Proteins turn over at different rates in plant tissues, and these have been quantified using stable isotope labeling of nitrogen and peptide mass spectrometry of leaf tissue from hydroponically grown barley.
Abstract
Protein turnover is a key component in cellular homeostasis; however, there is little quantitative information on degradation kinetics for individual plant proteins. We have used 15N labeling of barley (Hordeum vulgare) plants and gas chromatography-mass spectrometry analysis of free amino acids and liquid chromatography-mass spectrometry analysis of proteins to track the enrichment of 15N into the amino acid pools in barley leaves and then into tryptic peptides derived from newly synthesized proteins. Using information on the rate of growth of barley leaves combined with the rate of degradation of 14N-labeled proteins, we calculate the turnover rates of 508 different proteins in barley and show that they vary by more than 100-fold. There was approximately a 9-h lag from label application until 15N incorporation could be reliably quantified in extracted peptides. Using this information and assuming constant translation rates for proteins during the time course, we were able to quantify degradation rates for several proteins that exhibit half-lives on the order of hours. Our workflow, involving a stringent series of mass spectrometry filtering steps, demonstrates that 15N labeling can be used for large-scale liquid chromatography-mass spectrometry studies of protein turnover in plants. We identify a series of abundant proteins in photosynthesis, photorespiration, and specific subunits of chlorophyll biosynthesis that turn over significantly more rapidly than the average protein involved in these processes. We also highlight a series of proteins that turn over as rapidly as the well-known D1 subunit of photosystem II. While these proteins need further verification for rapid degradation in vivo, they cluster in chlorophyll and thiamine biosynthesis.
New proteins need to be synthesized while others must be degraded so that plants can respond to the daily requirements of cellular maintenance while also allowing progression through different developmental stages. Ubiquitinated proteins are targeted to the proteasome, which is responsible for a large part of the protein degradation process in both plants and animals (Vierstra, 2009). Its action is supplemented by vacuolar protein degradation during autophagy (Araújo et al., 2011) as well as compartment-specific ATP-dependent metalloproteases (Janska et al., 2013). The plant cell proteome differs most markedly from other eukaryotic cells by the abundance of chloroplasts, which can account for up to three-fourths of soluble protein in green plant tissues (Huber et al., 1976). Besides capturing energy from sunlight into fixed carbon, chloroplasts are also responsible for vitamin and cofactor generation as well as other anabolic processes needed to synthesize amino acids, lipids, and pigments (Rolland et al., 2012). Determining the turnover characteristics of chloroplast proteins relative to those in other parts of the plant cell will be important for understanding how metabolic functions are coordinated across plant organelles.
Over the life cycle of plants, the turnover of the chloroplast proteome is critical on several occasions. First, plastid protein degradation and the cytosolic ubiquitination system are known to be critical factors in deetiolation, the process of converting etioplasts to chloroplasts (Ling et al., 2012). Second, under carbon starvation induced by leaf shading or darkness, the catabolism of plastid proteins via autophagy is important in selectively degrading some proteins and tissues in order to sustain others (Araújo et al., 2011). Third, amino acids derived from degradation of the photosynthetic machinery are the primary source of nitrogen during the seed-filling process (Masclaux et al., 2000); therefore, plastid degeneration during natural senescence is central to the yield potential of crops. The protein content of chloroplasts is also modified on a shorter time scale in order to respond to environmental perturbations, to account for changing metabolic processes in diurnal cycles, and to renew the machinery required for the steady-state operations of the organelle. As a result, turnover of plastid proteins contributes to the significant energetic cost of protein synthesis required for cell maintenance (Penning De Vries, 1975). The most studied protein that exhibits fast turnover in plants is the D1 subunit protein of PSII in chloroplasts. The half-life of D1 is inversely correlated with light intensity, and environmental stress conditions are also associated with faster degradation of D1 (Aro et al., 1993; Marutani et al., 2012; Mittal et al., 2012). Plants generate their circadian rhythms by regulating gene expression and protein abundance over the diurnal cycle (McWatters and Devlin, 2011), and recently, these pathways have been shown to be interconnected with plastid proteins involved in thiamine metabolism (Bocobza et al., 2013). A deeper understanding of protein turnover dynamics within the plastid will enhance our understanding of plastid maintenance (van Wijk and Baginsky, 2011; Nevo et al., 2012) and also allow researchers to make more informed decisions regarding the energetic costs of genetic engineering strategies in the plastid compartment (Maliga and Bock, 2011).
The simultaneous measurement of degradation rate (Kd) and synthesis rate (Ks) for a range of proteins in parallel is a new proteomic tool available to biologists, resulting from improved mass spectrometry (MS) equipment, computational power, and algorithm development. Large data sets have been generated in yeast (Saccharomyces cerevisiae), mammalian cell cultures, as well as intact animals (Price et al., 2010; Cambridge et al., 2011; Schwanhäusser et al., 2011). Proteomic analyses of protein turnover are just beginning to gather momentum in plants. Several studies have assessed the utility of various metabolic labels (2H, 13C, and 15N) for this purpose (Yang et al., 2010; Chen et al., 2011; Li et al., 2012b). More focused reports have refined our understanding of the dynamics in the assembly of mitochondrial electron transport complexes using these tools (Li et al., 2012a, 2013). Most recently, we performed a shotgun study of mitochondrial proteins from Arabidopsis (Arabidopsis thaliana) cell culture, measured Kd values of 224 proteins, and assessed the turnover of several protein complexes by adaptation of this approach to assess larger scale liquid chromatography-tandem mass spectrometry (LC-MS/MS) data sets (Nelson et al., 2013). The important next step for the utility of this approach for plant biologists is to conduct and analyze whole-plant isotope labeling experiments in order to define the turnover of proteins in planta. Application of these techniques to a model species such as Arabidopsis will build upon the large body of information already available for this species (Lamesch et al., 2012). However, because there are significant differences between Arabidopsis and agriculturally relevant crop species, it is important to conduct studies in crops that can generate targets for geneticists and breeders.
Here, we assess the utility of 15N labeling to better understand protein turnover during the steady-state operation of plant leaves, with an emphasis on chloroplast proteins. We have analyzed leaves from barley (Hordeum vulgare), which has been a long-standing model for photosynthesis research, chloroplast biology, and genetic dissection of photosynthesis and photorespiration (Christopher and Mullet, 1994; Leegood et al., 1996; Dal Bosco et al., 2003; Rollins et al., 2013). Barley is also an important cereal crop for a variety of food and feed products (Schulte et al., 2009), and recent genome sequencing has scaffolded most of its expressed genes to a genetic map, opening new opportunities for in-depth proteome analysis of this species and linkage to quantitative trait loci of agronomic importance (Mayer et al., 2012). We have examined 15N labeling at the amino acid and protein levels and discuss the advantages and limitations of this method. We report Kd values for more than 500 protein groups, with most of these being localized to the chloroplast, and compare turnover rates for proteins across different organelles and functional categories. We observed a series of abundant protein subunits in photosynthesis, photorespiration, and cofactor synthesis pathways that are degraded much more rapidly than the average barley or plastid protein involved in these processes. We also highlight a series of proteins that turn over rapidly that have not previously been described in terms of their degradation kinetics, and we show that they cluster into certain metabolic pathways, specifically branches of chlorophyll and thiamine biosynthesis.
RESULTS
Labeling of Amino Acid Pools in Barley Leaves
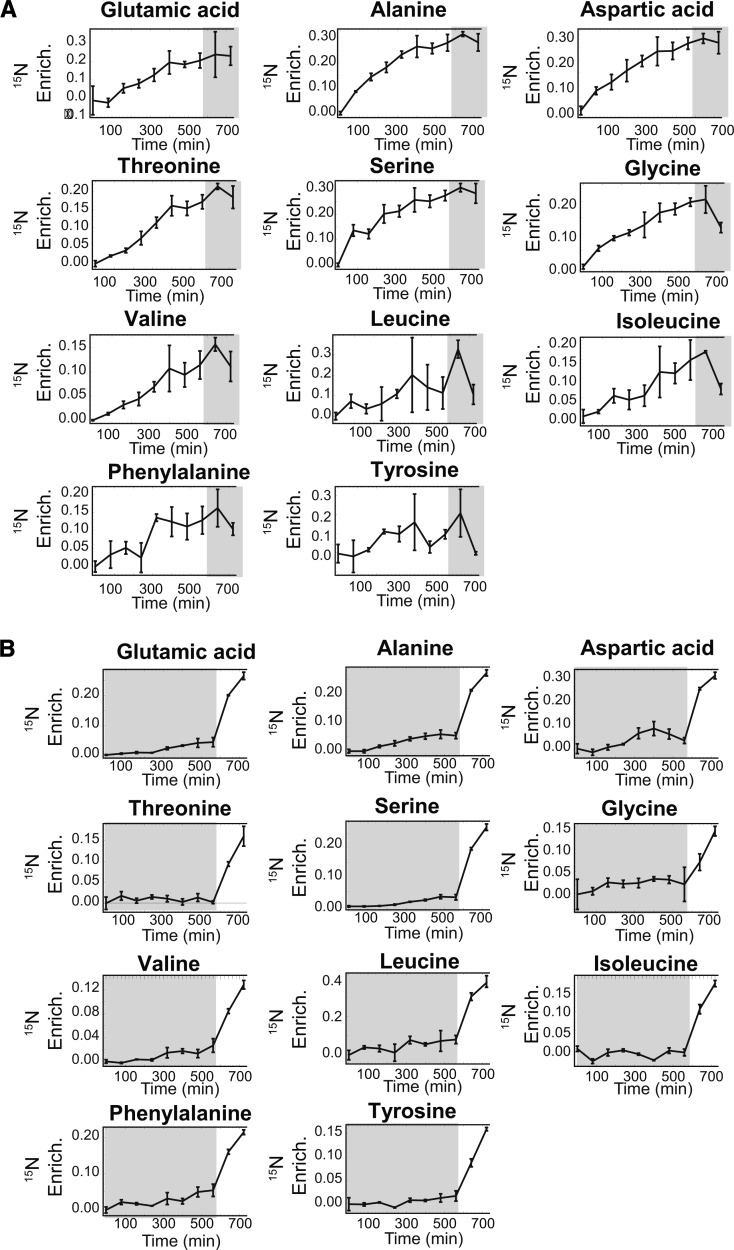
In order to determine the viability of 15N protein labeling in planta, we first examined the rate and consistency of label incorporation into amino acid pools in the leaves of hydroponically grown barley plants. After 25 d of plant growth and at 2.5 h into the light period, we swapped hydroponic medium containing KNO3 with natural abundance (NA) levels of 14N (99.6%) and 15N (0.4%) for medium containing K15NO3 (98% 15N) and began collecting whole plants for gas chromatography-mass spectrometry (GC-MS) amino acid analysis in leaves. As can be seen in Figure 1A, 15N labeling increased in an approximately linear fashion through the light period and then began to level off later in the photoperiod. Glu, which is one of the entry points for inorganic nitrogen, was rapidly labeled. Other abundant amino acids (Ala, Asp, Ser, and Thr) also incorporated the 15N label quickly. Other lower abundance amino acids such as Val and Ile were labeled more slowly. These results are in general agreement with prior data from metabolic labeling of barley plants (Thiele et al., 2008) and revealed that 15% to 30% of many of the major amino acid pools were 15N labeled in leaves after approximately 10 h of labeling. Alternatively, when the 15N label was applied 2.5 h after dark (Fig. 1B), very little label was incorporated into any of the amino acids. However, when the lights came on at 570 min, the label was rapidly incorporated into barley leaf amino acids. This observation is consistent with the reduced activity of nitrate reductase in the dark and with other observations in barley (Aslam and Huffaker, 1982; Gojon et al., 1986; Abdel-Latif et al., 2004).
Figure 1.
15N enrichment = Heavy/(Heavy + Light) ± se (n = 2 or 3) for the listed amino acids in 80-min intervals. A, 15N enrichment levels of amino acids in plants provided with 15N label at 2.5 h into the light period. B, 15N enrichment levels of amino acids in plants provided with 15N label at 2.5 h into the dark period. Light periods are marked in white and dark periods in gray.
