Abstract
Age-related macular degeneration (AMD) is a major cause of visual impairment in the western world. It is characterized by the presence of lipoproteinaceous deposits (drusen) in the inner layers of the retina. Immunohistochemistry studies identified deposition of complement proteins in the drusen as well as in the choroid. In the last decade, genetic studies have linked both common and rare variants in proteins of the complement system to increased risk of development of AMD. Here, we review the variants described to date and discuss the functional implications of dysregulation of the alternative pathway of complement in AMD.
Keywords: Age-related macular degeneration, complement system, alternative pathway, genetic variants
1. Introduction
Age-related macular degeneration (AMD) is the leading cause of vision loss in the developed world with approximately 50 million sufferers worldwide. Its prevalence is continuing to rise due to the increasing numbers of older individuals in the population (Lim et al., 2012; Sobrin and Seddon, 2014). AMD is a slow progressive, degenerative ophthalmologic disease, which normally occurs during or after the sixth decade of life. In populations of European ancestry, the prevalence of advanced AMD ranges from 1.4% at 70 years of age to 20% at 90 years of age (Rudnicka et al., 2012). The disease manifests itself with the loss of photoreceptor cells (the rod and cone cells) in the central region of the retina at the back of the eye (called the macula) (Figure 1). This leads to a loss of central vision, leaving patients dependent on the acuity of their peripheral vision.
Figure 1.
A cross-section diagram of the human eye (A) indicating the location of the retina and macula. In early AMD (B) accumulation of subretinal drusen can block nutrient uptake and cause damage in the photoreceptor cell layer, eventually leading to geographic atrophy (or `dry') AMD (C) with complete RPE cell loss and photoreceptor neurodegeneration. Neovascular or `wet' AMD (D) is characterized by invasion of abnormal, leaky blood vessels and macrophages in the retina, leading to photoreceptor cell degeneration.
Not only does the disease cause emotional hardship, it also imposes a large socioeconomic burden on healthcare services, patients and their caregivers. AMD affects reading and driving, greatly reducing the ability of patients to contribute to work-related activities. AMD is likely a syndrome with multiple environmental and genetic factors playing a role. While diet and environmental factors (i.e. smoking) are associated with the risk of AMD, it has become increasingly evident that AMD risk is driven by genetic factors as well (Cipriani et al., 2012; Fritsche et al., 2013; Sobrin and Seddon, 2014).
A number of genetic alterations are associated with increased risk of developing AMD and many reside in genes encoding the complement cascade (Klein et al., 2005; Maller et al., 2006; Hughes et al., 2006; Maller et al., 2007; Fritsche et al., 2010; Sofat et al., 2012). These variants span the allelic spectrum of disease from common variants that impart relatively low risk of disease to rare variants with nearly complete penetrance. This, together with the identification of a number of inflammatory mediators in drusen, the hallmark lesion of AMD, has led to the hypothesis that an inflammatory response, driven by an inadequately regulated complement cascade, significantly contributes to the progression of AMD (Anderson et al., 2010; Ambati et al., 2013).
Drusen are extracellular deposits containing cellular debris, lipids and various protein components including a number from the innate immune system (Johnson et al., 2000; Mullins et al., 2000; Crabb et al., 2002; Anderson et al., 2010). These drusen form in the extracellular matrix that separates the photoreceptor cells and the supporting retinal pigment epithelium (RPE) from the choroid and the posterior eye's blood supply (Figure 1). This disrupts the nutrient flow from the choroid to the RPE cells, leading to cell disruption and death, which subsequently affects the health of the adjacent photoreceptor cells. The consequences of drusen formation are also believed to contribute to excessive blood vessel growth from the choroid into the retina, bleeding, macrophage recruitment through the compromised Bruch's membrane, all of which leads to cell damage. The late-stage disease is commonly subdivided into two categories, neovascular (`wet') and atrophic (`dry') and albeit having different disease characteristics, both are usually preceded by formation of drusen and retinal pigment irregularities.
2. Common variants
2.1 Factor H and Factor H Related Proteins
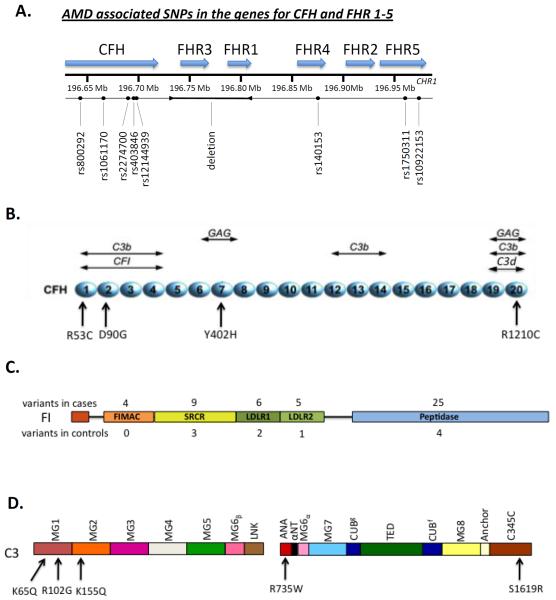
The implication that complement was somehow involved in AMD initiation and/or progression initially focused around the discovery of complement byproducts in drusen, including the main soluble regulator complement factor H (FH) (Hageman et al., 2001). Subsequent genetic studies identified a common single nucleotide polymorphism (SNP) in the CFH gene (rs1061170) (Figure 2A) that is associated with increased risk of developing AMD (Edwards et al., 2005; Hageman et al., 2005; Haines et al., 2005; Klein et al., 2005). This SNP results in a histidine residue replacing a tyrosine residue at position 402 (using the pro-protein sequence numbering; 384 in the mature protein) (Day et al., 1988). Heterozygous individuals for the Y402H polymorphism have a 2.3-fold increased risk of developing the disease, and homozygotes 5.2-fold (Sofat et al., 2012). Around 30% of people of European descent carry at least one copy of the 402H risk allele (Sofat et al., 2012).
Figure 2.
Schematic diagram of single nucleotide polymorphisms in Factor H and the Factor H related proteins (A). Factor H (B) is composed of 20 CCP repeats with multiple ligand binding sites. Locations of the variants associated with AMD are indicated. Factor I (C) is composed of two chains (heavy and light). The number of variants identified in each domain are indicated for the cases (above) and controls (below). C3 (D) is composed of two chains (α and β); variants associated with AMD in C3 are indicated.
