Abstract
In Rhizobium leguminosarum bv. viciae, quorum-sensing is regulated by CinR, which induces the cinIS operon. CinI synthesizes an AHL, whereas CinS inactivates PraR, a repressor. Mutation of praR enhanced biofilms in vitro. We developed a light (lux)-dependent assay of rhizobial attachment to roots and demonstrated that mutation of praR increased biofilms on pea roots. The praR mutant out-competed wild-type for infection of pea nodules in mixed inoculations. Analysis of gene expression by microarrays and promoter fusions revealed that PraR represses its own transcription and mutation of praR increased expression of several genes including those encoding secreted proteins (the adhesins RapA2, RapB and RapC, two cadherins and the glycanase PlyB), the polysaccharide regulator RosR, and another protein similar to PraR. PraR bound to the promoters of several of these genes indicating direct repression. Mutations in rapA2, rapB, rapC, plyB, the cadherins or rosR did not affect the enhanced root attachment or nodule competitiveness of the praR mutant. However combinations of mutations in rapA, rapB and rapC abolished the enhanced attachment and nodule competitiveness. We conclude that relief of PraR-mediated repression determines a lifestyle switch allowing the expression of genes that are important for biofilm formation on roots and the subsequent initiation of infection of legume roots.
Introduction
The infection of legume roots by rhizobia, leading to the formation of nitrogen-fixing nodules, is a clonal event and each individual bacterium that initiates an infection can grow rapidly, giving rise to over 106 progeny in nodules. It is somewhat of a lottery which individual soil Rhizobium will succeed in infecting roots and nodules, but critical to success is the ability of rhizobia to attach to legume roots and root hairs at potential infection sites (Downie, 2010). This attachment involves the secretion of both proteins that act as adhesins and different surface polysaccharides (Milner et al., 1992; Bittinger et al., 1997; Pobigaylo et al., 2008; Williams et al., 2008; Janczarek et al., 2009). Rhizobia attached to roots and root hairs are in an ideal position to detect plant-made signals such as flavonoids, which induce the production of nodulation signals (Nod-factors), that induce plant programmes for initiating infection and stimulating nodule morphogenesis (Oldroyd and Downie, 2008).
In view of the selective advantages of growing on exudates from root hairs and from infecting nodules, it is not surprising that rhizobia have multiple ways of attaching to roots and root hairs. This can involve different surface polysaccharides, secreted proteins and production of Nod-factor signalling molecules (Downie, 2010). Under slightly acidic conditions, a plant lectin expressed on root-hair tips promotes attachment of Rhizobium leguminosarum biovar viciae (R.l. viciae) to pea root hairs by binding to a polar-localized bacterial surface polysaccharide called glucomannan (Laus et al., 2006). Blocking glucomannan synthesis by the gmsA mutation did not block nodulation, but greatly reduced the ability of the mutants to compete with WT for nodule infection in competitive nodulation tests (Williams et al., 2008). Under slightly alkaline conditions, the lectin-glucomannan-mediated attachment is poor and a secreted calcium-binding protein called rhicadhesin becomes the dominant mechanism of attachment (Smit et al., 1987; 1989; Laus et al., 2006). The gene encoding rhicadhesin has not been identified, but a search for secreted proteins that attach to the surface of R. leguminosarum identified a group of rhizobial adhering proteins (Raps) that promote attachment and aggregation by rhizobia (Ausmees et al., 2001). These Rap proteins are calcium-binding lectins containing cadherin-like domains that bind to the acidic exopolysaccharide (Abdian et al., 2013), which is essential for attachment both in vitro and to root hairs (Russo et al., 2006; Williams et al., 2008). Increased expression of one of these proteins, RapA1, enhanced attachment to roots and increased infection competitiveness (Mongiardini et al., 2008; 2009). Cellulose fibrils play a role in biofilm growth (called cap formation) on root hairs after initial attachment, although a mutant unable to produce cellulose nodulated normally and apparently was not reduced for nodulation competitiveness (Smit et al., 1987; Laus et al., 2005; Williams et al., 2008). There are also other plant components such as an arabinogalactan protein, which can influence the attachment of R. leguminosarum to surfaces (Xie et al., 2012).
In previous work, we observed that mutations affecting the cinR–cinIS encoded quorum-sensing regulatory system affected biofilm formation in vitro (Edwards et al., 2009; Frederix et al., 2011). The cinS gene, co-transcribed with the AHL synthase gene cinI, encodes a small protein that acts as an anti-repressor of the transcriptional regulator PraR (Edwards et al., 2009; Frederix et al., 2011). In that work, PraR was identified because it bound to CinS, but PraR is highly conserved among the α proteobacteria (Akiba et al., 2010). PraR belongs to the HipB family of DNA-binding proteins and contains an N-terminal (residues 19–73) cro/C1-like helix–turn–helix domain and probably has a multimerization domain based on its pattern of binding to DNA (Frederix et al., 2011). In Escherichia coli HipB regulates the hipBA toxin-antitoxin operon (Gerdes and Maisonneuve, 2012); also belonging to this family is SinR a master regulator of Bacillus subtilis biofilms (Kearns et al., 2005). Repression mediated by HipB and SinR is relieved by their binding to the antirepressors HipA and SinI respectively. SinR has a terameric structure and in solution SinI destabilizes this tetramer by tightly binding individual SinR subunits forming a 1:1 dead-end complex, thereby reducing the ability of SinR to bind to DNA (Scott et al., 1999). We assume CinS destabilizes PraR in a similar manner, and indeed CinS is stabilized in the presence of PraR suggesting that a stable CinS-PraR complex is formed (Frederix et al., 2011).
In Azorhizobium caulinodans, PraR represses the expression of reb genes that are present in A. caulinodans, but absent from most rhizobia; the increased expression of the reb genes in a praR mutant caused decreased nitrogen fixation in nodules (Akiba et al., 2010). PraR is orthologous to PhrR from Sinorhizobium medicae, which was originally identified as a transcriptional regulator induced by low pH (Reeve et al., 1998), but was named praR in A. caulinodans and R. leguminosarum because induction by low pH was not observed in those strains (Akiba et al., 2010; Frederix et al., 2011).
In R.l. viciae, PraR represses the raiR and rhiR genes, which encode different LuxR-type quorum-sensing regulators. This repression is relieved as the population density increases and cinS expression is increased, and so the antirepressor CinS binds to soluble PraR relieving PraR-mediated repression (Edwards et al., 2009; Frederix et al., 2011). When induced by relief of repression, RhiR induces the expression of the AHL synthase encoded by rhiI and the consequent population-dependent induction of rhiABC genes plays a role in rhizosphere growth and nodulation (Cubo et al., 1992). The other regulator RaiR regulates the expression of the AHL synthase gene raiI, and in some strains, mutation of raiI increased nitrogen fixation in nodules (Rosemeyer et al., 1998). Not many strains have the raiI and raiR genes and genes regulated via the raiIR quorum-sensing system have not been identified.
In addition to directly repressing rhiR and raiR (Frederix et al., 2011), PraR may repress the expression of plyB in R.l. viciae; PlyB is one of three secreted polysaccharide glycanases that cleave the surface EPS, and the pattern of plyB expression mirrored that of raiR and rhiR in various quorum-sensing mutants (Edwards et al., 2009; Frederix et al., 2011). However, apart from the rhiR and raiR promoters, no other direct targets of PraR have been demonstrated.
In this work, we used microarray analysis, promoter gene fusions and promoter binding experiments to identify direct targets of PraR in R.l. viciae. Among the genes directly repressed by PraR, are praR, three rap genes, rosR, encoding a global regulator of polysaccharides and one gene similar to praR. We demonstrate that mutation of praR results in enhanced in vitro biofilm formation, enhanced attachment to root hairs and increased nodulation competitiveness primarily due to enhanced expression of Rap proteins.
Results
Mutation of praR increases biofilms and expression of genes encoding secreted attachment proteins
Mutation of praR in R.l. viciae strain 3841 enhanced biofilm formation both in polystyrene microtitre dishes (data not shown) and at the air–liquid interface on glass shake flasks (Fig. 1A). Quantification of the attachment using crystal violet staining confirmed that there was an increased biofilm with the praR mutant compared with WT (Fig. 1B). Since mutation of praR increased expression of rhiR (Frederix et al., 2011), we tested whether the increased biofilms were due to quorum sensing regulated by RhiR, by introducing a rhiR mutation into the praR mutant. Since the double mutant retained an enhanced biofilm (Fig. 1A and B), RhiR was not required. Strain 3841 lacks raiI and raiR and so the praR mutant phenotype could not be mediated via RaiR.
Fig. 1.
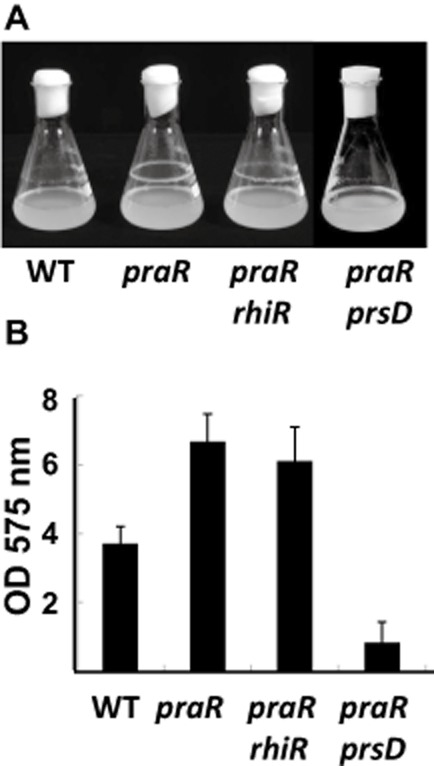
Mutation of praR enhances biofilm formation by R.l. viciae.A. Formation of rings of biofilm at the air–liquid interface after 5-day growth of 3841 (WT) and the mutants A963 (praR), A1149 (praR, rhiR) and A1161 (praR, prsD) in Y mannitol medium.B. The biofilms were quantified following staining with crystal violet and the data shown are averages (n = 10) ± SD. The mutants are all significantly different from WT (P = < 0.05 Student's t-test), whereas the praR, rhiR double mutant is not significantly different from the praR mutant.
