Abstract
Interphotoreceptor retinoid-binding protein (IRBP), which is critical to photoreceptor survival and function, is comprised of homologous tandem modules each ~300 amino acids, and contains 10 cysteines, possibly 8 as free thiols. Purification of IRBP has historically been difficult due to aggregation, denaturation and precipitation. Our observation that reducing agent 1,4-dithiothreitol dramatically prevents aggregation prompted investigation of possible functions for IRBP’s free thiols. Bovine IRBP (bIRBP) was purified from retina saline washes by a combination of concanavalin A, ion exchange and size exclusion chromatography. Antioxidant activity of the purified protein was measured by its ability to inhibit oxidation of 2,2'-azinobis [3-ethylbenzothiazoline-6-sulfonate] by metmyoglobin. Homology modeling predicted the relationship of the retinoid binding sites to cysteine residues. As a free radical scanvenger, bIRBP was more active than ovalbumin, thioredoxin, and vitamin E analog Trolox. Alkylation of free cysteines by N-ethylmaleimide inhibited bIRBP’s antioxidant activity, but not its ability to bind all-trans retinol. Structural modeling predicted that Cys 1051 is at the mouth of the module 4 hydrophobic ligand-binding site. Its free radical scavenging activity points to a new function for IRBP in defining the redox environment in the subretinal space.
Keywords: retina, retinoid binding protein, X-ray structure, crystallography, photoreceptors, interphotoreceptor-retinoid binding protein, interphotoreceptor matrix, visual cycle
1. Introduction
As the major soluble protein component of the interphotoreceptor matrix (IPM), interphotoreceptor retinoid-binding protein (IRBP) surrounds the individual rod and cone outer and inner segments occupying a critical interface between the retina and the RPE. Absence of IRBP results in photoreceptor degeneration in transgenic mice, and Asp1080Asn is associated with a form of recessive retinitis pigmentosa (den Hollander et al., 2009). Little is known about why IRBP is important to photoreceptor survival.
IRBP carries endogenous all-trans and 11-cis retinols, and 11-cis retinal in a light-dependent manner suggesting a role in facilitating the exchange of visual cycle retinoids between the rod, cone, RPE and Müller cell (Gonzalez-Fernandez and Ghosh, 2008; McBee et al., 2001). IRBP is known to promote outer segment release of all-trans retinol (Wu et al., 2007), and its delivery to the RPE. IRBP also enhances both the release of 11-cis retinal from the RPE (Carlson and Bok, 1992), and its return to the outer segments. Finally, an emerging concept is that IRBP may be important to protecting visual cycle retinoids from photodecomposition (Crouch et al., 1992; Parker et al., 2011; Tsin et al., 2013).
Visual cycle retinoids are vulnerable to photo-oxidation while crossing through the IPM. The primary alcoholic group and the double bonds in the side-chain or ring make 11-cis and all-trans retinol particularly susceptible to oxidative damage. Photo-degradation of vitamin A results in the formation of retinal, and a mixture of epoxy derivatives of vitamin A (Crank and Pardijanto, 1995; Failloux et al., 2003). The retina is particularly susceptible to oxidative stress, due to its high metabolic activity, oxygen tension, and concentration of readily oxidizable polyunsaturated fatty acids, and exposure to light (Chalam et al., 2011). Furthermore, although the RPE and Müller cells are known to have robust antioxidant systems (Bringmann et al., 2006; Cai et al., 2000; Plafker et al., 2012), the photoreceptors themselves and IPM are comparatively lacking in such protection. Although proteomic studies suggest that the IPM may contain neoruoprotective proteins including thioredoxin 5 (Hauck et al., 2005), immunohistochemical studies are needed to confirm the localization of these components to the IPM. It is therefore potentially significant that IRBP can protect visual cycle retinoids from photodecomposition. The mechanism of this protection is unknown.
Key to uncovering IRBP’s function will be elucidating the structure of the full-length protein. Bovine IRBP (bIRBP) has an overall elongated shape (Saari et al., 1985), and is composed of multiple homologous “modules” that may have diverse roles in the visual cycle. Functionally important structural changes appear to occur upon ligand binding (Adler et al., 1987). However, the preparation of pure bIRBP at concentrations required for crystallization trials proved to an insurmountable task owing to aggregation and precipitation of the protein, possibly through denaturation (Fong et al., 1984). Our early observation that 1,4-dithiothreitol (DTT) prevents aggregation permitted the purification of the pristine bIRBP, and prompted this investigation into the potential role of its free thiols.
2. Experimental Methods
This research was approved by the Buffalo Veterans Affairs Medical Center Research & Development Committee, and SUNY Buffalo and Upstate Medical University Biosafety Committee. All chemicals were of highest quality, and obtained from Sigma-Aldrich, unless otherwise specified.
2.1 Purification of Native Bovine IRBP
Our purification strategy is based methods previously established for the purfication of native bIRBP from bovine retina (Adler et al., 1990; Adler and Evans, 1983; Fong et al., 1986; Fong et al., 1984; Redmond et al., 1985; Saari et al., 1985). With the exception of (Saari et al., 1985) the use of a reducing agent during the purification of bIRBP is not described. As described below DTT was used throughout the purification.
IRBP is readily extracted from detached bovine retina by simple saline wash, which also affords an initial enrichment. Although some bIRBP remains adherent to the insoluble interphotoreceptor scaffold (Garlipp et al., 2012), at least 50% can be removed from the retina in this way. The extraction and purification were carried out at 4°C in the presence of 0.5 mM DTT, and protease inhibitors. Retinas were collected under dim red light by WL Lawson Co. (Lincoln, NE), and held at −80°C. For each purification, 200 retinas were thawed, and soaked for 15min in PBS (2 mM potassium phosphate, 7 mM sodium phosphate, 13.4 mM KCl, 136 mM NaCl, pH 7.4) containing 0.5 mM phenyl sulfonyl fluoride (PMSF), and centrifuged at 2,000×g for 5min. The supernatant was removed, and the retinas gently re-suspended in PBS, soaked for 10 min with gentle occasional agitation, and centrifuged for 10 min. at 3,000×g. The pooled washes were finally cleared at 20,000×g for 30min. A broad spectrum protease inhibitor cocktail was then added along with a 50% Concavalin A (ConA) Sepharose 4B slurry (GE Healthcare, Piscataway, NJ) in 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2 and 1 mM MnCl2. bIRBP was allowed to bind to the ConA for 4 hrs with continuous gentle agitation, and finally eluted overnight in 10% (w/v) methyl α-D-mannopyranoside, 50 mM Tris-HCl pH 7.5.
The ConA binding proteins were then loaded on a Q Sepharose high performance (QHP) column (1.6cm × 12cm) (GE Healthcare, Piscataway, NJ) equilibrated with 20 mM Tris-HCl pH 7.5, 50 mM NaCl on a Äkta Fast Protein Liquid Chromatography system. bIRBP was eluted at 600 mM NaCl with a linear gradient. The protein was concentrated to 5 mL using an Amicon centrifugal filter, and subjected to size exclusion using Sephacryl S-300HR in a 2.6cm × 100 cm column on a ÄKTA FPLC chromatography system (GE Healthcare, Piscataway, NJ). The running buffer was 20 mM Tris-HCl pH 7.5, and 100 mM NaCl. The pooled S300 bIRBP fractions were subjected to second QHP column (1.6cm ×12cm) and eluted with a NaCl gradient as before. The bIRBP containing fractions were pooled, and the concentration of the purified bIRBP determined by absorbance spectroscopy, and amino acid analysis. Purity was determined by SDS-PAGE analysis. The yield typically was 40 to 60mg of 98% pure bIRBP from 200 retinas.
