Associative transcriptomics in Brassica napus identified candidate genes for the control of variation in nitrate, phosphate, and sulfate contents.
Abstract
To assess the variation in nutrient homeostasis in oilseed rape and to identify the genes responsible for this variation, we determined foliar anion levels in a diversity panel of Brassica napus accessions, 84 of which had been genotyped previously using messenger RNA sequencing. We applied associative transcriptomics to identify sequence polymorphisms linked to variation in nitrate, phosphate, or sulfate in these accessions. The analysis identified several hundred significant associations for each anion. Using functional annotation of Arabidopsis (Arabidopsis thaliana) homologs and available microarray data, we identified 60 candidate genes for controlling variation in the anion contents. To verify that these genes function in the control of nutrient homeostasis, we obtained Arabidopsis transfer DNA insertion lines for these candidates and tested them for the accumulation of nitrate, phosphate, and sulfate. Fourteen lines differed significantly in levels of the corresponding anions. Several of these genes have been shown previously to affect the accumulation of the corresponding anions in Arabidopsis mutants. These results thus confirm the power of associative transcriptomics in dissection of the genetic control of complex traits and present a set of candidate genes for use in the improvement of efficiency of B. napus mineral nutrition.
Plants require 14 essential mineral nutrients (Marschner, 2012). The nutrients most commonly limiting plant growth are nitrogen, phosphorus, potassium, and sulfur, which are present in relatively large amounts in plant tissues and therefore named macronutrients (Maathuis, 2009; Marschner, 2012). Nitrogen, phosphorus, and sulfur are taken up as oxygenic anions and either stored in the vacuoles or assimilated into organic compounds. The availability of these minerals in soil is usually low, so for intensive agriculture it has to be improved by fertilization, adding significant monetary and environmental costs. A major target for crop improvement, therefore, is to relieve their dependence on high levels of mineral fertilizers and improve nutrient use efficiency (NUE; Parry and Hawkesford, 2012). NUE is defined as yield per unit of input (Good et al., 2004). NUE depends on the ability to efficiently take up the nutrient from the soil but also on transport, storage, mobilization, usage within the plant, and even the environment (Good et al., 2004; Rengel and Marschner, 2005). The partitioning of the nutrients between vacuolar storage and assimilation is thus an important contributing factor of their use efficiency. Several approaches have been taken to understand the genetic basis of nutrient homeostasis. First, the response of plants to nutrient deficiency stress has been explored to identify processes affected by such stress and regulatory networks (Hammond et al., 2003; Hirai et al., 2003, 2005; Wang et al., 2003, 2004; Wu et al., 2003; Nikiforova et al., 2005; Krouk et al., 2010).
Another major approach to dissect the control of complex traits, such as NUE, makes use of natural genetic variation (Loudet et al., 2003; Gallais and Hirel, 2004; Chardon et al., 2012; Weigel, 2012). These traits can be analyzed through quantitative trait locus (QTL) analysis (Loudet et al., 2003, 2007; Harada et al., 2004; Reymond et al., 2006; Habash et al., 2007; Ding et al., 2010) or genome-wide association studies (GWAS; Atwell et al., 2010; Chan et al., 2011; Harper et al., 2012). The usefulness of GWAS has been demonstrated by capturing numerous well-characterized candidate genes (Aranzana et al., 2005; Atwell et al., 2010). Traits connected with the accumulation of mineral elements have also been analyzed successfully using GWAS (Atwell et al., 2010; Chao et al., 2012). Understanding the control of nutrient homeostasis is particularly important for crop plants, as it may contribute to improving NUE and the reduction of fertilizer use. Both QTL and GWAS have been used not only in model species but also directly in crops, such as oilseed rape (Ding et al., 2010; Harper et al., 2012). Due to its polyploidy, Brassica napus presents a significant challenge for GWAS, which, however, has been successfully circumvented by using messenger RNA sequencing for the identification of the polymorphic molecular markers in an approach termed associative transcriptomics (AT; Harper et al., 2012). The validity of the approach was demonstrated by the identification of a polymorphism in a MYB28 gene, encoding a transcription factor controlling the synthesis of aliphatic glucosinolates, being responsible for accumulation of the corresponding glucosinolates in seeds (Harper et al., 2012).
Here, we show results of application of the AT approach to dissect the genetic control of variation in nitrate, phosphate, or sulfate in leaves of B. napus. Candidate genes indicated by GWAS were tested using Arabidopsis (Arabidopsis thaliana) transfer DNA (T-DNA) insertion lines, resulting in 14 lines affected in the accumulation of the corresponding anion. These genes include genes known to affect nutrient homeostasis as well as those that have never been associated with nutrition. This study thus confirms the power of AT and presents a set of interesting genes for detailed analysis of their roles in the control of plant mineral nutrition and, potentially, the improvement of NUE of crops.
