Abstract
Although there have been great strides in defining the mechanisms of RNA strand cleavage by 2′-O-transphosphorylation, long-standing questions remain. How do different catalytic modes such as acid/base and metal ion catalysis influence transition state charge distribution? Does the large rate enhancement characteristic of biological catalysis result in different transition states relative to solution reactions? Answering these questions is important for understanding biological catalysis in general, and revealing principles for designing small molecule inhibitors. Recent application of linear free energy relationships and kinetic isotope effects together with multi-scale computational simulations are providing tentative answers to these questions for this fundamentally important class of phosphoryl transfer reactions.
Introduction
Cleavage of RNA by nucleophilic attack of a ribose 2′-hydroxyl on the adjacent 3′5′, phosphodiester to generate cyclic 2′, 3′-cyclic phosphate and 5′-hydroxyl products is a fundamental reaction in biology [1–5]. Decades of detailed experimental and, more recently, computational analyses reveal a complex free energy landscape that includes both stepwise and concerted mechanisms. The specific pathway followed depends on interactions with acid, base and metal ion catalysts. Distinguishing between general and specific acid/base catalysis and the precise mode or modes of metal ion catalysis within these mechanisms is challenging. Nonetheless, such chemical detail is essential for understanding transition state stabilization, and designing new therapeutics that target phosphoryl transfer enzyme active sites [5–8].
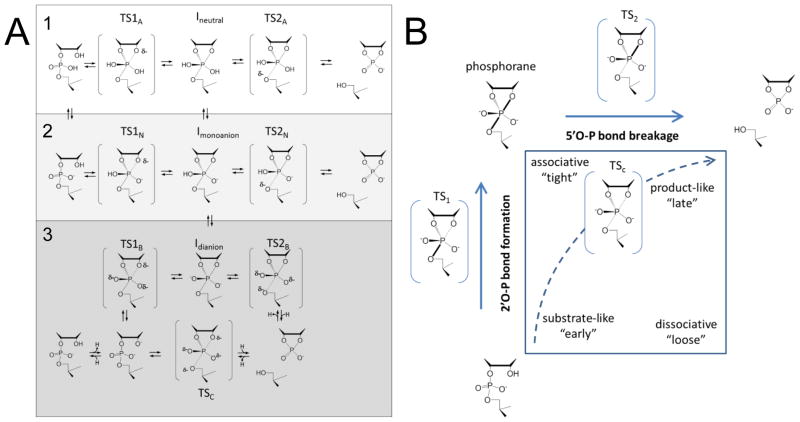
The overall mechanistic landscape for intramolecular transphosphorylation in solution depends on whether catalysis is by acid or base [1–3] (Figure 1A). At pH 0–5 both cleavage and isomerization (to a 2′,5′ phosphodiester) occur via a step-wise mechanism with a shared phosphorane intermediate. The increase in rate constant with decreasing pH is consistent with protonation of both the leaving group (pKa < 0) and one of the nonbridging oxygens (pKa ca. 2). At high pH (>8) the cleavage rate is log-linear with hydroxide concentration with an apparent pKa of ca. 13 reflecting deprotonation of the 2′-O nucleophile. Isomerization products are not formed at high pH suggesting a concerted mechanism, or short-lived dianionic phosphorane intermediate. A More-O’Ferrall/Jencks diagram illustrates the range of possible TSs (Figure 1B)[7–9]. For a stepwise mechanism involving a phosphorane intermediate the reaction proceeds first along the 2′O-P bond formation coordinate, and then along the 5′O-P bond cleavage coordinate. There are two separate transition states, TS1 and TS2, either of which can be rate limiting. TS1 and TS2 may be early or late along their respective reaction coordinate, and intermediates within the landscape that have partial 2′O-P and 5′O-P bonding are also possible. A fully concerted mechanism reacts via a single transition state (TSc) with partial 2′O-P and 5′O-P bonding.
Figure 1.
Reaction pathways and transition states for RNA 2′-O-transphosphorylation. (A). Overall reaction schemes for acid and base catalysis. Acid catalysis of the neutral diester (1) and monoanion (2) involve stepwise mechanisms via a phosphorane that may be neutral or monoanionic. Scheme 3 shows the potential stepwise (via a dianionic phosphorane) and concerted mechanisms for base catalysis. (B). More-O’Ferrall/Jencks diagram for RNA 2′-O-transphosphorylation showing the landscape for conversion of the substrate (bottom left) to products (top right). The reaction coordinate traverses the vertical 2′O-P bond formation and horizontal 5′O-P bond cleavage coordinates. A dotted arrow indicates the path for a hypothetical concerted mechanism with a single TSc.