2.2. Crystallization
Crystallization is generally accepted as a validation of integrity of the folded state and monodispersity (Ferre-D'Amare and Burley, 1997; Marmorstein, 1998; Weber, 1997). Bovine IRBP was exchanged into 50 mM Tris-HCl pH 7.5, 100 mM NaCl, and 4 mM DTT using YM-50 centricons and concentrated to 10mg/mL (0.07 mM). 0.5 mM oleic acid was then added, and the protein solution filtered to remove any particulate matter. Crystal screening was performed using various commercial screens from Hampton Research (San Diego, CA): Crystal Screen 1 and 2; Jena Biosciences (Jena Germany): JBS Membrane Screen 1–3; and Qiagen/Nextal (Montreal, Canada): Classics, SM4, SM5, Mb Class II Suite. The screening was performed at 23°C and 4°C in sitting drop plates (Hampton Research). Initial crystals were obtained from 0.1M MgCl2, 0.1M NaCl, 0.1M sodium citrate pH 5.5, 12% polyethylene glycol (PEG) 4000 at 23°C in a 1:1 protein:cocktail ratio using sitting drop vapor diffusion. This is a significant departure for the conditions used to grow crystal of recombinant IRBP (Loew and Gonzalez-Fernandez, 2002). Optimization of the initial growth conditions resulted in the procedure for routine growth of single crystals from a wide range of PEG concentrations and molecular weights. Single crystals were grown from 12mg/ml solutions of bIRBP in 0.1M MgCl2, 0.1M NaCl, 0.1M tri-Na citrate pH 5.5 to which the reservoir solution, 10 to 18% PEG 4K-35K in the same buffer cocktail, was added at 3:1 protein: reservoir volume ratio and vapor diffused in sitting drops against reservoirs at 23°C. The diffraction data presented here were gathered on a crystal grown against a reservoir of 18% PEG 35K.
Initial characterization of the crystals, and determination of space group and cell dimensions were performed at the Cornell High Energy Synchrotron Source (Ithaca, New York) by collecting low resolution (~5 Å) diffraction data sets at ambient temperature. Data processing routines MOSFLM (Powell, 1999), and HKL2000 (Otwinowski and Minor, 1995.) were used for this purpose.
2.3. Ligand-Binding Assays by Fluorescence Spectroscopy
The binding of all-trans retinol to bIRBP was characterized in titrations using a DM 45 scanning spectrofluorimeter (On-Line Instrument Systems, Inc., Bogart, GA) (Ward, 1985). Titrations monitoring enhancement of retinol fluorescence, quenching of the intrinsic protein fluorescence, and energy transfer were carried out as previously described (Baer et al., 1998). Enhancement of retinol fluorescence was followed by monitoring the increase in retinol fluorescence (excitation, 330 nm; emission, 480 nm). Assays following quenching of protein fluorescence used excitation, and emission wavelengths of 280 nm and 340 nm, respectively. In these experiments the inner filter effect was accounted for by graphical correction (Mertens and Kagi, 1979). The dissociation constant (Kd) and number of binding sites (N) was determined by nonlinear least square fit to a binding equation that assumes a single type of noninteracting site(s) as previously described (Baer et al., 1994).
2.4. Chemical Modification of bIRBP’s Cysteine Residues by Titration with N-ethylmaleimide (NEM)
NEM is widely used to probe the functional roles of protein free thiols (Carne, 1994; Chouchani et al., 2011). For example, it inhibits all cysteine peptidases, by alkylating the active site-thiol. Accessible thiols were modified by incubating 20 µM purified bIRBP with NEM at cysteine:NEM ratios of 10:10, 10:5, 10:2, 10:1 and 10:0 (bIRBP has 10 cysteines per polypeptide) in 10 mM sodium phosphate, pH 7.4, 120 mM NaCl, 3 mM KCl for 3 hrs at 22°C. Aliquoted NEM modified bIRBP was typically used in parallel fluorescence based all-trans retinol binding titrations, ABTS and retinol protection assays, and LC/MS/MS analysis. Fourier transform ion cyclotron resonance spectroscopy was used to identify NEM modified cysteines using a Bruker Apex 9.4T instrument (SUNY Upstate, NY). Following NEM alkylation, 4 mM DTT was used to quench unreacted residues. The protein was finally denatured in 8 M urea, and incubated with 40 mM iodoacetamide (IAA) overnight at 20°C in the dark to label unmodified cysteines prior to tryptic digestion.
2.5. LC/MS/MS of bIRBP Following Modification with NEM
Protein samples from three separate NEM-titration experiments at five cysteine:NEM molar ratios (see above) were digested with trypsin (Thermo Fisher Sci., Waltham, MA), and then analyzed by LC/MS/MS, liquid chromatography in line with tandem mass spectrometry. The molar ratio of 10:1 (and not 10:2) was used in one of the three experiments. An LC/MS/MS analysis was performed using a Surveyor MS Plus high-performance liquid chromatography (HPLC) system (with a flow splitter) in line with a LTQ Orbitrap XL mass spectrometer with a nanospray ionization source (Thermo Fisher Sci., Waltham, MA). Tryptic peptides (5µL) were separated by reverse-phase HPLC on a PicoFrit BioBasic C18 column (0.075 × 100 mm, New Objectives, Woburn, MA) at 0.3 µL/min flow rate. Water and acetonitrile with 0.1% formic acid were used as solvents A and B, respectively. The peptides were eluted by a linear gradient of solvent B (0–9 min 0% B, 9–20min 0–10% B, 20–80min 10–25% B, 80–90min 25–30% B, 90–110min 30–60% B, 110–120min 60–80% B). The eluted peptides were analyzed by tandem MS in data-dependent mode with dynamic exclusion. The peptides and the corresponding fragment ions were analyzed in the Orbitrap and the Linear Trap analyzer respectively. The database search was performed using SEQUEST algorithm within the mass informatics platform Proteome Discoverer 1.3 (version 1.3.0.339, Thermo Fisher Sci., Waltham, MA) and the non-redundant protein sequence database (nr.fasta, May 20, 2011) of the National Center for Biotechnology Information (NCBI). The validation of the search results was performed using Percolator algorithm within the same mass informatics platform. The spectra of identical peptides (precursor mass tolerance 10 ppm, retention time difference <0.75 min) were grouped. The following SEQUEST search parameters were used: digestion enzyme – trypsin, two missed cleavage sites were allowed, the precursor mass (mono-isotopic) tolerance – 10 ppm, fragment ion mass (mono-isotopic) tolerance – 0.8 Da, dynamic modifications – modification of Cys residues with N-ethylmaleimide or IAA, oxidation of Met residues, and phosphorylation of Ser, Thr, and Tyr residues. The database search was restricted to the proteins of bovine origin (56001 entries) and the corresponding tryptic peptides with the molecular masses of 500–5000. A decoy database with the reversed protein sequences was used for the validation of the search results; the validation was based on False Discovery Rate (FDR) or Posterior Error Probability (PEP). The peptide identification criteria: the peptide identification parameters exceeding the threshold value corresponding to FDR (q-Value) of 0.01; the additional requirement for peptides with modified cysteine residues was PEP error probability of ≤0,001. The protein identification criteria: identification of at least three peptides unique to a given protein. High confidence (P<0.01) cysteine-containing NEM-and IAA-modified peptides were tabulated.
2.6. ABTS assay
We took advantage of the green color of the stable free cationic radical of 2,2'-azino-bis[3-ethylbenzothiazoline-6-sulfonate] (ABTS•+) to assess IRBP’s antioxidant activity. The assay has been useful to characterize protein radical scavenging activity (Akerstrom et al., 2007). Reagents were obtained from Cayman Chemical Company (Ann Arbor, Michigan). Ferryl myoglobin radical formed from metmyoglobin and H2O2 oxidizes ABTS to the radical cation, ABTS•+. ABTS•+ is a green soluble chromogen is monitored at 750nm. Antioxidants act by scavenging the ABTS•+ radical cation (Miller and Rice-Evans, 1997). Selection of the appropriate incubation time can be important to the performance of the assay (Wang et al., 2004). In our system, 5 min provided an ideal interval. The radical scavenging activity of bIRBP was compared to buffer alone, synthetic water-soluble vitamin E analog Trolox™(6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid), thioredoxin, ovalbumin and NEM-modified bIRBP.
