Abstract
microRNAs (miRNAs) are enclosed within Argonaute proteins, the downstream effectors of small RNA-mediated gene silencing. As miRNAs mediate extensive networks of post-transcriptional control, cells have evolved multiple strategies to control their activity with precision. A growing theme of recent years regards how post-translational modifications of Argonaute proteins, such as prolyl-hydroxylation, phosphorylation, ubiquitination, and poly-ADP-ribosylation, alter miRNA activity at global or specific levels. In this review, we discuss recent advances on Argonaute modifications in mammalian cells, and emphasize how such alterations modulate small RNA function to coordinate appropriate downstream cellular responses. These findings provide a framework to understand how Argonaute protein modifications are linked to reorganization of post-transcriptional regulatory networks, enabling dynamic responses to diverse external stimuli and changing environmental conditions.
Keywords: Argonaute, microRNA, hydroxylation, phosphorylation, ubiquitination, PARylation
Overview of miRNA regulation
microRNAs (miRNAs) are ~22 nucleotide (nt) RNAs that mature from longer endogenous primary (pri-miRNA) transcripts [1]. Canonical pri-miRNAs are cleaved by the nuclear RNase III enzyme Drosha and its cofactor DGCR8 into pre-miRNA hairpins, which are then cleaved by the cytoplasmic RNase III enzyme Dicer into miRNA/miRNA* (star) duplexes. These are loaded into Argonaute (Ago) proteins, which preferentially retain the mature miRNA and eject the star strand. In addition, a subset of small RNAs that populate Ago proteins derive from non-canonical, Drosha-independent or Dicer-independent mechanisms [1].
Ago proteins associate with cofactors of the GW182/TNRC6 family, and are guided by miRNAs to target transcripts and mediate their destabilization and/or translational suppression [2]. Animal miRNA/Ago complexes recognize targets via complements to their 5′ ends, preferentially nts 2–8 [3–5]. Comparative genomic analyses indicate that a majority of human mRNAs are subject to conserved miRNA targeting, and conserved vertebrate miRNAs often have hundreds of conserved targets [6]. Moreover, additional target sites have non-canonical pairing features, are regulated via poorly-conserved target sites, or are regulated by evolutionarily young miRNAs.
Given the broad regulatory impact of miRNAs on gene expression, it follows that the activity and action of the miRNA pathway is subject to exquisite control. Perhaps the most straightforward strategy for this control lies at the transcriptional level, and certainly many miRNAs exhibit specific cell-type and/or cell-state expression patterns [7–9]. In this respect, miRNAs are subject to the transcriptional regulatory complexity that applies to mRNAs. However, as most tissues utilize miRNA regulation, with murine oocytes being a notable exception [10, 11], the miRNA machinery is expected to be expressed ubiquitously. While this mostly true, core miRNA pathway members are still subject to cell- or state-specific transcriptional modulation [12–14]. This has implications for global activity of the miRNA pathway in different tissues, as well as disease conditions including cancer.
Perhaps less intuitive is the possibility that the miRNA pathway might be regulated at post-transcriptional levels. However, this has recently emerged as a major aspect of miRNA regulation. Mechanisms described include: (1) diverse post-transcriptional modifications of small RNA intermediates, mature miRNAs, or the mRNAs encoding miRNA pathway factors; (2) post-translational modifications of miRNA pathway factors; (3) the action of ancillary proteins that modulate the core miRNA machinery [15, 16]. These strategies can either broadly influence miRNA activity, or selectively impact particular miRNAs. For example, phosphorylation of the Dicer cofactor TRBP by the MAP kinase ERK enhances miRNA biogenesis [17]. On the other hand, the RNA binding protein Lin-28 recruits the terminal uridyltransferases TUT4 and TUT7 to specifically modify the 3′ ends of pre-let-7 hairpins [18–20], which are subsequently degraded by the Dis3L2 exoribonuclease [21, 22].
While many modifications of small RNAs and their biogenesis factors are now recognized, the physiological utilities of these processes are less understood. In this review, we focus on the diversity of modifications reported for Ago proteins (Table 1). As the key downstream effectors of miRNAs and RNAi, Ago proteins are strategically positioned to receive state-specific modifications that allow for dynamic modulation of small RNA function. We consider the recent literature that addresses how post-translational control of Ago proteins can relay upstream stimuli to downstream gene regulatory responses, in contexts that range from hypoxia, cell differentiation to antiviral defense.
Table 1.
