SUMMARY
Lysine acetylation is a dynamic posttranslational modification with a well-defined role in regulating histones. The impact of acetylation on other cellular functions remains relatively uncharacterized. We explored the budding yeast acetylome with a functional genomics approach, assessing the effects of gene overexpression in the absence of lysine deacetylases (KDACs). We generated a network of 463 synthetic dosage lethal (SDL) interactions involving class I and II KDACs, revealing many cellular pathways regulated by different KDACs. A biochemical survey of genes interacting with the KDAC RPD3 identified 72 proteins acetylated in vivo. In-depth analysis of one of these proteins, Swi4, revealed a role for acetylation in G1-specific gene expression. Acetylation of Swi4 regulates interaction with its partner Swi6, both components of the SBF transcription factor. This study expands our view of the yeast acetylome, demonstrates the utility of functional genomic screens for exploring enzymatic pathways, and provides functional information that can be mined for future studies.
INTRODUCTION
Lysine acetylation influences gene expression (Robert et al., 2004), and the dynamic interplay between lysine acetyltransferases (KATs) and lysine deacetylases (KDACs) is required to maintain appropriate levels of histone acetylation to promote normal cell proliferation, growth, and differentiation. Consistent with a significant role for acetylation in regulating cell division, abnormal KAT/KDAC function results in disease states such as cancer (Archer and Hodin, 1999; Bradner et al., 2010; Chuang et al., 2009; Das and Kundu, 2005). The budding yeast genome encodes ten KDACs, which have been classified into three groups based on sequence homology (Gregoretti et al., 2004). Class I includes Rpd3, Hos1, and Hos2, and class II contains Hda1 and Hos3; these two classes are zinc-dependent KDACs. Class III enzymes, the sirtuins, use NAD+ as a cofactor for the deacetylase reaction (Shore, 2000; Smith et al., 2002).
Rpd3 exists in two large macromolecular complexes, Rpd3(L) and Rpd3(S), which share the core subunits Rpd3, Sin3, and Ume1 (Carrozza et al., 2005; Keogh et al., 2005; Shevchenko et al., 2008). Hda1 forms a heterotetrameric complex with two regulatory subunits, Hda2 and Hda3 (Wu et al., 2001). The HDA complex coregulates some genes with Rpd3, but also influences expression of a distinct group of genes (Bernstein et al., 2000). Although loss of either RPD3 or HDA1 is tolerated, deletion of both KDACs results in cell death (Lin et al., 2008; Rundlett et al., 1996). The other class I and II KDACs have received much less scrutiny. Hos2 appears to activate gene expression by restoring RNA polymerase-disrupted chromatin to a permissive state (Wang et al., 2002). Hos3 is the only class I/II KDAC that is insensitive to Tricostatin-A (TSA), an inhibitor of deacetylases (Carmen et al., 1999).
In the past decade, acetylation has been shown to regulate proteins other than histones (Glozak et al., 2005; Kurdistani and Grunstein, 2003). Proteomic studies identified ~2,500 acetylated proteins in mammalian cells, suggesting that acetylation may be as ubiquitous as phosphorylation (Choudhary et al., 2009; Glozak et al., 2005; Spange et al., 2009; Zhao et al., 2010). In yeast, the acetylome remains relatively unexplored, with only 28 nonhistone substrates identified to date (Basu et al., 2009; Beckouët et al., 2010; Borges et al., 2010; Choudhary et al., 2009; Heidinger-Pauli et al., 2009; Kim et al., 2010; Lin et al., 2008, 2009; Lu et al., 2011; Mitchell et al., 2011; Robert et al., 2011; VanDemark et al., 2007).
The genomic tools available in yeast provide an opportunity to systematically explore the acetylome and add functional information to our view of KAT/KDAC regulation. For example, synthetic genetic array (SGA) technology automates the analysis of genetic interactions in yeast and has been used to extensively map interactions between deletion alleles of nonessential genes (Baryshnikova et al., 2010; Costanzo et al., 2011). SGA has also been used to systematically assess synthetic dosage lethal (SDL) interactions (Sopko et al., 2006a). SDL interactions result when increased gene expression levels have little effect on the growth of a wild-type cell but produce a clear phenotype, such as lethality, in a specific mutant background (Kroll et al., 1996; Measday and Hieter, 2002; Sopko et al., 2006a). SDL interactions can identify enzyme-substrate relationships, and SDL screens have discovered targets of kinases (Huang et al., 2009; Sharifpoor et al., 2011; Sopko et al., 2006a; Zou et al., 2009), regulators of protein degradation (Liu et al., 2009), and lysine acetyltransferases (Mitchell et al., 2011). Hyperactivation of an opposing biological pathway or perturbation of protein complex stoichiometry may also result in SDL (Sopko et al., 2006b), and this can facilitate identification of new connections to a specific biological process (Measday et al., 2005).
Here, we describe the SDL interaction network for class I and II KDACs in yeast. Analysis of the network, combined with secondary biochemical tests, has allowed us to extend the list of acetylated proteins in yeast nearly 5-fold. To demonstrate the utility of this resource, we characterized one potential KDAC target identified in our rpd3Δ SDL screen in detail. Acetylation of the cell-cycle transcription factor Swi4 regulates its function in vivo, uncovering a new aspect of G1 transcription control in yeast. Our study expands the view of KDAC function and provides a valuable resource for predicting functional relationships in pathways regulated by KDACs. In addition, the data set captures functional information that cannot be revealed by biochemical surveys of the proteome.
RESULTS
Systematic Gene Overexpression Identifies 463 SDL Interactions for Class I and II KDACs
To perform whole-genome SDL screens, we introduced deletions of genes encoding class I and II KDACs into an arrayed collection of yeast strains, each conditionally overexpressing a unique gene (Sopko et al., 2006a). Overexpression phenotypes were measured with automated software using colony size as a proxy for cell fitness, an approach that we have validated in other large-scale genetic interaction screens (Baryshnikova et al., 2010; Costanzo et al., 2010; Sopko et al., 2006a). Genes whose overexpression resulted in a colony size reduction of > 20% compared to wild-type were independently confirmed (Figure S1 available online). A subset of interactions was confirmed in the presence of the KDAC inhibitor, TSA, showing that SDL interactions can result due to the loss of KDAC catalytic activity (Table S1). Our screens identified 463 SDL interactions (Figure 1A and Table S2) involving 374 unique genes, enriched for diverse processes including cell polarity and morphogenesis, protein sorting to the Golgi, endosome and vacuole, nuclear-cytoplasmic transport, peroxisome biogenesis, and drug/ion transport (Table 1). The SDL network includes genes encoding members of protein complexes (Figure 1A, black lines) and components of common pathways (Figure 1A, red lines), suggesting that acetylation may target protein complexes as well as coregulate many pathways, consistent with observations made in human acetylome studies (Choudhary et al., 2009) and SDL screens with NuA4 complex components in yeast (Mitchell et al., 2011).
Figure 1. Genetic Interactions Identified for Class I and II KDACs.
(A) Synthetic dosage lethal interactions for the catalytic subunits of the five class I and II KDACs are shown. Genes are grouped according to biological processes as annotated by Costanzo et al. (2010). The KDAC with which the SDL interaction occurs is indicated by a colored dot next to the gene. Lines connecting genes denote genes encoding proteins that either regulate the same biological pathway (red) or that are components of a common protein complex (black) as annotated in BioGRID. A complete list of SDL interactions is provided in Table S1.
(B) Bar graph showing enrichment of the rpd3Δ SDL interactions for biological process as defined by GO annotations (black columns) relative to the genome (gray bars).
(C) Enrichment in biological process categories for genes whose expression level changes in the absence of RPD3 (blue columns) relative to the genome (gray columns).
Table 1.
