Abstract
BACKGROUND
The recent revolutionary advances made in genome-wide sequencing technology have transformed biology and molecular diagnostics, allowing new sRNA (small RNA) classes to be discovered as potential disease-specific biological indicators. Cell-free microRNAs (miRNAs) have been shown to exist stably in a wide spectrum of body fluids and their expression profiles have been shown to reflect an assortment of physiological conditions, underscoring the utility of this new class of molecules to function as noninvasive biomarkers of disease.
CONTENT
We summarize information on the known mechanisms of miRNA protection and release into extracellular space and compile the current literature on extracellular miRNAs that have been investigated as biomarkers of 20 different cancers, 11 organ damage conditions and 10 diverse disease states. We also discuss the various strategies involved in the miRNA biomarker discovery workflow and provide a critical opinion on the impediments faced by this advancing field that need to be overcome in the laboratory.
SUMMARY
The field of miRNA-centered diagnostics is still in its infancy, and basic questions with regard to the exact role of miRNAs in the pathophysiology of diseases, and the mechanisms of their release from affected cells into biological fluids are yet to be completely understood. Nevertheless, these noninvasive micromarkers have immense potential in translational medicine not only for use in monitoring the efficacy and safety of therapeutic regimens but also to guide the diagnosis of diseases, to determine the risk of developing diseases or conditions, and more importantly, to inform treatment options.
It has been only 2 decades since the discovery of the first microRNA (miRNA),3 lin-4, as a heterochronic gene in the Caenorhabditis elegans developmental pathway (1). The sequence of let-7, which was discovered soon after (2), was found to be conserved across a wide range of phyla, being expressed at the later stages of development in all species, indicating temporal conservation as well (3). Since then, the research in this field has burgeoned, with over 21 000 sequences currently discovered in a total of 193 species. Today, we know miRNAs to be a family of small noncoding RNA molecules of approximately 22 nucleotides in length that regulate gene expression. In the human genome, 52% of miRNAs are located intergenically, and 40% and 8% are located within introns and exons, respectively (4).
miRNAs are transcribed in the nucleus by RNA polymerase II as primary transcripts called pri-miRNAs, which are further processed by the complex formed by RNase III endonuclease Drosha with DGCR8 protein (Pasha) into 70-nucleotide–long stem-looped precursor miRNAs (Fig. 1A) (5). The precursor miRNA (pre-miRNA) is exported to the cytoplasm via Exportin 5 in a Ran GTP– dependent manner, where it is cleaved by Dicer, another RNase III endonuclease which associates with trans-activation response (TAR) RNA-binding protein (TRBP) to produce an miRNA–miRNA* duplex comprising the functional fragment (miRNA) and a low-abundance fragment (miRNA*). Normally the functional miRNA that enters the RNA-induced silencing complex (RISC) originates from the strand with the least stable 5′ end, which lends asymmetry to the complex. The typical recovery ratio for miRNA and miRNA* from the miRNA–miRNA* duplex is about 100:1, except in the rare cases in which both arms of the precursor are equally stable and have similar recoveries (6).
Fig. 1. MicroRNA (miRNA) biogenesis and secretion into extracellular space.
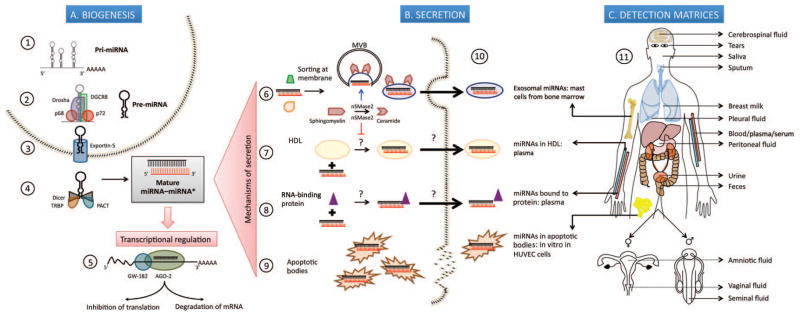
(A), Biogenesis. 1. Transcription of pri-miRNAs from genes in the nucleus by RNA polymerase II. 2. Processing of primary transcript into 70-nucleotide (nt)–long pre-miRNA by the Drosha-DGCR8 Microprocessor complex assisted by p68 (DDX5) and p72 (DDX17) DEAD-box RNA helicases that possibly act as scaffolding proteins to recruit cofactors. 3. Export into cytoplasm in a Ran-GTP– dependent manner through Exportin 5. 4. Cleavage of pre-miRNA by RNase III enzyme Dicer, into a 22-nt double-stranded RNA composed of the mature miRNA “guide” strand and the low-abundance miRNA* “passenger” strand; TRBP and protein activator of interferon-induced protein kinase PKR (PACT) are some of the molecules that regulate this step. 5. Incorporation of mature miRNA into the RISC, whose main components are AGO-2 and GW-182; partial complementarity of the seed region of miRNA to the 3′ untranslated region of target mRNAs causes translational repression while complete complementarity leads to degradation of the transcript. (B), Secretion of miRNAs into extracellular space by: 6. Packaging into multivesicular bodies (MVBs) that fuse with the plasma membrane and release as exosomes in a ceramide-dependent pathway positively regulated by nSMase2. 7. Encapsulation into HDL particles, a process which is repressed by nSMase2. 8. Binding to RNA-binding proteins, namely AGO-2 and nucleophosmin 1 (NPM1). 9. Incorporation into apoptotic bodies. 10. Pioneering studies describing exosomal miRNAs isolated from primary bone marrow derived mast cells; HDL-miRNA in human plasma; protein-bound miRNAs in human plasma (AGO-2) and human cell lines (NPM1 in HepG2 and A549); miR-126 from endothelial cell-derived apoptotic bodies in vitro in human umbilical vein endothelial cells (HUVEC). (C), Detection matrices. 11. Depiction of 13 different matrices where extracellular miRNAs have been discovered.
The functional miRNA is incorporated into the RISC assembled by Dicer/TRBP and an Argonaute protein (AGO-2), which functions as the catalytic endonuclease (5). Near perfect complementarity in the 3′ untranslated region of the mRNA to the seed sequence (2– 8 nucleotides) in the 5′ end of the miRNA leads to cleavage of the target mRNA. On the other hand, an imprecise complementarity assures translational repression of some form, which leaves the mRNA target intact (Fig. 1A). miRNAs are predicted to form pairing with nearly 60% of all protein-coding genes (7) and are known to impact diverse cellular processes, such as proliferation, differentiation, and cell death (5).
Mechanisms of miRNA Protection in Circulation
Recently, cell-free miRNAs have been discovered to exist in circulatory, secreted, and excreted body fluids. These extracellular miRNAs are physiologically functional and have been shown to exert gene silencing in downstream target cells (8). The surprising resistance of miRNAs to RNA-degrading enzymes in circulation is a result of several as yet unearthed mechanisms of protection (Fig. 1B). miRNAs in their naked form are not innately resistant to RNase activity owing to their size or structure, because artificially introduced synthetic Caenorhabditis elegans miRNAs are rapidly degraded in plasma, whereas endogenous miRNAs remain unaltered (8, 9). The mechanism that has been most investigated so far is the encapsulation of miRNAs into vesicular bodies such as exosomes (10–100 nm) and microvesicles (0.1–1 μm). The first report was in 2007 by Valadi et al., who showed that exosomes shed by human and mouse mast cells contain miRNAs that are deliverable in a functional form to recipient cells (10). Kosaka et al. showed in HEK293 cells that the packaging of miRNAs into exosomes is a ceramide-dependent mechanism under the regulation of neutral sphingomyelinase 2 (nSMase2) (8) and the process has been shown to be ATP dependent in a separate study (11). Exosomal miRNAs have been discovered in human saliva, breast milk, and urine apart from plasma (12–14). Another class of vesicles, apoptotic bodies (0.5–2 μm), have been demonstrated to carry miRNAs from apoptotic endothelial cells during atherosclerosis to neighboring cells to induce survival and growth signals (15). Experiments by Wang et al. on HepG2 and A549 cell lines indicate that following serum deprivation there is a sharp increase in the amount of extracellular miRNAs with a concomitant drop in intracellular concentration, which recovers after a few hours (11). This leads to the assumption that miRNAs to be exported are derived from a presynthesized pool. Apart from the vesicular fraction, miRNAs have also been identified in the supernatant after ultracentrifugation processes to isolate the vesicles (11, 16). This suggests that there are other forms of protection, and they have been shown to include conjugation with a variety of proteins, namely, nucleophosmin 1 (11), HDL (17), and AGO-2 (16, 18). Delivery of miRNAs conjugated with HDL to recipient cells is under the regulation of scavenger receptor class B type 1 (17). Interestingly, loading of HDL with miRNAs appears to be repressed by nSMase2, which indicates that exosomal secretion and HDL incorporation of miRNA may be opposing cellular mechanisms (17), although this has never been proven in the same cell line.
The clearance of miRNAs from circulation is another important mechanistic aspect that remains incompletely explored. Although the half-life of miRNAs has not been measured in extracellular fluids, it is likely that they persist for a long time because of the stable miRNA–protein/lipid complexes. Possible routes of clearance from circulation include miRNA-specific degradation pathways, filtration by the kidneys, or removal by the hepatic system. However, Weber et al. report that there is little correlation between the plasma and urinary profiles of miRNAs, indicating either that the kidneys are not involved in physiological clearance or that miRNAs filtered into the urine are degraded rapidly (19). Further evidence for the former possibility comes from a study which showed that decreased circulating concentrations of miRNAs in patients with chronic kidney disease was not associated with an increase in the urinary concentration (20).
