Abstract
Background
The multi-center cluster-randomized Strategies to Reduce Transmission of Antimicrobial Resistant Bacteria in Intensive Care Units (STAR*ICU) trial was carried out in 18 U.S. adult intensive care units (ICUs) and evaluated the effectiveness of infection control strategies in reducing transmission of methicillin-resistant Staphylococcus aureus (MRSA) colonization and/or infection. Our study objective was to examine the molecular epidemiology of MRSA and assess the prevalence and risk factors for community acquired (CA)-MRSA genotype nasal carriage at the time of ICU admission.
Methods
Selected MRSA isolates were subjected to molecular typing using pulsed-field gel electrophoresis.
Results
Among 5,512 ICU patient-admissions in the STAR*ICU trial during the intervention period, 626 (11%) had a positive nares culture for MRSA. 210/626 (34%) available isolates were selected by weighted random sampling for molecular typing. Of 210 patients, 123 (59%) were male; mean age was 63 years. Molecular typing revealed that 147 isolates (70%) were the USA100 clone; 26 (12%) USA300; 12 (6%) USA500; 8 (4%) USA800; 17 (8%) other. In multivariate analysis, patients with CA-MRSA genotype (USA300, USA400, or USA1000) colonization were less likely to have been hospitalized during the previous 12 months (PR=0.39; 95% C.I. 0.21–0.73) and less likely to have an older age (PR=0.97 per year; 0.95–0.98) compared to patients with a HA-MRSA genotype.
Conclusion
CA-MRSA genotypes have emerged as a cause of MRSA nares colonization among patients admitted to adult ICUs in the U.S. During the study period (2006), the predominant site of CA-MRSA genotype acquisition appeared to be in the community.
Keywords: MRSA, community-associated, healthcare-associated, ICU
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is widespread in hospitals, especially intensive care units [1] where MRSA accounts for >60% of S. aureus isolates in the U.S. hospital ICUs [2]. Risk factors for healthcare-associated MRSA (HA-MRSA) infections include recent surgery or hospitalization, prior antibiotic use, residence in a long-term care facility, dialysis, and the presence of or exposure to indwelling percutaneous catheters and other medical devices [3]. The presence of methicillin resistance results in greater lengths of stay, higher mortality [4], and increased costs [5, 6] compared to methicillin-susceptible S. aureus infections. However, MRSA is no longer only a nosocomial pathogen. Over the past decade, MRSA has emerged as an important cause of community-associated (CA) infections, particularly skin and soft tissue infections [7–9] among a variety of risk groups, including sports teams [10, 11], military recruits, correctional facilities [12], men who have sex with men and HIV-infected persons [13], children [14], Pacific Islanders, Alaskan Natives, Native Americans, as well as among healthy persons not in these identified risk groups [15].
Several different typing molecular methods have been used to study MRSA. Pulsed-field gel electrophoresis (PFGE) is typing method with high discriminatory power [16]. McDougal et al have defined 8 distinct clusters of MRSA genotypes through PFGE typing (USA100 through USA800) [17]. Of these, USA300 and USA400 were considered to be community-associated MRSA [17]. Recent studies have reported the presence of additional pulsed-field types [8, 18–20], including USA1000 and USA1100, which are also considered to be a community-associated strains of MRSA [8, 18].
The Strategies to Reduce Transmission of Antimicrobial Resistant Bacteria in Intensive Care Units (STAR*ICU) trial was a cluster randomized study that investigated whether a package of infection control measures were effective in reducing nosocomial transmission of MRSA [21]. The study was carried out at 18 geographically dispersed adult ICUs in the U.S. and provided a unique opportunity to study the molecular epidemiology of MRSA among patients admitted to these ICUs. The purpose of our study was to examine the molecular epidemiology of MRSA and assess the prevalence and risk factors for CA-MRSA genotype carriage among patients admitted to adult ICUs at the 18 study hospitals in the U.S. during the intervention period (March through August 2006) of the STAR*ICU trial.
Methods
Study design
The STAR*ICU study was a multi-center cluster-randomized trial conducted in 18 medical, surgical and medical/surgical ICUs in academic medical centers representing all regions of the country [21]. The trial was designed to evaluate the effectiveness of active culture-based surveillance for MRSA and vancomycin-resistant Enterococcus (VRE) and expanded use of barrier precautions on transmission of MRSA and VRE colonization/infection in the study ICUs [21]. Patients admitted into study ICUs with an expected length of stay of 3 days or longer had a nares surveillance culture performed for MRSA. Patients at institutions randomized to the control arm had a nares surveillance culture performed on admission as well but the “control” sites did not receive the results of the admission surveillance culture.