The sensitivity of this landscape to interactions with catalysts raises questions that are fundamentally important for understanding biological catalysis [8,10]. How do catalytic modes such as acid/base and metal ion catalysis influence TS structure and bonding? Do enzymes alter TSs in characteristic ways relative to solution reactions? Addressing these questions may provide insight into foundations of biological catalysis, and provide design principles for developing TS analogs as inhibitors. Experimental means for analyzing TSs include linear free energy relationship and kinetic isotope effect analyses. These data can constrain potential mechanisms, and serve as calibration points for multi-scale quantum modeling to provide an atomic-level interpretation of TS bonding. Characterization of the TS allows discrimination between free energy landscapes corresponding to different reaction channels, and, ultimately, reveals mechanistic detail. Here, we highlight recent progress using these approaches to understanding RNA transphosphorylation.
Transition states of solution RNA 2′-O-transphosphorylation
Linear free energy relationship (LFER) analyses compare the effect of changes in nucleophile or leaving group reactivity (pKa) on the reaction rate (βF for the forward reaction) calibrated against the effect of changing pKa on reaction equilibria (βEQ) in order to estimate charge development in the TS[8](and references therein). The Leffler parameter α = βF/βEQ is used to express the fraction of total charge development in the TS[11]. A βLG of −1.28 was measured for base catalyzed reaction of uridine-3′-alkylphosphodiesters indicating a late TS along the 5′O-P bond cleavage coordinate [12]. The βEQ for RNA transphosphorylation is unknown, however estimates of −1.6 to −1.7 [13,14] indicate α ≈ 0.7. For uridine-3′-arylphosphodiesters a βLG of −0.59 (α ≈ 0.34) was observed consistent with less charge accumulation on the leaving group for better aryl leaving groups [15]. These data suggest a change in mechanism with increasing leaving group pKa that may involve transition from a concerted to stepwise reaction [16]. Piccirilli and colleagues reported a βNUC of 0.75 for RNA transphosphorylation using a series of 2′-substituted analogs (α ≈ 0.5) [17] demonstrating the TS for RNA reactions is advanced along the 2′O-P bond formation coordinate. Significant βLG and βNUC values support a late, TS2-like transition state in either a concerted or stepwise mechanism. Although LFER provide a means to probe TS structure, the interpretation of these results is complicated by uncertainty in the accuracy of estimated βEQ values, an unclear understanding of the role of solvation, and ambiguity associated with kinetically equivalent mechanistic models within the framework of transition state theory [7,8]. Quantum mechanical calculations can greatly aid in establishing a connection between LFER data and TS bonding [18].
Kinetic isotope effects (KIEs) offer a complementary approach to investigate TS bonding. KIEs arise because heavier stable isotopes have lower zero point vibrational energies than their lighter counterparts [19–21]. Differences in bond stiffness between the ground state and TS result in differences in activation energy and consequently differences in rate constant (expressed as klight/kheavy). Decreasing bond stiffness in the TS favors the lighter isotope resulting in KIE >1 (referred to as “normal” effects), while increasing bond stiffness in the TS favors the heavier isotope (KIEs <1) resulting in an “inverse” KIE. TS structure can be inferred by comparing observed KIEs to equilibrium values. However, KIEs reflect changes in all vibrational modes involving the substituted atom, which can make them difficult to interpret unambiguously, although multi-scale quantum simulations can help to aid in theirin terpretation.
A large, normal KIE on the 5′O (18kLG) is observed for RNA 2′-O-transhosphorylation catalyzed by specific base (1.034)[22]. This result corresponds well to previous measurements by Cleland and colleagues for uridine-3′-m-nitrobenzyl phosphate (pKa 14.9) (18kLG = 1.027)[23] and the large βLG observed for alkyl leaving groups [12]. The 18kLG for uridine-3′-p-nitrophenyl phosphate (pKa 7.4) is small (18kLG = 1.006)[24] suggesting an early TS along the 5′O-P bond cleavage coordinate consistent with smaller βLG for aryl leaving groups.
Nucleophile KIEs (18kNUC) can range from normal to inverse for early versus late TSs because they reflect both participation in reaction coordinate motion as well as differences in bonding in the TS compared to the ground state [22,25]. A normal nucleophile KIE of 1.0327 is observed for hydroxypropyl-p-nitrophenol phosphate (HPpNP) reactions consistent with minimal contribution from 2′O-P bond formation. In contrast, the inverse 18kNUC (0.981) for RNA transphosphorylation (leaving group pKa ca. 14) indicates that 2′O-P bonding is advanced [22]. A lower 18kNUC for RNA is observed at pH values below the pKa of the 2′O (18kNUC = 0.996) due to a large normal contribution from loss of the 2′O-H stretching mode (ca. 1.027) to the observed KIE[22,26]. Thus, KIE and LFER analyses support base catalysis occurring by mechanisms with TS1- or TS2-like transition states depending on leaving group reactivity.
