Abstract
Runx2 is a developmentally regulated gene in vertebrates and is essential for bone formation and skeletal homeostasis. The induction of runx2-P1 isoform transcripts is a hallmark of early osteoblastogenesis. Although previous in vitro studies have defined a minimal Runx2-P1 promoter sequence with well-characterized functional elements, several lines of evidence suggest that transcription of the Runx2-P1 isoform relies on elements that extend beyond the previously defined P1 promoter boundaries. In this study, we examined Runx2-P1 transcriptional regulation in a cellular in vivo context during early osteoblastogenesis of MC3T3-E1 cultures and BMSCs induced towards the bone lineage by multi-layered analysis of the Runx2-P1 gene promoter using the following methodologies: 1) sequence homology among several mammalian species, 2) DNaseI hypersensitivity coupled with massively parallel sequencing (DNase-seq), and 3) chromatin immunoprecipitation of activating histone modifications coupled with massively parallel sequencing (ChIP-seq). These epigenetic features have allowed the demarcation of boundaries that redefine the minimal Runx2-P1 promoter to include a 336-bp sequence that mediates responsiveness to osteoblast differentiation. We also find that an additional level of control is contributed by a regulatory region in the 5′-UTR of Runx2-P1.
Keywords: osteoblast differentiation, Runx2-P1, gene regulation, DNase hypersensitivity, histone modification
1. Introduction
The Runt-related transcription factor 2 (RUNX2) is a known regulator of bone-related genes and a master regulator of osteoblast differentiation (Komori et al., 1997; Otto et al., 1997; Ducy et al., 1999; Lian et al., 2004). During development, Runx2 is highly expressed in the mesenchyme of the developing skeleton and endochondral bone. Mutations of RUNX2 in humans result in cleidocranial dysplasia (CCD) (Mundlos et al., 1997), and a reduction of as little as 30% of runx2 transcript levels results in a CCD-like phenotype in mice (Lou et al., 2009). Moreover, complete ablation of Runx2 function in mouse models results in lethality, which is marked by an absence of both intramembraneous and endochondral ossification (Otto et al., 1997; Choi et al., 2001).
The Runx2 gene locus produces two predominant isoforms (Lian et al., 2013). The Runx2 type-I isoform (regulated by the proximal P2 promoter) is expressed in both osseous and non-osseous mesenchyme, while the Runx2 type-II isoform (regulated by the distal P1 promoter) is expressed exclusively in osteo-progenitors, and is stimulated during bone formation (Harada et al., 1999). These alternative transcripts are also designated according to the promoters that drive them: runx2-P1 and runx2-P2. The runx2-P1 isoform is detected in pre-osteoblast cells, however upon differentiation, transcript levels increase several-fold. Importantly, specific loss of the runx2-P1 transcript causes severe developmental defects with CCD-like symptoms (Zhang et al., 2009a; Liu et al., 2011).
The Runx2-P1 promoter was previously characterized by a 0.6-kb region [−629 to −16 of the transcriptional start site (TSS)] that encompasses a DNaseI hypersensitive region containing several experimentally proven control elements including: C/EBPβ, Oct1, AP-1, Runx2, Msx2/Dlx3/Dlx5, ATF, HLH/Twist, VDRE, LEF/TCF, NKX, NF-1, Sp1, and Ets. (Drissi et al., 2000; Banerjee et al., 2001; Zambotti et al., 2002; Stein et al., 2004; Gaur et al., 2005; Lee et al., 2005; Lengner et al., 2005; Hassan et al., 2006; Hassan et al., 2007; Cruzat et al., 2009; Zhang et al., 2009b; Henriquez et al., 2011; Hovhannisyan et al., 2013). However, significant in vitro and in vivo evidence has accumulated to indicate that regulatory elements that drive promoter activity reside beyond the 0.6-kb region (Xiao et al., 2001; Lengner et al., 2002). Therefore, the complete cis-regulatory machinery that drives bone-specific Runx2-P1 expression remains incompletely described.
The development of next-generation sequencing technologies, and our improved understanding of epigenetic mechanisms, has aided our ability to predict and define regulatory sequences on a genome-wide scale (Bernstein et al., 2012). These methods have also made possible validation of the positions of experimentally discovered enhancer and promoter boundaries that were traditionally defined by promoter truncation, reporter gene assays, and comparative sequence analysis across diverse species. Although the ability to predict the locations of regulatory regions by querying sequence conservation alone was improved by the whole-genome sequencing of several vertebrate species, variations can mask critical cis-regulatory regions. Thus, the ability to also contrast sequence conservation with the profiling of epigenetic markers upon gene activation has allowed for the accurate prediction of critical regulatory regions responsible for transcriptional control.
In this study, our goal was to further characterize genomic segments of the endogenous Runx2 gene that are critical for the regulation of osteoblast differentiation-dependent Runx2-P1 transcription. We focused on the epigenetic composition of the Runx2-P1 promoter during early stages of differentiation to reveal that the promoter boundaries extend about 300 bp 5′ beyond the previously defined 0.6-kb promoter and about 400 bp 3′ beyond the TSS. To investigate the functional nature of the redefined 0.9-kb promoter, we designed luciferase reporter constructs that span from −956 to −16 of the TSS. We found that when this promoter was tested in pre-osteoblasts versus osteoblasts, it yielded an increase in activity during osteogenic differentiation, a function that is not observed for the previously characterized 0.6-kb P1 promoter. Of additional significance, this differentiation responsiveness is preserved with the inclusion of the 5′-UTR which acts to suppress P1 activity. Our findings further elucidate the transcriptional control of the osteogenic master regulator, Runx2.
2. Materials and methods
2.1. Cell culture and BMSC isolation
For genome-wide profiling assays and transcriptional activity studies, the MC3T3-E1 clone-4 pre-osteoblastic murine cell line (Wang et al., 1999) was used (American Type Culture Collection, Manassas, VA). Growth-phase cultures were maintained in α-MEM without ascorbic acid (Hyclone, Thermo Fisher Scientific, Rochester, NY) and supplemented with 1% penicillin-streptomycin (Gibco, Life Technologies, Grand Island, NY), 2 mM L-glutamine (Gibco, Life Technologies), and 10% Fetal Bovine Serum (Atlanta Biologicals, Lawrenceville, GA). When cultures reached ~90% confluency, differentiation was initiated by the addition of 142 μM ascorbic acid (Sigma-Aldrich, St. Louis, MO) to 10% FBS (Hyclone, Thermo Fisher Scientific) in α-MEM. After 2 days, the ascorbic acid concentration was increased to 280 μM and 5 mM β-Glycerophosphate (Sigma- Aldrich) was added (Quarles et al., 1992; Wang et al., 1999). Cultures were maintained at 37°C at 5% CO2, with fresh media changes every 2 days.
Primary Bone Marrow Stromal Cells (BMSCs) used in genome-wide profiling studies were isolated from 6–8 week old alpha smooth muscle actin (SMAA)-mCherry transgenic mice (gifted by Dr. David Rowe, University of Connecticut, Farmington, CT) following previously described methods (Kalajzic et al., 2008). BMSCs attached to the plate surface after 72 h, and were cultured for an additional four days with media changes every 2 days. SMAA-mCherry positive cells (definitive marker for BMSCs (Kalajzic et al., 2008)) were enriched using fluorescence-activated cell sorting (FACS), and plated at low-density (1500 cells/cm2) for expansion and maintenance in BMSC growth media. Passage 8 to 10 BMSCs were used for the experiments described. At 90% confluency, BMSCs were induced towards osteoblast differentiation with ascorbic acid-free α-MEM medium supplemented with 1% penicillin-streptomycin, 15% fetal bovine serum, 142 μM ascorbic acid, and 5 mM β-Glycerophosphate for up to 14 days with medium changes every 2–3 days. BMSCs at day 0 (non-differentiated), and days 7, and 14 (matrix-deposition) were harvested for experiments. All animal work was performed following an approved IACUC protocol at The University of Massachusetts Medical School, Worcester, MA.