Laboratory Methods
All cultures were processed at a single central laboratory (National Institutes of Health [NIH] Clinical Microbiology Laboratory). Nasal swabs were inoculated on to Mueller Hinton broth with 7% NaCl and 2µg/mL oxacillin and incubated at 35°C for 18–24 hours. Broth was used to inoculate mannitol salt agar plates supplemented with 4µg/mL oxacillin, which were incubated at 35°C and inspected at 18–24 hours and 42–48 hours of incubation. Isolates of S. aureus were tested for the mecA gene using the LightCycler MRSA detection test (Roche Applied Science, Indianapolis, IN).
Molecular Typing
A weighted random sample with a minimum of 7 MRSA isolates per each of 18 sites was selected for genotypic analysis from the pool of 626 MRSA isolates recovered from the nares cultures collected at the time of ICU admission (a total of 252 isolates). Isolates were stored in a Microbank (Pro Lab Diagnostics) system of beads in cryovials containing cryopreservative. After inoculation, the cryovials were stored at −70°C until removed to directly inoculate plate cultures. Molecular typing studies were performed on the available 210 (83%) MRSA isolates from the 252 randomly selected positive nares cultures using pulsed-field gel electrophoresis (PFGE) after restriction with SmaI (Roche Molecular Biochemicals) as previously described [22]. Digital photographs of the gels were saved as TIFF files for analysis with Bionumerics Software (Applied Maths, Austin, TX). Cluster analysis to determine strain relatedness was performed using unweighted pair-group methodology based on Dice coefficients where clusters were defined by a coefficient of similarity of >85%, allowing for either assignment of a pulsed-field type to one of the known MRSA USA types contained in the national CDC database [17] or identification of non-USA [19,20] or variant pulsed-field types.
Data analysis
Data analysis was performed using SAS software, version 9.1 (SAS Institute, Cary, NC). CA-MRSA genotype colonization was defined by a CA-MRSA pulsed-field type (i.e., USA300, USA400 or USA1000 genotypes). HA-MRSA colonization was defined by a HA-MRSA pulsed-field type (i.e., USA100, USA200, USA500, USA600, USA700, USA800 [17], eMRSA-15 [19], Brazilian [20], or any other non-CA MRSA genotypes). Potential risk factors were first assessed by univariate analysis. Prevalence ratios (PR) and the corresponding 95% confidence intervals (CI) were calculated. Multivariable log-binomial regression models included variables significantly associated with CA-MRSA genotype nasal colonization in univariate analysis, and based on biologic plausibility and epidemiological factors clinically felt to be associated with community-acquired MRSA. Variables considered for multivariable model included age, history of hospitalization in the past year, and documented history of MRSA or VRE colonization. Stepwise selection was used to arrive to the final model. A p-value of ≤.05 was defined as statistically significant.
Results
During the 6-month intervention period of the trial, there were a total of 5,512 ICU patient-admissions (5,133 unique patients); 626 (11.4%) of these patient-admissions had a positive nares culture for MRSA at the time of ICU admission (prevalent MRSA colonization); 252 (40.3%) of 626 isolates were selected for typing. A total of 210 (33.5%) of 626 isolates (or 83.3% from 252 selected for typing isolates) from a random weighted sample of these positive cultures were available for molecular typing (mean 12±3 isolates per site, range 7–17 isolates). Patients whose MRSA isolates were selected for molecular typing (n=252) were similar to those patients whose isolates were not selected (n=374) with the exception that selected patients more commonly had central venous catheter (55/252 [21.8%] vs. 54/374 [14.4%], p=.01), and had a history of solid organ transplantation (15/252 [6.0%] vs. 7/374 [1.9%], p=.006). Patients whose MRSA isolates were selected and available for molecular typing (n=210) were similar to those patients whose isolates were selected and not available for molecular typing (n=42) with the exception that the first group less commonly had black race (140/210 [66.7%] vs. 35/42 [83.3%], p=.03), and documented MRSA or VRE history (49/210 [23.3%] vs. 17/42 [40.5%], p=.03). Among the 210 patients from whom these MRSA isolates were recovered, 123 (59%) were male; 150 (72%) were Caucasian, 49 (23%) Black or African-American, and 11 (5%) had other race/ethnicity. The mean age of these 210 patients was 63 years. The mean length of stay in the ICU was 7.6 days (median – 4 days; range 0.5–74 days), and the mean time between hospital admission and admission to the ICU was 3.6 days (median – 0 days, range 0–70 days).