Gene Enrichments for KDAC SDL Screens
| Biological Processes | Fold Enrichment (p Value) |
|---|---|
| cell polarity/morphogenesis | 1.63 (5.8 × 10−3) |
| drug/ion transport | 1.85 (2.8 × 10−4) |
| Golgi/endosome/vacuole/sorting | 2.06 (1.4 × 10−4) |
| nuclear-cytoplasmic transport | 1.78 (4.8 × 10−2) |
| peroxisome biogenesis | 2.19 (2.4 × 10−2) |
Deletion of either RPD3 or HDA1 caused a significant fitness defect in standard conditions, and our screens in rpd3Δ and hda1Δ mutants produced the highest number of SDL interactions among KDAC mutants (244 and 168 interactions, respectively), consistent with other large-scale experiments showing an inverse relationship between degree of genetic interaction and fitness of a mutant strain (Costanzo et al., 2010). Because Rpd3 and Hda1 regulate a number of common genes (Bernstein et al., 2000), we expected some overlap between the interactions in the rpd3Δ and hda1Δ screens. Indeed, 57 SDL interactions were shared between the two KDACs (~34% of HDA1 and ~23% of RPD3 interactions [p < 2.54 × 10−34]). Consistent with significant nonoverlapping roles for each KDAC, the majority of SDL interactions appear to be unique to each deacetylase (Figure 1A). The SDL hits in each KDAC screen were enriched for distinct biological processes (Table S3), a property also observed in experiments assessing chromatin acetylation in the absence of KDACs (Robyr et al., 2002).
Given that KDACs regulate transcription through deacetylation of histones, a subset of SDL interactions may result from aberrant transcription in the absence of KDACs. For example, in some cases, overexpression of the gene from the GAL promoter together with the overexpression of the endogenous copy of the same gene due to histone hyperacetylation might lead to protein levels that are high enough to cause toxicity. To address this point, we compared our KDAC-SDL profiles to the gene expression profiles of rpd3Δ and hda1Δ mutants (Bernstein et al., 2000). Less than 4% of the genes that caused toxicity when overexpressed (12/244 genes in rpd3Δ and 6/168 genes in hda1Δ) were differentially regulated at the transcriptional level in the absence of the KDAC (Bernstein et al., 2000; Table S4, 2-fold cut-off]. Furthermore, SDL interactors of RPD3 and genes transcriptionally regulated by Rpd3 were enriched for distinct biological processes (Figures 1B and 1C). These results suggest that most SDL interactions do not reflect indirect effects resulting from defects in gene expression caused by the deletion of KDACs.
Gene Deletion and Gene Overexpression Uncover Distinct Genetic Interactions
Negative genetic interactions, such as synthetic sickness (SS) or lethality (SL), occur when the observed fitness of a double mutant is more severe than expected, given the fitness of the two single mutants (Mani et al., 2008). Our published SL/SS data set (Costanzo et al., 2010) includes 497 unique negative genetic interactions involving deletion alleles of class I and II KDACs. Of the 463 genes that were SDL in the absence of a KDAC, 14 were also SS/SL with the same KDAC (Table S5). Seven of these encode components of multiprotein complexes, consistent with the prediction that perturbation of stoichiometry gives rise to haploinsufficient phenotypes (Veitia, 2002). For example, Hda1 functions in a tetrameric complex (Carmen et al., 1996; Wu et al., 2001), and either overexpression or deletion of HDA1 caused lethality in the absence of RPD3; over-expression of HDA1 may mimic the deletion phenotype by disrupting HDA complex stoichiometry (Papp et al., 2003). The small overlap between SL and SDL data sets suggests that SL and SDL screens explore different facets of genetic interaction space (Kelley and Ideker, 2005; Measday et al., 2005; Sopko et al., 2006b; Tong et al., 2004).
The set of genetic interactions associated with mutation of a gene, the genetic interaction profile, can be used to construct correlation-based networks, allowing prediction of gene function, protein complexes, and biological pathways. To predict pathways that may be controlled by KDACs, we examined the correlation between the SDL profiles of the KDACs and available SL profiles (Costanzo et al. (2010). A correlation between a gene pair indicates that a set of genes that are toxic when overexpressed in a kdacΔ are also SL/SS when deleted in combination with the correlated gene (Figure 2). Our comparative analysis revealed that a correlation between genes may indicate involvement in a similar biological process. For example, two components of the SAGA acetyltransferase complex (SPT3 and TAF9) are SS/SL with a set of genes that exhibit an SDL phenotype in rpd3Δ mutants. This correlation profile is likely relevant because both Rpd3 and SAGA regulate transcription elongation (Carrozza et al., 2005; Daniel and Grant, 2007; Keogh et al., 2005; Li et al., 2007). Moreover, our data suggest that a correlation between a KDAC and several components of a protein complex with SL/SS correlations among them represents a strong biological connection (Figure 2, red lines). For example, the RPD3 SDL profile was correlated with the SL/ SS profiles of two genes required for autophagy, ATG13 and ATG4 (Funakoshi et al., 1997; Lang et al., 1998; Yorimitsu and Klionsky, 2005). Studies in cancer cells and in yeast show an increase in autophagy in the presence of KDAC inhibitors (Robert et al., 2011; Shao et al., 2004), and acetylation of some proteins promotes their degradation through autophagy (Choudhary et al., 2009; Jeong et al., 2009; Lin et al., 2009; Robert et al., 2011). The stability of Sae2, a protein involved in DNA damage repair, is regulated by the KDACs Rpd3 and Hda1 (Robert et al., 2011), highlighting the possibility that, in addition to Sae2, Rpd3 may be involved in negatively regulating other proteins that are required for autophagy. Thus, correlation networks may be useful for predicting nonchromatin substrates of KDACs.
Figure 2. Similarity Analysis of Synthetic Dosage Lethal and Synthetic Lethal/Sick Interaction Profiles for Yeast KDACs.

Similarity between the rpd3Δ and hda1Δ synthetic dosage lethal (SDL) profiles and the synthetic lethal/sick (SL/SS) profiles from Costanzo et al. (2010) was measured by computing Pearson correlation coefficients (PCCs). Gene pairs whose profile similarity exceeded a threshold PCC > 0.15 were connected in the network. Similarities between SDL and SL/SS profiles (black lines) as well as similarity between SL/SS and SL/SS profiles (red lines) of gene pairs are shown, and edge thickness is directly proportional to the degree of similarity. Nodes are colored according to the biological processes annotated by Costanzo et al. (2010).
SDL Screens Uncover Unique Roles for HDA Complex Subunits
The HDA complex is composed of a catalytic subunit, Hda1, and regulatory subunits, Hda2 and Hda3. Physical interactions among these subunits appear necessary for deacetylation activity of the complex in vivo and in vitro (Carmen et al., 1996; Wu et al., 2001). Because genes encoding proteins that are part of a complex generally have similar genetic interaction profiles (Collins et al., 2007; Tong et al., 2004), we reasoned that SDL experiments would provide an unbiased genetic approach to explore whether Hda1, Hda2, and Hda3 function only as part of the HDA complex or exhibit subunit-specific roles. We complemented our hda1Δ screen with two additional genome-wide screens for SDL interactions in hda2Δ and hda3Δ mutant strains. All interactions from each screen were cross-tested using serial spot dilution assays (Figure S1). Among the ~16 000 interactions tested, we identified 327 unique interactions for the HDA complex (Table S6). Surprisingly, only ~7% of the interactions were shared among all three of the components, whereas ~55% were specific to a single subunit (Figure 3A). The subunit-specific interactions were enriched for distinct biological processes, indicating that Hda2 and Hda3 may have unique functions (Figure 3B).
Figure 3. SDL Profiles for HDA Complex Subunits.
(A) Venn diagram highlighting the overlap of SDL interactions between HDA complex subunits. Numbers refer to shared SDL interactions, and a complete list of these interactions is provided in Table S5.