A near-unity non-bridging oxygen isotope effect (18kNPO) for specific base catalysis indicates similar bonding in the TS and ground state [20,22,27]. In contrast, during acid catalysis the stiffer bonding environment resulting from formation of a protonated phosphorane appears to be reflected in an observed inverse 18kNPO (0.991)[23,28]. The 18kLG for acid catalysis is small (1.005)[23,28], consistent with models involving rate-limiting 5′O-P bond cleavage with concomitant transfer of a proton from a phosphoryl oxygen to the 5′O[1]. Yet, the potential for offsetting contributions from O-P bond cleavage and leaving group protonation to observed KIEs make it difficult to pin down TS charge distribution unambiguously.
Multi-scale quantum simulations provide a wealth of atomic-level information about mechanism [29]. This approach involves performing molecular dynamics simulations using a potential that treats the local environment of the reacting atoms with quantum mechanical (QM) methods to describe the electronic structure needed to predict chemical bond formation and cleavage, and the remainder of the solvated macromolecular environment with molecular mechanics (MM) using simpler potential energy functions [30]. QM treatments range from computationally economical semi-empirical approaches [31] to more accurate and computationally expensive density functional and ab inito methods [32]. Development of both fast and accurate multi-scale methods has great potential to facilitate analysis of free energy surfaces for rigorous comparison with experimental measurements and for identification of alternative mechanisms of RNA transphosphorylation [33,34].
For a given mechanistic scenario (e.g., identification of general acid and base catalysis and specific metal ion binding modes), multi-dimensional free energy landscapes are determined along a set of appropriate reaction coordinates. These free energy landscapes provide the groundwork from which the most probable catalytic pathways can be determined. However, because these landscapes have been derived from models that are inherently approximate descriptions of Nature, they must be evaluated in light of experimental measurements, such as LFER analyses and KIE measurements.
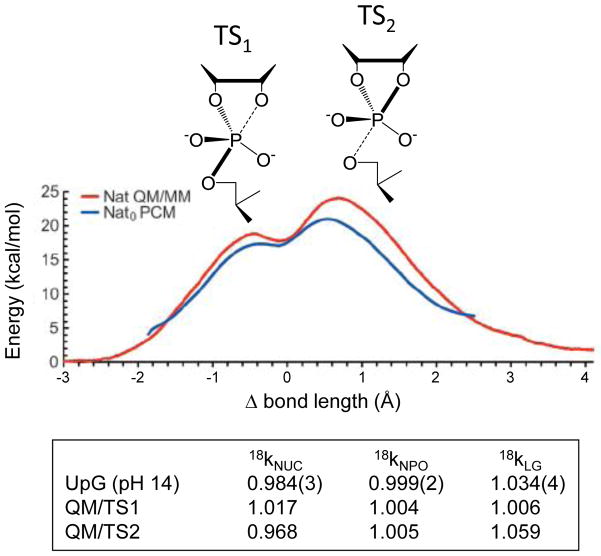
Recent density-functional calculations and QM/MM simulations reveal energy profiles for 2′-O-transphosphorylation showing an associative mechanism with distinct TS1 and TS2 separated by a metastable dianionic intermediate [27,35] (Figure 2). Calculated and measured free energies of activation match well (19.9–20.8 versus 19.9 kcal/mol) for a rate-limiting TS2; calculated 18kNUC and 18kLG also agree well with experiment (0.968 and 1.059, respectively; Figure 2)[27]. In this model for TS2 2′O-P bond formation is nearly complete (1.76 Å compared to 1.6 Å ideal length [36]), while 5′O departure is advanced (5′O-P of 2.3 Å). In contrast, TS1 was observed to have a normal 18kNUC (1.017) and near unity 18kLG (1.006), similar to the early TS proposed for reactions involving aryl leaving groups [26].
Figure 2.
Density-functional QM/MM free energy and adiabatic PCM profiles for the reaction of an RNA transphosphorylation model. Free energy is plotted as a function of the difference (Δ) in bond length between the 5′O-P and O2′-P bonds. The calculated density-functional free energy barrier is similar to experiment. The primary KIE values calculated for TS1 and TS2 by using an ab initio path-integral method are shown in the boxed table, below, compared to experimentally observed values.
Altered transition states for metal ion catalysis
Recent KIE and LFER analyses of catalysis by organometallic complexes reveal striking alterations in TS structure. Effects of organometallic complex catalysts on βLG values have been expertly covered by Mikkola and colleagues [37], and current reviews focusing on functional and design properties of organometallic catalysts are also available [38]. Here, we focus on examples where experimental and computational approaches are converging.
