Abstract
Gene variants found to associate with human longevity in one population rarely replicate in other populations. The lack of consistent findings may partly be explained by genetic heterogeneity among long-lived individuals due to cohort differences in survival probability. In most high-income countries the probability of reaching e.g. 100 years increases by 50–100% per decade, i.e. there is far less selection in more recent cohorts. Here we investigate the cohort specificity of variants in the APOE and FOXO3A genes by comparing the frequencies of the APOE ε4 allele and the minor alleles of two variants in FOXO3A at age 95+ and 100+ in 2,712 individuals from the genetically homogeneous Danish birth cohorts 1895–96, 1905, 1910–11, and 1915.
Generally, we find a decrease in the allele frequencies of the investigated APOE and FOXO3A variants in individuals from more recent birth cohorts. Assuming a recessive model, this negative trend is significant in 95+ year old individuals homozygous for the APOE ε4 allele (P = 0.026) or for the FOXO3A rs7762395 minor allele (P = 0.048). For the APOE ε4 allele, the significance is further strengthened when restricting to women (P = 0.006). Supportive, but non-significant, trends are found for two of the three tested variants in individuals older than 100 years.
Altogether, this indicates that cohort differences in selection pressure on survival to the highest ages are reflected in the prevalence of longevity gene variants. Although the effect seems to be moderate, our findings could have an impact on genetic studies of human longevity.
Keywords: Human Longevity, Genetics, Cohort Effects, Selection, Apolipoprotein E (APOE), Forkhead Box O3A (FOXO3A)
1. Introduction1
Only two genes have been consistently found to associate with human lifespan: the highly validated apolipoprotein E (APOE) gene, which repeatedly has been shown to associate with survival into old age (Schachter et al. 1994; Gerdes et al. 2000; Bathum et al. 2006; Jacobsen et al. 2010; Deelen et al. 2011; McKay et al. 2011; Nebel et al. 2011; Soerensen et al. 2013), and the forkhead box O3A (FOXO3A) gene, which more recently has been found to associate with human longevity in various populations (Willcox et al. 2008; Anselmi et al. 2009; Flachsbart et al. 2009; Li et al. 2009; Pawlikowska et al. 2009; Soerensen et al. 2010).
The limited number of genes known to associate with human longevity could potentially be explained by the complexity of the trait. Another possibility is that human longevity is likely to be affected by many small and low-frequent genetic effects (Christensen et al. 2006) as well as structural genetic variations and epigenetic changes (Murabito et al. 2012; Tan et al. 2013).
However, the lack of reliable and significant findings could also in part be due to heterogeneity among long-lived cases. Genetic studies of human longevity tend to focus on individuals having reached a given age, e.g. 90, 95 or 100 years, without considering the fact that the survival probability has changed dramatically over cohorts. During the past two centuries, record life expectancy in the industrialized countries has improved with a remarkable rate of 3 months per year (Oeppen & Vaupel 2002; Christensen et al. 2009), resulting in an increase of 50–100% per decade in the number of individuals surviving to extreme ages, e.g. 100 years, in many countries (Jeune & Kannisto 1997). Therefore, an interaction between birth cohort and the effect of longevity genes may be expected, and the reduced selection pressure on high age survival for more recent birth cohorts could be important to consider in genetic studies of long-lived individuals. A decrease in selection pressure for more recent birth cohorts could, on the one hand, be expected to increase the survival of persons carrying frailty genes, i.e. the frailty gene frequency would also increase (Vaupel et al. 1979). On the other hand, the reduction in mortality in later cohorts, e.g. due to improved living conditions and better health care, could imply that the effect of genetic factors as a cause of mortality would increase.
Here we explore the effect of changes in selection pressure over birth cohorts on the frequencies of the APOE ε4 allele and the minor alleles of two FOXO3A variants, rs7762395 and rs479744, previously reported to associate with longevity in Danish nonagenarians and centenarians (Soerensen et al. 2010), using cohorts of Danish long-lived individuals older than 95 years born in 1905 and 1915, and older than 100 years born in 1895–96, 1905 and 1910–11. The selection pressure has changed considerably over these birth cohorts, and together with the genetic homogeneity of the cohorts and the minimal immigration, this is an ideal setup for addressing the aspect of cohort differences in the prevalence of longevity-associated gene variants.
2. Materials and Methods
2.1. Study Population
The study was based on Danish Birth Cohort Study participants born in 1895–96, 1905, 1910, 1911 and 1915.