Structurally, the Y402H polymorphism occurs in the seventh of FH's twenty complement control protein (CCP) domains (Figure 2B) and does not alter the overall conformation of the protein (Herbert et al., 2007). Y402H, however, alters the binding of FH to a number of ligands (see (Clark et al., 2010a) and references within), most notably C-reactive protein (Sjöberg et al., 2007), Streptococcus M protein (Haapasalo et al., 2008) and sulfated polyanions (Clark et al., 2006), such as the glycosaminoglycan (GAG) chains of proteoglycans. Factor H anchors itself in part to the extracellular matrix and cell surface through interactions with GAGs (Clark et al., 2010b). The Y402H polymorphism perturbs the CCP 6–8 region of FH from binding GAG chains in Bruch's membrane (Clark et al., 2010b). Decreased localization to Bruch's membrane, the site of drusen deposition, would presumably lead to poorly controlled complement turnover and an excessive local, chronic, inflammatory response. Furthermore, the Y402H polymorphism reduces the binding of FH to malondialdehyde (MDA), a peroxidation product that accumulates in AMD as a result of oxidative stress (Weismann et al., 2011). It has been proposed that MDA-mediated FH recruitment inhibits complement activation in regions with drusen buildup, and therefore reduced binding of the 402H FH form would be proinflammatory (Weismann et al., 2011). A proposed explanation for the high prevalence of the Y402H polymorphism is that the 402H allele provides a survival advantage against streptococcal infections in early life (Haapasalo et al., 2008). The FH binding protein of streptococcus has a lower affinity for 402H than 402Y, which would lead to enhanced alternative pathway activation on these bacteria. Similarly, a second mechanism of positive selection was put forward by Dr. Robert Avery, relative to the bacterium Yersinia pestis. He hypothesized that less binding by 402H would prevent immune system evasion and confer protection from the Black Death, which killed 30–60% of the European population during the Middle-ages (Avery, 2010).
A number of other SNPs further downstream on chromosome 1 are associated with AMD and implicate the involvement of the factor H-related (FHR) proteins in disease pathogenesis (Figure 2A) (Hageman et al., 2006). Each of the five FHR proteins are encoded by their own gene and their exact function or role in immune homeostasis is still unclear. It has been postulated that some of the FHR proteins may well act as competitors for FH binding to various ligands and can even help form a novel C3 convertase by binding C3b (Hebecker and Józsi, 2012). Recently, studies have demonstrated that FHR-1, -2 and -5 circulate in the blood as dimers or higher oligomers (Goicoechea de Jorge et al., 2013; Tortajada et al., 2013). As such, it is likely that any competitive function they may possess would be accentuated by oligomers due to higher avidity.
Complete deletion of the genes for FHR-1 and FHR-3 was found to be protective against developing AMD (Hughes et al., 2006; Hageman et al., 2006; Fritsche et al., 2010). Other studies though have shown that the deletion of these genes is not independent of the intronic CFH SNP (rs 6677604) (Raychaudhuri et al., 2010; Ansari et al., 2013). The effect of the FHR-1/FHR-3 deletion and the intronic CFH SNP may be hard to separate out due to this linkage. With these recent discoveries and the presence of more rare SNPs in the FHR genes, this area of complement biology should continue to provide new insights into FHR protein contribution to immune regulation.
2.2 C3
C3 is the central component of complement and functional changes directly affect the downstream cascade. A common SNP (rs2230199) in C3 results in a R102G (pro-C3 numbering; R80G in the mature protein) substitution (Figure 2C) that is associated with risk of AMD (odds ratio 2.6) (Maller et al., 2007; Yates et al., 2007). The R102G polymorphism results in reduced FH binding to the 102G variant and subsequent decreased Factor I (FI) mediated cofactor activity. By extending the convertase lifetime, AP amplification is enhanced (Heurich et al., 2011). This effect on FH co-factor activity is specific as the C3 R102G polymorphism does not alter FH mediated decay accelerating activity nor does it affect the activity of either decay accelerating factor (DAF; CD55) or membrane cofactor protein (MCP; CD46) (Heurich et al., 2011). The R102G polymorphism is also associated with a number of other diseases such as the kidney condition dense deposit disease (Abrera-Abeleda et al., 2011).
2.3 Factor B
A common haplotype (set of variants that are highly correlated) spanning the genes of both complement factor B (FB) and C2 are associated with a decreased risk of AMD. The genes reside within 500 bp of each other in the major histocompatibility complex type III region of chromosome 6. The L9H variant in FB is in strong linkage disequilibrium with the E318D variant in C2, and the R32Q is in just as strong linkage disequilibrium with the intronic SNP in C2 (Gold et al., 2006). Both haplotypes are considered highly protective against developing AMD, although the current hypothesis is that the protection is mediated by the FB mutations (Gold et al., 2006; Maller et al., 2006). Factor B antigenic fragments are found in drusen at similar levels as FH and the R32Q mutation has been shown to result in a decreased potential to form convertase and amplify complement activation (Montes et al., 2009). The L9H mutation resides in the signal peptide for FB, and although it has to be fully tested, may well affect the secretion levels of FB. A recent meta-analysis of 19 separate studies has found that this locus lowers the risk of AMD in the general Caucasian population by up to 6% (Thakkinstian et al., 2012).
2.4 Factor I
A common polymorphism near the CFI gene has also been described (Fagerness et al., 2009) and was supported by other studies (Ennis et al., 2010; Kondo et al., 2010). This SNP (rs10033900) is 3' of the CFI gene. While alteration in FI expression or function would align with the pattern of defective regulation of complement in AMD, the specific functional effect of this variant remains to be elucidated.
3. Rare variants
3.1 Factor H
Since common variants in CFH had previously been associated with AMD, the CFH gene was interrogated for rare variants of large effect (Raychaudhuri et al., 2011). One mutation, R1210C (Figure 2B), was enriched in AMD cases compared to controls (1.4 % vs. <0.1 %, respectively; Table 1). The R1210C variant is highly penetrant and leads to an earlier age of onset compared to the overall AMD population (mean onset 65 vs. 71 yrs). This association of the R1210C variant with AMD has been confirmed by several groups (Zhan et al., 2013). Interestingly, this rare variant had previously been associated with atypical hemolytic uremic syndrome (aHUS) (Manuelian et al., 2003; Józsi et al., 2006). It is a missense mutation in the C-terminal region of the protein, which is responsible for FH localizing to sites of complement activation and polyanions. Functional studies of recombinantly produced R1210C protein, containing only CCPs 8–20, identified a defect in its binding to glycosaminoglycans (GAGs) on cell membranes (Manuelian et al., 2003; Józsi et al., 2006), although this has not yet been accomplished with the full length protein (i.e. in the presence of the other GAG binding domain in CCP 7). Also, western blot analysis under non-reducing conditions identified a higher mol. wt. band in the serum of R1210C carriers (210 vs the expected FH mol wt of 150 kDa), likely related to the presence of an extra cysteine in the 20th domain in this variant (Sánchez-Corral et al., 2002). Further analysis demonstrated that the R1210C protein forms a complex with albumin through formation of a disulphide bridge (Sánchez-Corral et al., 2002). The interaction of the R1210C FH-albumin heterodimer with immobilized C3b is also impaired. Thus, this variant in FH affects its complement regulatory functions, particularly on a membrane, and thereby would lead to increased AP activation at sites of debris deposition.
Table 1.
