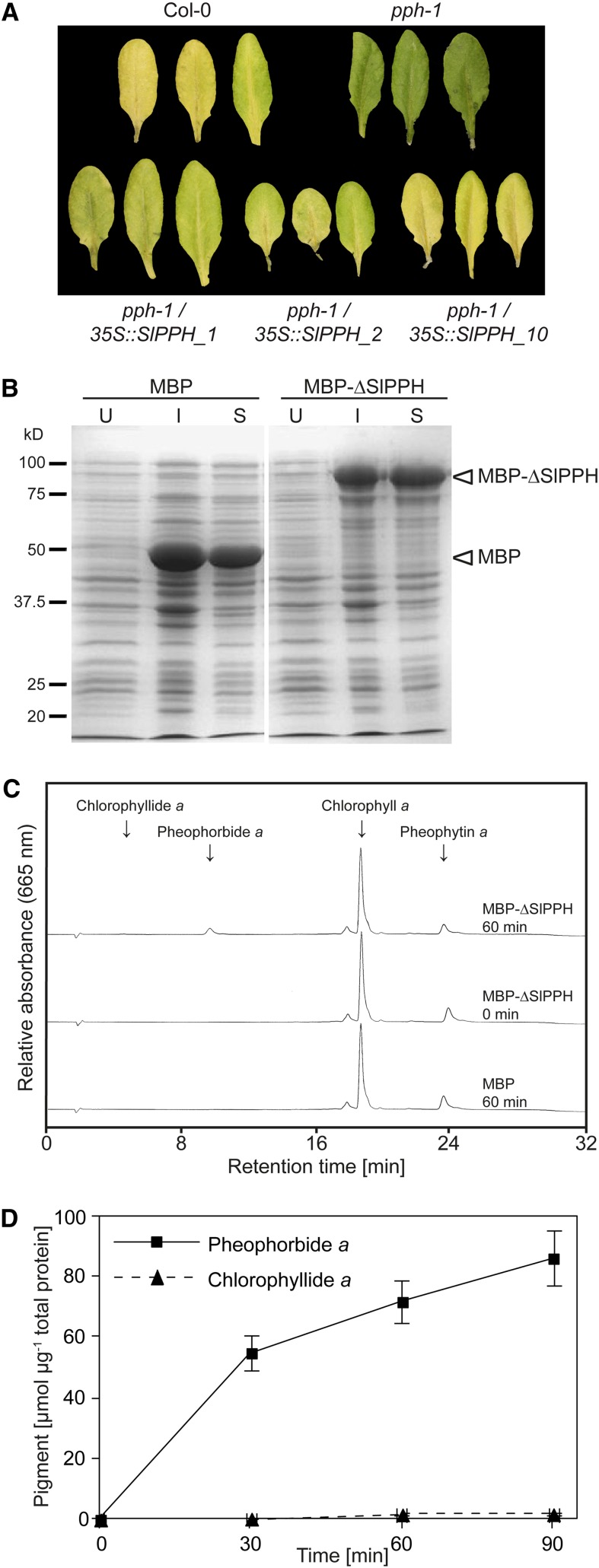
Phylogenetic analysis and sequence alignment of PPH homologs revealed the PPH motif including the proposed active-site Ser residue to be present in SlPPH (Fig. 3B). This indicated that SlPPH is a genuine PPH and, thus, an ortholog of Arabidopsis PPH (Schelbert et al., 2009). To confirm this, the Arabidopsis pph-1 mutant was complemented with an SlPPH cDNA construct (long version) under the control of the cauliflower mosaic virus (CaMV) 35S promoter. As shown earlier, pph-1 is impaired in chlorophyll breakdown and shows a stay-green phenotype (Schelbert et al., 2009). To induce senescence, detached T1 leaves of three independent complementation lines (pph-1/35S::SlPPH_1, pph-1/35S::SlPPH_2, and pph-1/35S::SlPPH_10) were dark incubated for 7 d. Indeed, ectopic expression of SlPPH complemented the pph-1 phenotype, and leaves of all three tested lines showed leaf yellowing comparable to the wild type (Fig. 5A). To further verify the function of SlPPH as PPH, we examined the enzymatic activity of a recombinant truncated version of SlPPH devoid of the predicted chloroplast transit peptide (ΔSlPPH). ΔSlPPH was expressed in Escherichia coli as an N-terminal maltose-binding protein fusion (MBP-ΔSlPPH). The recombinant fusion protein was highly stable and largely located in the soluble bacterial cell fraction (Fig. 5B). Using chlorophyll a or pheophytin a, or mixtures of both as substrate, we could confirm SlPPH to be highly specific for pheophytin a (Fig. 5, C and D), comparable to its Arabidopsis ortholog (Schelbert et al., 2009). These data strongly support the assumption that SlPPH acts as genuine PPH.
Figure 5.
Confirmation of SlPPH as a genuine PPH. A, Complementation of Arabidopsis pph-1 with SIPPH. Detached leaves of 4-week-old plants of three independent transformants (pph-1/35S::SlPPH_1, pph-1/35S::SlPPH_2, and pph-1/35S::SlPPH_10) in the T1 generation were dark incubated for 7 d. Col-0, Columbia-0. B to D, Analysis of recombinant SlPPH. B, Heterologous expression of MBP and MBP-ΔSlPPH fusion proteins in E. coli. U, Cells before induction with isopropylthio-β-galactoside; I, cells after isopropylthio-β-galactoside induction for 3 h; S, soluble cell fraction after lysis. Note that MBP-ΔSlPPH was largely retained in the soluble cell fraction. Molecular size markers (kD) are indicated on the left. C, HPLC analysis of 60-min assays employing soluble E. coli lysates expressing MBP-ΔSlPPH or MBP alone with mixtures of chlorophyll a and pheophytin a as substrate. Note that SlPPH specifically hydrolyzed pheophytin a to pheophorbide a, although chlorophyll a was present in excess. Arrows indicate HPLC retention times of substrates and the respective dephytylated products. D, Time-dependent formation of pheophorbide a and chlorophyllide a from pheophytin a and chlorophyll a, respectively, in assays with MBP-ΔSlPPH. Note that the activity of MBP-ΔSlPPH with chlorophyll a as substrate is marginal. Data are means ± sd of three assays.
SlPPH Catalyzes the Cleavage of Phytol in Senescing Tomato Leaves
To analyze whether SlPPH is required for in vivo chlorophyll breakdown in tomato, transgenic tomato plants were generated that harbored an SlPPH-silencing construct expressed under the control of the CaMV 35S promoter (SlPPHi). Levels of SlPPH expression of several independent transgenic tomato lines were determined in leaf and fruit tissues by semiquantitative RT-PCR and qPCR (Supplemental Fig. S1). Several independent RNA interference (RNAi) lines displayed strongly reduced SlPPH expression as compared with the wild type, and lines SlPPHi_17 and SlPPHi_27, with expression levels of less than 16% and 7%, respectively, in leaves and fruits at breaker + 1 d were chosen for further analysis.