3. Results
3.1. Stability and Integrity of Purified bIRBP
During the course of crystallization trials, we found that bIRBP typically aggregates above 10 mg/mL despite the presence of octyl-β-D-glucopyranoside (BOG). However, the presence of 4 mM DTT dramatically prevented aggregation. This allowed us to readily concentrate the protein sufficient for crystallization. Supplementary Materials Table 1 summarizes the four-step purification with yield and purity at each step. The method, which was modified from previously described protocols (Adler et al., 1990; Fong et al., 1986), yielded approximately 40% of the available bIRBP in the bovine retina.
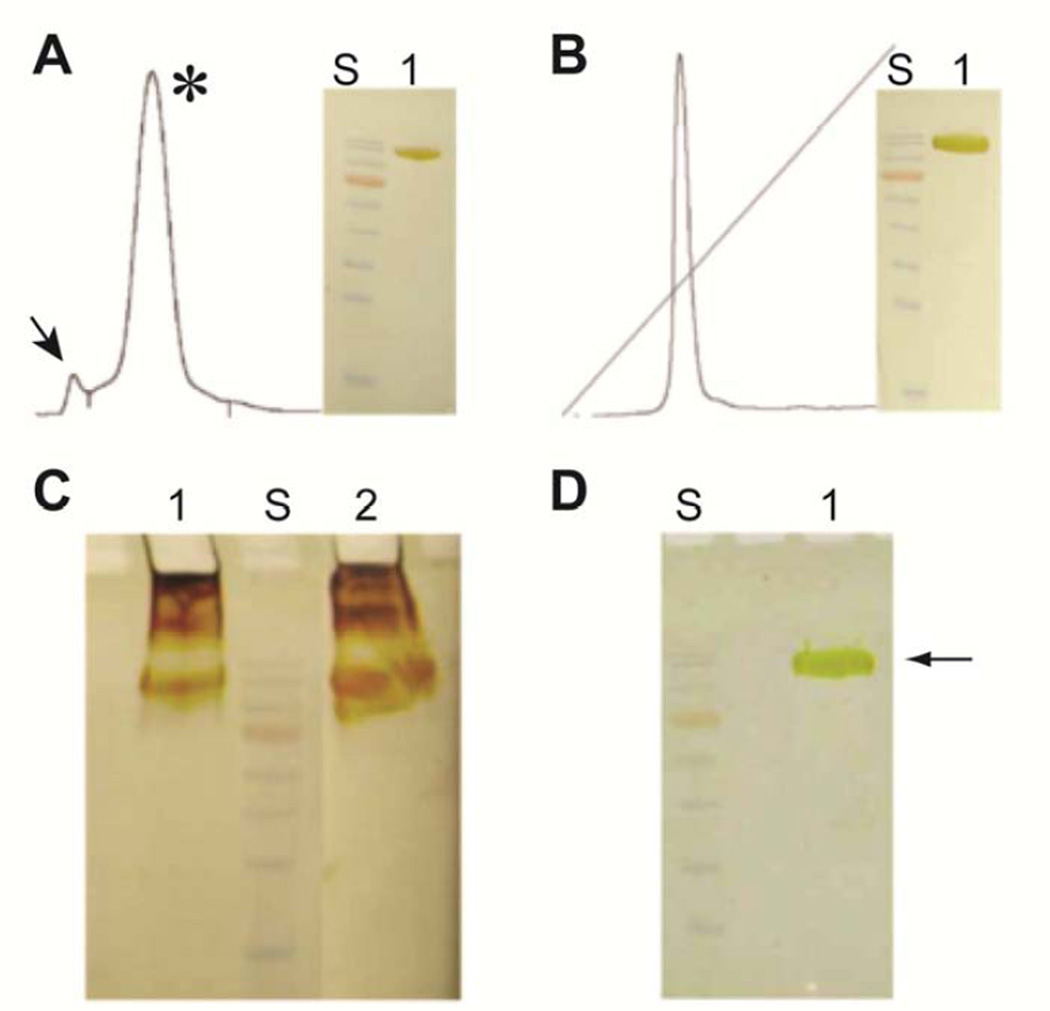
Chromatograms from steps 3 and 4 of the purification process, and associated gel electrophoresis results are shown in Fig. 1. At the end of step 4, about 60 mg of bIRBP was purified, essentially to homogeneity (Supplementary Materials Table 1). Fig. 1A is the S300 size exclusion elution profile showing a main peak of monodispersed bIRBP (asterisk), and leading minor peak representing aggregated bIRBP (arrow). The final ion exchange purification of bIRBP from the major S300 peak is shown in Fig. 1B. Without DTT, higher oligomeric states formed during concentration despite presence of BOG (Fig. 1C). Figure 1D shows that the presence of 4 mM DTT prevented oligomerization.
Fig. 1.
Aggregation of bovine IRBP is effectively prevented with reducing agent dithiothreitol (DTT). Aggregation typically occurs above 10 mg/ml even in the presence of detergents such as BOG. (A) S300 size exclusion elution profile showing a main peak of monodispersed bIRBP (asterisk), and leading minor peak representing aggregated bIRBP (arrow). (B) Final ion exchange purification of bIRBP from the major S300 peak. (C) Without DTT, higher oligomeric states form during concentration despite presence of BOG (1, precipitant; 2, supernatant). (D) The presence of 4 mM DTT prevented oligomerization. S, molecular weight standards (170, 130, 100, 72 (orange), 55, 40, 35, 25 kDa); Silver stained SDS-10%PAGE.
In sharp contrast to previous reports of bIRBP instability at high concentrations (Fong et al., 1984), the current protocol yielded protein that maintained its integrity, high solubility and activity for at least five weeks at 4°C, and could be concentrated beyond 10 mg/mL without having to use any detergents. In fact, although freshly purified preparations of bIRBP in the presence of 0.2% BOG were initially soluble and showed a single band in gel electrophoresis, the protein was highly unstable and precipitated on standing for 48 hr at 4°C at DTT concentrations of less than 0.5 mM, as evidenced by gel electrophoresis shown in Fig. 1C. The enhanced protein stability and sustained solubility are, therefore, directly attributable to the presence of molar excess of DTT in solution. Furthermore, the absence of BOG, or any other detergent was probably a stabilizing factor as well, since for soluble proteins, detergents could add to instability promoting gradual unfolding. Thus, under the reducing conditions of the purification experiments, highly pure bIRBP maintained its naturally folded state in a stable tertiary structure, and, therefore, was fully soluble and functionally active for a prolonged period of time.
The ability of the bIRBP to form crystals provides an indication the integrity of the folded purified, and concentrated polypeptide. Supplementary Materials Fig. 1A,B provides photomicrographs of crystals of bIRBP that grew from experiments conducted with PEG 35K as the precipitant, although the same crystals also grew from PEGs with lower molecular weights. A typical diffraction image gathered during initial characterization is shown in Supplementary Materials Fig. 1C. However, upon cryo-cooling with liquid nitrogen using a large number of cryo-protectants (combinations of various PEG solutions, 2-methyl-2, 4-pentanediol, glycerol, ethylene glycol etc), the crystals showed signs of high mosaicity and loss of long-range order. The resolution of the current crystals is roughly 5 Å. Gathering of diffraction data for determination of the structure awaits identification of the optimal cryo-cooling conditions and better diffraction-quality crystals. The space group is P43212 (or P41212), and cell dimensions of a=b=159.1Å, c=116.7Å, α=β=γ=90°, which is consistent with the bIRBP mass of 145 kDa and a specific volume of 2.6Å3/Da that is within the range for protein crystals. These observations indicate that a reducing environment is key to supporting the integrity, and monodispersity of purified bIRBP, and is allowing ongoing work toward determining the X-ray crystal structure of bIRBP.