The Danish 1895–96 Birth Cohort Study, also known as the Longitudinal Study of Danish Centenarians (LSDC), consists of all individuals who had reached an age of 100 years in the period from April 1st 1995 to May 31st 1996 (Andersen-Ranberg et al. 2001). A total of 276 eligible centenarians were identified through the Danish Civil Registration System (DCRS) (Pedersen et al. 2006) and of these, 207 (75.0%) chose to participate in the intake survey. Blood samples were collected from 154 (74.4%) individuals of whom 132 are included in this study.
The Danish 1905 Birth Cohort Study is an in-depth survey of all Danes born in 1905 and living in Denmark in 1998 (Nybo et al. 2001). The study was initiated in 1998 and follow-up studies of the participating survivors were carried out in years 2000, 2003 and 2005. At intake there was a total of 3,600 potential participants, of whom 2,262 (62.8%) agreed to take part in the study, and of these 1,651 (73.0%) provided a biological sample. To match the 1915 Birth Cohort (see below) only individuals who had reached an age of at least 95 years were included. This limited the number of potential participants to 2,205, of whom 1,432 (64.9%) participated in the study and 1,188 (83.0%) of these provided a biological sample. Of those who provided a biological sample, 1,169 are included in the present study. All the 1905 Birth Cohort Study participants included in the group of individuals older than 100 years are included in the group of individuals older than 95 years as well. The Danish 1910 Birth Cohort Study is a survey including all Danes born in 1910 and living in Denmark on September 1st 2010. A total of 400 individuals were identified through the DCRS and invited to participate in the survey, which 273 (68.3%) individuals agreed to (data not published). Blood samples were retrieved from 176 (64.5%) individuals, all of whom are included in the present study.
The Danish 1911–12 Birth Cohort Study is part of the international 5-Country Oldest Old Project (5-COOP), which intends to evaluate the health of the oldest old and make comparisons between the five participating countries, namely Denmark, France, Japan, Sweden and Switzerland (Robine et al. 2010). The study includes a random sample of 251 (48.5%) Danish individuals chosen from 518 individuals who had reached an age of 100 years in the period from April 1st 2011 to July 1st 2012 (data not published). Blood samples were collected from 202 (80.5%) individuals, of whom 130 (all born in 1911) are included in this study.
Due to the close proximity in birth year of the study population participants from the 1910 Birth Cohort Study and the 1911–12 Birth Cohort Study, these birth cohorts were grouped together (the 1910–11 cohort) in the present study. Prior to this grouping, the genotype distributions among the birth cohorts were compared (using a Chi2-test) to ensure that no bias would be introduced by the merging.
The Danish 1915 Birth Cohort Study includes all Danes born in 1915 and living in Denmark on September 1st 2010. A total of 2,509 individuals were identified though the DCRS as eligible participants, with 1,584 (63.1%) individuals participating in the study (Christensen et al. 2013). A biological sample was provided by 1,165 (73.5%) individuals, of whom 1,105 are included in this study.
Study approvals were received from the Danish National Committee on Biomedical Research Ethics.
2.2. Genotyping
DNA was isolated from whole blood or from blood spot cards using either the QIAamp DNA Mini and Micro Kits (Qiagen, Hilden, Germany), the Extract-N-Amp™ Blood PCR Kit (Sigma-Aldrich, St. Louis, MO, USA) or salting out applying a manual protocol or a semi-automated protocol based on the Autopure System (Qiagen, Hilden, Germany). For 336 of the samples, DNA was amplified using the GenomePlex® Complete Whole Genome Amplification Kit (Sigma-Aldrich, St. Louis, MO, USA) prior to genotyping.
Genotyping of the APOE variants rs429358 and rs7412 and the FOXO3A variants rs7762395 and rs479744 were primarily carried out using predesigned TaqMan® SNP Genotyping Assays (Applied Biosystems, Foster City, CA, USA) with genotyping efficiencies of between 96,8% and 99,7%. For 641 of the 1905 Birth Cohort Study participants, genotyping of the FOXO3A variants were performed as part of a previous study using the Illumina GoldenGate Technology (Illumina Inc, San Diego, CA, USA) as described by Soerensen et al. 2012 (Soerensen et al. 2012).
2.3. Statistical Analysis
Frequencies among individuals older than 95 years from the 1905 and the 1915 birth cohorts were compared using a two-sample z-test, whereas trend analyses of frequencies among individuals older than 100 years born in 1895–1896, 1905 and 1910–1911 were performed applying a Chi2-test for trend.