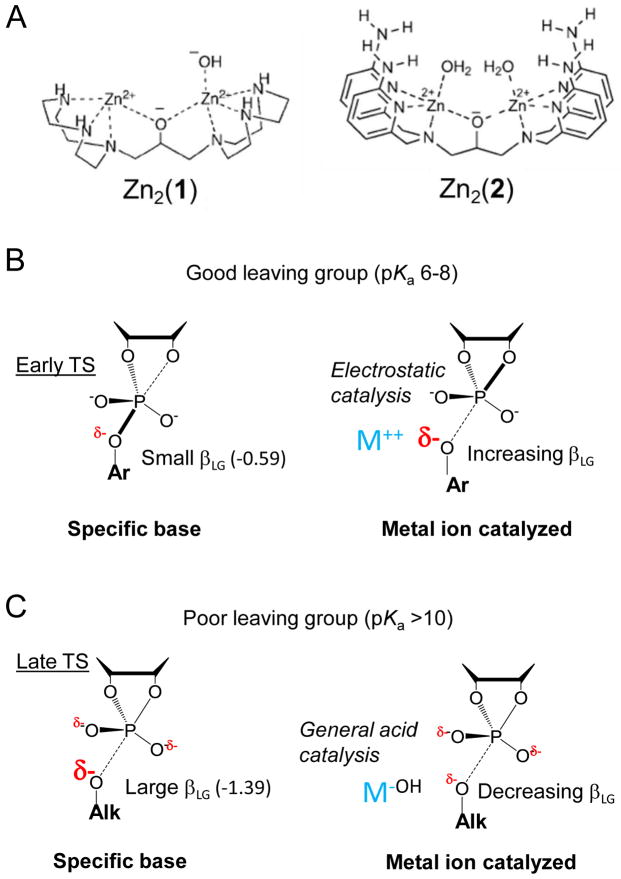
Two well-characterized examples involve dinuclear Zn2+ organometallic compounds (referred to here as Zn2(1) and (2)) [39] (Figure 3A). Consideration of pH dependence, solvent KIEs and inhibition by TS mimics indicates that promoting 2′OH deprotonation and indirect electrostatic charge stabilization, rather than direct inner sphere coordination, provide the primary modes of catalysis by Zn2(1). Interestingly, Zn2(1) catalysis of HPpNP transphosphorylation results in a more inverse 18kNUC (1.0079 vs. 0.9874), while the 18kLG becomes more normal (1.0064 versus 1.0113) consistent with a later TS[26] (Figure 3B). Recently reported QM calculations obtained assuming coordination of both non-bridging oxygens and the nucleophile by the two Zn2+ atoms in Zn2(1)[40] support a concerted mechanism with a later TS, although one that is more associative relative to the uncatalyzed TS, is consistent with the observed KIEs. In contrast, the distinct dinuclear Zn organometallic complex catalyst, Zn2(2) (Figure 3A) is proposed to act by stabilization of a phosphorane intermediate as evidenced by its ability to catalyze both cleavage and isomerization [37]. A βLG of −0.92 is observed for uridine-3′-alkylphosphate reactions catalyzed by Zn2(2)[41], compared to −1.28 for specific base catalysis (see above) indicating less charge development on the leaving group [41].
Figure 3.
Alteration of TS structure due to catalysis by metal ion and organometallic metal ion complexes. (A). Structure of dinuclear organometallic Zn complexes Zn2(1) (1,3-bis(1,4,7-triazacyclonon-1-yl)-2-hydroxypropane)[26] and the aminopyridine complex Zn2(2)[41]. Potential for different catalytic modes depending on leaving group reactivity. Differences in βLG values for catalysis by Zn2+ alone and by different dinuclear organometallic catalysts indicate different effects on transphosphorylation reactions with good (aryl) (B) versus poor (alkyl) (C) leaving groups [37,43].
Interestingly, catalysis of uridine-3′-alkylphosphate cleavage by Zn2+ ions alone also results in a significantly lower βLG (−0.43 to −0.32), which was interpreted as arising from general acid catalysis [41,42] (Figure 3C) although this and other mechanistic details remain to be confirmed. In contrast to the less negative βLG observed for alkyl phosphates, catalysis by Zn2+ aquo ions results in a more negative βLG for reactions of aryl leaving groups (−0.9), compared to specific base catalysis (−0.54), indicating greater charge accumulation in the TS compared to less reactive alkyl leaving groups. An emerging general feature appears to be that better (aryl) leaving groups depart as anions that can be stabilized by positive charge (resulting in a more negative βLG), but departure of less reactive (alkyl) groups like in RNA require greater charge stabilization offered by general acid catalysis (and a correspondingly less negative βLG)[43](Figure 3).