2.3. Antioxidant Activity, Detection of NEM Modification by LC/MS/MS and Effect of NEM Alkylation
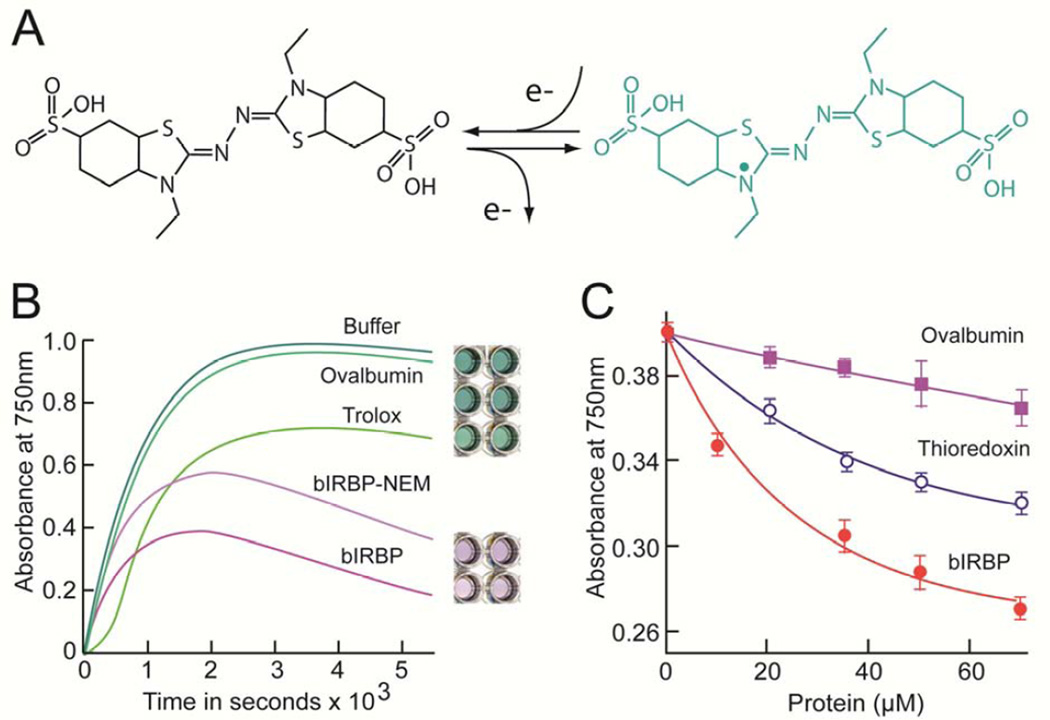
The antioxidant activity of IRBP was assessed by the ferryl myoglobin/ABTS assay (Fig. 2). This assay has proven useful in evaluating the radical scavenging activity of a variety of proteins including members of the lipocalin family (Akerstrom et al., 2007). The principle of the assay is that the ferryl myoglobin radical formed from metmyoglobin and H2O2 oxidizes ABTS to the radical cation, ABTS•+ (Fig. 2A). ABTS•+ is a green soluble chromogen that can be monitored spectrophotometrically at 750nm. Antioxidants act by scavenging the ABTS•+ radical cation, and not by inhibiting its formation through reduction of ferryl myoglobin, or its reaction with H2O2 (Miller and Rice-Evans, 1997). At the conclusion of incubations in the presence of bIRBP, the green color is often replaced by a light purple color that may represent a tyrosine-ABTS conjugation (Akerstrom et al., 2007). As the incubation time can be important to the performance of the assay (Wang et al., 2004), we conducted the time course study shown in Fig. 2B. Here, ABTS•+ formation in the presence of buffer alone, synthetic water-soluble vitamin E analog Trolox™, thioredoxin, ovalbumin and NEM-modified bIRBP were followed from 0 to 5,000 sec. Five min, which corresponds to the rising portion of the reaction curves, was selected to monitor assays at different concentrations of the test antioxidant (Fig. 2C). This panel compares the formation of ABTS•+ at 5 min for 0 to 60 µM ovalbumin, thioredoxin and bIRBP. At all concentrations tested, the free radical scavenging activity of bIRBP was greater than that of ovalbumin, or thioredoxin.
Fig. 2.
IRBP antioxidant activity assessed by the ferryl myoglobin/ABTS assay. (A) The assay monitors the ability to inhibit oxidation of 2,2'-azinobis [3-ethylbenzothiazoline-6-sulfonate] (ABTS; colorless) to the cationic radical ABTS•+ (green) by metmyoglobin. (B) Kinetics of ABTS•+ formation in the presence of buffer, ovalbumin, Trolox, bIRBP and NEM-modified bIRBP. Representative reaction wells at the conclusion of the incubation are shown from top to bottom for each condition respectively. (C) Dependence of ABTS•+ concentration on the concentration of ovalbumin, Trolox, and bIRBP. 750 nm absorbance measurements were taken at 5 min. Error bars, ± SE (N=3). Curves represent fits to first order exponential decay.
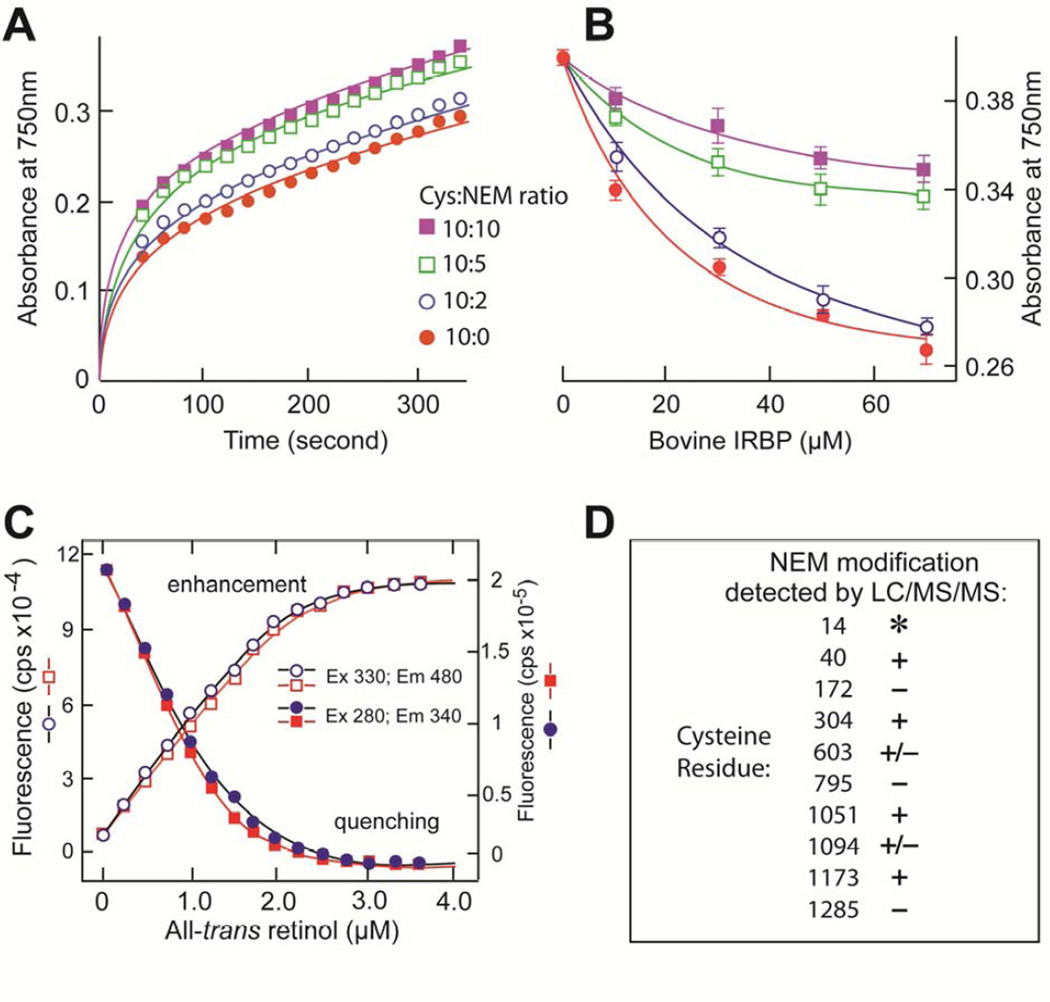
The effect of NEM on bIRBP’s antioxidant, and all-trans retinol binding activities are shown in Fig. 3A–C. In these experiments, bIRBP was treated with NEM at cysteine:NEM ratios 10:10, 10:5, 10:2 and 10:0. No discernable molar ratio dependence in NEM-labeling was evident in any of the three separate experiments. The combined result on alkylation of cysteine residues as characterized by LC/MS/MS in three separate experiments is shown in Fig. 3D. NEM inhibited the ability of IRBP to prevent the formation of ABTS•+ (Fig. 3A). The effect can be partly overcome by increasing protein concentration (Fig. 3B). To address whether the reduced antioxidant activity could be a consequence of disruption of IRBP’s putative ligand-binding site, we compared the effect of NEM modification on the binding of all-trans retinol to bIRBP and bIRBP-NEM at a cysteine:NEM ratio of 10:5. Titrations monitoring retinol fluorescence enhancement and quenching of intrinsic protein fluorescence by bound retinol did not detect a difference in retinol binding (Fig. 3C). The data indicates that cysteine residues 40, 304, 1051, and 1173 are candidates whose thiol groups could contribute to bIRBP’s antioxidant activity (Fig 3D). Cys 603 and Cys 1094 are also possible candidates since they were modified in one of the three experiments. Cys 14 resides in a trypsin peptide that is too short to be detected by our LC/MS/MS experiment, and was therefore not identified in any analysis; hence its role as a participant in antioxidant activity cannot be ascertained. The remaining cysteines could have a structural role as disulfides.
