SUMMARY
In Alzheimer’s disease (AD) brain exposure of axons to Aβ causes pathogenic changes that spread retrogradely by unknown mechanisms affecting the entire neuron. We found that locally applied Aβ1–42 initiates axonal synthesis of a defined set of proteins including the transcription factor ATF4. Inhibition of local translation and retrograde transport or knockdown of axonal Atf4 mRNA abolished Aβ-induced ATF4 transcriptional activity and cell loss. Aβ1–42 injection into the dentate gyrus (DG) of mice caused loss of forebrain neurons whose axons project to the DG. Protein synthesis and Atf4 mRNA were upregulated in these axons, and co-injection of Atf4 siRNA into the DG reduced the effects of Aβ1–42 in the forebrain. ATF4 protein and transcripts were found with greater frequency in axons in the brain of AD patients. These results reveal an active role for intra-axonal translation in neurodegeneration and identify ATF4 as a mediator for the spread of AD pathology.
INTRODUCTION
β-amyloid pathology is a central component of Alzheimer’s disease (AD) and Aβ1–42 is considered causative for most neurodegenerative alterations in AD (Hardy and Selkoe, 2002). Accumulation of soluble oligomeric forms of Aβ1–42 is positively correlated with the onset of cognitive decline in AD brain, and it elicits neurodegeneration in primary neurons. As axons and dendrites are generally much larger than their cell bodies and project over long distances in the brain, elevated Aβ1–42 levels will first be sensed by neurites. Consequently, pathogenic signaling mechanisms will initially be triggered within neurites. Several aspects of AD pathogenesis such as tau hyperphosphorylation or impaired transport are first apparent in axons (Iqbal et al., 2009; Perlson et al., 2010), and local application of Aβ1–42 is sufficient to induce neurite degeneration (Ivins et al., 1998) and to interfere with retrograde axonal trafficking (Poon et al., 2013). Indeed, pathogenic changes within axons may be primary events driving the development of the classical pathological changes (Krstic and Knuesel, 2013). For example, in AD brains with amyloid plaques restricted to the cortex, subcortical neurons with cortical projections degenerate suggesting that axonal exposure to Aβ1–42 is sufficient to induce neurodegeneration over long distances (Liu et al., 2008). Similarly, in AD patients’ brains monoaminergic neurodegeneration occurs in the locus coeruleus in the absence of local Aβ pathology (Marcyniuk et al., 1986). Therefore, in order to understand the pathogenesis of AD it is crucial to investigate the intra-axonal signaling pathways triggered by Aβ1–42 separately from its effects on soma and dendrites.
Compartmentalized signaling is especially important for neurons, the most morphologically polarized cells. In order to react to stimuli in a spatially and temporally acute manner, axons are able to synthesize a subset of proteins locally (Jung et al., 2014). During development intra-axonal protein synthesis is crucial for growth cone behavior, axonal pathfinding, axon maintenance, and retrograde signaling (Jung et al., 2014). After the developmental period, the composition of the axonally localized transcriptome changes (Gumy et al., 2011), overall levels of mRNAs and ribosomes are lower (Kleiman et al., 1994), and mature axons have long been thought to be incapable of protein synthesis. However, recent evidence shows that protein synthesis persists in post-developmental CNS axons in vivo (Dubacq et al., 2009; Kar et al., 2014; Willis et al., 2011; Yoon et al., 2012). Additionally, upon injury of mature axons, a specific set of mRNAs and translation machinery are rapidly recruited into axons, and proteins are locally synthesized within mature axons (Rishal and Fainzilber, 2014). In contrast to its well-established role during development and regeneration, the role of intra-axonal protein synthesis in the context of neurodegenerative disorders remains unexamined.
Here, we asked whether intra-axonal protein synthesis was activated in response to Aβ1–42 and functionally relevant for the retrograde transmission of neurodegenerative signals across brain regions. We report that axonal translation is activated in response to Aβ1–42. Axonal ATF4 synthesis is required for the retrograde spread of Aβ1–42-induced neurodegeneration, and axons in brains of AD patients show more frequent localization of ATF4 protein and mRNA.
RESULTS
Local exposure to Aβ1–42 oligomers induces intra-axonal protein synthesis in hippocampal neurons
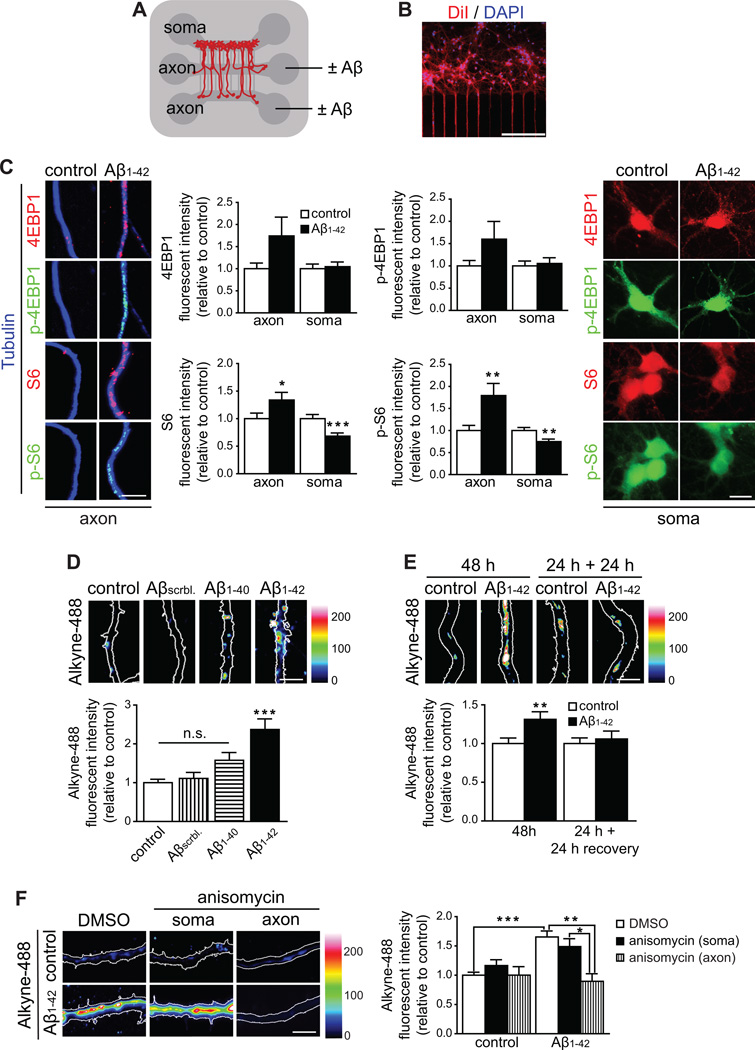
To investigate whether central nervous system (CNS) neurons locally synthesize proteins in axons in response to oligomeric Aβ1–42, rat embryonic hippocampal neurons were grown in tripartite microfluidic chambers which allow for the fluidic isolation of axons from cell bodies and dendrites (Figures 1A and 1B) (Hengst et al., 2009; Taylor et al., 2005). The small culture volume and the hydrophobicity of microfluidic chambers influences the effective concentrations of peptides (Toepke and Beebe, 2006). We used an Aβ1–42 concentration (3 µM) that is equivalent to ~250 nM in regular cultures (Figure S1A). Aβ concentrations in normal aging and AD brain range from ~2 pM to 2 µM, respectively (Wang et al., 1999). First we determined the axonal abundance of two molecular markers of translation: p-4EBP1 and p-S6. Levels for 4EBP1 and p-4EBP1 were non-significantly elevated, whereas S6 and p-S6 levels were significantly increased in axons upon Aβ1–42 treatment (Figure 1C). Cell body levels of 4EBP1 and p-4EBP1 did not change and levels of S6 and p-S6 were slightly reduced (Figures 1C). Aβ1–42 selectively applied to the cell body compartment caused an increase in both 4EBP1 and p-4EBP1 levels in the soma that did not propagate to the axonal compartment (Figures S1B and S1C). Next, we used bioorthogonal noncanonical amino acid tagging to detect newly synthesized proteins (Figure S1D). No local protein synthesis was detected in axons treated for 24 h with vehicle, a scrambled Aβ1–42 peptide or soluble oligomeric Aβ1–40 while Aβ1–42-treated axons exhibited a significant increase in L-azidohomoalanine (AHA) incorporation (Figure 1D). Protein synthesis was detected in axons exposed to Aβ1–42 for 48 h but not in axons treated for 24 h with Aβ1–42 followed by a 24 h recovery period, indicating that local protein synthesis does not persist after removal of Aβ1–42 (Figure 1E). AHA incorporation was prevented in the presence of the protein synthesis inhibitors anisomycin and emetine in the axonal but not the cell body compartment (Figures 1F and S1E). These results establish that axonally applied Aβ1–42 activates local protein synthesis within 24 h.
Figure 1. Locally applied Aβ1–42 oligomers induce intra-axonal protein synthesis.
(A) Scheme of a microfluidic chamber used to isolate axons of hippocampal neurons. Neurons were cultured in the upper compartment. Axons cross through two 200-µm-long microgroove barriers into the axonal compartments.
(B) Neuronal cell bodies were retrogradely labeled by applying DiI selectively to the axons. Typically between 40% (optical fields proximal to the microgrooves) and 30% (distal fields) of neurons were labeled indicating their axons had crossed the microgrooves. Scale bar, 200 µm.
(C) Hippocampal neurons were cultured in microfluidic chambers for 9–10 DIV and axons were treated with vehicle or Aβ1–42 for 24 h. Axons (left micrographs) and cell bodies (right micrographs) were immunostained for 4EBP1, p-4EBP1, S6 or p-S6. Mean ±SEM of 23–25 optical fields per condition (n=5 biological replicates per group). * p<0.05; **p<0.01; ***p<0.001. Scale bars, 5 µm (left micrographs), 20 µm (right micrographs).