Highly Penetrant Rare Variants of FH that Predispose to AMD
| VARIANT | FREQUENCY (%) | CCP | FUNCTIONAL IMPLICATIONS | REFERENCE(S) | ||
|---|---|---|---|---|---|---|
| C3b binding | DAA | CA | ||||
| R1210C | 1.4* | 20 | ↆ | Normal | Normal | (Raychaudhuri et al., 2011) |
| R53C | <0.01 | 1 | Normal | ↆ | ↆ | (Yu et al., in press) |
| D90G | <0.01 | 1 | Normal | Normal | ↆ | (Yu et al., in press) |
CCP, complement control protein repeat; DAA, decay accelerating activity; CA, cofactor activity
In AMD population. NHLBI ESP identified the frequency as <0.01% in 6,501 individuals.
More recently, two FH rare variants that exhibit high penetrance for AMD and an early onset of disease were identified through whole-exome sequencing of nine families with AMD. They were selected based on a high burden of disease but a low load of known genetic risk (Figure 2B; Table 1). In these families, the R53C and D90G missense variants segregated perfectly with AMD (Yu et al., 2014). Upon functional assessment, each had decreased regulatory activity. R53C, while binding to C3b normally by surface plasmon resonance, had reduced decay accelerating activity for the AP C3 convertase. Also, R53C and D90G displayed a defect in FH-mediated cofactor activity. Together, these rare variants (minor allele frequency <0.1%) support the concept that FH is key to maintaining a balance between complement activation and regulation in the posterior retina (Bradley et al., 2011).
3.2 Factor I
Following the identification of the rare, defective, highly penetrant R1210C FH variant, targeted deep sequencing was used to screen for other rare variants in 687 targeted genes including all 59 complement components and regulator genes in a cohort of 2,493 cases and controls. To our surprise, only FI variants had an unequivocal enrichment of rare variants in cases compared to controls. 7.8 % of cases vs 2.3 % of controls carried a variant (Seddon et al., 2013). 59 rare CFI variants were identified in 140 cases and 18 controls and these variants were confirmed by Sanger sequencing (Figure 2D). Factor I, a 100 kDa serine protease of plasma that inactivates C3b, is a two-chain protein with a 35 kDa catalytic domain located in the light chain. Notably, the excess of rare variants was largely present in the catalytic domain of the protein. Furthermore, a high proportion of the variants identified in cases were predicted to be deleterious by Polyphen-2 analysis. Perhaps not unexpected, a subset of these variants (13) had already been studied in the context of aHUS and resulted in either a protein with a secretion defect, or one that was defective in C3b and C4b cofactor activity (Seddon et al., 2013). Another study described two variants in FI, G119R and G188A, and in a replication cohort, G119R remained significant (van de Ven et al., 2013). Functional studies of recombinant G119R FI demonstrated that the mutant protein is secreted at a lower level than WT and had a decrease in FI mediated cleavage of C3b.
3.3 C3
A single rare variant in C3, K155Q, was reported to be associated with AMD risk by three separate groups in 2013 (OR=2.68–3.8; Table 2; Figure 2C). K155 in C3 is in close proximity to the FH binding site (Wu et al., 2009). In surface plasmon resonance experiments, binding of this mutant to FH was reduced compared to WT C3. Furthermore, FH is a cofactor for FI-mediated cleavage of C3b and fluid phase cofactor assays demonstrated a significant reduction in K155Q C3b cleavage compared to WT (Seddon et al., 2013). This variant was independent of the common risk variant R102G (rs2230199) and, in fact, was in phase with the protective allele; thus, its substantial risk apparently overpowered the weaker protective effect of the common polymorphism.
Table 2.
Minor Allele Frequency of C3 K155Q
| SEDDON, ET AL | HELGASON, ET AL | ZHAN, ET AL | |
|---|---|---|---|
| Cases (%) | 2.5 | 1.6 | 1.1 |
| Controls (%) | 0.7 | 0.4 | 0.4 |
| OR | 3.8 | 3.99 | 2.68 |
Subsequently, in a cohort of 84 AMD cases from the Netherlands, three rare C3 variants were identified: K155Q, which was previously reported, and R735W and S1619R (Figure 2C) (Duvvari et al., 2014). In two replication cohorts (from the EUGENDA and Rotterdam studies), the frequency of the rare variants R735W and S1619R was determined. Two variants previously identified in aHUS, K65Q and R161W were also included in this analysis. In the replication analysis, only K65Q remained significantly associated; a larger sample size will be required to rigorously determine if there is a significant association.
3.4 C9
The first C9 variant which was significant was P167S (Seddon et al., 2013). This variant was enriched approximately two-fold in AMD cases compared to controls and was significant (p= 6.5 × 10−7) and has been confirmed. While there is no functional data available regarding P167S, it was predicted to be damaging by PolyPhen-2 analysis. Interestingly, no missense mutations were observed in CD59 (the regulator of the membrane attack complex) in the 2,493 individuals sequenced (Seddon & Atkinson, unpublished data). Another C9 variant, a stop codon resulting in the change R95*, was reported to be associated with reduced risk of AMD in a small study, (Nishiguchi et al., 2012), but was nominally associated and has not been confirmed. These two variants suggest that the terminal complement pathway is playing a role in AMD pathogenesis. We have the hypothesis that the stop codon would lead to haploinsufficiency and therefore reduced C9 membrane perturbing activity and thus be protective in AMD while P167S increases activity through an unknown mechanism and increases risk.
4. Conclusions
Together, these common and rare variants in the AP establish the key role of this ancient innate immune system in the pathogenesis of AMD. The implication of these findings is that a balance must be reached between activation and regulation in clearing debris to avoid collateral tissue damage in the posterior retina. Specifically, several protein-protein and protein-target interactions must be in homeostasis. First, FH must bind to a damaged surface via exposed GAGs and/or C3b/iC3b/C3d fragments. Second, FI variants teach us that cofactor activity is the critical regulatory activity as it is in aHUS (Liszewski and Atkinson, 2011). Cofactor deficiency can be achieved in multiple ways: low FI levels or activity, low FH levels or activity, or mislocalization of FH not at critical sites. Decay accelerating activity does not appear to be as critical or limited. The discovery of two independent C9 variants (one protective and one risk) associated with AMD also points towards the membrane attack complex playing a role in mediating tissue injury.
The complement fragment(s) causing damage to the retina is (are) not known. The four major candidate fragments are outlined in Box 2. The two anaphylatoxins bind to their respective receptors to promote the local inflammatory response. The C3b fragment and its limited degradation products all serve as ligands for complement receptors (CR1, CR2, CR3, CR4). The membrane attack complex (MAC; C5b-9) can damage membranes. Any or all of these proinflammatory or cellular altering fragments could contribute to the disease.