Fig. 3.
Effect of N-ethylmaleimide (NEM) on bIRBP’s antioxidant and all-trans retinol binding activity, and identification of modified cysteine residues. (A,B) Time and concentration dependence respectively of ABTS•+ formation in the presence of IRBP, and NEM-modified bIRBP at cysteine:NEM molar ratios 10:10, 10:5, 10:2, 10:0. (C) Binding of all-trans retinol to bIRBP (red), and bIRBP-NEM (blue) (cysteine:NEM molar ratio = 10:5). Titrations monitored retinol fluorescence enhancement (excitation, 330 nm; emission, 480 nm), or quenching of intrinsic protein fluorescence by bound retinol (excitation, 280 nm; emission, 340 nm). (D) The cysteine residues modified with NEM were identified by LC/MS/MS. In three experiments, bIRBP was incubated with NEM at 10:10, 10:5 and 10:2 cysteine:NEM molar ratios. The total number of times an NEM modification was identified in these incubations was tabulated for each of IRBP’s cysteines. Cysteine residues listed as “+” were NEM-modified in all three experiments in at least one cysteine:NEM molar ratio. Those cysteine residues listed as “+/−” were NEM-modified in at least one of the three experiments in at least one of the NEM incubations. Cysteine residues listed as “−” were not NEM-modified at any cysteine:NEM molar ratio in any of the three experiments. Asterisk (*) indicates that a peptide containing cysteine 14 was not detected in any tryptic digest of any experiment.
4. Discussion
Stability and solubility results presented here for purified bIRBP demonstrate that under a reducing environment the protein maintains its structural and functional integrity for prolonged periods. In sharp contrast with the observation made by others (Fong et al., 1984), as well as our own earlier experience, in the presence of reducing agents bIRBP can be significantly concentrated without losses through precipitation. Although the presence of BOG affords a limited enhancement of this solubility, without reducing agents bIRBP slowly denatured and precipitated out of the solution even in the presence of such detergents (Fong et al., 1984).
The use of a thiol-based reducing agent in prevention of denaturation is significant since bIRBP, like many other IRBPs, contains 10 cysteines. Homology modeling of each module of bIRBP with the Xenopus module 2 structure (unpublished results) suggests that a large number of these residues, if not all, do not participate in disulfide formation and remain as free –SH groups exposed to the solvent. This is consistent with experimental data that eight of bIRBP’s thiols are free (Fedorovich et al., 2000). Our data suggests that bIRBP molecules aggregate, unfold, and denature irreversibly by cross-linking through their free sulfhydryl groups, a process that we have demonstrated is easily prevented by the addition of a thiol-based reducing agent, such as DTT (Fig. 1). Tris(2-carboxyethyl) phosphine (TCEP), a non-thiol based reducing agent, fails to provide similar protection (data not illustrated). Although the mechanism for this difference is not clear, it is generally recognized that selection of the optimum reductant is often application specific (Getz et al., 1999). TCEP has advantages of being orderless and active over a wider pH range. However, its nucleophilic phosphorus is more sterically crowded than DTT’s thiolate group, lowering the reactivity of the phosphine with protein disulfides compared to DTT in some cases (Cline et al., 2004). Although detergents such as BOG may initially help by solubilizing partially unfolded molecules, they ultimately aid in the unfolding and denaturation process. Interestingly, the Asp1080Asn mutation associated with a form of recessive retinitis pigmentosa results in the formation of insoluble high molecular weight complexes via disulfide bonds (Li et al., 2013).
What could be the function of IRBP’s free thiols? An earlier study noted that IRBP was able to protect the isomeric and oxidative state of all-trans retinol (Crouch et al., 1992). We wondered whether IRBP could function as a thiol-dependent antioxidant. We found that bIRBP is more active than thioredoxin, vitamin E analog trolox, and ovalbumin in inhibiting oxidation of ABTS, and that NEM inhibits the antioxidant capacity of bIRBP. However, a complete inhibition of the antioxidant effect was not achieved by equimolar titration, suggesting that more than one antioxidant mechanism may be present, or alternatively, not all of the functional cysteines may have been accessible to and/or modified by NEM. Denaturation of NEM-modified bIRBP followed by labeling with IAA identified additional cysteines not labeled by NEM alone. Although these residues could have been involved in disulfide bonds, the observation suggests that not all of the active thiols are accessible to NEM under the conditions used. Resolution of this question awaits defining the structure of the full-length bIRBP.