Differentiation was verified by the level of alkaline phosphatase (ALP) activity staining (Dean et al., 1994). ALP stained cultures were captured with a Leica microscope (M165FC) conjugated to a Leica digital color camera (DFC310FX, Leica Microsystems Inc., Buffalo Grove, IL), and acquired with LASv4.1 imaging software (Leica Microsystems Inc.). Phase contrast images were captured on a Nikon Eclipse TS100 inverted microscope (Nikon Instruments Inc., Melville, NY), conjugated to a SPOT RT3 CCD camera using SPOT imaging software v5.0 (Diagnostic Instruments Inc., Sterling Heights, MI).
2.2. Real-Time PCR
Total RNA from cultures was extracted with TRIzol (Invitrogen, Life Technologies, Grand Island, NY), followed by DNase treatment using the DNA-Free RNA Kit (Zymo Research, Irvine, CA) according to manufacturers’ instruction. cDNA was prepared using the SuperScriptIII First-Strand Synthesis System (Invitrogen). qPCR was performed with the iTaq SYBR Green Supermix with ROX (Bio-Rad, Hercules, CA) on the ViiA 7 Real Time PCR System (Applied Biosystems, Life Technologies, Grand Island, NY). Relative transcript levels were determined by the ΔΔCt method, normalized to gapdh. Primer sequences for runx2-P1, runx2-P2, bone gamma-carboxyglutamic acid-containing protein (bglap2), integrin-binding sialoprotein (ibsp), and glyceraldehyde 3-phosphate dehydrogenase (gapdh) are described elsewhere (Liu et al., 2011).
2.3. Chromatin immuno-precipitation and high-throughput sequencing
ChIP-seq analysis tracks presented here were obtained from (Wu et al., 2014) and an additional study that probes the genome-wide profiling of epigenetic marks during BMSC-osteoblast differentiation (Wu et al., 2013). Only tracks spanning the Runx2-P1 region and the muscle creatine kinase (Mck) locus were analyzed for this study. The following methods to obtain and analyze ChIP-seq libraries were used. Approximately 1×108 MC3T3-E1 cells or BMSCs were washed with PBS and fixed with 1% formaldehyde for 10 min at room temperature to crosslink DNA-protein complexes. Fixed cells were washed with ice-cold PBS, harvested, and pelleted. Extraction of nuclei was performed as described in (Wu et al., 2014). Isolated nuclei were subjected to shearing using a Misonix S-4000 Ultrasonic Processor (QSonica, Newton, CT) to obtain chromatin fragments ranging from 0.2 to 0.6 kb. Sheared chromatin was used for immunoprecipitation with antibodies against RUNX2 (M-70; Santa Cruz Biotechnology, Dallas, TX), H3K4me1 (ab8895, Abcam, Cambridge, MA), H3K4me3 (ab1012, Abcam), H3K9me3 (Ab8898, Abcam), H3K9ac (39137, Active Motif, Carlsbad, CA), H3K27me3 (07-449, Millipore, Billerica, MA), H3K27ac (07-360, Millipore), or immunoglobulin-G (IgG) (12-370; Millipore) followed by purification using Protein-G Dynabeads (Invitrogen). Crosslinks within precipitated chromatin were subsequently reversed at 65°C. DNA was recovered by phenol/chloroform/isoamyl alcohol (25:24:1 v/v) extraction and ethanol precipitation. High-throughput sequencing libraries were constructed by ligating Illumina SR adapters (Illumina, San Diego, CA) to purified DNA fragments following manufacturer’s recommendations. DNA libraries were size-selected (200 ± 50 bp), and subjected to single-end 36-base sequencing on an Illumina Genome Analyzer II (Deep Sequencing Core, University of Massachusetts Medical School). Base calls and sequence reads were generated by Illumina CASAVA software (version 1.6, Illumina). Two independent biological repeats of each antibody-specific ChIP-seq enrichment were prepared for each time point. For each cell type (BMSC and MC3T3-E1), two input ChIP-seq control libraries were prepared with sonicated total chromatin.
2.4. DNase-seq
DNase-seq tracks presented here were obtained from a genome-wide profiling study of DNase hypersensitivity throughout MC3T3-E1 osteoblastogenesis (Tai et al. manuscript in preparation). The following methods to obtain and analyze DNase-seq libraries were used. Genome-wide DNase hypersensitivity mapping of osteoblast cultures was performed by adapting a described DNase-seq protocol (Song and Crawford, 2010) with slight modifications. Approximately 40 × 106 growth-phase (day 0) or matrix-deposition stage (day 9) MC3T3-E1 clone-4 cells were harvested and subjected to 4, 12, and 40 U/132 μL of DNaseI digestion for 15 min at 37°C. Steps involving the isolation of chromatin embedded in agarose included a treatment with 10 U/mL β-agarase for 2 h at 37°C before extracting with phenol:chloroform:isoamyl alcohol (25:24:1 v/v) and ethanol precipitation. DNase-seq libraries were generated from pooled DNase1-treated chromatin. DNase-seq analyses were confirmed by two biological replicates, each constituting technical duplicates that pass ENCODE Consortium standards on significant peaks called by F-seq (Boyle et al., 2008) using IDR analysis (Li et al., 2011) and normalized using align2rawsignal (Kundaje A., http://code.google.com/p/align2rawsignal/). Representative peak signals from a single biological sample for each culture condition are shown.
2.5. Reporter constructs
The design and construction of the 3-kb (−2821 to −16) and 0.6-kb (−629 to −16) luciferase reporter plasmids have been described previously (Drissi et al., 2000). The Runx2-P1 0.9-kb-Luc construct was derived from the 3-kb luciferase construct by deleting sequence between −2821 and −966 using the quick-change method for large fragment deletion (Makarova et al., 2000). The −965 to +407 luciferase reporter construct was designed by PCR amplification from C57BL/6 genomic DNA and conventional cloning methods to insert the fragment into the pGL3 luciferase reporter plasmid (Promega, Madison, WI). Each construct was validated by Sanger sequencing. The TK-pGL3 and SV40-Renilla constructs were kind gifts from Dr. Stephen D. Hauschka (University of Washington, Seattle, WA).
2.6. Co-transfections and luciferase reporter assays
MC3T3-E1 cultures at >90% confluency were co-transfected with Firefly Luciferase reporter constructs and SV40-Renilla using Lipofectamine (Invitrogen) and Plus Reagent (Invitrogen) according to manufacturer’s instructions. The SV40-Renilla construct serves to control for slight variability in transfection efficiency between plates. A total of 2.5 μg of plasmid DNA (2 μg:0.5 μg, test: reference) was transfected per 60 mm plate. After 4 days post-differentiation, cultures were harvested and reporter activities were measured using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI) on a VICTOR X4 Multilabel Plate Reader (Perkin Elmer, Waltham, MA), according to manufacturers’ instructions. Each test condition described is represented by three biological replicates, each assayed in duplicate. Activities of test promoter constructs between pre-osteoblasts and differentiating osteoblasts were further normalized to the activity of the TK promoter construct that was transfected in parallel cultures. Statistical significance values assessed by Student’s t-test are reported where applicable.