(D) Axons were treated with vehicle, Aβscrambled, Aβ1–40 or Aβ1–42 for 24 h. 2 h prior to fixation, axons were sequentially incubated with AHA and 488-DIBO. Newly synthesized proteins were detected by the fluorescent signal (represented in pseudo color). Mean ±SEM of 25–35 optical fields per condition (n=5–7 biological replicates per group). ***p<0.001. Scale bars, 5 µm.
(E) Axons were treated with vehicle or Aβ1–42 for 48 h or for 48 h replacing the oligomer-containing medium with fresh 50% conditioned medium after 24 h. 2 h prior to sample processing axons were treated as in D. Mean ±SEM of 35–45 optical fields per condition (n=7–9 biological replicates per group). **p<0.01. Scale bar, 5 µm.
(F) Axons were treated with vehicle or Aβ1–42 for 24 h. 2 h and 30 min prior to fixation, axons were sequentially incubated with anisomycin or vehicle, and with AHA and 488-alkyne. Newly synthesized proteins were detected by their fluorescence signal (represented in pseudo color). Mean ±SEM of 25–65 optical fields per condition (5–13 biological replicates per group). *p<0.05; **p<0.01; ***p<0.001. Scale bar, 5 µm.
See also Figure S1.
Intra-axonal protein synthesis and retrograde transport are sequentially required for neurodegeneration triggered by axonal exposure to Aβ1–42
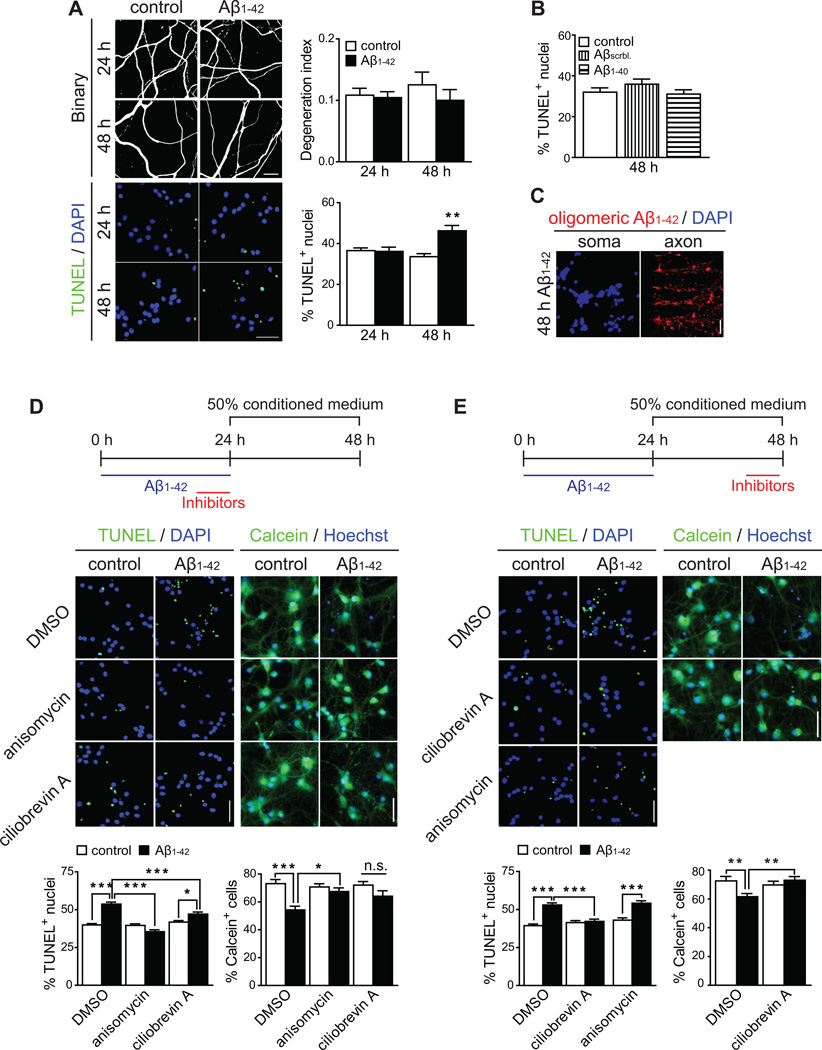
Application of Aβ1–42 to axons did not increase axonal fragmentation or cell death within 24 h, and after 48 h of Aβ1–42 exposure the number of TUNEL-positive neurons was significantly greater while axonal fragmentation was not induced (Figure 2A). This effect was specific for Aβ1–42 as neither the scrambled peptide nor Aβ1–40 had any effect on cell death (Figure 2B). The observed neurodegeneration at 48 h is the result of pathogenic changes originating in the axons as only extremely little Aβ1–42 was detected in the soma (Figure 2C).
Figure 2. Intra-axonal protein synthesis and retrograde transport are sequentially required for Aβ1–42-induced somatic degeneration.
(A) Axons were treated with vehicle or Aβ1–42 for 24 or 48 h. Fragmentation of axonal tubulin (upper micrographs) or nuclear TUNEL staining (lower micrographs) were measured. Mean ±SEM of 25–55 axonal fields per condition (upper graph, n=5–11 biological replicates per group) and 50–70 somatic fields per condition (lower graph, n=5–7 biological replicates per group). **p<0.01.
(B) Axons were treated with vehicle, Aβscrambled or Aβ1–40 for 48 h. TUNEL-positive nuclei were quantified. Mean ±SEM of 25–35 optical fields per condition (n=5–7 biological replicates).
(C) Immunostaining for Aβ1–42 on axons and cell bodies.
(D) Inhibitors were applied to axons during the last 6 h of the 24 h Aβ1–42 treatment period. The culture medium from the axonal compartments was then replaced with 50% conditioned medium and cells were allowed to recover. Cell death (left panels) or survival (right panels), were assessed by TUNEL and Calcein staining, respectively. Mean ±SEM of 50–70 somatic fields stained for TUNEL per condition (left graph) and 25–31 somatic fields stained for Calcein (right graphs) per condition (n=5–7 biological replicates per group). *p<0.05; ***p<0.001.
(E) Inhibitors were applied to axons during the last 6 h of the 48 h experimental period. Cell death and survival were assessed as before. Mean ±SEM of 50–100 somatic fields stained for TUNEL per condition (left graph) and 30 somatic fields stained for Calcein (right graphs) (n=5–10 biological replicates). *p<0.05; ***p<0.001.
Scale bars, 50 µm. See also Figure S2.
To test whether Aβ1–42-induced intra-axonal protein synthesis was required for the induction of cell death, axons were treated with vehicle or Aβ1–42 for 24 h in the absence or presence of anisomycin or emetine. To minimize toxic side effects of the protein synthesis inhibitors, axons were exposed to them only during the last 6 h of the Aβ1–42 treatment period. A significant increase in TUNEL-positive and corresponding decrease in Calcein-positive neurons were observed upon treatment of axons with Aβ1–42 (Figures 2D and S2A). Inhibition of intra-axonal protein synthesis completely abolished the effect of axonally applied Aβ1–42, demonstrating that intra-axonal protein synthesis is required for Aβ1–42-induced cell death.
To investigate whether transport from axons to soma was required for Aβ1–42-induced neurodegeneration we used the retrograde transport inhibitors ciliobrevin A and EHNA. Both inhibitors significantly reduced retrograde movement of axonal lysosomes in microfluidic chambers (Figures S2B). When applied during the last 6 h of the 24 h Aβ1–42 treatment period, ciliobrevin A only partially abolished Aβ1–42-mediated cell death while EHNA had no effect (Figures 2D and S2A). However, both inhibitors completely abolished Aβ1–42-dependent cell death when applied during the last 6 h of the 48 h experiment, while application of anisomycin at this time did not interfere with cell death (Figures 2E and S2C), consistent with our finding that axonal protein synthesis is not persistent after the removal of Aβ1–42 (Figure 1E). To ensure that the effect of the inhibitors was not due to alterations in the minute levels of Aβ1–42 transported to the cells bodies, axons were treated as before, and cell bodies were immunostained for Aβ1–42. No correlation was found between somatic Aβ1–42 levels and cell death (Figure S2D). These results establish that sequential intra-axonal protein synthesis and retrograde transport are required to transmit a neurodegenerative signal to the neuronal cell bodies in response to axonal Aβ1–42 application.