Several surface components could increase biofilm formation. To test if altered expression of exported surface proteins could be responsible, we mutated prsD in the praR mutant. The prsD–prsE operon encodes components of a Type I secretion system (Finnie et al., 1997) required for the secretion of several proteins including potential adhesins and rhizobial adhesion proteins (Raps) (Krehenbrink and Downie, 2008). The prsD mutation decreased biofilm formation compared to the praR mutant (Fig. 1). Isolation of proteins secreted by the WT, the praR mutant and a transductant of WT carrying the praR mutation identified a secreted protein present at a higher level in growth-medium supernatant of the praR mutants (Fig. 2). Mass spectrometry revealed the protein to be Rhizobium adhering protein RapA2, which is secreted via the prsDE-encoded Type I secretion system (Krehenbrink and Downie, 2008) and is similar (87% similarity, 83% identity) to the R. trifolii adhesin RapA1 that promotes attachment, aggregation and enhanced infection (Mongiardini et al., 2008; 2009).
Fig. 2.
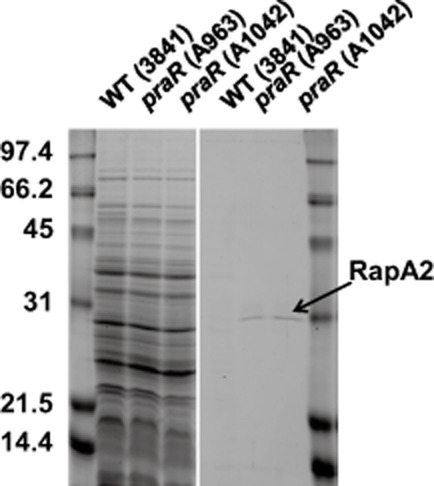
Mutation of praR enhances the level of the R.l. viciae secreted protein RapA2. Proteins from the growth-medium supernatants precipitated with trichloro-acetic acid (right panel) and cellular proteins (loading control) from the harvested bacteria (left panel) were separated by SDS-PAGE. The WT was 3841 and strain A1042 is a transductant of 3841 carrying the praR::Tn5 mutation from A963. The culture-supernatant protein arrowed was identified from the culture supernatant of A963 as the product of pRL100451 (RapA2) by MALDI-ToF mass spectrometry of a tryptic digest of the protein eluted from the gel; the statistical score calculated by the mascot program based on five matched fragments was 58 (a score above 50 indicates > 95% confidence). The bacteria were grown for 4 days in Y mannitol medium. The molecular weights of the protein standards in the flanking lanes are indicated.
We used microarrays to identify genes upregulated in the praR mutant compared with the WT. Thirty-seven genes showed an average induction of twofold or more compared with the WT (Table 1). One gene (RL0149) stood out in that it was strongly induced (over 11-fold) and this gene encodes a predicted transcriptional regulator with homology to PraR (55% similarity; 38% identity using clustalw over a full-length alignment). Most of the other genes apparently upregulated in the praR mutant have unknown functions, but three of the genes were of particular interest. One is praR, implying that PraR represses its own expression. As expected, one of the genes encodes RapA2, while another encodes RapC, which is related to RapA2. There are three functional rap genes in R.l. viciae 3841 (Krehenbrink and Downie, 2008) and although rapB (RL3911) did not appear in the genes induced more than twofold in the praR mutant, it was induced by an average of 1.7-fold (P value < 0.015). Therefore the three rap genes appear to be among those genes normally repressed by PraR and so it seemed possible that the increased expression of these rap genes could cause the enhanced biofilm formation in the praR mutant. However we also noted that the two genes, cadA (RL2961) and cadB (pRL100309) encoding predicted attachment proteins called cadherins (Krehenbrink and Downie, 2008), appeared to be slightly induced in the microarrays (1.8- and 1.4-fold respectively with P values of 0.021 and 0.009). Since these proteins are also secreted via the PrsDE system (Krehenbrink and Downie, 2008) and Rap proteins also contain cadherin-like domains (Abdian et al., 2013) we considered that cadA and cadB should be included in our analysis of biofilm formation. Although we focused in this work on genes likely to be repressed directly by PraR, it should be noted that expression of a few genes may be decreased in the praR mutant (Table 1); we did not independently confirm decreased expression of these genes, but presumably any decreased expression occurs as an indirect effect of the praR mutation.
Table 1.
Microarray analysis: genes expressed more than or less than twofold in praR mutant
| Gene ID | Fold up or down | Significance | Gene name | Predicted function of gene product |
|---|---|---|---|---|
| RL0149 | +11.1 | 1.92E-06 | PraR-like transcriptional regulator | |
| RL3192 | +3.3 | 0.017466 | Putative ATP-binding component of ABC transporter | |
| RL2937 | +3.0 | 9.63E-06 | Putative transmembrane protein | |
| pRL100451 | +2.9 | 5.41E-06 | rapA2 | Putative autoaggregation protein |
| pRL110037 | +2.7 | 0.004028 | Putative two-component fused sensor/regulator | |
| RL4665 | +2.7 | 0.03583 | Conserved hypothetical protein | |
| RL4097 | +2.7 | 0.038832 | Putative transmembrane cation transporter protein | |
| RL3074 | +2.7 | 0.015478 | rapC | Putative autoaggregation protein |
| RL3375 | +2.6 | 0.000292 | Conserved hypothetical protein | |
| RL2307 | +2.6 | 0.009037 | Conserved hypothetical protein | |
| RL4195 | +2.5 | 0.004917 | Putative transmembrane protein | |
| RL1338 | +2.5 | 0.008283 | pmtA | Putative phosphatidylethanolamine N-methyltransferase |
| RL3959 | +2.4 | 3.71E-05 | Conserved hypothetical protein | |
| RL4614 | +2.4 | 0.033157 | rpoH2 | Putative RNA polymerase sigma factor (heat shock) |
| RL3867 | +2.4 | 0.030485 | Conserved hypothetical protein | |
| RL3982 | +2.4 | 0.022785 | Conserved hypothetical protein | |
| pRL110129 | +2.4 | 0.021662 | Hypothetical protein | |
| RL0390 | +2.4 | 0.035746 | praR | PraR transcriptional regulator |
| pRL110586 | +2.3 | 0.029844 | Conserved hypothetical protein | |
| RL0610 | +2.3 | 0.013482 | Hypothetical exported protein | |
| pRL120342 | +2.3 | 5.75E-05 | hspD | Putative small heat shock protein |
| RL3701 | +2.3 | 0.00145 | Putative transmembrane protein | |
| RL1165 | +2.2 | 0.02779 | Conserved hypothetical protein | |
| RL1513 | +2.2 | 0.000623 | Putative FNR/CRP family transcriptional regulator | |
| RL1034 | +2.2 | 0.04639 | Conserved hypothetical protein | |
| RL3186 | +2.2 | 0.005128 | Putative transmembrane protein | |
| RL3702 | +2.2 | 0.000301 | Putative transmembrane protein | |
| RL0506 | +2.1 | 0.004262 | Conserved hypothetical protein | |
| pRL110257A | +2.1 | 0.003572 | Conserved hypothetical protein | |
| RL2415 | +2.1 | 0.000762 | Hypothetical protein | |
| RL4089 | +2.1 | 6.68E-13 | ibpA | Putative heat shock protein A hspA |
| RL3704 | +2.1 | 0.002125 | asfZ | Putative anti-sigma factor to RL3703 |
| RL4113 | +2.1 | 0.033118 | Hypothetical protein | |
| pRL110283 | +2.0 | 0.007827 | Putative DNA-binding protein | |
| RL4065 | +2.0 | 0.005843 | Conserved hypothetical protein | |
| RL3703 | +2.0 | 0.011679 | ecfZ | Putative RNA polymerase ECF sigma factor |
| RL4624 | +2.0 | 4.1E-06 | rpmF | Putative 50S ribosomal protein L32 |
| pRL110131 | −2.0 | 0.00376 | Hypothetical protein | |
| RL4371 | −2.1 | 0.034595 | Conserved hypothetical protein | |
| RL2877 | −2.1 | 0.018058 | Putative transmembrane transporter | |
| pRL120167 | −2.3 | 0.000351 | Putative MFS family transporter | |
| RL2588 | −2.3 | 0.021037 | tyrS1 | Putative tyrosyl-tRNA synthetase |
| pRL80130 | −2.5 | 0.030906 | Putative transmembrane protein | |
| RL1924 | −3.2 | 0.004394 | Conserved hypothetical exported protein | |
| RL1925 | −3.2 | 0.018171 | Conserved hypothetical protein | |
| RL3670 | −5.1 | 8.47E-05 | Conserved hypothetical protein |
Confirmation of PraR-repressed genes using promoter fusions
We made promoter fusion reporter plasmids for the rapA2, rapB, rapC, praR, RL0149, cadA and cadB genes identified from the microarrays as being potentially induced in the praR mutant. We also made and used other reporter fusions to test if the expression of other genes was affected by mutation of praR. Previously, (Edwards et al., 2009), plyB was shown to encode a secreted quorum-sensing-regulated glycanase that influences biofilm formation (Russo et al., 2006) and so we included reporter fusions with plyB and the closely related genes plyA and plyC (Finnie et al., 1998) in our analysis. Since polysaccharides influence biofilms, we also analysed expression of gene fusions with pssA and gmsA required for the acidic EPS and glucomannan respectively, both of which are important for biofilm formation on root hairs (Williams et al., 2008). We also assayed the expression of rosR encoding a global regulator of surface rhizobial polysaccharides (Janczarek and Urbanik-Sypniewska, 2013).