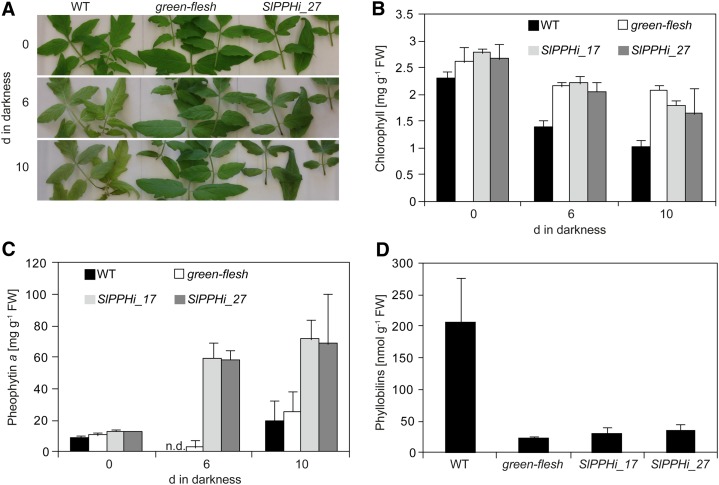
To elucidate whether the absence of SlPPH causes a stay-green phenotype during chlorophyll breakdown in leaves as described for Arabidopsis (Schelbert et al., 2009), senescence was initiated in detached leaves of the wild type, gf, SlPPHi_17, and SlPPHi_27 by dark incubation for up to 10 d in the presence of 1 mm ethephon, a precursor of ethylene. After 6 d, visual yellowing (Fig. 6A) and decrease of chlorophyll a and b (Fig. 6B) were observed in wild-type leaves, while leaves of gf and the two silencing lines still appeared green and chlorophyll degradation was significantly delayed. Thus, after 10 d, chlorophyll content was decreased to less than 50% in the wild type, whereas in gf, SlPPHi_17, and SlPPHi_27, approximately 70% of the initial chlorophyll was still present. In addition, HPLC analysis of pigment extracts showed that pheophytin accumulated in both analyzed RNAi lines after 6 and 10 d of dark incubation (Fig. 6C). By contrast, pheophytin was detected in only marginal amounts in wild-type and gf leaves. This was in agreement with the in vitro substrate specificity of SlPPH for pheophytin (Fig. 5) and comparable to the effect in the Arabidopsis pph-1 mutant (Schelbert et al., 2009).
Figure 6.
Silencing of SlPPH results in a stay-green phenotype in senescing tomato leaves. A, Leaf phenotype after 0, 6, and 10 d of ethylene-induced senescence in the dark. B to D, Pigment composition in senescing tomato leaves. B, Quantification of total chlorophyll. C, Quantification of pheophytin a. Note that pheophytin a was not detected (n.d.) in the wild type (WT) after 6 d of dark incubation. D, Quantification of phyllobilins after 10 d of dark incubation. All data are means of three biological replicates ± sd. FW, Fresh weight.
In Arabidopsis and many other species, nonfluorescent phyllobilins have been shown to constitute final catabolites of chlorophyll breakdown (Hörtensteiner and Kräutler, 2011; Kräutler and Hörtensteiner, 2013). Tomato wild-type leaves accumulated large quantities of phyllobilins after 10 d of dark incubation (Fig. 6D). By contrast, in SlPPHi_17 and SlPPHi_27 as well as in gf, phyllobilins did not accumulate to the same extent (Fig. 6D), confirming the impairment of chlorophyll degradation in these lines. In summary, we conclude that SlPPH is the core hydrolytic enzyme during chlorophyll breakdown in tomato leaves and that its absence blocks the overall process of chlorophyll degradation. As a consequence, chlorophyll is retained, pheophytin accumulates, and phyllobilin abundance is largely diminished.
SlPPH Is Active during Fruit Ripening, But Other Unknown Hydrolases Are Active in Parallel
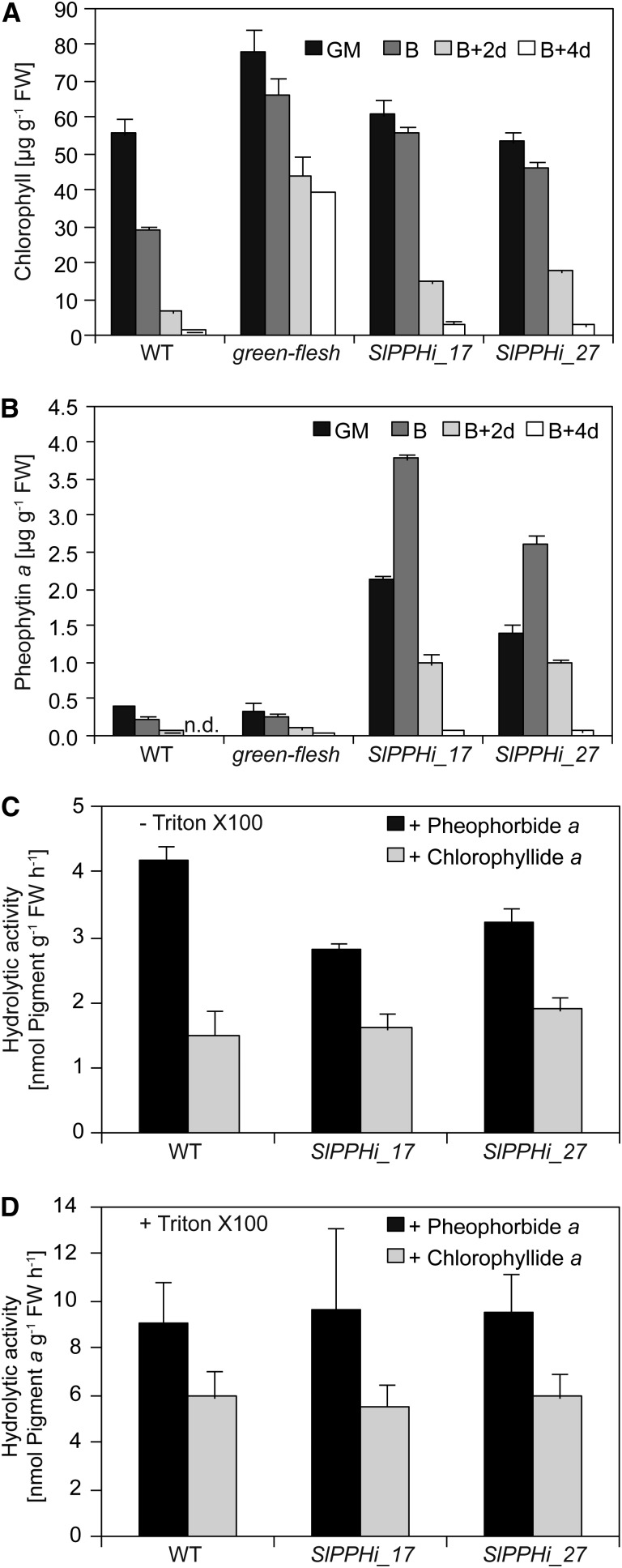
As shown in Figure 2, chlorophyll breakdown in tomato occurs during both leaf senescence and fruit ripening. Hence, we were interested in whether dephytylation in tomato fruits was also catalyzed by SlPPH, as shown for tomato leaves (Fig. 6). For this, we analyzed pigment composition in fruits of the wild type, gf, and the two RNAi lines SlPPHi_17 and SlPPHi_27 during the process of ripening at four different ripening stages: green mature, breaker, breaker + 2 d, and breaker + 4 d (Fig. 7). When compared with the wild type, the two SlPPH-silencing lines were retarded in chlorophyll breakdown and showed higher chlorophyll levels at the onset of ripening (breaker) and the half-ripe stage (breaker + 2 d). However, at the full-ripe stage (breaker + 4 d), the RNAi lines had lost chlorophyll comparable to the wild type. This indicated that the absence of SlPPH caused a transient retention of chlorophyll during fruit ripening but did not result in a true stay-green phenotype, as in gf fruits (Fig. 7A; Barry et al., 2008). The transient retardation of chlorophyll degradation in the silencing lines was accompanied by a transient accumulation of pheophytin a, the substrate of SlPPH, while wild-type and gf fruits did not accumulate pheophytin a at any stage of ripening (Fig. 7B). Thus, the RNAi lines accumulated up to 13-fold levels of pheophytin a at the breaker stage as compared with the controls. However, pheophytin a quantities were largely reduced at the breaker + 4 d stage in SlPPH-silencing fruits and were comparable to the wild type and gf (Fig. 7B). This transient accumulation of pheophytin a during the fruit ripening process implied an involvement of SlPPH in chlorophyll breakdown also during fruit ripening on the one hand; on the other hand, however, it indicated that other phytol hydrolytic activities may be involved and may compensate for the absence of PPH in the silencing lines. To address this, we performed in vitro activity assays using chromoplasts of wild-type and RNAi lines at the breaker + 2 d stage, thereby comparing pheophytin-specific activities in solubilized and nonsolubilized chromoplast membranes. For different plant species, including citrus fruits, membrane solubilization has been shown to be a prerequisite for the activation of CLHs (and possibly other dephytylating activities), which are present in membranes in a latent form (Amir-Shapira et al., 1986; Matile et al., 1999). Dephytylation of pheophytin was significantly reduced by about 25% in nonsolubilized chromoplasts of SlPPHi_17 and SlPPHi_27 when compared with the wild type (Fig. 7C). These differences likely reflect the absence of SlPPH in the RNAi lines; in addition, other dephytylating activities are present in chromoplasts. Furthermore, after solubilization, overall activity in the wild type was about twice that compared with nonsolubilized chromoplasts, but it was not different between the wild type and the silencing lines for both chlorophyll and pheophytin (Fig. 7D). This indeed supports the assumption that, besides PPH, major additional activities are present in ripening tomato fruit chromoplasts that are capable of dephytylation of either chlorophyll or pheophytin.