Labeling of Proteins and Calculation of Protein Kd in Barley Leaves
In order to track protein turnover using stable isotope labeling and peptide MS, the first step involves swapping unlabeled or NA amino acid pools for labeled or heavy (H) amino acids (Cambridge et al., 2011; Zhang et al., 2011; Trötschel et al., 2012; Li et al., 2012b). Following this label swap, proteins can be divided into two groups: NA (unlabeled) and H (labeled). The NA population contains proteins that were present prior to the label swap and only experience degradation. In contrast, the H population contains newly synthesized proteins and is subject to both synthesis and degradation. For our calculations, we define relative isotope abundance (RIA) as the proportion of the total population that carries the NA label: RIA = NA/(H + NA). As proteins are degraded over time, new copies of a protein are synthesized. This corresponds to a decrease in the proportional size of the NA population and a reciprocal increase in the proportional size of the H population. Protein Kd is calculated by tracking the decrease in the proportional size of the NA population over time using established procedures (Pratt et al., 2002; Cambridge et al., 2011; Zhang et al., 2011; Li et al., 2012b), with details provided in Supplemental Methods S1. Plant tissues also grow over time, which contributes to the H but not to the NA population; we account for this dilution effect by incorporating measurements of plant growth rates into our calculations, which are outlined in detail in Supplemental Methods S1.
Ideally, the label swap would be instantaneous, but in reality, when using whole plants, there is a delay in delivery of the label associated with the root uptake of 15NO3−, transport through the plant, and incorporation into amino acids by the nitrogen assimilation machinery. Thus, it is important to take this lag into account when choosing time points for sampling to ensure that the tissue of interest is adequately labeled, allowing reliable measurements of Kd. To make high-quality measurements, it is important to wait until the amino acid precursor pools reach a certain isotope enrichment threshold in order to maximize the chance that newly synthesized proteins will possess an isotopic label, or, alternatively stated, minimizes the amount of newly synthesized protein that possesses no label. The fraction of the labeled population that will have zero labeled nitrogens is equal to 14N enrichmentamino acidsnumber of nitrogens. For example, consider a typical tryptic peptide of 15 residues made of amino acids with an average 15N enrichment of 20%. In this case, less than 1% of peptides derived from a newly synthesized protein would have no labeled residues (Zhang et al., 2011). Therefore, based on our amino acid labeling data (Fig. 1), we selected 12 h as our first time point to assess protein turnover.
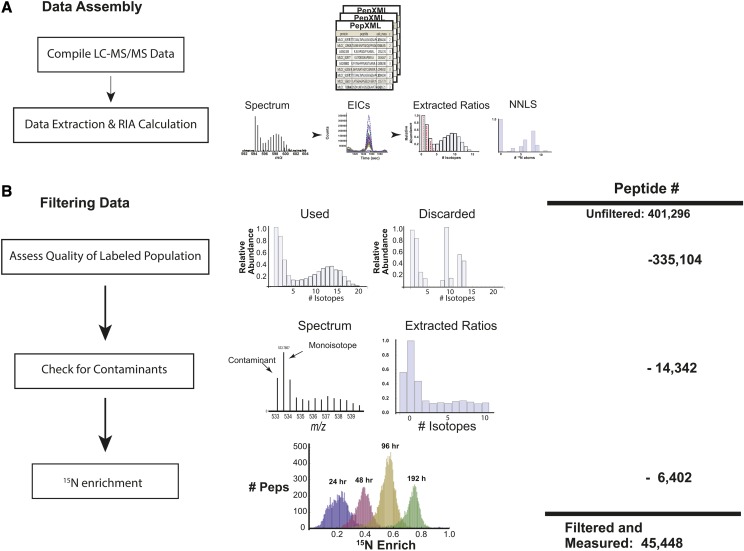
For our proteomic analysis of protein turnover rate in barley leaves, we also analyzed leaf tissue sampled at 24, 48, 72, 96, 144, and 192 h from the initiation of labeling. Images of barley plants from 25 to 33 d old are provided in Supplemental Figure S1. An example of our workflow is provided in Figure 2. Proteins were separated using SDS-PAGE, gels were cut into sequential fractions and proteolyzed with trypsin, and peptides were analyzed by LC-MS/MS. Isotopic labeling of proteins and peptides of an intermediate state of labeling has been reported to result in a loss of protein identifications (Whitelegge et al., 2004; Price et al., 2010). To counter this, we applied a modified approach whereby proteotypic peptides generated for barley leaf samples were cross extracted across all sample data files, so as not to suffer the negative consequences of reduced peptide and protein identifications from tandem mass spectrometry (MS/MS) experiments with partially labeled samples (Price et al., 2010; Trötschel et al., 2012). MS/MS data were searched against a list of barley proteins (Mayer et al., 2012) using the Mascot search algorithm (Perkins et al., 1999). The search results from all samples were then filtered and combined using the Trans Proteomic Pipeline (TPPL; Keller et al., 2005; Fig. 2A) in order to generate a list of proteotypic peptides (TPPL peptide P ≥ 0.8), which were then used to determine high-quality protein identifications with a protein P ≥ 0.95. Then, for each peptide and across all samples, extracted ion chromatograms (EICs) were generated for all relevant isotopes. Next, NA and H peptide populations were assigned using a previously described nonnegative least-squares algorithm, whereby the H population is not defined by one value that represents an average of all the labeled peptide but, rather, models the labeled peptides as several subpopulations based on the number of 15N atoms present in the peptide (Price et al., 2010). Because of the complex nature and wide dynamic range in abundance of proteins present in a crude lysate, we found that a significant portion of our peptide measurements were of low quality, due either to poor signal-to-noise ratio or chemical noise, which in many cases corresponded to similarly eluting peptides that were close in mass. In order to remove these poor measurements from the data set, we established a series of filters that removed low-fidelity measurements, as shown in Figure 2B and explained in detail in the Supplemental Methods S1.
Figure 2.
Workflow for processing of MS data. A, LC-MS data sets from all time points were compiled, and a list of proteotypic peptides characterized using mass, retention time, charge state, and gel fraction number was made. EICs were generated for isotopes of each peptide, the EICs were used to calculate RIA at each time point, and these values were used to calculate labeled and unlabeled populations via a nonnegative least-squares regression (NNLS). B, To reduce error in turnover calculations, a series of filters were applied to peptide level calculations. Peptide measurements were filtered first on the characteristics of measured isotopic ratios, such as the number of measurable ratios and their distribution. These filters are described more thoroughly in Supplemental Methods S1. Measurements not meeting these criteria were discarded. Shown are representative data that were kept and discarded. As a second filter, we measured the value of the isotope 1 D lighter (minus one peak) than the monoisotopic mass. If the ratio of the monoisotope to this minus one peak was less than 5:1, the measurement was discarded, as this was a good indicator of overlapping and contaminating peptides. As a third filter, we assessed 15N enrichment for all peptides from a given time point, and all peptides that fell outside 2 sd of the distribution were removed, as we found that these peptides were of low quality. Shown are 15N enrichment levels for peptides at 24, 48, 72, and 96 h. At the right is the total number of possible quantitation events across all experiments. Listed beside each filter is the number of quantitation events that were removed by the respective filter, with the final number of successful quantitation events listed below.
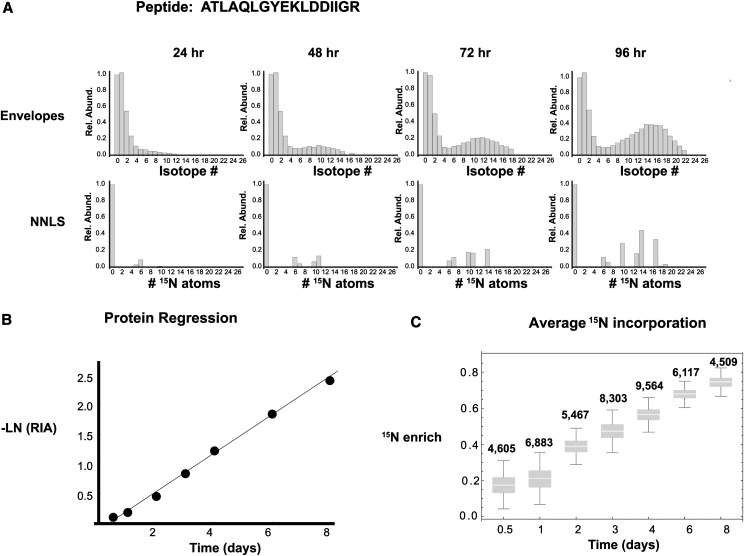
As an example, the ATLAQLGYEKLDDIIGR peptide from the ferredoxin-linked Gln oxoglutarate aminotransferase (MLOC_70866) is shown in Figure 3A. Examples of RIA are shown from samples at 24, 48, 72, and 96 h, with the modeled fits to the isotopic abundances immediately beneath. The negative of the natural log of RIA measurements for all measurements for a protein was used to determine the Kd for each protein (Fig. 3B), as detailed in Supplemental Methods S1. The appearance of peptide populations with a larger degree of 15N enrichment through time in the proteins would be consistent with enrichment in the translational amino acid pool as plants grow and are exposed to the isotopic label, consistent with studies in plants and mammals (Price et al., 2010; Li et al., 2012b). To more clearly demonstrate this, the average 15N enrichment levels for the labeled peptide fraction for all identified peptides at each sampling time were then calculated (Fig. 3C). At 12 h, the earliest time point, we had a median enrichment of 18%, and the enrichment then rose steadily to 75% in the plants sampled after 8 d of labeling. The 18% enrichment in the labeled peptide fraction at 12 h is consistent with the amino acid labeling profiles in Figure 1A.
Figure 3.
Example of protein turnover calculation. A, Top row, the isotopic envelope represented as isotopic ratios for the peptide ATLAQLGYEKLDDIIGR from Fd-GOGAT (MLOC_70866.1), at 24, 48, 72, and 96 h, with all isotopes normalized to the monoisotopic value. Bottom row, as determined by nonnegative least squares (NNLS), the NA form of the peptide, containing zero labeled 15N atoms, and the H form of the peptide, containing one or more 15N atoms. The RIA can then be measured as NA/(NA + H). B, The inverse of the natural log (−LN) of RIA for all time points for the protein was used to determine a degradation rate for the protein. C, Average 15N enrichment of labeled peptide populations at different time points through the course of the experiment.
A key assumption in the way we have conducted our calculation of protein turnover is that steady state (constant Kd and Ks) is a reasonable approximation for the duration of the experiment. We assessed the validity of this assumption in two ways. First, we looked at growth rates over the course of the experiment (Supplemental Fig. S2) and found that the values fit well to an exponential growth curve, with r2 = 0.94 determined via nonlinear regression. The growth constant (0.14, or 14% per day) is incorporated into our calculations of protein Kd to account for the dilution effect due to growth throughout the experiment. Second, we determined whether specific protein abundance levels varied during the experiment by measuring the abundance of proteins for which three or more peptides were observed in each time point. Our method for measuring protein abundance involved integrating the area under the EIC curve for the three most intense peptides derived from a given protein using established protocols (Silva et al., 2006; Ahrné et al., 2013), with a detailed explanation provided in Supplemental Methods S1. From our data set, we had 39 proteins that were observed by three or more peptides at all time points (Supplemental Table S1), with the abundances graphed in Supplemental Figure S3 and data provided in Supplemental Table S2. Additionally, for each protein, we conducted a Spearman rank correlation test for the relative abundance versus time, and after correcting for multiple testing, none of the proteins showed a significant trend in protein abundance relative to time. This suggests that the cellular proteome was quite consistent across the duration of the experiment. Therefore, both analyses support our assertion that plants were undergoing steady-state exponential growth, validating our choice of mathematical approaches to calculate protein Kd.