The rapA2, rapB, rapC, plyB, cadA and cadB genes encoding secreted proteins were induced in the praR mutant compared with WT (Fig. 3). The genes encoding the predicted regulators RL0149 and RosR were also increased in expression in the praR mutant. We also confirmed that PraR represses its own expression (Fig. 3). In contrast, mutation of praR did not significantly induce gmsA (required for glucomannan synthesis), or pssA, plyA and plyC (associated with the synthesis and cleavage of the acidic exopolysaccharide) (Fig. 3).
Fig. 3.
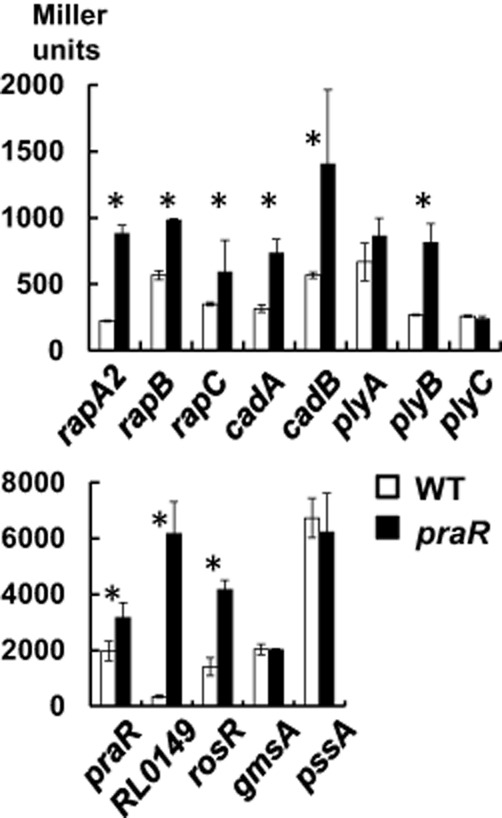
Expression of reporter gene fusions using the promoters of candidate genes potentially repressed by PraR. Levels of expression from the promoters were assayed in either 3841 (WT, open bars) or A963 (praR::Tn5, black bars) following growth for 48 h in Y mannitol medium. The plasmids containing cloned promoters were pIJ9651 (rapA2), pIJ11275 (rapB), pIJ11171 (rapC), pIJ9686 (cadA), pIJ9724 (cadB), pIJ11283 (plyA), pIJ9252 (plyB), pIJ11276 (plyC), pIJ11112 (praR), pIJ11114 (RL0149), pIJ11196 (rosR), pIJ11200 (gmsA) and pIJ11195 (pssA). Levels of expression of β-glucuronidase (rapA2 only) or β-galactosidase (all others) are expressed as averages (n = 6) ± SD and the asterisks mark those tests in which there is a significant (P < 0.05, Student's t-test) difference in expression in the praR mutant compared with WT. The data are shown in two histograms for simplicity of presentation due to the different levels of expression.
Identification of PraR-binding promoters
The enhanced expression of rapA2, rapB, rapC, cadA, cadB, plyB, rosR, praR and RL0149 in the praR mutant could be direct, due to release of repression by PraR, or indirect, e.g. due to the altered expression of a transcriptional regulator. At about 30–62 nM, PraR bound to the promoters of rapA2, rapB, rapC, rosR, praR and RL0149 (Fig. 4), reducing the mobility of radioactively labelled promoter fragments during electrophoresis as seen previously with the rhiR promoter (Frederix et al., 2011). This, together with the complete lack of binding of PraR to the plyC and cadB promoters (Fig. 4) demonstrates that PraR specifically interacts with the rapA2, rapB, rapC, rosR, praR and RL0149 promoters and so most probably directly represses their expression. There may be weaker binding of PraR to the cadA and plyB promoters, but this required higher levels of PraR (62–250 nM). We used MEME (Bailey and Elkan, 1994) to analyse the promoter fragments of rhiR, rapA2, rapB, rapC, rosR, praR and RL0149 for a consensus PraR binding site that was also present in the PraR-binding 20-nucleotide fragment identified previously (Frederix et al., 2011). This identified the consensus motif TTGCAA (Fig. S1). This conserved sequence was not present in the cadA, cadB, plyB or plyC promoters.
Fig. 4.
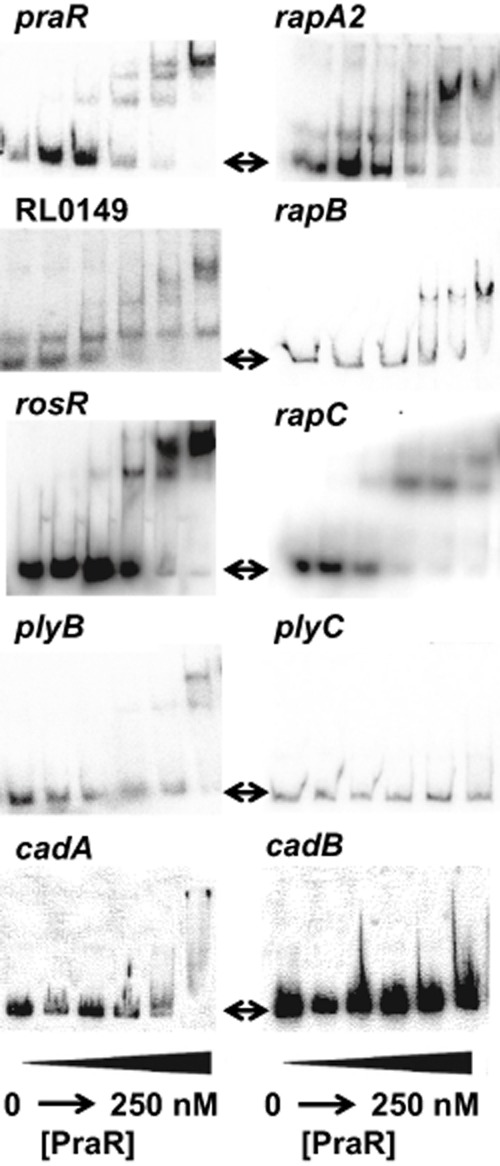
In vitro analysis of PraR binding to different promoters. Radioactively labelled promoters of the genes indicated were incubated with a purified PraR maltose-binding protein fusion (from left to right on each gel: 0, 16 nM, 32 nM, 64 nM, 125 nM and 250 nM protein). After the reactions the samples were separated by non-denaturing gel electrophoresis and the radioactively labelled bands were imaged using a phosphorimager as described previously (Frederix et al., 2011). The arrows indicate the sizes of unshifted fragments.
Since mutation of praR increased expression of most of the PraR-repressed genes only two- to threefold, PraR may repress expression of other genes not included in Table 1 because their expression increased less than twofold. As expected from the reporter fusion, PraR did not bind to the plyC promoter, but unexpectedly, PraR was not observed to bind to the cadB promoter (Fig. 4).
Previously (Frederix et al., 2011) we observed that the antirepressor CinS interacts with PraR, preventing PraR binding to the rhiR and raiR promoters. Since PraR and CinS can interact in the absence of cognate promoters, we expected that CinS would antagonize PraR binding to other promoters. We tested this using gel shift assays with the praR and rapA2 promoters and, as expected, CinS blocked PraR binding (Fig. S2).
Given the similarity between PraR and RL0149, we thought that there might be cross-regulation involving these two proteins. We made a mutation in the RL0149 gene and tested the expression of praR, rhiR, rosR and rapA2 in the mutant and saw no significant changes in gene expression or biofilm formation (data not shown). We also made a praR-RL0149 double mutant and saw no difference in the expression of praR or RL0149. Therefore the role of the RL0149 gene remains enigmatic.
A light emission (lux) assay shows that enhanced root attachment by the praR mutant requires Rap proteins and glucomannan
To examine whether the altered biofilm formation of the praR mutant was correlated with changes in root attachment, cells of the praR mutant and WT were labelled with 3H-leucine and then incubated with pea root segments; after several washes, attachment was then measured by determining the levels of radioactivity attached to the roots. This revealed that more of the praR mutant attached than the WT (Fig. 5A).
Fig. 5.
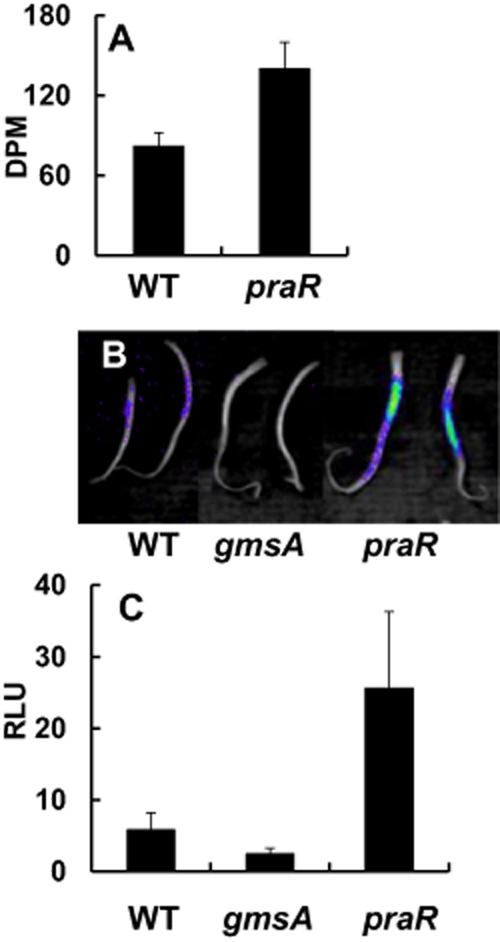
Enhanced root attachment by the praR mutant shown by radiolabelling and luminescence.A. R.l. viciae strains 3841 (WT) and A963 (praR::Tn5) were labelled with 3H-leucine and incubated with roots excised from pea seedlings. The roots were washed three times and then the bound radioactivity on each root was measured using a scintillation counter. The data show average counts determined using 10 roots (± SD).B. A similar attachment assay as in A was performed using strains 3841 (WT), A963 (praR) and A1042 (gmsA); instead of labelling with 3H-leucine, each strain carried the lux plasmid pIJ11282 expressing the luxCDABE operon (Fig. S1) and bacterial attachment was photographed using a NightOWL LB983 ultrasensitive light-imaging system. Strain A1042 (gmsA) was included to confirm that this method can document the observed decrease in attachment of the gmsA mutant (Williams et al., 2008).C. Attachment of 3841 (WT), A963 (praR) and A1042 (gmsA) all carrying pIJ11282 was done as in B, but quantified using a luminometer. Data shown are averages of Relative Light Units (RLU) based on separate measurements with 10 roots (± SD) and all are significantly different from each other (P = < 0.05, Student's t-test).
