SUMMARY
Clusters of circulating tumor cells (CTC-clusters) are present in the blood of patients with cancer but their contribution to metastasis is not well defined. Using mouse models with tagged mammary tumors, we demonstrate that CTC-clusters arise from oligoclonal tumor cell groupings and not from intravascular aggregation events. Although rare in the circulation compared with single CTCs, CTC-clusters have 23-50-fold increased metastatic potential. In patients with breast cancer, single-cell resolution RNA sequencing of CTC-clusters and single CTCs, matched within individual blood samples, identifies the cell junction component plakoglobin as highly differentially expressed. In mouse models, knockdown of plakoglobin abrogates CTC-cluster formation and suppresses lung metastases. In breast cancer patients, both abundance of CTC-clusters and high tumor plakoglobin levels denote adverse outcomes. Thus, CTC-clusters are derived from multicellular groupings of primary tumor cells held together through plakoglobin-dependent intercellular adhesion, and while rare, they greatly contribute to the metastatic spread of cancer.
INTRODUCTION
The metastatic spread of breast cancer, typically to bone, lung, liver and brain, accounts for the vast majority of cancer-related deaths (Nguyen et al., 2009). Our understanding of epithelial cancer metastasis is derived primarily from mouse models, and it is thought to involve a series of sequential steps: epithelial-to-mesenchymal transition (EMT) of individual cells within the primary tumor leading to their intravasation into the bloodstream, survival of such circulating tumor cells (CTCs) within the bloodstream, and finally their extravasation at distant sites, where mesenchymal-to-epithelial transition (MET) culminates in their proliferation as epithelial metastatic deposits (Hanahan and Weinberg, 2011). While EMT has indeed been demonstrated in human breast cancer cells in the circulation (Yu et al., 2013), the requirement for EMT to initiate metastasis has been debated (Ledford, 2011; Tarin et al., 2005). Alternative models proposed include tumor-derived microemboli that may break off from primary tumors, lodging into distal capillaries where they initiate metastatic growth (Fidler, 1973; Liotta et al., 1976; Molnar et al., 2001). Using diverse technological platforms, we and others have indeed detected clusters of CTCs, ranging from 2-50 cancer cells, within the circulation of patients with metastatic epithelial cancers (Cho et al., 2012; Fidler, 1973; Molnar et al., 2001; Stott et al., 2010; Yu et al., 2013).
Studies of cancer metastasis have emphasized the concept of “seed vs soil” as a key determinant of metastatic propensity (Fidler, 2003). This model matches the importance of mutated genetic drivers within tumor cells conferring proliferative and invasive properties, with that of the microenvironment of the distant organ or “niche”, which may facilitate metastatic growth. However, the physical characteristics of single CTCs and CTC-clusters may also contribute to metastatic propensity, especially as they impact the ability of epithelial tumor cells to survive the loss of cell adherence and shear forces in the blood stream, i.e. different survival signals among the cancer cell “seeds” may be important. For instance, in a mouse endogenous pancreatic cancer model, non-canonical Wnt signaling is elevated within CTCs, where it appears to suppress anoikis (Yu et al., 2012), while in a subcutaneous tumor xenograft model, the admixture of tumor and stromal cells within micro-emboli may contribute stromal-derived survival signals (Duda et al., 2010). In a recent study of human breast cancer, mesenchymal markers indicative of EMT were expressed within the cancer cells comprising CTC-clusters (Yu et al., 2013). Taken together, both human and mouse modeling studies point to the complexity of blood-borne metastasis, and the need to capture and characterize CTCs to better understand this process.
CTCs have been detected in the majority of epithelial cancers, where they represent cancer cells captured as they transit through the bloodstream (Alix-Panabieres and Pantel, 2013; Yu et al., 2011). As such, they hold the key to understanding critical pathways that mediate the blood-borne dissemination of cancer, which may not be readily evident through analyses of bulk primary or metastatic tumor populations. Factors leading to the generation of CTCs from a primary tumor are unknown, including the fraction derived from cancer cells that have actively intravasated into the bloodstream, versus those that are passively shed as a result of compromised tumor vasculature. Although exceedingly rare compared with normal blood cells, the number of CTCs in the bloodstream far exceeds the number of metastatic lesions in patients, indicating that the vast majority CTCs die in the bloodstream, with only a minor fraction representing viable metastatic precursors. Epithelial cells that have lost adhesion-dependent survival signals rapidly undergo anoikis, a fate likely to meet most CTCs in the bloodstream. It is in this context that either mesenchymal transformation, stromal-derived factors or persistent inter-epithelial cell junctions may provide survival signals that attenuate this apoptotic outcome (Duda et al., 2010; Mani et al., 2008; Robson et al., 2006; Yu et al., 2012). Dissecting the contributions of these various mechanisms to human cancer requires the ability to isolate individual CTCs from the bloodstream and subject these to detailed molecular analyses.
Multiple technologies have been developed for CTC capture, taking advantage of tumor-specific epitopes absent in normal blood cells, variations in their physical properties such as size, density and electromechanical characteristics, or by applying high throughput imaging to unpurified blood cell preparations (reviewed in (Yu et al., 2011). The fact that CTCs are extremely rare, even in patients with advanced metastatic cancers (estimated at one CTC/billion normal blood cells), and that they may be poised on the verge of apoptosis, has made their analysis contingent upon technological constraints. We have introduced a series of microfluidic devices, which have the advantage of low-shear, yet high throughput, interrogation of unprocessed whole blood, providing highly enriched and unfixed CTCs that are suitable for detailed molecular analysis (Nagrath et al., 2007; Ozkumur et al., 2013; Stott et al., 2010). Among these, the herringbone (HBCTC-Chip) makes use of grooves within the ceiling of the microfluidic chamber to generate turbulent microfluidic flow, directing cells against antibody-coated walls of the device, where CTCs are captured (Stott et al., 2010). This device, whose highly efficient design enabled our initial detection of large CTC-clusters, requires on-chip cell lysis for nucleic acid extraction and hence provides an enriched but heterogeneous CTC population for analysis (Yu et al., 2013; Yu et al., 2012). In contrast, our recently described negCTC-iChip achieves highly efficient depletion of erythrocytes and leukocytes from blood specimens, yielding untagged CTCs and small CTC-clusters in solution, where they can be micromanipulated for single cell RNA sequencing (Ozkumur et al., 2013). Here, we use both of these devices, along with in vivo flow cytometry and next generation RNA sequencing, to interrogate CTCs from both patients with metastatic breast cancer and mouse tumor models. We find, using mouse models, that CTC-clusters are derived from oligoclonal clumps of primary tumor cells and constitute a rare but very highly metastasis-competent subset of CTCs, compared with single circulating breast cancer cells. RNA sequencing of human breast CTC-clusters identifies plakoglobin as a key mediator of tumor cell clustering, which is expressed in a heterogeneous pattern within the primary tumor. Knockdown of plakoglobin expression in the mouse model suppresses CTC-cluster formation and reduces metastatic spread.