(B) Enrichment for the SDL interactions unique to each HDA complex subunit for biological process as annotated by Costanzo et al. (2010). The top panel shows fold enrichment for genes in a particular biological process, and the bottom panel shows the associated p values.
(C) Localization of a peroxisomal matrix marker in the absence of HDA complex components. Pot1-GFP was imaged in wild-type, hda1Δ, hda2Δ, and hda3Δ cells using confocal microscopy.
(D) Graph showing quantified data for the Pot1-GFP localization phenotype for the strains shown in (C). Error bars represent standard deviations from the mean generated from three independent experiments.
See also Figure S2.
Many genes involved in peroxisome biogenesis or maintenance were identified in the hda2Δ screen, suggesting a previously underappreciated role for the HDA complex in peroxisome biology. Of the 25 peroxisome-related genes that were present on our array, six were toxic in the absence of either HDA1 or HDA2, two were toxic only in the absence of HDA1, and eight were toxic only in the absence of HDA2, whereas only one was toxic in the absence of HDA3 (Table S6). We saw no difference in the number of peroxisomes between a wild-type strain and strains mutated for the three components of the HDA complex (Figure S2A), suggesting that the HDA complex has no role in regulating peroxisome number. However, we saw a reduction in translocation of the peroxisome matrix protein Pot1-GFP into the lumen of the peroxisome in the absence of HDA complex components (Figure 3C), with the hda2 mutant showing the most dramatic defect (33% of the cells had Pot1-GFP in the lumen of the peroxisome compared to 84% in a wild-type strain [Figure 3D]). These results suggest a potential role for the HDA complex in peroxisome protein import.
The SDL Data Set Is Enriched for In Vivo Acetylated Proteins
We next examined the proteins encoded by genes that had an SDL interaction with a single KDAC in greater detail. We chose the Rpd3 screen as a test case, as Rpd3 is the best-studied KDAC in S. cerevisiae, and its human homolog HDAC1 deacetylates multiple proteins in human cells (Glozak et al., 2005). We asked whether genes encoding acetylated proteins, which are probable KDAC targets, were enriched among the SDL interactions in our rpd3Δ screen (Figure 4A). Given the variability in epitope recognition for anti-acetyl lysine antibodies (Mitchell et al., 2011), it is possible that this assay has some false negatives. We tested 184 proteins whose overexpression was toxic in the absence of RPD3 and identified 72 in vivo acetylated proteins (40%; Figure 4B and Table S7). Only two, Yng2 and Rsc4, were previously shown to be acetylated (Choi et al., 2008; VanDemark et al., 2007). Our biochemical survey of RPD3 SDL hits expands the list of known acetylated proteins in yeast from 28 to 97 and identifies proteins involved in transcription, cell polarity and budding, growth and morphogenesis, vesicle fusion, and trans-membrane transport, consistent with the myriad roles of acetylated proteins in mammalian cells (Choudhary et al., 2009; Glozak et al., 2005; Zhao et al., 2010). Because we had seen effects of a KDAC on localization of peroxisomal proteins, we also tested our peroxisomal markers and found that Pot1, but not Pex15, was acetylated in vivo (Figure S2B). We also assayed a random set of 94 proteins and detected acetylation on ~20% of the proteins tested (Table S6). Thus, the KDAC-SDL roster is significantly enriched for acetylated proteins (~40% versus 20%; p < 1.5 × 10−14). We note that we have not been able to reproducibly assay acetylation in the absence of the KDAC because these proteins cause toxicity when overproduced in the absence of RPD3. However, consistent with direct regulation by the KDAC, 27/244 proteins encoded by genes SDL with RPD3 have protein-protein interactions (PPIs) with Rpd3 or an associated subunit (Sardiu et al., 2009; Stark et al., 2011; Table S2).
Figure 4. Analysis of Acetylation on Proteins Encoded by rpd3Δ SDL Genes.
(A) Western blot analysis of proteins using acetyl-lysine antibody. A wild-type strain was transformed with plasmids carrying genes identified in the rpd3Δ screen and the GST-proteins purified using glutathione Sepharose. The top panel shows a representative western blot with partially purified GST-tagged proteins probed with acetyl-lysine antibody. The bottom panel shows an anti-GST western blot to show protein levels in the pull-down experiment. Acetylation was clearly detected on known acetylation targets (Yng2, Rsc4) and on three of the five proteins tested (asterisk indicates acetylated proteins).
(B) Diagram showing proteins that were detectably acetylated in vivo encoded by genes that were also SDL in the rpd3Δ screen. Nodes are color coded according to biological processes as annotated by Costanzo et al. (2010). Yeast orthologs of mammalian proteins known to be acetylated are circled in black and are listed in Table S6.
See also Table S7.
A Cell Biological Screen Detects Proteins Mislocalized in the Absence of RPD3
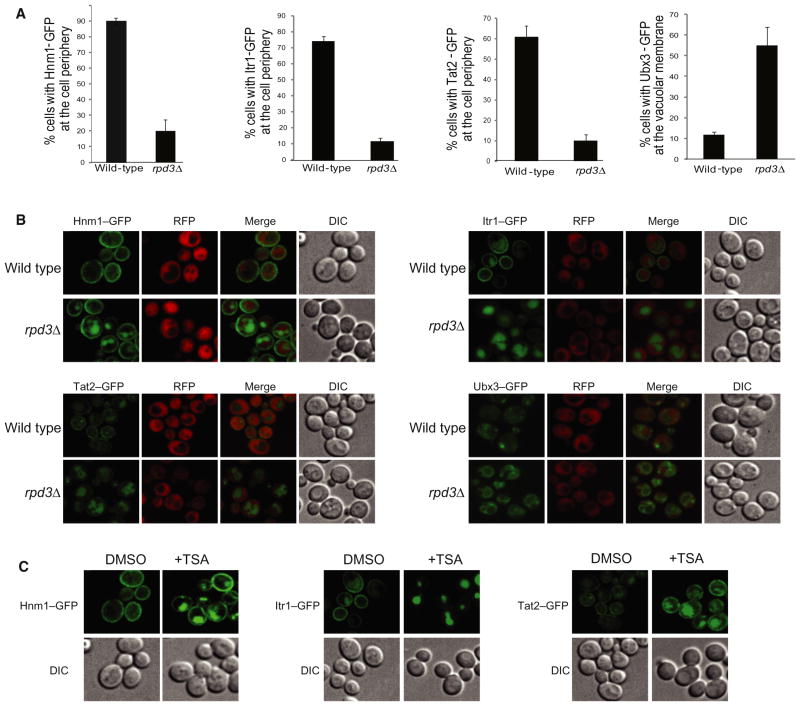
Similar to phosphorylation, acetylation can affect protein stability, protein-protein interactions, enzymatic activity, and protein localization (Kouzarides, 2000). To assess whether Rpd3 influences protein localization, we examined the subcellular localization of proteins identified in the rpd3Δ screen (Huh et al., 2003). Of the 187 proteins tested, we saw a defect in localization for four in the absence of RPD3: Ubx3, a ubiquitin regulatory X domain-containing protein, and three membrane proteins (Figure 5A). The group of genes most significantly enriched in the rpd3Δ SDL screen included membrane proteins and receptors with annotated roles in drug and ion transport (Table S3). Hnm1, a choline/ethanolamine transporter (Nikawa et al., 1986), Tat2, a tryptophan/tyrosine permease (Schmidt et al., 1994), and Itr1, a myo-inositol transporter (Nikawa et al., 1991), were mislocalized from their normal cell surface localization in wild-type cells to the vacuole in an rpd3Δ strain (Figure 5B). We saw a similar mislocalization phenotype when cells were treated with the KDAC inhibitor TSA (Figure 5C), suggesting that KDAC activity is required for proper transporter localization. We did not detect acetylation of any of these transporters, and thus it is possible that localization of some cell surface receptors in yeast may be regulated indirectly by acetylation.