3. Results
3.1. Runx2-P1 transcript levels increase several-fold during osteoblast differentiation
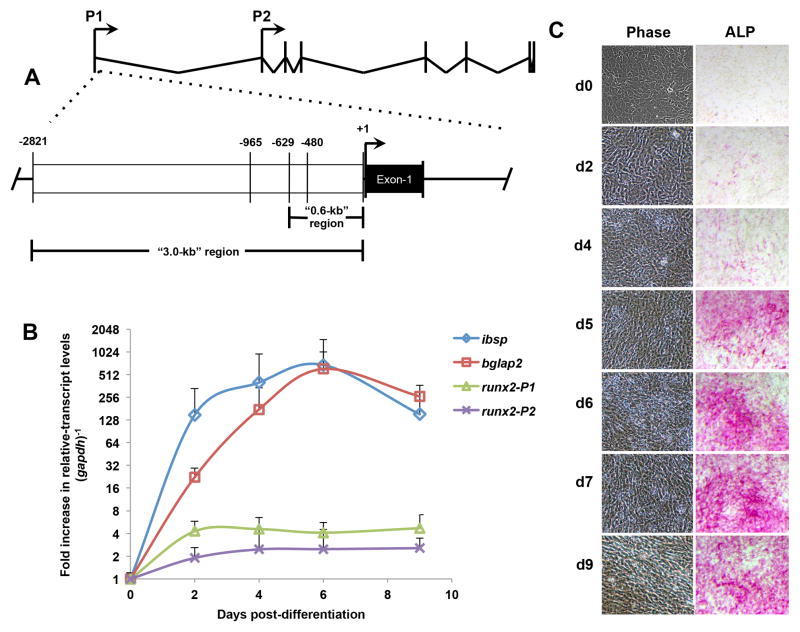
The murine Runx2 gene is located on chromosome 17 and spans a ~210-kb region. The osteogenic P1 promoter was previously described in vitro, in both rat and mouse osteoblast cultures, as a 0.6-kb region that encompasses a 0.4-kb hypersensitive region (Hovhannisyan et al., 2013) (Fig. 1A) and reviewed in Lian et al. (Lian et al., 2013). However, subsequent studies suggested that further elements beyond the 0.6-kb region could confer osteoblast-specific regulation (Xiao et al., 2001). To redefine the P1 promoter boundaries and activity, we chose the MC3T3-E1 pre-osteoblast model, which recapitulates the process of osteoblastogenesis, in vitro. We initially confirmed the differentiation parameters of the MC3T3-E1 cells by using RT-qPCR analysis to measure changes in expression of bone-specific genes (Fig. 1B). As expected, mRNA levels of bone-sialoprotein (ibsp) and osteocalcin (bglap2) reproducibly increase as observed in previous studies (Stein et al., 2004). In these specific cultures, ibsp and bglap2 levels reached maximal fold-change (>500-fold) between 5 and 6 days after induction with ascorbic acid and β-glycerophosphate treatment. As expected, the Runx2-P1 transcript demonstrates a ~4-fold higher expression level in cultures during early osteoblast differentiation (2 days post-osteogenic media) than in cultures maintained in growth-media (Fig. 1B). Steady-state levels of runx2 are maintained between 2 and 9 days after switching to osteogenic media. We also assayed cultures by alkaline phosphatase (ALP) activity. ALP is known to be induced by RUNX2 and is used as a definitive marker of osteogenic progression (Harada et al., 1999; Banerjee et al., 2001; Stein et al., 2004) (Fig. 1C). After switching to differentiation conditions, cultures predictably displayed increasing ALP activity (Fig. 1C), confirming a positive progression of osteoblast differentiation.
Fig. 1.
The runx2-P1 isoform is upregulated during MC3T3-E1 differentiation. (A) The Runx2 gene produces two predominant message isoforms transcribed from the P1 and P2 promoters. The Runx2-P1 promoter is defined by a previously discovered hypersensitive 0.4-kb “core” sequence (−480 to −1). The consensus ATG begins at +408 of Exon-1 (black region). (B) MC3T3 cultures show elevated bone-related transcript levels throughout the first 9 days post-switching to differentiation conditions as determined by qPCR. Relative levels at d0, d2, d4, d6, and d9 cultures are plotted bone-sialoprotein (ibsp, blue), osteocalcin (bglap2, red), runx2-P1 isoform (green), runx2-P2 isoform (purple). Error bars represent 1STD. (C) Osteoblast cultures display increasing alkaline phosphatase activity (ALP), a hallmark of cultures undergoing matrix-deposition. Select phase images (left column) of cultures throughout d0 to d9, accompanied by ALP stained parallel cultures (right column).
3.2. Sequence conservation and epigenetic landscape define the Runx2-P1 minimal promoter
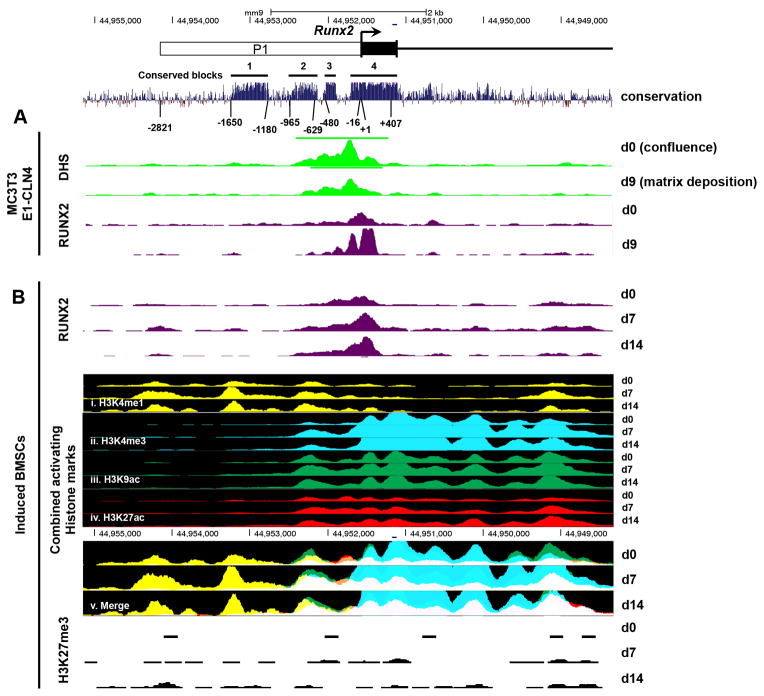
We applied the concept of phylogenetic footprinting to further establish regulatory elements within the Runx2-P1 region that are influenced by osteoblastic differentiation. This concept states that functionally important sequences, such as cis-regulatory regions, are highly conserved among different species (Tagle et al., 1988). Evolutionarily conserved sequence at the Runx2-P1 promoter were probed among several placental mammals (Fig. 2), as well as among six distantly related mammals (human, mouse, cow, horse, dog, and elephant) (Supplemental Fig. 1) via the publicly accessible UCSC Genome Bioinformatics resource (Kent et al., 2002). We observed high sequence conservation in four main blocks: block-1 (−1650 to −1180), block-2 (−960 to −540), block-3 (−480 to −310), and block-4 (−125 to +470) where blocks 3 and 4 are separated by an extended purine-rich region that is unique to the murine sequence (Drissi et al., 2000) (Supplemental Fig. 1). Conserved blocks 1 and 2 exist beyond the previously described 0.6-kb promoter, and suggest the presence of functional sequence. Conserved block-4 covers predominantly 5′-UTR sequence and exon-1. Although the conservation within this region may reflect selective pressure to promote mRNA stability or the post-transcriptional control of runx2-P1, the 5′-UTR has also been shown previously to contain several cis-acting elements that regulate Runx2-P1 expression (Drissi et al., 2000).
Fig. 2.