This radioactivity-based assay for attachment was inconvenient and so we devised an alternative strategy to measure attachment. We made a stably inherited plasmid (pIJ11282) constitutively expressing the lux operon (Fig. S3), transferred it into R.l. viciae 3841, and inoculated the resulting strain onto the roots of pea seedlings; light emission was then measured after washing off loosely attached bacteria, revealing that the bacteria were mostly attached to the region of the root above the tip, where young root-hairs are growing (Fig. 5B). In previous work (Williams et al., 2008), attachment of R.l. viciae to vetch root hairs was shown to require the surface glucomannan polysaccharide, which binds to a plant lectin expressed on root hairs (Laus et al., 2006). Blocking the production of the glucomannan by mutating the glucomannan synthase gene gmsA reduced attachment to pea roots measured using this lux-dependent light emission assay (Fig. 5B). In contrast, the praR mutant inoculated onto pea roots showed higher levels of light emission than WT (Fig. 5B); quantification using a luminometer confirmed that the praR mutant attached better than the WT (Fig. 5C). Colony counts of WT and praR mutant bacteria attached to pea roots also demonstrated that the praR mutant attaches at two- to threefold higher levels than WT, with the praR mutant attaching at 7.8 ± 0.19 × 105 cfu compared with the WT at 3.15 ± 0.07 × 105 cfu. Since the results with the lux-marked bacteria matched the enhanced attachment assayed by the radioactive assay, and by bacterial counts, we used the lux-based assay for further experiments.
We made a praR-gmsA double mutant to test whether increased expression of genes normally repressed by PraR could restore root attachment by the gmsA mutant. The praR-gmsA double mutant was reduced in attachment compared with the praR mutant (Fig. 6); this would fit with a model in which genes induced by mutation of praR enhance attachment following an initial lectin-mediated binding to the GmsA-determined glucomannan.
Fig. 6.
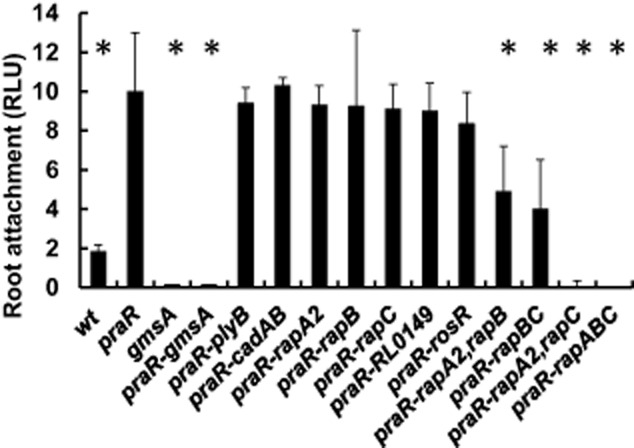
Enhanced root attachment by the praR mutant requires gmsA and rap genes. Attachment of the R.l. viciae strains was assayed using bacteria expressing the constitutively expressed lux operon on pIJ11282 following incubation of the bacteria with excised roots from pea seedlings and subsequent washing of the roots. The strains used were 300 (WT), A1345 (praR), A1208 (gmsA), A1367 (praR, gmsA), A1372 (praR, plyB), A1383 (praR, cadA, cadB), A1328 (praR, rapA2), A1416 (praR, rapB), A1374 (praR, rapC), A1385 (praR, rosR), A1427 (praR, rapA2, rapB), A1428 (praR, rapB, rapC), A1430 (praR, rapA2, rapC) and A1429 (praR, rapA2, rapB, rapC). Light emission of the attached bacteria was quantified using a luminometer. Data shown are averages of Relative Light Units (RLU) based on separate measurements with 10 roots (± SD) for each strain; averages significantly different (Student's t-test P = < 0.05) from the praR mutant are marked (*).
Since PraR represses expression of rapA2, rapB, rapC, cadA, cadB, plyB, rosR and R0149, we introduced mutations in each of these genes into the praR mutant to determine which gene(s) were required for the enhanced root attachment by the praR mutant. None of these mutations significantly decreased the enhanced attachment of the praR mutant (Fig. 6). Since the Rap proteins are structurally similar, we made all combinations of the rap gene mutations in the praR mutant and tested their attachment. Likewise since CadA and CadB are related we made a double cadA, cadB mutant. The combination of mutations in both rapA2 and rapC abolished the enhanced attachment of the praR mutant whereas the combination of mutations in rapA2 and rapB, or rapC and rapB decreased attachment (Fig. 6). Therefore we conclude that the enhanced attachment in the praR mutant is primarily due to increased levels of the secreted adhesins RapA2 and RapC, although RapB also plays a role. In contrast, the cadA, cadB double mutant was unaffected for attachment (Fig. 6).
The praR mutant out-competes the WT for nodule infection and this requires both the glucomannan and the Rap proteins
Root hair attachment can be related to competitiveness for nodule infection (Williams et al., 2008), and so we tested whether the praR mutant had a competitive advantage when co-inoculated with WT. To assay this, the praR::Tn5 mutation was transduced into strain 300, the streptomycin-sensitive parent of R.l. bv. viciae 3841, forming A1132. We first ascertained that strain 300 was equally competitive with strain 3841: when inoculated onto pea roots at a 1:1 ratio, the numbers of nodules formed by each strain was not significantly different from predicted (chi-squared test). Strain A1132 (praR), which grew with a similar doubling time as WT, was assayed for competitiveness in infection of pea nodules by co-inoculating A1132 with an equivalent amount of strain 3841 (WT) and then scoring bacteria from individual nodules for resistance to kanamycin (A1132) or streptomycin (3841). The praR mutant outcompeted the WT (Fig. 7) indicating that the increased attachment results in increased infection.
Fig. 7.
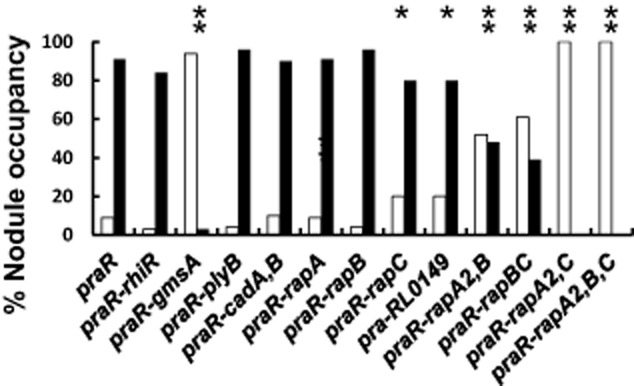
The R.l. viciae praR mutant has increased competitiveness for nodule infection and this requires the rap and gmsA genes. Strains A1345 (praR), A1208 (gmsA), A1367 (praR, gmsA), A1372 (praR, plyB), A1149 (praR, rhiR), A1383 (praR, cadA, cadB), A1328 (praR, rapA2), A1416 (praR, rapB), A1374 (praR, rapC), A1427 (praR, rapA2, rapB), A1428 (praR, rapB, rapC), A1430 (praR, rapA2, rapC) and A1429 (praR, rapA2, rapB, rapC) were each mixed in a 1:1 ratio with 3841 (WT) and inoculated onto peas. Bacteria were isolated from surface-sterilized individual nodules and the released bacteria were identified based on their antibiotic resistance. The graph shows the % of WT (open bars) or mutants (black bars) recovered from nodules; those results that were significantly different (chi-squared test) from A1345 (praR) + 3841 (WT) are marked (**P < 0.01; *P < 0.05). Each analysis is based on identification of mutant or WT bacteria isolated from at least 100 nodules from at least five separate plants and excludes potentially mixed infections.
To determine if the increased infection by the praR mutant required the glucomannan previously shown to be required for infection competitiveness (Williams et al., 2008), we tested the ability of the praR-gmsA double mutant to compete with WT. Since this mutant was a poor competitor (Fig. 7), we can conclude that the increased expression of rap genes cannot compensate for the loss of glucomannan in infection. The rapA or rapB mutations had no significant effect on the infection competitiveness of the praR mutant, whereas the rapC mutation slightly decreased its competitiveness (Fig. 7). The combination of mutations in rapA2 and rapB, or rapC and rapB further decreased competitiveness, while strains carrying combination of mutations in both rapA2 and rapC abolished the enhanced competitiveness of the praR mutant (Fig. 7). As with all the mutants tested (Fig. 7), the praR mutant carrying mutations in all three rap genes grew with the same doubling time as WT in planktonic culture in mannitol minimal medium and produced normal numbers of pink nodules when inoculated alone, showing that the effects of the mutations are primarily on the competitiveness, rather than effects on growth or nodule infection per se.
In contrast to these additive effects of rap mutations on competitiveness, the combination of the cadA and cadB mutations did not decrease nodulation competitiveness of the praR mutant. Likewise, introduction of a rhiR mutation into the praR mutant did not affect its increased nodule infection, demonstrating that the quorum-sensing genes induced by RhiR are not required for the increased infection potential (Fig. 7). Mutation of RL0149 slightly decreased competitiveness (Fig. 7), indicating that this gene may play a subtle role in competitive nodule infection.