Figure 5. Aberrant Localization of Proteins that Cause SDL in the Absence of RPD3.
(A) Yeast strains carrying C-terminally GFP-tagged proteins and cytosolic tdTomato under the control of the RPL39 promoter were imaged in wild-type and rpd3Δ backgrounds using confocal microscopy. Graphs show quantified localization changes for four proteins that changed localization in the absence of RPD3. Error bars represent standard deviation from the mean generated by three independent experiments.
(B) Increased vacuolar localization of transporters Hnm1, Itr1, and Tat2 in the absence of RPD3. Untagged tdTomato protein, which localizes to the cytosol and the nucleus but is excluded from the vacuole, was used to assess transporter localization in rpd3Δ mutants. Ubx3-GFP localization was punctate in wild-type but localized to vacuolar membrane in rpd3Δ cells.
(C) Strains carrying C-terminally GFP-tagged proteins were imaged in wild-type cells in the presence or absence of the KDAC inhibitor TSA using confocal microscopy.
Swi4 Is Regulated by Acetylation
To further study direct nonhistone KDAC targets, we compared our SDL screens to the results of high-sensitivity mass spectrometry experiments in mammalian cells (Choudhary et al., 2009; Kim et al., 2006; Zhao et al., 2010; Zhou et al., 2010). Thirty six percent (903) of the human proteins that are acetylated have yeast homologs (O’Brien et al., 2005), 52 of which were toxic when overexpressed in the absence of KDACs and 38 were specifically SDL in the absence of RPD3: 12 of these proteins were acetylated in vivo (Figure 4B, black circles). One of these proteins, Swi4, is the yeast analog of the mammalian transcription factor E2F, which is regulated by acetylation (Martínez-Balbás et al., 2000; Marzio et al., 2000). Both Swi4 and E2F activate G1-specific transcription via a regulatory pathway that is well conserved between budding yeast and higher eukaryotes (Costanzo et al., 2004; de Bruin et al., 2004; Schaefer and Breeden, 2004). Acetylation of E2F at sites adjacent to its DNA-binding domain augments DNA binding, increases stability, and stimulates transactivation activity (Martínez-Balbás et al., 2000; Marzio et al., 2000). Swi4 is the DNA-binding component of the transcription factor SBF and interacts with a heterodimeric partner Swi6 to regulate genes expressed in late G1 (Wittenberg and Reed, 2005). The repressor of SBF, Whi5, mediates repression in part through interaction with two KDACs, Hos3 and Rpd3 (Huang et al., 2009; Wang et al., 2009), and Rpd3 is recruited to G1 promoters, placing it near Swi4 (Robert et al., 2004; Takahata et al., 2009; Wang et al., 2009).
These data, together with the SDL interaction between RPD3 and SWI4 (Figure 6A), suggest that Swi4 may be directly regulated by acetylation. Consistent with an enzyme-substrate relationship between Rpd3 and Swi4, the levels of Swi4 acetylation increased in an rpd3Δ mutant (Figure 6B). Using mass spectrometry, we identified two acetylation sites, K1016 and K1066 (Figure S3), in the C-terminal domain of Swi4, which is required for interaction with its regulatory partner, Swi6 (Andrews and Moore, 1992). We mutated both residues to either arginine, which mimics constitutive deacetylation (Swi4-RR), or glutamine, which mimics constitutive acetylation (Swi4-QQ), at the endogenous SWI4 locus (Figure 6C). We then assessed the Swi4 mutant strains for phenotypes associated with defects in Swi4 function. The point mutations had little effect on cell growth (Figure S4A) or protein abundance (Figure S4B) and did not detectably alter the ability of Swi4 to bind to the CLN2 promoter in log-phase cells (Figure S4C). To assess the potential effects of defects in Swi4 acetylation on G1 transcription, we synchronized wild-type and swi4 mutant strains in G1 and then followed the expression of a Swi4 target gene, CLN2, using quantitative real-time PCR (Q-PCR) through the cell cycle. As expected, expression of CLN2 was induced during the G1-S phase transition in wild-type cells but was constitutively expressed at low levels in the swi4Δ strain (Figure 6D; Cross et al., 1994). Likewise, in cells expressing the constitutive deacetylation mimic, Swi4-RR, induction of CLN2 was dramatically reduced. In contrast, cells expressing the Swi4-QQ protein showed no defect in CLN2 expression. These results suggest that acetylation of Swi4 is important for its role in activating G1-specific transcription.
Figure 6. Regulation of the G1 Transcription Factor Swi4 by Acetylation In Vivo.
(A) Growth defect caused by overexpression of SWI4 in the absence of RPD3. Wild-type, rpd3Δ, hda1Δ, hos1Δ, hos2Δ, and hos3Δ strains bearing either vector or pGAL-GST-SWI4 were assessed for growth defects using liquid growth assays. The growth rate of all strains was measured relative to that of a wild-type strain transformed with vector (set at 100% fitness). Error bars represent standard deviation from the mean generated by three independent experiments.
(B) Acetylation of Swi4 in KDAC mutants. Endogenously produced Swi4-myc from wild-type and kdac mutant strains was immunoprecipitated and protein levels assessed using anti-myc antibody (bottom). Swi4 acetylation in the immunoprecipitates was assayed using an antibody against acetylated lysines.
(C) Schematic diagram of Swi4 showing location of relevant protein domains and the location of acetylated lysines (1016 and 1066; arrows).
(D) Cell-cycle regulation of G1 transcription in Swi4 acetylation site mutants. cDNA was prepared from strains containing wild-type Swi4, a point mutant mimicking constitutive acetylation, Swi4-QQ, a point mutant that mimics constitutive deacetylation, Swi4-RR, and a swi4Δ strain. CLN2 expression levels were normalized to transcript levels of ACT1. Error bars represent standard deviation from the mean generated by three independent experiments.
(E) Cross-linking of Swi6 and Swi4 to the CLN2 promoter in Swi4 acetylation site mutant strains. Swi6 association with the CLN2 promoter was detected using chromatin immunoprecipitation (ChIP) in both asynchronous samples and cells synchronized in G1 using α factor (15 min after release). Immunoprecipitations were performed using antibodies specific to Swi4 and Swi6. The presence of CLN2 promoter sequence was detected using quantitative RT-PCR. ChIP efficiency is shown as a ratio between Swi6 and Swi4. Error bars represent standard deviation from the mean generated by three independent experiments.
(F) Coimmunoprecipitation of Swi4 and Swi6 from KDAC mutant extracts. Endogenously tagged Swi4-myc was immunoprecipitated from wild-type, rpd3Δ, hda1Δ, and hos1Δ strains. The associated Swi6 was detected using an antibody specific to Swi6. Antibodies against the myc tag were used to test levels of immunoprecipitated Swi4.
See also Figures S3 and S4.
The acetylated residues of Swi4 reside in the C-terminal domain, which is required for interaction with Swi6. We therefore tested whether the Swi4-Swi6 interaction is regulated by Swi4 acetylation. First, we assessed binding of Swi6 and Swi4 to the CLN2 promoter. In both asynchronous cells and G1-synchronized cells, the ratio of Swi6 to Swi4-RR at the CLN2 promoter was reduced relative to both wild-type Swi4 and Swi4-QQ (Figure 6E), suggesting that acetylation of Swi4 may be needed for a stable association with Swi6 at G1 promoters. We also saw reduced association between Swi4-RR and Swi6 in a coimmuno-precipitation experiment (Figure S4D). Conversely, deletion of RPD3 increased the Swi4-Swi6 interaction (Figure 6F), suggesting that acetylation of Swi4 promotes association between Swi4 and Swi6. These results establish a role for Swi4 acetylation in regulating G1 transcription.