Figure 7.
Analysis of SlPPH during fruit ripening. A and B, Analysis of pigment composition during fruit ripening in SlPPH-silencing lines. A, Quantification of total chlorophyll. Note that silencing of SlPPH causes a transient delay of chlorophyll degradation. B, Quantification of pheophytin a. Note that SlPPH-silencing lines transiently accumulate pheophytin a. GM, Green mature; B, breaker. C and D, Phytol hydrolytic activities of tomato chromoplasts at the breaker + 2 d stage. Pheophytin a + b or chlorophyll a + b was used as substrate, and the formation of the respective products (pheophorbide a or chlorophyllide a) was analyzed by HPLC. Note that, because the b forms of substrates were present in only small quantities in the assays, their products were not quantified. C, Hydrolytic activities in nonsolubilized chromoplasts (−Triton X-100). D, Total hydrolytic activities in solubilized chromoplasts (+Triton X-100). Data are means of three biological replicates ± sd. FW, Fresh weight; WT, wild type.
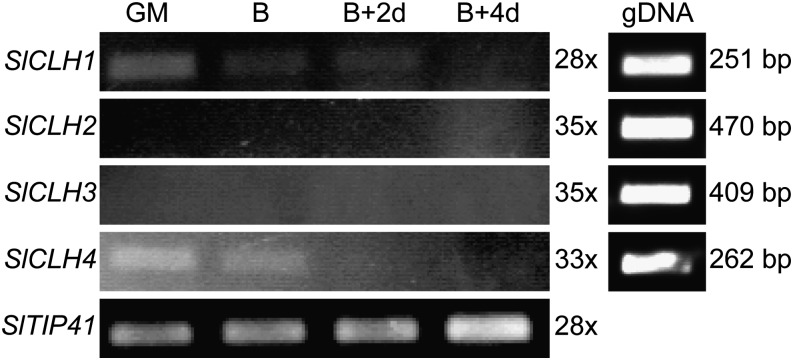
To test whether CLHs could be important, we analyzed tomato CLH (SlCLH) expression during leaf senescence and fruit ripening. The tomato genome contains four CLH genes. The deduced proteins of two of them (Solyc06g053980 = SlCLH1 and Solyc09g082600 = SlCLH3) clustered with Arabidopsis CLH2 in a phylogenetic tree, while Solyc09g06520 (SlCLH2) and Solyc12g005300 (SlCLH4) were more similar to AtCLH1 (Supplemental Fig. S2A; Lira et al., 2014). With the exception of a slight up-regulation of SlCLH1 during leaf senescence, the expression of none of the SlCLHs as analyzed by semiquantitative RT-PCR correlated with the progression of leaf senescence (Supplemental Fig. S2B) or fruit ripening (Fig. 8). Transcripts for SlCLH3 were hardly detectable. This confirmed published qPCR data on SlCLH expression (Lira et al., 2014). It is interesting that these results reflect the situation in Arabidopsis, where CLH1 expression decreases during leaf senescence (Zimmermann et al., 2004; Winter et al., 2007) and PPH represents the major dephytylating activity (Schelbert et al., 2009).
Figure 8.
Gene expression analyses of SlCLH1 to SlCLH4 during fruit ripening in wild-type tomato. SlTIP41 was used as a control (Expósito-Rodríguez et al., 2008). Expression was analyzed with the number of PCR cycles as indicated. PCR products were separated on agarose gels and visualized with ethidium bromide. PCR on genomic DNA (gDNA) was performed to test the efficacy of the primers used for gene expression analyses. The sizes of the fragments amplified with genomic DNA are indicated on the right. GM, Green mature; B, breaker.
DISCUSSION
The identification of pheophorbide a as an intermediate of chlorophyll breakdown (Hörtensteiner et al., 1995) demonstrated that dephytylation is an early step of breakdown and occurs within plastids. Phytol removal is important for two reasons: (1) it renders chlorophyll breakdown products water soluble (that is, a prerequisite for their ultimate storage in the vacuole as phyllobilins; Matile et al., 1988; Kräutler and Hörtensteiner, 2013); and (2) removal of phytol is regarded as a prerequisite for the degradation of chlorophyll-binding proteins during senescence. Thus, mutants that are incapable of phytol hydrolysis, such as Arabidopsis pph-1 and rice nonyellow coloring3 (nyc3), exhibit a stay-green phenotype during leaf senescence and retain large quantities of light-harvesting complex subunits (Morita et al., 2009; Schelbert et al., 2009). Likewise, mutations in steps upstream of dephytylation, such as SGR and NYC1 (that is, a CCE involved in chlorophyll b-to-chlorophyll a reduction), also result in stay-greenness coupled to apoprotein retention (Kusaba et al., 2007; Park et al., 2007; Aubry et al., 2008; Barry et al., 2008; Horie et al., 2009).
Pigment dephytylation was considered for more than a century to be catalyzed by CLHs (Willstätter and Stoll, 1913) that are able to hydrolyze both chlorophyll and pheophytin (Schelbert et al., 2009). However, their molecular identification in 1999 (Jacob-Wilk et al., 1999; Tsuchiya et al., 1999) was puzzling, since, in contrast with the predicted localization within plastid membranes, some of the cloned CLHs were suggested to localize extraplastidically and all of the identified genes encoded predicted soluble rather than membrane-localizing proteins (Takamiya et al., 2000; Hörtensteiner, 2006). Several reports that address the subcellular localization of CLHs have been published with conflicting results. Thus, the two Arabidopsis CLHs were shown to reside in the cytosol (Schenk et al., 2007), while the CLHs of citrus and Ginkgo biloba localize within plastids (Okazawa et al., 2006; Azoulay Shemer et al., 2008). The conflicting subcellular localization of CLHs prompted the hypothesis that additional extraplastidial breakdown pathways for chlorophyll may exist (Takamiya et al., 2000). However, demonstration that chloroplast-localized PAO, acting downstream of dephytylation, is involved in chlorophyll breakdown (Hörtensteiner et al., 1995; Sakuraba et al., 2012) and the finding that the absence of both Arabidopsis CLHs had only a marginal effect on chlorophyll breakdown (Schenk et al., 2007) challenged this idea and questioned whether CLHs are involved at all. The identification of PPH as a pheophytin-specific phytol hydrolase (Schelbert et al., 2009) supported this view, and now it is commonly accepted that PPHs rather than CLHs are responsible for leaf senescence-related chlorophyll breakdown (Tanaka et al., 2011), at least in Arabidopsis and rice. The results of this study extend this assumption to tomato, because, as in Arabidopsis pph mutants (Schelbert et al., 2009), leaf yellowing was largely blocked in SlPPH-silencing lines and significant amounts of pheophytin a accumulated upon senescence induction in the dark (Fig. 6). Furthermore, genes encoding highly conserved PPHs are commonly present in higher plants (Fig. 3), allowing the extrapolation that pheophytin-specific dephytylation by PPHs may be a common feature of chlorophyll breakdown during leaf senescence.