The Fastest Degrading Barley Proteins, and Calculations of Half-Lives in the Light
A list of the most rapidly degraded proteins is provided in Table I. The proteins came from several different functional categories, but all proteins with known localization were from the plastid. The fastest turning over protein, with a Kd of 1.65 d−1, was THI1, an enzyme involved in thiamine metabolism. The second and third most rapidly degraded proteins are involved in tetrapyrrole metabolism, these were magnesium (Mg)-chelatase (GUN5) and an iron-binding protein, CRD1. The D1 subunit of PSII was the fourth fastest in the data set. Because D1, THI1, and GUN5 appeared to be turning over so quickly and were difficult to measure beyond 24 h, we could not conduct a regression for these proteins. In order to account for the possibility that the abundance of these proteins varied over time, we also calculated Kd in an alternative fashion that considers potential changes in protein abundance; this is described thoroughly in Supplemental Methods S1 (Li et al., 2012b). As can be seen in Table I, the proteins with the fastest Kd values using the first method of calculation also exhibited the highest Kd values via the alternative calculation, supporting the argument that these proteins are indeed degraded quickly. Proteins from several other functional categories were also observed in the top 10, including an RNA-binding protein, a kinase, and a disulfide isomerase. Because the synthesis and turnover of several proteins on this list are known to be higher in the light (Tang et al., 2003; Pal et al., 2013), we were concerned that we were underestimating half-lives of this fastest set by considering data across all time points in our sampling regime, which encompassed long periods of darkness. To counteract this diurnal effect for the rapidly turning over subset, we also provide an estimate of the half-life for protein observations in the first 12-h time point, taking into account the lag in labeling time calculated from the average x intercept in the regression (Fig. 3B). The median x intercept from the regression values for all proteins was 8.6 h. As can be seen in Table I, based on these calculations, a number of the most rapidly labeled proteins possess predicted half-lives of around 2 h.
Table I. Rapid turnover protein Kd values versus protein half-life from 12-h measurements.
N.A. in the predicted half-life column indicates that no peptides were observed from this protein in either of the 12-h time points.
| Protein Identifier | Annotation | Kd, All Data | Predicted Half-Life in Light | Kd, Alternative |
|---|---|---|---|---|
| d−1 | h | d−1 | ||
| MLOC_11312.1 | THI1 | 1.65 | 1.8 | 1.21 |
| MLOC_82117.2 | Mg-chelatase (GUN5) | 1.59 | 1.9 | 2.18 |
| AK359887 | CRD1 | 1.1 | 2.4 | |
| MLOC_9279.1 | D1: PSBA | 0.94 | 2.5 | 1.28 |
| MLOC_43949.1 | THIC | 0.64 | 5.7 | |
| AK250295.1, MLOC_19141.1, MLOC_71436.1 | Lipid transfer protein4 | 0.58 | N.A. | |
| AK356209, AK360524 | RNA-binding family | 0.38 | 47.5 | |
| MLOC_65860.1 | CP12 domain containing | 0.37 | N.A. | |
| AK376220 | Asparate kinase-homoserine dehydrogenase II | 0.36 | N.A. | |
| MLOC_34830.1 | Disulfide isomerase-like protein | 0.33 | 10.3 |
Distribution of Kd Values for Barley Proteins across Functional Classes and Subcellular Locations
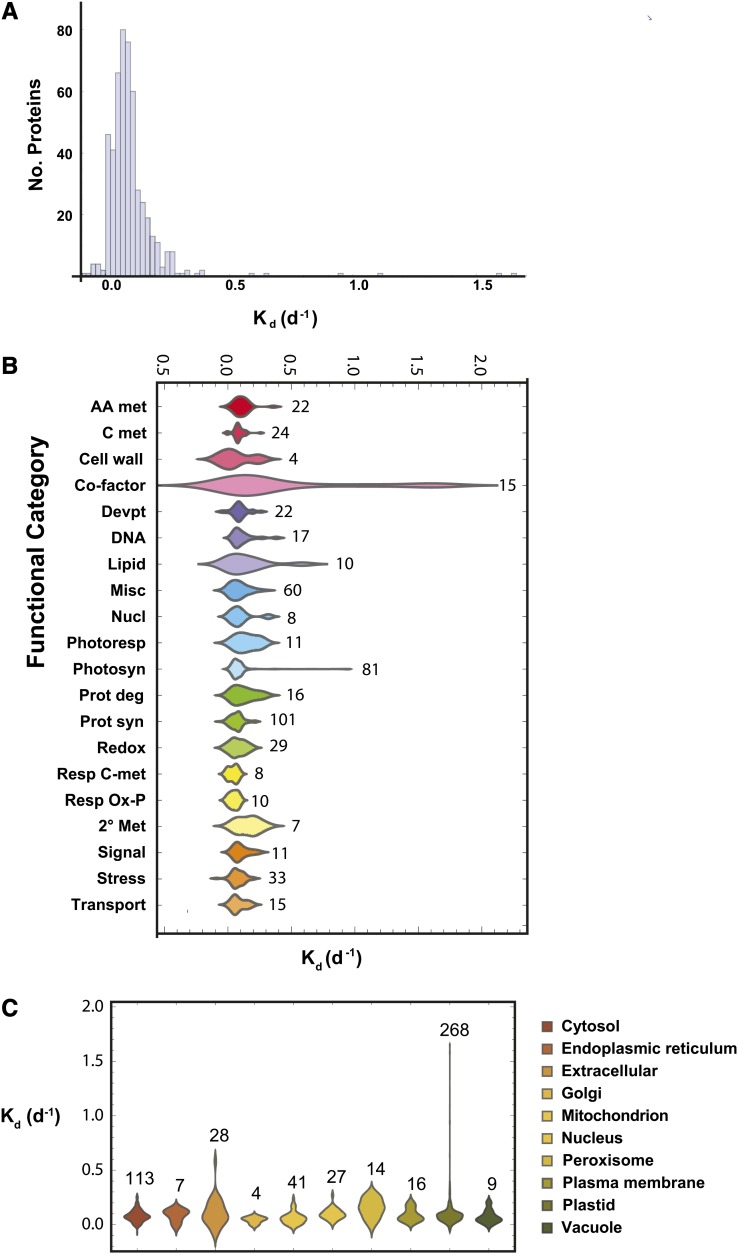
Considering peptides distinct for a protein group and with a TPP peptide P value of 0.8 or greater, we were able to measure Kd for 508 proteins when requiring three or more quantitation events. The full list of protein Kd values filtered with these thresholds is provided in Supplemental Table S2, with the list of measured peptides provided in Supplemental Table S3. A histogram of Kd values for all proteins is shown in Figure 4A. The median value for all proteins is 0.08 d−1, with most proteins falling between 0 and 0.2 d−1. Next, we wanted to determine what role biological function or subcellular localization might play in protein turnover rate. Because the Arabidopsis genome and its gene complement are well annotated, we used the BLAST algorithm (Altschul et al., 1990) to select the closest Arabidopsis homolog for each barley protein so that we could take advantage of the extensive resources that have been developed for Arabidopsis research. We obtained biological functional categorization from the MapCave Web site (Thimm et al., 2004; http://mapman.gabipd.org/web/guest/mapcave), with the data presented in Figure 4B. Next, we defined subcellular localizations for this set of proteins using the SUBAcon naïve Bayesian classifier (Tanz et al., 2013), and this information is provided in Figure 4C.
Figure 4.
A, Histogram of observed degradation rates. Values ranged over more than 2 orders of magnitude, with a median Kd of 0.08 d−1. B, Distribution of degradation rates for different protein functional categories. Using best BLAST matches from Arabidopsis, barley proteins were sorted into categories based on searches of the MapCave Web site (http://mapman.gabipd.org/web/guest/mapcave). The number of proteins in each category is marked to the right of each population. AA met, Amino acid metabolism; C met, carbon metabolism; Cell wall, cell wall metabolism; Co-factor, cofactor synthesis; Devpt, development and cellular organization; DNA, DNA and RNA metabolism; Lipid, lipid metabolism; Misc, miscellaneous; Nucl, nucleotide metabolism, Photoresp, photorespiration; Photosyn, photosynthesis; Prot deg, protein degradation; Prot syn, protein synthesis and assembly; Redox, redox metabolism; Resp C-met, respiratory carbon metabolism; Resp Ox-P, respiratory oxidative phosphorylation; 2° Met, secondary metabolism; Signal, signaling; Stress, stress response. C, Localizations were determined for Arabidopsis BLAST matches using the Suba Web site (http://suba.plantenergy.uwa.edu.au/), and organellar populations were plotted. The number of observed proteins for each organelle is marked above.
Based on functional annotation grouping, we observed that cell wall-associated proteins had the slowest turnover rate, followed by proteins involved in respiratory oxidative phosphorylation and protein synthesis. The fastest turnover rates were found in pigment and cofactor biosynthesis enzymes, followed somewhat distantly by secondary metabolism and photorespiratory components (Supplemental Table S2). When organellar Kd values were compared by ANOVA, there were no significant differences between groups.
Protein Turnover for Subunits of Protein Complexes
The coordinated assembly and operation of protein complexes makes it likely that most subunits of a complex will exhibit similar turnover characteristics. Exceptions to this rule might indicate (1) that individual subunits are being more rapidly damaged than other proteins in the complex and replaced, (2) that these subunits are peripheral to the complex and assembled separately, or (3) that these subunits are inducible and only present under certain conditions. Nine protein complexes with more than six subunits were represented in our data set. These included the plastid and mitochondrial ATP synthases, the cytosolic and plastidic ribosomal large and small subunit complexes, the photosynthetic electron transport chain PSI and PSII complexes, and a complex of Calvin cycle-related enzymes. These complexes showed average Kd values of 0.04 to 0.11 d−1; however, in some cases, there were outlier subunits that had Kd values more than 2 sd higher than the complex mean and were significantly different when assessed by an outlier test (Table II).
Table II. Turnover of protein complexes and outliers.
| Protein Complex Name | No. of Barley Proteins | No. of Quantified Observations | Average Kd | sd | Proteins Outside 2 sd |
|---|---|---|---|---|---|
| Mitochondrial ATP synthase | 8 | 72 | 0.04 | 0.04 | – |
| Cytosolic ribosome 60S | 24 | 272 | 0.04 | 0.04 | – |
| Cytosolic ribosome 40S | 17 | 174 | 0.06 | 0.04 | – |
| Chloroplast ribosome 50S | 22 | 414 | 0.08 | 0.06 | 3.5 sd: RPL33 (plastid encoded; Kd = 0.29) |
| Chloroplast ribosome 30S | 12 | 133 | 0.08 | 0.06 | 2.2 sd: S18 (plastid encoded; Kd = 0.22) |
| Chloroplast ATP synthase | 7 | 641 | 0.10 | 0.03 | – |
| PSII | 26 | 1,407 | 0.10 | 0.18 | 4.7 sd: PSBA D1 (plastid encoded; Kd = 0.94) |
| PSI | 15 | 764 | 0.04 | 0.02 | 2.2 sd: Subunit P of PSI/CURT1A (Kd = 0.09) |
| Calvin cycle | 8 | 767 | 0.11 | 0.11 | 2.4 sd: CP12 (Kd = 0.37) |
Cytosolic and Plastidic Ribosomes
We measured turnover rates for 75 ribosomal protein groups in barley across the cytosolic and plastidic ribosomes. Ribosome protein turnover was fairly slow, with median half-lives of approximately 11 d, which was one of the slower functional categories observed (Fig. 4A). This trend is consistent with other work from mice, where ribosomal proteins were turning over at slower rates compared with many other complexes (Price et al., 2010). In a study of mammalian cell culture, ribosomal subunits possessed different turnover rates (Cambridge et al., 2011), and the authors suggested that certain ribosomal proteins are generated in excess, with rapid degradation of proteins that are not stabilized by insertion into complexes. This view is supported by the observation that when proteasomal activity was chemically blocked in animal cells, ribosome subunits preferentially accumulated in the nucleolus (Lam et al., 2007). In the cytosolic 60S complex, we observed Kd values for 24 proteins, and from the 40S subunit, we observed values for 17 proteins. There were no proteins from either group that were noticeably different from their respective complexes. On the whole, barley chloroplast ribosome 50S and 30S subunits were turning over at a similar rate to their cytosolic counterparts. The two exceptions to this were ribosomal protein of the large subunit33 (RPL33) belonging to the 50S subunit, which was turning over 3.5 sd faster than the mean and was significantly different via Dixon Q test (P < < 0.001). Additionally, from the 30S complex, S18 was 2.2 sd from the mean for this complex but was not significantly different by Dixon Q test (P = 0.10). Both of these subunits are plastid encoded. Overall, six of the 35 chloroplast ribosomal subunits identified in this study were plastid encoded, and the other four plastid-encoded subunits had turnover rates within 40% of the mean of the nucleus-encoded subunits.