DISCUSSION
Exploration of the Yeast Lysine Acetylome Using Genetic Interactions
We report a systematic assessment of synthetic dosage interactions for yeast class I and II KDACs. Although a role for acetyl-transferases and deacetylases in regulating nonhistone proteins in higher eukaryotes has been previously appreciated, less information is available about the acetylation status of the S. cerevisiae proteome. Our comprehensive genetic data set provides a powerful functional counterpart to biochemical efforts to explore protein acetylation and links diverse biological pathways to lysine deacetylases.
Mechanisms for SDL
An SDL screen with the gene encoding the cyclin-dependent kinase Pho85 identified several genes encoding negatively regulated substrates of the kinase (Sopko et al., 2006a), suggesting that some SDL interactions may be caused by an accumulation of unmodified substrate (Sopko et al., 2007). We identified Yng2, a known target of Rpd3 (Lin et al., 2008), and Swi4, a novel target of Rpd3, in our SDL screen, both negatively regulated by deacetylation. Consistent with the possibility that SDL identifies direct targets of KDACs, the RPD3 SDL screen was enriched for genes encoding acetylated proteins, although the functional relevance of most of these acetylation events is unknown.
A second mechanism that could lead to SDL involves indirect effects of acetylation. Our rpd3Δ SDL screen was enriched for proteins involved in drug/ion transport. Although several of these transporters were mislocalized in the absence of RPD3 or in the presence of TSA, they did not appear to be acetylated. In this case, SDL may reflect defective regulation of components involved in protein transport. In human cells, the removal of KDAC6 results in increased degradation of EGFR (Deribe et al., 2009; Gao et al., 2010). However, it is the acetylation of α-tubulin, a nonhistone target of KDAC6, that is required for correct localization of EGFR as well as two other proteins: JNK-interaction protein 1 (Reed et al., 2006), and brain-derived neurotrophic factor (Dompierre et al., 2007).
A third mechanism that may result in SDL is hyperactivation of an opposing pathway. In this case, overexpression of a KAT in the absence of a cognate KDAC may produce SDL due to either hyperacetylation of histones and aberrant gene expression or the constitutive hyperacetylation of a common substrate. Consistent with this model, we identified two regulatory components of KATs, Hfi1 and Yng2, whose overproduction caused SDL in rpd3Δ. Multisubunit complexes such as KATs may require the overexpression of accessory subunits in addition to the overexpression of the catalytic subunit to produce a functional enzyme (Utley and Côté, 2003).
In a fourth mechanism, SDL may arise from perturbed stoichiometry of a multisubunit complex. Here, deletion and overexpression phenotypes are typically concordant, consistent with the balance hypothesis (Veitia, 2002). In this case, genes that result in SDL are likely to be corepressors that function in parallel with the given KDAC to produce the same biological outcome. One such example is Hda1, which is part of a tetrameric complex (Carmen et al., 1996; Wu et al., 2001). Although we highlight several mechanisms by which SDL may arise, additional mechanisms may be uncovered as the functional relevance of protein acetylation is characterized.
Novel Functions for the HDA Complex
Genes encoding proteins in the same complex are predicted to have similar genetic interactions (Collins et al., 2007; Tong et al., 2004); however, more than half of the dosage interactions identified for the HDA complex were unique to a given subunit. Most SL interactions and most PPIs described in the literature for hda1Δ, hda2Δ, and hda3Δ are also unique (Stark et al., 2011). Likewise, a dramatic difference in the number of SL and SDL interactions and a lack of overlap between complex components was observed for the NuA4 acetyltransferase complex (Mitchell et al., 2008; Mitchell et al., 2011). Because we cross-tested all interactions identified in our HDA complex screens, we have high confidence in our SDL data. HDA2 has several unique SDL interactions with genes whose products are involved in peroxisome biogenesis. A genome-wide study that examined genes involved in fatty acid metabolism revealed a growth defect for hda2Δ cells in myristic acid, a saturated fatty acid whose metabolism requires peroxisome function (Smith et al., 2006). Although peroxisome genes do not appear to be transcriptionally regulated by the HDA complex (Bernstein et al., 2000), we observed mislocalization of Pot1-GFP in the absence of HDA components. These data raise the possibility that the inability to metabolize fatty acids may be due to defective transport into the peroxisome. The enhanced Pot1 localization phenotype in the absence of HDA2, along with the defect in metabolizing myristic acid that was only observed in an hda2Δ mutant, suggests that Hda2 may be more important for peroxisome function than the other HDA components. These experiments highlight the importance of performing genetic screens with the regulatory subunits of multisubunit enzyme complexes.
Regulation of G1 Transcription Factors by Acetylation
We found that several genes identified in our RPD3 SDL screen encoded proteins that were acetylated in vivo and had clear human counterparts known to be regulated by acetylation. One of these genes, SWI4, is a G1 transcription factor analogous to the E2F in human cells. This is the first example of a transcription factor regulated by (de)acetylation in yeast. Our data suggest that acetylation of Swi4 is necessary for optimal interaction with Swi6 and binding of SBF to G1 promoters, as highlighted by a failure to induce G1 transcription in a swi4 mutant that mimics constitutive deacetylation. In early G1 phase, SBF is bound to promoters, but its activity is restrained by the repressor Whi5, a protein that is analogous to the Retinoblastoma protein that inhibits E2F in the G1 regulatory circuit in mammalian cells. Our data suggest that Rpd3 has a dual role in regulating G1 transcription in early G1, both by deacetylating histones to create repressive chromatin and by ensuring that Swi4 remains deacetylated (Figure 7). Phosphorylation and removal of Whi5 and Rpd3 by the CDKs, Cdc28 and Pho85, is necessary to initiate G1 gene expression (Huang et al., 2009; Wang et al., 2009), and the binding of SBF is facilitated by the recruitment of additional chromatin remodeling factors, including the Gcn5 KAT to G1 promoters (Cosma et al., 1999). Our data suggest that acetylation of Swi4 by a KAT strengthens the interaction between Swi4 and Swi6 and is necessary for the maximal induction of G1 transcription. Because Gcn5 is known to acetylate histones at G1 promoters, it may be the Swi4 acetyltransferase as well (Cosma, 2002; Cosma et al., 1999). The ortholog E2F is acetylated by two KATs, p300/CBP and P/CAF, and is deacetylated by HDAC1 (Martínez-Balbás et al., 2000; Marzio et al., 2000). The regulation of E2F and Swi4 by (de)acetylation is distinct from their regulation by the repressors Whi5 (for Swi4) and Rb (for E2F) and from the modulation of chromatin structure by KATs and KDACs. Our data provide evidence for acetylation modulating the protein-protein interaction between the two components of SBF and further extend the parallels between the G1 regulatory pathways between yeast and metazoans.
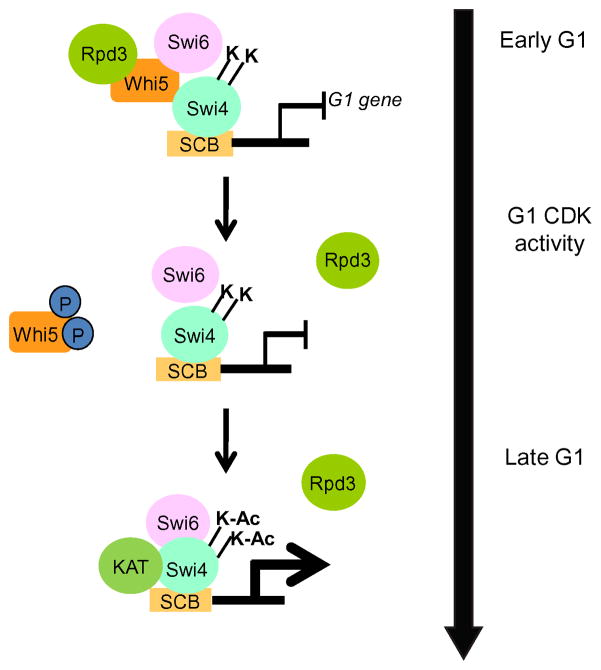
Figure 7. Model for Acetylation-Dependent Regulation of Swi4 and Transcriptional Induction during G1 Phase of the Cell Cycle.