RESULTS AND DISCUSSION
Anion Content in B. napus Varieties
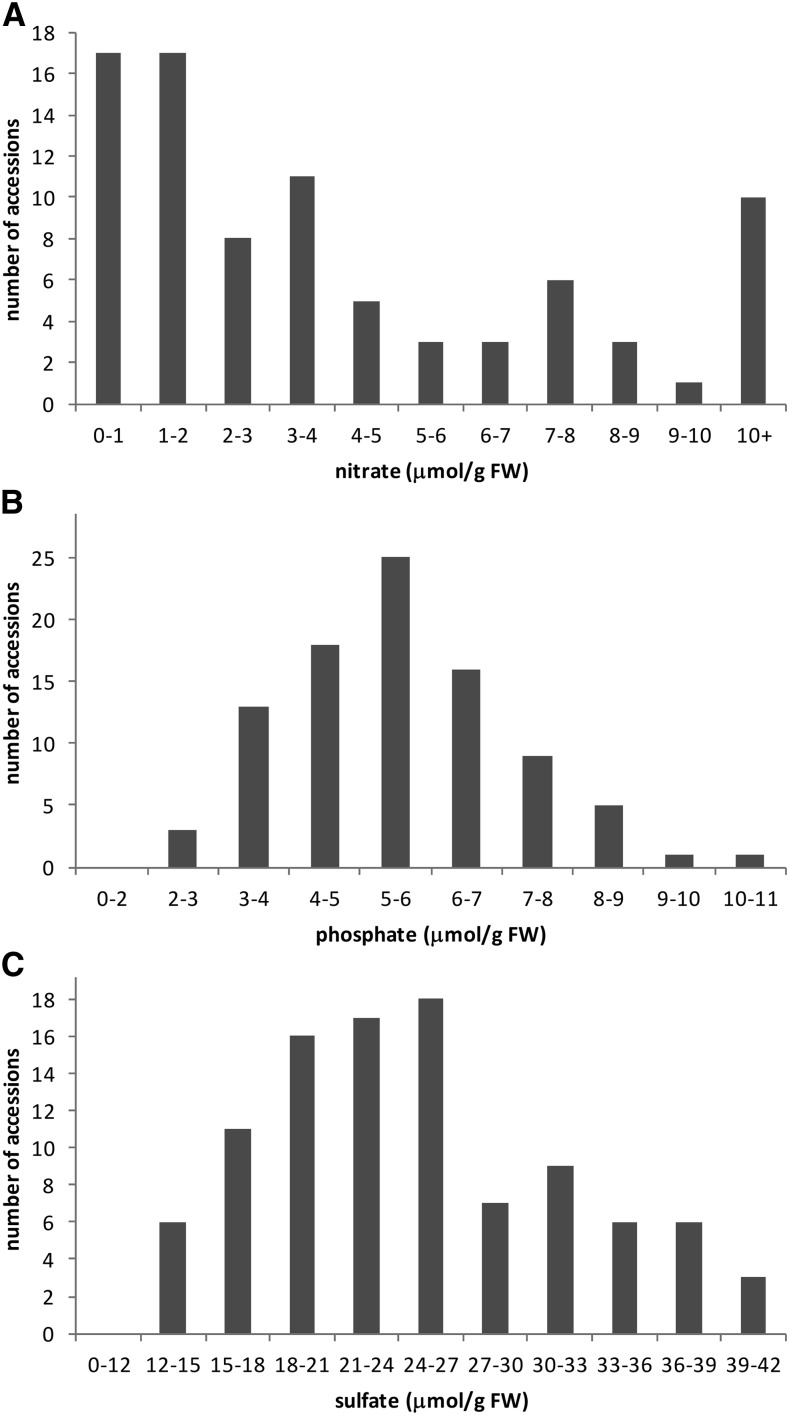
The B. napus core diversity set, comprising 99 varieties (Supplemental Table S1), was planted in a field at the John Innes Centre site. Two leaf discs from the youngest fully developed leaves of 8-week-old plants were sampled and used for the determination of nitrate, phosphate, and sulfate. The anion concentrations in the leaves varied substantially in the different varieties (Fig. 1). Nitrate was found to be the most variable anion, its levels varying from 0.31 to 25.7 μmol g−1 fresh weight, whereas phosphate was found in the range of 2.1 to 10.4 μmol g−1 fresh weight; sulfate levels were typically the highest, between 12.5 and 41.7 μmol g−1 fresh weight (Supplemental Table S1). Interestingly, whereas phosphate levels showed a normal distribution among the accessions and sulfate levels were also close to normal distribution, nitrate concentrations showed a very different pattern. More than one-third of the accessions contained very low nitrate levels, under 2 μmol g−1 fresh weight, whereas 10 accessions (i.e. 12%) contained more than 10 μmol g−1 fresh weight nitrate (Fig. 1). In most varieties, the most abundant anion was sulfate (Supplemental Table S1), which accumulates to high levels in various Brassica spp. (Blake-Kalff et al., 1998), and not nitrate, as typically observed in Arabidopsis (Fig. 2D; Mugford et al., 2009). The difference in nitrate concentration is remarkable. It is not linked to a specific group of B. napus, since varieties very low (less than 1 μmol g−1 fresh weight) and very high (more than 10 μmol g−1 fresh weight) are found among spring oilseed rapes, winter oilseed rapes, or swedes. Low nitrate content is a desirable trait, as it is connected with high nitrogen utilization efficiency; however, the significance of such contrasting accumulation of this anion is unknown and represents an intriguing question for further research.
Figure 1.
Distribution of anion contents among the B. napus varieties. Nitrate (A), phosphate (B), and sulfate (C) contents were determined in leaves of 99 B. napus varieties by HPLC. Shown is the frequency of anion levels among the 99 accessions. FW, Fresh weight.
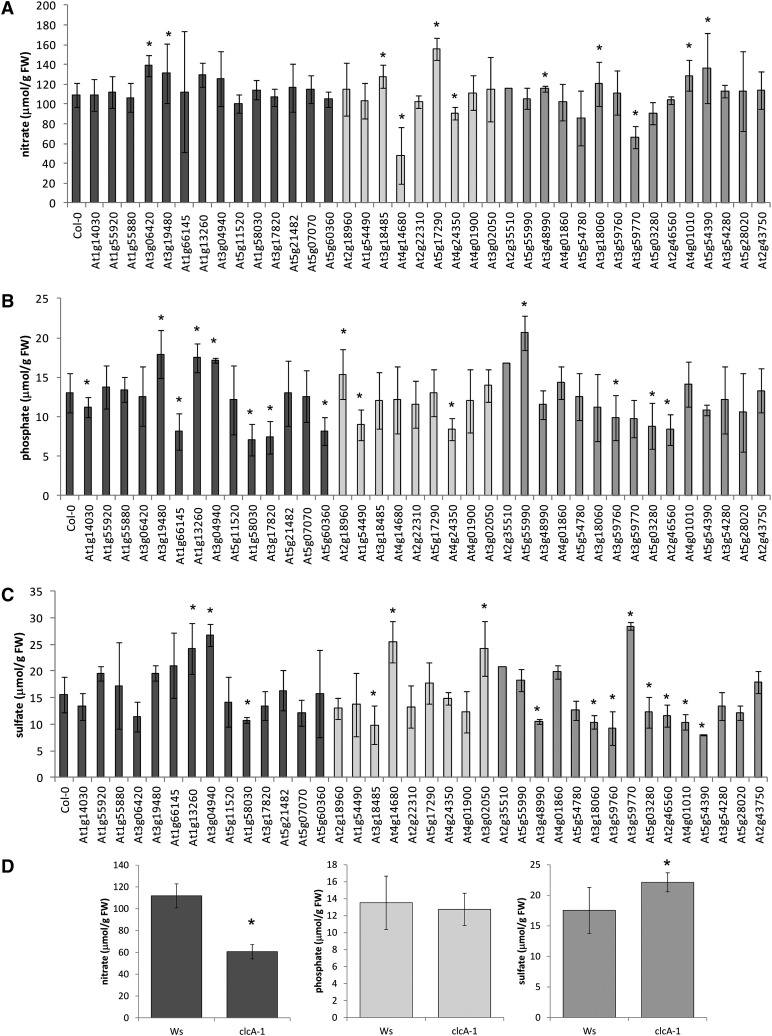
Figure 2.