RUNX2 enrichment and epigenetic landscape during the differentiation of osteoblast cultures at the Runx2-P1 locus. (A) The Runx2-P1 promoter, is defined by 4 blocks of high sequence conservation (black bars). A DNase hypersensitive region spanning a ~1.3-kb sequence as defined by F-seq peak caller (green bars and tracks) in growth phase and matrix deposition stages of MC3T3-E1-CLN1 cultures establishes the boundaries of an extended Runx2-P1 promoter. The enrichment of RUNX2 (purple tracks) during differentiation overlaps with DHS sequence. (B) RUNX2 enrichment in induced BMSC cultures. Increased signal throughout differentiation time-points d0, d7, and d14. Activating histone mark tracks: (i) H3K4me1 (yellow), (ii) H3K3me3 (cyan), (iii) H3K9ac (green), (iv) H3K27ac (red), and (v) merge of all activating histone marks demarcate the epigenetic landscape throughout differentiation. The repressive histone mark H3K27me3 (black track) is absent throughout this region. All scaling of tracks shown here are equal between each culturing time point and locus position. A 30-way alignment of sequence conservation among placental mammals is shown above each gene locus.
The high sequence conservation beyond the previously defined 0.6-kb promoter prompted us to determine whether these blocks coincide with chromatin accessible regions that house cis-regulatory elements. DNase hypersensitivity analysis permits the unbiased identification of chromatic regions that are either displaced or depleted of nucleosomes, due to factor or complex association. We therefore queried genome-wide DNase hypersensitivity coupled with deep sequencing (DNase-seq) analyses at sequence surrounding the Runx2-P1 promoter during two hallmark phases of osteoblastogenesis: pre-osteoblasts maintained in proliferation conditions at confluence 0 days (d0), and matrix deposition stage osteoblasts that were maintained in differentiation conditions for 9 days (d9) (Fig. 2A). Peak signals for DNase hypersensitivity indicate that significant chromatin accessibility is observed throughout the P1 region in both d0 and d9 cultures. F-seq analysis, an algorithm that generates a continuous tag sequence density estimation that allows for the identification of DNase hypersensitive sites (DHS) (Boyle et al., 2008), indicated that sequences ranging upstream of the TSS at −956 to downstream at +407 are associated with a DHS, These results suggest the occupancy of this region by transcription factors or complexes during both pre-osteoblast and differentiating osteoblast stages. Notably, this span extends beyond the previously established 0.6-kb promoter sequence. Although the highly conserved block-1 displays a low level of DHS peak signal in d0 cultures, this signal was not identified by F-seq analysis and likely represents either a small portion of cells that harbor a DHS at this position, or that the region has low DNaseI sensitivity. In either case, the signal detected is considered negligible.
We also addressed the occupancy of RUNX2 across the Runx2-P1 region using chromatin immunoprecipitation combined with massively parallel sequencing (ChIP-seq) (Wu et al., 2014). RUNX2 enrichment was observed in both d0 and d9 cultures, with a marked increase in enrichment in d9 cultures. We also found a greater enrichment of RUNX2 association within the 5′-UTR region that contains three RUNX2 binding sites centered at +31, +39, and +49 (Fig. 2A and Supplemental Fig. 1). These RUNX motifs have been shown in previous studies to suppress Runx2 expression as part of a negative feedback loop (Drissi et al., 2000). Additional RUNX2 peak signals overlap with the three previously identified RUNX2 binding sites centered at −336, −116, and −72 (Drissi et al., 2000; Hassan et al., 2006). Notably, the DHS region overlaps with RUNX2 enriched sequence in both pre-osteoblast and differentiating osteoblast cultures, coinciding with a “base-line” expression of Runx2-P1 in pre-osteoblasts that is increased upon differentiation.
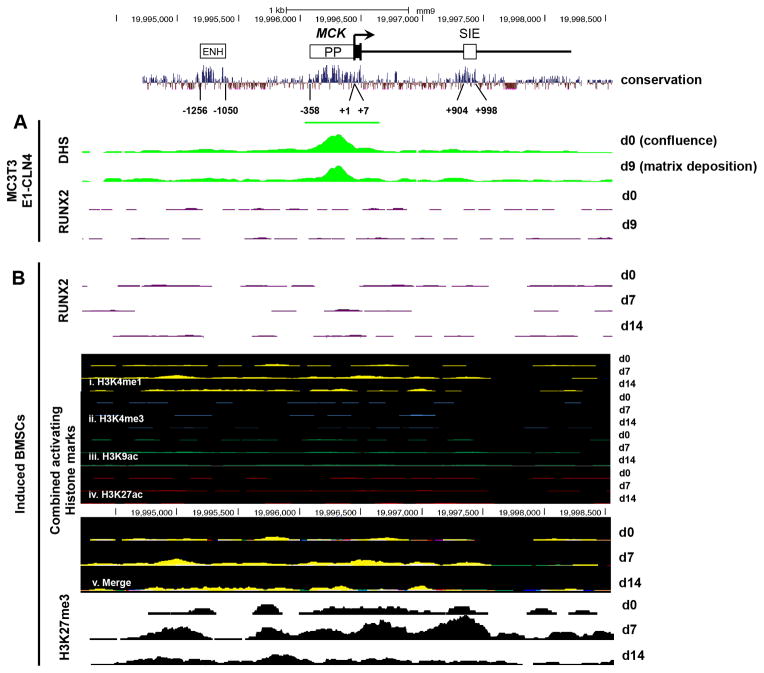
For comparison, we examined DNase hypersensitivity and RUNX2 enrichment surrounding the muscle creatine kinase gene (Mck), a tissue-specific gene expressed only in cells committed to the muscle lineage (Eppenberger et al., 1964). In mammals, Mck is controlled by three conserved and experimentally proven regulatory regions: an upstream 206-bp enhancer (−1256 to −1050), a 366-bp proximal promoter (−358 to +7), and a 995-bp intronic enhancer (+904 to +998) (Tai et al., 2011) (Fig. 3). Interestingly, the Mck promoter exhibits DNase-hypersensitivity in both d0 and d9 osteoblast cultures (Fig. 3A), indicating that although Mck is transcriptionally inactive, the promoter remains in an accessible conformation. As expected, the Mck regulatory regions show no evidence of enrichment for RUNX2, consistent with the absence of RUNX consensus binding motifs (Fig. 3A and B).
Fig. 3.
RUNX2 enrichment and epigenetic landscape during the differentiation of osteoblast cultures at the Mck locus. (A) The muscle creatine kinase (Mck) gene is transcriptionally controlled by three conserved regulatory regions: the upstream enhancer (−1256 to −1050), the proximal promoter (−358 to +7), and the small intronic enhancer (SIE, +904 to +998). In MC3T3-E1 cultures, the Mck locus displays hypersensitivity at the promoter region but not at the upstream or intronic enhancer regions. (B) Activating histone marks are absent within this region in induced BMSC cultures: (i) H3K4me1 (yellow tracks), (ii) H3K3me3 (blue tracks), (iii) H3K9ac (green tracks), and (iv) H3K27ac (red tracks) and (v) merge of all activating histone marks. The repressive H3K27me3 mark (black tracks) is dynamically enriched in differentiating cultures. All scaling of tracks shown here are equal between each culturing time point and locus position. A 30-way alignment of sequence conservation among placental mammals is shown above each gene locus.