The transcription factor ATF4 is locally synthesized in axons exposed to Aβ1–42
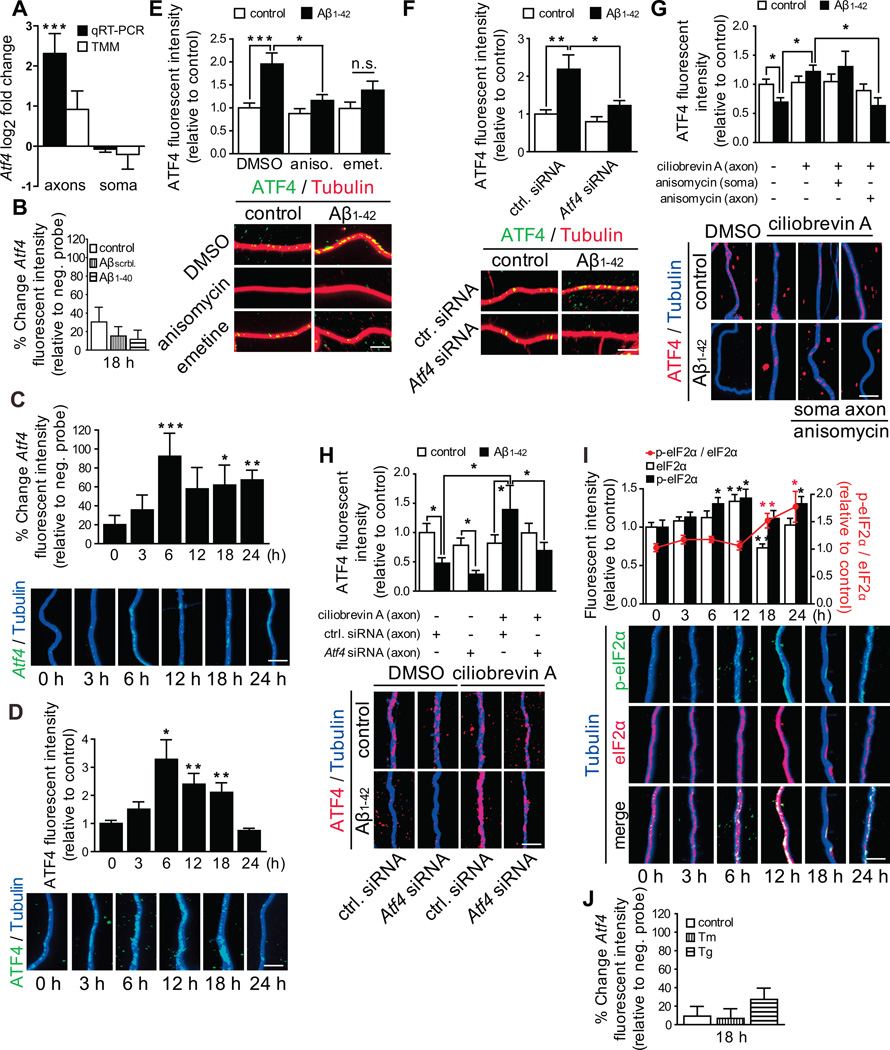
To identify proteins that might transmit the neurodegenerative signal from axons to the soma, we performed RNA sequencing on total RNA isolated from vehicle and Aβ1–42-treated axons and their cell bodies. Only mRNAs with higher expression levels than previously reported non-axonal transcripts were included in our analysis (Figure S3A). The axonal transcriptomes of control and treated axons show only partial overlap (Figure S3B), indicating that exposure of axons to Aβ1–42 triggers the recruitment of a specific cohort of mRNAs (Supplemental Table 1). Among the axonally recruited mRNAs was the transcript coding for activating transcription factor 4 (ATF4). As a transcription factor ATF4 is a prime candidate for a retrogradely transported protein, and it can suppress the transcription of memory related genes and activate the transcription of proapoptotic genes in response to intracellular stress (Ameri and Harris, 2008). Additionally, ATF4 is a key molecule of the unfolded protein response (UPR) pathway (Ron and Harding, 2012), which is activated in many neurodegenerative diseases, possibly including AD (Ma et al., 2013). Comparative analysis of the RNA-seq datasets and quantitative RT-PCR revealed an increase in axonal Atf4 abundance following Aβ1–42 treatment while levels in cell bodies were unchanged, indicating that the upregulation of Atf4 in axons is likely the result of increased axonal transport rather than transcription (Figures 3A). No increase in Atf4 was detected in axons treated with vehicle control, Aβscrambled, or Aβ1–40 for 18 h (Figure 3B).
Figure 3. Atf4mRNA is recruited into Aβ1–42-treated axons, and axonal ATF4 protein is locally synthesized and retrogradely transported.
(A) Log2 fold change for Atf4 mRNA as determined by real time RT-PCR and DESeq2 (TMM). ***p<0.001.
(B) Hippocampal neurons were cultured in microfluidic chamber for 9–10 DIV, axons were treated with vehicle, Aβscrambled or Aβ1–40 for 18 h, and axonal Atf4 mRNA levels were measured by quantitative FISH. Mean ±SEM of 25–30 optical fields per condition (n=5–6 biological replicates).
(C) Axons were treated with Aβ1–42 for the indicated times, and axonal Atf4 mRNA levels were measured by quantitative FISH. Mean ±SEM of 25–40 axonal fields per condition (n=5–8 biological replicates per group). The background fluorescence was determined using a non-targeting probe (neg. probe) and set to zero. *p<0.05; **p<0.01; ***p<0.001. Scale bar, 5 µm.
(D) Neurons were cultures and treated as in C. Axonal ATF4 protein levels were measured by quantitative immunofluorescence. Mean ±SEM of 20–40 axonal fields per condition (n=4–8 biological replicates per group). *p<0.05; **p<0.01. Scale bar, 5 µm.
(E) Hippocampal neurons were cultured and treated as in B. 3 h prior to sample processing axons were treated with DMSO, anisomycin or emetine. Axonal ATF4 protein levels were determined by quantitative immunofluorescence. Mean ±SEM of 25–35 axonal fields per condition (n=5–7 biological replicates per group). ***p<0.001: *p<0.05. Scale bar, 5 µm.
(F) Hippocampal neurons were cultured in microfluidic chambers for 8 DIV. Axons were transfected with a control (ctrl.) siRNA or a siRNA targeting Atf4. 24 h after transfection axons were treated with vehicle or Aβ1–42 for 18 h. ATF4 protein levels were measured by quantitative immunofluorescence. Mean ±SEM of 35–55 axonal fields per condition (n=7–11 biological replicates per group). **p<0.01: *p<0.05.
(G) Axons were treated with vehicle or Aβ1–42 for 24h, in the presence or absence of ciliobrevin A for 6h. Anisomycin was added to the cell body or the axonal compartment for 3 h. Axons were immunostained for ATF4 protein. Mean ±SEM of 30–40 axonal fields per condition (n=6–8 biological replicates per group). *p<0.05.
(H) Axons were transfected with a control siRNA or siRNAs targeting Atf4 mRNA and treated with Aβ1–42 and ciliobrevin A as in G. Axons were immunostained for ATF4 protein. Mean ±SEM of 30–40 axonal fields per condition (n=6–8 biological replicates per group). *p<0.05.
(I) Neurons were cultured and treated as in C. eIF2α and p-eIF2α levels were determined by quantitative immunofluorescence. Mean ±SEM of 20–35 axonal fields per condition (n=4–7 biological replicates per group).
(J) Neurons were cultured as in B. Axonal were treated for 18 h with tunicamycin (Tm) or thapsigargin (Tg) and Atf4 mRNA levels were determined by quantitative FISH. Mean ±SEM of 30 optical fields per condition (n=6 biological replicates).
Scale bars, 5 µm. See also Figure S3 and Supplemental Table S1.
Axonal Atf4 mRNA levels determined by quantitative fluorescent in situ hybridization (FISH) were significantly increased following 6 h of Aβ1–42 treatment and remained elevated until at least 24 h (Figure 3C). Similarly, ATF4 protein levels were significantly increased at 6, 12 and 18 h of Aβ1–42 treatment but dropped to lower than control levels at 24 h (Figure 3D). The increase in ATF4 at 18 h was abolished by the local application of protein synthesis inhibitors (Figure 3E) that did not affect Atf4 mRNA localization in Aβ1–42-treated axons (Figure S3C). To unambiguously demonstrate local translation of Atf4 in axons we transfected Atf4 targeting siRNAs into axons. The RNAi pathway is functional in axons, enabling knockdown of axonal mRNAs without affecting somato-dendritic mRNA levels (Hengst et al., 2006). Neither Atf4 siRNA significantly altered Atf4 levels in control axons but both blocked the increase of Atf4 in Aβ1–42-treated axons, with siRNA 1 decreasing Atf4 levels below control conditions (Figure S3D). In all subsequent experiments siRNA 1 was used. The siRNA’s effect was restricted to axons as ATF4 mRNA and protein levels were unchanged in cell bodies (Figures S3E and S3F). Selective knockdown of axonal Atf4 completely inhibited the increase in axonal ATF4 protein levels following 18 h of exposure to Aβ1–42 (Figure 3F).
To test whether the drop in axonal ATF4 abundance at 24 h of Aβ1–42 treatment was due to ATF4 transport to the soma, we applied retrograde transport inhibitors locally. Axonal ATF4 levels were significantly increased in axons after 24 h of Aβ1–42 treatment when retrograde transport was inhibited (Figure 3G, 3F and S3G), but axonal Atf4 mRNA levels were unchanged (Figure S3H). Inhibition of intra-axonal but not somatic protein synthesis completely abolished the Aβ1–42-dependent increase of axonal ATF4 in the presence of ciliobrevin A (Figure 3G). ATF4 protein levels were significantly decreased in control or Atf4 siRNA transfected axons exposed to Aβ1–42, and the accumulation of ATF4 in ciliobrevin A treated axons in response to Aβ1–42 was completely abolished in Atf4 siRNA transfected axons (Figure 3H). These results establish that local application of Aβ1–42 oligomers induces local ATF4 synthesis and its retrograde transport.
Aβ1–42 triggers moderate eIF2a activation
Atf4 belongs to a group of transcripts, whose translation is activated by phosphorylation of the translation initiation factor eIF2α (Ron and Harding, 2012). Total eIF2α levels in axons were significantly increased by 12 h of Aβ1–42 treatment but returned to control levels by 24 h (Figure 3I). p-eIF2α levels were significantly increased starting at 6 h, first due to the increase in total eIF2α and starting at 18 h due to an increase in the p-eIF2α/eIF2α ratio (Figure 3I). The increase in p-eIF2α was much lower than the increase in Atf4 mRNA levels indicating the increase in axonal ATF4 protein might be primarily driven by increased Atf4 localization. At 24 h, when we had observed strong upregulation of general protein synthesis in axons, we also detected a significant activation of eIF2α. There are four mammalian eIF2α-kinases, including the ER stress activated kinase PERK (Wek et al., 2006). Two activators of ER stress, tunicamycin and thapsigargin, did not trigger axonal recruitment of Atf4 mRNA at 18 h (Figure 3J), but both efficiently initiated ER-stress in neuronal cell bodies (Figure S4A), suggesting that local ER stress does not phenocopy the effect of Aβ1–42 oligomers on Atf4 mRNA recruitment.