Summary of known modifications to mammalian Ago proteins, and some of their reported consequences with respect to miRNA and/or siRNA activity.
| Argonaute Modification | Regulatory Impact | Refs |
|---|---|---|
| P700 prolyl 4-hydroxylation | increased Ago stability, increased miRNA levels/activity during hypoxia | [23, 24] |
| S253, T303, T307, S798 phosphorylation | unknown | [26] |
| S387 phosphorylation (MAPK or Akt3) | localization to processing bodies. increased translational repression vs target cleavage mode. |
[27, 28] |
| Y529 phosphorylation | impaired miRNA binding. transient relief of miRNA activity during macrophage activation. |
[26, 29] |
| Y393 phosphorylation (EGFR) | inhibited maturation of long-loop pre-miRNAs during hypoxia. increased cell survival/invasiveness. | [30] |
| ubiquitination | destabilization of unloaded Ago. also promotes turnover and remodeling of the miRISC pool during T cell activation. | [33, 39] [40, 49–51] |
| PARylation | suppression of RNA silencing | [53, 64] |
Prolyl 4-hydroxylation of Ago proteins
One of the first documented modifications of Ago proteins was hydroxylation. This was investigated following the recovery of α and β subunits of type I collagen prolyl-4-hydroxylase I [C-P4H(I)] in affinity purifications of human (h) Ago2 from HeLa cells, and both hAgo2 and hAgo4 associate with C-P4H [23]. Focusing on hAgo2, prolyl 4-hydroxylation of position P700 was shown to increase its stability (Figure 1). Evidence was shown that C-P4H positively influences siRNA-mediated silencing but not miRNA-mediated control [23]. However, as endogenous siRNAs appear to play limited roles in mammals, the functional impact of Ago hydroxylation remained mysterious.
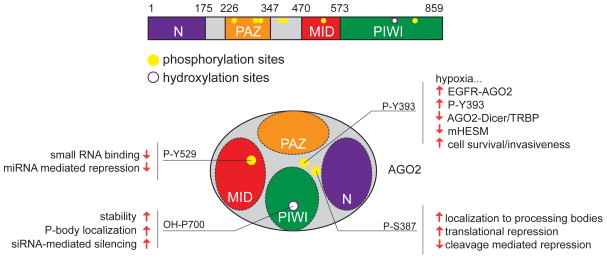
Figure 1.
Summary of site-specific modifications of Ago2. (Top) Positions of phosphorylation and hydroxylation in human Ago2. The main functional domains of Ago are noted. (Bottom) Schematic of the relative location of modified residues in Ago2, and some of the functional consequences ascribed to these phosphorylation and hydroxylation events.
More recently, hypoxia was found to increase C-P4H(I) levels in pulmonary artery smooth muscle cells (PASMCs). Hypoxia is a major contributing factor to pulmonary artery hypertension (PAH), and PASMCs undergo phenotypic alterations following hypoxia that model PAH, including lower contractility and elevated proliferation and migration. Elevated C-P4H(I) in hypoxic PASMCs stabilizes hAgo2 by increasing prolyl 4-hydroxylation at P700, and consequently increases general miRNA levels and miRNA activity [24]. Notably, hypoxia enhanced the capacity of PASMCs to mature miR-451 [24], a unique miRNA whose biogenesis absolutely depends on Slicer endonuclease activity of Ago2 [25]. Together, these findings demonstrate how a seemingly constitutive modification can be modulated in a context-specific manner to influence miRNA pathway activity.
An introduction to Ago phosphorylation events
The Ago modification studied most extensively has been phosphorylation [26] (Figure 2). Phosphorylation of hAgo2 at S387, mediated by p38 mitogen-activated protein kinase (MAPK) signaling, promotes its localization to processing bodies (P-bodies) [27]. The Akt3 kinase was recently shown to mediate S387 phosphorylation, an event proposed to alter Ago2 activity away from cleavage mode towards translational repression [28].
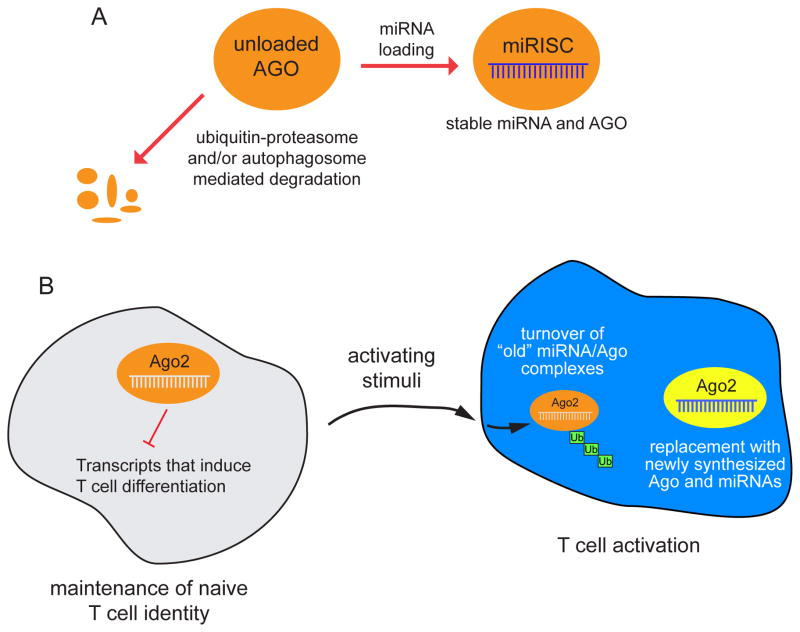
Figure 2.