Since DHS profiling seems to suggest that promoter regions can show accessibility without or perhaps prior to gene activation, we next asked whether profiling of the epigenetic landscape spanning the Runx2-P1 region might support our proposal that the Runx2-P1 promoter can be defined by the boundaries of the DHS region. To do so, we interrogated the genome-wide ChIP-seq profiling of activating histone modifications in lineage-committed osteogenic BMSCs (Wu et al., manuscripts in preparation) (Fig. 2B). We focused our analysis on the following modifications: H3 lysine 4 mono-methylation [H3K4me1], that marks active or poised enhancers; H3 lysine 4 tri-methylation [H3K4me3], that marks active or poised promoters; H3 lysine 9 acetylation [H3K9ac] that marks transcriptional activation; H3 lysine 27 acetylation [H3K27ac], a marker of active enhancers and promoters; and H3 lysine 27 tri-methylation [H3K27me3], a marker of transcriptionally silent chromatin. Three differentiation time points that reflect uncommitted BMSCs (d0) and matrix-deposition stages (d7 and d14) were examined. To demonstrate that the epigenetic profiles of induced BMSCs can be applied to our investigation of the Runx2-P1 region, we first examined the enrichment of RUNX2 at the Runx2-P1 locus between differentiating BMSCs and MC3T3-E1 cultures (Fig. 2A and B). For both cell types, the intensity of RUNX2 signal enrichment increased as differentiation progressed from growth to matrix deposition stages. The enrichment profiles in BMSCs are similar to MC3T3-E1 cultures, where peak signals are strongest within the 5′-UTR repressive region (Fig. 2B). There was no enrichment of RUNX2 at the Mck gene in BMSC cultures (Fig. 3B).
We asked whether the combined histone modification features could aide in establishing the Runx2-P1 border. Interestingly, enrichment profiles for the activating histone marks H3K4me3, H3K9ac, and H3K27ac exhibit periodicity downstream to the Runx2-P1 TSS, which likely reflects nucleosome positioning of this active region. It has previously been reported that the −1 and +1 nucleosomes flank a nucleosome-depleted region created by trans-acting factors recruited to active promoters (Hughes et al., 2012). The overall reduction in histone mark peak signals between −300 and −100 seems to reflect this. Importantly, H3K4me3, H3K9ac, and H3K27ac marks terminate near the −965-position, coinciding with the 5′ most border of the DHS region observed in the MC3T3-E1 cultures (Fig. 2A). H3K4me1, which marks active or poised enhancers, displays several areas of enrichment 5′ of the TSS, including a prominent peak signal that overlaps conserved block-1. However, conserved block-1 does not display a dominant DNase hypersensitivity peak signal (Fig. 2A). Interestingly, the region encompassing −965 to −310 (blocks 2 and 3) has overlapping H3K4me1 and H3K4me3 modifications, which is hallmark of activated cis-regulatory elements (Pekowska et al., 2011). We note that the repressive histone mark H3K27me3 is not enriched throughout this region (Fig. 2B). Overall, this comparative analysis suggests that the region spanning −965 to +407 demarcates regulatory sequence that is critical for expression of the Runx2-P1 promoter during osteoblast differentiation.
Activating histone marks are not observed at the Mck regulatory regions (Fig. 3B), in agreement with the absence of Mck expression in the osteoblast lineage (Eppenberger et al., 1964). The lack of activating histone marks contrasts with the presence of the DHS that overlaps the Mck promoter region in MC3T3-E1 cells (Fig. 3A). However, the repressive histone mark H3K27me3 is enriched throughout the Mck locus, where it is most prominent at the intronic enhancer (+904 to +998) in d7 cultures. These observations are suggestive of a mechanism for strong transcriptional repression of the Mck gene in d7 BMSCs in light of an “open” chromatin state at the silent promoter. Thus, although DNase hypersensitivity may predict the boundaries of promoter sequence, it alone cannot predict whether the DHS span contributes to the activation or repression of a gene.
3.3. Activity of the Runx2-P1 0.9-kb minimal promoter is responsive to osteoblast differentiation
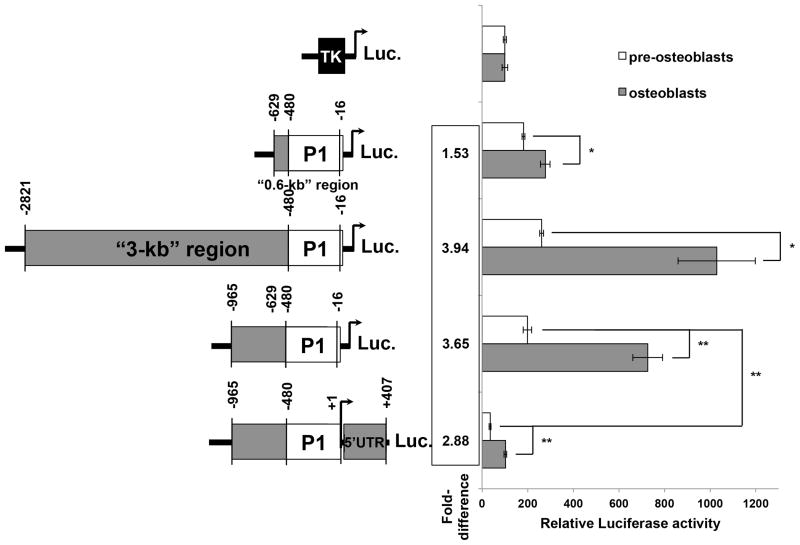
In previous studies, Drissi et al. demonstrated that two luciferase reporter constructs designed to encompass 0.6-kb (−629 to −16) and a larger a 3-kb (−2821 to −16) region of the Runx2-P1 promoter were equally efficient in driving reporter gene expression in non-differentiated rat osteosarcoma cells (ROS 17/2.8) (Drissi et al., 2000). This result suggested that the 0.6-kb construct encompasses all of the necessary regulatory elements to drive Runx2-P1 expression in vitro. However, our DHS and epigenetic profiling of the promoter region suggest that additional regulatory elements exist. Furthermore, differences in transcriptional activity between the 0.6-kb and 3-kb constructs during osteoblast differentiation had yet to be explored. We therefore examined how the Runx2-P1 promoter behaves in differentiating MC3T3-E1 osteoblasts.
To assay promoter activity of the region, identified in our DNase-seq and epigenetic profile queries, we constructed two luciferase reporter plasmids to test alongside the 0.6-kb promoter: a construct that spans from −965 to −16, and a construct that spans from −965 to +407. Again, the −965 to +407 construct encompasses sequence within the 5′-UTR. Although sequences within the Runx2-P1 5′-UTR have been experimentally proven to repress transcriptional activity (Drissi et al., 2000), it is unknown how the entire DHS defined promoter behaves during differentiation.
We transfected MC3T3-E1 pre-osteoblasts with the Runx2-P1 test constructs (Fig. 4) and luciferase activity was measured in non-differentiated cells or after four days of differentiation, when cultures reached plateauing expression of runx2-P1 mRNA (Fig. 1B). For comparison, parallel cultures subjected to the same conditions were also transfected with the 3-kb Runx2-P1 promoter described previously and the minimal thymidine kinase (TK) promoter driving luciferase. Comparisons to the TK promoter between pre-osteoblast cultures and cultures undergoing differentiation were performed to gauge the specific increase exclusive to Runx2 promoter sequence. The 0.6-kb, 0.9-kb, and 3-kb constructs produced similar reporter gene activities in pre-osteoblast cultures (Fig. 4), as predicted by previous studies. In cultures undergoing differentiation, the 0.6-kb construct demonstrated a ~30% greater transcriptional activity (p<0.05), indicating that the 0.6-kb construct activity remains relatively similar between pre-osteoblast and osteoblast stages. Because runx2-P1 mRNA levels, at least in vitro, are expected to increase ~4-fold after 2 days of differentiation, this result suggests that the 0.6-kb region lacks differentiation responsive control elements. Surprisingly, the 0.9-kb and the 3-kb Runx2-P1 promoter constructs produced nearly 4-fold more luciferase activity (p < 0.01) in cultures subjected to four days of differentiation, than in pre-osteoblast cultures (Fig. 4). These results indicate that both the 0.9-kb and 3-kb constructs contain differentiation responsive elements that are lacking in the 0.6-kb promoter.