Chlorophyll breakdown, however, not only occurs during leaf senescence but also, for example, during leaf desiccation in resurrection plants (Craterostigma pumilum and Xerophyta viscosa), during fruit ripening and seed maturation (Armstead et al., 2007; Delmas et al., 2013; Christ et al., 2014). Analysis of the dephytylation step in ripening fruits has been limited nearly exclusively to Citrus spp. (Amir-Shapira et al., 1987; Trebitsh et al., 1993; Jacob-Wilk et al., 1999; Azoulay Shemer et al., 2008), where leaf senescence-related chlorophyll breakdown has not been studied in detail (Katz et al., 2005). We chose tomato as a model because, besides a rather short life cycle, it offers established genetic tools as well as well-defined methods for fruit ripening and leaf senescence analysis (Akhtar et al., 1999; Barry et al., 2008) and, thus, allowed the simultaneous analysis of dephytylation during leaf senescence and fruit ripening (Figs. 6 and 7). With the SlPPH-silencing lines produced here, we are able to demonstrate that PPH surely participates in chlorophyll breakdown also during tomato fruit ripening, but its contribution is limited. Based on activity measurements on isolated chromoplast membranes (Fig. 7), we conclude that other phytol hydrolytic activities are present in ripening tomato fruits that either naturally participate in dephytylation as well or compensate for the absence of PPH in the silencing lines. The nature of these activities remains elusive; however, CLHs appeared as possible candidates. CLHs have been shown to dephytylate chlorophyll and pheophytin in vitro (Schelbert et al., 2009). Furthermore, CLHs exhibit an intriguing latency, which requires their in vitro activation by detergents or high concentrations of solvents (Amir-Shapira et al., 1986; Matile et al., 1999). In our assays, solubilization of chromoplasts with Triton X-100 increased the overall pheophytin hydrolytic activity by about 2-fold, indicating that CLHs contribute to the overall activity. This view that tomato CLHs may participate in dephytylation and/or may substitute for PPH seems to be in agreement with studies in citrus, where CLH was shown to play a major role in fruit ripening (Trebitsh et al., 1993; Brandis et al., 1996; Jacob-Wilk et al., 1999). Thus, citrus CLH was detected in chloroplasts by in situ immunofluorescence labeling. Furthermore, the enzyme is proteolytically processed at the N- and C termini, posttranslational modifications that are unrelated to chloroplast targeting but were shown to be important for activity (Harpaz-Saad et al., 2007; Azoulay Shemer et al., 2008; Azoulay-Shemer et al., 2011). Finally, citrus CLH is transcriptionally up-regulated during ethylene-induced citrus ripening (Jacob-Wilk et al., 1999). Because of the presence of four CLH genes in the tomato genome, analysis of CLH function during fruit ripening was beyond the scope of this work and needs to be addressed in a separate study in the future. Nevertheless, we analyzed CLH expression, but in contrast to PPH expression (Fig. 2), CLH transcript levels were rather low and did not correlate with the progression of fruit ripening or leaf senescence (Fig. 8; Supplemental Fig. S2B). We cannot exclude, however, the possibility that, also in tomato, CLHs may be regulated posttranscriptionally rather than at the expression level. Nevertheless, it is interesting that CLHs have not been identified in proteome analyses of tomato chromoplasts, in contrast to many CCEs, such as PPH, SGR, PAO and RCCR (Barsan et al., 2010, 2012; Wang et al., 2013), pointing to their presence, if at all, in rather low abundance.
Thus, despite the implication that CLHs may contribute to the overall phytol hydrolytic activity observed in tomato fruit chromoplasts, other explanations are possible as well. The genome of tomato, like other species (Schelbert et al., 2009), encodes several hundred α/β-hydrolases, many of which are predicted to localize to plastids. The common feature of such hydrolases is the presence of a catalytic triad with a conserved Ser residue (Tsuchiya et al., 2003), but they group into distinct protein families based on sequence similarity. As an example, both tomato PPH and CLHs belong to the α/β-hydrolases, but their overall sequence identity is below 27%. It is possible that one or several other, so far unidentified, plastid-localizing hydrolases are involved in dephytylation during chlorophyll breakdown in tomato fruits. These activities may also contribute to the remaining chlorophyll degradation activities observed in leaves of SlPPH-silencing lines (Fig. 6B) and Arabidopsis pph mutants (Schelbert et al., 2009).
This view is supported from investigations in Arabidopsis, where VITAMIN E5 (VTE5) has been shown to be responsible for the biosynthesis of 80% of α-tocopherol present in seeds (Valentin et al., 2006). VTE5 catalyzes the phosphorylation of phytol to phytyl phosphate (i.e. the first of two phosphorylation steps required to synthesize phytyl pyrophosphate for salvage into tocopherol; DellaPenna and Last, 2006; Ischebeck et al., 2006). It is commonly accepted that phytol hydrolysis of chlorophyll is a major source of phytol for tocopherol biosynthesis. Surprisingly, however, the absence of PPH, the two CLHs, or all three genes in a triple mutant does not affect seed tocopherol content in Arabidopsis (Zhang et al., 2014), pointing to a different phytol hydrolytic activity. Furthermore, triple pph-1 clh1 clh2 mutants do not show an embryo stay-green phenotype (Zhang et al., 2014), contrary to mutants deficient in SGR or NYC1 (Nakajima et al., 2012; Delmas et al., 2013). Thus, it appears that SGR and some CCEs, such as NYC1 and PAO, are commonly active during chlorophyll degradation in different plant tissues, while PPH is active in leaf senescence but plays only a minor role during fruit ripening and seed development.
MATERIALS AND METHODS
Plant Material and Senescence Induction
Seeds of tomato (Solanum lycopersicum) ecotype Ailsa Craig wild type and gf were obtained from Yoram Eyal (Volcani Center). For analysis of fruit ripening, plants were grown in soil under nutrient-sufficient conditions; plants were kept in small pots with limited nutrient supply to induce timely leaf senescence. Growth was under long-day conditions in a greenhouse with fluence rates of 100 to 200 µmol photons m−2 s−1 at 25°C and 60% humidity. Alternatively, sterilized seeds were placed on one-half-strength Murashige and Skoog (MS) medium (2.2 g L−1 MS basal salt mixture, 10 g L−1 Suc, and 0.6% [w/v] phyotagar), and plants were grown for 4 to 6 weeks at 80 µmol photons m−2 s−1 at 21°C. Plants were subsequently transferred to soil and grown for another 5 to 6 weeks in a phytotron (12-h/12-h light/dark cycle [40 to 50 µmol photons m−2 s−1], 60% humidity, and 22°C). For induction of senescence with ethylene, leaves of phytotron-grown plants were placed on filter paper soaked with 1 mm ethephon and incubated in the dark at room temperature. Likewise, leaves of Arabidopsis (Arabidopsis thaliana) Columbia-0 and pph-1 (Schelbert et al., 2009) were placed on wet filter paper and incubated in the dark.
Analysis of Chlorophyll and Catabolites
For the determination of chlorophyll and pheophytin concentrations, pigments were extracted from tomato leaf tissue and flavedo of fruits by homogenization in liquid nitrogen and subsequent extraction into 90% (v/v) acetone and 10% (v/v) 0.2 m Tris-HCl, pH 8 (Schelbert et al., 2009; Christ et al., 2012). After centrifugation (2 min, 16,000g, and 4°C), supernatants were used for spectrophotometric analysis (Strain et al., 1971) or for reverse-phase HPLC (C18 Hypersil ODS column [125 × 4.0 mm, 5 µm], Linear 206 PHD-diode array detector [365–700 nm], and ChromQuest version 2.51 software [Thermo Fisher Scientific]) as described (Langmeier et al., 1993). Phyllobilins were extracted and analyzed by HPLC as described (Christ et al., 2012).