Experimental and computational analysis of ribonuclease transition states
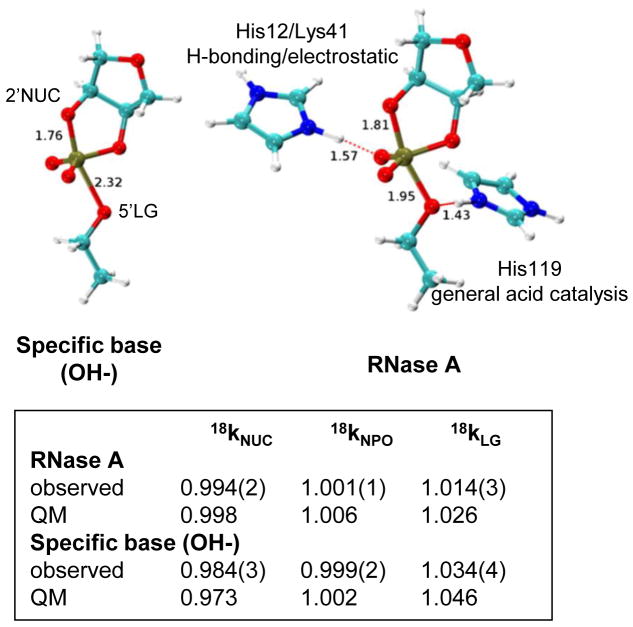
Ribonuclease A is a well-studied paradigm of enzyme catalysis in which active site His119 and His12 are thought to act as general acid and base, respectively. Yet, its mechanism and structure of the rate limiting TS remain ambiguous, making it an excellent system for merging theory and experiment to understand biological catalysis [6]. KIE results reveal intriguing differences in the enzyme TS compared to solution reactions. The 18kLG for RNA 2′-O-transphosphorylation by RNase A is less than the value observed for the solution reaction (1.014 vs. 1.037) [28,44], indicating a stiffer bonding environment for the 5′O on the enzyme. An inverse 18kNUC (0.994) indicates that, like specific base catalysis, 2′O-P bond formation is advanced [28]. A small 18kNPO and the lack of a thio effect [45] are most consistent with an dianionic TS rather than a monoanionic one involving protonation of a non-bridging oxygen.
MD simulations of RNase A with a dianionic phosphorane TS mimic provided a basis for a fully QM active site model [28,46]. His119 interacts with the departing 5′O consistent with a role as general acid. Interestingly, the calculations suggest a step-wise mechanism is possible in which a protonated His12 may hydrogen bond with a non-bridging phosphoryl oxygen, in addition to its role in nucleophilic activation. Alternative models involving a monoanionic TS and general acid catalysis alone were analyzed as well. Nonetheless, KIEs predicted from DFT calculations using the TS structure involving proton transfer from His119 to the 5′O, together with H-bonding between His12 and the anionic TS are most consistent with experimental values (Figure 4). The calculated 18kLG for the base-catalyzed reaction (1.048) was notably larger than the value for RNase A (1.026), consistent with the observed experimental trend (1.034 versus 1.017). The P-5′O appears therefore to retain a higher degree of covalent bond character, and proton transfer from the general acid (His119) further creates a stiffer bonding environment for stabilizing the leaving group.
Figure 4.
Transition state models for specific base catalysis (left) and ribonuclease A (right). The TS models used for the calculated KIE values are depicted with the 2′O-P and 5′O-P bond lengths indicated. For the RNase A model the distances between the His12 and His119 protons and the non-bridging oxygen or 5′O leaving group, respectively, are shown. The catalytic modes resulting in altered TS charge distribution for RNase A are indicated.
Recent analysis of the free energy landscape for RNase A compared the classic mechanism via a dianionic TS to one in which a non-bridging oxygen becomes protonated by His12 subsequent to nucleophilic attack. The stepwise mechanism was found to provide a lower free energy barrier, but TS1 was identified as the rate controlling TS[47], which appears inconsistent with a simple interpretation of the observed inverse 18kNUC and large 18kLG for this enzyme [28,44]. A fully triester-like mechanism involving complete proton transfer appears to conflicts with experimental KIE and thio-effect data [28,45]. However, phosphoryl oxygen vibrational modes are complex and additional computational and experimental effort are needed to achieve a consistent and chemically detailed understanding.
Conclusions
LFER and KIE are powerful because they fundamentally compare ground state and TS bonding, yet, there are inherent limits to their interpretation because of multiple contributions to bonding and kinetic ambiguity. Computational simulations can provide a complete theoretical picture of the reaction landscape and allow areas of mechanistic ambiguity to be identified or resolved, but need careful calibration to experimental data. In addition to gaining deeper insight into how ribonucleases work, an additional area calling for matched effort is in understanding metal ion catalysis where coordination, proton transfer and reaction coordinate effects appear to have large variable contributions to TS bonding depending on mechanism. Phosphoryl transferases have a variety of proposed active site metal geometries and distinct bonding environments compared to solution reactions. Thus, distinct transition states for these enzymes appear likely. In summary, results to date suggest that fundamental chemical constraints dominate solution and enzyme TS structures, however, interactions with active site residues can alter charge distribution in ways that reflect specific catalytic modes.
Highlights.
Long-standing questions remain regarding transition states for RNA cleavage reactions.
Different catalytic modes may profoundly alter transition state charge distribution.
Theory and experiment together can reveal how transition states are altered.