Fig. 4.
Runx2-P1 reporter construct activities in osteoblast cultures. The previously reported 0.6-kb (−629 to −16) and 3-kb (−2821 to −16) Runx2-P1 promoter constructs driving the luciferase reporter gene were transfected into MC3T3-E1-CLN4 cultures, and luciferase activity was assayed in either pre-osteoblasts (white bars) or osteoblasts maintained for 4 days in differentiation conditions (grey bars). The DNase hypersensitivity (DHS) and epigenetic topologically defined (−965 to −16) or (−965 to +407) sequences (Fig. 2) were cloned into similar luciferase constructs and tested in the same manner. Each construct activity is represented here as relative luciferase activity normalized to the co-transfected SV40 driven Renilla construct activity (see Materials and methods). Activities for each culturing condition were scaled to the activity of the thymidine kinase (TK) promoter construct in either pre-osteoblasts or osteoblasts to control for changes unrelated to Runx2-P1 promoter sequence. Fold-difference between conditions are displayed for each construct to the right of the bar graph. Error bars represent ± STD. Single asterisks (*) represent a p-value of < 0.05. Double asterisks (**) designate a p-value of < 0.01.
As anticipated, inclusion of the 5′-UTR region (−17 to +407) reduced the activity of the 0.9-kb promoter by ~5.6-fold in d0 cultures (p < 0.01) (Fig. 4). However, when the −965 to +407 construct was tested in differentiating cultures, luciferase activity was nearly 3-fold greater in d4 cultures than in d0 cultures (p < 0.01). Thus, although inclusion of the 5′-UTR repressive region reduces basal transcription levels, the 0.9-kb promoter remains responsive to the onset of differentiation. More importantly, our epigenetic landscape profiling was able to predict a functional role for the −965 to −629 region, which acts to promote differentiation responsiveness, while the −17 to +407 sequence acts to repress transcriptional activity.
4. Discussion
The transcriptional regulation of Runx2 is critical in mesenchymal tissue development and the permissive expression of Runx2 is concomitant with pathological consequences that include tumorigenesis and metastasis (Schroeder et al., 2005; Pratap et al., 2006). A better understanding of the regulatory regions that drive Runx2 expression will help to address how this critical factor is controlled during normal differentiation and in diseased states. In this study, we used several new approaches to define the regulatory boundaries that control transcription of the Runx2 gene. Analysis of the Runx2-P1 promoter using the UCSC genome browser revealed that highly conserved sequences are encompassed within a region from −1650 to +407 relative to the TSS (Fig. 2A). Querying DNase hypersensitivity and the positioning of activating histone marks offered supporting evidence that the minimal promoter should be redefined to include regions spanning from −0.9-kb to +0.4-kb. Although the principles by which the 0.6-kb region was originally defined in previous studies were also based on DNase-hypersensitivity analysis, it is intriguing that querying DNase-hypersensitivity on a genome-wide scale was able to reveal further accessible chromosomal sequence.
By designing luciferase reporter constructs to encompass this newly defined region, we were able to show that the region between −965 and −629 confers differentiation dependent transcription. These findings highlight the ability of a layered epigenetic analysis to redefine boundaries of regulatory regions. In essence, we were able to discover a region of sequence that imparts differentiation dependent responsiveness to the Runx2-P1 promoter.
Based on earlier studies by others, the −0.9-kb to −0.6-kb span contains consensus-binding motifs for Nkx2.5, Ik-2, SRY, Irf-2, GATA-1, CdxA, NF-kB and Oct-1 (Xiao et al., 2001) (Supplemental Fig. 1). Likely, a combination of these sites and possibly others act to define the differentiation responsiveness observed in our in vitro studies. Furthermore, recruitment of these factors may be intimately related to the overlapping H3K4me1 and H3K4me3 marks observed within this region (Fig. 2B). Whether H3K4 at this position is dynamically methylated and demethylated by histone modifying enzymes (Allis et al., 2007), or whether the nucleosomes associated with this region are marked by both modifications, remains to be addressed. A full understanding of how methylation status is established during differentiation may explain how this region responds to differentiation signals.
Interestingly, our study also demonstrates the presence of the H3K4me1 mark at the highly conserved sequence between −1650 and −1180 of the Runx2-P1 promoter (block-1, Fig. 2). Although this mark is strongly correlated with enhancers, this region lacks a strong DHS in both pre-osteoblasts and differentiating osteoblast cultures. This observation may indicate the presence of a poised enhancer region. Coincidently, a recent study reports that GATA-4 is recruited to two GATA-binding sites within −1664 to −1000 of Runx2-P1 to inhibit the binding of DLX5 to the promoter (Song et al., 2013). It is intriguing to hypothesize that the conserved sequence between −1650 and −1180 may modulate the activation of the 0.9-kb minimal promoter in cell-states that are beyond the osteoblastogenesis stages tested in our cell culture experiments.
Lastly, an earlier investigation demonstrated high-level expression of a 1.4-kb Runx2-P1 construct (−1403 to +14) in MC3T3 cultures (Xiao et al., 2001). However, this construct lacked the necessary elements to restrict expression to osteoblastic cells, as it was also highly expressed in C310T1/2, L929, C2C12, and NIH3T3 cells. It is plausible that the repressive DHS region defined in our study could aide in restricting or silencing expression in non-osseous tissues. We were able to show that inclusion of the 5′-UTR repression sequence still allowed the 0.9-kb construct to remain responsive to differentiation signals. This region may very well modulate P1 activity to restrict spurious transcription but allow other components of P1 to respond to stimulating signals to drive Runx2 expression. Notably, previous transgenic studies revealed the 3-kb region that spans from −2821 to −16, yields expression that is restricted to the axial skeleton (Lengner et al., 2002); these results suggest that additional far-distal regulatory elements controlling runx2-P1 transcription likely exist and are yet to be discovered. With the development of chromosomal conformation capture methodologies (Dekker et al., 2013), it is now feasible to query long-ranging interactions (e.g., enhancers, insulators, LCRs) that can modulate the transcriptional activity of the P1 promoter. As enhancer sequences are found to physically interact with promoter sequences and rely on these physical interactions for proper function (Williamson et al., 2011; Tai et al., 2014), any future investigations of enhancer regions acting to modulate P1 promoter activity should be tested within the context of the 0.9-kb promoter, rather than the traditional 0.6-kb construct.
5. Conclusions
Runx2-P1 gene transcription during the process of osteoblastogenesis shows complex regulation. Here we have demonstrated via multi-layered epigenetic profiling that Runx2-P1 transcription is defined by a bone-related promoter that encompasses a 965-bp sequence 5′ of the TSS. Additional regulatory regions remain elusive, and likely exist far-distal to this minimal promoter. The discovery of far-distal interacting regulatory regions (enhancers, silencers, LCRs, etc.) using methodologies such as 4C (circular chromosomal conformation capture) (Zhao et al., 2006) should therefore encompass the 0.9-kb promoter boundaries described in this study to fully understand how the Runx2 gene is regulated.
Supplementary Material
Six-way sequence alignment of the Runx2-P1 promoter. Select sequences of the human, mouse, cow, horse, dog, and elephant Runx2-P1 promoter region showing high-sequence similarity. Bases exhibiting high-sequence conservation across all species are in black and noted as asterisks (*) in the consensus sequence below each row. Bases that are semi-conserved are in gray. Gaps are designated as hyphens (−). The transcriptional start site (+1) is indicated by the black arrow. Currently known and experimentally proven control elements within the 0.9-kb minimal promoter are annotated and highlighted as colored texts of the associated transcription factors, and boxed to surround their respective motif sequences.
Highlights.