An additive model was applied as the baseline model. In addition, dominant and recessive models were applied for the APOE gene and a recessive model was applied for the FOXO3A gene.
Due to the a priori hypothesis of an association of the investigated genes with longevity, only uncorrected P-values are reported.
Data on changes in survival probabilities for the birth cohorts was retrieved from the Human Mortality Database (www.mortality.org).
3. Results
Characteristics of the study population are shown in Table 1. As expected, a higher number of long-lived women than men are present in the cohorts, with men comprising between 16.5% and 27.3%.
Table 1.
Characteristics of the study population.
| Birth Cohort | Mean Age | Age Range | N | % Men |
|---|---|---|---|---|
| 1895–96 | 100.1 | 99.0–100.4 | 132 | 22.7 |
| 1905 | 95 | - | 1169 | 24.5 |
| 1905 | 100 | - | 255 | 16.5 |
| 1910 | 100.2 | 99.7–100.9 | 176 | 24.4 |
| 1911 | 100.2 | 99.8–101.1 | 130 | 23.1 |
| 1915 | 95.3 | 94.7–95.9 | 1105 | 27.3 |
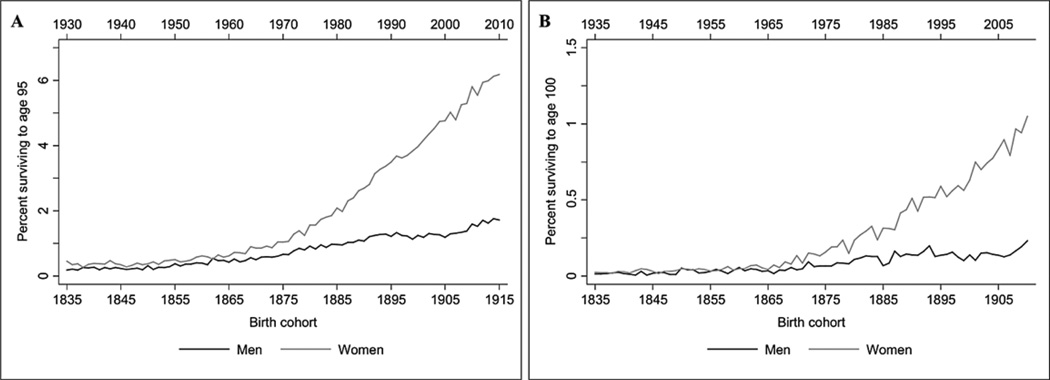
The probability of surviving from birth to age 95 (Figure 1A) and age 100 (Figure 1B) in Denmark is illustrated in Figure 1 (Human Mortality Database, www.mortality.org). Over the last four decades, the probability for both sexes combined of surviving to age 95 and 100 has increased from around 1% to 3.5%, and from around 0.2% to 0.6%, respectively. The increase has been much steeper for women than for men with the probability for reaching 95 years increasing from approximately 1.5% to 6% for women compared to an increase from 1% to 1.5% for men, and with the probability of reaching 100 years increasing from approximately 0.2% to 1% for women compared to an increase from 0.1% to 0.2% for men.
Figure 1.
The probability of surviving in Denmark from birth to age 95 in the period 1835 to 1915 (A) and from birth to age 100 in the period 1835 to 1910 (B).