Models for how ubiquitination can affect Ago proteins. (Top) Unloaded Ago proteins in many cellular contexts are susceptible to degradation by ubiquitin-proteasome and/or autophagosome-dependent pathways. These mechanisms adjust the level of Ago proteins in concert with miRNA biogenesis. (Bottom) During T cell activation, the ubiquitination-dependent turnover of Ago2 is associated with overall downregulation of miRNA accumulation. This may facilitate the re-setting of a different miRNA repertoire in activated T cells, in which the “old” miRNA/Ago pool is actively degraded and replaced with newly transcribed miRNAs and newly synthesized Ago proteins.
Systematic mapping of phosphorylation sites on hAgo2 identified 6 additional sites [26]. The impact of most of these sites awaits exploration, but Y529 is located within the MID domain that binds 5′ termini of small RNAs. An hAgo2 Y529E mutant, which mimics the negative charge of the phosphorylated residue, is impaired for small RNA binding. More recently, phosphorylation of Y529 upon macrophage interaction was reported to transiently relieve miRNA-mediated repression [29]. This facilitates translation of miRNA-regulated transcripts encoding pro-inflammatory cytokines.
EGFR phosphorylates Ago2 during hypoxia to selectively modulate miRNA biogenesis
Recent work proposes a mechanism by which Ago2 phosphorylation selectively regulates tumor suppressor miRNAs [30]. Mass spectrometry was applied to search for proteins that associate with the Epidermal Growth Factor Receptor (EGFR) kinase, and identified Ago2 as a novel candidate. This interaction was probed using a live sensor based on split YFP molecules, and confirmed using conventional co-immunoprecipitation techniques.
EGFR directly phosphorylates hAgo2 at Y393 (P-Y393), and the level of modified hAgo2 increased under hypoxia [30]. A consequence of P-Y393 is reduced association of hAgo2 with Dicer and its cofactor TRBP. However, testing of how EGFR and hypoxia affect miRNA biogenesis revealed that while transcription of many miRNAs is affected by these stimuli, a particular set of miRNAs exhibited an increased ratio of pre-miRNA:mature miRNA species (miRNAs regulated by hypoxia-dependent EGFR-suppressed maturation, or “mHESM” loci). These miRNAs were enriched in loci with annotated tumor-suppressor activities, and furthermore contained longer loop structures compared to non-mHESM loci.
Measurements of the mHESM loci mir-31, mir-192 and mir-193 showed that EGFR, but not catalytically inactive EGFR, inhibits mHESM maturation and their activity on synthetic reporters. In contrast, cells that expressed the non-phosphorylatable variant hAgo2-Y393F did not regulate mHESM loci, and this mutant Ago2 protein decreased cell survival and invasiveness in response to hypoxia. Knockdown of EGFR suppressed the phenotypic differences of wild type and mutant Ago2 proteins, suggesting the phosphorylation status underlies this effect, as opposed to an intrinsic functional difference of the Y393F variant. These data are of potential relevance to solid cancers, which are oxygen-deficient at their core. Indeed, greater immunoreactivity of a Y393 phosphospecific Ago2 antibody in breast cancer patient samples correlates with poorer overall survival.
Overall, these results support a mechanism by which an external stimulus can selectively modulate the biogenesis of cancer-relevant miRNAs (Figure 1). However, it is worth noting that some observations differ between the study introduced above [24]. For example, the Hung study observed reduced hAgo2 protein following hypoxic treatment [30], whereas a central conclusion of the Hata study was that hAgo2 was stabilized in hypoxic conditions via prolyl 4-hydroxylation [24]. Moreover, the Hung study used miR-21 as a control non-HESM locus unaffected by hypoxia, whereas Hata found that miR-21 was post-transcriptionally induced by hypoxia. Finally, the Hung study reported that hypoxia caused translocation of hAgo2 into Rab7+ late endosomes, whereas the Hata study observed that hypoxia induces translocation of hAgo2 into TIA-1-positive stress granules. These differences remain to be resolved, but in principle, they might be due to different experimental conditions or perhaps different cell types used (HeLa cells versus PASMCs). It would be important to appreciate if there are indeed substantial differences in the impact of hypoxia on the miRNA pathway in different cell types.