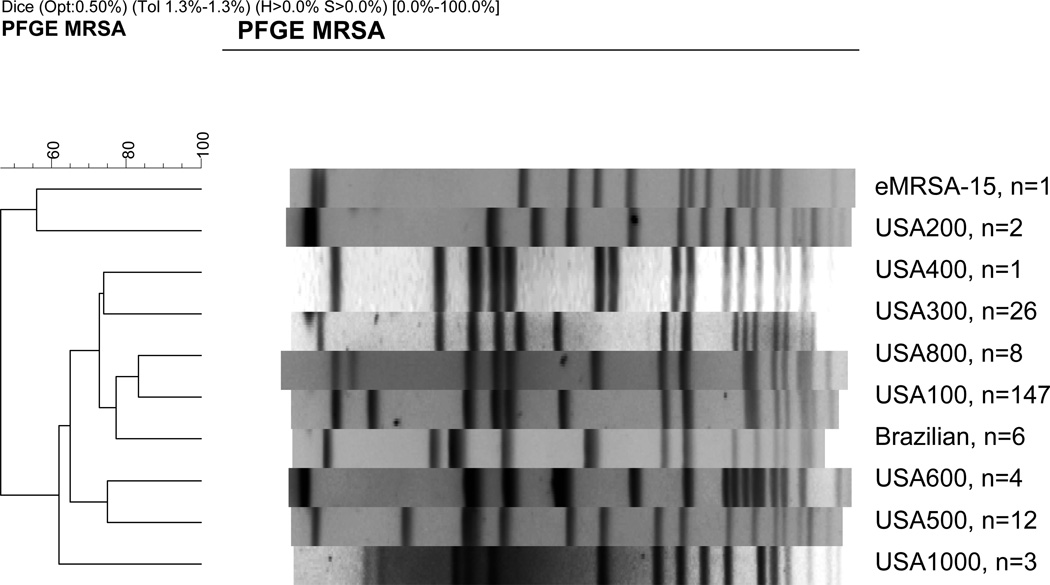
Molecular typing of the 210 MRSA isolates using pulsed-field gel electrophoresis (PFGE) revealed that 147 (70%) were the USA100 clone; 26 - USA300 (12%); 12 - USA500 (6%); 8 - USA800 (4%); 6 - Brazilian clone (3%); 4 - USA600 (2%); 3 - USA1000 (1%); 2 - USA200 (1%); 1 - USA400 (0.5%); and 1 was eMRSA-15 (0.5%). Overall, 30 (14%) patients had CA-MRSA genotype (USA300, USA400, or USA1000) colonization. Two non-USA type strains were present: the Brazilian strain and an eMRSA15 strain [19,20]. Representative PFGE types of MRSA strains are shown in Figure 1. The geographic distribution of pulsed-field types of MRSA isolates is shown in Table 1. All 6 Brazilian strains were seen in one hospital in Midwest, and the only eMRSA15 strain was seen in a hospital in East. There were no significant differences in frequency of CA-MRSA genotypes versus HA-MRSA genotypes by region.
Figure 1. Pulsed field gel electrophoresis (PFGE) of Methicillin-Resistant Staphylococcus aureus (MRSA) strains.
Representative pulsed-field types are shown. The number of isolates with each particular pulsed-field type is shown on the right of the figure. The coefficient of similarity is shown on the left of the figure.
Table 1.
Geographic distribution of pulsed-field types of Methicillin-Resistant Staphylococcus aureus (MRSA) strains.
| MRSA pulsed- field type |
East N=68 n (%) |
Midwest N=74 n (%) |
South N=42 n (%) |
West N=26 n (%) |
Total N=210 n (%) |
|---|---|---|---|---|---|
| USA100 | 48 | 53 | 25 | 21 | 147 |
| 70.6 | 71.6 | 59.5 | 80.8 | 70.0 | |
| USA1000 | 1 | 1 | 1 | 0 | 3 |
| 1.5 | 1.4 | 2.4 | 0.0 | 1.0 | |
| USA200 | 1 | 0 | 0 | 1 | 2 |
| 1.47 | 0 | 0 | 3.9 | 1.0 | |
| USA300 | 7 | 7 | 8 | 4 | 26 |
| 10.3 | 9.5 | 19.1 | 15.4 | 12.4 | |
| USA400 | 0 | 0 | 1 | 0 | 1 |
| 0.0 | 0.0 | 2.4 | 0.0 | 0.5 | |
| USA500 | 6 | 1 | 5 | 0 | 12 |
| 8.8 | 1.4 | 11.9 | 0.0 | 5.7 | |
| USA600 | 1 | 3 | 0 | 0 | 4 |
| 1.5 | 4.1 | 0.0 | 0.0 | 1.9 | |
| USA800 | 3 | 3 | 2 | 0 | 8 |
| 4.4 | 4.1 | 4.8 | 0.0 | 3.8 | |
| Brazil | 0 | 6 | 0 | 0 | 6 |
| 0.0 | 8.1 | 0.0 | 0.0 | 2.9 | |
| eMRSA15 | 1 | 0 | 0 | 0 | 1 |
| 1.5 | 0.0 | 0.0 | 0.0 | 0.5 | |
Note.
East: Connecticut, Maryland, Massachusetts, New Jersey, New York;
Midwest: Illinois, Iowa, Michigan, Minnesota, Ohio;
South: Alabama, Florida, Georgia;
West: Arizona.