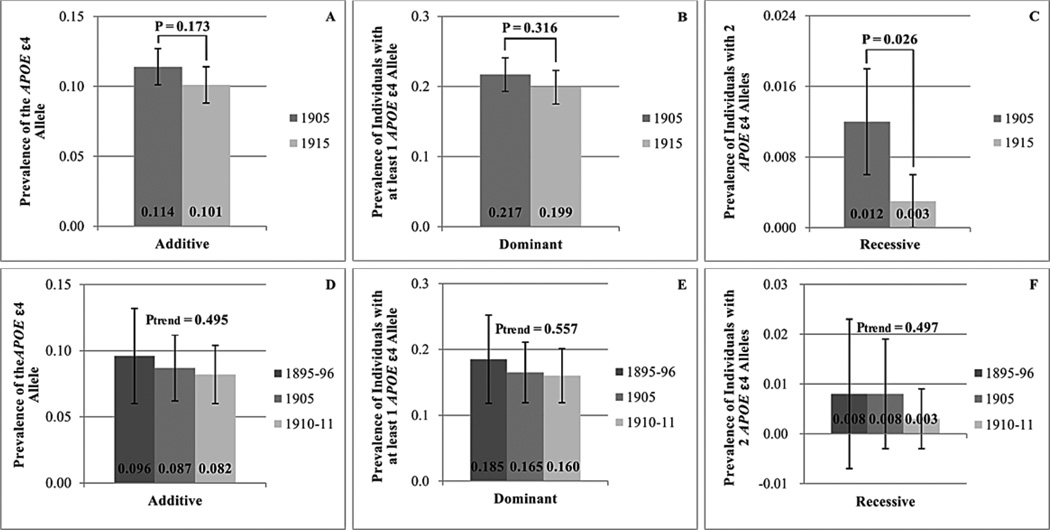
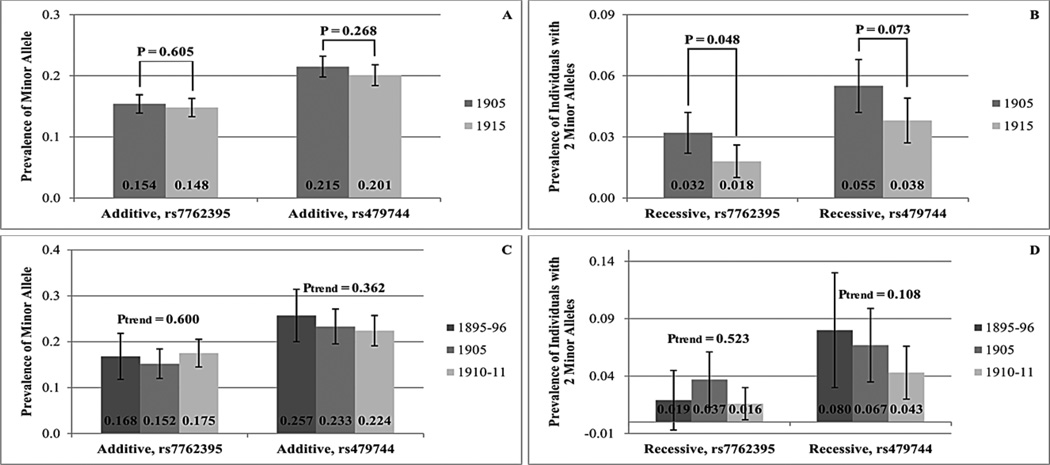
A comparison between the birth cohorts of allele frequencies and frequencies of carriers of the APOE ε4 allele and the minor alleles of the FOXO3A variants rs7762395 and rs479744 is seen in Figure 2 and 3. In individuals older than 95 years, the prevalence of the APOE ε4 allele (Figure 2A) and of APOE ε4 allele carriers (Figure 2B and 2C) is generally lower in individuals born in 1915 than in individuals born in 1905, although this tendency is only significant (P = 0.026) when assuming a recessive model for men and women combined (Figure 2C). When restricting to women, the negative trend of the APOE ε4 allele prevalence or of the prevalence of individuals homozygous for the APOE ε4 allele is significant, assuming an additive (P = 0.034) or a recessive (P = 0.006) model, respectively (separate results for women and men are shown in Supplementary Data, Supplementary Table 1 and 2). For the minor allele frequencies of the FOXO3A variants rs7762395 and rs479744 similar negative trends are seen, although these are not statistically significant (Figure 3A). When applying a recessive model (Figure 3B), the prevalence of rs7762395 minor allele homozygotes is significantly lower (P = 0.048) among individuals born in 1915 compared to individuals born in 1905, and rs479744 shows a similar borderline significant tendency (P = 0.073).
Figure 2.
The prevalence of the APOE ε4 allele and of APOE ε4 allele carriers in individuals older than 95 years born in 1905 and 1915 (A–C) and in individuals older than 100 years born in 1895–96, 1905 and 1910–11 (D–F) assuming an additive (A, D), a dominant (B, E) and a recessive (C, F) model. The 95% confidence interval is shown as well as the P-value for the significance of the difference between the birth cohorts (A–C) or of the significance of the trend (D–F).
Figure 3.
The prevalence of the FOXO3A rs7762395 and rs479744 minor alleles or of minor allele carriers in individuals older than 95 years born in 1905 and 1915 (A, B) and in individuals older than 100 years born in 1895–96, 1905 and 1910–11 (C, D) assuming an additive (A, C) and a recessive (B, D) model. The 95% confidence interval is shown as well as the P-value for the significance of the difference between the two birth cohorts (A, B) or of the significance of the trend (C, D).