In addition, questions remain regarding the underlying mechanism of how P-Y393 of Ago2 affects miRNA biogenesis. Although loop substitution experiments provided evidence that P-Y393 alters maturation of longer-looped pre-miRNAs [30], it is not clear how Ago proteins might affect Dicer activity, especially in a miRNA-selective fashion. As well, it was proposed that recruitment of Ago2 to the Dicer-TRBP complex facilitates loading of small RNA duplexes, but the importance of Dicer for loading or orientation of small RNA duplexes in mammalian Ago remains to be clarified [31, 32]. It is possible that hypoxia imposes biogenesis idiosyncrasies that are not representative of typical cell culture conditions. Overall, a better understanding of the mechanistic impact of P-Y393 Ago2 on long-loop miRNA biogenesis awaits further study.
An introduction to Ago ubiquitination events
Ubiquitination comprises another broad category of Ago protein modification. Previously, the ubiquitin ligase mLin-41 was reported to modify mAgo2 and inhibit its accumulation in mES cells [33]. Notably, C. elegans lin-41 emerged as one of the key targets of the let-7 miRNA during larval-to-adult transition [34], and this miRNA-target interaction is among the few conserved from invertebrates to vertebrates [35]. Although subsequent studies have not confirmed a functional impact of mLin-41 ubiquitination of Ago2, the association of these factors is reproducible [36–38].
Regulation of Ago protein levels by ubiquitination has also been documented in other contexts. Recently, studies in Drosophila and mammalian models showed that loss of miRNA biogenesis factors destabilize miRNA-class Ago proteins, whereas increased miRNA biogenesis could increase Ago levels [39]. This homeostatic control mechanism involves ubiquitination, because depletion of the Ubiquitin Activating Enzyme 1 stabilizes AGO1 in Drosophila, and treatment of dicer-knockout mouse embryonic fibroblasts or human HeLa or HEK cells with the proteasome inhibitor MG132 stabilizes Ago2 [39, 40] (Figure 2A). Currently it is not clear what E3 ubiquitin ligase provides specificity for this conserved process. As well, autophagy also restricts the stability of Ago proteins in some settings [41–43], indicating diverse strategies to control Ago protein accumulation.
Ago2 ubiquitination and degradation during T cell activation remodels the miRISC pool
It had previously been observed that T cells conditionally deleted for drosha, dgcr8 or dicer exhibit defects in proliferation and survival [44, 45]. Interestingly, cells lacking these miRNA factors undergo premature differentiation into IFN-γ-producing effector T cells. The miRNAs expressed in differentiated Th1 and Th2 cells are very different from those of naive T cells [46], suggesting that distinct miRNA cohorts contribute to the identity or function of these different cell types. However, reformatting the miRNA repertoire during T cell maturation might not be straightforward given that miRNA:Ago complexes seem to be long-lived, at least in some cell contexts [47, 48].
Recent work from Ansel and colleagues revealed global downregulation of miRNAs following T cell activation, concomitant with post-transcriptional loss of mAgo1 and mAgo2 proteins from two days post-activation onward [49]. This specifically affected miRNA effector proteins, since Dicer, Drosha and TRBP protein levels were elevated during the same time course. Deficiency of ago2 in naive T cells caused increased T cell proliferation and their premature differentiation into cytokine-producing effector cells. The authors reasoned that miRNAs in naive T cells prevent T cell differentiation until activating signals are received, while their removal permits differentiation [49].
Mechanistically, it appears that the proteasome clears mAgo2, since it is ubiquitinated upon T cell stimulation (Figure 2B) and treatment of activated T cells with MG132 rescued the accumulation of Ago2 protein. Interestingly, the same effect was observed when T cells were treated with different inhibitors of the mTOR signaling pathway (the PI3K inhibitor LY294002 or the mTOR inhibitor rapamycin), but not with inhibitors of other signaling pathways. This implies that mTOR signaling in activated T cells continuously promotes Ago ubiquitination and degradation. Overall, these data implicate ubiquitination of Ago proteins as a focal point between the activating stimulus and the downstream process of T cell differentiation.
The concept of using Ago ubiquitination to clear away state-specific small RNAs is analogous to a process that occurs during plant infection by certain viruses. As Argonaute-mediated silencing is antiviral, viruses have evolved diverse strategies to oppose small RNA activity, including Polerovirus P0, an F-box ubiquitin ligase that induces degradation of ARGONAUTE1 [50, 51]. In a similar vein, during late spermiogenesis in mouse testis, the anaphase promoting complex (APC), a multisubunit ubiquitin ligase, ubiquitinates the Piwi protein MIWI resulting in loss of its associated piRNAs [52]. A challenge for future studies of T cell maturation will be to identify a causal E3 ligase that mediates Ago degradation in this context. Such a factor might be predicted to be crucial for permitting T cell differentiation, and depending on its locations of expression, could potentially have implications for the global control of miRNA activity in other settings.