In univariate analysis, CA-MRSA genotype colonization (vs. HA-MRSA) was more common among those with Black race (PR =2.16, 95% C.I. 1.11–4.20); and less common among those with a documented prior history of MRSA or VRE colonization (PR=0.16, 95% C.I. 0.11–0.85), hospitalization during past 12 months (PR=0.37, 95% C.I. 0.19–0.70), and older patients (PR=0.96 per year, 95% C.I. 0.95–0.98) (Table 2). In multivariate analysis, patients with CA-MRSA genotype colonization were less likely to have been hospitalized during past 12 months (PR=0.39, 95% C.I. 0.21–0.73) and less likely to have an older age (PR=0.97 per year, 95% C.I. 0.95–0.98) compared to patients with HA-MRSA genotype colonization (Table 3).
Table 2.
Univariate analysis of the risk factors associated with community-associated (CA)-MRSA genotype nasal colonization in the STAR*ICU trial
| Risk factors | CA-MRSA N=30, n (%) |
HA-MRSA N=180, n (%) |
PR | 95% C.I. | P Value |
|---|---|---|---|---|---|
| Demographic data | |||||
| Age mean; median (range), years | 51; 48 (21–97) | 65; 66 (21–92) | 0.96 | 0.95–0.98 | <.001 |
| Female gender | 12 (40.0) | 75 (41.7) | 0.94 | 0.48–1.85 | .86 |
| Race | |||||
| White | 17 (56.7) | 133 (73.9) | 1.00 | ||
| Black | 12 (40.0) | 37 (20.5) | 2.16 | 1.11–4.20 | .02 |
| Other | 1 (3.3) | 10 (5.6) | 0.80 | 0.11–5.48 | .82 |
| Health care–associated risk factors | |||||
| Hospitalized in past year | 14 (46.7) | 134 (74.4) | 0.37 | 0.19–0.70 | .003 |
| Surgery in past year | 7 (23.3) | 70 (38.9) | 0.53 | 0.24–1.17 | .11 |
| Documented history of | |||||
| MRSA colonization | 4 (13.3) | 55 (30.6) | 0.39 | 0.14–1.08 | .07 |
| VRE colonization | 1 (3.3) | 24 (13.3) | 0.26 | 0.04–1.79 | .17 |
| MRSA or VRE colonization | 4 (13.3) | 66 (36.7) | 0.31 | 0.11–0.85 | .03 |
| Length of stay in hospital before ICU admission, mean, median (range), days | 4.2; 0 (0–70) | 3.5; 0 (0–49) | 1.01 | 0.97–1.05 | .66 |
| Device in place at ICU Admission | |||||
| Indwelling Device at ICU Admission | 15 (50.0) | 113 (62.8) | 0.64 | 0.33–1.24 | .19 |
| Gastrostomy Tube | 0 (0) | 15 (8.3) | undefined | undefined | 1.00 |
| Tracheostomy Tube | 3 (10.0) | 10 (5.6) | 1.68 | 0.59–4.82 | .33 |
| Indwelling Urinary Catheter | 12 (40.0) | 94 (52.2) | 0.65 | 0.33–1.29 | .22 |
| Central Venous Catheter | 5 (16.7) | 43 (23.9) | 0.68 | 0.27–1.67 | .39 |
| PICC | 1 (3.3) | 12 (6.7) | 0.52 | 0.08–3.54 | .51 |
| Medical history | |||||
| History of Solid Tumor | 4 (13.3) | 32 (17.8) | 0.74 | 0.28–2.00 | .56 |
| History of Hematologic Malignancy | 1 (3.3) | 6 (3.3) | 1.00 | 0.16–6.33 | 1.00 |
| History of Hematopoietic Stem Cell or Bone Marrow Transplant | 1 (3.3) | 2 (1.1) | 2.38 | 0.46–12.21 | .30 |
| History of Solid Organ Transplant | 2 (6.7) | 12 (6.7) | 1.00 | 0.27–3.77 | 1.00 |
| History of Chronic Hemodialysis | 1 (3.3) | 25 (13.9) | 0.24 | 0.03–1.72 | .16 |
| History of Chronic Hepatic Failure | 1 (3.3) | 16 (8.9) | 0.39 | 0.06–2.70 | .34 |
MRSA=methicillin-resistant Staphylococcus aureus
VRE=vancomycin-resistant Enterococcus
ICU=Intensive Care Unit
HA=healthcare-associated; CA=community-associated
PR=Prevalence Ratio
CI=Confidence Interval
PICC=peripherally inserted central catheter
Table 3.
Multivariate analysis of the risk factors associated with community-associated MRSA nasal colonization in the STAR*ICU trial
| Risk factor | PR | 95% C.I. | P Value |
|---|---|---|---|
| Age (per additional year) | 0.97 | 0.95–0.98 | <0.001 |
| Hospitalized in past 1 year | 0.39 | 0.21–0.73 | 0.003 |
MRSA=methicillin-resistant Staphylococcus aureus
PR=Prevalence Ratio
CI=Confidence Interval
Among the 210 patients with MRSA nasal colonization on ICU admission, 22 (10.5%) developed MRSA blood stream infection. There was no significant difference in rates of MRSA BSI among those with nasal colonization with CA-MRSA genotype compared to those with HA-MRSA colonization (3 [10.0%] of 30, and 19 [10.6%] of 180, respectively; PR=0.95, 95%CI 0.30–3.01; p=.93).