It is evident that those mutations that decreased root attachment by the praR mutant also decreased its ability to outcompete the WT for nodule infection. The enhanced attachment is not mediated via the RhiR regulon but requires the Rap proteins. Our observations are compatible with a model in which glucomannan-mediated attachment occurs first and that subsequent Rap-mediated adhesion (either between bacteria or enhancing bacterial attachment to roots) increases the infection potential of a rhizobial strain.
Although the rap genes are clearly required for competitive nodule infection and root attachment in a praR mutant, there is a possibility that this requirement is only seen in the praR mutant background. As proof of principle of the role of Rap proteins in competitive nodule infection we made a derivative of WT (3841) carrying mutations in rapA2 and rapC. This mutant (A1480) nodulated peas normally, but was completely defective for competitive nodule infection: when mixed 1:1 with strain 3841 all 192 nodules tested were occupied by 3841. There was also a complete loss of root attachment (0.05 ± 0.005 RLU) equivalent to that shown for the praR, rapA2, rapC mutant in Fig. 6. Therefore we conclude that the Rap proteins are essential for normal attachment and competitive nodule infection in the presence or absence of PraR.
The rapA2, rapB and rapC genes are differentially regulated
One apparent anomaly is that in addition to the praR mutation increasing biofilms, mutation of cinS also increased biofilms (Edwards et al., 2009; Frederix et al., 2011). Since CinS reduces PraR-mediated repression, mutation of cinS would be expected to enhance PraR-mediated repression and therefore decrease (rather than increase) biofilm attachment by reducing rap gene expression. Therefore we measured rapA2, rapB and rapC expression in a cinS mutant; as anticipated, there was a small decrease in rapC expression and a small decrease in rapA2 expression that is probably real but was just outside the significance range. Surprisingly there was a small increase in rapB expression (Fig. 8) possibly explaining the enhanced biofilm in the cinS mutant. Another potential regulator is ExpR, because mutation of expR also increased biofilm formation (Edwards et al., 2009); this might be due to by the increased rapA2 expression in the expR mutant (Fig. 8). One way in which differential regulation of the rap genes could occur could be for CinS to influence rap gene expression by interacting with another regulator (Fig. 9). To test this possibility, we introduced the cinS mutation into the praR mutant; the resulting increased rapA2 and rapB expression in the cinS, praR double mutant (Fig. 8) shows (as indicated in Fig. 9) that CinS must affect some regulator in addition to PraR (attenuating repression by another protein being the simplest explanation). Regulation of praR itself is complex, because mutation of cinS, praR or expR increased praR expression (Fig. 8). A working model for regulation of PraR expression and activity is shown in Fig. 9.
Fig. 8.
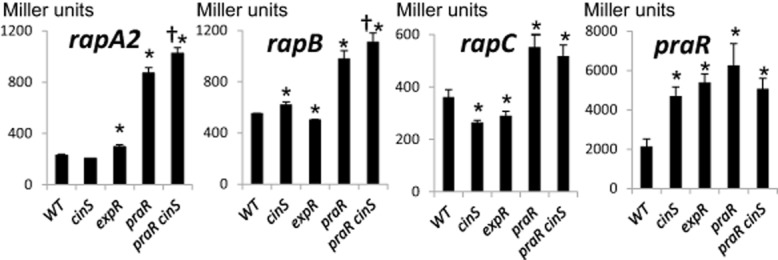
Differential regulation of rapA2, rapB, rapC and praR in cinS, expR and praR mutants. Levels of expression from the promoters were assayed in either 3841 (WT), A1229 (cinS), A1216 (expR), A963 (praR) or A1312 (praR, cinS) following growth for 48 h in Y mannitol medium. The plasmids containing cloned promoters were pIJ9651 (rapA2), pIJ11275 (rapB), pIJ11171 (rapC) or pIJ11112 (praR). Levels of expression of β-glucuronidase (rapA2 only) or β-galactosidase (all others) are expressed as averages (n = 6) ± SD in Miller units. Those tests in which there is a significant (P < 0.05, Student's t-test) difference in expression are marked: the asterisk (*) indicates significant differences compared with WT and the dagger (†) indicates significant differences in expression in A1312 (praR, cinS) compared with expression in A963 (praR). The reduced expression of rapA2 in the cinS mutant was just outside the range of significance (P = 0.0504).
Fig. 9.
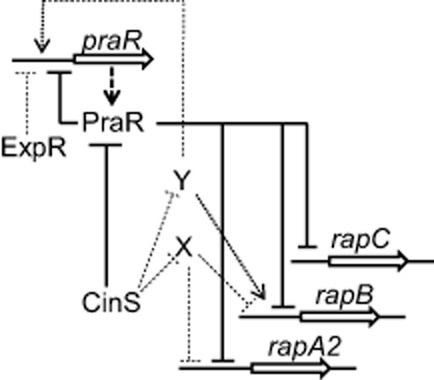
Model of regulation by PraR. Expression from the praR promoter is directly repressed by PraR (autoregulated) and since mutation of expR increased praR expression we speculate that ExpR also represses praR. PraR directly represses expression of rapA2, rapB, rapC (shown), rosR and RL0149 (not shown). CinS, which is expressed in a population-density-dependent manner via the CinR–CinI-regulated quorum-sensing system, binds to PraR attenuating its repression. Since expression of rapA2 and rapB increased in the cinS, praR mutant compared with the praR mutant (Fig. 8), CinS must interact with another protein or proteins. The increased rapA2 and rapB expression could be due to CinS binding to and inactivating an unknown repressor (X). The increased expression of both praR and rapB in the cinS mutant (Fig. 8) could be due to CinS attenuating the activity of an unknown positive transcriptional regulator (Y) as indicated; alternatively increased levels of repressor X in the cinS mutant could repress expression of another repressor of praR (not shown). Full arrows and lines are based on tested interactions; dotted arrows and lines are speculations that fit the gene expression patterns.
Discussion
The optimal mode of growth for many bacteria in nature is in biofilms, and this is particularly true for soil bacteria that can attach to the roots and root hairs of growing plants, which secrete nutrients into the rhizosphere. The switch between planktonic and biofilm growth is an important lifestyle switch and is coupled to changes in expression of many genes. There are two very good reasons why rhizobia attach to root hairs: one is to benefit from secreted nutrients and the other is to attach specifically to host legume root hairs so that by being well positioned they have a better chance of infecting roots and growing exponentially during infection and nodule development (Downie, 2010).
In R.l. viciae, we have shown that PraR is a regulator that modulates gene expression during adaptation to biofilm growth. PraR represses the expression of several genes including those encoding three rhizobial adhering proteins (RapA, RapB and RapC), a predicted cadherin (CadA), an EPS glycanase (PlyB) and at least two regulators, one of which (RL0149) is similar to PraR and the other of which (RosR) is a global regulator of production of surface polysaccharides (Janczarek and Urbanik-Sypniewska, 2013). Several other genes may be repressed by PraR, but since mutating praR causes only a small (two- to threefold) induction of those genes identified as direct targets, microarrays are not the best way to identify genes regulated by PraR; chromatin immunoprecipitation would identify a greater range of PraR regulated genes. Although the increase in expression of the rap genes in the praR mutant is relatively low, it is sufficient to initiate changes in attachment to roots, leading to increased nodulation competitiveness in mixed inoculations. Possibly other PraR-regulated genes including plyB, rosR and the cadherins may play a role in attachment/biofilm formation that would only be revealed under different test conditions. The fact that praR remains functional in R.l. viciae must mean that there is another stage in the lifestyle where it is important that the PraR-regulated genes are repressed. This could e.g. be explained by the requirement for rhizobia to move around in the rhizosphere and a lifestyle with too much attachment could hamper other aspects of their competitiveness in the wild.
PraR and its orthologue PhrR are well conserved within the α-proteobacteria (Akiba et al., 2010), but their regulation and the genes they regulate seem to be rather different, even among rhizobia. Low pH induced phrR in S. medicae, but praR appears not to be induced by low pH in R.l. viciae or A. caulinodans (Akiba et al., 2010; Frederix et al., 2011). PraR-regulated genes in A. caulinodans were identified using microarrays (Akiba et al., 2010), but none of the top 10 genes induced in the A. caulinodans praR mutant was induced by mutation of praR in R.l. viciae. PraR in A. caulinodans represses seven reb genes that are thought to determine the production of inclusion bodies; it is the enhanced expression of these reb genes that causes the Fix- phenotype of the A. caulinodans praR mutant (Akiba et al., 2010). In R.l. viciae there are no reb-gene homologues and the praR mutation does not cause a Fix- phenotype. One of the other genes (AZC-1189) that was most upregulated by mutation of praR in A. caulinodans also has no homologue in R.l. viciae and the homologues of the two induced flagellar genes (AZC-3379 and AZC-2699) did not appear to be enhanced in expression in the R.l. viciae praR mutant.
As illustrated (Fig. 9), the control of PraR activity in R.l. viciae is affected by quorum-sensing induction of CinS, which binds to PraR (Frederix et al., 2011), decreasing PraR-mediated repression; in this way PraR-repressed genes can be induced as the population density increases. However the regulation is complex because PraR represses its own expression (Fig. 9). There are parallels with the regulation of biofilm formation by SinR in B. subtilis; SinR represses expression of exopolysaccharide genes and a gene encoding a secreted protein that is required for normal biofilm formation (Kearns et al., 2005; Chu et al., 2006). Repression by SinR is relieved by SinI which binds to SinR, preventing SinR from acting as a repressor. CinS is not structurally similar to SinI, but by binding to PraR it can affect EPS production by increasing RosR expression and can allow increased expression of the three Rap proteins. As in B. subtilis, the regulation of R.l. viciae biofilm genes is complex, because another regulator, ExpR represses praR expression. Furthermore, mutations in expR and cinS have different effects on the regulation of the different rap genes (Fig. 9). It seems likely that differential expression of the rap genes can allow bacteria to attach at different stages of growth. For example rapB expression increased in the absence of CinS, a situation that would prevail at low population densities, whereas the expression of rapC is higher as CinS levels increase as seen at high population densities. However these interpretations are based on averages of gene expression across a population and this is unlikely to hold in biofilms. For example in B. subtilis biofilms, sinI was only expressed in a subpopulation of the cells whereas sinR was expressed in all cells (Chai et al., 2008).