An introduction to Ago PARylation
Poly(ADP-ribose), i.e. PAR, is a macromolecule whose regulatory effects are best-studied with respect to diverse chromosomal transactions. Beyond this, a systematic analysis showed that multiple PAR-polymerases and PAR-glycohydrolases are components of cytoplasmic stress granules [53]. Upon multiple paradigms of cellular stress, all four mammalian Ago proteins are recruited to stress granules and a portion of the Ago pool becomes PARylated, which correlates with inhibition of their activity [53, 54]. This provides a global mechanism for state-specific modulation of miRISC function.
More recently, cellular stress was reported to be accompanied by global remodeling of Ago2 binding across the transcriptome, including locations of release or enhanced binding [55]. Interestingly, the locations of Ago2 release were depleted of miRNA seed matches, whereas locations of stress-enhanced binding were enriched for sites of well-expressed miRNAs and correlated with increased translational suppression of these transcripts. While the basis of these seemingly different conclusions remains to be resolved, they may in principle involve differences in protocols and time points analyzed for stress responses.
More generally, other Ago modifications induced upon cellular stress have been associated with re-localization of mammalian Ago proteins to stress granules or P bodies [24, 27]. In addition, mitogenic signaling in Drosophila cells was recently reported to alter the composition and localization of distinct AGO1 complexes, and consequently increase the regulatory capacity of miRNAs [56]. It may be that the particular cell response by different post-translational and relocalization mechanisms of Ago proteins could differentially influence the aggregate effect on Ago activity.
Ago PARylation during viral infection relieves miRNA repression of the interferon-stimulated genes
While the importance of antiviral RNAi in plants and invertebrates is well established, the role of mammalian RNAi as a viral defense has been debated [57]. Viral replication is frequently associated with production of dsRNA, and most mammalian cells respond by inducing interferon signaling, which results in global translational suppression and mRNA degradation [58]. In addition, previous sequencing of small RNAs from diverse virus-infected mammalian cells did not yield a clear pattern of viral siRNAs [59], as is commonly seen in plants and invertebrates. Many DNA viruses clearly encode miRNAs, and in the best-studied cases confer benefits to the virus by repressing programs of host gene expression, or by providing control over viral genes [60]. It is unclear that host miRNAs can productively target viruses, since it should be relatively easy to evade regulation by selection of an appropriate mutation [61]. On the contrary, Eastern equine encephalitis virus takes advantage of miR-142 to restrict its tropism [62], and Hepatitis Virus C exploits host miR-122 to facilitate its replication [63].
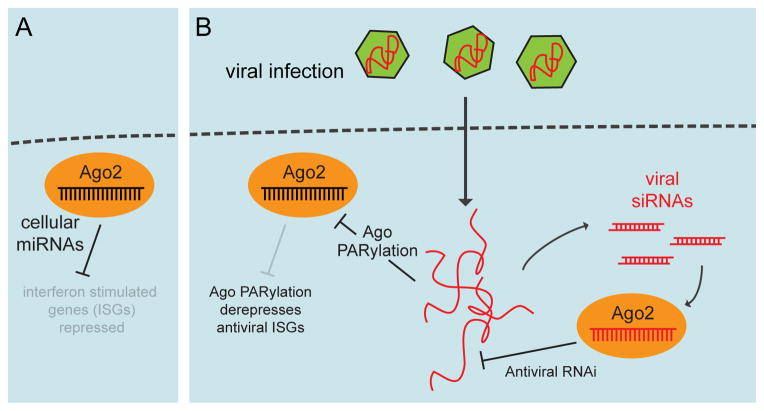
In a new twist, Sullivan and colleagues recently proposed that the mammalian miRNA pathway facilitates virus defense by regulating host gene expression [64] (Figure 3). They observed that miRNA/RNAi pathway activity was suppressed upon activation of interferon signaling by treatment with poly-I:C dsRNA, as well as following bona fide infection with herpesvirus HSV-1, Sendai virus, or influenza virus. Viral suppression of RNA silencing was accompanied by substantial PARylation of Ago2-associated proteins. Treatment with a pADP-r inhibitor partially relieved virus-induced suppression of miRNA/RNAi pathway activity, consistent with a functional role for this modification. In addition, full PARylation of Ago complexes depended on the antiviral factors MAVS and RNaseL, implicating a signaling pathway downstream of viral detection that leads to modification of Ago complexes [64].
Figure 3.
Opposing functions for small RNA pathway activity during the antiviral response. (A) In uninfected cells, endogenous miRNAs are proposed to repress many transcripts that encoded factors involved in viral defense (interferon-stimulated genes, ISGs). (B) Ago2 is PARylated following certain cellular stresses, including viral infection. As PARylation is thought to inhibit Ago function, this modification may broadly derepress ISGs in order to potentiate the antiviral interferon response. On the other hand, double-stranded RNA viral intermediates can also generate siRNAs that may in turn target the virus (right).