Anion contents in the candidate genes. A to C, Nitrate (A), phosphate (B), and sulfate (C) contents were determined in leaves of Arabidopsis T-DNA lines for genes potentially affecting anion homeostasis by HPLC. Lines originating from a nitrate screen are presented in black, those from the phosphate data set are in light gray, and lines from the sulfate screen are shown in dark gray. D, Anion levels in the clcA mutant and its corresponding wild type, Ws. Asterisks mark values significantly different from the wild type at P < 0.05 (Student’s t test). FW, Fresh weight.
AT
To determine the regions of a genome involved in the control of anion homeostasis, we made use of the AT approach recently developed for B. napus (Harper et al., 2012). This approach correlates variation in quantitative traits with markers derived from transcript sequencing. In a proof of concept, known QTLs for erucic acid content were correctly identified, thus showing the utility of the use of transcript-derived markers for GWAS (Harper et al., 2012). We used the diversity panel of 84 accessions for which the functional genotypes (i.e. single-nucleotide polymorphism [SNP] scores in expressed sequences) were reported by Harper et al. (2012). The analysis, using a linear mixed model to correct for population structure, revealed a large number of SNPs significantly associated with variation in the anion content. For nitrate, 151 SNPs were linked with significance of P < 0.005, whereas for phosphate and sulfate, the analysis revealed 146 and 168 significant markers, respectively (Supplemental Table S2).
Identification of Candidate Genes
Utilizing the colinearity between B. napus and Arabidopsis chromosomes, GWAS output identifies Arabidopsis orthologs of genes in which the SNPs are located (Harper et al., 2012). This allows immediate access to functional annotation of the genes and to a large variety of Arabidopsis resources. The Arabidopsis orthologs, therefore, were used as a basis to identify suitable candidate genes for causing the variation in anion content. Several candidate genes were immediately obvious. For example, among SNPs identified in the nitrate data set, the SNP with the fifth lowest P value is localized in a B. napus homolog of the nitrate transporter, calcium-activated chloride channel CLC-A (At5g40890; Supplemental Table S3), which was previously shown to contribute to the control of nitrate levels in Arabidopsis leaves (De Angeli et al., 2006). Also among the candidate genes containing significantly associated SNPs were genes for enzymes involved in nitrate assimilation, ASPARTATE AMINOTRANSFERASE5 (At4g31990) and GLUTAMINE SYNTHETASE2 (At5g35630). In the phosphate data set, genes for H+-ATPASE1 (At2g18960) and a hypothetical phosphate/phosphoenolpyruvate translocator (At5g33320) were found to contain the associated markers. Also in the data set based on sulfate levels, several clear candidates were identified directly, such as S-adenosyl-Met synthetase (At4g01850), Cys synthase C (At3g59760), or a 3(2),5-bisphosphate nucleotidase, SAL2 (At5g54390; Supplemental Table S3). However, the significant markers themselves are often not the causal polymorphisms affecting the traits but are only linked (Myles et al., 2009). Thus, the genes underlying the variations and containing the causal polymorphic SNPs are found within the linkage disequilibrium of the markers. For Arabidopsis, such linkage disequilibrium is about 10 kb (Kim et al., 2007), whereas for B. napus, it is 300 to 1,000 kb (Delourme et al., 2013). Therefore, we again exploited the colinearity of the Arabidopsis and B. napus genomes and continued the analysis on the Arabidopsis genome. We inspected Arabidopsis genes within ±15 kb from each significant marker for potential candidate genes controlling nutrient homeostasis using two complementary criteria. First, the functional annotations were used as a guide to search for genes involved in nutrient uptake or assimilation and signaling. Second, we hypothesized that genes controlling nutrient levels in the leaves are likely to be regulated by the deficiency of the corresponding nutrient and, therefore, assessed the expression of these genes in Arabidopsis microarray data for nitrate, phosphate, and sulfate deficiency (Wang et al., 2003; Wu et al., 2003; Maruyama-Nakashita et al., 2006). Together, these analyses resulted in 60 candidate genes, 21 of which directly included the SNPs used as GWAS markers (Supplemental Table S3). Thirty genes were identified in the nitrate data set, 11 in the phosphate data set, and 19 genes potentially control variation in sulfate content. The expression analysis contributed only six genes to the candidate list (Supplemental Table S3).
Analysis of Candidate Genes
The next step of the analysis was based on the hypothesis that mutants in genes controlling variation of anion content will be affected in the levels of such anions. We obtained T-DNA insertion lines for the candidate genes available from the Nottingham Arabidopsis Stock Centre and recovered homozygous mutants by standard PCR genotyping. Fourteen mutants were obtained for genes potentially controlling nitrate levels, and nine and 15 mutants were recovered from the phosphate and sulfate sets, respectively (Supplemental Table S4). For the CLC-A gene, a previously described clca-1 mutant in the Wassilewskija (Ws) background (Geelen et al., 2000) was provided by Sébastien Thomine at the Centre National de la Recherche Scientifique. These mutants were grown in controlled-environment conditions, and anion levels in leaves were determined (Fig. 2). From the 39 mutant lines tested, 12 showed significantly different nitrate levels and 15 each differed in phosphate and sulfate. Among these, two lines were simultaneously affected in nitrate and phosphate, eight lines in nitrate and sulfate, and six lines in phosphate and sulfate. Interestingly, when a second anion was affected in the lines with different phosphate levels, the changes were always in the same direction (i.e. for lines with higher phosphate levels, nitrate or sulfate also was higher). On the other hand, in the lines where both nitrate and sulfate were different from the wild type, the changes were complementary: increase in nitrate was accompanied by decrease in sulfate and vice versa. Since many of the candidate genes are involved in pathways important for general growth and development, it is possible that some of the nutrient effects, particularly when multiple nutrients are affected, are due to pleiotropic effects of the mutations.