RESULTS
Endogenous CTC-clusters have increased metastatic potential compared to single CTCs
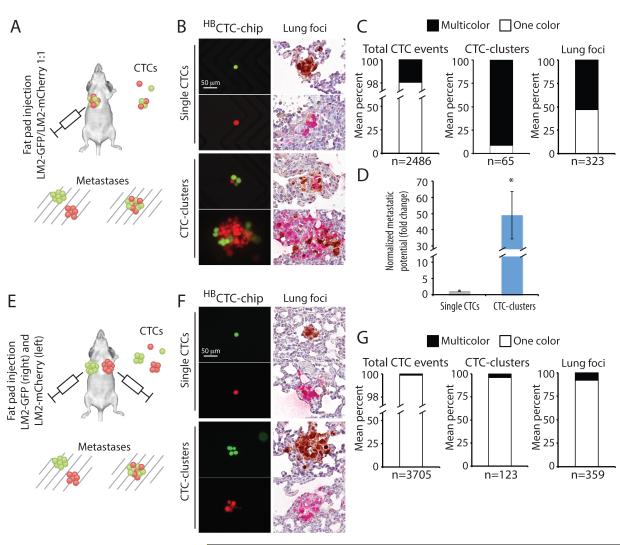
To define the origin and functional properties of CTC-clusters, compared with single CTCs, we made use of mouse models, where tumor cell composition, transit of CTCs through the bloodstream and metastatic deposits can be monitored and quantified. We first established a model to test the generation of endogenous CTCs and metastases from a primary orthotopic tumor xenograft. These experiments were designed both to test the metastatic propensity of CTC-clusters versus single CTCs, as well as to determine whether CTC-clusters originate from an oligoclonal grouping of primary tumor cells or from the clonal progeny of an individual tumor cell. MDA-MB-231-LM2 (LM2) cells, a lung-metastatic variant of MDA-MB-231 human breast cancer cells (Minn et al., 2005), were engineered to express either green fluorescent protein (LM2-GFP) or mCherry (LM2-mCherry), and a 1:1 mixture of these differentially tagged cells was injected into the mammary fat pad of immunodeficient (NSG) mice. As expected, overt primary breast tumors were observed after five weeks, and these retained an equal distribution of LM2-GFP and LM2-mCherry tagged cells, as confirmed by IHC staining (Fig. S1A and S1B). We sampled the blood of tumor-bearing animals for presence of single or clustered CTCs using a terminal bleed, and simultaneously harvested the lungs for analysis of metastatic deposits. In addition to enumeration of CTCs, we reasoned that clonally-derived CTC-clusters would uniformly express either GFP or mCherry, whereas aggregations of cells from the primary tumor would be heterogeneous for the two markers (Fig.1A). We observed a mean of 2,486 CTC events per mouse (n=5 mice), of which a mean of 65 (2.6%) were CTC-clusters and 2421 (97.4%) were single CTCs (Fig. 1B, 1C and S2A). Virtually all (91%) CTC-clusters were dual positives for GFP and mCherry. A mean of 5.6 (9%) CTC-clusters per mouse (with fewer than 3 cells per cluster) were comprised of cells expressing only one of the two markers, consistent with expected probabilities given a 1:1 mixture of GFP/mCherry expressing cells in the primary tumor (Fig. 1C and S2B). Thus, CTC-clusters do not result from the proliferation of a single tumor cell in the vasculature, instead they appear to represent the aggregation of neighboring cells, most likely within the primary tumor mass (see below).
Figure 1. CTC-clusters harbor increased metastatic potential compared to single CTCs.
A) Schematic of the experiment. MDA-MB-231-LM2 (LM2) cells expressing GFP (LM2-GFP) or mCherry (LM2-mCherry) cells were mixed at 1:1 ratio and injected in the right mammary gland of immunodeficient mice to generate one-color single CTCs and multicolor CTC-clusters. Accordingly, one-color metastatic foci are derived from a single CTC, while multicolor foci arise predominantly from a CTC-cluster. B) Representative images of single CTCs (GFP- or mCherry-positive) and CTC-clusters (GFP- and mCherry-positive) captured on the HBCTC-Chip (left). Lung metastatic foci derived from a single CTC (GFP- or mCherry-positive) or a CTC-cluster (GFP- and mCherry-positive) are shown (right). GFP (brown), mCherry (red). Blood samples and lung specimens were isolated five weeks after primary tumor development. n=5. C) Bar graphs showing the mean percentage of one-color versus multicolor CTC events captured by the HBCTC-Chip (left), the mean percentage of one-color versus multicolor CTC-clusters (middle), as well as the mean percentage of one-color versus multicolor lung foci (right). n=5. D) Bar graph showing the normalized metastatic potential of single CTCs and CTC-clusters. n=5, *p=0.031 by Student’s t test. E) Schematic of the experiment. LM2-GFP cells were injected in the right mammary gland while LM2-mCherry cells were injected in the left mammary gland of immunodeficient mice to generate tumors that give rise to one-color single CTCs and CTC-clusters, as well as rare multicolor CTC-clusters (resulting from aggregation events). Accordingly, one-color metastatic foci are derived from a single CTC or a CTC-cluster, while multicolor foci derive from CTC aggregates. F) Representative images of single CTCs (GFP- or mCherry-positive) and CTC-clusters (GFP- or mCherry-positive) captured on the HBCTC-Chip (left). Lung metastatic foci derived from a single CTC (GFP- or mCherry-positive) or a CTC-cluster (GFP- or mCherry-positive) are shown (right). GFP (brown), mCherry (red). Blood samples and lung specimens were isolated five weeks after primary tumor development. n=5. G) Bar graphs showing the mean percentage of one-color versus rare multicolor CTC events captured by the HBCTC-Chip (left), the mean percentage of one-color versus multicolor CTC-clusters (middle), as well as the mean percentage of one-color versus multicolor lung foci (right). n=5.
Metastatic deposits in the lungs were analyzed for both number and composition using anti-GFP and anti-mCherry antibodies, simultaneously with the CTC analyses (Fig. 1B, 1C and S2A). Given the distribution of GFP and mCherry staining in CTC-clusters, we reasoned that metastatic tumors derived from a single CTC would be positive for a single marker, while those derived from CTC-clusters would stain for both GFP and mCherry (Fig. 1A and 1B). A mean of 323 lung foci were identified per mouse (n=5 mice), of which 171 (53%) were multicolor, and therefore derived from CTC-clusters, versus 152 (47%) unicolor derivatives of single CTCs (Fig. 1C and S2A). Normalizing the number and distribution of lung metastases with that of single CTCs and CTC-clusters, we calculate that a CTC-cluster is approximately 50 times more likely to give rise to a metastatic deposit than a single CTC (Fig. 1D). Thus, while CTC-clusters are much more rare than single CTCs in this orthotopic mouse model of breast cancer, they contribute equally to the metastatic burden in the lung.
To further validate (1) that oligoclonal CTC-clusters arise from the fragmenting of primary tumor cell clumps into the vasculature, and not from intravascular aggregation of single CTCs and (2) that oligoclonal lung metastases arise from CTC-clusters and not from the reseeding of a metastatic site by multiple single CTCs, we undertook a second series of orthotopic mouse xenograft experiments, injecting LM2-GFP cells into the right mammary fat pad and LM2-mCherry cells in the left fat pad of immunodeficient mice (Fig. 1E). Five weeks after injection, mice harbored two independent and differentially tagged tumors, and we again simultaneously harvested the blood for analysis of CTCs and the lungs for enumeration of metastatic deposits. As expected, single CTCs in the circulation demonstrated equal contributions from the GFP and m-Cherry-tagged primary tumors. However, unlike the previous multi-tagged single tumor model, in mice with two independent individually-tagged tumors, the vast majority of CTC-clusters (96%) were of a single color, with equal contributions from GFP- or mCherry-positive primary tumors (Fig. 1F, 1G and S2A). Thus, the vast majority of CTC-clusters are derived from individual primary tumors, excluding intravascular aggregation of single CTCs as a significant source of CTC-clusters.
A very small fraction of CTC-clusters observed in the dual tumor-bearing mice were multicolor (4% of CTC-clusters, corresponding to 0.12% of total CTC events) (Fig. 1G and S2A). While extraordinarily rare, the presence of such CTC-clusters derived from two independent tumors may originate either from the uncommon intravascular aggregation of single CTCs or from a mixing of cancer cells within the two primary tumors, due to the previously reported “tumor reseeding” phenomenon (Kim et al., 2009) (Fig. S1B). Consistent with the latter hypothesis, we found that 3-5% of cells within the GFP-tagged primary tumor were positive for mCherry and 3-5% of cells within the mCherry-labeled tumor were positive for GFP (Fig. S1B). In addition to rare multicolor CTC-clusters, we observed a small fraction (8%) of multicolor tumors in the lung (Fig. 1G and S2A). These metastatic lesions could result either from the rare multicolor CTC-clusters or from the reseeding of metastatic lesions by multiple single CTCs.