Why might this process be implemented during viral infection? Since many interferon-stimulated genes (ISGs) are cytotoxic, they are subject to tight control. Inspection of 136 literature-annotated ISGs associated with cell death or proliferation showed that their 3′ UTRs contained about four times as many conserved miRNA target sites than background genes, and that many ISGs were upregulated in human colorectal cancer cells bearing a dicer hypomorphic allele relative to the DLD-1 parental cell line [64]. Amongst genes de-repressed by interferon treatment of these two cell lines, transcripts upregulated in the dicer mutant condition were enriched for target sites of the miR-17/93 family. Many of these target sites in ISGs were occupied by Ago, as measured in published transcriptome-wide sequencing of Ago binding sites [65, 66]. Moreover, forced expression of miR-17 or miR-93, but not control miRNA mimics, enhanced the production of HSV-1 and influenza virus possibly indicating that miRNAs, especially those of the miR-17/93 family, have “proviral” activity due to their default suppression of ISGs. Therefore, an effective cellular response to viral infection includes the downregulation of miRNA pathway activity via the PARylation of Ago2 complexes [64].
These results provide an interesting link to the earlier finding that PARylation can affect Ago proteins and dampen miRNA activity [53]. Interestingly, two contemporaneous studies provided evidence for an antiviral role for mammalian RNAi, at least in certain cell contexts [67, 68] (Figure 3). These studies identified a population of viral siRNAs loaded in Ago2, distinct from heterogeneous degradation fragments of viral sense strands, and provided evidence that suppression of RNAi by viral proteins contributes to productive infections of undifferentiated ES cells and mouse pups [67, 68]. The endogenous location of viral siRNA generation in intact animals was not identified, but ES cells lack an interferon response and harbor endogenous siRNAs [69, 70]. Notably, the capacity of ES cells to generate viral siRNAs was reduced upon differentiation, suggesting that antiviral RNAi is a property of an undifferentiated state [67]; however, it should be noted the other study utilized a kidney cell line to detect viral siRNAs [68].
Overall, these studies introduce new complexities to the cellular small RNA response to viral infection, including the opposing effects of antiviral RNAi that may directly restrict viruses, and suppression of proviral host miRNAs that may normally restrict cytotoxic interferon-responsive genes. It remains to be explored whether these processes occur simutaneously, or are temporally separated during viral response. They might even represent segregated processes that are preferentially executed according to cell differentiation status, or in response to specific stresses.
Concluding remarks
The study of post-translational modifications to mammalian Ago proteins not only highlights a multiplicity of strategies by which control over miRNA pathway activity can be achieved, but also illustrates how extracellular stimuli or environmental conditions can induce global or specific modulatory effects. Beyond all the strategies discussed, it remains to be seen whether Ago proteins are subject to methylation, as seen with Piwi-clade small RNA effectors [71], or perhaps acetylation, as shown for Drosha and DGCR8 [72, 73].
For many examples discussed, the relevant modifying enzymes are not yet identified (Table 1). Notably, Ago2-Y529 does not appear to be structurally accessible, although it is conceivable that Ago2 undergoes conformational changes. This question can be addressed upon identification of the relevant kinase. In addition, Ago destabilization following T cell activation is implied to be mediated by an E3 ubiquitin ligase. Identification of this factor would presumably be not only relevant to immunology, but such an E3 might be deployed in other situations that require resetting of miRNA repertoires.
Improved precision on tracking modified Ago proteins will depend on specific antibodies. Certain phospho-specific Ago2 antibodies exist, but such reagents are generally wanting for most known phosphorylated residues or for hydroxylated Ago2-P700. At this point, it is unclear which specific lysines are critical for any of the ubiquitin-dependent Ago turnover events reported, and it is not clear whether any antibody could specifically recognize PARylated Ago. Nevertheless, it is clear that additional modification-specific Ago antibodies would greatly aid profiling studies to understand how the distribution of Ago protein variants differs across cell types and tissues, during altered cell conditions, and/or in response to diverse extracellular stimuli. Moreover, while the majority of functional studies focused on Ago2, several of the documented modifications seem appended to multiple Ago proteins. Therefore, the “wishlist” should be extended to have modification-specific antibodies for each of the four mammalian Ago proteins.
A final consideration regards the specificity of most experiments regarding Ago modifications to date. Nearly all of the studies discussed utilized transfected constructs, knockdown conditions, and chemical treatments, but these methodologies have consequences in cells beyond the direct modulation of Ago activity. The ultimate “clean” experiment would be to knock-in point mutations of various Ago residues that are subject to modification. For example, slicing activity by Ago2 has been widely measured in cells and extracts, but firm demonstration that this is of substantial endogenous requirement only came with the generation of a mice bearing point mutation of a catalytic residue in ago2 [74]. Not only are these defective for slicing, they exhibit anemia-like defects that may involve loss of the erythroid-enriched, slicing-dependent mir-451 locus [25]. Nevertheless, as deletion of mir-451 does not phenocopy the loss of slicing capacity [25], there must be other genetic requirements for slicing.