However, while relatively high proportions of the selected lines, 31% or 38%, were indeed affected in anion contents, only a few lines showed changes in the same anion for which they were selected as candidates. Thus, among the 14 candidates for the control of nitrate content, only three differed in nitrate, while among the nine candidates for the regulation of phosphate homeostasis, only three were confirmed. On the other hand, in the set of 15 candidates for the control of sulfate content, eight showed correspondingly lower or higher sulfate levels. Altogether, the analysis resulted in the identification of 14 genes potentially responsible for variation in anion content in B. napus (Table I).
Table I. Candidate genes for the control of anion content.
P values in T-DNA experiments corresponding to the original dataset are printed in bold.
| Data Set | Gene | Annotation |
P Values Compared with Col-0 |
Anion Levels Compared with Col-0 | ||
|---|---|---|---|---|---|---|
| Nitrate | Phosphate | Sulfate | ||||
| Nitrate | At5g40890 | Nitrate transporter CLC-A | 0.000003 | 0.2699 | 0.0048 | − |
| Nitrate | At3g06420 | Autophagy-related protein | 0.0016 | 0.4297 | 0.0523 | + |
| Nitrate | At5g11520 | ASPARTATE AMINOTRANSFERASE3 | 0.0344 | 0.2792 | 0.3832 | − |
| Phosphate | At2g18960 | H+-ATPASE1 | 0.2424 | 0.0414 | 0.3039 | + |
| Phosphate | At1g54490 | EIN5 | 0.1843 | 0.0008 | 0.4803 | − |
| Phosphate | At4g24350 | Nutrient reservoir activity | 0.0003 | 0.0002 | 0.2098 | − |
| Sulfate | At3g48990 | Response to cadmium ion | 0.0385 | 0.1206 | 0.0265 | − |
| Sulfate | At3g18060 | CUL4 RING ubiquitin ligase complex | 0.0404 | 0.1091 | 0.0246 | − |
| Sulfate | At3g59760 | Cys synthase C | 0.3775 | 0.0139 | 0.0101 | − |
| Sulfate | At5g03280 | EIN2 | 0.1146 | 0.0059 | 0.0128 | − |
| Sulfate | At2g46560 | CUL4 RING ubiquitin ligase complex | 0.0517 | 0.0045 | 0.0406 | − |
| Sulfate | At4g01010 | CNGC13 | 0.0016 | 0.1984 | 0.0265 | − |
| Sulfate | At5g54390 | 3(2),5-Bisphosphate nucleotidase SAL2 | 0.0080 | 0.0969 | 0.0112 | − |
Genes Potentially Affecting Anion Homeostasis
The different levels of the respective anions in the 14 mutants were independently confirmed in 13 of them (Supplemental Fig. S1). The 13 genes include several genes for which evidence for or a clear link to causing variation in anion levels were described before as well as genes that were not previously associated with plant nutrition. Apart from CLC-A, which was shown to affect nitrate homeostasis in Arabidopsis (De Angeli et al., 2006), the 3(2),5-bisphosphate nucleotidase SAL2 also has a direct relevance for the control of anion content, as mutants of the closely related paralog FIERY1 possess significantly lower sulfate levels (Lee et al., 2012). From several candidate genes functioning in nitrate metabolism (Supplemental Table S4), only ASPARTATE AMINOTRANSFERASE3 has been confirmed to affect nitrate levels when disrupted. H+-ATPASE1 (At2g18960) may contribute to maintaining phosphate levels, as H+-ATPases were associated with the response to phosphate deficiency (Tomasi et al., 2009). Two genes with a direct relevance to sulfur metabolism were identified in the sulfate data set. Cys synthase C catalyzes the last step of primary sulfate assimilation, the synthesis of Cys (Heeg et al., 2008; Takahashi et al., 2011). The At3g48990 gene is annotated as functioning in response to cadmium, where sulfur compounds, such as phytochelatins, play a very important role (Sarry et al., 2006). Finally, the At3g06420 gene encodes one isoform of the ATG8 protein involved in autophagy (Thompson et al., 2005). Autophagy was recently revealed as an important process in nutrient homeostasis, particularly at nutrient limitation, as shown in studies with nitrate (Guiboileau et al., 2012) and sulfate (Álvarez et al., 2012). It is thus possible that it also contributes to nutrient homeostasis during vegetative growth, since the mutant in At3g06420 accumulated nitrate; however, the mechanisms of such contribution remain to be elucidated.
Other genes, however, do not have obvious functions in the control of plant nutrition. Particularly interesting is the identification of two genes involved in ethylene signaling, ETHYLENE INSENSITIVE2 (EIN2) and EIN5, associated with variation in sulfate and phosphate levels, respectively. Ethylene signaling is involved in several responses to phosphate deficiency: the regulation of gene expression, production of acid phosphatases, and accumulation of anthocyanins (Lei et al., 2011). Even more evidence is present on the role of ethylene in the regulation of the sulfate starvation response. The central regulator of the sulfate starvation response, SULFUR LIMITATION1, is a member of an EIN3-like family of transcription factors (Maruyama-Nakashita et al., 2006). Ethylene directly induces the activity of the key enzyme of sulfate assimilation, adenosine 5′-phosphosulfate reductase (APR), and ethylene signaling is important for the regulation of this enzyme by salt (Koprivova et al., 2008). Arabidopsis mutants in ethylene signaling are more sensitive to selenium, which primarily affects sulfate uptake and assimilation (Van Hoewyk et al., 2008). Ethylene production is affected by nitrate supply, and ethylene signaling is involved in the regulation of nitrate transporters (Zheng et al., 2013), showing a general role of this phytohormone in the control of plant nutrition. The findings of altered anion content in ein2 and ein5 mutants and the links between EIN2 and EIN5 and variation in sulfate and phosphate levels in the B. napus accessions thus support this conclusion and point to the necessity of more detailed investigations of the role of ethylene in the control of plant mineral nutrition.