The enhanced root attachment and increased infection competitiveness of the praR mutant appear to be due primarily to the increased expression of the rap genes (Fig. 9) and this is consistent with what was seen with increased expression of rapA1 on a plasmid in R.l. trifolii (Mongiardini et al., 2008). The enhanced root-hair attachment and infection of root hairs by the praR mutant was abolished by the gmsA mutation that prevents the formation of glucomannan. This polarly expressed polysaccharide is specifically recognized by a legume-specific root-hair-tip localized plant lectin and has been shown to be important for the initial step of rhizobial binding (Laus et al., 2006). In the absence of the glucomannan, infection can occur normally, but the bacteria are uncompetitive (Williams et al., 2008). In addition, the acidic EPS plays an essential role in root-hair infection. Since the Rap proteins bind to this EPS and are required for enhanced attachment to roots, we propose that after glucomannan-plant-lectin mediated attachment, Rap-mediated interactions between bacteria will permit an accumulation of bacteria on root hairs and that this promotes competitiveness. Bacterially made extracellular cellulose also promoted accumulation of on root hairs, but this cellulose-mediated effect did not significantly contribute to competitiveness (Williams et al., 2008). In fact cellulose production by R.l. viciae even appeared to inhibit initiation of infection in some circumstances (Laus et al., 2005). Based on these data, we propose that the glucomannan-plant lectin binding followed by the Rap-EPS interaction play key roles in competitive infection whereas the cellulose mediated stabilization of biofilms is important for bacterial growth on root-hairs.
Experimental procedures
Bacterial growth, assays of β-galactosidase and β-glucuronidase and competitive nodule infection
Rhizobium leguminosarum strains were grown at 28°C in TY medium (Beringer, 1974), AMS (minimal) medium (Poole et al., 1994) containing 10 mM NH4Cl and 30 mM glucose, or Y minimal medium (Sherwood, 1970) buffered with MOPS (Williams et al., 2008) and containing mannitol (0.2% w/v). E. coli was grown at 37°C in L medium (Sambrook et al., 1989). Antibiotics were added as appropriate to maintain selection for plasmids. β-Galactosidase was assayed as Miller units as described previously (Edwards et al., 2009) using at least three independent cultures. β-Glucuronidase was assayed using the same method except that the β-galactosidase substrate o-nitrophenol β-d-galactopyranoside was replaced with p-nitrophenol β-d-glucopyranoside. Conjugal matings using a helper plasmid and transductions using RL38 phage were done as described (Buchanan-Wollaston, 1979; Figurski and Helinski, 1979). DNA cloning, ligations and transformations were done using standard methods (Sambrook et al., 1989). Biofilm formation in flasks and microtitre plates was measured as described previously (Edwards et al., 2009). Competitive nodule infection experiments were carried out as described previously (Williams et al., 2008) using at least 100 nodules from a minimum of five separate plants in each test. Some crushed nodules (usually around 5% and always less than 20%) apparently contained two bacterial genotypes and these were excluded from the analyses; this co-infection of pea nodules may occur due to two infection threads infecting one nodule or the fusion of two nodules into what appears to be a single mutilobate nodule.
Construction of strains and plasmids
Strains, plasmids and primers used in this study are listed in Tables 2 and 3 and Tables S1 and S2. R. leguminosarum 3841 mutants with Tn5 insertions in cadA (RL2961), cadB (pRL100309), RL0149, rosR (RL1379), plyB (RL3023), or rapB (RL3911), were identified in pools of mutants from a Tn5 library using gene-specific and Tn5-specific PCR primers (Table S2) as previously described (Williams et al., 2008). To make A1206 (rapA2ΩSpecR), rapA2 (pRL100451) with 1 kb flanking regions was amplified by PCR using primers rapA2F and rapA2R (Table S2) and the product was digested with KpnI and SpeI and cloned into pJQ200KS; the spectinomycin resistance gene on pMP45Ω was then amplified (primers specF and specR) and cloned as an AccIII fragment into the AccIII site in rapA2 in pJQ200KS. The rapA2ΩspecR allele was then recombined into strain 3841 selecting for spectinomycin and sucrose-resistant transconjugants (Quandt and Hynes, 1993) and the rapA2ΩspecR allele in A1206 was confirmed by PCR. To make the rapC mutant A1362, an internal (347 bp) fragment of the rapC gene (RL3074) was amplified using PCR (primers rapCF and rapCR, Table S2) and cloned into pK19mob using the XhoI and HindIII restriction sites introduced on the primers. The resulting plasmid was conjugated into strain 3841 selecting on neomycin (400 μg ml−1) and a single-crossover integration into rapC was verified by PCR.
Table 2.
Bacterial strains
| Strain | Description | Source |
|---|---|---|
| 300 | Wild-type R. leguminosarum | Johnston and Beringer (1975) |
| 3841 | R. leguminosarum 300; strepR | Johnston and Beringer (1975) |
| A898 | 3841 prsD::Tn5ΩGent | Krehenbrink and Downie (2008) |
| A920 | 3841 rhiR::Tn5 | This work |
| A963 | 3841 praR::Tn5 | Frederix et al. (2011) |
| A1024 | 3841 rhiR::Tn5ΩGent | This work |
| A1042 | 3841 praR::Tn5 transduced from A963 | This work |
| A1045 | 3841 gmsA::Tn5ΩGent | Williams et al. (2008) |
| A1132 | 300 praR::Tn5 transduced from A963 | This work |
| A1149 | 3841 praR::Tn5, rhiR::Tn5ΩGent | This work |
| A1161 | 3841 praR::Tn5, prsD::Tn5ΩGent | This work |
| A1167 | 3841 praR:: Tn5ΩSpec | This work |
| A1176 | 3841 rosR::Tn5 | This work |
| A1206 | 3841 rapA2ΩSpecR | This work |
| A1208 | 300 gmsA::Tn5ΩGent | Williams et al. (2008) |
| A1216 | 3841 expRΩpK19mob | Frederix et al. (2011) |
| A1229 | 3841 cinSΩApra | Frederix et al. (2011) |
| A1253 | 3841 cadB::Tn5 | This work |
| A1254 | 3841 cadA::Tn5 | This work |
| A1261 | 300 cadB::Tn5 transduced from A1253 | This work |
| A1263 | 3841 cadB::Tn5ΩGent | This work |
| A1312 | 3841 praR::Tn5ΩSpec cinSΩApra | Frederix et al. (2011) |
| A1328 | 300 praR::Tn5 with rapA2ΩSpecR transduced from A1206 | This work |
| A1327 | 3841 RL0149::Tn5 | This work |
| A1340 | 300 RL0149::Tn5 transduced from A1327 | This work |
| A1345 | 300 praR::Tn5ΩSpec | This work |
| A1362 | 3841rapCΩpk19mob | This work |
| A1363 | 300 praR::Tn5ΩSpec cadA::Tn5 | This work |
| A1365 | 3841 plyB::Tn5 | This work |
| A1367 | 300 praR::Tn5ΩSpec gmsA::Tn5 | This work |
| A1368 | 300 praR::Tn5ΩSpec RL0149::Tn5 | This work |
| A1370 | 300 praR::Tn5ΩSpec rhiR::Tn5 | This work |
| A1372 | 300 praR::Tn5ΩSpec plyB::Tn5 | This work |
| A1374 | 300 praR::Tn5ΩSpec rapCΩpk19mob | This work |
| A1376 | 3841 rapB::Tn5 | This work |
| A1383 | 300 praR::Tn5ΩSpec cadA::Tn5 cadB::Tn5ΩGent | This work |
| A1384 | 300 praR::Tn5ΩSpec prsD::Tn5 | This work |
| A1385 | 300 praR::Tn5ΩSpec rosR::Tn5 | This work |
| A1416 | 300 praR::Tn5ΩSpec rapB::Tn5 | This work |
| A1417 | 300 rapB::Tn5 transduced from A1376 | This work |
| A1425 | 3841 rapB::Tn5ΩGen | This work |
| A1426 | 3841 rapC::Tn5ΩApra | This work |
| A1427 | 300 praR::Tn5 rapA2ΩSpec rapB::Tn5ΩGent | This work |
| A1428 | 300 praR::Tn5ΩSpec, rapB::Tn5ΩGent, rapCΩpk19mob | This work |
| A1429 | 300 praR::Tn5 rapA2ΩSpec rapB::Tn5ΩGen rapC::Tn5ΩApra | This work |
| A1430 | 300 praR::Tn5 rapA2ΩSpec rapC::Tn5ΩApra | This work |
| A1480 | 300 rapA2ΩSpec rapC::Tn5ΩApra | This work |
Table 3.