Overall mortality was 20% among those patients with MRSA colonization on ICU admission whose isolates were subjected to molecular typing (42 of 210 patients died); 27 (64.3%) of 42 died in an ICU, and 15 (35.7%) of 42 died in a non-ICU hospital setting.
Additional analyses were carried out to compare all patients with prevalent nasal MRSA colonization (i.e., positive nares culture for MRSA on ICU admission) to those patients without nasal MRSA colonization on ICU admission for all 5,133 patients admitted during the intervention period. Patients with prevalent nasal MRSA colonization were more likely to develop MRSA blood stream infection compared to those patients without nasal MRSA colonization on ICU admission (44 [7.4%] of 599 vs. 59 [1.3%] of 4,534 patients, PR=5.64, 95% CI 3.86–8.26, p<.001). There was no significant difference in mortality rates among those with prevalent nasal MRSA colonization and those with a negative nares culture at the time of ICU admission (120 [20.0%] of 599 vs. 851 [18.8%] of 4,534 patients, respectively, PR=1.07, 95% CI 0.90–1.27, p=0.46).
Discussion
MRSA has traditionally been a nosocomial pathogen. However, over the past decade MRSA has emerged as an important cause of community-onset infections, particularly those associated with skin and soft tissue [7–13, 23]. In our multicenter study which had a broad geographic distribution of sites in the U.S., we found that 14% of patients admitted to adult ICUs with MRSA nares colonization had strains that belonged to CA-MRSA genotypes (USA 300, USA 400, USA 1000). In multivariate analysis, patients with CA-MRSA genotype colonization were significantly more likely to be younger, and less likely to have a history of hospitalization during past 12 months compared to patients with HA-MRSA genotype colonization. These findings suggest that during the study period (2006), the community remained the predominant site where CA-MRSA genotype acquisition was taking place despite recent reports of nosocomial infections due to CA-MRSA USA300 (however, this might change over time and should continue to be studied). This is particularly of interest given that the lines between CA-MRSA and HA-MRSA have begun to blur. Seybold et al. [22] in a study carried out at Grady Memorial Hospital in Atlanta reported that the CA-MRSA USA300 genotype has emerged as a major cause of healthcare-associated MRSA bloodstream infections. Additional reports have also noted that CA-MRSA genotype USA300 has been introduced to healthcare settings and is causing nosocomial MRSA infections [24–27].
In our study, there was no significant difference in rates of MRSA BSI among those with nasal colonization with CA-MRSA genotypes compared to those with HA-MRSA genotype colonization on admission to the ICU. Overall crude in-hospital mortality was high among those patients included in the study (about 20%) and did not significantly differ among patients with MRSA colonization on admission compared to those without MRSA nares colonization.
The overall prevalence of MRSA colonization among patients admitted to 18 ICUs was approximately 11%. We found that that patients in the STAR*ICU study with MRSA colonization on ICU admission during the intervention period were significantly more likely to have a bacteremia due to MRSA compared to patients without nasal MRSA colonization at the time of ICU admission (PR=5.64, 95% CI 3.86–8.26). Our findings are consistent with previous reports which have noted that ICU patients (both adults and neonates) with MRSA nasal carriage are much more likely to develop a MRSA bacteremia compared to patients who lack nares colonization [28, 29]. Further data are needed to assess whether there is any benefit of nares or other decolonization strategies to try to reduce invasive MRSA infections among colonized patients in the ICU setting; this efficacy is needed as the number need to treat to prevent a single invasive infection is substantial (for example 12:1 based on study data assuming 100% efficacy) and the excessive use of mupirocin for nasal MRSA decolonization leads to development of mupirocin resistance. An alternative strategy which has demonstrated efficacy in reducing primary bloodstream infections in ICU patients, perhaps through decolonization or reduction in MRSA bacterial skin burden, is daily chlorhexidine baths [30].
The nomenclature surrounding MRSA molecular typing can be confusing given multiple different typing methods. In our study, we employed pulsed-field gel electrophoresis and characterized strain patterns using the USA typing system published by McDougal et al [17]. Most CA-MRSA genotypes in our study had the USA300 pulsed-field type. This is not surprising given that MRSA USA300 has also been reported to cause the overwhelming majority of community-acquired MRSA infections in the U.S. [7, 9, 31]. Overall in our study, HA-MRSA genotypes predominated with most HA-MRSA isolates (70%) belonging to the USA100 clone. Two non-USA type strains were found among the isolates we studied; this included the Brazilian strain and epidemic MRSA (EMRSA)-15 clone. EMRSA-15 and EMRSA-16 have emerged as the predominant MRSA clones recovered patients in hospitals in the United Kingdom [32] but remain uncommonly reported from the U.S. The Brazilian epidemic clone is the predominant clone in South America [20, 33]. It carries SCCmecIII.