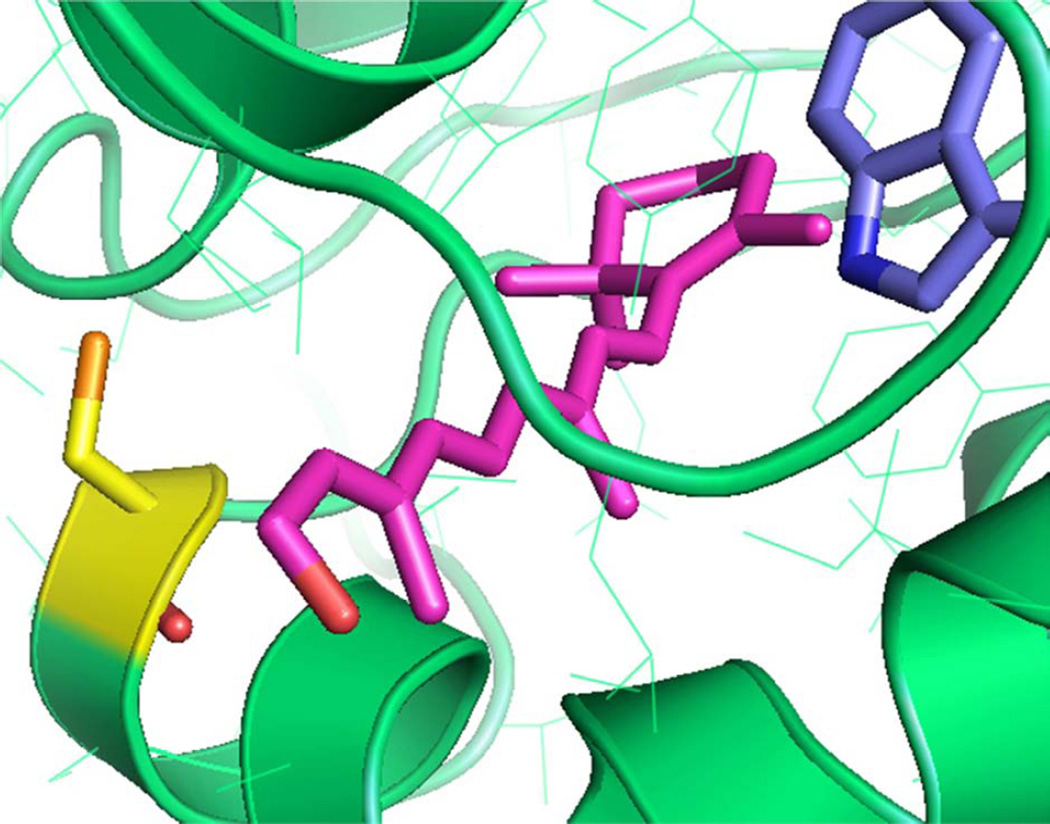
As the cysteine distribution is not uniform over the bIRBP’s four modules (modules 1 through 4 have 4, 1, 1 and 4 cysteines, respectively), the modules could have different reducing powers, and therefore be functionally distinct. Interestingly, the number of binding sites is determined to be less than 4 in the 4-module bIRBP, in agreement with previous reports (Chen et al., 1993). This data suggests that the retinol-binding site may be restricted to one, or two of the four modules, or intra-module sites defined by multiple modules, consistent with the notion that not all four modules are functionally equivalent. The idea that different homologous modules of a full-length IRBP polypeptide chain have different roles to play, and that all modules, although similar in the overall fold, are different in the atomistic details with regard to ligand binding or any other function, is supported by homology-modeling of the four-module Xenopus IRBP (Gonzalez-Fernandez et al., 2007; Nickerson et al., 1998). Perhaps some modules are tailored for antioxidant function, while others are masterly in retinoid or fatty acid capture and/or transport. An alignment highlighting the cysteines of bovine, human, Xenopus and zebrafish IRBPs is shown in Supplementary Fig. 2. The alignment shows that cysteines C40, C1094 and C1173 are conserved between each of these species. In fact, C40 is conserved through to lamprey IRBP (http://genomewiki.ucsc.edu/index.php/Opsin_evolution:_RBP3_(IRBP)). Three others (C14, C304 and C795) are conserved in human, bovine, zebrafish, but not Xenopus IRBP. C172 is conserved only between bovine and human IRBPs. C630, C1051 and C1285 are unique to bIRBP. The cysteines are not conserved with carboxy terminal processing proteases (CtpAs), a related protein family with proteolytic activity (Gonzalez-Fernandez, 2003; Gonzalez-Fernandez and Ghosh, 2008; Loew and Gonzalez-Fernandez, 2002) (for an alignment of CptAs from E. coli (tail-specific protease), Synechococcus, Bartonells bacilliformis and spinach see (Baer et al., 1998)). These observations suggest that IRBP’s cysteines have different functions, and that not all of these functions are conserved through to the CtpAs. Structural modeling suggests that module 4 of bIRBP is distinctive in that Cys 1051 sits at the mouth of the hydrophobic ligand-binding site (Fig. 5). A cysteine-mediated antioxidative retinol protection mechanism is expected to have these structural features.
While the quaternary association of the modules possibly defines the ligand-binding site, other regions of the polypeptide chain could serve to protect the fully reduced 11-cis and all-trans retinol, and fatty acids, particularly docosahexaenoic acid, from reactive oxidative species. The free –SH groups of bIRBP could then work as reducing agents, in a manner similar to peroxiredoxins (Wood et al., 2003) becoming themselves oxidized. In an oxidizing environment, these free –SH groups can act as antioxidants by reducing reactive oxidative species, thereby being oxidized themselves to sulfenic acid (–SOH). Unless they are over oxidized to sulfinic acid (–SO2H), other free thiol groups or other thiol-based reducing enzymes, such as thioredoxin or glutathione reductase, probably reduce them back to free –SH and the cycle continues. The reducing power of the then free –SH groups cannot be restored if they are over oxidized, in which case bIRBP molecules with over-oxidized cysteines exit the retinoid cycle being slated for degradation. This also explains the necessity of having a large polypeptide chain for transporting a small ligand, such as retinol, and the rapid turnover of IRBP in the eye (Cunningham and Gonzalez-Fernandez, 2000; Cunningham et al., 1999).
IRBP may have an important role not only in transporting, but also in protecting retinoids in the rod and cone photoreceptors (Gonzalez-Fernandez, 2003). However, the mechanism(s) remain a mystery, owing to the lack of structural insights into the nature of binding pockets, and correlation of the functional properties. The structure of the second Xenopus module (Loew and Gonzalez-Fernandez, 2002) has provided some clues to that effect. Nonetheless, since a functional full-length mammalian IRBP molecule contains four homologous modules, the structure of a mammalian protein in the ligand-bound form is likely to yield a more comprehensive understanding of the structure-function relationships. Our effort to investigate the structure of native bIRBP in complex with fatty acids, and retinoids is a step towards achieving that goal.
Little is known regarding how the redox state of the interphotoreceptor matrix is normally maintained, or altered in disease. Immunohistochemical studies have localized catalase and glutathione peroxidase to the photoreceptor inner segments and RPE rather than to the subretinal space (Atalla et al., 1990). Proteomic studies suggest that neuroprotective proteins may be present in porcine interphotoreceptor matrix extracts, including PEA-15, peroxiredoxin 5, alpha-B-crystallin, macrophage migration inhibitory factor, 78 kDa glucose-regulated protein (GRP78), protein disulfide-isomerase, and PEP-19 (Hauck et al., 2005). Of these, immunohistochemical evidence for a subretinal localization has been obtained only for GRP78. Thiobarbituric acid reacting substances have been found in patient subretinal fluid suggesting a role of lipid peroxidation in the pathogenesis of retinal detachment (Romero et al., 1998).
Further studies should evaluate the role of IRBP not only as a retinoid transport protein in the vitamin A cycle, but as a thiol-dependent antioxidant in the subretinal space. Given that IRBP is the major soluble protein component of the interphotoreceptor matrix, an antioxidant activity may have a significant protective effect in the retina. Furthermore, the results presented here may point to a role of this function in other processes beyond the vitamin A cycle such as protecting the outer retina from lipid peroxidation (Anderson et al., 1984; Catala, 2011). Ongoing studies are beginning to address this possibility in a physiological context (Sung et al., 2012).
Supplementary Material
Fig. 4.
Homology modeling of bIRBP module 4 with a molecule of all-trans retinol (purple) docked into the putative ligand-binding site (tryptophan residue is in blue). Structural modeling shown here predicts that cysteine 1051 (yellow) is located at the mouth of the hydrophobic ligand-binding site.
DTT prevents aggregation of IRBP allowing its crystallization.
IRBP is a thiol-dependent free radical scavenger.
IRBP may define the redox environment in the subretinal space.
Acknowledgments
We thank Jennifer Griswold and Molly Sprada for expert assistance in protein purification; Beth Lassman for her participation in the early phases of this work. Joshua Mimun and Brandi Betts-Obregon for assistance in performing the HPLC studies. The LC/MS/MS analysis was performed at the Proteomics Core Facility at SUNY Upstate Medical University. The contents of this report do not represent the views of the Department of Veterans Affairs or the United States Government.
Grant Support: This work was supported by NIH R01 EY09412 (D.G., F.G.-F), Merit Review Award I01BX007080 from the Biomedical Laboratory Research & Development Service of the Veterans Affairs Office of Research and Development (F.G.-F.), R24 EY 016662 core instrumentation grant; UTSA-Center for Research and Training in the Sciences (CRTS), the National Center for Research Resources (5 G12RR013646-12) and the National Institute on Minority Health and Health Disparities (G12MD007591) from the NIH; Unrestricted Research Grant from Research to Prevent Blindness to the Department of Ophthalmology at State University of New York at Buffalo.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Appendix A. Supplementary data
Supplementary Table 1. Summary of the purification of IRBP from bovine retina.