Two genes from the candidate list are part of the cullin4 (CUL4) RING ubiquitin ligase complex. The complex catalyzes the ubiquitination of a variety of proteins in different cellular pathways to control their degradation (Angers et al., 2006). The CUL4 RING ligase has been associated, for example, with DNA repair and the control of photomorphogenesis (Chen et al., 2006; Kapetanaki et al., 2006). The two genes identified in the study belong to DDB1 binding WD40 (DWD) proteins, substrate receptors of the CUL4 RING complex, which often regulate development (Lee et al., 2008). However, protein degradation leads to increased amino acid recycling and may so modulate the demand for reduced nitrogen or sulfur and, in this way, affect the homeostasis of the stored anions nitrate and sulfate. How the two DWD proteins are associated with plant nutrition, however, needs further study.
The At4g24350 gene encodes an uncharacterized protein belonging to the phosphorylase gene family with a predicted nutrient reservoir activity. However, the corresponding protein contains a nucleoside phosphorylase domain and so may be involved in the turnover of nucleotide phosphates and so affect phosphate homeostasis. The last gene from the list, At4g01010, encodes a cyclic nucleotide-gated channel, AtCNGC13. These calmodulin-binding channels have a plethora of functions in plants, from transport of potassium and other monovalent cations, to disease response signaling, to thermal sensing (Mäser et al., 2001; Ali et al., 2007; Finka et al., 2012). AtCNGC13 has not been characterized so far; however, reduced expression of its closest paralog, AtCNGC10, resulted in many developmental and growth defects, such as early flowering, starch accumulation, and growth retardation (Borsics et al., 2007). Given the number of functions members of this gene family have in plants, it is not possible to predict the function of AtCNGC13, but its interesting phenotype in sulfate accumulation encourages detailed investigation.
Power of AT
The number and nature of genes potentially controlling nutrient content confirm the value of AT in the genetic dissection of complex traits. First, previously known QTLs for erucic acid content were confirmed using this approach (Harper et al., 2012). Furthermore, analysis of association based on gene expression levels revealed that the seed glucosinolate content in B. napus is controlled by the expression of MYB28 (Harper et al., 2012). Interestingly, we observed no significant associations between gene expression levels and anion content at the selected high level of significance, 10−6, necessary to avoid false positives (Harper et al., 2012); all significant associations were found among the SNP markers. In the proof of concept experiments (Harper et al., 2012), the expression markers were identified with significance between 10−7 and 10−9, whereas the most highly significant SNP markers reached only P = 10−5 (Harper et al., 2012). The presence of CLC-A and SAL2 among the candidate genes showing significant effects on anion contents is further confirmation of the power of this approach. The analysis, however, revealed other gene candidates that were not linked to mineral nutrition before. The putative function of these genes was confirmed by an analysis of T-DNA lines, showing that AT has the potential to uncover new links between metabolic and/or regulatory pathways. In Arabidopsis, GWAS approaches also recovered previously known candidate genes (Aranzana et al., 2005; Atwell et al., 2010; Chan et al., 2011; Chao et al., 2012), but reports of new discoveries are still scarce (Angelovici et al., 2013; Meijón et al., 2014; Verslues et al., 2014).
The candidate genes were found in the analysis of 84 B. napus accessions, while a previous study showed a strong association based on only 53 varieties. First, Arabidopsis GWAS experiments were based on 96 accessions, but it has been calculated that at least 192 accessions are needed for a comprehensive sampling of genetic diversity (Atwell et al., 2010). The population structure of the B. napus diversity set thus might be more suitable for these studies, as fewer accessions are needed to obtain robust results. It should also be noted that the plant material used for AT originated from a field trial and not from controlled conditions. This confirms the robustness of the genome-wide methods even for nutrition traits that are naturally dependent on microenvironment. In addition, the AT approach was able to cope with the nitrate data set, which is far from a normal distribution of the values, since in the nitrate and phosphate data sets the number of significantly associated markers was similar and the number of confirmed candidates was identical. The prediction success, however, was not as high in nitrate (20%; three out of 15 tested) as for sulfate (47%) and phosphate (33%).
To test whether candidate genes from AT have a higher probability to be affected in nutrient content than genes of similar annotation but picked randomly, we performed an in silico experiment. Since no anion data set is publicly available, we used the total element data in the ionomics resources at www.ionomicshub.org (Baxter et al., 2007). We searched for data from T-DNA lines in genes between At1g60000 and At1g69000 with a similar annotation to the one we used for our selection and compared the concentrations of total phosphorus, selenium, as its distribution strongly correlates with sulfur, and iron in leaves. This interval was selected because it contains two genes with known effects on the accumulation of these elements, APR2, encoding an isoform of APR, for sulfur (and selenium) and the iron transporter IRT3. Indeed, these mutants showed significant effects on selenium and iron concentrations, respectively. From the 20 mutants analyzed, two were affected in phosphorus (P < 0.05), four in selenium, and three in iron (Supplemental Table S5). Thus, the success of the random set to identify genes affected in nutrient homeostasis was 10% to 20%, clearly significantly lower than the success rate of the candidates from AT analysis of sulfate and phosphate data sets and similar to the nitrate set.
From the three data sets analyzed in this study, most candidates were recovered for the control of sulfate levels. Interestingly, the two genes shown to control variation in sulfate levels in Arabidopsis have not been found among the candidate genes. These two genes were shown to underlie QTLs from an analysis of a Bay-0 × Shahdara population, and they encode isoforms of two consecutive enzymes of the sulfate assimilation pathway, ATP sulfurylase and APR (Loudet et al., 2007; Koprivova et al., 2013). The APR2 gene in Shahdara contained a unique SNP that resulted in an amino acid substitution near the active center and inactivation of the enzyme (Loudet et al., 2007). ATP sulfurylase (ATPS) affects sulfate levels due to variation in transcript levels of the main isoform, ATPS1, caused by a deletion in intron 1 found in a few accessions (Koprivova et al., 2013). Unique or very rare haplotypes, such as the Shahdara allele of APR2, are almost impossible to identify through GWAS, and it is thus no surprise that the gene was not recovered. For ATPS1, the number of accessions studied may not have been high enough to identify the small variation in expression levels observed in Arabidopsis. Thus, although the number of candidates for genes controlling trait variation is large, the list is still not complete, and other genes can be identified by other methods (e.g. QTL analysis).