Such findings illustrate the utility of analyzing Ago2 residues in cis. Therefore, it will be interesting to study ago2 alleles specifically mutated in lysine resides that are subject to different ubiquitination effects, or tyrosine/serine/threonine residues that are modified by different kinases. Such genetic reagents will enable stringent assessments of the endogenous requirements of various modified Ago2 residues, and potentially uncover novel small RNA biology.
Acknowledgments
We thank Ben TenOever, Ted Karginov, Katsutomo Okamura, Akiko Hata, Chris Sullivan, Mark Ansel and Gunter Meister for informative discussions. Work in E.C.L.’s group was supported by the Burroughs Wellcome Fund and the National Institute of Neurological Disorders and Stroke of the National Institutes of Health (1R01-NS083833). The content is solely the responsibility of the authors and does not represent the official views of these agencies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Molecular cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meister G. Argonaute proteins: functional insights and emerging roles. Nature reviews Genetics. 2013;14:447–459. doi: 10.1038/nrg3462. [DOI] [PubMed] [Google Scholar]
- 3.Brennecke J, et al. Principles of MicroRNA-Target Recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes & development. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai EC. microRNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nature genetics. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aboobaker AA, et al. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wienholds E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 10.Suh N, et al. MicroRNA Function Is Globally Suppressed in Mouse Oocytes and Early Embryos. Current Biology. 2010;20:271–277. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma J, et al. MicroRNA activity is suppressed in mouse oocytes. Curr Biol. 2010;20:265–270. doi: 10.1016/j.cub.2009.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su X, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467:986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy C, et al. Lineage-specific transcriptional regulation of DICER by MITF in melanocytes. Cell. 2010;141:994–1005. doi: 10.1016/j.cell.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, et al. c-Myc modulates microRNA processing via the transcriptional regulation of Drosha. Scientific reports. 2013;3:1942. doi: 10.1038/srep01942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim YK, et al. Modifications of small RNAs and their associated proteins. Cell. 2010;143:703–709. doi: 10.1016/j.cell.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 16.Heo I, Kim VN. Regulating the regulators: posttranslational modifications of RNA silencing factors. Cell. 2009;139:28–31. doi: 10.1016/j.cell.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 17.Paroo Z, et al. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heo I, et al. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 19.Hagan JP, et al. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nature structural & molecular biology. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thornton JE, et al. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) RNA. 2012;18:1875–1885. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang HM, et al. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013 doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ustianenko D, et al. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA. 2013;19:1632–1638. doi: 10.1261/rna.040055.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi HH, et al. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature. 2008;455:421–424. doi: 10.1038/nature07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu C, et al. Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Molecular and cellular biology. 2011;31:4760–4774. doi: 10.1128/MCB.05776-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang JS, Lai EC. Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates. Cell cycle. 2010;9:4455–4460. doi: 10.4161/cc.9.22.13958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudel S, et al. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic acids research. 2011;39:2330–2343. doi: 10.1093/nar/gkq1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng Y, et al. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J. 2008;413:429–436. doi: 10.1042/BJ20080599. [DOI] [PubMed] [Google Scholar]
- 28.Horman SR, et al. Akt-mediated phosphorylation of argonaute 2 downregulates cleavage and upregulates translational repression of MicroRNA targets. Molecular cell. 2013;50:356–367. doi: 10.1016/j.molcel.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazumder A, et al. A transient reversal of miRNA-mediated repression controls macrophage activation. EMBO reports. 2013;14:1008–1016. doi: 10.1038/embor.2013.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen J, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–387. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Betancur JG, Tomari Y. Dicer is dispensable for asymmetric RISC loading in mammals. RNA. 2012;18:24–30. doi: 10.1261/rna.029785.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noland CL, Doudna JA. Multiple sensors ensure guide strand selection in human RNAi pathways. RNA. 2013;19:639–648. doi: 10.1261/rna.037424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rybak A, et al. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nature cell biology. 2009;11:1411–1420. doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- 34.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 35.Ecsedi M, Grosshans H. LIN-41/TRIM71: emancipation of a miRNA target. Genes & development. 2013;27:581–589. doi: 10.1101/gad.207266.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang HM, et al. Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nature communications. 2012;3:923. doi: 10.1038/ncomms1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loedige I, et al. The mammalian TRIM-NHL protein TRIM71/LIN-41 is a repressor of mRNA function. Nucleic acids research. 2013;41:518–532. doi: 10.1093/nar/gks1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J, et al. The ubiquitin ligase mLin41 temporally promotes neural progenitor cell maintenance through FGF signaling. Genes & development. 