We confirmed the findings derived from the two LM2 mouse xenograft experiments with a second, mouse derived breast cancer cell line, 4T1 (Fig. S2C and S2D). Consistent with the LM2 results, a 1:1 mixture of 4T1-GFP and 4T1-mCherry cells within an orthotopic mammary tumor generated CTC-clusters that were overwhelmingly multicolored (90%), whereas two separate primary 4T1 tumors labeled either with GFP or mCherry produced CTC-clusters that were of a single color (87%). These observations further support that CTC-clusters arise as oligoclonal fragments derived from a single tumor (Fig. S2C). Normalizing the number and color distribution of 4T1-derived lung metastases relative to the prevalence of single CTCs and CTC-clusters, we calculated a 23-fold increase in metastatic competence for CTC-clusters versus single CTCs (Fig. S2D), an estimate that is comparable to the 50-fold increase derived from LM2 cell experiments. Taken together, these two mouse tumor models indicate that CTC-clusters constitute only 2-5% of all CTC events detected in the circulation, but their dramatically elevated metastatic potential (23-50 times that of single CTCs) contributes to approximately half of all metastatic lesions in orthotopic breast cancer models.
Clustered cancer cells are more resistant than single cells to apoptosis following dissemination to the lung
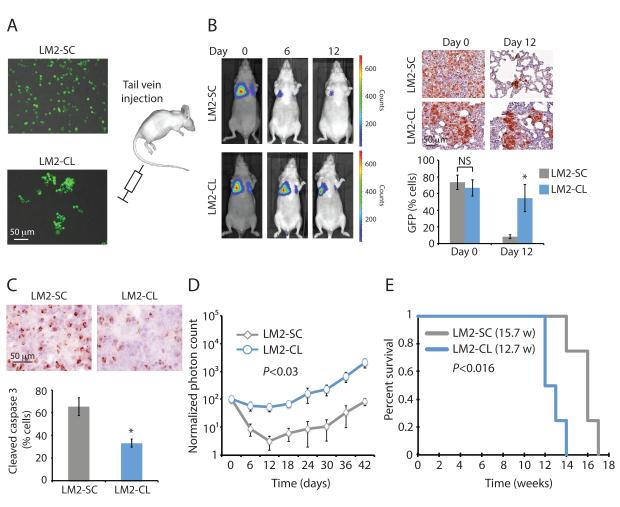
We generated an in vitro assay that allowed us to obtain a suspension of either single cells or clustered cells (2 to 30 cells) from cultures of GFP-Luciferase-tagged LM2 cells (see Supplementary Methods). We injected 200,000 LM2 cells prepared either as single cells (LM2-SC) or as clusters (LM2-CL) into the tail vein of immunodeficient mice, and subjected them to serial luciferase-based imaging (Fig. 2A and 2B). Both LM2-SC and LM2-CL cells reached the lungs with equal efficiency (day 0), as shown by both bioluminescence and GFP immunohistochemical (IHC) staining (Fig. 2B). However, over the following days, the LM2-SC lung signal progressively diminished as the cells underwent massive apoptosis, demonstrated by staining for cleaved caspase 3 (Fig. 2C). In contrast, the LM2-CL lung signal persisted following intravascular inoculation, with cells showing resistance to apoptosis and tumors expanding more rapidly (Fig. 2B, 2C and 2D). Lung tumors eventually grew in mice subjected to tail vein injection with either of the two LM2 derivatives, but injection of clustered cells resulted in reduced overall survival, with 12.7 weeks for LM2-CL versus 15.7 weeks for LM2-SC (p<0.016) (Fig. 2E). We confirmed the differential rate of apoptosis and metastatic growth in the lung for single versus clustered cancer cells using tail vein injection of two additional breast cancer cell lines, BT474 and 4T1 (Fig. S3A-D).
Figure 2. CTC-clusters are more resistant to apoptosis at distal metastatic sites.
A) Schematic showing MDA-MB-231-LM2-GFP-Luciferase (LM2) cells prepared as single cells (LM2-SC) or as clusters (LM2-CL) prior to injection into the tail vein of immunodeficient mice. 2×105 cells were injected as LM-SC or LM2-CL per mouse. B) Representative bioluminescence images of mice at 0, 6 and 12 days after tail vein injection with LM2-SC or LM2-CL cells (left). n=4. Representative images of GFP-stained sections of mouse lungs after injection with LM2-SC or LM2-CL cells (right). The bar graph shows the mean percentage of GFP-positive cells in lungs from LM2-SC-or LM2-CL-injected mice. n=4; NS=not significant, *p=0.03 by Student’s t test. C) Representative images of cleaved caspase 3-stained sections of mouse lungs 24 hours after injection with LM2-SC or LM2-CL cells. The bar graph shows the mean percentage of cleaved caspase 3-positive cells in lungs from LM2-SC- or LM2-CL-injected mice. n=4; *p<0.02 by Student’s t test. D) Lung metastasis growth curve from mice injected with LM2-SC or LM-CL. n=4; p<0.03 by Student’s t test. E) Kaplan-Meier survival plot showing survival rates for mice injected with LM2-SC or LM2-CL. n=4; p<0.016 by Log-rank test.
See also Figure S3.
Calculation of CTC-clusters and single CTC circulatory clearance rate using in vivo flow cytometry
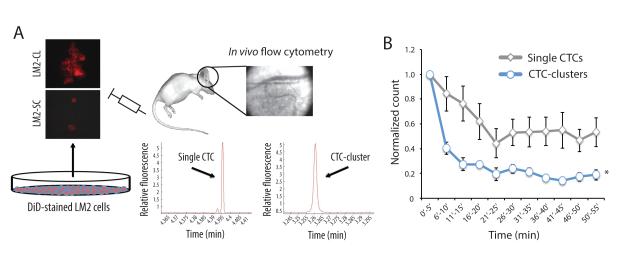
Clusters of tumor cells may exhibit considerable flexibility as they navigate through narrow channels, and capillary beds themselves may have uneven vessel diameters or bypass tracts that allow transit of large multi-cellular structures. However overall, CTC-clusters are more likely than single CTCs to be trapped in small capillaries of the lung and distal organs. Thus, the low steady state level of CTC-clusters in the circulation may reflect a considerably higher generation rate if their clearance rate is very high. To test if CTC-clusters indeed have a faster clearance rate from the bloodstream than single CTCs, we used in vivo flow cytometry (IVFC) to monitor LM2-SC and LM2-CL cells labeled with the lipophilic carbocyanine membrane dye DiD, following tail vein injection in immunodeficient mice (Fig. 3A). DiD was selected to achieve optimal detection of CTCs with our IVFC settings. Circulating DiD-labeled cells were detected in real time within the ear blood vessels for a total of 55 minutes in each mouse. Injected LM2-CL cells were cleared at least three times more rapidly than LM2-SC (half-life: 6-10 mins for LM2-CL versus 25-30 mins for LM2-SC) (Fig. 3B). Together, these observations define a circulating time for CTCs in the bloodstream: the shorter circulation half-life of CTC-clusters is consistent with their more rapid entrapment within capillaries of distal organs, where they may initiate metastatic growth (Liotta et al., 1976).
Figure 3. CTC-clusters demonstrate faster clearance rate from the bloodstream.