CONCLUSION
We have performed GWAS of leaf anion content in B. napus using AT. The analysis resulted in the identification of 13 genes, potentially involved in the control of sulfate, nitrate, and phosphate levels. First, genes within linkage disequilibrium of the significant markers were inspected. Genes annotated as involved in nutrient uptake and metabolism, signaling networks, and transcription factors were marked for further analysis, as were genes affected in expression by the particular nutrient deficiency. Homozygous T-DNA insertion lines for these genes were obtained and analyzed for anion content. This pipeline generally can be adopted to facilitate the identification of new genes controlling trait variation from GWAS results not only in Arabidopsis but also in B. napus. The presence of several genes known to affect anion levels in the resulting gene list confirms the suitability of the approach. The mechanisms of action and the causal polymorphisms of these candidates will be subjects of future more detailed studies.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The plant material for initial anion content measurements was derived from a field-grown diversity set of 99 Brassica napus lines, as listed in Supplemental Table S1. The seeds were germinated and grown in long-day glasshouse conditions (16-h photoperiod) at 15°C (400-W HQI metal halide lamps). Plants were pricked out after 11 d and transferred into a field at the John Innes Centre site, arranged into a four block, one-way randomized design with one plant of each of accession per block and randomized within each block. Leaf discs were cut from mature leaves of 8-week-old plants approximately 4 h into the light period and immediately frozen in liquid nitrogen.
For functional analysis of the candidate genes, Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 (Col-0) was used as the wild type, except for the clcA-1 mutant (FLAG_171A06), obtained from Sébastien Thomine at the Centre National de la Recherche Scientifique, which is in the Ws background. T-DNA lines disrupting the candidate genes were obtained from the Nottingham Arabidopsis Stock Centre and genotyped by PCR (for accession numbers and primers, see Supplemental Table S4) to obtain homozygous mutants. Plants for analysis were grown for 5 weeks in a controlled-environment room under a short-day 10-h-light/14-h-dark cycle at a constant temperature of 22°C, 60% relative humidity, and light intensity of 160 µE s–1 m–2. Four individual plants from each genotype were analyzed, and the experiment was independently repeated.
Measurement of Anion Content
For anion measurements, approximately 50 mg of frozen plant material was homogenized in 1 mL of deionized water, and the anions, nitrate, phosphate, and sulfate, were separated by HPLC on an IC-PAK ion-exchange column as described (Scheerer et al., 2010).
AT
The previously developed SNP data set (Supplemental Data File 6 in Harper et al., 2012) had been entered previously into the program STRUCTURE 2.3.3 (Pritchard et al., 2000) for Bayesian population structure analysis. An admixture model with independent allele frequencies was used, and K was set between 1 and 5, each repeated three times with a burn-in length of 100,000, followed by 100,000 iterations of the Monte Carlo Markov Chain algorithm. The method of Evanno et al. (2005) was employed to estimate the number of clusters that best represents the data set. Once the optimal number of K populations was established, K-1 Q matrix scores for each individual could be used as a fixed effect in the subsequent association analysis.
The trait data, STRUCTURE Q matrix, and SNP data were entered into the program TASSEL V3.0 (Bradbury et al., 2007). Minor allele states below 0.05 were removed from the SNP data set, leaving 62,980 SNPs (Harper et al., 2012), and a kinship matrix was calculated to estimate the pairwise relatedness between individuals. These data sets were entered into a mixed linear model with optimum compression and Population Parameters Previously Determined variance component estimation to decrease computing time for the large data set.
Using methods and scripts described by Bancroft et al. (2011) and Higgins et al. (2012) along with sequence data sets submitted previously to sequence reads archive (under accession nos. ERA122949, ERA036824, and ERA063602), transcript quantification was undertaken for the B. napus accessions used in this study. The sequence reads were mapped to the cured reference described by Harper et al. (2012), which comprised the A and C genome versions of each unigene (189,116 unigene sequences in all). Transcript abundance was quantified and normalized as reads per kilobase per million aligned reads separately for the A and C genome versions of each unigene. The relationship between gene expression and the trait values was calculated by linear regression using R (http://www.R-project.org/). For each unigene, previously calculated reads per kilobase per million aligned reads values (Harper et al., 2012) were regressed as the dependent variable on the trait value as the independent variable, and r2 and significance values were calculated for each unigene.
Statistical Analysis
The normality of the distribution of anions in B. napus varieties was tested using the Kolmogorov-Smirnov test (http://www.physics.csbsju.edu/stats/KS-test.html). Significant differences between the wild type and the T-DNA lines were analyzed according to Student’s t test for P < 0.05.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Confirmation of changed anion contents in leaves of mutants in candidate genes.
Supplemental Table S1. Anion contents in leaves of B. napus varieties.
Supplemental Table S2. Markers significantly associated with variation in anion levels in B. napus.
Supplemental Table S3. Candidate genes for analysis of T-DNA lines.
Supplemental Table S4. T-DNA lines used for analysis.
Supplemental Table S5. Variation in the content of phosphorus, selenium, and iron in leaves of randomly selected T-DNA from www.ionomicshub.org.