Box 2. AP Complement Activation: Who is causing the damage in AMD?
| Effectors: C3a | Anaphylatoxin |
| *C3b/iC3b/C3dg/C3d | Ligand for receptors |
| C5a | Anaphylatoxin |
| C5b-C9 | Membrane perturbation/Cell Lysis |
| * These C3 fragments would all be covalently bound to the target via an ester linkage at the site of the thioester bond in the C3d portion of the α-chain. | |
The initial observation of FH, C3b and about 20 other components (including other complement proteins) being deposited in drusen and surrounding structures was largely unappreciated as to its biologic significance (Hageman et al., 2001). However, the subsequent and now overwhelming genetic evidence conclusively shows that AMD is strongly related to an overactive AP. The hyperinflammatory complement phenotype has been implicated in other diseases (Richards et al., 2003; Kavanagh et al., 2008; Lachmann, 2009). An instructive example is atypical hemolytic uremic syndrome (aHUS). Atypical HUS is characterized by the triad of microangiopathic hemolytic anemia, thrombocytopenia and acute renal failure induced by thrombosis in the capillaries of the glomerulus. Excessive activation of the AP is central to the development of aHUS and rare variants with large effects on risk have been identified in FH, FI, MCP, C3 and FB (Frémeaux-Bacchi et al., 2008; Goicoechea de Jorge et al., 2007; Kavanagh et al., 2005; Manuelian et al., 2003; Noris et al., 2003; Richards et al., 2001). Functional characterization of these variants has provided insight relative to the mechanism of disease. The variants in the inhibitory proteins (FH, FI and MCP) result in decreased regulatory activity: either a defect in protein secretion (Type I mutation) or a secreted protein that has reduced regulatory activity for C3b via cofactor activity (Type II mutation). In contrast, mutations in C3 and FB lead to a secondary gain of function and are generally resistant to the regulation. In several cases, the variant is located in the binding site for FH or MCP. In others, the missense mutation interferes with FI mediated inactivation of C3b. Using aHUS as an example of an hyperinflammatory complement disease, “complementopathy” (Lachmann, 2009), together with the variants described thus far in AMD, we have gained enormous insight into the pathogenesis of these diseases.
Remarkably, one is an acute injury state predominantly affecting young children involving glomerular endothelial cells, while the other is a chronic disease of the aged retina primarily involving epithelial cells and debris handling. On first glance, it is hard to imagine two more distinct and contrasting disease states in which pathogenesis is due to the same innate immune system overreacting to an injured state. AMD and aHUS also point out how little we know about immune responses in most specialized tissues and organs. A major question now relates to whether inhibition of C5 that has been successfully used in aHUS or inhibition of another AP target will be a successful therapeutic intervention in AMD.
A number of complement inhibitors are under evaluation for the treatment of AMD (Ambati et al., 2013). They target the complement cascade at several levels in the AP. POT-4 (Alcon) is a peptide that prevents activation of C3 and is currently in Phase II trials. Eculizumab (Alexion) is a monoclonal antibody that prevents cleavage of C5 by a C5 convertase, and is FDA approved for use in aHUS. A combined Phase I/II trial was recently completed evaluating the effect of eculizumab on the growth of geographic atrophy in AMD patients (Yehoshua et al., 2014). In this placebo- controlld randomized trial, Eculizmab or placebo was delivered intravenously to thirty patients. No differences were seen in the growth of geographic atrophy between the treatment group and placebo. However, this was a small study and perhaps a different delivery method would be more effective (e.g. intravital injection). An anti-FD antibody that blocks cleavage of the C3bB proconvertase by FD is also currently in clinical trials, but no results are available to date (Genentech). Based on the genetic evidence for a role of the AP in AMD, manipulation of complement activity systemically or locally is a promising therapeutic avenue to continue to pursue.
Highlights.
Age-related macular degeneration is a leading cause of visual impairment in the elderly.
Common and rare variants in the complement system are associated with risk of AMD.
Functional studies of a subset of variants demonstrate over-activation of the AP.
Box 1. Factor I Variants in AMD.
7.8% AMD cases vs. 2.3% controls (OR=3.6), p=2×10−8
59 distinct variants; 140 total rare varia nts in cases (137 confirmed by Sanger sequencing)
- Variants in cases versus controls were:
- Predicted to be loss of function
- Observed in aHUS (13 cases, 0 controls)
- Enriched in the catalytic domain
- About 50% had antigenic FI levels <30 ug/ml
- Independent of common risk allele (rs4698775)
Class IV and V AMD patients: 5% carry a rare FI variant and ~50% of those have low FI levels (haploinsufficiency).
Acknowledgements
This work was supported by US National Institutes of Health (NIH) grants (R01 AI041592 and P30AR48335 to JPA and R01-EY11309 to JMS); The Edward N and Della L Thome Memorial Foundation (JPA); Massachusetts Lions Eye Research Fund, Inc.(JMS); the Foundation Fighting Blindness (JMS); the Macular Vision Research Foundation (JMS); Research to Prevent Blindness Challenge Grant to the New England Eye Center, Department of Ophthalmology, Tufts University School of Medicine; American Macular Degeneration Foundation (JMS) and the Macular Degeneration Research Fund of the Ophthalmic Epidemiology and Genetics Service, New England Eye Center, Tufts Medical Center, Tufts University School of Medicine. SJC is supported by MRC Career Development Fellowship (MR/K024418/1). MPT was supported by the F30 National Heart Lung and Blood Institute Ruth L Kirschstein National Research Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrera-Abeleda MA, Nishimura C, Frees K, Jones M, Maga T, Katz LM, Zhang Y, Smith RJH. Allelic variants of complement genes associated with dense deposit disease. J. Am. Soc. Nephrol. JASN. 2011;22:1551–1559. doi: 10.1681/ASN.2010080795. doi:10.1681/ASN.2010080795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambati J, Atkinson JP, Gelfand BD. Immunology of age-related macular degeneration. Nat. Rev. Immunol. 