Our study was subject to some limitations. In our study, only nasal swabs for MRSA colonization were performed. The study did not include surveillance cultures at other body sites (e.g., no throat, inguinal, rectal/perianal, wound or tracheal aspirate surveillance cultures were performed). This may have led to an underestimation of the prevalence of MRSA colonization, and differentially impact detection of colonization with CA-MRSA genotypes, as these may be relatively more likely (compared to HA-MRSA genotypes) to colonize non-nasal body sites (such as the groin). In addition, while participating centers were geographically dispersed, only academic medical centers were included and thus our patient population many not be representative of all patients entering ICUs. We did not perform molecular typing on all 626 MRSA isolates recovered (only on 210 MRSA isolates), so selection bias cannot be excluded. However, we randomly selected the isolates to perform molecular typing on. The genotype distribution of the 22 MRSA strains causing nosocomial bacteremia among the 210 patients with MRSA nasal colonization whose isolates underwent molecular typing was unknown (as collection of bloodstream isolates was not part of the study protocol); thus we could not ascertain if bacteremic episodes were of an endogenous nature, i.e. same genotype as nasal isolate, or were possibly due to cross-infection in the ICU setting.
Conclusion
Our study findings have implications with regard to targeted approaches to screening, since some targeted approaches include a history of prior hospitalization as an indication to screen, which may miss people with CA-MRSA who are less likely to have history of prior hospitalizations. We also found in our study that patients with nares MRSA colonization on admission to the ICU were significantly more likely to develop an MRSA BSI than patients without MRSA colonization, but there were no differences in risk of developing BSI based on MRSA genotype (i.e., CA-MRSA genotypes compared to those with HA-MRSA).
Acknowledgments
Funding. Supported in part by the National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases (NIAID) [Contract Numbers N01 AI-15440 and N01 AI-15441] including the NIAID Bacterial and Mycoses Study Group, the NIH/National Center for Research Resources (NCRR) [UL1 RR025008 (Atlanta Clinical and Translational Science Institute), M01-RR-00585, and UL1 RR024150 (Mayo Clinic Center for Translational Science Activities)] as well as Merck and Co., Inc., Elan Pharmaceuticals, Inc, Roche Diagnostics, and Kimberly Clark.
We thank the STAR*ICU trial study coordinators, ICU physician and nurse directors, and all other ICU staff at the participating sites for their assistance in conducting the study and the technologists at the NIH Clinical Center Microbiology Laboratory (Laura Ediger, Robin Greb Odom, Amy Kominsky, Muthoni Rwenji and Liliana Rao) for their work processing the surveillance cultures.
Participating sites
The investigators (listed alphabetically), their participating centers, and their academic affiliations were: Harper University Hospital, Wayne State University, Detroit, Michigan – G. Alangaden; University of Alabama at Birmingham Hospital, University of Alabama at Birmingham, Birmingham, Alabama – J. Baddley; Mayo Clinic Arizona, Phoenix, Arizona and College of Medicine, Mayo Clinic, Rochester, Minnesota – J. Blair; R. Adams Cowley Shock Trauma Center, University of Maryland, Baltimore, Maryland – G. Bochicchio; Grady Memorial Hospital, Emory University, Atlanta, Georgia - H. Blumberg; Beth Israel Medical Center, Yeshiva University, New York, New York – N. Schulhof*, B. Koll; University Hospital, University of Michigan, Ann Arbor, Michigan – C. Chenoweth; University Medical Center, University of Arizona, Tucson, Arizona – C. Glasby; University Hospitals of Cleveland, Case Western Reserve University, Cleveland, Ohio – R. Hejal; Mayo Clinic Jacksonville, Jacksonville, Florida and College of Medicine, Mayo Clinic, Rochester, Minnesota – W. Hellinger; University of Iowa Hospitals and Clinics, University of Iowa Carver College of Medicine, Iowa City, Iowa – L. Herwaldt; Mayo Clinic Rochester, College of Medicine, Mayo Clinic, Rochester, Minnesota - W. Huskins; Yale-New Haven Medical Center, Yale University, New Haven, Connecticut – L. Kaplan, H. Frankel †; Jackson Memorial Hospital, Miller School of Medicine at the University of Miami, Miami, Florida – D. Kett; Cooper University Hospital, University of Medicine and Dentistry of New Jersey, Camden, New Jersey – A. Reboli; Oregon Health and Sciences University Hospital, Oregon Health and Sciences University, Portland, Oregon – R. Taplitz ‡; University of Chicago Medical Center, University of Chicago, Chicago, Illinois – S. Weber; Beth Israel Deaconess Medical Center, Harvard Medical School, Boston, Massachusetts – S. Wright; Massachusetts General Hospital, Harvard Medical School, Boston, Massachusetts – K. Zachary.