Circulating miRNAs as Biomarkers
miRNAs have not only been shown to play critical regulatory roles in health and disease, but have also been investigated as biomarkers for detecting and predicting disease progression, as well as for therapeutic intervention. Abundant expression, lower complexity, stability in various detection matrices, and amplifiable signals are some qualities that make extracellular miRNAs attractive candidates as biomarkers reflecting a variety of pathophysiological conditions (21–24) (Table 1). Although proteins have significantly advanced our understanding of biomarker science, there are several drawbacks concerning methods of discovery and detection, availability and stability. Classical immunoaffinity-based methods for measurement of protein biomarkers detect antigen–antibody complex formation in various platforms. These techniques, which include ELISA, rely heavily on the specificity and concentration of the antibody used (25). Platforms that provide multiplexing to measure several antigens in one assay run into problems of diverse range of abundance resulting in the inability for uniform amplification. Posttranslational modifications like glycosylation, acetylation, and lipidation occur frequently and further increase the complexity of the proteome (26). The presence of proteolytic enzymes in nearly all biological samples used for biomarker analysis renders the need for careful collection and preservation of samples. Detection of proteins from preserved specimens such as formalin-fixed paraffin-embedded (FFPE) sections requires extensive antigen retrieval and extraction treatments (27).
Table 1.
Salient features of miRNAs making them ideal biomarkers.
| Sensitive: immediate release before mRNA transcription and protein translation |
| Specific: tissue- and disease-specific regulation |
| Dynamic range: approximately 50,000 copies of miRNA/cell |
| Translational potential: highly conserved with a high degree of inter- and intraspecies homology |
| Stability: more resistant to degradation than mRNA and proteins and able to be detected in FFPE tissues (although DNA is more stable, it is less tissue specific) |
| Accessible in a plethora of biological fluids, allowing noninvasive detection |
| Quantitative by PCR amplification |
Since the discovery of stably expressed miRNAs in serum in 2008 (28), these small molecules have been discovered in a plethora of other biological fluids such as urine, saliva, breast milk, amniotic fluid, tears, feces, seminal and vaginal fluid, sputum, and cerebrospinal fluid (CSF) (Fig. 1C). The most striking advantage of miRNAs over all other molecules that are in use as biomarkers is their stability across a wide range of physiological and storage conditions. miRNAs in serum have been shown to resist degradation due to extreme temperature and pH fluctuations, freeze–thaw cycles, extended storage of samples, and RNase A digestion (9, 24, 29). Several groups have demonstrated successful isolation of miRNAs from a wide range of sample types including FFPE sections (30), bone marrow aspirate slides (31), and bone marrow core biopsies (32). miRNAs are ubiquitously present and are conserved in mature sequence across several organisms (33), and yet within a species, their expression levels are reproducibly consistent among individuals. The absence of post-transcriptional modification of miRNAs is another attractive feature that confers uniformity and lowers the complexity of the system. Several miRNAs have unique cellular or tissue localization or disease-specific expression that lends specificity to their roles as biomarkers.
Pioneering studies characterizing miRNAs as fluid-based biomarkers have typically used an individual miRNA or a panel of miRNAs to distinguish between cohorts, and statistical measures such as the areas under ROC curves indicate that they have diagnostic accuracies ranging from 70% to 95% (34, 35). These proof-of-concept experiments have laid the foundation on which future miRNA research has developed, taking forward the theme of characterizing entire miRNA profiles to specific diseases (36). An miRNA-based marker will ensure specificity because by virtue of sheer numbers, unique expression signatures can be identified for disease initiation, progression, prognosis, and classification. The wide batteries of tests now being conducted for the same purpose involve measurement of antigens, proteins, lipids, and enzymes and are hindered by shortcomings such as high detection thresholds, low throughput, delayed expression, and variable sensitivities. Although a universally standardized procedure of RNA isolation and miRNA detection is awaited, amplification-based techniques such as quantitative PCR (qPCR) have been shown to be simple, fast, reproducible, and cost-effective.
Some studies have examined the variation of miRNAs in the healthy population, because this is important in interpreting the results from different disease states and biological fluids. Weber et al. measured the miRNA composition of 12 different body fluids in 5 healthy samples and determined that saliva, breast milk, and seminal fluid have larger numbers of detectable miRNA species in comparison with urine or CSF (19). On the basis of commonly expressed miRNAs, these investigators were able to cluster the fluids into 2 major groups, with plasma having a profile distinct from most other fluid types. We analyzed the temporal variation in a select number of candidate miRNAs in the urine samples of 29 healthy individuals (36) and observed that although there was considerable background variation in the healthy samples, the miRNA concentrations were significantly increased in disease conditions. This ubiquitous and variant dispersion of the miRNA population suggests their importance in maintaining normal physiological functions in a variety of organ systems.
miRNAS AS BIOMARKERS OF CANCER
The knowledge that miRNAs are released outside the cell and exist stably in various body fluids has received considerable attention from the field of cancer biology, where the gold standards of detection are usually invasive, and the necessity for extracellular and noninvasive sources of biomarkers is growing urgent (Table 2). Most of the studies have been performed on serum/ plasma, but some groups have examined other fluids relevant to diseases, such as saliva for oral carcinoma (37), urine for renal (38) and bladder cancer (39), sputum for lung cancer (40), and CSF for lymphoma of the central nervous system (41) and glioblastoma (42). One of the first attempts to identify miRNA biomarkers in a noninvasive body fluid was made in 2008, by comparing the concentrations of 3 miRNAs known to be associated with tumors in the serum of patients with diffuse large B-cell lymphoma with concentrations in healthy controls (28). It was found that miRNA concentrations are higher in the patient sera and specifically that high miR-21 concentrations indicated relapse-free survival. Around the same time, another group showed that miRNAs are present in a very stable and detectable form in human plasma and can distinguish prostate cancer xenograft mice from controls (9).
Table 2.
Extracellular microRNAs as biomarkers of cancer.a
| Disease | Body fluid | Candidate miRNAs | Cohort used | Normalizer | Reference | ||
|---|---|---|---|---|---|---|---|
| Discovery | Confirmation | Evaluation | |||||
| Prostate cancer (PC) | Serum | miR-141 | 1 Control | 25 PC, 25 controls | — | C. elegans synthetic miRNA | Mitchell et al. (9) |
| Serum | Panel of 15 miRNAs | 5 PC, 8 controls | — | — | z-Score normalization | Sita-Lumsden et al. (94) | |
| Serum | miR-375, -141 | 7 Metastatic, 14 localized PC | 42 PC, 3 metastatic | 71 Primary PC | C. elegans synthetic miRNA | Sita-Lumsden et al. (94) | |
| Diffuse large B-cell lymphoma (DLBCL) | Serum | miR-155, -210, -21 | 60 DLBCL, 43 controls | — | — | miR-16 | Lawrie al. (28) |
| Serum | miR-15a, -16–1, -29c, -155 | 20 DLBCL, 20 controls | 75 DLBCL, 77 controls | — | C. elegans miR-39 | Fang et al. (95) | |
| Leukemia | Plasma | miR-92a, -638 | 2 Acute myeloid leukemia (AML), 7 controls | 54 AML, 7 Acute lymphoblastic leukemia, 16 controls | — | Raw data | Tanaka et al. (96) |
| Pancreatic cancer (PnC) | Plasma | miR-210 | 11 PnC, 14 controls | 11 PnC, 11 controls | — | C. elegans miR-54 | Ho et al. (97) |
| Serum | miR-20a, -21, -24, -25, -99a, -185, -191 | Pooled 25 PnC, 25 controls | 25 PnC, 25 controls; 95 PnC, 81 controls | 82 Chronic pancreatitis (CP); 37 PnC, 32 controls; 55 retrospective | Serum volume | Liu et al. (98) | |
| Plasma | miR-21, -221, -200b, -200c, -146a, let 7b, let 7d | Pooled 50 PnC, 10 controls | 32 PnC, 10 controls | — | miR-16 | Ali et al. (99) | |
| Pancreatic ductal adenocarcinoma (PDAC) | Plasma | miR-21, -210, -155, -196a | 28 PDAC, 19 controls | — | — | miR-16 | Wang et al. (100) |
| Serum | miR-196a, -21, -155, -210 | 35 PDAC, 15 CP, 15 controls | — | — | Raw data | Kong et al. (50) | |
| Breast cancer (BC) | Whole blood | miR-195, let 7a | 83 BC, 44 controls | 29 BC after surgery | — | miR-16 | Heneghan et al. (101) |
| Serum | miR-10b, -34a, -155 | 4 BC cell lines | 59 Primary BC, 30 metastatic, 29 controls | — | miR-16 | Roth et al. (47) | |
| Serum | miR-21 | 14 BC and normal tissue | 40 BC (10 each stage), 10 controls | 102 BC different stages, 20 controls | miR-16 | Asaga et al. (43) | |
| Plasma | miR-16, -21, -145, -451 | 5 BC and control plasma, BC and normal tissue | 15 BC before and after surgery, 15 controls | 170 BC, 95 different cancers, 100 controls; 70 BC, 50 controls | RNU6B | Ng et al. (102) | |