Acknowledgments
The work in the author’s laboratories is funded by NIH GM096000 to MEH, AI081987 to JAP and GM062248 to DMY. The authors apologize to colleagues who have fundamentally advanced our understanding of the RNA reactions reviewed here whose advances could not be directly discussed due to limitations of space and scope.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Oivanen M, Kuusela S, Lonnberg H. Kinetics and mechanism for the cleavage and isomerization of phosphodiester bonds of RNA by bronsted acids and bases. Chemical reviews. 1998;98 (3):961–990. doi: 10.1021/cr960425x. [DOI] [PubMed] [Google Scholar]
- 2.Perreault DM, Anslyn EV. Unifying the current data on the mechanism of cleavage–transesterification of rna. Angewandte Chemie International Edition in English. 1997;36(5):432–450. [Google Scholar]
- 3.Emilsson GM, Nakamura S, Roth A, Breaker RR. Ribozyme speed limits. RNA. 2003;9(8):907–918. doi: 10.1261/rna.5680603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang W. Nucleases: Diversity of structure, function and mechanism. Quarterly Reviews of Biophysics. 2011;44(01):1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilcox JL, Ahluwalia AK, Bevilacqua PC. Charged nucleobases and their potential for RNA catalysis. Acc Chem Res. 2011;44(12):1270–1279. doi: 10.1021/ar2000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuchillo CM, Nogues MV, Raines RT. Bovine pancreatic ribonuclease: Fifty years of the first enzymatic reaction mechanism. Biochemistry. 2011;50(37):7835–7841. doi: 10.1021/bi201075b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **7.Kamerlin SC, Sharma PK, Prasad RB, Warshel A. Why nature really chose phosphate. Quarterly reviews of biophysics. 2013;46(1):1–132. doi: 10.1017/S0033583512000157. A wide-ranging review of phosphoryl transfer including basic chemistry to complex enzyme systems stressing the importance of understanding the entire free energy landscape using computational methods. Overarching themes include the importance of kinetic stability of phosphoesters due to electrostatic repulsion between the nucleophile and phosphoryl group and the ability to regulate reactivity via electrostatic interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **8.Lassila JK, Zalatan JG, Herschlag D. Biological phosphoryl-transfer reactions: Understanding mechanism and catalysis. Annu Rev Biochem. 2011;80:669–702. doi: 10.1146/annurev-biochem-060409-092741. A comprehensive and authoritative (while still student-friendly) introduction to the conceptual and experimental background for understanding the chemistry and catalysis of phosphoryl transfer reactions. The authors illustrate the application of these principles in several examples from their work and present important future challenges for understanding phosphoryl transfer mechanism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jencks WP. General acid-base catalysis of complex reactions in water. Chemical reviews. 1972;72 (6):705–718. [Google Scholar]
- 10.Jencks WP. Binding energy, specificity, and enzymic catalysis: The circe effect. Adv Enzymol Relat Areas Mol Biol. 1975;43:219–410. doi: 10.1002/9780470122884.ch4. [DOI] [PubMed] [Google Scholar]
- 11.Leffler JE. Parameters for the description of transition states. Science. 1953;117(3039):340–341. doi: 10.1126/science.117.3039.340. [DOI] [PubMed] [Google Scholar]
- 12.Kosonen M, Youseti-Salakdeh E, Stromberg R, Lonnberg H. Mutual isomerization of uridine 2′- and 3′-alkylphosphates and cleavage to a 2′,3′-cyclic phosphate: The effect of the alkyl group on the hydronium- and hydroxide-ion-catalyzed reactions. Journal of the Chemical Society, Perkin Transactions. 1997;2(12):2661–2666. [Google Scholar]
- 13.Bourne N, Williams A. Effective charge on oxygen in phosphoryl group transfer from an oxygen donor. The Journal of Organic Chemistry. 1984;49(7):1200–1204. [Google Scholar]
- 14.Ye JD, Barth CD, Anjaneyulu PS, Tuschl T, Piccirilli JA. Reactions of phosphate and phosphorothiolate diesters with nucleophiles: Comparison of transition state structures. Org Biomol Chem. 2007;5(15):2491–2497. doi: 10.1039/b707205h. [DOI] [PubMed] [Google Scholar]
- 15.Davis AM, Hall AD, Williams A. Charge description of base-catalyzed alcoholysis of aryl phosphodiesters: A ribonuclease model. J Am Chem Soc. 1988;110:5105–5108. [Google Scholar]
- 16.Lonnberg H, Stromberg R, Williams A. Compelling evidence for a stepwise mechanism of the alkaline cyclisation of uridine 3′-phosphate esters. Org Biomol Chem. 2004;2(15):2165–2167. doi: 10.1039/b406926a. [DOI] [PubMed] [Google Scholar]
- 17.Ye JD, Li NS, Dai Q, Piccirilli JA. The mechanism of rna strand scission: An experimental measure of the bronsted coefficient, βnuc. Angew Chem Int Ed Engl. 2007;46(20):3714–3717. doi: 10.1002/anie.200605124. [DOI] [PubMed] [Google Scholar]