Axonally synthesized ATF4 induces gene expression in cell bodies and mediates retrograde somatic degeneration via CHOP
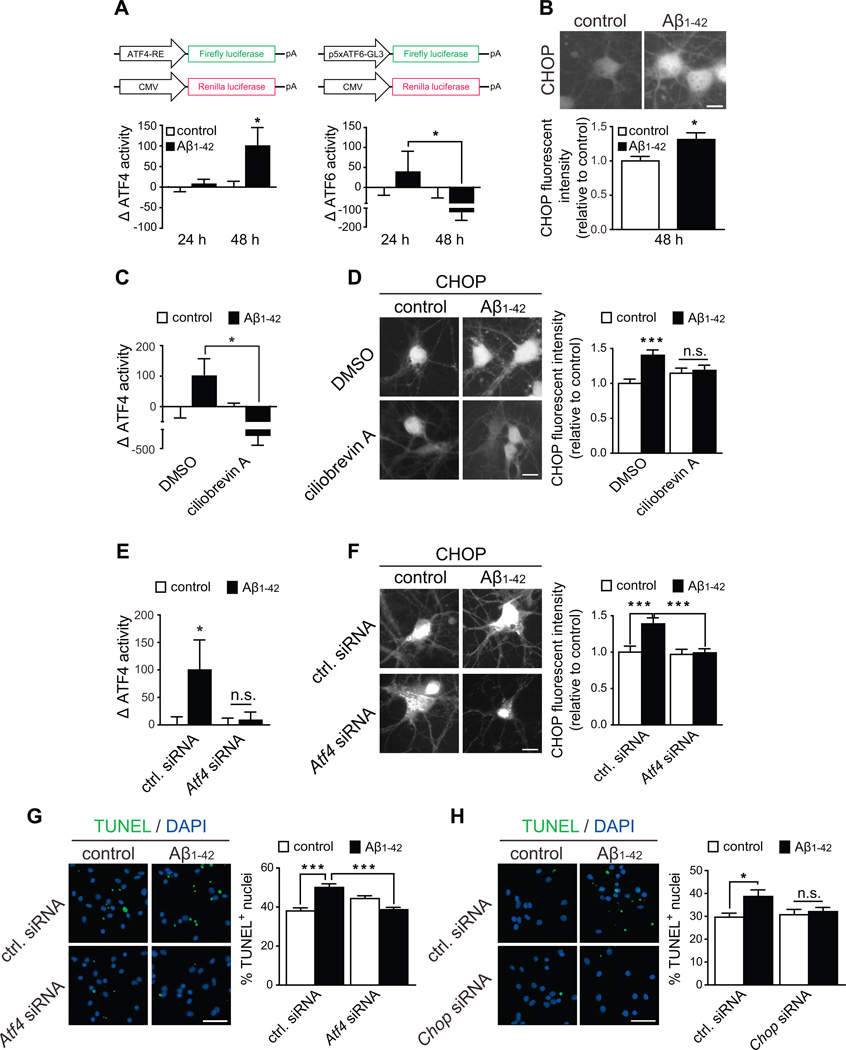
Next we asked if axonally derived ATF4 would function as a transcription factor in response to local application of Aβ1–42 using an ATF4 firefly luciferase reporter gene construct. We also included an ATF6 luciferase reporter (Wang et al., 2000) to investigate whether Aβ1–42 causes local ER stress leading to the activation of the ATF6 arm of the UPR. ATF4- or ATF6-dependent luciferase transcription was efficiently detected upon treatment of cell bodies with tunicamycin or thapsigargin for 24 h (Figure S4A). No firefly luciferase activity of either construct could be detected when axons were treated with vehicle, or following 24 h of Aβ1–42 treatment (Figure 4A). However, 48 h after Aβ1–42 exposure a significant increase in cell body ATF4 abundance (Figure S4B) and transcriptional activity was observed (Figure 4A, left graph) while ATF6 activity remained undetectable (Figure 4A, right graph). Thus, axonal exposure to Aβ1–42 induces ATF4-but not ATF6-dependent transcription. Next we analyzed somatic expression of CHOP, a transcriptional target of ATF4 (Averous et al., 2004), following 48 h of Aβ1–42 treatment. CHOP expression was significantly increased in cell bodies in response to axonal Aβ1–42 but not Aβscrambled or Aβ1–40 exposure (Figures 4B and S4C).
Figure 4. Axonally synthesized ATF4 induces ATF4-dependent gene expression in the nucleus and leads to retrograde somatic degeneration via CHOP.
(A) Neurons were grown in microfluidic chambers and cell bodies were transfected with the reporter gene constructs 24 h before local exposure of axons to Aβ1–42. Luciferase activities were measured in cell lysates 24 and 48 h after axons had been treated with vehicle or Aβ1–42. Data are plotted as the ratio Firefly(RLU)/Renilla(RLU) and normalized to vehicle. The maximum increase in Firefly(RLU) activity per experiment was set to 100%. Mean ±SEM of 7–12 biological replicates per condition. *p<0.05.
(B) CHOP levels were measured in cell bodies by quantitative immunofluorescence after 48 h of local application of Aβ1–42 to axons. Mean ±SEM of 30–40 microscopy fields per condition (n=6–8 biological replicates per group). *p<0.05. Scale bar, 20 µm.
(C) Neurons were cultured as in A and axons were exposed to Aβ1–42 oligomers for 48 h. 6 h prior to luciferase measurement axons were exposed to vehicle or ciliobrevin A. Mean ±SEM of 6–10 biological replicates per condition. *p<0.05.
(D) Axons were treated as in C. CHOP levels were measured in cell bodies by quantitative immunofluorescence. Mean ±SEM of 35–45 optical fields per condition (n=7–9 biological replicates per group). ***p<0.001. Scale bar, 20 µm.
(E) Neurons were cultured as in A and axons were transfected with control or Atf4 siRNA 24 h before Aβ1–42 treatment. Luciferase activities were measured and represented as in A. Mean ±SEM of 10–12 biological replicates per condition. *p<0.05.
(F) Axons were treated as in E. CHOP levels in cell bodies were measured by quantitative immunofluorescence. Mean ±SEM of 30–40 microscopy fields (n=6–8 biological replicates per group). ***p<0.001. Scale bar, 20 µm.
(G) Neurons were cultured and treated as in E. Cell bodies were processed for TUNEL staining. Mean ±SEM of 70–90 microscopy fields (n=7–9 biological replicates per group). ***p<0.001. Scale bar, 50 µm.
(H) Neurons were cultured as in A and cell bodies were transfected with control or Chop siRNA 24 h before Aβ1–42 treatment. Cell bodies were processed for TUNEL staining after 48 h of Aβ1–42 application to axons. Mean ±SEM of 60 microscopy fields (n=6 biological replicates per group). *p<0.05. Scale bar, 50 µm. See also Figure S4.
We then asked whether activation of ATF4-dependent gene expression was mediated by axonally synthesized ATF4. The ATF4 increase in cell bodies after Aβ1–42 exposure was fully blocked by axonally applied anisomycin and partially blocked by ciliobrevin A (Figure S4D). Thus, we treated axons with Aβ1–42 for 48 h, adding ciliobrevin A 6 h prior to sample processing and assessed ATF4 activity via luciferase and CHOP expression assays. In both assays inhibition of retrograde transport completely abolished the effect of axonal Aβ1–42 (Figures 4C and 4D), and knockdown of axonal Atf4 prevented Aβ1–42-dependent transcription of luciferase, CHOP expression, or increase of ATF4 in cell bodies (Figures 4E, 4F and S4E), demonstrating that axonally synthesized ATF4 is required for ATF4-dependent gene expression after axonal Aβ1–42 treatment.
Prolonged CHOP expression leads to cell death (Zinszner et al., 1998), and therefore, we asked if Aβ1–42-dependent neurodegeneration was mediated by axonally synthesized ATF4. A significant induction of apoptosis and corresponding decrease in Calcein staining was found when control siRNA transfected axons were treated with Aβ1–42, whereas depletion of axonal Atf4 mRNA fully rescued the cells (Figures 4G and S4F). Additionally, Aβ1–42 significantly increased the amount of TUNEL-positive nuclei in cell bodies transfected with control siRNA, but Chop knockdown blocked Aβ1–42-mediated neurodegeneration (Figure 4H).
These results reveal that local application of Aβ1–42 triggers the intra-axonal synthesis and retrograde transport of ATF4, and these events are required for ATF4-dependent transcription leading to CHOP-dependent cell loss.
Atf4 is locally translated in cholinergic axons in the mouse brain in response to Aβ
Next, we used a mouse model of semi-acute amyloidopathy by intra-hippocampal injection of Aβ1–42 oligomers to analyze the in vivo relevance of our in vitro findings (Sotthibundhu et al., 2008). In contrast to the more widely used transgenic mouse models for Aβ1–42 amyloidopathy, this model allows the spatially restricted and temporally acute exposure of axons to elevated Aβ1–42 levels. Intra-hippocampal injection of oligomeric Aβ1–42 induces neurodegeneration of basal forebrain cholinergic neurons (BFCNs) within 2 weeks post-injection (Sotthibundhu et al., 2008). BFCNs project their axons ipsi-laterally to the hippocampus (Leranth and Frotscher, 1989), allowing the contra-lateral injection of vehicle to be utilized as a control in the same animal. Also, with the exception of very few cholinergic neuronal cell bodies in the dentate hilus, which can easily be avoided, choline acetyltransferase (ChAT) immunoreactivity in the dentate gyrus (DG) is a specific marker for BFCN axons (Leranth and Frotscher, 1989).