Photosystems
In PSII, one protein was turning over 4.7 sd faster than the mean for the complex (P < < 0.001 via Dixon Q test). This subunit was PSBA, also known as D1, and is the best known and most studied rapid turnover protein in plants. The oxidative damage, removal, and replacement of D1 in PSII have been measured repeatedly (Aro et al., 1993; Christopher and Mullet, 1994; Tyystjärvi et al., 1994). Based on our calculations using only the 12-h time point data, it has a half-life in the light of as little as 2.4 h (Table I). Much slower than D1 but still double the average Kd of PSII and significantly different (P < < 0.001 via Dixon Q test) was PSBC, alternatively named CP43. This observation is similar to prior observations made in algae (Yao et al., 2012) and land plants (Christopher and Mullet, 1994) and is consistent with the close proximity of D1 and CP43 in the PSII structure (Kato and Sakamoto, 2009). In PSI, there was one faster turnover subunit, PSIP, that has a Kd of 0.09 compared with the complex mean of 0.05, but its turnover rate was not significantly different by Dixon Q test. Interestingly, the homolog of this protein in Arabidopsis has recently been shown to have a role entirely independent of PSI as a curvature thylakoid protein (CURT1A; Armbruster et al., 2013), providing a reason why it would not have the same turnover characteristics as other PSI subunits.
ATP Synthases
The structure of both the plastid and mitochondrial ATP synthases has been studied extensively (Hamasur and Glaser, 1992; Yoshida et al., 2001; Sunamura et al., 2012). We have shown previously that the subunits from both complexes turn over at comparable rates in Arabidopsis cell culture (Nelson et al., 2013). In barley leaves, no subunit’s turnover rate was more than 2 sd from the mean for each complex or significantly different by Dixon Q test, suggesting that the complexes turn over largely as intact units in planta.
Calvin Cycle
Reports over several decades have noted different combinations of Calvin cycle protein complexes, including early reports of associations between Rubisco with other Calvin cycle enzymes and ferredoxin reductase (Suss et al., 1993). Additionally, there are detailed analyses of the role of CP12 in complexing with and modifying the activity of the Calvin cycle enzymes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and phosphoribulokinase (Wedel et al., 1997; Carmo-Silva et al., 2011; Gontero and Maberly, 2012) as well as a recent report of megadalton complexes of GAPDH with small and large subunits of Rubisco and 400-kD complexes of Rubisco, GAPDH, and phosphoribulokinase (Behrens et al., 2013). Combining this larger group of enzymes, we noted a similar turnover rate of 0.11 d−1, with the notable exception of CP12 (Kd = 0.37 d−1), which was 2.3 sd faster than the mean for the Calvin cycle group and significantly different via Dixon Q test (P = 0.002).
Fast Turnover of Enzymes in Select Metabolic Pathways
Analysis of functional groups (Fig. 4) and individual protein turnover rates (Supplemental Table S2) revealed certain metabolic pathways that were overrepresented among fast turnover proteins. Multiple enzymes of thiamine and tetrapyrrole biosynthesis were found to possess high turnover rates, along with peroxisomal enzymes of the photorespiratory cycle. We examined these pathways more closely to determine if turnover rates might be informative in understanding the regulation or function of these pathways (Fig. 5).
Figure 5.
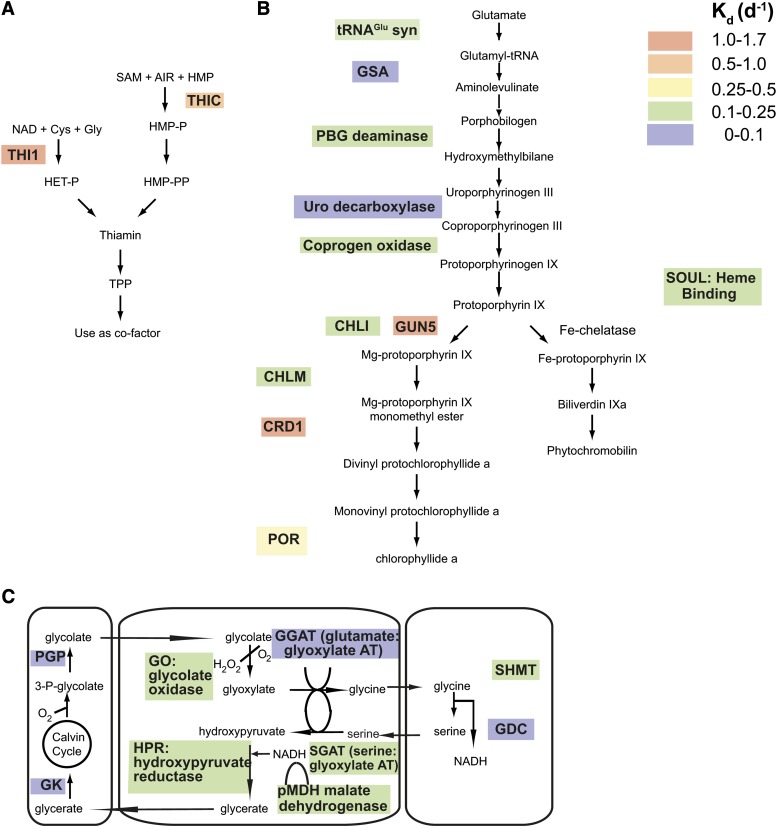
Turnover of proteins in metabolic pathways in barley leaves. A, Biosynthetic pathway for the cofactor TPP. NAD (NAD+), Cys, and Gly are converted to 4-methyl-5-(β-hydroxyethyl) thiazole phosphate (HET-P) via the rapidly degraded THI1. In the other branch of the pathway, another fast turnover enzyme, THIC, catalyzes the conversion of S-adenosyl Met (SAM), 5-aminoimidazole ribonucleotide (AIR), and hydroxymethylpyrimidine (HMP) to HMP phosphate (HMP-P), which is then converted to HMP pyrophosphate (HMP-PP). HET-P and HMP-PP are then converted to thiamine phosphate (Thiamine) and subsequently to TPP for use as an enzymatic cofactor. B, The tetrapyrrole biosynthetic pathway. In the top portion of the pathway, five proteins (glutamyl tRNA synthetase [tRNAGlu syn], Glu-1-semialdehyde aminomutase [GSA], porphobilinogen [PBG] deaminase, uroporyphrinogen [Uro] decarboxylase, and coprogen oxidase) all had fairly typical degradation rates. In the Mg2+ branch of the pathway, leading to chlorophyll biosynthesis, two of the most rapidly degraded proteins in the data set were observed, an Mg-chelatase subunit (GUN5) and MgMPPE cyclase (CRD1), as well as POR. Also observed in this branch were a CHLM protein from the Mg-protoporphyrin IX methyltransferase complex and another subunit of the magnesium chelatase complex (CHLI). One protein was classified as a heme-binding protein (SOUL) and involved in tetrapyrrole metabolism but has no clearly assigned function. C, Photorespiratory pathway. The oxygenic reaction of Rubisco leads to the formation of 2-phosphogycolate, which is converted to glycolate that is exported to the cytosol and enters peroxisomes, where it is oxidized to glyoxylate and aminated to Gly by the fast turnover of glycolate oxidase and Glu:glyoxylate aminotransferase (AT). After decarboxylation of Gly and the synthesis of Ser in mitochondria by the combined action of Gly decarboxylase (GDC) and Ser hydroxymethyltransferase (SHMT), the Ser reenters peroxisomes, where it is deaminated to hydroxypyruvate and reduced to glycerate by two rapidly degrading enzymes, Ser glyoxylate aminotransferase (SGAT) and hydroxypyruvate reductase (HPR). Glycerate is then returned to the chloroplast, where it is phosphorylated by glycerate kinase (GK) to reenter the Calvin cycle as 3-P-glycolate and then converted to glycolate by phosphoglycolate phosphatase (PGP).
Thiamine Synthesis
THIC and THI1 turn over very quickly and are involved in thiamine biosynthesis, which is ultimately used to make thiamine pyrophosphate (TPP), an important enzymatic cofactor (Fig. 5A). The protein with the largest Kd value in the entire data set was THI1, which converts Gly and NAD+ to ADP 5-(2-hydroxyethyl)-4-methylthiazole-2-carboxylic acid, a precursor to thiazole. Mutants for this protein require thiamine supplementation for growth (Papini-Terzi et al., 2003). Using the modified method of calculating Kd in the light, the half-life of this protein would be 1.9 h during illumination (Table I). To our knowledge, there are no reports of turnover for this protein in plants. Additionally, we observed a Kd of 0.69 d−1 for THIC, which converts S-adenosyl Met and 5-aminoimidazole ribonucleotide to hydroxymethylpyrimidine phosphate and is the rate-limiting step in thiamine biosynthesis (Bocobza et al., 2013). These plastid enzymes were the only two enzymes in this pathway for which we obtained data, but these results clearly indicate that enzymes in both branches leading to thiamine synthesis are turned over several times each day.
Tetrapyrrole Biosynthesis
We measured degradation rates for several enzymes involved in tetrapyrrole biosynthesis, which bifurcates into a branch for chlorophyll biosynthesis and a branch for heme biosynthesis (Fig. 5B). In the upper portion of the pathway prior to the split, we have made estimates of turnover for glutamyl tRNA synthetase, Glu semialdehyde aminotransferase, porphobilinogen deaminase, uroporyphrinogen decarboxylase, and coprogen oxidase, with Kd values of 0.11, 0.03, 0.17, −0.04, and 0.16 d−1, respectively. Although there were no measurements of Kd for proteins with clearly defined enzymatic functions in the heme branch, we measured unusually fast turnover rates for several enzymes involved in the Mg2+ branch of tetrapyrrole biosynthesis, which is the branch associated with chlorophyll biosynthesis. Most prominent is the rapid turnover of Mg-chelatase H subunit GUN5, which was the second fastest degraded protein in the data set, with a predicted half-life of 2 h during illumination. GUN5 is one of three subunits in the Mg-chelatase complex that converts protoporphyrin IX to Mg-protoporphyrin IX in the first step in the chlorophyll branch of tetrapyrrole biosynthesis, and it is a key player in chloroplast retrograde signaling (Kindgren et al., 2012). Another subunit of this complex, CHLI, had a more typical degradation rate of 0.11 d−1. In the next step of the pathway, Mg-protoporphyrin IX methyl transferase (CHLM) catalyzes the methylation of Mg-protoporphyrin IX to Mg-protoporphyrin IX monomethyl ester (MgMPPE). CHLM was degraded at a faster rate than the median protein in our data set, but it had a more typical rate in comparison with other enzymes in this branch of tetrapyrrole biosynthesis (Kd = 0.19 d−1). The MgMPPE is then converted to protochlorophyllide via MgMPPE cyclase, which contains at least one soluble subunit and two membranous subunits (Rzeznicka et al., 2005). CRD1 is one of the membranous subunits of this complex and was the third most rapidly degraded protein in our data set, with an estimated half-life of 2.4 h during illumination. In the last step of chlorophyll biosynthesis, NADPH:protochlorophyllide oxidoreductase (POR) is responsible for the catalysis of protochlorophyllide to chlorophyllide and is composed of two subunits in dicots (PORA and PORB) but only one subunit in monocots. We observed POR as turning over rapidly, with a Kd of 0.27 d−1, which is three times the median decay rate for all observed barley leaf proteins. Finally, there was one heme-binding protein (SOUL) classified as involved with tetrapyrrole metabolism, but it had no known specific function and had a Kd of 0.15 d−1.