2013;13:438–451. doi: 10.1038/nri3459. doi:10.1038/nri3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DH, Radeke MJ, Gallo NB, Chapin EA, Johnson PT, Curletti CR, Hancox LS, Hu J, Ebright JN, Malek G, Hauser MA, Rickman CB, Bok D, Hageman GS, Johnson LV. The pivotal role of the complement system in aging and age-related macular degeneration: hypothesis re-visited. Prog. Retin. Eye Res. 2010;29:95–112. doi: 10.1016/j.preteyeres.2009.11.003. doi:10.1016/j.preteyeres.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari M, McKeigue PM, Skerka C, Hayward C, Rudan I, Vitart V, Polasek O, Armbrecht A-M, Yates JRW, Vatavuk Z, Bencic G, Kolcic I, Oostra BA, Van Duijn CM, Campbell S, Stanton CM, Huffman J, Shu X, Khan JC, Shahid H, Harding SP, Bishop PN, Deary IJ, Moore AT, Dhillon B, Rudan P, Zipfel PF, Sim RB, Hastie ND, Campbell H, Wright AF. Genetic influences on plasma CFH and CFHR1 concentrations and their role in susceptibility to age-related macular degeneration. Hum. Mol. Genet. 2013;22:4857–4869. doi: 10.1093/hmg/ddt336. doi:10.1093/hmg/ddt336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery RL. The plague and macular degeneration. Ophthalmology. 2010;117:2442. doi: 10.1016/j.ophtha.2010.08.052. doi:10.1016/j.ophtha.2010.08.052. [DOI] [PubMed] [Google Scholar]
- Bradley DT, Zipfel PF, Hughes AE. Complement in age-related macular degeneration: a focus on function. Eye Lond. Engl. 2011;25:683–693. doi: 10.1038/eye.2011.37. doi:10.1038/eye.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipriani V, Leung H-T, Plagnol V, Bunce C, Khan JC, Shahid H, Moore AT, Harding SP, Bishop PN, Hayward C, Campbell S, Armbrecht AM, Dhillon B, Deary IJ, Campbell H, Dunlop M, Dominiczak AF, Mann SS, Jenkins SA, Webster AR, Bird AC, Lathrop M, Zelenika D, Souied EH, Sahel J-A, Léveillard T, French AMD Investigators. Cree AJ, Gibson J, Ennis S, Lotery AJ, Wright AF, Clayton DG, Yates JRW. Genome-wide association study of age-related macular degeneration identifies associated variants in the TNXB-FKBPL-NOTCH4 region of chromosome 6p21.3. Hum. Mol. Genet. 2012;21:4138–4150. doi: 10.1093/hmg/dds225. doi:10.1093/hmg/dds225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Bishop PN, Day AJ. Complement factor H and age-related macular degeneration: the role of glycosaminoglycan recognition in disease pathology. Biochem. Soc. Trans. 2010a;38:1342–1348. doi: 10.1042/BST0381342. doi:10.1042/BST0381342. [DOI] [PubMed] [Google Scholar]
- Clark SJ, Higman VA, Mulloy B, Perkins SJ, Lea SM, Sim RB, Day AJ. His-384 allotypic variant of factor H associated with age-related macular degeneration has different heparin binding properties from the non-disease-associated form. J. Biol. Chem. 2006;281:24713–24720. doi: 10.1074/jbc.M605083200. doi:10.1074/jbc.M605083200. [DOI] [PubMed] [Google Scholar]
- Clark SJ, Perveen R, Hakobyan S, Morgan BP, Sim RB, Bishop PN, Day AJ. Impaired binding of the age-related macular degeneration-associated complement factor H 402H allotype to Bruch's membrane in human retina. J. Biol. Chem. 2010b;285:30192–30202. doi: 10.1074/jbc.M110.103986. doi:10.1074/jbc.M110.103986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabb JW, Miyagi M, Gu X, Shadrach K, West KA, Sakaguchi H, Kamei M, Hasan A, Yan L, Rayborn ME, Salomon RG, Hollyfield JG. Drusen proteome analysis: an approach to the etiology of age-related macular degeneration. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14682–14687. doi: 10.1073/pnas.222551899. doi:10.1073/pnas.222551899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day AJ, Willis AC, Ripoche J, Sim RB. Sequence polymorphism of human complement factor H. Immunogenetics. 1988;27:211–214. doi: 10.1007/BF00346588. [DOI] [PubMed] [Google Scholar]
- Duvvari MR, Paun CC, Buitendijk GHS, Saksens NTM, Volokhina EB, Ristau T, Schoenmaker-Koller FE, van de Ven JPH, Groenewoud JMM, van den Heuvel LPWJ, Hofman A, Fauser S, Uitterlinden AG, Klaver CCW, Hoyng CB, de Jong EK, den Hollander AI. Analysis of rare variants in the c3 gene in patients with age-related macular degeneration. PloS One. 2014;9:e94165. doi: 10.1371/journal.pone.0094165. doi:10.1371/journal.pone.0094165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards AO, Ritter R, 3rd, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. doi:10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- Ennis S, Gibson J, Cree AJ, Collins A, Lotery AJ. Support for the involvement of complement factor I in age-related macular degeneration. Eur. J. Hum. Genet. 2010;18:15–16. doi: 10.1038/ejhg.2009.113. doi:10.1038/ejhg.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagerness JA, Maller JB, Neale BM, Reynolds RC, Daly MJ, Seddon JM. Variation near complement factor I is associated with risk of advanced AMD. Eur. J. Hum. Genet. 2009;17:100–104. doi: 10.1038/ejhg.2008.140. doi:10.1038/ejhg.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frémeaux-Bacchi V, Miller EC, Liszewski MK, Strain L, Blouin J, Brown AL, Moghal N, Kaplan BS, Weiss RA, Lhotta K, Kapur G, Mattoo T, Nivet H, Wong W, Gie S, Hurault de Ligny B, Fischbach M, Gupta R, Hauhart R, Meunier V, Loirat C, Dragon-Durey M-A, Fridman WH, Janssen BJC, Goodship THJ, Atkinson JP. Mutations in complement C3 predispose to development of atypical hemolytic uremic syndrome. Blood. 