The trends of decreasing prevalence with more recent birth cohort seen in individuals older than 95 years are generally mirrored in individuals older than 100 years (Figures 2D–F and 3C–D), at least for the APOE ε4 allele and the FOXO3A rs479744 variant, although none of them reach statistical significance.
No significant differences in the prevalence of the APOE ε2 allele or of APOE ε2 allele carriers are found across the cohorts at age 95+ or 100+ (data not shown). However, the tendency of a higher prevalence in the more recent cohort seen in the larger sample of nonagenarians is consistent with an overall conclusion of the APOE gene becoming more important for mortality in more recent cohorts.
4. Discussion
The aim of this study was to explore whether the pronounced improvement in survival probability and the related reduction in selection pressure seen over recent cohorts of long-lived Danes were reflected in the prevalence of variants in known longevity-associated genes.
Generally, we found that the prevalence of the investigated variants in the APOE and FOXO3A genes was differing between cohorts born over a 20-year period from 1895 to 1915. A comparison of individuals older than 95 years born in 1905 and 1915 revealed a lower prevalence in the 1915 cohort, both of the APOE ε4 allele and of the minor alleles of the FOXO3A variants rs7762395 and rs479744. This was particularly evident when applying a recessive model and the trend was generally supported when we compared individuals older than 100 years born with an interval of 5–10 years from 1895 to 1911. None of the comparisons in the 100+ year olds reached statistical significance, probably due to lower power as a consequence of fewer participants compared to the group of 95+ year olds.
The APOE ε4 allele has previously been found to associate with increased mortality (Christensen et al. 2006), and the lower prevalence seen in the more recent birth cohorts is consistent with ε4 noncarriers having an increasingly larger survival advantage compared to ε4 carriers. It can be speculated that this could cause the mortality of individuals to a greater extent to be dependent on their APOE genotype, since competing causes of death have been eliminated due to improved living conditions and better medical treatment in the more recent birth cohorts (Oeppen & Vaupel 2002; Christensen et al. 2009). This line of thought is, however, opposite to what would be expected from the heterogeneity hypothesis that a selection pressure in older cohorts would decrease the effect of frailty gene variants such as the APOE ε4 allele when compared to younger cohorts (Vaupel et al. 1979). This is supported by the findings in a meta-study by Ewbank suggesting that the mortality by APOE genotype diminished at the oldest ages (Ewbank 2007). Also, a decline in the risk over age for carriers of the ε4 allele was found when using a statistical model incorporating heterogeneity (Ewbank 2002). The lack of concordance between the effect of the APOE gene and the heterogeneity hypothesis seen in the present study has previously been suggested in a longitudinal study of 92+ year olds examining the survival of APOE ε4 carriers versus non-carriers (Jacobsen et al. 2010). One possible explanation for the discrepancy between Ewbank (Ewbank 2002; Ewbank 2007), and the present study and Jacobsen et al. 2010 may be the different age groups studied, since APOE may have different effects on survival at different ages (Jacobsen et al. 2010).
For the FOXO3A minor alleles, which associate with longevity and not mortality, we found a decrease in minor allele frequency in the more recent birth cohorts. This is what we would expect from the heterogeneity hypothesis, leaving alive persons with the frailer version of the FOXO3A genotype and with other factors compensating for this. Since the role of FOXO3A in longevity is still largely unknown, more studies are needed to explain the pattern seen for this gene in the present study.
Since the probability of surviving to extreme ages has increased much faster in women than in men (see Figure 1), sex-stratified analyses were carried out (see Supplementary Data, Supplementary Table 1 and 2). Significant decreases were found for the prevalence of the APOE ε4 allele or of individuals carrying two ε4 alleles when we compared 95+ year old women born in 1905 and 1915 and assumed an additive or a recessive model. In contrast, non-significant increases were seen in men. These sex-stratified results are as predicted by the theory presented above, with a bigger decrease in selection pressure resulting in a larger influence of the APOE genotype as a cause of death, which is exactly what is seen for women. For the FOXO3A variants, no general differences between women and men are seen. There is a vague tendency for bigger differences between the birth cohorts in men, but inference is difficult due to the relatively small number of men.