Genome-wide analyses define the epigenetic landscape of the Runx2-P1 promoter
Activating histone marks demarcate the boundaries of the active Runx2-P1 promoter
The redefined 0.9-kb promoter responds to differentiation queues
Acknowledgments
Funding
This work was supported by National Institutes of Health (P01 CA082834 and P01 AR48818 to G.S.S. R01 AR039588 to G.S.S. and J.B.L., and R37 DE012528 to J.B.L.).
We thank the University of Vermont Advanced Genome Technologies Core and the UMASS Medical School Deep Sequencing Core for assistance with analysis. Special thanks to Dr. L. Song and Dr. G. Crawford for advise on the DNase-seq methodology, and S. Tighe, T. Hunter, Dr. J. Dragon, Dr. J. Bond, Dr. E. Kittler, Dr. M. Zapp, and E. Carr.
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
Contributor Information
Phillip W. L. Tai, Email: phillip.tai@uvm.edu.
Hai Wu, Email: hai.wu@uvm.edu.
Jonathan A. R. Gordon, Email: jonathan.a.gordon@uvm.edu.
Troy W. Whitfield, Email: troy.whitfield@umassmed.edu.
A. Rasim Barutcu, Email: rasim.barutcu@umassmed.edu.
André J. van Wijnen, Email: vanwijnen.andre@mayo.edu.
Jane B. Lian, Email: jane.lian@uvm.edu.
Gary S. Stein, Email: gary.stein@uvm.edu.
Janet L. Stein, Email: janet.stein@uvm.edu.
References
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, Shilatifard A, Workman J, Zhang Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Banerjee C, Javed A, Choi JY, Green J, Rosen V, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Differential regulation of the two principal Runx2/Cbfa1 n-terminal isoforms in response to bone morphogenetic protein-2 during development of the osteoblast phenotype. Endocrinology. 2001;142:4026–4039. doi: 10.1210/endo.142.9.8367. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Guinney J, Crawford GE, Furey TS. F-Seq: a feature density estimator for high-throughput sequence tags. Bioinformatics. 2008;24:2537–2538. doi: 10.1093/bioinformatics/btn480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Pratap J, Javed A, Zaidi SK, Xing L, Balint E, Dalamangas S, Boyce B, van Wijnen AJ, Lian JB, Stein JL, Jones SN, Stein GS. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:8650–8655. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruzat F, Henriquez B, Villagra A, Hepp M, Lian JB, van Wijnen AJ, Stein JL, Imbalzano AN, Stein GS, Montecino M. SWI/SNF-independent nuclease hypersensitivity and an increased level of histone acetylation at the P1 promoter accompany active transcription of the bone master gene Runx2. Biochemistry. 2009;48:7287–7295. doi: 10.1021/bi9004792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean DD, Schwartz Z, Bonewald L, Muniz OE, Morales S, Gomez R, Brooks BP, Qiao M, Howell DS, Boyan BD. Matrix vesicles produced by osteoblast-like cells in culture become significantly enriched in proteoglycan-degrading metalloproteinases after addition of beta-glycerophosphate and ascorbic acid. Calcified Tissue International. 1994;54:399–408. doi: 10.1007/BF00305527. [DOI] [PubMed] [Google Scholar]
- Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nature Reviews Genetics. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissi H, Luc Q, Shakoori R, Chuva De Sousa Lopes S, Choi JY, Terry A, Hu M, Jones S, Neil JC, Lian JB, Stein JL, Van Wijnen AJ, Stein GS. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. Journal of Cellular Physiology. 2000;184:341–350. doi: 10.1002/1097-4652(200009)184:3<341::AID-JCP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Ducy P, Starbuck M, Priemel M, Shen J, Pinero G, Geoffroy V, Amling M, Karsenty G. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes & Development. 1999;13:1025–1036. doi: 10.1101/gad.13.8.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppenberger HM, Eppenberger M, Richterich R, Aebi H. The Ontogeny of Creatine Kinase Isozymes. Developmental Biology. 1964;10:1–16. doi: 10.1016/0012-1606(64)90002-8. [DOI] [PubMed] [Google Scholar]
- Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PV, Komm BS, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. Journal of Biological Chemistry. 2005;280:33132–33140. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- Harada H, Tagashira S, Fujiwara M, Ogawa S, Katsumata T, Yamaguchi A, Komori T, Nakatsuka M. Cbfa1 isoforms exert functional differences in osteoblast differentiation. Journal of Biological Chemistry. 1999;274:6972–6978. doi: 10.1074/jbc.274.11.6972. [DOI] [PubMed] [Google Scholar]
- Hassan MQ, Tare R, Lee SH, Mandeville M, Weiner B, Montecino M, van Wijnen AJ, Stein JL, Stein GS, Lian JB. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Molecular and Cellular Biology. 2007;27:3337–3352. doi: 10.1128/MCB.01544-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MQ, Tare RS, Lee SH, Mandeville M, Morasso MI, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB. BMP2 commitment to the osteogenic lineage involves activation of Runx2 by DLX3 and a homeodomain transcriptional network. Journal of Biological Chemistry. 2006;281:40515–40526. doi: 10.1074/jbc.M604508200. [DOI] [PubMed] [Google Scholar]
- Henriquez B, Hepp M, Merino P, Sepulveda H, van Wijnen AJ, Lian JB, Stein GS, Stein JL, Montecino M. C/EBPbeta binds the P1 promoter of the Runx2 gene and up-regulates Runx2 transcription in osteoblastic cells. Journal of Cellular Physiology. 2011;226:3043–3052. doi: 10.1002/jcp.22652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovhannisyan H, Zhang Y, Hassan MQ, Wu H, Glackin C, Lian JB, Stein JL, Montecino M, Stein GS, van Wijnen AJ. Genomic occupancy of HLH, AP1 and Runx2 motifs within a nuclease sensitive site of the Runx2 gene. Journal of Cellular Physiology. 2013;228:313–321. doi: 10.1002/jcp.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes AL, Jin Y, Rando OJ, Struhl K. A functional evolutionary approach to identify determinants of nucleosome positioning: a unifying model for establishing the genome-wide pattern. Molecular Cell. 2012;48:5–15. doi: 10.1016/j.molcel.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalajzic Z, Li H, Wang LP, Jiang X, Lamothe K, Adams DJ, Aguila HL, Rowe DW, Kalajzic I. Use of an alpha-smooth muscle actin GFP reporter to identify an osteoprogenitor population. Bone. 2008;43:501–510. doi: 10.1016/j.bone.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Lee MH, Kim YJ, Yoon WJ, Kim JI, Kim BG, Hwang YS, Wozney JM, Chi XZ, Bae SC, Choi KY, Cho JY, Choi JY, Ryoo HM. Dlx5 specifically regulates Runx2 type II expression by binding to homeodomain-response elements in the Runx2 distal promoter. Journal of Biological Chemistry. 2005;280:35579–35587. doi: 10.1074/jbc.M502267200. [DOI] [PubMed] [Google Scholar]
- Lengner CJ, Drissi H, Choi JY, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Activation of the bone-related Runx2/Cbfa1 promoter in mesenchymal condensations and developing chondrocytes of the axial skeleton. Mechanisms of Development. 2002;114:167–170. doi: 10.1016/s0925-4773(02)00050-3. [DOI] [PubMed] [Google Scholar]
- Lengner CJ, Hassan MQ, Serra RW, Lepper C, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Nkx3.2-mediated repression of Runx2 promotes chondrogenic differentiation. Journal of Biological Chemistry. 2005;280:15872–15879. doi: 10.1074/jbc.M411144200. [DOI] [PubMed] [Google Scholar]