Supplementary Fig. 1. (A,B) Photomicrographs of bIRBP crystals formed under the reducing environment described. The crystals typically ranged in size from 100 to 20 µm. (C) X-ray diffraction pattern with ~5 Å resolution recorded from a single bIRBP crystal. The formation of diffractable crystals such as these, indicates that the purified protein is correctly folded, and exists in a monodisperse state. This advance is allowing studies to move forward toward solving the X-ray crystal structure of IRBP.
Supplementary Fig. 2. Multisequence alignment of bovine, human, Xenopus and zebrafish IRBPs. Three cysteines (C40, C1094 and C1173) are conserved between each species; C14, C304 and C795 are conserved in each of these species except Xenopus. C172 is conserved only between bovine and human IRBPs. C630, C1051 and C1285 are unique to bIRBP. The cysteines are not conserved with CtpAs. Symbols: “*”, identical residue; “:”, conserved substitutions, "." semi-conserved substitutions.
References
- Adler AJ, Chader GJ, Wiggert B. Purification and assay of interphotoreceptor retinoid-binding protein from the eye. Methods Enzymol. 1990;189:213–223. doi: 10.1016/0076-6879(90)89292-p. [DOI] [PubMed] [Google Scholar]
- Adler AJ, Evans CD. Rapid isolation of bovine interphotoreceptor retinol-binding protein. Biochim. Biophys. Acta. 1983;761:217–222. doi: 10.1016/0304-4165(83)90068-5. [DOI] [PubMed] [Google Scholar]
- Adler AJ, Stafford WF, III, Slayter HS. Size and shape of bovine interphotoreceptor retinoid-binding protein by electron microscopy and hydrodynamic analysis. J. Biol. Chem. 1987;262:13198–13203. [PubMed] [Google Scholar]
- Akerstrom B, Maghzal GJ, Winterbourn CC, Kettle AJ. The lipocalin alpha1-microglobulin has radical scavenging activity. J. Biol. Chem. 2007;282:31493–31503. doi: 10.1074/jbc.M702624200. [DOI] [PubMed] [Google Scholar]
- Anderson RE, Rapp LM, Wiegand RD. Lipid peroxidation and retinal degeneration. Curr.Eye Res. 1984;3:223–227. doi: 10.3109/02713688408997203. [DOI] [PubMed] [Google Scholar]
- Atalla LR, Sevanian A, Rao NA. Immunohistochemical localization of peroxidative enzymes in ocular tissue. CLAO J. 1990;16:S30–33. [PubMed] [Google Scholar]
- Baer CA, Kittredge KL, Klinger AL, Briercheck DM, Braiman MS, Gonzalez-Fernandez F. Expression and characterization of the fourth repeat of Xenopus interphotoreceptor retinoid-binding protein in E. coli. Curr. Eye Res. 1994;13:391–400. doi: 10.3109/02713689408999866. [DOI] [PubMed] [Google Scholar]
- Baer CA, Retief JD, Van Niel E, Braiman MS, Gonzalez-Fernandez F. Soluble expression in E. coli of a functional interphotoreceptor retinoid-binding protein module fused to thioredoxin: correlation of vitamin A binding regions with conserved domains of C-terminal processing proteases. Exp. Eye Res. 1998;66:249–262. doi: 10.1006/exer.1997.0418. [DOI] [PubMed] [Google Scholar]
- Bringmann A, Pannicke T, Grosche J, Francke M, Wiedemann P, Skatchkov SN, Osborne NN, Reichenbach A. Muller cells in the healthy and diseased retina. Prog. Retin. Eye Res. 2006;25:397–424. doi: 10.1016/j.preteyeres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Cai J, Nelson KC, Wu M, Sternberg P, Jr., Jones DP. Oxidative damage and protection of the RPE. Prog. Retin. Eye Res. 2000;19:205–221. doi: 10.1016/s1350-9462(99)00009-9. [DOI] [PubMed] [Google Scholar]
- Carlson A, Bok D. Promotion of the release of 11-cis-retinal from cultured retinal pigment epithelium by interphotoreceptor retinoid-binding protein. Biochemistry. 1992;31:9056–9062. doi: 10.1021/bi00152a049. [DOI] [PubMed] [Google Scholar]
- Carne AF. Chemical modification of proteins. Methods Mol. Biol. 1994;32:311–320. doi: 10.1385/0-89603-268-X:311. [DOI] [PubMed] [Google Scholar]
- Catala A. Lipid peroxidation of membrane phospholipids in the vertebrate retina. Front Biosci (Schol Ed) 2011;3:52–60. doi: 10.2741/s131. [DOI] [PubMed] [Google Scholar]
- Chalam KV, Khetpal V, Rusovici R, Balaiya S. A review: role of ultraviolet radiation in age-related macular degeneration. Eye Contact Lens. 2011;37:225–232. doi: 10.1097/ICL.0b013e31821fbd3e. [DOI] [PubMed] [Google Scholar]
- Chen Y, Saari JC, Noy N. Interactions of all-trans-retinol and long-chain fatty acids with interphotoreceptor retinoid-binding protein. Biochemistry. 1993;32:11311–11318. doi: 10.1021/bi00093a007. [DOI] [PubMed] [Google Scholar]
- Chouchani ET, James AM, Fearnley IM, Lilley KS, Murphy MP. Proteomic approaches to the characterization of protein thiol modification. Curr. Opin. Chem. Biol. 2011;15:120–128. doi: 10.1016/j.cbpa.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline DJ, Redding SE, Brohawn SG, Psathas JN, Schneider JP, Thorpe C. New water-soluble phosphines as reductants of peptide and protein disulfide bonds: reactivity and membrane permeability. Biochemistry. 2004;43:15195–15203. doi: 10.1021/bi048329a. [DOI] [PubMed] [Google Scholar]
- Crank G, Pardijanto MS. Photo-oxidations and photosensitized oxidations of vitamin A and its palmitate ester. J Photochem Photobiol. 1995;85:93–100. [Google Scholar]
- Crouch RK, Hazard ES, Lind T, Wiggert B, Chader G, Corson DW. Interphotoreceptor retinoid-binding protein and alpha-tocopherol preserve the isomeric and oxidation state of retinol. Photochem. Photobiol. 1992;56:251–255. doi: 10.1111/j.1751-1097.1992.tb02154.x. [DOI] [PubMed] [Google Scholar]
- Cunningham LL, Gonzalez-Fernandez F. Coordination between production and turnover of interphotoreceptor retinoid-binding protein in zebrafish. Invest Ophthalmol.Vis.Sci. 2000;41:3590–3599. [PubMed] [Google Scholar]
- Cunningham LL, Yang L, Gonzalez-Fernandez F. Interphotoreceptor retinoidbinding protein (IRBP) is rapidly cleared from the Xenopus interphotoreceptor matrix. Exp. Eye Res. 1999;68:399–410. doi: 10.1006/exer.1998.0633. [DOI] [PubMed] [Google Scholar]
- den Hollander AI, McGee TL, Ziviello C, Banfi S, Dryja TP, Gonzalez-Fernandez F, Ghosh D, Berson EL. A homozygous missense mutation in the IRBP gene (RBP3) associated with autosomal recessive retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci. 2009;50:1864–1872. doi: 10.1167/iovs.08-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Failloux N, Bonnet I, Baron MH, Perrier E. Quantitative analysis of vitamin A degradation by Raman spectroscopy. Appl Spectrosc. 2003;57:1117–1122. doi: 10.1366/00037020360695973. [DOI] [PubMed] [Google Scholar]
- Fedorovich IB, Semenova EM, Grant K, Converse CA, Ostrovsky MA. Photosensitized light-induced damage of IRBP (interphotoreceptor retinoid-binding protein): effects on binding properties. Curr. Eye Res. 2000;21:975–980. doi: 10.1076/ceyr.21.6.975.6984. [DOI] [PubMed] [Google Scholar]
- Ferre-D'Amare AR, Burley SK. Dynamic light scattering in evaluating crystallizability of macromolecules. In: Carter CW, Sweet RM, editors. Macromolecular Crystallography. San Diego: Academic Press; 1997. pp. 157–166. [DOI] [PubMed] [Google Scholar]