Plasmids
| Plasmid | Description | Reference |
|---|---|---|
| pIJ9252 | plyB′-lacZ | Edwards et al. (2009) |
| pIJ9651 | rapA2′-gus | This work |
| pIJ9686 | cadA′-lacZ | This work |
| pIJ9724 | cadB′-lacZ | This work |
| pIJ11112 | praR′ -lacZ | This work |
| pIJ11114 | RL0149′-lacZ | This work |
| pIJ11171 | rapC′-lacZ | This work |
| pIJ11195 | pssA′-lacZ | This work |
| pIJ11196 | rosR′-lacZ | This work |
| pIJ11200 | gmsA′-lacZ | This work |
| pIJ11275 | rapB′-lacZ | This work |
| pIJ11276 | plyC′-lacZ | This work |
| pIJ11283 | plyA′-lacZ | This work |
| pIJ11268 | Broad host range; promoterless luxCDABE TetR, AmpR | This work |
| pIJ11282 | Derivative of pIJ11268 with nptII promoter | This work |
| pIJ11304 | For exchanging KanR in Tn5 to ApraR | This work |
Mutations in strain 300 were transduced from the appropriate 3841 strains containing single mutations. To facilitate construction of strains carrying multiple mutations, the nptII gene on Tn5 in some mutants was replaced by the spectinomycin or gentamicin resistance genes on plasmids pJQ173 and pJQ175 respectively as described previously (Quandt et al., 2004). Also, an apramycin-resistance plasmid was made by amplifying the apramycin resistance gene on pIJ733 using the primers pApra and aac5 (Table S2) and cloning it as a BamHI fragment into BamHI-cut pJQ173 to give plasmid pIJ11304 which was used in the same way as pJQ173. The praR::Tn5 mutation was transduced into 3841 (WT) to make A1042; to make praR mutants with mutations in additional genes, A1042 (praR::Tn5) or A1345 (praR::Tn5ΩSpec) were transduced using phage RL39 propagated on strains carrying the appropriate single mutations, selecting with appropriate antibiotics. One of these mutants, A1363 (praR, cadA) was then transduced with phage from A1261 to make the praR, cadA, cadB mutant A1383. A1374 (praR, rapC) was transduced with phage from A1425 to make the praR, rapB, rapC mutant A1428. The rapA2, praR double mutant A1328 was made by transduction of A1132 (praR::Tn5) using phage from A1206. The rapB::Tn5ΩGent and rapCTn5ΩApra alleles were introduced into A1328 to make A1427 and A130 respectively, by transduction using phage propagated on A1425 or A1426. The praR, rapA2, rapB, rapC mutant A1429 was generated by transduction of A1427 with phage from A1426. A1480 a derivative of 300 carrying mutations in rapA2 and rapC was made by sequential transduction with phage from A1206 and A1426.
Promoter fusion reporter plasmids were constructed by cloning PCR-amplified promoters into the lacZ reporter plasmid pMP220 (Spaink et al., 1987) or the gus reporter plasmid pRU1156 (Karunakaran et al., 2005). Promoter amplification was carried out using the gene-specific primers listed in Table S2. Either restriction sites (underlined) were introduced during PCR or the amplification product was first cloned into pGEM T-easy and then excised as an EcoRI fragment. The correct orientation of the insert was determined by PCR using a vector and gene-specific primers.
Protein work
Protein electrophoresis and MALDI-ToF were done as described (Krehenbrink and Downie, 2008). Gel shift assays were done as described previously (Frederix et al., 2011) with the same promoter fragments amplified to make the reporter-plasmid fusions described above. These fragments were amplified using the primers listed in Table S2 and then end-labelled using γ32P-ATP. Each 20 microlitres of reaction buffer (20 mM TrisHCL, 100 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 0.5 mM dithiothreitol and 8% glycerol by volume), contained 0.1 nM of each labelled promoter fragment, 200 ng salmon sperm DNA and varying amounts of the maltose-binding-PraR fusion protein (MBP-PraR) as described previously (Frederix et al., 2011). As indicated in Fig. S2, different amounts of CinS-His5 purified as described previously (Frederix et al., 2011) were added after the addition of 250 nM MBP-PraR.
RNA purification and microarrays
Three independent Rhizobium cultures were grown in AMS minimal medium to exponential phase (OD600 0.7–0.8). Samples (12 ml) were rapidly mixed with 24 ml RNAlater (Qiagen) and stored until further use. RNA was purified from Rhizobium cultures using the RNeasy Mini kit (Qiagen) as described in the manufacturer's instructions. Labelling and microarray analysis was performed as described previously using custom-designed arrays with unique 70-mer oligonucleotides representing 7344 genes of R.l. viciae strain 3841 as described previously (Karunakaran et al., 2009). Duplicates or triplicates of the 70mers were printed on the arrays in a random pattern such that each array included technical replicates and data were analysed using Genespring 7.2 (Silicon Genetics, Redwood, CA). After subtracting local background values from the intensity of each spot, a Lowess normalization was applied. Dye swaps were done on different biological repeats and the normalized expression ratios were calculated as described previously (Karunakaran et al., 2009).
Radioactivity and bacterial count-based root attachment assays
Pea seeds were surface-sterilized by washing with 70% ethanol and incubating in 1% hypochlorite for 5 min then germinated on water agar plates in the dark at room temperature for 4–5 days. Bacteria were grown overnight in Y mannitol MOPS containing 185 kBq of 3H-leucine to OD600 0.5. Cells were pelleted washed twice and then resuspended in an equal volume of 25 mM phosphate buffer pH 6.0. Aliquots were counted showing that 23.7 kBq had been incorporated and that there was less than 2% difference of incorporation in the two samples. Aliquots (100 μl) were added to 50 ml Falcon tubes containing 10 ml FP medium and 10 pea roots (2 cm lengths from the root tip). Tubes were incubated on a rocking platform for 2 h at room temperature. The roots were then washed three times with 20 ml of FP medium. The roots were then put into separate scintillation vials containing HiSafe2 scintillation fluid (Perkin Elmer) and counted in a Perkin Elmer Scintillation Counter.
In parallel experiments performed without radioactivity labelling, after incubation with bacteria and washing of the roots, attached bacteria were released by extensive vortexing (30 min) of the roots in phosphate buffer containing 0.5 mM EDTA and the released bacteria were counted by plating.
Luminescence-based root attachment arrays
A stably inherited plasmid conferring luminescence was constructed using the pJP2 replicon (Prell et al., 2002). To generate a fragment with the luxCDABE operon containing appropriate upstream cloning sites, pBlueLux (Brackman et al., 2008) was first linearized by digestion with XhoI and EcoRI and then recircularized, ligating in a linker made by annealing the oligonucleotides 5′- TCGAGGGTACCCTCGAGGGATCCGTTTAAACG-3′ and 5′-AATTCGTTTAAACGGATCCCTCGAGGGTACCC-3′); this introduced KpnI, XhoI, BamHI, PmeI and EcoRI sites upstream of luxC and there is a PstI site downstream of luxE. The 6 kb KpnI–PstI fragment containing the luxCDABE operon was then cloned into KpnI- and PstI-digested pJP2 to generate pIJ11268, in which the β-glucuronaidase has been replaced by the lux operon with unique KpnI, XhoI, BamHI and PmeI sites upstream of luxC (Fig. S3). To generate pIJ11282, in which the lux operon is expressed under the control of the nptII promoter from Tn5 (Fig. S3), the nptII promoter was amplified as a 400 bp fragment (using the primers 5′-TTTGGTACCAGGCCTGAATCGCCCCATC-3′ and 5′-CTTCGAATTCGAGCTCCCGGGTAC-3′, cut with KpnI and EcoRI and cloned into the same sites upstream of luxC in pIJ11268.
Plasmid pIJ11282 was then transferred by conjugation into R.l. viciae strains and these strains were grown, resuspended in phosphate buffer and added to pea roots as described above. The roots were then washed as described above and attached bacteria were measured by luminescence visualized using a NightOWL LB 983 in vivo Imaging System (Berthold Technologies, Bad Wildbad Germany); luminescence from individual roots was quantified using an FB12 Luminometer (Berthold Technologies). Luminescence is scored as relative light units; data shown in individual figures were all collected under identical conditions, and so are directly comparable. The RLU values may vary between experiments but the ratios of the signals remained very similar within experiments.
Acknowledgments
We thank Rachel Duffy for assisting with the initial trials of the lux system and the Nuffield Trust for supporting her visit. We thank Alison East for her advice and Gerhard Saalbach for MALDI-ToF and Q-ToF analysis. This work was supported by the Biotechnology and Biological Sciences Research Council by a Grant-in-Aid, Grant BB/E017045/1 (to JAD), and a studentship (to AW). MF was supported in part by a Marie-Curie short-term EST fellowship (019727) and JAD was supported in part by the John Innes Foundation.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web-site.