* current affiliation: Mount Sinai Medical Center, Mount Sinai School of Medicine, New York, New York
† current affiliation: University of Texas Southwestern Medical Center, University of Texas Southwestern Medical School, Dallas, Texas
‡ current affiliation: Moores Cancer Center, University of California, San Diego, La Jolla, California
Footnotes
Conflict of Interest. Dr. Huskins reports serving on an advisory board for Glaxo Smith Kline. The other authors declare no conflict of interest.
IRB approval. The study was approved by Emory University Institutional Review Board (IRB) and IRBs at all participating sites.
References
- 1.Boucher HW, Corey GR. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 2008 Jun 1;46(Suppl 5):S344–S349. doi: 10.1086/533590. [DOI] [PubMed] [Google Scholar]
- 2.National Nosocomial Infections Surveillance (NNIS) System Report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control. 2004 Dec;32(8):470–485. doi: 10.1016/S0196655304005425. [DOI] [PubMed] [Google Scholar]
- 3.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998 Aug 20;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 4.Cosgrove SE, Sakoulas G, Perencevich EN, Schwaber MJ, Karchmer AW, Carmeli Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis. 2003 Jan 1;36(1):53–59. doi: 10.1086/345476. [DOI] [PubMed] [Google Scholar]
- 5.Cosgrove SE, Qi Y, Kaye KS, Harbarth S, Karchmer AW, Carmeli Y. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infect Control Hosp Epidemiol. 2005 Feb;26(2):166–174. doi: 10.1086/502522. [DOI] [PubMed] [Google Scholar]
- 6.Engemann JJ, Carmeli Y, Cosgrove SE, et al. Adverse clinical and economic outcomes attributable to methicillin resistance among patients with Staphylococcus aureus surgical site infection. Clin Infect Dis. 2003 Mar 1;36(5):592–598. doi: 10.1086/367653. [DOI] [PubMed] [Google Scholar]
- 7.King MD, Humphrey BJ, Wang YF, Kourbatova EV, Ray SM, Blumberg HM. Emergence of community-acquired methicillin-resistant Staphylococcus aureus USA 300 clone as the predominant cause of skin and soft-tissue infections. Ann Intern Med. 2006 Mar 7;144(5):309–317. doi: 10.7326/0003-4819-144-5-200603070-00005. [DOI] [PubMed] [Google Scholar]
- 8.Klevens RM, Morrison MA, Nadle J, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007 Oct 17;298(15):1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 9.Moran GJ, Krishnadasan A, Gorwitz RJ, et al. Methicillin-resistant S. aureus infections among patients in the emergency department. N Engl J Med. 2006 Aug 17;355(7):666–674. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- 10.Methicillin-resistant staphylococcus aureus infections among competitive sports participants--Colorado, Indiana, Pennsylvania, and Los Angeles County, 2000–2003. MMWR Morb Mortal Wkly Rep. 2003 Aug 22;52(33):793–795. [PubMed] [Google Scholar]
- 11.Begier EM, Frenette K, Barrett NL, et al. A high-morbidity outbreak of methicillin-resistant Staphylococcus aureus among players on a college football team, facilitated by cosmetic body shaving and turf burns. Clin Infect Dis. 2004 Nov 15;39(10):1446–1453. doi: 10.1086/425313. [DOI] [PubMed] [Google Scholar]
- 12.Outbreaks of community-associated methicillin-resistant Staphylococcus aureus skin infections--Los Angeles County, California, 2002–2003. MMWR Morb Mortal Wkly Rep. 2003 Feb 7;52(5):88. [PubMed] [Google Scholar]
- 13.Diep BA, Chambers HF, Graber CJ, et al. Emergence of multidrug-resistant, community-associated, methicillin-resistant Staphylococcus aureus clone USA300 in men who have sex with men. Ann Intern Med. 2008 Feb 19;148(4):249–257. doi: 10.7326/0003-4819-148-4-200802190-00204. [DOI] [PubMed] [Google Scholar]
- 14.Adcock PM, Pastor P, Medley F, Patterson JE, Murphy TV. Methicillin-resistant Staphylococcus aureus in two child care centers. J Infect Dis. 1998 Aug;178(2):577–580. doi: 10.1086/517478. [DOI] [PubMed] [Google Scholar]
- 15.Herold BC, Immergluck LC, Maranan MC, et al. Community-acquired methicillin-resistant Staphylococcus aureus in children with no identified predisposing risk. JAMA. 1998 Feb 25;279(8):593–598. doi: 10.1001/jama.279.8.593. [DOI] [PubMed] [Google Scholar]