Photorespiration
The most rapidly degraded protein in this pathway was an isoform of the peroxisomal glycolate oxidase (Kd of 0.27 d−1; Fig. 5C), with two other isoforms having somewhat lower Kd values (0.24 and 0.12 d−1). This is closely followed by three other peroxisome-located enzymes involved in the pathway: the Ser/glyoxylate aminotransferase (Kd of 0.26 d−4), the peroxisomal malate dehydrogenase (Kd of 0.26 d−1), and the hydroxypyruvate reductase (Kd of 0.16 d−1). This is in contrast with the mitochondrial photorespiratory components (average Kd of 0.12 d−1) and the plastid-localized enzymes, glycerate kinase and phosphogylcolate phosphatase, which respectively initiate and complete the cycle (average Kd of 0.04 d−1).
DISCUSSION
Protein turnover is integral in maintaining cellular function, particularly in response to environmental stress (Araújo et al., 2011). In addition, the cost of protein synthesis/degradation is known to be a significant energetic cost for plants (Penning De Vries, 1975; Hachiya et al., 2007). Measurement of protein turnover kinetics as a proteomic technique is just beginning in planta, with prior large-scale work conducted in plant cell culture (Li et al., 2012b; Nelson et al., 2013). Assuming that Ks and Kd are constant across the duration of our experiment, we used MS to acquire steady-state measurements of degradation rates for a selection of more than 500 of the most abundant leaf proteins in the major crop species barley under control conditions and show more than 100-fold differences between their half-lives. This highlights the highly differential cost of maintaining the major components of the proteome at steady-state levels. For example, we have documented the investment required to maintain the abundance of rapidly degraded proteins such as GUN5 and THI1. While it is interesting to consider these fast turning over proteins on the list, the total cost of synthesis is also a function of the protein’s abundance. While the Kd values for the large and small subunits of Rubisco were close to the median value for all proteins, the abundance of these proteins in the cell will dominate the associated costs of protein maintenance. However, there are various improvements that can be made and challenges to be addressed in future protein turnover studies in intact plants to maximize the value and accuracy of calculations, especially for proteins possessing faster turnover rates. These include measurement of lag in labeling, determining diurnal patterns of amino acid and protein synthesis, alternative label options for proteins turning over at different rates, biological interpretation of aberrant turnover rates in pathways and protein complexes, and non-steady-state analysis of changes in turnover characteristics.
Defining Lag and Diurnal Effects in Labeling to Extend Dynamic Range
When applying isotopic labels to intact plants for quantifying protein turnover, one of the primary concerns is the lag from label application to incorporation into amino acids and proteins. In a prior study of 2H2O labeling in Arabidopsis seedlings, proteins incorporated the isotopic label quickly, but high levels of deuterium were problematic due to the resulting physiological changes (Yang et al., 2010). In an assessment of a pulse-chase 13CO2 labeling approach in leaves from Arabidopsis during which plants were grown in 13CO2 and the gas label was exchanged for 12CO2, a measurable but diffuse isotopic envelope was observed after 24 h of label swap. To our knowledge, there are no proteomic studies in intact plants that have assessed the utility of 15N for protein turnover studies. Based on results from amino acid data and x intercept data from regression studies of protein measurements, we see approximately a 9-h lag between label application and robust protein labeling of 25-d-old barley plants. This value is comparable to or less than the delays reported for labeling of tissues from mice (Price et al., 2010; Zhang et al., 2011).
The lag in label incorporation into proteins is probably due to several reasons. First, the 15NO3− must be taken up in the root, transported to the leaves via transpiration, reduced to ammonia, and assimilated into amino acids. Second, the labeling of translational amino acid pools with 15N is potentially further retarded by competing with NH3 generated by photorespiration in leaves for reassimilation into amino acids. Earlier work reported that reassimilation of photorespiratory NH3 was significantly greater than new primary NH3 acquisition (Keys et al., 1978). Interestingly, photorespiration is required for efficient reduction of NO3− to NH3 (Rachmilevitch et al., 2004; Bloom et al., 2010). In barley, while there is extensive exchange of reduced nitrogen between tissues (Gojon et al., 1986; Abdel-Latif et al., 2004), studies suggest that leaves account for the vast majority of nitrate reduction under normal conditions, with most of this reduction occurring in the light (Aslam and Huffaker, 1982; Gojon et al., 1986; Abdel-Latif et al., 2004).
It is also interesting that for many of the amino acids observed in our study, there was a drop in the RIA for the H isotope after the lights went off at 570 min after label swap (Fig. 1). While nitrate assimilation is known to occur at slower rates in the dark compared with illuminated conditions (Aslam and Huffaker, 1982), this does not provide an obvious explanation for the decrease in amino acid 15N incorporation during the dark period. A possible explanation is the reported decrease in the amino acid pool sizes in the dark in both barley and Arabidopsis (Winter et al., 1993; Tschoep et al., 2009). This effect was most clearly demonstrated by Gly (Fig. 1), which is also reported to be significantly reduced in abundance in the dark due to the cessation of photorespiration in darkness (Winter et al., 1993; Tschoep et al., 2009). Also worth considering is that plant cells contain multiple pools of amino acids that differ in their metabolic activity (Holleman and Key, 1967), with large inert storage pools of many amino acids being sequestered in the vacuole (Tohge et al., 2011). Therefore, this decrease in the incorporation of whole tissue extracts during the dark period may represent both the cessation of nitrogen assimilation and a relative decrease in the size of metabolically active amino acid pools, which would have been more highly labeled relative to the metabolically inert pools. While this dynamic nature of amino acid pool labeling shows the complexity of the labeling process, the protein incorporation and turnover calculations using steady-state assumptions remain straightforward as long as the average 15N enrichment of utilized amino acid pools is 0.15 or greater in newly synthesized proteins.
The D1 protein of PSII is well documented as degrading quickly in leaf tissue and, therefore, is a good example to consider the applicability of our 15N labeling for the measurement of rapidly degraded proteins with this technique. Reports vary from 1 to 20 h in terms of half-life for this protein across different species (Wilson and Greenberg, 1993; Jansen et al., 1999; Booij-James et al., 2000). When calculated via regression, D1 was the fourth fastest turning over protein, with a Kd of 0.94 d−1, or, in other words, a half-life of 15 h, and thus is within the range of reported values for D1, albeit on the slower side. Because of the challenges that contribute to 15N lag and our desire to make accurate measurements of Kd for all proteins, we consider whether other considerations in calculation would offer viable alternatives for rapid turnover proteins. We know that translation of D1 is strongly dependent on light intensity, with the translation rate being much faster in the light than in the dark (Kim and Mayfield, 1997; Barnes and Mayfield, 2003). Our calculation of D1 Kd incorporates data from both 12- and 24-h time points, and this time series encompasses a long period of darkness. So to account for the light-induced variation in synthesis, we modified our calculation of Kd, using only data from the 12-h time point. This results in an estimated half-life of 8.5 h, which is well within the reported measurements mentioned above. However, this calculation does not account for the lag in label uptake and reduction. We further modified our calculation of D1 half-life, accounting for the lag in label delivery based on the x intercept data from the regressions so that we could allow for the lag in label incorporation. This provided a D1 half-life as short as 2.5 h, which is comparable to some of the fastest observations in the literature (Aro et al., 1993; Tyystjärvi et al., 1994). We believe that the 2.5-h half-life estimate is probably the most accurate value, as it factors in the limitations of our system and is quite close to values reported by other methods. So while it can be complicated to obtain precise measurements of rapidly degraded leaf proteins using 15N, our data set illustrates the power of 15N labeling and MS for obtaining accurate measurements of Kd for hundreds of proteins, placing D1 in the top percentile.
Label Selection for Protein Turnover Studies
The ideal label would be one that is rapidly incorporated into amino acid pools with minimal lag time and that can accurately measure Kd over a wide dynamic range (i.e. hours to weeks). In addition, a preferred label would be present in most peptides, and the organism being analyzed would be auxotrophic for the label (Hinkson and Elias, 2011). Plants can synthesize all of the standard 20 amino acids, which would suggest that providing exogenous labeled amino acids may not work. One prior work using stable isotope labeling by amino acids (SILAC) in Arabidopsis cell culture confirmed that amino acid labeling would be problematic due to incomplete labeling (Gruhler et al., 2005), while another investigation of SILAC for proteomic applications in seedlings showed that the strategy may be viable, with high incorporations of labeled amino acids into proteins (Lewandowska et al., 2013). But even if this strategy is viable in some defined scenarios, given the high cost of SILAC reagents, this strategy will be of limited use in hydroponic systems and would not scale well to the measurement of older plants and larger species of agricultural importance. Given the autotrophic nature of plants, this would leave the elements in peptides that have multiple stable isotopes (sulfur, hydrogen, carbon, nitrogen, and oxygen). Given the variable fashion in which different elemental labels are assimilated by plants, there probably is no single best solution for all plant tissues and for all experimental conditions. As has already been established, deuterium at high levels is toxic (Kushner et al., 1999; Yang et al., 2010). Sulfur is present in only a fraction of peptides, so it is probably not a good choice as a label. Oxygen has been used as a proteomic tracer in bacteria (Bernlohr, 1972), but given the price of 18O water, this strategy may be cost prohibitive. Nitrogen is easy to apply in a controlled fashion as a salt, which is then taken up by root tissue; however, lag before delivery to aerial tissues limits measurements in the first few hours following labeling. It may be possible to shorten the lag by applying the label as reduced NH4+, but there are two shortcomings with this strategy. In many agricultural scenarios, nitrogen fertilizer may be provided in a reduced form such as urea or NH4+, but this nitrogen is rapidly oxidized to nitrate via the action of bacteria in the soil (Skiba et al., 2011); also, different crop species have varying sensitivities to ammonium toxicity (Britto and Kronzucker, 2002). The final element to consider is carbon, which is challenging because the label, 13CO2, must be supplied as a gas and therefore is more difficult to regulate. However, given the rapid incorporation of this label into amino acids of illuminated leaves (Szecowka et al., 2013), it would be anticipated that the lag in robust labeling of proteins might be reduced to only minutes for aerial tissues. For more slowly degraded proteins, extended labeling periods may be required. While 13C conceivably could be used to measure slowly turning over proteins, this strategy would be expensive and difficult due to the challenges associated with a gas-phase label. A combination of labels, 13C for proteins that have fast turnover rates and 15N for slower turning over proteins, is probably the optimal compromise.
Reasons for Rapid Protein Degradation
Proteins could be rapidly degraded for one of two reasons: either the protein serves a regulatory role and rapid turnover is required to respond rapidly to some environmental perturbation, or the protein is damaged through its functioning. The canonical rapidly degraded protein, D1, is turned over due to light-induced damage in PSII. In addition to our observations of D1, we observed several other rapidly degraded proteins in our data set, with two of these proteins, THI1 and THIC, being involved in thiamine biosynthesis. In tobacco (Nicotiana tabacum), THI1 transcript abundance and polysome loading of this mRNA are high in the light and decrease in the dark (Tang et al., 2003), which would be consistent with our observations of the daily regeneration of THI1 protein. Recent work with the yeast THI1 ortholog THI4p reported that the sulfur atom necessary for thiazole synthesis was transferred from an internal Cys residue, leaving behind a dehydro-Ala residue so that this protein was classified as a suicide enzyme, due to the irreversible nature of this modification (Chatterjee et al., 2011). When the authors examined data on THI1 crystal structures, they concluded that a dehydro-Ala residue was present in the analogous Cys residue, suggesting that this enzymatic mechanism may be conserved across eukaryotic taxa (Chatterjee et al., 2011). This would mean that organisms would need to synthesize a new copy of the THI1 protein for each newly synthesized thiamine molecule, which is surprising given the abundance of THI1 and the associated energetic costs. Even if THI1 is not a suicide enzyme, but instead is susceptible to consuming the internal Cys residue during catalysis, it still could be expected that this enzyme would need to be recycled frequently. The other rapidly turning over protein involved in thiamine metabolism was THIC, which is the rate-limiting step in thiamine biosynthesis (Bocobza et al., 2013). Interestingly, this protein is the only known riboswitch in plants, whereby the THIC mRNA binds TPP resulting in a shift to transcription of an alternative, less stable transcript that is rapidly degraded (Bocobza et al., 2013). Perhaps rapid turnover of the THIC protein represents an additional level of control.