| Ovarian cancer (OvC) | Serum | miR-21, -92, -93, -126, -29a, -155, -127, -99b | 9 OvC, 4 controls | 19 OvC, 11 controls | — | miR-142–3p | Resnick et al. (103) |
| Serum exosome | miR-21, -141, -200a, -200b, -200c, -203, -205, -214 | Exosome derived vs tumor derived profile | 50 OvC different stages, 10 benign, 10 controls | — | Ambion control miRNA | Taylor and Gercel-Taylor (104) | |
| Gastric cancer (GC) | Plasma | miR-17–5p, -21, -106a, -106b, let-7a | 34 GC, 15 controls; 8 GC in plasma and FFPE | 10 GC before and after surgery | 69 GC different stages | RNU6B | Wang et al. (105) |
| Serum | miR-1, -20a, -27a, -34, -423, -5p | Pooled 20 metastatic GC, 20 controls | 22 GC, 22 controls | 142 GC, 105 controls | Standard curve calibration | Wang et al. (105) | |
| Plasma | miR-223, -21, -218 | 10 GC, 10 controls | 8 GC plasma and tissue | 60 GC, 60 controls | RNU6B | Li et al. (106) | |
| Serum | miR-221, -376c, -744 | Pooled and individual 14 GC, 14 controls | 68 GC, 68 controls; 42 dysplasia, 42 controls | 58 GC retrospective | ath-miR-159a, profiling; C. elegans miR-39, qPCR | Wang et al. (105) | |
| Serum | miR-378 | 7 GC, 7 CRC, 10 controls | 10 GC, 10 CRC, 10 controls | 40 GC, 41 controls; 4 GC tissue | RNU6B | Wang et al. (105) | |
| Colorectal cancer (CRC) | Plasma | miR-92, -17–3p | 5 CRC and control plasma; CRC and normal tissue | 25 CRC, 20 controls; 10 CRC before and 7 days after surgery | 90 CRC, 20 GC, 20 inflammatory bowel diseases, 50 controls | RNU6B | Menendez et al. (107) |
| Plasma | miR-29a, -92a | 20 CRC, 20 controls | 80 CRC, 37 adenomas, 39 controls | 20 CRC before and after surgery | miR-16 | Menendez et al. (107) | |
| Stool | miR-92a, -21 | 88 CRC, 57 polyps, 101 controls | 40 CRC and normal tissue | 9 and 10 CRC before and after surgery | Standard curve calibration | Wu et al. (108) | |
| Stool | miR-144* | 15 CRC and controls | 35 CRC and 40 controls | 15 CRC and normal tissue | miR-378 | Kalimutho et al. (109) | |
| Plasma | miR-141 | 74 CRC different stages, 28 controls | 108 CRC, 48 controls | 21 Metastatic, 24 localized tumors | C. elegans miR-39 | Menendez et al. (107) | |
| Non–small cell lung carcinoma (NSCLC) | Serum | miR-21 | 6 NSCLC and normal tissue | 70 NSCLC, 44 controls | — | RNU6B | Liu et al. (44) |
| Lung adenocarcinoma (LA) | Sputum | miR-21, -486, -375, -200b | 20 LA and normal tissue | 36 LA, 36 controls | 64 LA, 58 controls | RNU6B | Yu et al. (110) |
| Squamous lung cell carcinoma (LC) | Sputum | miR-205, -210, -708 | 15 LC and normal tissue | 48 LC, 48 controls | 67 LC, 55 controls | RNU6B | Xing et al. (40) |
| Hepatocellular carcinoma (HCC) | Serum | miR-500 | Mouse tissue; 6 liver cancer cell lines | 40 HCC and normal tissues | 3 HCC before and after surgery | RNU6B, tissue; miR-1, serum | Yamamoto et al. (46) |
| Serum | miR-25, -375, let-7f | 30 HCC, 30 controls | 30 HCC, 30 controls | 55 HCC, 50 controls | Serum volume | Li et al. (111) | |
| Urine | miR-618, -650 | Pooled 32 HCC post–hepatitis C virus (HCV), 74 HCV, 12 controls | Individual validation | — | 5S rRNA | Abdalla and Haj-Ahmad (112) | |
| Plasma | miR-21 | 126 HCC, 30 hepatitis, 50 controls | 10 HCC before and after surgery | — | miR-16 | Tomimaru et al. (113) | |
| Primary central nervous system lymphoma (PCL) | CSF | miR-21, -19b, -92 | 23 PCL, 10 other neurological problems | 23 PCL, 30 other neurological problems | — | miR-24 | Baraniskin et al. (41) |
| Glioblastoma (GBM) | CSF | miR-10b, -21 | 19 GBM, 16 breast to brain, 26 breast leptomeningeal metastasis (LM), 28 lung to brain, 4 lung LM, 15 controls | 1 GBM, 2 NSCLC temporal study | — | miR-24 | Teplyuk et al. (42) |
| Bladder cancer (BCa) | Urine | miR-143, -222, -452 | 37 BCa tissue and urine | 37 BCa, 37 benign urinary diseases, 20 controls | — | miR-16 | Puerta-Gil et al. (39) |
| Urine | miR-1224–3p, -135b, -15b | 68 BCa, 58 benign urinary diseases | — | — | RNU44, RNU48 | Miah et al. (114) | |
| Urine | miR-126, -152, -182 | 9 low BCa, 9 high BCa, 9 urinary tract infection (UTI), 9 controls | 11 low BCa, 18 high BCa, 7 UTI, 11 controls | — | RNU6B | Hanke et al. (51) | |
| Malignant melanoma (MM) | Serum | miR-221 | 90 different types of MM, 20 controls | 8 MM before, after surgery, at recurrence | — | C. elegans miR-54 | Kanemaru et al. (115) |
| Serum | miR-103, -221, -222, -423, -5p, 199a-5p, -33a, -424 | 80 MM (discovery) and 50 MM (validation) | 10 MM before and after surgery | 17 MM before and after recurrence | Raw data | Friedman et al. (49) | |
| Oral squamous cell carcinoma (OSCC) | Plasma | miR-24 | 43 OSCC and normal tissue | 33 OSCC, 10 controls | — | RNU6B, let7a | Lin et al. (116) |
| Plasma | miR-31 | 43 OSCC before and after surgery, 21 controls | 9 OSCC - saliva before and after surgery | — | miR-16 | Liu et al. (117) | |
| Saliva | miR-125a, -200a | 12 Controls | 12 OSCC, 12 controls | 38 OSCC, 38 controls | RNU6B | Park et al. (37) | |
| Saliva | miR-31 | 45 OSCC, 10 Oral verrucous leukoplakia, 24 controls | 22 OSCC before and after surgery | 28 OSCC, 17 controls, plasma and saliva | miR-16 | Liu et al. (48) | |
| Esophageal squamous cell carcinoma (ESCC) | Serum | miR-10a, -22, -100, -148b, -223, -133a, -127–3p | Pooled 86 nonmetastatic, 55 metastatic ESCC, 40 controls | 36 ESCC, 30 controls | 113 ESCC, 67 controls | Serum volume | Zhang et al. (45) |
| Plasma | miR-21, -375 | 20 ESCC, 10 controls | 8 ESCC before and after surgery | 50 ESCC, 20 controls | RNU6B | Komatsu et al. (118) | |
| Serum | miR-31 | 45 ESCC and normal tissue; 120 ESCC, 121 controls | 64 ESCC before and after surgery | 81 ESCC, 120 different tumors, 81 controls | miR-16 | Zhang et al. (119) | |
| Renal cell carcinoma (RCC) | Urine | miR-15a | 23 RCC and 5 controls tissue | 10 RCC before and after surgery | 10 RCC, 35 different tumors | 5S rRNA | von Brandenstein et al. (38) |
| Serum | miR-26a-2*, -191, - 337–3p, -378 | 25 RCC, 25 controls | 117 RCC, 14 benign renal tumors, 123 controls | — | C. elegans miR-39 | Hauser et al. (120) | |
| Serum | miR-378, -451 | Pooled 15 RCC, 12 controls | 90 RCC, 35 controls | — | miR-16 | Redova et al. (121) | |
List of 20 different cancer types in which miRNAs have been evaluated as biomarkers. Each row describes the detection matrix of, study design, and normalization method used. Discovery and confirmation steps were typically performed in a cohort with a smaller sample size to select/verify candidate miRNAs, and evaluation was conducted in an expanded cross-sectional or longitudinal cohort to test the specificity, sensitivity, or early diagnostic capability.
Studies were conducted using human biospecimens unless otherwise stated.
One of the most common study designs that many groups have followed is to screen for differentially regulated miRNAs in paired cancerous and noncancerous tissues. Thereby, individual miRNAs or a panel is selected and further examined in the chosen fluid matrix (43, 44). Another widely used approach is to perform the initial screen in pooled samples and then confirm the results in individual samples (45). A few groups have studied the dysregulation of miRNAs in mouse models and translated the results to humans (46) or have performed in vitro experiments in cancerous cell lines before expanding the study to patient samples (47). An appropriate inclusion in the study design is to evaluate the miRNA expression before and after tumor removal as a prognostic indicator of disease regression (48). Friedman et al. used serum miRNAs to predict the recurrence of melanoma after surgical resection of the tumor (49).
Several studies highlight the specificity of candidate miRNAs as biomarkers of cancer by comparing their concentrations in closely related cancer types or disease states. For example, while identifying potential serum miRNA biomarkers for pancreatic ductal adenocarcinoma, Kong et al. used chronic pancreatic cancer samples to validate the specificity of miR-196a (50). Hanke’s group compared urine samples from patients with low- and high-grade bladder cancer to noncancerous urinary tract infection in their study (51). Some studies highlight the prognostic ability of miRNAs to classify different grades or stages of cancer. Roth et al. compared the concentrations of serum miRNAs in primary vs metastatic breast cancer patients (47), while a study by Asaga et al. demonstrated that patients with advanced stages of breast cancer have significantly higher circulating miR-21 than those in earlier stages (43).
miRNAS AS BIOMARKERS OF ORGAN DAMAGE CONDITIONS
miRNA-based diagnostics have also been investigated in various organ damage conditions, some of which are summarized in Table 3. The spectrum of cardiovascular diseases has been a prime focus of miRNA bio-marker research. miR-1 and -499 have been proposed and verified as biomarkers of acute myocardial infarction (AMI) that are comparable to conventional standards (52). Cheng’s group employed a rat model of AMI and found serum miR-1 to increase early, peaking at 6 h, and to be well correlated with the size of the infarct (52). On translating their findings to humans, they observed serum miR-1 patterns to correlate with the conventional serum creatine kinase-MB. In both rats and humans, urinary miR-1 was found to increase significantly at early time-points after AMI as well, peaking at 24 h (53). Heart failure, coronary artery disease, and hypertension are other heart diseases that have been extensively worked on (52).
Table 3.