A schematic depicting how acetylation of Swi4 may facilitate the interaction between Swi4 and Swi6 to promote timely activation of G1 transcription. Early in G1, Rpd3 is recruited to promoters by the repressor Whi5, where it de-acetylates Swi4, leading to a weaker interaction with Swi6, preventing transcription. Phosphorylation and removal of Whi5 by G1 CDKs allows the recruitment of a KAT, which acetylates Swi4, strengthening the interaction between Swi4 and Swi6. Together, these events ensure that SBF targets are appropriately activated in late G1 phase prior to DNA replication.
Conclusions
The results presented here show that synthetic dosage lethal screens can provide a powerful counterpart to biochemical efforts in systematically exploring protein acetylation. KDAC overexpression is linked to poor prognosis in many cancers (Wang et al., 2001), and several KDAC inhibitors are currently being used as chemotherapies (Kavanaugh et al., 2010) and as treatments for neurodegenerative diseases (D’Mello, 2009; Dietz and Casaccia, 2010). Little information is available about changes in acetylation patterns of the proteome (Spange et al., 2009); thus, studies linking specific acetylation events to cognate KATs/KDACs coupled with an examination of their biological effects are important. Though these types of high-throughput genome-wide screens remain technically difficult in mammalian cells, KDACs and their regulatory pathways are highly conserved (Bradner et al., 2010; Yang and Seto, 2008). Systematic genetic studies in yeast can enhance our understanding of the global relationships between acetylation events and the propensity of acetylation networks to lapse into malign states in diseased cells.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Conditions
S. cerevisiae strains and growth conditions are described in Extended Experimental Procedures.
SDL Screens and Confirmations
Screens were performed as previously described (Sopko et al., 2006a). For details, refer to Extended Experimental Procedures.
Cell Biology
For the vacuolar internalization experiments, GFP-tagged proteins from the yeast GFP collection (Huh et al., 2003) were imaged in wild-type and rpd3Δ backgrounds, and for the peroxisome experiments, GFP-tagged proteins were imaged in wild-type, hda1Δ, hda2Δ, and hda3Δ backgrounds as previously described (Youn et al., 2010). Cells were treated with 10 μM TSA or DMSO for 6 hr before imaging. Peroxisome number was determined by eye in three independent experiments. For details of the cell biology screen and quantification, refer to Extended Experimental Procedures.
Pull-Down of GST-Tagged Proteins and Anti-Acetylation Western Blots
Pull-downs were performed as previously described (Huang et al., 2009). Anti-acetyl lysine antibodies are described in detail in the Extended Experimental Procedures.
Mass Spectrometry
Protein purification and mass spectrometry analysis of acetylation sites on Swi4-TAP was performed as previously described (Lambert et al., 2009).
Cell-Cycle Synchronization, Quantitative PCR, and Gene Expression Analysis
Cultures were grown to midlog phase in YPD at 30°C, and their cell cycle was arrested by incubating with 5 μM α factor (GenScript) for 2–3 hr. Cells were then washed with cold YP and resuspended in fresh YPD medium. RNA extraction, cDNA preparation, and qPCR reactions were performed as previously described (Fillingham et al., 2009). FACS analysis for the cell-cycle-synchronized samples was performed as previously described (Huang et al., 2009).
Chromatin Immunoprecipitations
Formaldehyde (Sigma) cross-linking and whole-cell extracts were prepared as previously described (Kim et al., 2004). Immunoprecipitations were performed using 1:200 dilution of α-myc monoclonal antibody (9E10) and α-Swi6 or α-Swi4 polyclonal antibodies (Andrews and Herskowitz, 1989; Ogas et al., 1991). Enrichment of the CLN2 promoter sequence was quantified with real-time PCR, using a dual fluorogenic reporter TaqMan assay in an ABI PRISM 7500HT Sequence Detection System as previously described (Costanzo et al., 2004).
Supplementary Material
Acknowledgments
We thank members of the Andrews group for helpful discussions. This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) to B.A. and Tim Hughes (MOP-11206). The automated cell biology screens were supported by CIHR grant MOP97939 to B.A. and Charles Boone. Operation of our automated yeast genetics platform is supported by grants from the CIHR (MOP-102629) and the National Institutes of Health (US-R01HG005853-01). S.K.D. was the recipient of an Ontario Graduate Scholarship.
Footnotes
Supplemental Information includes Extended Experimental Procedures, four figures, and eight tables and can be found with this article online at doi:10.1016/j.cell.2012.02.064.
References
- Andrews BJ, Herskowitz I. The yeast SWI4 protein contains a motif present in developmental regulators and is part of a complex involved in cell-cycle-dependent transcription. Nature. 1989;342:830–833. doi: 10.1038/342830a0. [DOI] [PubMed] [Google Scholar]
- Andrews BJ, Moore LA. Interaction of the yeast Swi4 and Swi6 cell cycle regulatory proteins in vitro. Proc Natl Acad Sci USA. 1992;89:11852–11856. doi: 10.1073/pnas.89.24.11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SY, Hodin RA. Histone acetylation and cancer. Curr Opin Genet Dev. 1999;9:171–174. doi: 10.1016/s0959-437x(99)80026-4. [DOI] [PubMed] [Google Scholar]
- Baryshnikova A, Costanzo M, Kim Y, Ding H, Koh J, Toufighi K, Youn JY, Ou J, San Luis BJ, Bandyopadhyay S, et al. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat Methods. 2010;7:1017–1024. doi: 10.1038/nmeth.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Rose KL, Zhang J, Beavis RC, Ueberheide B, Garcia BA, Chait B, Zhao Y, Hunt DF, Segal E, et al. Proteome-wide prediction of acetylation substrates. Proc Natl Acad Sci USA. 2009;106:13785–13790. doi: 10.1073/pnas.0906801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckouët F, Hu B, Roig MB, Sutani T, Komata M, Uluocak P, Katis VL, Shirahige K, Nasmyth K. An Smc3 acetylation cycle is essential for establishment of sister chromatid cohesion. Mol Cell. 2010;39:689–699. doi: 10.1016/j.molcel.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Tong JK, Schreiber SL. Genomewide studies of histone deacetylase function in yeast. Proc Natl Acad Sci USA. 2000;97:13708–13713. doi: 10.1073/pnas.250477697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges V, Lehane C, Lopez-Serra L, Flynn H, Skehel M, Rolef Ben-Shahar T, Uhlmann F. Hos1 deacetylates Smc3 to close the cohesin acetylation cycle. Mol Cell. 2010;39:677–688. doi: 10.1016/j.molcel.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, Mazitschek R. Chemical phylogenetics of histone deacetylases. Nat Chem Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmen AA, Rundlett SE, Grunstein M. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- Carmen AA, Griffin PR, Calaycay JR, Rundlett SE, Suka Y, Grunstein M. Yeast HOS3 forms a novel trichostatin A-insensitive homodimer with intrinsic histone deacetylase activity. Proc Natl Acad Sci USA. 1999;96:12356–12361. doi: 10.1073/pnas.96.22.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Choi JK, Grimes DE, Rowe KM, Howe LJ. Acetylation of Rsc4p by Gcn5p is essential in the absence of histone H3 acetylation. Mol Cell Biol. 2008;28:6967–6972. doi: 10.1128/MCB.00570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Chuang DM, Leng Y, Marinova Z, Kim HJ, Chiu CT. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 2009;32:591–601. doi: 10.1016/j.tins.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Cosma MP. Ordered recruitment: gene-specific mechanism of transcription activation. Mol Cell. 2002;10:227–236. doi: 10.1016/s1097-2765(02)00604-4. [DOI] [PubMed] [Google Scholar]
- Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Nishikawa JL, Tang X, Millman JS, Schub O, Breitkreuz K, Dewar D, Rupes I, Andrews B, Tyers M. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117:899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science. 2010;327:425–431. doi: 10.1126/science.1180823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, Baryshnikova A, Myers CL, Andrews B, Boone C. Charting the genetic interaction map of a cell. Curr Opin Biotechnol. 2011;22:66–74. doi: 10.1016/j.copbio.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Cross FR, Hoek M, McKinney JD, Tinkelenberg AH. Role of Swi4 in cell cycle regulation of CLN2 expression. Mol Cell Biol. 1994;14:4779–4787. doi: 10.1128/mcb.14.7.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Mello SR. Histone deacetylases as targets for the treatment of human neurodegenerative diseases. Drug News Perspect. 2009;22:513–524. doi: 10.1358/dnp.2009.9.1428871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Grant PA. Multi-tasking on chromatin with the SAGA coactivator complexes. Mutat Res. 2007;618:135–148. doi: 10.1016/j.mrfmmm.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C, Kundu TK. Transcriptional regulation by the acetylation of nonhistone proteins in humans — a new target for therapeutics. IUBMB Life. 2005;57:137–149. doi: 10.1080/15216540500090629. [DOI] [PubMed] [Google Scholar]
- de Bruin RA, McDonald WH, Kalashnikova TI, Yates J, III, Wittenberg C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell. 2004;117:887–898. doi: 10.1016/j.cell.2004.05.025. [DOI] [PubMed] [Google Scholar]
- Deribe YL, Wild P, Chandrashaker A, Curak J, Schmidt MH, Kalaidzidis Y, Milutinovic N, Kratchmarova I, Buerkle L, Fetchko MJ, et al. Regulation of epidermal growth factor receptor trafficking by lysine deacetylase HDAC6. Sci Signal. 2009;2:ra84. doi: 10.1126/scisignal.2000576. [DOI] [PubMed] [Google Scholar]
- Dietz KC, Casaccia P. HDAC inhibitors and neurodegeneration: at the edge between protection and damage. Pharmacol Res. 2010;62:11–17. doi: 10.1016/j.phrs.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dompierre JP, Godin JD, Charrin BC, Cordelières FP, King SJ, Humbert S, Saudou F. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington’s disease by increasing tubulin acetylation. J Neurosci. 2007;27:3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham J, Kainth P, Lambert JP, van Bakel H, Tsui K, Peña-Castillo L, Nislow C, Figeys D, Hughes TR, Greenblatt J, Andrews BJ. Two-color cell array screen reveals interdependent roles for histone chaperones and a chromatin boundary regulator in histone gene repression. Mol. Cell. 2009;35:340–351. doi: 10.1016/j.molcel.2009.06.023. [DOI] [PubMed] [Google Scholar]
- Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997;192:207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- Gao YS, Hubbert CC, Yao TP. The microtubule-associated histone deacetylase 6 (HDAC6) regulates epidermal growth factor receptor (EGFR) endocytic trafficking and degradation. J Biol Chem. 2010;285:11219–11226. doi: 10.1074/jbc.M109.042754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Unal E, Koshland D. Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol Cell. 2009;34:311–321. doi: 10.1016/j.molcel.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Kaluarachchi S, van Dyk D, Friesen H, Sopko R, Ye W, Bastajian N, Moffat J, Sassi H, Costanzo M, Andrews BJ. Dual regulation by pairs of cyclin-dependent protein kinases and histone deacetylases controls G1 transcription in budding yeast. PLoS Biol. 2009;7:e1000188. doi: 10.1371/journal.pbio.1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Jeong H, Then F, Melia TJ, Jr, Mazzulli JR, Cui L, Savas JN, Voisine C, Paganetti P, Tanese N, Hart AC, et al. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 2009;137:60–72. doi: 10.1016/j.cell.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh SM, White LA, Kolesar JM. Vorinostat: A novel therapy for the treatment of cutaneous T-cell lymphoma. Am J Health Syst Pharm. 2010;67:793–797. doi: 10.2146/ajhp090247. [DOI] [PubMed] [Google Scholar]
- Kelley R, Ideker T. Systematic interpretation of genetic interactions using protein networks. Nat Biotechnol. 2005;23:561–566. doi: 10.1038/nbt1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kim M, Ahn SH, Krogan NJ, Greenblatt JF, Buratowski S. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 2004;23:354–364. doi: 10.1038/sj.emboj.7600053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Kim JH, Saraf A, Florens L, Washburn M, Workman JL. Gcn5 regulates the dissociation of SWI/SNF from chromatin by acetylation of Swi2/Snf2. Genes Dev. 2010;24:2766–2771. doi: 10.1101/gad.1979710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll ES, Hyland KM, Hieter P, Li JJ. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics. 1996;143:95–102. doi: 10.1093/genetics/143.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- Lambert JP, Mitchell L, Rudner A, Baetz K, Figeys D. A novel proteomics approach for the discovery of chromatin-associated protein networks. Mol Cell Proteomics. 2009;8:870–882. doi: 10.1074/mcp.M800447-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang T, Schaeffeler E, Bernreuther D, Bredschneider M, Wolf DH, Thumm M. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 1998;17:3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- Lin YY, Qi Y, Lu JY, Pan X, Yuan DS, Zhao Y, Bader JS, Boeke JD. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev. 2008;22:2062–2074. doi: 10.1101/gad.1679508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YY, Lu JY, Zhang J, Walter W, Dang W, Wan J, Tao SC, Qian J, Zhao Y, Boeke JD, et al. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell. 2009;136:1073–1084. doi: 10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, van Dyk D, Li Y, Andrews B, Rao H. A genome-wide synthetic dosage lethality screen reveals multiple pathways that require the functioning of ubiquitin-binding proteins Rad23 and Dsk2. BMC Biol. 2009;7:75. doi: 10.1186/1741-7007-7-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JY, Lin YY, Sheu JC, Wu JT, Lee FJ, Chen Y, Lin MI, Chiang FT, Tai TY, Berger SL, et al. Acetylation of yeast AMPK controls intrinsic aging independently of caloric restriction. Cell. 2011;146:969–979. doi: 10.1016/j.cell.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani R, St Onge RP, Hartman JLt, Giaever G, Roth FP. Defining genetic interaction. Proc Nat Acad Sci USA. 2008;105:3461–3466. doi: 10.1073/pnas.0712255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Balbás MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzio G, Wagener C, Gutierrez MI, Cartwright P, Helin K, Giacca M. E2F family members are differentially regulated by reversible acetylation. J Biol Chem. 2000;275:10887–10892. doi: 10.1074/jbc.275.15.10887. [DOI] [PubMed] [Google Scholar]
- Measday V, Hieter P. Synthetic dosage lethality. Methods Enzymol. 2002;350:316–326. doi: 10.1016/s0076-6879(02)50971-x. [DOI] [PubMed] [Google Scholar]