Several enzymes involved in chlorophyll biosynthesis, belonging to the Mg2+ branch of tetrapyrrole metabolism, were observed, and many of these were turned over rapidly. Flux through this pathway is tightly regulated due to the toxic nature of the intermediates. Two of the fastest degrading proteins in our data set were GUN5 and CRD1. We are not aware of any reports of protein turnover measurements for either of these gene products; however, GUN5 is known to function in retrograde signaling between the chloroplast and the nucleus (Mochizuki et al., 2001). Another report established that GUN5 is regulated in a circadian manner by the clock gene, timing of CAB expression1 (TOC1; Legnaioli et al., 2009). Rapid degradation for GUN5 would be consistent with its signaling role as well as its links with circadian oscillations. Other proteins that turned over rapidly in our data set were POR and CHLM. While there are no other precise quantitative measurements of Kd for these proteins in the literature, in one study from barley leaf discs the POR protein, as assessed through western blotting, showed rapid degradation in illuminated leaf discs (Richter et al., 2010). In total, these observations suggest that protein turnover plays an important role in regulating the Mg2+ branch of tetrapyrrole biosynthesis.
There also were differences within the compartmental components of photorespiration. Specifically, we observed that some peroxisome-localized enzymes were significantly different from their mitochondrial and plastid-localized partners. Hydrogen peroxide produced by glycolate oxidase in peroxisomes could lead to its damage during high photorespiratory flux in the light, which may also affect neighboring enzymes. Metabolic channeling of photorespiratory intermediates (NADH, glyoxylate, and hydroxypyruvate) between these enzymes does occur, even when the peroxisomal membrane is disrupted, suggesting close proximity or protein complex formation (Heupel et al., 1991). The turnover of these peroxisomal enzymes of photorespiration is unlikely to be a result of bulk peroxisome turnover, as one of the major components of peroxisomes, catalase, has a Kd of 0.06 d−1, close to the cellular median.
Extension to Non-Steady-State Analysis of Protein Turnover
One obvious extension of these methodologies in intact plants will be the analysis of non-steady-state systems, such as the initial responses to biotic and abiotic stresses, as well as developmental transitions, such as germination, senescence, tillering, and seed filling. These types of studies would help us understand the role of protein degradation in scenarios associated with agricultural productivity. Such analyses could be integrated with biochemical data that quantify the energetic requirements of growth, metabolism, and cellular maintenance (Cheung et al., 2013; Poolman et al., 2013). This would allow us to measure the changing cost of protein turnover and estimate the proportion of cellular energy that is expended on polypeptide synthesis and degradation in different scenarios. This information would greatly further our understanding of the energy budgeting of plants, enhancing strategies that attempt to rationally engineer the partitioning of resources between maintenance, growth, and other energy-consuming processes for optimal resource use efficiency in plants.
CONCLUSION
By tracking the decay of unlabeled proteins following a switch to 15N-labeled medium and using an assumption of constant Ks, we present, to our knowledge, the first large-scale liquid chromatography-mass spectrometry (LC-MS) report of steady-state Kd values in leaves from intact plants. These data give insights into the dynamics of protein complexes and identify a number of previously undocumented fast turnover proteins, some of which are control points that regulate the metabolic response to environmental cues. This work serves as a foundation for future studies investigating the role of protein turnover in plant energy budgeting across different scenarios.
MATERIALS AND METHODS
Plant Growth
Barley (Hordeum vulgare variety Baudin) seed was sterilized for 5 min with 6% (v/v) bleach and rinsed several times prior to placing seeds on wetted filter paper. After allowing 4 d for germination, seedlings were transferred to hydroponic tubs in medium containing 3 mm KNO3, 0.5 mm KH2PO4, 0.5 mm MgSO4, 0.5 mm CaCl2, 0.1 mm Fe-EDTA, 0.05 mm KCl, 0.025 mm H3BO3, 0.005 mm Na2MoO4, 0.002 mm MnSO4, 0.002 mm ZnSO4, and 0.0005 mm CuSO4, adjusted to pH 5.9 with KOH. Roots were flooded and drained with nutrient solution every 30 min, according to the design of earlier work (Munns and James, 2003). Plants were grown for 25 d on a 12 h/12-h light/dark schedule and a 23°C/18°C temperature regime, with 500 µmol m−2 s−1 light intensity and 65% relative humidity. Medium was changed twice weekly. From days 21 to 24, the nitrogen was checked and adjusted daily. After 25 d of growth, the nitrogen salt in the nutrient solution was switched to K15NO3.
Amino Acid Analysis
For amino acid analysis, plants were snap frozen immediately at label application and every 80 min thereafter for the first 12 h of labeling. Samples were ground with a mortar and pestle in liquid nitrogen. To 25 mg of ground tissue, 500 μL of prechilled (−20°C) metabolite extraction buffer (90% [v/v] methanol and 0.008 mg mL−1 ribitol) was added. Samples were heated to 65°C and shaken at maximum speed for 30 min in a thermomixer. Tubes were then centrifuged at 20,000g, and 60 μL of the supernatant was dried in a Centrivap (Labconco) and stored at −80°C until analysis. To dried samples, 20 μL of methoxyamine in pyridine (20 mg mL−1) was added and shaken at 750 rpm for 90 min at 37°C. Next, 30 μL of N-Methyl-N-(trimethylsilyl) trifluoroacetamide (Pierce) was added and shaken for 60 min at 750 rpm and incubated at 37°C. After 60 min of additional incubation without shaking, GC-MS analysis was conducted in a manner similar to previous work (Howell et al., 2009) with the following gas chromatography oven conditions: initial conditions were 70°C, held for 1 min, increased to 76°C at 1°C min−1, from 76°C to 325°C at 6°C min−1, held at 325°C for 8 min, and then returned to 70°C for a total run time of 60 min. The ribitol spike was used as a retention time marker, and samples were aligned using Enhanced Chemstation Data Analysis version E.02.02.1431 (Agilent). Chromatographic peak areas were determined for amino acids and their 15N-labeled counterparts, making corrections for the NA of H atomic isotopes that are mainly derived from 13C. Quantifier ions for amino acids and their 15N-labeled forms are provided in Supplemental Table S4.
Proteomic Analysis
For protein analysis, tissue samples from leaves were harvested immediately after label application and at 12, 24, 48, 72, 96, 144, and 192 h for the assessment of peptide 15N incorporation and protein turnover rates. Samples were snap frozen in liquid nitrogen and stored at −80°C until preparation for proteomic analysis. Frozen leaves from two samples at each time point were ground with a mortar and pestle sitting on dry ice, and whole-cell protein was extracted using a modified chloroform-methanol protocol (Wessel and Flügge, 1984). Briefly, 400 μL of extraction buffer (125 mm Tris-HCl, 7% [w/v] SDS, and 10% [v/v] β-mercaptoethanol, pH 7) was added to 250 mg of ground tissue, with samples then placed in ice on a rocking platform for 10 min, with intermittent inversion of the samples to facilitate thawing. Samples were then centrifuged at 10,000g for 5 min at 4°C, with 200 μL of the resulting supernatant transferred to a new Eppendorf tube. To the supernatant, 800 μL of precooled methanol (4°C), 200 μL of precooled chloroform (4°C), and 500 μL of precooled double distilled water (ddH2O; 4°C) were added and the sample was vortexed. The solutions were then centrifuged at 10,000g for 5 min at 4°C in order to form an aqueous/organic phase separation, with precipitated protein present at the interface. The upper aqueous phase was removed, and 500 μL of methanol was added before vortexing again. Samples were centrifuged at 9,000g for 10 min at 4°C, with the supernatant removed and discarded. To the remaining pellet, 1 mL of precooled acetone (−20°C) was added and vortexed vigorously. Samples were incubated for 1 h at −20°C and then centrifuged at 10,000g for 10 min at 4°C. The supernatant was discarded, and the protein pellets were left to air dry for approximately 15 min at room temperature.
To precipitated protein, sample buffer (2% [w/v] SDS, 50 mm Tris, pH 8.5, and 10 mm dithiothreitol) was added and incubated at 55°C for 30 min while gently shaking with an orbital mixer. Samples were then cooled to room temperature. Protein concentrations were determined using the Amido Black protein assay (Schweikl et al., 1989). To 150 μg of resolubilized protein, 0.5 m iodoacetamide was added to each sample to a final concentration of 25 mm and incubated for 30 min at room temperature in the dark, with the reaction quenched by the addition of dithiothreitol to a final concentration of 30 mm. The protein concentrations of the samples were adjusted to 2.5 mg mL−1 by the addition of modified sample buffer (2% [w/v] SDS, 50 mm Tris-HCl, pH 8.5, and 20% [v/v] glycerol), and 20 μL of sample was loaded onto three adjacent lanes of a precast AnyKD gel (Bio-Rad). Proteins were electrophoresed at 300 V for 20 min. Following separation, gels were placed in gel fix solution (40% [v/v] methanol and 10% [v/v] acetic acid) for 60 min. Gel fix solution was then removed and replaced with three parts Coomassie Brilliant Blue stain (80 mL of methanol, 0.6 g of Coomassie Brilliant Blue powder, 9.4 mL of 85% [v/v] phosphoric acid, and ddH2O to a final volume of 320 mL) and one part ammonium persulfate buffer (50%, w/v). Gels were left to stain overnight before being destained with ddH2O water several times as required.
Gel samples were then cut into 12 fractions grouped by Mr, and each fraction was sliced into approximately 1-mm cubes with a razor blade. Gel pieces were placed in microfuge tubes and destained in 1 mL of 100 mm (NH4)HCO3/50% (v/v) methanol for 10 min while being vigorously shaken at maximum with an orbital mixer for 10 min at room temperature. Supernatants were discarded, and the destaining process was repeated. Samples were then dehydrated using 1 mL of 10 mm (NH4)HCO3/50% (v/v) acetonitrile for 2 min. Supernatant was removed, and samples were further dehydrated with 100% acetonitrile for 30 s. Supernatant was removed, and samples were dried in a Centrivap (Labconco) for 15 min. Trypsin [5 ng μL−1 in 10 mm (NH4)HCO3 and 3% (v/v) acetonitrile] was added to each sample until gel pieces were covered. Additional 10 mm (NH4)HCO3 was added as necessary to keep the gel pieces immersed. Samples were then incubated at 37°C and left to digest overnight. Following overnight digestion, digested proteins were extracted using 700 μL of HPLC-grade water and 1% (v/v) trifluoroacetic acid by vortexing at room temperature in an orbital shaker for 10 min at high speed. Supernatant was transferred to a new 1.5-mL Eppendorf tube. An additional extraction was performed using 700 μL of 70% (v/v) acetonitrile, 25% (v/v) ddH2O water, and 5% (v/v) trifluoroacetic acid. The supernatant was removed and added to the first extraction, and samples were then dried in the Centrivap.
MS
Digested peptides were resuspended in 20 μL of 2% (v/v) acetonitrile and 0.1% (v/v) formic acid, and 5 μL of this resuspension was loaded onto a C18 high-capacity nano LC chip (Agilent) in 98% buffer A (0.1% [v/v] formic acid in Optima grade water [Fisher]) and 2% buffer B (0.1% [v/v] formic acid in Optima grade acetonitrile [Fisher]) using a 1200 series capillary pump (Agilent). Following loading, samples were eluted from the C18 column and into an inline 6550 Series quadrupole time-of-flight mass spectrometer (Agilent) with a 1200 series nano pump (Agilent) using the following gradient: 2% to 45% B in 26 min, 45% to 60% B in 3 min, and 60% to 100% B in 1 min. Fractions from a given sample were analyzed in sequence, and a blank was run between samples. The quadrupole time-of-flight was operated in a data-dependent fashion with an MS spectrum collected prior to the three most abundant ions subjected to MS/MS from doubly, triply, and higher charge states. Ions were dynamically excluded for 0.4 min following fragmentation. MS data were collected at eight spectra per second, while MS/MS spectra were collected at three spectra per second, with a minimum threshold of 10,000 counts and a target of 25,000 ions per MS/MS event.