A) Schematic showing the experimental setup for measuring the clearance time of single CTCs and CTC-clusters. Briefly, DiD-stained LM2 cells were prepared as LM2-SC or LM2-CL and injected into the tail vein of immunodeficient mice. In vivo flow cytometry was applied to the ear blood vessels to detect single CTCs and CTC-clusters over a 55 minutes period after injection. Graphs show representative fluorescence peaks corresponding to the transit of a single CTC or CTC-cluster through the ear blood vessel. B) Graph showing single CTCs and CTC-clusters clearance curves. n=5 for single CTCs and n=4 for CTC-clusters, *p<0.01 by two-way ANOVA.
The presence of CTC-clusters in patients with breast and prostate cancer correlates with poor prognosis
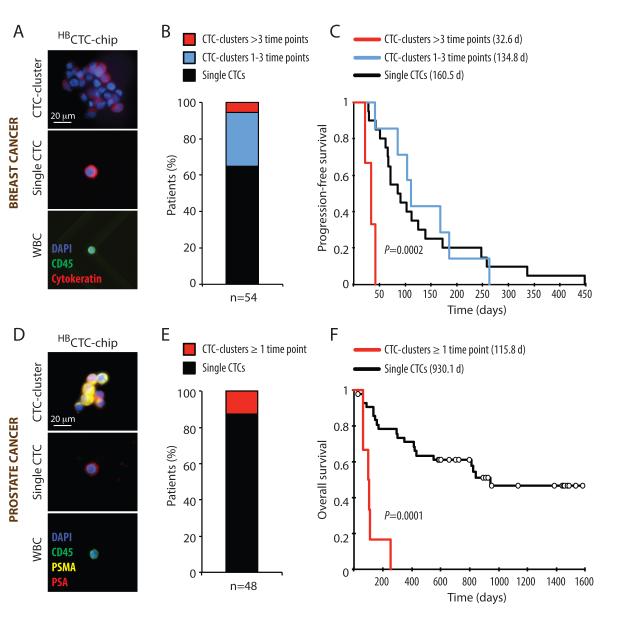
Having characterized the origin and metastatic potential of CTC-clusters in mouse models, we undertook to study their properties in patients with cancer. To first test the clinical significance of CTC-clusters in the blood of patients with progressing metastatic breast cancer, we measured their presence in blood specimens from a total of 79 patients, drawn at multiple time points over a period of 19 months. Patients were recruited to an IRB-approved study at the Massachusetts General Hospital Cancer Center, including women with estrogen receptor positive (n=49), HER2 positive (n=13), triple negative (n=17) subtypes of breast cancer (total: 265 data points). For these experiments, we made use of the HBCTC-Chip, which is highly efficient in capturing both large and small CTC-clusters (Stott et al., 2010). We coated the microfluidic chamber with a combination of antibodies, targeting the Epithelial Cell Adhesion Molecule (EpCAM), as well as the lineage markers Epithelial Growth Factor Receptor (EGFR) and Human Epithelial growth factor Receptor 2 (HER2/ErbB2), which together efficiently capture both epithelial and mesenchymal breast CTCs (Yu et al., 2013). After processing 3ml of whole blood from patients with breast cancer, the CTCs captured on the chip were stained with antibodies against wide spectrum cytokeratin (CK) to identify CTCs, and against the leukocyte marker CD45 to assess white blood cell (WBC) contamination (Fig. 4A). CTCs were identified in 54 out of 79 patients (68%). Among patients with CTCs, 3 (5.6%) had CTC-clusters evident across more than three time points, while 16 (29.6%) had CTC-clusters during one to three time points and 35 (64.8%) had no detectable clusters (Fig. 4B). We correlated the presence of CTC-clusters with progression-free survival (PFS) for all patients where such data were available (n=30) (Table S1). Of note, PFS was calculated as time from initiation of therapy to discontinuation by the treating clinician (blinded to the CTC results), and PFS data analysis was performed only when clinical measurements bracketed the CTCs isolation time frame. Patients with CTC-clusters across more than three time points had a mean progression-free survival time of 32.6 days, compared with 134.8 days for patients where CTC-clusters were found during one to three time points and 160.5 days for patients with single CTCs only (p=0.0002) (Fig. 4C and Table S1). Thus, even among patients with advanced metastatic breast cancer, the continuous presence of CTC-clusters is associated with an adverse clinical outcome.
Figure 4. The presence of CTC-clusters in patients with cancer correlates with poor prognosis.
A) Representative images of a CTC-cluster, a single CTC and a white blood cell (WBC) isolated from a breast cancer patient using the HBCTC-Chip and stained with wide-spectrum cytokeratin (CK, red), CD45 (green) and DAPI (nuclei, blue). B) A total of 79 breast cancer patients (corresponding to 265 timepoints) were analyzed for the presence of CTCs, with 54 of the 79 patients scoring positive for CTCs. The bar graph shows the percentage of CTC-positive patients having CTC-clusters during more than three time points (red), CTC-clusters across one to three time points (blue) or single CTCs only (black). C) Kaplan-Meier progression-free survival plot showing progression rates for breast cancer patients having CTC-clusters during more than three time points (red), CTC-clusters across one to three time points (blue) or single CTCs only (black). The mean progression-free survival time for each group is given in parentheses. p=0.0002 by Log-rank test. D) Representative images of a CTC-cluster, a single CTC and a white blood cell (WBC) isolated from a prostate cancer patient using the HBCTC-Chip and stained with prostate-specific antigen (PSA, red), prostate-specific membrane antigen (PSMA, yellow), CD45 (green) and DAPI (nuclei, blue). B) A total of 64 prostate cancer patients (corresponding to 202 timepoints) were analyzed for the presence of CTCs, with 48 of the 64 patients scoring positive for CTCs. The bar graph shows the percentage of CTC-positive patients having CTC-clusters during at least one time point (red) or single CTCs only (black). C) Kaplan-Meier overall survival plot showing progression rates for prostate cancer patients having CTC-clusters during at least one time point (red) or single CTCs only (black). The mean overall survival time for each group is given in parentheses. p=0.0001 by Log-rank test.
See also Table S1.
Given the relatively short time to progression in patients with advanced breast cancer, we sought to test the correlation between CTC-clusters and adverse prognosis in patients with a longer clinical course. We measured the number of CTCs in a total of 64 patients with prostate cancer using blood specimens drawn at multiple time points over a period of 53 months (total: 202 data points). CTCs in prostate cancer patients were visualized by staining with a cocktail of antibodies against prostate-specific antigen (PSA) and prostate-specific membrane antigen (PSMA); anti-CD45 staining was used to exclude white blood cells (Miyamoto et al., 2012). CTCs were detected in 48/64 patients (75%). CTC-clusters were present in in 6/48 samples (12.5%) (Figure 4D and 4E). In this cohort, the presence of CTC-clusters during at least one time point strongly correlated with a dramatically shorter overall survival time (mean survival time was 115.8 days for patients with CTC-clusters versus 930.1 days for patients with single CTCs; p=0.00001) (Fig. 4F and Table S1). While further studies will be required to ascertain the clinical utility of CTC-clusters versus single CTCs as prognostic determinants in either breast or prostate cancer, these initial results point to the potential relevance of CTC-clusters in the progression of human cancer.