Traditionally, it has been difficult to replicate primary genetic association findings in aging research, and only a few genes have been found to associate with longevity across populations. The results of this study suggest that birth year and population-dependent differences in selection pressure may be part of the explanation for this general lack of replication, although our data points to only small genetic differences between the investigated birth cohorts. However, the genetic variations related to longevity are currently expected to be rare and/or have small effects, and therefore even modest cohort effects could, when unaccounted for, confound results and leave true associations undiscovered.
A potential problem with this study might be bias due to differences in participation rate, e.g. if fewer sick or disabled individuals are included in a study, it could reduce the APOE ε4 frequency. Thus, it cannot be excluded that part of the genetic difference seen between the cohorts is due to cohort differences in the participation rate. However, the significant cohort differences in the prevalence of the investigated alleles are seen in individuals older than 95 years from the 1905 and 1915 birth cohorts, which have only very small differences in participation rates (Christensen et al. 2013).
The genetic differences over cohorts found in this study are substantial considering that the birth year intervals are less than two decades. Also, Denmark is among the high-income countries with the smallest increase in survival among the oldest-old and thus larger cohort differences in the allele distribution of longevity-associated genes may be expected in countries with a more pronounced increase in the number of nonagenarians and centenarians.
Altogether, our results point toward the need to consider the birth cohort of long-lived individuals in future genetic studies of human longevity. The results presented in this study are based on a unique collection of samples. Most other longevity samples contain individuals from a wide range of birth cohorts, and therefore the results of this study may be difficult to replicate directly. Studies of longevity genes may, however, benefit from being stratified into decades of birth year.
Supplementary Material
Highlights.
The chance of surviving to age 90+ has increased dramatically over the last decades
We studied 2,712 long-lived individuals from four Danish cohorts born 1895–1915
Allele frequencies of variants in the longevity genes APOE and FOXO3A were compared
Moderate cohort differences in allele frequencies were observed for both genes
This indicates an interaction between birth cohort and longevity gene effects
Acknowledgments
This study was financially supported by the INTERREG 4 A programme Syddanmark-Schleswig-K.E.R.N (with EU funds from the European Regional Development Fund), the VELUX Foundation, the European Union's Seventh Framework Programme (FP7/2007–2011) under grant agreement n° 259679, the Max-Planck Institute for Demographic Research (Rostock, Germany), the Danish Interdisciplinary Research Council, the Danish National Research Foundation, the US National Institutes of Health - National Institute on Aging (grant number P01 AG08761), the AXA Research Fund, and the Danish Agency for Science, Technology and Innovation (grant number. 09-070081).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: APOE, apolipoprotein E; FOXO3A, forkhead box O3A; LSDC, Longitudinal Study of Danish Centenarians; DCRS, Danish Civil Registration System; 5-COOP, 5-Country Oldest Old Project.
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Marianne Nygaard, Email: mnygaard@health.sdu.dk.
Rune Lindahl-Jacobsen, Email: rjacobsen@health.sdu.dk.
Mette Soerensen, Email: msoerensen@health.sdu.dk.
Jonas Mengel-From, Email: jmengel-from@health.sdu.dk.
Karen Andersen-Ranberg, Email: KARanberg@health.sdu.dk.
Bernard Jeune, Email: BJeune@health.sdu.dk.
James W. Vaupel, Email: jwv@demogr.mpg.de.
Qihua Tan, Email: qtan@health.sdu.dk.
Lene Christiansen, Email: LChristiansen@health.sdu.dk.
Kaare Christensen, Email: KChristensen@health.sdu.dk.