2008;112:4948–4952. doi: 10.1182/blood-2008-01-133702. doi:10.1182/blood-2008-01-133702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Chen W, Schu M, Yaspan BL, Yu Y, Thorleifsson G, Zack DJ, Arakawa S, Cipriani V, Ripke S, Igo RP, Jr, Buitendijk GHS, Sim X, Weeks DE, Guymer RH, Merriam JE, Francis PJ, Hannum G, Agarwal A, Armbrecht AM, Audo I, Aung T, Barile GR, Benchaboune M, Bird AC, Bishop PN, Branham KE, Brooks M, Brucker AJ, Cade WH, Cain MS, Campochiaro PA, Chan C-C, Cheng C-Y, Chew EY, Chin KA, Chowers I, Clayton DG, Cojocaru R, Conley YP, Cornes BK, Daly MJ, Dhillon B, Edwards AO, Evangelou E, Fagerness J, Ferreyra HA, Friedman JS, Geirsdottir A, George RJ, Gieger C, Gupta N, Hagstrom SA, Harding SP, Haritoglou C, Heckenlively JR, Holz FG, Hughes G, Ioannidis JPA, Ishibashi T, Joseph P, Jun G, Kamatani Y, Katsanis N, N Keilhauer C, Khan JC, Kim IK, Kiyohara Y, Klein BEK, Klein R, Kovach JL, Kozak I, Lee CJ, Lee KE, Lichtner P, Lotery AJ, Meitinger T, Mitchell P, Mohand-Saïd S, Moore AT, Morgan DJ, Morrison MA, Myers CE, Naj AC, Nakamura Y, Okada Y, Orlin A, Ortube MC, Othman MI, Pappas C, Park KH, Pauer GJT, Peachey NS, Poch O, Priya RR, Reynolds R, Richardson AJ, Ripp R, Rudolph G, Ryu E, Sahel J-A, Schaumberg DA, Scholl HPN, Schwartz SG, Scott WK, Shahid H, Sigurdsson H, Silvestri G, Sivakumaran TA, Smith RT, Sobrin L, Souied EH, Stambolian DE, Stefansson H, Sturgill-Short GM, Takahashi A, Tosakulwong N, Truitt BJ, Tsironi EE, Uitterlinden AG, van Duijn CM, Vijaya L, Vingerling JR, Vithana EN, Webster AR, Wichmann H-E, Winkler TW, Wong TY, Wright AF, Zelenika D, Zhang M, Zhao L, Zhang K, Klein ML, Hageman GS, Lathrop GM, Stefansson K, Allikmets R, Baird PN, Gorin MB, Wang JJ, Klaver CCW, Seddon JM, Pericak-Vance MA, Iyengar SK, Yates JRW, Swaroop A, Weber BHF, Kubo M, Deangelis MM, Léveillard T, Thorsteinsdottir U, Haines JL, Farrer LA, Heid IM, Abecasis GR, AMD Gene Consortium Seven new loci associated with age-related macular degeneration. Nat. Genet. 2013;45:433–439. 439e1–2. doi: 10.1038/ng.2578. doi:10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Lauer N, Hartmann A, Stippa S, Keilhauer CN, Oppermann M, Pandey MK, Köhl J, Zipfel PF, Weber BHF, Skerka C. An imbalance of human complement regulatory proteins CFHR1, CFHR3 and factor H influences risk for age-related macular degeneration (AMD) Hum. Mol. Genet. 2010;19:4694–4704. doi: 10.1093/hmg/ddq399. doi:10.1093/hmg/ddq399. [DOI] [PubMed] [Google Scholar]
- Goicoechea de Jorge E, Caesar JJE, Malik TH, Patel M, Colledge M, Johnson S, Hakobyan S, Morgan BP, Harris CL, Pickering MC, Lea SM. Dimerization of complement factor H-related proteins modulates complement activation in vivo. Proc. Natl. Acad. Sci. U. S. A. 2013;110:4685–4690. doi: 10.1073/pnas.1219260110. doi:10.1073/pnas.1219260110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goicoechea de Jorge E, Harris CL, Esparza-Gordillo J, Carreras L, Arranz EA, Garrido CA, López-Trascasa M, Sánchez-Corral P, Morgan BP, Rodríguez de Córdoba S. Gain-of-function mutations in complement factor B are associated with atypical hemolytic uremic syndrome. Proc. Natl. Acad. Sci. U. S. A. 2007;104:240–245. doi: 10.1073/pnas.0603420103. doi:10.1073/pnas.0603420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold B, Merriam JE, Zernant J, Hancox LS, Taiber AJ, Gehrs K, Cramer K, Neel J, Bergeron J, Barile GR, Smith RT, AMD Genetics Clinical Study Group. Hageman GS, Dean M, Allikmets R. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat. Genet. 2006;38:458–462. doi: 10.1038/ng1750. doi:10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haapasalo K, Jarva H, Siljander T, Tewodros W, Vuopio-Varkila J, Jokiranta TS. Complement factor H allotype 402H is associated with increased C3b opsonization and phagocytosis of Streptococcus pyogenes. Mol. Microbiol. 2008;70:583–594. doi: 10.1111/j.1365-2958.2008.06347.x. doi:10.1111/j.1365-2958.2008.06347.x. [DOI] [PubMed] [Google Scholar]
- Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, Smith RJH, Silvestri G, Russell SR, Klaver CCW, Barbazetto I, Chang S, Yannuzzi LA, Barile GR, Merriam JC, Smith RT, Olsh AK, Bergeron J, Zernant J, Merriam JE, Gold B, Dean M, Allikmets R. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc. Natl. Acad. Sci. U. S. A. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. doi:10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hageman GS, Hancox LS, Taiber AJ, Gehrs KM, Anderson DH, Johnson LV, Radeke MJ, Kavanagh D, Richards A, Atkinson J, Meri S, Bergeron J, Zernant J, Merriam J, Gold B, Allikmets R, Dean M, AMD Clinical Study Group Extended haplotypes in the complement factor H (CFH) and CFH-related (CFHR) family of genes protect against age-related macular degeneration: characterization, ethnic distribution and evolutionary implications. Ann. Med. 2006;38:592–604. [PMC free article] [PubMed] [Google Scholar]
- Hageman GS, Luthert PJ, Victor Chong NH, Johnson LV, Anderson DH, Mullins RF. An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch's membrane interface in aging and age-related macular degeneration. Prog. Retin. Eye Res. 2001;20:705–732. doi: 10.1016/s1350-9462(01)00010-6. [DOI] [PubMed] [Google Scholar]
- Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. doi:10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- Hebecker M, Józsi M. Factor H-related protein 4 activates complement by serving as a platform for the assembly of alternative pathway C3 convertase via its interaction with C3b protein. J. Biol. Chem. 2012;287:19528–19536. doi: 10.1074/jbc.M112.364471. doi:10.1074/jbc.M112.364471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert AP, Deakin JA, Schmidt CQ, Blaum BS, Egan C, Ferreira VP, Pangburn MK, Lyon M, Uhrín D, Barlow PN. Structure shows that a glycosaminoglycan and protein recognition site in factor H is perturbed by age-related macular degeneration-linked single nucleotide polymorphism. J. Biol. Chem. 2007;282:18960–18968. doi: 10.1074/jbc.M609636200. doi:10.1074/jbc.M609636200. [DOI] [PubMed] [Google Scholar]
- Heurich M, Martínez-Barricarte R, Francis NJ, Roberts DL, Rodríguez de Córdoba S, Morgan BP, Harris CL. Common polymorphisms in C3, factor B, and factor H collaborate to determine systemic complement activity and disease risk. Proc. Natl. Acad. Sci. U. S. A. 2011;108:8761–8766. doi: 10.1073/pnas.1019338108. doi:10.1073/pnas.1019338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AE, Orr N, Esfandiary H, Diaz-Torres M, Goodship T, Chakravarthy U. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat. Genet. 2006;38:1173–1177. doi: 10.1038/ng1890. doi:10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- Johnson LV, Ozaki S, Staples MK, Erickson PA, Anderson DH. A potential role for immune complex pathogenesis in drusen formation. Exp. Eye Res. 2000;70:441–449. doi: 10.1006/exer.1999.0798. doi:10.1006/exer.1999.0798. [DOI] [PubMed] [Google Scholar]
- Józsi M, Heinen S, Hartmann A, Ostrowicz CW, Hälbich S, Richter H, Kunert A, Licht C, Saunders RE, Perkins SJ, Zipfel PF, Skerka C. Factor H and atypical hemolytic uremic syndrome: mutations in the C-terminus cause structural changes and defective recognition functions. J. Am. Soc. Nephrol. JASN. 2006;17:170–177. doi: 10.1681/ASN.2005080868. doi:10.1681/ASN.2005080868. [DOI] [PubMed] [Google Scholar]