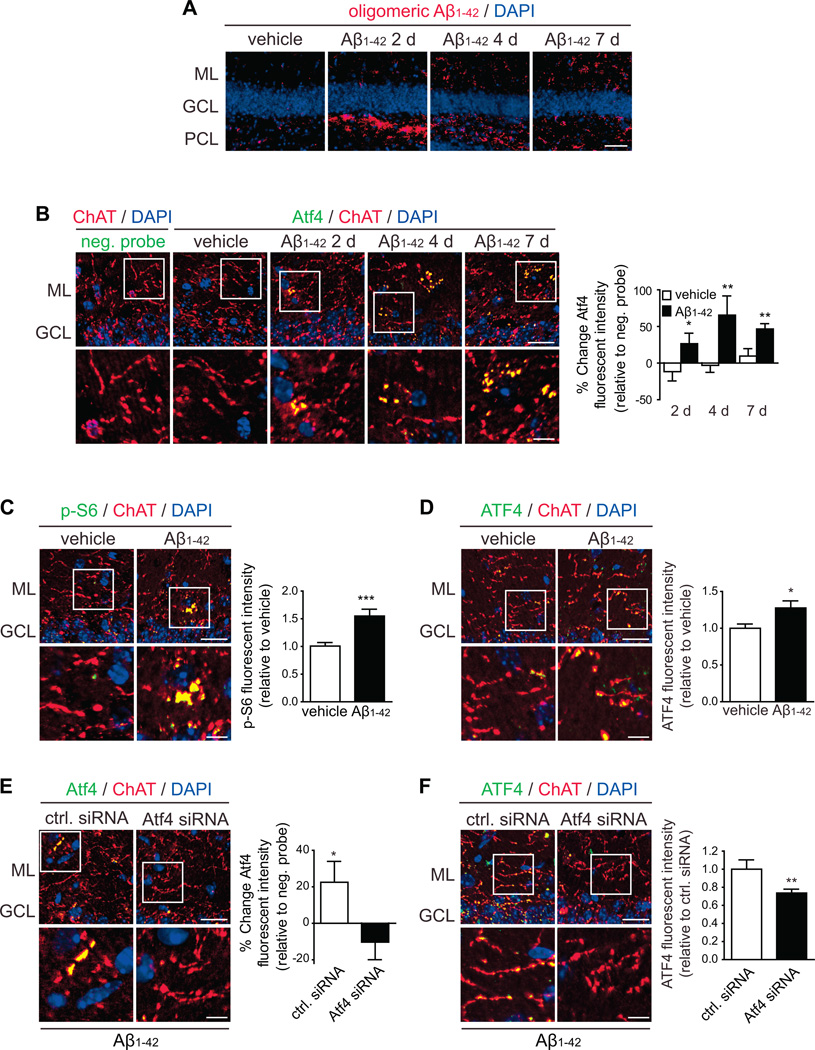
We injected Aβ1–42 into the DG and analyzed brain sections 2 to 7 days post injection (DPI) at sites adjacent to the injection where the DG layers were intact (Figures 5A, S5A and S5B). First, we confirmed the presence of oligomeric Aβ1–42 in these sites at 2, 4 and 7 DPI (Figure 5A). Atf4 mRNA was readily detectable above background levels in cholinergic axons in all layers of the DG 2, 4 and 7 DPI in Aβ1–42- but not vehicle-injected hemispheres (Figures 5B). ChAT staining appeared to be more punctate in the vicinity of cell bodies, especially in the granule cell layer (GCL) and co-localized with synaptophysin staining in control hemispheres (Figure S5B), consistent with the known termination pattern of BFCN axons. Puncta were more evident over time in Aβ1–42-injected hemispheres suggesting synaptic/neuritic retraction. Atf4 granules were frequently found in these puncta, possibly indicating their localization to synaptic terminals and/or retracting synapses. However, no reduction in ChAT-positive features was seen in Aβ1–42-injected hemispheres even 7 DPI (Figure S5C). Also, no Atf4 above background was observed in granule cell bodies under any condition (Figure S5D).
Figure 5. Intra-hippocampal injection of Aβ1–42 induces synthesis of ATF4 in BFCN axons.
(A) Presence of Aβ1–42 in the DG of mice injected with vehicle and Aβ1–42 oligomers 2 to 7 DPI. 4 to 5 mice were analyzed per condition. ML, molecular layer; GCL, granule cell layer; PCL, polymorphic cell layer. Scale bar, 50 µm.
(B) FISH for Atf4 mRNA in the DG of mice injected with vehicle and Aβ1–42. BFCN axons were identified by ChAT immunostaining. Cell bodies were counterstained with DAPI. Mean ±SEM of measurements performed in 3–4 brain slices per mouse (n=4 mice per group). Background fluorescence was determined non-targeting probe signal and set to zero. *p<0.05; **p<0.01. Scale bars, 20 µm, 5 µm (insets).
(C) Phosphorylation levels of ribosomal protein S6 within ChAT-positive axons were measured by quantitative immunofluorescence on brain sections 7 DPI. Mean ±SEM of measurements typically performed in 4 brain slices per mouse (n=4 mice). ***p<0.001.Scale bars, 20 µm, (insets, 5 µm).
(D) ATF4 protein levels within ChAT-positive axons were measured by quantitative immunofluorescence on brain sections 7 DPI. Mean ±SEM of measurements typically performed in 4 brain slices per mouse (n=4 mice). *p<0.05. Scale bars, 20 µm, (insets, 5 µm).
(E) Mice were injected with Aβ1–42 oligomers in both hemispheres of the brain. The left hemisphere was co-injected with a control (ctrl.) siRNA and the right hemisphere with an Atf4 siRNA. The presence of Atf4 mRNA within ChAT-positive axons was analyzed by FISH 7 DPI. Mean ±SEM of measurements typically performed in 3 brain slices per mouse (n=3 mice). Background fluorescence was determined non-targeting probe signal and set to zero. *p<0.05. Scale bars, 20 µm, (insets, 5 µm).
(F) Mice were injected as in E. ATF4 protein levels within ChAT-positive axons were measured by quantitative immunofluorescence on brain sections 7 DPI. Mean ±SEM of measurements typically performed in 4 brain slices per animal (n=4 mice). **p<0.01. Scale bars, 20 µm, (insets, 5 µm).
See also Figure S5.
p-S6 and ATF4 levels were significantly increased within ChAT-positive axons in the Aβ1–42-injected side 7 DPI (Figures 5C and 5D). In granule cells, a moderate increase in p-S6 and a strong upregulation of ATF4 were detected (Figures S5E and S5F), indicating that both axons and cell bodies respond to Aβ1–42 by increasing ATF4 levels. To confirm synthesis of ATF4 within BFCN axons, both hemispheres of the brain were injected with Aβ1–42, and either a control siRNA or an Atf4 siRNA. At 7 DPI, Atf4 siRNA caused a completed knockdown of axonal Atf4 mRNA and significant reduction of ATF4 protein (Figures 5E and 5F) without causing axonal loss (Figure S5I). ATF4 protein was significantly reduced, and Atf4 mRNA remained undetectable in granule cells (Figures S5G and S5H). These results demonstrate that axons in the mature mammalian brain synthesize ATF4 and likely other proteins in response to Aβ1–42.
Axonally synthesized ATF4 is required to transmit a neurodegenerative signal from the DG to BFCNs
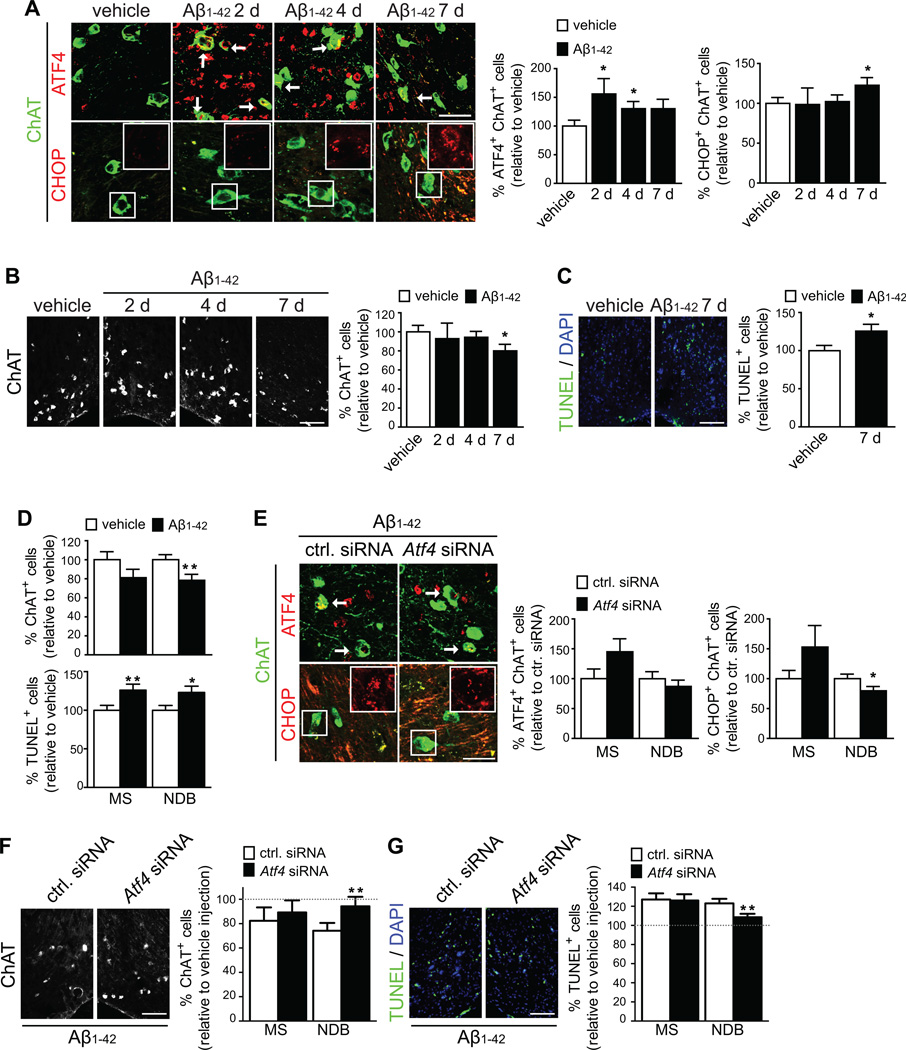
Next we investigated if ATF4-dependent gene expression was induced in BFCNs. Fluorogold was co-injected into both hemispheres of the brain to define the region of the basal forebrain from which axons close to the injection site originated. The unaffected detection of retrogradely transported fluorogold in the basal forebrains of all mice (Figure S6A and S6B) suggests that cholinergic afferents were functional and capable of retrograde transport. ATF4 levels in BFCNs were significantly increased at 2 and 4 DPI with a non-significant increase at 7 DPI (Figure 6A, upper panels and left graph). ATF4 induction was evident in cholinergic neurons but not in all neurons present in the basal forebrain (Figures S5C). CHOP positive cholinergic neurons were significantly increased at 7 DPI (Figure 6A, lower panels and right graph), indicating that ATF4-dependent gene expression was induced in the basal forebrain.