- 16.Deurenberg RH, Stobberingh EE. The evolution of Staphylococcus aureus. Infect Genet Evol. 2008 Dec 8;(6):747–763. doi: 10.1016/j.meegid.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 17.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003 Nov;41(11):5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boyle-Vavra S, Ereshefsky B, Wang CC, Daum RS. Successful multiresistant community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel Staphylococcal chromosome cassette mec (SCCmec) type VT or SCCmec type IV. J Clin Microbiol. 2005 Sep;43(9):4719–4730. doi: 10.1128/JCM.43.9.4719-4730.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson AP, Aucken HM, Cavendish S, et al. Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS) J Antimicrob Chemother. 2001 Jul;48(1):143–144. doi: 10.1093/jac/48.1.143. [DOI] [PubMed] [Google Scholar]
- 20.Vandenesch F, Naimi T, Enright MC, et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis. 2003 Aug 9;(8):978–984. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huskins WC, Huckabee CM, O'Grady NP, Murray P, Kopetskie H, Zimmer L, Walker ME, Sinkowitz-Cochran RL, Jernigan JA, Samore M, Wallace D, Goldmann DA. STAR*ICU Trial Investigators. Intervention to reduce transmission of resistant bacteria in intensive care. N Engl J Med. 2011;364:1407–1418. doi: 10.1056/NEJMoa1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seybold U, Kourbatova EV, Johnson JG, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin Infect Dis. 2006 Mar 1;42(5):647–656. doi: 10.1086/499815. [DOI] [PubMed] [Google Scholar]
- 23.Zetola N, Francis JS, Nuermberger EL, Bishai WR. Community-acquired meticillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect Dis. 2005 May 5;(5):275–286. doi: 10.1016/S1473-3099(05)70112-2. [DOI] [PubMed] [Google Scholar]
- 24.Kourbatova EV, Halvosa JS, King MD, Ray SM, White N, Blumberg HM. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA 300 clone as a cause of health care-associated infections among patients with prosthetic joint infections. Am J Infect Control. 2005 Sep;33(7):385–391. doi: 10.1016/j.ajic.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Otter JA, French GL. Nosocomial transmission of community-associated methicillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect Dis. 2006 Dec 6;(12):753–755. doi: 10.1016/S1473-3099(06)70636-3. [DOI] [PubMed] [Google Scholar]
- 26.Patel M, Kumar RA, Stamm AM, Hoesley CJ, Moser SA, Waites KB. USA300 genotype community-associated methicillin-resistant Staphylococcus aureus as a cause of surgical site infections. J Clin Microbiol. 2007 Oct;45(10):3431–3433. doi: 10.1128/JCM.00902-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Said-Salim B, Mathema B, Kreiswirth BN. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging pathogen. Infect Control Hosp Epidemiol. 2003 Jun;24(6):451–455. doi: 10.1086/502231. [DOI] [PubMed] [Google Scholar]
- 28.Corbella X, Dominguez MA, Pujol M, et al. Staphylococcus aureus nasal carriage as a marker for subsequent staphylococcal infections in intensive care unit patients. Eur J Clin Microbiol Infect Dis. 1997 May;16(5):351–357. doi: 10.1007/BF01726362. [DOI] [PubMed] [Google Scholar]
- 29.Seybold U, Halvosa JS, White N, Voris V, Ray SM, Blumberg HM. Emergence of and risk factors for methicillin-resistant Staphylococcus aureus of community origin in intensive care nurseries. Pediatrics. 2008 Nov;122(5):1039–1046. doi: 10.1542/peds.2007-3161. [DOI] [PubMed] [Google Scholar]
- 30.Bleasdale SC, Trick WE, Gonzalez IM, Lyles RD, Hayden MK, Weinstein RA. Effectiveness of chlorhexidine bathing to reduce catheter-associated bloodstream infections in medical intensive care unit patients. Arch Intern Med. 2007 Oct 22;167(19):2073–2079. doi: 10.1001/archinte.167.19.2073. [DOI] [PubMed] [Google Scholar]
- 31.Como-Sabetti K, Harriman KH, Buck JM, Glennen A, Boxrud DJ, Lynfield R. Community-associated methicillin-resistant Staphylococcus aureus: trends in case and isolate characteristics from six years of prospective surveillance. Public Health Rep. 2009 May-Jun;124(3):427–435. doi: 10.1177/003335490912400312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson AP, Pearson A, Duckworth G. Surveillance and epidemiology of MRSA bacteraemia in the UK. J Antimicrob Chemother. 2005 Sep;56(3):455–462. doi: 10.1093/jac/dki266. [DOI] [PubMed] [Google Scholar]
- 33.Da Silva Coimbra MV, Teixeira LA, Ramos RL, et al. Spread of the Brazilian epidemic clone of a multiresistant MRSA in two cities in Argentina. J Med Microbiol. 2000 Feb;49(2):187–192. doi: 10.1099/0022-1317-49-2-187. [DOI] [PubMed] [Google Scholar]