LC-MS Data Analysis
Agilent .D files were converted to mzML using the Msconvert package (version 2.2.2973) from the Proteowizard project, and mzML files were subsequently converted to Mascot generic files using the mzxml2search tool from the TPPL version 4.6.2. Mascot generic files peak lists were searched against protein sequences released for barley (ftp://ftpmips.helmholtz-muenchen.de/plants/barley/public_data/) using Mascot 2.3 running on an in-house server, allowing for variable Met oxidation, alkylation of Cys, N-terminal acetylation of the protein, 100 ppm mass tolerance for the parent ion, and 0.5 D tolerance on MS/MS peaks. Results were downloaded from the Mascot server as .dat files and then converted to pep.xml files using the ToPepXML tool in the TPPL. Results from LC-MS/MS analysis were then assessed using the PeptideProphet tool with the accurate mass binning option selected and then processed further using ProteinProphet. Further details on the calculation of protein turnover rates are provided in Supplemental Methods S1.
Statistical Analysis of Organelles and Protein Complexes
Proteins were grouped by known subcellular localization of the Arabidopsis (Arabidopsis thaliana) homolog according to the SUBAcon call in the SUBA3 database (suba.plantenergy.uwa.edu.au), and an ANOVA was applied across compartments to identify organelle protein sets that varied significantly. For protein complexes, protein subunit outliers relative to their complex were determined by applying a Dixon Q test (Dixon, 1950; Rorabacher, 1991). All analyses were conducted with R (http://www.R-project.org).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Representative images of plant growth across the experiment.
Supplemental Figure S2. Leaf growth in mass over the course of the experiment.
Supplemental Figure S3. Normalized abundance of barley proteins by the top three ion approach.
Supplemental Table S1. Relative abundance of major barley proteins.
Supplemental Table S2. Information on barley proteins for which Kd was measured.
Supplemental Table S3. Peptides used to quantify proteins.
Supplemental Table S4. Information on fragment ions used to measure 14N and 15N forms of amino acids measured by GC-MS.
Supplemental Methods S1. Details of MS methods and data analysis for protein turnover rate determination.
Acknowledgments
We thank Dorothee Hahne for assistance with GC-MS analyses performed by the University of Western Australia.
Glossary
- Kd
degradation rate
- Ks
synthesis rate
- MS
mass spectrometry
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- GC-MS
gas chromatography-mass spectrometry
- NA
natural abundance
- H
heavy
- RIA
relative isotope abundance
- TPP
Trans Proteomic Pipeline
- EIC
extracted ion chromatogram
- Mg
magnesium
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- MgMPPE
magnesium-protoporphyrin IX monomethyl ester
- POR
protochlorophyllide oxidoreductase
- SILAC
stable isotope labeling by amino acids
- LC-MS
liquid chromatography-mass spectrometry
- MS/MS
tandem mass spectrometry
Footnotes
This work was supported by the Australian Research Council Centre of Excellence in Plant Energy Biology (grant no. CE140100008) and by the Australian Research Council (Linkage grant no. LP120200102, Future Fellow grant no. FT110100242 to A.H.M., and Superscience Fellow grant no. FS100100022 to R.A.) The University of Western Australia Centre for Metabolomics was supported by infrastructure funding from the Western Australian State Government in partnership with the Australian Federal Government, through the National Collaborative Research Infrastructure Strategy.
The online version of this article contains Web-only data.
Articles can be viewed online without a subscription.
References
- Abdel-Latif S, Kawachi T, Fujikake H, Ohtake N, Ohyama T, Sueyoshi K. (2004) Contribution of shoots and roots to in vivo nitrate reduction in NADII-specific nitrate reductase-deficient mutant seedlings of barley (Hordeum vulgare L.). Soil Sci Plant Nutr 50: 527–535 [Google Scholar]
- Ahrné E, Molzahn L, Glatter T, Schmidt A. (2013) Critical assessment of proteome-wide label-free absolute abundance estimation strategies. Proteomics 13: 2567–2578 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Araújo WL, Tohge T, Ishizaki K, Leaver CJ, Fernie AR. (2011) Protein degradation: an alternative respiratory substrate for stressed plants. Trends Plant Sci 16: 489–498 [DOI] [PubMed] [Google Scholar]
- Armbruster U, Rühle T, Kreller R, Strotbek C, Zühlke J, Tadini L, Blunder T, Hertle AP, Qi Y, Rengstl B, et al. (2013) The photosynthesis affected mutant68-like protein evolved from a PSII assembly factor to mediate assembly of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell 25: 3926–3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro EM, McCaffery S, Anderson JM. (1993) Photoinhibition and D1 protein degradation in peas acclimated to different growth irradiances. Plant Physiol 103: 835–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam M, Huffaker RC. (1982) In vivo nitrate reduction in roots and shoots of barley (Hordeum vulgare L.) seedlings in light and darkness. Plant Physiol 70: 1009–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes D, Mayfield SP. (2003) Redox control of posttranscriptional processes in the chloroplast. Antioxid Redox Signal 5: 89–94 [DOI] [PubMed] [Google Scholar]
- Behrens C, Blume C, Senkler M, Eubel H, Peterhänsel C, Braun HP. (2013) The ‘protein complex proteome’ of chloroplasts in Arabidopsis thaliana. J Proteomics 91: 73–83 [DOI] [PubMed] [Google Scholar]
- Bernlohr RW. (1972) 18Oxygen probes of protein turnover, amino acid transport, and protein synthesis in Bacillus licheniformis. J Biol Chem 247: 4893–4899 [PubMed] [Google Scholar]
- Bloom AJ, Burger M, Rubio Asensio JS, Cousins AB. (2010) Carbon dioxide enrichment inhibits nitrate assimilation in wheat and Arabidopsis. Science 328: 899–903 [DOI] [PubMed] [Google Scholar]
- Bocobza SE, Malitsky S, Araújo WL, Nunes-Nesi A, Meir S, Shapira M, Fernie AR, Aharoni A. (2013) Orchestration of thiamin biosynthesis and central metabolism by combined action of the thiamin pyrophosphate riboswitch and the circadian clock in Arabidopsis. Plant Cell 25: 288–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booij-James IS, Dube SK, Jansen MA, Edelman M, Mattoo AK. (2000) Ultraviolet-B radiation impacts light-mediated turnover of the photosystem II reaction center heterodimer in Arabidopsis mutants altered in phenolic metabolism. Plant Physiol 124: 1275–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britto DT, Kronzucker HJ. (2002) NH4+ toxicity in higher plants: a critical review. J Plant Physiol 159: 567–584 [Google Scholar]
- Cambridge SB, Gnad F, Nguyen C, Bermejo JL, Krüger M, Mann M. (2011) Systems-wide proteomic analysis in mammalian cells reveals conserved, functional protein turnover. J Proteome Res 10: 5275–5284 [DOI] [PubMed] [Google Scholar]
- Carmo-Silva AE, Marri L, Sparla F, Salvucci ME. (2011) Isolation and compositional analysis of a CP12-associated complex of Calvin cycle enzymes from Nicotiana tabacum. Protein Pept Lett 18: 618–624 [DOI] [PubMed] [Google Scholar]
- Chatterjee A, Abeydeera ND, Bale S, Pai PJ, Dorrestein PC, Russell DH, Ealick SE, Begley TP. (2011) Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase. Nature 478: 542–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WP, Yang XY, Harms GL, Gray WM, Hegeman AD, Cohen JD. (2011) An automated growth enclosure for metabolic labeling of Arabidopsis thaliana with 13C-carbon dioxide: an in vivo labeling system for proteomics and metabolomics research. Proteome Sci 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung CY, Williams TC, Poolman MG, Fell DA, Ratcliffe RG, Sweetlove LJ. (2013) A method for accounting for maintenance costs in flux balance analysis improves the prediction of plant cell metabolic phenotypes under stress conditions. Plant J 75: 1050–1061 [DOI] [PubMed] [Google Scholar]
- Christopher DA, Mullet JE. (1994) Separate photosensory pathways co-regulate blue light/ultraviolet-A-activated psbD-psbC transcription and light-induced D2 and CP43 degradation in barley (Hordeum vulgare) chloroplasts. Plant Physiol 104: 1119–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Bosco C, Busconi M, Govoni C, Baldi P, Stanca AM, Crosatti C, Bassi R, Cattivelli L. (2003) cor gene expression in barley mutants affected in chloroplast development and photosynthetic electron transport. Plant Physiol 131: 793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon WJ. (1950) Analysis of extreme values. Ann Math Stat 21: 488–506 [Google Scholar]
- Gojon A, Soussana JF, Passama L, Robin P. (1986) Nitrate reduction in roots and shoots of barley (Hordeum vulgare L.) and corn (Zea mays L.) seedlings. I. N study. Plant Physiol 82: 254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontero B, Maberly SC. (2012) An intrinsically disordered protein, CP12: jack of all trades and master of the Calvin cycle. Biochem Soc Trans 40: 995–999 [DOI] [PubMed] [Google Scholar]
- Gruhler A, Schulze WX, Matthiesen R, Mann M, Jensen ON. (2005) Stable isotope labeling of Arabidopsis thaliana cells and quantitative proteomics by mass spectrometry. Mol Cell Proteomics 4: 1697–1709 [DOI] [PubMed] [Google Scholar]
- Hachiya T, Terashima I, Noguchi K. (2007) Increase in respiratory cost at high growth temperature is attributed to high protein turnover cost in Petunia × hybrida petals. Plant Cell Environ 30: 1269–1283 [DOI] [PubMed] [Google Scholar]
- Hamasur B, Glaser E. (1992) Plant mitochondrial F0F1 ATP synthase: identification of the individual subunits and properties of the purified spinach leaf mitochondrial ATP synthase. Eur J Biochem 205: 409–416 [DOI] [PubMed] [Google Scholar]
- Heupel R, Markgraf T, Robinson DG, Heldt HW. (1991) Compartmentation studies on spinach leaf peroxisomes: evidence for channeling of photorespiratory metabolites in peroxisomes devoid of intact boundary membrane. Plant Physiol 96: 971–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinkson IV, Elias JE. (2011) The dynamic state of protein turnover: it’s about time. Trends Cell Biol 21: 293–303 [DOI] [PubMed] [Google Scholar]
- Holleman JM, Key JL. (1967) Inactive and protein precursor pools of amino acids in the soybean hypocotyl. Plant Physiol 42: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell KA, Narsai R, Carroll A, Ivanova A, Lohse M, Usadel B, Millar AH, Whelan J. (2009) Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol 149: 961–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SC, Hall TC, Edwards GE. (1976) Differential localization of fraction I protein between chloroplast types. Plant Physiol 57: 730–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen MA, Mattoo AK, Edelman M. (1999) D1-D2 protein degradation in the chloroplast: complex light saturation kinetics. Eur J Biochem 260: 527–532 [DOI] [PubMed] [Google Scholar]
- Janska H, Kwasniak M, Szczepanowska J. (2013) Protein quality control in organelles: AAA/FtsH story. Biochim Biophys Acta 1833: 381–387 [DOI] [PubMed] [Google Scholar]
- Kato Y, Sakamoto W. (2009) Protein quality control in chloroplasts: a current model of D1 protein degradation in the photosystem II repair cycle. J Biochem 146: 463–469 [DOI] [PubMed] [Google Scholar]
- Keller A, Eng J, Zhang N, Li XJ, Aebersold R. (2005) A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol 1: 0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keys AJ, Bird IF, Cornelius MJ, Lea PJ, Wallsgrove RM, Miflin BJ. (1978) Photorespiratory nitrogen cycle. Nature 275: 741–743 [Google Scholar]
- Kim J, Mayfield SP. (1997) Protein disulfide isomerase as a regulator of chloroplast translational activation. Science 278: 1954–1957 [DOI] [PubMed] [Google Scholar]
- Kindgren P, Norén L, López JdeD, Shaikhali J, Strand A. (2012) Interplay between Heat Shock Protein 90 and HY5 controls PhANG expression in response to the GUN5 plastid signal. Mol Plant 5: 901–913 [DOI] [PubMed] [Google Scholar]