Extracellular miRNAs as biomarkers of organ damage.a
| Disease | Body fluid | Candidate miRNAs | Cohorts used | Normalizer | Reference | ||
|---|---|---|---|---|---|---|---|
| Discovery | Confirmation | Evaluation | |||||
| AMI | Plasma | miR-1 | 93 AMI, 66 controls | 83 AMI temporal study | — | RNU6B | Creemers et al. (52) |
| Serum | miR-1 | 8 AMI, 8 controls rats temporal study | 12 AMI, 12 controls rats | 31 AMI, 20 controls humans | Blood volume | Creemers et al. (52) | |
| Plasma | miR-122, -375 | 6 ST-segment elevation MI (STEMI), 6 controls | 25 STEMI, 17 controls | 8 STEMI temporal study | Self-normalization | D’Alessandra et al. (122) | |
| Plasma | miR-208b, -499 | 36 AMI, 36 controls | 14 Acute viral myocarditis (VM), 20 post VM, 20 controls | 33 Acute heart failure, 20 controls | C. elegans miR-39 | Creemers et al. (52) | |
| Plasma | miR-208a | 4 Controls | 33 AMI, 16 coronary heart disease, 17 different cardiovascular diseases, 30 controls | 5 AMI before and after treatment | Raw data | Wang et al. (123) | |
| Plasma | miR-499 | 14 Acute coronary syndromes, 15 congestive heart failure, 10 controls | — | — | Synthetic small RNA | Creemers et al. (52) | |
| Urine | miR-1 | 8 AMI, 8 controls rats | 8 AMI, 8 controls rats, temporal study, serum, and urine | 20 AMI, 20 controls humans | Standard curve calibration | Cheng et al. (53) | |
| Acute kidney injury (AKI) | Plasma | miR-210 | Pooled 5 AKI, 5 controls | 77 AKI, 18 AMI, 30 controls | — | C. elegans miR-54 | Lorenzen et al. (34) |
| Urine | miR-21, -200c, - 423, -4640 | Pooled 6 AKI, 6 controls | 6 AKI and 6 healthy controls | 98 AKI, 97 controls | miR-1287 | Ramachandran et al. (36) | |
| Chronic kidney disease (CKD) | Plasma and Urine | miR-16, -155, -210, -638 | 33 CKD, 22 controls - plasma | 12 CKD, 7 controls - urine | — | Raw data | Neal et al. (20) |
| Acute allograft rejection | Urine | miR-10a, -10b, -210 | Pooled 5 rejection, 5 stable transplant | 68 Rejection, 20 stable transplant, 13 UTI after transplant | 19 Before and after rejection | C. elegans miR-39 | Lorenzen et al. (54) |
| Chronic allograft dysfunction (CAD) | Urine | miR-142–3p, -204, -211 | 13 CAD, 5 controls tissue | 7 CAD, 7 controls | 36 Transplant recipients temporal study | RNU48 | Scian et al. (124) |
| ESRD | Plasma | miR-638, -21, -155, -210, -16 | 20 ESRD, 33 CKD, 22 controls | — | — | Raw data | Neal et al. (20) |
| Acetaminophen (APAP)- induced acute liver injury (ALI) | Serum | miR-122 | 6 APAP–no ALI, 11 no APAP–ALI, 22 CKD, 53 APAP–ALI, 25 controls, | 53 APAP–ALI prospective study | 11 APAP–ALI temporal study, 3 APAP–ALI before and after transplant | RNU6B | Starkey Lewis et al. (56) |
| Liver fibrosis | Serum | miR-29a | 5 Fibrotic, 5 control mouse tissue | 11 Fibrotic, 9 controls human tissue | 67 Fibrosis, 17 controls | RNU6B | Roderburg et al. (125) |
| Chronic hepatitis C (CHC) and nonalcoholic fatty liver disease (NAFLD) | Serum | miR-34a, -122, -16 | 18 CHC, 19 controls | 35 CHC | 34 NAFLD, 19 controls | C. elegans miR- 238 | Cermelli et al. (57) |
| Active pulmonary tuberculosis (TB) | Sputum | miR-3179, -147, 19b-2* | Pooled 58 TB, 32 controls | 30 TB, 30 controls | - | RNU6B | Yi et al. (126) |
| Serum | miR-29a | Pooled 30 TB, 30 controls | 30 TB, 30 controls | 30 TB; 30 controls, sputum | RNU6B | Fu et al. (127) | |
| Biliary atresia (BA) | Serum | miR-200b, -429 | 5 BA, 5 controls | 24 BA, 24 controls | — | C.el-miR-54, -238 | Zahm et al. (128) |
List of 11 different types of organ damages in which microRNAs have been evaluated as biomarkers. Each row describes the detection matrix, study design, and normalization method used. Discovery and confirmation steps were typically performed in a cohort with a smaller sample size to select/verify candidate miRNAs, and evaluation was conducted in an expanded cross-sectional or longitudinal cohort to test the specificity, sensitivity, or early diagnostic capability. Studies were conducted using human biospecimens unless otherwise stated.
Urine has been one of the most widely used matrices for biomarker discovery in the context of kidney diseases. miR-210 has been proposed as a plasma and urinary biomarker for acute and chronic forms of injury and end-stage renal disease (ESRD) (20, 34). The urinary form could predict acute allograft rejection and has the potential to be a noninvasive monitor of graft function and susceptibility to medication (54). miR-155 deregulation in urine and plasma has also been studied in acute and chronic injury and ESRD (20, 55).
One of the most common forms of liver damage, acetaminophen toxicity, was found to upregulate miR-122 in the plasma in several rodent and human studies (21, 56). miR-122 and miR-192, both liver-enriched species, were shown to have an earlier and dose-dependent response to drug administration and are translatable, making them very suitable candidates for toxicity monitoring of large-scale drug and compound screening (21). miR-122 has also been shown to be immensely deregulated in nonalcoholic fatty liver disease and chronic hepatitis C (57).
miRNAS AS BIOMARKERS OF DISEASE CONDITIONS
Apart from cancer and organ damage conditions, there are many other disease states in which miRNAs have been investigated as biomarkers, and a few such studies are listed in Table 4. Zampetaki et al. discovered that the concentrations of 5 miRNAs in the plasma of patients with diabetes differed from those in plasma from healthy individuals. This dysregulation was then confirmed in about 200 samples from individuals with or without diabetes, and miR-126 evaluation was further extended to 822 different samples (58). Another clinically relevant condition that necessitates rapid and early diagnosis and lacks an ideal biomarker is sepsis. It was shown that plasma miR-150 is significantly lower in sepsis patients and that the concentrations correlate with the severity of the disease (59). Serum miR-146a and -223 concentrations were both found to be lower in sepsis patients than in healthy controls (60).
Table 4.
Extracellular microRNAs as biomarkers of disease states.a
| Disease | Body fluid | Candidate miRNAs | Cohorts used | Normalizer | Reference | ||
|---|---|---|---|---|---|---|---|
| Discovery | Confirmation | Evaluation | |||||
| Sepsis | Plasma | miR-150 | 8 Sepsis, 8 control leukocytes | 10 Sepsis, 12 control leukocytes | 24 Sepsis, 32 controls | RNU6B, leukocytes; miR-192, plasma | Vasilescu et al. (59) |
| Serum | miR-15a, -16, -223, -499– 5p, -122, -193b* | 166 Sepsis, 32 systemic inflammatory response syndrome (SIRS), 24 controls | 43 Mild, 123 severe sepsis, 24 controls | — | RNU6B | Wang et al. (129) | |
| Serum | miR-146a, -223 | 50 Sepsis, 30 SIRS, 20 controls | — | — | miR-295 | Wang et al. (60) | |
| Systemic lupus erythematosus (SLE) | Serum and urine | miR-200a, -200b, -141, -200c, -429, -205, -192 | 40 SLE, 30 controls– serum | 40 SLE, 30 controls– urine | — | RNU48 | Wang et al. (130) |
| Urine | miR-146a, -155 | 40 SLE, 13 controls | 40 SLE at time 0, 3 and 6 months after treatment | — | RNU48 | Wang et al. (131) | |
| Diabetes mellitus type 2 (DM) | Plasma | miR-126, -15a, -29b, -223, -28–3p | Pooled 2 DM, 6 controls | 99 DM, 99 controls | 822 Different samples | miR-454, RNU6B | Zampetaki et al. (58) |
| Hypertension (Hyp) | Plasma | Human cytomegalovirus miR-UL112, miR-296– 5p, let7e | 13 Hyp, 5 controls | 24 Hyp, 22 controls | 194 Hyp, 67 controls | RNU6B | Li et al. (132) |
| Pulmonary arterial hypertension (PAH) | Plasma | miR-150 | 8 PAH, 8 controls | 145 PAH, 10 controls | 30 PAH | C. elegans miR-39 | Rhodes et al. (133) |
| Multiple sclerosis (MS) | CSF | miR-181c, -633, -922 | Pooled 10 MS, 10 other neurological diseases | 53 MS, 39 other neurological diseases | — | C. elegans miR-39 | Haghikia et al. (61) |
| Plasma | miR-92a-1*, -454, -30e, -22, -210, -574–3p, -135a, -140–3p, let-7a | 9 Secondary progressive MS (SPMS), 10 RRMS, 9 controls | 51 SPMS, 50 relapsing remitting MS (RRMS), 32 controls | 15 Amyotrophic lateral sclerosis | RNA amount | Gandhi et al. (134) | |
| HIV encephalitis (HIVE) | CSF | miR-1224–3p, -204, -484, -720, -934, -937 | 5 HIV+, 4 HIVE, 10 HIV− | — | — | miR-622, -1266 | Pacifici et al. (62) |
| Pregnancy | Serum | miR-520d-5p, -526a, -527 | 20 Pregnant temporal study, 10 controls | — | — | Panel of 6 miRNAs | Gilad et al. (135) |
| Bipolar disorder | Plasma | miR-134 | 21 Bipolar disorder temporal study, 21 controls | — | — | C. elegans lin-4 | Rong et al. (63) |
| Mild cognitive impairment (MCI) | Plasma | Panel of 8 miRNAs | 10 MCI, 10 controls | 20 MCI, 20 Alzheimer disease, 20 controls | 20 young and 20 old controls; 19 retrospective | Ratio of miRNA pairs | Sheinerman et al. (136) |
List of 10 different disease conditions in which microRNAs have been evaluated as biomarkers. Each row describes the detection matrix, study design, and normalization method used. Discovery and confirmation steps were typically performed in a cohort with a smaller sample size to select/verify candidate miRNAs, and evaluation was conducted in an expanded cross-sectional or longitudinal cohort to test the specificity, sensitivity, or early diagnostic capability.