- Kavanagh D, Kemp EJ, Mayland E, Winney RJ, Duffield JS, Warwick G, Richards A, Ward R, Goodship JA, Goodship THJ. Mutations in complement factor I predispose to development of atypical hemolytic uremic syndrome. J. Am. Soc. Nephrol. JASN. 2005;16:2150–2155. doi: 10.1681/ASN.2005010103. doi:10.1681/ASN.2005010103. [DOI] [PubMed] [Google Scholar]
- Kavanagh D, Richards A, Atkinson J. Complement Regulatory Genes and Hemolytic Uremic Syndromes. Annu. Rev. Med. 2008;59:293–309. doi: 10.1146/annurev.med.59.060106.185110. doi:10.1146/annurev.med.59.060106.185110. [DOI] [PubMed] [Google Scholar]
- Klein RJ, Zeiss C, Chew EY, Tsai J-Y, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. doi:10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo N, Bessho H, Honda S, Negi A. Additional evidence to support the role of a common variant near the complement factor I gene in susceptibility to age-related macular degeneration. Eur. J. Hum. Genet. 2010;18:634–635. doi: 10.1038/ejhg.2009.243. doi:10.1038/ejhg.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachmann PJ. The amplification loop of the complement pathways. Adv. Immunol. 2009;104:115–149. doi: 10.1016/S0065-2776(08)04004-2. doi:10.1016/S0065-2776(08)04004-2. [DOI] [PubMed] [Google Scholar]
- Lim LS, Mitchell P, Seddon JM, Holz FG, Wong TY. Age-related macular degeneration. Lancet. 2012;379:1728–1738. doi: 10.1016/S0140-6736(12)60282-7. doi:10.1016/S0140-6736(12)60282-7. [DOI] [PubMed] [Google Scholar]
- Liszewski MK, Atkinson JP. Too much of a good thing at the site of tissue injury: the instructive example of the complement system predisposing to thrombotic microangiopathy. Hematol. Educ. Program Am. Soc. Hematol. Am. Soc. Hematol. Educ. Program. 2011;2011:9–14. doi: 10.1182/asheducation-2011.1.9. doi:10.1182/asheducation-2011.1.9. [DOI] [PubMed] [Google Scholar]
- Maller J, George S, Purcell S, Fagerness J, Altshuler D, Daly MJ, Seddon JM. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat. Genet. 2006;38:1055–1059. doi: 10.1038/ng1873. doi:10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- Maller JB, Fagerness JA, Reynolds RC, Neale BM, Daly MJ, Seddon JM. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat. Genet. 2007;39:1200–1201. doi: 10.1038/ng2131. doi:10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- Manuelian T, Hellwage J, Meri S, Caprioli J, Noris M, Heinen S, Jozsi M, Neumann HPH, Remuzzi G, Zipfel PF. Mutations in factor H reduce binding affinity to C3b and heparin and surface attachment to endothelial cells in hemolytic uremic syndrome. J. Clin. Invest. 2003;111:1181–1190. doi: 10.1172/JCI16651. doi:10.1172/JCI16651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes T, Tortajada A, Morgan BP, Rodríguez de Córdoba S, Harris CL. Functional basis of protection against age-related macular degeneration conferred by a common polymorphism in complement factor B. Proc. Natl. Acad. Sci. U. S. A. 2009;106:4366–4371. doi: 10.1073/pnas.0812584106. doi:10.1073/pnas.0812584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RF, Russell SR, Anderson DH, Hageman GS. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2000;14:835–846. [PubMed] [Google Scholar]
- Nishiguchi KM, Yasuma TR, Tomida D, Nakamura M, Ishikawa K, Kikuchi M, Ohmi Y, Niwa T, Hamajima N, Furukawa K, Terasaki H. C9-R95X polymorphism in patients with neovascular age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 2012;53:508–512. doi: 10.1167/iovs.11-8425. doi:10.1167/iovs.11-8425. [DOI] [PubMed] [Google Scholar]
- Noris M, Brioschi S, Caprioli J, Todeschini M, Bresin E, Porrati F, Gamba S, Remuzzi G. Familial haemolytic uraemic syndrome and an MCP mutation. Lancet. 2003;362:1542–1547. doi: 10.1016/S0140-6736(03)14742-3. doi:10.1016/S0140-6736(03)14742-3. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Iartchouk O, Chin K, Tan PL, Tai AK, Ripke S, Gowrisankar S, Vemuri S, Montgomery K, Yu Y, Reynolds R, Zack DJ, Campochiaro B, Campochiaro P, Katsanis N, Daly MJ, Seddon JM. A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nat. Genet. 2011;43:1232–1236. doi: 10.1038/ng.976. doi:10.1038/ng.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S, Ripke S, Li M, Neale BM, Fagerness J, Reynolds R, Sobrin L, Swaroop A, Abecasis G, Seddon JM, Daly MJ. Associations of CFHR1-CFHR3 deletion and a CFH SNP to age-related macular degeneration are not independent. Nat. Genet. 2010;42:553–555. doi: 10.1038/ng0710-553. author reply 555–556. doi:10.1038/ng0710-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards A, Buddles MR, Donne RL, Kaplan BS, Kirk E, Venning MC, Tielemans CL, Goodship JA, Goodship TH. Factor H mutations in hemolytic uremic syndrome cluster in exons 18–20, a domain important for host cell recognition. Am. J. Hum. Genet. 2001;68:485–490. doi: 10.1086/318203. doi:10.1086/318203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards A, Kemp EJ, Liszewski MK, Goodship JA, Lampe AK, Decorte R, Müslümanoğlu MH, Kavukcu S, Filler G, Pirson Y, Wen LS, Atkinson JP, Goodship THJ. Mutations in human complement regulator, membrane cofactor protein (CD46), predispose to development of familial hemolytic uremic syndrome. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12966–12971. doi: 10.1073/pnas.2135497100. doi:10.1073/pnas.2135497100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudnicka AR, Jarrar Z, Wormald R, Cook DG, Fletcher A, Owen CG. Age and gender variations in age-related macular degeneration prevalence in populations of European ancestry: a meta-analysis. Ophthalmology. 2012;119:571–580. doi: 10.1016/j.ophtha.2011.09.027. doi:10.1016/j.ophtha.2011.09.027. [DOI] [PubMed] [Google Scholar]