- Kushner DJ, Baker A, Dunstall TG. (1999) Pharmacological uses and perspectives of heavy water and deuterated compounds. Can J Physiol Pharmacol 77: 79–88 [PubMed] [Google Scholar]
- Lam YW, Lamond AI, Mann M, Andersen JS. (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol 17: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegood RC, Lea PJ, Häusler RE. (1996) Use of barley mutants to study the control of photorespiratory metabolism. Biochem Soc Trans 24: 757–761 [DOI] [PubMed] [Google Scholar]
- Legnaioli T, Cuevas J, Mas P. (2009) TOC1 functions as a molecular switch connecting the circadian clock with plant responses to drought. EMBO J 28: 3745–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowska D, ten Have S, Hodge K, Tillemans V, Lamond AI, Brown JW. (2013) Plant SILAC: stable-isotope labelling with amino acids of Arabidopsis seedlings for quantitative proteomics. PLoS ONE 8: e72207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Carrie C, Nelson C, Whelan J, Millar AH. (2012a) Accumulation of newly synthesized F1 in vivo in Arabidopsis mitochondria provides evidence for modular assembly of the plant F1Fo ATP synthase. J Biol Chem 287: 25749–25757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Nelson CJ, Carrie C, Gawryluk RM, Solheim C, Gray MW, Whelan J, Millar AH. (2013) Subcomplexes of ancestral respiratory complex I subunits rapidly turn over in vivo as productive assembly intermediates in Arabidopsis. J Biol Chem 288: 5707–5717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Nelson CJ, Solheim C, Whelan J, Millar AH. (2012b) Determining degradation and synthesis rates of Arabidopsis proteins using the kinetics of progressive 15N labeling of two-dimensional gel-separated protein spots. Mol Cell Proteomics 11: 010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Q, Huang W, Baldwin A, Jarvis P. (2012) Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338: 655–659 [DOI] [PubMed] [Google Scholar]
- Maliga P, Bock R. (2011) Plastid biotechnology: food, fuel, and medicine for the 21st century. Plant Physiol 155: 1501–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marutani Y, Yamauchi Y, Kimura Y, Mizutani M, Sugimoto Y. (2012) Damage to photosystem II due to heat stress without light-driven electron flow: involvement of enhanced introduction of reducing power into thylakoid membranes. Planta 236: 753–761 [DOI] [PubMed] [Google Scholar]
- Masclaux C, Valadier MH, Brugière N, Morot-Gaudry JF, Hirel B. (2000) Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211: 510–518 [DOI] [PubMed] [Google Scholar]
- Mayer KF, Waugh R, Brown JW, Schulman A, Langridge P, Platzer M, Fincher GB, Muehlbauer GJ, Sato K, Close TJ, et al. (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716 [DOI] [PubMed] [Google Scholar]
- McWatters HG, Devlin PF. (2011) Timing in plants: a rhythmic arrangement. FEBS Lett 585: 1474–1484 [DOI] [PubMed] [Google Scholar]
- Mittal S, Kumari N, Sharma V. (2012) Differential response of salt stress on Brassica juncea: photosynthetic performance, pigment, proline, D1 and antioxidant enzymes. Plant Physiol Biochem 54: 17–26 [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. (2001) Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc Natl Acad Sci USA 98: 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, James RA. (2003) Screening methods for salinity tolerance: a case study with tetraploid wheat. Plant Soil 253: 201–218 [Google Scholar]
- Nelson CJ, Li L, Jacoby RP, Millar AH. (2013) Degradation rate of mitochondrial proteins in Arabidopsis thaliana cells. J Proteome Res 12: 3449–3459 [DOI] [PubMed] [Google Scholar]
- Nevo R, Charuvi D, Tsabari O, Reich Z. (2012) Composition, architecture and dynamics of the photosynthetic apparatus in higher plants. Plant J 70: 157–176 [DOI] [PubMed] [Google Scholar]
- Pal SK, Liput M, Piques M, Ishihara H, Obata T, Martins MC, Sulpice R, van Dongen JT, Fernie AR, Yadav UP, et al. (2013) Diurnal changes of polysome loading track sucrose content in the rosette of wild-type Arabidopsis and the starchless pgm mutant. Plant Physiol 162: 1246–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papini-Terzi FS, Galhardo RS, Farias LP, Menck CF, Van Sluys MA. (2003) Point mutation is responsible for Arabidopsis tz-201 mutant phenotype affecting thiamin biosynthesis. Plant Cell Physiol 44: 856–860 [DOI] [PubMed] [Google Scholar]
- Penning De Vries FWT. (1975) Cost of maintenance processes in plant cells. Ann Bot (Lond) 39: 77–92 [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Poolman MG, Kundu S, Shaw R, Fell DA. (2013) Responses to light intensity in a genome-scale model of rice metabolism. Plant Physiol 162: 1060–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt JM, Robertson DH, Gaskell SJ, Riba-Garcia I, Hubbard SJ, Sidhu K, Oliver SG, Butler P, Hayes A, Petty J, et al. (2002) Stable isotope labelling in vivo as an aid to protein identification in peptide mass fingerprinting. Proteomics 2: 157–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JC, Guan S, Burlingame A, Prusiner SB, Ghaemmaghami S. (2010) Analysis of proteome dynamics in the mouse brain. Proc Natl Acad Sci USA 107: 14508–14513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachmilevitch S, Cousins AB, Bloom AJ. (2004) Nitrate assimilation in plant shoots depends on photorespiration. Proc Natl Acad Sci USA 101: 11506–11510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A, Peter E, Pörs Y, Lorenzen S, Grimm B, Czarnecki O. (2010) Rapid dark repression of 5-aminolevulinic acid synthesis in green barley leaves. Plant Cell Physiol 51: 670–681 [DOI] [PubMed] [Google Scholar]
- Rolland N, Curien G, Finazzi G, Kuntz M, Maréchal E, Matringe M, Ravanel S, Seigneurin-Berny D. (2012) The biosynthetic capacities of the plastids and integration between cytoplasmic and chloroplast processes. Annu Rev Genet 46: 233–264 [DOI] [PubMed] [Google Scholar]
- Rollins JA, Habte E, Templer SE, Colby T, Schmidt J, von Korff M. (2013) Leaf proteome alterations in the context of physiological and morphological responses to drought and heat stress in barley (Hordeum vulgare L.). J Exp Bot 64: 3201–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorabacher DB. (1991) Statistical treatment for rejection of deviant values: critical values of Dixon’s “Q” parameter and related subrange ratios at the 95% confidence level. Anal Chem 63: 139–146 [Google Scholar]
- Rzeznicka K, Walker CJ, Westergren T, Kannangara CG, von Wettstein D, Merchant S, Gough SP, Hansson M. (2005) Xantha-l encodes a membrane subunit of the aerobic Mg-protoporphyrin IX monomethyl ester cyclase involved in chlorophyll biosynthesis. Proc Natl Acad Sci USA 102: 5886–5891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte D, Close TJ, Graner A, Langridge P, Matsumoto T, Muehlbauer G, Sato K, Schulman AH, Waugh R, Wise RP, et al. (2009) The International Barley Sequencing Consortium: at the threshold of efficient access to the barley genome. Plant Physiol 149: 142–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473: 337–342 [DOI] [PubMed] [Google Scholar]
- Schweikl H, Klein U, Schindlbeck M, Wieczorek H. (1989) A vacuolar-type ATPase, partially purified from potassium transporting plasma membranes of tobacco hornworm midgut. J Biol Chem 264: 11136–11142 [PubMed] [Google Scholar]
- Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics 5: 144–156 [DOI] [PubMed] [Google Scholar]
- Skiba MW, George TS, Baggs EM, Daniell TJ. (2011) Plant influence on nitrification. Biochem Soc Trans 39: 275–278 [DOI] [PubMed] [Google Scholar]
- Sunamura E, Konno H, Imashimizu M, Mochimaru M, Hisabori T. (2012) A conformational change of the γ subunit indirectly regulates the activity of cyanobacterial F1-ATPase. J Biol Chem 287: 38695–38704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suss KH, Arkona C, Manteuffel R, Adler K. (1993) Calvin cycle multienzyme complexes are bound to chloroplast thylakoid membranes of higher plants in situ. Proc Natl Acad Sci USA 90: 5514–5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szecowka M, Heise R, Tohge T, Nunes-Nesi A, Vosloh D, Huege J, Feil R, Lunn J, Nikoloski Z, Stitt M, et al. (2013) Metabolic fluxes in an illuminated Arabidopsis rosette. Plant Cell 25: 694–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Bhat S, Petracek ME. (2003) Light control of nuclear gene mRNA abundance and translation in tobacco. Plant Physiol 133: 1979–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanz SK, Castleden I, Hooper CM, Vacher M, Small I, Millar HA. (2013) SUBA3: a database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Res 41: D1185–D1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele B, Füllner K, Stein N, Oldiges M, Kuhn AJ, Hofmann D. (2008) Analysis of amino acids without derivatization in barley extracts by LC-MS-MS. Anal Bioanal Chem 391: 2663–2672 [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37: 914–939 [DOI] [PubMed] [Google Scholar]
- Tohge T, Ramos MS, Nunes-Nesi A, Mutwil M, Giavalisco P, Steinhauser D, Schellenberg M, Willmitzer L, Persson S, Martinoia E, et al. (2011) Toward the storage metabolome: profiling the barley vacuole. Plant Physiol 157: 1469–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trötschel C, Albaum SP, Wolff D, Schröder S, Goesmann A, Nattkemper TW, Poetsch A. (2012) Protein turnover quantification in a multilabeling approach: from data calculation to evaluation. Mol Cell Proteomics 11: 512–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschoep H, Gibon Y, Carillo P, Armengaud P, Szecowka M, Nunes-Nesi A, Fernie AR, Koehl K, Stitt M. (2009) Adjustment of growth and central metabolism to a mild but sustained nitrogen-limitation in Arabidopsis. Plant Cell Environ 32: 300–318 [DOI] [PubMed] [Google Scholar]
- Tyystjärvi E, Mäenpää P, Aro EM. (1994) Mathematical modelling of photoinhibition and photosystem II repair cycle. I. Photoinhibition and D1 protein degradation in vitro and in the absence of chloroplast protein synthesis in vivo. Photosynth Res 41: 439–449 [DOI] [PubMed] [Google Scholar]
- van Wijk KJ, Baginsky S. (2011) Plastid proteomics in higher plants: current state and future goals. Plant Physiol 155: 1578–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wedel N, Soll J, Paap BK. (1997) CP12 provides a new mode of light regulation of Calvin cycle activity in higher plants. Proc Natl Acad Sci USA 94: 10479–10484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D, Flügge UI. (1984) A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Whitelegge JP, Katz JE, Pihakari KA, Hale R, Aguilera R, Gómez SM, Faull KF, Vavilin D, Vermaas W. (2004) Subtle modification of isotope ratio proteomics: an integrated strategy for expression proteomics. Phytochemistry 65: 1507–1515 [DOI] [PubMed] [Google Scholar]
- Wilson MI, Greenberg BM. (1993) Protection of the D1 photosystem II reaction center protein from degradation in ultraviolet radiation following adaptation of Brassica napus L to growth in ultraviolet B. Photochem Photobiol 57: 556–563 [Google Scholar]
- Winter H, Robinson DG, Heldt HW. (1993) Subcellular volumes and metabolite concentrations in barley leaves. Planta 191: 180–190 [Google Scholar]
- Yang XY, Chen WP, Rendahl AK, Hegeman AD, Gray WM, Cohen JD. (2010) Measuring the turnover rates of Arabidopsis proteins using deuterium oxide: an auxin signaling case study. Plant J 63: 680–695 [DOI] [PubMed] [Google Scholar]
- Yao DC, Brune DC, Vermaas WF. (2012) Lifetimes of photosystem I and II proteins in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett 586: 169–173 [DOI] [PubMed] [Google Scholar]
- Yoshida M, Muneyuki E, Hisabori T. (2001) ATP synthase: a marvellous rotary engine of the cell. Nat Rev Mol Cell Biol 2: 669–677 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Reckow S, Webhofer C, Boehme M, Gormanns P, Egge-Jacobsen WM, Turck CW. (2011) Proteome scale turnover analysis in live animals using stable isotope metabolic labeling. Anal Chem 83: 1665–1672 [DOI] [PubMed] [Google Scholar]