Studies were conducted using human biospecimens unless otherwise stated.
There have been studies of other disease conditions, including multiple sclerosis (61), HIV encephalitis (62), and biopolar disorder (63) for the use of miRNAs in various body fluids as novel and noninvasive biomarkers.
Tools and Techniques Used in miRNA Biomarker Research
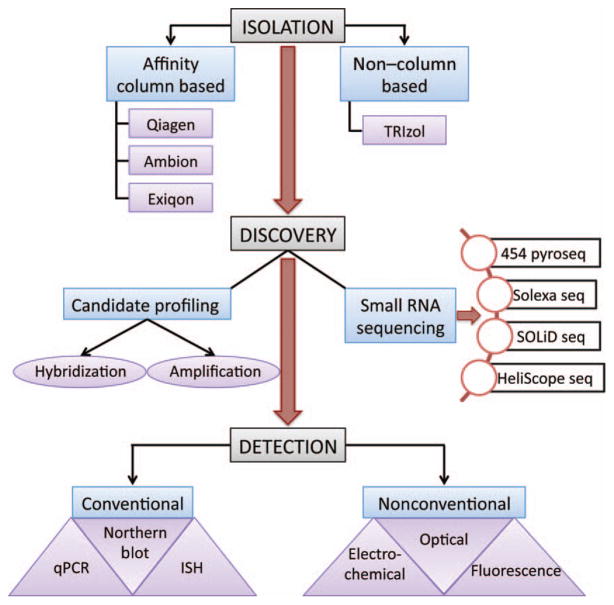
ISOLATION
The conventional methods of RNA isolation include phenol– chloroform extraction with commercial reagents or glass–fiber column– based extraction from various manufacturers (Fig. 2). Techniques for exosome isolation and preconcentration include commercial precipitation reagents, ultracentrifugation, chromatography, and Dynabeads (13, 64). Yoo et al. have described an innovative method to isolate miRNAs directly from exosomes using immunoaffinity magnetic beads (65). Quantification of the extracted RNA is usually performed by spectophotometric analysis on the Nanodrop. Agilent’s microfluidics platform can be used to assess the quantity and quality of total RNA as well as low molecular weight species separately. Studies that specifically isolate exosomes use transmission electron microscopy (TEM) to study the vesicle structure and integrity or perform immunoblot analysis for exosome-specific proteins such as CD63 and TSG101 (14).
Fig. 2.
Tools and techniques used in miRNA bio-marker discovery in the RNA isolation, candidate discovery, and verification stages. Seq, sequencing.
DISCOVERY
Profiling
Numerous companies offer miRNA profiling using amplification-based as well as hybridization-based platforms (Fig. 2). Although hybridization systems work well for profiling in tissues and avoid the amplification bias, the low RNA yield from extra-cellular fluids poses a problem in this context. Thus, amplification-based profiling techniques that provide an additional option of preamplification may be the best choice for systems such as urine, which have a very low RNA abundance to begin with. In addition to commercially available platforms, several groups have developed other techniques for expression profiling (66).
Sequencing
Next generation sequencing of nucleic acids is increasingly used to identify new species-specific and tissue-specific miRNAs. The technologies that are most commonly used are 454 pyrosequencing, Solexa sequencing, and ABI solid sequencing. HeliScope single-molecule sequencing has also been demonstrated for the sequencing of small RNAs (Fig. 2). There are comprehensive reviews detailing the complete workflow of deep sequencing and the advantages and drawbacks of each method (67).
DETECTION
The detection of single or multiple miRNAs obtained after profiling/sequencing is usually performed by qPCR, Northern blotting, or in situ hybridizations. Traditional qPCR is performed by using stem-looped RT primers (like Taqman), an miRNA-specific forward primer, and a universal reverse primer for PCR. Another strategy is to add a polyA tag to the RNA and use an oligo-dT primer for RT and specific forward and universal reverse primers for PCR (like Qiagen). Improvisations include polyuridylating and using a polyA stem-loop primer during RT for better specificity and convenience (68). In situ hybridization is another commonly used method of detection, which is improved in selectivity by using locked nucleic acid probes (69). De Planell-Saguer et al. have developed a unique technique to simultaneously detect miRNAs and proteins from FFPE sections and cell cultures (70). This could be especially useful in miRNA–target interaction studies.
Several novel and original methods of detection have been developed, many of them having a fluorescence-, electrochemical-, or optical-based detection step. These protocols aim at expanding the utility of miRNAs into diagnostic tools that can be efficiently incorporated into a clinical setting. Many of these assays allow for rapid quantification, have improved sensitivity and specificity of detection, and reduce the number of amplification steps required before detection. For instance, a scheme put forth by Schoch et al. isolates, concentrates, and quantifies small RNAs from cell lysates using on-chip isotachophoresis (71). Strategies of detection vary from using simple molecular beacons (72), graphene-oxide– based fluorescence quenching (73), and enzymatic luminescence (74) to different nanoparticle-based probes like silver (75), gold (76), and carbon (77). More complex methods involve different forms of electrophoresis, like isotachophoresis (78) and capillary electrophoresis (79), circular exponential amplification (80), or isothermal amplification (81) and various forms of hybridization (82).
Challenges
Although the use of miRNAs as biomarkers has been proposed and verified in various disease models, there are several impediments that need to be overcome before they can be scaled up into clinical translational markers.
ORIGINS
The origins of extracellular miRNAs remain obscure and could potentially affect the significance of the results. For example, it has been shown that tumors specifically secrete exosomes that carry miRNAs into the circulation (83), which indicates that extracellular miRNAs have an associative role in the initiation, progression, or metastasis of the tumor, making them relevant choices for biomarkers. A recent study indicated that miRNAs present in serum and saliva are for the most part concentrated in exosomes (14). However, studies by Arroyo et al. and Turchinovich et al. assert that miRNAs in circulation are principally found in association with protein complexes and not vesicles (16, 18), raising the possibility that the detected miRNAs are remnants of dead cells protected by stable protein complexes (16).
BIOMATERIAL SELECTION
It is not clear whether choosing serum or plasma contributes any difference to the results. Although Mitchell’s group reported that there is no difference in abundance between serum and plasma for selected miRNAs (9), a recent study showed that the total concentration of RNAs as well as detectable miRNA abundance is higher in serum than in plasma (84). The group suggests that coagulation may trigger the release or secretion of miRNAs, or RNA extrusion due to cell lysis may contribute to the effect. Another important factor to take into consideration while dealing with clinical plasma/serum samples is the extent of hemolysis. Kirschner et al. demonstrate that miR-16 and -451, commonly used as normalizing factors, are present in substantial amounts in red blood cells and the extent of hemolysis causes considerable variation in their concentrations between samples (85).
ISOLATION TECHNIQUES
Differences in RNA stabilization systems for blood may cause differences in yields and purity of the RNA, although they do not seem to affect downstream qPCR applications (86). Isolation of RNA is a crucial step, and yield differs with technique, introducing considerable variability in qPCR and microarray-based results (87). Several studies have been performed that compare TRIzol extraction to column-based isolation and the use of exosome precipitation protocols to ultracentrifugation in a wide variety of samples (29, 87). The discrepancies in the conclusions indicate that sample type, protocol, reagent variation, and quantification methods influence the quality and quantity of RNA isolated. As with mRNA analysis, different microarray platforms used for quantification of miRNAs have inconsistencies in their results, and there is variation between microarray and qPCR/Northern blot validation data (88). Even with deep sequencing strategies, the protocols for library preparation and selection of the sequencing platform were found to influence the findings (89).
NORMALIZATION
The lack of a standardized housekeeping miRNA that is consistent between different tissues and extracellular fluids and between disease states is a crippling factor in their putative use as biomarkers (90). Alternative means of normalizing include synthetic miRNA spiked into the sample, which serves as a good technical control, or the use of equal amounts of starting total RNA as quantified on a spectrophotometer. However, a spiked miRNA will not account for endogenous differences in miRNA concentrations due to disease conditions, and there are studies that indicate that the correlation between total RNA amounts and miRNA concentrations is low (84). Meyer et al. provide an analysis of the utility of 7 different methods of normalization for microarray data (91). The most commonly used mode of normalization for microarray data is to select a panel of endogenous invariant miRNAs across the cases and controls for a particular disease condition and profiling platform (92).
Conclusions
The field of miRNA-centered diagnostics is still in its infancy and these micromolecules have immense potential not only to be developed into sensitive, specific, and robust markers of organ damage and disease conditions but also to monitor disease progression and efficacy of treatment. The National Center for Advancing Translational Sciences, which is a part of NIH, has recently funded a program on extracellular RNA communication that includes miRNAs. Their initiatives are to assess the clinical utility of these molecules as both biomarkers and therapeutic agents and to create healthy reference profiles of extracellular RNAs present in various body fluids. An industry consortium comprising Pfizer and Eli Lilly and overseen by the Predictive Safety Testing Consortium (PSTC) at the Critical Path Institute is examining circulating miRNAs as markers of testicular damage in response to toxicity (93). However, basic questions with regard to their exact roles in the pathophysiology of disease and the mechanisms of their release from affected cells into biological fluids are yet to be answered. miRNAs have been launched as a new generation of biomarkers, and once the various technical and biological hurdles can be overcome, they hold the promise of traversing the biomarker pipeline all the way from preclinical and clinical studies to emerge into the market for consumer use.
Footnotes
Nonstandard abbreviations: miRNA, microRNA; pri-miRNA, primary miRNA transcripts; pre-miRNA, precursor miRNA; TAR, trans-activation response; TRBP, TAR binding protein; RISC, RNA- induced silencing complex; AGO-2, Argonaute protein; nSMase2, neutral sphingomyelinase 2; FFPE, formalin-fixed paraffin-embedded; CSF, cerebrospinal fluid; qPCR, quantitative PCR; AMI. acute myocardial infarction; ESRD, end-stage renal disease.