Figure 6. Intra-axonal synthesis of ATF4 leads to neurodegeneration in the adult mouse brain.
(A) Mice were injected with vehicle in the left hemisphere of the brain and with Aβ1–42 in the contralateral hemisphere. Sections of the basal forebrain were immunostained for ChAT and ATF4 or CHOP 2 to 7 DPI. Mean ±SEM of positive cells relative to vehicle in ~8 brain slices per animal (n=4–5 mice per condition). *p<0.05. Scale bar, 50 µm.
(B) ChAT-positive neurons in the basal forebrain of injected mice. Mean ±SEM of ChAT-positive neurons relative to the vehicle injected side in ~8 brain slices per animal (n=4–5 mice per condition). *p<0.05. Scale bar, 100 µm.
(C) TUNEL-positive cells in the basal forebrain of injected mice 7 DPI. Mean ±SEM of TUNEL-positive cells relative to the vehicle injected side in ~8 brain slices per mouse (n=5 mice). *p<0.05. Scale bar, 100 µm.
(D) Comparison of the effect of Aβ1–42 injection on ChAT- and TUNEL-positive cells in the MS and NDB 7 DPI. Mean ±SEM of positive cells in ~8 brain slices per mouse (n=5 mice). *p<0.05; **p<0.01.
(E) Aβ1–42 injections were performed in both hemispheres of the brain. A control (ctrl.) siRNA was co-injected into the left hemisphere and an Atf4 siRNA was co-injected in the right hemisphere. Basal forebrain sections were immunostained for CHAT and ATF4 or CHOP. ATF4- and CHOP-positive cholinergic neurons were quantified in the MS and NDB. Mean ±SEM of double-positive cells relative to ctrl. siRNA in ~8 brain sections per animal (n=5 mice). *p<0.05. Scale bar, 50 µm.
(F) Mice were injected as in E. ChAT-positive neurons in the basal forebrain of injected mice were quantified in the MS and NDB. Mean ±SEM of ChAT-positive neurons relative to ctrl. siRNA in ~8 brain slices per animal (n=5 mice per condition). **p<0.01. Scale bar, 100 µm.
(G) TUNEL-positive cells in the forebrain of injected mice. Mean ±SEM of TUNEL-positive cells relative to ctrl. siRNA in ~8 brain slices per mouse (n=5 mice). **p<0.01. Scale bar, 100 µm.
See also Figure S6 and Supplemental Table S2.
Next we quantified the number of ChAT-positive neurons to determine if Aβ1–42 injected in the hippocampus was sufficient to induce neurodegeneration of BFCNs at some point between 2 and 7 days. Aβ1–42 injection did not change the number of ChAT-positive neurons in the forebrain at 2 or 4 DPI, but caused a significant ~20% reduction at 7 DPI (Figure 6B). Conversely, no overall decrease was seen in NeuN-positive neurons (Figure S6C, right graph) as expected considering that not only BFCNs resides in the basal forebrain. We confirmed these results using stereology as a complimentary approach (Figure S6D and S6E, left graphs). A significant ~24% increase in TUNEL-positive cells was found in the Aβ1–42-injected hemisphere compared to the control hemisphere (Figure 6C). These results demonstrate that Aβ1–42 injection into the hippocampus induces retrograde degeneration of BFCNs.
We had observed that ATF4 and CHOP induction was uneven across the basal forebrain suggesting a greater response of BFCNs in the nucleus in the diagonal band (NDB) than in the medial septum (MS). Indeed, a significant decrease in BFCNs was apparent only in the NDB, whereas cell death affected both nuclei to a similar extent (Figures 6D and S6D, right graph), suggesting that cells other than BFCNs degenerate in the MS in response to Aβ1–42 injection. Next we determined whether ATF4-dependent signaling in the basal forebrain required Aβ1–42-dependent Atf4 synthesis in cholinergic axons in the hippocampus. Consistent with our previous observations that ATF4 protein was not significantly induced in the basal forebrain at 7 DPI, no reduction was detected in Atf4 siRNA injected hemispheres (Figure 6E, upper panels, left graph). However, Aβ1–42-dependent CHOP induction was significantly reduced by Atf4 siRNA in the NDB (Figure 6E, lower panels, right graph). Thus, synthesis of ATF4 in the hippocampus induces ATF4-dependent signaling in BFCNs.
Finally, we sought to determine whether axonally derived ATF4 was required for the loss of BFCNs. Co-injection of Atf4 siRNA blocked the decrease in density of BFCNs in the NDB, in contrast to the MS, which remained unaffected (Figures 6F and S6F, Supplemental Table S2). When compared to non-siRNA conditions (dashed lines in Figures 6F and 6G), Atf4 siRNA reduced the number of TUNEL-positive cells in the NDB by ~63% but restored the number of ChAT-positive neurons to normal levels. This discrepancy indicates that other cells in the forebrain die as well, but only BFCNs die in an Atf4-dependent manner. Additionally axonally synthesized ATF4 might cause a loss of cholinergic phenotype in BFCNs, as is suggested by the fact the number of NeuN positive cells does not decrease significantly in the forebrain upon Aβ1–42 injection.
We observed a significant thinning of the GCL and increased cell death in DG exposed to Aβ1–42 (Figure S6G), but Atf4 siRNA had no effect on the thickness of the GCL (Figure S6H, left graph), and far from rescuing dying cells, Atf4 siRNA exacerbated cell death in the DG (Figure S6H, right graph). Thus, the decrease in BFCNs was not caused by neurodegeneration in the hippocampus.
Atf4 mRNA granules and ATF4 protein are present in processes in human AD brains
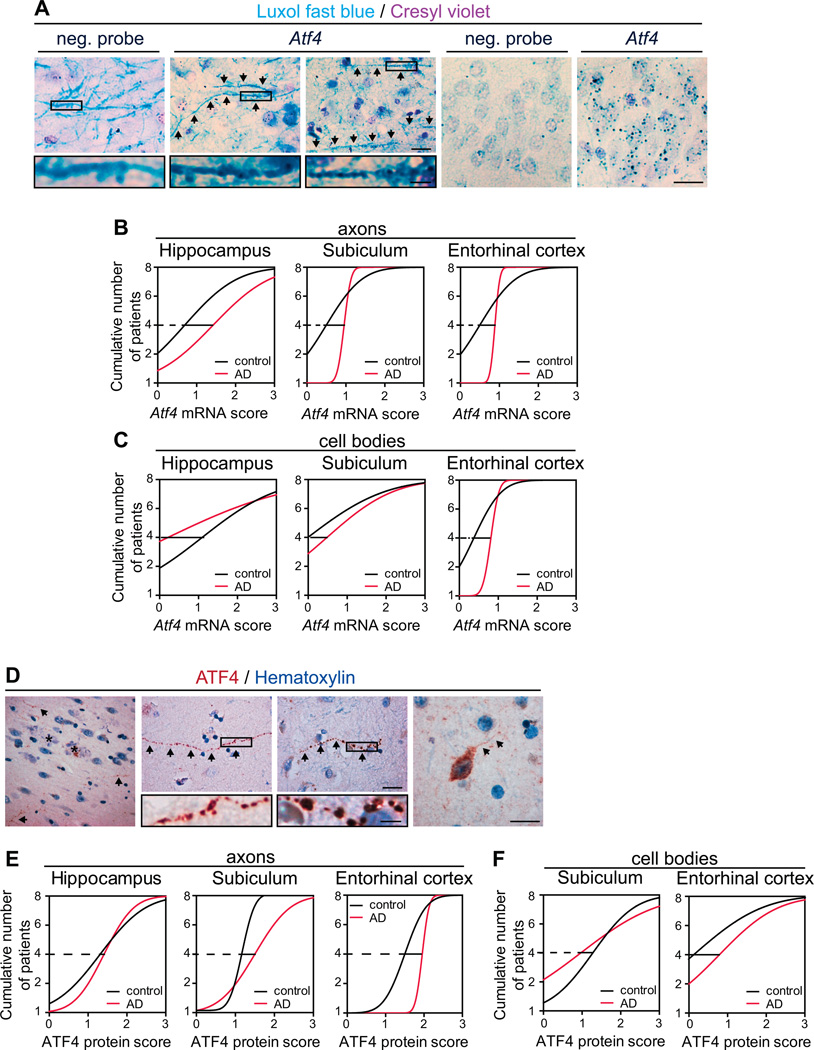
Finally, we analyzed the presence of ATF4 mRNA and protein in post mortem brain samples of 8 AD patients and 8 age-matched controls. Axons and cell bodies containing Atf4 mRNA granules were found in the hippocampal formation in all cases (Figure 7A). However, AD brains exhibited a higher frequency of Atf4-containing axons in the hippocampus, the subiculum, and the entorhinal cortex (Figure 7B). A decrease was observed in Atf4-positive cell bodies in the hippocampus of AD brains, but a higher frequency was found in the subiculum and entorhinal cortex (Figure 7C). In AD brains, ATF4-positive processes could be observed in the vicinity of amyloid plaques (Figure 7D). ATF4 was found in relatively intact processes and in beaded neurites (Figure 7D). More ATF4-positive axonal structures were found in the subiculum and the entorhinal cortex but not the hippocampus of AD brains (Figure 7E). ATF4-positive cell bodies (Figure 7D) were generally restricted to the subiculum and entorhinal cortex for both control and AD cases, with a higher frequency in the entorhinal cortex for AD cases (Figure 7F). The increased frequencies of ATF4 mRNA and protein in axons in the subiculum and entorhinal cortex of AD patients are highly suggestive of intra-axonal ATF4 synthesis in those regions of the brain that are especially vulnerable in AD (Khan et al., 2014). The results from human brain samples, although correlative, closely mirror our findings in hippocampal neurons and in the adult mouse brain providing evidence for the pathophysiological significance of our proposed model (Figure S6I).
Figure 7. Presence of Atf4 mRNA granules and ATF4 protein in axons and axonal-like structures in the AD brain.