- Li QH, Brown JB, Huang HY, Bickel PJ. Measuring Reproducibility of High-Throughput Experiments. Annals of Applied Statistics. 2011;5:1752–1779. [Google Scholar]
- Lian JB, Gordon JA, Stein GS. Redefining the activity of a bone-specific transcription factor: novel insights for understanding bone formation. Journal of Bone and Mineral Research. 2013;28:2060–2063. doi: 10.1002/jbmr.2076. [DOI] [PubMed] [Google Scholar]
- Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen AJ, Stein JL, Stein GS. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Critical Reviews in Eukaryotic Gene Expression. 2004;14:1–41. [PubMed] [Google Scholar]
- Liu JC, Lengner CJ, Gaur T, Lou Y, Hussain S, Jones MD, Borodic B, Colby JL, Steinman HA, van Wijnen AJ, Stein JL, Jones SN, Stein GS, Lian JB. Runx2 protein expression utilizes the Runx2 P1 promoter to establish osteoprogenitor cell number for normal bone formation. Journal of Biological Chemistry. 2011;286:30057–30070. doi: 10.1074/jbc.M111.241505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y, Javed A, Hussain S, Colby J, Frederick D, Pratap J, Xie R, Gaur T, van Wijnen AJ, Jones SN, Stein GS, Lian JB, Stein JL. A Runx2 threshold for the cleidocranial dysplasia phenotype. Human Molecular Genetics. 2009;18:556–568. doi: 10.1093/hmg/ddn383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova O, Kamberov E, Margolis B. Generation of deletion and point mutations with one primer in a single cloning step. BioTechniques. 2000;29:970–972. doi: 10.2144/00295bm08. [DOI] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, Owen MJ, Mertelsmann R, Zabel BU, Olsen BR. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Pekowska A, Benoukraf T, Zacarias-Cabeza J, Belhocine M, Koch F, Holota H, Imbert J, Andrau JC, Ferrier P, Spicuglia S. H3K4 tri-methylation provides an epigenetic signature of active enhancers. The EMBO Journal. 2011;30:4198–4210. doi: 10.1038/emboj.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratap J, Lian JB, Javed A, Barnes GL, van Wijnen AJ, Stein JL, Stein GS. Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone. Cancer Metastasis Reviews. 2006;25:589–600. doi: 10.1007/s10555-006-9032-0. [DOI] [PubMed] [Google Scholar]
- Quarles LD, Yohay DA, Lever LW, Caton R, Wenstrup RJ. Distinct proliferative and differentiated stages of murine MC3T3-E1 cells in culture: an in vitro model of osteoblast development. Journal of Bone and Mineral Research. 1992;7:683–692. doi: 10.1002/jbmr.5650070613. [DOI] [PubMed] [Google Scholar]
- Schroeder TM, Jensen ED, Westendorf JJ. Runx2: a master organizer of gene transcription in developing and maturing osteoblasts. Birth Defects Research Part C, Embryo Today: Reviews. 2005;75:213–225. doi: 10.1002/bdrc.20043. [DOI] [PubMed] [Google Scholar]
- Song I, Kim K, Kim JH, Lee YK, Jung HJ, Byun HO, Yoon G, Kim N. GATA4 negatively regulates osteoblast differentiation by downregulation of Runx2. BMB Reports. 2013 doi: 10.5483/BMBRep.2014.47.8.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harbor Protocols. 2010;2010 doi: 10.1101/pdb.prot5384. pdb prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein GS, Lian JB, van Wijnen AJ, Stein JL, Montecino M, Javed A, Zaidi SK, Young DW, Choi JY, Pockwinse SM. Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene. 2004;23:4315–4329. doi: 10.1038/sj.onc.1207676. [DOI] [PubMed] [Google Scholar]
- Tagle DA, Koop BF, Goodman M, Slightom JL, Hess DL, Jones RT. Embryonic epsilon and gamma globin genes of a prosimian primate (Galago crassicaudatus). Nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. Journal of Molecular Biology. 1988;203:439–455. doi: 10.1016/0022-2836(88)90011-3. [DOI] [PubMed] [Google Scholar]
- Tai PW, Fisher-Aylor KI, Himeda CL, Smith CL, Mackenzie AP, Helterline DL, Angello JC, Welikson RE, Wold BJ, Hauschka SD. Differentiation and fiber type-specific activity of a muscle creatine kinase intronic enhancer. Skeletal Muscle. 2011;1:25. doi: 10.1186/2044-5040-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai PW, Zaidi SK, Wu H, Grandy RA, Montecino M, van Wijnen AJ, Lian JB, Stein GS, Stein JL. The dynamic architectural and epigenetic nuclear landscape: developing the genomic almanac of biology and disease. Journal of Cellular Physiology. 2014;229:711–727. doi: 10.1002/jcp.24508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Christensen K, Chawla K, Xiao G, Krebsbach PH, Franceschi RT. Isolation and characterization of MC3T3-E1 preosteoblast subclones with distinct in vitro and in vivo differentiation/mineralization potential. Journal of Bone and Mineral Research. 1999;14:893–903. doi: 10.1359/jbmr.1999.14.6.893. [DOI] [PubMed] [Google Scholar]
- Williamson I, Hill RE, Bickmore WA. Enhancers: from developmental genetics to the genetics of common human disease. Developmental Cell. 2011;21:17–19. doi: 10.1016/j.devcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Wu H, Whitfield TW, Gordon JA, Dobson JR, Tai PW, van Wijnen AJ, Stein JL, Lian JB, Stein GS. In: Compston J, editor. Genomic occupancy of Runx2 combined with global expression profiling identifies novel mechanisms regulating osteoblastogenesis; American Society for Bone and Mineral Research Annual Meeting; 2013; Baltimore, Maryland: American Society for Bone and Mineral Research; 2013. p. S396. [Google Scholar]
- Wu H, Whitfield TW, Gordon JA, Dobson JR, Tai PW, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome Biology. 2014;15:R52. doi: 10.1186/gb-2014-15-3-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao ZS, Liu SG, Hinson TK, Quarles LD. Characterization of the upstream mouse Cbfa1/Runx2 promoter. Journal of Cellular Biochemistry. 2001;82:647–659. doi: 10.1002/jcb.1192. [DOI] [PubMed] [Google Scholar]
- Zambotti A, Makhluf H, Shen J, Ducy P. Characterization of an osteoblast-specific enhancer element in the CBFA1 gene. Journal of Biological Chemistry. 2002;277:41497–41506. doi: 10.1074/jbc.M204271200. [DOI] [PubMed] [Google Scholar]
- Zhang S, Xiao Z, Luo J, He N, Mahlios J, Quarles LD. Dose-dependent effects of Runx2 on bone development. Journal of Bone and Mineral Research. 2009a;24:1889–1904. doi: 10.1359/JBMR.090502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Hassan MQ, Xie RL, Hawse JR, Spelsberg TC, Montecino M, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Co-stimulation of the bone-related Runx2 P1 promoter in mesenchymal cells by SP1 and ETS transcription factors at polymorphic purine-rich DNA sequences (Y-repeats) Journal of Biological Chemistry. 2009b;284:3125–3135. doi: 10.1074/jbc.M807466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nature Genetics. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Six-way sequence alignment of the Runx2-P1 promoter. Select sequences of the human, mouse, cow, horse, dog, and elephant Runx2-P1 promoter region showing high-sequence similarity. Bases exhibiting high-sequence conservation across all species are in black and noted as asterisks (*) in the consensus sequence below each row. Bases that are semi-conserved are in gray. Gaps are designated as hyphens (−). The transcriptional start site (+1) is indicated by the black arrow. Currently known and experimentally proven control elements within the 0.9-kb minimal promoter are annotated and highlighted as colored texts of the associated transcription factors, and boxed to surround their respective motif sequences.