- Sánchez-Corral P, Pérez-Caballero D, Huarte O, Simckes AM, Goicoechea E, López-Trascasa M, de Córdoba SR. Structural and functional characterization of factor H mutations associated with atypical hemolytic uremic syndrome. Am. J. Hum. Genet. 2002;71:1285–1295. doi: 10.1086/344515. doi:10.1086/344515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seddon JM, Yu Y, Miller EC, Reynolds R, Tan PL, Gowrisankar S, Goldstein JI, Triebwasser M, Anderson HE, Zerbib J, Kavanagh D, Souied E, Katsanis N, Daly MJ, Atkinson JP, Raychaudhuri S. Rare variants in CFI, C3 and C9 are associated with high risk of advanced age-related macular degeneration. Nat. Genet. 2013;45:1366–1370. doi: 10.1038/ng.2741. doi:10.1038/ng.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöberg AP, Trouw LA, Clark SJ, Sjölander J, Heinegård D, Sim RB, Day AJ, Blom AM. The factor H variant associated with age-related macular degeneration (His-384) and the non-disease-associated form bind differentially to C-reactive protein, fibromodulin, DNA, and necrotic cells. J. Biol. Chem. 2007;282:10894–10900. doi: 10.1074/jbc.M610256200. doi:10.1074/jbc.M610256200. [DOI] [PubMed] [Google Scholar]
- Sobrin L, Seddon JM. Nature and nurture- genes and environment- predict onset and progression of macular degeneration. Prog. Retin. Eye Res. 2014;40C:1–15. doi: 10.1016/j.preteyeres.2013.12.004. doi:10.1016/j.preteyeres.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofat R, Casas JP, Webster AR, Bird AC, Mann SS, Yates JRW, Moore AT, Sepp T, Cipriani V, Bunce C, Khan JC, Shahid H, Swaroop A, Abecasis G, Branham KEH, Zareparsi S, Bergen AA, Klaver CCW, Baas DC, Zhang K, Chen Y, Gibbs D, Weber BHF, Keilhauer CN, Fritsche LG, Lotery A, Cree AJ, Griffiths HL, Bhattacharya SS, Chen LL, Jenkins SA, Peto T, Lathrop M, Leveillard T, Gorin MB, Weeks DE, Ortube MC, Ferrell RE, Jakobsdottir J, Conley YP, Rahu M, Seland JH, Soubrane G, Topouzis F, Vioque J, Tomazzoli L, Young I, Whittaker J, Chakravarthy U, de Jong PTVM, Smeeth L, Fletcher A, Hingorani AD. Complement factor H genetic variant and age-related macular degeneration: effect size, modifiers and relationship to disease subtype. Int. J. Epidemiol. 2012;41:250–262. doi: 10.1093/ije/dyr204. doi:10.1093/ije/dyr204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer KL, Olson LM, Anderson BM, Schnetz-Boutaud N, Scott WK, Gallins P, Agarwal A, Postel EA, Pericak-Vance MA, Haines JL. C3 R102G polymorphism increases risk of age-related macular degeneration. Hum. Mol. Genet. 2008;17:1821–1824. doi: 10.1093/hmg/ddn075. doi:10.1093/hmg/ddn075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakkinstian A, McEvoy M, Chakravarthy U, Chakrabarti S, McKay GJ, Ryu E, Silvestri G, Kaur I, Francis P, Iwata T, Akahori M, Arning A, Edwards AO, Seddon JM, Attia J. The association between complement component 2/complement factor B polymorphisms and age-related macular degeneration: a HuGE review and meta-analysis. Am. J. Epidemiol. 2012;176:361–372. doi: 10.1093/aje/kws031. doi:10.1093/aje/kws031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortajada A, Yébenes H, Abarrategui-Garrido C, Anter J, García-Fernández JM, Martínez-Barricarte R, Alba-Domínguez M, Malik TH, Bedoya R, Cabrera Pérez R, López Trascasa M, Pickering MC, Harris CL, Sánchez-Corral P, Llorca O, Rodríguez de Córdoba S. C3 glomerulopathy-associated CFHR1 mutation alters FHR oligomerization and complement regulation. J. Clin. Invest. 2013;123:2434–2446. doi: 10.1172/JCI68280. doi:10.1172/JCI68280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Ven JPH, Nilsson SC, Tan PL, Buitendijk GHS, Ristau T, Mohlin FC, Nabuurs SB, Schoenmaker-Koller FE, Smailhodzic D, Campochiaro PA, Zack DJ, Duvvari MR, Bakker B, Paun CC, Boon CJF, Uitterlinden AG, Liakopoulos S, Klevering BJ, Fauser S, Daha MR, Katsanis N, Klaver CCW, Blom AM, Hoyng CB, den Hollander AI. A functional variant in the CFI gene confers a high risk of age-related macular degeneration. Nat. Genet. 2013;45:813–817. doi: 10.1038/ng.2640. doi:10.1038/ng.2640. [DOI] [PubMed] [Google Scholar]
- Weismann D, Hartvigsen K, Lauer N, Bennett KL, Scholl HPN, Charbel Issa P, Cano M, Brandstätter H, Tsimikas S, Skerka C, Superti-Furga G, Handa JT, Zipfel PF, Witztum JL, Binder CJ. Complement factor H binds malondialdehyde epitopes and protects from oxidative stress. Nature. 2011;478:76–81. doi: 10.1038/nature10449. doi:10.1038/nature10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wu Y-Q, Ricklin D, Janssen BJC, Lambris JD, Gros P. Structure of complement fragment C3b-factor H and implications for host protection by complement regulators. Nat. Immunol. 2009;10:728–733. doi: 10.1038/ni.1755. doi:10.1038/ni.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates JRW, Sepp T, Matharu BK, Khan JC, Thurlby DA, Shahid H, Clayton DG, Hayward C, Morgan J, Wright AF, Armbrecht AM, Dhillon B, Deary IJ, Redmond E, Bird AC, Moore AT, Genetic Factors in AMD Study Group Complement C3 variant and the risk of age-related macular degeneration. N. Engl. J. Med. 2007;357:553–561. doi: 10.1056/NEJMoa072618. doi:10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- Yehoshua Z, Alexandre de Amorim Garcia Filho C, Nunes RP, Gregori G, Penha FM, Moshfeghi AA, Zhang K, Sadda S, Feuer W, Rosenfeld PJ. Systemic Complement Inhibition with Eculizumab for Geographic Atrophy in Age-Related Macular Degeneration. Ophthalmology. 2014;121:693–701. doi: 10.1016/j.ophtha.2013.09.044. doi:10.1016/j.ophtha.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Triebwasser MP, Wong EKS, Schramm EC, Thomas B, Reynolds R, Mardis ER, Atkinson JP, Daly M, Raychaudhuri S, Kavanagh D, Seddon JM. Whole-exome sequencing identifies rare, functional CFH variants in families with macular degeneration. Hum. Mol. Genet. 2014 doi: 10.1093/hmg/ddu226. doi:10.1093/hmg/ddu226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Larson DE, Wang C, Koboldt DC, Sergeev YV, Fulton RS, Fulton LL, Fronick CC, Branham KE, Bragg-Gresham J, Jun G, Hu Y, Kang HM, Liu D, Othman M, Brooks M, Ratnapriya R, Boleda A, Grassmann F, von Strachwitz C, Olson LM, Buitendijk GHS, Hofman A, van Duijn CM, Cipriani V, Moore AT, Shahid H, Jiang Y, Conley YP, Morgan DJ, Kim IK, Johnson MP, Cantsilieris S, Richardson AJ, Guymer RH, Luo H, Ouyang H, Licht C, Pluthero FG, Zhang MM, Zhang K, Baird PN, Blangero J, Klein ML, Farrer LA, Deangelis MM, Weeks DE, Gorin MB, Yates JRW, Klaver CCW, Pericak-Vance MA, Haines JL, Weber BHF, Wilson RK, Heckenlively JR, Chew EY, Stambolian D, Mardis ER, Swaroop A, Abecasis GR. Identification of a rare coding variant in complement 3 associated with age-related macular degeneration. Nat. Genet. 2013;45:1375–1379. doi: 10.1038/ng.2758. doi:10.1038/ng.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]