(A) Representative micrographs of Atf4 mRNA granules in axons and cell bodies in human brain samples. Panels 1–3: axons stained with luxol fast blue and a negative probe or an Atf4-targeting probe. Atf4-containing axons are indicated with arrows. Panels 4–5: examples of granule cells stained with cresyl violet and a negative or Atf4-targeting probe. Scale bars, 20 µm (Insets, 5 µm).
(B) Cumulative frequency distributions of Atf4-containing axons in the hippocampus, the subiculum, and the entorhinal cortex of control and AD cases (n=8 brains per condition).
(C) Cumulative frequency distributions of Atf4-containing cell bodies in the hippocampus, the subiculum, and the entorhinal cortex of control and AD cases (n=8 brains per condition).
(D) Representative micrographs of ATF4 protein in processes and cell bodies in human brain samples. First panel: an ATF4-positive process (arrows) in the vicinity of amyloid plaques (asterisks). Second panel: a relatively intact ATF4-positive process. Third panel: a beaded process. Fourth panel: A positive cell body and neurite (arrows). Scale bars, 20 µm (insets, 5 µm).
(E) Cumulative frequency distributions of ATF4-positive processes axons in the hippocampus, the subiculum, and the entorhinal cortex of control and AD cases (n=8 brains per condition).
(F) Cumulative frequency distributions of ATF4-positive cell bodies in the subiculum and the entorhinal cortex of control and AD cases (n=8 brains per condition).
DISCUSSION
Several prior studies have demonstrated the importance of local translation for axon maintenance (Yoon et al., 2012), mitochondrial function (Kar et al., 2014) and survival (Cox et al., 2008), and suppression of local translation of lb2 mRNA causes neurodegeneration in vivo (Yoon et al., 2012). Here we report another dimension of local protein synthesis: in response to a physiologically relevant neurodegenerative stimulus axonal protein synthesis plays an active role in the transmission of neurodegeneration. Rather than acting solely as a factor in cellular homeostasis, local protein synthesis can be a major component of neuronal dyshomeostasis under pathological conditions.
Our finding that oligomeric Aβ1–42 application to distal axons triggers the rapid recruitment and local translation of a distinct set of mRNAs is reminiscent of the activation of local translation upon nerve injury (Rishal and Fainzilber, 2014). However, the changes to the axonal transcriptome appear to be unique to the exposure of distal axons to oligomeric Aβ1–42. For example we find that the transcriptome of Aβ1–42-treated axons contains mRNAs of many AD related genes, including transcripts for 4 out of the current list of 20 AD susceptibility loci (Lambert et al., 2013): APP, ApoE, Clu, and FERMT2. These proteins function in Aβ1–42 production (APP) and metabolism (Apoe, Clu), and have been implicated in tau pathology (FERMT2) (Shulman et al., 2014). The post-transcriptional regulation of these genes by Aβ1–42 suggests that these proteins might function in feedback mechanisms downstream of amyloid-pathology in AD.
Transcriptional changes in AD brain or in response to Aβ1–42 have been extensively studied in various experimental settings (Miller and Geschwind, 2010). While these studies have provided valuable insight into the signaling pathways affected in Aβ1–42 pathology, many of the mRNAs we identified as regulated by Aβ1–42 in axons have never before been described to be changed in response to Aβ1–42. This is likely due to the fact that they are post-transcriptionally regulated, rather than by increased promoter activity; in fact we did not observe an overall up- or down-regulation for the vast majority of the axonally localized mRNAs. Our study is thus a demonstration that post-transcriptional mechanisms of gene expression must be taken into account when investigating changes in gene expression. Especially in morphologically polarized cells such as neurons, mRNA localization can be as functionally relevant as transcriptional regulation, and disorders of the nervous system cannot be completely understood without the consideration of translational mechanisms.
We found that the increase of ATF4-positive BFCNs is greater than the observed cell loss, suggesting a model in which ATF4 is not directly leading to the transcription of pro-apoptotic genes but rather triggers the expression of a variety of genes whose functions cause pathogenic changes in the neurons, leading to cell death as a secondary effect. The finding that Atf4 siRNA is more efficient in rescuing the loss of ChAT-positive BCFNs than in preventing apoptosis supports this model. Our finding that BFCNs in the MS and NDB react differentially to Aβ1–42 injection into the DG indicates that the exact transcriptional response to axonally derived ATF4 differs between cell types. In fact, depending on the context, ATF4 in neurons has variously been described as pro-apoptotic, pro-survival, or memory suppressing (Ameri and Harris, 2008). It is possible that in response to low-levels of eIF2α phosphorylation, as has been seen in AD patients’ brains and AD model mice (Ma et al., 2013), ATF4 acts mainly in a neuroprotective and memory suppressing manner while upon prolonged exposure to Aβ1–42 it can contribute to cell death.
Our study adds to a growing body of evidence that some transcription factors are axonally synthesized (Ji and Jaffrey, 2014). It remains an unanswered question what might be the advantage of synthesizing a transcription factor in axons. In the case of ATF4 an appealing idea is that local synthesis might favor dimerization with an otherwise outcompeted binding partner. ATF4 binds promoter sequences either as a homodimer or a heterodimer (Ameri and Harris, 2008). The relative abundance of potential binding partners in axons could favor the formation of other heterodimers in axons than in cell bodies leading to differential transcriptional activities.
AD progression is characterized by the spread of pathology throughout the brain. Interfering with the spread would be an ideal approach to slow the decline of cognitive function that is characteristic of AD. Our results unravel a mechanism for the spread of disease that is based on the retrograde transport of ATF4. In this model, the exposure of axons to pathological levels of Aβ1–42 leads to neuron-wide pathogenic changes due to pathogenic alterations in gene expression. Our finding that siRNA-mediated knockdown of Atf4 mRNA in axons alone is sufficient to prevent neurodegeneration in response to acutely applied Aβ1–42 in vivo indicates a an unexpected target for a future therapy. Indeed, small molecules exist that could be used to repress ATF4 expression in the brain (Moreno et al., 2013; Sidrauski et al., 2013).
In conclusion, we describe a pathway through which a neurodegenerative signal is transmitted from the periphery of neurons to the soma across macroscopic distances in the brain. Our findings provide a mechanistic explanation for the spread of parts of the pathological changes in AD brain and potentially indicate new avenues for the development of therapeutic interventions for AD.
EXPERIMENTAL PROCEDURES
Extended Experimental Procedures can be found in Supplemental Information.
Axon specific treatment in vitro
To apply peptides, inhibitors or siRNA specifically to axons, rat embryonic hippocampal neurons were grown in tripartite microfluidic chambers with two 200-µm-long microgrooves barriers (Taylor et al., 2005). Synthetic Aβ1–42 peptides were oligomerized (Stine et al., 2003) and applied to the axonal compartment at 3 µM at 9–10 DIV. Whenever stated, the axonal or cell body compartments were treated with 10 µM anisomycin, 500 nM emetine, 30 µM ciliobrevin A, 10 µM EHNA, 10 µg ml−1 tunicamycin, or 1 µM thapsigargin, or transfected with siRNA using NeuroPORTER (Genlantis, San Diego, CA).
RNA-seq Analysis
Axons were exposed to Aβ1–42 or vehicle for 24 h. Total RNA was purified from the cell bodies and axons using the PrepEase RNA isolation kit (Affymetrix, Santa Clara, CA). cDNA libraries were created using the TruSeq RNA Sample Preparation Kit (Illumina, San Diego, CA). Sequencing was performed on an Illumina MiSeq instrument (paired-end, 2× 150 bp) with biological replicates. Reads were aligned to the rat genome (Rn5) and counted using DESeq2.
Real time RT-PCR
Total RNA from the cell bodies and the axonal compartments was isolated as above, reverse transcribed, pre-amplified with the TaqMan PreAmp Kit (Life Technologies, Carlsbad, CA) and real time RT-PCR was performed with TaqMan Gene Expression master mix and the Atf4 gene expression set (Rn00824644_g1). Gene expression was normalized to input RNA.
Fluorescent In Situ Hybridization (FISH)
Atf4 mRNA in hippocampal neurons was detected by quantitative FISH using a mixture of in vitro transcribed, digoxigenin-labeled riboprobes following establish protocols (Hengst et al., 2009). FISH on sections of mouse and human brain was performed with RNAscope Multiplex Fluorescent Reagent Kit (Advanced Cell Diagnostics, Hayward, CA) according to manufacturer’s instructions.
Luciferase Assay
Cell bodies of hippocampal neurons were transfected using NeuroPORTER with an ATF4-(Promega) or ATF6-firefly reporter (Addgene, Cambridge, MA) and a Renilla luciferase construct (Promega). Firefly luciferase activity was measured 24 or 48 h after Aβ1–42 treatment using the Dual-Luciferase Reporter Assay System (Promega).
Aβ1–42 injection experiments
Stereotaxic were performed following Sotthibundhu et al. (2008). 9–12-month-old C57Bl/6J mice were anesthetized, and placed in a stereotaxic frame (Stoelting, Wood Dale, IL). Stereotaxic injections were conducted using convection-enhanced delivery at a rate of 0.5 µl min−1 using the Quintessential Stereotaxic Injector (Stoelting) (coordinates from bregma: anterior-posterior, −2.00 mm; medial-lateral, ±1.3 mm; dorsal-ventral, −2.2 mm) resulting in an estimated Aβ1–42 concentration in the DG of ~30 nM. Guidelines for the care and use of laboratory animals were followed for all mouse experimentation.
Brain samples
Post mortem brain samples of AD patients and age-matched controls were obtained from the New York Brain Bank. 8 µm paraffin embedded sections were analyzed histochemically for the presence of ATF4 protein or by RNAscope for Atf4 mRNA.
Statistical Analyses
When comparing multiple groups, one-way ANOVA followed by Bonferroni post-hoc test was performed. To compare two groups, t-tests were used.
Supplementary Material
HIGHLIGHTS.
Locally applied Aβ1–42 triggers recruitment of mRNAs into axons and local translation.
ATF4 is locally synthesized and retrogradely transported in response to Aβ1–42.
Knockdown of axonal Atf4 mRNA reduces Aβ1–42-induced neurodegeneration in vivo.