Author Contributions: All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership: V.S. Vaidya, Harvard Program in Therapeutic Sciences.
Consultant or Advisory Role: None declared.
Stock Ownership: None declared.
Honoraria: None declared.
Research Funding: V.S. Vaidya, National Institutes of Health/National Institutes of Environmental Health Sciences–Outstanding New Environmental Scientist Award (ES017543) and an Innovation in Regulatory Science Award from the Burroughs Wellcome Fund.
Expert Testimony: None declared.
Patents: None declared.
References
- 1.Lee R, Feinbaum R, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Reinhart B, Slack F, Basson M, Pasquinelli A, Bettinger J, Rougvie A, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 3.Pasquinelli A, Reinhart B, Slack F, Martindale M, Kuroda M, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 4.Hsu P, Huang H-D, Hsu S-D, Lin L-Z, Tsou A-P, Tseng C-P, et al. MiRNAmap: genomic maps of microRNA genes and their target genes in mammalian genomes. Nucleic Acids Res. 2006;34:D135–9. doi: 10.1093/nar/gkj135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 6.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 7.Friedman R, Farh K, Burge C, Bartel D. Most mammalian mRNAs are conserved targets of microRNAs. Genome research. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–52. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–9. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 11.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38:7248–59. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q, et al. Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci. 2012;8:118–23. doi: 10.7150/ijbs.8.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rani S, O’Brien K, Kelleher FC, Corcoran C, Germano S, Radomski MW, et al. Isolation of exosomes for subsequent mRNA, microRNA, and protein profiling. Methods Mol Biol. 2011;784:181–95. doi: 10.1007/978-1-61779-289-2_13. [DOI] [PubMed] [Google Scholar]
- 14.Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7:e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 16.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–33. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–8. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, et al. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56:1733–41. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neal CS, Michael MZ, Pimlott LK, Yong TY, Li JY, Gleadle JM. Circulating microRNA expression is reduced in chronic kidney disease. Nephrol Dial Transplant. 2011;26:3794– 802. doi: 10.1093/ndt/gfr485. [DOI] [PubMed] [Google Scholar]
- 21.Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106:4402–7. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sharma CM, Vogel J. Experimental approaches for the discovery and characterization of regulatory small RNA. Curr Opin Microbiol. 2009;12:536– 46. doi: 10.1016/j.mib.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Hede K. Small RNAs are raising big expectations. J Natl Cancer Inst. 2009;101:840–1. doi: 10.1093/jnci/djp172. [DOI] [PubMed] [Google Scholar]
- 24.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 25.Oellerich M. Enzyme-immunoassay: a review. J Clin Chem Clin Biochem. 1984;22:895–904. [PubMed] [Google Scholar]
- 26.Jensen O. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr Opin Chem Biol. 2004;8:33–41. doi: 10.1016/j.cbpa.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 27.Addis MF, Tanca A, Pagnozzi D, Crobu S, Fanciulli G, Cossu-Rocca P, Uzzau S. Generation of high-quality protein extracts from formalin-fixed, paraffin-embedded tissues. Proteomics. 2009;9:3815–23. doi: 10.1002/pmic.200800971. [DOI] [PubMed] [Google Scholar]
- 28.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–5. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 29.Mraz M, Malinova K, Mayer J, Pospisilova S. MicroRNA isolation and stability in stored RNA samples. Biochem Biophys Res Commun. 2009;390:1–4. doi: 10.1016/j.bbrc.2009.09.061. [DOI] [PubMed] [Google Scholar]
- 30.Siebolts U, Varnholt H, Drebber U, Dienes HP, Wickenhauser C, Odenthal M. Tissues from routine pathology archives are suitable for microRNA analyses by quantitative PCR. J Clin Pathol. 2009;62:84– 8. doi: 10.1136/jcp.2008.058339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morenos L, Saffery R, Mechinaud F, Ashley D, Elwood N, Craig JM, Wong NC. Evaluation of microRNA expression in patient bone marrow aspirate slides. PLoS One. 2012;7:e42951. doi: 10.1371/journal.pone.0042951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borze I, Guled M, Musse S, Raunio A, Elonen E, Saarinen-Pihkala U, et al. MicroRNA microarrays on archive bone marrow core biopsies of leukemias–method validation. Leuk Res. 2011;35:188–95. doi: 10.1016/j.leukres.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Lee CT, Risom T, Strauss W. Evolutionary conservation of microRNA regulatory circuits: an examination of microRNA gene complexity and conserved microRNA-target interactions through metazoan phylogeny. DNA Cell Biol. 2007;26:209–18. doi: 10.1089/dna.2006.0545. [DOI] [PubMed] [Google Scholar]
- 34.Lorenzen JM, Kielstein JT, Hafer C, Gupta SK, Kumpers P, Faulhaber-Walter R, et al. Circulating miR-210 predicts survival in critically ill patients with acute kidney injury. Clin J Am Soc Nephrol. 2011;6:1540– 6. doi: 10.2215/CJN.00430111. [DOI] [PubMed] [Google Scholar]
- 35.Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly MB, et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci U S A. 2007;104:19971–6. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramachandran K, Saikumar J, Bijol V, Koyner JL, Qian J, Betensky RA, et al. Human miRNome profiling identifies microRNAs differentially present in the urine after kidney injury. Clin Chem. 2013;59:1742–52. doi: 10.1373/clinchem.2013.210245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, Wong DT. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin Cancer Res. 2009;15:5473–7. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Brandenstein M, Pandarakalam JJ, Kroon L, Loeser H, Herden J, Braun G, et al. MicroRNA 15a, inversely correlated to PKCα, is a potential marker to differentiate between benign and malignant renal tumors in biopsy and urine samples. Am J Pathol. 2012;180:1787–97. doi: 10.1016/j.ajpath.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 39.Puerta-Gil P, Garcia-Baquero R, Jia AY, Ocana S, Alvarez-Mugica M, Alvarez-Ossorio JL, et al. mir-143, mir-222, and mir-452 are useful as tumor stratification and noninvasive diagnostic biomarkers for bladder cancer. Am J Pathol. 2012;180:1808–15. doi: 10.1016/j.ajpath.2012.01.034. [DOI] [PubMed] [Google Scholar]
- 40.Xing L, Todd NW, Yu L, Fang H, Jiang F. Early detection of squamous cell lung cancer in sputum by a panel of microRNA markers. Mod Pathol. 2010;23:1157–64. doi: 10.1038/modpathol.2010.111. [DOI] [PubMed] [Google Scholar]
- 41.Baraniskin A, Kuhnhenn J, Schlegel U, Chan A, Deckert M, Gold R, et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood. 2011;117:3140– 6. doi: 10.1182/blood-2010-09-308684. [DOI] [PubMed] [Google Scholar]
- 42.Teplyuk NM, Mollenhauer B, Gabriely G, Giese A, Kim E, Smolsky M, et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol. 2012;14:689–700. doi: 10.1093/neuonc/nos074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asaga S, Kuo C, Nguyen T, Terpenning M, Giuliano AE, Hoon DS. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin Chem. 2011;57:84–91. doi: 10.1373/clinchem.2010.151845. [DOI] [PubMed] [Google Scholar]
- 44.Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ, Wang YK, et al. High expression of serum mir-21 and tumor mir-200c associated with poor prognosis in patients with lung cancer. Med Oncol. 2012;29:618–26. doi: 10.1007/s12032-011-9923-y. [DOI] [PubMed] [Google Scholar]
- 45.Zhang C, Wang C, Chen X, Yang C, Li K, Wang J, et al. Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem. 2010;56:1871–9. doi: 10.1373/clinchem.2010.147553. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto Y, Kosaka N, Tanaka M, Koizumi F, Kanai Y, Mizutani T, et al. MicroRNA-500 as a potential diagnostic marker for hepatocellular carcinoma. Biomarkers. 2009;14:529–38. doi: 10.3109/13547500903150771. [DOI] [PubMed] [Google Scholar]
- 47.Roth C, Rack B, Muller V, Janni W, Pantel K, Schwarzenbach H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12:R90. doi: 10.1186/bcr2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu CJ, Lin SC, Yang CC, Cheng HW, Chang KW. Exploiting salivary mir-31 as a clinical biomarker of oral squamous cell carcinoma. Head Neck. 2012;34:219–24. doi: 10.1002/hed.21713. [DOI] [PubMed] [Google Scholar]
- 49.Friedman EB, Shang S, de Miera EV, Fog JU, Teilum MW, Ma MW, et al. Serum microRNAs as biomarkers for recurrence in melanoma. J Transl Med. 2012;10:155. doi: 10.1186/1479-5876-10-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kong X, Du Y, Wang G, Gao J, Gong Y, Li L, et al. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: mir-196a could be a potential marker for poor prognosis. Dig Dis Sci. 2011;56:602–9. doi: 10.1007/s10620-010-1285-3. [DOI] [PubMed] [Google Scholar]
- 51.Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, et al. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 2010;28:655–61. doi: 10.1016/j.urolonc.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 52.Creemers E, Tijsen A, Pinto Y. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483–95. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 53.Cheng Y, Wang X, Yang J, Duan X, Yao Y, Shi X, et al. A translational study of urine miRNAs in acute myocardial infarction. J Mol Cell Cardiol. 2012;53:668–76. doi: 10.1016/j.yjmcc.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lorenzen JM, Volkmann I, Fiedler J, Schmidt M, Scheffner I, Haller H, et al. Urinary miR-210 as a mediator of acute T-cell mediated rejection in renal allograft recipients. Am J Transplant. 2011;11:2221–7. doi: 10.1111/j.1600-6143.2011.03679.x. [DOI] [PubMed] [Google Scholar]