- **18.Huang M, York DM. Linear free energy relationships in rna transesterification: Theoretical models to aid experimental interpretations. Phys Chem Chem Phys. 2014 doi: 10.1039/c4cp01050g. submitted A key challenge for this field is the integration of LFER and KIE data into computational simulations. This study presents a detailed set of simulations of the effect of changes in leaving group pKa on RNA transphosphorylation models reproducing the breakpoint at high pKa observed experimentally. The results support a unifying perspective on the transition from concerted to stepwise mechanisms with decreasing leaving group reactivity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bigeleisen J, Wolfsberg M. Theoretical and experimental aspects of isotope effects in chemical kinetics. Adv Chem Phys. 1958;1(1):15–76. [Google Scholar]
- 20.Hengge AC. Isotope effects in the study of phosphoryl and sulfuryl transfer reactions. Acc Chem Res. 2002;35(2):105–112. doi: 10.1021/ar000143q. [DOI] [PubMed] [Google Scholar]
- 21.Cook PF, Cleland WW. Enzyme kinetics and mechanism. Garland Science; New York: 2007. Isotope effects as a probe of mechanism; pp. 253–324. [Google Scholar]
- 22.Harris ME, Dai Q, Gu H, Kellerman DL, Piccirilli JA, Anderson VE. Kinetic isotope effects for RNA cleavage by 2′-O-transphosphorylation: Nucleophilic activation by specific base. J Am Chem Soc. 2010;132(33):11613–11621. doi: 10.1021/ja103550e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerratana B, Sowa GA, Cleland WW. Characterization of the transition state structures and mechanisms for the isomerization and cleavage reactions of 3′-m-nitrobenzyl phosphate. J Am Chem Soc. 2000;122(51):12615–12621. [Google Scholar]
- 24.Hengge AC, Bruzik KS, Tobin AE, Cleland WW, Tsai MD. Kinetic isotope effects and stereochemical studies on a ribonuclease model: Hydrolysis reactions of uridine 3′-nitrophenyl phosphate. Bio-Organic Chemistry. 2000;28(3):119–133. doi: 10.1006/bioo.2000.1170. [DOI] [PubMed] [Google Scholar]
- 25.Iyer S, Hengge AC. The effects of sulfur substitution for the nucleophile and bridging oxygen atoms in reactions of hydroxyalkyl phosphate esters. J Org Chem. 2008;73(13):4819–4829. doi: 10.1021/jo8002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Humphry T, Iyer S, Iranzo O, Morrow JR, Richard JP, Paneth P, Hengge AC. Altered transition state for the reaction of an RNA model catalyzed by a dinuclear zinc(ii) catalyst. J Am Chem Soc. 2008;130(52):17858–17866. doi: 10.1021/ja8059864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **27.Wong KY, Gu H, Zhang S, Piccirilli JA, Harris ME, York DM. Characterization of the reaction path and transition states for RNA transphosphorylation models from theory and experiment. Angew Chem Int Ed Engl. 2012;51(3):647–651. doi: 10.1002/anie.201104147. The nucleophile, leaving group and nonbridging oxygen KIEs for a model RNA cleavage transesterification were calculated and compared with experimental measurements for specific base catalysis in this paper. The results support two distinct TSs and an overall rate limiting TS for departure of the leaving group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **28.Gu H, Zhang S, Wong KY, Radak BK, Dissanayake T, Kellerman DL, Dai Q, Miyagi M, Anderson VE, York DM, Piccirilli JA, et al. Experimental and computational analysis of the transition state for ribonuclease A-catalyzed rna 2′-O-transphosphorylation. Proc Natl Acad Sci U S A. 2013;110(32):13002–13007. doi: 10.1073/pnas.1215086110. Despite its well-studied active site the mechanism of RNase A is still ambiguous and this paper describes combined computational and experimental analyses of this classic enzyme system. The results demonstrate an altered transition state consistent with general acid stabilization of the leaving group and suggest stabilizing interactions with the non-bridging oxygens as well. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee TS, Giambasu GM, Moser A, Nam K, Silva Lopez C, Guerra F, Nieto Faza O, Giese TJ, Gao J, York DM. Unraveling the mechanisms of ribozyme catalysis with multi-scale simulations. In: Lee TS, York DM, editors. Multi-scale quantum models for biocatalysis. Springer; 2009. [Google Scholar]
- 30.van der Kamp MW, Mulholland AJ. Combined quantum mechanics/molecular mechanics (QM/MM) methods in computational enzymology. Biochemistry. 2013;52(16):2708–2728. doi: 10.1021/bi400215w. [DOI] [PubMed] [Google Scholar]