Biocomputational Methods and Phylogenetic Analysis
SlPPH (Solyc01g088090.2) and SlCLHs (SlCLH1, Solyc06g053980.2; SlCLH2, Solyc09g065620.2; SlCLH4, Solyc12g005300.1; and SlCLH3, Solyc09g082600.1) were identified by BLASTP searches (Altschul et al., 1997) with the Sol Genomics Network database (http://solgenomics.net/) using Arabidopsis PPH (AtPPH) and CLH1 (AtCLH1), respectively, as queries. Full-length protein sequences of PPH homologs from other species were identified by BLASTP searches at the National Center for Biotechnology Information (http://ncbi.nlm.nih.gov/). Phylogenetic trees (Fig. 3A; Supplemental Fig. S2A) were estimated using the maximum likelihood method (http://phylogeny.fr; Dereeper et al., 2008). Branch support values of the phylogram are based on 100 nonparametric bootstrap replicates. The sequence alignment between SlPPH and AtPPH (Fig. 3B) was performed using the program DIALIGN (http://bibiserv.techfak.uni-bielefeld.de/dialign/submission.html; Morgenstern, 2004).
Generation of Transgenic Tomato Lines and pph-1 Complementation
cDNA derived from mature green tomato fruits was obtained from Yoram Eyal and was used to clone the full-length sequence of SlPPH [SlPPH(long)]. For silencing of SlPPH by RNAi, a 400-bp cDNA sense and antisense fragment of SlPPH was amplified using Pfu polymerase (Promega) with gene-specific primers as listed in Supplemental Table S1 and cloned in the silencing vector pHannibal (Wesley et al., 2001). A NotI fragment containing the silencing construct between the CaMV 35S promoter and an OCTOPINE SYNTHASE terminator was excised and subcloned into pGreen0029 (Hellens et al., 2000). For ectopic complementation of pph-1, full-SlPPH(long) was cloned in a pGreen0029-derived vector (pGr-At-RCCR; Pružinská et al., 2007) that harbors a CaMV 35S promoter and a CaMV poly(A) terminator. For that, the NdeI/EcoRI insert of pGr-At-RCCR was replaced with a PCR-amplified (for primers, see Supplemental Table S1), NdeI/EcoRI-restricted fragment containing SlPPH(long). After verifying the inserts by sequencing, both constructs were transformed into Agrobacterium tumefaciens strain GV3101 together with pSOUP (Hellens et al., 2000). Arabidopsis pph-1 mutants were transformed by the floral dip method (Clough and Bent, 1998). Transformants were selected on kanamycin, and plants of the T1 generation were used for further experiments.
To generate SlPPH-silencing tomato lines, seeds were sterilized with 1.2% (v/v) sodium hypochlorite for 15 min. Seeds were rinsed three times with sterile water and placed on medium (one-half-strength MS, 1.5% [w/v] Suc, and 0.8% [w/v] phytagar) in 10-cm-high sterile glass pots. After 9 to 12 d of growth under long-day conditions in a culture room at 80 µmol photons m−2 s−1 at 21°C, cotyledons were excised by removing 2 to 3 mm of the leaf blades from both the proximal and distal ends. Cotyledons were placed upside down in petri dishes containing D1 medium (4.4 g L−1 MS salts including B5 vitamins, 30 g L−1 Glc, 1 mg L−1 zeatin, 0.1 mg L−1 naphthyl acetic acid, 1 mg L−1 folic acid, 2 mm MES-KOH, pH 5.6–5.7, and 8 g L−1 phytagar) and incubated in the culture room for 2 d. A. tumefaciens cells harboring the silencing construct were grown overnight at 28°C. Cells of a 20-mL culture were collected by centrifugation (6,000g for 15 min), and the pellet was resuspended in MSO-KOH, pH 5.6 (4.4 g L−1 MS salts including B5 vitamins and 20 g L−1 Suc) to an optical density at 600 nm of 0.4 to 0.5. Acetosyringone (100 µm) was added, and the culture was grown for another 2 h at 28°C. For transformation, cotyledons were incubated with the bacterial culture for 2 h in the dark. After another 2 to 3 d of cultivation on D1 medium, the cotyledons were transferred to D1 medium containing kanamycin (75 mg L−1) and timenten (100 mg L−1). Shoot regeneration was detected after about 30 d, and respective plantlets were then transferred to DL medium (4.4 g L−1 MS salts including B5 vitamins, 20 g L−1 Glc, 2 mg L−1 indole-3-butyric acid, 1 mg L−1 folic acid, 2 mm MES-KOH, pH 5.6–5.7, and 8 g L−1 agar) for root induction. Rescued transformants were transferred to soil.
GFP Fusion Protein Analysis
Both SlPPH cDNA varieties, SlPPH(long) and SlPPH(short), were amplified using PCR Extender polymerase (5Prime) with the gene-specific primers listed in Supplemental Table S1. After restriction digestion with XmaI, the fragment was cloned into the corresponding site of pUC18-spGFP6 (Meyer et al., 2006), thereby producing C-terminal fusions of SlPPH with GFP (SlPPH-GFP). Sequence accuracy was confirmed by sequencing. Mesophyll protoplasts were isolated from leaves of Arabidopsis (Columbia-0) grown under short-day conditions according to published procedures (Endler et al., 2006). Leaves were incubated in the dark for 3 d prior to protoplast isolation. Cell numbers were quantified with a Neubauer chamber, and density was adjusted to 2 × 106 protoplasts mL−1. Transformation of protoplasts was performed with 20% (w/v) polyethylene glycol as published (Meyer et al., 2006). Transformed protoplasts were incubated in the dark at room temperature for 24 to 48 h prior to confocal laser-scanning microscopic analysis (Leica TCS SP5; Leica Microsystems). GFP fluorescence was imaged at an excitation wavelength of 488 nm, and the emission signal was detected between 495 and 530 nm for GFP and between 643 and 730 nm for chlorophyll autofluorescence.
RNA Isolation, Semiquantitative RT-PCR, and qPCR
For semiquantitative RT-PCR, total RNA was extracted from leaf tissues or the flavedo of fruits using TRIzol according to the manufacturer’s instructions (Life Technologies). Polyvinylpolypyrrolidone was added to ground tissue for extraction. After DNA digestion with RQ1 DNase (Promega), first-strand cDNA was synthesized from total RNA using either the RETROscript kit (Life Technologies) or Moloney murine leukemia virus reverse transcriptase (Promega) and oligo(dT)15 primers (Promega). PCR was performed with gene-specific primers as listed in Supplemental Table S1. To control primer suitability for RT-PCR analysis, PCR was run with genomic DNA extracted from tomato fruits. Tomato type 2A-interacting protein41 (SlTIP41) (Solyc10g049850.1) was used as the control gene (Expósito-Rodríguez et al., 2008).
RNA extraction for qPCR analysis and qPCR were performed as described (Quadrana et al., 2013). The PCR primers used are listed in Supplemental Table S1. All reactions were performed with two technical replicates and at least three biological replicates. mRNA levels were quantified using the 7500 Real-Time PCR system (Applied Biosystems) and SYBR Green Master Mix (Applied Biosystems). Data were analyzed with LinRegPCR software (Ruijter et al., 2009) to obtain cycle threshold values and to calculate primer efficiency. Expression values were normalized to the mean of two constitutively expressed genes, TIP41 and EXPRESSED (Solyc07g025390.2.1; Expósito-Rodríguez et al., 2008). A permutation test, which lacks sample distribution assumptions (Pfaffl et al., 2002), was used to detect statistical (P < 0.05) differences in expression levels between samples using the algorithms in the fgStatistics software (http://sites.google.com/site/fgStatistics/).