Single cell resolution RNA sequencing of matched CTC-clusters and single CTCs purified from patients with breast cancer
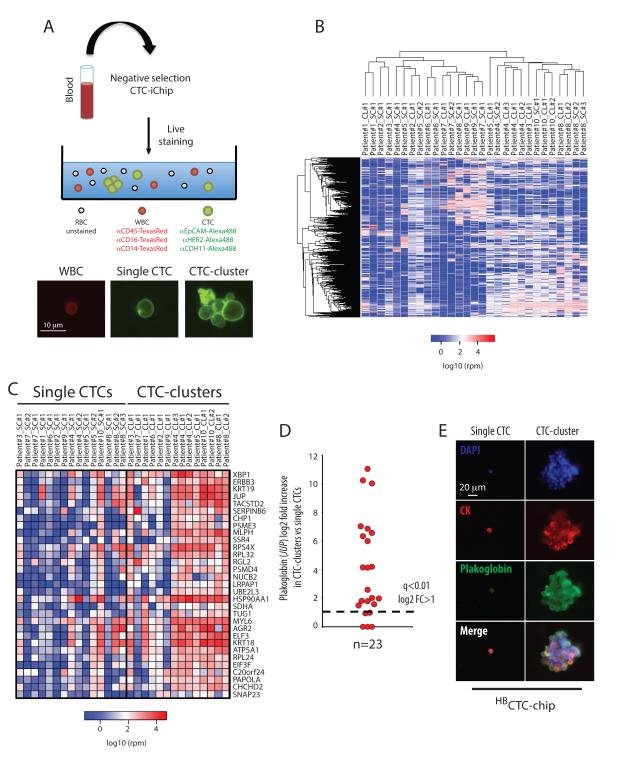
The ability to capture both single CTCs and CTC-clusters from the same blood specimen made it possible to undertake single cell resolution RNA sequencing, searching for differences in expression profiles matched to individual patients. For these experiments, we applied the negCTC-iChip, which enables isolation and single cell manipulation of untagged CTCs, together with an optimized protocol for next generation RNA sequencing from minute amounts of template (Ozkumur et al., 2013; Tang et al., 2010). Blood specimens from 10 patients with metastatic breast cancer were subjected to microfluidic depletion of RBCs and CD45- and CD66b-positive WBCs, leaving untagged single CTCs and small CTC-clusters in the final product (Ozkumur et al., 2013). Unfixed tumor cells were stained for cell surface expression of EpCAM, HER2 and the mesenchymal marker CDH11 (Alexa488-conjugated), and counterstained with antibodies against CD45, CD14 and CD16 to identify contaminating leukocytes (TexasRedconjugated) (Fig. 5A). Individual CTC-clusters (median of 3 cells per cluster) were isolated using a micromanipulator and compared with numerically matched pools of single CTCs from the same specimen, followed by next generation RNA sequencing (SOLiD 5500XL) (Fig. 5A). We derived normalized expression profiles for a total of 29 samples (15 pools of single CTCs and 14 CTC-clusters) isolated from 10 breast cancer patients.
Figure 5. RNA sequencing of CTC-clusters and single CTCs reveals a CTC-clusters-associated gene set.
A) Illustration of the experimental setup (top). Representative images of a labeled white blood cell (WBC, red), a single CTC and a CTC-cluster (green) (bottom). B) Heatmap showing unsupervised hierarchical clustering of 15 single CTCs pools and matched 14 CTC-clusters isolated from 10 breast cancer patients (SC: single CTCs, CL: CTC-cluster). C) Heatmap showing the top 31 transcripts upregulated in CTC-clusters. n=15 for single CTCs and n=14 for CTC-clusters; q<0.01, log2 fold change (FC) >1 in more than 70% intra-patient comparisons (SC: single CTCs, CL: CTC-cluster). D) Graph showing log2 fold increase in plakoglobin for each comparison between matched CTC-clusters versus single CTCs. The threshold line represents a q<0.01 and log2 fold increase >1. E) Representative images of a single CTC and a CTC-cluster captured on the HBCTC-Chip from a breast cancer patient and stained with wide-spectrum cytokeratin (CK, red), plakoglobin (green) and DAPI (nuclei, blue).
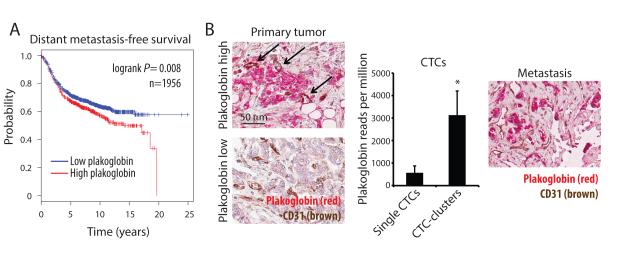
Unsupervised hierarchical clustering of RNA sequencing data showed no obvious distinctions at the global gene expression level between single CTCs and CTC-clusters, with both of these clustering closely by patient of origin (Fig. 5B). Consistent with the microscopic appearance of CTC-clusters as primarily tumor cell derived, we did not identify RNA signatures of other cell types, including T cells, B cells, dendritic cells, natural killer cells, hematopoietic stem cells, macrophages/monocytes, granulocytes, endothelial cells or fibroblasts (Fig. S4). Markers for platelets were present in both single CTCs and CTC-clusters, consistent with their known adherence to cancer cells in the circulation. For each patient, we compared gene expression data of CTC-clusters versus single-CTCs, generating a list of 31 CTC-cluster-associated genes shared across different patients (q<0.01, log2FC>1, in more than 70% of all intra-patient comparisons) (Fig. 5C, 5D and Table S2). To identify potential drivers of metastasis among CTC-cluster enriched genes, we tested for correlation between their overexpression in primary tumor specimens and clinical outcomes in a cohort of 1,956 patients with ER-positive, HER2-positive and triple-negative breast cancers. Among the candidate CTC-cluster genes, plakoglobin was unique in its high level of overexpression in CTC-clusters compared with single CTCs (219-fold) and the fact that its expression in primary tumors associated with a significantly reduced distant metastasis-free survival (p=0.008) (Fig. 5D, 6A and S5). We therefore selected plakoglobin as a CTC-cluster enriched transcript for more detailed analysis.
Figure 6. Plakoglobin expression correlates with decreased distant metastasis-free survival.
A) Kaplan-Meier distant metastasis-free survival plot showing progression rates for patients whose primary tumor expressed either “low plakoglobin” or “high plakoglobin” transcript. n=1956; p=0.008 by Log-rank test. B) Representative images of plakoglobin (red) and CD31 (blood vessels, brown)-stained tissue sections of matched primary tumor (left) and bone metastasis (right) from a hormone receptor-positive breast cancer patient with high CTC-cluster counts. Arrows indicate blood vessels in “high plakoglobin” regions. Nuclei are stained with hematoxylin. The bar graph (middle) shows plakoglobin reads per million in matched single CTCs and CTC-clusters isolated from the same patient. n=3; *p=0.031.
Plakoglobin (JUP) is a member of the Armadillo family of proteins and an important component of desmosomes and adherence junctions (Aktary and Pasdar, 2012), which has been reported to have both positive and negative roles in diverse malignancies (Hakimelahi et al., 2000; Kolligs et al., 2000; Shiina et al., 2005). Along with upregulation of plakoglobin RNA, multiple components of both desmosomes and adherence junctions were significantly enriched in CTC-clusters (Fig. S6A-E). Consistent with the RNA sequencing results, we confirmed plakoglobin protein expression in multiple CTC-clusters, but not in matched single CTCs from a breast cancer patient (Fig. 5E). While CTC-clusters express epithelial cell junction components, including plakoglobin and E-cadherin, we have previously shown that some mesenchymal markers may also be upregulated in such clusters, an effect that may be associated with adherence in the bloodstream with TGFβ-rich platelets (Labelle et al., 2011; Yu et al., 2013). Matched primary and metastatic tumors biopsies were available from this patient: plakoglobin expression was remarkably heterogeneous in both the primary and metastatic breast tumors, with foci of high expression interspersed with regions without detectable protein (Fig. 6B). Thus, while plakoglobin is a key component of intercellular junctions, its variable expression levels within primary tumors raises the possibility that it might demarcate tightly adherent groups of cells that may constitute precursors to CTC-clusters.