References
- Andersen-Ranberg K, Schroll M, Jeune B. Healthy centenarians do not exist, but autonomous centenarians do: a population-based study of morbidity among Danish centenarians. J Am Geriatr Soc. 2001;49:900–908. doi: 10.1046/j.1532-5415.2001.49180.x. [DOI] [PubMed] [Google Scholar]
- Anselmi CV, Malovini A, Roncarati R, Novelli V, Villa F, Condorelli G, Bellazzi R, Puca AA. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 2009;12:95–104. doi: 10.1089/rej.2008.0827. [DOI] [PubMed] [Google Scholar]
- Bathum L, Christiansen L, Jeune B, Vaupel J, McGue M, Christensen K. Apolipoprotein e genotypes: relationship to cognitive functioning, cognitive decline, and survival in nonagenarians. J Am Geriatr Soc. 2006;54:654–658. doi: 10.1111/j.1532-5415.2005.53554.x. [DOI] [PubMed] [Google Scholar]
- Christensen K, Johnson TE, Vaupel JW. The quest for genetic determinants of human longevity: challenges and insights. Nat Rev Genet. 2006;7:436–448. doi: 10.1038/nrg1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K, Doblhammer G, Rau R, Vaupel JW. Ageing populations: the challenges ahead. Lancet. 2009;374:1196–1208. doi: 10.1016/S0140-6736(09)61460-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K, Thinggaard M, Oksuzyan A, Steenstrup T, Andersen-Ranberg K, Jeune B, McGue M, Vaupel JW. Physical and cognitive functioning of people older than 90 years: a comparison of two Danish cohorts born 10 years apart. Lancet. 2013 doi: 10.1016/S0140-6736(13)60777-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen J, Beekman M, Uh HW, Helmer Q, Kuningas M, Christiansen L, Kremer D, van der Breggen R, Suchiman HE, Lakenberg N, van den Akker EB, Passtoors WM, Tiemeier H, van Heemst D, de Craen AJ, Rivadeneira F, de Geus EJ, Perola M, van der Ouderaa FJ, Gunn DA, Boomsma DI, Uitterlinden AG, Christensen K, van Duijn CM, Heijmans BT, Houwing-Duistermaat JJ, Westendorp RG, Slagboom PE. Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell. 2011;10:686–698. doi: 10.1111/j.1474-9726.2011.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewbank DC. Mortality differences by APOE genotype estimated from demographic synthesis. Genetic epidemiology. 2002;22:146–155. doi: 10.1002/gepi.0164. [DOI] [PubMed] [Google Scholar]
- Ewbank DC. Differences in the association between apolipoprotein E genotype and mortality across populations. The journals of gerontology. Series A, Biological sciences and medical sciences. 2007;62:899–907. doi: 10.1093/gerona/62.8.899. [DOI] [PubMed] [Google Scholar]
- Flachsbart F, Caliebe A, Kleindorp R, Blanche H, von Eller-Eberstein H, Nikolaus S, Schreiber S, Nebel A. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A. 2009;106:2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes LU, Jeune B, Ranberg KA, Nybo H, Vaupel JW. Estimation of apolipoprotein E genotype-specific relative mortality risks from the distribution of genotypes in centenarians and middle-aged men: apolipoprotein E gene is a "frailty gene," not a "longevity gene". Genetic epidemiology. 2000;19:202–210. doi: 10.1002/1098-2272(200010)19:3<202::AID-GEPI2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Jacobsen R, Martinussen T, Christiansen L, Jeune B, Andersen-Ranberg K, Vaupel JW, Christensen K. Increased effect of the ApoE gene on survival at advanced age in healthy and long-lived Danes: two nationwide cohort studies. Aging Cell. 2010;9:1004–1009. doi: 10.1111/j.1474-9726.2010.00626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeune B, Kannisto V. Emergence of Centenarians and Super-centenarians. In: Robine J-M, Vaupel J, Jeune B, Allard M, editors. Longevity: To the Limits and Beyond. Berlin Heidelberg: Springer; 1997. pp. 77–89. [Google Scholar]
- Li Y, Wang WJ, Cao H, Lu J, Wu C, Hu FY, Guo J, Zhao L, Yang F, Zhang YX, Li W, Zheng GY, Cui H, Chen X, Zhu Z, He H, Dong B, Mo X, Zeng Y, Tian XL. Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum Mol Genet. 2009;18:4897–4904. doi: 10.1093/hmg/ddp459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay GJ, Silvestri G, Chakravarthy U, Dasari S, Fritsche LG, Weber BH, Keilhauer CN, Klein ML, Francis PJ, Klaver CC, Vingerling JR, Ho L, De Jong PT, Dean M, Sawitzke J, Baird PN, Guymer RH, Stambolian D, Orlin A, Seddon JM, Peter I, Wright AF, Hayward C, Lotery AJ, Ennis S, Gorin MB, Weeks DE, Kuo CL, Hingorani AD, Sofat R, Cipriani V, Swaroop A, Othman M, Kanda A, Chen W, Abecasis GR, Yates JR, Webster AR, Moore AT, Seland JH, Rahu M, Soubrane G, Tomazzoli L, Topouzis F, Vioque J, Young IS, Fletcher AE, Patterson CC. Variations in apolipoprotein E frequency with age in a pooled analysis of a large group of older people. American journal of epidemiology. 