Analysis of Recombinant SlPPH
For heterologous expression of SlPPH in Escherichia coli, a truncated cDNA fragment, lacking the 61 5′-terminal amino acids encoding the likely chloroplast transit peptide, was produced by PCR using Extender polymerase (5Prime) with primers as listed in Supplemental Table S1. After restriction digestion with EcoRI, the fragment was cloned into pMal_c2 (New England Biolabs), producing a truncated MBP-SlPPH fusion (MBP-ΔSlPPH). After verifying the insert by sequencing, the construct was transformed into E. coli BL21(DE3). Recombinant SlPPH protein was expressed and cells were lysed as described (Schelbert et al., 2009). PPH activity assays (300 µL) were performed with 15 µL of crude protein extract (approximately 130 µg of soluble protein), 0.1 mm pheophytin a and/or chlorophyll a (final acetone concentration, 6.7% [v/v]), and 0.1 m HEPES-KOH, pH 8, containing 1 mm EDTA. In assays with substrate mixtures, pheophytin a and chlorophyll a were present at concentrations of 35 and 65 µm, respectively. After incubation at 34°C for various time periods, reactions were stopped by adding 2 volumes of acetone and analyzed by reverse-phase HPLC as described (Schelbert et al., 2009). Pheophytin a was produced from pure chlorophyll a (LivChem) by acidification as described (Schelbert et al., 2009).
Chromoplast Isolation and Activity Measurements
Chromoplasts of tomato mesocarp tissue at the breaker + 2 d stage were isolated as published for red pepper (Capsicum annuum; Christ et al., 2012) with some modifications. Mesocarp tissue was blended in a Sorvall mixer three times for 5 s with isolation buffer (1 mL g−1 fresh weight) containing 400 mm Suc, 50 mm Tris-MES, pH 8, 2 mm EDTA, 10 mm polyethylene glycol 4000, 5 mm dithiothreitol, and 5 mm L(+)-ascorbic acid. Subsequently, the suspension was filtered through two layers of gauze and centrifuged (10 min at 12,000g). The pellet was carefully resuspended in isolation buffer (1 mL g−1 fresh weight). After repeating the centrifugation step, chromoplasts were resuspended in Tris-MES buffer (0.05 mL g−1 fresh weight) containing 25 mm Tris-MES, pH 8, and 5 mm L(+)-ascorbic acid. Isolated chromoplasts were divided into two fractions and either supplemented with 0.1 volume of Tris-MES buffer containing 10% (v/v) Triton X-100 to obtain a final Triton X-100 concentration of 1% (v/v) (+Triton X-100) or chromoplasts were supplemented with 0.1 volume of Tris-MES buffer (−Triton X-100). Both chromoplast fractions were incubated with rotation in the dark at 4°C for 30 min. Aliquots of isolated chromoplasts were frozen in liquid nitrogen and stored at −80°C. Phytol hydrolysis assays (total volume of 100 µL) consisted of 10 µL of chromoplasts (corresponding to 0.2 g fresh weight), 70 µm pheophytin a/b or chlorophyll a/b, with about 10-fold excess of the a pigment in both cases (3% [v/v] final acetone concentration) and reaction buffer (0.1 m HEPES-KOH, pH 8, and 1 mm EDTA). After incubation at 34°C for 45 min, reactions were stopped by adding 2 volumes of acetone. After centrifugation (16,000g for 2 min), samples were analyzed by reverse-phase HPLC as described (Langmeier et al., 1993). Substrate production and quantification were performed as described (Schelbert et al., 2009).
GenBank or Sol Genomics Network (http://solgenomics.net/) identification numbers for the DNA/protein sequences used in this work are as follows. PPH sequences: Aegilops tauschii, 475611823; Amborella trichopoda, 548840076; Arabidopsis lyrata, 297811489; Arabidopsis, 15240707 (AtPPH, At5g13800); Brachypodium distachyon, 357123819; Capsella rubella, 565459260; Cicer arietinum, 502127590; Citrus clementina, 567892823; Citrus sinensis, 568858818; Cucumis sativus, 449436343; Eutrema salsugineum, 567173584; Fragaria vesca, 470134497; Genlisea aurea, 527208569; soybean, 356539136 (Glyma1), 356531629 (Glyma2), 356542875 (Glyma3); barley, 326498881; Lotus japonicus, 388497996; Medicago truncatula, 357458507; Nicotiana tabacum, 156763846; Oryza brachyantha, 573959173; rice, 115467988; common bean, 561022305 (Phavu1), 561004436 (Phavu2); Populus trichocarpa, 224106163; Prunus persica, 462415467; Setaria italica, 514804304; tomato, 460367643 (SlPPH, Solyc01g088090.2); Solanum tuberosum, 565357100; Sorghum bicolor, 242060434; Theobroma cacao, 508704687; Triticum urartu, 473998920; Vitis vinifera, 225449963; and Zea mays, 226530215. Additional sequences for Arabidopsis: AtCLH1, 30912637 (At1g19670); AtCLH2, 30912739 (At5g43860); SGR, 75100772 (At4g22920); and PAO, 41688605 (At3g44880). Additional sequences for tomato: SlCLH1, 460390857 (Solyc06g053980.2); SlCLH2, 460403437 (Solyc09g065620.2); SlCLH4, 460412186 (Solyc12g005300.1); SlCLH3 (Solyc09g082600.1); SlTIP41, 460406627 (Solyc10g049850.1); and EXPRESSED, 460394765 (Solyc07g025390.2.1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Expression analysis of SlPPH in SlPPH-silencing lines.
Supplemental Figure S2. Analysis of tomato CLHs.
Supplemental Table S1. List of primers used in this study.
Acknowledgments
We thank Yoram Eyal for providing tomato seeds and cDNA, and gardeners Christian Frey and Kari Huwiler, for taking care of the plants.
Glossary
- pFCC
primary fluorescent chlorophyll catabolite
- CCE
chlorophyll catabolic enzyme
- RT
reverse transcription
- qPCR
quantitative PCR
- cDNA
complementary DNA
- CaMV
cauliflower mosaic virus
- RNAi
RNA interference
- MS
Murashige and Skoog
Footnotes
This work was supported by the Swiss National Science Foundation (grant no. 31003A–132603 to S.H.).