Plakoglobin is required for CTC-cluster formation and contributes to breast cancer metastasis
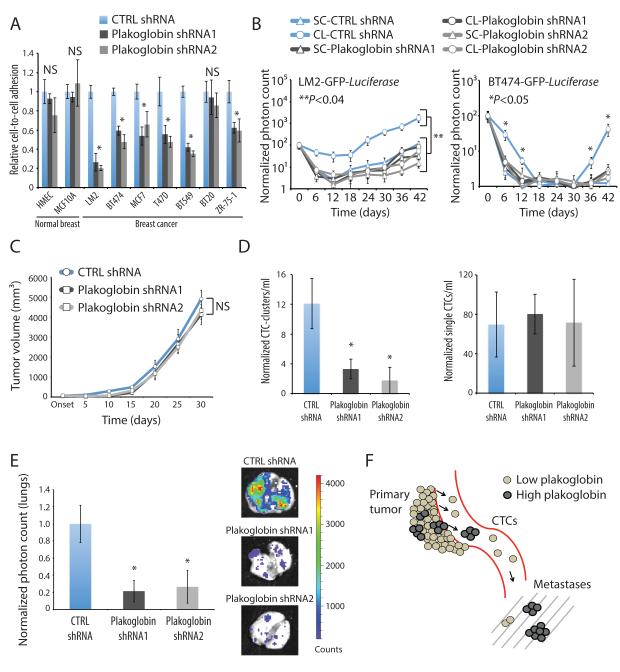
To define the functional consequences of plakoglobin expression in the context of CTC-clusters, we first applied an in vitro assay (Vybrant™), which utilizes a fluorogenic dye to measure cell-to-cell adhesion under a variety of culture conditions (El Khoury et al., 1996). We compared seven breast cancer cell lines (MDA-MB-231-LM2, BT474, MCF7, T47D, BT549, BT20 and ZR-75-1) with two non-transformed human mammary epithelial cells (HMEC and MCF10A), following stable lentiviral-mediated plakoglobin knockdown. shRNA-mediated plakoglobin suppression triggered disruption of cell-cell contacts in 6/7 breast cancer lines grown as a monolayer, while it had no detectable effect in either of the two non-transformed breast epithelial cells (p<0.04) (Fig. 7A, S7A and S7B). Thus, breast cancer cells may be more dependent on plakoglobin-mediated cell junctions than normal epithelial cells, which may benefit from additional or alternative pathways in forming intercellular connections (Alford and Taylor-Papadimitriou, 1996; Cavallaro and Christofori, 2004).
Figure 7. Plakoglobin is required for CTC-cluster formation and lung metastasis.
A) Bar graph showing the relative cell-to-cell adhesion in a panel of mammary epithelial cells and breast cancer cell lines grown in the presence or absence of plakoglobin. n=5; *p<0.04. B) Lung metastasis growth curves from mice injected with LM2-GFP-Luciferase (left) or BT474-GFP-Luciferase (right) cells expressing control or plakoglobin shRNAs and prepared as single cells (SC) or clusters (CL). n=4; *p<0.05, **p<0.04 by Student’s t test. C) LM2-GFP-Luciferase tumor growth curves in the presence or absence of plakoglobin. n=4; NS=not significant. D) Bar graphs showing the normalized number of CTC-clusters (left) and single CTCs (right) per ml of blood. Blood samples were isolated 4 weeks after primary tumor development and processed with the HBCTC-Chip. n=4; *p<0.05 by Student’s t test. E) Bar graph showing normalized lung photon counts from mice bearing a LM2-GFP-Luciferase control or plakoglobin knockdown primary tumor for 4 weeks. n=4; *p<0.045 by Student’s t test. F) Schematic showing that “high plakoglobin” regions in the primary tumor are likely to generate CTC-clusters with increased metastatic potential.
See also Figure S7.
To extend these observations in vivo, we introduced either plakoglobin shRNAs or non-target controls into GFP-Luciferase-tagged LM2 and BT474 cells, and prepared these as single cells (SC) or clusters (CL) for tail vein injection into immunosuppressed mice (Fig. S7C). Consistent with our previous results, both LM2 and BT474 cells expressing control shRNAs showed dramatically increased persistence in the lung when prepared under CL versus SC conditions. In contrast, despite CL conditions, plakoglobin knockdown in both LM2 and BT474 cells dissociated clusters into single cells, consistent with the requirement for plakoglobin for inter-cellular adhesion in these cells. Following plakoglobin knockdown, tail vein inoculation of CL and SC preparations of both LM2 and BT474 were comparable in producing a reduced number of lung metastases (Fig. 7B and S7C). Thus, plakoglobin knockdown abrogates intercellular interactions required to generate clustered cancer cells, thereby reducing their potential to produce lung foci after direct intravascular injection.
Finally, we generated orthotopic xenografts, injecting LM2-GFP-Luciferase cells expressing either control or plakoglobin shRNAs into the mammary fat pad of immunodeficient mice and measuring tumor growth as well as tumor-derived CTCs. Plakoglobin knockdown did not alter the primary tumor growth rate, measured for up to 30 days (Fig. 7C and S7D), nor did it affect the total number of single CTCs derived from the primary tumor (Fig. 7D). Remarkably, the number of tumor-derived CTC-clusters was significantly reduced in mice bearing LM2 plakoglobin shRNA-expressing tumors compared to control mice (Fig. 7D). In parallel, bioluminescence imaging of mouse lungs demonstrated a striking 80% reduction in lung nodules for mice bearing plakoglobin-suppressed primary tumors (Fig. 7E).
Together, our data suggest a model whereby plakoglobin-expressing regions within a primary tumor produce aggregated tumor cells, i.e. CTC-clusters, that are shed into the bloodstream, where they demonstrate rapid clearing at distant sites and enhanced metastatic potential (Fig. 7F). This mechanism of metastatic spread and the possibility that CTC-clusters may be targeted therapeutically through disruption of cell-cell junctions provide an opportunity for novel strategies to reduce the metastatic spread of breast cancer.
DISCUSSION
By applying microfluidic CTC isolation technologies to both patients with breast cancer and mouse models, we have characterized CTC-clusters, a striking but poorly understood feature of blood-borne metastasis. CTC-clusters have been observed in patients with cancers of different origin and using multiple technologies (Cho et al., 2012; Fidler, 1973; Liotta et al., 1976; Molnar et al., 2001; Stott et al., 2010; Yu et al., 2013). While most clusters are relatively small, some comprise dozens of tumor cells, raising the question of how they navigate through normal capillaries. Our in vivo flow cytometry studies indicate that clusters are more rapidly cleared from the circulation than single CTCs. Nonetheless, both the structural deformability of the aggregated cells within these clusters and the presence of vascular shunts within the circulation may allow a subset of these to circulate. The rapid clearance of clusters within distal tissues, together with their potentially increased cellular viability may underlie their dramatically enhanced metastatic potential. The increased metastatic propensity of CTC-clusters in reconstituted mouse models, together with the adverse prognosis of breast and prostate cancer patients with abundant CTC-clusters, support an important role for these cellular aggregates in the blood-borne spread of cancer.
Based on cellular tagging and mixing studies in the mouse, almost all CTC-clusters appear to be of oligoclonal origin, rather than being derived from the progeny of a single migratory cell. Our studies exclude intravascular aggregation of CTCs as a significant cause for CTC-clusters, demonstrating instead that they originate from a single tumor. We cannot determine whether their entry into the vascular space results from “grouped migration”, an active invasive process that has been described for epithelial cell masses (Friedl and Gilmour, 2009), or passive shedding into compromised tumor vasculature. Interestingly, the high expression of plakoglobin within foci of cells within the primary tumor raises the possibility that these demarcate the origin of clusters that ultimately enter the circulation. In mouse reconstitution models, plakoglobin knockdown in cells that constitute the primary tumor does not suppress tumorigenesis itself, but it abrogates the generation of CTC-clusters in the circulation and greatly reduces the number of metastatic deposits in the lung.