- 55.Saikumar J, Hoffmann D, Kim TM, Gonzalez VR, Zhang Q, Goering PL, et al. Expression, circulation, and excretion profile of microRNA-21, -155, and -18a following acute kidney injury. Toxicol Sci. 2012;129:256– 67. doi: 10.1093/toxsci/kfs210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Starkey Lewis PJ, Dear J, Platt V, Simpson KJ, Craig DG, Antoine DJ, et al. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology. 2011;54:1767–76. doi: 10.1002/hep.24538. [DOI] [PubMed] [Google Scholar]
- 57.Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PloS One. 2011;6:e23937. doi: 10.1371/journal.pone.0023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107:810–7. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 59.Vasilescu C, Rossi S, Shimizu M, Tudor S, Veronese A, Ferracin M, et al. microRNA fingerprints identify mir-150 as a plasma prognostic marker in patients with sepsis. PloS One. 2009;4:e7405. doi: 10.1371/journal.pone.0007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang JF, Yu ML, Yu G, Bian JJ, Deng XM, Wan XJ, Zhu KM. Serum miR-146a and miR-223 as potential new biomarkers for sepsis. Biochem Biophys Res Commun. 2010;394:184– 8. doi: 10.1016/j.bbrc.2010.02.145. [DOI] [PubMed] [Google Scholar]
- 61.Haghikia A, Haghikia A, Hellwig K, Baraniskin A, Holzmann A, Decard BF, et al. Regulated microRNAs in the CSF of patients with multiple sclerosis: a case-control study. Neurology. 2012;79:2166–70. doi: 10.1212/WNL.0b013e3182759621. [DOI] [PubMed] [Google Scholar]
- 62.Pacifici M, Delbue S, Ferrante P, Jeansonne D, Kadri F, Nelson S, et al. Cerebrospinal fluid miRNA profile in HIV-encephalitis. J Cell Physiol. 2013;228:1070–5. doi: 10.1002/jcp.24254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rong H, Liu TB, Yang KJ, Yang HC, Wu DH, Liao CP, et al. MicroRNA-134 plasma levels before and after treatment for bipolar mania. J Psychiatr Res. 2011;45:92–5. doi: 10.1016/j.jpsychires.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 64.Taylor DD, Zacharias W, Gercel-Taylor C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol Biol. 2011;728:235–46. doi: 10.1007/978-1-61779-068-3_15. [DOI] [PubMed] [Google Scholar]
- 65.Yoo CE, Kim G, Kim M, Park D, Kang HJ, Lee M, Huh N. A direct extraction method for microRNAs from exosomes captured by immunoaffinity beads. Anal Biochem. 2012;431:96– 8. doi: 10.1016/j.ab.2012.09.008. [DOI] [PubMed] [Google Scholar]
- 66.Duan D, Zheng K-x, Shen Y, Cao R, Jiang L, Lu Z, et al. Label-free high-throughput microRNA expression profiling from total RNA. Nucleic Acids Res. 2011;39:e154. doi: 10.1093/nar/gkr774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Creighton CJ, Reid JG, Gunaratne PH. Expression profiling of microRNAs by deep sequencing. Brief Bioinform. 2009;10:490–7. doi: 10.1093/bib/bbp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mei Q, Li X, Meng Y, Wu Z, Guo M, Zhao Y, et al. A facile and specific assay for quantifying microRNA by an optimized RT-qPCR approach. PLoS One. 2012;7:e46890. doi: 10.1371/journal.pone.0046890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song R, Ro S, Yan W. In situ hybridization detection of microRNAs. Methods Mol Biol. 2010;629:287–94. doi: 10.1007/978-1-60761-657-3_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.de Planell-Saguer M, Rodicio MC, Mourelatos Z. Rapid in situ codetection of noncoding RNAs and proteins in cells and formalin-fixed paraffin-embedded tissue sections without protease treatment. Nat Protoc. 2010;5:1061–73. doi: 10.1038/nprot.2010.62. [DOI] [PubMed] [Google Scholar]
- 71.Schoch R, Ronaghi M, Santiago J. Rapid and selective extraction, isolation, preconcentration, and quantitation of small RNAs from cell lysate using on-chip isotachophoresis. Lab Chip. 2009;9:2145–52. doi: 10.1039/b903542g. [DOI] [PubMed] [Google Scholar]
- 72.Baker M, Bao G, Searles C. In vitro quantification of specific microRNA using molecular beacons. Nucleic Acids Res. 2012;40:e13. doi: 10.1093/nar/gkr1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dong H, Zhang J, Ju H, Lu H, Wang S, Jin S, et al. Highly sensitive multiple microRNA detection based on fluorescence quenching of graphene oxide and isothermal strand-displacement polymerase reaction. Anal Chem. 2012;84:4587–93. doi: 10.1021/ac300721u. [DOI] [PubMed] [Google Scholar]
- 74.Sun Y, Gregory K, Chen N, Golovlev V. Rapid and direct microRNA quantification by an enzymatic luminescence assay. Analytical biochemistry. 2012;429:11–7. doi: 10.1016/j.ab.2012.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dong H, Jin S, Ju H, Hao K, Xu LP, Lu H, Zhang X. Trace and label-free microRNA detection using oligonucleotide encapsulated silver nano-clusters as probes. Anal Chem. 2012;84:8670– 4. doi: 10.1021/ac301860v. [DOI] [PubMed] [Google Scholar]
- 76.Tu Y, Wu P, Zhang H, Cai C. Fluorescence quenching of gold nanoparticles integrating with a conformation-switched hairpin oligonucleotide probe for microRNA detection. Chem Commun. 2012;48:10718–20. doi: 10.1039/c2cc35564g. [DOI] [PubMed] [Google Scholar]
- 77.Wang L, Cheng Y, Wang H, Li Z. A homogeneous fluorescence sensing platform with water-soluble carbon nanoparticles for detection of microRNA and nuclease activity. Analyst. 2012;137:3667–72. doi: 10.1039/c2an35396b. [DOI] [PubMed] [Google Scholar]
- 78.Garcia-Schwarz G, Santiago J. Integration of on-chip isotachophoresis and functionalized hydrogels for enhanced-sensitivity nucleic acid detection. Anal Chem. 2012;84:6366–9. doi: 10.1021/ac301586q. [DOI] [PubMed] [Google Scholar]
- 79.Jiang RM, Chang YS, Chen SJ, Chen JH, Chen HC, Chang PL. Multiplexed microRNA detection by capillary electrophoresis with laser-induced fluorescence. J Chromatogr A. 2011;1218:2604–10. doi: 10.1016/j.chroma.2011.02.061. [DOI] [PubMed] [Google Scholar]
- 80.Wang GL, Zhang CY. Sensitive detection of microRNAs with hairpin probe-based circular exponential amplification assay. Anal Chem. 2012;84:7037–42. doi: 10.1021/ac3012544. [DOI] [PubMed] [Google Scholar]
- 81.Harcourt EM, Kool ET. Amplified microRNA detection by templated chemistry. Nucleic Acids Res. 2012;40:e65. doi: 10.1093/nar/gkr1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pohlmann C, Sprinzl M. Electrochemical detection of microRNAs via gap hybridization assay. Anal Chem. 2010;82:4434– 40. doi: 10.1021/ac100186p. [DOI] [PubMed] [Google Scholar]
- 83.Palma J, Yaddanapudi SC, Pigati L, Havens MA, Jeong S, Weiner GA, et al. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 2012;40:9125–38. doi: 10.1093/nar/gks656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang K, Yuan Y, Cho JH, McClarty S, Baxter D, Galas DJ. Comparing the microRNA spectrum between serum and plasma. PLoS One. 2012;7:e41561. doi: 10.1371/journal.pone.0041561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, van Zandwijk N, Reid G. Haemolysis during sample preparation alters microRNA content of plasma. PLoS One. 2011;6:e24145. doi: 10.1371/journal.pone.0024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weber DG, Casjens S, Rozynek P, Lehnert M, Zilch-Schoneweis S, Bryk O, et al. Assessment of mRNA and microRNA stabilization in peripheral human blood for multicenter studies and bio-banks. Biomark Insights. 2010;5:95–102. doi: 10.4137/bmi.s5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Eldh M, Lotvall J, Malmhall C, Ekstrom K. Importance of RNA isolation methods for analysis of exosomal RNA: evaluation of different methods. Mol Immunol. 2012;50:278– 86. doi: 10.1016/j.molimm.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 88.Jensen SG, Lamy P, Rasmussen MH, Ostenfeld MS, Dyrskjot L, Orntoft TF, Andersen CL. Evaluation of two commercial global miRNA expression profiling platforms for detection of less abundant miRNAs. BMC Genomics. 2011;12:435. doi: 10.1186/1471-2164-12-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Toedling J, Servant N, Ciaudo C, Farinelli L, Voinnet O, Heard E, Barillot E. Deep-sequencing protocols influence the results obtained in small-RNA sequencing. PLoS One. 2012;7:e32724. doi: 10.1371/journal.pone.0032724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ajit SK. Circulating microRNAs as biomarkers, therapeutic targets, and signaling molecules. Sensors. 2012;12:3359– 69. doi: 10.3390/s120303359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meyer SU, Kaiser S, Wagner C, Thirion C, Pfaffl MW. Profound effect of profiling platform and normalization strategy on detection of differentially expressed microRNAs–a comparative study. PLoS One. 2012;7:e38946. doi: 10.1371/journal.pone.0038946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Latham GJ. Normalization of microRNA quantitative RT-PCR data in reduced scale experimental designs. Methods Mol Biol. 2010;667:19–31. doi: 10.1007/978-1-60761-811-9_2. [DOI] [PubMed] [Google Scholar]
- 93.Dere E, Anderson LM, Coulson M, McIntyre BS, Boekelheide K, Chapin RE. SOT symposium highlight: translatable indicators of testicular toxicity: inhibin b, microRNAs, and sperm signatures. Toxicol Sci. 2013;136:265–73. doi: 10.1093/toxsci/kft207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sita-Lumsden A, Dart DA, Waxman J, Bevan CL. Circulating microRNAs as potential new bio-markers for prostate cancer. Br J Cancer. 2013;108:1925–30. doi: 10.1038/bjc.2013.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fang C, Zhu DX, Dong HJ, Zhou ZJ, Wang YH, Liu L, et al. Serum microRNAs are promising novel biomarkers for diffuse large B cell lymphoma. Ann Hematol. 2012;91:553–9. doi: 10.1007/s00277-011-1350-9. [DOI] [PubMed] [Google Scholar]