Supporting information
References
- Abdian PL, Caramelo JJ, Ausmees N, Zorreguieta A. RapA2 is a calcium-binding lectin composed of two highly conserved cadherin-like domains that specifically recognize Rhizobium leguminosarum acidic exopolysaccharides. J Biol Chem. 2013;288:2893–2904. doi: 10.1074/jbc.M112.411769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiba N, Aono T, Toyazaki H, Sato S, Oyaizu H. A phrR-like gene, praR, of Azorhizobium caulinodans ORS571 is essential for symbiosis with Sesbania rostrata, and is involved in the expression of reb genes. Appl Environ Microbiol. 2010;76:3475–3485. doi: 10.1128/AEM.00238-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausmees N, Jacobsson K, Lindberg M. A unipolarly located, cell-surface-associated agglutinin, RapA, belongs to a family of Rhizobium-adhering proteins (Rap) in Rhizobium leguminosarum bv. trifolii. Microbiology. 2001;147:549–559. doi: 10.1099/00221287-147-3-549. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximisation to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Beringer JE. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- Bittinger MA, Milner JL, Saville BJ, Handelsman J. rosR, a determinant of nodulation competitiveness in Rhizobium etli. Mol Plant Microbe Interact. 1997;10:180–186. doi: 10.1094/MPMI.1997.10.2.180. [DOI] [PubMed] [Google Scholar]
- Brackman G, Defoirdt T, Miyamoto C, Bossier P, Van Calenbergh S, Nelis H, et al. Cinnamaldehyde and cinnamaldehyde derivatives reduce virulence in Vibrio spp. by decreasing the DNA-binding activity of the quorum sensing response regulator LuxR. BMC Microbiol. 2008;8:149. doi: 10.1186/1471-2180-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston AV. Generalized transduction in Rhizobium leguminosarum. J Gen Microbiol. 1979;112:135–142. [Google Scholar]
- Chai Y, Chu F, Kolter R, Losick R. Bistability and biofilm formation in Bacillus subtilis. Mol Microbiol. 2008;67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F, Kearns DB, Branda SS, Kolter R, Losick R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol. 2006;59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- Cubo MT, Economou A, Murphy G, Johnston AW, Downie JA. Molecular characterization and regulation of the rhizosphere-expressed genes rhiABCR that can influence nodulation by Rhizobium leguminosarum biovar viciae. J Bacteriol. 1992;174:4026–4035. doi: 10.1128/jb.174.12.4026-4035.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie JA. The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol Rev. 2010;34:150–170. doi: 10.1111/j.1574-6976.2009.00205.x. [DOI] [PubMed] [Google Scholar]
- Edwards A, Frederix M, Wisniewski-Dye F, Jones J, Zorreguieta A, Downie JA. The cin and rai quorum-sensing regulatory systems in Rhizobium leguminosarum are coordinated by ExpR and CinS, a small regulatory protein coexpressed with CinI. J Bacteriol. 2009;191:3059–3067. doi: 10.1128/JB.01650-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnie C, Hartley NM, Findlay KC, Downie JA. The Rhizobium leguminosarum prsDE genes are required for secretion of several proteins, some of which influence nodulation, symbiotic nitrogen fixation and exopolysaccharide modification. Mol Microbiol. 1997;25:135–146. doi: 10.1046/j.1365-2958.1997.4471803.x. [DOI] [PubMed] [Google Scholar]
- Finnie C, Zorreguieta A, Hartley NM, Downie JA. Characterization of Rhizobium leguminosarum exopolysaccharide glycanases that are secreted via a type I exporter and have a novel heptapeptide repeat motif. J Bacteriol. 1998;180:1691–1699. doi: 10.1128/jb.180.7.1691-1699.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederix M, Edwards A, McAnulla C, Downie JA. Co-ordination of quorum-sensing regulation in Rhizobium leguminosarum by induction of an anti-repressor. Mol Microbiol. 2011;81:994–1007. doi: 10.1111/j.1365-2958.2011.07738.x. [DOI] [PubMed] [Google Scholar]
- Gerdes K, Maisonneuve E. Bacterial persistence and toxin-antitoxin loci. Annu Rev Microbiol. 2012;66:103–123. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- Janczarek M, Urbanik-Sypniewska T. Expression of the Rhizobium leguminosarum bv. trifolii pssA gene, involved in exoplolysaccharide synthesis, is regulated by RosR, phosphate and the carbon source. J Bacteriol. 2013;195:3412–3423. doi: 10.1128/JB.02213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janczarek M, Jaroszuk-Scisel J, Skorupska A. Multiple copies of rosR and pssA genes enhance exopolysaccharide production, symbiotic competitiveness and clover nodulation in Rhizobium leguminosarum bv. trifolii. Antonie Van Leeuwenhoek. 2009;96:471–486. doi: 10.1007/s10482-009-9362-3. [DOI] [PubMed] [Google Scholar]
- Johnston AW, Beringer J. Identification of the Rhizobium strains in pea root nodules using genetic markers. J Gen Microbiol. 1975;87:343–350. doi: 10.1099/00221287-87-2-343. [DOI] [PubMed] [Google Scholar]
- Karunakaran R, Mauchline TH, Hosie AH, Poole PS. A family of promoter probe vectors incorporating autofluorescent and chromogenic reporter proteins for studying gene expression in Gram-negative bacteria. Microbiology. 2005;151:3249–3256. doi: 10.1099/mic.0.28311-0. [DOI] [PubMed] [Google Scholar]
- Karunakaran R, Ramachandran VK, Seaman JC, East AK, Mouhsine B, Mauchline TH, et al. Transcriptomic analysis of Rhizobium leguminosarum biovar viciae in symbiosis with host plants Pisum sativum and Vicia cracca. J Bacteriol. 2009;191:4002–4014. doi: 10.1128/JB.00165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm production by Bacillus subtilis. Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- Krehenbrink M, Downie JA. Identification of protein secretion systems and novel secreted proteins in Rhizobium leguminosarum bv. viciae. BMC Genomics. 2008;9:55. doi: 10.1186/1471-2164-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laus MC, van Brussel AA, Kijne JW. Role of cellulose fibrils and exopolysaccharides of Rhizobium leguminosarum in attachment to and infection of Vicia sativa root hairs. Mol Plant Microbe Interact. 2005;18:533–538. doi: 10.1094/MPMI-18-0533. [DOI] [PubMed] [Google Scholar]
- Laus MC, Logman TJ, Lamers GE, Van Brussel AA, Carlson RW, Kijne JW. A novel polar surface polysaccharide from Rhizobium leguminosarum binds host plant lectin. Mol Microbiol. 2006;59:1704–1713. doi: 10.1111/j.1365-2958.2006.05057.x. [DOI] [PubMed] [Google Scholar]
- Milner JL, Araujo RS, Handelsman J. Molecular and symbiotic characterization of exopolysaccharide-deficient mutants of Rhizobium tropici strain CIAT899. Mol Microbiol. 1992;6:3137–3147. doi: 10.1111/j.1365-2958.1992.tb01770.x. [DOI] [PubMed] [Google Scholar]
- Mongiardini EJ, Ausmees N, Pérez-Giménez J, Julia Althabegoiti M, Ignacio Quelas J, López-Garcia SL, et al. The rhizobial adhesion protein RapA1 is involved in adsorption of rhizobia to plant roots but not in nodulation. FEMS Microbiol Ecol. 2008;65:279–288. doi: 10.1111/j.1574-6941.2008.00467.x. [DOI] [PubMed] [Google Scholar]
- Mongiardini EJ, Pérez-giménez J, Althabegoiti MJ, Covelli J, Quelas JI, López-Garcia SL, et al. Overproduction of the rhizobial adhesin RapA1 increases competitiveness for nodulation. Soil Biol Biochem. 2009;41:2017–2020. [Google Scholar]
- Oldroyd GE, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- Pobigaylo N, Szymczak S, Nattkemper TW, Becker A. Identification of genes relevant to symbiosis and competitiveness in Sinorhizobium meliloti using signature-tagged mutants. Mol Plant Microbe Interact. 2008;21:219–231. doi: 10.1094/MPMI-21-2-0219. [DOI] [PubMed] [Google Scholar]
- Poole PS, Schofield NA, Reid CJ, Drew EM, Walshaw DL. Identification of chromosomal genes located downstream of dctD that affect the requirement for calcium and the lipopolysaccharide layer of Rhizobium leguminosarum. Microbiology. 1994;140:2797–2809. doi: 10.1099/00221287-140-10-2797. [DOI] [PubMed] [Google Scholar]
- Prell J, Boesten B, Poole P, Priefer UB. The Rhizobium leguminosarum bv. viciae VF39 gamma-aminobutyrate (GABA) aminotransferase gene (gabT) is induced by GABA and highly expressed in bacteroids. Microbiology. 2002;148:615–623. doi: 10.1099/00221287-148-2-615. [DOI] [PubMed] [Google Scholar]
- Quandt J, Hynes MF. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- Quandt J, Clark RG, Venter AP, Clark SR, Twelker S, Hynes MF. Modified RP4 and Tn5-Mob derivatives for facilitated manipulation of large plasmids in Gram-negative bacteria. Plasmid. 2004;52:1–12. doi: 10.1016/j.plasmid.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Reeve WG, Tiwari RP, Wong CM, Dilworth MJ, Glenn AR. The transcriptional regulator gene phrR in Sinorhizobium meliloti WSM419 is regulated by low pH and other stresses. Microbiology. 1998;144:3335–3342. doi: 10.1099/00221287-144-12-3335. [DOI] [PubMed] [Google Scholar]
- Rosemeyer V, Michiels J, Verreth C, Vanderleyden J. luxI- and luxR-homologous genes of Rhizobium etli CNPAF512 contribute to synthesis of autoinducer molecules and nodulation of Phaseolus vulgaris. J Bacteriol. 1998;180:815–821. doi: 10.1128/jb.180.4.815-821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo DM, Williams A, Edwards A, Posadas DM, Finnie C, Dankert M, et al. Proteins exported via the PrsD–PrsE type I secretion system and the acidic exopolysaccharide are involved in biofilm formation by Rhizobium leguminosarum. J Bacteriol. 2006;188:4474–4486. doi: 10.1128/JB.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Scott DJ, Leejeerajumnean S, Brannigan JA, Lewis RJ, Wilkinson AJ, Hoggett JG. Quaternary re-arrangement analysed by spectral enhancement: the interaction of a sporulation repressor with its antagonist. J Mol Biol. 1999;293:997–1004. doi: 10.1006/jmbi.1999.3221. [DOI] [PubMed] [Google Scholar]
- Sherwood MT. Improved synthetic medium for the growth of Rhizobium. J Appl Bacteriol. 1970;33:708–713. doi: 10.1111/j.1365-2672.1970.tb02253.x. [DOI] [PubMed] [Google Scholar]
- Smit G, Kijne JW, Lugtenberg BJ. Involvement of both cellulose fibrils and a Ca2+-dependent adhesin in the attachment of Rhizobium leguminosarum to pea root hair tips. J Bacteriol. 1987;169:4294–4301. doi: 10.1128/jb.169.9.4294-4301.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit G, Kijne JW, Lugtenberg BJ. Roles of flagella, lipopolysaccharide, and a Ca2+-dependent cell surface protein in attachment of Rhizobium leguminosarum biovar viciae to pea root hair tips. J Bacteriol. 1989;171:569–572. doi: 10.1128/jb.171.1.569-572.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaink H, Okker RJ, Wijffelman CA, Pees E, Lugtenberg B. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]
- Williams A, Wilkinson A, Krehenbrink M, Russo DM, Zorreguieta A, Downie JA. Glucomannan-mediated attachment of Rhizobium leguminosarum to pea root hairs is required for competitive nodule infection. J Bacteriol. 2008;190:4706–4715. doi: 10.1128/JB.01694-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Williams A, Edwards A, Downie JA. A plant arabinogalactan-like glycoprotein promotes a novel type of polar surface attachment by Rhizobium leguminosarum. Mol Plant Microbe Interact. 2012;25:250–258. doi: 10.1094/MPMI-08-11-0211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information