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Akhtar MS, Goldschmidt EE, John I, Rodoni S, Matile P, Grierson D. (1999) Altered patterns of senescence and ripening in gf, a stay-green mutant of tomato (Lycopersicon esculentum Mill.). J Exp Bot 50: 1115–1122 [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir-Shapira D, Goldschmidt E, Altman A. (1986) Autolysis of chlorophyll in aqueous and detergent suspensions of chloroplast fragments. Plant Sci 43: 201–206 [Google Scholar]
- Amir-Shapira D, Goldschmidt EE, Altman A. (1987) Chlorophyll catabolism in senescing plant tissues: in vivo breakdown intermediates suggest different degradative pathways for Citrus fruit and parsley leaves. Proc Natl Acad Sci USA 84: 1901–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstead I, Donnison I, Aubry S, Harper J, Hörtensteiner S, James C, Mani J, Moffet M, Ougham H, Roberts L, et al. (2007) Cross-species identification of Mendel’s I locus. Science 315: 73. [DOI] [PubMed] [Google Scholar]
- Aubry S, Mani J, Hörtensteiner S. (2008) Stay-green protein, defective in Mendel’s green cotyledon mutant, acts independent and upstream of pheophorbide a oxygenase in the chlorophyll catabolic pathway. Plant Mol Biol 67: 243–256 [DOI] [PubMed] [Google Scholar]
- Azoulay Shemer T, Harpaz-Saad S, Belausov E, Lovat N, Krokhin O, Spicer V, Standing KG, Goldschmidt EE, Eyal Y. (2008) Citrus chlorophyllase dynamics at ethylene-induced fruit color-break: a study of chlorophyllase expression, posttranslational processing kinetics, and in situ intracellular localization. Plant Physiol 148: 108–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azoulay-Shemer T, Harpaz-Saad S, Cohen-Peer R, Mett A, Spicer V, Lovat N, Krokhin O, Brand A, Gidoni D, Standing KG, et al. (2011) Dual N- and C-terminal processing of citrus chlorophyllase precursor within the plastid membranes leads to the mature enzyme. Plant Cell Physiol 52: 70–83 [DOI] [PubMed] [Google Scholar]
- Barry CS. (2009) The stay-green revolution: recent progress in deciphering the mechanisms of chlorophyll degradation in higher plants. Plant Sci 176: 325–333 [Google Scholar]
- Barry CS, McQuinn RP, Chung MY, Besuden A, Giovannoni JJ. (2008) Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol 147: 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsan C, Sanchez-Bel P, Rombaldi C, Egea I, Rossignol M, Kuntz M, Zouine M, Latché A, Bouzayen M, Pech JC. (2010) Characteristics of the tomato chromoplast revealed by proteomic analysis. J Exp Bot 61: 2413–2431 [DOI] [PubMed] [Google Scholar]
- Barsan C, Zouine M, Maza E, Bian W, Egea I, Rossignol M, Bouyssie D, Pichereaux C, Purgatto E, Bouzayen M, et al. (2012) Proteomic analysis of chloroplast-to-chromoplast transition in tomato reveals metabolic shifts coupled with disrupted thylakoid biogenesis machinery and elevated energy-production components. Plant Physiol 160: 708–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovsky Y, Paran I. (2008) Chlorophyll breakdown during pepper fruit ripening in the chlorophyll retainer mutation is impaired at the homolog of the senescence-inducible stay-green gene. Theor Appl Genet 117: 235–240 [DOI] [PubMed] [Google Scholar]
- Brandis A, Vainstein A, Goldschmidt EE. (1996) Distribution of chlorophyllase among components of chloroplast membranes in Citrus sinensis organs. Plant Physiol Biochem 34: 49–54 [Google Scholar]
- Chen LFO, Lin CH, Kelkar SM, Chang YM, Shaw JF. (2008) Transgenic broccoli (Brassica oleracea var. italicia) with antisense chlorophyllase (BoCLH1) delays postharvest yellowing. Plant Sci 174: 25–31 [Google Scholar]
- Christ B, Egert A, Süssenbacher I, Kräutler B, Bartels D, Peters S, Hörtensteiner S. (2014) Water deficit induces chlorophyll degradation via the ‘PAO/phyllobilin’ pathway in leaves of homoio- (Craterostigma pumilum) and poikilochlorophyllous (Xerophyta viscosa) resurrection plants. Plant Cell Environ (in press) [DOI] [PubMed] [Google Scholar]
- Christ B, Hörtensteiner S. (2014) Mechanism and significance of chlorophyll breakdown. J Plant Growth Regul 33: 4–20 [Google Scholar]
- Christ B, Schelbert S, Aubry S, Süssenbacher I, Müller T, Kräutler B, Hörtensteiner S. (2012) MES16, a member of the methylesterase protein family, specifically demethylates fluorescent chlorophyll catabolites during chlorophyll breakdown in Arabidopsis. Plant Physiol 158: 628–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- DellaPenna D, Last RL. (2006) Progress in the dissection and manipulation of plant vitamin E biosynthesis. Physiol Plant 126: 356–368 [Google Scholar]
- Delmas F, Sankaranarayanan S, Deb S, Widdup E, Bournonville C, Bollier N, Northey JG, McCourt P, Samuel MA. (2013) ABI3 controls embryo degreening through Mendel’s I locus. Proc Natl Acad Sci USA 110: E3888–E3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard J-F, Guindon S, Lefort V, Lescot M, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucl Acids Res 36: W465–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea I, Barsan C, Bian W, Purgatto E, Latché A, Chervin C, Bouzayen M, Pech J-C. (2010) Chromoplast differentiation: current status and perspectives. Plant Cell Physiol 51: 1601–1611 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A, Meyer S, Schelbert S, Schneider T, Weschke W, Peters SW, Keller F, Baginsky S, Martinoia E, Schmidt UG. (2006) Identification of a vacuolar sucrose transporter in barley and Arabidopsis mesophyll cells by a tonoplast proteomic approach. Plant Physiol 141: 196–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA. (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8: 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpaz-Saad S, Azoulay T, Arazi T, Ben-Yaakov E, Mett A, Shiboleth YM, Hörtensteiner S, Gidoni D, Gal-On A, Goldschmidt EE, et al. (2007) Chlorophyllase is a rate-limiting enzyme in chlorophyll catabolism and is posttranslationally regulated. Plant Cell 19: 1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens R, Edwards EA, Leyland NR, Bean S, Mullineaux PM. (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Hendry GAF, Houghton JD, Brown SB. (1987) The degradation of chlorophyll: a biological enigma. New Phytol 107: 255–302 [DOI] [PubMed] [Google Scholar]
- Horie Y, Ito H, Kusaba M, Tanaka R, Tanaka A. (2009) Participation of chlorophyll b reductase in the initial step of the degradation of light-harvesting chlorophyll a/b-protein complexes in Arabidopsis. J Biol Chem 284: 17449–17456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörtensteiner S. (2006) Chlorophyll degradation during senescence. Annu Rev Plant Biol 57: 55–77 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S. (2009) Stay-green regulates chlorophyll and chlorophyll-binding protein degradation during senescence. Trends Plant Sci 14: 155–162 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S. (2013) Update on the biochemistry of chlorophyll breakdown. Plant Mol Biol 82: 505–517 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S, Kräutler B. (2011) Chlorophyll breakdown in higher plants. Biochim Biophys Acta 1807: 977–988 [DOI] [PubMed] [Google Scholar]
- Hörtensteiner S, Vicentini F, Matile P. (1995) Chlorophyll breakdown in senescent cotyledons of rape, Brassica napus L.: enzymatic cleavage of phaeophorbide a in vitro. New Phytol 129: 237–246 [DOI] [PubMed] [Google Scholar]
- Ischebeck T, Zbierzak AM, Kanwischer M, Dörmann P. (2006) A salvage pathway for phytol metabolism in Arabidopsis. J Biol Chem 281: 2470–2477 [DOI] [PubMed] [Google Scholar]
- Jacob-Wilk D, Holland D, Goldschmidt EE, Riov J, Eyal Y. (1999) Chlorophyll breakdown by chlorophyllase: isolation and functional expression of the Chlase1 gene from ethylene-treated Citrus fruit and its regulation during development. Plant J 20: 653–661 [DOI] [PubMed] [Google Scholar]
- Katz E, Riov J, Weiss D, Goldschmidt EE. (2005) The climacteric-like behaviour of young, mature and wounded citrus leaves. J Exp Bot 56: 1359–1367 [DOI] [PubMed] [Google Scholar]
- Kräutler B. (2008) Chlorophyll breakdown and chlorophyll catabolites in leaves and fruit. Photochem Photobiol Sci 7: 1114–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kräutler B, Hörtensteiner S (2013) Chlorophyll breakdown: chemistry, biochemistry and biology. In GC Ferreira, KM Kadish, KM Smith, R Guilard, eds, Handbook of Porphyrin Science, Vol 28. World Scientific Publishing, Hackensack, NJ, pp 117–185 [Google Scholar]