- 96.Tanaka M, Oikawa K, Takanashi M, Kudo M, Ohyashiki J, Ohyashiki K, Kuroda M. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PloS One. 2009;4:e5532. doi: 10.1371/journal.pone.0005532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ho AS, Huang X, Cao H, Christman-Skieller C, Bennewith K, Le QT, Koong AC. Circulating miR-210 as a novel hypoxia marker in pancreatic cancer. Transl Oncol. 2010;3:109–13. doi: 10.1593/tlo.09256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu R, Chen X, Du Y, Yao W, Shen L, Wang C, et al. Serum microRNA expression profile as a bio-marker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58:610– 8. doi: 10.1373/clinchem.2011.172767. [DOI] [PubMed] [Google Scholar]
- 99.Ali S, Almhanna K, Chen W, Philip PA, Sarkar FH. Differentially expressed miRNAs in the plasma may provide a molecular signature for aggressive pancreatic cancer. Am J Transl Res. 2010;3:28– 47. [PMC free article] [PubMed] [Google Scholar]
- 100.Wang J, Chen J, Chang P, LeBlanc A, Li D, Abbruzzesse JL, et al. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev Res. 2009;2:807–13. doi: 10.1158/1940-6207.CAPR-09-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Heneghan HM, Miller N, Lowery AJ, Sweeney KJ, Newell J, Kerin MJ. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251:499–505. doi: 10.1097/SLA.0b013e3181cc939f. [DOI] [PubMed] [Google Scholar]
- 102.Ng EK, Li R, Shin VY, Jin HC, Leung CP, Ma ES, et al. Circulating microRNAs as specific bio-markers for breast cancer detection. PloS One. 2013;8:e53141. doi: 10.1371/journal.pone.0053141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112:55–9. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 104.Taylor D, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic bio-markers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 105.Wang F, Sun GP, Zou YF, Hao JQ, Zhong F, Ren WJ. MicroRNAs as promising biomarkers for gastric cancer. Cancer Biomark. 2012;11:259–67. doi: 10.3233/CBM-2012-00284. [DOI] [PubMed] [Google Scholar]
- 106.Li BS, Zhao YL, Guo G, Li W, Zhu ED, Luo X, et al. Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS One. 2012;7:e41629. doi: 10.1371/journal.pone.0041629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Menendez P, Villarejo P, Padilla D, Menendez JM, Montes JA. Diagnostic and prognostic significance of serum microRNAs in colorectal cancer. J Surg Oncol. 2013;107:217–20. doi: 10.1002/jso.23245. [DOI] [PubMed] [Google Scholar]
- 108.Wu CW, Ng SS, Dong YJ, Ng SC, Leung WW, Lee CW, et al. Detection of miR-92a and miR-21 in stool samples as potential screening biomarkers for colorectal cancer and polyps. Gut. 2012;61:739– 45. doi: 10.1136/gut.2011.239236. [DOI] [PubMed] [Google Scholar]
- 109.Kalimutho M, Del Vecchio Blanco G, Di Cecilia S, Sileri P, Cretella M, Pallone F, et al. Differential expression of miR-144* as a novel fecal-based diagnostic marker for colorectal cancer. J Gastroenterol. 2011;46:1391–402. doi: 10.1007/s00535-011-0456-0. [DOI] [PubMed] [Google Scholar]
- 110.Yu L, Todd NW, Xing L, Xie Y, Zhang H, Liu Z, et al. Early detection of lung adenocarcinoma in sputum by a panel of microRNA markers. Int J Cancer. 2010;127:2870– 8. doi: 10.1002/ijc.25289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, et al. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70:9798– 807. doi: 10.1158/0008-5472.CAN-10-1001. [DOI] [PubMed] [Google Scholar]
- 112.Abdalla MA, Haj-Ahmad Y. Promising candidate urinary microRNA biomarkers for the early detection of hepatocellular carcinoma among high-risk hepatitis C virus Egyptian patients. J Cancer. 2012;3:19–31. doi: 10.7150/jca.3.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tomimaru Y, Eguchi H, Nagano H, Wada H, Kobayashi S, Marubashi S, et al. Circulating microRNA-21 as a novel biomarker for hepato-cellular carcinoma. J Hepatol. 2012;56:167–75. doi: 10.1016/j.jhep.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 114.Miah S, Dudziec E, Drayton RM, Zlotta AR, Morgan SL, Rosario DJ, et al. An evaluation of urinary microRNA reveals a high sensitivity for bladder cancer. Br J Cancer. 2012;107:123–8. doi: 10.1038/bjc.2012.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kanemaru H, Fukushima S, Yamashita J, Honda N, Oyama R, Kakimoto A, et al. The circulating microRNA-221 level in patients with malignant melanoma as a new tumor marker. J Dermatol Sci. 2011;61:187–93. doi: 10.1016/j.jdermsci.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 116.Lin SC, Liu CJ, Lin JA, Chiang WF, Hung PS, Chang KW. miR-24 up-regulation in oral carcinoma: positive association from clinical and in vitro analysis. Oral Oncol. 2010;46:204– 8. doi: 10.1016/j.oraloncology.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 117.Liu CJ, Kao SY, Tu HF, Tsai MM, Chang KW, Lin SC. Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis. 2010;16:360– 4. doi: 10.1111/j.1601-0825.2009.01646.x. [DOI] [PubMed] [Google Scholar]
- 118.Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Morimura R, Nagata H, et al. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer. 2011;105:104–11. doi: 10.1038/bjc.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang T, Wang Q, Zhao D, Cui Y, Cao B, Guo L, Lu SH. The oncogenetic role of microRNA-31 as a potential biomarker in oesophageal squamous cell carcinoma. Clin Sci. 2011;121:437–47. doi: 10.1042/CS20110207. [DOI] [PubMed] [Google Scholar]
- 120.Hauser S, Wulfken LM, Holdenrieder S, Moritz R, Ohlmann CH, Jung V, et al. Analysis of serum microRNAs (miR-26a-2*, miR-191, miR-337–3p and miR-378) as potential biomarkers in renal cell carcinoma. Cancer Epidemiol. 2012;36:391–4. doi: 10.1016/j.canep.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 121.Redova M, Poprach A, Nekvindova J, Iliev R, Radova L, Lakomy R, et al. Circulating miR-378 and miR-451 in serum are potential biomarkers for renal cell carcinoma. J Transl Med. 2012;10:55. doi: 10.1186/1479-5876-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.D’Alessandra Y, Devanna P, Limana F, Straino S, Di Carlo A, Brambilla PG, et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31:2765–73. doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, et al. Circulating microRNA: a novel potential bio-marker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–66. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 124.Scian MJ, Maluf DG, David KG, Archer KJ, Suh JL, Wolen AR, et al. MicroRNA profiles in allograft tissues and paired urines associate with chronic allograft dysfunction with IF/TA. Am J Transplant. 2011;11:2110–22. doi: 10.1111/j.1600-6143.2011.03666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–18. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 126.Yi Z, Fu Y, Ji R, Li R, Guan Z. Altered microRNA signatures in sputum of patients with active pulmonary tuberculosis. PloS one. 2012;7:e43184. doi: 10.1371/journal.pone.0043184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fu Y, Yi Z, Wu X, Li J, Xu F. Circulating microRNAs in patients with active pulmonary tuberculosis. J Clin Microbiol. 2011;49:4246–51. doi: 10.1128/JCM.05459-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zahm AM, Hand NJ, Boateng LA, Friedman JR. Circulating microRNA is a biomarker of biliary atresia. J Pediatr Gastroenterol Nutr. 2012;55:366–9. doi: 10.1097/MPG.0b013e318264e648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang HJ, Zhang PJ, Chen WJ, Feng D, Jia YH, Xie LX. Four serum microRNAs identified as diagnostic biomarkers of sepsis. J Trauma Acute Care Surg. 2012;73:850– 4. doi: 10.1097/TA.0b013e31825a7560. [DOI] [PubMed] [Google Scholar]
- 130.Wang G, Tam LS, Li EK, Kwan BC, Chow KM, Luk CC, et al. Serum and urinary free microRNA level in patients with systemic lupus erythematosus. Lupus. 2011;20:493–500. doi: 10.1177/0961203310389841. [DOI] [PubMed] [Google Scholar]
- 131.Wang G, Tam LS, Kwan BC, Li EK, Chow KM, Luk CC, et al. Expression of miR-146a and miR-155 in the urinary sediment of systemic lupus erythematosus. Clin Rheumatol. 2012;31:435–40. doi: 10.1007/s10067-011-1857-4. [DOI] [PubMed] [Google Scholar]
- 132.Li S, Zhu J, Zhang W, Chen Y, Zhang K, Popescu LM, et al. Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection. Circulation. 2011;124:175–84. doi: 10.1161/CIRCULATIONAHA.110.012237. [DOI] [PubMed] [Google Scholar]
- 133.Rhodes CJ, Wharton J, Boon RA, Roexe T, Tsang H, Wojciak-Stothard B, et al. Reduced microRNA-150 is associated with poor survival in pulmonary arterial hypertension. Am J Respir Crit Care Med. 2013;187:294–302. doi: 10.1164/rccm.201205-0839OC. [DOI] [PubMed] [Google Scholar]
- 134.Gandhi R, Healy B, Gholipour T, Egorova S, Musallam A, Shuja M, et al. Circulating microRNAs as biomarkers for disease staging in multiple sclerosis. Ann Neurol. 2013;73:729– 40. doi: 10.1002/ana.23880. [DOI] [PubMed] [Google Scholar]
- 135.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, et al. Serum microRNAs are promising novel biomarkers. PloS One. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sheinerman KS, Tsivinsky VG, Crawford F, Mullan MJ, Abdullah L, Umansky SR. Plasma microRNA biomarkers for detection of mild cognitive impairment. Aging. 2012;4:590– 605. doi: 10.18632/aging.100486. [DOI] [PMC free article] [PubMed] [Google Scholar]