2011;173:1357–1364. doi: 10.1093/aje/kwr015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murabito JM, Yuan R, Lunetta KL. The search for longevity and healthy aging genes: insights from epidemiological studies and samples of long-lived individuals. The journals of gerontology. Series A, Biological sciences and medical sciences. 2012;67:470–479. doi: 10.1093/gerona/gls089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebel A, Kleindorp R, Caliebe A, Nothnagel M, Blanche H, Junge O, Wittig M, Ellinghaus D, Flachsbart F, Wichmann HE, Meitinger T, Nikolaus S, Franke A, Krawczak M, Lathrop M, Schreiber S. A genome-wide association study confirms APOE as the major gene influencing survival in long-lived individuals. Mech Ageing Dev. 2011;132:324–330. doi: 10.1016/j.mad.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Nybo H, Gaist D, Jeune B, Bathum L, McGue M, Vaupel JW, Christensen K. The Danish 1905 cohort: a genetic-epidemiological nationwide survey. J Aging Health. 2001;13:32–46. doi: 10.1177/089826430101300102. [DOI] [PubMed] [Google Scholar]
- Oeppen J, Vaupel JW. Demography. Broken limits to life expectancy. Science. 2002;296:1029–1031. doi: 10.1126/science.1069675. [DOI] [PubMed] [Google Scholar]
- Pawlikowska L, Hu D, Huntsman S, Sung A, Chu C, Chen J, Joyner AH, Schork NJ, Hsueh WC, Reiner AP, Psaty BM, Atzmon G, Barzilai N, Cummings SR, Browner WS, Kwok PY, Ziv E. Association of common genetic variation in the insulin/IGF1 signaling pathway with human longevity. Aging Cell. 2009;8:460–472. doi: 10.1111/j.1474-9726.2009.00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen CB, Gotzsche H, Moller JO, Mortensen PB. The Danish Civil Registration System. A cohort of eight million persons. Dan Med Bull. 2006;53:441–449. [PubMed] [Google Scholar]
- Robine JM, Cheung SL, Saito Y, Jeune B, Parker MG, Herrmann FR. Centenarians Today: New Insights on Selection from the 5-COOP Study. Current gerontology and geriatrics research. 2010;2010:120354. doi: 10.1155/2010/120354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter F, Faure-Delanef L, Guenot F, Rouger H, Froguel P, Lesueur-Ginot L, Cohen D. Genetic associations with human longevity at the APOE and ACE loci. Nature genetics. 1994;6:29–32. doi: 10.1038/ng0194-29. [DOI] [PubMed] [Google Scholar]
- Soerensen M, Dato S, Christensen K, McGue M, Stevnsner T, Bohr VA, Christiansen L. Replication of an association of variation in the FOXO3A gene with human longevity using both case-control and longitudinal data. Aging Cell. 2010;9:1010–1017. doi: 10.1111/j.1474-9726.2010.00627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soerensen M, Dato S, Tan Q, Thinggaard M, Kleindorp R, Beekman M, Jacobsen R, Suchiman HE, de Craen AJ, Westendorp RG, Schreiber S, Stevnsner T, Bohr VA, Slagboom PE, Nebel A, Vaupel JW, Christensen K, McGue M, Christiansen L. Human longevity and variation in GH/IGF-1/insulin signaling, DNA damage signaling and repair and pro/antioxidant pathway genes: Cross sectional and longitudinal studies. Exp Gerontol. 2012;47:379–387. doi: 10.1016/j.exger.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soerensen M, Dato S, Tan Q, Thinggaard M, Kleindorp R, Beekman M, Suchiman HE, Jacobsen R, McGue M, Stevnsner T, Bohr VA, de Craen AJ, Westendorp RG, Schreiber S, Slagboom PE, Nebel A, Vaupel JW, Christensen K, Christiansen L. Evidence from case-control and longitudinal studies supports associations of genetic variation in APOE, CETP, and IL6 with human longevity. Age (Dordr) 2013;35:487–500. doi: 10.1007/s11357-011-9373-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Q, Christiansen L, Thomassen M, Kruse TA, Christensen K. Twins for epigenetic studies of human aging and development. Ageing Res Rev. 2013;12:182–187. doi: 10.1016/j.arr.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaupel JW, Manton KG, Stallard E. The impact of heterogeneity in individual frailty on the dynamics of mortality. Demography. 1979;16:439–454. [PubMed] [Google Scholar]
- Willcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.