- 31.Giese TJ, Chen H, Dissanayake T, Giambaşu GM, Heldenbrand H, Huang M, Kuechler ER, Lee T-S, Panteva MT, Radak BK, York DM. A variational linear-scaling framework to build practical, efficient next-generation orbital-based quantum force fields. Journal of Chemical Theory and Computation. 2013;9(3):1417–1427. doi: 10.1021/ct3010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Šponer J, Šponer JE, Mládek A, Banaáš P, Jurečka P, Otyepka M. How to understand quantum chemical computations on DNA and RNA systems? A practical guide for non-specialists. Methods. 2013;64(1):3–11. doi: 10.1016/j.ymeth.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 33.Lee TS, Radak BK, Huang M, Wong KY, York DM. Roadmaps through free energy landscapes calculated using the multi-dimensional VFPE approach. J Chem Theory Comput. 2014;10(1):24–34. doi: 10.1021/ct400691f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee TS, Radak BK, Pabis A, York DM. A new maximum likelihood approach for free energy profile construction from molecular simulations. J Chem Theory Comput. 2013;9(1):153–164. doi: 10.1021/ct300703z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radak BK, Harris ME, York DM. Molecular simulations of RNA 2′-O-transesterification reaction models in solution. J Phys Chem B. 2013;117(1):94–103. doi: 10.1021/jp3084277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lide D. CRC handbook of chemistry and physics. 88. CRC Press; 2007. [Google Scholar]
- 37.Korhonen H, Williams NH, Mikkola S. βlg values in mechanistic studies on the transesterification of RNA models and their application in a metal ion complex promoted transesterification. Journal of Physical Organic Chemistry. 2013;26(2):182–186. [Google Scholar]
- 38.Niittymaki T, Lonnberg H. Artificial ribonucleases. Org Biomol Chem. 2006;4(1):15–25. doi: 10.1039/b509022a. [DOI] [PubMed] [Google Scholar]
- 39.Morrow JR, Amyes TL, Richard JP. Phosphate binding energy and catalysis by small and large molecules. Acc Chem Res. 2008;41(4):539–548. doi: 10.1021/ar7002013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao H, Ke Z, DeYonker NJ, Wang J, Xu H, Mao ZW, Phillips DL, Zhao C. Dinuclear Zn(ii) complex catalyzed phosphodiester cleavage proceeds via a concerted mechanism: A density functional theory study. J Am Chem Soc. 2011;133(9):2904–2915. doi: 10.1021/ja106456u. [DOI] [PubMed] [Google Scholar]
- **41.Korhonen H, Mikkola S, Williams NH. The mechanism of cleavage and isomerisation of RNA promoted by an efficient dinuclear Zn2+ complex. Chemistry. 2012;18(2):659–670. doi: 10.1002/chem.201100721. This study reports the catalysis of both cleavage and, remarkably, isomerisation of uridine 3′-alkylphosphates by a dinuclear Zn(2+) complex (termed Zn2(2) in the text). The authors propose a model in which the compound acts as an electrophilic catalyst to neutralize negative charge on the phosphate and stabilizing a dianionic phosphorane intermediate. [DOI] [PubMed] [Google Scholar]
- 42.Mikkola S, Stenman E, Nurmi K, Yousefi-Salakdeh E, Stromberg R, Lonnberg H. The mechanism of the metal ion promoted cleavage of RNA phosphodiester bonds involves a general acid catalysis by the metal aquo ion on the departure of the leaving group. J Chem Soc, Perkin Trans. 1999;2:1619–1625. [Google Scholar]
- **43.Korhonen H, Koivusalo T, Toivola S, Mikkola S. There is no universal mechanism for the cleavage of RNA model compounds in the presence of metal ion catalysts. Org Biomol Chem. 2013;11(48):8324–8339. doi: 10.1039/c3ob41554f. The authors review LFER analyses of the transesterification of uridine 3′-phosphodiesters catalyzed by both monometallic and bimetallic complexes. A general framework is proposed in which the catalytic mechanism, either concerted or stepwise, depends on the acidity of the catalyst and leaving group reactivity and there is an overall change from general base catalysis to general acid catalysis as leaving group reactivity decreases. [DOI] [PubMed] [Google Scholar]
- 44.Sowa GA, Hengge AC, Cleland WW. 18O isotope effects support a concerted mechanism for ribonuclease A. J Am Chem Soc. 1997;119:2319–2320. [Google Scholar]
- 45.Herschlag D. Ribonuclease revisited: Catalysis via the classical general acid-base mechanism or a triester-like mechanism? Journal of the American Chemical Society. 1994;116(26):11631–11635. [Google Scholar]
- 46.Formoso E, Matxain JM, Lopez X, York DM. Molecular dynamics simulation of bovine pancreatic ribonuclease A-CpA and transition state-like complexes. J Phys Chem B. 2010;114(21):7371–7382. doi: 10.1021/jp909004y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **47.Elsässer B, Fels G, Weare JH. QM/MM simulation (B3LYP) of the RNase A cleavage-transesterification reaction supports a triester An + Dn associative mechanism with an O2′ H internal proton transfer. Journal of the American Chemical Society. 2013;136(3):927–936. doi: 10.1021/ja406122c. This paper reports QM/MM free energy calculations for a dianionic TS mechanism and a pathway involving proton transfer from the 2′OH to a non-bridging oxygen. A lower free energy is calculated for the trimester mechanism as opposed to the commonly proposed general acid/base mechanism. [DOI] [PubMed] [Google Scholar]