ATF4 transcript and protein levels are increased in axons in the brain of AD patients.
ACKNOWLEDGMENTS
This work was supported by the Alzheimer’s Association (NIRG-10-171721, U.H.), NINDS (NS081333, C.M.T.), and pilot study awards from the NIA-funded ADRC at Columbia University (AG008702, J.B. and Y.Y.J) that also supports the New York Brain Bank. We thank S. Cano and staff in the Personalized Genomic Medicine laboratory for their contribution; H. Moore for assistance with stereology; J.P. Vonsattel for access to human samples; and members of the Hengst group for comments and discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.B. with C.A.W. performed and analyzed most experiments. C.M.T proposed and established the in vivo model, Y.Y.J. performed the Aβ1–42 injections. P.L.N. and J.B. performed the RNA-sequencing. J.F.C. and J.B. performed and analyzed the experiments on human samples. U.H. conceived the project; U.H. and J.B. designed the experiments, analyzed the RNA sequencing data with assistance from P.L.N., and wrote the manuscript.
REFERENCES
- Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol. 2008;40:14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- Averous J, Bruhat A, Jousse C, Carraro V, Thiel G, Fafournoux P. Induction of CHOP expression by amino acid limitation requires both ATF4 expression and ATF2 phosphorylation. J Biol Chem. 2004;279:5288–5297. doi: 10.1074/jbc.M311862200. [DOI] [PubMed] [Google Scholar]
- Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10:149–159. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubacq C, Jamet S, Trembleau A. Evidence for developmentally regulated local translation of odorant receptor mRNAs in the axons of olfactory sensory neurons. J Neurosci. 2009;29:10184–10190. doi: 10.1523/JNEUROSCI.2443-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy LF, Yeo GS, Tung YC, Zivraj KH, Willis D, Coppola G, Lam BY, Twiss JL, Holt CE, Fawcett JW. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17:85–98. doi: 10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- Hengst U, Cox LJ, Macosko EZ, Jaffrey SR. Functional and selective RNA interference in developing axons and growth cones. J Neurosci. 2006;26:5727–5732. doi: 10.1523/JNEUROSCI.5229-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengst U, Deglincerti A, Kim HJ, Jeon NL, Jaffrey SR. Axonal elongation triggered by stimulus-induced local translation of a polarity complex protein. Nat Cell Biol. 2009;11:1024–1030. doi: 10.1038/ncb1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Liu F, Gong CX, Alonso Adel C, Grundke-Iqbal I. Mechanisms of tau-induced neurodegeneration. Acta Neuropathol. 2009;118:53–69. doi: 10.1007/s00401-009-0486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivins KJ, Bui ET, Cotman CW. β-amyloid induces local neurite degeneration in cultured hippocampal neurons: evidence for neuritic apoptosis. Neurobiol Dis. 1998;5:365–378. doi: 10.1006/nbdi.1998.0228. [DOI] [PubMed] [Google Scholar]
- Ji SJ, Jaffrey SR. Axonal transcription factors: novel regulators of growth cone-to-nucleus signaling. Dev Neurobiol. 2014;74:245–258. doi: 10.1002/dneu.22112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Gkogkas CG, Sonenberg N, Holt CE. Remote control of gene function by local translation. Cell. 2014;157:26–40. doi: 10.1016/j.cell.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar AN, Sun CY, Reichard K, Gervasi NM, Pickel J, Nakazawa K, Gioio AE, Kaplan BB. Dysregulation of the axonal trafficking of nuclear-encoded mitochondrial mRNA alters neuronal mitochondrial activity and mouse behavior. Dev Neurobiol. 2014;74:333–350. doi: 10.1002/dneu.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan UA, Liu L, Provenzano FA, Berman DE, Profaci CP, Sloan R, Mayeux R, Duff KE, Small SA. Molecular drivers and cortical spread of lateral entorhinal cortex dysfunction in preclinical Alzheimer's disease. Nat Neurosci. 2014;17:304–311. doi: 10.1038/nn.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiman R, Banker G, Steward O. Development of subcellular mRNA compartmentation in hippocampal neurons in culture. J Neurosci. 1994;14:1130–1140. doi: 10.1523/JNEUROSCI.14-03-01130.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krstic D, Knuesel I. Deciphering the mechanism underlying late-onset Alzheimer disease. Nat Rev Neurol. 2013;9:25–34. doi: 10.1038/nrneurol.2012.236. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, Destefano AL, Bis JC, Beecham GW, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leranth C, Frotscher M. Organization of the septal region in the rat brain: cholinergic-GABAergic interconnections and the termination of hippocampo-septal fibers. J Comp Neurol. 1989;289:304–314. doi: 10.1002/cne.902890210. [DOI] [PubMed] [Google Scholar]
- Liu Y, Yoo MJ, Savonenko A, Stirling W, Price DL, Borchelt DR, Mamounas L, Lyons WE, Blue ME, Lee MK. Amyloid pathology is associated with progressive monoaminergic neurodegeneration in a transgenic mouse model of Alzheimer's disease. J Neurosci. 2008;28:13805–13814. doi: 10.1523/JNEUROSCI.4218-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Trinh MA, Wexler AJ, Bourbon C, Gatti E, Pierre P, Cavener DR, Klann E. Suppression of eIF2α kinases alleviates Alzheimer's disease-related plasticity and memory deficits. Nat Neurosci. 2013;16:1299–1305. doi: 10.1038/nn.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcyniuk B, Mann DM, Yates PO. The topography of cell loss from locus caeruleus in Alzheimer's disease. J Neurol Sci. 1986;76:335–345. doi: 10.1016/0022-510x(86)90179-6. [DOI] [PubMed] [Google Scholar]
- Miller JA, Geschwind DH. Transcriptional Changes in Alzheimer’s Disease. In: Choi S, editor. In Systems Biology for Signaling Networks. New York: Springer; 2010. pp. 611–643. [Google Scholar]
- Moreno JA, Halliday M, Molloy C, Radford H, Verity N, Axten JM, Ortori CA, Willis AE, Fischer PM, Barrett DA, et al. Oral Treatment Targeting the Unfolded Protein Response Prevents Neurodegeneration and Clinical Disease in Prion-Infected Mice. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3006767. 206ra138. [DOI] [PubMed] [Google Scholar]
- Perlson E, Maday S, Fu MM, Moughamian AJ, Holzbaur EL. Retrograde axonal transport: pathways to cell death? Trends Neurosci. 2010;33:335–344. doi: 10.1016/j.tins.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon WW, Carlos AJ, Aguilar BL, Berchtold NC, Kawano CK, Zograbyan V, Yaopruke T, Shelanski M, Cotman CW. β-Amyloid (Aβ) oligomers impair brain-derived neurotrophic factor retrograde trafficking by down-regulating ubiquitin C-terminal hydrolase, UCH-L1. J Biol Chem. 2013;288:16937–16948. doi: 10.1074/jbc.M113.463711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rishal I, Fainzilber M. Axon-soma communication in neuronal injury. Nat Rev Neurosci. 2014;15:32–42. doi: 10.1038/nrn3609. [DOI] [PubMed] [Google Scholar]
- Ron D, Harding HP. Protein-folding homeostasis in the endoplasmic reticulum and nutritional regulation. Cold Spring Harb Perspect Biol. 2012;4:a013177. doi: 10.1101/cshperspect.a013177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman JM, Imboywa S, Giagtzoglou N, Powers MP, Hu Y, Devenport D, Chipendo P, Chibnik LB, Diamond A, Perrimon N, et al. Functional screening in Drosophila identifies Alzheimer's disease susceptibility genes and implicates Tau-mediated mechanisms. Hum Mol Genet. 2014;23:870–877. doi: 10.1093/hmg/ddt478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C, Acosta-Alvear D, Khoutorsky A, Vedantham P, Hearn BR, Li H, Gamache K, Gallagher CM, Ang KK, Wilson C, et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. eLife. 2013;2:e00498. doi: 10.7554/eLife.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotthibundhu A, Sykes AM, Fox B, Underwood CK, Thangnipon W, Coulson EJ. β-amyloid1–42 induces neuronal death through the p75 neurotrophin receptor. J Neurosci. 2008;28:3941–3946. doi: 10.1523/JNEUROSCI.0350-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stine WB, Jr, Dahlgren KN, Krafft GA, LaDu MJ. In vitro characterization of conditions for amyloid-β peptide oligomerization and fibrillogenesis. J Biol Chem. 2003;278:11612–11622. doi: 10.1074/jbc.M210207200. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Blurton-Jones M, Rhee SW, Cribbs DH, Cotman CW, Jeon NL. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat Methods. 2005;2:599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toepke MW, Beebe DJ. PDMS absorption of small molecules and consequences in microfluidic applications. Lab on a Chip. 2006;6:1484–1486. doi: 10.1039/b612140c. [DOI] [PubMed] [Google Scholar]
- Wang J, Dickson DW, Trojanowski JQ, Lee VM. The levels of soluble versus insoluble brain Aβ distinguish Alzheimer's disease from normal and pathologic aging. Exp Neurol. 1999;158:328–337. doi: 10.1006/exnr.1999.7085. [DOI] [PubMed] [Google Scholar]
- Wang Y, Shen J, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J Biol Chem. 2000;275:27013–27020. doi: 10.1074/jbc.M003322200. [DOI] [PubMed] [Google Scholar]
- Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem Soc Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- Willis DE, Xu M, Donnelly CJ, Tep C, Kendall M, Erenstheyn M, English AW, Schanen NC, Kirn-Safran CB, Yoon SO, et al. Axonal Localization of transgene mRNA in mature PNS and CNS neurons. J Neurosci. 2011;31:14481–14487. doi: 10.1523/JNEUROSCI.2950-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon BC, Jung H, Dwivedy A, O'Hare CM, Zivraj KH, Holt CE. Local translation of extranuclear lamin B promotes axon maintenance. Cell. 2012;148:752–764. doi: 10.1016/j.cell.2011.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinszner H, Kuroda M, Wang X, Batchvarova N, Lightfoot RT, Remotti H, Stevens JL, Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.