- Measday V, Baetz K, Guzzo J, Yuen K, Kwok T, Sheikh B, Ding H, Ueta R, Hoac T, Cheng B, et al. Systematic yeast synthetic lethal and synthetic dosage lethal screens identify genes required for chromosome segregation. Proc Natl Acad Sci USA. 2005;102:13956–13961. doi: 10.1073/pnas.0503504102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell L, Lambert JP, Gerdes M, Al-Madhoun AS, Skerjanc IS, Figeys D, Baetz K. Functional dissection of the NuA4 histone acetyltransferase reveals its role as a genetic hub and that Eaf1 is essential for complex integrity. Mol Cell Biol. 2008;28:2244–2256. doi: 10.1128/MCB.01653-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell L, Lau A, Lambert JP, Zhou H, Fong Y, Couture JF, Figeys D, Baetz K. Regulation of septin dynamics by the Saccharomyces cerevisiae lysine acetyltransferase NuA4. PLoS ONE. 2011;6:e25336. doi: 10.1371/journal.pone.0025336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J, Tsukagoshi Y, Yamashita S. Cloning of a gene encoding choline transport in Saccharomyces cerevisiae. J Bacteriol. 1986;166:328–330. doi: 10.1128/jb.166.1.328-330.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa J, Tsukagoshi Y, Yamashita S. Isolation and characterization of two distinct myo-inositol transporter genes of Saccharomyces cerevisiae. J Biol Chem. 1991;266:11184–11191. [PubMed] [Google Scholar]
- O’Brien KP, Remm M, Sonnhammer EL. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 2005;33:D476–D480. doi: 10.1093/nar/gki107. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J, Andrews BJ, Herskowitz I. Transcriptional activation of CLN1, CLN2, and a putative new G1 cyclin (HCS26) by SWI4, a positive regulator of G1-specific transcription. Cell. 1991;66:1015–1026. doi: 10.1016/0092-8674(91)90445-5. [DOI] [PubMed] [Google Scholar]
- Papp B, Pál C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ. Microtubule acetylation promotes kinesin-1 binding and transport. Curr Biol. 2006;16:2166–2172. doi: 10.1016/j.cub.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Robert F, Pokholok DK, Hannett NM, Rinaldi NJ, Chandy M, Rolfe A, Workman JL, Gifford DK, Young RA. Global position and recruitment of HATs and HDACs in the yeast genome. Mol Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, Botrugno OA, Parazzoli D, Oldani A, Minucci S, Foiani M. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature. 2011;471:74–79. doi: 10.1038/nature09803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, Grunstein M. Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell. 2002;109:437–446. doi: 10.1016/s0092-8674(02)00746-8. [DOI] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Nat Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiu ME, Gilmore JM, Carrozza MJ, Li B, Workman JL, Florens L, Washburn MP. Determining protein complex connectivity using a probabilistic deletion network derived from quantitative proteomics. PLoS ONE. 2009;4:e7310. doi: 10.1371/journal.pone.0007310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer JB, Breeden LL. RB from a bud’s eye view. Cell. 2004;117:849–850. doi: 10.1016/j.cell.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Hall MN, Koller A. Two FK506 resistance-conferring genes in Saccharomyces cerevisiae, TAT1 and TAT2, encode amino acid permeases mediating tyrosine and tryptophan uptake. Mol Cell Biol. 1994;14:6597–6606. doi: 10.1128/mcb.14.10.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Gao Z, Marks PA, Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci USA. 2004;101:18030–18035. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifpoor S, Nguyen Ba AN, Young JY, van Dyk D, Friesen H, Douglas AC, Kurat CF, Chong YT, Founk K, Moses AM, Andrews BJ. A quantitative literature-curated gold standard for kinase-substrate pairs. Genome Biol. 2011;12:R39. doi: 10.1186/gb-2011-12-4-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Roguev A, Schaft D, Buchanan L, Habermann B, Sakalar C, Thomas H, Krogan NJ, Shevchenko A, Stewart AF. Chromatin Central: towards the comparative proteome by accurate mapping of the yeast proteomic environment. Genome Biol. 2008;9:R167. doi: 10.1186/gb-2008-9-11-r167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D. The Sir2 protein family: A novel deacetylase for gene silencing and more. Proc Natl Acad Sci USA. 2000;97:14030–14032. doi: 10.1073/pnas.011506198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Avalos J, Celic I, Muhammad S, Wolberger C, Boeke JD. SIR2 family of NAD(+)-dependent protein deacetylases. Methods Enzymol. 2002;353:282–300. doi: 10.1016/s0076-6879(02)53056-1. [DOI] [PubMed] [Google Scholar]
- Smith JJ, Sydorskyy Y, Marelli M, Hwang D, Bolouri H, Rachubinski RA, Aitchison JD. Expression and functional profiling reveal distinct gene classes involved in fatty acid metabolism. Mol Syst Biol. 2006;2:2006.0009. doi: 10.1038/msb4100051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol Cell. 2006a;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Sopko R, Papp B, Oliver SG, Andrews BJ. Phenotypic activation to discover biological pathways and kinase substrates. Cell Cycle. 2006b;5:1397–1402. doi: 10.4161/cc.5.13.2922. [DOI] [PubMed] [Google Scholar]
- Sopko R, Huang D, Smith JC, Figeys D, Andrews BJ. Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast. EMBO J. 2007;26:4487–4500. doi: 10.1038/sj.emboj.7601847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spange S, Wagner T, Heinzel T, Krämer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–D704. doi: 10.1093/nar/gkq1116. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata S, Yu Y, Stillman DJ. The E2F functional analogue SBF recruits the Rpd3(L) HDAC, via Whi5 and Stb1, and the FACT chromatin reorganizer, to yeast G1 cyclin promoters. EMBO J. 2009;28:3378–3389. doi: 10.1038/emboj.2009.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Utley RT, Côté J. The MYST family of histone acetyltransferases. Curr Top Microbiol Immunol. 2003;274:203–236. doi: 10.1007/978-3-642-55747-7_8. [DOI] [PubMed] [Google Scholar]
- VanDemark AP, Kasten MM, Ferris E, Heroux A, Hill CP, Cairns BR. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol Cell. 2007;27:817–828. doi: 10.1016/j.molcel.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia RA. Exploring the etiology of haploinsufficiency. Bioessays. 2002;24:175–184. doi: 10.1002/bies.10023. [DOI] [PubMed] [Google Scholar]
- Wang C, Fu M, Mani S, Wadler S, Senderowicz AM, Pestell RG. Histone acetylation and the cell-cycle in cancer. Front Biosci. 2001;6:D610–D629. doi: 10.2741/1wang1. [DOI] [PubMed] [Google Scholar]
- Wang A, Kurdistani SK, Grunstein M. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science. 2002;298:1412–1414. doi: 10.1126/science.1077790. [DOI] [PubMed] [Google Scholar]
- Wang H, Carey LB, Cai Y, Wijnen H, Futcher B. Recruitment of Cln3 cyclin to promoters controls cell cycle entry via histone deacetylase and other targets. PLoS Biol. 2009;7:e1000189. doi: 10.1371/journal.pbio.1000189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberg C, Reed SI. Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes. Oncogene. 2005;24:2746–2755. doi: 10.1038/sj.onc.1208606. [DOI] [PubMed] [Google Scholar]
- Wu J, Carmen AA, Kobayashi R, Suka N, Grunstein M. HDA2 and HDA3 are related proteins that interact with and are essential for the activity of the yeast histone deacetylase HDA1. Proc Natl Acad Sci USA. 2001;98:4391–4396. doi: 10.1073/pnas.081560698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JY, Friesen H, Kishimoto T, Henne WM, Kurat CF, Ye W, Ceccarelli DF, Sicheri F, Kohlwein SD, McMahon HT, Andrews BJ. Dissecting BAR domain function in the yeast Amphiphysins Rvs161 and Rvs167 during endocytosis. Mol Biol Cell. 2010;21:3054–3069. doi: 10.1091/mbc.E10-03-0181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Chaerkady R, Shaw PG, Kensler TW, Pandey A, Davidson NE. Screening for therapeutic targets of vorinostat by SILAC-based proteomic analysis in human breast cancer cells. Proteomics. 2010;10:1029–1039. doi: 10.1002/pmic.200900602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Friesen H, Larson J, Huang D, Cox M, Tatchell K, Andrews B. Regulation of cell polarity through phosphorylation of Bni4 by Pho85 G1 cyclin-dependent kinases in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20:3239–3250. doi: 10.1091/mbc.E08-12-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.