The identification of specific transcripts that enhance the metastatic potential of tumor cells may enable novel therapeutic strategies to suppress the blood-borne spread of cancer, a critical although challenging goal. To date, candidate metastasis genes have been derived primarily from mouse tumor models. Some, like inducers of EMT, alter the migratory properties of breast cancer epithelial cells and confer stem-like properties (Mani et al., 2008). In human breast cancer CTCs, we recently documented marked enrichment for mesenchymal transcripts in CTCs, using quantitative RNA-in-situ hybridization (Yu et al., 2013). In addition to generalized migratory properties associated with EMT, tissue-specific tropism studies in the mouse have identified subsets of genes involved in breast cancer metastases to lung (e.g. Epiregulin, CXCL1, SPARC and MMP2) (Minn et al., 2005), brain (e.g. COX2, HBEGF and ST6GALNAC5) (Bos et al., 2009), and bone (mainly driven by Src activation) (Zhang et al., 2009). A recent study interrogating candidate genes in breast CTCs derived from a patient with breast cancer has suggested that coexpression of EpCAM, CD44, CD47 and MET identifies a subset with increased metastatic capacity (Baccelli et al., 2013). While these candidate metastasis genes were not upregulated in CTC-clusters compared with single CTCs, our study was not designed to compare CTCs with their matched primary tumors. Finally, we note that our RNA expression studies focused on clusters comprised of tumor cells, since the negCTC-iChip favors isolation of small CTC-clusters. We therefore cannot exclude additional contributions from non-malignant cells within larger micro-emboli (Duda et al., 2010; Labelle et al., 2011; Stott et al., 2010), including potential stromal-derived tropism signals (Zhang et al., 2013).
Our study identifies novel mediators of metastasis by comparing two distinct populations of circulating tumor cells, one with very high metastatic potential (CTC-clusters) compared with the other (single CTCs). The development of advanced microfluidic CTC isolation technology (Ozkumur et al., 2013) enabled us to undertake such a detailed study of human breast cancer cells as they transiently circulate in the bloodstream of patients with metastatic disease. Single cell-resolution RNA sequencing demonstrated a very high level of concordance in expression patterns between matched CTC-clusters and single CTCs from individual breast cancer patients. We identified only a small number of candidate genes with significantly divergent expression (Table S2), including transcriptional regulators (XBP1), signaling molecules (AGR2 and HER3) and plakoglobin. While we focused this study on the functional characterization of plakoglobin due to the clinical association between high plakoglobin expression and adverse outcome in patients with breast cancer, additional CTC-cluster associated genes may be involved in their generation and their metastatic potential. The striking consequences of plakoglobin knockdown, suppressing both CTC-cluster generation and metastatic tumor formation in mouse models, points to this gene product being a major determinant of tumor dissemination. Plakoglobin contributes to both adherens junctions and desmosomes: in adherens junctions, the C-terminal intracellular domain of E-cadherin interacts in a mutually exclusive manner with either β-catenin or plakoglobin, which in turn associates with the actin-binding protein α-catenin (Harris and Tepass, 2010). At desmosomes, the intracellular domains of desmocolin and desmoglein interact with plakophilin and plakoglobin, which in turn binds the intermediate filament binding protein desmoplakin (Garrod and Chidgey, 2008). Thus, plakoglobin is a critical constituent of both adherens junctions and desmosomes, a role that may underlie its unique contribution to cell-to-cell adhesion in tumor cells. While plakoglobin has been implicated as both oncogene and tumor suppressor in different contexts (Hakimelahi et al., 2000; Kolligs et al., 2000; Shiina et al., 2005), it is neither in the model proposed here, functioning instead as an intercellular tether that confers added metastatic potential to tumor cells as they break off into the circulation. Interestingly, plakoglobin knockdown has far less impact on intercellular connections of non-transformed breast epithelial cells, which may benefit from additional adhesion mechanisms. This differential effect may offer an opportunity for therapeutic intervention.
In summary, our studies of CTCs in both breast and prostate cancer patients and mouse models point to CTC-clusters as critical mediators of cancer metastasis. These coexist with single migratory CTCs, making a contribution to the metastatic burden that far exceeds their comparatively small numbers in the circulation. The ability of tumor cell aggregates to detach from a primary tumor and maintain their cohesion as they survive in the bloodstream may identify a novel and potentially targetable step in the blood-borne dissemination of cancer.
EXPERIMENTAL PROCEDURES
CTC capture and identification
Blood specimens for CTC analysis were obtained after informed patient consent, per IRB protocol (05-300), at the Massachusetts General Hospital. A maximum of 20 ml of blood was drawn in EDTA vacutainers. Within four hours from blood draw, approximately 3 ml of blood was processed through the HBCTC-Chip or 6-12 ml of blood was processed through the negCTC-iChip. For mouse studies, blood was retrieved via cardiac puncture and approximately 1 ml of blood was processed through the HBCTC-Chip.
HBCTC-Chips were manufactured on site at the Massachusetts General Hospital Cancer Center/BioMEMS Resource Facility. For patient samples and mouse xenografts, chips were functionalized as previously described (Yu et al., 2013) with a cocktail of 10 μg/ml each of biotinylated antibodies against EpCAM (R&D Systems), EGFR (Cetuximab, Lilly) and HER2 (R&D Systems). For 4T1 mouse mammary tumor cells, chips were functionalized with a cocktail of antibodies against mouse EpCAM (BioLegend) and EGFR (Cetuximab, Lilly). Samples from patients with prostate cancer were processed as described (Miyamoto et al., 2012). negCTC-iChips were designed and fabricated as previously described (Ozkumur et al., 2013) (see Supplementary Methods).
Tumorigenesis assays
All mouse experiments were carried out in compliance with institutional guidelines. For tail vein experiments, NOD SCID Gamma (NSG) mice (Jackson Labs) were injected with 2×105 LM2 cells, 4×105 BT474 cells or 2×105 4T1 cells and monitored with IVIS® Lumina II (Caliper LifeSciences). For CTC-clusters metastatic potential assessment, 2×106 LM2-GFP (or 4T1-GFP) and 2×106 LM2-mCherry (or 4T1-mCherry) cells were prepared separately or mixed 1:1, suspended in 100 μl of 50% Basement Membrane Matix Phenol Red-free (BD Biosciences) in PBS and injected orthotopically in NSG mice. Blood draw for CTCs enumeration was performed five weeks after tumor onset. For plakoglobin knockdown experiments, 1×106 LM2-CTRL or LM2-Plakoglobin shRNA cells were suspended in 100 μl of 50% Basement Membrane Matrix Phenol Red-free in PBS and injected orthotopically in NSG mice. Blood draw for CTCs enumeration and lung metastasis analysis were performed four weeks after tumor onset.
Analysis of RNA Sequencing data
Determination of reads-per-million (rpm): color space reads were aligned using tophat and bowtie1 with the no-novel-juncs argument set with human genome version hg19 and transcriptome defined by the hg19 knownGene table from genome.ucsc.edu. Reads that did not align or aligned to multiple locations in the genome were discarded. The hg19 table knownToLocusLink from genome.ucsc.edu was used to map, if possible, each aligned read to the gene whose exons the read had aligned to. The reads count for each gene was the number of reads that were so mapped to that gene. This count was divided by the total number of reads that were mapped to any gene and multiplied by one million to form the reads-per-million (rpm) count. We used rpm rather than rpkm because we noted a 3′ bias in the alignments. Clustering algorithms are described under Supplementary Methods.
Supplementary Material
HIGHLIGHTS.