- Kräutler B, Jaun B, Bortlik KH, Schellenberg M, Matile P. (1991) On the enigma of chlorophyll degradation: the constitution of a secoporphinoid catabolite. Angew Chem Int Ed Engl 30: 1315–1318 [Google Scholar]
- Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M, et al. (2007) Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19: 1362–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmeier M, Ginsburg S, Matile P. (1993) Chlorophyll breakdown in senescent leaves: demonstration of Mg-dechelatase activity. Physiol Plant 89: 347–353 [Google Scholar]
- Lira BS, de Setta N, Rosado D, Almeida J, Freschi L, Rossi M. (2014) Plant degreening: evolution and expression of tomato (Solanum lycopersicum) dephytylation enzymes. Gene 546: 359–366 [DOI] [PubMed] [Google Scholar]
- Luo ZD, Zhang JH, Li JH, Yang CX, Wang TT, Ouyang B, Li HX, Giovannoni J, Ye ZB. (2013) A STAY-GREEN protein SlSGR1 regulates lycopene and beta-carotene accumulation by interacting directly with SlPSY1 during ripening processes in tomato. New Phytologist 198: 442–452 [DOI] [PubMed] [Google Scholar]
- Matile P, Ginsburg S, Schellenberg M, Thomas H. (1988) Catabolites of chlorophyll in senescing barley leaves are localized in the vacuoles of mesophyll cells. Proc Natl Acad Sci USA 85: 9529–9532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matile P, Hörtensteiner S, Thomas H. (1999) Chlorophyll degradation. Annu Rev Plant Physiol Plant Mol Biol 50: 67–95 [DOI] [PubMed] [Google Scholar]
- Meyer A, Eskandari S, Grallath S, Rentsch D. (2006) AtGAT1, a high affinity transporter for g-aminobutyric acid in Arabidopsis thaliana. J Biol Chem 281: 7197–7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgenstern B. (2004) DIALIGN: multiple DNA and protein sequence alignment at BiBiServ. Nucleic Acids Res 32: W33–W36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita R, Sato Y, Masuda Y, Nishimura M, Kusaba M. (2009) Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J 59: 940–952 [DOI] [PubMed] [Google Scholar]
- Moser D, Matile P. (1997) Chlorophyll breakdown in ripening fruits of Capsicum annuum. J Plant Physiol 150: 759–761 [Google Scholar]
- Moser S, Müller T, Holzinger A, Lütz C, Jockusch S, Turro NJ, Kräutler B. (2009) Fluorescent chlorophyll catabolites in bananas light up blue halos of cell death. Proc Natl Acad Sci USA 106: 15538–15543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlecker W, Ongania KH, Kräutler B, Matile P, Hörtensteiner S. (1997) Tracking down chlorophyll breakdown in plants: elucidation of the constitution of a ‘fluorescent’ chlorophyll catabolite. Angew Chem Int Ed Engl 36: 401–404 [Google Scholar]
- Nakajima S, Ito H, Tanaka R, Tanaka A. (2012) Chlorophyll b reductase plays an essential role in maturation and storability of Arabidopsis seeds. Plant Physiol 160: 261–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazawa A, Tango L, Itoh Y, Fukusaki E, Kobayashi A. (2006) Characterization and subcellular localization of chlorophyllase from Ginkgo biloba. Z Naturforsch C 61: 111–117 [DOI] [PubMed] [Google Scholar]
- Park SY, Yu JW, Park JS, Li J, Yoo SC, Lee NY, Lee SK, Jeong SW, Seo HS, Koh HJ, et al. (2007) The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 19: 1649–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruzinská A, Anders I, Aubry S, Schenk N, Tapernoux-Lüthi E, Müller T, Kräutler B, Hörtensteiner S. (2007) In vivo participation of red chlorophyll catabolite reductase in chlorophyll breakdown. Plant Cell 19: 369–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrana L, Almeida J, Otaiza SN, Duffy T, Corrêa da Silva JV, de Godoy F, Asís R, Bermúdez L, Fernie AR, Carrari F, et al. (2013) Transcriptional regulation of tocopherol biosynthesis in tomato. Plant Mol Biol 81: 309–325 [DOI] [PubMed] [Google Scholar]
- Ren G, An K, Liao Y, Zhou X, Cao Y, Zhao H, Ge X, Kuai B. (2007) Identification of a novel chloroplast protein AtNYE1 regulating chlorophyll degradation during leaf senescence in Arabidopsis. Plant Physiol 144: 1429–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Zhou Q, Wu S, Zhang Y, Zhang L, Huang J, Sun Z, Kuai B. (2010) Reverse genetic identification of CRN1 and its distinctive role in chlorophyll degradation in Arabidopsis. J Integr Plant Biol 52: 496–504 [DOI] [PubMed] [Google Scholar]
- Rodoni S, Mühlecker W, Anderl M, Kräutler B, Moser D, Thomas H, Matile P, Hörtensteiner S. (1997) Chlorophyll breakdown in senescent chloroplasts: cleavage of pheophorbide a in two enzymic steps. Plant Physiol 115: 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AFM. (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuraba Y, Schelbert S, Park SY, Han SH, Lee BD, Andrès CB, Kessler F, Hörtensteiner S, Paek NC. (2012) STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell 24: 507–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelbert S, Aubry S, Burla B, Agne B, Kessler F, Krupinska K, Hörtensteiner S. (2009) Pheophytin pheophorbide hydrolase (pheophytinase) is involved in chlorophyll breakdown during leaf senescence in Arabidopsis. Plant Cell 21: 767–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk N, Schelbert S, Kanwischer M, Goldschmidt EE, Dörmann P, Hörtensteiner S. (2007) The chlorophyllases AtCLH1 and AtCLH2 are not essential for senescence-related chlorophyll breakdown in Arabidopsis thaliana. FEBS Lett 581: 5517–5525 [DOI] [PubMed] [Google Scholar]
- Strain HH, Cope BT, Svec WA. (1971) Analytical procedures for the isolation, identification, estimation and investigation of the chlorophylls. Methods Enzymol 23: 452–476 [Google Scholar]
- Takamiya KI, Tsuchiya T, Ohta H. (2000) Degradation pathway(s) of chlorophyll: what has gene cloning revealed? Trends Plant Sci 5: 426–431 [DOI] [PubMed] [Google Scholar]
- Tanaka A, Tanaka R. (2006) Chlorophyll metabolism. Curr Opin Plant Biol 9: 248–255 [DOI] [PubMed] [Google Scholar]
- Tanaka R, Kobayashi K, Masuda T. (2011) Tetrapyrrole metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0145, DOI: 10.1199/tab.0145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh T, Goldschmidt EE, Riov J. (1993) Ethylene induces de novo synthesis of chlorophyllase, a chlorophyll degrading enzyme, in Citrus fruit peel. Proc Natl Acad Sci USA 90: 9441–9445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T, Ohta H, Okawa K, Iwamatsu A, Shimada H, Masuda T, Takamiya K. (1999) Cloning of chlorophyllase, the key enzyme in chlorophyll degradation: finding of a lipase motif and the induction by methyl jasmonate. Proc Natl Acad Sci USA 96: 15362–15367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya T, Suzuki T, Yamada T, Shimada H, Masuda T, Ohta H, Takamiya K. (2003) Chlorophyllase as a serine hydrolase: identification of a putative catalytic triad. Plant Cell Physiol 44: 96–101 [DOI] [PubMed] [Google Scholar]
- Valentin HE, Lincoln K, Moshiri F, Jensen PK, Qi Q, Venkatesh TV, Karunanandaa B, Baszis SR, Norris SR, Savidge B, et al. (2006) The Arabidopsis vitamin E pathway gene5-1 mutant reveals a critical role for phytol kinase in seed tocopherol biosynthesis. Plant Cell 18: 212–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YQ, Yang Y, Fei Z, Yuan H, Fish T, Thannhauser TW, Mazourek M, Kochian LV, Wang X, Li L. (2013) Proteomic analysis of chromoplasts from six crop species reveals insights into chromoplast function and development. J Exp Bot 64: 949–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang M, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbot D, Stoutjesdijk PA, et al. (2001) Construct design for efficient, effective and high-throuput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Willstätter R, Stoll A (1913) Die Wirkungen der Chlorophyllase. In R Willstätter, A Stoll, eds, Untersuchungen über Chlorophyll. Verlag Julius Springer, Berlin, pp 172–187 [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Liu TQ, Ren GD, Hörtensteiner S, Zhou YM, Cahoon EB, Zhang CY. (2014) Chlorophyll degradation: the tocopherol biosynthesis related phytol hydrolase in Arabidopsis seeds is still missing. Plant Physiol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136: 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Han L, Pislariu C, Nakashima J, Fu C, Jiang Q, Quan L, Blancaflor EB, Tang Y, Bouton JH, et al. (2011) From model to crop: functional analysis of a STAY-GREEN gene in the model legume Medicago truncatula and effective use of the gene for alfalfa improvement. Plant Physiol 157: 1483–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]