Glossary
- NUE
nutrient use efficiency
- QTL
quantitative trait locus
- GWAS
genome-wide association studies
- AT
associative transcriptomics
- T-DNA
transfer DNA
- SNP
single-nucleotide polymorphism
- Ws
Wassilewskija
- Col-0
Columbia-0
Footnotes
This work was supported by the Biotechnology and Biological Sciences Research Council (grant nos. BB/H004351/1, BB/J004561/1, and BB/L002124/1) and the John Innes Foundation.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Ali R, Ma W, Lemtiri-Chlieh F, Tsaltas D, Leng Q, von Bodman S, Berkowitz GA. (2007) Death don’t have no mercy and neither does calcium: Arabidopsis CYCLIC NUCLEOTIDE GATED CHANNEL2 and innate immunity. Plant Cell 19: 1081–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez C, García I, Moreno I, Pérez-Pérez ME, Crespo JL, Romero LC, Gotor C. (2012) Cysteine-generated sulfide in the cytosol negatively regulates autophagy and modulates the transcriptional profile in Arabidopsis. Plant Cell 24: 4621–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Lipka AE, Deason N, Gonzalez-Jorge S, Lin H, Cepela J, Buell R, Gore MA, Dellapenna D. (2013) Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. Plant Cell 25: 4827–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angers S, Li T, Yi X, MacCoss MJ, Moon RT, Zheng N. (2006) Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443: 590–593 [DOI] [PubMed] [Google Scholar]
- Aranzana MJ, Kim S, Zhao K, Bakker E, Horton M, Jakob K, Lister C, Molitor J, Shindo C, Tang C, et al. (2005) Genome-wide association mapping in Arabidopsis identifies previously known flowering time and pathogen resistance genes. PLoS Genet 1: e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft I, Morgan C, Fraser F, Higgins J, Wells R, Clissold L, Baker D, Long Y, Meng J, Wang X, et al. (2011) Dissecting the genome of the polyploid crop oilseed rape by transcriptome sequencing. Nat Biotechnol 29: 762–766 [DOI] [PubMed] [Google Scholar]
- Baxter I, Ouzzani M, Orcun S, Kennedy B, Jandhyala SS, Salt DE. (2007) Purdue Ionomics Information Management System: an integrated functional genomics platform. Plant Physiol 143: 600–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake-Kalff MM, Harrison KR, Hawkesford MJ, Zhao FJ, McGrath SP. (1998) Distribution of sulfur within oilseed rape leaves in response to sulfur deficiency during vegetative growth. Plant Physiol 118: 1337–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsics T, Webb D, Andeme-Ondzighi C, Staehelin LA, Christopher DA. (2007) The cyclic nucleotide-gated calmodulin-binding channel AtCNGC10 localizes to the plasma membrane and influences numerous growth responses and starch accumulation in Arabidopsis thaliana. Planta 225: 563–573 [DOI] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 [DOI] [PubMed] [Google Scholar]
- Chan EK, Rowe HC, Corwin JA, Joseph B, Kliebenstein DJ. (2011) Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol 9: e1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao DY, Silva A, Baxter I, Huang YS, Nordborg M, Danku J, Lahner B, Yakubova E, Salt DE. (2012) Genome-wide association studies identify heavy metal ATPase3 as the primary determinant of natural variation in leaf cadmium in Arabidopsis thaliana. PLoS Genet 8: e1002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardon F, Noël V, Masclaux-Daubresse C. (2012) Exploring NUE in crops and in Arabidopsis ideotypes to improve yield and seed quality. J Exp Bot 63: 3401–3412 [DOI] [PubMed] [Google Scholar]
- Chen H, Shen Y, Tang X, Yu L, Wang J, Guo L, Zhang Y, Zhang H, Feng S, Strickland E, et al. (2006) Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angeli A, Monachello D, Ephritikhine G, Frachisse JM, Thomine S, Gambale F, Barbier-Brygoo H. (2006) The nitrate/proton antiporter AtCLCa mediates nitrate accumulation in plant vacuoles. Nature 442: 939–942 [DOI] [PubMed] [Google Scholar]
- Delourme R, Falentin C, Fomeju BF, Boillot M, Lassalle G, André I, Duarte J, Gauthier V, Lucante N, Marty A, et al. (2013) High-density SNP-based genetic map development and linkage disequilibrium assessment in Brassica napus L. BMC Genomics 14: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding G, Yang M, Hu Y, Liao Y, Shi L, Xu F, Meng J. (2010) Quantitative trait loci affecting seed mineral concentrations in Brassica napus grown with contrasting phosphorus supplies. Ann Bot (Lond) 105: 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14: 2611–2620 [DOI] [PubMed] [Google Scholar]
- Finka A, Cuendet AFH, Maathuis FJM, Saidi Y, Goloubinoff P. (2012) Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell 24: 3333–3348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallais A, Hirel B. (2004) An approach to the genetics of nitrogen use efficiency in maize. J Exp Bot 55: 295–306 [DOI] [PubMed] [Google Scholar]
- Geelen D, Lurin C, Bouchez D, Frachisse JM, Lelièvre F, Courtial B, Barbier-Brygoo H, Maurel C. (2000) Disruption of putative anion channel gene AtCLC-a in Arabidopsis suggests a role in the regulation of nitrate content. Plant J 21: 259–267 [DOI] [PubMed] [Google Scholar]
- Good AG, Shrawat AK, Muench DG. (2004) Can less yield more? Is reducing nutrient input into the environment compatible with maintaining crop production? Trends Plant Sci 9: 597–605 [DOI] [PubMed] [Google Scholar]
- Guiboileau A, Yoshimoto K, Soulay F, Bataillé MP, Avice JC, Masclaux-Daubresse C. (2012) Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol 194: 732–740 [DOI] [PubMed] [Google Scholar]
- Habash DZ, Bernard S, Schondelmaier J, Weyen J, Quarrie SA. (2007) The genetics of nitrogen use in hexaploid wheat: N utilisation, development and yield. Theor Appl Genet 114: 403–419 [DOI] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ. (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132: 578–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Kuromori T, Hirayama T, Shinozaki K, Leigh RA. (2004) Quantitative trait loci analysis of nitrate storage in Arabidopsis leading to an investigation of the contribution of the anion channel gene, AtCLC-c, to variation in nitrate levels. J Exp Bot 55: 2005–2014 [DOI] [PubMed] [Google Scholar]
- Harper AL, Trick M, Higgins J, Fraser F, Clissold L, Wells R, Hattori C, Werner P, Bancroft I. (2012) Associative transcriptomics of traits in the polyploid crop species Brassica napus. Nat Biotechnol 30: 798–802 [DOI] [PubMed] [Google Scholar]
- Heeg C, Kruse C, Jost R, Gutensohn M, Ruppert T, Wirtz M, Hell R. (2008) Analysis of the Arabidopsis O-acetylserine(thiol)lyase gene family demonstrates compartment-specific differences in the regulation of cysteine synthesis. Plant Cell 20: 168–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins J, Magusin A, Trick M, Fraser F, Bancroft I. (2012) Use of mRNA-seq to discriminate contributions to the transcriptome from the constituent genomes of the polyploid crop species Brassica napus. BMC Genomics 13: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K. (2003) Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J 33: 651–663 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Klein M, Fujikawa Y, Yano M, Goodenowe DB, Yamazaki Y, Kanaya S, Nakamura Y, Kitayama M, Suzuki H, et al. (2005) Elucidation of gene-to-gene and metabolite-to-gene networks in Arabidopsis by integration of metabolomics and transcriptomics. J Biol Chem 280: 25590–25595 [DOI] [PubMed] [Google Scholar]
- Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapić-Otrin V, Levine AS. (2006) The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci USA 103: 2588–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M. (2007) Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet 39: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Koprivova A, Giovannetti M, Baraniecka P, Lee BR, Grondin C, Loudet O, Kopriva S. (2013) Natural variation in the ATPS1 isoform of ATP sulfurylase contributes to the control of sulfate levels in Arabidopsis. Plant Physiol 163: 1133–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, North KA, Kopriva S. (2008) Complex signaling network in regulation of adenosine 5′-phosphosulfate reductase by salt stress in Arabidopsis roots. Plant Physiol 146: 1408–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Mirowski P, LeCun Y, Shasha DE, Coruzzi GM. (2010) Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol 11: R123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BR, Huseby S, Koprivova A, Chételat A, Wirtz M, Mugford ST, Navid E, Brearley C, Saha S, Mithen R, et al. (2012) Effects of fou8/fry1 mutation on sulfur metabolism: is decreased internal sulfate the trigger of sulfate starvation response? PLoS ONE 7: e39425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Terzaghi W, Gusmaroli G, Charron JB, Yoon HJ, Chen H, He YJ, Xiong Y, Deng XW. (2008) Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20: 152–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M, Zhu C, Liu Y, Karthikeyan AS, Bressan RA, Raghothama KG, Liu D. (2011) Ethylene signalling is involved in regulation of phosphate starvation-induced gene expression and production of acid phosphatases and anthocyanin in Arabidopsis. New Phytol 189: 1084–1095 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Merigout P, Talbotec J, Daniel-Vedele F. (2003) Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis. Plant Physiol 131: 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudet O, Saliba-Colombani V, Camilleri C, Calenge F, Gaudon V, Koprivova A, North KA, Kopriva S, Daniel-Vedele F. (2007) Natural variation for sulfate content in Arabidopsis thaliana is highly controlled by APR2. Nat Genet 39: 896–900 [DOI] [PubMed] [Google Scholar]
- Maathuis FJ. (2009) Physiological functions of mineral macronutrients. Curr Opin Plant Biol 12: 250–258 [DOI] [PubMed] [Google Scholar]
- Marschner H (2012) Mineral Nutrition of Higher Plants, Ed 3. Academic Press, London [Google Scholar]
- Maruyama-Nakashita A, Nakamura Y, Tohge T, Saito K, Takahashi H. (2006) Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18: 3235–3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser P, Thomine S, Schroeder JI, Ward JM, Hirschi K, Sze H, Talke IN, Amtmann A, Maathuis FJM, Sanders D, et al. (2001) Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol 126: 1646–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijón M, Satbhai SB, Tsuchimatsu T, Busch W. (2014) Genome-wide association study using cellular traits identifies a new regulator of root development in Arabidopsis. Nat Genet 46: 77–81 [DOI] [PubMed] [Google Scholar]
- Mugford SG, Yoshimoto N, Reichelt M, Wirtz M, Hill L, Mugford ST, Nakazato Y, Noji M, Takahashi H, Kramell R, et al. (2009) Disruption of adenosine-5′-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. Plant Cell 21: 910–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costich DE, Buckler ES. (2009) Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell 21: 2194–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R. (2005) Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol 138: 304–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry MA, Hawkesford MJ. (2012) An integrated approach to crop genetic improvement. J Integr Plant Biol 54: 250–259 [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. (2000) Inference of population structure using multilocus genotype data. Genetics 155: 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengel Z, Marschner P. (2005) Nutrient availability and management in the rhizosphere: exploiting genotypic differences. New Phytol 168: 305–312 [DOI] [PubMed] [Google Scholar]
- Reymond M, Svistoonoff S, Loudet O, Nussaume L, Desnos T. (2006) Identification of QTL controlling root growth response to phosphate starvation in Arabidopsis thaliana. Plant Cell Environ 29: 115–125 [DOI] [PubMed] [Google Scholar]
- Sarry JE, Kuhn L, Ducruix C, Lafaye A, Junot C, Hugouvieux V, Jourdain A, Bastien O, Fievet JB, Vailhen D, et al. (2006) The early responses of Arabidopsis thaliana cells to cadmium exposure explored by protein and metabolite profiling analyses. Proteomics 6: 2180–2198 [DOI] [PubMed] [Google Scholar]
- Scheerer U, Haensch R, Mendel RR, Kopriva S, Rennenberg H, Herschbach C. (2010) Sulphur flux through the sulphate assimilation pathway is differently controlled by adenosine 5′-phosphosulphate reductase under stress and in transgenic poplar plants overexpressing gamma-ECS, SO, or APR. J Exp Bot 61: 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kopriva S, Giordano M, Saito K, Hell R. (2011) Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol 62: 157–184 [DOI] [PubMed] [Google Scholar]
- Thompson AR, Doelling JH, Suttangkakul A, Vierstra RD. (2005) Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol 138: 2097–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi N, Kretzschmar T, Espen L, Weisskopf L, Fuglsang AT, Palmgren MG, Neumann G, Varanini Z, Pinton R, Martinoia E, et al. (2009) Plasma membrane H-ATPase-dependent citrate exudation from cluster roots of phosphate-deficient white lupin. Plant Cell Environ 32: 465–475 [DOI] [PubMed] [Google Scholar]
- Van Hoewyk D, Takahashi H, Inoue E, Hess A, Tamaoki M, Pilon-Smits EA. (2008) Transcriptome analyses give insights into selenium-stress responses and selenium tolerance mechanisms in Arabidopsis. Physiol Plant 132: 236–253 [DOI] [PubMed] [Google Scholar]
- Verslues PE, Lasky JR, Juenger TE, Liu TW, Kumar MN. (2014) Genome-wide association mapping combined with reverse genetics identifies new effectors of low water potential-induced proline accumulation in Arabidopsis. Plant Physiol 164: 144–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM. (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tischner R, Gutiérrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM. (2004) Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol 136: 2512–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D. (2012) Natural variation in Arabidopsis: from molecular genetics to ecological genomics. Plant Physiol 158: 2–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW. (2003) Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol 132: 1260–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Han X, An Y, Guo H, Xia X, Yin W. (2013) The nitrate transporter NRT2.1 functions in the ethylene response to nitrate deficiency in Arabidopsis. Plant Cell Environ 36: 1328–1337 [DOI] [PubMed] [Google Scholar]