- Fong SL, Liou GI, Bridges CD. Purification of interstitital retinol-binding protein from the eye. Methods Enzymol. 1986;189:102–111. doi: 10.1016/s0076-6879(86)23014-1. [DOI] [PubMed] [Google Scholar]
- Fong SL, Liou GI, Landers RA, Alvarez RA, Bridges CD. Purification and characterization of a retinol-binding glycoprotein synthesized and secreted by bovine neural retina. J. Biol. Chem. 1984;259:6534–6542. [PubMed] [Google Scholar]
- Garlipp MA, Nowak KR, Gonzalez-Fernandez F. Cone outer segment extracellular matrix as binding domain for interphotoreceptor retinoid-binding protein. J. Comp. Neurol. 2012;520:756–769. doi: 10.1002/cne.22773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getz EB, Xiao M, Chakrabarty T, Cooke R, Selvin PR. A comparison between the sulfhydryl reductants tris(2-carboxyethyl)phosphine and dithiothreitol for use in protein biochemistry. Anal. Biochem. 1999;273:73–80. doi: 10.1006/abio.1999.4203. [DOI] [PubMed] [Google Scholar]
- Ghosh D, Haswell K, Sprada M, Gonzaez-Fernandez F. Structure of zebrafish IRBP reveals its fatty acid-binding sites and retention of protease fold. Biochemistry. (first paper in Appendix 3). [Google Scholar]
- Gonzalez-Fernandez F. Interphotoreceptor retinoid-binding protein--an old gene for new eyes. Vision Res. 2003;43:3021–3036. doi: 10.1016/j.visres.2003.09.019. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Fernandez F, Baer CA, Ghosh D. Module structure of interphotoreceptor retinoid-binding protein (IRBP) may provide bases for its complex role in the visual cycle -structure / function study of Xenopus IRBP. BMC Biochem. 2007;8:15. doi: 10.1186/1471-2091-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Fernandez F, Ghosh D. Focus on Molecules: interphotoreceptor retinoidbinding protein (IRBP) Exp. Eye Res. 2008;86:169–170. doi: 10.1016/j.exer.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Hauck SM, Schoeffmann S, Deeg CA, Gloeckner CJ, Swiatek-de Lange M, Ueffing M. Proteomic analysis of the porcine interphotoreceptor matrix. Proteomics. 2005;5:4637. doi: 10.1002/pmic.200401223. [DOI] [PubMed] [Google Scholar]
- Li S, Yang Z, Hu J, Gordon WC, Bazan NG, Haas AL, Bok D, Jin M. Secretory defect and cytotoxicity: the potential disease mechanisms for the retinitis pigmentosa (RP)-associated interphotoreceptor retinoid-binding protein (IRBP) J. Biol. Chem. 2013 doi: 10.1074/jbc.M112.418251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loew A, Gonzalez-Fernandez F. Crystal structure of the functional unit of interphotoreceptor retinoid binding protein. Structure. 2002;10:43–49. doi: 10.1016/s0969-2126(01)00698-0. [DOI] [PubMed] [Google Scholar]
- Marmorstein R. Crystallization of soluble macromolecules. In: Coligan JE, Dunn BM, Ploegh HL, Speicher DW, Wingfield PT, editors. Current Protocols in Protein Science. John Wiley & Sons; 1998. pp. 17.14.11–17.14.17. [Google Scholar]
- McBee JK, Palczewski K, Baehr W, Pepperberg DR. Confronting complexity: the interlink of phototransduction and retinoid metabolism in the vertebrate retina. Prog. Retin. Eye Res. 2001;20:469–529. doi: 10.1016/s1350-9462(01)00002-7. [DOI] [PubMed] [Google Scholar]
- Mertens ML, Kagi JH. A graphical correction procedure for inner filter effect in fluorescence quenching titrations. Anal. Biochem. 1979;96:448–455. doi: 10.1016/0003-2697(79)90605-5. [DOI] [PubMed] [Google Scholar]
- Miller NJ, Rice-Evans CA. Factors influencing the antioxidant activity determined by the ABTS.+ radical cation assay. Free Radic. Res. 1997;26:195–199. doi: 10.3109/10715769709097799. [DOI] [PubMed] [Google Scholar]
- Nickerson JM, Li GR, Lin ZY, Takizawa N, Si JS, Gross EA. Structure-function relationships in the four repeats of human interphotoreceptor retinoid-binding protein (IRBP) Mol. Vis. 1998;4:33. [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. The HKL Program Suit, HKL Manual. New Haven, CT: Yale University Press; 1995. [Google Scholar]
- Parker R, Wang JS, Kefalov VJ, Crouch RK. Interphotoreceptor Retinoid-Binding Protein as the Physiologically Relevant Carrier of 11-cis-Retinol in the Cone Visual Cycle. J. Neurosci. 2011;31:4714–4719. doi: 10.1523/JNEUROSCI.3722-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plafker SM, O'Mealey GB, Szweda LI. Mechanisms for countering oxidative stress and damage in retinal pigment epithelium. Int Rev Cell Mol Biol. 2012;298:135–177. doi: 10.1016/B978-0-12-394309-5.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell HR. The Rossmann Fourier autoindexing algorithm in MOSFLM. Acta CrystallogrDBiol. Crystallogr. 1999;55:1690–1695. doi: 10.1107/s0907444999009506. [DOI] [PubMed] [Google Scholar]
- Redmond TM, Wiggert B, Robey FA, Nguyen NY, Lewis MS, Lee L, Chader GJ. Isolation and characterization of monkey interphotoreceptor retinoid-binding protein, a unique extracellular matrix component of the retina. Biochemistry. 1985;24:787–793. doi: 10.1021/bi00324a038. [DOI] [PubMed] [Google Scholar]
- Romero FJ, Bosch-Morell F, Romero MJ, Jareno EJ, Romero B, Marin N, Roma J. Lipid peroxidation products and antioxidants in human disease. Environ. Health Perspect. 1998;106(Suppl 5):1229–1234. doi: 10.1289/ehp.98106s51229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saari JC, Teller DC, Crabb JW, Bredberg L. Properties of an interphotoreceptor retinoid-binding protein from bovine retina. J. Biol. Chem. 1985;260:195–201. [PubMed] [Google Scholar]
- Sung D, Sprada M, Haswell K, Ghosh D. Interphotoreceptor retinoid binding protein (IRBP) - a thiol based antioxidant in the subretinal space. Invest Ophthal Vis Sci. 2012 ARVO abstract. [Google Scholar]
- Tsin AT, Mimun JM, Gonzaez-Fernandez F. Interphotoreceptor Retinoid Binding Protein Protects and Delivers Retinoids in the Cone Visual Cycle. , ARVO Abstract Invest. Ophthal. Vis. Sci. 2013 3765-A0104. [Google Scholar]
- Wang CC, Chu CY, Chu KO, Choy KW, Khaw KS, Rogers MS, Pang CP. Troloxequivalent antioxidant capacity assay versus oxygen radical absorbance capacity assay in plasma. Clin. Chem. 2004;50:952–954. doi: 10.1373/clinchem.2004.031526. [DOI] [PubMed] [Google Scholar]
- Ward LD. Measurement of ligand binding to protein by fluorescence spectroscopy. In: Hirs CHW, Timasheff SN, editors. Methods Enzymol. New York: Academic press; 1985. pp. 400–414. [DOI] [PubMed] [Google Scholar]
- Weber PC. Overview of protein crystallization methods. In: Carter CW, Sweet RS, editors. Macromolecular Crystallography. San Diego: Academic Press; 1997. pp. 13–22. [Google Scholar]
- Wood ZA, Schroder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem. Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- Wu Q, Blakeley LR, Cornwall MC, Crouch RK, Wiggert BN, Koutalos Y. Interphotoreceptor Retinoid-Binding Protein Is the Physiologically Relevant Carrier That Removes Retinol from Rod Photoreceptor Outer Segments. Biochemistry. 2007;46:8669–8679. doi: 10.1021/bi7004619. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.