CTC-clusters originate as oligoclonal groups of cells from the primary tumor
CTC-clusters exhibit increased metastatic propensity compared to single CTCs
Abundance of CTC-clusters in patients denotes adverse outcome
Plakoglobin mediates CTC-cluster formation, enhancing metastatic spread
ACKNOWLEDGMENTS
We express our gratitude to all the patients who participated in this study. We thank C. Hart, A. McGovern, L.C. Davis and the Massachusetts General Hospital (MGH) clinical research coordinators for help with the clinical studies; Drs. P.S. Spuhler and T.A. Barber for support with the CTC-iChip technology; L. Libby and J. Brockmann for excellent technical support. This work was supported by grants for Breast Cancer Research Foundation (D.A.H), Stand Up to Cancer (D.A.H., M.T., S.M.), National Foundation for Cancer Research (D.A.H.), Howard Hughes Medical Institute (D.A.H.), NIH CA129933 (D.A.H.), NIBIB EB008047 (M.T., D.A.H.), Susan G. Komen for the Cure KG09042 (S.M.), NCI Federal Share Program and Income (S.M.), NIH P41 E8015903-02S1 (C.P.L.), NIH P50 CA086355-12 (C.P.L.) and the MGH-Johnson & Johnson Center for Excellence in CTCs (M.T., S.M.). N. Aceto is a fellow of the Human Frontiers Science Program, the Swiss National Science Foundation, and the Swiss Foundation for Grants in Biology and Medicine. Sequencing data have been deposited in the Gene Expression Omnibus database (accession no. GSE51827).
Footnotes
AUTHOR CONTRIBUTIONS N.A., D.A.H. and S.M. designed and performed the experiments, analyzed the data and wrote the manuscript. A.B. and D.T.M. provided clinical samples and analyzed clinical data. M.C.D., M.Y., A.E., H.Z. and B.W.B. performed immunofluorescence staining, CTCs isolation from patients and mouse blood samples, RNA amplification and library preparation. B.S.W. and S.R. analyzed the RNA sequencing data. J.A.S., A.P. and C.P.L. performed the in vivo flow cytometry. R.K., S.L.S., T.S., D.T.T. and M.T. provided the CTC isolation technology and the single cell RNA sequencing platform.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aktary Z, Pasdar M. Plakoglobin: role in tumorigenesis and metastasis. International journal of cell biology. 2012;2012:189521. doi: 10.1155/2012/189521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alford D, Taylor-Papadimitriou J. Cell adhesion molecules in the normal and cancerous mammary gland. Journal of mammary gland biology and neoplasia. 1996;1:207–218. doi: 10.1007/BF02013644. [DOI] [PubMed] [Google Scholar]
- Alix-Panabieres C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clinical chemistry. 2013;59:110–118. doi: 10.1373/clinchem.2012.194258. [DOI] [PubMed] [Google Scholar]
- Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, Klein C, Saini M, Bauerle T, Wallwiener M, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nature biotechnology. 2013;31:539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavallaro U, Christofori G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nature reviews Cancer. 2004;4:118–132. doi: 10.1038/nrc1276. [DOI] [PubMed] [Google Scholar]
- Cho EH, Wendel M, Luttgen M, Yoshioka C, Marrinucci D, Lazar D, Schram E, Nieva J, Bazhenova L, Morgan A, et al. Characterization of circulating tumor cell aggregates identified in patients with epithelial tumors. Physical biology. 2012;9:016001. doi: 10.1088/1478-3975/9/1/016001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DG, Duyverman AM, Kohno M, Snuderl M, Steller EJ, Fukumura D, Jain RK. Malignant cells facilitate lung metastasis by bringing their own soil. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21677–21682. doi: 10.1073/pnas.1016234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Khoury J, Hickman SE, Thomas CA, Cao L, Silverstein SC, Loike JD. Scavenger receptor-mediated adhesion of microglia to beta-amyloid fibrils. Nature. 1996;382:716–719. doi: 10.1038/382716a0. [DOI] [PubMed] [Google Scholar]
- Fidler IJ. The relationship of embolic homogeneity, number, size and viability to the incidence of experimental metastasis. European journal of cancer. 1973;9:223–227. doi: 10.1016/s0014-2964(73)80022-2. [DOI] [PubMed] [Google Scholar]
- Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nature reviews Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nature reviews Molecular cell biology. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- Garrod D, Chidgey M. Desmosome structure, composition and function. Biochimica et biophysica acta. 2008;1778:572–587. doi: 10.1016/j.bbamem.2007.07.014. [DOI] [PubMed] [Google Scholar]
- Hakimelahi S, Parker HR, Gilchrist AJ, Barry M, Li Z, Bleackley RC, Pasdar M. Plakoglobin regulates the expression of the anti-apoptotic protein BCL-2. The Journal of biological chemistry. 2000;275:10905–10911. doi: 10.1074/jbc.275.15.10905. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Harris TJ, Tepass U. Adherens junctions: from molecules to morphogenesis. Nature reviews Molecular cell biology. 2010;11:502–514. doi: 10.1038/nrm2927. [DOI] [PubMed] [Google Scholar]
- Kim MY, Oskarsson T, Acharyya S, Nguyen DX, Zhang XH, Norton L, Massague J. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolligs FT, Kolligs B, Hajra KM, Hu G, Tani M, Cho KR, Fearon ER. gamma-catenin is regulated by the APC tumor suppressor and its oncogenic activity is distinct from that of beta-catenin. Genes & development. 2000;14:1319–1331. [PMC free article] [PubMed] [Google Scholar]
- Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer cell. 2011;20:576–590. doi: 10.1016/j.ccr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledford H. Cancer theory faces doubts. Nature. 2011;472:273. doi: 10.1038/472273a. [DOI] [PubMed] [Google Scholar]
- Liotta LA, Saidel MG, Kleinerman J. The significance of hematogenous tumor cell clumps in the metastatic process. Cancer research. 1976;36:889–894. [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massague J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto DT, Lee RJ, Stott SL, Ting DT, Wittner BS, Ulman M, Smas ME, Lord JB, Brannigan BW, Trautwein J, et al. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer discovery. 2012;2:995–1003. doi: 10.1158/2159-8290.CD-12-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar B, Ladanyi A, Tanko L, Sreter L, Tulassay Z. Circulating tumor cell clusters in the peripheral blood of colorectal cancer patients. Clinical cancer research : an official journal of the American Association for Cancer Research. 2001;7:4080–4085. [PubMed] [Google Scholar]
- Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nature reviews Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen PI, Morgan B, Trautwein J, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Science translational medicine. 2013;5:179ra147. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson EJ, Khaled WT, Abell K, Watson CJ. Epithelial-to mesenchymal transition confers resistance to apoptosis in three murine mammary epithelial cell lines. Differentiation; research in biological diversity. 2006;74:254–264. doi: 10.1111/j.1432-0436.2006.00075.x. [DOI] [PubMed] [Google Scholar]
- Shiina H, Breault JE, Basset WW, Enokida H, Urakami S, Li LC, Okino ST, Deguchi M, Kaneuchi M, Terashima M, et al. Functional Loss of the gamma-catenin gene through epigenetic and genetic pathways in human prostate cancer. Cancer research. 2005;65:2130–2138. doi: 10.1158/0008-5472.CAN-04-3398. [DOI] [PubMed] [Google Scholar]
- Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Nordman E, Li B, Xu N, Bashkirov VI, Lao K, Surani MA. RNA-Seq analysis to capture the transcriptome landscape of a single cell. Nature protocols. 2010;5:516–535. doi: 10.1038/nprot.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarin D, Thompson EW, Newgreen DF. The fallacy of epithelial mesenchymal transition in neoplasia. Cancer research. 2005;65:5996–6000. doi: 10.1158/0008-5472.CAN-05-0699. discussion 6000-5991. [DOI] [PubMed] [Google Scholar]
- Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. The Journal of cell biology. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, Ciciliano JC, Smas ME, Winokur D, Gilman AJ, et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012;487:510–513. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Jin X, Malladi S, Zou Y, Wen YH, Brogi E, Smid M, Foekens JA, Massague J. Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell. 2013;154:1060–1073. doi: 10.1016/j.cell.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Wang Q, Gerald W, Hudis CA, Norton L, Smid M, Foekens JA, Massague J. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer cell. 2009;16:67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.