2012;26:803–815. doi: 10.1101/gad.187641.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smibert P, et al. Homeostatic control of Argonaute stability by microRNA availability. Nature structural & molecular biology. 2013 doi: 10.1038/nsmb.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnston M, et al. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Mol Biol Cell. 2010;21:1462–1469. doi: 10.1091/mbc.E09-10-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez NJ, Gregory RI. Argonaute2 expression is post-transcriptionally coupled to microRNA abundance. RNA. 2013;19:605–612. doi: 10.1261/rna.036434.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang P, Zhang H. Autophagy modulates miRNA-mediated gene silencing and selectively degrades AIN-1/GW182 in C. elegans. EMBO reports. 2013;14:568–576. doi: 10.1038/embor.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibbings D, et al. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nature cell biology. 2012;14:1314–1321. doi: 10.1038/ncb2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chong MM, et al. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205:2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muljo SA, et al. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005 doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monticelli S, et al. MicroRNA profiling of the murine hematopoietic system. Genome biology. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olejniczak SH, et al. Long-lived microRNA-Argonaute complexes in quiescent cells can be activated to regulate mitogenic responses. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:157–162. doi: 10.1073/pnas.1219958110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Rooij E, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 49.Bronevetsky Y, et al. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med. 2013;210:417–432. doi: 10.1084/jem.20111717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baumberger N, et al. The Polerovirus silencing suppressor P0 targets ARGONAUTE proteins for degradation. Curr Biol. 2007;17:1609–1614. doi: 10.1016/j.cub.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 51.Bortolamiol D, et al. The Polerovirus F box protein P0 targets ARGONAUTE1 to suppress RNA silencing. Curr Biol. 2007;17:1615–1621. doi: 10.1016/j.cub.2007.07.061. [DOI] [PubMed] [Google Scholar]
- 52.Zhao S, et al. piRNA-Triggered MIWI Ubiquitination and Removal by APC/C in Late Spermatogenesis. Developmental cell. 2013;24:13–25. doi: 10.1016/j.devcel.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 53.Leung AK, et al. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Molecular cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leung AK, et al. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Karginov FV, Hannon GJ. Remodeling of Ago2-mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes & development. 2013;27:1624–1632. doi: 10.1101/gad.215939.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu PH, et al. Functionally diverse microRNA effector complexes are regulated by extracellular signaling. Molecular cell. 2013;52:113–123. doi: 10.1016/j.molcel.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cullen BR, et al. Is RNA interference a physiologically relevant innate antiviral immune response in mammals? Cell host & microbe. 2013;14:374–378. doi: 10.1016/j.chom.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 58.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nature reviews Immunology. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parameswaran P, et al. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS pathogens. 2010;6:e1000764. doi: 10.1371/journal.ppat.1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cullen BR. Viral and cellular messenger RNA targets of viral microRNAs. Nature. 2009;457:421–425. doi: 10.1038/nature07757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yeung ML, et al. siRNA, miRNA and HIV: promises and challenges. Cell Res. 2005;15:935–946. doi: 10.1038/sj.cr.7290371. [DOI] [PubMed] [Google Scholar]
- 62.Trobaugh DW, et al. RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature. 2013 doi: 10.1038/nature12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jopling CL, et al. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 64.Seo GJ, et al. Reciprocal Inhibition between Intracellular Antiviral Signaling and the RNAi Machinery in Mammalian Cells. Cell host & microbe. 2013;14:435–445. doi: 10.1016/j.chom.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gottwein E, et al. Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines. Cell host & microbe. 2011;10:515–526. doi: 10.1016/j.chom.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Skalsky RL, et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS pathogens. 2012;8:e1002484. doi: 10.1371/journal.ppat.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maillard PV, et al. Antiviral RNA interference in mammalian cells. Science. 2013;342:235–238. doi: 10.1126/science.1241930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Y, et al. RNA interference functions as an antiviral immunity mechanism in mammals. Science. 2013;342:231–234. doi: 10.1126/science.1241911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Babiarz JE, et al. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes & development. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flemr M, et al. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell. 2013;155:807–816. doi: 10.1016/j.cell.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 71.Vagin VV, et al. Arginine methylation as a molecular signature of the Piwi small RNA pathway. Cell cycle. 2009;8 doi: 10.4161/cc.8.24.10146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wada T, et al. Histone deacetylase 1 enhances microRNA processing via deacetylation of DGCR8. EMBO reports. 2012;13:142–149. doi: 10.1038/embor.2011.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang X, et al. Acetylation of drosha on the N-terminus inhibits its degradation by ubiquitination. PloS one. 2013;8:e72503. doi: 10.1371/journal.pone.0072503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cheloufi S, et al. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]